- 1Department of Viroscience, Postgraduate School of Molecular Medicine, Erasmus MC, University Medical Centre Rotterdam, Rotterdam, Netherlands
- 2Medical Library, Erasmus MC, University Medical Centre Rotterdam, Rotterdam, Netherlands
Respiratory tract infections (RTI) are a major cause of morbidity and mortality in humans. A large number of RTIs is caused by viruses, often resulting in more severe disease in infants, elderly and the immunocompromised. Upon viral infection, most individuals experience common cold-like symptoms associated with an upper RTI. However, in some cases a severe and sometimes life-threatening lower RTI may develop. Reproducible and scalable in vitro culture models that accurately reflect the human respiratory tract are needed to study interactions between respiratory viruses and the host, and to test novel therapeutic interventions. Multiple in vitro respiratory cell culture systems have been described, but the majority of these are based on immortalized cell lines. Although useful for studying certain aspects of viral infections, such monomorphic, unicellular systems fall short in creating an understanding of the processes that occur at an integrated tissue level. Novel in vitro models involving primary human airway epithelial cells and, more recently, human airway organoids, are now in use. In this review, we describe the evolution of in vitro cell culture systems and their characteristics in the context of viral RTIs, starting from advances after immortalized cell cultures to more recently developed organoid systems. Furthermore, we describe how these models are used in studying virus-host interactions, e.g. tropism and receptor studies as well as interactions with the innate immune system. Finally, we provide an outlook for future developments in this field, including co-factors that mimic the microenvironment in the respiratory tract.
Introduction
Respiratory tract infections (RTIs) are a major source of morbidity and mortality in humans (1). Almost 300 million episodes of lower RTIs (e.g. pneumoniae and bronchitis) occurred in 2015 and about 3 million people die each year. This places RTIs amongst the leading causes of death worldwide (2, 3). A large fraction of RTIs is caused by viruses (50 to 90%) (4). Respiratory viruses include human rhinovirus (HRV), influenza A and B virus (IAV and IBV), human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), human coronavirus (HCoV), human parainfluenzavirus (HPIV), and human adenovirus (HAdV). Most respiratory viruses have single-stranded RNA genomes, with the exception of adenoviruses that have double-stranded DNA genomes. The highest morbidity and mortality due to RTIs is seen in infants, elderly and immunocompromised individuals, but also healthy individuals without underlying risk factors can be affected (5). Furthermore, RTIs have been associated with exacerbations of asthma or chronic obstructive pulmonary disease (COPD) (6, 7).
Recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has captured global headlines as the causative agent in the coronavirus disease 2019 (COVID-19) pandemic (8), highlighting the impact of respiratory viruses on global health. As this pandemic has shown, options for prophylaxis and treatment of viral respiratory infections are limited. For some viruses, such as influenza virus or adenovirus, vaccines are available but both efficacy and coverage are suboptimal. Antiviral drugs against acute respiratory virus infections often have limited efficacy. Therefore, development of novel and improved antiviral drugs and vaccines remains of high priority to improve global health.
The human respiratory tract (RT) is the primary site where respiratory viruses enter, replicate, disseminate and cause disease. These viruses are transmitted by aerosols and/or droplets. RTIs in most cases start by infection of airway epithelial cells in the upper respiratory tract (URT) (nasal cavity, pharynx, larynx), and are associated with common cold-like symptoms, including rhinitis, sore throat, runny nose, and nasal congestion. During the course of infection, the lower respiratory tract (LRT) (trachea, primary bronchi, lungs) can become involved, causing more severe disease, such as pneumonia or bronchitis (9). How severe lower RTIs develop and why only some individuals are affected could be a stochastic process related to dose and route of the inoculum. However, the development of severe LRTIs remains a black box and is an important topic of investigation. Many of these studies are performed in vivo, which provides important data on the pathogenesis of respiratory viruses [reviewed elsewhere (10–14)] but has some downsides, such as differences between animal or human host factors, ethical concerns and practical challenges. In vitro models have as advantages that they originate from the relevant host species, express the relevant host factors, are of less ethical concern and easier to work with than animal models. Therefore, a reproducible and scalable in vitro culture system that accurately represents the human RT would be valuable to study respiratory virus infections and test new treatments. Here we investigate the differences between commonly used in vitro primary airway cell culture models and how these models are used for studying respiratory virus infections. The first part of this review describes several characteristics of the RT, then we will review the evolution of in vitro cell culture systems in the context of viral RTIs, starting from advances after immortalized cell lines to recently developed stem cell (SC)-based organoid cultures. Finally, an outlook is provided for future developments in this field.
Methodology and Study Design
This systematic review was prepared in accordance with PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) guidelines (15). A systematic literature search, on the 21st of January of 2021, was conducted using title, abstract, index term and author keyword fields for respiratory viruses and human airway epithelial cells (HAEC) or their variations. Of note, primary olfactory epithelial cells were not included, since there are described previously (16). Embase, Medline (Ovid), and Web of Science Core Collection databases (Supplementary Material 1) were searched from inception until 21st January 2021. Search results were restricted to English and excluded case-reports. Articles were imported and deduplicated in EndNote™. Two researchers independently screened title and abstract for study eligibility criteria (17). Consensus was achieved after discussion upon disagreement. The electronic search was performed in Embase, Medline and WoS (Supplementary Material 2).
Results
943 references were obtained and screened on basis of title and abstract (Figure 1). After the first screening, studies using only animal material, material of diseased patients, or immortalized cell lines were excluded. In addition, studies performed with respiratory bacteria (e.g. Heamophilus influenzae) and where adenoviruses were used as a vector were excluded. For the second screening full-text articles were assessed for eligibility and divided into seven topics: pathogenesis (N=165), signal transduction (N=93), co-cultures (N=21), zoonosis (N=11), reviews (N=32), drug/molecule testing (N=145), and disease (e.g. cystic fibrosis and chronic obstructive pulmonary disease) (N=28). Studies regarding signal transduction, drug/molecule testing and diseases such as chronic obstructive pulmonary disease and cystic fibrosis were excluded at this stage (n=504). Finally, 151 studies were selected for this systematic review. To provide sufficient background information we also included 69 additional papers based on relevant content for this review, resulting in a final selection of 220 papers.
Discussion
The Respiratory Tract
The RT can be divided into separate sections, either based on physiology (URT versus LRT, Figure 2A) or the type of respiratory epithelium (Figure 2B). The URT includes the nasal cavity, mouth, larynx and pharynx (throat), and beginning of the trachea (18). In the nasal cavity inspired air is warmed and humidified before it travels down the RT. Additionally, the nasal cavity houses the olfactory receptors that bind molecules resulting in impulses to the brain that enable smell. The nasal cavities drain into the nasopharynx that is in turn attached to the trachea (19). The trachea connects the throat to the LRT, starting at the primary bronchial branches (20, 21). The two bronchial branches bifurcate further into smaller tubes called the bronchi and bronchioles, which end in the alveoli; tiny air sacs facilitating gas exchange (22, 23). The entire RT is covered by epithelium and it is estimated that an adult of 176 cm or an infant of 60 cm have a lung epithelial surface of 78 m2 or 4 m2, respectively (24). The airway epithelium has a crucial barrier and immune functions. The mucus covering the airway epithelium is the first barrier that respiratory viruses have to pass to reach the airway epithelial cells. The airway epithelium itself has essential inflammatory, immune and regenerative capacities to combat these viruses.
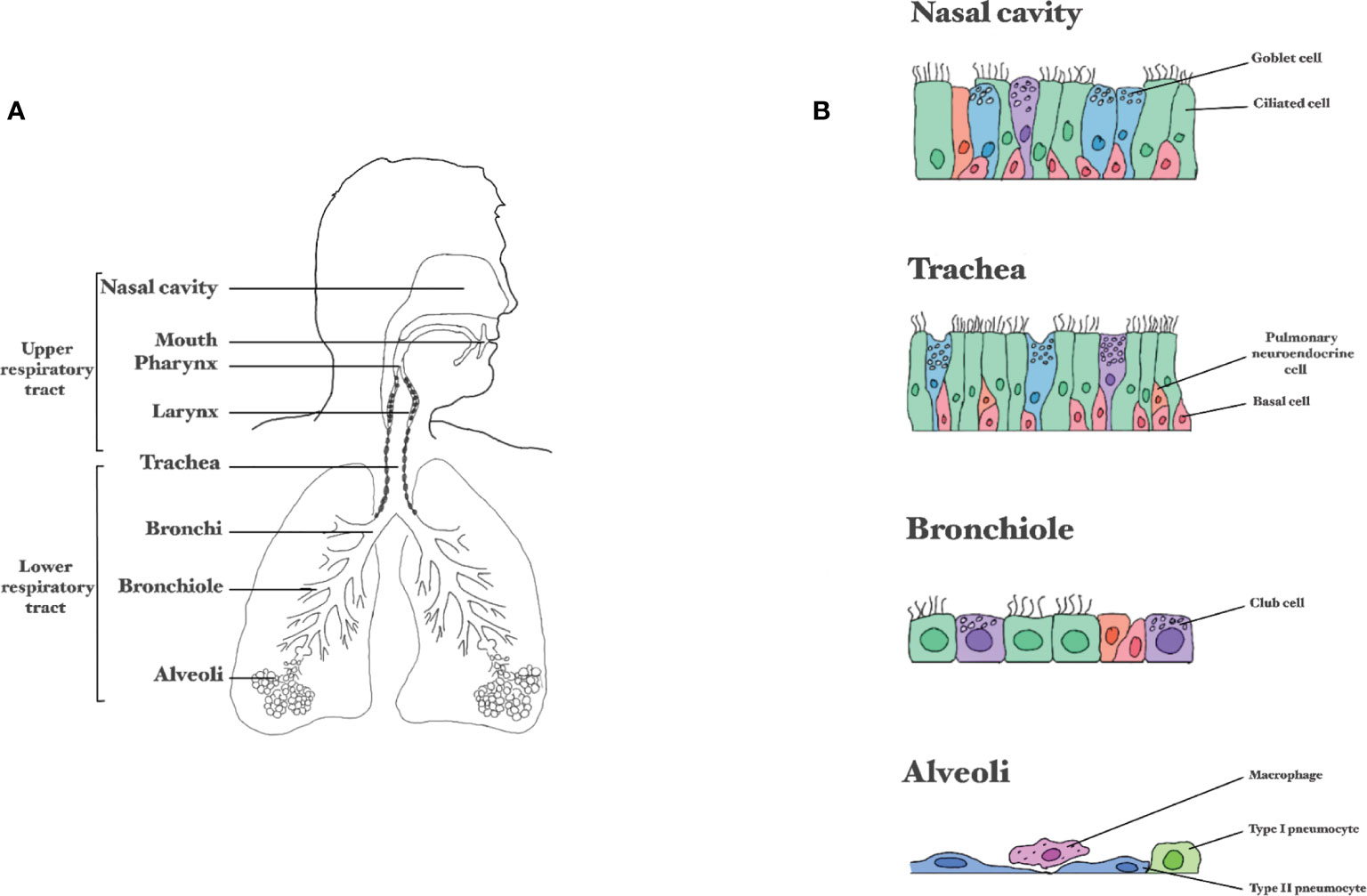
Figure 2 Composition of the respiratory tract. (A) Division between the upper and lower respiratory tract. (B) Schematic representation of epithelial layer in the different parts of the respiratory tract.
The airway epithelium is composed of several cell types held together by tight junctions and adherens junctions that form a barrier against invading pathogens. The nasal cavity is lined with two distinct kinds of epithelium: the olfactory epithelium (1-2% of the nasal epithelium) and the respiratory epithelium (>98% of the nasal epithelium) (19). The neurons in the olfactory epithelium in the nose are directly connected to the central nervous system via the olfactory nerve. Viruses that infect the olfactory epithelium can use this nerve as a shortcut to reach the central nervous system (25). The epithelial layer in the nasal atrium consists of multi-layered keratinized squamous epithelium and further down the nose this becomes multi-row cylindrical epithelium. Many respiratory epithelial cells are also ciliated, facilitating mucus transport towards the pharynx and trachea (19). From the trachea downwards, the RT is lined with ciliated pseudostratified columnar epithelium (20). In the bronchioles, the epithelium gradually changes from pseudostratified into simple cuboidal and in the alveolar ducts and alveoli there is simple squamous epithelium (mainly type alveolar type I cells) (20).
Ciliated cells account for 50% to 80% of the airway epithelial cells, and have 200-300 motile cilia on their surface to displace mucus, enabling muco-ciliary clearance (26). The cells producing mucus are goblet cells, comprising around 9% of the respiratory epithelial cells (27). Next to goblet cells there are club cells; cuboidal non-ciliated non-mucous secretory cells. Club cells secrete extracellular matrix components, and can serve as progenitor cells for themselves and for ciliated cells (28). These cells are not very abundant in the URT, but in the terminal bronchioles account for approximately 11-22% of the cells (28). Epithelial and mucus-producing cells are supported by airway basal cells. Whereas the epithelial and mucus-producing cells are terminally differentiated and cannot renew, basal cells possess stem-cell like properties (29, 30). Basal cells occupy 31% of the respiratory epithelial cell population, the density of basal cells decreases when descending into the small airways (31). Another important cell type of the airways is the pulmonary neuroendocrine cell, comprising 1 out of 2500 epithelial cells from the trachea onto the alveolar ducts (32), forming the lung self-renewing SC niche relevant in airway epithelial regeneration (33). The alveolar epithelium consists mostly of two cell types, alveolar type I and type II cells (ATI and ATII). ATI are very thin, squamous cells, accounting for over 90% of the surface area in the lungs and provide an efficient barrier for air exchange (34). About 7% of the surface area comprises ATII, which are smaller and cuboidal, and function mostly in the production and uptake of lung surfactant (35). ATIIs can differentiate into ATI (36). In addition to ATI and ATII, also immune cells, like alveolar macrophages, are present in the alveoli.
Besides a barrier function, respiratory epithelial cells have inherent innate immunity functions. They recognize respiratory viruses via pattern recognition receptors, eventually leading to the production of cytokines and chemokines that render the cells and their neighboring cells in an antiviral state (37). The bridge between this innate response and adaptive immunity is formed by dendritic cells (DCs). There are tissue-resident DCs, that form an integrated network within the respiratory epithelium. Upon activation, for instance by a virus infection, they can travel to the lymph nodes to initiate an adaptive immune response. There are also migratory DCs that can be attracted to the site of infection and aid here in the local immune response. Besides DCs, natural killer cells, innate lymphoid cells, T cells and B cells orchestrate the immune response in the respiratory epithelium (38). Eventually, all elements contribute to an effective integrated immune response.
In Vitro Models
Multiple in vitro respiratory cell culture systems have been developed to study the interaction between the different airway cell types and respiratory viruses. To date, the majority of these models is based on immortalized cell lines. Although useful in the study of direct viral infection and replication mechanisms at a cellular and molecular level, such monomorphic and unicellular systems fall short in creating an understanding of the processes that occur at an integrated tissue level (39, 40). We need more advanced models to better understand infection dynamics in relation to intrinsic cellular resistance mechanisms, including the innate immune response, and other factors such as microbiome and immune cells. New in vitro models have been proposed and used for some time, especially involving the use of primary HAEC. HAEC are derived from surgical material or brushings (nose or throat) that are subsequently cultured in the laboratory. More recently, 3D-cultures such as airway organoids (AO), formed from SCs, have been developed and hold promise as a useful tool to study host-pathogen interactions. However, this technique is still in its infancy, laborious and expensive. To perform in-depth studies into interactions between the host airway and respiratory viruses, the ultimate goal is to create a reproducible, scalable, feasible and economic in vitro culture system that faithfully recapitulates the architecture of the RT as well as the dynamics of infection.
Primary Respiratory Epithelial Cells
HAEC are obtained from human respiratory tissue, which can originate from different anatomical sites of the RT (41). Respiratory tissue is now obtained in different ways: during lung transplantation, during tissue resection in cancer patients, during other surgical procedures (e.g. turbinoplasty/-ectomy or nasal polypectomy) or from cadaveric explants (41). In addition, nasal and bronchial brushings, which are less invasive, can be performed to obtain HAEC (42). HAEC from healthy donors are now commercially available (43). Additionally, material from, for instance, COPD patients and/or smokers is also available (44, 45). Acquiring HAEC from various donors allows us to study and compare the RT of both healthy and diseased individuals.
Undifferentiated Primary Respiratory Epithelial Cells
Primary undifferentiated HAEC (HAECun) are relatively easy to culture, but can only be passaged a few times. After obtaining tissue, the primary airway epithelial cells are directly isolated and cultured. For the isolation of primary airway epithelial cells from lung transplants or biopsies, the tissue is cut into smaller pieces and then dissociated via the addition of a protease-containing digestion cocktail, followed by generation of uniform single-cell suspensions (46, 47). Tissue obtained from brushings can be cultured directly (47). After isolation, HAEC can be used in this undifferentiated form for experiments. In addition to normal culture flasks or plates, HAEC can also be seeded on a collagen-coated semi-permeable membrane (transwell) (48). In this transwell system, medium is present on both the apical and the basolateral side. When a 100% confluent monolayer has formed to separate the apical and basolateral compartments, the HAECun can be used for experiments. These HAECun are not polarized and these cultures do not have ciliated cells or goblet cells, and therefore lack important characteristics of the airways.
Differentiated Primary Respiratory Epithelial Cells
Although more challenging to culture, differentiated HAEC (HAECdif) represent the RT more faithfully than their undifferentiated counterparts. To differentiate primary respiratory epithelial cells in the transwell system, medium is removed from the apical compartment after the confluent monolayer has formed, creating an air-liquid interface (ALI). This, in combination with specific growth factors, induces differentiation of these cells over a timeframe of 3-4 weeks (48). Eventually, a polarized, pseudostratified respiratory epithelium is formed, containing basal cells, ciliated cells, and goblet cells or club cells (depending on the anatomical location) (48, 49). These HAECdif resemble the human RT anatomically (50, 51) but a continuous airflow, blood flow, and the presence of immune cells are still lacking.
Considerations for Use of Primary Cells
Comparing studies using primary HAEC is difficult, because of differences in donor variability (1), anatomic source of the cells (2), culture methods (e.g. medium and growth factors) (3) and the use of undifferentiated versus differentiated cells (4) (Figure 3).
1. Variability in culturability, morphology and phenotype between primary HAEC obtained from different donors has been described. This donor variability can depend on multiple factors, like age, gender, smoking history, or obesity (52–56). Variation in infection and replication levels of viruses between donors was regularly observed (57–60). Explanations could be differences in: receptor presence or distribution, cytokine responses, donors and environmental factors (61, 62). It is dependent on the research question how important these dissimilarities are.
2. In addition to donor variability, the source of the cells (ranging from nose to alveoli) can make comparison of studies difficult (63, 64), because of variations in susceptibility to viral infections and virus-host interactions (9). The preferential anatomical site should be determined by the research question (e.g. study URT or LRT) and comparing cells of multiple regions of the RT for susceptibility, virus replication and host responses.
3. The importance of culture medium is highlighted in a HRSV infection study, where two different media for differentiation of pediatric HAEC (nasal) were used: PneumaCultTM and PromocellTM. The former led to an overall greater total number of cells, but the proportion of ciliated and goblet cells was similar. The replication kinetics of HRSV was similar in both cultures, but in the culture with PneumoCultTM more ciliated cells were infected. HRSV infection of the PneumoCultTM cultures led to a higher IFNλ secretion, which could be due to the increased infection of ciliated cells (65).
4. Several studies describe different expression patterns of cellular receptors that can mediate virus entry, resulting in varying infection percentages or different immune response between HAECun and HAECdif (66–69).
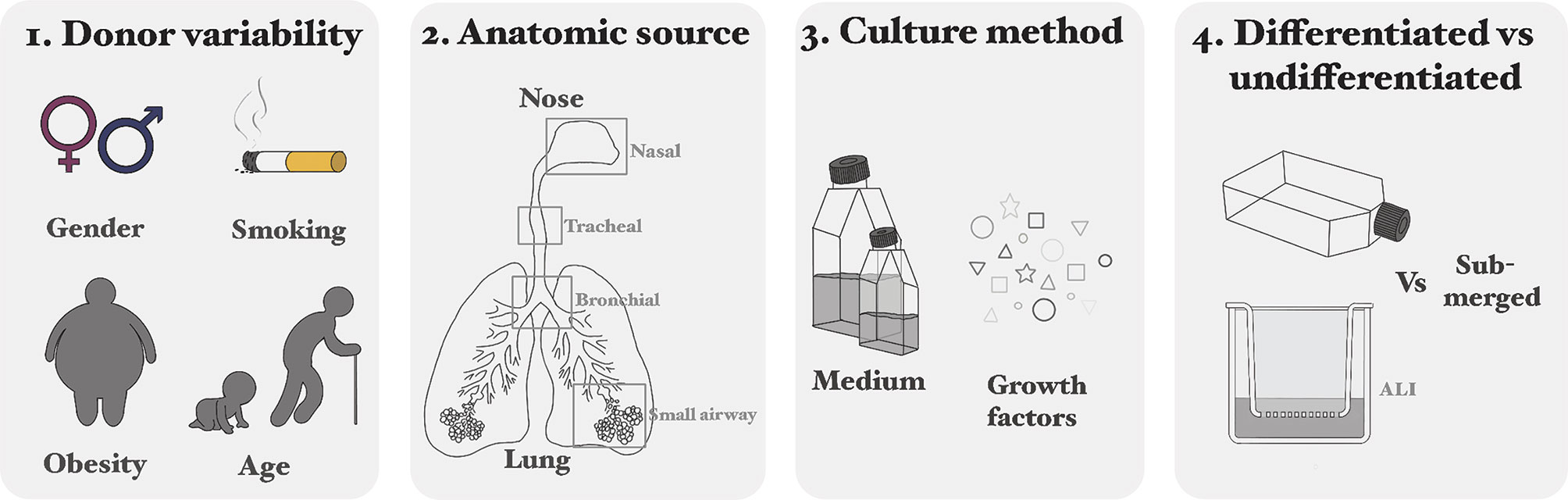
Figure 3 Considerations for using primary cells: differences in donor variability (1), anatomic source of the cells (2), culture methods (e.g. medium and growth factors) (3) and the use of undifferentiated versus differentiated cells (4).
HAECun can be passaged approximately three times prior to differentiation, after which loss of epithelial integrity, differential gene expression, or senescence occurs (70–72). This restricts the number of experiments that can be performed. That is why immortalized variants of HAEC have been created. Jonsdottir and colleagues (2019) described that insertion of the Rho-associated protein kinase (ROCK) inhibitor Y-27632 via a lentiviral vector increased the longevity of the primary respiratory epithelial cells (73). Alternatively, exogenous induction of human telomerase reverse transcriptase (hTERT) can be used to prolong primary cell life (74). Immortalization resolves the issue of the restricted number of experiments, but it remains unknown if all the characteristics of the respiratory epithelial cells remain the same. And, even in these immortalized variants, senescence can occur after a few passages (75, 76). Genetic engineering of HAEC is challenging and may compromise translational aspects of the model.
In conclusion, primary HAEC can be extracted from all parts of the airways and differentiated into a pseudostratified monolayer resembling the in vivo situation. Although several variables and parameters must be considered, primary HAEC are a promising culture system to study RTIs.
Stem Cell-Based Models
SC-based models have the potential to overcome the problems associated with the limited lifespan of primary HAEC. SCs can be used to make organoids, which are three dimensional cultures that can self-organize and renew (77). Organoid culture is a novel and innovative technology and was first described in 2009 with organoids from the gut (78). In 2012, the technique to culture AO was developed (79). Until now, human AO are mostly made from SCs obtained from adult lungs. Adults tissues are more accessible than embryonic SCs or induced pluripotent (iPSCs) and have less ethical restraints. Nevertheless, AO from embryonic SCs are also used (80, 81). Although most studies describe AO obtained from the lung, AO can be made from almost all parts of the RT, and are thus a suitable alternative to primary HAEC (82). Additionally, genetic modifications are also more feasible in AO than in primary HAEC.
Different SCs can be used to generate AO. First, embryonic SCs obtained from the inner cell mass of an early staged embryo have the potential to form every cell of the human body (83). However, the use of these cells remains ethically controversial. Second, iPSCs can also differentiate into almost every cell type of the human body (84). To obtain these iPSCs, somatic cells (for example skin cells) are reprogrammed to the progenitor state by addition of several transcription factors. However, specific factors, including growth factors and cytokines, are needed for differentiation into respiratory epithelial cells (85), which has so far not been successful. The challenges to find the optimal factors to differentiate these iPSCs into airway epithelial cells can be overcome by using organ-specific adult SCs. These cells can differentiate into the cell types of the respective organ (86). In this case, airway epithelial SCs can be isolated from tissue or URT brushings (87). The isolated basal cells can subsequently be used for the formation of 3D undifferentiated AO, using Matrigel and medium with specific growth factors. These AO, cultured either in 3D in matrigel or on transwell filters at ALI, can then be differentiated into cultures that represent the cells of the RT, such as basal cells, goblet cells, ciliated cells or alveolar cells (81, 87–92) (Figure 4A). In a differentiated spheroid AO, the basal cells are on the outside of the sphere, the goblet cells excrete their mucus into the lumen while the cilia move the mucus around (see Figure 4B). Similar to primary cell cultures, there are indications that culture medium influences organoid morphology and behavior, which is something that has to be studied in more detail (92, 93).
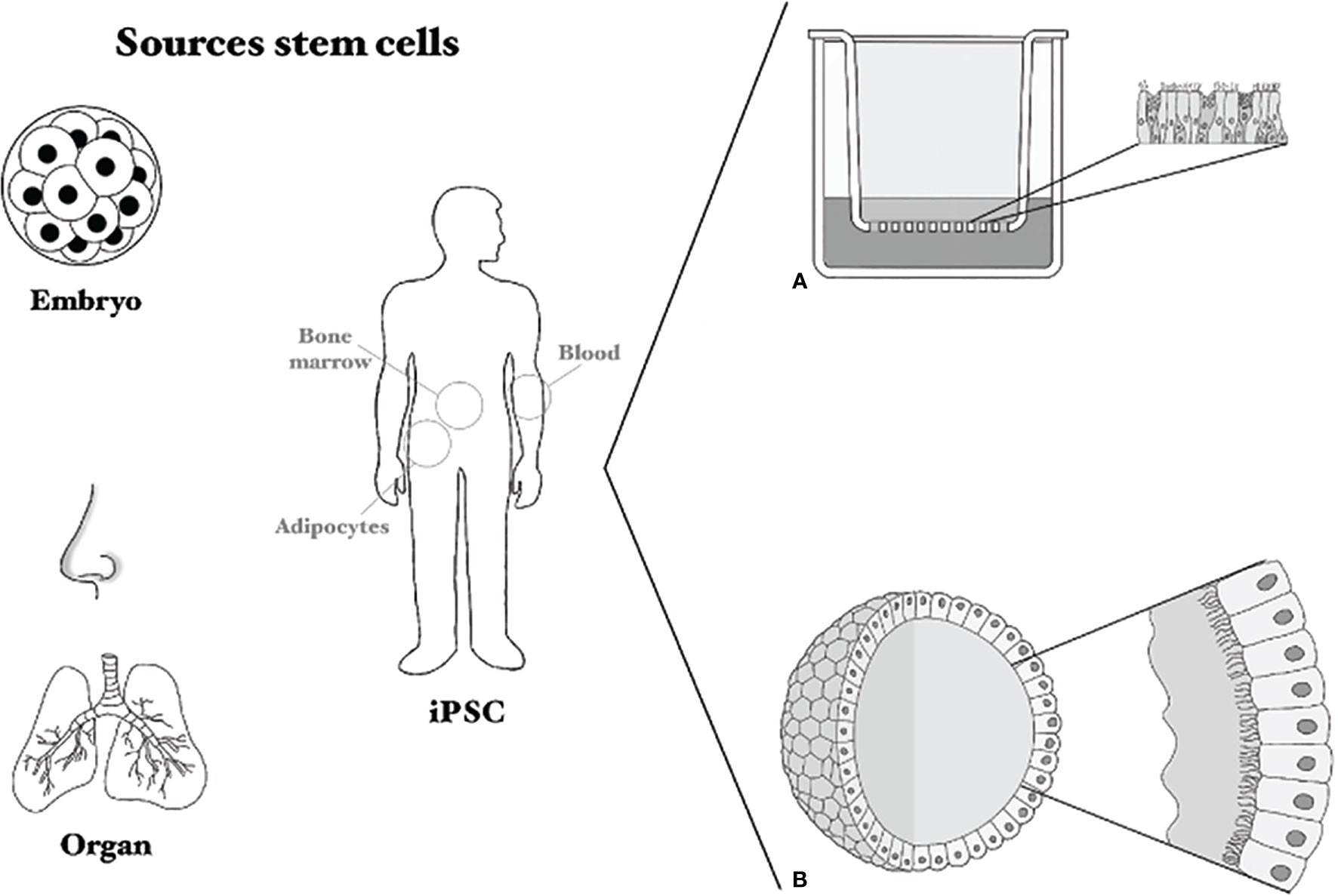
Figure 4 Sources of stem cells and culture methods. Several sources for stem cells are shown: embryonic stem cells, induced pluripotent stem cells (iPSC) and organ-derived or nasal brushing-derived. (A) Shows a Transwell system on which primary cells or stem-cell based cells can be cultured, (B) shows an airway organoid sphere with cilia and mucus on the inside.
AO appear to be a promising model mimicking the human RT and once established can be maintained for a long period of time. Limited studies have been performed with AO and respiratory viruses so far due to the novelty of the technique. However, the studies that have been performed show great promise for studying virus-host interaction and drug screening studies in this model.
Studying Respiratory Virus Infections
To understand the pathogenesis of respiratory virus infections, it is important to study (1) tropism, (2) receptor usage, (3) immune response, (4) cytopathic effects and damage, and (5) tissue regeneration after clearance. Primary cell culture models are crucial in these studies, because viruses rapidly adapt to culture in immortalized cell lines (81, 94, 95). In this second part of the review, we summarize how these properties of respiratory viruses have been investigated in light of the advantages of using primary HAEC and AO (Table 1).
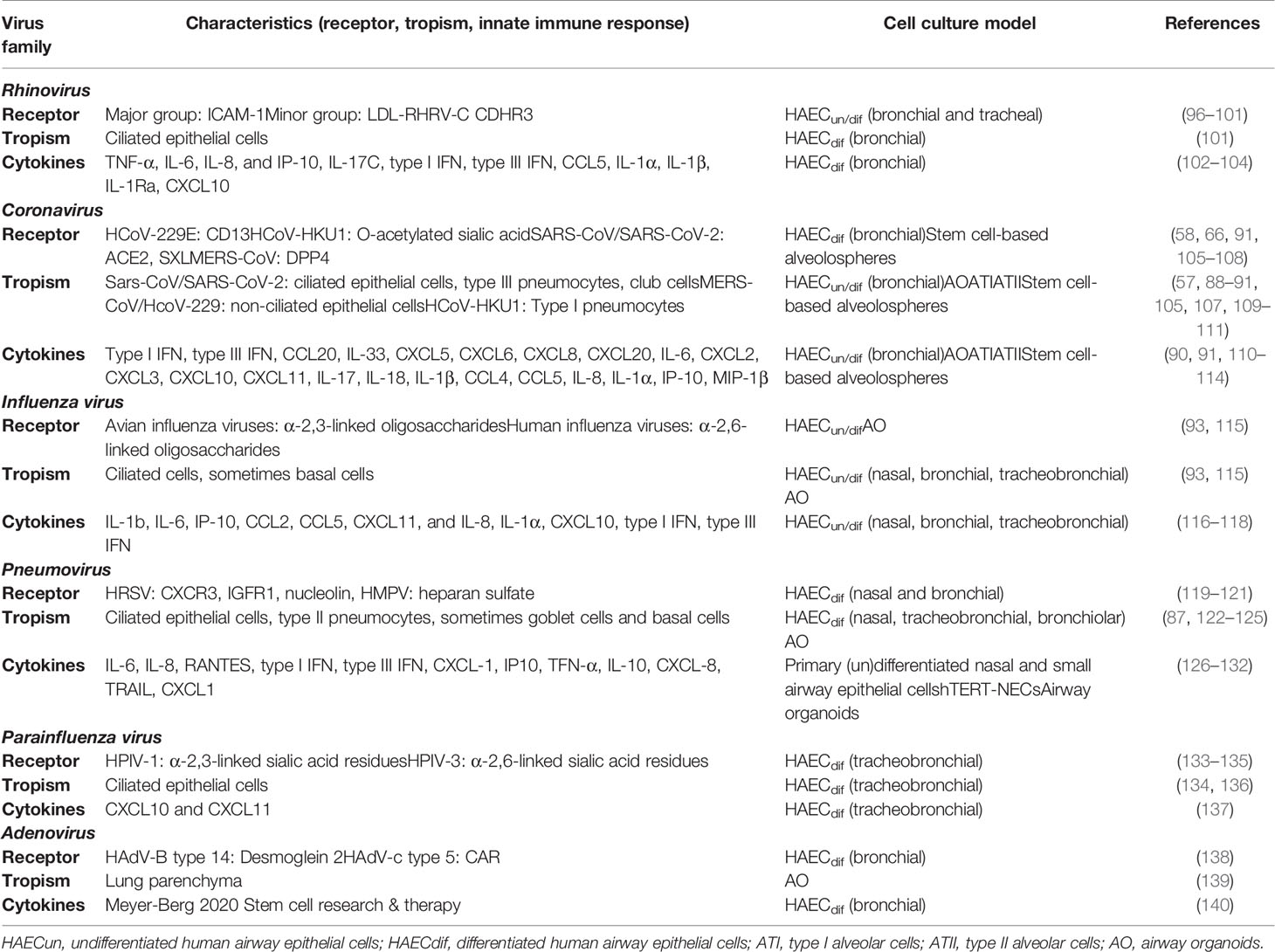
Table 1 Summary table of the studies on respiratory viruses performed in primary respiratory epithelial cells and stem cell-based models.
Receptor Studies
One of the mechanisms by which viruses enter a host cell is receptor-mediated entry; the appropriate receptor needs to be present on the cell for the virus to enter. Immortalized cell lines are often used to identify such a cellular receptor, for example by genomic- and interactome- based approaches (141, 142). Primary HAEC mimic the natural targets cells of respiratory viruses better than continuous cell lines and are thus important for receptor identification, as illustrated in the following examples.
For most HCoVs, receptors were initially identified in cell lines, followed by confirmation and verification of these receptors in primary HAEC (58, 66, 91, 105–107, 143–148). An example of the importance of using primary HAEC was seen in the search for a model for HCoV-HKU1. This virus only replicates in primary differentiated tracheobronchial epithelial cells. Through studies in these cells, the HCoV-HKU1 receptor (O-acetylated sialic acid) was ultimately identified (108, 149). Differential binding of avian and human influenza viruses to their receptors was investigated in primary HAECdif (bronchial), which express both α-2,3- and α-2,6-linked oligosaccharides. Infections with avian and human influenza viruses confirmed that viruses with avian origin preferably bind to α-2,3-linked oligosaccharides, whereas viruses from human origin bind to α-2,6-linked oligosaccharides (133). For most HRV subtypes their receptor was identified in immortalized cell lines and validated in HAEC, except for HRV-C (96–99, 150–153). For HRV-C the receptor was found in primary differentiated bronchial epithelial cells and in ex vivo organ transplants, since this HRV subtype does not replicate in continuous cell lines (101, 154).
Many HRSV receptor candidates have been proposed in literature, but arguably the relevant in vivo receptor has not yet been identified. Ciliated epithelial cells are the main target cell of HRSV (155), therefore primary HAECdif are the most suitable model to perform receptor studies. CXC3R1, insulin-like growth factor-1 (IGFR1) and nucleolin have been identified as potential cellular receptors for HRSV. CXC3R1 would probably not have been found if it were not for primary HAEC, since this receptor is absent (unless transfected) on immortalized cell lines (119, 120). The other two receptors have been confirmed and validated in primary HAEC (121, 156). Although closely related to HRSV, the proposed receptors for HMPV are not the same as for HRSV, but studies with primary respiratory epithelial cells from different locations in the RT have proposed integrins and heparin sulfate as receptors (126, 127, 157, 158).
Lastly, using primary HAECdif (tracheobronchial), it was confirmed that HPIV-3 uses α-2,6-linked sialic acid residues and HPIV-1 uses α-2,3-linked sialic acid residues as cellular receptors (133, 134). Primary HAEC have also been used to identify or verify the receptors for HAdV (138, 159).
All the above-mentioned examples clearly illustrate that HAEC are important for identifying the cellular entry receptor for respiratory viruses. Although initially the receptor is often found in cell lines, primary cell models are essential in confirming and validating this receptor and these results translate better to the in vivo situation.
Tropism
Regarding viral tropism, in vivo models and ex vivo culture models provide valuable information. However, it has to be kept in mind that in in vivo studies the animal is often not the natural host of the virus, introducing a possible bias. Human post-mortem material from either healthy or infected individuals is also useful for binding studies, but this material is not always available. Cell lines are immortalized and often express unique gene expression patterns that are not representative of in vivo cell types. Primary airway models can overcome these hurdles and are a thus powerful tool to investigate viral tropism, since most cell types from the RT are represented in these models.
In the SARS-CoV-2 pandemic, differentiated AO (AOdif) and primary HAEC rapidly helped elucidate that SARS-CoV-2 mainly infects ciliated cells, club cells and ATII, but not goblet cells (88–91, 109, 110). For other coronaviruses, similar models were used to determine their respective tropism, for instance HCoV-229E and MERS-CoV infect non-ciliated cells and HCoV-HKU1 predominantly infects ATII (105, 107, 160, 161). Multiple studies have been performed with influenza viruses in HAEC or AO. It was found that, for example, seasonal H1N1 was able to infect ciliated cells, goblet cells and alveolar cells, whereas pandemic H1N1 was also able to infect club cells (89, 93, 115). HRV is a common cause of respiratory infections in humans and both HRV-A and HRV-B can be cultured in conventional cell models, while this is not the case for HRV-C. However, by using 3D Matrigel cultures with primary HAECdif HRV-C can be propagated (159). For all three HRV subtypes the tropism could be determined in primary HAECdif (bronchial): HRV-A and HRV-B infect basal cells and HRV-C infects ciliated epithelial cells (101, 162). It was also confirmed that HRV can replicate in cells from both the URT (adenoids) and LRT (bronchus) (163). Clinical characteristics of HRV are also recapitulated in primary airway models. For instance, HRV-B infections are associated with less severe infections and studies in primary HAECdif (nasal and bronchial) reported that HRV-B replicates slower and leads to less cytotoxicity than HRV-A and HRV-C (164, 165). For both HRSV and HMPV it was confirmed in HAECdif and AOdif mainly targets the ciliated epithelial cells (122, 123, 128, 166–168). One study showed that basal cells could also be infected by HRSV after epithelial injury, using commercially obtained primary HAECdif (bronchial) (124).
In conclusion, with primary airway cultures or SC-based cultures it is possible to culture viruses that are difficult to culture in immortalized cell lines and these models can provide reliable information on the viral tropism.
Disease Modelling
Studying pathogenesis is challenging in vitro, both in continuous and primary cell lines. A single or limited number of cell types are present in in vitro cultures, not resembling a full organism. As alternative, cytopathic effect (CPE) can be studied. CPE in primary HAECdif often resembles the natural situation more closely compared to continuous cells. HRSV offers a good example: in continuous cell lines it forms big syncytia (hence the name), but in primary HAEC HRSV forms little to no syncytia, which is in line with observations in vivo (both animal models and humans) (48, 122, 129, 168–170). A clinical observation in HRSV disease is neutrophil inflammation, which is involved in severe HRSV disease. This clinical phenomenon has been mimicked in HRSV-infected primary HAECdif (nasal) that were co-cultured with neutrophils (171). AO have significantly improved disease modelling opportunities. HRSV, HMPV and HPIV have been investigated in AOdif: and for example for HRSV epithelial cell sloughing was shown, which was absent in HPIV infections. These observations both fit with clinical observations. Other hallmarks of HRSV and HMPV disease, such as infection of ciliated epithelial cells and mucus production were also recapitulated in AO and in primary HAEC (80, 87, 125, 155, 172). These hallmarks are impossible to mimic in 2D cell lines and thus highlight the usefulness of 3D AO to study respiratory virus pathogenesis.
Innate Immune Responses
When a virus enters a cell, this cell will directly mount an innate immune response, both in the infected cell and in surrounding cells. This usually includes the production of several cytokines and chemokines to attract more immune cells and activate the adaptive immune system. Studying the innate immune system in primary HAEC has as an advantage over continuous cell lines, because they potentially produce more clinically relevant cytokines that would also be produced by the natural target cells. For instance, for SARS-CoV it was described that ATII are less susceptible than ATI, which was linked to a robust innate immune response (161). This pronounced innate immune response was not found in primary HAECdif (bronchial, chemically differentiated) after MERS-CoV, SARS-CoV or HCoV-229E infection, indicating immune evasion (173). It appears that each corona-virus induces specific innate immune responses, with few cytokines and ISGs overlapping (90, 91, 110, 112–114). In response to influenza virus infection, similar cytokines, mainly IL-1β, IL-6 and IL-8, were produced in HAECdif from different parts of the RT (114, 116–118). For HRSV and HMPV infections it has also been shown that similar cytokines, predominantly type I and/or type III IFN, are produced in HAECdif (nasal, bronchial and small airway) (123, 126–132, 168).
Different subtypes of HPIV are associated with certain clinical observations: HPIV1 can stay undetected for days, whereas HPIV2 is associated with mild disease and HPIV3 with more severe respiratory disease. These clinical observations have been linked to cytokine profiles that were found in primary HAECdif (tracheobronchial). It was shown that for HPIV1 there was no early innate immune response, whereas for HPIV2 innate cytokines were produced at early time points and for HPIV3 cytokines increased overtime (61).
It was observed that HRV induced an innate immune response in 3D matrigel cultures with primary HAECdif that inhibited viral replication, but that this was only short-lived because a second peak of viral replication was measured. This correlated with the production of the cytokines TNF-α, IL-6, Il-8, and IP-10. Based on these results, the authors postulated that the second peak appeared due to the production of new virus, after the eclipse caused by the innate immune system (102). Similar replication kinetics and cytokines were seen in other studies, including type I and type III IFN (103, 104, 174, 175). In one study it was shown that upon HRV infection IL-17C was produced in the basolateral compartment and induced CXLC1, which is a neutrophil attractant and may contribute to exacerbations of lower airway disease (103).
To summarize, many studies underline the importance of using primary HAEC and AO when studying innate immune responses. Although these models seem to be a good proxy for clinically relevant cytokines in vivo, studies directly comparing the innate cytokine responses between continuous cell lines and primary cell models are lacking. Nevertheless, complex cell systems that mimic the in vivo target cells of viruses probably model the antiviral innate immune response better than cell lines.
Other Applications of Primary Respiratory Epithelial Cells
Primary HAEC can mimic host factors related to lifestyle, such as smoking history and obesity. Smoking history might predispose to more severe SARS-CoV-2 disease, possibly to the upregulation of the ACE2 receptor, which was found in formalin-fixed paraffin-embedded human lung tissues and in brushings of the airway epithelium (176–178). This difference was not observed in primary HAECdif (bronchial), but the number of donors was limited (179). Obesity is related to more severe influenza symptoms, which has been shown in primary ATII, where cells from obese donors were more susceptible to a pandemic IAV than non-obese donors (52).
Primary HAEC are also useful for studying stability of viruses under environmental conditions. For example, primary HAECdif (bronchial) were used to study the impact of relative humidity on the stability of a pandemic influenza A virus in aerosols and droplets. Aerosols and droplets were created and supplemented with extracellular matrix material harvested from the apical surface of the HAEC and then the amount of virus was determined in MDKC cells (180). None of the experimental humidity conditions affected the stability of IAV H1N1 when aerosols and droplets were supplemented with extracellular matrix, but without supplementation, a humidity-dependent decay of virus infectivity was observed (180). This provided evidence that extracellular matrix can protect influenza viruses from decay and thereby promote spreading (181, 182).
Primary HAECdif have also been proposed as a screening model for evaluating and monitoring the infectivity, pathogenicity and antigenicity of virus variants, for instance during an outbreak. In the current SARS-CoV-2 pandemic many new, and supposedly more infectious, variants are arising. Information on these variants is important, so that interventions can be tailored. Using primary HAEC, it was found that SARS-CoV-2 variant (D614G) replicates better than the original wild-type strain in the nasal and proximal airways, which was not observed in VeroE6 cells. Increased replication in the URT might lead to more transmission (183). During influenza seasons of 2013-2014 and 2015-2016 the vaccine effectiveness against the circulating influenza strains was reduced. The reason for this was a reduction in sustained multi-cycle replication. This was found in primary HAECdif (nasal), but not in MDCK cells (182). These important data regarding novel virus variants would not have been available without primary airway models and are important for managing pandemics and developing interventions.
In this section, we illustrated that primary cell models may serve as an authentic model for in vitro respiratory viral pathogenesis studies, recapitulating viral infection in the host. They are useful for identifying viral receptors, determining viral tropism, mimicking disease and assessing innate immune responses in respiratory epithelial cells. Additionally, some viruses that are difficult to culture can be propagated in primary HAEC and often not in cell lines (184). Although continuous cell lines can give seminal insights into virus-host interactions, they also render selective pressure on viruses, leading to culture adaptations. One of the advantages of continuous cell lines is the possibility for genetic modification, which is thus far impossible in primary respiratory cell cultures. This hurdle can be overcome in the future by using AO derived from human SCs, which could be engineered to express desired host features.
Co-Cultures
As described above, in vitro primary cell models can be used to investigate different aspects of virus-host interactions. However, still a big part of the micro-environment of the cells is not reflected in these models. (1) The lungs fill up with air and deflate during breathing, creating an airflow and stretching of the epithelium. (2) To supply the lungs with sufficient nutrients and to take up incoming oxygen, endothelial blood vessels are connected to the respiratory epithelial cells. (3) The airways are also normally colonized with all different kinds of micro-organisms, referred to as the microbiome. (4) Also, several types of immune cells are present to fight potential pathogens. These four features are usually not recapitulated in HAEs nor AOs, but are important for an integrated in vitro airway model.
Endothelial Cells
Endothelial blood vessels are connected to the airway epithelial cells in vivo and therefore important to also include in an in vitro airway model. Co-cultures with endothelium have been performed, with primary HAEC placed on a chip and microvascular endothelial cells on the opposite side of the porous membrane, with a fluid flow underneath (185). This small airway-on-a-chip model recapitulates tissue-tissue interactions, physicochemical microenvironments, and vascular perfusion of the RT (185, 186). Unfortunately, these models are costly and often not compatible with a biosafety level (BSL)-II or BSL-III laboratory environment required for performing pathogenic virus infections. A way to mimic air flow has been addressed by applying mechanical forces to the airway-on-a-chip model, recreating a breathing movement (187). In another study, primary HAEC were cultured at the apical side of transwell filters and primary microvascular endothelial cells at the basolateral side to create an alveolar-capillary system. This system was used to study influenza virus and staphylococcus aureus (SA) co-infections (188). Thus, although progress is being made, the use of transwell filters and airway-on-a-chip models to model the epithelial-endothelial barrier to study respiratory virus pathogenesis is still limited.
Microbiome
Next to co-cultures with endothelial cells, co-culture models with bacteria and/or viruses to mimic co-infections or the microbiome are being developed. One option is an infection with a bacterium preceding viral infection. In HAECdif (alveolar), co-cultured with primary microvascular endothelial cells, it was found that methicillin-resistant SA (MRSA) dysregulated the host immune response and decreased the barrier function (188). Another option is a viral infection preceding an infection with a bacterium, for instance initial IAV infection resulted in increased replication of S. pneumonia in primary HAECdif (bronchial). In contrast, in primary HAEC it was then found that pre-exposure to HRV reduced the viral titers of subsequent influenza virus exposure, which was supported by clinical data (189). Co-culturing cells with bacteria poses a challenge, since these micro-organisms replicate fast and rapidly overgrow the respiratory epithelial cells, often leading to cell death. In some of these studies, bacteria were inactivated to overcome this problem, but it is questionable if this still mimics the real-life situation since several features of the bacteria are lost (190). However, it has been shown that inactivated bacteria can influence respiratory virus infections. For instance, UV-inactivated nontypable Heamophilus influenzae (NTHI) enhanced the susceptibility of human bronchial epithelial cells to HPIV, likely due to upregulation of ICAM-1, a cellular receptor for HPIV (191, 192). Another way around the toxicity problem could be only co-culturing bacteria for a short time. Obviously, this does not mimic the microbiome or a co-infection (193–196). Nonetheless, this still allowed investigation of bacterial adherence to the (infected) primary HAEC or transmigration of bacteria. Alternatively, co-cultures could be regularly washed. Using this method researchers were able to culture NTHI with commercially available primary HAEC for 10 days (197). Combining co-cultures of bacteria and AO have, to our knowledge, not been described yet, but lessons can be learned from co-cultures of bacteria and intestinal organoids or skin organoids (197, 198).
It is important to study the effect of bacteria and viruses on the innate immune state of the HAEC. Certain innate immune profiles influence the outcome of viral infections, for instance for HRSV. It was described, in humans, that neutrophilic inflammation at the time of viral challenge predisposed individuals to symptomatic HRSV infection (199). These data suggest that co-cultures with, for instance, neutrophils might be required to fully mimic the microenvironment of the RT. In conclusion, although there is only limited data available on this topic, more studies are being performed and we are confident it is feasible to create an in vitro airway model combined with the microbiome.
Immune Cells
Another important feature of the airways is the (intra-epithelial) presence of immune cells that are often involved in the defense against respiratory virus infections. Co-cultures of neutrophils with primary HAECun (trachea) that were infected with HPIV were already performed in the 1990s (200, 201). These studies showed enhanced adhesion of neutrophils to HPIV-infected cells. This has later been shown in AO infected with HRSV, where it was even possible to visualize the preferential neutrophil movement to the infected AO. Co-cultures of primary HAEC (nasal or bronchial) with DCs or T cells have also been described, either with virus present prior to adding the cells, or after. In these studies, researchers investigated both the effect on the immune responses as well as viral replication (87, 202, 203). They showed that the presence of DCs or T cells enhanced antiviral and inflammatory responses and inhibited viral replication. A triple co-culture has also been described, in which primary HAECdif (bronchial) were co-cultured with monocyte-derived DCs (moDCs) and macrophages (204). Here, the moDCs were infected with HRSV and added apically to a layer of HAEC in a transwell system. Uninfected moDCs were simultaneously added to the basolateral membrane of the insert. This led to the transmission of HRSV to the HAEC, but not to the moDCs located basolaterally. However, when macrophages were added to the apical surface of this co-culture, the basolateral moDCs were infected too, indicating some type of trans-epithelial transport mechanism. This study shows the importance of having multiple factors in a culture to understand the infection process (205). All in all, combining primary HAECdif with endothelial cells, airflow, the microbiome and immune cells would be an ideal model to mimic our RT.
Conclusion and Future Directions
We need scalable, feasible, affordable and reproducible in vitro models that adequately reflect the complexity of our RT and the cascade of events that occur during viral infections. We illustrated in this review that primary HAEC and AO are promising models for the RT and for studying respiratory viral pathogenesis. Since the field of primary epithelial cell models and AO is rapidly progressing, future applications of these models are endless. Currently these models are used for testing and developing effective treatments for SARS-CoV-2 (206). Another potential future application of primary HAEC is using species-specific cells to study the zoonotic potential of viruses. With these models it is possible to test the infectivity of the viruses isolated from the natural hosts in primary human respiratory epithelial cells and vice versa (207–210).
Furthermore, primary HAEC and AO can be used for drug screening studies, to test the potential of a drug, small molecule or antibody to inhibit virus replication in the context of a more representative tissue environment. There are many papers describing drug studies performed in HAEC (211). In the future, the use of AO for drug screening studies can be of great value as well (212).
The ultimate goal is to design a model that reflects the respiratory environment in all critical aspects to understand respiratory virus infections and host interaction in detail. Therefore, the usage of self-organized AO models would be preferable over the use of primary HAEC, either in 2D at ALI or 3D. Although initially more expensive and laborious to set up, such AO cultures can be maintained for long periods and are expected to generate the most reproducible results. However, primary HAEC are still a good model to use, especially for rapid screening of inter-individual variation of different donors in response to respiratory virus infections, or fast initial drug screening.
A state-of-the-art development is the lung-on-a-chip model. This model allows co-culture of primary respiratory epithelial cells, for instance primary HAEC or AO, with endothelial cells and mimics breathing by using an airflow and stretching. This model might recapitulate the human airways most faithfully, but there are still many challenges to be overcome before this can be used widely. This model is expensive, and requires specific expertise and equipment. One of the biggest hurdles is using the lung-on-a-chip model in a BSL-II or higher laboratory environment. However, the first study using a lung-on-a-chip model in combination with a respiratory virus (Influenza A) has been published recently showing the potential of this model (213).
In conclusion, primary HAEC are better suited for investigating basic virus-host interactions, such as receptor use and tropism of a respiratory virus than cell lines. However, for investigating the more complex interplay between virus, target cells and immune responses, AO appear to be more suitable. Further improving this model by introducing additional system factors, such endothelium and airflow (as has been done for lung-on-a-chip models), commensal bacteria and immune cells are required to even more closely mimic the micro-environment present in the RT. This culture system will help us understand viral respiratory infections and host responses, and to develop effective therapies to cure these infections.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
LR, LD, RV, and RS have been involved in conceptualization, project administration, and visualization. ME was responsible for the methodology with direct involvement of LR and LD. LR and LD were responsible for data curation, investigation, formal analysis, and wrote the original manuscript. All authors contributed to the article and approved the submitted version. RV and RS have been responsible for the supervision. In addition to RV and RS, ME was also involved in providing the resources.
Funding
Funds were received for open access publication by the Erasmus University Library Data and Publication support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.683002/full#supplementary-material
Supplementary material 1 | Core collection Erasmus MC.
Supplementary material 2 | Electronic search terms for Embase, Medline and Web of Science.
References
1. GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Lower Respiratory Infections in 195 Countries, 1990-2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis (2018) 18(11):1191–210. doi: 10.1016/S1473-3099(18)30310-4
2. Shi T, McAllister DA, O’Brien KL, Simoes EAF, Madhi SA, Gessner BD, et al. Global, Regional, and National Disease Burden Estimates of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children in 2015: A Systematic Review and Modelling Study. Lancet (2017) 390(10098):946–58. doi: 10.1016/S0140-6736(17)30938-8
3. Who. The Top 10 Causes of Death (2018). Available at: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
4. Henrickson KJ. Viral Pneumonia in Children. Semin Pediatr Infect Dis (1998) 9(3):217–33. doi: 10.1016/S1045-1870(98)80035-6
5. Greenberg SB. Respiratory Viral Infections in Adults. Curr Opin Pulm Med (2002) 8(3):201–8. doi: 10.1097/00063198-200205000-00009
6. Busse WW, Lemanske RF, Gern JE. Role of Viral Respiratory Infections in Asthma and Asthma Exacerbations. Lancet (2010) 376(9743):826–34. doi: 10.1016/S0140-6736(10)61380-3
7. Nicholson KG, Kent J, Ireland DC. Respiratory Viruses and Exacerbations of Asthma in Adults. Bmj (1993) 307(6910):982–6. doi: 10.1136/bmj.307.6910.982
8. Zheng J. SARS-CoV-2: An Emerging Coronavirus That Causes a Global Threat. Int J Biol Sci (2020) 16(10):1678–85. doi: 10.7150/ijbs.45053
10. Altamirano-Lagos MJ, Díaz FE, Mansilla MA, Rivera-Pérez D, Soto D, McGill JL, et al. Current Animal Models for Understanding the Pathology Caused by the Respiratory Syncytial Virus. Front Microbiol (2019) 10:873. doi: 10.3389/fmicb.2019.00873
11. X M, P NG. Cellular and Animals Models for Rhinovirus Infection in Asthma. Contrib Microbiol (2007) 14:33–41. doi: 10.1159/000107053
12. Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell P-S, Riveros-Balta AX, Albrecht RA, et al. Animal Models for COVID-19. Nat (2020) 586(7830):509–15. doi: 10.1159/000107053
13. N TQ, R R, C YK. Animal Models for Influenza Research: Strengths and Weaknesses. Viruses (2021) 13(6):1011. doi: 10.3390/v13061011
14. MacPhail M, Schickli JH, Tang RS, Kaur J, Robinson C, Fouchier RAM, et al. Identification of Small-Animal and Primate Models for Evaluation of Vaccine Candidates for Human Metapneumovirus (hMPV) and Implications for hMPV Vaccine Design. J Gen Virol (2004) 85(6):1655–63. doi: 10.1099/vir.0.79805-0
15. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Vol. 339, BMJ. Tavistock Square, London, UK: British Medical Journal Publishing Group (2009). p. 332–6.
16. Rawson NE, Ozdener MH. Primary Culture of the Human Olfactory Neuroepithelium. Methods Mol Biol (2013) 945:81–93. doi: 10.1007/978-1-62703-125-7_6
17. Bramer WM, Milic J, Mast F. Reviewing Retrieved References for Inclusion in Systematic Reviews Using Endnote. Vol. 105, Journal of the Medical Library Association. Erasmus MC, Rotterdam, The Netherlands: Medical Library Association (2017). p. 84–7.
18. Morris IR. Functional Anatomy of the Upper Airway. Emerg Med Clin North Am (1988) 6(4):639–69. doi: 10.1016/S0733-8627(20)30518-6
19. Scherzad A, Hagen R, Hackenberg S. Current Understanding of Nasal Epithelial Cell Mis-Differentiation. J Inflammation Res (2019) 12:309–17. doi: 10.2147/JIR.S180853
20. Breeze R, Turk M. Cellular Structure, Function and Organization in the Lower Respiratory Tract. Env Heal Perspect (1984) 55:3–24. doi: 10.1289/ehp.84553
21. Brand-Saberi BEM, Schäfer T. Trachea: Anatomy and Physiology. Thorac Surg Clin (2014) 24(1):1–5. doi: 10.1016/j.thorsurg.2013.09.004
22. Davies A, Moores C. Structure of the Respiratory System, Related to Function. Respir Syst (2010) 11–28. doi: 10.1016/B978-0-7020-3370-4.00002-5
23. Ranga V, Kleinerman J. Structure and Function of Small Airways in Health and Disease. Arch Pathol Lab Med (1978) 102(12):609–17.
24. Guha S, Hariharan P, Myers MR. Enhancement of ICRP’s Lung Deposition Model for Pathogenic Bioaerosols. Aerosol Sci Technol (2014) 48(12):1226–35. doi: 10.1080/02786826.2014.975334
25. Riel DV, Verdijk R, Kuiken T. The Olfactory Nerve: A Shortcut for Influenza and Other Viral Diseases Into the Central Nervous System. J Pathol (2015) 235(2):277–87. doi: 10.1002/path.4461
26. Yaghi A, Dolovich MB. Airway Epithelial Cell Cilia and Obstructive Lung Disease. Cells (2016) 5(4):40. doi: 10.3390/cells5040040
27. Harkema JR, Plopper CG, Pinkerton KE. Comparative Structure of the Respiratory Tract: Airway Architecture in Humans and Animals. In: Cohen MD, Zelikoff JT, Schlesinger RB, editors. Pulmonary Immunotoxicology. Boston, MA: Springer US (2000). p. 1–59.
28. Rokicki W, Rokicki M, Wojtacha J, Dżeljijli A. The Role and Importance of Club Cells (Clara Cells) in the Pathogenesis of Some Respiratory Diseases. Kardiochir Torakochirurgia Pol (2016) 13(1):26–30. doi: 10.5114/kitp.2016.58961
29. Hajj R, Baranek T, Le Naour R, Lesimple P, Puchelle E, Coraux C. Basal Cells of the Human Adult Airway Surface Epithelium Retain Transit-Amplifying Cell Properties. Stem Cells (2007) 25(1):139–48. doi: 10.1634/stemcells.2006-0288
30. Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In Vivo Differentiation Potential of Tracheal Basal Cells: 666evidence for Multipotent and Unipotent Subpopulations. Am J Physiol Lung Cell Mol Physiol (2004) 286(4):L643–9. doi: 10.1152/ajplung.00155.2003
31. Boers JE, Ambergen AW, Thunnissen FB. Number and Proliferation of Basal and Parabasal Cells in Normal Human Airway Epithelium. Am J Respir Crit Care Med (1998) 157:2000–6. doi: 10.1164/ajrccm.157.6.9707011
32. Gosney JR, Sissons MC, Allibone RO. Neuroendocrine Cell Populations in Normal Human Lungs: A Quantitative Study. Thorax (1988) 43(11):878–82. doi: 10.1136/thx.43.11.878
33. Cutz E, Yeger H, Pan J. Pulmonary Neuroendocrine Cell System in Pediatric Lung Disease-Recent Advances. Pediatr Dev Pathol (2007) 10(6):419–35. doi: 10.2350/07-04-0267.1
34. Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell Number and Cell Characteristics of the Normal Human Lung. Am Rev Respir Dis (1982) 126(2):332–7.
35. Daniels CB, Orgeig S. Pulmonary Surfactant: The Key to the Evolution of Air Breathing. News Physiol Sci (2003) 18:151–7. doi: 10.1152/nips.01438.2003
36. Adamson IY, Bowden DH. The Type 2 Cell as Progenitor of Alveolar Epithelial Regeneration. A Cytodynamic Study in Mice After Exposure to Oxygen. Lab Invest (1974) 30(1):35–42.
37. Vareille M, Kieninger E, Edwards MR, Regamey N. The Airway Epithelium: Soldier in the Fight Against Respiratory Viruses. Clin Microbiol Rev (2011) 24(1):210–29. doi: 10.1128/CMR.00014-10
38. Zhang JM, An J. Cytokines, Inflammation, and Pain. Int Anesth Clin (2007) 45(2):27–37. doi: 10.1097/AIA.0b013e318034194e
39. Ferreira Lopes S, Vacher G, Ciarlo E, Savova-Bianchi D, Roger T, Niculita-Hirzel H. Primary and Immortalized Human Respiratory Cells Display Different Patterns of Cytotoxicity and Cytokine Release Upon Exposure to Deoxynivalenol, Nivalenol and Fusarenon-X. Toxins (Basel) (2017) 9(11):337. doi: 10.3390/toxins9110337
40. Swain RJ, Kemp SJ, Goldstraw P, Tetley TD, Stevens MM. Assessment of Cell Line Models of Primary Human Cells by Raman Spectral Phenotyping. Biophys J (2010) 98(8):1703–11. doi: 10.1016/j.bpj.2009.12.4289
41. Forrest IA, Murphy DM, Ward C, Jones D, Johnson GE, Archer L, et al. Primary Airway Epithelial Cell Culture From Lung Transplant Recipients. Eur Respir J (2005) 26(6):1080–5. doi: 10.1183/09031936.05.00141404
42. Lopez-Souza N, Avila PC, Widdicombe JH. Polarized Cultures of Human Airway Epithelium From Nasal Scrapings and Bronchial Brushings. Vitr Cell Dev Biol - Anim (2003) 39(7):266–9. doi: 10.1290/1543-706X(2003)039<0266:PCOHAE>2.0.CO;2
43. Müller L, Brighton LE, Carson JL, Fischer 2WA, Jaspers I. Culturing of Human Nasal Epithelial Cells at the Air Liquid Interface. J Vis Exp (2013) 80):50646. doi: 10.3791/50646
44. Awatade NT, Wong SL, Hewson CK, Fawcett LK, Kicic A, Jaffe A, et al. Human Primary Epithelial Cell Models: Promising Tools in the Era of Cystic Fibrosis Personalized Medicine. Front Pharmacol (2018) 9:1429. doi: 10.3389/fphar.2018.01429
45. Horvath KM, Brighton LE, Zhang W, Carson JL, Jaspers I. Epithelial Cells From Smokers Modify Dendritic Cell Responses in the Context of Influenza Infection. Am J Respir Cell Mol Biol (2011) 45(2):237–45. doi: 10.1165/rcmb.2010-0190OC
46. Bandyopadhyay G, Huyck HL, Misra RS, Bhattacharya S, Wang Q, Mereness J, et al. Dissociation, Cellular Isolation, and Initial Molecular Characterization of Neonatal and Pediatric Human Lung Tissues. Am J Physiol Lung Cell Mol Physiol (2018) 315(4):L576–l583. doi: 10.1152/ajplung.00041.2018
47. Gowers KHC, Hynds RE, Thakrar RM, Carroll B, Birchall MA, Janes SM. Optimized Isolation and Expansion of Human Airway Epithelial Basal Cells From Endobronchial Biopsy Samples. J Tissue Eng Regener Med (2018) 12(1):e313–7. doi: 10.1002/term.2466
48. Villenave R, Shields MD, Power UF. Respiratory Syncytial Virus Interaction With Human Airway Epithelium. Trends Microbiol (2013) 21(5):238–44. doi: 10.1016/j.tim.2013.02.004
49. Broadbent L, Villenave R, Guo-Parke H, Douglas I, Shields MD, Power UF. In Vitro Modeling of RSV Infection and Cytopathogenesis in Well-Differentiated Human Primary Airway Epithelial Cells (WD-PAECs). Methods Mol Biol (2016) 1442:119–39. doi: 10.1007/978-1-4939-3687-8_9
50. Pezzulo AA, Starner TD, Scheetz TE, Traver GL, Tilley AE, Harvey BG, et al. The Air-Liquid Interface and Use of Primary Cell Cultures are Important to Recapitulate the Transcriptional Profile of In Vivo Airway Epithelia. Am J Physiol Lung Cell Mol Physiol (2011) 300(1):L25–31. doi: 10.1152/ajplung.00256.2010
51. Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. Do Airway Epithelium Air-Liquid Cultures Represent the In Vivo Airway Epithelium Transcriptome? Am J Respir Cell Mol Biol (2011) 44(4):465–73. doi: 10.1165/rcmb.2009-0453OC
52. Travanty E, Zhou B, Zhang H, Peter Di Y, Alcorn JF, Wentworth DE, et al. Differential Susceptibilities of Human Lung Primary Cells to H1N1 Influenza Viruses. J Virol (2015) 89(23):11935–44. doi: 10.1128/JVI.01792-15
53. Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, et al. Gene Expression and in Situ Protein Profiling of Candidate SARS-CoV-2 Receptors in Human Airway Epithelial Cells and Lung Tissue. Eur Respir J (2020) 56(3):2001123. doi: 10.1183/13993003.01123-2020
54. Smith JC, Sausville EL, Girish V, Yuan ML, Vasudevan A, John KM, et al. Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Dev Cell (2020) 53(5):514–29.e3. doi: 10.1016/j.devcel.2020.05.012
55. Peretz J, Pekosz A, Lane AP, Klein SL. Estrogenic Compounds Reduce Influenza a Virus Replication in Primary Human Nasal Epithelial Cells Derived From Female, But Not Male, Donors. Am J Physiol Lung Cell Mol Physiol (2016) 310(5):L415–25. doi: 10.1152/ajplung.00398.2015
56. Salka K, Arroyo M, Chorvinsky E, Abutaleb K, Perez GF, Wolf S, et al. Innate IFN-Lambda Responses to dsRNA in the Human Infant Airway Epithelium and Clinical Regulatory Factors During Viral Respiratory Infections in Early Life. Clin Exp Allergy (2020) 50(9):1044–54. doi: 10.1111/cea.13701
57. Dominguez SR, Shrivastava S, Berglund A, Qian Z, Góes LGB, Halpin RA, et al. Isolation, Propagation, Genome Analysis and Epidemiology of HKU1 Betacoronaviruses. J Gen Virol (2014) 95(PART 4):836–48. doi: 10.1099/vir.0.059832-0
58. Sims AC, Burkett SE, Yount B, Pickles RJ. SARS-CoV Replication and Pathogenesis in an In Vitro Model of the Human Conducting Airway Epithelium. Virus Res (2008) 133(1):33–44. doi: 10.1016/j.virusres.2007.03.013
59. Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon 3KH, et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell (2020) 182(2):429–46.e14. doi: 10.1016/j.cell.2020.05.042
60. Pickles RJ. Human Airway Epithelial Cell Cultures for Modeling Respiratory Syncytial Virus Infection. Curr Top Microbiol Immunol (2013) 372:371–87. doi: 10.1007/978-3-642-38919-1_19
61. Schaap-Nutt A, Liesman R, Bartlett EJ, Scull MA, Collins PL, Pickles RJ, et al. Human Parainfluenza Virus Serotypes Differ in Their Kinetics of Replication and Cytokine Secretion in Human Tracheobronchial Airway Epithelium. Virology (2012) 433(2):320–8. doi: 10.1016/j.virol.2012.08.027
62. Gillen AE, Yang R, Cotton CU, Perez A, Randell SH, Leir SH, et al. Molecular Characterization of Gene Regulatory Networks in Primary Human Tracheal and Bronchial Epithelial Cells. J Cyst Fibros (2018) 17(4):444–53. doi: 10.1016/j.jcf.2018.01.009
63. Mihaylova VT, Kong Y, Fedorova O, Sharma L, Dela Cruz CS, Pyle AM, et al. Regional Differences in Airway Epithelial Cells Reveal Tradeoff Between Defense Against Oxidative Stress and Defense Against Rhinovirus. Cell Rep (2018) 24(11):3000–7.e3. doi: 10.1016/j.celrep.2018.08.033
64. Zhao Y, Jamaluddin M, Zhang Y, Sun H, Ivanciuc T, Garofalo RP, et al. Systematic Analysis of Cell-Type Differences in the Epithelial Secretome Reveals Insights Into the Pathogenesis of Respiratory Syncytial Virus-Induced Lower Respiratory Tract Infections. J Immunol (2017) 198(8):3345–64. doi: 10.4049/jimmunol.1601291
65. Broadbent L, Manzoor S, Zarcone MC, Barabas J, Shields MD, Saglani S, et al. Comparative Primary Paediatric Nasal Epithelial Cell Culture Differentiation and RSV-Induced Cytopathogenesis Following Culture in Two Commercial Media. Tripp RA, Editor. PloS One (2020) 15(3):e0228229. doi: 10.1371/journal.pone.0228229
66. Jia HP, Look DC, Shi L, Hickey M, Pewe L, Netland J, et al. ACE2 Receptor Expression and Severe Acute Respiratory Syndrome Coronavirus Infection Depend on Differentiation of Human Airway Epithelia. J Virol (2005) 79(23):14614–21. doi: 10.1128/JVI.79.23.14614-14621.2005
67. Chan MC, Chan RW, Tsao GS, Peiris JS. Replication and Pathogenesis of Avian Influenza A (H5N1) Virus Infection in Polarised Human Bronchial and Alveolar Epithelium. Hong Kong Med J (2013) 19 Suppl 4:24–8.
68. Lopez-Souza N, Dolganov G, Dubin R, Sachs LA, Sassina L, Sporer H, et al. Resistance of Differentiated Human Airway Epithelium to Infection by Rhinovirus. Am J Physiol Lung Cell Mol Physiol (2004) 286(2 30-2):L373–81. doi: 10.1152/ajplung.00300.2003
69. Chan RWY, Yuen KM, Yu WCL, Ho CCC, Nicholls JM, Malik Peiris JS, et al. Influenza H5N1 and H1N1 Virus Replication and Innate Immune Responses in Bronchial Epithelial Cells are Influenced by the State of Differentiation. PloS One (2010) 5(1):e8713. doi: 10.1371/journal.pone.0008713
70. Reeves SR, Barrow KA, White MP, Rich LM, Naushab M, Debley JS. Stability of Gene Expression by Primary Bronchial Epithelial Cells Over Increasing Passage Number. BMC Pulm Med (2018) 18(1):91. doi: 10.1186/s12890-018-0652-2
71. Gray TE, Guzman K, Davis CW, Abdullah LH, Nettesheim P. Mucociliary Differentiation of Serially Passaged Normal Human Tracheobronchial Epithelial Cells. Am J Respir Cell Mol Biol (1996) 14(1):104–12. doi: 10.1165/ajrcmb.14.1.8534481
72. Gardner DE. Toxicology of the Lung. 4th edition. Boca Raton, Florida, United States: CRC Press (2005). p. 124.
73. Jonsdottir HR, Marti S, Geerts D, Rodriguez R, Thiel V, Dijkman R. Establishment of Primary Transgenic Human Airway Epithelial Cell Cultures to Study Respiratory Virus–Host Interactions. Viruses (2019) 11(8):747. doi: 10.3390/v11080747
74. Kurose M, Kojima T, Koizumi J, Kamekura R, Ninomiya T, Murata M, et al. Induction of Claudins in Passaged hTERT-Transfected Human Nasal Epithelial Cells With an Extended Life Span. Cell Tissue Res (2007) 330(1):63–74. doi: 10.1007/s00441-007-0453-z
75. Toouli CD, Huschtscha LI, Neumann AA, Noble JR, Colgin LM, Hukku B, et al. Comparison of Human Mammary Epithelial Cells Immortalized by Simian Virus 40 T-Antigen or by the Telomerase Catalytic Subunit. Oncogene (2002) 21(1):128–39. doi: 10.1038/sj.onc.1205014
76. Wu X, Wang S, Li M, Li J, Shen J, Zhao Y, et al. Conditional Reprogramming: Next Generation Cell Culture. Acta Pharm Sin B (2020) 10(8):1360–81. doi: 10.1016/j.apsb.2020.01.011
77. Lehmann R, Lee CM, Shugart EC, Benedetti M, Charo RA, Gartner Z, et al. Human Organoids: A New Dimension in Cell Biology. Mol Biol Cell (2019) 30(10):1129–37. doi: 10.1091/mbc.E19-03-0135
78. Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro Without a Mesenchymal Niche. Nature (2009) 459(7244):262–5. doi: 10.1038/nature07935
79. Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, et al. Directed Differentiation of Human Pluripotent Stem Cells Into Mature Airway Epithelia Expressing Functional CFTR Protein. Nat Biotechnol (2012) 30(9):876–82. doi: 10.1038/nbt.2328
80. Chen YW, Huang SX, De Carvalho ALRT, Ho SH, Islam MN, Volpi S, et al. A Three-Dimensional Model of Human Lung Development and Disease From Pluripotent Stem Cells. Nat Cell Biol (2017) 19(5):542–9. doi: 10.1038/ncb3510
81. Porotto M, Ferren M, Chen YW, Siu Y, Makhsous N, Rima B, et al. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. MBio (2019) 10(3):1360–81. doi: 10.1128/mBio.00723-19
82. Evans KV, Lee JH. Alveolar Wars: The Rise of In Vitro Models to Understand Human Lung Alveolar Maintenance, Regeneration, and Disease. Stem Cells Transl Med (2020) 9(8):867–81. doi: 10.1002/sctm.19-0433
83. Vazin T, Freed WJ. Human Embryonic Stem Cells: Derivation, Culture, and Differentiation: A Review. Restor Neurol Neurosci (2010) 28(4):589–603. doi: 10.3233/RNN-2010-0543
84. Medvedev SP, Shevchenko AI, Zakian SM. Induced Pluripotent Stem Cells: Problems and Advantages When Applying Them in Regenerative Medicine. Acta Naturae (2010) 2(2):18–28. doi: 10.32607/20758251-2010-2-2-18-27
85. Huang SX, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, et al. Efficient Generation of Lung and Airway Epithelial Cells From Human Pluripotent Stem Cells. Nat Biotechnol (2014) 32(1):84–91. doi: 10.1038/nbt.2754
86. Hombach-Klonisch S, Panigrahi S, Rashedi I, Seifert A, Alberti E, Pocar P, et al. Adult Stem Cells and Their Trans-Differentiation Potential–Perspectives and Therapeutic Applications. J Mol Med (2008) 86(12):1301–14. doi: 10.1007/s00109-008-0383-6
87. Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Böttinger L, Klay D, et al. Long-Term Expanding Human Airway Organoids for Disease Modeling. EMBO J (2019) 38(4):e100300. doi: 10.15252/embj.2018100300
88. Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, et al. An Organoid-Derived Bronchioalveolar Model for SARS-CoV-2 Infection of Human Alveolar Type II-Like Cells. EMBO J (2021) 40(5):e105912. doi: 10.15252/embj.2020105912
89. Salahudeen AA, Choi SS, Rustagi A, Zhu J, van Unen V, de la O SM, et al. Progenitor Identification and SARS-CoV-2 Infection in Human Distal Lung Organoids. Nature (2020) 588(7839):670–5. doi: 10.1038/s41586-020-3014-1
90. Youk J, Kim T, Evans KV, Jeong YI, Hur Y, Hong SP, et al. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell (2020) 27(6):905–19.e10. doi: 10.1016/j.stem.2020.10.004
91. Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights Into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell (2020) 27(6):890–904.e8. doi: 10.1016/j.stem.2020.10.005
92. Zhou J, Li C, Sachs N, Chiu MC, Wong BHY, Chu H, et al. Differentiated Human Airway Organoids to Assess Infectivity of Emerging Influenza Virus. Proc Natl Acad Sci USA (2018) 115(26):6822–7. doi: 10.1073/pnas.1806308115
93. Hui KPY, Ching RHH, Chan SKH, Nicholls JM, Sachs N, Clevers H, et al. Tropism, Replication Competence, and Innate Immune Responses of Influenza Virus: An Analysis of Human Airway Organoids and Ex-Vivo Bronchus Cultures. Lancet Respir Med (2018) 6(11):846–54. doi: 10.1016/S2213-2600(18)30236-4
94. Villenave R, O’Donoghue D, Thavagnanam S, Touzelet O, Skibinski G, Heaney LG, et al. Differential Cytopathogenesis of Respiratory Syncytial Virus Prototypic and Clinical Isolates in Primary Pediatric Bronchial Epithelial Cells. Virol J (2011) 8:43. doi: 10.1186/1743-422X-8-43
95. Iketani S, Shean RC, Ferren M, Makhsous N, Aquino DB, des Georges A, et al. Viral Entry Properties Required for Fitness in Humans Are Lost Through Rapid Genomic Change During Viral Isolation. MBio (2018) 9(4):e00898–18. doi: 10.1128/mBio.00898-18
96. Terajima M, Yamaya M, Sekizawa K, Okinaga S, Suzuki T, Yamada N, et al. Rhinovirus Infection of Primary Cultures of Human Tracheal Epithelium: Role of ICAM-1 and IL-1β. Am J Physiol Lung Cell Mol Physiol (1997) 273(4 17-4):L749–59. doi: 10.1152/ajplung.1997.273.4.L749
97. Schroth MK, Grimm E, Frindt P, Galagan DM, Konno SI, Love R, et al. Rhinovirus Replication Causes RANTES Production in Primary Bronchial Epithelial Cells. Am J Respir Cell Mol Biol (1999) 20(6):1220–8. doi: 10.1165/ajrcmb.20.6.3261
98. Papi A, Stanciu LA, Papadopoulos NG, Teran LM, Holgate ST, Johnston SL. Rhinovirus Infection Induces Major Histocompatibility Complex Class I and Costimulatory Molecule Upregulation on Respiratory Epithelial Cells. J Infect Dis (2000) 181(5):1780–4. doi: 10.1086/315463
99. Papadopoulos NG, Bates PJ, Bardin PG, Papi A, Leir SH, Fraenkel DJ, et al. Rhinoviruses Infect the Lower Airways. J Infect Dis (2000) 181(6):1875–84. doi: 10.1086/315513
100. Suzuki T, Yamaya M, Sekizawa K, Hosoda M, Yamada N, Ishizuka S, et al. Erythromycin Inhibits Rhinovirus Infection in Cultured Human Tracheal Epithelial Cells. Am J Respir Crit Care Med (2002) 165(8):1113–8. doi: 10.1164/ajrccm.165.8.2103094
101. Griggs TF, Bochkov YA, Basnet S, Pasic TR, Brockman-Schneider RA, Palmenberg AC, et al. Rhinovirus C Targets Ciliated Airway Epithelial Cells. Respir Res (2017) 18(1):84. doi: 10.1186/s12931-017-0567-0
102. Chen YX, Xie GC, Pan D, Du YR, Pang LL, Song JD, et al. Three-Dimensional Culture of Human Airway Epithelium in Matrigel for Evaluation of Human Rhinovirus C and Bocavirus Infections. BioMed Env Sci (2018) 31(2):136–45. doi: 10.3967/bes2018.016
103. Jamieson KC, Wiehler S, Michi AN, Proud D. Rhinovirus Induces Basolateral Release of IL-17C in Highly Differentiated Airway Epithelial Cells. Front Cell Infect Microbiol (2020) 10:103. doi: 10.3389/fcimb.2020.00103
104. Veerati PC, Troy NM, Reid AT, Li NF, Nichol KS, Kaur P, et al. Airway Epithelial Cell Immunity Is Delayed During Rhinovirus Infection in Asthma and COPD. Front Immunol (2020) 11:974. doi: 10.3389/fimmu.2020.00974
105. Raj VS, Mou H, Smits SL, Dekkers DHW, Müller MA, Dijkman R, et al. Dipeptidyl Peptidase 4 is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature (2013) 495(7440):251–4. doi: 10.1038/nature12005
106. Wang G, Deering C, Macke M, Shao J, Burns R, Blau DM, et al. Human Coronavirus 229E Infects Polarized Airway Epithelia From the Apical Surface. J Virol (2000) 74(19):9234–9. doi: 10.1128/JVI.74.19.9234-9239.2000
107. Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jonsdottir HR, Molenkamp R, et al. Isolation and Characterization of Current Human Coronavirus Strains in Primary Human Epithelial Cell Cultures Reveal Differences in Target Cell Tropism. J Virol (2013) 87(11):6081–90. doi: 10.1128/JVI.03368-12
108. Huang X, Dong W, Milewska A, Golda A, Qi Y, Zhu QK, et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J Virol (2015) 89(14):7202–13. doi: 10.1128/JVI.00854-15
109. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 Productively Infects Human Gut Enterocytes. Science (2020) 369(6499):50–4. doi: 10.1126/science.abc1669
110. Pei R, Feng J, Zhang Y, Sun H, Li L, Yang X, et al. Host Metabolism Dysregulation and Cell Tropism Identification in Human Airway and Alveolar Organoids Upon SARS-CoV-2 Infection. Protein Cell (2020) 12:1–17. doi: 10.1007/s13238-020-00811-w
111. Qian Z, Ou X, Góes LGB, Osborne C, Castano A, Holmes KV, et al. Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1. J Virol (2015) 89(17):8816–27. doi: 10.1128/JVI.03737-14
112. Jang Y, Seo SH. Gene Expression Pattern Differences in Primary Human Pulmonary Epithelial Cells Infected With MERS-CoV or SARS-CoV-2. Arch Virol (2020) 165(10):2205–11. doi: 10.1007/s00705-020-04730-3
113. Loo SL, Wark PAB, Esneau C, Nichol KS, Hsu ACY, Bartlett NW. Human Coronaviruses 229E and OC43 Replicate and Induce Distinct Antiviral Responses in Differentiated Primary Human Bronchial Epithelial Cells. Am J Physiol Lung Cell Mol Physiol (2020) 319(6):L926–31. doi: 10.1152/ajplung.00374.2020
114. Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol (2020) 94(19):e00985–20. doi: 10.1101/2020.05.19.105437
115. Bui CHT, Chan RWY, Ng MMT, Cheung MC, Ng KC, Chan MPK, et al. Tropism of Influenza B Viruses in Human Respiratory Tract Explants and Airway Organoids. Eur Respir J (2019) 54(2):1900008. doi: 10.1183/13993003.00008-2019
116. Bhowmick R, Derakhshan T, Liang Y, Ritchey J, Liu L. Gappa-Fahlenkamp H. A Three-Dimensional Human Tissue-Engineered Lung Model to Study Influenza A Infection. Tissue Eng Part A (2018) 24(19–20):1468–80. doi: 10.1089/ten.tea.2017.0449
117. Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, et al. Plasticity and Virus Specificity of the Airway Epithelial Cell Immune Response During Respiratory Virus Infection. J Virol (2012) 86(10):5422–36. doi: 10.1128/JVI.06757-11
118. Lin YT, Lin CF, Yeh TH. Influenza A Virus Infection Induces Indoleamine 2,3-Dioxygenase (IDO) Expression and Modulates Subsequent Inflammatory Mediators in Nasal Epithelial Cells. Acta Otolaryngol (2020) 140(2):149–56. doi: 10.1080/00016489.2019.1700304
119. Jeong KI, Piepenhagen PA, Kishko M, DiNapoli JM, Groppo RP, Zhang L, et al. CX3CR1 Is Expressed in Differentiated Human Ciliated Airway Cells and Co-Localizes With Respiratory Syncytial Virus on Cilia in a G Protein-Dependent Manner. PloS One (2015) 10(6):e0130517. doi: 10.1371/journal.pone.0130517
120. Johnson SM, McNally BA, Ioannidis I, Flano E, Teng MN, Oomens AG, et al. Respiratory Syncytial Virus Uses CX3CR1 as a Receptor on Primary Human Airway Epithelial Cultures. PLoS Pathog (2015) 11(12):e1005318. doi: 10.1371/journal.ppat.1005318
121. Griffiths CD, Bilawchuk LM, McDonough JE, Jamieson KC, Elawar F, Cen Y, et al. IGF1R Is an Entry Receptor for Respiratory Syncytial Virus. Nature (2020) 583(7817):615–9. doi: 10.1038/s41586-020-2369-7
122. Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory Syncytial Virus Infection of Human Airway Epithelial Cells is Polarized, Specific to Ciliated Cells, and Without Obvious Cytopathology. J Virol (2002) 76(11):5654–66. doi: 10.1128/JVI.76.11.5654-5666.2002
123. Hong GP, Canning P, Douglas I, Villenave R, Heaney LG, Coyle PV, et al. Relative Respiratory Syncytial Virus Cytopathogenesis in Upper and Lower Respiratory Tract Epithelium. Am J Respir Crit Care Med (2013) 188(7):842–51. doi: 10.1164/rccm.201304-0750OC
124. Persson BD, Jaffe AB, Fearns R, Danahay H. Respiratory Syncytial Virus can Infect Basal Cells and Alter Human Airway Epithelial Differentiation. PloS One (2014) 9(7):e102368. doi: 10.1371/journal.pone.0102368
125. Porotto M, Ferren M, Chen YW, Siu Y, Makhsous N, Rima B, et al. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. MBio (2019) 10(3):e00723–19. doi: 10.1128/mBio.00723-19
126. Loevenich S, Malmo J, Liberg AM, Sherstova T, Li Y, Rian K, et al. Cell-Type-Specific Transcription of Innate Immune Regulators in Response to Hmpv Infection. Mediat Inflammation (2019) 4964239. doi: 10.1155/2019/4964239
127. Bao X, Liu T, Spetch L, Kolli D, Garofalo RP, Casola A. Airway Epithelial Cell Response to Human Metapneumovirus Infection. Virology (2007) 368(1):91–101. doi: 10.1016/j.virol.2007.06.023
128. Yu X, Lakerveld AJ, Imholz S, Hendriks M, ten Brink SCA, Mulder HL, et al. Antibody and Local Cytokine Response to Respiratory Syncytial Virus Infection in Community-Dwelling Older Adults. mSphere (2020) 5(5):e00577–20. doi: 10.1128/mSphere.00577-20
129. Villenave R, Thavagnanam S, Sarlang S, Parker J, Douglas I, Skibinski G, et al. In Vitro Modeling of Respiratory Syncytial Virus Infection of Pediatric Bronchial Epithelium, the Primary Target of Infection In Vivo. Proc Natl Acad Sci USA (2012) 109(13):5040–5. doi: 10.1073/pnas.1110203109
130. Villenave R, Broadbent L, Douglas I, Lyons JD, Coyle PV, Teng MN, et al. Induction and Antagonism of Antiviral Responses in Respiratory Syncytial Virus-Infected Pediatric Airway Epithelium. J Virol (2015) 89(24):12309–18. doi: 10.1128/JVI.02119-15
131. Das S, Palmer OP, Leight WD, Surowitz JB, Pickles RJ, Randell SH, et al. Cytokine Amplification by Respiratory Syncytial Virus Infection in Human Nasal Epithelial Cells. Laryngoscope (2005) 115(5):764–8. doi: 10.1097/01.MLG.0000159527.76949.93
132. Okabayashi T, Kojima T, Masaki T, Yokota S-I, Imaizumi T, Tsutsumi H, et al. Type-III Interferon, Not Type-I, is the Predominant Interferon Induced by Respiratory Viruses in Nasal Epithelial Cells. Virus Res (2011) 160:360–6. doi: 10.1016/j.virusres.2011.07.011
133. Kogure T, Suzuki T, Takahashi T, Miyamoto D, Hidari KIPJ, Guo CT, et al. Human Trachea Primary Epithelial Cells Express Both Sialyl(α2-3)Gal Receptor for Human Parainfluenza Virus Type 1 and Avian Influenza Viruses, and Sialyl(α2-6)Gal Receptor for Human Influenza Viruses. Glycoconjugate J (2006) 23(1–2):101–6. doi: 10.1007/s10719-006-5442-z
134. Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, et al. Infection of Ciliated Cells by Human Parainfluenza Virus Type 3 in an In Vitro Model of Human Airway Epithelium. J Virol (2005) 79(2):1113–24. doi: 10.1128/JVI.79.2.1113-1124.2005
135. Zhang H, Newman DR, Sannes PL. HSULF-1 Inhibits ERK and AKT Signaling and Decreases Cell Viability In Vitro in Human Lung Epithelial Cells. Respir Res (2012) 13:69. doi: 10.1186/1465-9921-13-69
136. Zhang L, Collins PL, Lamb RA, Pickles RJ. Comparison of Differing Cytopathic Effects in Human Airway Epithelium of Parainfluenza Virus 5 (W3A), Parainfluenza Virus Type 3, and Respiratory Syncytial Virus. Virology (2011) 421(1):67–77. doi: 10.1016/j.virol.2011.08.020
137. Schaap-Nutt A, D’Angelo C, Scull MA, Amaro-Carambot E, Nishio M, Pickles RJ, et al. Human Parainfluenza Virus Type 2 V Protein Inhibits Interferon Production and Signaling and Is Required for Replication in non-Human Primates. Virology (2010) 397(2):285–98. doi: 10.1016/j.virol.2009.11.018
138. Lam E, Ramke M, Warnecke G, Schrepfer S, Kopfnagel V, Dobner T, et al. Effective Apical Infection of Differentiated Human Bronchial Epithelial Cells and Induction of Proinflammatory Chemokines by the Highly Pneumotropic Human Adenovirus Type 14p1. PloS One (2015) 10(7):e0131201. doi: 10.1371/journal.pone.0131201
139. Meyer-Berg H, Zhou Yang L, Pilar de Lucas M, Zambrano A, Hyde SC, Gill DR. Identification of AAV Serotypes for Lung Gene Therapy in Human Embryonic Stem Cell-Derived Lung Organoids. Stem Cell Res Ther (2020) 11(1):488. doi: 10.1186/s13287-020-01950-x
140. Yang TI, Li WL, Chang TH, Lu CY, Chen JM, Lee PI, et al. Adenovirus Replication and Host Innate Response in Primary Human Airway Epithelial Cells. J Microbiol Immunol Infect (2019) 52(2):207–14. doi: 10.1016/j.jmii.2018.08.010
141. Raj VS, Lamers MM, Smits SL, Demmers JA, Mou H, Bosch BJ, et al. Identification of Protein Receptors for Coronaviruses by Mass Spectrometry. Methods Mol Biol (2015) 1282:165–82. doi: 10.1007/978-1-4939-2438-7_15
142. Barrass SV, Butcher SJ. Advances in High-Throughput Methods for the Identification of Virus Receptors. Vol. 209, Medical Microbiology and Immunology. Springer; (2020) 209(3):309–23. doi: 10.1007/s00430-019-00653-2
143. Wang S, Qiu Z, Hou Y, Deng X, Xu W, Zheng T, et al. AXL is a Candidate Receptor for SARS-CoV-2 That Promotes Infection of Pulmonary and Bronchial Epithelial Cells. Cell Res (2021) 31(2):126–40. doi: 10.1038/s41422-020-00460-y
144. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, et al. Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature (1992) 357(6377):420–2. doi: 10.1038/357420a0
145. Vlasak R, Luytjes W, Spaan W, Palese P. Human and Bovine Coronaviruses Recognize Sialic Acid-Containing Receptors Similar to Those of Influenza C Viruses. Proc Natl Acad Sci USA (1988) 85(12):4526–9. doi: 10.1073/pnas.85.12.4526
146. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-Converting Enzyme 2 is a Functional Receptor for the SARS Coronavirus. Nature (2003) 426(6965):450–4. doi: 10.1038/nature02145
147. Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. Human Coronavirus NL63 Employs the Severe Acute Respiratory Syndrome Coronavirus Receptor for Cellular Entry. Proc Natl Acad Sci USA (2005) 102(22):7988–93. doi: 10.1073/pnas.0409465102
148. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell (2020) 181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052
149. Pyrc K, Sims AC, Dijkman R, Jebbink M, Long C, Deming D, et al. Culturing the Unculturable: Human Coronavirus HKU1 Infects, Replicates, and Produces Progeny Virions in Human Ciliated Airway Epithelial Cell Cultures. J Virol (2010) 84(21):11255–63. doi: 10.1128/JVI.00947-10
150. Staunton DE, Merluzzi VJ, Rothlein R, Barton R, Marlin SD, Springer TA. A Cell Adhesion Molecule, ICAM-1, is the Major Surface Receptor for Rhinoviruses. Cell (1989) 56(5):849–53. doi: 10.1016/0092-8674(89)90689-2
151. Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, et al. Members of the Low Density Lipoprotein Receptor Family Mediate Cell Entry of a Minor-Group Common Cold Virus. Proc Natl Acad Sci USA (1994) 91(5):1839–42. doi: 10.1073/pnas.91.5.1839
152. Marlovits TC, Abrahamsberg C, Blaas D. Very-Low-Density Lipoprotein Receptor Fragment Shed From HeLa Cells Inhibits Human Rhinovirus Infection. J Virol (1998) 72(12):10246–50. doi: 10.1128/JVI.72.12.10246-10250.1998
153. Suzuki T, Yamaya M, Kamanaka M, Jia YX, Nakayama K, Hosoda M, et al. Type 2 Rhinovirus Infection of Cultured Human Tracheal Epithelial Cells: Role of LDL Receptor. Am J Physiol Lung Cell Mol Physiol (2001) 280(3 24-3):L409–20. doi: 10.1152/ajplung.2001.280.3.L409
154. Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, et al. Molecular Modeling, Organ Culture and Reverse Genetics for a Newly Identified Human Rhinovirus C. Nat Med (2011) 17(5):627–32. doi: 10.1038/nm.2358
155. Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The Histopathology of Fatal Untreated Human Respiratory Syncytial Virus Infection. Mod Pathol (2007) 20(1):108–19. doi: 10.1038/modpathol.3800725
156. Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. Identification of Nucleolin as a Cellular Receptor for Human Respiratory Syncytial Virus. Nat Med (2011) 17(9):1132–5. doi: 10.1038/nm.2444
157. Chang A, Masante C, Buchholz UJ, Dutch RE. Human Metapneumovirus (HMPV) Binding and Infection Are Mediated by Interactions Between the HMPV Fusion Protein and Heparan Sulfate. J Virol (2012) 86(6):3230–43. doi: 10.1128/JVI.06706-11
158. Cox RG, Erickson JJ, Hastings AK, Becker JC, Johnson M, Craven RE, et al. Human Metapneumovirus Virus-Like Particles Induce Protective B and T Cell Responses in a Mouse Model. J Virol (2014) 88(11):6368–79. doi: 10.1128/JVI.00332-14
159. Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, et al. Isolation of a Common Receptor for Coxsackie B Viruses and Adenoviruses 2 and 5. Science (1997) 275(5304):1320–3. doi: 10.1126/science.275.5304.1320
160. Dominguez SR, Travanty EA, Qian Z, Mason RJ. Human Coronavirus HKU1 Infection of Primary Human Type II Alveolar Epithelial Cells: Cytopathic Effects and Innate Immune Response. PloS One (2013) 8(7):e70129. doi: 10.1371/journal.pone.0070129
161. Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, et al. Innate Immune Response of Human Alveolar Type II Cells Infected With Severe Acute Respiratory Syndrome-Coronavirus. Am J Resp Cell Mol Biol (2013) 48(6):742–8. doi: 10.1165/rcmb.2012-0339OC
162. Jakiela B, Brockman-Schneider R, Amineva S, Lee WM, Gern JE. Basal Cells of Differentiated Bronchial Epithelium Are More Susceptible to Rhinovirus Infection. Am J Respir Cell Mol Biol (2008) 38(5):517–23. doi: 10.1165/rcmb.2007-0050OC
163. Mosser AG, Brockman-Schneider R, Amineva S, Burchell L, Sedgwick JB, Busse WW, et al. Similar Frequencyaa of Rhinovirus-Infectible Cellsasas in Upper and Lower Airway Epithelium. J Infect Dis (2002) 185(6):734–43xxzx. doi: 10.1086/339339
164. Nakagome K, Bochkov YA, Ashraf S, Brockman-Schneider RA, Evans MD, Pasic TR, et al. Effects of Rhinovirus Species on Viral Replication and Cytokine Production. J Allergy Clin Immunol (2014) 134(2):332–41.e10. doi: 10.1016/j.jaci.2014.01.029
165. Yeo NK, Jang YJ. Rhinovirus Infection-Induced Alteration of Tight Junction and Adherens Junction Components in Human Nasal Epithelial Cells. Laryngoscope (2010) 120(2):346–52. doi: 10.1002/lary.20764
166. Teng MN, Whitehead SS, Collins PL. Contribution of the Respiratory Syncytial Virus G Glycoprotein and its Secreted and Membrane-Bound Forms to Virus Replication In Vitro and In Vivo. Virology (2001) 289(2):283–96. doi: 10.1006/viro.2001.1138
167. de Graaf M, Herfst S, Aarbiou J, Burgers PC, Zaaraoui-Boutahar F, Bijl M, et al. Small Hydrophobic Protein of Human Metapneumovirus Does Not Affect Virus Replication and Host Gene Expression In Vitro. PloS One (2013) 8(3):e58572. doi: 10.1371/journal.pone.0058572
168. Geiser J, Boivin G, Huang S, Constant S, Kaiser L, Tapparel C, et al. RSV and HMPV Infections in 3D Tissue Cultures: Mechanisms Involved in Virus-Host and Virus-Virus Interactions. Viruses (2021) 13(1):139. doi: 10.3390/v13010139
169. Rijsbergen LC, Rennick LJ, Laksono BM, van Run PRWA, Kuiken T, Duprex WP, et al. In Vivo Comparison of a Laboratory-Adapted and Clinical-Isolate-Based Recombinant Human Respiratory Syncytial Virus. J Gen Virol (2020) 101(10):1037–46. doi: 10.1099/jgv.0.001468
170. Liesman RM, Buchholz UJ, Luongo CL, Yang L, Proia AD, DeVincenzo JP, et al. RSV-Encoded NS2 Promotes Epithelial Cell Shedding and Distal Airway Obstruction. J Clin Invest (2014) 124(5):2219–33. doi: 10.1172/JCI72948
171. Deng Y, Herbert JA, Robinson E, Ren L, Smyth RL, Smith CM. Neutrophil-Airway Epithelial Interactions Result in Increased Epithelial Damage and Viral Clearance During Respiratory Syncytial Virus Infection. J Virol (2020) 94(13):e2161–19. doi: 10.1128/JVI.02161-19
172. Piedimonte G, Perez MK. Respiratory Syncytial Virus Infection and Bronchiolitis. Pediatr Rev (2014) 35(12):519–30. doi: 10.1542/pir.35-12-519
173. Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, et al. Efficient Replication of the Novel Human Betacoronavirus EMC on Primary Human Epithelium Highlights its Zoonotic Potential. MBio (2013) 4(1):e00611–12. doi: 10.1128/mBio.00611-12
174. Montgomery ST, Frey DL, Mall MA, Stick SM, Kicic A. Rhinovirus Infection Is Associated With Airway Epithelial Cell Necrosis and Inflammation via Interleukin-1 in Young Children With Cystic Fibrosis. Front Immunol (2020) 11:596. doi: 10.3389/fimmu.2020.00596
175. Montgomery ST, Stick SM, Kicic A. An Adapted Novel Flow Cytometry Methodology to Delineate Types of Cell Death in Airway Epithelial Cells. J Biol Methods (2020) 7(4):e139. doi: 10.14440/jbm.2020.336
176. Liu A, Zhang X, Li R, Zheng M, Yang S, Dai L, et al. Overexpression of the SARS-CoV-2 Receptor ACE2 is Induced by Cigarette Smoke in Bronchial and Alveolar Epithelia. J Pathol (2021) 253(1):17–30. doi: 10.1002/path.5555
177. Radzikowska U, Ding M, Tan G, Zhakparov D, Peng Y, Wawrzyniak P, et al. Distribution of ACE2, CD147, CD26, and Other SARS-CoV-2 Associated Molecules in Tissues and Immune Cells in Health and in Asthma, COPD, Obesity, Hypertension, and COVID-19 Risk Factors. Allergy Eur J Allergy Clin Immunol (2020) 75(11):2829–45. doi: 10.1111/all.14429
178. Zhang H, Rostami MR, Leopold PL, Mezey JG, O’Beirne SL, Strulovici-Barel Y, et al. Expression of the SARS-CoV-2 ACE2 Receptor in the Human Airway Epithelium. Am J Respir Crit Care Med (2020) 202(2):219–29. doi: 10.1164/rccm.202003-0541OC
179. Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, et al. SARS-CoV-2 Receptor ACE2 and TMPRSS2 Are Primarily Expressed in Bronchial Transient Secretory Cells. EMBO J (2020) 39(10):e105114. doi: 10.15252/embj.20105114
180. Kormuth KA, Lin K, Prussin AJ, Vejerano EP, Tiwari AJ, Cox SS, et al. Influenza Virus Infectivity Is Retained in Aerosols and Droplets Independent of Relative Humidity. J Infect Dis (2018) 218(5):739–47. doi: 10.1093/infdis/jiy221
181. Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, et al. Comparative Study of Influenza Virus Replication in MDCK Cells and in Primary Cells Derived From Adenoids and Airway Epithelium. J Virol (2012) 86(21):11725–34. doi: 10.1128/JVI.01477-12
182. Hawksworth A, Lockhart R, Crowe J, Maeso R, Ritter L, Dibben O, et al. Replication of Live Attenuated Influenza Vaccine Viruses in Human Nasal Epithelial Cells Is Associated With H1N1 Vaccine Effectiveness. Vaccine (2020) 38(26):4209–18. doi: 10.1016/j.vaccine.2020.04.004
183. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, et al. SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission In Vivo. Science (2020) 370(6523):1464–8. doi: 10.1101/2020.09.28.317685
184. Komabayashi K, Matoba Y, Seto J, Ikeda Y, Tanaka W, Aoki Y, et al. Isolation of Human Coronaviruses OC43, HKU1, NL63, and 229E in Yamagata, Japan, Using Primary Human Airway Epithelium Cells Cultured by Employing an Air-Liquid Interface Culture. Jpn J Infect Dis (2020) 74(4):285–92. doi: 10.7883/yoken.JJID.2020.776
185. Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee H-H, et al. Small Airway-on-a-Chip Enables Analysis of Human Lung Inflammation and Drug Responses In Vitro. Nat Methods (2016) 13(2):151–7. doi: 10.1038/nmeth.3697
186. Bhatia SN, Ingber DE. Microfluidic Organs-on-Chips. Nat Biotechnol (2014) 32(8):760–72. doi: 10.1038/nbt.2989
187. Kaarj K, Yoon JY. Methods of Delivering Mechanical Stimuli to Organ-On-a-Chip. Micromachines (Basel) (2019) 10(10):700. doi: 10.3390/mi10100700
188. Nickol ME, Lyle SM, Dennehy B, Kindrachuk J. Dysregulated Host Responses Underlie 2009 Pandemic Influenza-Methicillin Resistant Staphylococcus Aureus Coinfection Pathogenesis at the Alveolar-Capillary Barrier. Cells (2020) 9(11):1–19. doi: 10.3390/cells9112472
189. Wu A, Mihaylova VT, Landry ML, Foxman EF. Interference Between Rhinovirus and Influenza A Virus: A Clinical Data Analysis and Experimental Infection Study. Lancet Microbe (2020) 1(6):e254–62. doi: 10.1016/S2666-5247(20)30114-2
190. Cebrián G, Condón S, Mañas P. Physiology of the Inactivation of Vegetative Bacteria by Thermal Treatments: Mode of Action, Influence of Environmental Factors and Inactivation Kinetics. Foods (2017) 6(12):107. doi: 10.3390/foods6120107
191. Gulraiz F, Bellinghausen C, Dentener MA, Reynaert NL, Gaajetaan GR, Beuken EV, et al. Efficacy of IFN-λ1 to Protect Human Airway Epithelial Cells Against Human Rhinovirus 1B Infection. PloS One (2014) 9(4):e95134. doi: 10.1371/journal.pone.0095134
192. Bellinghausen C, Gulraiz F, Heinzmann AC, Dentener MA, Savelkoul PH, Wouters EF, et al. Exposure to Common Respiratory Bacteria Alters the Airway Epithelial Response to Subsequent Viral Infection. Respir Res (2016) 17(1):68. doi: 10.1186/s12931-016-0382-z
193. Sajjan U, Wang Q, Zhao Y, Gruenert DC, Hershenson MB. Rhinovirus Disrupts the Barrier Function of Polarized Airway Epithelial Cells. Am J Respir Crit Care Med (2008) 178(12):1271–81. doi: 10.1164/rccm.200801-136OC
194. Wang JH, Kwon HJ, Jang YJ. Rhinovirus Enhances Various Bacterial Adhesions to Nasal Epithelial Cells Simultaneously. Laryngoscope (2009) 119(7):1406–11. doi: 10.1002/lary.20498
195. Golda A, Malek N, Dudek B, Zeglen S, Wojarski J, Ochman M, et al. Infection With Human Coronavirus NL63 Enhances Streptococcal Adherence to Epithelial Cells. J Gen Virol (2011) 92(6):1358–68. doi: 10.1099/vir.0.028381-0
196. Avadhanula V, Rodriguez CA, De Vincenzo JP, Wang Y, Webby RJ, Ulett GC, et al. Respiratory Viruses Augment the Adhesion of Bacterial Pathogens to Respiratory Epithelium in a Viral Species- and Cell Type-Dependent Manner. J Virol (2006) 80(4):1629–36. doi: 10.1128/JVI.80.4.1629-1636.2006
197. Min S, Kim S, Cho SW. Gastrointestinal Tract Modeling Using Organoids Engineered With Cellular and Microbiota Niches. Exp Mol Med (2020) 52(2):227–37. doi: 10.1038/s12276-020-0386-0
198. Choi KG, Wu BC, Lee AH, Baquir B, Hancock REW. Utilizing Organoid and Air-Liquid Interface Models as a Screening Method in the Development of New Host Defense Peptides. Front Cell Infect Microbiol (2020) 10:228. doi: 10.3389/fcimb.2020.00228
199. Habibi MS, Thwaites RS, Chang M, Jozwik A, Paras A, Kirsebom F, et al. Neutrophilic Inflammation in the Respiratory Mucosa Predisposes to RSV Infection. Science (2020) 370(6513):eaba:9301. doi: 10.1126/science.aba9301
200. Stark JM, Van Egmond AWA, Zimmerman JJ, Carabell SK, Tosi MF. Detection of Enhanced Neutrophil Adhesion to Parainfluenza-Infected Airway Epithelial Cells Using a Modified Myeloperoxidase Assay in a Microtiter Format. J Virol Methods (1992) 40(2):225–42. doi: 10.1016/0166-0934(92)90071-K
201. Tosi MF, Stark JM, Hamedani A, Smith CW, Gruenert DC, Huang YT. Intercellular Adhesion Molecule-1 (ICAM-1)-Dependent and ICAM-1- Independent Adhesive Interactions Between Polymorphonuclear Leukocytes and Human Airway Epithelial Cells Infected With Parainfluenza Virus Type 2. J Immunol (1992) 149(10):3345–9.
202. Jornot L, Cordey S, Caruso A, Gerber C, Vukicevic M, Tapparel C, et al. T Lymphocytes Promote the Antiviral and Inflammatory Responses of Airway Epithelial Cells. PloS One (2011) 6(10):e29293. doi: 10.1371/journal.pone.0026293
203. Rahmatpanah F, Agrawal S, Jaiswal N, Ngyuen HM, McClelland M, Agrawal A. Airway Epithelial Cells Prime Plasmacytoid Dendritic Cells to Respond to Pathogens via Secretion of Growth Factors. Mucosal Immunol (2019) 12(1):77–84. doi: 10.1038/s41385-018-0097-1
204. Ugonna K, Bingle CD, Plant K, Wilson K, Everard ML. Macrophages are Required for Dendritic Cell Uptake of Respiratory Syncytial Virus From an Infected Epithelium. PloS One (2014) 9(3):e91855. doi: 10.1371/journal.pone.0091855
205. Papazian D, Würtzen PA, Hansen SW. Polarized Airway Epithelial Models for Immunological Co-Culture Studies. Int Arch Allergy Immunol (2016) 170(1):1–21. doi: 10.1159/000445833
206. van der V J, L MM, H BL, C H. Advancing Lung Organoids for COVID-19 Research. Dis Model Mech (2021) 14(6):dmm049060. doi: 10.1242/dmm.049060
207. Suksatu A, Sangsawad W, Thitithanyanont A, Smittipat N, Fukuda MM, Ubol S. Characteristics of Stork Feces-Derived H5N1 Viruses That are Preferentially Transmitted to Primary Human Airway Epithelial Cells. Microbiol Immunol (2009) 53(12):675–84. doi: 10.1111/j.1348-0421.2009.00177.x
208. Kim J-S, Heo P, Yang T-J, Lee K-S, Cho D-H, Kim BT, et al. Selective Killing of Bacterial Persisters by a Single Chemical Compound Without Affecting Normal Antibiotic-Sensitive Cells. Antimicrob Agents Chemother (2011) 55(11):5380–3. doi: 10.1128/AAC.00708-11
209. Huang DTN, Lu CY, Chi YH, Li WL, Chang LY, Lai MJ, et al. Adaptation of Influenza A (H7N9) Virus in Primary Human Airway Epithelial Cells. Sci Rep (2017) 7(1):11300. doi: 10.1038/s41598-017-10749-5
210. Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. MBio (2017) 8(2):e00373–17. doi: 10.1128/mBio.00373-17
211. Forbes B, Ehrhardt C. Human Respiratory Epithelial Cell Culture for Drug Delivery Applications. Eur J Pharm Biopharm (2005) 60(2):193–205. doi: 10.1016/j.ejpb.2005.02.010
212. Fang Y, Eglen RM. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discovery (2017) 22(5):456–72. doi: 10.1177/1087057117696795
Keywords: respiratory viral diseases, primary airway epithelium culture, organoid culture, co-culture, airway modeling
Citation: Rijsbergen LC, van Dijk LLA, Engel MFM, de Vries RD and de Swart RL (2021) In Vitro Modelling of Respiratory Virus Infections in Human Airway Epithelial Cells – A Systematic Review. Front. Immunol. 12:683002. doi: 10.3389/fimmu.2021.683002
Received: 19 March 2021; Accepted: 30 July 2021;
Published: 18 August 2021.
Edited by:
Nabila Seddiki, Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), FranceReviewed by:
Anass Bouchnita, University of Texas at Austin, United StatesAntonella Casola, University of Texas Medical Branch at Galveston, United States
Copyright © 2021 Rijsbergen, van Dijk, Engel, de Vries and de Swart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rik L. de Swart, ci5kZXN3YXJ0QGVyYXNtdXNtYy5ubA==
 Laurine C. Rijsbergen
Laurine C. Rijsbergen Laura L. A. van Dijk1
Laura L. A. van Dijk1 Rory D. de Vries
Rory D. de Vries Rik L. de Swart
Rik L. de Swart