- Department of Biology, University of Texas at Arlington, Arlington, TX, United States
Pattern recognition receptors (PRRs) are evolutionarily ancient and crucial components of innate immunity, recognizing danger-associated molecular patterns (DAMPs) and activating host defenses. Basal non-bilaterian animals such as cnidarians must rely solely on innate immunity to defend themselves from pathogens. By investigating cnidarian PRR repertoires we can gain insight into the evolution of innate immunity in these basal animals. Here we utilize the increasing amount of available genomic resources within Cnidaria to survey the PRR repertoires and downstream immune pathway completeness within 15 cnidarian species spanning two major cnidarian clades, Anthozoa and Medusozoa. Overall, we find that anthozoans possess prototypical PRRs, while medusozoans appear to lack these immune proteins. Additionally, anthozoans consistently had higher numbers of PRRs across all four classes relative to medusozoans, a trend largely driven by expansions in NOD-like receptors and C-type lectins. Symbiotic, sessile, and colonial cnidarians also have expanded PRR repertoires relative to their non-symbiotic, mobile, and solitary counterparts. Interestingly, cnidarians seem to lack key components of mammalian innate immune pathways, though similar to PRR numbers, anthozoans possess more complete immune pathways than medusozoans. Together, our data indicate that anthozoans have greater immune specificity than medusozoans, which we hypothesize to be due to life history traits common within Anthozoa. Overall, this investigation reveals important insights into the evolution of innate immune proteins within these basal animals.
Introduction
Animals sense and interact with microbes through their immune system (1). The majority of knowledge about immune system function stems from studies in vertebrates, which interact with microbes through both an innate and an adaptive immune system (2, 3). In contrast, invertebrates rely on innate immunity to detect and respond to microbes. Pattern recognition receptors (PRRs) are key components of innate immunity that detect both danger associated molecular patterns (DAMPs) and microbe associated molecular patterns (MAMPs), activating downstream signaling pathways following this recognition (4). PRRs are drivers of immune specificity in invertebrates, as diverse repertoires of receptors are needed in order to generate a microbe specific immune response (5).
The four best studied families of PRRs are Toll-like receptors (TLRs), retinoic acid-inducible gene I-like receptors (RLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), and C-type lectins (CTLs) (6, 7). While it was originally thought that many of these PRR families first arose in vertebrates, studies following the advent of next generation sequencing revealed that TLRs, RLRs, NLRs, and CTLs are present in basal metazoans and thus are evolutionarily ancient (8–12). Studying PRRs in basal taxa informs our understanding of PRR evolution and more broadly immune evolution.
Cnidaria is a basal phylum sister to Bilateria with over 10,000 species spanning three major clades: Anthozoa, Medusozoa, and Endocnidozoa which span a diverse array of life history strategies ranging from sedentary to planktonic to parasitic (13). Despite their primitive morphology, cnidarian genomes are complex and contain a repertoire of innate immune genes unexpectedly similar to mammals (14, 15). Due to this, cnidarians are exceptional candidates for investigations into innate immune evolution as they are not only basal, but also have immune repertoires are not reduced like the nematode model Caenorhabditis elegans or derived in function like the arthropod model Drosophila melanogaster (15–17). Additionally, common life history traits within Cnidaria have been linked to immunity. Several cnidarian taxa in Anthozoa and Medusozoa form intracellular nutritional symbiosis with algae in the family Symbiodiniaceae, the maintenance of which has been linked to several different PRRs as well as NFκB signaling (18). Similarly, many cnidarians have colonial body plans, and likely have a larger need for allorecognition and thus immune specificity (13, 19, 20). Therefore, the wide variety of life history traits found within cnidaria has the potential to create a gradient of selective pressures for immune specificity, which likely is reflected in their PRR repertoires.
Toll-like receptors (TLRs) are transmembrane PRRs that are capable of recognizing a wide variety of ligands including bacterial cell wall components, viral RNA, and developmental cues (21–23). Canonically, TLRs consist of extracellular leucine rich repeats (LRRs) which bind to DAMPs and MAMPs and an intracellular Toll/interleukin-1 receptor (TIR) domain that activates signal transduction through protein-protein interactions with other TIR domain-containing proteins (21, 22, 24). Several pathways can be activated by TLRs following ligand engagement, including the MAPK, IFN, and NFκB pathways (22, 24). Within Cnidaria, prototypical TLRs have been identified in several anthozoan species, and functional studies indicate that the Nematostella vectensis TLR functions in pathogen recognition, activation of NFκB signaling, and development (25, 26). Additionally, a medusozoan species lacking prototypical TLRs, Hydra vulgaris, is still capable of TLR-NFκB signaling through TLR-like proteins containing a transmembrane domain and an intracellular TIR domain that appear to interact with transmembrane proteins with extracellular leucine rich repeats to perform the function of the prototypical TLR (27).
Retinoic acid-inducible gene I-like receptors (RLRs) are cytosolic PRRs that detect intracellular viral RNA (28–30). Mammals have three RLRs: RIG-I, MDA5, and LGP2. All RLRs have a central ATPase containing DExD/H box helicase domain and a C-terminal regulatory domain (28, 30). RIG-I and MDA5 also contain CARD domains which in mammals interact with the CARD domain of signaling adaptor MAVS to initiate downstream signaling, activating transcription factors IRF3 and NFκB and ultimately resulting in an antiviral response (31, 32). Additionally, RIG-I and LGP2 have a repressor domain (RD) within the C-terminal regulatory domain (28, 30). As LGP2 lacks CARD domains, it is unable to initiate antiviral signaling and instead likely acts as a concentration dependent biphasic switch in mammals, positively regulating MDA5 at low concentrations and negatively regulating RIG-I and MDA5 at high concentrations (33–35). Some RLRs with sequences most similar to RIG-I have been identified in the N. vectensis genome (9).
Nucleotide-binding oligomerization domain-like receptors (NLRs) are intracellular PRRs capable of recognizing a wide array of DAMPs and MAMPs including reactive oxygen species (ROS) (36), organelle calcium efflux (37), Lipopolysaccharide (38), peptidoglycan (39), and viral RNA (40). Prototypically NLRs contain an N terminal effector domain, a central NACHT/nucleotide binding domain, and C terminal leucine rich repeats. NLRs can activate several innate immune pathways following ligand engagement, including the NFκB, MAPK, interferon and inflammasome assembly pathways (36, 39, 41). H. vulgaris and two stony coral species have been shown to have large NLR repertoires, containing unique domain combinations that are not seen in mammalian NLRs (10, 42, 43).
C-type lectins (CTLs) are a very diverse protein family that can act as either soluble or transmembrane PRRs (5, 44). They are characterized by the C-type lectin domain (CTLD) which is most well-known for calcium dependent carbohydrate binding but is also capable of binding to proteins, lipids, and inorganic compounds (44). CTLs can activate NFκB as well as the lectin complement pathway (45, 46). A bioinformatic study of N. vectensis CTLs also found a large repertoire that could not be categorized by the mammalian CTL classification system (47).
Our study aims to build upon the current base of knowledge on cnidarian PRRs by expanding to investigate four PRR types in a phylogenetically diverse group of cnidarians. Previous studies are heavily concentrated in two classes, the anthozoan class Hexacorillia (stony corals and anemones), and two model systems within the medusozoan class Hydrozoa: H. vulgaris and Hydractinia symbiolongicarpus (8, 9, 15, 26, 27, 42, 47, 48). To date, PRRs have not been investigated in cnidarians with a free-swimming adult medusae form, meaning that current studies also lack diversity in terms of life history strategies. Furthermore, only one anemone species, N. vectensis, has studies of all four PRR types (9, 25, 43, 47). Thus, we lack knowledge of the number and structure of PRRs in the remaining classes of the phylum and the full PRR repertoires of even the well-studied species.
Within the past couple of years, a wealth of cnidarian genomic resources has become available, particularly in the medusozoan clade, making it possible to investigate PRRs in a far more diverse set of cnidarian species (nine anthozoans and six medusozoans), with strong potential to provide a more detailed picture of PRR and innate immune evolution (49–53). Therefore, we surveyed the proteomes of 15 cnidarians, nine anthozoans and six medusozoans, for putative TLRs, RLRs, NLRs, and CTLs with the hypothesis that medusozoans would have less diverse PRR repertoires. Next, because TLRs, RLRs, NLRs, and CTLs are all capable of activating NFκB signaling in mammals, we investigated the PRR to NFκB pathways in all 15 cnidarian species, as well as the lectin complement pathway to determine if there is a disparity in downstream immune pathway completeness between the anthozoan and medusozoan clades.
Materials and Methods
PRR Survey
The proteomes of Acropora millepora (54), Actinia tenebrosa (53), Aurelia sp. (50), Calavadosia cruxmelitensis (51), Cassiopea xamachana (55), Clytia hemisphaerica (49), Dendronephyta gigantea (56), Exaiptasia daiphana (previously Exaiptaisa pallida) (57), Hydra vulgaris (14), Montipora capitata (58), Morbakka virulenta (59), Nematostella vectensis (60), Orbicella faveolata (61), Pocillopora damicornis (62), Xenia sp. (52), and sponge Amphimedon queenslandica (63) were surveyed for TLRs, RLRs, NLRs, and CTLs using HMMR (64). A summary the clade, class, life history traits, genome assembly size, and predicted proteins for each species can be found in Supplementary Table 1. All proteomes used were genome based with the exception of N. vectensis. This was due to a failure to find the complete prototypical TLR in the genome-based proteome of N. vectensis, despite several previous studies reporting its presence in the genome (25). The complete prototypical TLR was also absent in smallest transcriptome shotgun assembly available (NCBI GenBank: HADO000000000.1), so the second smallest transcriptome shotgun assembly (NCBI GenBank : HADN000000000.1) was filtered and used (60). Transdecoder was used to extract the longest open reading frame of each contig and translate it into a predicted peptide sequence (65). Then CD-HIT was used to collapse sequences with a similarity level of 0.85 to limit the number of splice isoforms in the assembly (66). This resulted in a proteome with 42,379 contigs, 14,000 higher than the average number of predicted proteins in the genome-based proteomes used in this study (Supplementary Table 1) (67). However, PRR numbers are similar across the genome based proteome of N. vectensis, NCBI GenBank: HADO000000000.1, and GenBank : HADN000000000.1 (Supplementary Table 2) so excess contigs have minimal impact on results.
Queries were made by using Clustal Omega to make an alignment of all human TLRs, RLRs, NLRs, and CTLs respectively (68, 69). All sequences that had an E-value less than 10-4.9 following the HMMR search were then run through Pfam to predict protein domains using the batch search tool (70). Non-repeat domains with an individual E-value of less than 10-4.9 were counted and repeat domains were counted if they had an individual E-value less than 10-3. TMHMM was used to predict transmembrane domains in TLRs and CTLs (71).
TLRs were classified as prototypical if the predicted protein had both a TIR domain, LRR, and transmembrane domain that met our inclusion threshold. Proteins with a TIR domain and a transmembrane domain were classified as TLR-like proteins. Proteins with a DExD/H box helicase, CARD, and C terminal RIG regulatory domain were classified as RIG-I/MDA5-like receptors. Proteins were classified as LGP2-like receptors if they had a DExD/H box helicase, and RIG-I repressor domain (Figure 1). In several proteins the DExD/H box helicase domains had E-values meeting our inclusion threshold but were not considered as a member of the Pfam domain clan following post processing. However, given that Cnidaria is a phylogenetically distant and basal phyla, these domains were counted given the context of the surrounding domains which always included the RIG-I repressor domain. NLRs were classified as prototypical if the predicted protein contained at a minimum both the NACHT domain and LRRs and as NLR-like if they contained at a minimum the NACHT domain. CTLs were divided into 3 groups. Proteins with the CTLD and a transmembrane domain, proteins with CLTD and a Pfam domain indicative of extracellular localization, and proteins with the CTLD and no additional domains indicative of where they may be localized (Figure 1). Pfam domains considered to be indicative of extracellular localization included cysteine rich secretory domain (72), CUB (73), F5/8 (74), ShK (75), Von Willebrand factors (76), thrombospondin (77), trefoil (78), Fibrinogen β/γ terminal globular domain (79), NIDO (80), PKD (81), coagulation factor Xa inhibitory domain (82), U-PAR/Ly6 (83), complement Clr like EGF (84), and Xlink (85).
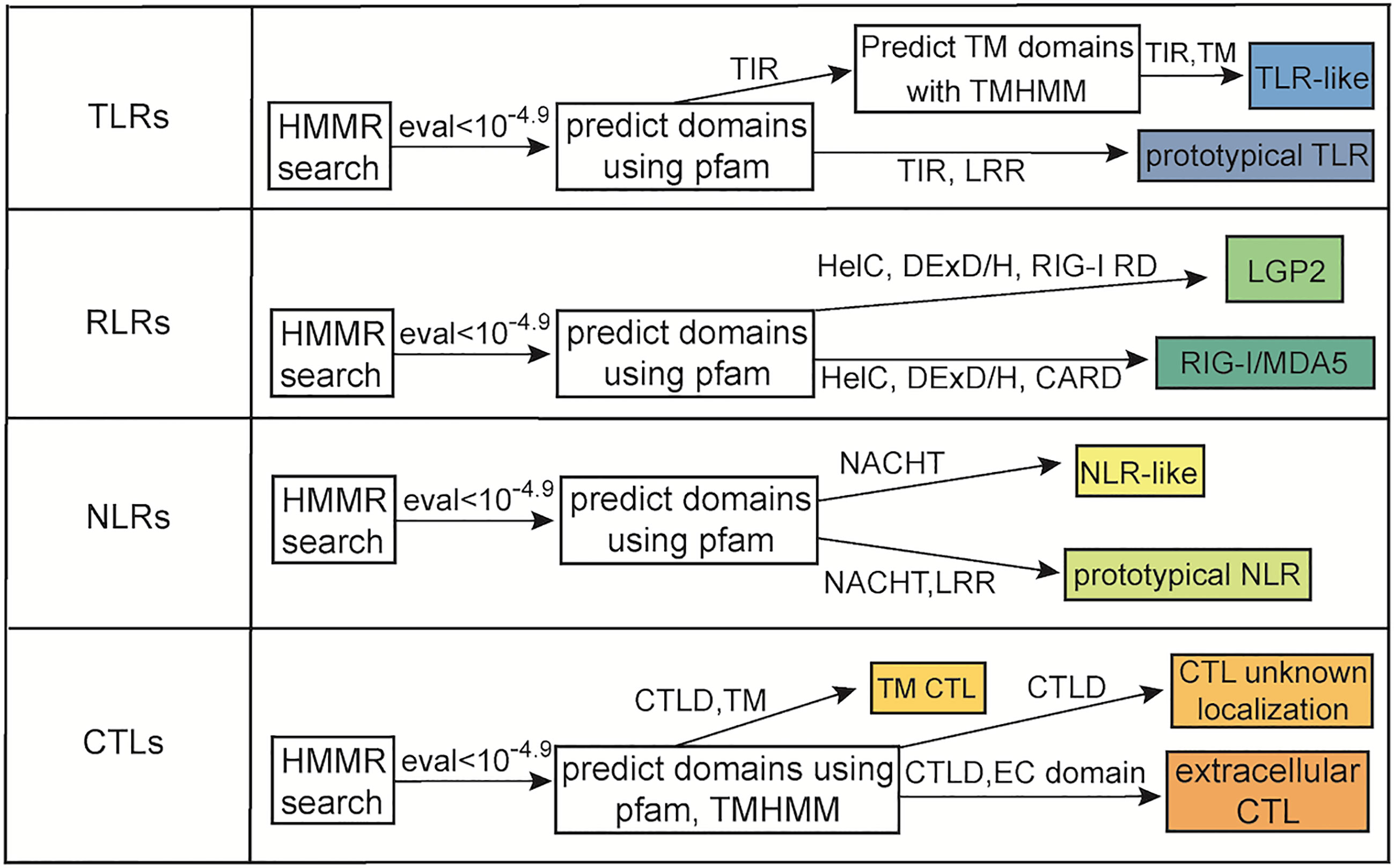
Figure 1 Flow chart of methodology used to identify PRRs. Shown is the minimum domain requirements for a protein to be classified in the various categories as well as homology requirements based upon E-value.
The total number of TLRs, RLRs, NLRs and CTLs in each species were then used as input for ancestral state reconstructions. The phylogenetic tree used for this analysis was created using Orthofinder (86). Ancestral state reconstructions based on maximum likelihood (ML) were made using the fastAnc function in the R package phytools. Phytools was also used to visualize the ML ancestral state reconstructions (87).
Relationship to Clade and Life History Traits
Principal component analysis was run in R and visualized with the R package ggfortify (88). The input of the PCA analysis was the total number of PRRs in each PRR type per species. To test for associations between total PRR number and clade, intracellular algal symbiosis, coloniality, and mobility, generalized linear models were run in R. Because there is considerable overlap between anthozoan species, symbiotic species, colonial species, and sedentary species, five individual models were tested (total PRRs~trait, family = quasipoisson) (Supplementary Table 1).
Downstream Immune Pathway Completeness
NFκB
BLASTp was used to identify NFκB and IKBA in all species included in the study (89). Human sequences of p100, p105, and IKBA were used as queries (69). The top 5 best hits to each query were then run through Pfam to predict domains using the same inclusion thresholds as the PRRs. Because the N. vectensis NFκB is known to be truncated only the Rel DNA binding domain and Rel dimerization domain were required for a cnidarian protein to be considered NFκB (90). Multiple ankyrin repeats were required for a cnidarian protein to be considered IKBA.
PRR to NFκB Pathway
Cnidarian proteomes were searched for members of the PRR to NFκB KEGG pathways using BLASTp and a human query (69, 89, 91). The cnidarian protein with the highest E-value to each query was then blasted against the human proteome (GCA_000001405) (69). If the human query and best human hit following reciprocal blast were the same protein, the protein was counted as present in the cnidarian.
Lectin Complement Pathway
Because complement consists 5 mosaic protein families that likely did not expand until teleosts, we used pBLAST to search for representatives of the C3, Factor B/C2, MASP, and C6 families (89, 92). Human sequences for all members of a given family were used as queries (69). Each protein family has a unique domain composition so the top 5 best hits to each query were run through Pfam using the batch search option to predict domains (70). Proteins were considered members of the C3 family if they contained multiple macroglobulin domains, CUB, and C345c. Proteins were considered members of the Factor B/C2 family if they contained sushi repeats, von Willebrand factors, and a serine protease. Proteins were considered members of the MASP family if they contained 2 CUB domains, sushi repeats, and serine protease. Proteins were considered members of the C6 family if they contained TSP, low density lipoprotein receptor domain class A, MACPF, and sushi repeats (92).
Results
PRR Survey
The number of prototypical TLRs and TLR-like proteins vary across the species surveyed from 0 to 24. No prototypical TLRs were found in any of the six medusozoan species and in two anthozoans, E. daiphana and Xenia sp. (Figure 2). In contrast, two anthozoans have expansions in prototypical TLRs, D. gigantea and A. millepora, with four prototypical TLRs found in D. gigantea and ten in A. millepora. TLR-like proteins with a TIR domain and a transmembrane domain were found in all species except for C. hemisphaerica and M. virulenta (Figure 2 and Supplementary Table 3). Based upon ancestral state reconstruction, one TLR was present in the common ancestor shared by medusozoans and anthozoans (Figure 3A). Relative to this ancestral cnidarian and other anthozoans, stony corals (A. millepora, M. capitata, P. damicornis, O. faveolata) have expansions in TLRs, specifically TLR-like proteins (Figures 2, 3A).
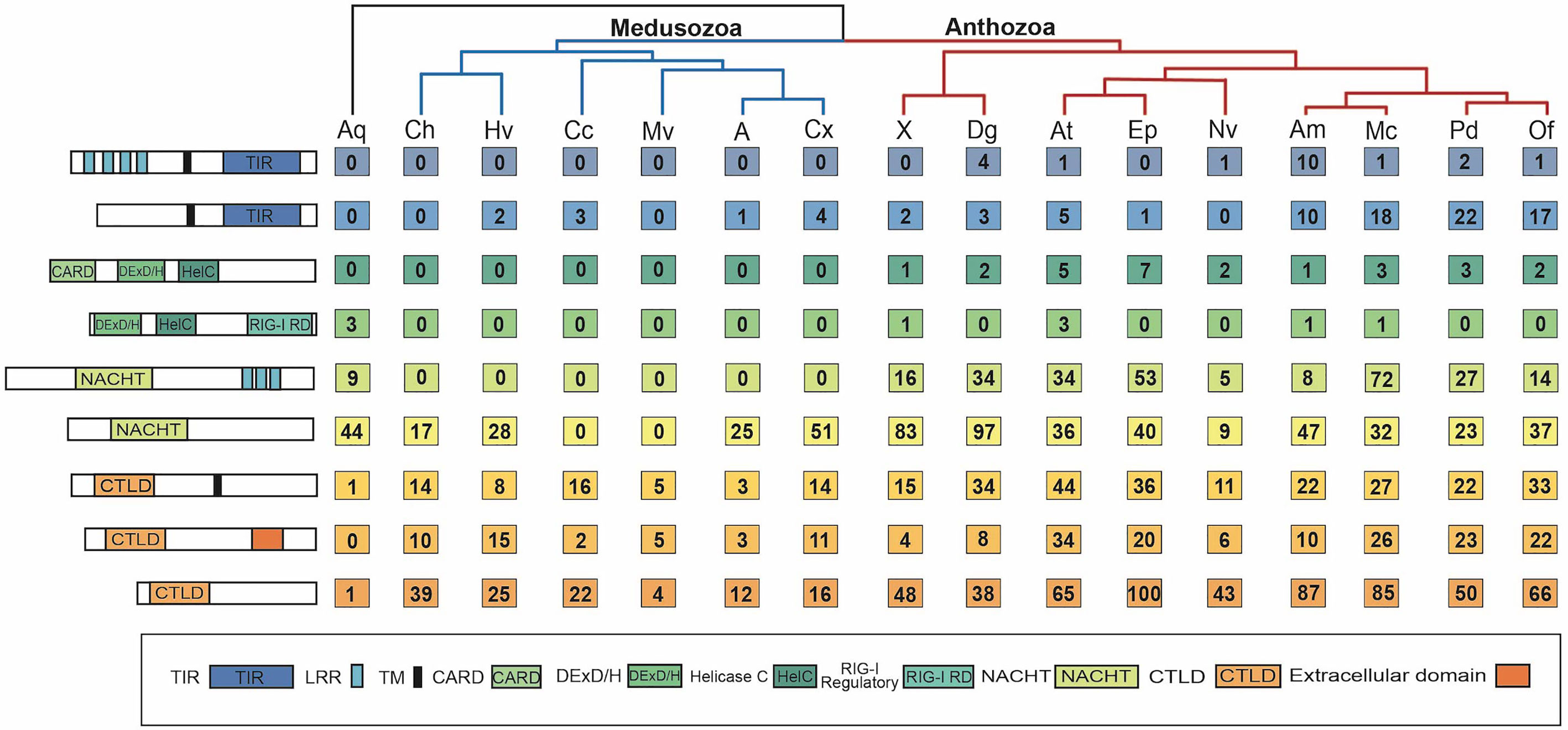
Figure 2 Number of PRRs of each type found in each species. Far left shows schematics of the minimum domain composition for each PRR type, from top to bottom: prototypical TLR, TLR-like, RIG-I/MDA5-like, LGP2-like, prototypical NLR, NLR-like, transmembrane CTL, extracellular CTL, CTL with unknown localization. To the right of the schematics is the corresponding number of each PRR type found in each species’ proteome. Species are grouped by phylogeny, with anthozoans in red, medusozoans in blue, and the sponge outgroup in black. Aq, Amphimedon queenslandica; Ch, Clytia hemisphaerica; Hv, Hydra vulgaris; Cc, Calavadosia cruxmelitensis; Mv, Morbakka virulenta; A, Aurelia sp.; Cx, Cassiopea xamachana; X, Xenia sp.; Dg, Dendonephthya gigantea; At, Actinia tenebrosa; Ep, Exaiptasia daiphana; Nv, Nematostella vectensis; Am, Acropora millepora; Mc, Montipora capitata; Pd, Pocillopora damicornis.
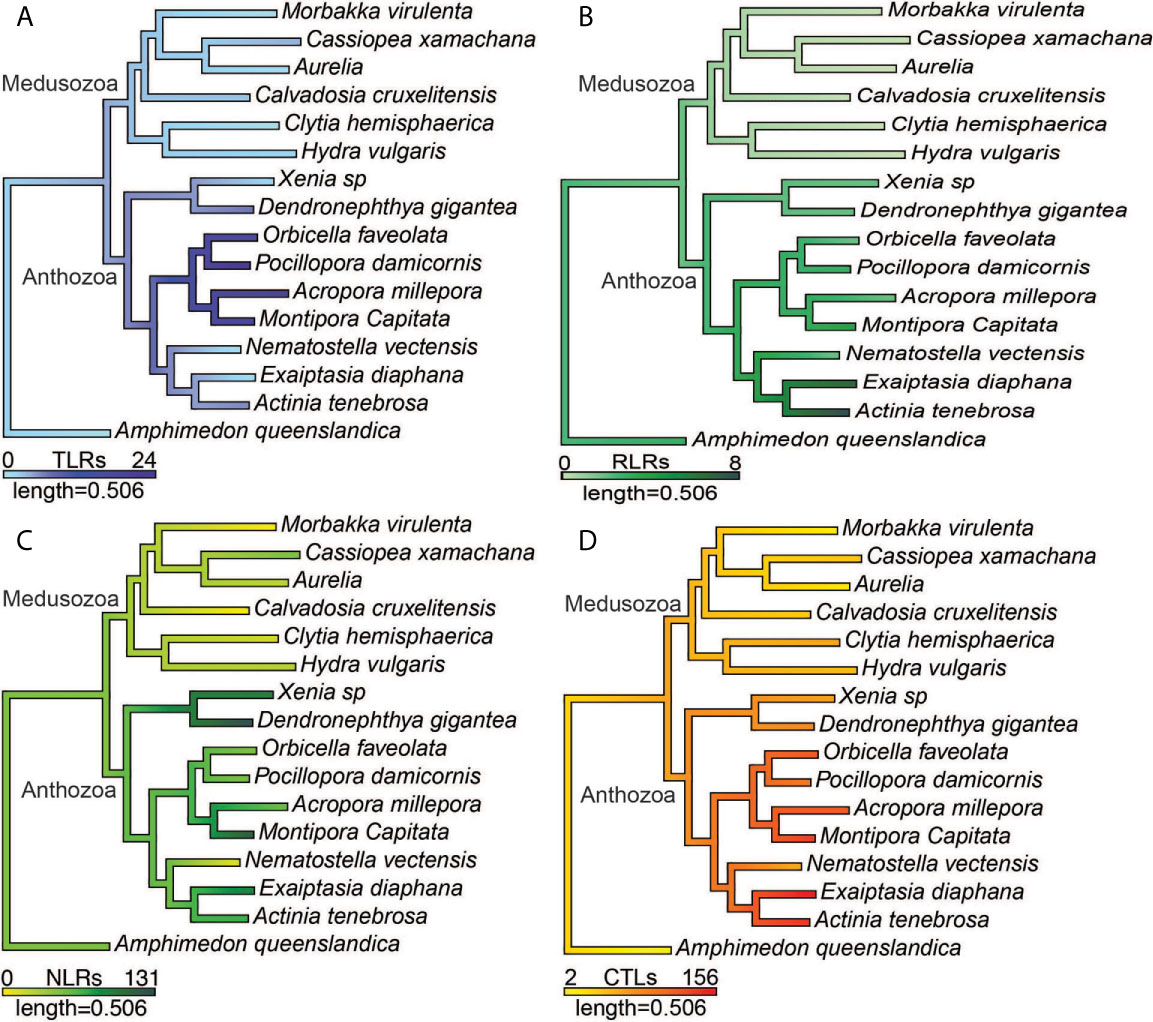
Figure 3 Ancestral state reconstructions of the number of (A) prototypical TLRs and TLR-like proteins, (B) RIG-I/MDA5-like receptors and LGP2-like receptors (C) prototypical NLRs and NLR-like proteins (D) CTLs.
RIG-I/MDA5-like receptors were found in the proteomes of all nine anthozoan species (Figure 2). These proteins all contained both the C terminal regulatory domain and the RIG-I C terminal repressor domain (Supplementary Table 4). Two species of anemone, A. tenebrosa and E. daiphana, have expansions in RLRs relative to both other anthozoans and the common ancestor shared by medusozoans and anthozoans (Figure 3B). These expansions are largely due to the high number of RIG-I/MDA5-like receptors in these species (Figure 2). A. queenslandica, the sponge outgroup, Xenia sp., A. tenebrosa, A. millepora, and M. capitata all contained proteins with domains characteristic of LGP2 (Figure 2). In A. millepora and M. capitata, these proteins had an RNA dependent RNA polymerase domain in addition to the canonical LGP2 domain organization (Supplementary Table 4). No RLRs were found in any of the six medusozoan species (Figure 2).
Diverse arrays of NACHT containing proteins were found in the majority of cnidarian species surveyed. Prototypical NLRs containing NACHT and LRR domains were present in the sponge A. queenslandica and all nine anthozoan species but absent in all medusozoan species (Figure 2). No NACHT containing proteins were found in two medusozoan species, the Staurozoan C. cruxmelitensis and the Cubozoan M. virulenta (Figure 2). The majority of anthozoan species were found to have expanded NLR repertoires relative to medusozoans. However, only a smaller subset of anthozoan NLR repertoires are expanded relative to the common ancestor shared by medusozoans and anthozoans (Figure 3C).
Anthozoan NLR repertoires are not only generally larger (Figure 3C), but also more diverse in terms of domain composition (Supplementary Table 5). Excluding LRRs 35 different Pfam domains were found in combination with NACHT across the 15 cnidarian species. Several of these domains are associated with immunity, including Dzip3/hRUL138-like HEPN nuclease (93), caspase recruitment domain (CARD) (94), death domain (DD) (95), ZU5 (96), glycosyl transferase (97), NB-ARC (98), WD-40 repeats (99), and Toll/interleukin receptor (TIR) domain (100). Excluding LRRs, the Dzip3/hRUL138- like HEPN domain was the most common domain found in combination with NACHT in C. hemisphaerica, Xenia sp., D. gigantea, A. tenebrosa, E. daiphana, A. millepora, M. capitata, P. damicornis, and O. faveolata. Anthozoans show large expansions of proteins with Dzip3/hRUL138- like HEPN domain fused to NACHT, which ranged from 17 proteins in O. faveolata to 83 in D. gigantea. TIR was the most common domain found with NACHT in C. xamachana and DD was most common domain found with NACHT in H. vulgaris. Domains associated with transposable elements were found with NACHT in two species, H. vulgaris and M. capitata. A protein model with NACHT and the hAT family C terminal dimerization domain was found in H. vulgaris while M. capitata’s proteome contains a protein with NACHT and an endonuclease reverse transcriptase domain and a protein with NACHT and integrase (Supplementary Table 5).
All of the cnidarians in this study have expanded repertoires of CTLs relative to the sponge outgroup (Figures 2, 3D). Two medusozoan species that are planktonic as adults, M. virulenta and Aurelia sp., had the fewest CTLs of the cnidarian species (Figures 2, 3D). Additionally, anthozoans have expanded CTL repertoires relative to both medusozoans and the common ancestor shared by medusozoans and anthozoans (Figure 3D). The species with the most predicted CTLs were A. tenebrosa and E. daiphana, two closely related sea anemones (Figures 2, 3D).
Across all 15 cnidarians, 70 different protein domains were found in combination with CTLD. The two clades differed in the domains most commonly found with CTLD. In six out of the nine anthozoan species the epidermal growth factor domain (EGF) was the most common domain found in conjunction with CTLD. The concanavalin A-like lectin domain was most commonly found with CTLD in four of the six medusozoans and two of the nine anthozoans. In the remaining medusozoans, C. hemisphaerica and M. virulenta, Von Willebrand factors and cysteine rich secretory domains respectively were most commonly found in conjunction with CTLD. Other common domains fused to CTLD were immunoglobulin, fibronectin, MAM, CUB, cysteine rich scavenger receptor, Kazal-type serine protease inhibitor, and PAN. Proteins with the domain organization of mannose binding lectin (MBL) (CTLD, collagen) were found in C. xamachana and C. hemisphaerica. As with the NLRs, the CTL search yielded surprising domain combinations. Reverse transcriptase domains were found in combination with CTLD in four species, M. virulenta, H. vulgaris, E. daiphana and M. capitata, and integrase and CTLD were found in M. capitata. Additionally, M. capitata and P. damicornis had predicted proteins with the MAC/Perforin domain in addition to CTLD (Supplementary Table 6).
Relationship to Clade and Life History Traits
Our results indicate the presence of a divide between the two cnidarian clades in PRR number across all four PRR types. Principle component analysis resulted in the medusozoans and a single anthozoan, N. vectensis, grouping tightly across both PC1 and PC2. The anthozoans excluding N. vectensis group relatively tightly across PC1, driven largely by CTLs and NLRs, but show more variance across PC2. The separation of the anthozoans across PC2 is most likely due to the expansions in prototypical TLRs and TLR-like proteins found in stony corals and the expansion of RIG-I/MDA5-like proteins in A. tenebrosa and E. daiphana (Figure 4A). A generalized linear model found significant associations between the total PRR number and clade (p = 0.0001). Total PRR number was also associated with ability to host intracellular algal symbionts (p=0.038), colonial animals (p = 0.033), and sedentary animals (p=0.014) (Figures 4B–E).
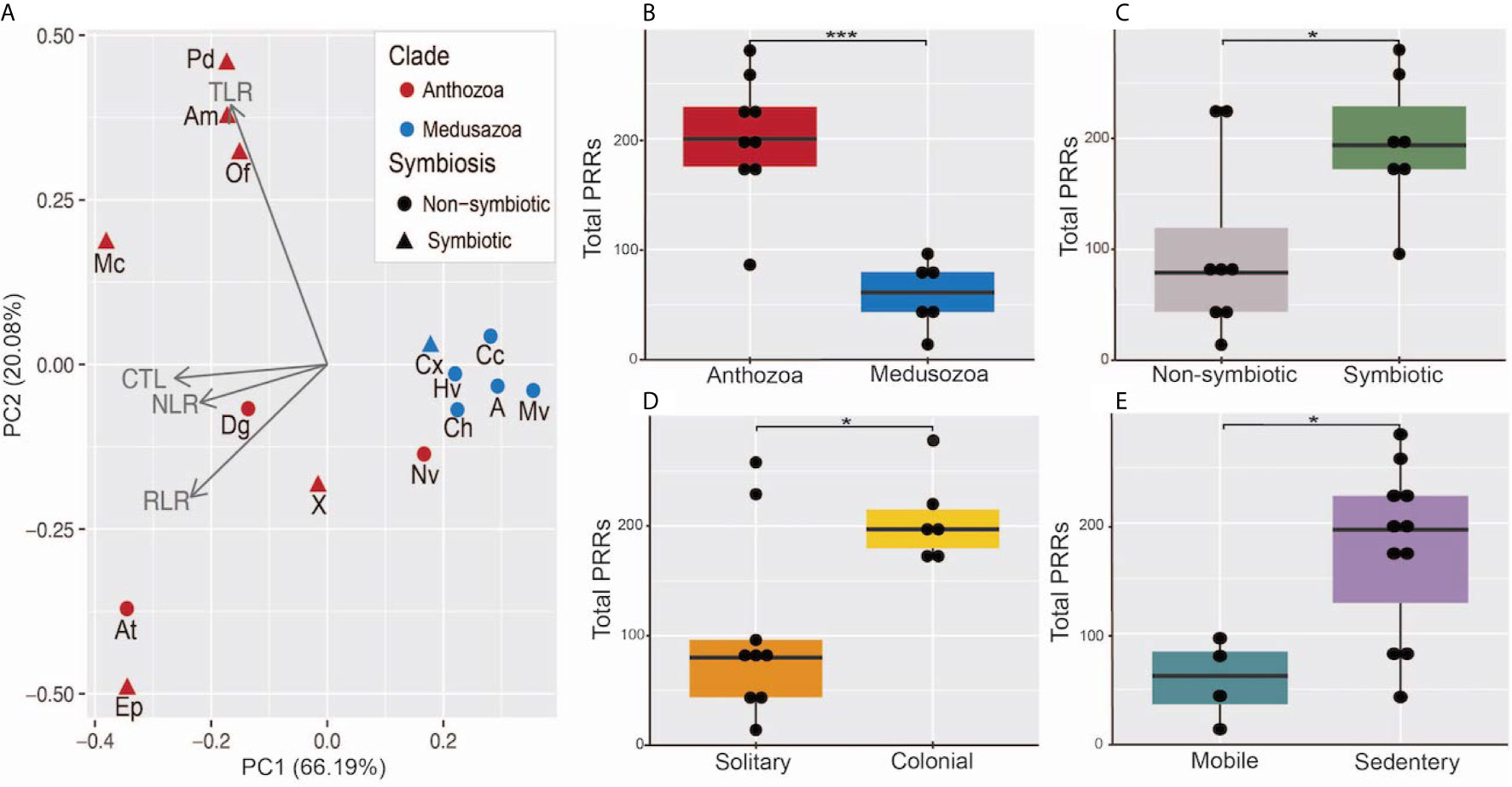
Figure 4 (A) Principal component analysis of total number of PRRs in each category, red points indicate anthozoan species, blue points indicate medusozoan species, triangular points indicate symbiotic species, and circular points represent non-symbiotic species. Aq, Amphimedon queenslandica; Ch, Clytia hemisphaerica; Hv, Hydra vulgaris; Cc, Calavadosi cruxmelitensis; Mv, Morbakka virulenta; A, Aurelia sp.; Cx, Cassiopea xamachana; X, Xenia sp.; Dg, Dendonephthya gigantea; At, Actinia tenebrosa; Ep, Exaiptasia daiphana; Nv, Nematostella vectensis; Am, Acropora millepora; Mc, Montipora capitata; Pd, Pocillopora damicornis. (B–E) Boxplots of total PRR number separated by (B) Clade, (C) Symbiotic status, (D) Colonial status, and (E) motility. *** indicates a p-value less < 0.001 and * indicates a p-value < 0.05 resulting from a generalized linear model. Points represent individual species’ total PRR numbers binned into groups of 10.
Downstream Immune Pathway Completeness
Given the differences in PRR numbers across the two cnidarian clades, we then investigated the completeness of the pathways downstream of TLRs, RLRs, NLRs, and CTLs that lead to master immune regulator NFκB (Figure 5). All species’ proteomes contained at least one homolog of NFκB. Stony corals and sponge outgroup A. queenslandica contain NFκB homologs most similar in domain composition to the mammalian NFκB queries (p100, p105), containing the Rel binding domain, Rel dimerization domain, ankyrin repeats, and the death domain (Figure 5C). Anthozoans E. daiphana and A. tenebrosa, have seemingly lost the death domain in their NFκB, which contains the Rel binding domain, Rel dimerization domain, and ankyrin repeats (Figure 5C). Notably, anthozoans N. vectensis, D. gigantea, Xenia sp., and all of the medusozoan species have NFκB homologs that have lost ankyrin repeats and the death domain (Figure 5C).
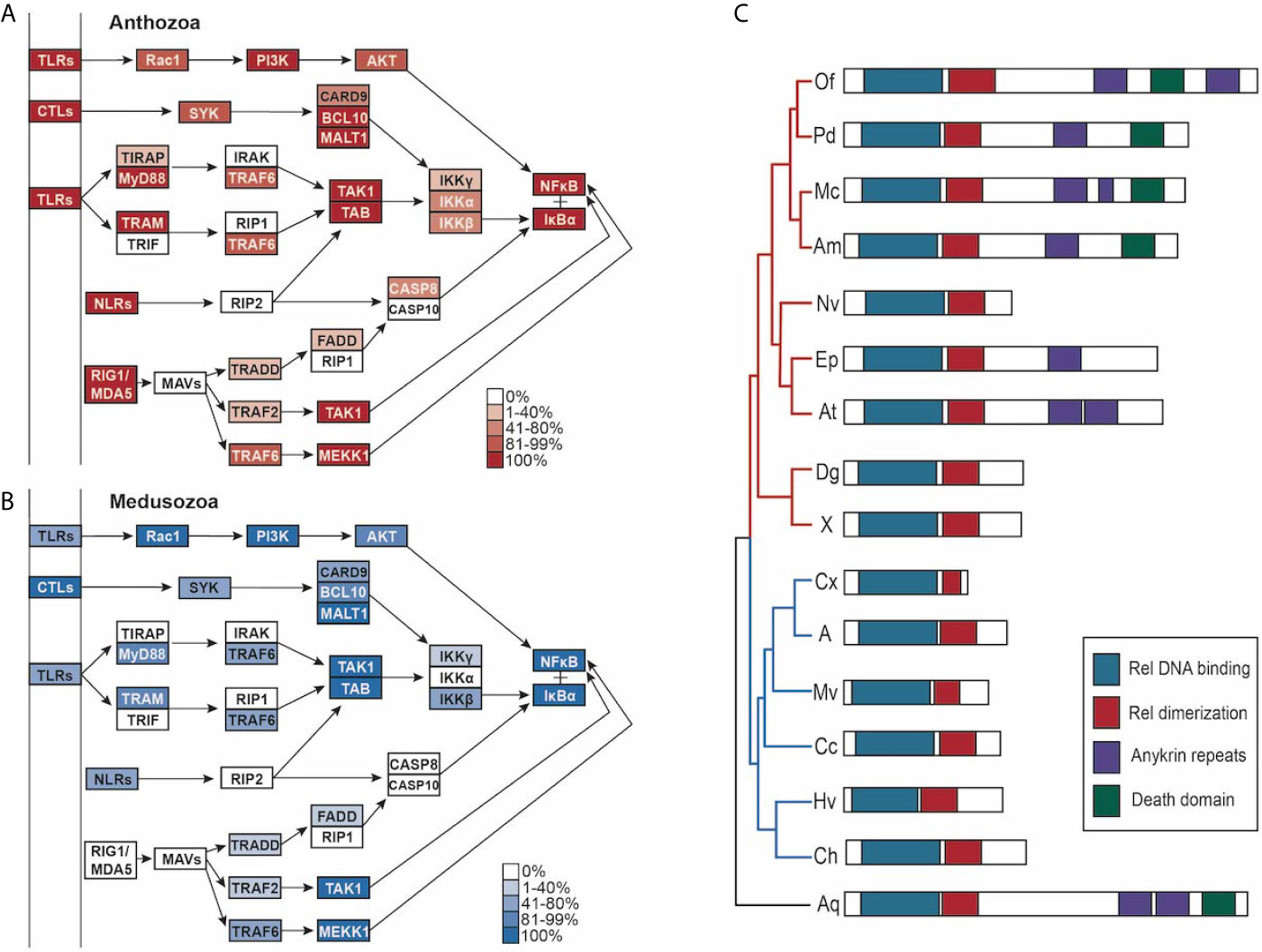
Figure 5 (A, B) PRR-NFκB pathways modified from Kegg pathway. Opacity of the boxes represents the percentage of (A) Anthozoans or (B) Medusozoans with that pathway component. (C) schematic of the domains found in NFκB in each species, grouped by phylogeny. Medusozoan species are indicated by the blue phylogenetic tree branches. Anthozoan species indicated by the red phylogenetic tree branches.
In addition to lacking RIG1/MDA5, prototypical NLRs and prototypical TLRs, medusozoans have slightly less complete PRR to NFκB pathways when compared to anthozoans (Figures 5A, B). Notably, IKKγ (NEMO) is absent in the majority of species, although the majority of species. contained at least one IKK (Supplementary Table 7). IRAK1/4, TRIF, RIP1, RIP2, MAVs, and CASP10 were missing in all cnidarian species (Figures 5A, B). In the case of CASP8, CASP10, and CARD9 the best reciprocal best blast hit was a slightly different caspase or caspase recruitment protein (Supplementary Table 7).
Homologs of MASP, C2, and C3 were found in the majority of species in this study (Figure 6). Two medusozoans, C. cruxmelitensis and H. vulgaris lacked MASP homologs. Notably, all anthozoans had proteins with the domain structure characteristic of the C2/Bf family and C3 family while C. hemisphaerica and H. vulgaris lacked C2/Bf proteins and Aurelia sp. and C. xamachana lacked proteins in the C3 family. However, both the Aurelia sp. and C. xamachana proteomes contained proteins with all of the characteristic C3 family domains except for the C34c domain (Supplementary Table 8). No proteins matching the C6-9 protein family’s domain structure were found in any of the species (Figure 6).
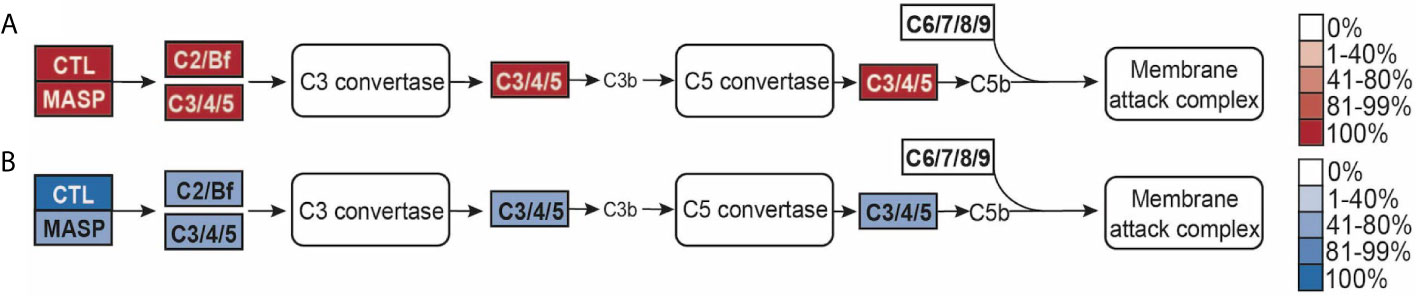
Figure 6 (A, B) Lectin-complement pathway modified from Kegg pathway. Opacity of the boxes represents the percentage of (A) Anthozoans or (B) Medusozoans with that pathway component.
Discussion
PRR Survey
Our study builds upon previous studies of cnidarian PRRs to give a higher resolution picture of PRR evolution within the phylum. We show a clear split in PRR number between two major cnidarian clades, Anthozoa and Medusozoa. Most notably medusozoans lack prototypical TLRs, all RLRs, and prototypical NLRs, while anthozoans possess prototypical PRRs. Further, we show that within Anthozoa extensive expansions of NLRs and CTLs are present in soft corals as well as what has been previously described in stony corals and anemones. In some cases, our results vary slightly from previous studies in the exact number of PRRs identified, due to differences in methodology including queries and search algorithms used, inclusion thresholds, genomic resources, and treatment of splice isoforms (10, 26, 43, 47, 101). These slight differences do not impact the overall findings of this study, including expanded PRR repertoires in the majority of anthozoans relative to medusozoans and a lack of prototypical PRRs in medusozoans not previously reported outside of Hydrozoa (27).
The presence of prototypical TLRs in two anthozoan classes, Octocorallia and Hexacorallia, and the absence of prototypical TLRs in medusozoans, sponges (102, 103), ctenophores (104), and placozoans (105) suggests that within Metazoa prototypical TLRs first appear in the ancestral anthozoan and have been secondarily lost in Xenia sp. and E. daiphana (57). Anthozoan prototypical TLRs likely are similar in function to the N. vectensis TLR which has been shown to play a role in pathogen recognition, NFkB signal transduction, and development (25). Despite their lack of prototypical TLRs, it is likely that Xenia sp., E. daiphana, and medusozoans are capable of TLR-NFκB signaling through TLR-like proteins, as studies in H. vulgaris have indicated (27).
The full mechanism by which transmembrane LRR proteins interact with TLR-like proteins is unknown; however, if multiple transmembrane LRRs are capable of interacting with a single TLR-like protein this pathogen recognition system could provide a versatile source of immune specificity in cnidarians. This study provides further support to previous findings of a lineage specific expansion in TLR-like proteins in stony coral species. This expansion is hypothesized to be involved in distinguishing mutualistic or commensal microorganisms from parasitic microorganisms within the coral holobiont and mounting an appropriate immune response (8, 26, 106). This potential function of TLR-like proteins is particularly vital to understand in corals as microbiome composition is thought to influence resistance to both disease and thermal stress, two existential threats to coral reefs (107, 108)
The presence of the C terminal RIG-I repressor domain indicates that the RIG-I/MDA5-like receptors in anthozoans are more similar to RIG-I than MDA5. This supports previous findings that N. vectensis RIG-I/MDA5-like receptors group more closely with vertebrate RIG-I than MDA5 (9). The presence of LGP2-like proteins in A. queenslandica, Xenia sp., A. tenebrosa, A. millepora, and M. capitata contradicts the hypothesis that RIG-I is the most evolutionarily ancient RLR and supports the hypothesis that LGP2 is the first RLR to emerge (9, 109). The fact that LGP2 is not consistently present in anthozoans indicates either that it is commonly secondarily lost due to low selective pressure or that it has evolved independently several times. Additionally, It is unclear what function LGP2 has in the absence of RIG-I and MDA5; however, the presence of an RNA dependent RNA polymerase domain in the A. millepora and M. capitata LGP2-like receptors indicates that their LGP2-like receptors most possibly amplify dsRNA targets for RNA interference and do not function like vertebrate LGP2 (34, 35, 110).
The presence of prototypical NLRs in A. queenslandica and all nine anthozoan species indicates that prototypical NLRs have been secondarily lost in Medusozoa. However, LRRs appear to not be necessary for DAMP recognition in medusozoan NLRs, as an NLR in H. vulgaris was shown to be upregulated in response to both lipopolysaccharide and flagellin (10). Thus, NLR-like proteins may still be a source of immune specificity in cnidarians. As in previous studies, we found expansions of NLRs within anthozoan species, indicating that NLRs are a substantial source of immune specificity within the clade (42, 43). Gene expression studies indicate that NLRs are upregulated in response to immune stressors and modulate apoptosis and immunity (42, 111, 112). Our data support the link between cnidarian NLRs and apoptosis as many of the effector domains found in cnidarian NLRs including CARD (94), ZU5 (96), DD (95), NBARC (98), and WD-40 repeats (99) are associated with apoptotic signal transduction.
Because NLRs are involved not only in pathogen recognition but also in self/altered-self/non-self-recognition, traits such as coloniality and ability to form nutritional algal symbiosis may be linked to expansions in NLRs (18, 113). A recent study indicates that in E. daiphana, a symbiotic anemone, microalgae are taken into the cell largely indiscriminately and the decision to retain these microalgae as symbionts or expel them likely occurs intracellularly (114). As intracellular PRRs, NLRs are great candidates for modulating interactions between the algal symbionts and the immune system of their cnidarian hosts during the establishment of symbiosis, a hypothesis with some support from transcriptomic studies (115).
Currently, knowledge of cnidarian NLRs stems from both bioinformatic and gene expression studies (10, 42, 43, 111, 112) as this family of PRRs has yet to be functionally studied within the phylum. Functional studies of cnidarian NLRs are needed, as our results and previous studies indicate that anthozoans have invested in large and diverse NLR repertoires (42, 43).
The expansion of CTLs across all 15 cnidarian species relative to the sponge outgroup aligns with previous findings of diverse and large CTLD containing protein repertoires in invertebrate species (5, 47). Despite this diversity and the potential for cnidarian CTLs to greatly contribute to immune specificity, functional studies have only been conducted on homologs of mannose binding lectin (MBL), the activator of the lectin complement pathway (11, 116). These studies indicate that MBL homologs in two corals, P. damicornis (116) and A millepora (11) are capable of binding to both bacteria and Symbiodiniaceae leading to the hypothesis that lectin/glycan interactions are a mechanism of recognition during symbiont infection. This hypothesis has found some support from transcriptomic studies (117, 118). The ability of cnidarian lectins to interact with MASP and activate the lectin-complement pathway has yet to be investigated. Both of the cnidarian lectins for which we have functional studies and the majority of cnidarian species in this study lack CTLs with the collagen helix domain characteristic of MBL (11, 116). However, it is possible that cnidarians are capable of lectin-complement activation despite lacking collagen domains as studies have shown that CTLs lacking collagen domains are still able to interact with MASP (119, 120).
CTLs in cnidarians are potentially a large source of immune specificity which to date has been understudied. Based on their large and diverse CTL repertoires, cnidarians are likely utilizing CTLD containing proteins for a variety of functions. Transcriptomic and proteomic studies indicate that CTLs are involved in coral wound healing (121) and disease response (122, 123) in addition to their hypothesized role in mediating symbiosis with Symbiodiniaceae (11, 116–118). Thus, further functional studies of cnidarian CTLs are warranted and should focus both on highly conserved CTLs and on novel cnidarian CTLs with the potential to shed light on disease processes.
Relationship to Clade and Life History Traits
There is a divide in number of PRRs and downstream immune pathway completeness between Medusozoa and Anthozoa. There are several life history traits common in the anthozoan clade that could explain a greater need for immune specificity and thus this division (13). Three of these life history traits, intracellular algal symbiosis, coloniality, and sedentary lifestyle, we found to have a significant association with total number of PRRs. While intracellular algal symbiosis occurs in both Medusozoa and Anthozoa, there are far more symbiotic anthozoan genera (18, 124). The establishment and maintenance of intracellular algal symbionts is a complex process, with many cnidarian species hosting several species of algae, and thus it likely requires immune specificity (18). Similar to intracellular algal symbiosis, there are colonial organisms in both Medusozoa and Anthozoa, but they are far more common in Anthozoa. Colonial invertebrates require allorecognition systems to distinguish self-tissues from conspecific tissues, which may necessitate a more diverse repertoire of PRRs (19, 20). Animals that spend the majority of their lifespan motile are far more common in the medusozoan clade. In contrast, anthozoan species are largely sedentary, often residing in microbe rich environments like estuaries and coral reefs (13). As sedentary animals are unable to avoid antigen accumulation through movement, they likely also have a greater need for immune specificity (125).
There are several other life history traits that may be associated with immune specificity in cnidarians that we were unable to test due to either a lack of data or low sample size. Number of mutualistic and commensal bacterial symbioses is almost certainly factor in the amount of immune specificity a given cnidarian has, however this information is not available for many of the species in this study (126). It is also possible that anthozoans and medusozoans have a similar need for immune specificity and simply employ different methods to meet this demand. Medusozoans could rely more heavily on other pattern recognition receptor types, such scavenger receptors, rather than TLRs, RLRs, NLRs, and CTLs to provide their immune specificity (127). Other possible sources of immune specificity not reflected in PRR number include increased substitution rates and post translational modifications (128, 129). With future studies and increased resolution of genomic and proteomic resources we hope that this study can be used as a basis for linking life history to mechanisms of immune specificity.
Downstream Immune Pathway Completeness
Our study indicates a complex history of NFκB within Cnidaria, as ankyrin repeats appear to have been secondarily lost at least three times within the phylum. This C-terminal inhibitory domain prevents NFκB from trafficking to the nucleus and must be removed via proteolysis in order for NFκB to bind to DNA. This regulatory function may not be under strong selective pressure within Cnidaria. There are several functional studies on cnidarian NFκB proteins, including N. vectensis Nv-NFκB, which is truncated and does not include ankyrin repeats (90, 130). These studies show that E. daiphana’s NFκB binding specificity is more similar to both human NFκB p50 and Nv-NFκB than c-Rel and RelA, despite the Nv-NFκB having a similar domain organization to c-Rel and RelA (130). Because ankyrin repeats appear to have been independently lost, it is unclear if soft coral and medusozoan NFκB share similar binding specificity to Ep-NFκB, Nv-NFκB, and human NFκB p50. However, several studies in both medusozoans and anthozoans show that NFκB is responsive to pathogen exposure (27, 111, 112, 130).
Consistent with previous studies, we found that while cnidarians have the majority of proteins in PRR to NFκB pathways they are missing some components found in mammalian PRR to NFκB pathways (15, 130). However, the absence of these components does not mean that cnidarians are incapable of PRR to NFκB signaling (27, 130). Cnidarians can compensate for missing pathway components either by proteins upstream of the missing component interacting directly with proteins further downstream or through proteins that are not homologous to mammalian pathway members but are able to functionally replace them (10, 131).
While cnidarians likely retain PRR to NFκB signaling through these mechanisms, these missing proteins still indicate potential fundamental differences between cnidarian and mammalian innate immune pathways. Functional replacements may not be regulated or regulate immune pathways in the same manner as their mammalian counterparts. For example, although it appears as though cnidarians have a functional replacement for RIPs, this functional replacement likely is not regulated in the same manner as RIPs because the adaptor proteins RIPs interact with in the decision to promote pro-life NFκB signaling or cell death signaling are also absent (10, 132). The lack of RIPs in cnidarians could result in a greater propensity for cell death signaling over pro-life NFκB signaling, a hypothesis that could explain disease phenotypes in white syndrome coral diseases (133). The absence of MAVs from the RLR-NFκB pathway is notable, as it is unclear how RLR antiviral signaling occurs in the absence of this key adaptor protein, however interactions with other CARD domain containing proteins may mediate RLR signaling in cnidarians (31, 32).
No proteins in the C6 family were found in any of the 15 cnidarian species in this study, consistent with previous findings in N. vectensis, E. daiphana and reef building corals (57, 92, 118). This indicates that cnidarians are unable to form the membrane attack complex and instead use complement for opsonization through C3 (92). While opsonization of microbes by C3 has not been directly shown in cnidarians, transcriptomic studies in anthozoans show that complement signaling is responsive to bacterial pathogens (112, 118). Scyphozoans C. xamachana and Aurelia sp. lack the complete domain structure of C3. However, because the domain they are missing, C345c mainly functions in interacting with C6 family proteins, this protein may still be able to opsonize (134).
Conclusions
As a whole, our data indicate that anthozoans have greater immune specificity than medusozoans, with expansions of NLRs and CTLs providing the majority of this specificity. We hypothesize that a greater immune specificity in anthozoans is needed due to life history traits common within the clade, such as being sedentary, having a colonial body plan, and hosting a complex microbiota that includes intracellular algal symbionts. More broadly, our data indicate that studying cnidarian PRRs can give insight not only into where within Metazoa prototypical PRRs arose but also how basal prototypical PRRs function and the systems by which DAMPS and MAMPs were recognized prior to the emergence of these prototypical PRRs. Further investigations into medusozoan immunity would likely provide a greater understanding of non-prototypical pattern recognition systems. The ecological threat coral diseases pose has led to a wealth of knowledge on anthozoan immune responses (135). Placing these studies in an evolutionary context could give further information as to how basal prototypical PRRs function and more broadly how innate immunity evolved.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
ME and BD conceived of the project. Data collection and bioinformatic analysis was conducted by ME. ME, BD, and LM wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding was provided by NSF grant no. OCE 1712134 and OCE 1928771 to LM, as well as support from UTA Research Enhancement Program and UTA College of Science for support for ME.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors would like to thank Mydlarz lab members Nicholas MacKnight, Kelsey Bevers, and Emily Van Buren for their input and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.689463/full#supplementary-material
Supplementary Table 1 | Life history traits and genome assembly statistics of species included in this study.
Supplementary Table 2 | Comparison of N. vectensis proteomes.
Supplementary Table 3 | TLR domains and sequence IDs.
Supplementary Table 4 | RLR domains and sequence IDs.
Supplementary Table 5 | NLR domains and sequence IDs.
Supplementary Table 6 | CTL domains and sequence IDs.
Supplementary Table 7 | PRR-NFkB pathways sequence IDs.
Supplementary Table 8 | Lectin complement pathway domains and sequence IDs.
References
1. Buchmann K. Evolution of Innate Immunity: Clues From Invertebrates Via Fish to Mammals. Front Immunol (2014) 5:1–8. doi: 10.3389/fimmu.2014.00459/full
2. oehm. Evolution of Vertebrate Immunity | Elsevier Enhanced Reader (2012). Available at: https://reader.elsevier.com/reader/sd/pii/S0960982212007944?token=4197AD9ED7F9C49EE933D95310DF08D581B5F0156E3454BA83FE996D6C38C58EF6623C84F271890CBC42DE3B9E2C1E2C.
3. Litman GW, Rast JP, Fugmann SD. The Origins of Vertebrate Adaptive Immunity. Nat Rev Immunol (2010) 10(8):543–53. doi: 10.1038/nri2807
4. Janeway CA, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
5. Pees B, Yang W, Zárate-Potes A, Schulenburg H, Dierking K. High Innate Immune Specificity Through Diversified C-Type Lectin-Like Domain Proteins in Invertebrates. J Innate Immun (2016) 8(2):129–42. doi: 10.1159/000441475
6. Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front Immunol (2018) 9:1–12. doi: 10.3389/fimmu.2018.02379/full
7. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu Rev Immunol (2015) 33(1):257–90. doi: 10.1146/annurev-immunol-032414-112240
8. Brennan JJ, Gilmore TD. Evolutionary Origins of Toll-like Receptor Signaling. Mol Biol Evol (2018) 35(7):1576–87. doi: 10.1093/molbev/msy050
9. Zou J, Chang M, Nie P, Secombes CJ. Origin and Evolution of the RIG-I Like RNA Helicase Gene Family. BMC Evo Bio (2009) 9:1–14. doi: 10.1186/1471-2148-9-85
10. Lange C, Hemmrich G, Klostermeier UC, López-Quintero JA, Miller DJ, Rahn T, et al. Defining the Origins of the NOD-Like Receptor System at the Base of Animal Evolution. Mol Biol Evol (2011) 28(5):1687–702. doi: 10.1093/molbev/msq349
11. Kvennefors ECE, Leggat W, Hoegh-Guldberg O, Degnan BM, Barnes AC. An Ancient and Variable Mannose-Binding Lectin From the Coral Acropora Millepora Binds Both Pathogens and Symbionts. Dev Comp Immunol (2008) 32(12):1582–92. doi: 10.1016/j.dci.2008.05.010
12. Ting JP-Y, Davis BK. Caterpiller: A Novel Gene Family Important in Immunity, Cell Death, and Diseases. Annu Rev Immunol (2004) 23(1):387–414. doi: 10.1146/annurev.immunol.23.021704.115616
13. Kayal E, Bentlage B, Sabrina Pankey M, Ohdera AH, Medina M, Plachetzki DC, et al. Phylogenomics Provides a Robust Topology of the Major Cnidarian Lineages and Insights on the Origins of Key Organismal Traits. BMC Evol Biol (2018) 18(1):68. doi: 10.1186/s12862-018-1142-0
14. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The Dynamic Genome of Hydra. Nature (2010) 464(7288):592–6. doi: 10.1038/nature08830
15. Miller DJ, Hemmrich G, Ball EE, Hayward DC, Khalturin K, Funayama N, et al. The Innate Immune Repertoire in Cnidaria - Ancestral Complexity and Stochastic Gene Loss. Genome Biol (2007) 8(4):R59. doi: 10.1186/gb-2007-8-4-r59
16. Schulenburg H, Kurz CL, Ewbank JJ. Evolution of the Innate Immune System: The Worm Perspective. Immunol Rev (2004) 198(1):36–58. doi: 10.1111/j.0105-2896.2004.0125.x
17. Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic Evolution of the Innate Immune System in Drosophila. Nat Genet (2007) 39(12):1461–8. doi: 10.1038/ng.2007.60
18. Mansfield KM, Gilmore TD. Innate Immunity and cnidarian-Symbiodiniaceae Mutualism. Dev Comp Immunol (2019) 90:199–209. doi: 10.1016/j.dci.2018.09.020
19. Nydam ML, De Tomaso T. Creation and Maintenance of Variation in Allorecognition Loci. Front Immunol (2011) 2:1–6. doi: 10.3389/fimmu.2011.00079/full
20. Oberbarnscheidt MH, Lakkis FG. Innate Allorecognition. Immunol Rev (2014) 258(1):145–9. doi: 10.1111/imr.12153
21. Medzhitov R. Toll-Like Receptors and Innate Immunity. Nat Rev Immunol (2001) 1(2):135–45. doi: 10.1038/35100529
22. Akira S, Takeda K. Toll-Like Receptor Signalling. Nat Rev Immunol (2004) 4(7):499–511. doi: 10.1038/nri1391
23. Anthoney N, Foldi I, Hidalgo A. Toll and Toll-like Receptor Signalling in Development. Development (2018) 145(9):dev156018. doi: 10.1242/dev.156018
24. Vasselon T, Detmers PA. Toll Receptors: A Central Element in Innate Immune Responses. Infect Immun (2002) 70(3):1033–41. doi: 10.1128/IAI.70.3.1033-1041.2002
25. Brennan JJ, Messerschmidt JL, Williams LM, Matthews BJ, Reynoso M, Gilmore TD. Sea Anemone Model has a Single Toll-like Receptor That can Function in Pathogen Detection, NF-κb Signal Transduction, and Development. Proc Natl Acad Sci (2017) 114(47):E10122–31. doi: 10.1073/pnas.1711530114
26. Poole AZ, Weis VM. TIR-Domain-Containing Protein Repertoire of Nine Anthozoan Species Reveals Coral–Specific Expansions and Uncharacterized Proteins. Dev Comp Immunol (2014) 46(2):480–8. doi: 10.1016/j.dci.2014.06.002
27. Bosch TCG, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, et al. Uncovering the Evolutionary History of Innate Immunity: The Simple Metazoan Hydra Uses Epithelial Cells for Host Defence. Dev Comp Immunol (2009) 33(4):559–69. doi: 10.1016/j.dci.2008.10.004
28. Dixit E, Kagan JC. Intracellular Pathogen Detection by RIG-I-Like Receptors. In: Advances in Immunology. Cambridge Massachusetts: Elsevier (2013). p. 99–125. Available at: https://linkinghub.elsevier.com/retrieve/pii/B9780124105249000049.
29. Rehwinkel J, Gack MU. Rig-I-like Receptors: Their Regulation and Roles in RNA Sensing. Nat Rev Immunol (2020) 20(9):537–51. doi: 10.1038/s41577-020-0288-3
30. Loo Y-M, Gale M. Immune Signaling by RIG-I-like Receptors. Immunity (2011) 34(5):680–92. doi: 10.1016/j.immuni.2011.05.003
31. Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein That Activates Nf-κb and IRF3. Cell (2005) 122(5):669–82. doi: 10.1016/j.cell.2005.08.012
32. Potter JA, Randall RE, Taylor GL. Crystal Structure of Human IPS-1/MAVS/VISA/Cardif Caspase Activation Recruitment Domain. BMC Struct Biol (2008) 8(1):11. doi: 10.1186/1472-6807-8-11
33. Bruns AM, Horvath CM. LGP2 Synergy With MDA5 in RLR-mediated RNA Recognition and Antiviral Signaling. Cytokine (2015) 74(2):198–206. doi: 10.1016/j.cyto.2015.02.010
34. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a Positive Regulator of RIG-I- and MDA5-mediated Antiviral Responses. Proc Natl Acad Sci (2010) 107(4):1512–7. doi: 10.1073/pnas.0912986107
35. Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J Virol (2014) 88(15):8194–200. doi: 10.1128/JVI.00640-14
36. Martinon F, Mayor A, Tschopp J. The Inflammasomes: Guardians of the Body. Annu Rev Immunol (2009) 27(1):229–65. doi: 10.1146/annurev.immunol.021908.132715
37. Lee G-S, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, et al. The Calcium-Sensing Receptor Regulates the NLRP3 Inflammasome Through Ca 2+ and Camp. Nature (2012) 492(7427):123–7. doi: 10.1038/nature11588
38. Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin Activates the Inflammasome in Response to Toxins and ATP. Nature (2006) 440(7081):228–32. doi: 10.1038/nature04515
39. Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-Like Proteins in Immunity, Inflammation and Disease. Nat Immunol (2006) 7(12):1250–7. doi: 10.1038/ni1412
40. Kanneganti T-D, Body-Malapel M, Amer A, Park J-H, Whitfield J, Franchi L, et al. Critical Role for Cryopyrin/Nalp3 in Activation of Caspase-1 in Response to Viral Infection and Double-stranded Rna*. J Biol Chem (2006) 281(48):36560–8. doi: 10.1074/jbc.M607594200
41. Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the Antimicrobial Response by NLR Proteins. Immunity (2011) 34(5):665–79. doi: 10.1016/j.immuni.2011.05.007
42. Dimos BA, Butler CC, Ricci CA, MacKnight NJ, Mydlarz LD. Responding to Threats Both Foreign and Domestic: Nod-Like Receptors in Corals. Integr Comp Biol (2019) 59(4):819–29. doi: 10.1093/icb/icz111
43. Hamada M, Shoguchi E, Shinzato C, Kawashima T, Miller DJ, Satoh N. The Complex Nod-Like Receptor Repertoire of the Coral Acropora Digitifera Includes Novel Domain Combinations. Mol Biol Evol (2013) 30(1):167–76. doi: 10.1093/molbev/mss213
44. Zelensky AN, Gready JE. The C-type Lectin-Like Domain Superfamily. FEBS J (2005) 272(24):6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
45. Geijtenbeek TBH, Gringhuis SI. Signalling Through C-type Lectin Receptors: Shaping Immune Responses. Nat Rev Immunol (2009) 9(7):465–79. doi: 10.1038/nri2569
46. Fujita T. Evolution of the Lectin–Complement Pathway and its Role in Innate Immunity. Nat Rev Immunol (2002) 2(5):346–53. doi: 10.1038/nri800
47. Wood-Charlson EM, Weis VM. The Diversity of C-type Lectins in the Genome of a Basal Metazoan, Nematostella Vectensis. Dev Comp Immunol (2009) 33(8):881–9. doi: 10.1016/j.dci.2009.01.008
48. Zárate-Potes A, Ocampo ID, Cadavid LF. The Putative Immune Recognition Repertoire of the Model Cnidarian Hydractinia Symbiolongicarpus is Large and Diverse. Gene (2019) 684:104–17. doi: 10.1016/j.gene.2018.10.068
49. Leclère L, Horin C, Chevalier S, Lapébie P, Dru P, Peron S, et al. The Genome of the Jellyfish Clytia Hemisphaerica and the Evolution of the Cnidarian Life-Cycle. Nat Ecol Evol (2019) 3(5):801–10. doi: 10.1038/s41559-019-0833-2
50. Gold DA, Katsuki T, Li Y, Yan X, Regulski M, Ibberson D, et al. The Genome of the Jellyfish Aurelia and the Evolution of Animal Complexity. Nat Ecol Evol (2019) 3(1):96–104. doi: 10.1038/s41559-018-0719-8
51. Ohdera A, Ames CL, Dikow RB, Kayal E, Chiodin M, Busby B, et al. Box, Stalked, and Upside-Down? Draft Genomes From Diverse Jellyfish (Cnidaria, Acraspeda) Lineages: Alatina Alata (Cubozoa), Calvadosia Cruxmelitensis (Staurozoa), and Cassiopea Xamachana (Scyphozoa). GigaScience (2019) 8(7):1–15. doi: 10.1093/gigascience/giz069
52. Hu M, Zheng X, Fan C-M, Zheng Y. Lineage Dynamics of the Endosymbiotic Cell Type in the Soft Coral Xenia. Nature (2020) 582(7813):534–8. doi: 10.1038/s41586-020-2385-7
53. Surm JM, Stewart ZK, Papanicolaou A, Pavasovic A, Prentis PJ. The Draft Genome of Actinia Tenebrosa Reveals Insights Into Toxin Evolution. Ecol Evol (2019) 9(19):11314–28. doi: 10.1002/ece3.5633
54. Fuller ZL, Mocellin VJL, Morris LA, Cantin N, Shepherd J, Sarre L, et al. Population Genetics of the Coral Acropora Millepora: Toward Genomic Prediction of Bleaching. Science (2020) 369(6501):eaba474. doi: 10.1126/science.aba4674
55. Home - Cassiopea xamachana. Available at: https://mycocosm.jgi.doe.gov/Casxa1/Casxa1.home.html.
56. Jeon Y, Park SG, Lee N, Weber JA, Kim H-S, Hwang S-J, et al. The Draft Genome of an Octocoral, Dendronephthya Gigantea. Genome Biol Evol (2019) 11(3):949–53. doi: 10.1093/gbe/evz043
57. Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, et al. The Genome of Aiptasia, a Sea Anemone Model for Coral Symbiosis. Proc Natl Acad Sci (2015) 112(38):11893–8. doi: 10.1073/pnas.1513318112
58. Shumaker A, Putnam HM, Qiu H, Price DC, Zelzion E, Harel A, et al. Genome Analysis of the Rice Coral Montipora Capitata. Sci Rep (2019) 9(1):2571. doi: 10.1038/s41598-019-39274-3
59. Khalturin K, Shinzato C, Khalturina M, Hamada M, Fujie M, Koyanagi R, et al. Medusozoan Genomes Inform the Evolution of the Jellyfish Body Plan. Nat Ecol Evol (2019) 3(5):811–22. doi: 10.1038/s41559-019-0853-y
60. Babonis LS, Martindale MQ, Ryan JF. Do Novel Genes Drive Morphological Novelty? An Investigation of the Nematosomes in the Sea Anemone Nematostella Vectensis. BMC Evol Biol (2016) 16(1):114. doi: 10.1186/s12862-016-0683-3
61. Prada C, Hanna B, Budd AF, Woodley CM, Schmutz J, Grimwood J, et al. Empty Niches After Extinctions Increase Population Sizes of Modern Corals. Curr Biol (2016) 26(23):3190–4. doi: 10.1016/j.cub.2016.09.039
62. Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N. Comparative Analysis of the Pocillopora Damicornis Genome Highlights Role of Immune System in Coral Evolution. Sci Rep (2018) 8(1):16134. doi: 10.1038/s41598-018-34459-8
63. Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, et al. The Amphimedon Queenslandica Genome and the Evolution of Animal Complexity. Nature (2010) 466(7307):720–6. doi: 10.1038/nature09201
64. Eddy SR. Accelerated Profile Hmm Searches. PloS Comput Biol (2011) 7(10):e1002195. doi: 10.1371/journal.pcbi.1002195
65. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De Novo Transcript Sequence Reconstruction From RNA-Seq: Reference Generation and Analysis With Trinity. Nat Protoc (2013) 8(8):1494–512. doi: 10.1038/nprot.2013.084
66. Fu L, Niu B, Zhu Z, Wu S, Li W. Cd-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinforma Oxf Engl (2012) 28(23):3150–2. doi: 10.1093/bioinformatics/bts565
67. Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, et al. Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Science (2007) 317(5834):86–94. doi: 10.1126/science.1139158
68. Madeira F, Park Y, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res (2019) 47(W1):W636–41. doi: 10.1093/nar/gkz268
69. Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, et al. Evaluation of GRCh38 and De Novo Haploid Genome Assemblies Demonstrates the Enduring Quality of the Reference Assembly. Genome Res (2017) 27(5):849–64. doi: 10.1101/gr.213611.116
70. Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res (2021) 49(D1):D412–9. doi: 10.1093/nar/gkaa913
71. Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting Transmembrane Protein Topology With a Hidden Markov Model: Application to Complete genomes11Edited by F. Cohen J Mol Biol (2001) 305(3):567–80. doi: 10.1006/jmbi.2000.4315
72. Gibbs GM, Roelants K, O’Bryan MK. The CAP Superfamily: Cysteine-Rich Secretory Proteins, Antigen 5, and Pathogenesis-Related 1 Proteins–Roles in Reproduction, Cancer, and Immune Defense. Endocr Rev (2008) 29(7):865–97. doi: 10.1210/er.2008-0032
73. Bork P, Beckmann G. The CUB Domain: A Widespread Module in Developmentally Regulated Proteins. J Mol Biol (1993) 231(2):539–45. doi: 10.1006/jmbi.1993.1305
74. Johnson JD, Edman JC, Rutter WJ. A Receptor Tyrosine Kinase Found in Breast Carcinoma Cells has an Extracellular Discoidin I-like Domain. Proc Natl Acad Sci U S A (1993) 90(12):5677–81. doi: 10.1073/pnas.90.12.5677
75. Rangaraju S, Khoo KK, Feng Z-P, Crossley G, Nugent D, Khaytin I, et al. Potassium Channel Modulation by a Toxin Domain in Matrix Metalloprotease 23*. J Biol Chem (2010) 285(12):9124–36. doi: 10.1074/jbc.M109.071266
76. Colombatti A, Bonaldo P, Doliana R. Type A Modules: Interacting Domains Found in Several non-Fibrillar Collagens and in Other Extracellular Matrix Proteins. Matrix Stuttg Ger (1993) 13(4):297–306. doi: 10.1016/S0934-8832(11)80025-9
77. Adams JC. Thrombospondins: Multifunctional Regulators of Cell Interactions. Annu Rev Cell Dev Biol (2001) 17:25–51. doi: 10.1146/annurev.cellbio.17.1.25
78. Hoffmann W, Hauser F. The P-domain or Trefoil Motif: A Role in Renewal and Pathology of Mucous Epithelia? Trends Biochem Sci (1993) 18(7):239–43. doi: 10.1016/0968-0004(93)90170-R
79. Matsushita M, Fujita T. The Role of Ficolins in Innate Immunity. Immunobiology (2002) 205(4–5):490–7. doi: 10.1078/0171-2985-00149
80. Ciccarelli FD, Doerks T, Bork P. AMOP, a Protein Module Alternatively Spliced in Cancer Cells. Trends Biochem Sci (2002) 27(3):113–5. doi: 10.1016/S0968-0004(01)02049-7
81. Bycroft M, Bateman A, Clarke J, Hamill SJ, Sandford R, Thomas RL, et al. The Structure of a PKD Domain From polycystin-1: Implications for Polycystic Kidney Disease. EMBO J (1999) 18(2):297–305. doi: 10.1093/emboj/18.2.297
82. Fujimoto T, Imaeda Y, Konishi N, Hiroe K, Kawamura M, Textor GP, et al. Discovery of a Tetrahydropyrimidin-2(1H)-one Derivative (Tak-442) as a Potent, Selective, and Orally Active Factor Xa Inhibitor. J Med Chem (2010) 53(9):3517–31. doi: 10.1021/jm901699j
83. Behrendt N, Ploug M, Patthy L, Houen G, Blasi F, Danø K. The Ligand-Binding Domain of the Cell Surface Receptor for Urokinase-Type Plasminogen Activator. J Biol Chem (1991) 266(12):7842–7. doi: 10.1016/S0021-9258(20)89526-X
84. Wouters MA, Rigoutsos I, Chu CK, Feng LL, Sparrow DB, Dunwoodie SL. Evolution of Distinct EGF Domains With Specific Functions. Protein Sci Publ Protein Soc (2005) 14(4):1091–103. doi: 10.1110/ps.041207005
85. Kohda D, Morton CJ, Parkar AA, Hatanaka H, Inagaki FM, Campbell ID, et al. Solution Structure of the Link Module: A Hyaluronan-Binding Domain Involved in Extracellular Matrix Stability and Cell Migration. Cell (1996) 86(5):767–75. doi: 10.1016/S0092-8674(00)80151-8
86. Emms DM, Kelly S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol (2019) 20(1):238. doi: 10.1186/s13059-019-1832-y
87. Revell LJ. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol Evol (2012) 3(2):217–23. doi: 10.1111/j.2041-210X.2011.00169.x
88. Horikoshi M, Tang [aut Y, Dickey A, Grenié M, Thompson R, et al. Ggfortify: Data Visualization Tools for Statistical Analysis Results (2020). Available at: https://CRAN.R-project.org/package=ggfortify.
89. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol (1990) 215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2
90. Wolenski FS, Garbati MR, Lubinski TJ, Traylor-Knowles N, Dresselhaus E, Stefanik DJ, et al. Characterization of the Core Elements of the NF-κb Signaling Pathway of the Sea Anemone Nematostella Vectensis. Mol Cell Biol (2011) 31(5):1076–87. doi: 10.1128/MCB.00927-10
91. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res (2016) 44(D1):D457–62. doi: 10.1093/nar/gkv1070
92. Kimura A, Sakaguchi E, Nonaka M. Multi-Component Complement System of Cnidaria: C3, Bf, and MASP Genes Expressed in the Endodermal Tissues of a Sea Anemone, Nematostella Vectensis. Immunobiology (2009) 214(3):165–78. doi: 10.1016/j.imbio.2009.01.003
93. Anantharaman V, Makarova KS, Burroughs AM, Koonin EV, Aravind L. Comprehensive Analysis of the HEPN Superfamily: Identification of Novel Roles in Intra-Genomic Conflicts, Defense, Pathogenesis and RNA Processing. Biol Direct (2013) 8(1):15. doi: 10.1186/1745-6150-8-15
94. Bouchier-Hayes L, Martin SJ. CARD Games in Apoptosis and Immunity. EMBO Rep (2002) 3(7):616–21. doi: 10.1093/embo-reports/kvf139
95. Sartorius U, Schmitz I, Krammer PH. Molecular Mechanisms of Death-Receptor-Mediated Apoptosis. ChemBioChem (2001) 2(1):20–9. doi: 10.1002/1439-7633(20010105)2:1<20::AID-CBIC20>3.0.CO;2-X
96. Williams ME, Strickland P, Watanabe K, Hinck L. Unc5h1 Induces Apoptosis Via Its Juxtamembrane Region Through an Interaction With NRAGE*. J Biol Chem (2003) 278(19):17483–90. doi: 10.1074/jbc.M300415200
97. Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell (2006) 126(5):855–67. doi: 10.1016/j.cell.2006.08.019
98. van der Biezen EA, Jones JDG. The NB-ARC Domain: A Novel Signalling Motif Shared by Plant Resistance Gene Products and Regulators of Cell Death in Animals. Curr Biol (1998) 8(7):R226–8. doi: 10.1016/S0960-9822(98)70145-9
99. Jain BP, Pandey S. Wd40 Repeat Proteins: Signalling Scaffold With Diverse Functions. Protein J (2018) 37(5):391–406. doi: 10.1007/s10930-018-9785-7
100. O’Neill LAJ, Bowie AG. The Family of Five: TIR-domain-containing Adaptors in Toll-like Receptor Signalling. Nat Rev Immunol (2007) 7(5):353–64. doi: 10.1038/nri2079
101. Yuen B, Bayes JM, Degnan SM. The Characterization of Sponge Nlrs Provides Insight Into the Origin and Evolution of This Innate Immune Gene Family in Animals. Mol Biol Evol (2014) 31(1):106–20. doi: 10.1093/molbev/mst174
102. Wiens M, Korzhev M, Perovic-Ottstadt S, Luthringer B, Brandt D, Klein S, et al. Toll-Like Receptors are Part of the Innate Immune Defense System of Sponges (Demospongiae: Porifera). Mol Biol Evol (2007) 24(3):792–804. doi: 10.1093/molbev/msl208
103. Gauthier MEA, Pasquier LD, Degnan BM. The Genome of the Sponge Amphimedon Queenslandica Provides New Perspectives Into the Origin of Toll-like and Interleukin 1 Receptor Pathways. Evol Dev (2010) 12(5):519–33. doi: 10.1111/j.1525-142X.2010.00436.x
104. Traylor-Knowles N, Vandepas LE, Browne WE. Still Enigmatic: Innate Immunity in the Ctenophore Mnemiopsis Leidyi. Integr Comp Biol (2019) 59(4):811–8. doi: 10.1093/icb/icz116
105. Kamm K, Schierwater B, DeSalle R. Innate Immunity in the Simplest Animals – Placozoans. BMC Genomics (2019) 20(1):5. doi: 10.1186/s12864-018-5377-3
106. Thompson JR, Rivera HE, Closek CJ, Medina M. Microbes in the Coral Holobiont: Partners Through Evolution, Development, and Ecological Interactions. Front Cell Infect Microbiol (2015) 4:1–20. doi: 10.3389/fcimb.2014.00176
107. Rosales SM, Miller MW, Williams DE, Traylor-Knowles N, Young B, Serrano XM. Microbiome Differences in Disease-Resistant vs. Susceptible Acropora Corals Subjected to Disease Challenge Assays. Sci Rep (2019) 9(1):18279. doi: 10.1038/s41598-019-54855-y
108. Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. Bacterial Community Dynamics are Linked to Patterns of Coral Heat Tolerance. Nat Commun (2017) 8(1):14213. doi: 10.1038/ncomms14213
109. Sarkar D, Desalle R, Fisher PB. Evolution of MDA-5/RIG-I-dependent Innate Immunity: Independent Evolution by Domain Grafting. Proc Natl Acad Sci U S A (2008) 105(44):17040–5. doi: 10.1073/pnas.0804956105
110. Venkataraman S, Prasad BVLS, Selvarajan R. Rna Dependent Rna Polymerases: Insights From Structure, Function and Evolution. Viruses (2018) 10(2):76. doi: 10.3390/v10020076
111. Fuess LE, Pinzón C JH, Weil E, Grinshpon RD, Mydlarz LD. Life or Death: Disease-Tolerant Coral Species Activate Autophagy Following Immune Challenge. Proc R Soc B Biol Sci (2017), 284(1856):20170771. doi: 10.1098/rspb.2017.0771
112. Roesel CL, Vollmer SV. Differential Gene Expression Analysis of Symbiotic and Aposymbiotic Exaiptasia Anemones Under Immune Challenge With Vibrio Coralliilyticus. Ecol Evol (2019) 9(14):8279–93. doi: 10.1002/ece3.5403
113. Maekawa T, Kufer TA, Schulze-Lefert P. NLR Functions in Plant and Animal Immune Systems: So Far and Yet So Close. Nat Immunol (2011) 12(9):817–26. doi: 10.1038/ni.2083
114. Jacobovitz MR, Rupp S, Voss PA, Maegele I, Gornik SG, Guse A. Dinoflagellate Symbionts Escape Vomocytosis by Host Cell Immune Suppression. Nat Microbiol (2021) 6:1–14. doi: 10.1038/s41564-021-00897-w
115. Mohamed AR, Andrade N, Moya A, Chan CX, Negri AP, Bourne DG, et al. Dual RNA-sequencing Analyses of a Coral and its Native Symbiont During the Establishment of Symbiosis. Mol Ecol (2020) 29(20):3921–37. doi: 10.1111/mec.15612
116. Zhou Z, Zhao S, Ni J, Su Y, Wang L, Xu Y. Effects of Environmental Factors on C-type Lectin Recognition to Zooxanthellae in the Stony Coral Pocillopora Damicornis. Fish Shellfish Immunol (2018) 179:228–33. doi: 10.1016/j.fsi.2018.05.026
117. Wolfowicz I, Baumgarten S, Voss PA, Hambleton EA, Voolstra CR, Hatta M, et al. Aiptasia Sp. Larvae as a Model to Reveal Mechanisms of Symbiont Selection in Cnidarians. Sci Rep (2016) 6(1):32366. doi: 10.1038/srep32366
118. Kvennefors ECE, Leggat W, Kerr CC, Ainsworth TD, Hoegh-Guldberg O, Barnes AC. Analysis of Evolutionarily Conserved Innate Immune Components in Coral Links Immunity and Symbiosis. Dev Comp Immunol (2010) 34(11):1219–29. doi: 10.1016/j.dci.2010.06.016
119. Li H, Zhang H, Jiang S, Wang W, Xin L, Wang H, et al. A Single-CRD C-type Lectin From Oyster Crassostrea Gigas Mediates Immune Recognition and Pathogen Elimination With a Potential Role in the Activation of Complement System. Fish Shellfish Immunol (2015) 44(2):566–75. doi: 10.1016/j.fsi.2015.03.011
120. Sekine H, Kenjo A, Azumi K, Ohi G, Takahashi M, Kasukawa R, et al. An Ancient Lectin-Dependent Complement System in an Ascidian: Novel Lectin Isolated From the Plasma of the Solitary Ascidian, Halocynthia Roretzi. J Immunol (2001) 167(8):4504–10. doi: 10.4049/jimmunol.167.8.4504
121. Lõhelaid H, Teder T, Tõldsepp K, Ekins M, Samel N. Up-Regulated Expression of AOS-LOXa and Increased Eicosanoid Synthesis in Response to Coral Wounding. PloS One (2014) 9(2):e892115. doi: 10.1371/journal.pone.0089215
122. Libro S, Kaluziak ST, Vollmer SV. RNA-Seq Profiles of Immune Related Genes in the Staghorn Coral Acropora Cervicornis Infected With White Band Disease. PloS One (2013) 8(11):e81821. doi: 10.1371/journal.pone.0081821
123. Ricci CA, Kamal AHM, Chakrabarty JK, Fuess LE, Mann WT, Jinks LR, et al. Proteomic Investigation of a Diseased Gorgonian Coral Indicates Disruption of Essential Cell Function and Investment in Inflammatory and Other Immune Processes. Integr Comp Biol (2019) 59(4):830–44. doi: 10.1093/icb/icz107
124. Djeghri N, Pondaven P, Stibor H, Dawson MN. Review of the Diversity, Traits, and Ecology of Zooxanthellate Jellyfishes. Mar Biol (2019) 166(11):147. doi: 10.1007/s00227-019-3581-6
125. Takeuchi T, Koyanagi R, Gyoja F, Kanda M, Hisata K, Fujie M, et al. Bivalve-Specific Gene Expansion in the Pearl Oyster Genome: Implications of Adaptation to a Sessile Lifestyle. Zool Lett (2016) 2(1):1–13. doi: 10.1186/s40851-016-0039-2
126. van Oppen MJH, Blackall LL. Coral Microbiome Dynamics, Functions and Design in a Changing World. Nat Rev Microbiol (2019) 17(9):557–67. doi: 10.1038/s41579-019-0223-4
127. Canton J, Neculai D, Grinstein S. Scavenger Receptors in Homeostasis and Immunity. Nat Rev Immunol (2013) 13(9):621–34. doi: 10.1038/nri3515
128. Pinheiro A, Águeda-Pinto A, Melo-Ferreira J, Neves F, Abrantes J, Esteves PJ. Analysis of Substitution Rates Showed That TLR5 is Evolving at Different Rates Among Mammalian Groups. BMC Evol Biol (2019) 19(1):221. doi: 10.1186/s12862-019-1547-4
129. Cao X. Self-Regulation and Cross-Regulation of Pattern-Recognition Receptor Signalling in Health and Disease. Nat Rev Immunol (2016) 16(1):35–50. doi: 10.1038/nri.2015.8
130. Williams LM, Fuess LE, Brennan JJ, Mansfield KM, Salas-Rodriguez E, Welsh J, et al. A Conserved Toll-like Receptor-to-NF-κb Signaling Pathway in the Endangered Coral Orbicella Faveolata. Dev Comp Immunol (2018) 79:128–36. doi: 10.1016/j.dci.2017.10.016
131. Franzenburg S, Fraune S, Kunzel S, Baines JF, Domazet-Loso T, Bosch TCG. Myd88-Deficient Hydra Reveal an Ancient Function of TLR Signaling in Sensing Bacterial Colonizers. Proc Natl Acad Sci (2012) 109(47):19374–9. doi: 10.1073/pnas.1213110109
132. Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-κb Signaling Pathways. Nat Immunol (2011) 12(8):695–708. doi: 10.1038/ni.2065
133. Meiling SS, Muller EM, Lasseigne D, Rossin A, Veglia AJ, MacKnight N, et al. Variable Species Responses to Experimental Stony Coral Tissue Loss Disease (Sctld) Exposure. Front Mar Sci (2021) 8:670829. doi: 10.3389/fmars.2021.670829
134. Thai C-T, Ogata RT. Expression and Characterization of the C345C/NTR Domains of Complement Components C3 and C5. J Immunol (2003) 171(12):6565–73. doi: 10.4049/jimmunol.171.12.6565
Keywords: innate immunity, non-bilaterian, pattern recognition receptors, Toll-like receptor, NOD-like receptor, RIG-I like receptor, C-type lectin
Citation: Emery MA, Dimos BA and Mydlarz LD (2021) Cnidarian Pattern Recognition Receptor Repertoires Reflect Both Phylogeny and Life History Traits. Front. Immunol. 12:689463. doi: 10.3389/fimmu.2021.689463
Received: 31 March 2021; Accepted: 07 June 2021;
Published: 23 June 2021.
Edited by:
Katherine Buckley, Auburn University, United StatesReviewed by:
Sabyasachi Das, Emory University, United StatesAlex Cordoba-Aguilar, National Autonomous University of Mexico, Mexico
Copyright © 2021 Emery, Dimos and Mydlarz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura D. Mydlarz, bXlkbGFyekB1dGEuZWR1
 Madison A. Emery
Madison A. Emery Bradford A. Dimos
Bradford A. Dimos Laura D. Mydlarz
Laura D. Mydlarz