- 1Key Laboratory of South China Sea Fishery Resources Exploitation & Utilization, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 2Key Laboratory of Maricultural Organism Disease Control, Ministry of Agriculture and Rural Affairs, Qingdao Key Laboratory of Mariculture Epidemiology and Biosecurity, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, China
- 3Key Laboratory of Aquatic Product Processing, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
In recent years, more and more studies have shown that early pathogenic bacterial infection in invertebrates can enhance immunity and significantly reduce mortality when reinfected with the same pathogen. There are mechanisms to explain this phenomenon, but they are relatively few. In addition, dose-dependent primary infection is also associated with increased immunity. In the present study, the initial infection dose and mortality of abalone Haliotis diversicolor after reinfection with Vibrio harveyi were recorded, and the mechanism of immune enhancement was investigated by the transcriptomic response of abalone after two successive stimuli with V. harveyi. Priming with different concentrations of pathogen can enhance immunity; however, higher concentration is not always better. Compared with the first exposure, more genes were up-regulated after the second exposure. Among the commonly expressed genes, the immune related genes were significantly or persistently highly expressed after two infections and included pattern recognition receptors as well as immune effectors, such as toll-like receptors, perlucin 4, scavenger receptor class B-like protein, cytochrome P450 1B1-like, glutathione S-transferase 6, lysozyme and so on; in addition, these immune-related genes were mainly distributed in the pathways related to phagocytosis and calcium signaling. Among the specifically expressed genes, compared with the first infection, more genes were involved in the immune, metabolic and digestive pathways after the second infection, which would be more conducive to preventing the invasion of pathogens. This study outlined the mechanism of immune enhancement in abalone after secondary infection at the global molecular level, which is helpful for a comprehensive understanding of the mechanism of immune priming in invertebrates.
Introduction
More and more studies have shown that the innate immune system has a memory similar to the adaptive immune system, which endows the organism with a stronger and more effective resistance to reinfection, and can be found in a variety of organisms (1, 2). This characteristic is found in plants (3), bacteria (4, 5) and viruses (6). And, this phenomenon is defined in different terms depending on the species studied, for example “trained immunity” in vertebrates (7–9), “immune priming” in invertebrates (10, 11) and “Systemic Acquired Resistance” (SAR) in plants (12, 13). Based on molecular, immunological and evolutionary arguments, Netea et al. (9) proposed that innate immune memory is a primitive form of immune memory, while adaptive immune memory is an advanced form of immune memory. It is necessary to develop an immune memory as it is of great advantage in improving the organism’s survival rate in an unfavorable environment.
Invertebrates include a wide variety of species, accounting for more than 95% of the animal kingdom (14). Invertebrate immunology has attracted more and more attention, and with the further study of invertebrate immune priming, some mechanisms have been identified. It is generally believed that improved resistance after reinfection is mainly related to the up-regulated expression of pattern recognition receptors (PRRs) and the enhancement of phagocytic activity. For example, fibrinogen-related proteins (FREPs) were over represented in the Vector Snail following a secondary challenge (15). Following re-exposure to the pathogen, C-lectins and peptidoglycan recognition protein-S1 were up-regulated in scallop (16, 17). Increased phagocytosis was found in silkworm (18), Drosophila (19) and Pacific oyster (20) after homologous exposure. In addition, other mechanisms such as DNA synthesis (21) and RNA methylation (22) have also been used to explain immune priming. Moreover, initial priming doses can affect the immune priming outcome. A positive correlation between increased resistance and priming dose was found in Galleria mellonella larvae (23). However, immune priming is not universal. Immune memory failed to be detected in damselflies (24) and ants (25).
Abalone is an important economic shellfish in China, and plays a pivotal role in the aquaculture of marine shellfish. Compared with 2018, production in 2019 increased by 9.85%; the annual growth rate far exceeds that of other farmed shellfish. Although abalone farming is generally on the rise in China, the frequent occurrence of disease has seriously affected the rapid development of abalone aquaculture. The cultivation and promotion of new varieties of abalone have promoted the revitalization and development of the abalone breeding industry to a certain extent, but diseases still occur from time to time 3–5 years after the breeding of new varieties. Vibrio harveyi is a Gram-negative bacterium, which is widely distributed in various waters and has high pathogenicity in abalone. In previous studies, we confirmed that improved survival rate could be obtained during re-infection when Haliotis diversicolor was primed with V. harveyi. However, it is not clear whether the initial priming dose is associated with increased resistance in re-infection. Although mechanisms of immune priming at the cytological level have been studied in abalone (26), molecular studies have not been reported. To fully understand the basis of innate immune memory generation, a global molecular approach is needed. The development of high-throughput sequencing technology has accelerated the study of non-model organisms and made it possible to investigate immune priming mechanisms at the overall molecular level.
In order to determine whether different infection doses affect resistance and address the mechanism of immune enhancement after abalone secondary infection, in this study, two different experiments were conducted. In the first experiment, H. diversicolor were exposed to different concentrations of V. harveyi in the first infection. However, the same concentration was used in the second infection to detect the relationship between the infection dose and immune resistance via survival rate changes. In the second experiment, we examined the alterations in mRNA expression of immune-related genes during one and two infections of abalone, respectively, using a transcriptomic approach. In this study, we hoped to understand the mechanism of improved immunity during re-infection.
Materials and Methods
Abalone and Microbes
The abalone (mean shell length 41.35 ± 0.23 mm) used in this study were obtained from our aquaculture base in Jieyang, Guangdong Province, China. The animals were acclimated in experimental barrels with continuous oxygenation and a flow-through sea water supply for 2 weeks. Filtered seawater and salinity were maintained at 28°C and 30, respectively.
V. harveyi isolated from the hepatopancreas of moribund abalone, was used in these experiments. The bacteria were incubated in LB medium at 28°C for 20 h, and harvested by centrifugation at 7,000 × g at 25°C for 5 min. The pellet was resuspended in sterile sea water (SSW) which was filtered using a 0.2 μm Millipore filter, and the final concentration was adjusted according to experimental requirements.
Immune Challenge and Sample Collection
For observation of immune priming, the abalone received two immune challenges in total. A schematic diagram of the experimental design is shown in Figure 1. For the primary immune challenge, abalone were divided into five treatment groups designated as V3, V4, V5, V6 and V7, and were injected with 50 μL of different concentrations of V. harveyi as follows: 1.42 × 103 CFU mL-1, 1.42 × 104 CFU mL-1, 1.42 × 105 CFU mL-1, 1.42 × 106 CFU mL-1 and 1.42 × 107 CFU mL-1, respectively. Abalone in a control group received an injection of 50 μL SSW as a control (designated the V0 group). Fifteen days later, the secondary immune challenge was performed. Five treatment groups designated as V3 + 6, V4 + 6, V5 + 6, V6 + 6 and V7 + 6, corresponding to the five primary immune challenge groups were each injected with 50 μL V. harveyi at a concentration of 1.58 × 106 CFU mL-1 for the secondary immune challenge. In addition, the SSW group was divided into two subgroups, designated as V0 + 0 (receiving an injection of 50 μL SSW) and V0 + 6 (receiving an injection of 50 μL V. harveyi at the concentration of 1.58 × 106 CFU mL-1). During the first injection, 360 abalone were used in each group. During the second infection, 90 abalone in each group were used for mortality monitoring, and at least 30 abalone were used in each of the sampling groups. All experiments were performed in triplicate with abalone in 500 L plastic tanks. In these experiments, dead animals were removed in a timely manner to avoid affecting the water quality.
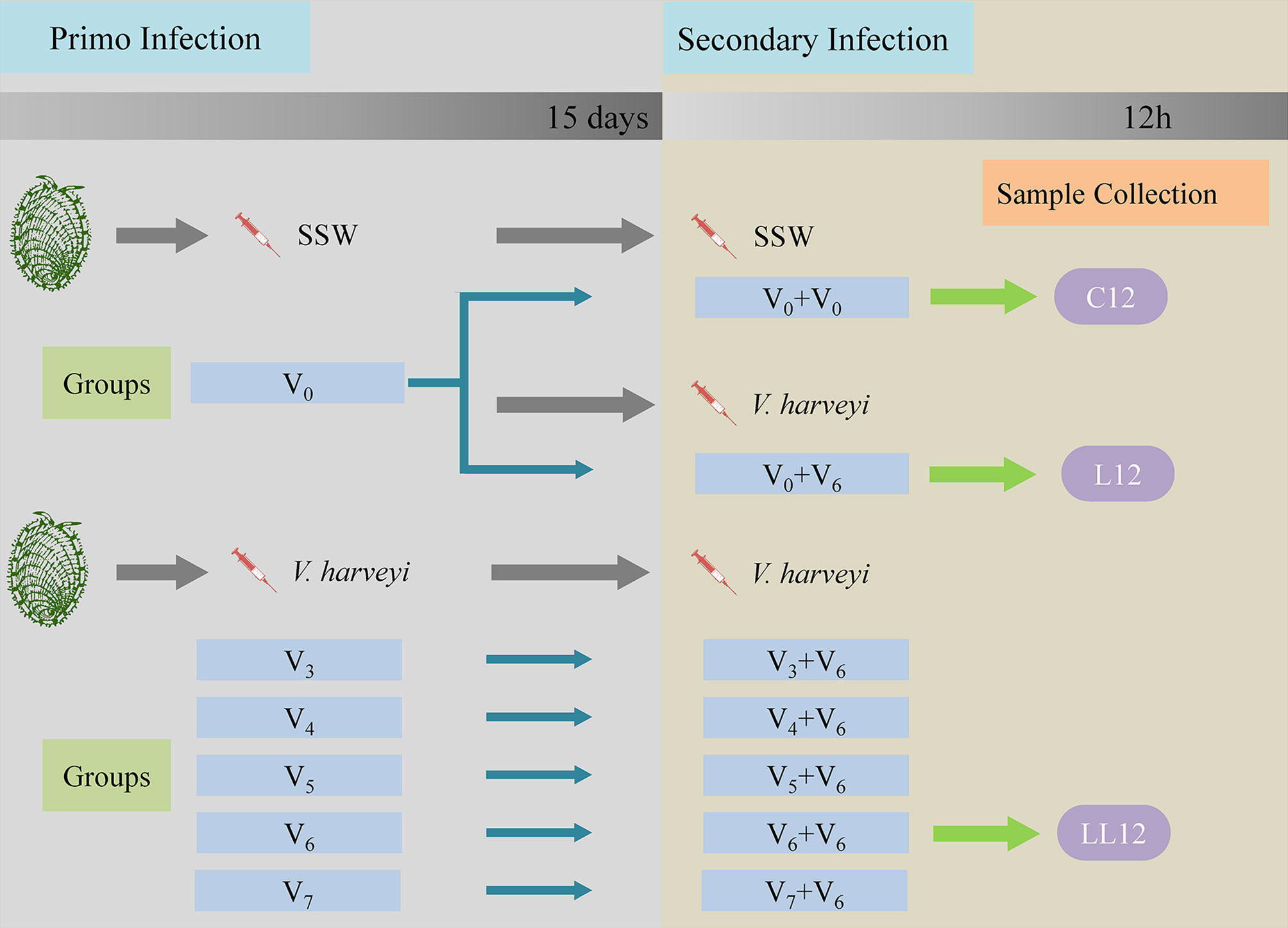
Figure 1 Schematic diagram of the experimental design. Each experiment consisted of two successive infection challenges. For the primary immune challenge, abalone from the same batch were injected with either sterile sea water (SSW, group V0) or different concentrations of V. harveyi: 1.42 × 103 CFU mL-1, 1.42 × 104 CFU mL-1, 1.42 × 105 CFU mL-1, 1.42 × 106 CFU mL-1 and 1.42 × 107 CFU mL-1, designated as V3, V4, V5, V6 and V7, respectively. Fifteen days later, the five treatment groups and the V0 group received an injection of V. harveyi at a concentration of 1.58 × 106 CFU mL-1 (designated as V3 + 6, V4 + 6, V5 + 6, V6 + 6 V7 + 6 and V0 + 6). The V0 group was also injected with SSW as a control for the secondary infection (V0 + 0). During each experimental infection, mortalities were monitored daily post-challenge. The abalone hepatopancreases were sampled 12 h after the second stimulation in the V0 + 0, V0 + 6 and V6 + 6 groups, and named C12, L12 and LL12 groups, respectively. Gene expression levels in the hepatopancreas were then analyzed by transcriptomics.
To study the correlation between initial infection dose and degree of protection, daily mortality rates in each group were monitored. To study the molecular mechanism of immune priming, the hepatopancreases of live abalone were randomly sampled at the time point of 12 h after the second stimulation from the V0 + 0, V0 + 6 and V6 + 6 groups, and were named the C12, L12 and LL12 groups, respectively. A total of nine abalone were sacrificed in each group, and three hepatopancreases from each replicate were pooled together as one sample. These samples were immediately frozen in liquid nitrogen and stored separately at -80°C for subsequent transcriptome sequencing. The survival rate was calculated with a Kaplan-Meier estimate followed by a log-rank test in SPSS 25. Significant differences were set at P < 0.05.
RNA Extraction, Library Construction, and Sequencing
Total RNA was extracted using the TRIzol kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA samples were then digested with DNase I (Ambion, Thermo Scientific, Waltham, MA, USA) to remove potential genomic DNA contamination. RNA degradation and contamination were monitored in 1% agarose gels. RNA quantity and integrity were measured using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively. The resultant RNA samples were then used for RNA-seq.
The poly (A) mRNA was enriched using poly-T oligo-attached magnetic beads (Dynabeads; Invitrogen, Carlsbad, CA, USA) and then cleaved into fragments in the NEB proprietary fragmentation buffer. Following first strand and complementary strand synthesis, the resultant RNA was ligated with sequencing adapters. Then PCR amplification was performed to generate the RNA-seq library. The library preparations were sequenced on an Illumina Novaseq 6000 platform (Illumina, San Diego, CA, USA) and 150 bp paired-end reads were generated.
Bioinformatics Analysis
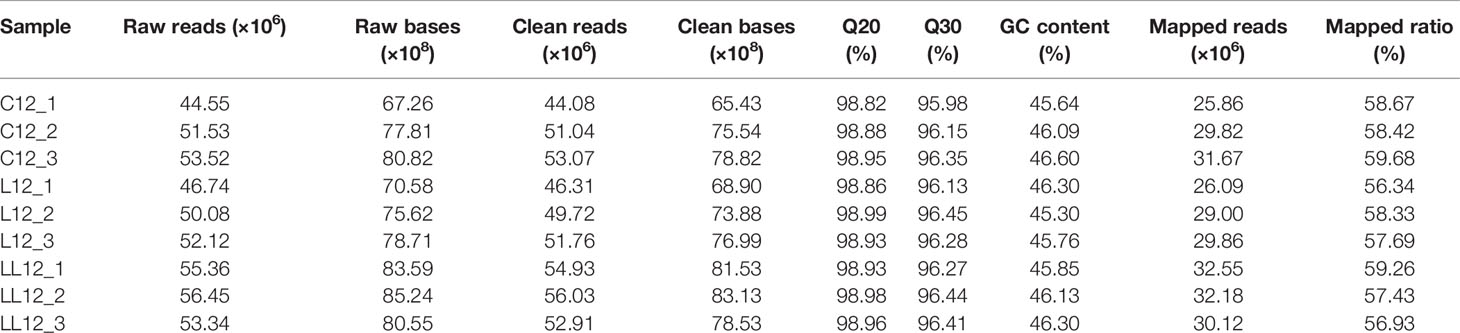
Clean reads were obtained from the raw reads (149,598,910, 148,942,646 and 165,147,008 reads in C12, L12 and LL12 groups, respectively) by removing adapter reads, unknown reads (with ‘N’ ratios > 10%), low quality reads (with quality value ≤ 20) and short reads (with length < 30 bp). After this processing, 148,180,604, 147,780,808 and 163,874,166 clean reads were obtained from the C12, L12 and LL12 groups, respectively (Table 1). The resultant reads were then assembled into a transcriptome using Trinity software with default settings (27). BlastX was used to obtain functional annotation of all expressed genes, by comparing with six databases, including the NCBI nonredundant protein (NR), Swiss-Prot, Protein family (Pfam), Gene Ontology (GO), Cluster of Orthologous Groups (COG) and Kyoto Encyclopedia of Genes and Genome (KEGG) databases, with a cut-off E-value of < 10-5. GO annotations were analyzed with the Blast2GO program (28) utilizing default parameters. After treatment, the mapping rates ranged from 56.34% to 59.68% for all groups (Table 1). The expression abundance of H. diversicolor was calculated using RSEM software (29) and gene expression levels were measured using the FPKM (Fragments Per Kilobase of transcript per Million fragments) method. Differentially expressed genes (DEGs) in the different groups were detected using the DESeq package (30). Significant differential expression was defined by setting absolute log2Flodchange > 1 and FDR (false discovery rate) p-value (q-value) < 0.05 as the threshold. Finally, the obtained DEGs were included in the GO and KEGG enrichment analysis, based on the Hypergeometric distribution model.
Gene Expression Validation
Twelve DEGs, including toll-like receptor (TLR), TLR2, TLR8, scavenger receptor class B-like protein (SR‐BI), X-box binding protein (XBP), cathepsin B (CatB), tumor necrosis factor ligand superfamily member 6-like (TNFSF6), legumain (Lgmn), Zinc transporter ZIP10 (ZIP10), peptidoglycan-recognition protein SC2 isoform X1 (PGRP-SC2), pannexin 5 (Px5) and tumor necrosis factor ligand superfamily member 6 isoform X2 (TNFSF6-X2) were selected to validate Illumina sequencing data by real-time qRT-PCR analysis, and qPCR was performed on different individuals exposed to the same conditions. All primers were acquired with Primer Premier 5.0 based on the reference transcriptome sequences and β-actin was selected as the reference gene (Table 2). RNA preparation was carried out as above. The first strand cDNA was synthesized from approximately 1 μg of total RNA using the PrimeScript™ RT Reagent Kit (Takara, Dalian, China) in a 20 μL reaction system following the manufacturer’s protocol. The qRT-PCR amplifications were performed with a LightCycler® 480 system (Roche Biochemicals, Indianapolis, IN, USA) using TB Green® Premix Ex Taq™ II (Takara, Dalian, China) in a 10 μL reaction system. The thermal cycling parameters were 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Three biological replicates and three technical replicates were included in these experiments and the data were obtained using the 2−ΔΔCT method (31). The data were expressed as the mean ± SD (n = 3).
Results
Survival Rates After First and Second Challenge With V. harveyi
To investigate the relationship between immune protection and initial dose of infection, two consecutive V. harveyi infection experiments were performed. In this study, five different concentrations of V. harveyi diluents were used. After the first immune stimulation, the survival rate decreased with increased infection concentration (Figure 2A). Among them, the survival rate in the V6 group was 48.9%. Thus, a bacterial concentration of 106 CFU mL-1 was selected for the second infection experiment. After the second infection, the log-rank test showed that the survival rates in all groups receiving the first immune stimulation were significantly higher than that in group V0 + 6 (Figure 2B), but there were no significant differences among groups V3 + 6, V4 + 6, V5 + 6, V6 + 6 and V7 + 6 (P < 0.05). This implies that, the previous infection protected the abalone against a secondary infection. However, there was no positive correlation between immune protection and the initial dose of infection, as group V7+6 did not have the highest survival rate.
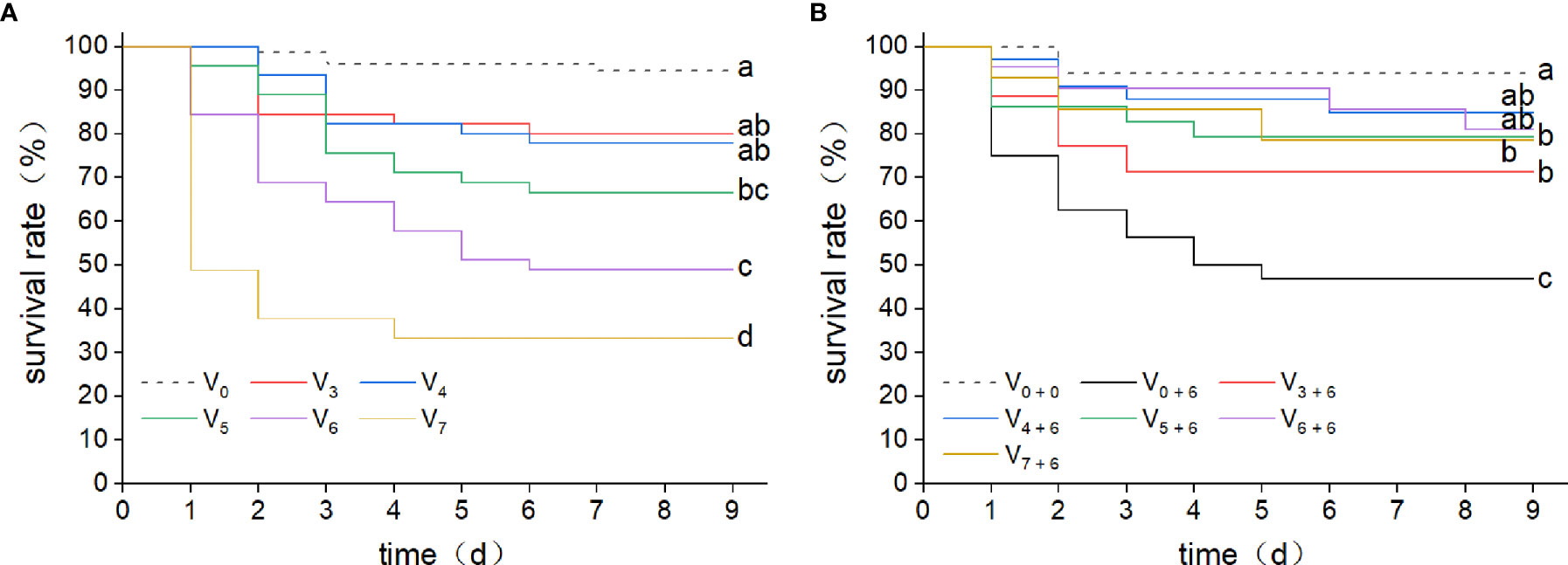
Figure 2 Kaplan-Meier survival curves for abalone after two consecutive infections with V. harveyi. (A) Kaplan-Meier survival curves were generated from abalone following a first injection with V. harveyi at concentrations of either 1.42 × 103 CFU mL-1 (V3), 1.42 × 104 CFU mL-1 (V4), 1.42 × 105 CFU mL-1 (V5), 1.42 × 106 CFU mL-1 (V6) or 1.42 × 107 CFU mL-1 (V7), or with SSW as control (V0). A total of 360 abalone were used in each group (120 per tank). Mortalities were monitored for nine days after infection. Different letter labels next to the graph lines indicate statistically significant differences among treatments (p < 0.05, log-rank test, n = 360). (B) Kaplan-Meier survival curves were generated from abalone primed by injection with V. harveyi at five different concentrations, or with SSW, followed by a second injection with V. harveyi at 1.58 × 106 CFU mL-1 (V3 + 6, V4 + 6, V5 + 6, V6 + 6 V7 + 6 and V0 + 6) or with a second SSW injection as control (V0 + 0). A total of 90 abalone were used in each group (30 per tank). Mortalities were monitored for nine days after infection. Different letter labels next to the graph lines indicate statistically significant differences among treatments (p < 0.05, log-rank test, n = 90).
Analysis of Sequenced Data Quality
Nine cDNA libraries were constructed for Illumina sequencing and the data processing results are summarized in Table 1. After assembly, the length of these transcripts ranged from 201 to 16,691 bp with an average length of 885 bp, and the N50 length was 1,311 bp. The values of the Q30 were all > 95.9% and the GC percentage of the clean reads in the nine libraries ranged from 45.30% to 46.60%, suggesting a good assembled quality and sufficient for subsequent analysis. Raw sequencing data were archived under the accession ID SRR13931757-SRR13931765 in the NCBI Sequence Read Archive.
Cluster Analysis and Pairwise Comparisons
In order to comprehensively understand the distribution of different genes, a hierarchical cluster analysis was performed for all DEGs (Supplementary File 1). As shown in Figure 3A, the stimulus group sample, including L12 and LL12, formed one cluster, and were then grouped with C12. When compared with C12, 1092 DEGs were identified in L12 (289 up-regulated and 803 down-regulated genes) and 1,035 DEGs were identified in LL12 (415 up-regulated and 620 down-regulated genes). In addition, 304 DEGs were detected between L12 and LL12, including 226 up-regulated and 78 down-regulated genes (Figure 3B).
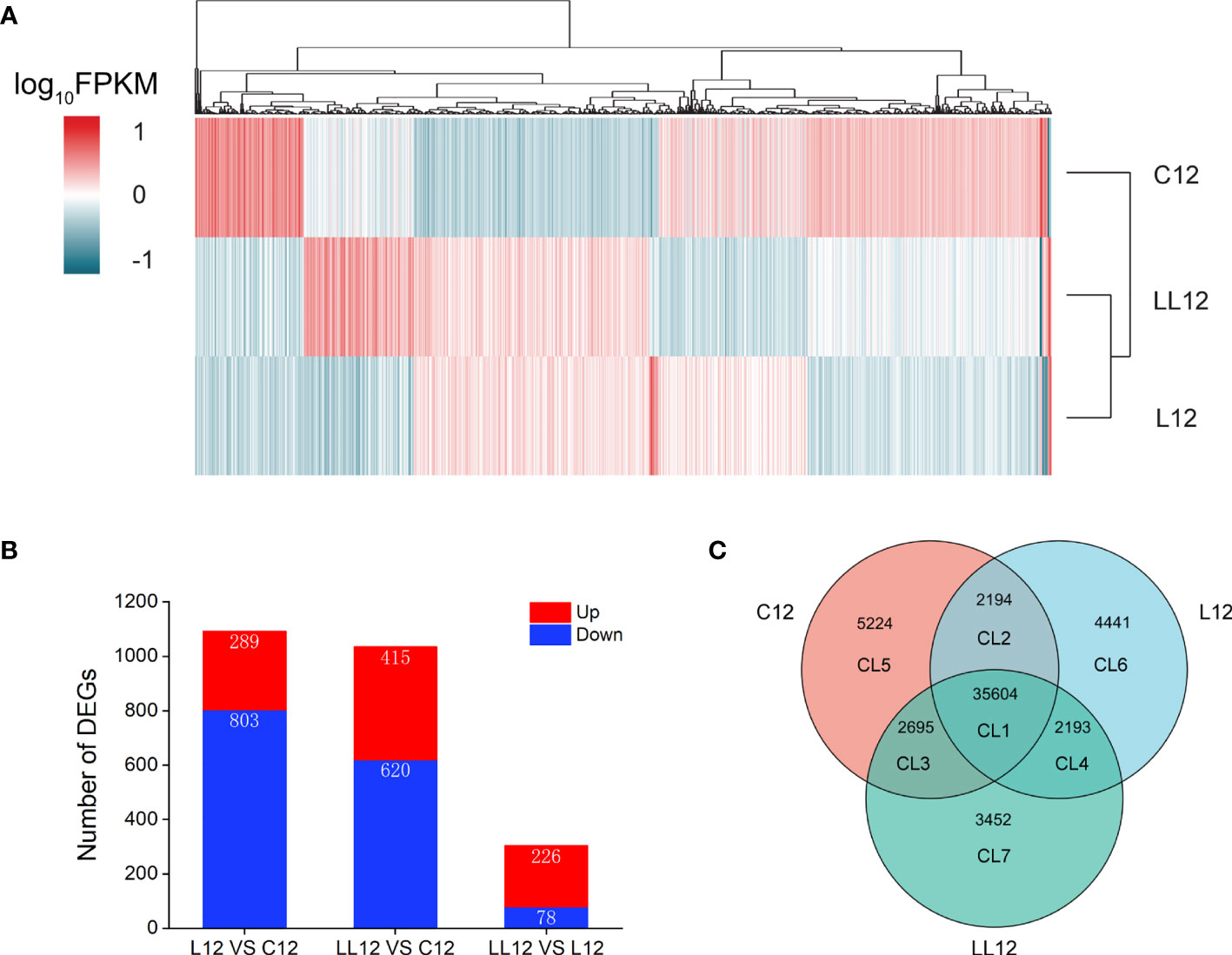
Figure 3 Overview of the expression analysis for C12, L12 and LL12. (A) Hierarchical clustering heat map constructed based on DEGs. Each row represents a group, each column represents a DEG. The expression level of each DEG is shown as the log10(FPKM) value. Red and blue gradients indicate increased and decreased transcript abundance, respectively. (B) Number of DEGs in the different groups. (C) Venn diagram showing the number of commonly and uniquely expressed genes among different groups.
Venn Analysis of Expressed Genes Among Different Groups
In order to better study the DEGs, a Venn diagram was constructed base on unigenes of different groups, and 7 clusters (CLs) were generated (Figure 3C). CL1 showed co-expressed genes in all groups. CL2, CL3 and CL4 showed co-expressed genes between groups. CL5, CL6 and CL7 showed specific expressed genes in each group. DEGs were identified in CL1, CL2, CL3 and CL4. A total of 1174 DEGs were identified in CL1 and they were further studied (Supplementary File 2). In CL2, four nonimmune related DEGs were identified. Thirty DEGs were generated in CL3; however, only one was an immune-related gene; thus, no further analysis was performed. Finally, no DEGs were found in CL4.
Patterns of DEGs in CL1
The DEGs in CL1 from two consecutive V. harveyi infections were divided into 6 subclusters based on a K-means clustering algorithm. As shown in Figure 4A, similar patterns in gene expression were clustered together. In subcluster 1 and 4, genes were continuously down-regulated after the first and second infection. In subcluster 6, genes decreased after the first infection and increased after the second infection; however, the expression level was lower than that in the control group. Subcluster 2 and 5 represented a class of genes that were persistently highly expressed. In subcluster 3, genes were down-regulated after one infection; however, in the second infection, the expression level increased and exceeded the control group. In view of the expression pattern, the genes in subclasses 2, 3 and 5 appeared to be the most promising possible sources of immune priming, and they were analyzed in depth.
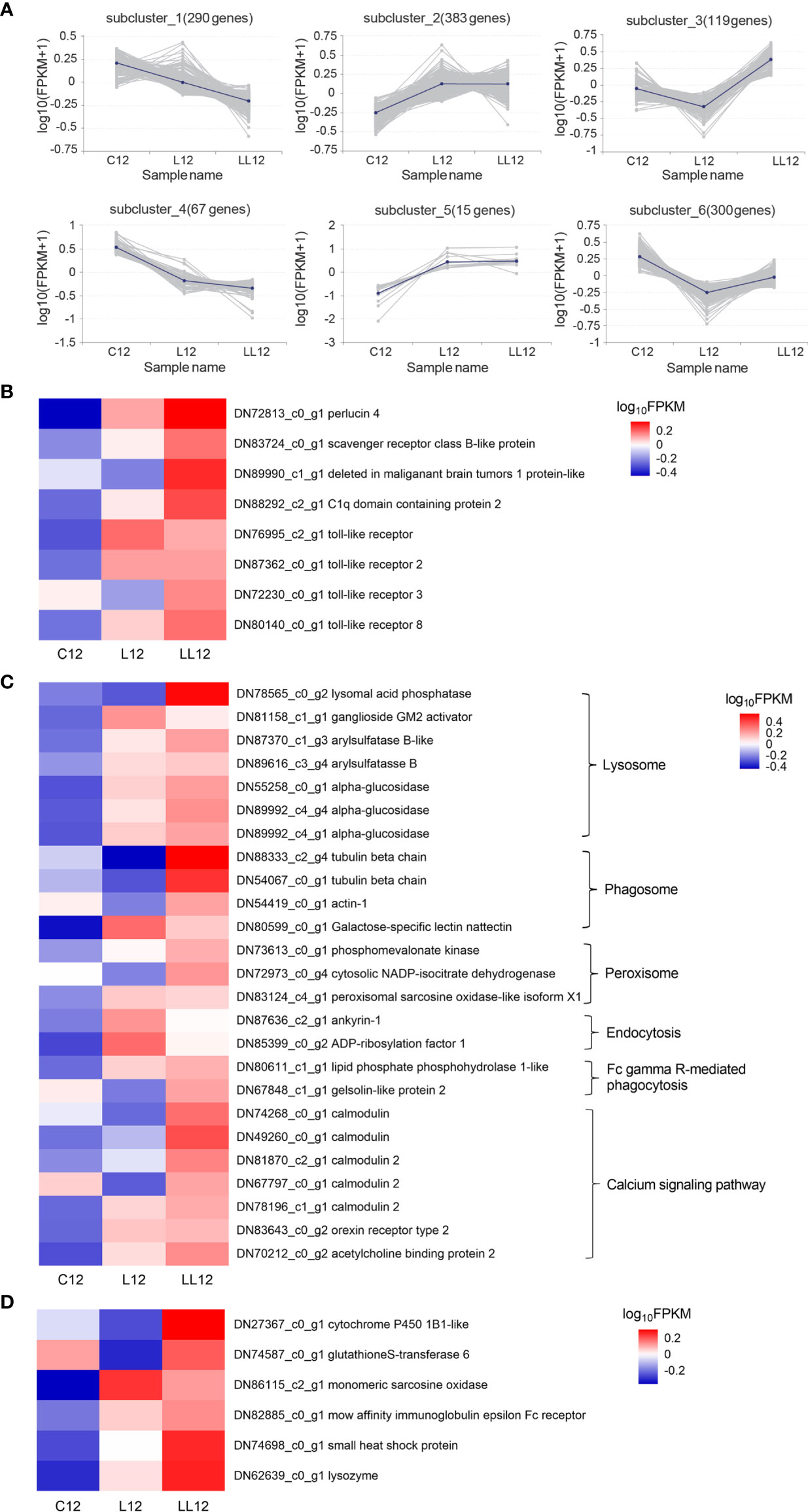
Figure 4 Analysis of differentially expressed genes. (A) Patterns of DEGs in CL1 by K-means clustering analysis. The lines represent the expression tendency of DEGs. The number of genes represented by each pattern is shown above the graphs. (B) The DEGs involved in the PRRs; (C) The DEGs involved in the immune-related KEGG. (D) The DEGs involved in the immune effectors. The heatmap shows log10 (FPKM) values of DEGs among different groups.
Immune Priming Related Genes in CL1
PRRs can bind to pathogen-associated molecular patterns (PAMPs) and are thought to be able to establish immunological memory. Eight PRRs were identified as up-regulated after the second immune challenge, including perlucin 4, C1q domain containing protein 2, scavenger receptor class B-like protein, deleted in malignant brain tumors 1 protein-like, TLR, TLR2, TLR3 and TLR8 (Figure 4B). Some immune-related pathways including the calcium signaling pathway and pathways associated with pathogen clearance (lysosome, phagosome, peroxisome, and Fc gamma R-mediated phagocytosis) were also activated when subjected to a continuous stimulus (Figure 4C). In addition, some immune effector factors including cytochrome P450 1B1-like, glutathione S-transferase 6, monomeric sarcosine oxidase, low affinity immunoglobulin epsilon Fc receptor, and lysozyme were also increased in the second infection (Figure 4D).
Pathway Analysis of CL6 and CL7
All genes collected in CL6 and CL7 were subjected to pathway analysis, respectively. As shown in Figure 5, six categories of the pathways were divided into metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems and human diseases. There were different gene numbers within the same pathway in CL6 and CL7. Some pathways such as carbohydrate metabolism, lipid metabolism, signal transduction, digestive system and immune system had more gene members in CL7 than in CL6. Furthermore, immune-related pathways were analyzed in depth. As shown in Table 3, some pathways associated with pathogen clearance had more gene numbers in CL7 than in CL6, such as lysosome, phagosome, ubiquitin-mediated proteolysis, and autophagy-animal. Other signaling pathways such as the Ras signaling pathway, NOD-like receptor signaling pathway, MAPK signaling pathway, Toll-like receptor signaling pathway and Calcium signaling pathway also showed consistent results.
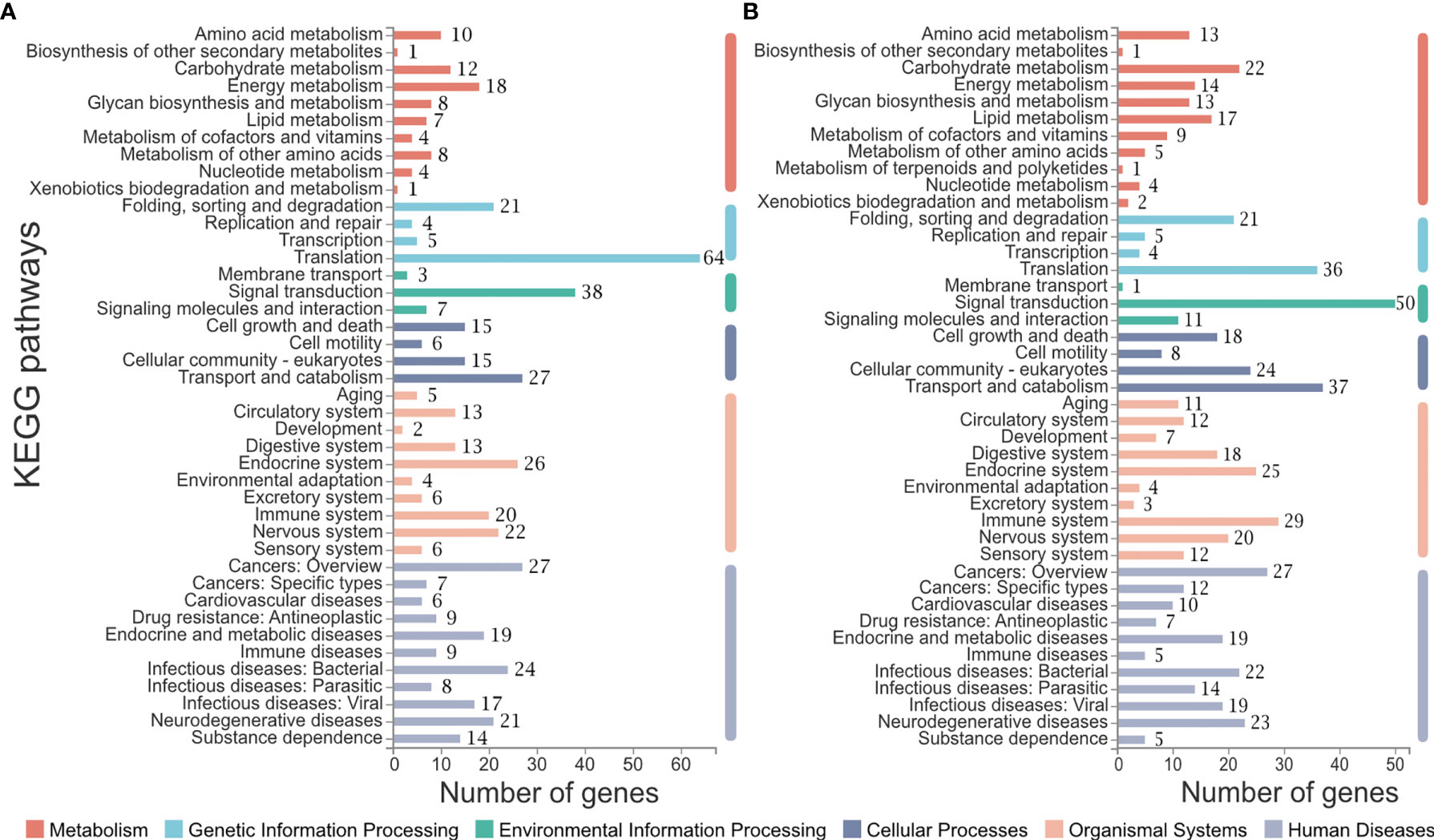
Figure 5 Pathway analysis of CL6 and CL7. (A) Pathways constituted in CL6; (B) Pathways constituted in CL7. The number of genes belonging to each pathway are labeled on the right of the bar.
Validation of RNA-Seq Data by qPCR
In order to evaluate the accuracy of the RNA-seq results, 12 genes including TLR, TLR2, TLR8, SR-BI, XBP, CatB, TNFSF6, Lgmn, ZIP10, PGRP-SC2, Px5 and TNFSF6-x2 from DEG libraries were analyzed by qRT-PCR. Fold changes were compared with the DEG analysis results. As shown in Figure 6, the expression trends of selective genes by qRT-PCR were consistent with the DEG analysis results, and the Spearman’s correlation was 0.875 (p value < 0.001), which indicated the reliability of the transcriptome data.
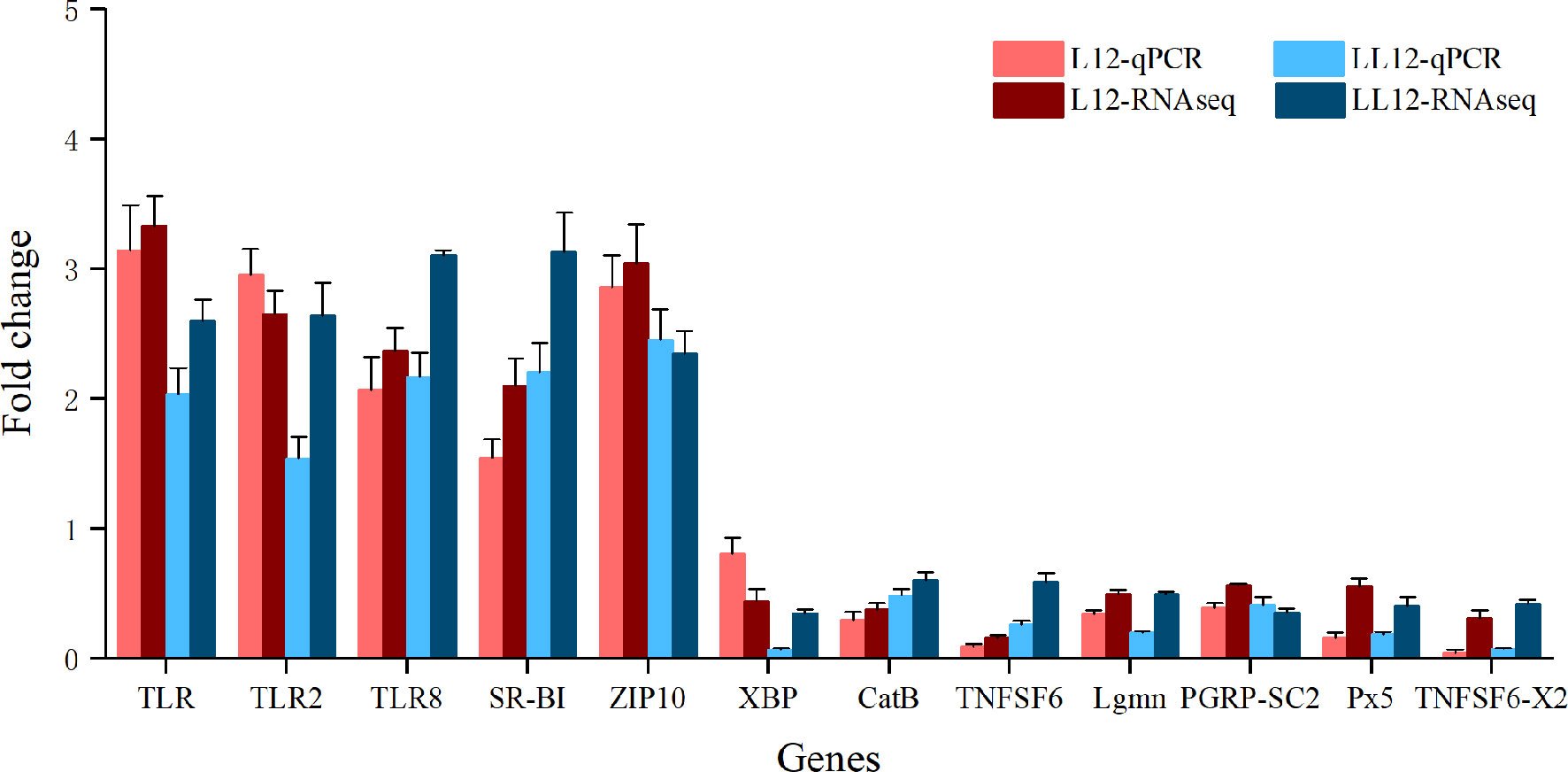
Figure 6 Quantitative RT-PCR validation of 12 genes that are differentially expressed after two successive infections. The x-axis is the gene name and the y-axis represents the fold change in gene expression compared to the control. Data are represented as mean ± SD for three biological replicates.
Discussion
It was reported that immune priming was related to exposure dose and time post priming (2), only certain concentrations were able to induce a primed response, and low dose may lead to less obvious effects (23, 32). In the present study, the abalone were first injected with five different concentrations of V. harveyi diluents, which conferred the infected abalone with higher immune protection during subsequent re-exposure. We observed that different initial infection doses of V. harveyi provided different levels of protection; i.e., priming with the lowest initial infection dose provided the lowest immune protection, although the highest initial infection dose did not provide the highest immune protection (minimal immune protection was observed after priming with 1.42 × 103 CFU mL-1 of V. harveyi, but 1.42 × 107 CFU mL-1 did not offer maximum immune protection). Although enhanced immune protection has been found in many invertebrates, persistent immune protection time varied according to the invertebrate and pathogen examined (33–35). In mollusks, infection with V. anguillarum can offer Chlamys farreri immune protection for up to one week (17, 36). In Biomphalaria glabrata, immunological memory to Schistosoma mansoni was maintained for the rest of the animal’s lifespan (37, 38). In poly(I:C) primed Crassostrea gigas, resistance to Ostreid herpesvirus 1 (OsHV-1) could last for more than 5 months (39); and this protection was even transmitted to offspring (40, 41). In the current study, the time interval between priming and re-exposure was 15 days. Whether enhanced immune protection can last for longer time periods requires further research. However, in other species of abalone, it was reported that improved antibacterial response was induced within 25 days after injection of V. fluvialis in H. discus hannai (42). The appearance of a natural resistant population of the European abalone indicated that immune priming of abalone has strong plasticity, and functions within and across generations (26).
Previous studies have suggested that the formation of immune memory is related to the diversity of PRRs, the synergistic interactions between receptors and the expression level of receptors (43). In this study, PRRs in CL1 including perlucin 4, C1q domain containing protein 2, scavenger receptor class B-like protein, deleted in malignant brain tumors 1 protein-like, and 4 TLRs showed increased expression after re-exposure to V. harveyi. C1q domain containing proteins can bind to LPS, PGN, polyI:C, beta-glucan, mannan, and yeast-glycan, giving them a broader bacterial agglutinating spectrum, and powerful function in the recognition strategy. In addition, they can act as opsonins to enhance phagocytic activity (44–46). Scavenger receptors (SRs) are major endocytic receptors that can bind Gram-negative and Gram-positive bacteria to promote hemocyte phagocytosis (47, 48). Perlucin is a typical C-type lectin, which can directly agglutinate bacterial pathogens to restrict their spread in plasma, and is involved in regulating the innate immunity by regulating phagocytosis and AMP expression (49). Toll and toll-like receptors are involved in pathogen recognition in plants, invertebrates and vertebrates (50). In Drosophila, the toll pathway was reported to regulate hemocyte proliferation (51), antimicrobial peptide expression (52), and initiate a systemic response in which hemocytes are mobilized and activated (53); more importantly, specific immune priming requires the toll pathway (19). In mollusks, a number of TLRs have been identified in various species, including hard clam (54), mussels (55, 56) and oysters (57, 58). TLRs were highly expressed in molluscan hemocytes and the TLR pathway was suggested to play a central role in initiating the cellular response to infection (59). It has been reported that TLR signaling is involved in enhanced immune protection of oysters against pathogen re-infection (60). In this study, the increased expression of PRRs suggested their contribution to improved immune protection during the re-exposure of abalone. Moreover, synergistic interactions may exist between different PRRs to enhance pathogen clearance.
In invertebrates, cellular immune responses are performed by hemocytes. Phagocytosis is the core defense mechanism for hemocytes to eliminate external invaders (14, 61). Many studies have shown that phagocytosis is necessary for the evaluation of immune memory in invertebrates. Studies have reported that specific primed immune responses of Drosophila (19) and silkworm (18, 62) were dependent on phagocytes. In mollusks, the total hemocyte count and phagocytic activity were increased after secondary infection in snails (63), scallop (36) and oysters (20, 64). In the present study, we demonstrated the importance of phagocytosis from a molecular perspective, as some genes in CL1 involved in lysosome, phagosome, endocytosis and Fc gamma R-mediated phagocytosis had higher expression levels in secondary infection. In addition, among the specifically expressed genes, CL7 contained more gene annotations to phagocytosis-related pathways than CL6. In oyster, sequencing results also showed that Fc gamma-mediated phagocytosis and ubiquitin-mediated proteolysis were significantly enriched, and phagocytosis was suggested to have a key role in second immune protection (60). In addition, the activation of immune-related pathways in CL4 also plays an important role in pathogen clearance (Supplementary File 3). With the exception of the classical immune-related pathway, some genes involved in the calcium signaling pathway were also significantly up-regulated in the present study. Calcium signaling is critical for diverse biological processes including fertilization, differentiation, proliferation and gene transcription (65). Calcium signaling also plays a vital role in immune function (66). Weavers et al. (67) demonstrated that calcium bursts mediate molecular memory generated by corpse engulfment by Drosophila macrophages. In addition, C1q domain containing proteins, scavenger receptors and perlucin functionally promote phagocytosis. Thus, it can be speculated that phagocytosis plays an important role in preventing re-infection of the same pathogen in abalone.
Effector factors are the most basic molecules, and are the main agents involved in the elimination of pathogens. Of these factors, antimicrobial peptides (AMPs) constitute the first line of host defense against pathogen infection, and are crucial components of the innate immune system in mollusks (68). To date, several types of AMPs have been identified and characterized from mollusks, including mytilins, myticins, mytimycins, mytimacins, defensins, big defensins, histones, lysozymes, abhisin and molluscidin (69–75). In marine mollusks, AMPs can bind the bacterial membrane directly and kill the invading bacterial pathogens by membrane disruption (76). In this study, only lysozyme in CL1 was found to be notably up-regulated in secondary infection. This may have been due to the sampling time points, as not all AMPs transcripts were increased 12 h after pathogen infection (77–79). However, AMPs are only effectors of the immune response and have a spectrum of antimicrobial characteristics, which can be induced indiscriminately, and the AMP transcripts quickly return to the baseline state after infection (19). Therefore, the AMPs are considered unable to establish a primed response (1, 18). We also found that a range of immune effectors associated with detoxification and antioxidant stress in CL1 were significantly increased during re-infection, including cytochrome P450 1B1-like, glutathione S-transferase 6 and small heat shock protein (80–82). Our sequencing results showed that the enhanced immune protection after secondary infection was not only due to the role of AMPs, but also involves the synergistic effect of multiple immune effectors.
In mammals, there is increasing evidence that trained immunity involves metabolic regulation such as glycolysis, oxidative phosphorylation, fatty acid and amino acid metabolism, which endows innate immune cells with the ability to respond more strongly to a second stimulus (83). Glycolysis, an alternative form of glucose metabolism, can produce ATP faster and it is thought to play a key role in immunity (84). Induction of glycolysis has recently been shown to be crucial for the initiation of trained immunity in human volunteers after BCG vaccination (85). Epigenetic modification also plays an important role in the establishment of innate immune memory. Several metabolites of glycolysis and the TCA cycle are also co-factors for epigenetic enzymes (86). Fatty acids can activate innate immune pathways, amino acids are the basic chemical building blocks during biogenesis, lipid metabolism and amino acid metabolism, and were proved to be necessary for the induction of innate immune memory (83, 87). In this study, we also found that many genes involved in glycolysis, fatty acid and amino acid metabolism were significantly up-regulated in CL1 (Supplementary File 4), and more genes were found in CL7 than in CL6 (Figure 5; Supplementary File 5). This suggested that metabolic pathways also played a role in abalone immune priming. In addition, it was found that innate memory in macrophages could polarize other neighboring cells in ways that drive antibacterial, Th17 and M1 responses (88). This suggested that intercellular signal transduction played an important role in bacterial clearance in re-infections. The results in Table 3 and Figure 5 also demonstrated this, as more genes were annotated to signal transduction in CL7.
In summary, in the present study we demonstrated that an infection enhanced abalone immunity to secondary infection with the same pathogen, although this protection was not linearly correlated with the initial infection dose. Comparative transcriptome analysis has improved our understanding of the mechanism of enhanced immune protection in abalone. Increased immunity in abalone was due to the synergistic effect of the recognition of a variety of pattern recognition receptors, phagocytosis of hemocytes, detoxification and anti-oxidation of immune effectors, the enhancement of metabolism, and so on. The study on the mechanism of immune protection enhancement of abalone carried out in this study will enrich the content of invertebrate immunology, and contribute to a deeper understanding of the diversity of invertebrate immune priming mechanisms and the evolutionary process of the invertebrate immune system.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Short Read Archive database under the accession ID SRR13931757-SRR13931765.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Ethics Committee of South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences.
Author Contributions
TY and LY conceived, designed the experiment, and wrote the manuscript. TY and JL performed the experiments. CB and ZX analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Key R&D Program of China (2019YFD0900105), the Science and Technology Planning Project of Guangzhou (202002030488), the China Agriculture Research System of MOF and MARA, the Central Public-interest Scientific Institution Basal Research Fund, CAFS (2020TD42 and 2021SD05), the Science and Technology Planning Project of Jieyang (2019029 and sxm029), the Professorial and Doctoral Scientific Research Foundation of Huizhou University (2020JB065), the Shellfish and Large Algae Industry Innovation Team Project of Guangdong Province (2020KJ146) and the Guangdong Rural Revitalization Strategy Special Funds (Fishery Industry Development) (YueCaiNong[2020]4).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.685896/full#supplementary-material
References
1. Cooper D, Eleftherianos I. Memory and Specificity in the Insect Immune System: Current Perspectives and Future Challenges. Front Immunol (2017) 8:539. doi: 10.3389/fimmu.2017.00539
2. Milutinović B, Kurtz J. Immune Memory in Invertebrates. Semin Immunol (2016) 28:328–42. doi: 10.1016/j.smim.2016.05.004
3. Reimer-Michalski E-M, Conrath U. Innate Immune Memory in Plants. Semin Immunol (2016) 28:319–27. doi: 10.1016/j.smim.2016.05.006
4. Glass Z, Lee M, Li Y, Xu Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol (2018) 36:173–85. doi: 10.1016/j.tibtech.2017.11.006
5. Barrangou R, Marraffini LA. CRISPR-Cas Systems: Prokaryotes Upgrade to Adaptive Immunity. Mol Cell (2014) 54:234–44. doi: 10.1016/j.molcel.2014.03.011
6. Levasseur A, Bekliz M, Chabrière E, Pontarotti P, La Scola B, Raoult D. MIMIVIRE Is a Defence System in Mimivirus That Confers Resistance to Virophage. Nature (2016) 531:249–52. doi: 10.1038/nature17146
7. Netea MG. Training Innate Immunity: The Changing Concept of Immunological Memory in Innate Host Defence. Eur J Clin Invest (2013) 43:881–4. doi: 10.1111/eci.12132
8. Netea MG, Quintin J, van der Meer JWM. Trained Immunity: A Memory for Innate Host Defense. Cell Host Microbe (2011) 9:355–61. doi: 10.1016/j.chom.2011.04.006
9. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe (2019) 25:13–26. doi: 10.1016/j.chom.2018.12.006
10. Kurtz J. Specific Memory Within Innate Immune Systems. Trends Immunol (2005) 26:186–92. doi: 10.1016/j.it.2005.02.001
11. Little TJ, Kraaijeveld AR. Ecological and Evolutionary Implications of Immunological Priming in Invertebrates. Trends Ecol Evol (2004) 19:58–60. doi: 10.1016/j.tree.2003.11.011
12. Kachroo A, Robin GP. Systemic Signaling During Plant Defense. Curr Opin Plant Biol (2013) 16:527–33. doi: 10.1016/j.pbi.2013.06.019
13. Jaskiewicz M, Conrath U, Peterhänsel C. Chromatin Modification Acts as a Memory for Systemic Acquired Resistance in the Plant Stress Response. EMBO Rep (2011) 12:50–5. doi: 10.1038/embor.2010.186
14. Syed Musthaq SK, Kwang J. Reprint of “Evolution of Specific Immunity in Shrimp – A Vaccination Perspective Against White Spot Syndrome Virus.” Dev Comp Immunol (2015) 48:342–53. doi: 10.1016/j.dci.2014.07.016
15. Pinaud S, Portela J, Duval D, Nowacki FC, Olive M-A, Allienne J-F, et al. A Shift From Cellular to Humoral Responses Contributes to Innate Immune Memory in the Vector Snail Biomphalaria glabrata. PLoS Pathog (2016) 12:e1005361. doi: 10.1371/journal.ppat.1005361
16. Cong M, Song L, Qiu L, Li C, Wang B, Zhang H, et al. The Expression of Peptidoglycan Recognition Protein-S1 Gene in the Scallop Chlamys farreri Was Enhanced After a Second Challenge by Listonella anguillarum. J Invertebr Pathol (2009) 100:120–2. doi: 10.1016/j.jip.2008.10.004
17. Wang J, Wang L, Yang C, Jiang Q, Zhang H, Yue F, et al. The Response of mRNA Expression Upon Secondary Challenge With Vibrio anguillarum Suggests the Involvement of C-Lectins in the Immune Priming of Scallop Chlamys farreri. Dev Comp Immunol (2013) 40:142–7. doi: 10.1016/j.dci.2013.02.003
18. Wu G, Li M, Liu Y, Ding Y, Yi Y. The Specificity of Immune Priming in Silkworm, Bombyx mori, Is Mediated by the Phagocytic Ability of Granular Cells. J Insect Physiol (2015) 81:60–8. doi: 10.1016/j.jinsphys.2015.07.004
19. Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A Specific Primed Immune Response in Drosophila Is Dependent on Phagocytes. PLoS Pathog (2007) 3:e26. doi: 10.1371/journal.ppat.0030026
20. Li Y, Song X, Wang W, Wang L, Yi Q, Jiang S, et al. The Hematopoiesis in Gill and Its Role in the Immune Response of Pacific Oyster Crassostrea gigas Against Secondary Challenge With Vibrio splendidus. Dev Comp Immunol (2017) 71:59–69. doi: 10.1016/j.dci.2017.01.024
21. Cime-Castillo J, Arts RJW, Vargas-Ponce de León V, Moreno-Torres R, Hernández-Martínez S, Recio-Totoro B, et al. DNA Synthesis Is Activated in Mosquitoes and Human Monocytes During the Induction of Innate Immune Memory. Front Immunol (2018) 9:2834. doi: 10.3389/fimmu.2018.02834
22. Castro-Vargas C, Linares-López C, López-Torres A, Wrobel K, Torres-Guzmán JC, Hernández GAG, et al. Methylation on RNA: A Potential Mechanism Related to Immune Priming Within But Not Across Generations. Front Microbiol (2017) 8:473. doi: 10.3389/fmicb.2017.00473
23. Wu G, Yi Y, Lv Y, Li M, Wang J, Qiu L. The Lipopolysaccharide (LPS) of Photorhabdus luminescens TT01 Can Elicit Dose- and Time-Dependent Immune Priming in Galleria mellonella larvae. J Invertebr Pathol (2015) 127:63–72. doi: 10.1016/j.jip.2015.03.007
24. González-Tokman DM, González-Santoyo I, Lanz-Mendoza H, Córdoba Aguilar A. Territorial Damselflies Do Not Show Immunological Priming in the Wild. Physiol Entomol (2010) 35:364–72. doi: 10.1111/j.1365-3032.2010.00752.x
25. Reber A, Chapuisat M. No Evidence for Immune Priming in Ants Exposed to a Fungal Pathogen. PLoS One (2012) 7:e35372. doi: 10.1371/journal.pone.0035372
26. Dubief B, Nunes FLD, Basuyaux O, Paillard C. Immune Priming and Portal of Entry Effectors Improve Response to Vibrio Infection in a Resistant Population of the European Abalone. Fish Shellfish Immunol (2017) 60:255–64. doi: 10.1016/j.fsi.2016.11.017
27. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-Length Transcriptome Assembly From RNA-Seq Data Without a Reference Genome. Nat Biotechnol (2011) 29:644–52. doi: 10.1038/nbt.1883
28. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
29. Li B, Dewey CN. RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf (2011) 12:323. doi: 10.1186/1471-2105-12-323
30. Anders S, Huber W. Differential Expression Analysis for Sequence Count Data. Genome Biol (2010) 11:R106. doi: 10.1186/gb-2010-11-10-r106
31. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔct Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
32. Wu G, Zhao Z, Liu C, Qiu L. Priming Galleria mellonella (Lepidoptera: Pyralidae) Larvae With Heat-Killed Bacterial Cells Induced an Enhanced Immune Protection Against Photorhabdus luminescens TT01 and the Role of Innate Immunity in the Process. J Econ Entomol (2014) 107:559–69. doi: 10.1603/EC13455
33. Thomas AM, Rudolf VHW. Challenges of Metamorphosis in Invertebrate Hosts: Maintaining Parasite Resistance Across Life-History Stages. Ecol Entomol (2010) 35:200–5. doi: 10.1111/j.1365-2311.2009.01169.x
34. Haine ER, Moret Y, Siva-Jothy MT, Rolff J. Antimicrobial Defense and Persistent Infection in Insects. Science (2008) 322:1257–9. doi: 10.1126/science.1165265
35. Moret Y, Siva-Jothy MT. Adaptive Innate Immunity? Responsive-Mode Prophylaxis in the Mealworm Beetle, Tenebrio molitor. Proc R Soc London Ser B Biol Sci (2003) 270:2475–80. doi: 10.1098/rspb.2003.2511
36. Cong M, Song L, Wang L, Zhao J, Qiu L, Li L, et al. The Enhanced Immune Protection of Zhikong Scallop Chlamys farreri on the Secondary Encounter with Listonella anguillarum. Comp Biochem Physiol Part B Biochem Mol Biol (2008) 151:191–6. doi: 10.1016/j.cbpb.2008.06.014
37. Pinaud S, Portet A, Allienne JF, Belmudes L, Saint-Beat C, Arancibia N, et al. Molecular Characterisation of Immunological Memory Following Homologous or Heterologous Challenges in the Schistosomiasis Vector Snail, Biomphalaria glabrata. Dev Comp Immunol (2019) 92:238–52. doi: 10.1016/j.dci.2018.12.001
38. Portela J, Duval D, Rognon A, Galinier R, Boissier J, Coustau C, et al. Evidence for Specific Genotype-Dependent Immune Priming in the Lophotrochozoan Biomphalaria glabrata Snail. J Innate Immun (2013) 5:261–76. doi: 10.1159/000345909
39. Lafont M, Petton B, Vergnes A, Pauletto M, Segarra A, Gourbal B, et al. Long-Lasting Antiviral Innate Immune Priming in the Lophotrochozoan Pacific Oyster, Crassostrea gigas. Sci Rep (2017) 7:13143. doi: 10.1038/s41598-017-13564-0
40. Lafont M, Goncalves P, Guo X, Montagnani C, Raftos D, Green T. Transgenerational Plasticity and Antiviral Immunity in the Pacific Oyster (Crassostrea Gigas) Against Ostreid Herpesvirus 1 (OsHV-1). Dev Comp Immunol (2019) 91:17–25. doi: 10.1016/j.dci.2018.09.022
41. Green TJ, Helbig K, Speck P, Raftos DA. Primed for Success: Oyster Parents Treated With Poly(I:C) Produce Offspring With Enhanced Protection Against Ostreid Herpesvirus Type I Infection. Mol Immunol (2016) 78:113–20. doi: 10.1016/j.molimm.2016.09.002
42. Li T, Ding M, Xiang J, Liu R. Immunnological Studies on Haliotis discus hannai With Vibrio Fluvialis-II. Oceanol Limnol Sin (1997) 28:27–32. doi: CNKI:SUN:HYFZ.0.1997-01-004
43. Schulenburg H, Boehnisch C, Michiels NK. How do Invertebrates Generate a Highly Specific Innate Immune Response? Mol Immunol (2007) 44:3338–44. doi: 10.1016/j.molimm.2007.02.019
44. Wang L, Wang L, Kong P, Yang J, Zhang H, Wang M, et al. A Novel C1qDC Protein Acting as Pattern Recognition Receptor in Scallop Argopecten irradians. Fish Shellfish Immunol (2012) 33:427–35. doi: 10.1016/j.fsi.2012.05.032
45. Kong P, Zhang H, Wang L, Zhou Z, Yang J, Zhang Y, et al. AiC1qDC-1, A Novel Gc1q-Domain-Containing Protein From Bay Scallop Argopecten irradians With Fungi Agglutinating Activity. Dev Comp Immunol (2010) 34:837–46. doi: 10.1016/j.dci.2010.03.006
46. Kishore U, Ghai R, Greenhough TJ, Shrive AK, Bonifati DM, Gadjeva MG, et al. Structural and Functional Anatomy of the Globular Domain of Complement Protein C1q. Immunol Lett (2004) 95:113–28. doi: 10.1016/j.imlet.2004.06.015
47. Tang M, Li X, Yang L, Wang Q, Li W. A Class B Scavenger Receptor Mediates Antimicrobial Peptide Secretion and Phagocytosis in Chinese Mitten Crab (Eriocheir Sinensis). Dev Comp Immunol (2020) 103:103496. doi: 10.1016/j.dci.2019.103496
48. Wu YM, Yang L, Li XJ, Li L, Wang Q, Li WW. A Class B Scavenger Receptor From Eriocheir sinensis (EsSR-B1) Restricts Bacteria Proliferation by Promoting Phagocytosis. Fish Shellfish Immunol (2017) 70:426–36. doi: 10.1016/j.fsi.2017.09.034
49. Bi J, Ning M, Xie X, Fan W, Huang Y, Gu W, et al. A Typical C-Type Lectin, Perlucin-Like Protein, Is Involved in the Innate Immune Defense of Whiteleg Shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2020) 103:293–301. doi: 10.1016/j.fsi.2020.05.046
50. Bell JK, Mullen GED, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-Rich Repeats and Pathogen Recognition in Toll-Like Receptors. Trends Immunol (2003) 24:528–33. doi: 10.1016/S1471-4906(03)00242-4
51. Qiu P, Pan PC, Govind S. A Role for the Drosophila Toll/Cactus Pathway in Larval Hematopoiesis. Development (1998) 125:1909–20. doi: 10.1007/s004290050153
52. Tzou P, Reichhart J-M, Lemaitre B. Constitutive Expression of a Single Antimicrobial Peptide Can Restore Wild-Type Resistance to Infection in Immunodeficient Drosophila Mutants. Proc Natl Acad Sci (2002) 99:2152–7. doi: 10.1073/pnas.042411999
53. Yang H, Hultmark D. Tissue Communication in a Systemic Immune Response of Drosophila. Fly (Austin) (2016) 10:115–22. doi: 10.1080/19336934.2016.1182269
54. Perrigault M, Tanguy A, Allam B. Identification and Expression of Differentially Expressed Genes in the Hard Clam, Mercenaria mercenaria, in Response to Quahog Parasite Unknown (QPX). BMC Genomics (2009) 10:377. doi: 10.1186/1471-2164-10-377
55. Philipp EER, Kraemer L, Melzner F, Poustka AJ, Thieme S, Findeisen U, et al. Massively Parallel RNA Sequencing Identifies a Complex Immune Gene Repertoire in the Lophotrochozoan Mytilus edulis. PLoS One (2012) 7:e33091. doi: 10.1371/journal.pone.0033091
56. Toubiana M, Gerdol M, Rosani U, Pallavicini A, Venier P, Roch P. Toll-Like Receptors and MyD88 Adaptors in Mytilus: Complete Cds and Gene Expression Levels. Dev Comp Immunol (2013) 40:158–66. doi: 10.1016/j.dci.2013.02.006
57. Tanguy A, Guo X, Ford SE. Discovery of Genes Expressed in Response to Perkinsus marinus Challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) Oysters. Gene (2004) 338:121–31. doi: 10.1016/j.gene.2004.05.019
58. Wang L, Zhang H, Wang M, Zhou Z, Wang W, Liu R, et al. The Transcriptomic Expression of Pattern Recognition Receptors: Insight Into Molecular Recognition of Various Invading Pathogens in Oyster Crassostrea gigas. Dev Comp Immunol (2019) 91:1–7. doi: 10.1016/j.dci.2018.09.021
59. Allam B, Raftos D. Immune Responses to Infectious Diseases in Bivalves. J Invertebr Pathol (2015) 131:121–36. doi: 10.1016/j.jip.2015.05.005
60. Wang W, Wang L, Liu Z, Song X, Yi Q, Yang C, et al. The Involvement of TLR Signaling and Anti-Bacterial Effectors in Enhanced Immune Protection of Oysters After Vibrio splendidus Pre-Exposure. Dev Comp Immunol (2020) 103:103498. doi: 10.1016/j.dci.2019.103498
61. Melillo D, Marino R, Italiani P, Boraschi D. Innate Immune Memory in Invertebrate Metazoans: A Critical Appraisal. Front Immunol (2018) 9:1915. doi: 10.3389/fimmu.2018.01915
62. Yi Y, Xu H, Li M, Wu G. RNA-Seq Profiles of Putative Genes Involved in Specific Immune Priming in Bombyx mori Haemocytes. Infect Genet Evol (2019) 74:103921. doi: 10.1016/j.meegid.2019.103921
63. de Melo ES, Brayner FA, Junior NCP, França IRS, Alves LC. Investigation of Defense Response and Immune Priming in Biomphalaria glabrata and Biomphalaria straminea, Two Species With Different Susceptibility to Schistosoma mansoni. Parasitol Res (2020) 119:189–201. doi: 10.1007/s00436-019-06495-4
64. Zhang T, Qiu L, Sun Z, Wang L, Zhou Z, Liu R, et al. The Specifically Enhanced Cellular Immune Responses in Pacific Oyster (Crassostrea Gigas) Against Secondary Challenge With Vibrio splendidus. Dev Comp Immunol (2014) 45:141–50. doi: 10.1016/j.dci.2014.02.015
65. Berridge MJ, Lipp P, Bootman MD. The Versatility and Universality of Calcium Signalling. Nat Rev Mol Cell Biol (2000) 1:11–21. doi: 10.1038/35036035
66. Rao A, Hogan PG. Calcium Signaling in Cells of the Immune and Hematopoietic Systems. Immunol Rev (2009) 231:5–9. doi: 10.1111/j.1600-065X.2009.00823.x
67. Weavers H, Evans IR, Martin P, Wood W. Corpse Engulfment Generates a Molecular Memory That Primes the Macrophage Inflammatory Response. Cell (2016) 165:1658–71. doi: 10.1016/j.cell.2016.04.049
68. Liao Z, Wang X, Liu H, Fan M, Sun J, Shen W. Molecular Characterization of a Novel Antimicrobial Peptide From Mytilus coruscus. Fish Shellfish Immunol (2013) 34:610–6. doi: 10.1016/j.fsi.2012.11.030
69. De Zoysa M, Nikapitiya C, Whang I, Lee JS, Lee J. Abhisin: A Potential Antimicrobial Peptide Derived From Histone H2A of Disk Abalone (Haliotis Discus Discus). Fish Shellfish Immunol (2009) 27:639–46. doi: 10.1016/j.fsi.2009.08.007
70. Li H, Parisi M-G, Toubiana M, Cammarata M, Roch P. Lysozyme Gene Expression and Hemocyte Behaviour in the Mediterranean Mussel, Mytilus galloprovincialis, After Injection of Various Bacteria or Temperature Stresses. Fish Shellfish Immunol (2008) 25:143–52. doi: 10.1016/j.fsi.2008.04.001
71. Seo JK, Lee MJ, Nam BH, Park NG. Cgmolluscidin, a Novel Dibasic Residue Repeat Rich Antimicrobial Peptide, Purified From the Gill of the Pacific Oyster, Crassostrea gigas. Fish Shellfish Immunol (2013) 35:480–8. doi: 10.1016/j.fsi.2013.05.010
72. Dorrington T, Villamil L, Gómez-chiarri M. Upregulation in Response to Infection and Antibacterial Activity of Oyster Histone H4. Fish Shellfish Immunol (2011) 30:94–101. doi: 10.1016/j.fsi.2010.09.006
73. Venier P, Varotto L, Rosani U, Millino C, Celegato B, Bernante F, et al. Insights Into the Innate Immunity of the Mediterranean Mussel Mytilus galloprovincialis. BMC Genomics (2011) 12:69. doi: 10.1186/1471-2164-12-69
74. Yao T, Lu J, Ye L, Wang J. Molecular Characterization and Immune Analysis of a Defensin From Small Abalone, Haliotis diversicolor. Comp Biochem Physiol Part B Biochem Mol Biol (2019) 235:1–7. doi: 10.1016/j.cbpb.2019.05.004
75. Gerdol M, De Moro G, Manfrin C, Venier P, Pallavicini A. Big Defensins and Mytimacins, New AMP Families of the Mediterranean Mussel Mytilus galloprovincialis. Dev Comp Immunol (2012) 36:390–9. doi: 10.1016/j.dci.2011.08.003
76. Zhang L, Yang D, Wang Q, Yuan Z, Wu H, Pei D, et al. A Defensin From Clam Venerupis philippinarum: Molecular Characterization, Localization, Antibacterial Activity, and Mechanism of Action. Dev Comp Immunol (2015) 51:29–38. doi: 10.1016/j.dci.2015.02.009
77. Yang J, Luo J, Zheng H, Lu Y, Zhang H. Cloning of a Big Defensin Gene and Its Response to Vibrio parahaemolyticus Challenge in the Noble Scallop Chlamys nobilis (Bivalve: Pectinidae). Fish Shellfish Immunol (2016) 56:445–9. doi: 10.1016/j.fsi.2016.07.030
78. De Zoysa M, Whang I, Lee Y, Lee S, Lee J-S, Lee J. Defensin From Disk Abalone Haliotis discus discus: Molecular Cloning, Sequence Characterization and Immune Response Against Bacterial Infection. Fish Shellfish Immunol (2010) 28:261–6. doi: 10.1016/j.fsi.2009.11.005
79. Muñoz M, Vandenbulcke F, Saulnier D, Bachère E. Expression and Distribution of Penaeidin Antimicrobial Peptides Are Regulated by Haemocyte Reactions in Microbial Challenged Shrimp. Eur J Biochem (2002) 269:2678–89. doi: 10.1046/j.1432-1033.2002.02934.x
80. Kao C-H, Sun C-N. In Vitro Degradation of Some Organophosphorus Insecticides by Susceptible and Resistant Diamondback Moth. Pestic Biochem Physiol (1991) 41:132–41. doi: 10.1016/0048-3575(91)90067-V
81. Wagner MA, Khanna P, Jorns MS. Structure of the Flavocoenzyme of Two Homologous Amine Oxidases: Monomeric Sarcosine Oxidase and N -Methyltryptophan Oxidase †. Biochemistry (1999) 38:5588–95. doi: 10.1021/bi982955o
82. Song L, Wang L, Zhang H, Wang M. The Immune System and Its Modulation Mechanism in Scallop. Fish Shellfish Immunol (2015) 46:65–78. doi: 10.1016/j.fsi.2015.03.013
83. Domínguez-Andrés J, Joosten LA, Netea MG. Induction of Innate Immune Memory: The Role of Cellular Metabolism. Curr Opin Immunol (2019) 56:10–6. doi: 10.1016/j.coi.2018.09.001
84. Van den Bossche J, Baardman J, Otto NA, van der Velden S, Neele AE, van den Berg SM, et al. Mitochondrial Dysfunction Prevents Repolarization of Inflammatory Macrophages. Cell Rep (2016) 17:684–96. doi: 10.1016/j.celrep.2016.09.008
85. Arts RJW, Moorlag SJCFM, Novakovic B, Li Y, Wang S-Y, Oosting M, et al. BCG Vaccination Protects Against Experimental Viral Infection in Humans Through the Induction of Cytokines Associated With Trained Immunity. Cell Host Microbe (2018) 23:89–100.e5. doi: 10.1016/j.chom.2017.12.010
86. Etchegaray J-P, Mostoslavsky R. Interplay Between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell (2016) 62:695–711. doi: 10.1016/j.molcel.2016.05.029
87. Pérez-Vázquez D, Contreras-Castillo E, Licona-Limón P. Innate Immune Memory, the Missing Piece of the Immunological Response. TIP Rev Espec en Cienc Químico-Biológicas (2018) 21:112–23. doi: 10.22201/fesz.23958723e.2018.0.151
Keywords: Haliotis diversicolor, Vibrio harveyi, enhanced immune protection, secondary infection, immune priming
Citation: Yao T, Lu J, Bai C, Xie Z and Ye L (2021) The Enhanced Immune Protection in Small Abalone Haliotis diversicolor Against a Secondary Infection With Vibrio harveyi. Front. Immunol. 12:685896. doi: 10.3389/fimmu.2021.685896
Received: 26 March 2021; Accepted: 22 June 2021;
Published: 06 July 2021.
Edited by:
Osamu Takeuchi, Kyoto University, JapanReviewed by:
Surya Pandey, University of Chicago, United StatesXiaocui He, La Jolla Institute for Immunology (LJI), United States
Copyright © 2021 Yao, Lu, Bai, Xie and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingtong Ye, bGluZ3RvbmcyNzUzQDEyNi5jb20=
 Tuo Yao
Tuo Yao Jie Lu
Jie Lu Changming Bai2
Changming Bai2 Lingtong Ye
Lingtong Ye