- 1Department of Cardiothoracic Surgery, Taihe Hospital, Hubei University of Medicine, Shiyan, China
- 2Department of Oncology, Huanggang Central Hospital, Huanggang, China
- 3School of Basic Medicine, Fourth Military Medical University, Xi’an, China
- 4Department of Respiratory, Xinchang People’s Hospital, Xinchang, China
Background: The occurrence and development of cancer could be promoted by abnormally competing endogenous RNAs (ceRNA) network. This article aims to determine the prognostic biomarker of ceRNA for non-small-cell lung cancer (NSCLC) prognosis.
Methods: The expression and clinical significance of LINC00973 in NSCLC tissues were analyzed via the The Cancer Genome Atlas (TCGA), Gene Expression Profiling Interactive Analysis (GEPIA), lnCAR, and clinical samples in Taihe Hospital. The biological functions and signaling pathways involved in target genes of ceRNA network were analyzed via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Survival analysis, univariate and multivariate Cox regression analysis were used for prognostic-related mRNA.
Results: Expression of LINC00973 was increased in NSCLC tissues. High expression of LINC00973 was associated with poor prognosis of NSCLC patients. There were 15 miRNA and 238 differential mRNA in the INC00973-miRNA-mRNA ceRNA network, involving cell migration, endothelial cell proliferation, tumor growth factor (TGF)-β, cellular senescence, phosphatidylinositol 3-hydroxy kinase (PI3K)-Akt, Hippo, Rap1, mitogen-activated protein kinase (MAPK), cell cycle signaling pathway, etc. The expression levels of RTKN2, NFIX, PTX3, BMP2 and LOXL2 were independent risk factors for the poor prognosis of NSCLC patients.
Conclusions: LINC00973-miRNA-mRNA ceRNA network might be the basis for determining pivotal post-translational regulatory mechanisms in the progression of NSCLC. BMP2, LOXL2, NFIX, PTX3 and RTKN2 might be valuable prognostic markers and potential therapeutic targets.
Introduction
According to Global Cancer Statistics 2020, with an estimated 2.2 million new cancer cases and 1.8 million deaths, lung cancer was the second most commonly diagnosed cancer and the leading cause of cancer death in 2020, representing approximately one in 10 (11.4%) cancers diagnosed and one in 5 (18.0%) deaths (1–4). Lung cancer included non-small cell carcinoma (NSCLC) and small cell carcinoma. At present, surgery was the first choice for NSCLC patients at early-stage, and combined therapy was the main way for NSCLC patients at middle and advanced stage (5). With the improvement of treatment methods, the prognosis of cancer patients was improved, but the overall survival (OS) of NSCLC patients remained still frustrating. Recently, the researches had provided new insights into the molecular mechanisms of NSCLC in genomics and transcriptomics. For example, LCAT1, a member of long non-coding RNA (lncRNA), was an upregulated marker in lung cancer tissues. Elevated LCAT1 expression level was closely associated with poor prognosis of cancer patients (6). LncRNA MNX1-AS1 was upregulated in lung cancer tissues, and the prognosis of lung cancer patients with overexpression of MNX1-AS1 was often terrible. MNX1-AS1 promoted cell proliferation, migration and invasion of lung cancer. The MNX1-AS1/miR-527/BRF2 signaling axis was involved in the occurrence and development of lung cancer (7). The expression of LncRNA AFAP1-AS1 was increased in NSCLC tissues, and related to the TNM stage and tumor size of NSCLC patients. Interfering with the expression of AFAP1-AS1 could inhibit the growth of NSCLC cells in vitro and in vivo (8). Therefore, lncRNA played an essential role in the progression of lung cancer.
Studies have confirmed that non-coding RNA (ncRNA) played important roles in tumorigenesis and metastasis (6–8). microRNA (miRNA) and lncRNA belong to ncRNA, and both of which have miRNA recognition elements (MRE) (9, 10). miRNA could trigger degradation of downstream target genes by recognizing the MRE in the 3’non-transcribed region (11, 12). Therefore, when lncRNA and mRNA had the same MRE, both of them could competitively bind to miRNA, forming a competitive endogenous RNA (ceRNA) regulatory network (13, 14). When miRNA bound to the target mRNA, the stability of mRNA decreased and its translation was hindered, thereby affecting gene expression. When lncRNA bound to miRNA, the stability of competitive mRNA increases instead, so that transcription and translation were well performed. This was a competitive two-way gene expression regulation mechanism composed of endogenous RNA (15, 16).
The ceRNA network mechanism presents a significant role in the occurrence and development of cancer. For example, the expression of circ_0025033 and small nuclear ribonucleoprotein Sm-like4 (LSM4) increased in ovarian cancer tissues and cells. Interfered with circ_0025033 or LSM4 expression could inhibit colony formation, migration, invasion and glycolytic metabolism of ovarian cancer cells. Circ_0025033 acted as a ceRNA to regulate LSM4 expression by targeting miR-184. Overexpression of LSM4 promoted the expression of circ_0025033, thereby inducing colony formation, migration, invasion and glycolysis (17). This indicated that there were abnormalities in the ceRNA network mechanism in the development of cancer. It was recently discovered that lncRNA LINC00973 had a vital effect in cancer. For example, chemotherapy could upregulate the expression level of LINC00973 in normal cells and cancer cells, which might be related to the activation of DNA damage response pathways or mitotic arrest. LINC00973 could reduce p21 levels, activate cancer cell proliferation, and reduce the lethality of drugs (18). Siglec-15 was a tumor immunosuppressive molecule. LINC00973 was highly expressed in Siglec-15 positive clear cell renal cell carcinoma, and it could positively regulate the expression of Siglec-15, which was the direct target molecule of miR-7109. LINC00973 could regulate the expression of Siglec-15 by affecting the role of sponge miR-7109 and forming ceRNA. Therefore, the LINC00973-miR-7109-Siglec-15 ceRNA mechanism was involved in regulating the progression of renal cell carcinoma (19). Nowadays, there was no related literature report about the regulation mechanism of ceRNA network composed of LINC00973 in the progress of NSCLC. Therefore, this study aims to explore the role and potential value of LINC00973-miRNA-mRNA ceRNA in the progression of NSCLC, and to provide new target molecules and potential regulatory mechanisms for NSCLC diagnosis and treatment.
Materials and Methods
NSCLC Tissue Sample
Cancer and normal adjacent tissues of 25 patients with NSCLC undergoing surgical treatment were collected at the Department of Thoracic Surgery, Taihe Hospital from December 2019 to April 2020. Inclusion criteria: (1) NSCLC was diagnosed pathologically; (2) Neoadjuvant chemotherapy or radiotherapy was not performed before surgery; (3) Immunosuppressant therapy, biological therapy, or targeted therapy were not performed. Exclusion criteria: 1) SCLC; 2) Patients voluntarily withdraw. This study was approved by the ethics committee of the Taihe Hospital, Hubei University of Medicine, and the patient has signed an informed consent. Among them, 11 cases were lung adenocarcinoma (LUAD) and 14 cases were lung squamous cell carcinoma (LUSC).
TCGA Database
In October 2020, 1145 samples were downloaded from the TCGA (https://portal.gdc.cancer.gov/) website (108 cases of normal lung tissue, including 59 cases of normal lung tissue from LUAD and 49 cases of normal lung tissue from LUSC; 1037 cases of NSCLC tissue samples, including 535 samples of LUAD Cases and 522 cases of LUSC samples) HTSeq-FPKM transcriptome data and TCGA clinical data of 1026 NSCLC patients. The clinical parameters and prognostic information of NSCLC patients was sorted out. Patients with unknown or incomplete parameters to analyze the relationship between the expression level of LINC00973 and the prognosis was excluded. The limma package was used to analyze the differential expression genes in the tissues of NSCLC patients, and the screening conditions: fold change > 1 and P < 0.05.
Quantitative Real-Time PCR
According to the RNA kit (Invitrogen, USA) instructions, total RNA was extracted from NSCLC tissue, and the qualified RNA concentration was detected. Copying DNA (cDNA) was synthesized using a reverse transcription kit (Takara, Japan), and the expression of LINC00973 was detected by quantitative real-time PCR (qRT-PCR). Primers was supplied by Sangong Co Ltd. GAPDH was used as an internal control. Primer sequence (5’-3’): LINC00973: TTGAAGGCTTCCTGGTCTGAG (Forward), AGGCTTACATTCCAGCTGTGT (Reverse), GAPDH: AACGGATTTGGTCGTATTG (Forward), GGAAGATGGTGATGGGATT (Reverse). Experiments were performed in triplicate independently.
LNCAR and GEPIA Database
The LNCAR (http://lncar.renlab.org/) database data were acquired from the GEO database, which contained the cancer-related lncRNA expression database. The expression of LINC00973 in lung cancer tissues and its potential clinical value were analyzed in the LNCAR database. The screening criteria: P < 0.05. The GEPIA (http://gepia.cancer-pku.cn/) database was a reanalysis online database based on the TCGA and GTEx database data. The expression of LINC00973 and biomarkers in ceRNA network were explored. And the relationship between expression of LINC00973 and NSCLC overall survival (OS) and disease-free survival (DFS) were analyzed in the GEPIA database.
ceRNA Network
The miRNA of LINC00973 were predicted in the LncBase Predicted v.2 database, and the screening criteria: binding coefficient ≥ 0.6. In addition, the miRNA that predicting the differentially expressed genes of TCGA database were performed via the PITA, RNA22, miRmap, microT, miRanda, PicTar, and TargetScan databases, and the screening criteria: the number of databases with targeting relationships needed to be greater than 3. Screening for overlapping miRNA in the LINC00973 target miRNA and miRNA that differentially expressed genes of TCGA database, and the ceRNA network signaling mechanism was constructed via Cytoscape 3.6.1 software.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
LncRNA competitively inhibited the mRNA binding to miRNA, and could maintain the stability of mRNA and promotes RNA transcription when LncRNA binds to miRNA. Therefore, the biological functions and signaling mechanisms involved in the ceRNA network was explored via the R clusterProfiler package. GO annotation included the biological process (BP), molecular function (MF) and cellular component (CC). The screening criteria: P < 0.05.
Prognostic Survival Analysis
Kaplan-Meier survival analysis was used to evaluate the role of differentially expressed genes in the prognosis of NSCLC patients to understand the important target molecules in the ceRNA mechanism. The screening criteria was P < 0.05. On this basis, univariate Cox and multivariate Cox regression analysis were performed, and a nomogram was constructed.
The Value of Poor Prognostic Factors in NSCLC Patients Were Verified
The GEPIA database was used to verify the expression of poor prognostic factors in the ceRNA network in NSCLC tissues. In addition, the Cbioportal database (http://www.cbioportal.org) was used to explore the relationship between poor prognostic factors and clinical pathological characteristics of patients with NSCLC. PrognoScan (http://www.abren.net/PrognoScan/) and Kaplan-Meier Plotter (http://kmplot.com/analysis/) databases were used to verify the value of poor prognostic factors in the prognosis of NSCLC patients, and the grouping standard was according to the best cutoff value.
Statistical Analysis
Data processing and statistical analysis adopt perl and R language. The expression level and prognostic value of LINC00973 and mRNA in the ceRNA network were analyzed by the wilcoxon signed-rank test and Kaplan-Meier survival analysis. In addition, the role of differentially expressed genes in NSCLC prognosis in the ceRNA network were explored via the univariate and multivariate Cox regression analysis. P < 0.05 was considered statistically significant.
Results
LINC00973 Was Upregulated in NSCLC Tissues
In the TCGA database, the expression of LINC00973 was increased in unpaired NSCLC tissues (Figure 1A). The expression of LINC00973 was increased in unpaired LUAD and LUSC tissues (Figures 1B, C). The expression of LINC00973 in 106 paired NSCLC tissues was increased significantly (Figure 1D). The expression of LINC00973 was also increased in 57 paired LUAD and 49 paired LUSC significantly (Figures 1E, F).
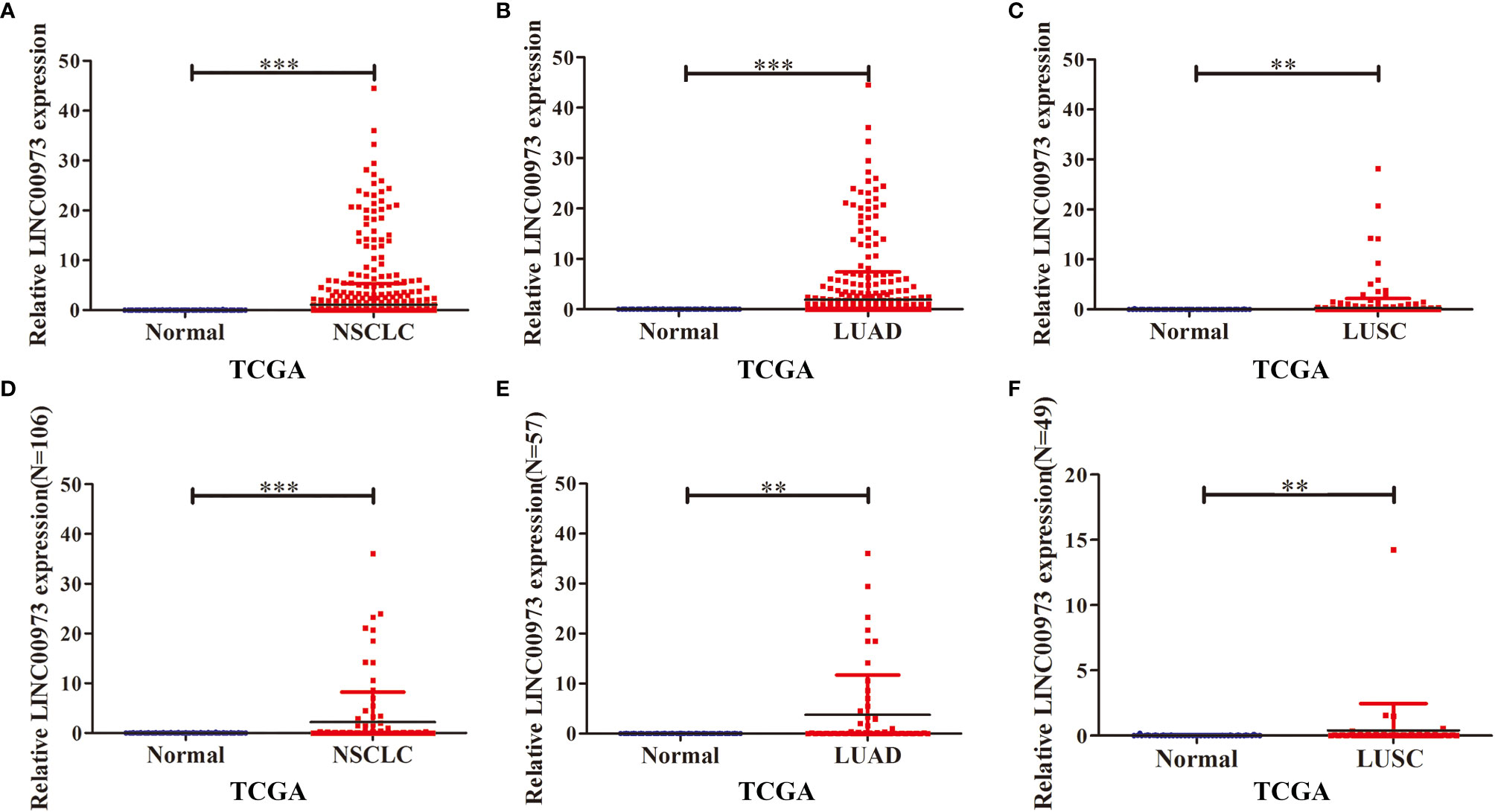
Figure 1 LINC00973 expression was elevated in NSCLC tissue in the TCGA database. (A–C) Expression of LINC00973 in unpaired samples of NSCLC, LUAD and LUSC; (D–F) Expression of LINC00973 in paired samples of NSCLC, LUAD and LUSC. NSCLC, Non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; Normal, normal tissues; TCGA, The Cancer Genome Atlas; **P < 0.01; ***P < 0.001.
In addition, the expression of LINC00973 was increased in NSCLC tissues in LNCAR database (Figures 2A–F). The GSE27262 and GSE89039 datasets showed that the expression of LINC00973 (LC_S148 and LC_S39) in lung cancer tissues was elevated, significantly (Figures 2A, B). The GSE101929 dataset revealed that the expression of LINC00973 (LC_S3) in NSCLC tissues was elevated (Figure 2C). The GSE40791 and GSE33532 datasets showed that the expression of LINC00973 (LC_S257 and LC_S216) in LUAD tissues was increased, and it was statistically significant (Figures 2D, E). The GSE33532 dataset showed that the expression of LINC00973 (LC_S218) in LUSC tissues was increased. However, it was not statistically significant (Figure 2E).
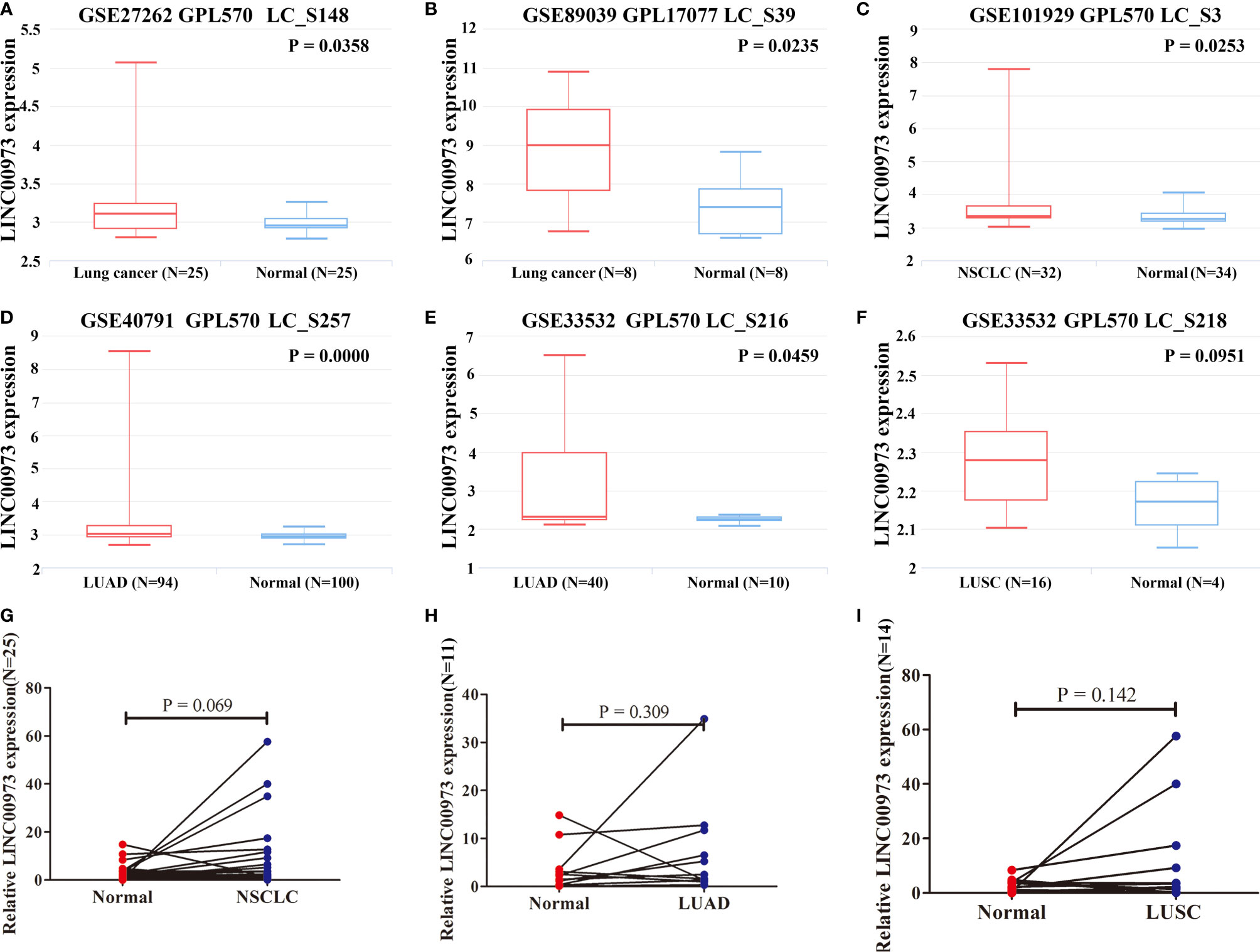
Figure 2 LINC00973 expression was upregulated in the LNCAR database and clinical NSCLC tissues. (A) GSE27262 (LC_S148); (B) GSE89039 (LC_S39); (C) GSE101929 (LC_S3); (D) GSE40791 (LC_S257); (E) GSE33532 (LC_S216); (F) GSE33532 (LC_S218); (G) Clinical NSCLC samples; (H) Clinical LUSD samples; (I) Clinical LUSCsamples. NSCLC, Non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; Normal, normal tissues.
Moreover, we collected 25 clinical NSCLC tissues and adjacent normal lung tissues. We found that the expression of LINC00973 was increased in NSCLC and its subtypes via qRT-PCR. However, it was not statistically significant (Figures 2G–I).
Elevated LINC00973 Expression Was Correlated With Poor Prognosis in NSCLC Patients
In the TCGA database, Kaplan-Meier survival curve suggested that increasing level of LINC00973 was closely associated with poor prognosis of NSCLC patients (Figure 3A). Subgroup analysis found that the prognosis of LUAD and LUSC patients with elevated LINC00973 expression levels were with poor prognosis, whereas there was no statistical significance between the LINC00973 expression levels and the prognosis of LUSC patients (Figures 3B, C). In addition, we found that increasing the expression levels of LINC00973 were with poor prognosis of NSCLC patients in the GEPIA database (Figures 3D–F). In detail, increasing levels of LINC00973 were negatively interrelated with OS and DFS in NSCLC patients, which indicated that LINC00973 might be a carcinogen. A consistent conclusion was reached in the LNCAR database. We found that anti-cancer drugs could inhibit the expression of LINC00973 in NSCLC PC9 and HCC827 cells (Figure 4). In detail, compared with the control group, GSE67051,GSE80316, and GSE51212 datasets showed that the expression of LINC00973 in the erlotinib treatment group was significantly decreased. The GSE38302 dataset showed that the expression of LINC00973 in the gefitinib group was significantly decreased.
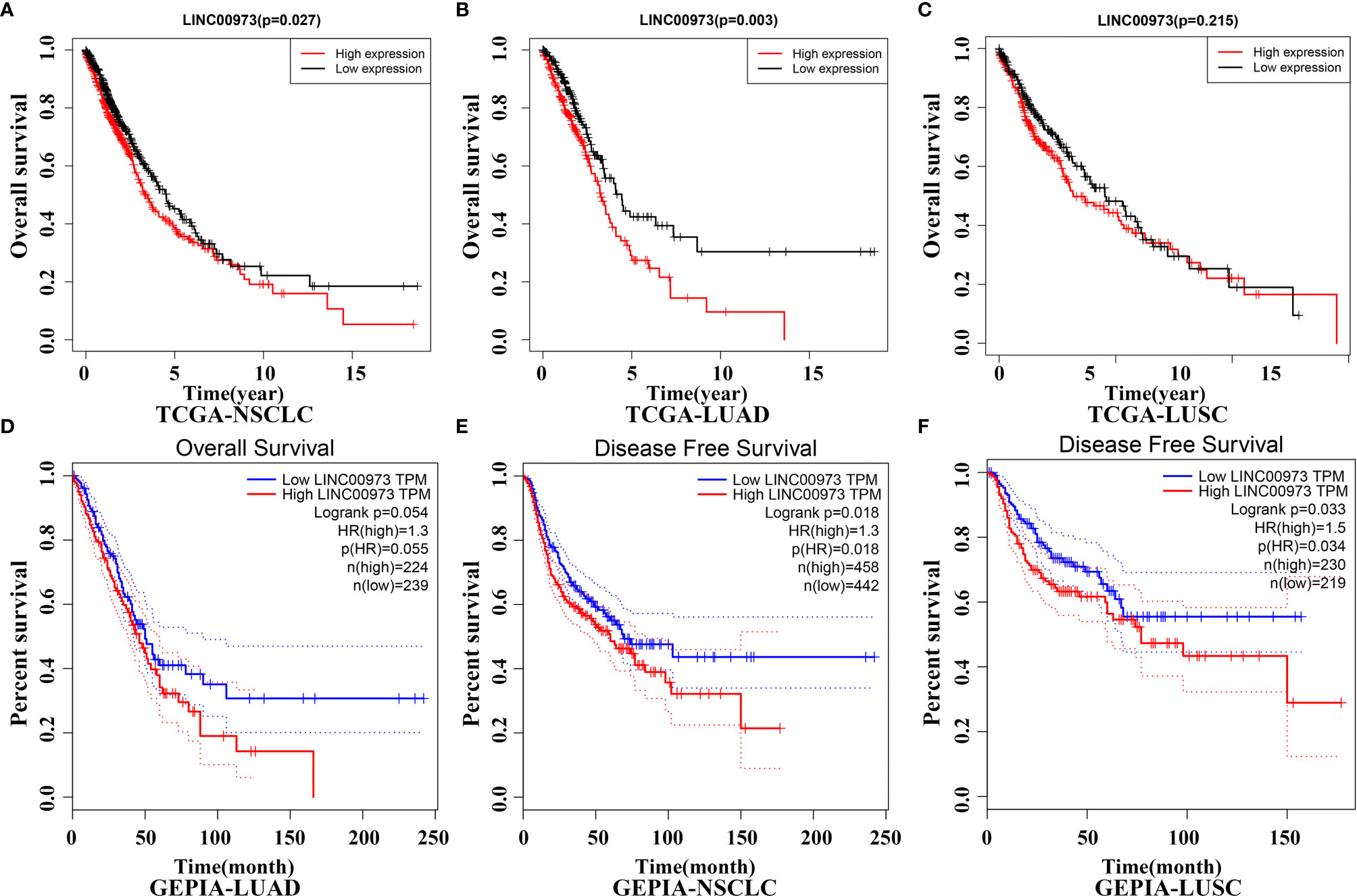
Figure 3 NSCLC patients with elevated lncRNA expression have poor prognosis in TCGA and GEPIA databases. (A) TCGA-NSCLC-OS; (B) TCGA-LUAD-OS; (C) TCGA-LUSC-OS; (D) GEPIA-LUAD-OS; (E) GEPIA-NSCLC-DFS; (F) GEPIA-LUSC-DFS. NSCLC, Non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas; GEPIA, Gene Expression Profiling Interactive Analysis.
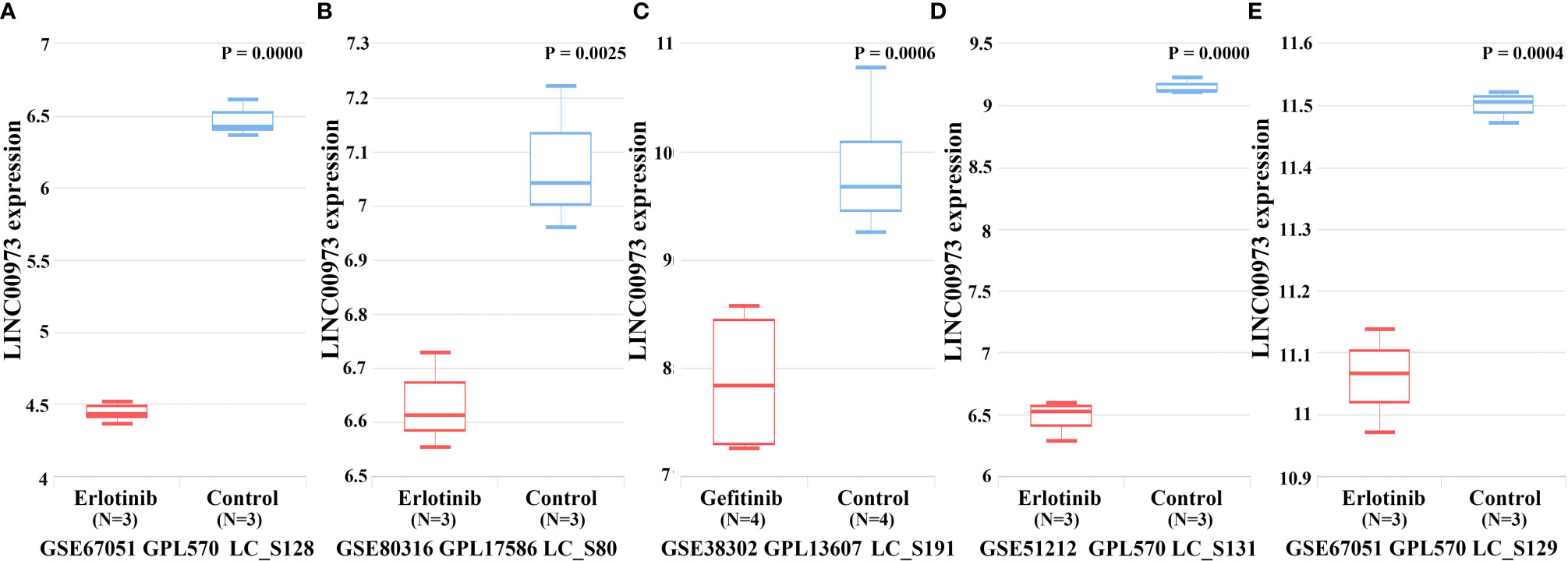
Figure 4 Anti-cancer drugs could down-regulate the expression of LINC00973. (A) GSE67051-erlotinib; (B) GSE80316-erlotinib; (C) GSE38302-gefitinib; (D) GSE51212-erlotinib; (E) GSE67051-erlotinib.
Construction of LINC00973-miRNA-mRNA ceRNA Network
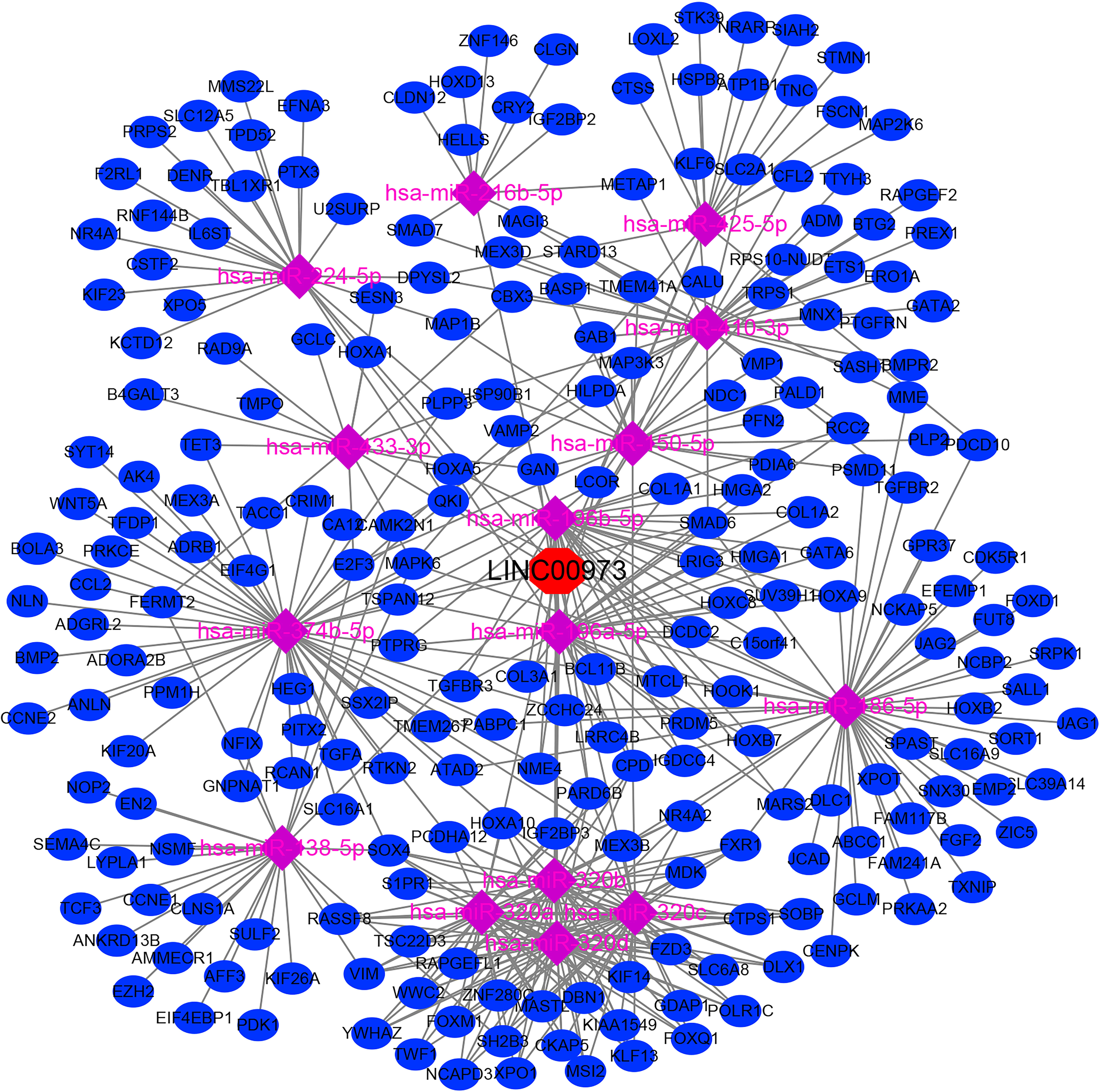
In the LncBase Predicted v.2 database, a total of 210 miRNA were screened with the binding coefficient ≥ 0.6 (Table 1). In the TCGA database, 5367 differentially expressed genes were found in NSCLC tissues. We use heat map and violin map to show the top 20 differentially expressed genes in TCGA via fold changes (Figures S1A, B). We predicted that there were 232 miRNAs for differentially expressed genes (Table S1). Further comparison suggested that there were 15 miRNA that were competitively bound, namely hsa-miR-374b-5p, hsa-miR-216b-5p, hsa-miR-196a-5p, hsa-miR-320c, hsa-miR-138 -5p, hsa-miR-150-5p, hsa-miR-196b-5p, hsa-miR-224-5p, hsa-miR-320a, hsa-miR-186-5p, hsa-miR-320b, hsa-miR -425-5p, hsa-miR-410-3p, hsa-miR-320d and hsa-miR-433-3p (Figure S2). There were 238 target genes for miRNA in the ceRNA network (Table S2), and the LINC0097-miRNA-mRNA ceRNA network signaling mechanism was constructed (Figure 5).
The Biological Functions and Signaling Pathways Involved in the ceRNA Network Mechanism
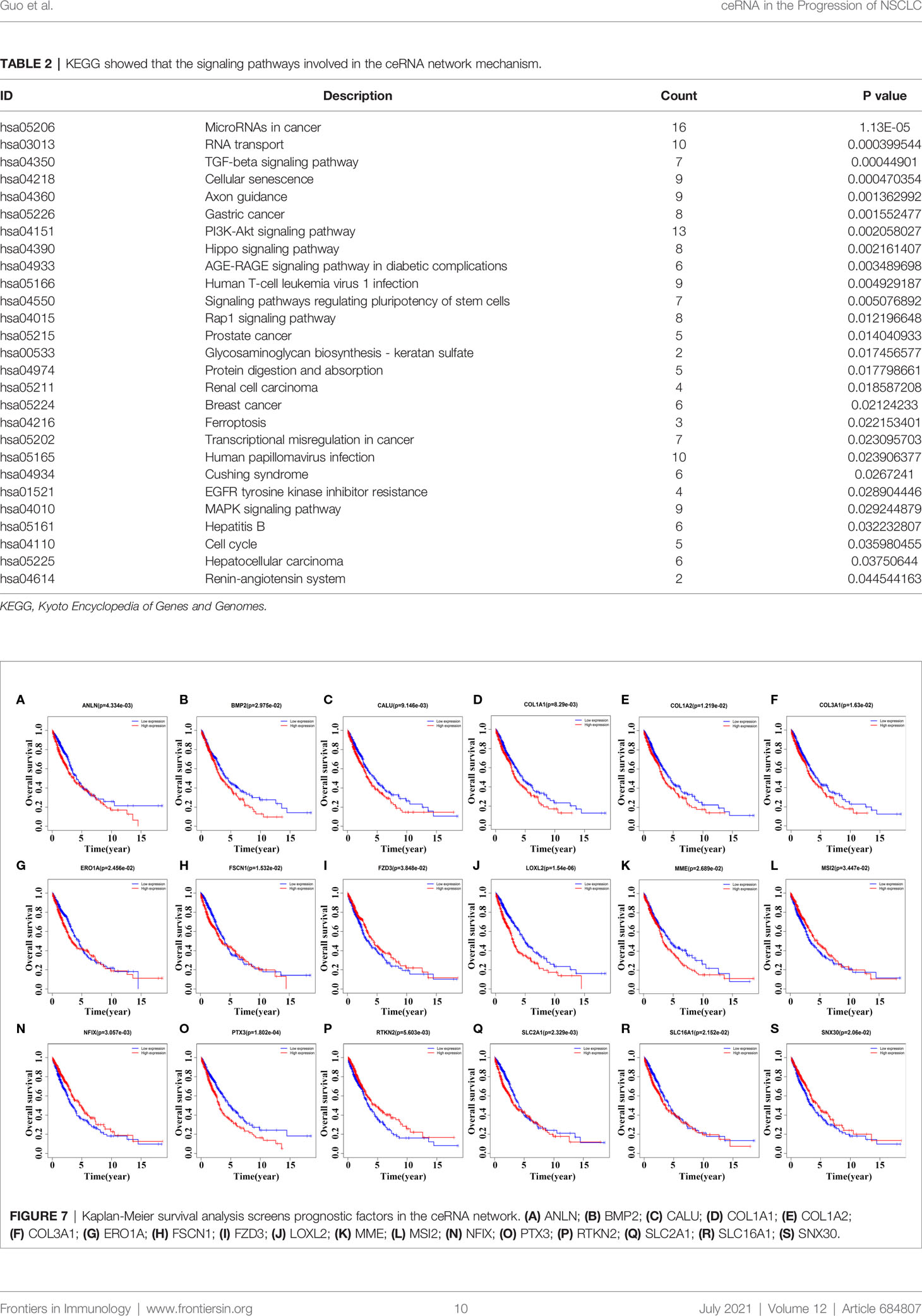
We performed GO annotation and KEGG on 238 mRNAs in the ceRNA network to explore the biological roles of LINC00973 of NSCLC. The GO annotation showed that LINC00973 might be involved in biological processes such as cell migration, response to oxygen levels, positive regulation of cell cycle process, and endothelial cell proliferation (Figures 6A–C and Table S3). KEGG showed that LINC00973 might be involved in the regulation of MicroRNAs in cancer, RNA transport, TGF β signaling pathway, Cellular senescence, PI3K-Akt signaling pathway, Hippo signaling pathway, Rap1 signaling pathway, Transcriptional misregulation in cancer, EGFR tyrosine kinase inhibitor resistance, MAPK signaling pathway, cell cycle and renin-angiotensin system and other signaling pathways (Figure 6D and Table 2).
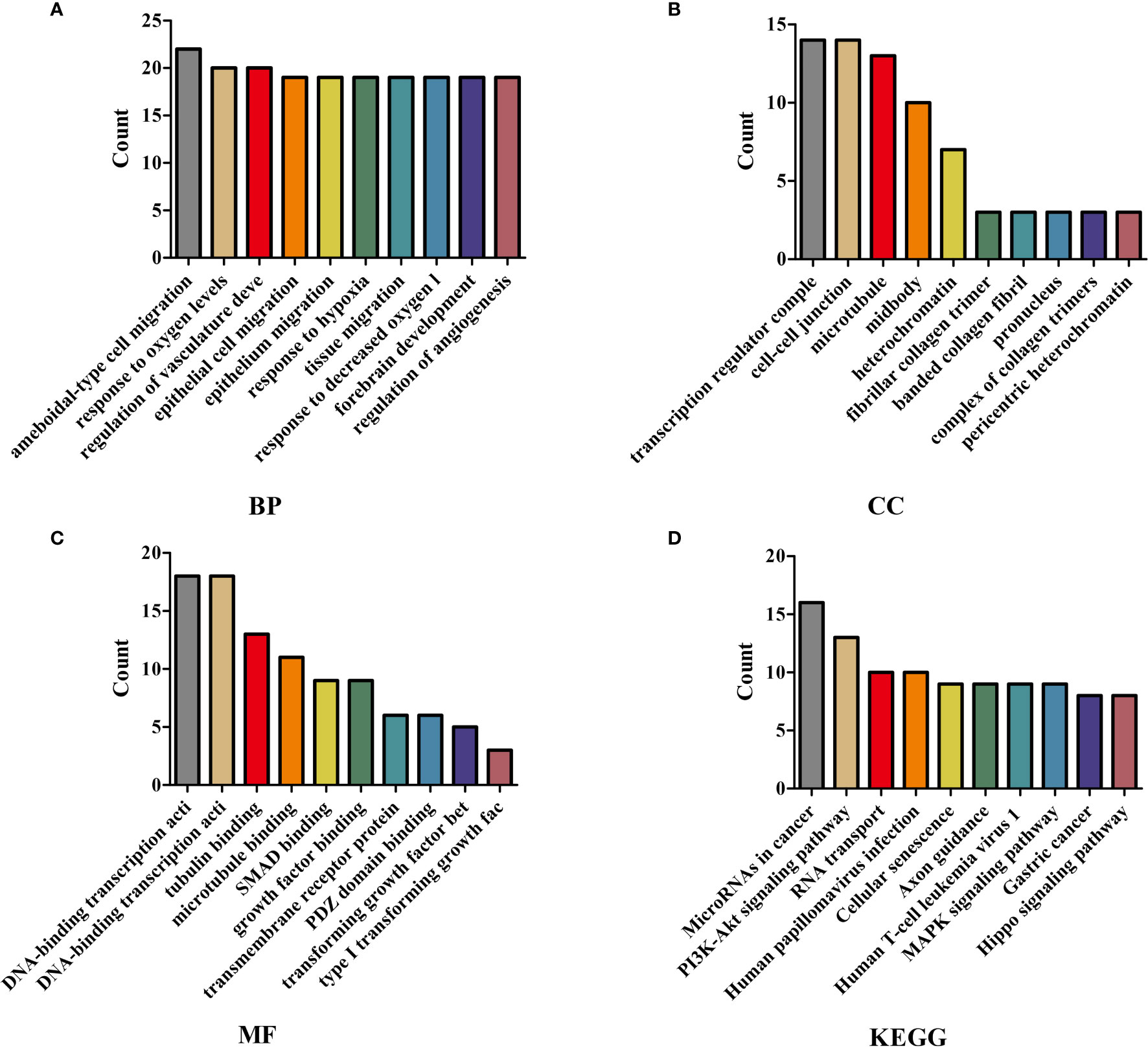
Figure 6 GO and KEGG showed that the biological functions and signaling pathways involved in the ceRNA network mechanism. (A) BP; (B) CC; (C) MF; (D) KEGG. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, Biological process; CC, Cellular components; MF, Molecular function.
Kaplan-Meier Survival Analysis Screens Prognostic Genes in the ceRNA Network
Kaplan-Meier survival analysis revealed that the expression levels of ANLN, BMP2, CALU, COL1A1, COL1A2, COL3A1, ERO1A, FSCN1, FZD3, LOXL2, MME, MSI2, NFIX, PTX3, RTKN2, SLC2A1, SLC16A1, and SNX30 were relevant with poor prognosis in NSCLC patients (Figure 7). In detail, NSCLC patients with elevated expression levels of ANLN, BMP2, CALU, COL1A1, COL1A2, COL3A1, LOXL2, MME, PTX3 and SLC2A1 had a poor prognosis. The prognosis of NSCLC patients with reduced expression levels of FZD3, MSI2, NFIX, RTKN2 and SNX30 were poor, respectively (P < 0.05).
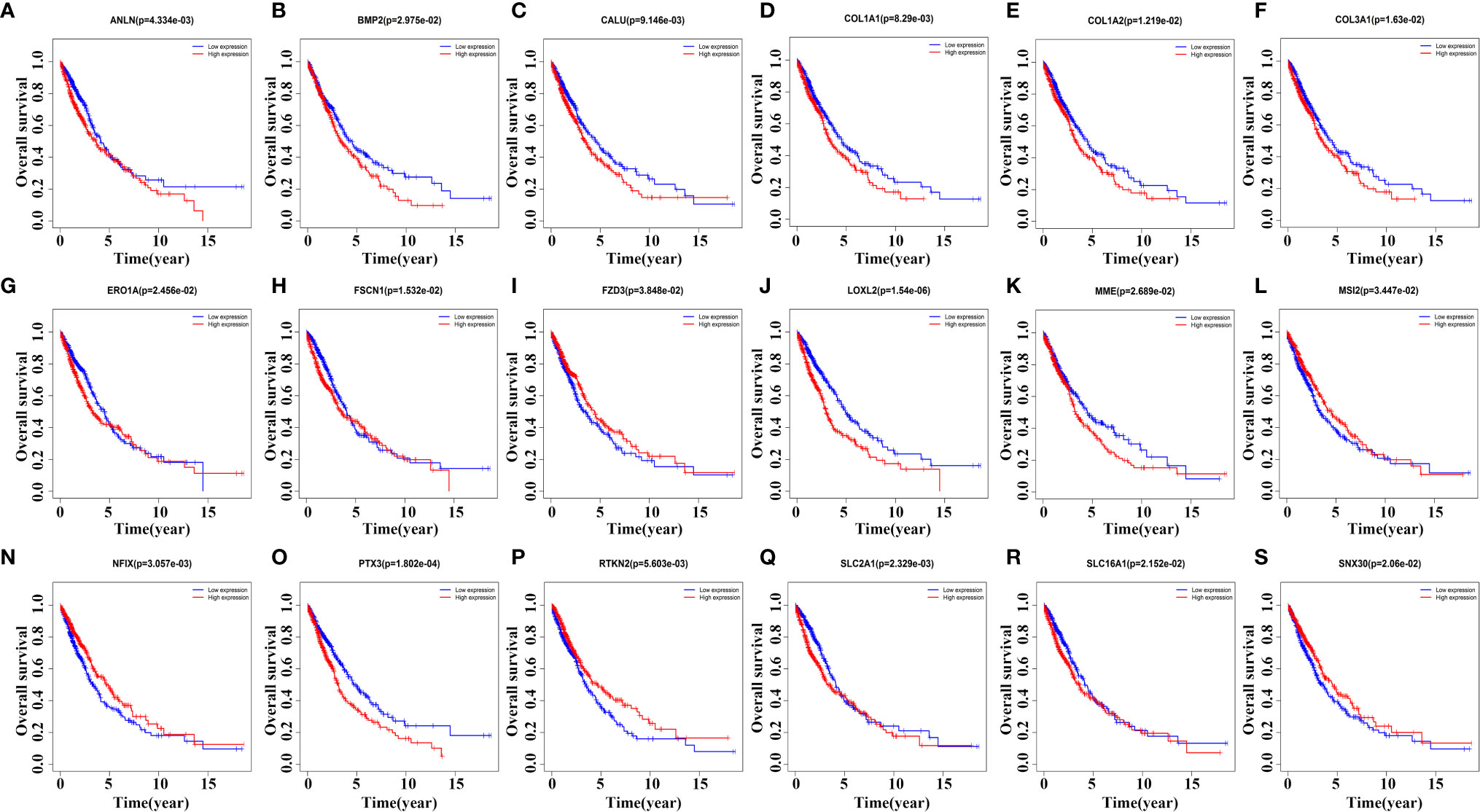
Figure 7 Kaplan-Meier survival analysis screens prognostic factors in the ceRNA network. (A) ANLN; (B) BMP2; (C) CALU; (D) COL1A1; (E) COL1A2; (F) COL3A1; (G) ERO1A; (H) FSCN1; (I) FZD3; (J) LOXL2; (K) MME; (L) MSI2; (N) NFIX; (O) PTX3; (P) RTKN2; (Q) SLC2A1; (R) SLC16A1; (S) SNX30.
Univariate and Multivariate Cox Regression Analysis to Screen Prognostic Factors in ceRNA Network
Univariate Cox regression analysis to screen 18 prognostic-related factors, it was found that LOXL2, PTX3, SLC2A1, RTKN2, ANLN, NFIX, CALU, ERO1A, BMP2, FSCN1, COL1A2, SLC16A1, COL3A1, MME, SNX30, COL1A1, and MSI2 might be prognostic risk factors of NSCLC patients (Table 3). Multivariate Cox regression analysis suggested that RTKN2, NFIX, PTX3, BMP2 and LOXL2 were independent risk factors for poor prognosis of NSCLC patients (Figure 8).
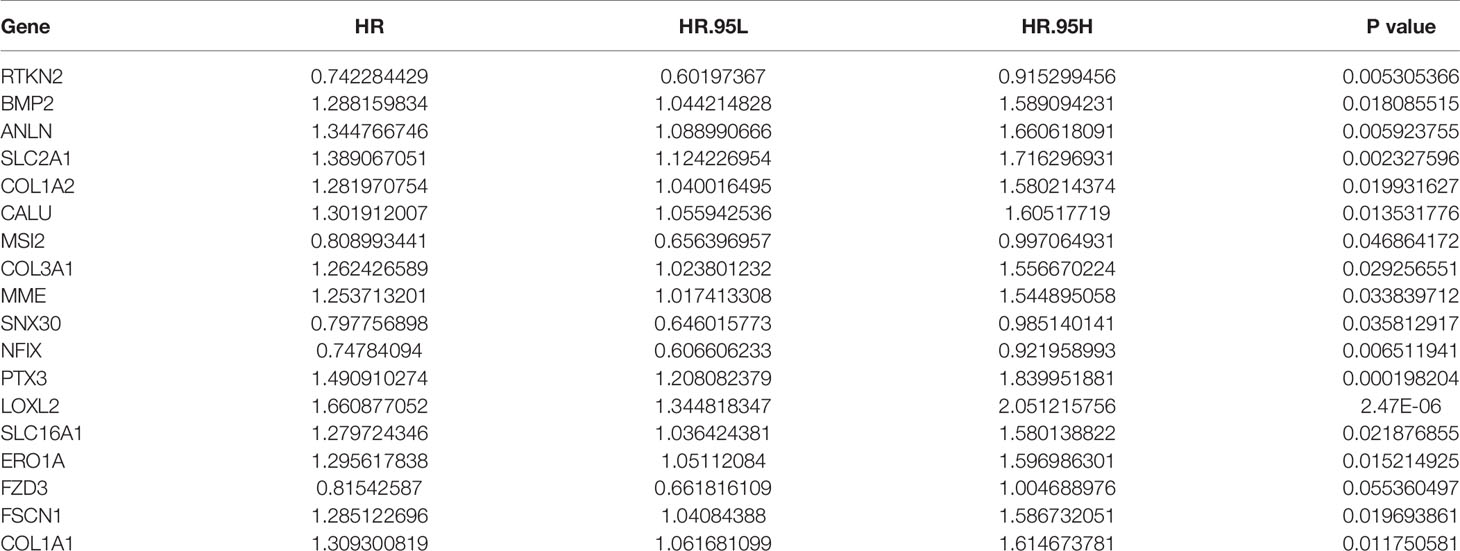
Table 3 Univariate Cox regression analysis showed that the prognostic factors of NSCLC patients in ceRNA. network.
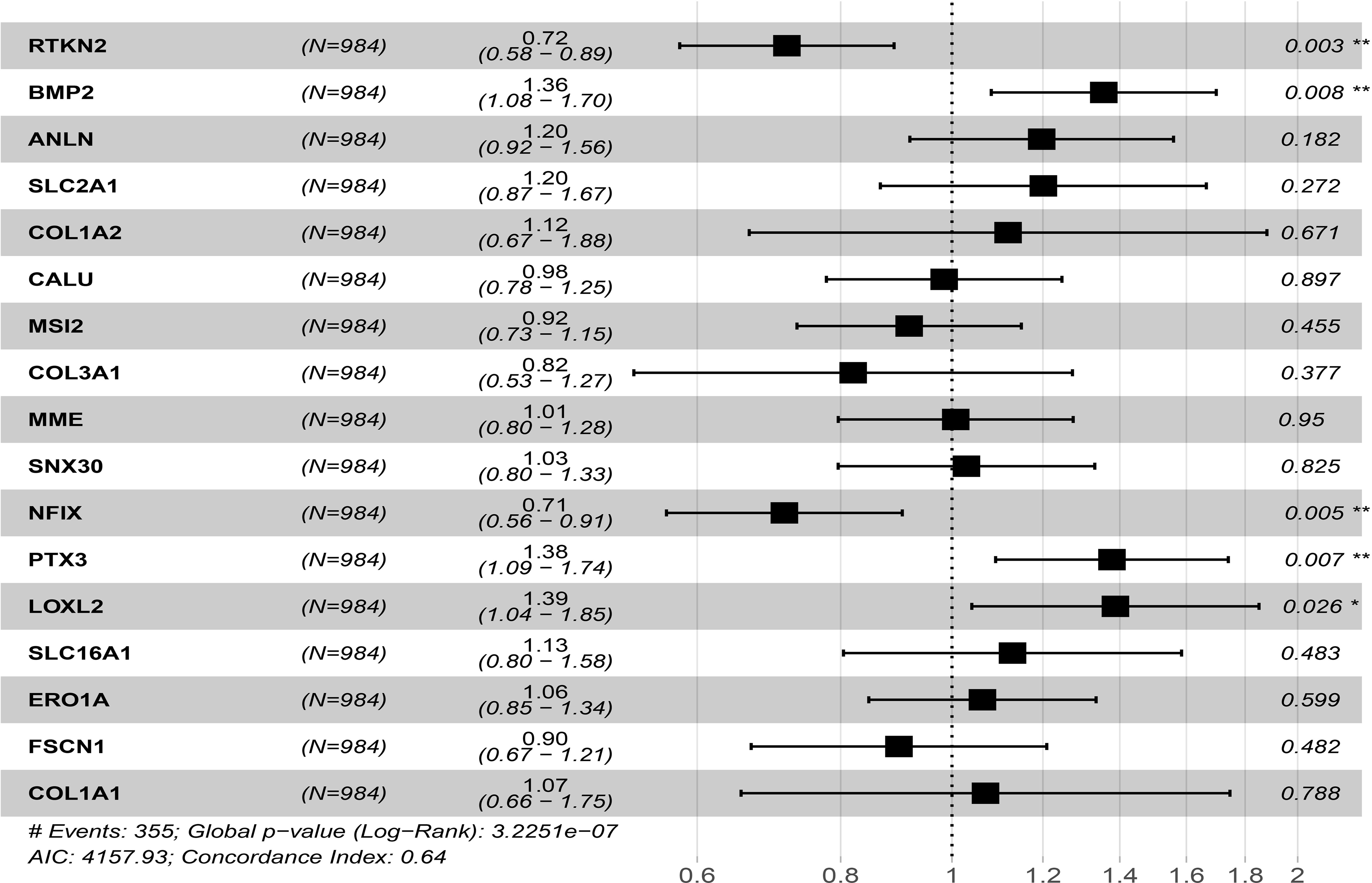
Figure 8 Univariate and multivariate cox regression analysis showed that prognostic factors of NSCLC patients in ceRNA network. *P < 0.05; **P < 0.01.
Verified the Expression and Potential Clinical Value of Independent Risk Factors in NSCLC Tissues
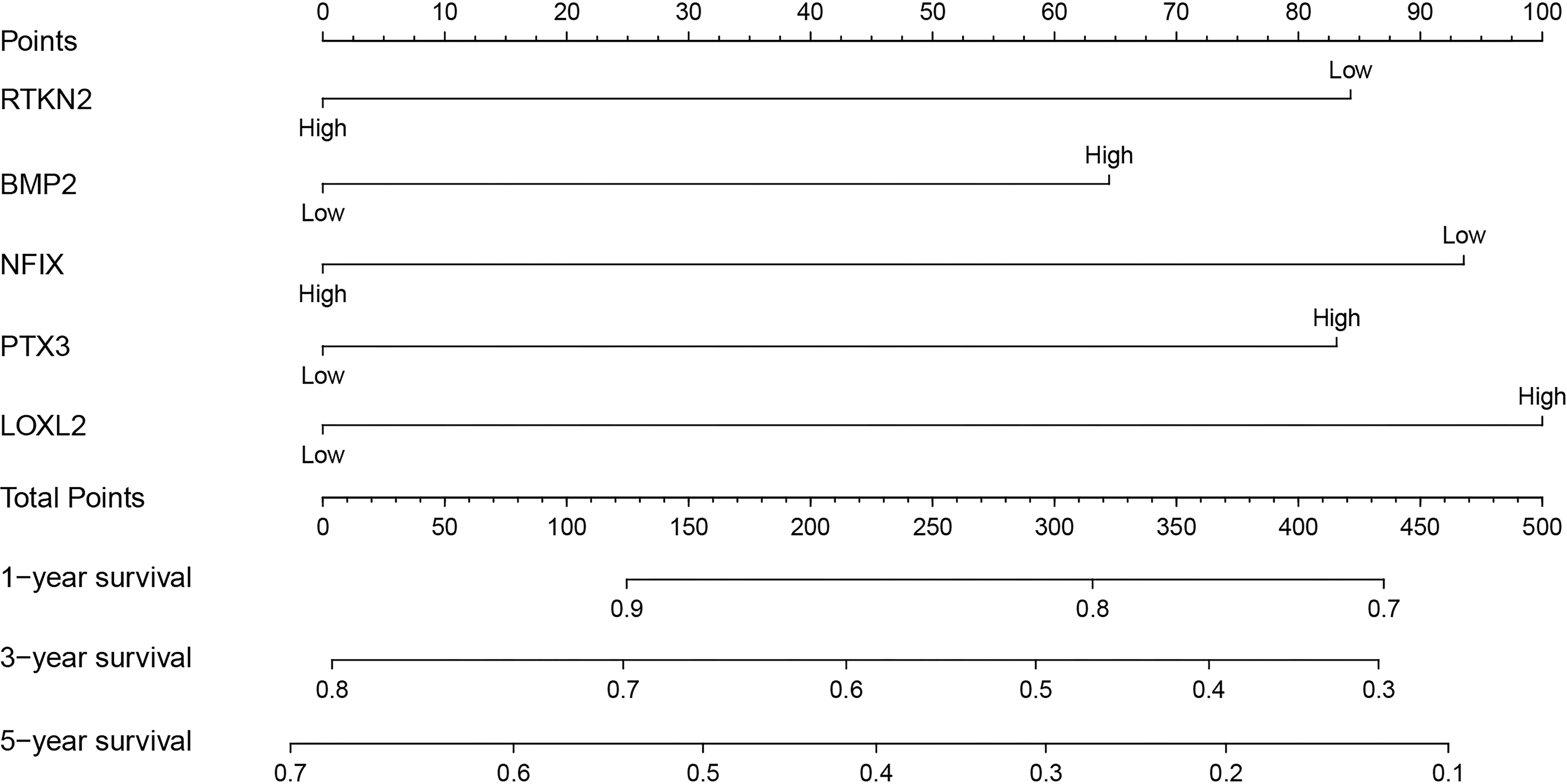
In the GEPIA database, we found that the expression levels of RTKN2, NFIX, PTX3 and BMP2 in NSCLC subtypes LUAD and LUSC tissues were significantly declined, while the expression of LOXL2 in NSCLC tissues increased (Figure S3). Figure S4 showed the relationship between RTKN2, NFIX, PTX3, BMP2, and LOXL2 mutation status and clinicopathological characteristics of NSCLC patients. In addition, the expression levels of RTKN2, NFIX, PTX3, BMP2 and LOXL2 were correlated with poor prognosis of NSCLC patients in the PrognoScan and Kaplan-Meier Plotter databases (Figure S5 and Table 4). Therefore, we grouped the median values of RTKN2, NFIX, PTX3, BMP2, and LOXL2 expression to construct a nomogram to assess the prognosis of patients (Figure 9).
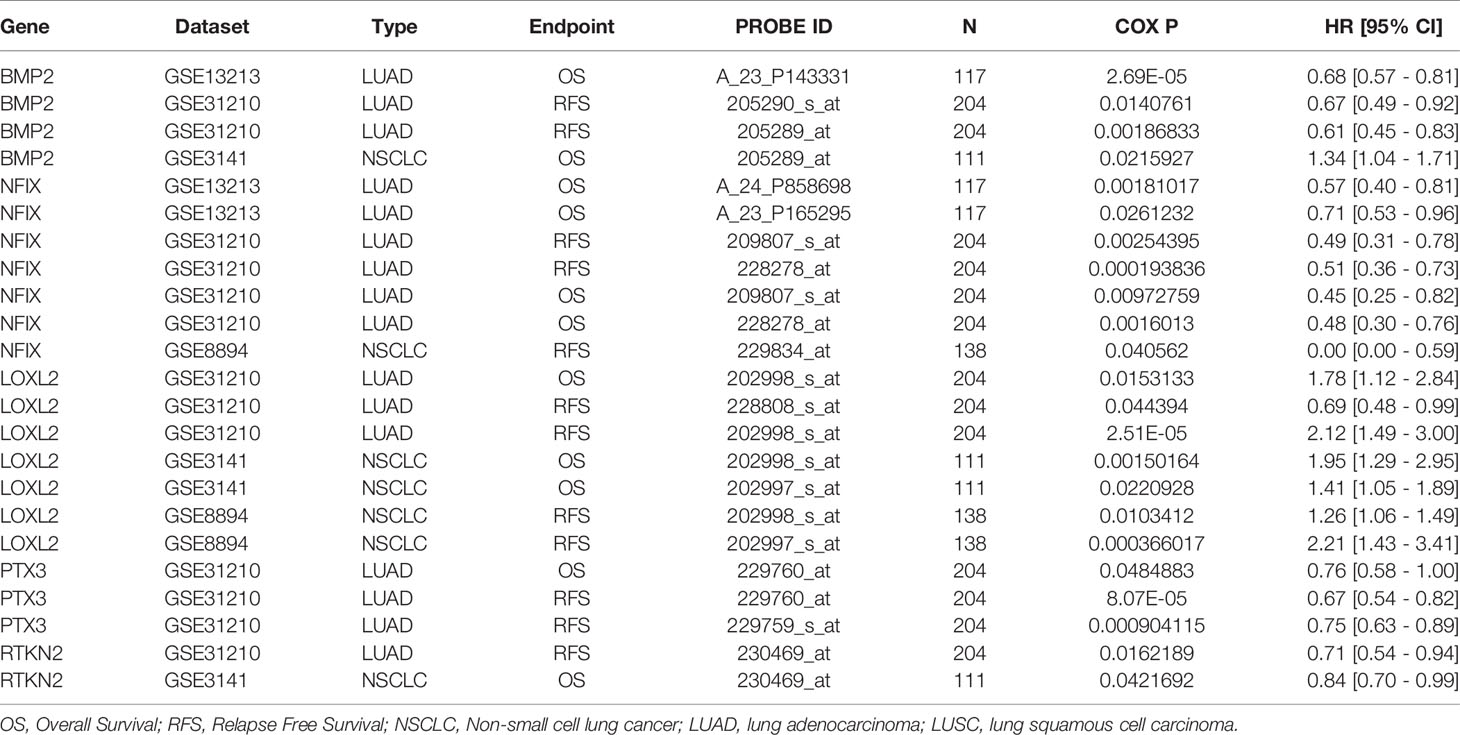
Table 4 The expression levels of RTKN2, NFIX, PTX3, BMP2 and LOXL2 were correlated with poor prognosis of NSCLC patients in the PrognoScan databases.
Discussion
Currently, it was urgent for us to improve the prognosis of NSCLC patients by finding novel and improved therapies. More and more evidences have showed that a large number of lncRNAs played extremely pivotal biological roles in the progression of NSCLC (20). For example, LncRNA KTN1-AS1, CASC9, ABHD11-AS1, LINC00467 could participate in the progress of NSCLC (20–23). Studies have reported that LINC00973 was related to cancer progression (18, 19, 24, 25). The expression level of LINC00973 was upregulated in cetuximab-resistant colorectal cancer H508/CR cells. Interfering the expression of LINC00973 decreased cell viability, increased cell apoptosis, and reduced glucose consumption and lactate secretion, which indicated that LINC00973 acted as a carcinogen (25). In addition, studies have reported that different doses of 5-fluorouracil, oxaliplatin, and irinotecan could impact the expression of LINC00973 in HT-29 and HCT-116 colon cancer cells. Combination medication increased the expression of LINC00973 in colon cancer cells (24). In our results, the expression of LINC00973 was elevated in NSCLC tissues. The high expression of LINC00973 was associated with poor prognosis in NSCLC patients, which indicated that LINC00973 might act as a carcinogen in the progression of NSCLC. As we all know, gefitinib and erlotinib were common anti-cancer drugs for lung cancer (26, 27), and Gefitinib and erlotinib could reduce the expression of carcinogenic LINC00973, which was expected to improve the prognosis of lung cancer patients. Therefore, LINC00973 might be a promising target for the development of new therapies for NSCLC.
lncRNAs could regulate the growth and migration behavior of cancer cells through a variety of mechanisms, including regulate expression of mRNA by ceRNA network, which could lead to its degradation (28, 29). For example, Cui et al. reported that the expression of lncRNA TRPM2-AS was significantly upregulated in NSCLC tissues and cells. Interfering with TRPM2-AS expression could inhibit cell proliferation, migration and invasion, and induce cell apoptosis. miR138-5p was the target downstream molecule of TRPM2-AS, and they showed a negative correlation. Interfering with TRPM2-AS expression could inhibit tumor formation, reduce the expression of EGFR and Ki67, and promote tumor cell apoptosis (28). At present, miR-374b-5p, miR-196a-5p, miR-138-5p, miR-150-5p, miR-196b-5p, miR-320a, miR-186-5p, etc. were related to the progress of NSCLC (30–38). For example, miR-138-5p could reduce the growth of NSCLC cells and increase the number of tumor-infiltrating dendritic cells (DCs). miR-138-5p down-regulates the expression of cyclin D3, CCD20, Ki67 and MCM in A549/3LL cells (37). The expression level of TSPAN12 was down-regulated in NSCLC tissues, and it was negatively correlated with the expression level of miR-196b-5p (35). However, LINC00973 was compatible with miR-374b-5p, miR-216b-5p, miR-196a-5p, miR-320c, miR-138-5p, miR-150-5p, miR-196b-5p, miR-224-5p. The relationship between miR-320a, miR-186-5p, miR-320b, miR-425-5p, miR-410-3p, miR-320d and miR-433-3p has not been reported yet, and we still need to conduct cell research to confirm.
RTKN2, NFIX, PTX3, BMP2 and LOXL2 of the ceRNA network play an important role in cancer progression (39–43). For instance, Ji et al. found that the expression level of RTKN2 in NSCLC tissues and cells was upregulated. Interfered with RTKN2 expression could induce NSCLC cell apoptosis and inhibit cell proliferation by increasing Bax levels and downregulating Bcl-2 levels. Interfered with RTKN2 expression could inhibit the migration and invasion of NSCLC cells by upregulating the expression of matrix metalloproteinase 9 (MMP9) and matrix metalloproteinase 2 (MMP2) (43). Huang et al. reported that elevated the levels of BMP2 were associated with short OS in NSCLC patients. BMP signaling was activated in the metastatic bone tumor of Lewis lung cancer in mice, and BMP2 signaling activation could enhance the bone metastasis of Lewis lung cancer, leading to poor prognosis. BMP2 secreted by mesenchymal fibroblasts could promote the migration and invasion of NSCLC cells (42). The expression of miR-504 in NSCLC tissues was significantly downregulated. The downregulation of miR-504 levels were positively correlated with lymph node metastasis and TNM stage. The overexpression of miR-504 significantly inhibited the proliferation, invasion and EMT process of NSCLC cells. miR-504 could bind to the 3’UTR region of LOXL2 and regulate its expression. Overexpression of LOXL2 could rescue the miR-504-induced inhibition of NSCLC cell proliferation and invasion (41). In our study, Univariate and multivariate Cox analysis found that the target molecules RTKN2, NFIX, PTX3, BMP2 and LOXL2 in the ceRNA network were independent risk factors for poor prognosis of NSCLC patients. This further showed that they had an important biological role in the progression of NSCLC.
In our study, we found that the expression of LINC00973 in NSCLC tissues was increased, and that the increased expression levels of LINC00973 were associated with the poor prognosis of NSCLC patients, and constructed the LINC00973-miRNA-mRNA ceRNA network competition mechanism. Most of the miRNA and mRNA in the competitive ceRNA network have been confirmed to have important biological roles in the progression of NSCLC. There were few studies on LINC00973 in cancer, but we first formally formalize the expression of LINC00973 in NSCLC tissues, and explore the ceRNA network regulation mechanism of LINC00973 to provide new candidate markers for the treatment of NSCLC. Then, our research needs to be verified by in vivo and in vitro experiments to confirm the role and value of LINC00973-miRNA-mRNA ceRNA in the progression of NSCLC.
Conclusion
This study found that LINC00973-miRNA-mRNA ceRNA network might be the basis for determining important post-translational regulatory mechanisms in the progression of NSCLC. BMP2, LOXL2, NFIX, PTX3 and RTKN2 might be valuable prognostic markers and potential therapeutic targets in the progression of NSCLC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics committee of the Affiliated Taihe Hospital of Hubei University of Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HL, TL, and XW conceived the research topic, made the research plan and directed the implementation of the whole research. QG drafted the manuscript together and processed the data. DL assisted to collect and analyze the data. XL and YY performed data checking and language polishing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Home for Researchers editorial team (www.home-for-researchers.com) for language editing service.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.684807/full#supplementary-material
Abbreviation
NSCLC, Non-small cell lung cancer; miRNA, microRNAs; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; lncRNA, long non-coding RNA; MRE, miRNA recognition elements; OS, overall survival.
References
1. Khan P, Siddiqui JA, Maurya SK, Lakshmanan I, Jain M, Ganti AK, et al. Epigenetic Landscape of Small Cell Lung Cancer: Small Image of a Giant Recalcitrant Disease. Semin Cancer Biol (2020). doi: 10.1016/j.semcancer.2020.11.006
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Woodman C, Vundu G, George A, Wilson CM. Applications and Strategies in Nanodiagnosis and Nanotherapy in Lung Cancer. Semin Cancer Biol (2021) 69:349–64. doi: 10.1016/j.semcancer.2020.02.009
4. Guo Q, Ke XX, Fang SX, Gao WL, Song YX, Chen C, et al. PAQR3 Inhibits Non-Small Cell Lung Cancer Growth by Regulating the NF-κb/P53/Bax Axis. Front Cell Dev Biol (2020) 8:581919. doi: 10.3389/fcell.2020.581919
5. Harrison PT, Vyse S, Huang PH. Rare Epidermal Growth Factor Receptor (EGFR) Mutations in Non-Small Cell Lung Cancer. Semin Cancer Biol (2020) 61:167–79. doi: 10.1016/j.semcancer.2019.09.015
6. Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, et al. Long Noncoding RNA LCAT1 Functions as a ceRNA to Regulate RAC1 Function by Sponging miR-4715-5p in Lung Cancer. Mol Cancer (2019) 18:171. doi: 10.1186/s12943-019-1107-y
7. Liu H, Han L, Liu Z, Gao N. Long Noncoding RNA MNX1-AS1 Contributes to Lung Cancer Progression Through the miR-527/BRF2 Pathway. J Cell Physiol (2019) 234:13843–50. doi: 10.1002/jcp.28064
8. Yin D, Lu X, Su J, He X, De W, Yang J, et al. Long Noncoding RNA AFAP1-AS1 Predicts a Poor Prognosis and Regulates Non-Small Cell Lung Cancer Cell Proliferation by Epigenetically Repressing P21 Expression. Mol Cancer (2018) 17:92. doi: 10.1186/s12943-018-0836-7
9. McCabe EM, Rasmussen TP. lncRNA Involvement in Cancer Stem Cell Function and Epithelial-Mesenchymal Transitions. Semin Cancer Biol (2020). doi: 10.1016/j.semcancer.2020.12.012
10. Ferragut Cardoso AP, Banerjee M, Nail AN, Lykoudi A, States JC. miRNA Dysregulation Is an Emerging Modulator of Genomic Instability. Semin Cancer Biol (2021). doi: 10.1016/j.semcancer.2021.05.004
11. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA Translation and Stability by microRNAs. Annu Rev Biochem (2010) 79:351–79. doi: 10.1146/annurev-biochem-060308-103103
12. Jin X, Guan Y, Zhang Z, Wang H. Microarray Data Analysis on Gene and miRNA Expression to Identify Biomarkers in Non-Small Cell Lung Cancer. BMC Cancer (2020) 20:329. doi: 10.1186/s12885-020-06829-x
13. Yu H, Cheng Y, Li W, Li Z, Wu P, Qiu S, et al. A Novel lncRNA-miRNA-mRNA Competitive Endogenous RNA Network for Uveal Melanoma Prognosis Constructed by Weighted Gene Co-Expression Network Analysis. Life Sci (2020) 260:118409. doi: 10.1016/j.lfs.2020.118409
14. Majed SO, Mustafa SA. MACE-Seq-Based Coding RNA and TrueQuant-Based Small RNA Profile in Breast Cancer: Tumor-Suppressive miRNA-1275 Identified as a Novel Marker. BMC Cancer (2021) 21:473. doi: 10.1186/s12885-021-08218-4
15. Smillie CL, Sirey T, Ponting CP. Complexities of Post-Transcriptional Regulation and the Modeling of ceRNA Crosstalk. Crit Rev Biochem Mol Biol (2018) 53:231–45. doi: 10.1080/10409238.2018.1447542
16. Cheng Y, Su Y, Wang S, Liu Y, Jin L, Wan Q, et al. Identification of circRNA-lncRNA-miRNA-mRNA Competitive Endogenous RNA Network as Novel Prognostic Markers for Acute Myeloid Leukemia. Genes (Basel) (2020) 11:8. doi: 10.3390/genes11080868
17. Hou W, Zhang Y. Circ_0025033 Promotes the Progression of Ovarian Cancer by Activating the Expression of LSM4 via Targeting miR-184. Pathol Res Pract (2021) 217:153275. doi: 10.1016/j.prp.2020.153275
18. Karpov DS, Spirin PV, Zheltukhin AO, Tutyaeva VV, Zinovieva OL, Grineva EN, et al. LINC00973 Induces Proliferation Arrest of Drug-Treated Cancer Cells by Preventing P21 Degradation. Int J Mol Sci (2020) 21:21. doi: 10.3390/ijms21218322
19. Liu Y, Li X, Zhang C, Zhang H, Huang Y. LINC00973 Is Involved in Cancer Immune Suppression Through Positive Regulation of Siglec-15 in Clear-Cell Renal Cell Carcinoma. Cancer Sci (2020) 111:3693–704. doi: 10.1111/cas.14611
20. Chen Z, Chen Q, Cheng Z, Gu J, Feng W, Lei T, et al. Long Non-Coding RNA CASC9 Promotes Gefitinib Resistance in NSCLC by Epigenetic Repression of DUSP1. Cell Death Dis (2020) 11:858. doi: 10.1038/s41419-020-03047-y
21. Li C, Zhao W, Pan X, Li X, Yan F, Liu S, et al. LncRNA KTN1-AS1 Promotes the Progression of Non-Small Cell Lung Cancer via Sponging of miR-130a-5p and Activation of PDPK1. Oncogene (2020) 39:6157–71. doi: 10.1038/s41388-020-01427-4
22. Zhu Y, Li J, Bo H, He D, Xiao M, Xiang L, et al. LINC00467 Is Up-Regulated by TDG-Mediated Acetylation in Non-Small Cell Lung Cancer and Promotes Tumor Progression. Oncogene (2020) 39:6071–84. doi: 10.1038/s41388-020-01421-w
23. Xue L, Li J, Lin Y, Liu D, Yang Q, Jian J, et al. M(6) A Transferase METTL3-Induced lncRNA ABHD11-AS1 Promotes the Warburg Effect of Non-Small-Cell Lung Cancer. J Cell Physiol (2021) 236:2649–58. doi: 10.1002/jcp.30023
24. Zinovieva OL, Grineva EN, Prokofjeva MM, Karpov DS, Zheltukhin AO, Krasnov GS, et al. Expression of Long Non-Coding RNA LINC00973 Is Consistently Increased Upon Treatment of Colon Cancer Cells With Different Chemotherapeutic Drugs. Biochimie (2018) 151:67–72. doi: 10.1016/j.biochi.2018.05.021
25. Jing C, Ma R, Cao H, Wang Z, Liu S, Chen D, et al. Long Noncoding RNA and mRNA Profiling in Cetuximab-Resistant Colorectal Cancer Cells by RNA Sequencing Analysis. Cancer Med (2019) 8:1641–51. doi: 10.1002/cam4.2004
26. Thomas P, Vincent B, George C, Joshua JM, Pavithran K, Vijayan M. A Comparative Study on Erlotinib & Gefitinib Therapy in Non-Small Cell Lung Carcinoma Patients. Indian J Med Res (2019) 150:67–72. doi: 10.4103/ijmr.IJMR_1896_17
27. Zhang Y, Fu F, Hu H, Wang S, Li Y, Hu H, et al. Gefitinib as Neoadjuvant Therapy for Resectable Stage II-IIIA Non-Small Cell Lung Cancer: A Phase II Study. J Thorac Cardiovasc Surg (2021) 161:434–442.e2. doi: 10.1016/j.jtcvs.2020.02.131
28. Cui D, Feng Y, Shi K, Zhang H, Qian R. Long Non-Coding RNA TRPM2-AS Sponges microRNA-138-5p to Activate Epidermal Growth Factor Receptor and PI3K/AKT Signaling in Non-Small Cell Lung Cancer. Ann Transl Med (2020) 8:1313. doi: 10.21037/atm-20-6331
29. Ren XY, Han YD, Lin Q. Long Non-Coding RNA MIR155HG Knockdown Suppresses Cell Proliferation, Migration and Invasion in NSCLC by Upregulating TP53INP1 Directly Targeted by miR-155-3p and miR-155-5p. Eur Rev Med Pharmacol Sci (2020) 24:4822–35. doi: 10.26355/eurrev_202005_21171
30. Wang J, Shi C, Wang J, Cao L, Zhong L, Wang D. MicroRNA-320a Is Downregulated in Non-Small Cell Lung Cancer and Suppresses Tumor Cell Growth and Invasion by Directly Targeting Insulin-Like Growth Factor 1 Receptor. Oncol Lett (2017) 13:3247–52. doi: 10.3892/ol.2017.5863
31. Bao M, Pan S, Yang W, Chen S, Shan Y, Shi H. Serum miR-10a-5p and miR-196a-5p as Non-Invasive Biomarkers in Non-Small Cell Lung Cancer. Int J Clin Exp Pathol (2018) 11(2):773–80.
32. Zhao W, Sun Q, Yu Z, Mao S, Jin Y, Li J, et al. MiR-320a-3p/ELF3 Axis Regulates Cell Metastasis and Invasion in Non-Small Cell Lung Cancer via PI3K/Akt Pathway. Gene (2018) 670:31–7. doi: 10.1016/j.gene.2018.05.100
33. Zeng Z, Zhao G, Rao C, Hua G, Yang M, Miao X, et al. Knockdown of lncRNA ZFAS1-Suppressed Non-Small Cell Lung Cancer Progression via Targeting the miR-150-5p/HMGA2 Signaling. J Cell Biochem (2019). doi: 10.1002/jcb.29542
34. Li J, Zhang X, Tang J, Gong C. MicroRNA-374b-5p Functions as a Tumor Suppressor in Non-Small Cell Lung Cancer by Targeting FOXP1 and Predicts Prognosis of Cancer Patients. Oncol Targets Ther (2020) 13:4229–37. doi: 10.2147/ott.S243221
35. Liang G, Meng W, Huang X, Zhu W, Yin C, Wang C, et al. miR-196b-5p-Mediated Downregulation of TSPAN12 and GATA6 Promotes Tumor Progression in Non-Small Cell Lung Cancer. Proc Natl Acad Sci USA (2020) 117:4347–57. doi: 10.1073/pnas.1917531117
36. Liu X, Zhou X, Chen Y, Huang Y, He J, Luo H. miR-186-5p Targeting SIX1 Inhibits Cisplatin Resistance in Non-Small-Cell Lung Cancer Cells (NSCLCs). Neoplasma (2020) 67:147–57. doi: 10.4149/neo_2019_190511N420
37. Song N, Li P, Song P, Li Y, Zhou S, Su Q, et al. MicroRNA-138-5p Suppresses Non-Small Cell Lung Cancer Cells by Targeting PD-L1/PD-1 to Regulate Tumor Microenvironment. Front Cell Dev Biol (2020) 8:540. doi: 10.3389/fcell.2020.00540
38. Wu Z, Li W, Li J, Zhang Y, Zhang X, Xu Y, et al. Higher Expression of miR-150-5p Promotes Tumorigenesis by Suppressing LKB1 in Non-Small Cell Lung Cancer. Pathol Res Pract (2020) 216:153145. doi: 10.1016/j.prp.2020.153145
39. Zhan P, Shen XK, Qian Q, Zhu JP, Zhang Y, Xie HY, et al. Down-Regulation of Lysyl Oxidase-Like 2 (LOXL2) Is Associated With Disease Progression in Lung Adenocarcinomas. Med Oncol (2012) 29:648–55. doi: 10.1007/s12032-011-9959-z
40. Infante M, Allavena P, Garlanda C, Nebuloni M, Morenghi E, Rahal D, et al. Prognostic and Diagnostic Potential of Local and Circulating Levels of Pentraxin 3 in Lung Cancer Patients. Int J Cancer (2016) 138:983–91. doi: 10.1002/ijc.29822
41. Ye MF, Zhang JG, Guo TX, Pan XJ. MiR-504 Inhibits Cell Proliferation and Invasion by Targeting LOXL2 in Non Small Cell Lung Cancer. BioMed Pharmacother (2018) 97:1289–95. doi: 10.1016/j.biopha.2017.11.005
42. Huang F, Cao Y, Wu G, Chen J, Lin W, Lan R, et al. BMP2 Signalling Activation Enhances Bone Metastases of Non-Small Cell Lung Cancer. J Cell Mol Med (2020) 24:10768–84. doi: 10.1111/jcmm.15702
Keywords: LINC00973, non-small cell lung cancer, prognosis, biomarker, ceRNA
Citation: Guo Q, Li D, Luo X, Yuan Y, Li T, Liu H and Wang X (2021) The Regulatory Network and Potential Role of LINC00973-miRNA-mRNA ceRNA in the Progression of Non-Small-Cell Lung Cancer. Front. Immunol. 12:684807. doi: 10.3389/fimmu.2021.684807
Received: 31 March 2021; Accepted: 25 June 2021;
Published: 29 July 2021.
Edited by:
Hubing Shi, Sichuan University, ChinaReviewed by:
Yanyang Liu, Sichuan University, ChinaVirginia Tirino, Università della Campania Luigi Vanvitelli, Italy
Copyright © 2021 Guo, Li, Luo, Yuan, Li, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huasong Liu, aHVhc29uZzgxOUBxcS5jb20=; Xinju Wang, eGN3eGowMDZAb3V0bG9vay5jb20=; Tian Li, Zm1tdWx0QGZveG1haWwuY29t
 Qiang Guo
Qiang Guo Dan Li2
Dan Li2 Tian Li
Tian Li