Corrigendum: Long Non-Coding RNAs in the Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential
- 1Department of Gynecology, Harbin Medical University Cancer Hospital, Harbin, China
- 2Department of Gynecology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 3Institute of Cancer Neuroscience, Medical Frontier Innovation Research Center, The First Hospital of Lanzhou University, The First Clinical Medical College of Lanzhou University, Lanzhou, China
Cancer immunotherapy (CIT) is considered a revolutionary advance in the fight against cancer. The complexity of the immune microenvironment determines the success or failure of CIT. Long non-coding RNA (lncRNA) is an extremely versatile molecule that can interact with RNA, DNA, or proteins to promote or inhibit the expression of protein-coding genes. LncRNAs are expressed in many different types of immune cells and regulate both innate and adaptive immunity. Recent studies have shown that the discovery of lncRNAs provides a novel perspective for studying the regulation of the tumor immune microenvironment (TIME). Tumor cells and the associated microenvironment can change to escape recognition and elimination by the immune system. LncRNA induces the formation of an immunosuppressive microenvironment through related pathways, thereby controlling the escape of tumors from immune surveillance and promoting the development of metastasis and drug resistance. Using lncRNA as a therapeutic target provides a strategy for studying and improving the efficacy of immunotherapy.
Introduction
Antitumor therapy is based on two fundamental principles: 1) direct killing of tumor cells or 2) regulation of the tumor microenvironment. To achieve the goal of tumor eradication, cancer biology cannot be understood based on the characteristics of tumor cells alone, but must also include the effect of the tumor microenvironment (TME) on the tumor (1). Therefore, there are some cases in which treatment methods that directly target tumor cells fail to achieve the expected efficacy in clinical application (1). Although Stephen Paget first proposed the “seed and soil” hypothesis in 1889, the subsequent renewed understanding of the TME has made it an important target for tumor research and therapy (2–4). The TME is a complex and dynamic network structure composed of tumor cells and the surrounding region (including tumor-associated immune cells, fibroblasts, vascular endothelial cells, adipocytes and extracellular matrix, as well as secreted cytokines and chemokines) (5). The immune microenvironment, hypoxic niche, metabolism microenvironment, acidic niche and innervated niche microenvironment, and other microenvironments are interconnected, which significantly contributes to the complexity and heterogeneity of the TME (6–12). The immune microenvironment is considered to be a critical specialized microenvironment that can reprogram cancer biology, and is closely related to cancer prognosis and response to treatment (7, 13).
The immune microenvironment is primarily composed of myeloid cells [i.e., macrophages, myeloid inhibitory cells (MDSCs), and neutrophils], lymphocytes [i.e., CD4+ T helper cells (Th), regulatory T cells (Tregs), CD8+ cytotoxic T cells (CTLs), B cells, natural killer (NK) cells, and dendritic cells (DCs)]. The composition and status of immune cells varies between different types of tumors and between patients with the same tumor (14). Both activated and suppressive immune phenotypes have been found in the TME based on the infiltration of immune cells (1). Moreover, high resolution single-cell RNA sequencing, flow cytometry, and immunoscore techniques have been applied in an effort to further understand the density and diversity of tumor-infiltrating immune cells (14–18). While these methods can help explain how immunotherapy-based strategies improve clinical outcomes, the therapeutic responses of immunotherapy-based strategies are limited to a small number of patients who have significantly improved patient-specific clinical outcomes (19, 20). In addition, reversing immunosuppressive strategies can improve the efficacy of immunotherapy (21). Immune escape and therapy resistance are the two major obstacles associated with radical tumor therapy, which are also primarily mediated by an immunosuppressive microenvironment (22, 23). Therefore, immune microenvironment reprogramming represents the key to improving the antitumor response and is a powerful target for CIT.
LncRNA is a type of non-coding RNA (ncRNA) longer than 200 nucleotides (24). LncRNAs have been found to be associated with multiple types of cancer [e.g., breast (25), lung (26), and liver (27) cancer], as well as resistance to chemotherapy and immunotherapy (28, 29). LncRNAs do not directly encode proteins involved in the innate or adaptive immune response; however, they can regulate the differentiation and function of immune cells (30). LncRNAs can also facilitate the escape of tumor cells from immune surveillance by promoting the formation of an immunosuppressive microenvironment and other mechanisms (31). For example, the lncRNA NKILA can induce the apoptosis of tumor-specific T cells so that they cannot penetrate the tumor (32). Recently, lncRNA has been considered a potential target for immunotherapy, and has attracted extensive attention in the field of cancer therapy research. This review primarily focuses on lncRNA-mediated reprogramming of the tumor immune microenvironment (TIME). In particular, we describe the mechanism by which lncRNA inhibits the generation of the microenvironment, inducing immune escape and immune checkpoints to promote resistance. Next, we summarize the potential application of lncRNA as a target for tumor immunotherapy.
LncRNA: Molecular Features and Biological Mechanisms
In the human genome, approximately 93% of DNA can be transcribed into RNA, of which only 2% is protein-coding mRNA and the remaining 98% is termed non-coding RNA (33). LncRNAs lack protein-coding ability, can be spliced, capped, and/or polyadenylated, and are localized in the nucleus or cytoplasm (34). Based on their localization and the length between protein coding target mRNAs, lncRNAs can be roughly divided into intronic, intergenic, sense, antisense, bidirectional, and enhancer lncRNAs (35). Since the advent of the genomic era in the 2000s, significant progress has been made in understanding the biogenesis and function of different types of lncRNAs that are found ubiquitously across species (36, 37). The lncRNA is no longer considered to be “transcriptional noise”, but rather a highly efficient RNA factor (38, 39) that functions through epigenetic control and transcription, translation, RNA metabolism, and other mechanisms (40, 41). LncRNAs act as competing endogenous RNA (ceRNA) to competitively bind to miRNAs, thereby preventing miRNAs from binding to target mRNA (42–44). LncRNAs are directly involved in the epigenetic regulation of cancer by interacting with key histone modification enzymes, as well as chromatin modification, direct transcriptional regulation, and post-transcriptional functions (e.g., splicing, editing, localization, translation, and degradation) (45–47). Additionally, in gastric cancer, lncRNA SNHG17 has been shown to promote cancer progression by epigenetically silencing p15 and p57 (48). Recent studies have also suggested that cis-regulatory elements associated with the specific chromatin architecture are formed by epigenetic factors, endowing innate immune cells with specific phenotypes and unique functions by establishing cell-specific gene expression patterns (49). Moreover, lncRNAs can be used as immune modulators to regulate the immune response at the epigenetic level. Several studies have shown that lncRNAs are dysregulated in cancer and play a role in tumor proliferation, angiogenesis, apoptosis, and metastasis (50). In addition, lncRNAs are also closely related to the regulation of the TIME and antitumor immunity (50).
LncRNA Regulation of the TIME: Focus on Immune Escape
LncRNAs play a regulatory role in the immune system. Immune regulation is achieved primarily through the processes of RNA/protein binding or RNA/DNA base pairing, and both lncRNA and mRNA use a common promoter region to conduct bidirectional transcription (51, 52). In addition, lncRNAs can regulate the immune response through several different pathways, including NF-κB/MAPK and JAK/STAT (53). MYC-regulated NEAT1 was found to promote diffuse large B cell lymphoma (DLBCL) proliferation via the miR-34b-5p-GLI1 pathway (54). It has also been reported that some immune-related lncRNAs control the differentiation, development, and effector function of these cells (55). Moreover, lncRNA can mediate the activation and inhibition of immune response genes. In a breakthrough study, lncRNA-DC was found to be expressed only in human DCs, directly bind to STAT3 in the cytoplasm, and promote STAT3 phosphorylation on tyrosine-705 by preventing the binding and dephosphorylation of STAT3 to SHP1. An Lnc-DC knockout was demonstrated to impair DC differentiation in human monocytes in vitro and mouse bone marrow cells in vivo, which decreased the ability of DCs to stimulate T cell activation (56). These results indicate that lncRNAs are key immunomodulators.
Tumor cells can evade immune recognition and elimination by changing their phenotype or the microenvironment (57). The activation of immunosuppressive cells and factors [e.g., MDSCs, tumor-associated macrophage (TAMs) subsets], abnormal antitumor immune cells (e.g., DC, NK, and T cells), and Tregs represent important features of the microenvironment that promote tumor immune escape (58, 59). At the microenvironmental level, lncRNAs are involved in mediating and controlling various immune and cancer cell interactions and other important mechanisms of the immune response (Table 1). Various studies have confirmed that lncRNAs induce the formation of an immunosuppressive microenvironment through related pathways, thereby contributing to tumor escape of immune surveillance, as well as the development of metastasis and drug resistance (Figure 1).
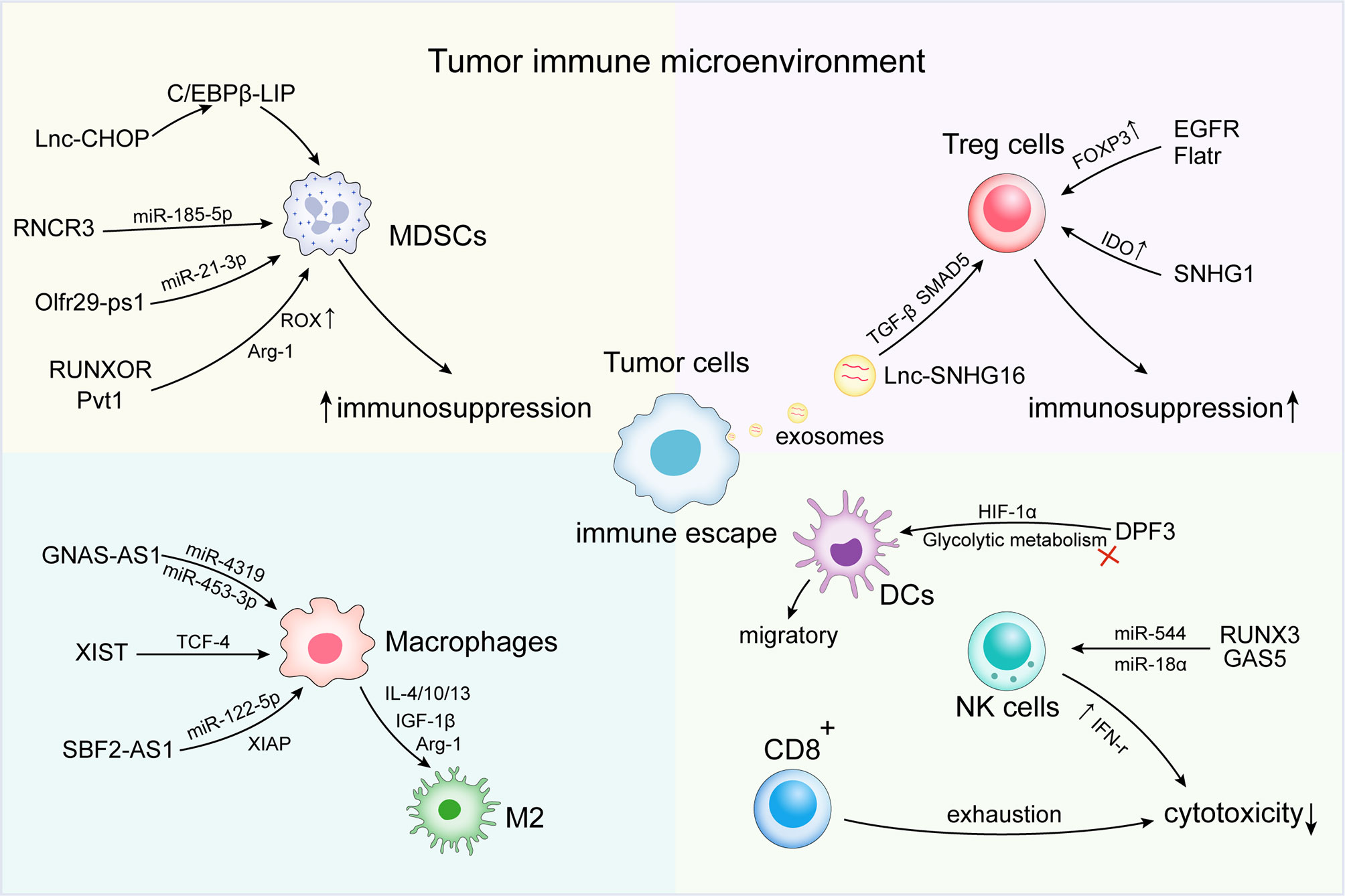
Figure 1 Long non-coding RNAs (lncRNAs) regulate immune escape in the tumor immune microenvironment (TIME). At the microenvironmental level, lncRNAs are involved in mediating and controlling various immune and cancer cell interactions, promoting the activation of immunosuppressive cells and factors [e.g., myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophage (TAMs) subsets]. Abnormal antitumor immune cells [e.g., dendritic cell (DC), natural killer (NK) cells, and T cells] and regulatory cells T cells (Tregs) induce the formation of an immunosuppressive microenvironment, thus contributing to the immune escape of tumor cells.
LncRNA Regulation of MDSCs
LncRNAs can inhibit the immune response by regulating the activity of immunosuppressive cells. Under pathological conditions, extramedullary bone marrow generates MDSCs (112). MDSCs play a central role in cancer progression by mediating immunosuppression in the TME through a variety of mechanisms, including the production of inducible nitric oxide synthase (iNOS), arginase-1 (Arg1), oxygen free radicals (ROS), and nitric oxide (NO) (113, 114). Recent studies suggest that lncRNAs play an important role in the immunosuppressive functions of MDSCs. In particular, lnc-CHOP and RNCR3 can positively regulate the growth and inhibitory function of MDSCs (60, 61). Moreover, lnc-chop may interact with CHOP and the C/EBPB isoform, LIP, to encourage C/EBPB activation. C/EBPB is associated with the differentiation of MDSCs. Therefore, lnc-chop may affect the differentiation of MDSCs and activate the expression of immunosuppressive genes. Furthermore, the combination of lnc-chop and CHOP in MDSCs may have important significance for the control of tumor growth, since increased CHOP expression in tumor-associated MDSCs has been observed in a variety of tumor models (115). Similarly, the expression of the lncRNA, RNCR3, in MDSCs is upregulated by both inflammatory and tumor-associated factors. In addition, an RNCR3 knockout was found to result in suppressed MDSC differentiation and function both in vitro and in vivo (61). Another study found that the lncRNA, pseudogene Olfr29-ps1, is expressed in MDSCs. Pseudogene Olfr29-ps1 has been shown to regulate the differentiation and immunosuppressive function of MDSCs via the N6-methyladenosine (M6A) modified Olfr29-ps1/miR-214-3p/MyD88 regulatory network (62, 116). The lncRNAs, Pvt1, MALAT1, HOTAIRM1, RUNXOR, and others have also strongly confirmed the regulatory effect of lncRNA on MDSC activity (63–66).
LncRNA Regulation of Tregs
Tregs are an immunosuppressive subset of CD4+ T cells (117). The depletion of CD4+CD25+ regulatory T cells promotes a tumor-specific immune response in the pancreas cancer-bearing mice (118). Tumor-infiltrating Tregs may also interfere with host antitumor responses by inhibiting tumor-specific immune effector cells. Multiple studies have shown that lncRNAs [e.g., lnc epidermal growth factor receptor (lnc-EGFR), lncRNA SNHG1, Flicr, and Flatr] can regulate the biological function of Tregs (30, 69–71). The upregulation of lnc-EGFR in Tregs was positively correlated with tumor size and EGFR/Foxp3 expression. Lnc-EGFR functions by activating the downstream AP-1/NF-AT1 axis and inducing EGFR expression. Moreover, lnc-EGFR has also been shown to stimulate Treg differentiation, inhibit CTL activity, and promote hepatocellular carcinoma (HCC) growth (69).
The lncRNA, Flatr, is a part of the upstream cascade that leads to enhanced differentiation, FOXP3 expression, and immunosuppressive function in Tregs (71). Breast cancer cells promote the expression of SMAD5 in γδT cells through the transfer of the lncRNA, SNHG16, in exosomes, which functions as a ceRNA through miR-16-5p, thereby enhancing the TGF-β1/Smad5 pathway and upregulating CD73 expression (72). The study by Pei et al. showed that interference with SNHG1 promoted miR-448 expression, reduced the level of indoleamine 2,3-dioxygenase (IDO), and inhibited Treg differentiation, thereby impeding tumor immune escape (30).
LncRNA Effects Macrophage Differentiation in Immune Escape
In the immune microenvironment, macrophages are classified as proinflammatory, antitumorigenic M1, and anti-inflammatory protumorigenic M2 phenotypes (119). TAMs function by directly or indirectly inhibiting effector T cells (120). Multiple studies have shown that lncRNA can affect the immune escape of tumor cells by regulating M2 macrophage polarization. LncRNA GNAS-AS1 expression is significantly enhanced in TAM non-small cell lung cancer (NSCLC) cell lines and clinical tumor tissues in lung cancer, and is negatively correlated to the overall survival of NSCLC patients. Moreover, lncRNA GNAS-AS1 promotes tumor progression in NSCLC by altering macrophage polarization through the GNAS-AS1/MIR4319/NECAB3 axis (76). LncRNA-XIST is regulated by TCF-4, which also plays a role in promoting M2 macrophage polarization (78). However, some lncRNAs can negatively regulate TAM M2 polarization. In endometrial cancer, NIFK-AS1 inhibited the M2-like polarization of macrophages by targeting miR-146a, thereby reducing the proliferation, migration, and invasion of estrogen-induced endometrial cancer cells (79). Zhou et al. co-incubated a mouse liver cell line (HEPAL6 cells) and a liver cancer cell line (HepG2 cells) with M1 or M2 macrophages, and found that lncRNA COX-2 expression was higher in M1 macrophages than in M2 macrophages. LncRNA COX-2 inhibits HCC immune escape and tumor growth by inhibiting M2 macrophage polarization (80). In pancreatic cancer, studies have shown that the blocking Sbf2-AS1 in M2 macrophage-derived exosomes inhibited XIAP expression through the negative regulation of miR-122-5p, and played a role in reducing the oncogenic ability of tumor cells (81). In the report by Zhang et al., RNA sequencing and other methods were used to identify differentially expressed miRNAs and lncRNAs in MΦ-CM co-cultured osteosarcoma cells and the corresponding control group, which confirmed that lncRNA LIFR-AS1 was upregulated in MΦ-CM co-cultured osteosarcoma cells (97). In addition, LIFR-AS1 can be transmitted from macrophages to osteosarcoma cells via exosomes, and promote tumor progression via spongy transfection of miR-29a (97). These findings show that abnormally expressed lncRNAs can be used as potential biological targets for cancer therapy.
LncRNA Modulation of Antitumor Immune Cells
DCs are associated with overall survival in cancer patients, reflecting the unique ability of humans to initiate CD8+ T cell responses (121). However, the TIME often interferes with the normal function of DCs to avoid immune surveillance (122, 123). LncRNAs can regulate DC infiltration, differentiation, and metabolism, as well as influence other immune cells, including T cells, to modify the local immune environment. Lnc-DC was found to promote DC maturation and inhibit trophoblast invasion without the involvement of CD4+ T cells. In addition, lnc-DC controlled the immune response by reducing the concentration of TNF-α, IL-6, IL-12, and IFN-γ secretion, as well as increasing IL-1β production by DCs (99, 100). Lnc-DPF3 inhibits DC migration by directly binding to HIF1A and inhibiting HIF1A activity via the HRE motif to suppress glycolysis (98).
The first line of immune defense includes NK cells, which are cytotoxic immune cells that can directly kill cancer cells (124). Recently, Zhang et al. measured the expression profile of lncRNAs in human primary lymphocytes, and found that NK-specific lncRNAs are closely associated with the differentiation and function of NK cells. The expression of the NK-specific lncRNA, lnc-CD56, was found to be a positive regulator of CD56 (104). In addition, the lncRNA, GAS5, could regulate the killing effect of NK cells in several types of cancer (105, 106). These results demonstrate the importance of lncRNAs in NK cell function and the antitumor immune response. LncRNA can regulate the function of CD8+ T cells in the TME through a variety of mechanisms to alter the immune response. LNC-TIM3 was found to be upregulated in tumor-infiltrating CD8 T cells from HCC patients and negatively correlated with the level of IFN-γ and IL-2 production. LNC-TIM3 specifically binds to Tim-3 and blocks its interaction with BAT3, thereby inhibiting the downstream LCK/NFAT1/AP-1 signaling pathway, and plays a key role in promoting CD8 T suppression (109). In another study, both NEAT1 and TIM-3 expression were upregulated in the PBMCs of liver cancer patients compared with healthy subjects. The downregulation of NEAT1 can inhibit the apoptosis of CD8+ T cells through the miR-155/Tim-3 pathway, enhance cell lysis activity, and inhibit tumor growth in mice with HCC (110).
LncRNA Impacts Resistance to Immune Checkpoint Therapy
Immunotherapy, primarily represented by PD-1/PD-L1 inhibitors, has made substantial breakthroughs for the treatment of multi-solid tumors (125–127). Thus, immunotherapy has become a popular form of cancer treatment. However, after experiencing an initial response to immune checkpoint inhibitor (ICIS) therapy, most patients develop secondary resistance. The mechanism by which secondary resistance develops remains largely uncertain (128, 129). ICIS therapy functions by relieving the immunosuppression of tumor cells or the associated microenvironment. Several factors can impact the efficacy of ICIS, including antigen presentation, tumor mutation burden, and T cell infiltration (128, 130).
The mechanism of immune escape is dominated by the formation of an immunosuppressive microenvironment, which can be regulated by lncRNAs. Some lncRNAs also promote the generation of drug resistance through the PD-1/PD-L1 axis and the presentation of inhibitory antigens. For example, the lncRNA, MALAT1, can regulate tumor immunity by indirectly upregulating the expression of PD-L1 through miR-195 and miR-200a-3 (131, 132). In addition, the SNHG14/miR-5590-3p/ZEB1 positive feedback loop was found to promote the progression and immune escape of DLBCL by regulating the PD-1/PD-L1 checkpoint, suggesting that targeting SNHG14 is a potential method of improving the efficacy of DLBCL immunotherapy (133). More importantly, silencing LINC00473 resulted in increased expression of Bcl-2 X-related proteins (Burlington), interferon (IFN)-γ, and IL-4, but reduced the expression of B cell lymphoma-2 (Bcl-2), matrix metalloproteinase (MMP-2), MMP-9, and IL-10, thereby inducing enhanced apoptosis and inhibiting proliferation of DLBCL. In addition, silencing LINC00473 or elevating miR-195-5p was found to increase the number of activated CD8+ T cells (134). In contrast, NKX2-1-AS1 has been shown to aid in inhibiting immune escape by negatively regulating PD-L1 (135). LncRNA can also regulate antigen presentation, as Link-A has been shown to inactivate tumor suppressor pathways and downregulate antigen presentation through inactivating the PKA pathway. Therapy with Link-A locked nucleic acid or a GPCR antagonist has been found stabilize the PLC components, Rb and p53, and sensitize breast tumors to immune checkpoint blockers. Elevated Link-A levels were also confirmed in patients with programmed cell death protein 1 (PD-1)-blocking triple-negative breast cancer (TNBC) (31). Therefore, the regulation of lncRNA plays a key role in resistance to ICIS therapy.
Potential Therapeutic Approaches of lncRNA as Important Targets
From a clinical perspective, lncRNA-mediated regulation of the immune microenvironment represents a highly promising target for immunotherapy. There are many therapeutic strategies targeting lncRNAs, including small molecule inhibitors, antisense oligonucleotides (ASOs), RNA interference (RNAi) technology, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 genome editing (136). Small molecule inhibitors mainly bind to the higher structural regions of lncRNAs that are similar to protein targets (137). Screening and identification of small molecule compounds that may inhibit RNA can be achieved by high-throughput sequencing. ASO belongs to a class of drugs that bind to the lncRNA transcriptome via base pairing (138). Gapmer was developed based on this mechanism, and uses RNA nucleotides with extra covalent bonds to 2 ‘-O and 4’ -C nucleotide rings to specifically bind to RNA targets and recruit the RNA-H enzyme to induce target degradation (139). RNAi is a biological process of inducing a specific gene knockout by neutralizing targets with exogenous double-stranded RNA, including both short interfering RNAs (siRNAs; with high specificity and short effects) and short hairpin RNAs (shRNAs; with long-lasting and stable effects) (140). Moreover, the CRISPR/Cas9 system can be used to silence or knock-out lncRNA-expressing loci (141). After the CRISPR/CAS system enters the cell, gRNAs guide the CAS enzyme to locate specific DNA sequences on PAM complementary to the gRNA, after which the CAS enzyme will cut the DNA double strand, changing it or inducing a mutation through a frameshift, which finally leads to the silencing of the edited gene (142). Off-target effects represent the main difficulties associated with CRISPR/Cas9 gene therapy.
The key to treatment is to optimize target delivery. As therapeutic carriers, nanomaterials and exosomes can protect against drug degradation or aggregation and are associated with good targeting. As such, the safe and efficient intracellular delivery of CRISPR/Cas9 is critical for effective therapeutic genome editing. The study by He et al. showed that the use of epithelial cell-derived microvesicles (MVS) as a carrier to deliver CRISPR/Cas9 components to cancer cells showed strong anticancer effects against xenograft tumors, and this may become a safe CRISPR/Cas9 delivery platform for cancer patients (143). Furthermore, the application of nanotechnology can maximize the advantages of using lncRNA in combination with immunotherapy.
Nanoparticles
Over the past decade, the development of nanoparticle platforms has yielded promising prospects for their application in RNA therapy and cancer immunotherapy (144). Nanoparticles are granular dispersions or solid particles ranging in size from 10 nm to 1000 nm. Therapeutics can be delivered using nanoparticles to achieve enhanced permeability and retention (EPR). Typical nanoparticles include liposomes, polymer nanoparticles (NPs), inorganic NPs, and exosomes (145–149). Nanocarriers also typically exhibit good biocompatibility and stability. Moreover, nanoparticles can be customized via unique physical properties (e.g., dimensional charge and surface chemistry), to enable specific tissue or tumor targeting. On their own, nano pharmaceuticals can enhance cellular interactions, stimulate the immune system, and sustain an antitumor response (150). Therefore, the use of nanoparticles as a lncRNA-targeted therapy carrier combined with immunotherapy represents a multi-effect strategy. Gong et al. successfully constructed MALAT1-specific ASO and nucleo-targeted Tat peptide synergized Au nanoparticles (i.e., ASO-Au-Tat NPS), which could stabilize fragile ASOs, enhance nuclear internalization, and demonstrate good biocompatibility. Following treatment with ASO-Au-Tat NPS, the level of MALAT1 expression in A549 lung cancer cells was significantly reduced. In addition, ASO-Au-Tat NPS has been found to significantly reduce the formation of metastatic tumor nodules in vivo (151). Another study demonstrated that RGD-peg-ECO/siDANCR nanoparticle treatment of MDA-MB-231 cells and BT549, siRNA could be effectively passed to the cell, and continue to silence targeted nanoparticles. In addition, combined treatment significantly reduced TNBC cell survival, proliferation and tumor globular form of migration (152). In a recent study, researchers designed a novel type of polymer nanoparticle, which simultaneously targeted T cell immunoreceptor with Ig and ITIM domains (TIGIT)/polio virus receptors (PVRs), T cell immune receptors, and long non-coding RNA antisense non-coding RNA in the INK4 locus (lncRNA ANRIL) to suppress liver cancer. DTTP/3NP/siANRIL have a good antitumor effect against liver cancer, and inhibition of miR-203a and its downstream gene expression increases the percentage of NK cells and T cells (153). Nanoparticle-based delivery systems not only deliver high-dose therapeutic payloads to target cells, but also exhibit the same regulatory function in immunotherapy with RNA therapy. At the same time, combining lncRNA-mediated nanotherapy with existing immunotherapy provides an opportunity to improve the efficacy of cancer treatment. However, relatively few studies have investigated the use of this delivery method, and it will take some time before this application can be used in the clinic.
Exosomes
Exosomes are extracellular nanovesicles (30–150 nm in diameter) of endocytic origin that are secreted by most mammalian cell types. Exosomes are present in a wide range of bodily fluids (154). Moreover, exosomes are now recognized as important intercellular signaling messengers that encapsulate and transfer versatile molecular cargo to recipient cells. Exosomes naturally possess sophisticated specificity and are capable of passing through most biological barriers in vivo (155). Exosomes engineered to deliver specific small interfering RNA (siRNA) payloads can be protected from degradation by blood-derived ribonuclease (156). In addition, the surfaces of exosomes can be designed with required ligands to increase targeting efficiency (157). For example, exosomes can effectively deliver microRNAs (miRNAs) to breast cancer cells expressing epidermal growth factor receptor (EGFR) (158). Furthermore, exosomes themselves can regulate innate and acquired immunity, as well as the TME (119).
Exosomes have the unique advantages of improving cancer therapeutic indicators. They can also be engineered into therapeutic exosomes that improve the efficiency and targeting ability of antitumor drugs. Exosomes with siRNA targeting KRASG12D were shown to reduce KRAS GTPase activity and the downstream activation of RAF-MEK-ERK or PI3K-AKT-mTOR signaling, inhibit cancer cell proliferation, and increase pancreatic cancer cell apoptosis (159, 160). Recently, a trial using exosome vectors as a means of siRNA delivery was conducted in breast cancer cells. These exosomes were able to specifically bind to HER2/Neu and were capable of delivering siRNA molecules against the TPD52 gene into a SKBR3 cell line, which downregulated TPD52 gene expression by up to 70% (161). In addition, exosomal AFAP1-AS1 was found to induce trastuzumab resistance through associating with AUF1 and promoting ERBB2 translation (162).
Since lncRNAs play an important role in tumor immune escape and immunotherapy resistance, targeted lncRNA drugs combined with immunotherapy may provide an effective strategy for the treatment of cancer. For example, link-A may represent a potential therapeutic target for increasing ICIS sensitivity (31). Moreover, NKILA silencing in metastatic tumor infiltrating lymphocytes and CAR T cells can overcome tumor immune escape and improve the efficacy of adoptive T cell therapy in cancer treatment (32). Thus, nanoparticle or exosome-loaded lncRNA targeted therapy combined with immunotherapy has broad applicability in the field of cancer therapy.
Challenges and Future Perspectives
This article mainly reviews the reprogramming of the TIME mediated by lncRNAs. The role and mechanism of lncRNA in the formation of an inhibitory microenvironment and in inducing tumor cells to escape immune surveillance have been described in detail. The TME is highly complex. Although the importance of lncRNA in the regulation of the TIME has been demonstrated, a clear mechanism remains to be elucidated. Next, we discussed relevant strategies for targeting lncRNA therapy, which can improve therapeutic efficacy and accelerate clinical application by optimizing the targeted delivery of vectors. LncRNA is both a potential therapeutic target for cancer, as well as a predictor of the survival and treatment response. Tu et al. found that MSC derivatives induced the expression of LINC01119 in adjacent TNBC cells and accelerated the growth of cancer cells in vitro. LINC01119 is a strong prognostic indicator for poor prognosis in patients with TNBC (163). Additionally, lncRNA-based therapy is a promising approach in the field of cancer immunotherapy (164). In a recent cohort study, overall survival (OS) with immunofunctional lncRNA features and high CTL infiltration benefited the most. At the same time, a multiomics panel based on lncRNA score has been designed as a useful biomarker for cancer immunotherapy (165). Another study demonstrated that lncRNA miR155 was closely associated with the OS of different tumor types, immune cell infiltration, and immune checkpoint molecule expression, and also provided great value for predicting the efficacy of immune checkpoint inhibitor therapy (166). Thus, lncRNA-based immune subtypes are associated with survival and response to cancer immunotherapy.
Recent findings offer novel insight into lncRNA-based cancer treatment and draw attention to areas that require further research; however, there are some problems that remain to be solved. The application of lncRNA as a therapeutic target is associated with several challenges, the most important of which is the method by which specific molecules can be delivered to target cells. Next, problems with molecular delivery and off-target effects may cause safety concerns during treatment. The application of nanoparticles and exosome carriers are the key to solving these problems, and these platforms have good targeting. However, there are drawbacks regarding material selection and application (e.g., toxicity of nanomaterials, as well as the storage and large-scale preparation of exosomes). Although the goal of all studies is to facilitate clinical application, additional work must be performed before the clinical transformation of lncRNA-targeted therapy can be achieved. To date, there have been no clinical trials on the independent use of lncRNAs as a cancer treatment. Thus, studies involving organoids and patient-derived xenografts (PDX) may accelerate this process.
Conclusion
In summary, lncRNA molecules play a significant role in remodeling the TIME and regulating the immune escape of tumor cells. Thus, lncRNA-based targeted cancer immunotherapy has a promising future. Despite the continued problems associated with the application of lncRNA-based therapy, as research progresses and becomes optimized, the use of lncRNA as a therapeutic target will contribute to the development of novel therapeutic strategies for cancer.
Author Contributions
B-RX, GL, and W-LJ designed the manuscript. Y-NP wrote the manuscript. Y-NP and W-CQ drew the figures and tables. B-RX, GL, and W-LJ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81872507), Nn10 Program of Harbin Medical University Cancer Hospital (Nn10py2017-01), and HaiYan fund of Harbin Medical University Cancer Hospital (No. JJZD2017-01) to GL; National Key Research and Development Program of China (No. 2017FYA0205302) to W-LJ; and the National Natural Science Foundation of China (No.81872430), Special Fund in China Postdoctoral Science Foundation (No. 2019T120281, 2019M661304) and Heilongjiang Province Postdoctoral Science Foundation (No. LBH-Z18109) to B-RX.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ming-Zhu Jin for her guidance.
References
1. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
2. Akhtar M, Haider A, Rashid S, Al-Nabet A. Paget’s “Seed and Soil” Theory of Cancer Metastasis: An Idea Whose Time has Come. Adv Anat Pathol (2019) 26(1):69–74. doi: 10.1097/PAP.0000000000000219
3. Maman S, Witz IP. A History of Exploring Cancer in Context. Nat Rev Cancer (2018) 18(6):359–76. doi: 10.1038/s41568-018-0006-7
4. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
5. Hui L, Chen Y. Tumor Microenvironment: Sanctuary of the Devil. Cancer Lett (2015) 368(1):7–13. doi: 10.1016/j.canlet.2015.07.039
6. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune Cells Within the Tumor Microenvironment: Biological Functions and Roles in Cancer Immunotherapy. Cancer Lett (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
7. Jin MZ, Jin WL. The Updated Landscape of Tumor Microenvironment and Drug Repurposing. Signal Transduction Targeted Ther (2020) 5(1):166. doi: 10.1038/s41392-020-00280-x
8. Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the Tumor in the Hypoxic Niche: The Emerging Concept and Associated Therapeutic Strategies. Trends Pharmacol Sci (2017) 38(8):669–86. doi: 10.1016/j.tips.2017.05.002
9. Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell (2020) 78(6):1019–33. doi: 10.1016/j.molcel.2020.05.034
10. Paolini L, Adam C, Beauvillain C, Preisser L, Blanchard S, Pignon P, et al. Lactic Acidosis Together With GM-CSF and M-CSF Induces Human Macrophages Toward an Inflammatory Protumor Phenotype. Cancer Immunol Res (2020) 8(3):383–95. doi: 10.1158/2326-6066.CIR-18-0749
11. Shurin MR, Shurin GV, Zlotnikov SB, Bunimovich YL. The Neuroimmune Axis in the Tumor Microenvironment. J Immunol (2020) 204(2):280–5. doi: 10.4049/jimmunol.1900828
12. Zahalka AH, Frenette PS. Nerves in Cancer. Nat Rev Cancer (2020) 20(3):143–57. doi: 10.1038/s41568-019-0237-2
13. Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The Immune Contexture in Human Tumours: Impact on Clinical Outcome. Nat Rev Cancer (2012) 12(4):298–306. doi: 10.1038/nrc3245
14. Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal Dynamics of Intratumoral Immune Cells Reveal the Immune Landscape in Human Cancer. Immunity (2013) 39(4):782–95. doi: 10.1016/j.immuni.2013.10.003
15. Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PloS Med (2016) 13(12):e1002194. doi: 10.1371/journal.pmed.1002194
16. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust Enumeration of Cell Subsets From Tissue Expression Profiles. Nat Methods (2015) 12(5):453–7. doi: 10.1038/nmeth.3337
17. Aran D, Hu Z, Butte AJ. xCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol (2017) 18(1):220. doi: 10.1186/s13059-017-1349-1
18. Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity (2016) 44(3):698–711. doi: 10.1016/j.immuni.2016.02.025
19. Saleh R, Elkord E. Treg-Mediated Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Lett (2019) 457:168–79. doi: 10.1016/j.canlet.2019.05.003
20. Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy B, et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity (2016) 44(6):1255–69. doi: 10.1016/j.immuni.2016.06.001
21. Liu Z, Han C, Fu YX. Targeting Innate Sensing in the Tumor Microenvironment to Improve Immunotherapy. Cell Mol Immunol (2020) 17(1):13–26. doi: 10.1038/s41423-019-0341-y
22. Kamimura N, Wolf AM, Iwai Y. Development of Cancer Immunotherapy Targeting the PD-1 Pathway. J Nippon Med Sch (2019) 86(1):10–4. doi: 10.1272/jnms.JNMS.2019_86-2
23. Passarelli A, Mannavola F, Stucci LS, Tucci M, Silvestris F. Immune System and Melanoma Biology: A Balance Between Immunosurveillance and Immune Escape. Oncotarget (2017) 8(62):106132–42. doi: 10.18632/oncotarget.22190
24. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-Coding RNAs and Cancer: A New Frontier of Translational Research? Oncogene (2012) 31(43):4577–87. doi: 10.1038/onc.2011.621
25. Zhang T, Hu H, Yan G, Wu T, Liu S, Chen W, et al. Long Non-Coding RNA and Breast Cancer. Technol Cancer Res Treat (2019) 18:1533033819843889. doi: 10.1177/1533033819843889
26. Lu T, Wang Y, Chen D, Liu J, Jiao W. Potential Clinical Application of lncRNAs in non-Small Cell Lung Cancer. OncoTargets Ther (2018) 11:8045–52. doi: 10.2147/OTT.S178431
27. O’Brien A, Zhou T, Tan C, Alpini G, Glaser S. Role of Non-Coding Rnas in the Progression of Liver Cancer: Evidence From Experimental Models. Cancers (Basel) (2019) 11(11). doi: 10.3390/cancers11111652
28. Jiang W, Xia J, Xie S, Zou R, Pan S, Wang ZW, et al. Long non-Coding RNAs as a Determinant of Cancer Drug Resistance: Towards the Overcoming of Chemoresistance Via Modulation of Lncrnas. Drug Resist Updates: Rev Commentaries Antimicrob Anticancer Chemother (2020) 50:100683. doi: 10.1016/j.drup.2020.100683
29. Zhang HD, Jiang LH, Zhong SL, Li J, Sun DW, Hou JC, et al. The Role of Long non-Coding RNAs in Drug Resistance of Cancer. Clin Genet (2021) 99(1):84–92. doi: 10.1111/cge.13800
30. Pei X, Wang X, Li H. Lncrna SNHG1 Regulates the Differentiation of Treg Cells and Affects the Immune Escape of Breast Cancer Via Regulating Mir-448/IDO. Int J Biol Macromol (2018) 118(Pt A):24–30. doi: 10.1016/j.ijbiomac.2018.06.033
31. Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncRNA Downregulates Cancer Cell Antigen Presentation and Intrinsic Tumor Suppression. Nat Immunol (2019) 20(7):835–51. doi: 10.1038/s41590-019-0400-7
32. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. Nkila lncRNA Promotes Tumor Immune Evasion by Sensitizing T Cells to Activation-Induced Cell Death. Nat Immunol (2018) 19(10):1112–25. doi: 10.1038/s41590-018-0207-y
33. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of Transcription in Human Cells. Nature (2012) 489(7414):101–8. doi: 10.1038/nature11233
34. Fatica A, Bozzoni I. Long non-Coding RNAs: New Players in Cell Differentiation and Development. Nat Rev Genet (2014) 15(1):7–21. doi: 10.1038/nrg3606
35. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding Rnas. Cell (2009) 136(4):629–41. doi: 10.1016/j.cell.2009.02.006
36. Yang L, Froberg JE, Lee JT. Long Noncoding RNAs: Fresh Perspectives Into the RNA World. Trends Biochem Sci (2014) 39(1):35–43. doi: 10.1016/j.tibs.2013.10.002
37. Wu H, Yang L, Chen LL. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet: TIG (2017) 33(8):540–52. doi: 10.1016/j.tig.2017.05.004
38. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22(9):1775–89. doi: 10.1101/gr.132159.111
39. Ransohoff JD, Wei Y, Khavari PA. The Functions and Unique Features of Long Intergenic non-Coding RNA. Nat Rev Mol Cell Biol (2018) 19(3):143–57. doi: 10.1038/nrm.2017.104
40. McDonel P, Guttman M. Approaches for Understanding the Mechanisms of Long Noncoding RNA Regulation of Gene Expression. Cold Spring Harb Perspect Biol (2019) 11(12). doi: 10.1101/cshperspect.a032151
41. Chen LL. Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci (2016) 41(9):761–72. doi: 10.1016/j.tibs.2016.07.003
42. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell (2011) 146(3):353–8. doi: 10.1016/j.cell.2011.07.014
43. Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-induced Mouse Model of Melanoma. Cell (2011) 147(2):382–95. doi: 10.1016/j.cell.2011.09.032
44. Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, et al. Long Noncoding Rna LINC01234 Functions as a Competing Endogenous RNA to Regulate Cbfb Expression by Sponging miR-204-5p in Gastric Cancer. Clin Cancer Res: an Off J Am Assoc Cancer Res (2018) 24(8):2002–14. doi: 10.1158/1078-0432.CCR-17-2376
45. Wilusz JE, Sunwoo H, Spector DL. Long Noncoding RNAs: Functional Surprises From the RNA World. Genes Dev (2009) 23(13):1494–504. doi: 10.1101/gad.1800909
46. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding Rnas. Cell (2007) 129(7):1311–23. doi: 10.1016/j.cell.2007.05.022
47. Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing Through Chromatin-Level Regulation. Mol Cell (2008) 32(2):232–46. doi: 10.1016/j.molcel.2008.08.022
48. Aguilo F, Zhou MM, Walsh MJ. Long Noncoding RNA, Polycomb, and the Ghosts Haunting INK4b-ARF-INK4a Expression. Cancer Res (2011) 71(16):5365–9. doi: 10.1158/0008-5472.CAN-10-4379
49. Zhang Q, Cao X. Epigenetic Remodeling in Innate Immunity and Inflammation. Annu Rev Immunol (2021) 39:279–311. doi: 10.1146/annurev-immunol-093019-123619
50. Wei B, Kong W, Mou X, Wang S. Comprehensive Analysis of Tumor Immune Infiltration Associated With Endogenous Competitive RNA Networks in Lung Adenocarcinoma. Pathol Res Pract (2019) 215(1):159–70. doi: 10.1016/j.prp.2018.10.032
51. Turner M, Galloway A, Vigorito E. Noncoding RNA and its Associated Proteins as Regulatory Elements of the Immune System. Nat Immunol (2014) 15(6):484–91. doi: 10.1038/ni.2887
52. Bonasio R, Shiekhattar R. Regulation of Transcription by Long Noncoding Rnas. Annu Rev Genet (2014) 48:433–55. doi: 10.1146/annurev-genet-120213-092323
53. Mathy NW, Chen XM. Long non-Coding RNAs (lncRNAs) and Their Transcriptional Control of Inflammatory Responses. J Biol Chem (2017) 292(30):12375–82. doi: 10.1074/jbc.R116.760884
54. Qian CS, Li LJ, Huang HW, Yang HF, Wu DP. MYC-Regulated Lncrna NEAT1 Promotes B Cell Proliferation and Lymphomagenesis Via the miR-34b-5p-GLI1 Pathway in Diffuse Large B-Cell Lymphoma. Cancer Cell Int (2020) 20:87. doi: 10.1186/s12935-020-1158-6
55. Mumtaz PT, Bhat SA, Ahmad SM, Dar MA, Ahmed R, Urwat U, et al. LncRNAs and Immunity: Watchdogs for Host Pathogen Interactions. Biol Proced Online (2017) 19:3. doi: 10.1186/s12575-017-0052-7
56. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding Long Noncoding RNA lnc-DC Controls Human Dendritic Cell Differentiation. Science (2014) 344(6181):310–3. doi: 10.1126/science.1251456
57. Martin JD, Cabral H, Stylianopoulos T, Jain RK. Improving Cancer Immunotherapy Using Nanomedicines: Progress, Opportunities and Challenges. Nat Rev Clin Oncol (2020) 17(4):251–66. doi: 10.1038/s41571-41019-40308-z
58. Tang S, Ning Q, Yang L, Mo Z, Tang S. Mechanisms of Immune Escape in the Cancer Immune Cycle. Int Immunopharmacol (2020) 86:106700. doi: 10.1016/j.intimp.2020.106700
59. Simiczyjew A, Dratkiewicz E, Mazurkiewicz J, Ziętek M, Matkowski R, Nowak D. The Influence of Tumor Microenvironment on Immune Escape of Melanoma. Int J Mol Sci (2020) 21(21). doi: 10.3390/ijms21218359
60. Gao Y, Wang T, Li Y, Zhang Y, Yang R. Lnc-Chop Promotes Immunosuppressive Function of Myeloid-Derived Suppressor Cells in Tumor and Inflammatory Environments. J Immunol (2018) 200(8):2603–14. doi: 10.4049/jimmunol.1701721
61. Shang W, Tang Z, Gao Y, Qi H, Su X, Zhang Y, et al. Lncrna RNCR3 Promotes Chop Expression by Sponging miR-185-5p During MDSC Differentiation. Oncotarget (2017) 8(67):111754–69. doi: 10.18632/oncotarget.22906
62. Shang W, Gao Y, Tang Z, Zhang Y, Yang R. The Pseudogene Olfr29-Ps1 Promotes the Suppressive Function and Differentiation of Monocytic Mdscs. Cancer Immunol Res (2019) 7(5):813–27. doi: 10.1158/2326-6066.CIR-18-0443
63. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long Noncoding RNA Pvt1 Regulates the Immunosuppression Activity of Granulocytic Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. Mol Cancer (2019) 18(1):61. doi: 10.1186/s12943-019-0978-2
64. Zhou Q, Tang X, Tian X, Tian J, Zhang Y, Ma J, et al. Lncrna MALAT1 Negatively Regulates MDSCs in Patients With Lung Cancer. J Cancer (2018) 9(14):2436–42. doi: 10.7150/jca.24796
65. Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Long Non-Coding Rna HOXA Transcript Antisense Rna Myeloid-Specific 1-Hoxa1 Axis Downregulates the Immunosuppressive Activity of Myeloid-Derived Suppressor Cells in Lung Cancer. Front Immunol (2018) 9:473. doi: 10.3389/fimmu.2018.00473
66. Tian X, Ma J, Wang T, Tian J, Zheng Y, Peng R, et al. Long non-Coding RNA RUNXOR Accelerates MDSC-Mediated Immunosuppression in Lung Cancer. BMC Cancer (2018) 18(1):660. doi: 10.1186/s12885-018-4564-6
67. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/Ebpβ Negatively Regulates the Suppressive Function of Myeloid-Derived Suppressor Cells. Cancer Immunol Res (2018) 6(11):1352–63. doi: 10.1158/2326-6066.CIR-18-0108
68. Gao Y, Shang W, Zhang D, Zhang S, Zhang X, Zhang Y, et al. Lnc-C/Ebpβ Modulates Differentiation of MDSCs Through Downregulating Il4i1 With C/Ebpβ LIP and WDR5. Front Immunol (2019) 10:1661. doi: 10.3389/fimmu.2019.01661
69. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The Long Noncoding RNA lnc-EGFR Stimulates T-Regulatory Cells Differentiation Thus Promoting Hepatocellular Carcinoma Immune Evasion. Nat Commun (2017) 8:15129. doi: 10.1038/ncomms15129
70. Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C. Flicr, a Long Noncoding RNA, Modulates Foxp3 Expression and Autoimmunity. Proc Natl Acad Sci USA (2017) 114(17):E3472–80. doi: 10.1073/pnas.1700946114
71. Brajic A, Franckaert D, Burton O, Bornschein S, Calvanese AL, Demeyer S, et al. The Long non-Coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Front Immunol (2018) 9:1989. doi: 10.3389/fimmu.2018.01989
72. Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J, et al. Breast Cancer-Derived Exosomes Transmit Lncrna SNHG16 to Induce CD73+γδ1 Treg Cells. Signal Transduction Targeted Ther (2020) 5(1):41. doi: 10.1038/s41392-020-0129-7
73. Xiong G, Yang L, Chen Y, Fan Z. Linc-POU3F3 Promotes Cell Proliferation in Gastric Cancer Via Increasing T-Reg Distribution. Am J Trans Res (2015) 7(11):2262–9.
74. Wang J, Huang F, Shi Y, Zhang Q, Xu S, Yao Y, et al. Rp11-323N12.5 Promotes the Malignancy and Immunosuppression of Human Gastric Cancer by Increasing YAP1 Transcription. Gastric Cancer: Off J Int Gastric Cancer Assoc Jpn Gastric Cancer Assoc (2021) 24(1):85–102. doi: 10.1007/s10120-020-01099-9
75. Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long Non-coding Rna FENDRR Acts as a Mir-423-5p Sponge to Suppress the Treg-Mediated Immune Escape of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids (2019) 17:516–29. doi: 10.1016/j.omtn.2019.05.027
76. Li Z, Feng C, Guo J, Hu X, Xie D. Gnas-AS1/miR-4319/NECAB3 Axis Promotes Migration and Invasion of non-Small Cell Lung Cancer Cells by Altering Macrophage Polarization. Funct Integr Genomics (2020) 20(1):17–28. doi: 10.1007/s10142-019-00696-x
77. Liu SQ, Zhou ZY, Dong X, Guo L, Zhang KJ. Lncrna GNAS-AS1 Facilitates ER+ Breast Cancer Cells Progression by Promoting M2 Macrophage Polarization Via Regulating miR-433-3p/GATA3 Axis. Biosci Rep (2020) 40(7). doi: 10.1042/BSR20200626
78. Sun Y, Xu J. Tcf-4 Regulated Lncrna-XIST Promotes M2 Polarization of Macrophages and Is Associated With Lung Cancer. OncoTargets Ther (2019) 12:8055–62. doi: 10.2147/OTT.S210952
79. Zhou YX, Zhao W, Mao LW, Wang YL, Xia LQ, Cao M, et al. Long non-Coding RNA Nifk-AS1 Inhibits M2 Polarization of Macrophages in Endometrial Cancer Through Targeting Mir-146a. Int J Biochem Cell Biol (2018) 104:25–33. doi: 10.1016/j.biocel.2018.08.017
80. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-Coding RNA Cox-2 Prevents Immune Evasion and Metastasis of Hepatocellular Carcinoma by Altering M1/M2 Macrophage Polarization. J Cell Biochem (2018) 119(3):2951–63. doi: 10.1002/jcb.26509
81. Yin Z, Zhou Y, Ma T, Chen S, Shi N, Zou Y, et al. Down-Regulated Lncrna SBF2-AS1 in M2 Macrophage-Derived Exosomes Elevates miR-122-5p to Restrict XIAP, Thereby Limiting Pancreatic Cancer Development. J Cell Mol Med (2020) 24(9):5028–38. doi: 10.1111/jcmm.15125
82. Liu J, Ding D, Jiang Z, Du T, Liu J, Kong Z. Long non-Coding RNA CCAT1/Mir-148a/Pkcζ Prevents Cell Migration of Prostate Cancer by Altering Macrophage Polarization. Prostate (2019) 79(1):105–12. doi: 10.1002/pros.23716
83. Zhou L, Tian Y, Guo F, Yu B, Li J, Xu H, et al. LincRNA-p21 Knockdown Reversed Tumor-Associated Macrophages Function by Promoting MDM2 to Antagonize* p53 Activation and Alleviate Breast Cancer Development. Cancer Immunol Immunother: CII (2020) 69(5):835–46. doi: 10.1007/s00262-020-02511-0
84. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. Lncrna BCRT1 Promotes Breast Cancer Progression by Targeting miR-1303/PTBP3 Axis. Mol Cancer (2020) 19(1):85. doi: 10.1186/s12943-020-01206-5
85. Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, et al. Long Noncoding RNA LINC00662 Promotes M2 Macrophage Polarization and Hepatocellular Carcinoma Progression Via Activating Wnt/β-Catenin Signaling. Mol Oncol (2020) 14(2):462–83. doi: 10.1002/1878-0261.12606
86. Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, et al. Lncrna-MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor-Associated Macrophage Fgf2 Protein Secretion. J Cell Biochem (2017) 118(12):4821–30. doi: 10.1002/jcb.26153
87. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-Coding RNA MALAT1 Promotes Angiogenesis and Immunosuppressive Properties of HCC Cells by Sponging Mir-140. Am J Physiol Cell Physiol (2020) 318(3):C649–63. doi: 10.1152/ajpcell.00510.2018
88. Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA Tuc339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer (2013) 4(7-8):261–72. doi: 10.1177/1947601913499020
89. Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC-Derived Exosomal lncRNA Tuc339. Int J Mol Sci (2018) 19(10). doi: 10.3390/ijms19102958
90. Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, et al. Lncrna RPPH1 Promotes Colorectal Cancer Metastasis by Interacting With TUBB3 and by Promoting Exosomes-Mediated Macrophage M2 Polarization. Cell Death Dis (2019) 10(11):829. doi: 10.1038/s41419-019-2077-0
91. Cao J, Dong R, Jiang L, Gong Y, Yuan M, You J, et al. Lncrna-MM2P Identified as a Modulator of Macrophage M2 Polarization. Cancer Immunol Res (2019) 7(2):292–305. doi: 10.1158/2326-6066.CIR-18-0145
92. Yang D, Liu K, Fan L, Liang W, Xu T, Jiang W, et al. Lncrna RP11-361F15.2 Promotes Osteosarcoma Tumorigenesis by Inhibiting M2-Like Polarization of Tumor-Associated Macrophages of CPEB4. Cancer Lett (2020) 473:33–49. doi: 10.1016/j.canlet.2019.12.041
93. Xie C, Guo Y, Lou S. Lncrna ANCR Promotes Invasion and Migration of Gastric Cancer by Regulating Foxo1 Expression to Inhibit Macrophage M1 Polarization. Digestive Dis Sci (2020) 65(10):2863–72. doi: 10.1007/s10620-019-06019-1
94. Liu A, Liu L, Lu H. Lncrna XIST Facilitates Proliferation and Epithelial-Mesenchymal Transition of Colorectal Cancer Cells Through Targeting miR-486-5p and Promoting Neuropilin-2. J Cell Physiol (2019) 234(8):13747–61. doi: 10.1002/jcp.28054
95. Zhang Y, Feng J, Fu H, Liu C, Yu Z, Sun Y, et al. Coagulation Factor X Regulated by CASC2c Recruited Macrophages and Induced M2 Polarization in Glioblastoma Multiforme. Front Immunol (2018) 9:1557. doi: 10.3389/fimmu.2018.01557
96. Wang B, Li X, Hu W, Zhou Y, Din Y. Silencing of Lncrna SNHG20 Delays the Progression of Nonalcoholic Fatty Liver Disease to Hepatocellular Carcinoma Via Regulating Liver Kupffer Cells Polarization. IUBMB Life (2019) 71(12):1952–61. doi: 10.1002/iub.2137
97. Zhang H, Yu Y, Wang J, Han Y, Ren T, Huang Y, et al. Macrophages-Derived Exosomal LncRNA LIFR-AS1 Promotes Osteosarcoma Cell Progression Via miR-29a/NFIA Axis. Cancer Cell Int (2021) 21(1):192. doi: 10.1186/s12935-021-01893-0
98. Liu J, Zhang X, Chen K, Cheng Y, Liu S, Xia M, et al. Ccr7 Chemokine Receptor-Inducible lnc-Dpf3 Restrains Dendritic Cell Migration by Inhibiting Hif-1α-Mediated Glycolysis. Immunity (2019) 50(3):600–615.e615. doi: 10.1016/j.immuni.2019.01.021
99. Zhang W, Yang M, Yu L, Hu Y, Deng Y, Liu Y, et al. Long non-Coding RNA lnc-DC in Dendritic Cells Regulates Trophoblast Invasion Via P-STAT3-Mediated TIMP/MMP Expression. Am J Reprod Immunol (New York NY: 1989) (2020) 83(6):e13239. doi: 10.1111/aji.13239
100. Zhuang L, Tian J, Zhang X, Wang H, Huang C. Lnc-DC Regulates Cellular Turnover and the HBV-Induced Immune Response by TLR9/STAT3 Signaling in Dendritic Cells. Cell Mol Biol Lett (2018) 23:43. doi: 10.1186/s11658-018-0108-y
101. Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X, et al. Knockdown of NEAT1 Induces Tolerogenic Phenotype in Dendritic Cells by Inhibiting Activation of NLRP3 Inflammasome. Theranostics (2019) 9(12):3425–42. doi: 10.7150/thno.33178
102. Xin J, Li J, Feng Y, Wang L, Zhang Y, Yang R. Downregulation of Long Noncoding RNA HOTAIRM1 Promotes Monocyte/Dendritic Cell Differentiation Through Competitively Binding to Endogenous Mir-3960. OncoTargets Ther (2017) 10:1307–15. doi: 10.2147/OTT.S124201
103. Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu YJ, et al. Chemokine (C-C Motif) Ligand 5 is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression Through non-Coding RNA Malat-1. J Cell Physiol (2015) 230(8):1883–94. doi: 10.1002/jcp.24918
104. Zhang R, Ni F, Fu B, Wu Y, Sun R, Tian Z, et al. A Long Noncoding RNA Positively Regulates CD56 in Human Natural Killer Cells. Oncotarget (2016) 7(45):72546–58. doi: 10.18632/oncotarget.12466
105. Fang P, Xiang L, Chen W, Li S, Huang S, Li J, et al. Lncrna GAS5 Enhanced the Killing Effect of NK Cell on Liver Cancer Through Regulating Mir-544/RUNX3. Innate Immun (2019) 25(2):99–109. doi: 10.1177/1753425919827632
106. Wei MF, Gu ZS, Zheng LL, Zhao MX, Wang XJ. Long non-Coding RNA GAS5 Promotes Natural Killer Cell Cytotoxicity Against Gastric Cancer by Regulating Mir-18a. Neoplasma (2020) 67(5):1085–93. doi: 10.4149/neo_2020_191014N1034
107. Stein N, Berhani O, Schmiedel D, Duev-Cohen A, Seidel E, Kol I, et al. Ifng-As1 Enhances Interferon Gamma Production in Human Natural Killer Cells. iScience (2019) 11:466–73. doi: 10.1016/j.isci.2018.12.034
108. Li S, Zhu A, Ren K, Li S, Chen L. Ifnβ-Induced Exosomal linc-EPHA6-1 Promotes Cytotoxicity of NK Cells by Acting as a ceRNA for hsa-miR-4485-5p to Up-Regulate NKp46 Expression. Life Sci (2020) 257:118064. doi: 10.1016/j.lfs.2020.118064
109. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-Coding RNA lnc-Tim3 Exacerbates CD8 T Cell Exhaustion Via Binding to Tim-3 and Inducing Nuclear Translocation of Bat3 in HCC. Cell Death Dis (2018) 9(5):478. doi: 10.1038/s41419-018-0528-7
110. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of Lncrna NEAT1 Enhances the Antitumor Activity of CD8(+)T Cells Against Hepatocellular Carcinoma Via Regulating Mir-155/Tim-3. Int J Biochem Cell Biol (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
111. Wu K, Zhao Z, Liu K, Zhang J, Li G, Wang L. Long Noncoding RNA lnc-Sox5 Modulates CRC Tumorigenesis by Unbalancing Tumor Microenvironment. Cell Cycle (Georgetown Tex) (2017) 16(13):1295–301. doi: 10.1080/15384101.2017.1317416
112. Veglia F, Perego M, Gabrilovich D. Myeloid-Derived Suppressor Cells Coming of Age. Nat Immunol (2018) 19(2):108–19. doi: 10.1038/s41590-017-0022-x
113. Zhang S, Ma X, Zhu C, Liu L, Wang G, Yuan X. The Role of Myeloid-Derived Suppressor Cells in Patients With Solid Tumors: A Meta-Analysis. PloS One (2016) 11(10):e0164514. doi: 10.1371/journal.pone.0164514
114. Tesi RJ. MDSC; the Most Important Cell You Have Never Heard of. Trends Pharmacol Sci (2019) 40(1):4–7. doi: 10.1016/j.tips.2018.10.008
115. Thevenot PT, Sierra RA, Raber PL, Al-Khami AA, Trillo-Tinoco J, Zarreii P, et al. The Stress-Response Sensor Chop Regulates the Function and Accumulation of Myeloid-Derived Suppressor Cells in Tumors. Immunity (2014) 41(3):389–401. doi: 10.1016/j.immuni.2014.08.015
116. Delano MJ, Scumpia PO, Weinstein JS, Coco D, Nagaraj S, Kelly-Scumpia KM, et al. MyD88-dependent Expansion of an Immature GR-1(+)CD11b(+) Population Induces T Cell Suppression and Th2 Polarization in Sepsis. J Exp Med (2007) 204(6):1463–74. doi: 10.1084/jem.20062602
117. Ohue Y, Nishikawa H. Regulatory T (Treg) Cells in Cancer: Can Treg Cells be a New Therapeutic Target? Cancer Sci (2019) 110(7):2080–9. doi: 10.1111/cas.14069
118. Viehl CT, Moore TT, Liyanage UK, Frey DM, Ehlers JP, Eberlein TJ, et al. Depletion of CD4+CD25+ Regulatory T Cells Promotes a Tumor-Specific Immune Response in Pancreas Cancer-Bearing Mice. Ann Surg Oncol (2006) 13(9):1252–8. doi: 10.1245/s10434-006-9015-y
119. Huang Y, Liu K, Li Q, Yao Y, Wang Y. Exosomes Function in Tumor Immune Microenvironment. Adv Exp Med Biol (2018) 1056:109–22. doi: 10.1007/978-3-319-74470-4_7
120. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-Associated Macrophages as Treatment Targets in Oncology. Nat Rev Clin Oncol (2017) 14(7):399–416. doi: 10.1038/nrclinonc.2016.217
121. Sánchez-Paulete AR, Teijeira Á, Quetglas JI, Rodríguez-Ruiz ME, Sánchez-Arráez Á, Labiano S, et al. Intratumoral Immunotherapy With XCL1 and Sflt3l Encoded in Recombinant Semliki Forest Virus-Derived Vectors Fosters Dendritic Cell-Mediated T-cell Cross-Priming. Cancer Res (2018) 78(23):6643–54. doi: 10.1158/0008-5472.CAN-18-0933
122. Giovanelli P, Sandoval TA, Cubillos-Ruiz JR. Dendritic Cell Metabolism and Function in Tumors. Trends Immunol (2019) 40(8):699–718. doi: 10.1016/j.it.2019.06.004
123. Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, et al. A Conserved Dendritic-Cell Regulatory Program Limits Antitumour Immunity. Nature (2020) 580(7802):257–62. doi: 10.1038/s41586-020-2134-y
124. Shimasaki N, Jain A, Campana D. NK Cells for Cancer Immunotherapy. Nat Rev Drug Discov (2020) 19(3):200–18. doi: 10.1038/s41573-019-0052-1
125. Kato K, Cho BC, Takahashi M, Okada M, Lin CY, Chin K, et al. Nivolumab Versus Chemotherapy in Patients With Advanced Oesophageal Squamous Cell Carcinoma Refractory or Intolerant to Previous Chemotherapy (ATTRACTION-3): A Multicentre, Randomised, Open-Label, Phase 3 Trial. Lancet Oncol (2019) 20(11):1506–17. doi: 10.1016/S1470-2045(19)30626-6
126. Motzer RJ, Rini BI, McDermott DF, Arén Frontera O, Hammers HJ, Carducci MA, et al. Nivolumab Plus Ipilimumab Versus Sunitinib in First-Line Treatment for Advanced Renal Cell Carcinoma: Extended Follow-Up of Efficacy and Safety Results From a Randomised, Controlled, Phase 3 Trial. Lancet Oncol (2019) 20(10):1370–85. doi: 10.1016/S1470-2045(19)30413-9
127. Hellmann MD, Paz-Ares L, Bernabe Caro R, Zurawski B, Kim SW, Carcereny Costa E, et al. Nivolumab Plus Ipilimumab in Advanced non-Small-Cell Lung Cancer. N Engl J Med (2019) 381(21):2020–31. doi: 10.1056/NEJMoa1910231
128. Schoenfeld AJ, Hellmann MD. Acquired Resistance to Immune Checkpoint Inhibitors. Cancer Cell (2020) 37(4):443–55. doi: 10.1016/j.ccell.2020.03.017
129. Saleh R, Elkord E. Acquired Resistance to Cancer Immunotherapy: Role of Tumor-Mediated Immunosuppression. Semin Cancer Biol (2020) 65:13–27. doi: 10.1016/j.semcancer.2019.07.017
130. Fares CM, Van Allen EM, Drake CG, Allison JP, Hu-Lieskovan S. Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Annu Meeting (2019) 39:147–64. doi: 10.1200/EDBK_240837
131. Wang QM, Lian GY, Song Y, Huang YF, Gong Y. Lncrna MALAT1 Promotes Tumorigenesis and Immune Escape of Diffuse Large B Cell Lymphoma by Sponging Mir-195. Life Sci (2019) 231:116335. doi: 10.1016/j.lfs.2019.03.040
132. Wei S, Wang K, Huang X, Zhao Z, Zhao Z. Lncrna MALAT1 Contributes to non-Small Cell Lung Cancer Progression Via Modulating miR-200a-3p/programmed Death-Ligand 1 Axis. Int J Immunopathol Pharmacol (2019) 33:2058738419859699. doi: 10.1177/2058738419859699
133. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. Lncrna SNHG14/miR-5590-3p/ZEB1 Positive Feedback Loop Promoted Diffuse Large B Cell Lymphoma Progression and Immune Evasion Through Regulating PD-1/PD-L1 Checkpoint. Cell Death Dis (2019) 10(10):731. doi: 10.1038/s41419-019-1886-5
134. Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long Noncoding RNA LINC00473 Drives the Progression of Pancreatic Cancer Via Upregulating Programmed Death-Ligand 1 by Sponging Microrna-195-5p. J Cell Physiol (2019) 234(12):23176–89. doi: 10.1002/jcp.28884
135. Kathuria H, Millien G, McNally L, Gower AC, Tagne JB, Cao Y, et al. Nkx2-1-AS1 Negatively Regulates CD274/PD-L1, Cell-Cell Interaction Genes, and Limits Human Lung Carcinoma Cell Migration. Sci Rep (2018) 8(1):14418. doi: 10.1038/s41598-018-32793-5
136. Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and Clinical Development of siRNA-Based Therapeutics. Adv Drug Deliv Rev (2015) 87:108–19. doi: 10.1016/j.addr.2015.01.007
137. Abulwerdi FA, Xu W, Ageeli AA, Yonkunas MJ, Arun G, Nam H, et al. Selective Small-Molecule Targeting of a Triple Helix Encoded by the Long Noncoding RNA, Malat1. ACS Chem Biol (2019) 14(2):223–35. doi: 10.1021/acschembio.8b00807
138. Lee JS, Mendell JT. Antisense-Mediated Transcript Knockdown Triggers Premature Transcription Termination. Mol Cell (2020) 77(5):1044–1054.e1043. doi: 10.1016/j.molcel.2019.12.011
139. Lennox KA, Behlke MA. Tips for Successful Lncrna Knockdown Using Gapmers. Methods Mol Biol (Clifton NJ) (2020) 2176:121–40. doi: 10.1007/978-1-0716-0771-8_9
140. Watanabe C, Cuellar TL, Haley B. Quantitative Evaluation of First, Second, and Third Generation Hairpin Systems Reveals the Limit of Mammalian Vector-Based Rnai. RNA Biol (2016) 13(1):25–33. doi: 10.1080/15476286.2015.1128062
141. Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. Crispri-Based Genome-Scale Identification of Functional Long Noncoding RNA Loci in Human Cells. Science (2017) 355(6320). doi: 10.1126/science.aah7111
142. Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary Classification of CRISPR-Cas Systems: A Burst of Class 2 and Derived Variants. Nat Rev Microbiol (2020) 18(2):67–83. doi: 10.1038/s41579-019-0299-x
143. He C, Jaffar Ali D, Xu H, Kumaravel S, Si K, Li Y, et al. Epithelial Cell -Derived Microvesicles: A Safe Delivery Platform of CRISPR/Cas9 Conferring Synergistic Anti-Tumor Effect With Sorafenib. Exp Cell Res (2020) 392(2):112040. doi: 10.1016/j.yexcr.2020.112040
144. Lin YX, Wang Y, Blake S, Yu M, Mei L, Wang H, et al. RNA Nanotechnology-Mediated Cancer Immunotherapy. Theranostics (2020) 10(1):281–99. doi: 10.7150/thno.35568
145. Allen TM, Cullis PR. Liposomal Drug Delivery Systems: From Concept to Clinical Applications. Adv Drug Deliv Rev (2013) 65(1):36–48. doi: 10.1016/j.addr.2012.09.037
146. Samal SK, Dash M, Van Vlierberghe S, Kaplan DL, Chiellini E, van Blitterswijk C, et al. Cationic Polymers and Their Therapeutic Potential. Chem Soc Rev (2012) 41(21):7147–94. doi: 10.1039/c2cs35094g
147. Zhang S, Zhao B, Jiang H, Wang B, Ma B. Cationic Lipids and Polymers Mediated Vectors for Delivery of Sirna. J Controlled Release: Off J Controlled Release Soc (2007) 123(1):1–10. doi: 10.1016/j.jconrel.2007.07.016
148. Lin YX, Gao YJ, Wang Y, Qiao ZY, Fan G, Qiao SL, et al. Ph-Sensitive Polymeric Nanoparticles With Gold(I) Compound Payloads Synergistically Induce Cancer Cell Death Through Modulation of Autophagy. Mol Pharm (2015) 12(8):2869–78. doi: 10.1021/acs.molpharmaceut.5b00060
149. Shen J, Zhang W, Qi R, Mao ZW, Shen H. Engineering Functional Inorganic-Organic Hybrid Systems: Advances in siRNA Therapeutics. Chem Soc Rev (2018) 47(6):1969–95. doi: 10.1039/C7CS00479F
150. Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer Nanomedicine: Progress, Challenges and Opportunities. Nat Rev Cancer (2017) 17(1):20–37. doi: 10.1038/nrc.2016.108
151. Gong N, Teng X, Li J, Liang XJ. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting Lncrna MALAT1 Inhibits Cancer Metastasis. ACS Appl Mater Interfaces (2019) 11(1):37–42. doi: 10.1021/acsami.8b18288
152. Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X, Jiang H, et al. Systemic Delivery of Tumor-Targeting Sirna Nanoparticles Against an Oncogenic Lncrna Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjugate Chem (2019) 30(3):907–19. doi: 10.1021/acs.bioconjchem.9b00028
153. Wang T, Li P, Wan T, Tu B, Li J, Huang F. TIGIT/PVR and LncRNA ANRIL Dual-Targetable PAMAM Polymeric Nanoparticles Efficiently Inhibited the Hepatoma Carcinoma by Combination of Immunotherapy and Gene Therapy. J Drug Target (2021) 1–9. doi: 10.1080/1061186X.2021.1879088
154. Kalluri R, LeBleu VS. The Biology, Function, and Biomedical Applications of Exosomes. Science (2020) 367(6478). doi: 10.1126/science.aau6977
155. Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, et al. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm Res (2015) 32(6):2003–14. doi: 10.1007/s11095-014-1593-y
156. Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes Provide a Protective and Enriched Source of miRNA for Biomarker Profiling Compared to Intracellular and Cell-Free Blood. J Extracell Vesicles (2014) 3. doi: 10.3402/jev.v3.23743
157. Mentkowski KI, Snitzer JD, Rusnak S, Lang JK. Therapeutic Potential of Engineered Extracellular Vesicles. AAPS J (2018) 20(3):50. doi: 10.1208/s12248-018-0211-z
158. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically Injected Exosomes Targeted to EGFR Deliver Antitumor microRNA to Breast Cancer Cells. Mol Ther: J Am Soc Gene Ther (2013) 21(1):185–91. doi: 10.1038/mt.2012.180
159. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature (2017) 546(7659):498–503. doi: 10.1038/nature22341
160. Buscail L. Pancreatic Cancer: Exosomes for Targeting KRAS in the Treatment of Pancreatic Cancer. Nat Rev Gastroenterol Hepatol (2017) 14(11):636–8. doi: 10.1038/nrgastro.2017.113
161. Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered Exosomes for Targeted Transfer of siRNA to HER2 Positive Breast Cancer Cells. Appl Biochem Biotechnol (2019) 187(1):352–64. doi: 10.1007/s12010-018-2813-4
162. Han M, Gu Y, Lu P, Li J, Cao H, Li X, et al. Exosome-Mediated Lncrna AFAP1-AS1 Promotes Trastuzumab Resistance Through Binding With AUF1 and Activating ERBB2 Translation. Mol Cancer (2020) 19(1):26. doi: 10.1186/s12943-020-1145-5
163. Tu Z, Schmoellerl J, Mariani O, Zheng Y, Hu Y, Vincent-Salomon A, et al. The LINC01119-SOCS5 Axis as a Critical Theranostic in Triple-Negative Breast Cancer. NPJ Breast Cancer (2021) 7(1):69. doi: 10.1038/s41523-021-00259-z
164. Di Martino MT, Riillo C, Scionti F, Grillone K, Polerà N, Caracciolo D, et al. miRNAs and lncRNAs as Novel Therapeutic Targets to Improve Cancer Immunotherapy. Cancers (Basel) (2021) 13(7). doi: 10.3390/cancers13071587
165. Yu Y, Zhang W, Li A, Chen Y, Ou Q, He Z, et al. Association of Long Noncoding RNA Biomarkers With Clinical Immune Subtype and Prediction of Immunotherapy Response in Patients With Cancer. JAMA Network Open (2020) 3(4):e202149. doi: 10.1001/jamanetworkopen.2020.2149
Keywords: LncRNA, tumor microenvironment, immunosuppression, immune escape, therapeutic target
Citation: Pi Y-N, Qi W-C, Xia B-R, Lou G and Jin W-L (2021) Long Non-Coding RNAs in the Tumor Immune Microenvironment: Biological Properties and Therapeutic Potential. Front. Immunol. 12:697083. doi: 10.3389/fimmu.2021.697083
Received: 19 April 2021; Accepted: 23 June 2021;
Published: 06 July 2021.
Edited by:
Susanna Mandruzzato, University Hospital of Padua, ItalyReviewed by:
Tibor Bakacs, Alfred Renyi Institute of Mathematics, HungaryChandrasekhar Kanduri, University of Gothenburg, Sweden
Copyright © 2021 Pi, Qi, Xia, Lou and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bai-Rong Xia, eGlhYmFpcm9uZ0B1c3RjLmVkdS5jbg==; Ge Lou, bG91Z2VAZW1zLmhyYm11LmVkdS5jbg==; Wei-Lin Jin, bGR5eV9qaW53bEBsenUuZWR1LmNu; d2VpbGluamluQHlhaG9vLmNvbQ==
 Ya-Nan Pi1
Ya-Nan Pi1 Bai-Rong Xia
Bai-Rong Xia Ge Lou
Ge Lou Wei-Lin Jin
Wei-Lin Jin