
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 18 June 2021
Sec. Alloimmunity and Transplantation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.692225
This article is part of the Research TopicNovel and improved methods for the prevention and treatment of Graft-Versus-Host Disease (GVHD)View all 13 articles
 Vitor Heidrich1,2†
Vitor Heidrich1,2† Julia S. Bruno1†
Julia S. Bruno1† Franciele H. Knebel1†
Franciele H. Knebel1† Vinícius C. de Molla3,4†
Vinícius C. de Molla3,4† Wanessa Miranda-Silva1
Wanessa Miranda-Silva1 Paula F. Asprino1
Paula F. Asprino1 Luciana Tucunduva3
Luciana Tucunduva3 Vanderson Rocha3,5,6
Vanderson Rocha3,5,6 Yana Novis3
Yana Novis3 Celso Arrais-Rodrigues3,4
Celso Arrais-Rodrigues3,4 Eduardo R. Fregnani3
Eduardo R. Fregnani3 Anamaria A. Camargo1*
Anamaria A. Camargo1*Acute graft-versus-host disease (aGVHD) is one of the major causes of death after allogeneic hematopoietic stem cell transplantation (allo-HSCT). Recently, aGVHD onset was linked to intestinal microbiota (IM) dysbiosis. However, other bacterial-rich gastrointestinal sites, such as the mouth, which hosts several distinctive microbiotas, may also impact the risk of GVHD. The dental biofilm microbiota (DBM) is highly diverse and, like the IM, interacts with host cells and modulates immune homeostasis. We characterized changes in the DBM of patients during allo-HSCT and evaluated whether the DBM could be associated with the risk of aGVHD. DBM dysbiosis during allo-HSCT was marked by a gradual loss of bacterial diversity and changes in DBM genera composition, with commensal genera reductions and potentially pathogenic bacteria overgrowths. High Streptococcus and high Corynebacterium relative abundance at preconditioning were associated with a higher risk of aGVHD (67% vs. 33%; HR = 2.89, P = 0.04 and 73% vs. 37%; HR = 2.74, P = 0.04, respectively), while high Veillonella relative abundance was associated with a lower risk of aGVHD (27% vs. 73%; HR = 0.24, P < 0.01). Enterococcus faecalis bloom during allo-HSCT was observed in 17% of allo-HSCT recipients and was associated with a higher risk of aGVHD (100% vs. 40%; HR = 4.07, P < 0.001) and severe aGVHD (60% vs. 12%; HR = 6.82, P = 0.01). To the best of our knowledge, this is the first study demonstrating that DBM dysbiosis is associated with the aGVHD risk after allo-HSCT.
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the only curative treatment for several hematologic diseases. However, allo-HSCT recipients may experience potentially fatal complications, such as infections and graft-versus-host disease (GVHD) (1).
Acute GVHD (aGVHD) is a clinical syndrome characterized by maculopapular rash, hyperbilirubinemia, anorexia, diarrhea and abdominal pain (2). The incidence of aGVHD grade II-IV is 30-40% at day 100 (3). During transplantation, chemotherapy, radiotherapy, and infection can damage host cells, releasing sterile damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) into the extracellular milieu. DAMPs and PAMPs activate donor T cells leading to a proinflammatory state. Simultaneously, donor regulatory T cells, myeloid-derived suppressor cells and tolerogenic dendritic cells are activated, counterbalancing the inflammation as an anti-inflammatory response. An imbalance in these events towards the proinflammatory state may result in aGVHD (4).
In addition to the graft source and the intensity of the conditioning regimen (4), the intestinal microbiota (IM) composition was shown to be associated with the risk and intensity of aGVHD. Loss of IM diversity has been observed during the pre- and post-transplantation period (5), and low microbiota diversity at the time of stem cell engraftment has been associated with a higher risk of severe aGVHD (5) and transplant-related death (6).
Two non-exclusive ecological events can explain the link between loss of bacterial diversity and aGVHD risk: absence or loss of protective commensal bacterial species and sudden expansion (also known as bloom) of opportunistic pathogenic bacteria. Both events have been independently linked to aGVHD development. For instance, a higher abundance of commensal bacteria from the Blautia genus in the IM after allo-HSCT has been associated with reduced GVHD-related mortality and improved overall survival (7, 8). On the other hand, a shift in IM leading to the dominance of bacteria from the Enterococcus genus occurs more prominently in allo-HSCT recipients developing aGVHD (9), and it is associated with increased GVHD-related mortality (10).
Recent studies have shown that bacteria inhabiting the oral cavity can translocate to the gut (11) and drive IM dysbiosis (12). However, direct evaluation of the effect of allo-HSCT on the oral microbiota (OM) and the influence of OM dysbiosis on aGVHD risk have not been performed. To further understand the impact of gastrointestinal bacterial communities on aGVHD development following allo-HSCT, it would be crucial to extend the scope of these analyses to the OM.
The OM comprises over 700 bacterial species that stick to surfaces of the mouth, forming biofilms (13). The dental biofilm microbiota (DBM), in particular, is among the richest and most diverse and, like the IM, interacts with host cells and modulates immune homeostasis (14). In this study, we characterized changes of the DBM in patients during allo-HSCT and evaluated whether alterations in DBM diversity and composition could be associated with the risk of aGVHD.
Supragingival biofilm samples were collected from patients who underwent allo-HSCT. Samples were collected with sterile swabs at three phases during allo-HSCT: before the conditioning regimen (preconditioning), at aplasia and at engraftment. All patients were requested not to perform oral hygiene for at least 6h before sample collection. All patients were examined by an oral medicine specialist for potential infections and followed the same protocol for oral mucositis prophylaxis with photobiomodulation and oral hygiene with fluoride toothpaste and 0.12% chlorhexidine mouthwash. Informed consent was obtained from all participants prior to sample collection. The study was approved by the Institutional Ethics Committee (Protocol #1.414.217), in line with the Declaration of Helsinki.
Bacterial cells were recovered from swabs by vortexing in TE buffer supplemented with PureLink RNAse A (Thermo Fisher Scientific, Waltham, MA, USA). DNA was extracted using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Next, 12.5 ng of total DNA and pre-validated primers (15) were used to amplify 16S rRNA hypervariable regions V3–V4. Amplicons were sequenced as described elsewhere (16) on the MiSeq platform (Illumina, San Diego, CA, USA).
Reads were demultiplexed and primer sequences were removed using the MiSeq Reporter software. Read processing was carried out within the QIIME 2 (Quantitative Insights Into Microbial Ecology 2) framework (17). Briefly, forward and reverse sequences were filtered for quality and bimeras, denoised, and merged into consensus sequences with the DADA2 pipeline (18), generating unique amplicon sequencing variants (ASVs). ASVs were further filtered for chimeric sequences using the SILVA database (19) and UCHIME (20), resulting in a total of 6 434 516 high-quality 16S rRNA sequences, with the median number of sequences obtained per sample being 58 867 (range: 2 153 - 240 734). Afterwards, ASVs were taxonomically assigned using the SILVA database and VSEARCH tool (21).
As determined by per sample alpha diversity rarefaction curves, <12 500 reads samples were considered defective and excluded. To adjust for differences in library sizes, the remaining samples were rarefied to 14 157 reads before calculating alpha diversity indexes (Shannon and Gini-Simpson indexes and the number of observed ASVs as a proxy for species richness) with the QIIME 2 q2-diversity plugin. Alpha diversity across transplantation phases was compared with the Mann-Whitney U test. The relative abundance of each genus was calculated with the QIIME 2 q2-taxa plugin. Differentially abundant genera across transplantation phases were identified using ANCOM (22). ANCOM W represents the proportion of null hypotheses rejected when subtesting the differential abundance of a genus normalized by the abundance of each one of the genera in the dataset. W > 0.7 was considered as statistically significant. Cumulative incidence (CMI) rates for aGVHD (grade II to IV) and severe aGVHD (grade III and IV) were calculated with death as a competing event. Relative risks for developing aGVHD and severe aGVHD were estimated using the Fine-Gray risk regression model and adjusted for graft source and intensity of the conditioning regimen. Relative risks are presented as hazard ratios with 95% CIs and two-tailed P-values. R software (version 3.6.2) and the statistical package cmprsk (version 2.2.9) were used for statistical analyses.
A total of 30 patients who underwent allo-HSCT for hematologic disorders at Hospital Sírio-Libanês between January 2016 and April 2018 were consecutively enrolled in our study. Patient clinical characteristics are summarized in Table 1. The most common underlying disease was acute leukemia (60%). The majority of patients received reduced-intensity conditioning (60%) and grafts from peripheral blood (67%).
The standard antimicrobial prophylaxis in our institution included oral levofloxacin, antiviral prophylaxis with acyclovir or valacyclovir, and antifungal prophylaxis with echinocandins or azoles according to the patient’s risk of fungal infection. In addition, cephalosporin and antibiotics for anaerobic bacteria (metronidazole, meropenem or piperacillin/tazobactam) were administered to 70% and 57% of patients, respectively.
aGVHD was diagnosed and classified according to the Glucksberg grading system (23). Fifteen patients developed grade II-IV aGVHD and, of those, 6 developed severe aGVHD (grade III-IV). None of this cohort’s clinical characteristics, including graft source, conditioning regimen, GVHD prophylaxis and antibiotics usage, was significantly associated with the risk of aGVHD (Table S1).
Supragingival biofilm samples were collected for bacterial profiling at preconditioning, aplasia, and engraftment to characterize changes in DBM during allo-HSCT. Three engraftment samples were excluded from downstream analyses due to insufficient high-quality reads.
DBM alpha diversity was assessed using the Shannon index. We observed a statistically significant decrease in DBM alpha diversity during allo-HSCT, with engraftment samples presenting the lowest overall bacterial diversity (median at each collection phase: 4.15, 3.39, and 2.75, respectively; Figure 1A). A similar decrease in alpha diversity was observed when using the Gini-Simpson index (Figure S1A) or the number of observed ASVs as a proxy for species richness (Figure S1B).
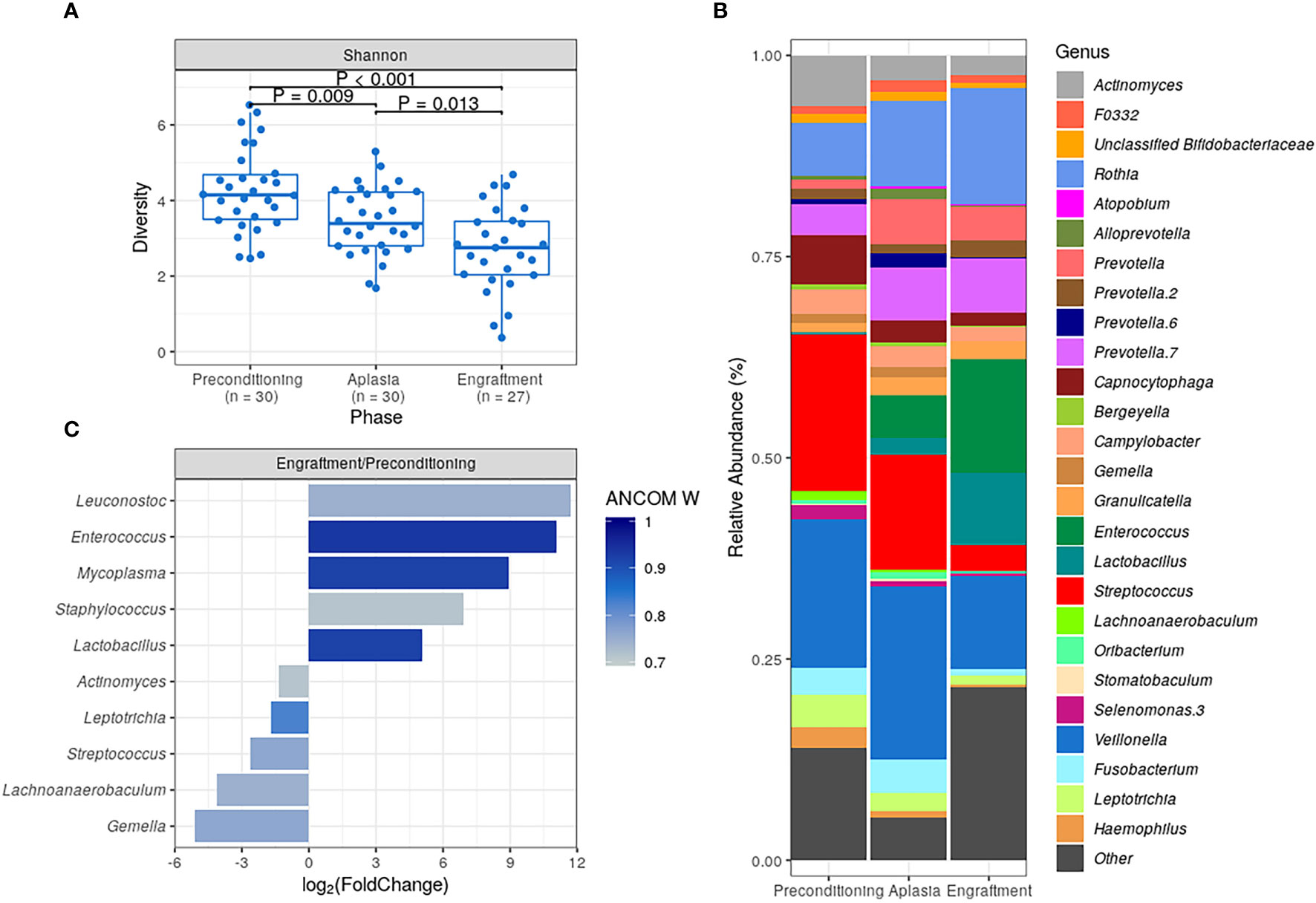
Figure 1 Characterization of dental biofilm microbiota (DBM) during allogeneic hematopoietic stem cell transplantation. (A) DBM alpha diversity (Shannon) boxplots at preconditioning (n = 30), aplasia (n = 30) and engraftment (n = 27). Mann-Whitney U test was used with the preconditioning as the reference for comparisons. The boxes highlight the median value and cover the 25th and 75th percentiles, with whiskers extending to the more extreme value within 1.5 times the length of the box. (B) Average DBM genera relative abundance composition across transplantation phases. Only genera with at least 0.1% relative abundance in at least 25% study samples are shown. Taxa are sorted based on taxonomic relatedness. (C) Significant genera relative abundance variations from preconditioning to engraftment according to ANCOM test (W > 0.7). Log2(Fold Change) for the average relative abundance variation (Engraftment/Preconditioning) is shown.
Marked changes in DBM genera composition were observed for all patients during allo-HSCT (Figure S2). As expected, several dental biofilm commensal genera were detected at a high average relative abundance at preconditioning, including Streptococcus (19.5%), Veillonella (18.4%), Actinomyces (6.3%), and Capnocytophaga (6.1%) (Figure 1B). However, their average relative abundance decreased during allo-HSCT. Likewise, we observed an increase in the average relative abundance of potentially pathogenic genera, such as Enterococcus and Lactobacillus (Figure 1B).
For a more quantitative assessment of DBM changes during allo-HSCT, we compared genera abundances at preconditioning and engraftment using the ANCOM test (Figure 1C). The most statistically significant differences in abundance were observed for Enterococcus, Lactobacillus, and Mycoplasma, confirming the expansion of these potentially pathogenic genera in DBM during allo-HSCT. We also observed statistically significant (although less pronounced in terms of relative abundance change) decreases in commensal genera (Figure 1C).
Patients were stratified into two equal-sized groups (high and low-diversity groups) by the entire cohort’s median alpha diversity value to evaluate the association between DBM diversity and aGVHD risk. Using the Shannon diversity index, DBM diversity showed no association with the risk of aGVHD at preconditioning, aplasia, or engraftment (Figures 2A–C and Table 2). Similar results were obtained when using the Gini-Simpson diversity index or the number of observed ASVs as a proxy for species richness (Figure S3).
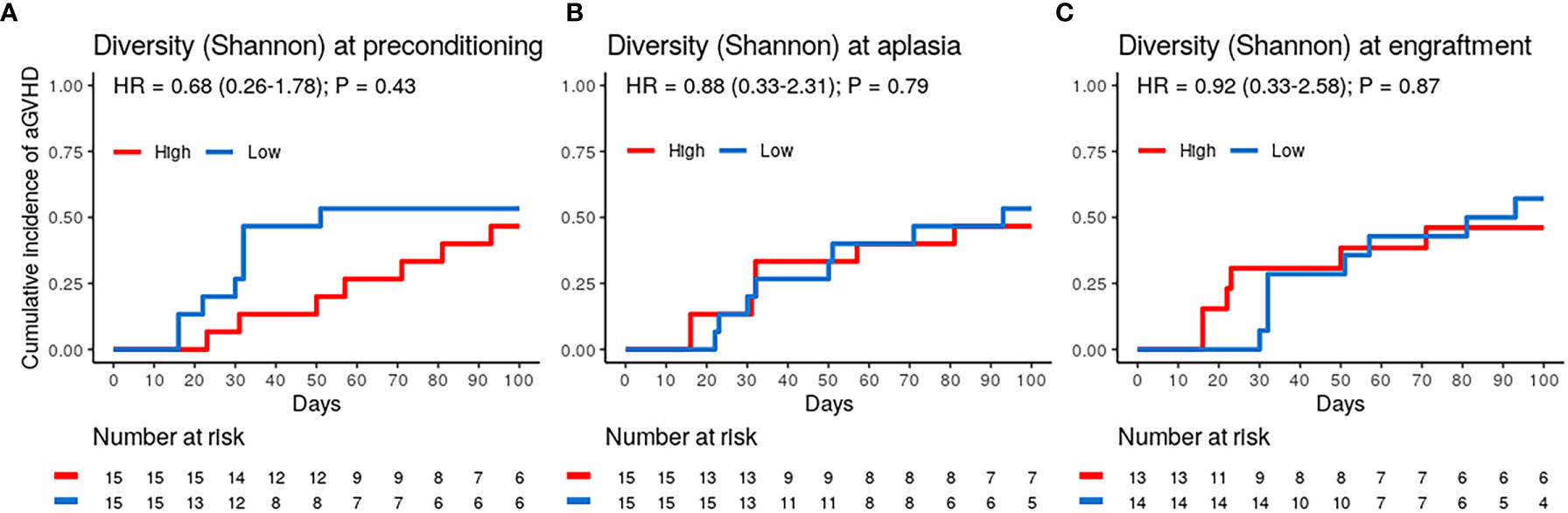
Figure 2 Dental biofilm microbiota alpha diversity is not associated with the risk of acute graft-versus-host disease (aGVHD). (A–C) Cumulative incidence of aGVHD with patients stratified by Shannon diversity index (High vs. Low) at preconditioning (A; n = 30), aplasia (B; n = 30) or engraftment (C; n = 27).
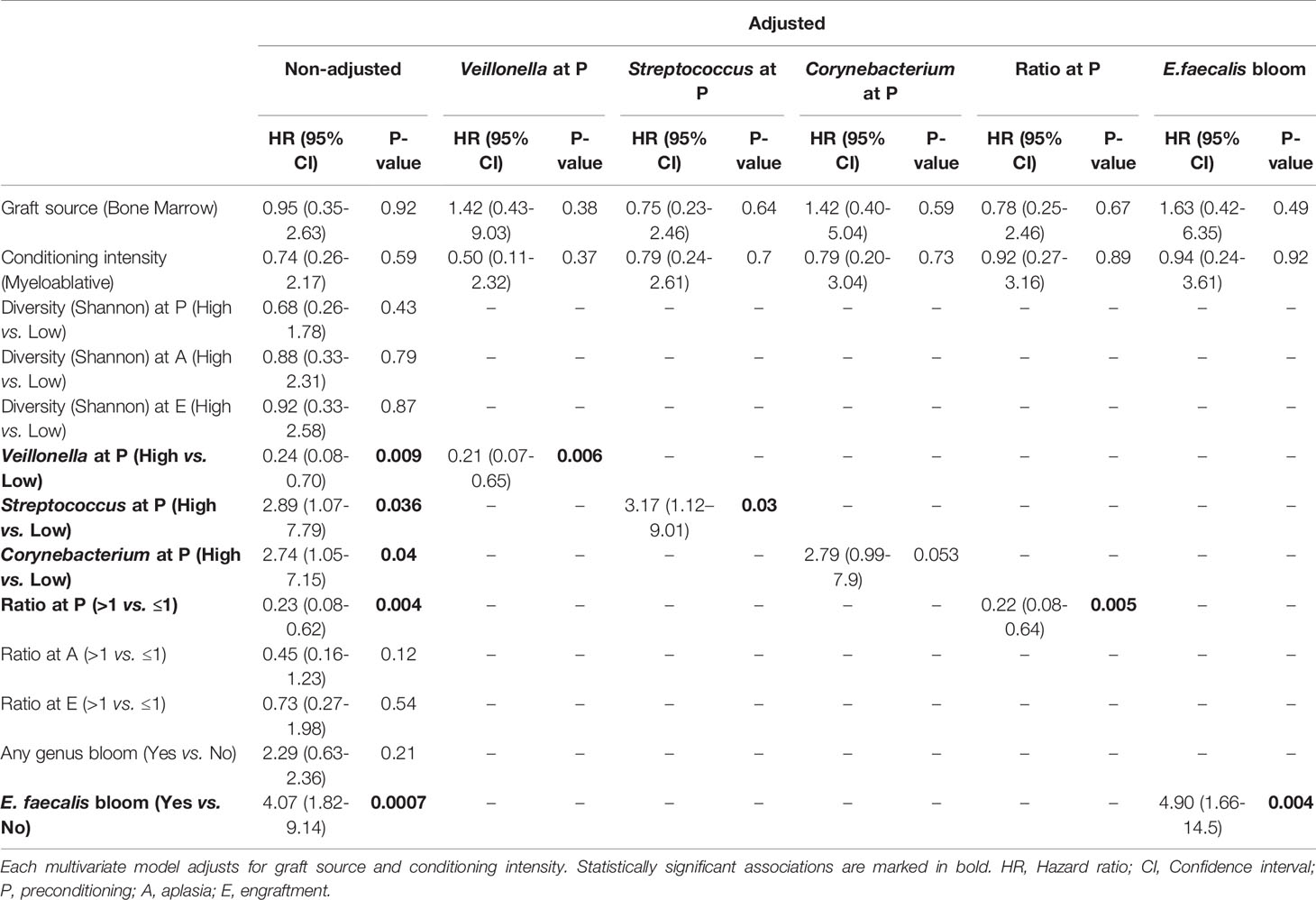
Table 2 Univariate (non-adjusted) and adjusted competing risk analyses for the association of acute graft-versus-host disease with relevant microbiota variables.
We then evaluated whether the abundance of specific genera at preconditioning, aplasia, or engraftment was associated with the risk of aGVHD (Figure 3). Only genera present at relative abundance ≥ 0.1% in at least 25% of the samples were considered for these analyses. Patients were stratified into two equal-sized groups (high and low relative abundance groups) by the median relative abundance observed in the entire cohort of each genus. Veillonella, Streptococcus, and Corynebacterium at preconditioning were significantly associated with the risk of aGVHD. We did not observe a similar association between the relative abundance of these or any other genus with the risk of aGVHD at aplasia or engraftment (Figure 3A).
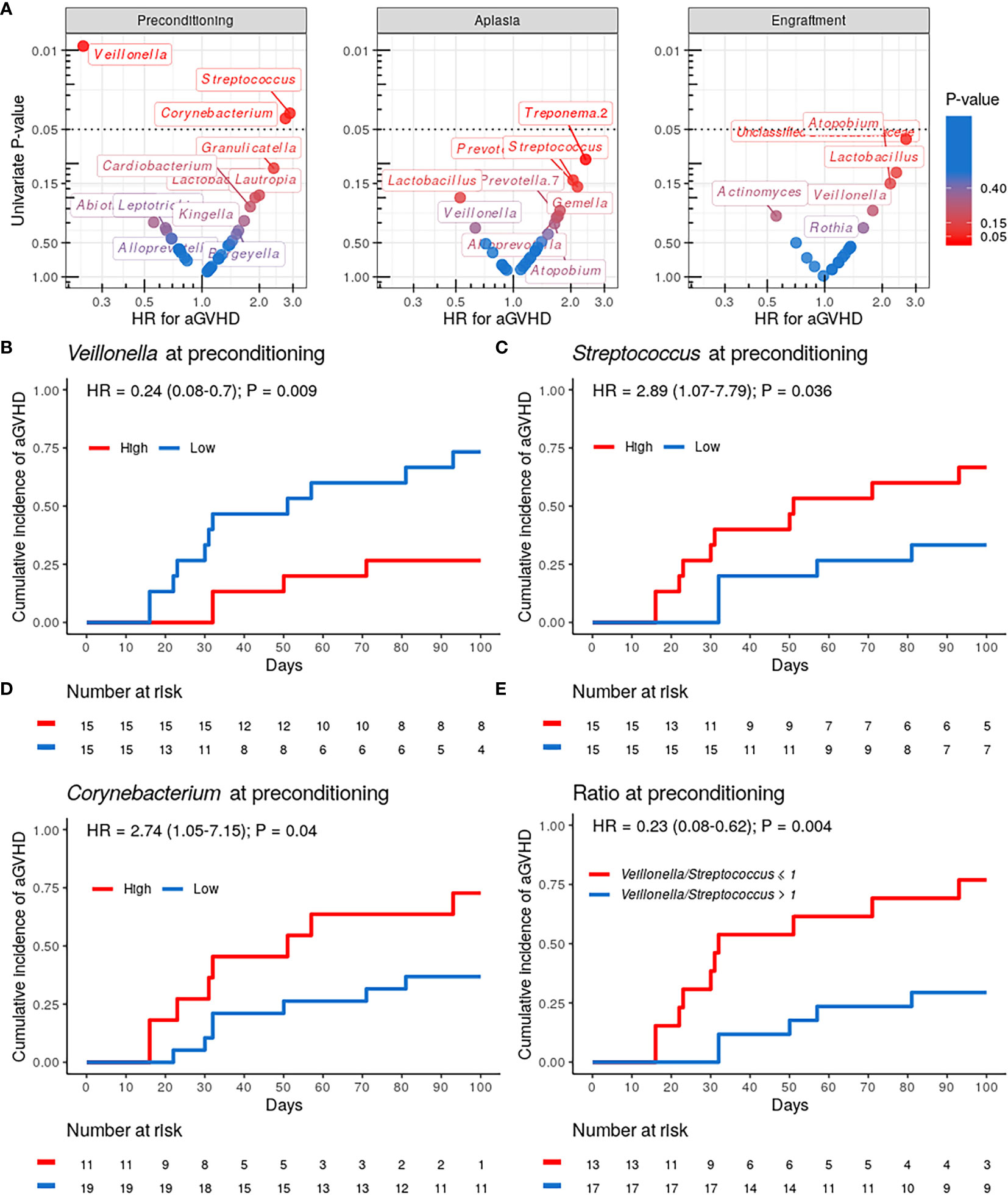
Figure 3 Specific genera relative abundance at preconditioning are associated with the risk of acute graft-versus-host disease (aGVHD). (A) Volcano plot for the univariate competing risk analysis for the association of aGVHD with genera relative abundance (hazard ratio vs. P-value) at preconditioning (left), aplasia (center) and engraftment (right). Only genera with ≥0.1% relative abundance in at least 25% of samples at a given phase were evaluated. Genera with P-value < 0.4 for the association are indicated explicitly. (B–D) Cumulative incidence of aGVHD with patients (n = 30) stratified by either Veillonella (B), Streptococcus (C) or Corynebacterium (D) relative abundance at preconditioning (High vs. Low). (E) Cumulative incidence of aGVHD with patients (n = 30) stratified by Veillonella/Streptococcus relative abundance ratio at preconditioning (>1 vs. ≤1).
Patients with high Veillonella relative abundance at preconditioning had a lower CMI of aGVHD (27% vs. 73%; HR = 0.24, 95% CI: 0.08–0.7, P = 0.009; Figure 3B and Table 2). This association remained significant after adjusting for graft source and intensity of the conditioning regimen (adjusted-HR = 0.21, 95% CI: 0.07–0.65, P = 0.006, Table 2). Patients with high Streptococcus or Corynebacterium relative abundance at preconditioning had a higher CMI of aGVHD (67% vs. 33%; HR = 2.89, 95% CI: 1.07–7.79, P = 0.036 and 73% vs. 37%; HR = 2.74, 95% CI: 1.05–7.15, P = 0.04, respectively; Figures 3C, D and Table 2). However, only Streptococcus remained significantly associated with the risk of aGVHD after adjusting for graft source and intensity of the conditioning regimen (adjusted-HR = 3.17, 95% CI: 1.12–9.01, P = 0.03, Table 2).
Veillonella and Streptococcus showed the highest average relative abundance at preconditioning (Figure 1B). Given their overall high relative abundance and an inverse association with the risk of aGVHD, we next evaluated whether the Veillonella/Streptococcus ratio at preconditioning was associated with the risk of aGVHD. Patients with a Veillonella/Streptococcus ratio >1 at preconditioning had a lower CMI of aGVHD (29% vs. 77%; HR = 0.23, 95% CI: 0.08–0.62, P = 0.004; Figure 3E and Table 2). Interestingly, the association between the Veillonella/Streptococcus ratio at preconditioning and aGVHD risk was stronger than the association observed for each genus separately and remained significant after adjusting for graft source and intensity of the conditioning regimen (adjusted-HR = 0.22, 95% CI: 0.08–0.64, P=0.005, Table 2). The Veillonella/Streptococcus ratio at aplasia or engraftment was not associated with the risk of aGVHD (Table 2).
Finally, we analyzed whether the blooming of potentially pathogenic genera observed during allo-HSCT was associated with the risk of aGVHD. For these analyses, bloom was defined as the sudden expansion of a particular genus from near absence (relative abundance <1% at preconditioning) to dominance (relative abundance ≥30% at aplasia or engraftment). Analyzing variations in genera relative abundance during allo-HSCT, we observed 23 blooms, involving 12 different genera and affecting a total of 20 patients. Three patients experienced more than one blooming event (Figure S4). Patients experiencing any genus bloom (n = 20) did not have altered aGVHD risk (Table 2). Enterococcus bloom was the most frequent event (Figure 4A), observed in 20% of the patients undergoing allo-HSCT. For all patients experiencing Enterococcus bloom except one, the phenomenon was attributed exclusively to Enterococcus faecalis expansion (Figure 4B). There was no association between E. faecalis bloom and cephalosporin (Fisher’s exact test, P = 0.29) or antibiotic for anaerobic bacteria usage (Fisher’s exact test, P = 1).
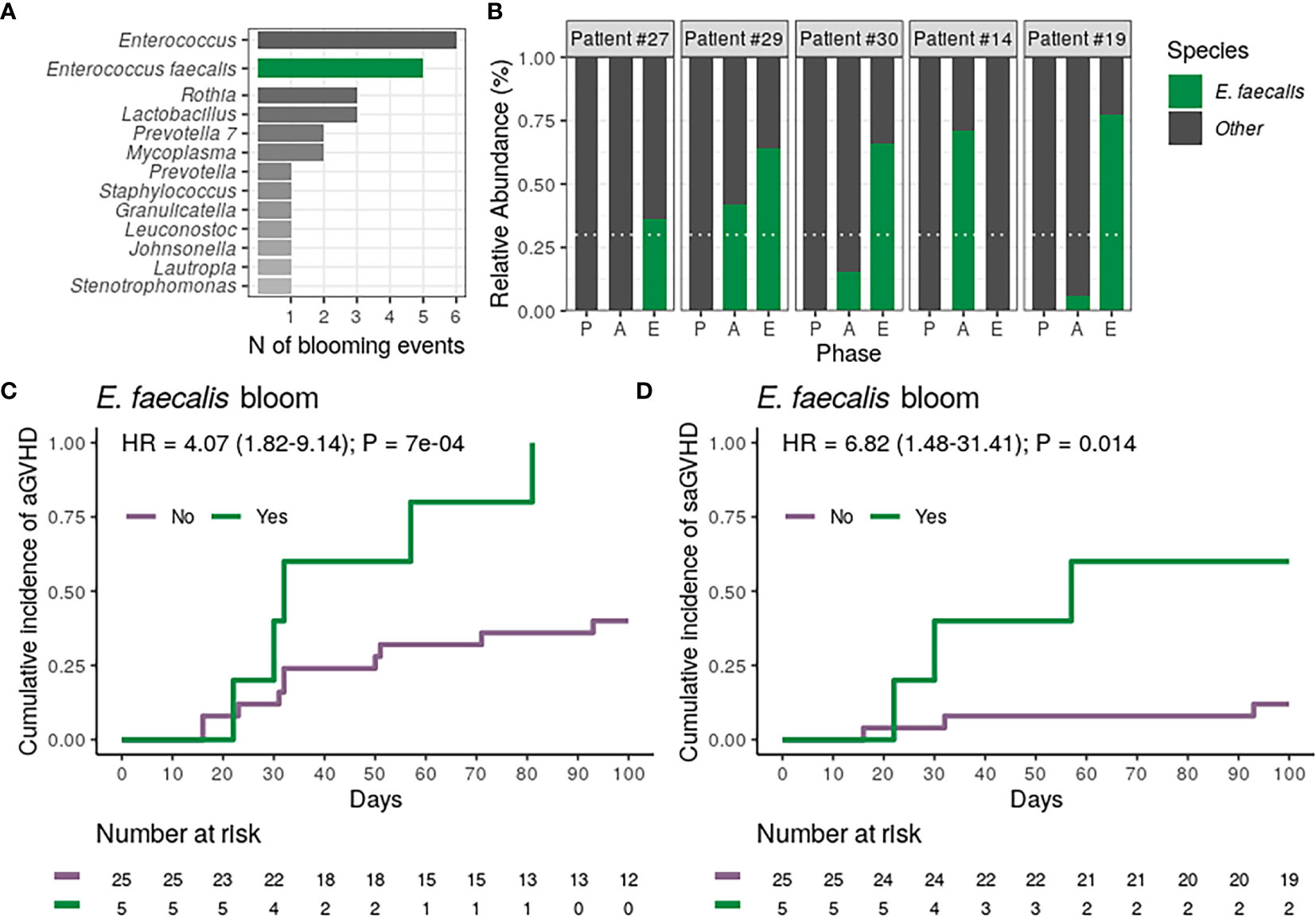
Figure 4 Dental biofilm Enterococcus faecalis bloom during allogeneic hematopoietic stem cell transplantation is associated with a higher risk of acute graft-versus-host disease (aGVHD) and severe aGVHD (saGVHD). (A) Number of observed blooming events per genera in all patients (n = 30). The number of Enterococcus blooms caused exclusively by Enterococcus faecalis is indicated. (B) Relative abundance of Enterococcus faecalis across transplantation phases for all patients experiencing Enterococcus faecalis bloom (n = 5). Patients are sorted based on the highest Enterococcus faecalis relative abundance observed per patient. White horizontal dashed line indicates dominance threshold. P, Preconditioning; A, Aplasia; E, Engraftment. (C, D) Cumulative incidence of aGVHD (C) or saGVHD (D) with patients (n = 30) stratified by Enterococcus faecalis bloom occurrence (No vs. Yes).
We next tested whether the occurrence of E. faecalis bloom was associated with the risk of aGVHD. All patients experiencing E. faecalis bloom developed aGVHD, and E. faecalis bloom was strongly associated with a higher CMI of aGVHD (100% vs. 40%; HR = 4.07, 95% CI: 1.82–9.14, P = 0.0007; Figure 4C and Table 2). This association remained significant after adjusting for graft source and intensity of the conditioning regimen (adjusted-HR = 4.90, 95% CI: 1.66–14.50, P = 0.004, Table 2). Notably, CMI of severe aGVHD (grade III-IV) was higher in patients experiencing E. faecalis bloom (60% vs. 12%; HR = 6.82, 95% CI: 1.48–31.41, P = 0.014; Figure 4D; Table 2), revealing a direct association between DBM E. faecalis bloom and aGVHD risk and grade.
In our study, we describe, for the first time using high-throughput 16S rRNA sequencing, changes in DBM diversity and composition in 30 patients undergoing allo-HSCT. As observed for IM, DBM dysbiosis during allo-HSCT was marked by a gradual loss of bacterial diversity, with engraftment samples presenting the lowest overall bacterial diversity. Like for the IM, we also observed significant changes in DBM genera composition, with a decrease in the abundance of commensal core DBM genera, such as Streptococcus and Actinomyces (the only genera that can adhere to the tooth surface to start ordinary DB formation) (24), and overgrowths of potentially pathogenic bacteria, such as Enterococcus, Lactobacillus, and Mycoplasma. Most importantly, we observed that DBM genera relative abundance at preconditioning and changes in DBM composition during allo-HSCT (namely, E. faecalis bloom) were both predictive of aGVHD risk after allo-HSCT. There was no association between these aGVHD-associated microbiota variables and other allo-HSCT outcomes, including chronic GVHD (Table S2), as diagnosed in accordance with the NIH 2014 consensus (25).
aGVHD is a major cause of non-relapse mortality following allo-HSCT, with a one-year survival rate for patients developing severe aGVHD of only 40% (26). First-line therapy for aGVHD is based on corticosteroids, with response rates that vary between 40 and 70% (27). In this scenario, identifying biomarkers capable of predicting aGVHD risk and developing preventive therapies are critical.
Recently, the IM composition has been analyzed as a biomarker for clinical outcomes in allo-HSCT recipients, including the development of aGVHD (5, 7). Moreover, microbiota-based therapeutic interventions, including microbiota-driven antibiotics selection, alternative dietary regimens (including probiotics/prebiotics usage) and fecal microbiota transplantation have been proposed to prevent and treat aGVHD (28–32).
Like the IM, the OM plays an essential role in maintaining local and systemic health. Dental biofilm (DB) bacteria, as opposed to other shedding surface-living bacteria in the oral cavity, can adhere to hard surfaces and coaggregate (33), allowing the assembly of an organized three-dimensional structure, which confers DBM its distinctive ecological properties. The DBM interacts directly with host immune cells and modulates immune homeostasis (14). Moreover, DBM can also act as a microbial reservoir for systemic diseases. DBM dysbiosis can trigger local inflammation, destruction of surrounding periodontal tissue, and systemic translocation of oral microbes (24). The influence of the OM in systemic diseases such as colorectal cancer (34) and arthritis (35) has been increasingly studied. However, in the allo-HSCT context, studies are still limited and have focused mainly on the saliva and the tongue microbiota (36–39).
Loss of bacterial diversity in the salivary microbiota of patients undergoing allo-HSCT has been previously described and associated with oral mucositis (36). Likewise, a steep decline in the tongue microbiota diversity was observed in severe aplastic anemia patients from preconditioning to the day of transplantation (37). On the other hand, no appreciable changes in OM during allo-HSCT were observed in an additional study evaluating 4 different oral sites (buccal mucosa, saliva, tongue, and DB) (38). However, this latter study used a low-resolution methodology (microarray) for microbiota characterization in a small number of patients (n = 11). Noteworthy, a single study evaluated the association between OM and allo-HSCT outcomes (39). Allo-HSCT recipients showed a less diverse and distinct tongue microbiota on the day of transplantation than that of community-dwelling adults. In this study, the presence of the non-commensal bacteria Staphylococcus haemolyticus and/or Ralstonia pickettii in the tongue microbiota was significantly associated with lower overall survival after allo-HSCT, but not with aGVHD.
Out of the many allo-HSCT outcomes evaluated so far (40), aGVHD onset has the clearest causal connection to the IM (28, 29, 40). Briefly, it has been shown that the loss of commensal bacteria (especially SCFA-producing Clostridia species) during the conditioning regimen reduces the intestinal concentration of butyrate and indole-3-aldehyde (41, 42). Low levels of these metabolites compromise mucosal integrity (42, 43), promoting extravasation of bacterial lipopolysaccharide and activation of donor reactive T cells (40). Additionally, Enterococcus faecalis might contribute to aGVHD development via production of metalloproteases that impair barrier function (44) and by stimulating macrophages to secrete TNF (45). Accordingly, low IM diversity at the time of stem cell engraftment (6, 7), low abundance of commensal bacteria from Clostridia class (7, 8), and intestinal enterococci dominance during allo-HSCT (10) have been all associated with worsened aGVHD-related outcomes in studies evaluating stool specimens from allo-HSCT recipients (28, 29, 40).
In our study, DBM diversity was not associated with the risk of aGVHD in any transplantation phase evaluated, which is in line with a recent IM study that did not find differences in IM diversity between aGVHD groups neither pre- nor post-transplantation (46). Also, despite the presence (as expected (47)) of many Clostridia genera in DBM (such as Oribacterium), we did not find DBM Clostridia class members significantly associated with the risk of aGVHD. However, as for the IM, we observed a decrease in the relative abundance of several DB commensal genera during allo-HSCT, such as Streptococcus, Veillonella, Actinomyces, and Capnocytophaga, and an increase in the relative abundance of potentially pathogenic genera such as Enterococcus and Lactobacillus. Most importantly, high Streptococcus and high Corynebacterium relative abundance at preconditioning were associated with a higher risk of aGVHD, while high Veillonella relative abundance at preconditioning was associated with a lower risk of aGVHD.
Streptococci, corynebacteria, and veillonellae are part of the core DBM (48) and represent the 1st, 2nd and 10th most important genera in terms of relative abundance in healthy volunteers DBM, respectively (47). In our study, streptococci and veillonellae showed the highest average relative abundance at preconditioning and were both associated with the risk of aGVHD. Given their overall high relative abundance and the relative nature of the data, higher Veillonella relative abundance imposes lower Streptococcus relative abundance and vice versa. Hence, it is not possible to determine whether both genera are genuinely associated with the risk of aGVHD. Interestingly, the association between the Veillonella/Streptococcus ratio at preconditioning and aGVHD risk, independently of the conditioning regimen and graft source, was stronger than the association observed for each genus separately, suggesting a partial role for both genera in the observed effect.
During DB formation, bacterial early colonizers, after adhering to teeth salivary pellicles, coaggregate with other early and late colonizers, and a repeatable microbial succession takes place on the tooth surface (33). Streptococci are the most abundant microbe in DB, representing a predominant early colonizer with broad coaggregation partnerships. Streptococci and veillonellae are in close physical contact during the early phases of DB maturation (33, 49) and can grow together in a metabolic cooperation-dependent manner (33, 49). Since this interaction occurs in the early phases of DB formation (and therefore are instrumental for DB maturation), the ratio Veillonella/Streptococcus might be a marker of early DBM disruption associated with a higher risk of aGVHD.
Corynebacteria bridge the early biofilm members to late colonizers (48). In contradiction with the documented in the aforementioned healthy volunteers study (47), we did not observe a high corynebacteria average relative abundance in any of the allo-HSCT phases evaluated. It is possible that the overall lower relative abundance of corynebacteria in detriment of early colonizers (such as streptococci and veillonellae) in our study may be indicative of a basal DBM disruption afflicting all allo-HSCT recipients. Alternatively, the lower relative abundance of corynebacteria may be explained by the stricter oral hygiene protocol recommended to our patients.
Finally, in our study, E. faecalis bloom in the DBM was observed in 17% of allo-HSCT recipients and was significantly associated with a higher risk of aGVHD and saGVHD. Noteworthy, despite recent in vitro evidence suggesting that high-dose of cephalosporin may promote E. faecalis biofilm formation (50), there was no association between cephalosporin usage and DBM E. faecalis bloom in the evaluated cohort.
During allo-HSCT, intestinal enterococci expansion is well documented and is linked to both aGVHD development (10) and subsequent bacteremia (51). Notably, E. faecalis alone exacerbates aGVHD severity in gnotobiotic mouse models (10). Our study reveals an additional site with enterococci expansion that might have systemic impacts after allo-HSCT. We can speculate that, during allo-HSCT, the dysbiotic DBM may act as an enterococci reservoir, triggering translocation to the gut and intestinal enterococci domination. This possibility is corroborated by the fact that there is intense oral bacteria translocation to the gut in hepatic cirrhosis patients (52) and that such translocations in colorectal cancer patients are negatively correlated with intestinal Clostridia bacteria presence (34). Indeed, oral bacteria translocation to the gut has been described in allo-HSCT recipients, and the presence of oral Actinobacteria and oral Firmicutes in stool samples of these patients was positively correlated with subsequent aGVHD development (5). Alternatively, DBM enterococci may have an intestinal origin, since the injury to Goblet cells during conditioning regimen was shown to induce dissemination of dominant intestinal bacteria (28). Further studies evaluating synchronously IM and DBM are necessary to decipher whether IM and DBM enterococci bloom are linked and which event precedes the other. Importantly, enterococci are present in small amounts in the healthy OM (47) but may overgrow in pathogenic/dysbiotic settings, including after solid organ transplantation (53), in a biofilm-dependent manner (54). This may explain why previous microbiota studies on soft oral sites have not reported the expansion of Enterococcus in allo-HSCT recipients.
Our study has many limitations. As a pioneering and exploratory work, it is single-centered and has a limited sample size. Besides, the study patients analyzed are heterogeneous and encompass several underlying diseases. Therefore, validation cohorts and multicentric prospective studies are needed to confirm our findings. We also emphasize that the associations reported herein are correlative, so that further studies on DBM during allo-HSCT that include synchronous fecal sampling and metabolomics analyses are needed to associate DBM dysbiosis with aGVHD pathophysiology.
Although patients usually receive rigorous oral health care during allo-HSCT (55), OM dysbiosis has been overlooked. Common oral care protocols already used in allo-HSCT patients to prevent and counteract oral health decay can also be used to directly (e.g. chlorhexidine mouthwash) or indirectly (e.g. photobiomodulation) modulate the OM. However, as the role of oral microbes in allo-HSCT outcomes become more prominent, complementary odontologic/pharmacologic interventions targeting specific sites and bacteria of the OM will be necessary. For instance, DBM dysbiosis could be managed by antimicrobial photodynamic therapy, which can eliminate pathogens with no risk of the emergence of drug-resistant strains (56). DBM dysbiosis could also be countervailed with the use of nanoparticles that alters DBM composition by interfering in fundamental biofilm properties such as adhesion and quorum-sensing (57, 58). These innovative approaches will be instrumental to evaluate whether early interventions to correct DBM dysbiosis can prevent aGVHD onset.
In conclusion, to our knowledge, this is the first study evaluating the DBM during allo-HSCT using a high-resolution technique. We identified markers of DBM dysbiosis during allo-HSCT. Most importantly, we showed that DBM composition during allo-HSCT may be predictive of aGVHD onset after transplantation, providing a simple and reproducible protocol for collection and analysis of allo-HSCT recipients microbiota before transplantation that may substitute fecal sampling when evaluating gastrointestinal dysbiosis and Enterococcus bloom.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: European Nucleotide Archive (ENA), PRJEB42862.
The studies involving human participants were reviewed and approved by the Ethics Committee of Hospital Sírio-Libanês (Protocol #1.414.217). The patients/participants provided their written informed consent to participate in this study.
AC and EF designed the study. FK, PA, VH, JB, and WM-S performed the sequencing. VH and AC developed the bioinformatics pipeline. VH, JB, VM, CA-R, EF, and AC contributed to the analysis and interpretation of data. VH, JB, VM, CA-R, and AC contributed to writing the manuscript. All authors contributed to the article and approved the submitted version.
VH was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process no. 13996-0/2018). VM was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, process no. 141575/2018-2). JB was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, process no. 001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
A manuscript regarding this work has been previously submitted to medRxiv as a preprint (59).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.692225/full#supplementary-material
1. Styczyński J, Tridello G, Koster L, Iacobelli S, van Biezen A, van der Werf S, et al. Death After Hematopoietic Stem Cell Transplantation: Changes Over Calendar Year Time, Infections and Associated Factors. Bone Marrow Transpl (2020) 55(1):126–36. doi: 10.1038/s41409-019-0624-z
2. Schoemans HM, Lee SJ, Ferrara JL, Wolff D, Levine JE, Schultz KR, et al. EBMT-NIH-CIBMTR Task Force Position Statement on Standardized Terminology & Guidance for Graft-Versus-Host Disease Assessment. Bone Marrow Transpl (2018) 53(11):1401–15. doi: 10.1038/s41409-018-0204-7
3. Greinix HT, Eikema D-J, Koster L, Penack O, Yakoub-Agha I, Montoto S, et al. Incidence of Acute Graft-Versus-Host Disease and Survival After Allogeneic Hematopoietic Cell Transplantation Over Time: A Study From the Transplant Complications and Chronic Malignancies Working Party of the EBMT. Blood (2018) 132(Supplement 1):2120–0. doi: 10.1182/blood-2018-99-111764
4. Zeiser R, Blazar BR. Acute Graft-versus-Host Disease - Biologic Process, Prevention, and Therapy. N Engl J Med (2017) 377(22):2167–79. doi: 10.1056/NEJMra1609337
5. Golob JL, Pergam SA, Srinivasan S, Fiedler TL, Liu C, Garcia K, et al. Stool Microbiota at Neutrophil Recovery Is Predictive for Severe Acute Graft vs Host Disease After Hematopoietic Cell Transplantation. Clin Infect Dis Off Publ Infect Dis Soc Am (2017) 65(12):1984–91. doi: 10.1093/cid/cix699
6. Taur Y, Jenq RR, Perales M-A, Littmann ER, Morjaria S, Ling L, et al. The Effects of Intestinal Tract Bacterial Diversity on Mortality Following Allogeneic Hematopoietic Stem Cell Transplantation. Blood (2014) 124(7):1174–82. doi: 10.1182/blood-2014-02-554725
7. Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, et al. Intestinal Blautia Is Associated With Reduced Death From Graft-Versus-Host Disease. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transpl (2015) 21(8):1373–83. doi: 10.1016/j.bbmt.2015.04.016
8. Payen M, Nicolis I, Robin M, Michonneau D, Delannoye J, Mayeur C, et al. Functional and Phylogenetic Alterations in Gut Microbiome Are Linked to Graft-Versus-Host Disease Severity. Blood Adv (2020) 4(9):1824–32. doi: 10.1182/bloodadvances.2020001531
9. Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, et al. Metagenomic Analysis of the Stool Microbiome in Patients Receiving Allogeneic Stem Cell Transplantation: Loss of Diversity Is Associated With Use of Systemic Antibiotics and More Pronounced in Gastrointestinal Graft-Versus-Host Disease. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transpl (2014) 20(5):640–5. doi: 10.1016/j.bbmt.2014.01.030
10. Stein-Thoeringer CK, Nichols KB, Lazrak A, Docampo MD, Slingerland AE, Slingerland JB, et al. Lactose Drives Enterococcus Expansion to Promote Graft-Versus-Host Disease. Science (2019) 366(6469):1143–9. doi: 10.1126/science.aax3760
11. Olsen I, Yamazaki K. Can Oral Bacteria Affect the Microbiome of the Gut? J Oral Microbiol (2019) 11(1):1586422. doi: 10.1080/20002297.2019.1586422
12. Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic Colonization of Oral Bacteria in the Intestine Drives TH1 Cell Induction and Inflammation. Science (2017) 358(6361):359–65. doi: 10.1126/science.aan4526
13. Sharma N, Bhatia S, Sodhi AS, Batra N. Oral Microbiome and Health. AIMS Microbiol (2018) 4(1):42–66. doi: 10.3934/microbiol.2018.1.42
14. Moutsopoulos NM, Konkel JE. Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends Immunol (2018) 39(4):276–87. doi: 10.1016/j.it.2017.08.005
15. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res (2013) 41(1):e1. doi: 10.1093/nar/gks808
16. Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast Tissue, Oral and Urinary Microbiomes in Breast Cancer. Oncotarget (2017) 8(50):88122–38. doi: 10.18632/oncotarget.21490
17. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat Biotechnol (2019) 37(8):852–7. doi: 10.1038/s41587-019-0209-9
18. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-Resolution Sample Inference From Illumina Amplicon Data. Nat Methods (2016) 13(7):581–3. doi: 10.1038/nmeth.3869
19. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res (2013) 41(Database issue):D590–596. doi: 10.1093/nar/gks1219
20. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinforma Oxf Engl (2011) 27(16):2194–200. doi: 10.1093/bioinformatics/btr381
21. Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ (2016) 4:e2584. doi: 10.7717/peerj.2584
22. Kaul A, Mandal S, Davidov O, Peddada SD. Analysis of Microbiome Data in the Presence of Excess Zeros. Front Microbiol (2017) 8:2114. doi: 10.3389/fmicb.2017.02114
23. Glucksberg H, Storb R, Fefer A, Buckner CD, Neiman PE, Clift RA, et al. Clinical Manifestations of Graft-Versus-Host Disease in Human Recipients of Marrow From HL-A-Matched Sibling Donors. Transplantation (1974) 18(4):295–304. doi: 10.1097/00007890-197410000-00001
24. Vieira Colombo AP, Magalhães CB, Hartenbach FARR, Martins do Souto R, Maciel da Silva-Boghossian C. Periodontal-Disease-Associated Biofilm: A Reservoir for Pathogens of Medical Importance. Microb Pathog (2016) 94:27–34. doi: 10.1016/j.micpath.2015.09.009
25. Jagasia MH, Greinix HT, Arora M, Williams KM, Wolff D, Cowen EW, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-Versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group Report. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transpl (2015) 21(3):389–401. doi: 10.1016/j.bbmt.2015.05.004
26. Khoury HJ, Wang T, Hemmer MT, Couriel D, Alousi A, Cutler C, et al. Improved Survival After Acute Graft-Versus-Host Disease Diagnosis in the Modern Era. Haematologica (2017) 102(5):958–66. doi: 10.3324/haematol.2016.156356
27. Garnett C, Apperley JF, Pavlů J. Treatment and Management of Graft-Versus-Host Disease: Improving Response and Survival. Ther Adv Hematol (2013) 4(6):366–78. doi: 10.1177/2040620713489842
28. Kumari R, Palaniyandi S, Hildebrandt GC. Microbiome: An Emerging New Frontier in Graft-Versus-Host Disease. Dig Dis Sci (2019) 64(3):669–77. doi: 10.1007/s10620-018-5369-9
29. Staffas A, Burgos da Silva M, van den Brink MRM. The Intestinal Microbiota in Allogeneic Hematopoietic Cell Transplant and Graft-Versus-Host Disease. Blood (2017) 129(8):927–33. doi: 10.1182/blood-2016-09-691394
30. Taur Y, Coyte K, Schluter J, Robilotti E, Figueroa C, Gjonbalaj M, et al. Reconstitution of the Gut Microbiota of Antibiotic-Treated Patients by Autologous Fecal Microbiota Transplant. Sci Transl Med (2018) 10(460):eaap9489. doi: 10.1126/scitranslmed.aap9489
31. Spindelboeck W, Schulz E, Uhl B, Kashofer K, Aigelsreiter A, Zinke-Cerwenka W, et al. Repeated Fecal Microbiota Transplantations Attenuate Diarrhea and Lead to Sustained Changes in the Fecal Microbiota in Acute, Refractory Gastrointestinal Graft-Versus-Host-Disease. Haematologica (2017) 102(5):e210–3. doi: 10.3324/haematol.2016.154351
32. van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor Fecal Microbiota Transplantation Ameliorates Intestinal Graft-Versus-Host Disease in Allogeneic Hematopoietic Cell Transplant Recipients. Sci Transl Med (2020) 12(556):eaaz8926. doi: 10.1126/scitranslmed.aaz8926
33. Kolenbrander PE, Palmer RJ, Periasamy S, Jakubovics NS. Oral Multispecies Biofilm Development and the Key Role of Cell-Cell Distance. Nat Rev Microbiol (2010) 8(7):471–80. doi: 10.1038/nrmicro2381
34. Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, et al. The Oral Microbiota in Colorectal Cancer Is Distinctive and Predictive. Gut (2018) 67(8):1454–63. doi: 10.1136/gutjnl-2017-314814
35. Lorenzo D, GianVincenzo Z, Carlo Luca R, Karan G, Jorge V, Roberto M, et al. Oral-Gut Microbiota and Arthritis: Is There an Evidence-Based Axis? J Clin Med (2019) 8(10):1753. doi: 10.3390/jcm8101753
36. Shouval R, Eshel A, Dubovski B, Kuperman AA, Danylesko I, Fein JA, et al. Patterns of Salivary Microbiota Injury and Oral Mucositis in Recipients of Allogeneic Hematopoietic Stem Cell Transplantation. Blood Adv (2020) 4(13):2912–7. doi: 10.1182/bloodadvances.2020001827
37. Ames NJ, Barb JJ, Ranucci A, Kim H, Mudra SE, Cashion AK, et al. The Oral Microbiome of Patients Undergoing Treatment for Severe Aplastic Anemia: A Pilot Study. Ann Hematol (2019) 98(6):1351–65. doi: 10.1007/s00277-019-03599-w
38. Ames NJ, Sulima P, Ngo T, Barb J, Munson PJ, Paster BJ, et al. A Characterization of the Oral Microbiome in Allogeneic Stem Cell Transplant Patients. PloS One (2012) 7(10):e47628. doi: 10.1371/journal.pone.0047628
39. Oku S, Takeshita T, Futatsuki T, Kageyama S, Asakawa M, Mori Y, et al. Disrupted Tongue Microbiota and Detection of Nonindigenous Bacteria on the Day of Allogeneic Hematopoietic Stem Cell Transplantation. PloS Pathog (2020) 16(3):e1008348. doi: 10.1371/journal.ppat.1008348
40. Shono Y, van den Brink MRM. Gut Microbiota Injury in Allogeneic Haematopoietic Stem Cell Transplantation. Nat Rev Cancer (2018) 18(5):283–95. doi: 10.1038/nrc.2018.10
41. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and Receptors for Short-Chain Fatty Acids as the Molecular Link Between Colonic Bacteria and the Host. Curr Opin Pharmacol (2013) 13(6):869–74. doi: 10.1016/j.coph.2013.08.006
42. Ferrara JLM, Chaudhry MS. GVHD: Biology Matters. Blood Adv (2018) 2(22):3411–7. doi: 10.1182/bloodadvances.2018020214
43. Mathewson ND, Jenq R, Mathew AV, Koenigsknecht M, Hanash A, Toubai T, et al. Gut Microbiome-Derived Metabolites Modulate Intestinal Epithelial Cell Damage and Mitigate Graft-Versus-Host Disease. Nat Immunol (2016) 17(5):505–13. doi: 10.1038/ni.3400
44. Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, et al. Enterococcus Faecalis Metalloprotease Compromises Epithelial Barrier and Contributes to Intestinal Inflammation. Gastroenterology (2011) 141(3):959–71. doi: 10.1053/j.gastro.2011.05.035
45. Kim SO, Sheikh HI, Ha S-D, Martins A, Reid G. G-CSF-mediated Inhibition of JNK Is a Key Mechanism for Lactobacillus Rhamnosus-Induced Suppression of TNF Production in Macrophages. Cell Microbiol (2006) 8(12):1958–71. doi: 10.1111/j.1462-5822.2006.00763.x
46. Ilett EE, Jørgensen M, Noguera-Julian M, Nørgaard JC, Daugaard G, Helleberg M, et al. Associations of the Gut Microbiome and Clinical Factors With Acute GVHD in Allogeneic HSCT Recipients. Blood Adv (2020) 4(22):5797–809. doi: 10.1182/bloodadvances.2020002677
47. Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the Adult Digestive Tract Bacterial Microbiome Based on Seven Mouth Surfaces, Tonsils, Throat and Stool Samples. Genome Biol (2012) 13(6):R42. doi: 10.1186/gb-2012-13-6-r42
48. Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a Human Oral Microbiome at the Micron Scale. Proc Natl Acad Sci USA (2016) 113(6):E791–800. doi: 10.1073/pnas.1522149113
49. Mashima I, Nakazawa F. The Interaction Between Streptococcus Spp. and Veillonella Tobetsuensis in the Early Stages of Oral Biofilm Formation. J Bacteriol (2015) 197(3):2104–11. doi: 10.1128/JB.02512-14
50. Thieme L, Klinger-Strobel M, Hartung A, Stein C, Makarewicz O, Pletz MW. In Vitro Synergism and Anti-Biofilm Activity of Ampicillin, Gentamicin, Ceftaroline and Ceftriaxone Against Enterococcus Faecalis. J Antimicrob Chemother (2018) 73(6):1553–61. doi: 10.1093/jac/dky051
51. Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin Infect Dis Off Publ Infect Dis Soc Am (2012) 55(7):905–14. doi: 10.1093/cid/cis580
52. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the Human Gut Microbiome in Liver Cirrhosis. Nature (2014) 513(7516):59–64. doi: 10.1038/nature13568
53. Diaz PI, Hong B-Y, Frias-Lopez J, Dupuy AK, Angeloni M, Abusleme L, et al. Transplantation-Associated Long-Term Immunosuppression Promotes Oral Colonization by Potentially Opportunistic Pathogens Without Impacting Other Members of the Salivary Bacteriome. Clin Vaccine Immunol CVI (2013) 20(6):920–30. doi: 10.1128/CVI.00734-12
54. Ch’ng J-H, Chong KKL, Lam LN, Wong JJ, Kline KA. Biofilm-Associated Infection by Enterococci. Nat Rev Microbiol (2019) 17(2):82–94. doi: 10.1038/s41579-018-0107-z
55. Current Practice of Oral Care for Hematopoietic Stem Cell Transplant Patients: A Survey of the Eastern Mediterranean Blood and Marrow Transplantation Group - Pubmed. Available at: https://pubmed.ncbi.nlm.nih.gov/33631114/.
56. Hu X, Huang Y-Y, Wang Y, Wang X, Hamblin MR. Antimicrobial Photodynamic Therapy to Control Clinically Relevant Biofilm Infections. Front Microbiol (2018) 9:1299. doi: 10.3389/fmicb.2018.01299
57. Zhang T, Kalimuthu S, Rajasekar V, Xu F, Yiu YC, Hui TKC, et al. Biofilm Inhibition in Oral Pathogens by Nanodiamonds. Biomater Sci (2021). doi: 10.1039/D1BM00608H
58. Shah S, Gaikwad S, Nagar S, Kulshrestha S, Vaidya V, Nawani N, et al. Biofilm Inhibition and Anti-Quorum Sensing Activity of Phytosynthesized Silver Nanoparticles Against the Nosocomial Pathogen Pseudomonas Aeruginosa. Biofouling (2019) 35(1):34–49. doi: 10.1080/08927014.2018.1563686
59. Heidrich V, Bruno JS, Knebel FH, Molla VC de, Miranda-Silva W, Asprino PF, et al. Dental Biofilm Microbiota Dysbiosis Is Associated With the Risk of Acute Graft-Versus-Host Disease After Allogeneic Hematopoietic Stem Cell Transplantation. medRxiv (2021). 2021.02.04.21251019. doi: 10.1101/2021.02.04.21251019
Keywords: oral microbiota, supragingival plaque, microbiome dysbiosis, acute GVHD, allogeneic HSCT, bone marrow transplant
Citation: Heidrich V, Bruno JS, Knebel FH, de Molla VC, Miranda-Silva W, Asprino PF, Tucunduva L, Rocha V, Novis Y, Arrais-Rodrigues C, Fregnani ER and Camargo AA (2021) Dental Biofilm Microbiota Dysbiosis Is Associated With the Risk of Acute Graft-Versus-Host Disease After Allogeneic Hematopoietic Stem Cell Transplantation. Front. Immunol. 12:692225. doi: 10.3389/fimmu.2021.692225
Received: 13 April 2021; Accepted: 02 June 2021;
Published: 18 June 2021.
Edited by:
Evelyn Ullrich, Goethe University Frankfurt, GermanyReviewed by:
Michele Malagola, University of Brescia, ItalyCopyright © 2021 Heidrich, Bruno, Knebel, de Molla, Miranda-Silva, Asprino, Tucunduva, Rocha, Novis, Arrais-Rodrigues, Fregnani and Camargo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anamaria A. Camargo, YW5hbWFyaWEuYWNhbWFyZ29AaHNsLm9yZy5icg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.