
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 15 July 2021
Sec. Viral Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.690976
This article is part of the Research TopicImmune Evasion Mechanisms by RNA VirusesView all 32 articles
 Betsy Yang1
Betsy Yang1 Kuender D. Yang2,3,4*
Kuender D. Yang2,3,4*Different emerging viral infections may emerge in different regions of the world and pose a global pandemic threat with high fatality. Clarification of the immunopathogenesis of different emerging viral infections can provide a plan for the crisis management and prevention of emerging infections. This perspective article describes how an emerging viral infection evolves from microbial mutation, zoonotic and/or vector-borne transmission that progresses to a fatal infection due to overt viremia, tissue-specific cytotropic damage or/and immunopathology. We classified immunopathogenesis of common emerging viral infections into 4 categories: 1) deficient immunity with disseminated viremia (e.g., Ebola); 2) pneumocytotropism with/without later hyperinflammation (e.g., COVID-19); 3) augmented immunopathology (e.g., Hanta); and 4) antibody-dependent enhancement of infection with altered immunity (e.g., Dengue). A practical guide to early blocking of viral evasion, limiting viral load and identifying the fatal mechanism of an emerging viral infection is provided to prevent and reduce the transmission, and to do rapid diagnoses followed by the early treatment of virus neutralization for reduction of morbidity and mortality of an emerging viral infection such as COVID-19.
Whether an RNA virus causes an endemic, epidemic, or pandemic is determined by the interactions among microorganism, host immunity, and environment. “Death or Survival” in an emerging viral infection depends largely on host immune responses because some patients are succumbed to death but most of the patients survive from the emerging infection. It is necessary to clarify varied immunopathogenesis of different emerging viral infections to prevent infection, morbidities, and mortality. Based on literature and our experiences with different emerging viral infections including enterovirus 71 encephalitis, dengue hemorrhagic fever, severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1), novel influenza A(H1N1), and SARS-CoV-2 in the past 2 decades, this perspective article describes the evasion and evolution of an emerging infection among different aspects of RNA virus, environment and host, including microbial evasion and evolution (mutation, deletion, and recombination), changes of ecosystems (season, climate, and urbanization), host susceptibility, and herd immunity. “Know thyself and thy adversary to win a hundred battles”, each emerging viral infection requires individualized strategies to prevent infection and to avoid post-infectious immunopathology and fatality. A series of stepwise practical guides to infection and immunity controls are provided to prevent evasion, morbidity, and fatality of emerging viral infections.
An emerging infection is usually caused by the naive immunity of human beings encountering a novel pathogen arising from microbial mutation, vector-borne, or/and zoonotic transmission (Table 1) (1–12). Most of the common emerging infections are mediated by RNA viruses, which pose a higher rate of genetic mutation, sequence deletion, recombination, and reassortment of RNA virus codes (1, 13). As shown in Table 1, severe acute respiratory syndrome (SARS), avian flu, swine flu, and enterovirus 71 (EV71) are known to emerge from sequence mutation, deletion, recombination, and/or reassortment of RNA segments (1–6). Vector-borne diseases such as yellow fever, dengue hemorrhagic fever, and West Nile virus encephalitis are transmitted by mosquitos (Table 1) and affected by weather, global warming, and herd immunity (5–9). Zoonotic diseases such as Ebola, Lassa, and Hantavirus infections are affected by culture, movement of animals, and exploitation of forests (8–12).
Many emerging infections are caused by single-stranded RNA viruses. RNA viruses pose a higher mutation rate because its RNA-dependent polymerases usually lack a 3’-exonuclease that is present in DNA-dependent polymerases to provide proofreading ability for the genome stability during replication. RNA polymerases can produce one mutation over 10,000 replications (1, 13) and DNA viruses can produce only one mutation in 106 ~ 108 replications (14). The higher mutation rates of RNA viruses pose challenges to many emerging infections in human beings. For instance, influenza viruses, which are single-stranded negative RNA viruses, frequently have a nucleotide mutation causing an antigen drift responsible for periodic seasonal flu within the same host species (15). Similarly, EV71, which is a single-stranded positive RNA virus reveals varied genomic sequences in the isolates with different phenotypes (16). In an animal model, point mutations in different regions of EV71 have been attributed to different tissue tropism and fatality (17). Human severe acute respiratory syndrome coronavirus (SARS-CoV-1) caused an epidemic in Asia in 2003. SARS-CoV-1 virus mutation was estimated to be low at 0.1 per genome, similar to common single-stranded RNA viruses (18). However, SARS-CoV-2 appears to have an average of 7.23 mutations per sample (19). Single nucleotide transitions have been recognized as the major mutation of SARS-CoV-2 worldwide (19–23). The SARS-CoV-2 variant B.1.1.7 with mutation of N501Y and P681H reported from United Kingdom showed a 61% more virulence, and the variant B.1.617 with point mutations of E484Q, L452R and P681R from India revealed a 160% higher transmission rate (Ro > 5.0) (20, 21). More importantly, the variant B.1.351 with mutations of N501Y, K417N, and E484K from South Africa tended to cause breakthrough of COVID-19 vaccines showing a significant reduction in neutralization of antibodies raised by different vaccines (22, 23), potentially contributing to re-infection after natural infection or vaccination.
It is believed that the 1918 Pandemic Spanish flu that killed millions of people originated from the reassortment of cross-species virus genetic segments among avian, swine, horse, and humans (24). Flu viruses from different host animals recognize different sugar residues on respiratory and/or gastrointestinal epithelium. For instance, avian flu viruses recognize sialic acid alpha 2,3 galactose as a receptor and human flu viruses recognize sialic acid alpha 2,6 galactose as a receptor. A mutation or RNA segment reassortment of avian flu virus codes can change its hemagglutinin and recognize sialic acid alpha 2,6 galactose, thus expanding its host range to humans (25). Avian influenza uses segment reassortment of the genome to promote its cell surface binding, expand its host ranges and pose an epidemic or pandemic threat (26). These studies supported the cross-species adaptation of flu viruses through a series of reassortment events in mammals over a period of years before a pandemic outbreak (24–26), suggesting continual surveillance strategies for detection of flu viruses with cross-species genetic codes may alert to pandemics in advance.
Coronavirus, which possesses a 3’-exonulcease to maintain a relative larger RNA genome, uses recombination and deletion to expand to and adapt in human beings (27–29). The SARS-CoV-1 likely originated in civet cats and raccoon dogs, with precursor SARS-like viruses potentially circulating in live-animal markets, and later transmitted to and adapted in humans by certain nucleotide sequence deletion (30). SARS-CoV-2 is also believed to have jumped from bats to pangolins and humans via a recombination of the genome in the cell binding region of spike glycoprotein (31, 32). Similarly, MERS-CoV jumped from bats to camels and humans through a series of recombinations among coronaviruses of bats, civet cats, and camels (33). Deletion of certain nucleotides in the open reading frame 8 (ORF8) has been found in SARS-CoV-2 isolates, which potentially contribute to milder infections in humans (34). Another pattern of gene deletion involved in the emerging infection of a zoonotic disease are the vaccinia-like viruses Aracatuba and Cantagalo viruses, which have been isolated from diary workers and cattle (35). The viruses have a 99% homology to the vaccinia virus but show an 18-nucleotide deletion in the A56R hemagglutinin gene (35, 36).
In addition to virus mutation, temperature and humidity are known to affect human-human transmission of emerging infections. Aerosol, droplet and vector-born transmissions are affected by extreme climate changes and global transportation, and zoonotic infections are affected by urbanization, moving of animals, and exploitation of forests.
Human seasonal influenza is usually prevalent during the winter season in which lower temperature and humidity enhance droplet and aerosol transmission. Enteroviruses are prone to outbreaks during the summer season when higher humidity enhances oral-fecal route transmission. An experimental study showed that higher temperatures and humidity block droplet and aerosol routes of influenza transmission but not close contact transmission (37). In contrast to seasonal patterns of influenza and enterovirus infection, the SARS-CoV-2 pandemic widely spread to over 180 countries in both hemispheres at the same time, suggesting that this pandemic could be related to a micro-organism that is relatively insensitive to warmer temperature and/or humidity, and can survive for a longer time on fomites, such as surfaces of handles and/or handrails. The fact that mandated face-covering, and regional or national lockdowns, even in a region of SARS-CoV-2 variant with a high reproduction number, accounts for the significantly reduced number of infections in different countries, suggests aerosol transmission as the dominant route for the SARS-CoV-2 infection (21, 38).
Warming temperatures and precipitation (humidity) may decrease aerosol transmission of influenza infections, but increase mosquito-borne diseases, such as Dengue fever (DF), Zika fever, Yellow fever, and Chikungunya infections which have emerged in Western and Eastern countries (39). Global transportation and urbanization may also enhance mosquito-transmitted emerging infections. These emerging RNA viruses are primarily transmitted by the mosquito Aedes aegypti, which originated in Africa and breeds in fresh water such as tree holes or standing water, and is now responsible for outbreaks of urban Yellow fever, dengue, and Zika fever, following the movement of larva or eggs of Ae. aegypti through slave trade from Africa to the New World (39–41). The relatively cold-hardy Ae. albopictus has moved even further north with global warming (40, 41). More than 100 countries in Africa, the Americas, the Eastern Mediterranean, South-East Asia and the Western Pacific are seriously affected by DF, with Asia representing approximately 70% of the global burden (42). Dengue fever, caused by 4 different serotypes, used to present a benign febrile illness for a century until the 1950s when a severe form of dengue called dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) was reported in the Philippines (41). DHF/DSS spread to South America in 1981 and currently threatens countries in East Asia and South America (42). The reasons for the transition of benign DF to life-threatening DFH/DSS may be related to vector adaptation, climate change (warming and precipitation), and/or prevalence of heterotypic serotype infections (43). Like dengue, Zika virus with a mutation of NS1, transmitted by Ae. aegypti, has spread worldwide with a recent introduction from African and Asian lineages to the Americas (44). Zika virus causes intrauterine infection, especially in the first trimester, which can lead to congenital anomalies, particularly microcephaly, intrauterine growth restriction, and eye diseases (45).
Transition of jungle Yellow fever to endemic and epidemic urban Yellow fever is largely due to environmental changes, particularly industrialization and urbanization which enhance contacts between humans and the virus vectors in forests, as well as contacts between humans and urban virus vectors after urbanization (46). Although an effective live attenuated vaccine is available for Yellow fever, recent outbreaks in Africa and South America, where urbanization has promoted the Yellow fever virus to circulate from a jungle cycle (jungle mosquito-nonhuman primate) into an urban cycle (human-urban mosquito, Ae. aegypti), pose a risk to an estimated 400-500 million unvaccinated people living in at-risk areas (47). West Nile virus (WNV) is transmitted between avian hosts. The virus is transmitted by Culex spp. mosquitos that are infected from feeding on birds. The virus has however, expanded its geographic range from Africa, Europe, and the Mid-East to the Americas through global commerce and ecological changes (48). West Nile virus is not transmitted by a human-to-human or human-to-mosquito transmission, but rather by bird-to-mosquito-to-human transmission in which humans are the dead-end host; most of the infections are subclinical, but some can develop into severe neurological diseases, including fatal encephalitis and meningitis, particularly in older or immunocompromised patients (49). The spread of WNV north to Canada and south to Argentina indicates the growing burden of WNV in the world (50). A similar situation also occurs in Japanese encephalitis virus (JEV) transmission. JEV is an emerging flavivirus infection, transmitted by Culex spp. mosquitos in the Asia-Pacific region (51). JEV was initially reported in Africa and is now prevalent in the Asia-pacific region. Recently, both Aedes and Culex spp. have been shown to carry JEV in Europe (52), posing a great concern over its further spread in Northern Hemisphere countries.
Moreover, increased precipitation is associated with prevalence of Hantavirus hemorrhagic fever. Hantavirus hemorrhagic fever is transmitted by secretions of rodents and does not cause human-human transmission. The virus is found in urine and body secretion of rodents in large quantities and causes infection in humans by aerosol transmission. Hantavirus infection can lead to massive vascular damage causing “hemorrhagic fever with renal syndrome” (HFRS). HFRS was initially reported in Korea in 1950s and is now prevalent in China and Europe (53–55). The other hemorrhagic fever called “hantavirus cardio-pulmonary syndrome” (HCPS) is prevalent in the New World in North and South America (56). Recently certain overlapping hemorrhagic manifestations between HFRS and HCPS are increasingly observed (55). The HFRS has a relatively low fatality rate at about 1-3%, and the HCPS has a higher fatality rate of about 15-45%, depending on different outbreaks (53–56).
Herd immunity is another key factor that determines the endemic or epidemic spread of an emerging infection. Seasonal flu is usually involved in a community where less than 10% of the population has immunity to a mutant influenza virus. Each year, human seasonal flu emerges with a certain serotype of a mutant with antigen drift resulting in an endemic or epidemic depending on herd immunity and immunization coverage. The seasonal flu, whether endemic or epidemic, usually occurs in autumn and winter when humans live in an atmosphere with a closer social distance, and lower temperature and humidity. The flu epidemic can be limited by herd immunity and/or mass vaccination that is selected and prepared from the emergence of seasonal influenza in the previous years. This is an example of the balance between virus mutation and herd immunity (57). A seasonal flu usually has a reproduction number (Ro) about 1.2~1.3, which can be controlled by herd immunity or vaccination if coverage is over 25% of the population (1 - 1/Ro = 1 - 1/1.3 = 25%). A flu pandemic is different and is usually caused by a series of antigenic reassortments (shifts) among cross-species flu viruses, which is novel to a population without immunity and causes a potential pandemic and fatal transmission. A novel cross-species flu virus usually causes a pandemic involving about 30-50% of the population in the initial years because almost all humans are susceptible to the novel influenza virus (58). This pandemic could re-emerge after a period of several years or decades; approximately 36 years (58), depending on the evolution and adaptation of a cross-species flu virus among avian, swine, and human hosts, and on the control of school closures, vaccination, facemask use, and isolation (59, 60). Another hypothesis for pandemic re-emergence is related to introduction of a dominant flu subtype virus into a population where the kinetic balance between virus virulence and human immunity is broken. Once a novel strain of flu virus can cause human-human transmission, it usually has a Ro value around 1.8 in the first wave of the epidemic and an attack rate of 10-30%. Second and/or 3rd waves will follow until herd immunity of over 60% is reached (60). The novel strain virus eventually transforms into a dominant subtype of the influenza epidemic and affects most of the population, particularly children who become infected with the pandemic strain over several years. This will confer some level of protection to older individuals and protect them from morbidity and mortality of influenza until the next pandemic (60). In a simulation model, the Ro of a novel influenza virus transmission among human-human transmission is around 1.3~1.8 (61), and that of the SARS-CoV-2 is around 2.3 (62), respectively. To control the pandemic requires infection or immunization rates of 33% (1 – 1/1.5) and 57% (1 – 1/2.3), respectively, based on the equation, 1 - 1/Ro, to cease the pandemic (63). Although a number of SARS-CoV-2 vaccines have been shown effectiveness on controlling the outbreaks with different SARS-CoV-2 variants, certain variants cause higher virulence, higher reproduction number, and/or breakthrough of COVID-19 vaccines (20–23), potentially contributing to re-infection after natural infection or vaccination. Whether the novel SARS-CoV-2 pandemic might also cause periodic waves of epidemics remains a great concern (64).
Genetic polymorphisms of immunity genes and virus receptors also affect infectivity and fatality of an emerging infection. Polymorphisms of CCR5, CCR2, CX3CR1, and SDF1 have been shown to influence HIV susceptibility and treatment responses (65). Polymorphisms of human leukocyte antigen (HLA), MBL2, CD209, and vitamin D receptor genes were associated with development of TB in HIV patients (66). We have found that a combination of TGFβ and CTLA-4 genotypes was significantly associated with the susceptibility to DHF (67). We (68) and others (69) have shown that a promoter polymorphism of CD209, a C-type lectin, was significantly associated with DHF. Recently, we found that the L-SIGN (CD299) polymorphism at the neck region of 9-tandem repeats was associated with susceptibility to DHF and correlated to virus replication and immune response (unpublished data). Similarly, the nine-repeat of CD299 isoform was associated with increased HIV viral load and HIV sexual transmission (70). ACE2 is a receptor for SARS-CoV-1 and SARS-CoV-2 infection, but the polymorphism of ACE2 was not associated with severity of infection (71). In contrast, glycosylation of Spike antigen is critically involved in recognition and binding of coronavirus (72) and affects binding affinity of host antibodies (73). TLR7 genetic variants cause predisposition to severe COVID-19 infections (74). Genetic variants in IL6R, TLR3, and DC-SIGN genes were associated with susceptibility and/or severity of DF (75). Genetic polymorphisms of DC-SIGN, TLR3 and TNF-α genes are also risk factors for the susceptibility and disease progression of Chikungunya infection (76). Interferon-inducible transmembrane protein 3 (IFITM3) gene is associated with susceptibility to severe influenza (77), and the variant with higher TMPRSS2 expression confers a higher risk to susceptibility of human A(H7N9) influenza and severity of A(H1N1)09 influenza (78). Ran Binding Protein 2 (RANBP2) gene mutations increase the susceptibility to recurrent episodes of necrotizing encephalitis with respiratory viral infections, particularly influenza infection (79).
Culture, occupation, and socioeconomic status also affect the spread of emerging infections. A patient with hemorrhagic fever and symptoms of bloody diarrhea, bleeding gums and skin, hemorrhagic eyes or urine, should be traced back to the suspected contact of Ebola virus or Marburg virus through a dead or sick animal (for Ebola virus) or a mine or cave with bat colonies (for Marburg virus) (80). Ebola and Marburg hemorrhagic fevers usually begin as an exposure to affected animals followed by human-human transmission (80). Travelers who visit the endemic area of Africa may spread the filoviruses worldwide (81). Health caregivers or people in diagnostic laboratories who come into contact with tissue fluid samples may become infected through human-human transmission of Ebola virus because the virus shedding time in tissue fluids can persist for 30 to 60 days (82). Ebola outbreaks are also related to cultural funeral ceremonies, including washing and touching the corpse and close contact during funeral ceremonies (83). Another cultural issue that influences emerging infections is wet markets in Asia. SARS coronaviruses and avian influenza viruses can be identified in live poultry markets (84), posing a need for virological and serological monitoring of viruses and hosts in live poultry markets which are still popular in Asian countries.
An emerging infection can rapidly lead to a pandemic with high fatality rates. Each individual emerging infection has its unique pattern of infectivity related to virus-host interactions underlying ligation of pathogen-associated molecular pattern (PAMP) to pattern recognition receptor (PRR) for the signaling of immune responses toward proper defense or morbidity. It is always debatable whether the high fatality of an emerging infection is related to viral virulence, immune deficiency, or immunopathology.
We studied immune responses to enterovirus 71 (EV71) (85–88), dengue (67, 68, 89–96), SARS-CoV-1 (30, 97–100), and influenza A (H1N1) 2009 infections (101–104) employing a real time simultaneous detection of viral load and immune responses (Figure 1). A TaqMan qRT-PCR was used to replace classical time-consuming plaque-forming unit assay of viral load, and cell cytometers were used to measure quantity and quality of leukocyte counts and activation. Based upon our studies and others’ studies, we have classified common immunopathogeneses of different emerging infections into 4 categories in Table 2: 1) Deficient immunity with disseminated viremia; 2) Pneumocytotropism with/without later hyperinflammation; 3) Augmented immunopathology; and 4) Antibody-dependent enhancement of infection with altered immunity.
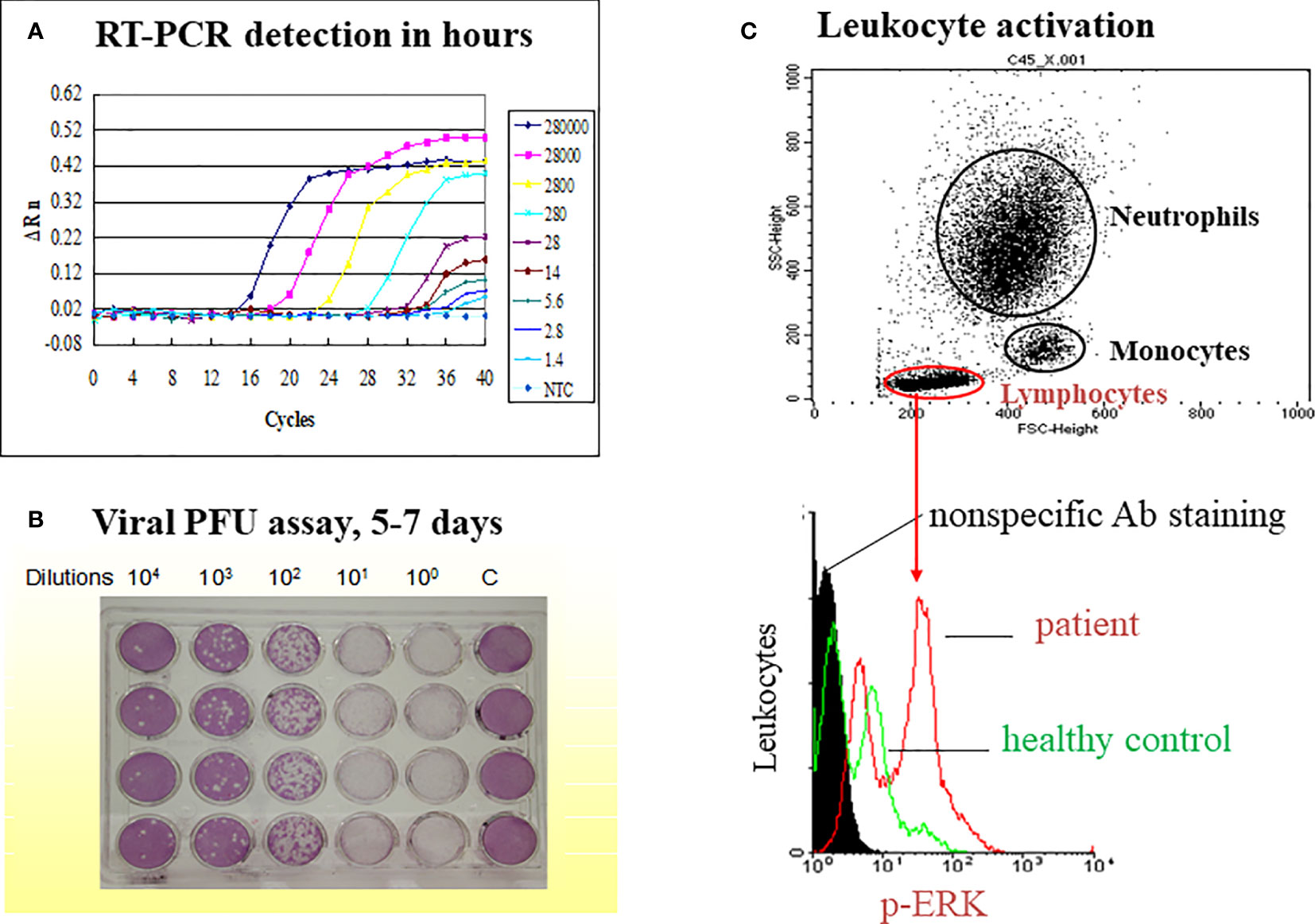
Figure 1 A model on simultaneous measurement of viral load and immune responses in an emerging infectious disease such as dengue fever. The study model presented is derived from our previous publication (Yeh, et al. FEMS Immunol Med Microbiol. 2006;48 (1):84-90). (A) A TaqMan qRT-PCR is used to replace classical time-consuming plaque-forming unit assay of viral load (B). The limit of detection (LOD) is 14 copies of the dengue 2 virus while cutting off the PCR cycle at 35, and the LOD is 1.4 copies of the dengue virus while cutting off the PCR cycle at 40. (C) A flow cytometric assay is used to gate different populations of leukocytes (neutrophil, monocyte and lymphocyte) for the activation assay such as ERK activation in lymphocytes as indicated.
Emerging infections that fit into this category include Ebola, Lassa fever, West Nile virus (WNV) encephalitis, and EV71 encephalitis (Table 2). A study with Ebola and Lassa viruses showed that Ebola and Lassa virus infection could compromise monocyte-derived dendritic cell function resulting in impaired adaptive immunity (105). Patients with fatal Ebola infection tended to have an impaired humoral response associated with 100% detectable viremia (106, 107). Lassa fever with fatal outcome was related to impaired T cell reaction associated with overt viremia and disseminated vascular insults (108, 109). For WNV encephalitis, the virus tended to infect immunocompromised hosts, especially those with B cell defect, causing higher mortality (110).
Our study on the immunopathogenesis of EV71 encephalitis also demonstrated that younger children with impaired T cell activation of CD40L were associated with EV71 infection complicated by encephalitis (85). Patients with EV71 encephalitis tended to have higher IL-8 and IL-2 levels than those without (86). Patients with encephalitis associated with neurogenic pulmonary syndrome had augmented IL-6 and TNFα levels in their blood (87). Further studies showed sialylated glycans as a receptor and inhibitor of EV71 infection to DLD-1 intestinal cells (88). The blood viral load in EV71 encephalitis patients was significantly higher than in those without encephalitis (Figure 2A). In contrast, the blood viral load in patients with dengue hemorrhagic fever (DHF) was not significantly different from patients with dengue fever (Figure 2B). Taken together, the severity of Ebola, Lassa fever, West Nile encephalitis and EV71 encephalitis is correlated to immune deficiency with disseminated viremia. Detection of definite impaired immunity and/or viremia in these infections alerts to the seriousness and calls for emergent medical assistance.
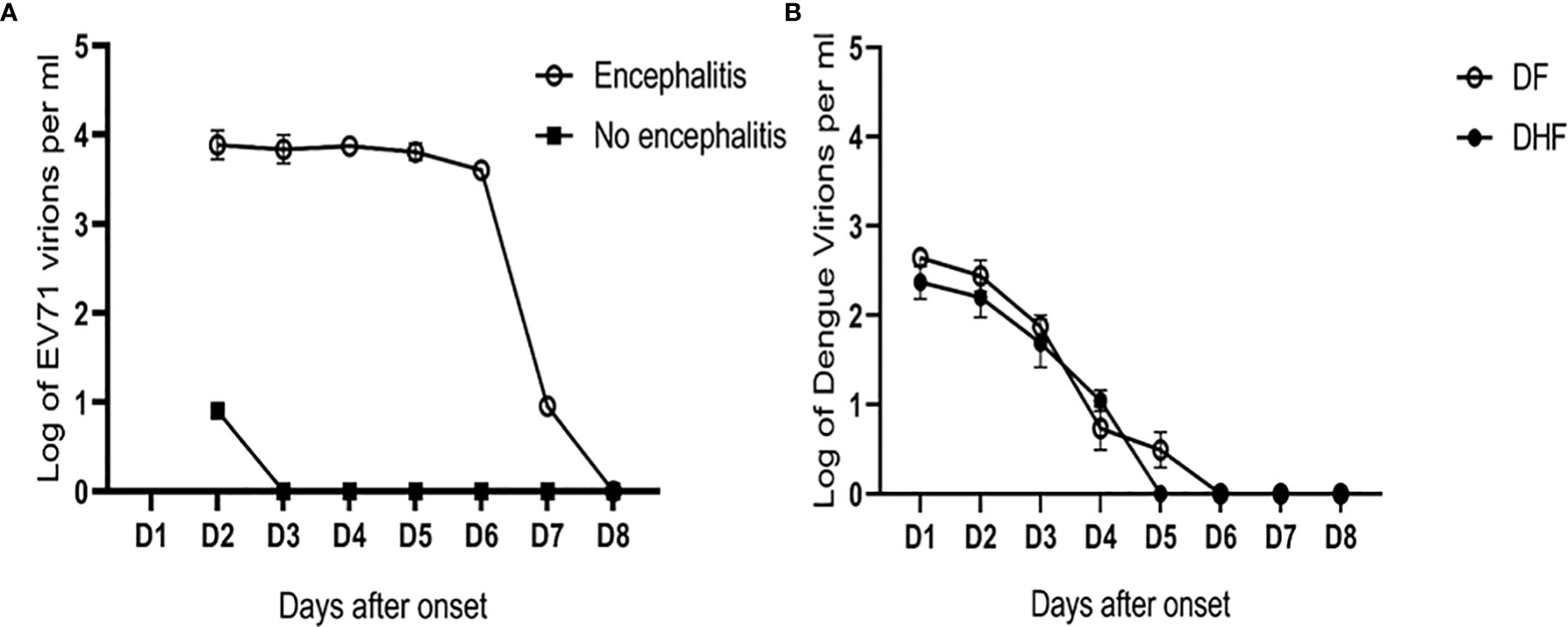
Figure 2 Different patterns of viral load in EV71 and dengue infections. (A) The blood viral loads in EV71 encephalitis patients were significantly higher than those with no encephalitis (data derived from 12 pairs of case-control samples). Based on the unit of one milliliter blood (ml), the limit of detection (LOD) is 9 copies/ml in patients with EV71. In contrast, (B) the blood viral loads in patients with dengue hemorrhagic fever (DHF) were not significantly different from those in dengue fever (DF). The LOD is 3 copies/ml in patients with dengue infection (the representative graph is derived from the publication (Chen, et al. FEMS Immunol Med Microbiol. 2005;44 (1):43-50).
Emerging infections fit into this category include SARS-CoV-1, SARS-CoV-2, and swine influenza A(H1N1)2009 which bind and fuse into the cells of respiratory tract and cause proinflammatory reaction in the lungs, called pneumocytotropism (Table 2). SARS-CoV-1 and SARS-CoV-2 are believed to infect the human respiratory tract by binding to angiotensin-converting enzyme 2 (ACE2) (111), and influenza A virus recognizes sialic acid alpha 2,6 galactose on respiratory epithelial cells as a receptor (23). The viruses enter the lung epithelial cells and induce innate immunity with production of interferons which limit viral replication before adaptive immunity. In different virus-host interactions, the virulent antigen(s) of the viruses (112), or host genetic variants (74–78), could impair the innate immune response and cause proinflammation or immunosuppression, followed by altered hyperinflammation with skewed Th17 reaction (113, 114). The viral RNA of SARS-CoV-1 and SARS-CoV-2 cannot only be detected in respiratory secretions but also in urine, feces, tears, and blood (115, 116). Virus shedding is not apparent during the incubation period in SARS-CoV-1 but can persist for 15-20 days after illness onset (115). However, virus shedding of SARS-CoV-2 virus is found in nasopharyngeal swabs before symptom onset and can persist for at least 3 weeks (116). The RNA virus was found in blood and urine of SARS-CoV-2 patients, but the urine or blood samples never yielded the virus from culture (116). In SARS-CoV-2 infections, the predominant pattern of lung lesions in autopsy is ARDS, similar to the findings in other two coronavirus, SARS-CoV-1 and MERS-CoV, showing capillary congestion, hyaline membrane, interstitial edema, pneumocyte hyperplasia and platelet-fibrin thrombi, associated with infiltration of macrophages in alveolar lumens, and lymphocytes (117, 118). Electron microscopy revealed viral particles in cytoplasmic vacuoles of pneumocytes. Pathogenesis of the deaths in cardiopulmonary events of COVID-19 patients are not due to pneumonia with ARDS at all, but some sort of thrombosis or disseminated intravascular coagulopathy (DIC) which occurred before death (119). Patients with severe COVID-19 have a hyperinflammation with higher plasma IL-2, IL-7, IL-10, G-SCF, IP-10, MCP-1, MIP1A, and TNFα levels, particularly in elders showing “inflamed-aging” (120). Elders with SARS-CoV-1 or SARS-CoV-2 infections had a higher mortality in an age-dependent correlation, and in an association with co-morbidities (121–123).
We found that one-third of SARS patients had detectable blood SARS-CoV-1 RNA, although the viremia was unlikely related to the outcome of the disease (97). Patients with SARS-CoV-1 infection had a significant higher IL-8 level associated with augmented phosphorylated p38 expression of CD14 cells and depressed phosphorylated p38 expression of CD8 T cells in early stage (<7 days) but higher IL-2 levels in late stage (>7 days) (97, 98). One of the 15 SARS patients studied had a late exacerbation of ARDS with a surge of p-ERK expression of CD8 T cells requiring steroid pulse therapy, which reversed the hyperactivation of p-ERK expression after the steroid pulse therapy (Figure 3). An exposure history and an early progression of chest X-rays in SARS-CoV-1 patients was associated with poor outcomes (99). SARS patients tended to have lymphopenia and thrombocytopenia which was caused by cell apoptosis associated with higher sFasL levels, and vascular sequestration associated with increased sVCAM-1 levels (100). Taken together, these results suggest uncontrolled regional pneumocytotropic lung damage, but not viremia responsible for the poor outcome of SARS-CoV-1. Some SARS-CoV-1 patients revealed a secondary exacerbation between the second and third weeks of infection in which CD8 T cell activation with higher IL-2 production was found (Figure 3).
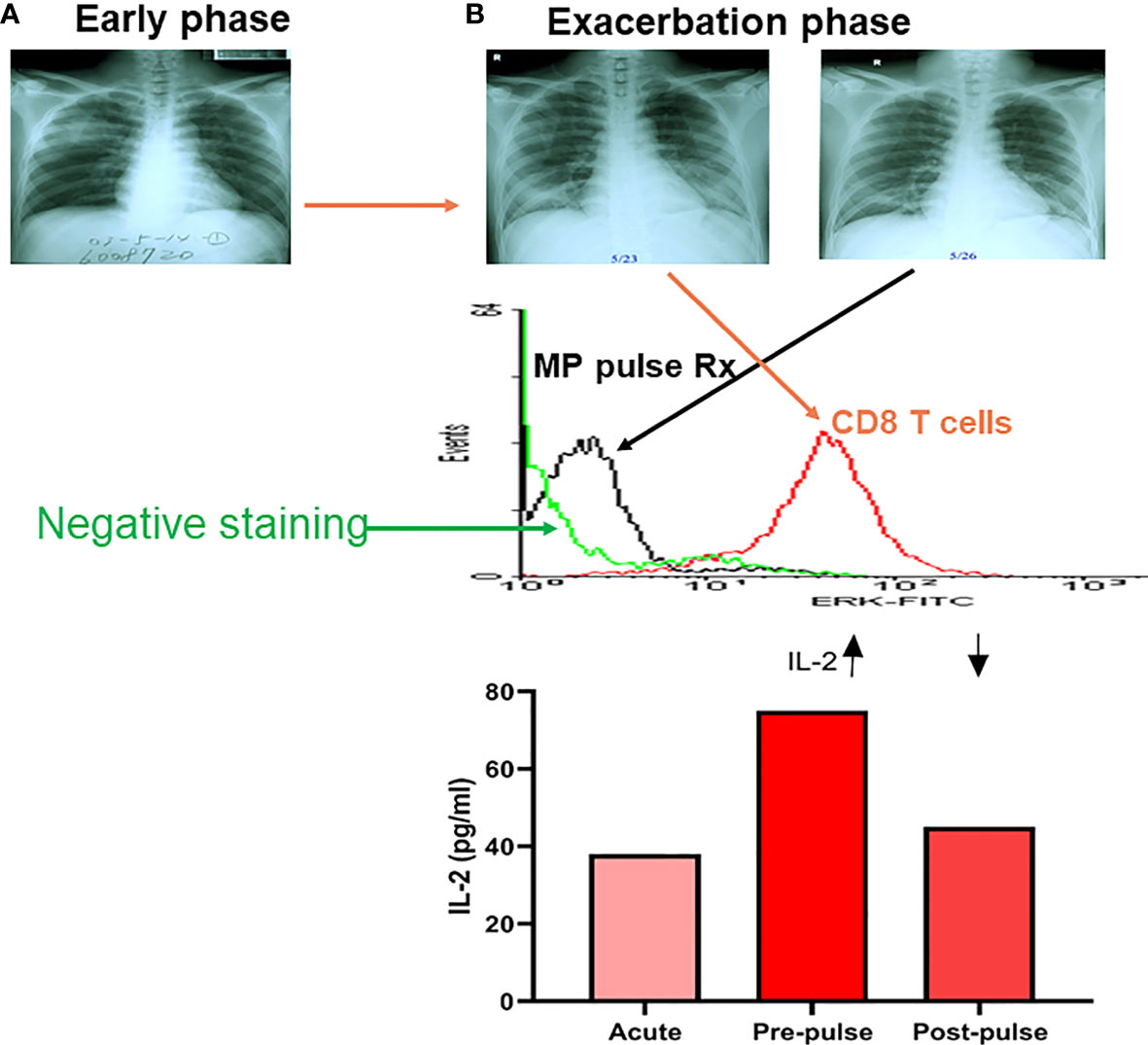
Figure 3 A kinetic tracing of chest X-rays, intracellular p-ERK and plasma IL-2 levels in a patient with late exacerbation of SARS-CoV-1 before and after methylprednisolone pulse treatment (MP pulse Rx). An early phase X-rays film in a SARS patient (A), who developed late phase exacerbation (B) showing a high intracellular p-ERK level of CD8 T cells (flow cytometric analysis of intracellular phosphorylated ERK levels) in exacerbation, and dramatically decreased after a 3-day course of methylprednisolone (MP; 1 gm/day) pulse Rx, associated with a decrease in plasma IL-2 level after the MP pulse Rx. (Data presented are derived from Li & Yang, et al. J Immunol. 2004;172 (12):7841-7).
In the swine influenza A (H1N1) 2009 outbreak, we found different clinical features between children and adults (101–103), and the younger children had a longer viral shedding time (102), and characteristic early lymphopenia and lower C-reactive protein levels (103). The influenza A (H1N1)2009 infection was associated with depressed NK cell function (Figures 4A, B). In other words, a higher initial viral uptake and/or suppressed immunity determines whether there is overwhelming regional lung damage and complication or not.
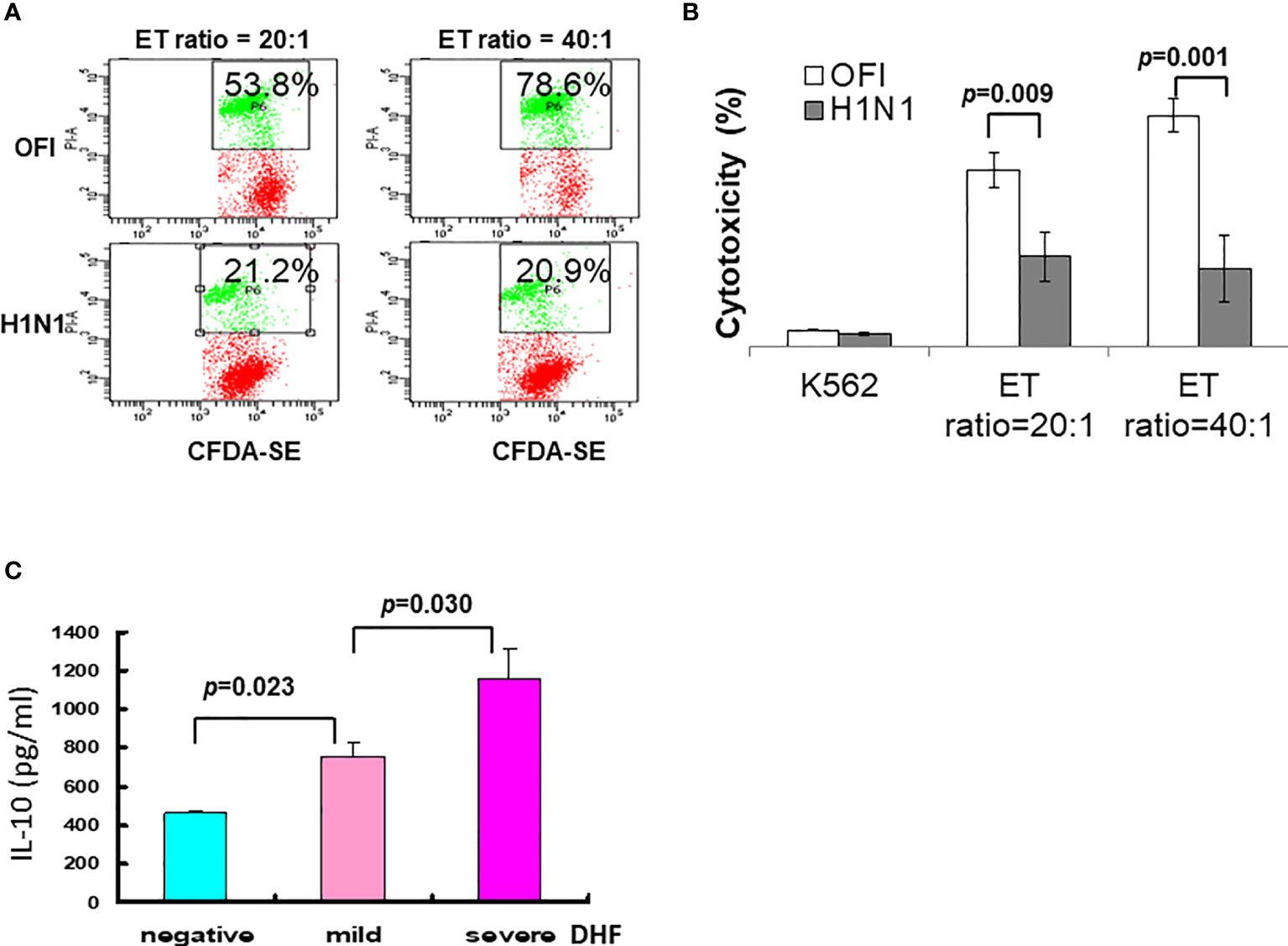
Figure 4 Immune alteration of influenza A (H1N1) 2009 and dengue infection. (A) Depressed NK cell activity in influenza A (H1N1) 2009 infection. Employing K562 cells labeled with CFDA dye, the cell cytotoxicity was detected by propidium iodide (PI). (B) The NK cell cytotoxic activities in patients with influenza A (H1N1) 2009 infection were significantly depressed either in effector-target (ET) ratio at 20:1 or 40:1 in comparison to those in patients with other febrile illness (OFI). (C) A significant association of IL-10 levels among dengue infections with mild or severe DHF. In a dengue outbreak, a cohort of patients with dengue fever, and dengue hemorrhagic fever mild or severe showed a significant higher level of IL-10 toward severe DHF (data derived from Chen RF, Yang KD, et al. Trans R Soc Trop Med Hyg. 2007;101 (11):1106-13).
Certain viruses do not cause systemic virus dissemination in the blood, but cause a systemic immune response with cytokine storm, or indirectly assault vessels by augmented immune reactions resulting in hemorrhage or vascular leakage (124–126). Emerging infections fit into this category, including Avian flu and Hanta viruses (Table 2). Patients with fatal H5N1 infections had a cytokine storm with low peripheral blood T-lymphocyte counts, associated with pharyngeal viral loads (127). Patients with fatal Hantavirus fever renal syndrome (HFRS) or Hanta cardiopulmonary syndrome (HCPS) had varied cytokine storms without viremia (128). Currently, H5N1 avian flu virus infects humans via the bird-to-human transmission and likely by the oral-fecal route, but not via aerosol transmission (129). However, avian H5H1 flu virus RNA was detected by RT-PCR in the lungs, intestines, and spleen. Active viral replication was limited to the lungs and intestine. This is compatible with clinical symptoms of pneumonia and diarrhea associated with altered immunity with circulating thrombocytopenia, cytokine storm and hemophagocytic syndrome (130). This suggests that regional unlimited viral replication due to depressed immunity, which is associated with uncontrolled proinflammatory cytokine production, is involved in the immunopathogenesis. An appropriate treatment may require not only an anti-viral agent (e.g. Tamiflu for avian flu within 3 days), but also immunomodulation of cytokine storm (e.g. anti-IL6 for COVID-19) as early as possible.
Hantavirus infects humans exposed to secretions of reservoir hosts (e.g., rats), resulting in a dead-end infection in humans with a long incubation period between 2-4 weeks. The hantavirus replicates in endothelial cells without cytopathic effect (CPE) but induces vascular leakage by a mechanism related to anti-viral mediators of endothelial cells, or cell immunity directed against infected cells by different cytokine storms in blood and affected tissues (56, 131). Infections occurring in the lungs are called HCPS (55, 56), and those occurring in the kidneys are called HFRS (53, 54). Hantaviruses infect endothelial cells via the β3 integrins which induce hyperresponsive to the permeability of endothelial cells by VEGF (131).
The human immune system can discriminate non-self-microbes and raise a memory immune reaction after the infection. The memory immune response produces neutralizing antibodies for immuno-surveillance of the same microbes and/or cross-reactive protection of similar microbial infections. Unfortunately, certain emerging infections that raises antibodies may cause cross-enhancement of infections as seen in dengue fever and Ross River viral infections (Table 2). Patients with secondary dengue fever are more susceptible to complications of DHF and dengue shock syndrome (DSS) (132–134). The antibodies raised in primary dengue infection can circulate in the blood for years or even decades, providing protection from the same serotype of dengue infection, but cause cross-enhancement of secondary heterotypic dengue infections, in which subneutralizing antibodies enhance heterotypic dengue virus infection (89–92), and alter immune response shifting type 1 T helper (Th1) response to Th2 response with dominant IL-10 in patients with DHF (89, 92, 94). The first implication for DHF was the observation that over 85% of children with DHF had high dengue heterotypic cross-reactive antibody titers in a Bangkok outbreak of DHF (43, 132), suggesting an antibody-dependent enhancement (ADE) of dengue infection in the pathogenesis. This hinders dengue vaccine development because of antibody-dependent enhancement (ADE) of dengue infections due to vaccine-induced heterotypic antibodies. In contrast to the DHF, which more frequently occurs to children in East Asia, our studies found that elders with comorbidities are more susceptible to DHF (90, 91, 95), and patients carrying certain genotypes were significantly associated with DHF (67, 75). We also found that previous subclinical dengue infections are more frequently associated with DHF (92, 94, 96), and elders with comorbidity or concurrent bacteremia have a higher mortality (91, 95). To explore the biomarker for early detection of DHF, we found that blood IL-10 levels were significantly associated with severity of DHF (Figure 4C). In addition to the ADE of dengue infections (43, 89, 92), another example of ADE was demonstrated in Ross River viral infections with polyarthritis, in which the presence of antibody enhances viral infection by macrophages (135). Like ADE of dengue, antibodies of COVID-19 infections have been proposed to induce augmented immune response by Fcγ-receptor mediated enhancement (136).
Mechanisms of the fatality in various emerging infections are different so that protection from fatality of each emerging infection requires an advanced deployment on early detection of the fatal pathogenesis among viral dissemination, immune deficiency, and immunopathology to develop a proper strategy to prevent or decrease fatality. For those with disseminated viremia, anti-viral agents such as interferons, inhibition of RNA replication with drugs such as remdesivir or favipiravir, and/or agents that block viral shedding, such as silmitasertib can be applied (137–139). Those with immune deficiency or with high viral load require earlier supplementation of hyperimmune immunoglobulins, neutralizing MoAbs, or convalescent plasma from convalescent patients (140–142). Those with immunopathology such as cytokine storm require administration of cytokine antagonist, inhibition of complement cascade, or adsorption of circulating cytokines (143–145). Those with infection-associated hemophagocytosis, also called secondary hemophagocytic lymphohistiocytosis or macrophage activation syndrome, require administration of IVIG, cyclosporin-A, corticosteroids, and/or anti-cytokine therapy (146, 147). As shown in Table 3, we have summarized how to differentiate fatal mechanisms and early signature markers for crisis management of early recognition and prevention, based on pathogenic mechanisms of overt viremia, tissue-specific organ failure, cytokine storm, and iatrogenic insults.
An emerging infection that causes impaired or delayed cell immunity or production of neutralizing antibodies can raise systemic viremia or immunopathology that causes a high fatality with hemorrhagic fever (coagulopathy), respiratory failure, and/or encephalitis. Patients with fatal Ebola infection tend to have 100% detectable viremia (106, 107). The early recognition of infection, viremia or antigenemia could promote not only an early administration of neutralizing antibodies (MoAbs or convalescent plasma) for reducing viral load and fatality, but also a timely interruption of the transmission of the emerging infection (Table 3.1). For instance, a rapid diagnostic test (RDT) for the Lassa viral antigen by a point of care test of immunochromatography can alert for systemic viremia (148). Detection of an early systemic viral load of Lassa fever in blood will raise the warning sign for early intervention (149) with administration of ribavirin (anti-viral agent), and/or convalescent plasma. These early interventions were shown to significantly reduced fatality (150, 151). Our study on the immunopathogenesis of enterovirus 71 encephalitis also demonstrates that younger children with impaired T cell reaction are associated with delayed CD40L expression and viremia (85, 86). In a simulation for Ebola containment based on a Ro value of 2.0, it is estimated that a rapid blood test reduces the attack rate from 80% to nearly zero, and the average diagnostic time from 5 days to 1 day in 60% of Ebola virus-infected patients (152). More importantly, in an Ebola outbreak, the early diagnosis would also promote the efficacy of ring vaccination by rVSV-ZEBOV which provided 100% vaccine efficacy (0/4539 vs. 39/4557 cases) in the immediate vaccinees after known exposure compared to the delayed group vaccinated 21 days after exposure (153). Administration of MoAbs or convalescent plasma in early stage of infection has also been shown effectiveness on the limitation of disease progression in Ebola (140), SARS-CoV-1 (141), MERS-CoV (142), and SARS-CoV-2 (154).
Rapid diagnoses of emerging viral infections using point of care tests (POCT) for detection of specific antigen or nucleotide are made available in recent years, particularly during the COVID-19 pandemic (155). The paper-based POCT can be done in 15 minutes by detecting antigen-antibody reaction in secretion of upper respiratory tract or blood (155). The early detection within 3 days may be followed by early treatment of neutralizing antibodies to reduce viral load of the lung and reduce complication (140, 154). Some emerging infections can cause tissue-specific cytotropism; for instance, SARS-CoV-2 and Avian influenza virus can cause respiratory distress syndrome and Hanta virus can cause renal failure. The emergence of Avian flu and Hantavirus syndrome did not cause systemic viral dissemination, but assaulted vascular endothelium by augmented immune reactions, resulting in hemorrhage, pulmonary edema, or renal failure (125, 126). As shown in Table 3.3, kinetic monitoring of lung and kidney functions is mandatory to prevent Hantavirus-induced organ failure. This can be accomplished through ventilation support, continuous renal replacement therapy (CRRT), and/or extracorporeal membrane oxygenation (ECMO) support (156). In addition, Icatibant which blocks the binding of bradykinin has been used to treat hantavirus infection with complement activation and coagulopathy (157). For patients with a fulminant or a treatment resistant course, strategies to identify host genetic variants that compromise defense, or to identify viral virulent factors that induce immunosuppression are required. For instance, a respiratory tract infection with repeated influenza infections or fulminant (necrotizing) encephalitis should be screened for genetic mutations at Ran Binding Protein 2 (RANBP2) (79) or interferon-inducible transmembrane protein 3 (IFITM3) (77), respectively, and anti-virus treatment (e.g. Tamiflu) should be initiated as early as possible.
Certain emerging infections can cause altered immunity which results in the release of untoward cytokines causing cytokine storm of immunopathology. Because organ failure is related to inflammatory insults, anti-inflammatory regimens are necessary (Table 3.3). The cytokine storms in different emerging infections are frequently associated with augmented levels of IL-6, IL-1β, IL-8, TNFα, and/or IP-10 (87, 89, 92, 97, 120, 158). Anti-IL6R and/or anti-IL1 antibodies are indicated in the treatment of cytokine storm of COVID-19 (159). Moreover, the cytokine profiles induced by coronavirus infections are related to T helper cell type 17 (Th17) reactions (97, 114, 120, 158, 159), to which immunoregulatory therapies have been proposed (113, 160). For cases complicated by abnormal complement cascade and coagulopathy (higher D-dimer and lower platelets), a combined therapy with anti-C5a antibody and Jak1 inhibitor may be needed (161). In addition, some patients may require utilization of heparin, ECMO and/or CRRT treatment (162). Avian flu with cytokine storm might be associated with augmented immune responses such as hemophagocytosis showing anemia, thrombocytopenia, hyperferritinemia, hypertriglyceridemia, and adult type respiratory distress syndrome (ARDS) without detectable viremia, which may require a combination of IVIG with steroids, and cyclosporin A or etoposide (163, 164).
An emerging infection can cause high fatality when conditions such as sepsis and complications due to comorbidities or malpractices are superimposed (Table 3.4). Strategies to alert to these superimposed conditions will promote integrated therapies including anti-virus, anti-bacteria, anti-inflammation, and ventilation or renal support. Many patients with an emerging infection die of sepsis because of virus-induced immunosuppression (95, 165). In these cases mass spectrometry fingerprinting of blood culture is necessary to early detect bacteremia, identify antibiotic resistance, and prevent sepsis. New drugs or crisis management may result in novel toxicity or unexpected drug interactions in patients with comorbidities. Certain emerging infections, particularly those prone to nosocomial infections such as Ebola and SARS, can impact not only the general population but also health care providers and medical institutions. Containment of nosocomial and emerging infections in health care centers and long-term care facilities where elders are frequently bedridden with multiple comorbidities is especially important, since co-morbid patients are usually super-spreaders and succumb to higher morbidity and mortality, requiring early RDTs and reduction in viral load by MoAbs or convalescent plasma. Overtreatment or undertreatment of an emerging infection may cause iatrogenic morbidity and mortality. For instance, early mechanical ventilation or late use of neutralizing antibodies may increase morbidity and mortality. Shortage of medical resources or shortages of health providers could also increase potential complications. Continuing education with advanced deployment and use of computer simulation can be used to reduce iatrogenic side effects.
Because each individual emerging infection has its own evolutionary trait, transmission route, and immunopathogenesis, each emerging infection requires individualized strategies to prevent infection, morbidity and mortality. However, “stones from other hills may serve to polish the jade of this one,” advance deployment may be made for mitigating a pandemic and reducing fatality. A stepwise guideline for infection and immunity controls to prevent an emerging infection may be possible (166). As shown in Figure 5, there are 5 check points of infection controls to prevent infection, morbidity and fatality:
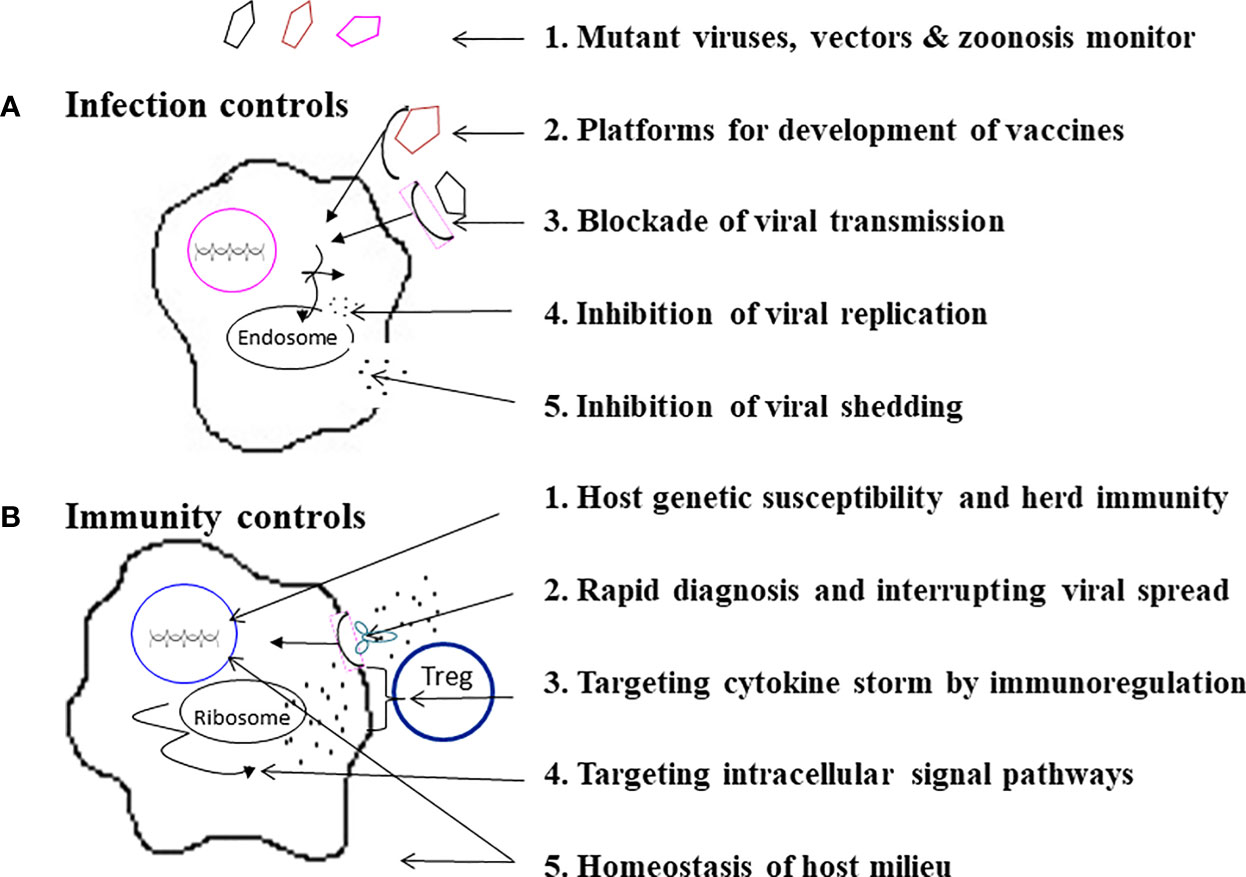
Figure 5 A stepwise practical guide to do infection controls and immunity controls. As specified, (A) Infection controls can be made by a 5-step program, and (B) Immunity controls can be approached by another 5-step program.
(1) Monitoring for mutant viruses, vectors & zoonosis. The best way to prevent pandemics and fatalities due to an emerging infection is to monitor potential emerging microbials in mutations, vectors, and zoonosis before and during pandemics (1–13, 27–30, 84). In this era of a global village and changes of ecosystems, early prediction, recognition, and elimination of an emerging infection is not guaranteed. Preparedness of mass vaccination, convalescent plasma and specific anti-virus agents is also important.
(2) Platforms for development of vaccines. A couple of new platforms for rapid development of vaccines by avirulent virus vectors with DNA, mRNA vaccine and recombinant protein technologies that are safe and efficacious have been made possible (167–170). For instance, the fast pipelines of vaccines for an emerging infection such as COVID-19 were made available within one year (169, 170).
(3) Blockade of viral transmission. Before a vaccine is available for an emerging infection, it is important to encourage wearing of facemasks, keeping social distance and doing surface disinfection. These measures may not only have an effect on blocking transmission of the emerging infection, but may also have collateral benefits by decreasing other upper respiratory tract infections (171, 172).
(4) Inhibition of viral replication. The inhibition of viral replication could be made by antiviral agents directed against virus-cell fusion, virus and host proteases, and RNA synthetase (137–139).
(5) Inhibition of viral shedding. In SARS-CoV-2 infections activations of casein kinases (CK2) and protein kinases (MAPK) have been demonstrated (137). Inhibitors of CK2 and protein kinases which have demonstrated safety data in human trials have been proposed to re-purposing of the FDA-approved kinases inhibitors for the treatment of COVID-19 (137, 173, 174). A combination of anti-viral replication and shedding may provide a synergistic effect on mitigation of viral transmission
There are 5 other check points for immunity controls of an emerging infection:
(1) Host genetic susceptibility and herd immunity. In different emerging infections mortality ranges from 1% to 60%. Many humans survive because of host immunity and herd immunity. For patients who experience a fulminant disease course or treatment resistance, it is necessary to survey for host genetic susceptibility. For instances, deletion or mutation of TLR7 has been attributed to severity of COVID-19 in young adults (74), in which protection or early administration of MoAbs (REGN-CoV-2) may limit morbidity and mortality. Similarly, Ran Binding Protein 2 (RANBP2) mutation has been associated with fulminant necrotizing encephalitis of influenza (79), in which early prophylactic use of Tamiflu may prevent complication and fatality.
(2) Rapid diagnosis and interrupting viral spread. Rapid diagnostic tests have made early detection and interruption of disease progression and viral transmission possible (148, 149, 152, 153, 155). In an Ebola outbreak, an RDT made a ring vaccination possible that provided 100% vaccine efficacy in the immediate vaccinees (152, 153). Early administration of MoAbs or convalescent plasma has also shown effectiveness on the limitation of disease progression in Ebola (140), SARS-CoV-1 (141), MERS-CoV (142), and SARS-CoV-2 (154, 175–177). It is also postulated that a combination of neutralizing MoAbs and anti-virus agent may induce a synergistic effect (178). The early diagnosis followed by early treatment with MoAbs or convalescent plasma in 72 hours has been shown to reduce viral load, hospitalization and disease progression of COVID-19 (175–177).
(3) Targeting cytokine storm by immunoregulation. Different emerging infections may induce variant types of cytokine storm (87, 92, 97, 120, 158) to which immunotherapies with anti-cytokine and/or immune regulatory therapies have been proposed to rescue the patients with cytokine storm (114, 159–161). It is, however, postulated that aiming at a single target of one cytokine action may be ineffective, and sequential targeting may be required for eliminating the cytokine storm (178). A combined regimen with circulating supports by ECMO and eliminating cytokines by CRRT (156, 157, 162) may be beneficial.
(4) Targeting intracellular signal pathways. Hyperactivation of MAPK pathway and CK2 (casein kinase 2)-phosphorylation have been associated with SARS-CoV-1 and SARS-CoV-2 infections (97, 137), and inhibition of p38 activation or CK2 activation has been shown to decrease viral replication and cytokine induction (137, 174).
(5) Homeostasis of host milieu. The abnormal virus-host interactions for fulminant inflammation on emerging infections may not only depend on viral mutation and host genetic variants, but also host milieu: interior environment, such as imbalances of vitamins and microbiota, and external environment, such as temperature, humidity and protection equipment. For instance, maintenance of host interior homeostasis on vitamins (e.g. vitamin D, retinoids, vitamin K2) and metabolites of microbiota, which provide anti-virus properties and/or better Treg responses for anti-inflammatory reactions (179–184), may regulate immunity and reduce mortality of an emerging infection.
BY completed the literature review, performed several studies on immunopathogenesis of enterovirus 71, and drafted the manuscript. KY made the scheme for writing this perspective article and revised the manuscript for final submission. All authors contributed to the article and approved the submitted version.
This study was supported in part by a grant MMH109-005 from Mackay Memorial Hospital, Taiwan; and a grant MOST 109-2811-B195-503 from Ministry of Science and Technology, Taiwan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the SARS team of Chang Gung Memorial Hospital at Kaohsiung for their collection of samples for parts of the study. The authors also thank Ms. Susan Wright for her critical review of the manuscript.
1. Hui EK. Reasons for the Increase in Emerging and Re-Emerging Viral Infectious Diseases. Microbes Infect (2006) 8:905–16. doi: 10.1016/j.micinf.2005.06.032
2. Clark LK, Green TJ, Petit CM. Structure of Nonstructural Protein 1 From SARS-Cov-2. J Virol (2021) 95(4):e02019–20. doi: 10.1101/2020.11.03.366757
3. Jester BJ, Uyeki TM, Jernigan DB. Fifty Years of Influenza A(H3N2) Following the Pandemic of 1968. Am J Public Health (2020) 110(5):669–76. doi: 10.2105/AJPH.2019.305557
4. Lowen A, Palese P. Transmission of Influenza Virus in Temperate Zones is Predominantly by Aerosol, in the Tropics by Contact: A Hypothesis. PloS Curr (2009) 1:RRN1002. doi: 10.1371/currents.rrn1002
5. Phillips ML. Dengue Reborn: Widespread Resurgence of a Resilient Vector. Environ Health Perspect (2008) 116:A382–8. doi: 10.1289/ehp.116-a382
6. Johnson B, Chambers TV, Crabtree MB, Filippis AM, Vilarinhos PT, Resende MC, et al. Vector Competence of Brazilian Aedes Aegypti and Ae. Albopictus for a Brazilian Yellow Fever Virus Isolate. Trans R Soc Trop Med Hyg (2002) 96:611–3. doi: 10.1016/s0035-9203(02)90326-3
7. Brault AC. Changing Patterns of West Nile Virus Transmission: Altered Vector Competence and Host Susceptibility. Vet Res (2009) 40:43. doi: 10.1051/vetres/2009026
8. Zeier M, Handermann M, Bahr U, Rensch B, Müller S, Kehm R, et al. New Ecological Aspects of Hantavirus Infection: A Change of a Paradigm and a Challenge of Prevention–a Review. Virus Genes (2005) 30:157–80. doi: 10.1007/s11262-004-5625-2
9. Chae JS, Adjemian JZ, Kim HC, Ko S, Klein TA, Foley J. Predicting the Emergence of Tick-Borne Infections Based on Climatic Changes in Korea. Vector Borne Zoonotic Dis (2008) 8:265–75. doi: 10.1089/vbz.2007.0190
10. Zhang YZ, Dong X, Li X, Ma C, Xiong HP, Yan GJ, et al. Seoul Virus and Hantavirus Disease, Shenyang, People’s Republic of China. Emerg Infect Dis (2009) 15:200–6. doi: 10.3201/eid1502.080291
11. Letko M, Seifert SN, Olival KJ, Plowright RK, Munster VJ. Bat-Borne Virus Diversity, Spillover and Emergence. Nat Rev Microbiol (2020) 18(8):461–71. doi: 10.1038/s41579-020-0394-z
12. Leffel EK, Reed DS. Marburg and Ebola Viruses as Aerosol Threats. Biosecur Bioterror (2004) 2:186–91. doi: 10.1089/bsp.2004.2.186
13. Domingo E, Holland JJ. RNA Virus Mutations and Fitness for Survival. Ann Rev Microbiol (1997) 51:151–78. doi: 10.1146/annurev.micro.51.1.151
14. Sanjuán R, Domingo-Calap P. Mechanisms of Viral Mutatio. Cell Mol Life Sci (2016) 73:4433–48. doi: 10.1007/s00018-016-2299-6
15. Bedford T, Riley S, Barr IG, Broor S, Chadha M, et al. Global Circulation Patterns of Seasonal Influenza Viruses Vary With Antigenic Drift. Nature (2015) 523:217–20. doi: 10.1038/nature14460
16. Wen HL, Si LY, Yuan XJ, Hao SB, Gao F, Chu F, et al. Complete Genome Sequencing and Analysis of Six Enterovirus 71 Strains With Different Clinical Phenotypes. Virol J (2013) 10:115. doi: 10.1186/1743-422X-10-115
17. Chang CK, Wu SR, Chen YC, Lee KJ, Chung NH, Lu YJ, et al. Mutations in VP1 and 5’-UTR Affect Enterovirus 71 Virulence. Sci Rep (2018) 8:6688. doi: 10.1038/s41598-018-25091-7
18. Yeh SH, Wang HY, Tsai CY, Kao CL, Yang JY, Liu HW, et al. Characterization of Severe Acute Respiratory Syndrome Coronavirus Genomes in Taiwan: Molecular Epidemiology and Genome Evolution. Proc Natl Acad Sci U S A (2004) 101:2542–7. doi: 10.1073/pnas.0307904100
19. Mercatelli D, Giorgi FM. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front Microbiol (2020) 11:1800. doi: 10.3389/fmicb.2020.01800
20. Wikipedia. Variants of SASR-CoV-2. Retrieved on May, 26, 2021 (2021). Available at: https://en.wikipedia.org/wiki/Variants_of_SARS-CoV-2#cite_note-28.
21. Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, et al. Changes in Symptomatology, Reinfection, and Transmissibility Associated With the SARS-CoV-2 Variant B.1.1.7: An Ecological Study. Lancet Public Health (2021) 6(5):e335–45.doi: 10.1016/S2468-2667(21)00055-4
22. Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, et al. Sensitivity of Infectious SARS-CoV-2 B.1.1.7 and B.1.351 Variants to Neutralizing Antibodies. Nat Med (2021) 27(5):917–24. doi: 10.1038/s41591-021-01318-5
23. Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Porrot F, et al. Vaccine Breakthrough Infections With SARS-CoV-2 Variants. N Engl J Med (2021) NEJMoa2105000. doi: 10.1056/NEJMoa2105000
24. He CQ, He M, He HB, Wang HM, Ding NZ. The Matrix Segment of the “Spanish Fl” Virus Originated From Intragenic Recombination Between Avian and Human Influenza A Viruses. Transbound Emerg Dis (2019) 66:2188–95. doi: 10.1111/tbed.13282
25. Kumlin U, Olofsson S, Dimock K, Arnberg N. Sialic Acid Tissue Distribution and Influenza Virus Tropism. Influenza Other Respir Viruses (2008) 2:147–54. doi: 10.1111/j.1750-2659.2008.00051.x
26. Essere B, Yver M, Gavazzi C, Terrier O, Isel C, Fournier E, et al. Critical Role of Segment-Specific Packaging Signals in Genetic Reassortment of Influenza A Viruses. Proc Natl Acad Sci U S A (2013) 110:E3840–8. doi: 10.1073/pnas.1308649110
27. Smith EC, Denison MR. Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity. PloS Pathog (2013) 9:e1003760. doi: 10.1371/journal.ppat.1003760
28. Cagliani R, Forni D, Clerici M, Sironi M. Computational Inference of Selection Underlying the Evolution of the Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2. J Virol (2020) 94:e00411–20. doi: 10.1128/JVI.00411-20
29. Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Origin and Animal Reservoir. Virol J (2016) 13:87. doi: 10.1186/s12985-016-0544-0
30. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus From Animals in Southern China. Science (2003) 302:276–8. doi: 10.1126/science.1087139
31. Li X, Giorgi EE, Marichann MH, Foley B, Xiao C, Kong XP, et al. Emergence of SARS-CoV-2 Through Recombination and Strong Purifying Selection. Sci Adv (2020) 6:eabb9153. doi: 10.1126/sciadv.abb9153
32. Zhu Z, Meng K, Meng G. Genomic Recombination Events may Reveal the Evolution of Coronavirus and the Origin of SARS-Cov-2. Sci Rep (2020) 10(1):21617. doi: 10.1038/s41598-020-78703-6
33. Zhang Z, Shen L, Gu X. Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission. Sci Rep (2016) 6:25049. doi: 10.1038/srep25049
34. Gong YN, Tsao KC, Hsiao MJ, Huang CG, Huang PN, Huang PW, et al. Sars-CoV-2 Genomic Surveillance in Taiwan Revealed Novel ORF8-Deletion Mutant and Clade Possibly Associated With Infections in Middle East. Emerg Microb Infect (2020) 9:1457–66. doi: 10.1080/22221751.2020.1782271
35. de Souza Trindade G, da Fonseca FG, Marques JT, Lacerda Nogueira M, Nogueira Mendes LC, Secorun Borges A, et al. Araçatuba Virus: A Vaccinia Like Virus Associated With Infection in Humans and Cattle. Emerg Infect Dis (2003) 9:155–60. doi: 10.3201/eid0902.020244
36. Damaso CR, Reis SA, Jesus DM, Lima PS, Moussatché N. A PCR-Based Assay for Detection of Emerging Vaccinia-Like Viruses Isolated in Brazil. Diagn Microbiol Infect Dis (2007) 57(1):39–46. doi: 10.1016/j.diagmicrobio.2006.07.012
37. Lowen AC, Steel J, Mubareka S, Palese P. High Temperature (30°C) Blocks Aerosol But Not Contact Transmission of Influenza Virus. J Virol (2008) 82:5650–2. doi: 10.1128/JVI.00325-08
38. Zhang R, Li Y, Zhang AL, Wang Y, Molina MJ. Identifying Airborne Transmission as the Dominant Route for the Spread of COVID-19. Proc Natl Acad Sci USA (2020) 117:14857–63. doi: 10.1073/pnas.2009637117
39. Servadio JL, Rosenthal SR, Carlson L, Bauer C. Climate Patterns and Mosquito-Borne Disease Outbreaks in South and Southeast Asia. J Infect Public Health (2018) 11:566–71. doi: 10.1016/j.jiph.2017.12.006
40. Kotsakiozi P, Evans BR, Gloria-Soria A, Kamgang B, Mayanja M, Lutwama J, et al. Population Structure of a Vector of Human Diseases: Aedes Aegypti in its Ancestral Range, Africa. Ecol Evol (2018) 8:7835–48. doi: 10.1002/ece3.4278
42. WHO. Dengue and Severe Dengue. Retrieved on March 13, 2021 (2020). Available at: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
43. Halstead S. Recent Advances in Understanding Dengue. F1000Res (2019) 8:F1000 Faculty Rev–1279. doi: 10.12688/f1000research.19197.1
44. Liu Y, Liu J, Du S, Shan C, Nie K, Zhang R, et al. Evolutionary Enhancement of Zika Virus Infectivity in Aedes Aegypti Mosquitoes. Nature (2017) 545:482–6. doi: 10.1038/nature22365
45. Platt DJ, Miner JJ. Consequences of Congenital Zika Virus Infection. Curr Opin Virol (2017) 27:1–7. doi: 10.1016/j.coviro.2017.09.005
46. Gardner CL, Ryman KD. Yellow Fever: A Reemerging Threat. Clin Lab Med (2010) 30:237–60. doi: 10.1016/j.cll.2010.01.001
47. Douam F, Ploss A. Yellow Fever Virus: Knowledge Gaps Impeding the Fight Against an Old Foe. Trends Microbiol (2018) 26:913–28. doi: 10.1016/j.tim.2018.05.012
48. Roehrig JT. West Nile Virus in the United States - a Historical Perspective. Viruses (2013) 5:3088–108. doi: 10.3390/v5123088
49. Hadfield J, Brito AF, Swetnam DM, Vogels CBF, Tokarz RE, Andersen KG, et al. Twenty Years of West Nile Virus Spread and Evolution in the Americas Visualized by Nextstrain. PloS Pathog (2019) 15:e1008042. doi: 10.1371/journal.ppat.1008042
50. Sejvar JJ. Clinical Manifestations and Outcomes of West Nile Virus Infection. Viruses (2014) 6:606–23. doi: 10.3390/v6020606
51. Filgueira L, Lannes N. Review of Emerging Japanese Encephalitis Virus: New Aspects and Concepts About Entry Into the Brain and Inter-Cellular Spreading. Pathogens (2019) 8:111. doi: 10.3390/pathogens8030111
52. de Wispelaere M, Desprès P, Choumet V. European Aedes Albopictus and Culex Pipiens are Competent Vectors for Japanese Encephalitis Virus. PloS Negl Trop Dis (2017) 11:e0005294. doi: 10.1371/journal.pntd.0005294
53. Zhang YZ, Zou Y, Fu ZF, Plyusnin A. Hantavirus Infections in Humans and Animals, China. Emerg Infect Dis (2010) 16:1195–203. doi: 10.3201/eid1608.090470
54. de Oliveira SV, Faccini-Martínez ÁA. Hantavirus Infection and the Renal Syndrom. In: Bezerra da Silva Junior G, De Francesco Daher E, Barros E, editors. Tropical Nephrology (2020) 175–92. doi: 10.1007/978-3-030-44500-3_14
55. Clement J, LeDuc JW, Lloyd G, Reynes JM, McElhinney L, Van Ranst M, et al. Wild Rats, Laboratory Rats, Pet Rats: Global Seoul Hantavirus Disease Revisited. Viruses (2019) 11:652. doi: 10.3390/v11070652
56. López R, Pérez-Araos R, Salazar Á, Ulloa AL, Vial C, Vial PA, et al. Hemodynamic and Pulmonary Permeability Characterization of Hantavirus Cardiopulmonary Syndrome by Transpulmonary Thermodilution. Viruses (2019) 11:900. doi: 10.3390/v11100900
57. Brooke CB. Population Diversity and Collective Interactions During Influenza Virus Infection. J Virol (2017) 91:e01164–17. doi: 10.1128/JVI.01164-17
58. Morans DM, Taubenberger JK. Influenza: The Once and Future Pandemic. Public Health Rep (2010) 125(Suppl 3):16–26.
59. Taubenberger JK, Kash JC. Influenza Virus Evolution, Host Adaptation and Pandemic Formation. Cell Host Microbe (2010) 7:440–51. doi: 10.1016/j.chom.2010.05.009
60. Reperant LA, Moesker FM, Osterhaus ADME. Influenza: From Zoonosis to Pandemic. ERJ Open Res (2016) 2(1):00013–2016. doi: 10.1183/23120541.00013-2016
61. Biggerstaff M, Cauchemez S, Reed C, Gambhir M, Finelli L. Estimates of the Reproduction Number for Seasonal, Pandemic, and Zoonotic Influenza: A Systematic Review of the Literature. BMC Infect Dis (2014) 14:480.doi: 10.1186/1471-2334-14-480
62. Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the Reproductive Number of Novel Coronavirus (COVID-19) and the Probable Outbreak Size on the Diamond Princess Cruise Ship: A Data-Driven Analysis. Int J Infect Dis (2020) 93:201–4. doi: 10.1016/j.ijid.2020.02.033
63. Chang HJ. Estimation of Basic Reproduction Number of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) During the Outbreak in South Korea, 2015. Biomed Eng Online (2017) 16(1):79. doi: 10.1186/s12938-017-0370-7
64. D’Souza GA, Dowdy D. What is Herd Immunity and How can We Achieve it With COVID-19? Johns Hopkins Bloomberg School of Public Health (2020). (10 April 2020). Available at: https://www.jhsph.edu/covid-19/articles/achieving-herd-immunity-with-covid19.html.
65. Puissant B, Roubinet F, Massip P, Sandres-Saune K, Apoil PA, et al. Analysis of CCR5, CCR2, CX3CR1, and SDF1 Polymorphisms in HIV-Positive Treated Patients: Impact on Response to HAART and on Peripheral T Lymphocyte Counts. AIDS Res Hum Retroviruses (2006) 22:153–62. doi: 10.1089/aid.2006.22.153
66. Raghavan S, Alagarasu K, Selvaraj P. Immunogenetics of HIV and HIV Associated Tuberculosis. Tuberculosis (Edinb) (2012) 92:18–30. doi: 10.1016/j.tube.2011.08.004
67. Chen RF, Wang L, Cheng JT, Chuang H, Chang JC, Liu JW, et al. Combination of CTLA-4 and TGFbeta1 Gene Polymorphisms Associated With Dengue Hemorrhagic Fever and Virus Load in a Dengue-2 Outbreak. Clin Immunol (2009) 131:404–9. doi: 10.1016/j.clim.2009.01.015
68. Wang L, Chang WP, Wang L, Lin YJ, Liang CD, Kuo HC, et al. Dc-SIGN (Cd209) Promoter -336 a/G Polymorphism Is Associated With Dengue Hemorrhagic Fever and Correlated to DC-SIGN Expression and Immune Augmentation. PloS Negl Trop Dis (2011) 5:e934. doi: 10.1371/journal.pntd.0000934
69. Sakuntabhai A, Turbpaiboon C, Casadémont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, et al. A Variant in the CD209 Promoter Is Associated With Severity of Dengue Disease. Nat Genet (2005) 37:507–13. doi: 10.1038/ng1550
70. Xu L, Li Q, Ye H, Zhang Q, Chen H, Huang F, et al. The Nine-Repeat DC-SIGNR Isoform Is Associated With Increased HIV-RNA Loads and HIV Sexual Transmission. J Clin Immunol (2010) 30:402–7. doi: 10.1007/s10875-010-9376-7
71. Chiu RW, Tang NL, Hui DS, Chung GT, Chim SS, Chan KCA, et al. ACE2 Gene Polymorphisms Do Not Affect Outcome of Severe Acute Respiratory Syndrome. Clin Chem (2004) 50:1683–6. doi: 10.1373/clinchem.2004.035436
72. Grant OC, Montgomery D, Ito K, Woods RJ. Analysis of the SARS-CoV-2 Spike Protein Glycan Shield Reveals Implications for Immune Recognition. Sci Rep (2020) 10:14991. doi: 10.1038/s41598-020-71748-7
73. Zhou D, Tian X, Qi R, Peng C, Zhang W. Identification of 22 N-Glycosites on Spike Glycoprotein of SARS-CoV-2 and Accessible Surface Glycopeptide Motifs: Implications for Vaccination and Antibody Therapeutics. Glycobiology (2020) 2020:cwaa052. doi: 10.1093/glycob/cwaa052
74. van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA (2020) 324:663–73. doi: 10.1001/jama.2020.13719
75. Avendaño-Tamayo E, Rúa A, Parra-Marín MV, Rojas W, Campo O, Agudelo-Flórez P, et al. Evaluation of Variants in IL6R, TLR3, and DC-SIGN Genes Associated With Dengue in Sampled Colombian Population. Biomedica (2019) 39:88–101. doi: 10.7705/biomedica.v39i1.4029
76. Bucardo F, Reyes Y, Morales M, Briceño R, González F, Lundkvist Å, et al. Genetic Polymorphisms in DC-SIGN, TLR3 and TNFa Genes and the Lewis-Negative Phenotype Are Associated With Chikungunya Infection and Disease in Nicaragua. J Infect Dis (2020) 2020:jiaa364. doi: 10.1093/infdis/jiaa364
77. Prabhu SS, Chakraborty TT, Kumar N, Banerjee I. Association Between IFITM3 Rs12252 Polymorphism and Influenza Susceptibility and Severity: A Meta-Analysi. Gene (2018) 674:70–9. doi: 10.1016/j.gene.2018.06.070
78. Cheng Z, Zhou J, To KK, Chu H, Li C, Wang D, et al. Identification of TMPRSS2 as a Susceptibility Gene for Severe 2009 Pandemic A(H1N1) Influenza and A(H7N9) Influenza. J Infect Dis (2015) 212:1214–21. doi: 10.1093/infdis/jiv246
79. Howard A, Uyeki TM, Fergie J. Influenza-Associated Acute Necrotizing Encephalopathy in Siblings. J Pediatr Infect Dis Soc (2018) 7:e172–7. doi: 10.1093/jpids/piy033
80. Languon S, Quaye O. Filovirus Disease Outbreaks: A Chronological Overview. Virol (Auckl) (2019) 10:1178122X19849927. doi: 10.1177/1178122X19849927
81. Bogoch II, Creatore MI, Cetron MS, Brownstein JS, Pesik N, Miniota J, et al. Assessment of the Potential for International Dissemination of Ebola Virus Via Commercial Air Travel During the 2014 West African Outbreak. Lancet (2015) 385:29–35. doi: 10.1016/S0140-6736(14)61828-6
82. Vetter P, Fischer WA 2nd, Schibler M, Jacobs M, Bausch DG, Kaiser L. Ebola Virus Shedding and Transmission: Review of Current Evidence. J Infect Dis (2016) 214(Suppl 3):S177–84. doi: 10.1093/infdis/jiw254
83. Victory KR, Coronado F, Ifono SO, Soropogui T, Dahl BA. Ebola Transmission Linked to a Single Traditional Funeral Ceremony — Kissidougou, Guinea, December, 2014–January 2015. MMWR Morb Mortal Wkly Rep (2015) 64:386–8.
84. Webster RG. Wet Markets—A Continuing Source of Severe Acute Respiratory Syndrome and Influenza? Lancet (2004) 363:234–6. doi: 10.1016/S0140-6736(03)15329-9
85. Yang KD, Yang MY, Li CC, Lin SF, Chong MC, Wang CL, et al. Altered Cellular But Not Humoral Reactions in Children With Complicated Enterovirus 71 Infections in Taiwan. J Infect Dis (2001) 183:850–6.
86. Li CC, Yang MY, Chen RF, Lin TY, Tsao KC, Ning HC, et al. Clinical Manifestations and Laboratory Assessment in an Enterovirus 71 Outbreak in Southern Taiwan. Scand J Infect Dis (2002) 34:104–9. doi: 10.1080/00365540110077119
87. Lin TY, Chang LY, Huang YC, Hsu KH, Chiu CH, Yang KD. Different Proinflammatory Reactions in Fatal and Non-Fatal Enterovirus 71 Infections: Implications for Early Recognition and Therapy. Acta Paediatr (2002) 91(6):632–5. doi: 10.1080/080352502760069016
88. Yang B, Chuang H, Yang KD. Sialylated Glycans as Receptor and Inhibitor of Enterovirus 71 Infection to DLD-1 Intestinal Cells. Virol J (2009) 6:141. doi: 10.1186/1743-422X-6-141
89. Yang KD, Yeh WT, Yang MY, Chen RF, Shaio MF. Antibody-Dependent Enhancement of Heterotypic Dengue Infections Involved in Suppression of IFNgamma Production. J Med Virol (2001) 63:150–7.
90. Liu JW, Lee IK, Wang L, Chen RF, Yang KD. The Usefulness of Clinical-Practice-Based Laboratory Data in Facilitating the Diagnosis of Dengue Illness. Biomed Res Int (2013) 2013:198797. doi: 10.1155/2013/198797
91. Lee IK, Liu JW, Yang KD. Fatal Dengue Hemorrhagic Fever in Adults: Emphasizing the Evolutionary Pre-Fatal Clinical and Laboratory Manifestations. PloS Negl Trop Dis (2012) 6:e1532. doi: 10.1371/journal.pntd.0001532
92. Yeh WT, Chen RF, Wang L, Liu JW, Shaio MF, Yang KD. Implications of Previous Subclinical Dengue Infection But Not Virus Load in Dengue Hemorrhagic Fever. FEMS Immunol Med Microbiol (2006) 48:84–90. doi: 10.1111/j.1574-695X.2006.00127.x
93. Chen RF, Wang L, Cheng JT, Yang KD. Induction of Ifnα or IL-12 Depends on Differentiation of THP-1 Cells in Dengue Infections Without and With Antibody Enhancement. BMC Infect Dis (2012) 12:340. doi: 10.1186/1471-2334-12-340
94. Lee IK, Hsieh CJ, Chen RF, Yang ZS, Wang L, Chen CM, et al. Increased Production of Interleukin-4, Interleukin-10, and Granulocyte-Macrophage Colony-Stimulating Factor by Type 2 Diabetes’ Mononuclear Cells Infected With Dengue Virus, But Not Increased Intracellular Viral Multiplication. Biomed Res Int (2013) 2013:965853. doi: 10.1155/2013/965853
95. Lee IK, Liu JW, Yang KD. Clinical Characteristics and Risk Factors for Concurrent Bacteremia in Adults With Dengue Hemorrhagic Fever. Am J Trop Med Hyg (2005) 72:221–6.
96. Lee IK, Liu JW, Yang KD. Clinical and Laboratory Characteristics and Risk Factors for Fatality in Elderly Patients With Dengue Hemorrhagic Fever. Am J Trop Med Hyg (2008) 79:149–53.
97. Lee CH, Chen RF, Liu JW, Yeh WT, Chang JC, Eng HL, et al. Altered P38 Mitogen-Activated Protein Kinase Expression in Different Leukocytes With Increment of Immunosuppressive Mediators in Patients With Severe Acute Respiratory Syndrome. J Immunol (2004) 172:7841–7. doi: 10.1016/j.micinf.2005.06.007
98. Lee YS, Chen CH, Chao A, Chen ES, Wei ML, Chen LK, et al. Molecular Signature of Clinical Severity in Recovering Patients With Severe Acute Respiratory Syndrome Coronavirus (SARS-Co). BMC Genomics (2005) 6:132. doi: 10.1186/1471-2164-6-132
99. Ko SF, Lee TY, Huang CC, Cheng YF, Ng SH, Kuo YL, et al. Severe Acute Respiratory Syndrome: Prognostic Implications of Chest Radiographic Findings in 52 Patients. Radiology (2004) 233:173–81. doi: 10.1148/radiol.2323031547
100. Chen RF, Chang JC, Yeh WT, Lee CH, Liu JW, Eng HL, et al. Role of Vascular Cell Adhesion Molecules and Leukocyte Apoptosis in the Lymphopenia and Thrombocytopenia of Patients With Severe Acute Respiratory Syndrome (SAR). Microbes Infect (2006) 8:122–7. doi: 10.1016/j.micinf.2005.06.007.
101. Chu TP, Li CC, Wang L, Hsu LW, Eng HL, You HL, et al. A Surveillance System to Reduce Transmission of Pandemic H1N1 (2009) Influenza in a 2600-Bed Medical Center. PloS One (2012) 7:e32731. doi: 10.1371/journal.pone.0032731
102. Lee IK, Liu JW, Wang L, Yang KD, Li CC, Eng HL. 2009 Pandemic Influenza A (H1N1): Clinical and Laboratory Characteristics in Pediatric and Adult Patients and in Patients With Pulmonary Involvement. Influenza Other Respir Viruses (2012) 6:e152–61. doi: 10.1111/j.1750-2659.2012.00410.x
103. Wang L, Chang LS, Lee IK, Tang KS, Li CC, Eng HL, et al. Clinical Diagnosis of Pandemic A(H1N1) 2009 Influenza in Children With Negative Rapid Influenza Diagnostic Test by Lymphopenia and Lower C-Reactive Protein Levels. Influenza Other Respir Viruses (2014) 8:91–8. doi: 10.1111/irv.12182
104. Li CC, Wang L, Eng HL, You HL, Chang LS, Tang KS, et al. Correlation of Pandemic (H1N1) 2009 Viral Load With Disease Severity and Prolonged Viral Shedding in Children. Emerg Infect Dis (2010) 16:1265–72. doi: 10.3201/eid1608.091918
105. Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, Pulendran B. Cutting Edge: Impairment of Dendritic Cells and Adaptive Immunity by Ebola and Lassa Viruses. J Immunol (2003) 170:2797–801. doi: 10.4049/jimmunol.170.6.2797
106. Leroy EM, Baize S, Lu CY, McCormick JB, Georges AJ, Georges-Courbot MC, et al. Diagnosis of Ebola Haemorrhagic Fever by RT-PCR in an Epidemic Setting. J Med Virol (2000) 60:463–7.
107. Baize S, Leroy EM, Georges-Courbot MC, Capron M, Lansoud-Soukate J, Debré P, et al. Defective Humoral Responses and Extensive Intravascular Apoptosis Are Associated With Fatal Outcome in Ebola Virus-Infected Patients. Nat Med (1999) 5:423–6. doi: 10.1038/7422
108. Johnson KM, McCormick JB, Webb PA, Smith ES, Elliott LH, King IJ. Clinical Virology of Lassa Fever in Hospitalized Patients. J Infect Dis (1987) 155:456–64. doi: 10.1093/infdis/155.3.456
109. Chen JP, Cosgriff TM. Hemorrhagic Fever Virus-Induced Changes in Hemostasis and Vascular Biology. Blood Coag Fibrin (2000) 11:461–83. doi: 10.1097/00001721-200007000-00010
110. Diamond MS, Sitati EM, Friend LD, Higgs S, Shrestha B, Engle M. A Critical Role for Induced IgM in the Protection Against West Nile Virus Infection. J Exp Med (2003) 198:1853–62. doi: 10.1084/jem.20031223
111. Davidson AM, Wysocki J, Batlle D. Interaction of SARS-CoV-2 and Other Coronavirus With ACE (Angiotensin-Converting Enzyme)-2 as Their Main Receptor: Therapeutic Implications. Hypertension (2020) 76:1339–49. doi: 10.1161/HYPERTENSIONAHA.120.15256
112. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: Current State of the Science. Immunity (2020) 52:910–41. doi: 10.1016/j.immuni.2020.05.002
113. Wu D, Yang XO. TH17 Responses in Cytokine Storm of COVID-19: An Emerging Target of JAK2 Inhibitor Fedratinib. J Microbiol Immunol Infect (2020) 53:368–70. doi: 10.1016/j.jmii.2020.03.005
114. De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, et al. Marked T Cell Activation, Senescence, Exhaustion and Skewing Towards TH17 in Patients With COVID-19 Pneumonia. Nat Commun (2020) 11:3434. doi: 10.1038/s41467-020-17292-4
115. Peiris JS, Chu CM, Cheng VCC, Chan KS, Hung IFN, Poon LLM, et al. Clinical Progression and Viral Load in a Community Outbreak of Coronavirus-Associated SARS Pneumonia: A Prospective Study. Lancet (2003) 361:1767–72. doi: 10.1016/s0140-6736(03)13412-5
116. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological Assessment of Hospitalized Patients With COVID-2019. Nature (2020) 581:465–9. doi: 10.1038/s41586-020-2196-x
117. Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann Intern Med (2020) 173:268–77. doi: 10.7326/M20-2003
118. Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, et al. Lung Pathology of Fatal Severe Acute Respiratory Syndrome. Lancet (2003) 361:1773–8. doi: 10.1016/s0140-6736(03)13413-7
119. Merrill JT, Erkan D, Winakur J, James JA. Emerging Evidence of a COVID-19 Thrombotic Syndrome has Treatment Implications. Nat Rev Rheumatol (2020) 16:581–9. doi: 10.1038/s41584-020-0474-5
120. Meftahi GH, Jangravi, Sahraei H, Bahari Z. The Possible Pathophysiology Mechanism of Cytokine Storm in Elderly Adults With COVID-19 Infection: The Contribution of “Inflame-Aging”. Inflamm Res (2020) 69:825–89. doi: 10.1007/s00011-020-01372-8
121. WHO. Update 49 - SARS Case Fatality Ratio, Incubation Period. Available at: https://www.who.int/csr/sarsarchive/2003_05_07a/en/.
122. Reilev M, Kristensen KB, Pottegaard A. Characteristics and Predictors of Hospitalization and Death in the First 11,122 Cases With a Positive RT-PCR Test for SARS-CoV-2 in Denmark: A Nationwide Cohort. Int J Epidemiol (2020) 49:1468–81. doi: 10.1093/ije/dyaa140
123. Williamson E, Walker AJ, Bhaskaran KJ. Factors Associated With COVID-19-Related Death Using OpenSAFELY. Nature (2020) 584:430–6. doi: 10.1016/j.micinf.2005.06.032
124. Kurane I, Innis BL, Nimmannitya S, Nisalak A, Meager A, Janus J, et al. Activation of T Lymphocytes in Dengue Virus Infections. High Levels of Soluble Interleukin 2 Receptor, Soluble CD4, Soluble CD8, Interleukin 2, and Interferon-Gamma in Sera of Children With Dengue. J Clin Invest (1991) 88:1473–80. doi: 10.1172/JCI115457
125. Khaiboullina SF, St Jeor SC. Hantavirus Immunology. Viral Immunol (2002) 15:609–25. doi: 10.1089/088282402320914548
126. To KF, Chan PK, Chan KF, Lee WK, Lam WY, Wong KF, et al. Pathology of Fatal Human Infection Associated With Avian Influenza A H5N1 Virus. J Med Virol (2001) 63:242–6. doi: 10.1002/1096-9071(200103)63:3<242::aid-jmv1007>3.0.co;2-n
127. de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TNB, et al. Fatal Outcome of Human Influenza A (H5N1) Is Associated With High Viral Load and Hypercytokinemia. Nat Med (2006) 12:1203–7. doi: 10.1038/nm1477
128. Khaiboullina SF, Levis S, Morzunov SP, Martynova EV, Anokhin VA, Gusev OA, et al. Serum Cytokine Profiles Differentiating Hemorrhagic Fever With Renal Syndrome and Hantavirus Pulmonary Syndrome. Front Immunol (2017) 8:567. doi: 10.3389/fimmu.2017.00567
129. WHO. Review of Latest Available Evidence on Potential Transmission of Avian Influenza (lrm;H5N1)lrm; Through Water and Sewage and Ways to Reduce the Risks to Human Health. Available at: https://apps.who.int/iris/handle/10665/204275.
130. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the Eye of the Cytokine Storm. Microbiol Mol Biol Rev (2012) 76:16–32. doi: 10.1128/MMBR.05015-11
131. Gavrilovskaya I, Gorbunova E, Koster F, Mackow E. Elevated VEGF Levels in Pulmonary Edema Fluid and PBMCs From Patients With Acute Hantavirus Pulmonary Syndrome. Adv Virol (2012) 2012:674360. doi: 10.1155/2012/674360
132. Kongsomboon K, Singhasivanon P, Kaewkungwal J, Nimmannitya S, Mammen MP Jr., Nisalak A, et al. Temporal Trends of Dengue Fever/Dengue Hemorrhagic Fever in Bangkok, Thailand From 1981 to 2000: An Age-Period-Cohort Analysis. Southeast Asian J Trop Med Public Health (2004) 35:913–7.
133. Welsh RM, Selin LK, Szomolanyi-Tsuda E. Immunological Memory to Viral Infections. Ann Rev Immunol (2004) 22:711–43. doi: 10.1146/annurev.immunol.22.012703.104527
134. Chaturvedi UC, Agarwal R, Elbishbishi EA, Mustafa AS. Cytokine Cascade in Dengue Hemorrhagic Fever: Implications for Pathogenesis. FEMS Immunol Med Microbiol (2000) 28:183–8. doi: 10.1111/j.1574-695X.2000.tb01474.x
135. Mahalingam S, Lidbury BA. Suppression of Lipopolysaccharide-Induced Antiviral Transcription Factor (STAT-1 and NF-Kappa B) Complexes by Antibody-Dependent Enhancement of Macrophage Infection by Ross River Virus. Proc Natl Acad Sci U S A (2002) 99:13819–24. doi: 10.1073/pnas.202415999
136. Ulrich H, Pillat MM, Tárnok A. Dengue Fever, COVID-19 (Sars-CoV-2), and Antibody-Dependent Enhancement (ADE): A Perspective. Cytometry A (2020) 97:662–7. doi: 10.1002/cyto.a.24047
137. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell (2020) 182:685–712.e19. doi: 10.1016/j.cell.2020.06.034
138. Consiglio CR, Cotugno N, Sardh F, Pou C, Amodio D, Rodriguez L, et al. The Immunology of Multisystem Inflammatory Syndrome in Children With COVID-19. Cell (2020) 183:1–14. doi: 10.1016/j.cell.2020.09.016
139. Fu L, Ye F, Feng Y, Yu F, Wang Q, Wu Y, et al. Both Boceprevir and GC376 Efficaciously Inhibit SARS-CoV-2 by Targeting Its Main Protease. Nat Commun (2020) 11:4417. doi: 10.1038/s41467-020-18233-x
140. Mulangu S, Dodd LE, Davey RT Jr, Tshiani Mbaya O, Proschan M, Mukadi D, et al. A Randomized, Controlled Trial of Ebola Virus Disease Therapeutics. N Engl J Med (2019) 381:2293–303. doi: 10.1056/NEJMoa1910993
141. Cheng Y, Wong R, Soo YO, Wong WS, Lee CK, Ng MHL, et al. Use of Convalescent Plasma Therapy in SARS Patients in Hong Kong. Eu J Clin Microbiol Infect Dis (2005) 24:44–6. doi: 10.1007/s10096-004-1271-9
142. Arabi YM, Hajeer AH, Luke T, Raviprakash K, Balkhy H, Johani S, et al. Feasibility of Using Convalescent Plasma Immunotherapy for MERS-CoV Infection, Saudi Arabia. Emerging Infect Dis (2016) 22:1554–61. doi: 10.3201/eid2209.151164
143. Xu X, Han M, Li T, Sun W, Wang D, Fu B, et al. Effective Treatment of Severe COVID-19 Patients With Tocilizumab. Proc Natl Acad Sci U S A (2020) 117:10970–5. doi: 10.1073/pnas.2005615117
144. Nasonov E, Samsonov M. The Role of Interleukin 6 Inhibitors in Therapy of Severe COVID-19. Biomed Pharmacother (2020) 131:110698. doi: 10.1016/j.biopha.2020.110698
145. Carvelli J, Demaria O, Vély F, Batista L, Chouaki Benmansour N, Fares J, et al. Association of COVID-19 Inflammation With Activation of the C5a-C5aR1 Axis. Nature (2020) 588:146–50. doi: 10.1038/s41586-020-2600-6
146. Morimoto A, Nakazawa Y, Ishii E. 016. Hemophagocytic Lymphohistiocytosis: Pathogenesis, Diagnosis, and Management. Pediatr Int 58:817–25. doi: 10.1111/ped.13064
147. McGonagle D, Sharif K, O’Regan A, Bridgewood C. The Role of Cytokines Including Interleukin-6 in COVID-19 Induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun Rev (2020) 19:102537. doi: 10.1016/j.autrev.2020.102537
148. Boisen ML, Hartnett JN, Shaffer JG, Goba A, Momoh M, Sandi JD, et al. Field Validation of Recombinant Antigen Immunoassays for Diagnosis of Lassa Fever. Sci Rep (2018) 8:5939. doi: 10.1038/s41598-018-24246-w
149. Takah NF, Brangel P, Shrestha P, Peeling R. Sensitivity and Specificity of Diagnostic Tests for Lassa Fever: A Systematic Review. BMC Infect Dis (2019) 19:647. doi: 10.1186/s12879-019-4242-6
150. McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, et al. Lassa Fever. Effective Therapy With Ribavirin. N Engl J Med (1986) 314:20–6. doi: 10.1056/NEJM198601023140104
151. Frame JD, Verbrugge GP, Gill RG, Pinneo L. The Use of Lassa Fever Convalescent Plasma in Nigeria. Trans R Soc Trop Med Hyg 78:319–24. doi: 10.1016/0035-9203(84)90107-x
152. Jacob ST, Crozier I, Fischer WA 2nd, Hewlett A, Kraft CS, Vega MA, et al. Ebola Virus Disease. Nat Rev Dis Primers (2020) 6:13. doi: 10.1038/s41572-020-0147-3
153. Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, et al. Efficacy and Effectiveness of an rVSV-Vectored Vaccine in Preventing Ebola Virus Disease: Final Results From the Guinea Ring Vaccination, Open-Label, Cluster-Randomised Trial (Ebola Ça Suffi)! Lancet (2017) 389:505–18. doi: 10.1016/S0140-6736(16)32621-6
154. Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. Regn-COV2, A Neutralizing Antibody Cocktail, in Outpatients With Covid-19. N Engl J Med (2021) 384(3):238–51. doi: 10.1056/NEJMoa2035002
155. CDC. Interim Guidance for Antigen Testing for SARS-CoV-2. Retrieved on May 25, 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
156. Brown RA, Murthy J, Manian P, Rumbaoa B, Connolly T. An Early Aggressive Strategy for the Treatment of Hanta Virus Cardiopulmonary Syndrome: A Perspective From an Extracorporeal Membrane Oxygenation Center. Clin Infect Dis (2014) 59:458–9. doi: 10.1093/cid/ciu295
157. Vaheri A, Strandin T, Jääskeläinen AJ, Vapalahti O, Jarva H, Lokki M, et al. Pathophysiology of a Severe Case of Puumala Hantavirus Infection Successfully Treated With Bradykinin Receptor Antagonist Icatibant. Antiviral Res (2014) 111:23–5. doi: 10.1016/j.antiviral.2014.08.007
158. Wong JP, Viswanathan S, Wang M, Sun LQ, Clark GC, D’Elia RV. Current and Future Developments in the Treatment of Virus-Induced Hypercytokinemia. Future Med Chem (2017) 9:169–78. doi: 10.4155/fmc-2016-0181
159. Buckley LF, Wohlford GF, Ting C, Alahmed A, Van Tassell BW, Abbate A, et al. Role for Anti-Cytokine Therapies in Severe Coronavirus Disease 2019. Crit Care Explor (2020) 2:e0178. doi: 10.1097/CCE.0000000000000178
160. Sadeghi A, Tahmasebi S, Mahmood A, Kuznetsova M, Valizadeh H, Nazemiyeh M, et al. Th17 and T Reg Cells Function in SARS-CoV2 Patients Compared With Healthy Controls. J Cell Physiol (2020). doi: 10.1002/jcp.30047
161. Giudice V, Pagliano P, Vatrella A, Masullo A, Poto S, Polverino BM, et al. Combination of Ruxolitinib and Eculizumab for Treatment of Severe SARS-CoV-2-Related Acute Respiratory Distress Syndrome: A Controlled Study. Front Pharmacol (2020) 11:857. doi: 10.3389/fphar.2020.00857
162. Streng AS, Delnoij TSR, Mulder MMG, Sels JWEM, Wetzels RJH, Verhezen PWM, et al. Monitoring of Unfractionated Heparin in Severe COVID-19: An Observational Study of Patients on CRRT and ECMO. TH Open (2020) 4:e365–75. doi: 10.1055/s-0040-1719083
163. Ando M, Miyazaki E, Hiroshige S, Ashihara Y, Okubo T, Ueo M, et al. Virus Associated Hemophagocytic Syndrome Accompanied by Acute Respiratory Failure Caused by Influenza A (H3n2). Intern Med (2006) 45:1183–6. doi: 10.2169/internalmedicine.45.1736
164. Trujillo H, Miyamura T, Takahama S, Sonomoto K, Nakamura M, Lumbreras C. Influenza-Associated Hemophagocytic Syndrome in Adults: Case Report and Review. J Antivir Antiretrovir (2018) 10:1–5. doi: 10.4172/1948-5964.1000172
165. Kalil AC, Thomas PG. Influenza Virus-Related Critical Illness: Pathophysiology and Epidemiology. Crit Care (2019) 23:258. doi: 10.1186/s13054-019-2539-x
166. Yang KD. A Stepwise Infection and Immunity Strategies to Prevent and Treat an Emerging Infection. Am J Biomed Sci Res (2020) 11:AJBSR.MS.ID.001602. doi: 10.34297/AJBSR.2020.11.001602
167. Ollmann Saphire E. A Vaccine Against Ebola Virus. Cell (2020) 181:6. doi: 10.1016/j.cell.2020.03.011
168. Suschak JJ, Schmaljohn CS. Vaccines Against Ebola Virus and Marburg Virus: Recent Advances and Promising Candidates. Hum Vaccin Immunother (2019) 15:2359–77. doi: 10.1080/21645515.2019.1651140
169. Pardi N, Hogan MJ, Weissman D. Recent Advances in mRNA Vaccine Technology. Curr Opin Immunol (2020) 65:14–20. doi: 10.1016/j.coi.2020.01.008
170. van Riel D, de Wit E. Next-Generation Vaccine Platforms for COVID-19. Nat Materials (2020) 19:810–2. doi: 10.1038/s41563-020-0746-0
171. Kuo SC, Shih SM, Chien LH, Hsiung CA. Collateral Benefit of COVID-19 Control Measures on Influenza Activity, Taiwan. Emerg Infect Dis (2020) 26(8):1928–30. doi: 10.3201/eid2608.201192
172. Yang CJ, Chen TC, Kuo SH, Hsieh MH, Chen YH. Severe Complicated Influenza Declined During the Prevention of COVID-19 in Taiwan. Infect Control Hosp Epidemiol (2020) 2020:1–2. doi: 10.1017/ice.2020.272
173. Totura AL, Bavari S. Broad-Spectrum Coronavirus Antiviral Drug Discovery. Expert Opin Drug Discov (2019) 14:397–412. doi: 10.1080/17460441.2019.1581171
174. Weisberg E, Parent A, Yang PL, Sattler M, Liu Q, Liu Q, et al. Repurposing of Kinase Inhibitors for Treatment of COVID-19. Pharm Res (2020) 37:167. doi: 10.1007/s11095-020-02851-7
175. Cohen MS. Monoclonal Antibodies to Disrupt Progression of Early Covid-19 Infection. N Engl J Med (2021) 384(3):289–91. doi: 10.1056/NEJMe2034495
176. Liu STH, Lin HM, Baine I, Wajnberg A, Gumprecht JP, Rahman F, et al. Convalescent Plasma Treatment of Severe COVID-19: A Propensity Score-Matched Control Study. Nat Med (2020) 26:1708–13. doi: 10.1038/s41591-020-1088-9
177. Libster R, Pérez Marc G, Wappner D, Coviello S, Bianchi A, Braem V, et al. Early High-Titer Plasma Therapy to Prevent Severe COVID-19 in Older Adults. N Engl J Med (2021) 384(7):610–8. doi: 10.1056/NEJMoa2033700
178. Chen MR, Kuo HC, Lee YJ, Chi H, Li SC, Lee HC, et al. Phenotype, Susceptibility, Autoimmunity and Immunotherapy Between Kawasaki Disease (KD) and COVID-19 Associated Multisystem Inflammatory Syndrome in Children (MIS-C). Front Immunol (2021) 12:632890. doi: 10.3389/fimmu.2021.632890
179. Rhodes JM, Subramanian S, Laird E, Griffin G, Kenny RA. Perspective: Vitamin D Deficiency and COVID-19 Severity - Plausibly Linked by Latitude, Ethnicity, Impacts on Cytokines, ACE2, and Thrombosis. J Intern Med (2020) 289:97–115. doi: 10.1111/joim.13149
180. Mohammad S, Mishra A, Ashraf MZ. Emerging Role of Vitamin D and its Associated Molecules in Pathways Related to Pathogenesis of Thrombosis. Biomolecules (2019) 9:649. doi: 10.3390/biom9110649
181. Shoemark KD, Colenso CK, Toelzer C, Gupta K, Sessions RB, Davidson AD, et al. Molecular Simulations Suggest Vitamins, Retinoids and Steroids as Ligands Binding the Free Fatty Acid Pocket of SARS-CoV-2 Spike Protein. Angew Chem Int Ed (2020). doi: 10.1002/anie.202015639
182. Castellani ML, Shaik-Dasthagirisaheb YB, Tripodi D, Anogeianaki A, Felaco P, Toniato E, et al. Interrelationship Between Vitamins and Cytokines in Immunity. J Biol Regul Homeost Agents (2010) 24:385–90.
183. Goddek S. Vitamin D3 and K2 and Their Potential Contribution to Reducing the COVID-19 Mortality Rate. Int J Infect Dis (2020) 99:286–90. doi: 10.1016/j.ijid.2020.07.080
Keywords: emerging viral infections, immunopathogenesis, evasion, fatality, prevention, early diagnosis, early treatment
Citation: Yang B and Yang KD (2021) Immunopathogenesis of Different Emerging Viral Infections: Evasion, Fatal Mechanism, and Prevention. Front. Immunol. 12:690976. doi: 10.3389/fimmu.2021.690976
Received: 05 April 2021; Accepted: 14 June 2021;
Published: 15 July 2021.
Edited by:
R. Brad Jones, Cornell University, United StatesReviewed by:
Jianzhong Zhu, Yangzhou University, ChinaCopyright © 2021 Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuender D. Yang, eWFuZ2tkLnllaEBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.