
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 04 June 2021
Sec. B Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.682397
This article is part of the Research Topic Multi-Dimensional Genomic Structures in the Immune System View all 12 articles
Regulation of gene expression in time, space and quantity is orchestrated by the functional interplay of cis-acting elements and trans-acting factors. Our current view postulates that transcription factors recognize enhancer DNA and read the transcriptional regulatory code by cooperative DNA binding to specific DNA motifs, thus instructing the recruitment of transcriptional regulatory complexes forming a plethora of higher-ordered multi-protein-DNA and protein-protein complexes. Here, we reviewed the formation of multi-dimensional chromatin assemblies implicated in gene expression with emphasis on the regulatory role of enhancer hubs as coordinators of stochastic gene expression. Enhancer hubs contain many interacting regulatory elements and represent a remarkably dynamic and heterogeneous network of multivalent interactions. A functional consequence of such complex interaction networks could be that individual enhancers function synergistically to ensure coordination, tight control and robustness in regulation of expression of spatially connected genes. In this review, we discuss fundamental paradigms of such inter- and intra- chromosomal associations both in the context of immune-related genes and beyond.
Regulation of gene expression in time, space and quantity is orchestrated by the functional interplay of cis-acting regulatory elements and trans-acting factors. The establishment of cell identity during development and the response to environmental cues such as pathogens requires transcriptional reprogramming (1, 2). Cells respond to signals by engaging various receptors that recognize and interpret the environment by initiating signal transduction cascades, culminating in the activation of transcription factors (TFs), which bind to specific regulatory DNA sequences to control the expression of genes, thus orchestrating a dynamic interplay between genome form and function (3–6). Inappropriate regulation of gene expression during development or adult life can cause abnormal embryogenesis and disease (7, 8). Enhancers and promoters represent the major classes of DNA regulatory elements responsible for warranting the execution of precise gene expression programs by functioning as information processing units interpreting the extra- and intra-cellular signals in the form of transcription factor binding events, which are followed by the recruitment of various transcriptional regulatory proteins such as cofactors, chromatin modifiers and the transcriptional apparatus at the core promoter (9–15). Core promoters are DNA regions, surrounding the start site of transcription (TSS), directing the accurate initiation of transcription by interacting with the various components of the general transcription machinery, including RNA Pol II (16). In addition, the DNA region (100-200 bp) located immediately upstream of the TSS often bears critical transcription factor binding sites and it is required to increase the rate of transcription (17, 18). In some cases, upstream promoter elements can function in a cell-type specific manner to mediate the interactions with distal regulatory elements like the enhancers (19, 20).
Enhancers activate transcription of target genes independent of their distance, position and orientation relative to the promoter of the affected gene (10, 21, 22). In certain cases, enhancers can act synergistically to ensure for proper gene activation (23). Our current model envisions that TFs recognize enhancer DNA in a combinatorial manner by reading the transcriptional regulatory code through the interplay of nucleotide sequence recognition, sequence context, 3D DNA structure (shape), DNA binding cooperativity, interacting cofactors and nucleosome occupancy (24–29). Establishment and maintenance of cell identity and function arise from integrated actions of transcription factors with chromatin–associated proteins, coactivators, and with the spatially organized genome (30). Combinatorial TF DNA binding leads to a synchronized targeting of enhancers by multifunctional transcriptional regulatory proteins such as coactivators, corepressors, chromatin modifiers and remodelers through direct and indirect physical associations, to assemble a plethora of high-order multi-protein-DNA and protein-protein complexes required for regulation of transcription (4, 9, 14). TFs belong to a diverse class of DNA binding protein families with distinct structural characteristics and individual members from the same family can bind similar DNA motifs, yet carry out distinct functions, in vivo (31–33). In addition, as the number of DNA binding sites in the genome for most TFs is greater than the number of the corresponding protein molecules per cell, it appears that only some TF molecules occupy most of their functional relevant sites in physiological conditions (34–38). TFs typically recognize and bind to short 6-12bp-long degenerate DNA motifs. Consistently, TFs bind to thousands of genomic sites across chromatin landscapes, but the number of genes changing their expression is frequently limited to a few hundred (39–42). Cells, at least in part, resolve this apparent paradox through combinatorial strategies where TFs must work together when bound to enhancers and promoters to achieve the specificity required for the selection of the appropriate genes to become activated. Cooperative DNA binding of TFs to low-affinity sites provides the means to target the correct genes for expression in time, space and in response to signals and is mediated by direct or indirect DNA-and/or chromatin-facilitating contacts between the proteins (33). Interestingly, high throughput ChIP-seq studies identified that binding of TFs to certain genomic regions is highly clustered, each cluster is composed of both functionally related and unrelated TFs, representing strong binding of one or a few TFs, and weaker binding to more degenerate motifs by many other TFs, implying a substantial degree of cooperation between individual motifs (43, 44). Large genomic regions with an unusually high degree of enrichment for lineage-specific TFs, cofactors (e.g. Med1) and histone modifications (H3K27ac) have been recently identified and characterized as super-enhancers (SEs), different from classical enhancers. These clusters of closely spaced enhancer elements often regulate the expression of genes determining cell identity and fate and driving oncogenic transcription (45–48). In general, super-enhancers are involved in strong intradomain interactions that is, between its individual enhancer constituents, which are stronger compared to typical enhancer-promoter interactions (15, 49). As super-enhancers consist of multiple enhancer elements, we assume that there will be complex functional relationships among the different constituents of any given super-enhancer working, depending on the biological context, either in a synergistic or additive manner or a combination thereof, thus resembling the arrangement and functional relationships of TF motifs building a typical enhancer.
One of the best-characterized examples of assembled super-enhancers is provided after stimulation of endothelial cells with TNFα, which leads to the formation of super-enhancers specific for inflammatory genes (50). Major players for the inducible assembly of these super-enhancers are the transcription factor NF-κB and the coactivator BRD4, a member of the bromodomain and extraterminal domain (BET) family of factors that is typically recruited to enhancers and super-enhancers. In brief, stimulation of endothelial cells with TNFα promotes the rapid redistribution of NF-κB across diverse genomic loci and subsequently the assembly of inflammatory SE chromatin configurations. This is accompanied by striking colocalization of the NF-κB p65 subunit and BRD4 in regulatory sequences marked by histone acetylation (H3K27ac). Thus, NF-κB efficiently targets its binding sites and promotes a pronounced recruitment of BRD4 across inflammatory SEs, which coordinate the development of a pro-inflammatory gene expression program (50).
The blood cell-specific SE cluster, known as BENC, is located 1.7 MB downstream of the Myc locus, and consists of multiple enhancer modules working in a combinatorial manner. BENC is bound by the transcription factors GFI1b, RUNX1 and MYB (51) and is essential for the regulation of Myc expression in hematopoietic and leukemic stem cells critical for hematopoietic malignancies. BENC deletion in hematopoietic stem cells closely mimics the conditional deletion of the Myc gene. Thus, overlapping, adjacent and/or partially redundant regulatory units arranged in clusters and bound by a variety of transcription factors functioning in a combinatorial manner, are essential for the generation of cell-type specific transcriptional regulatory outputs.
The application of high-throughput sequencing-based chromatin profiling (e.g. ChIP-seq, DNAseI-seq), ideal for measuring the potential regulatory activity, indicates that the human genome contains about a million candidate different enhancers (52, 53) interspersed at regions located proximally, distantly, or within genes, and that in each cell type a fraction of these putative enhancers is active. Enhancers regulate gene expression in quite diverse manners. For example, enhancers do not necessarily regulate their closest gene (10), each gene may be regulated by more than one enhancer (54, 55), and a single enhancer can regulate multiple genes (56). The above mechanistic principles suggest the existence of complex and highly dynamic enhancer-promoter networks and diverse modes of enhancer action. The multiplicity of the enhancer-promoter functional interactions is highly regulated by transcription factors and cofactors associated with them to direct the re-configuration and the spatial folding of the 3D chromatin interaction, thus bridging remotely located transcriptional regulatory elements to a functional assembly. Although there is strong evidence that enhancer-promoter interactions are usually established concurrently with gene activation, and thus comprising an integral part of global genomic regulation, it is not yet proven whether enhancer-promoter contacts are the cause or the consequence of regulated gene expression. Some enhancer-promoter paired configurations are formed in the absence of any transcriptional events, thus suggesting that, simply, their identification does not necessarily have functional implications (57). However, forced chromatin looping between an enhancer and a promoter led to induction of transcription at high levels (58), thus strongly suggesting the requirement for establishing functional 3D structures connecting enhancers and promoters that could be integrated into a higher order condensed multi-genic 3D transcriptionally-active hubs at specific nuclear positions to coordinate gene expression of functionally related genes (59, 60).
Furthermore, although the identification of specific enhancer-promoter contacts provides important information regarding the assignment of enhancers to the correct target genes, these interactions are not necessarily predictive of functional regulation. For example, recent studies have shown that sonic hedgehog (Shh) expression in the developing embryo is regulated by multiple enhancer elements. Using a combination of 3D-DNA FISH and chromosome conformation capture approaches, Benabdallah et al., demonstrated that Shh expression is incompatible with the classic enhancer-promoter looping model. The authors found that, practically, there is no spatial proximity between the enhancer and the promoter during the differentiation of embryonic stem cells to progenitor neural cells (61). In contrast, however, Shh expression in the zone of polarized activity in the developing limb in mice, which is controlled by a different enhancer called ZRS, is characterized by the highest enhancer-promoter proximity as compared to other non-expressing tissues, a finding consistent with the formation of a tight chromosomal loop between the enhancer and the promoter (62). Similar to the findings of Shh expression in ES cells, live-cell imaging experiments revealed that the expression of the key pluripotency transcription factor Sox2 in ES cells is not controlled by the spatial proximity between its control region SCR and the promoter, despite the fact that 3C assays have identified enriched contacts between the enhancer and the promoter (63). Although it is not yet fully understood how a spatially remote enhancer could affect the activity of its target promoter, we speculate that the assembly of condensates composed of transcriptional regulatory proteins could trigger the formation of large macromolecular bridges or hubs, which may alter the configuration of the intervening chromatin structure, thus leading to increased enhancer-promoter spatial distances, despite the fact that these regulatory sequences are functionally linked.
Essential questions related to the formation of dense enhancer-promoter or enhancer-enhancer networks are how they are established, maintained, or dissolved. We speculate that it would be difficult or nearly impossible for any distal enhancer to locate its target promoter if the genome was largely homogeneously structured, thus lacking an internal higher order organization. We now know that, in general, the hierarchical organization of chromatin involves the A (euchromatin) and B (heterochromatin) compartments, with compartment B marked by a more compact chromatin packaging as compared to compartment A (64), which is characterized by increased chromatin accessibility and higher gene expression levels (65). Chromatin contacts within each compartment are specifically enriched, but contacts between genomic regions present in the A and B compartments are not favored. Each chromatin compartment is further substructured to additional hierarchical layers termed Topological Associated Domains (TADs) (66, 67), where regions within the same TAD tend to interact more frequently with each other than with regions belonging to different TADs. Thus, within TADs, numerous genomic interactions (loops) are formed connecting regulatory elements such as enhancers and promoters, with a significantly smaller number of interactions spanning across the boundaries of adjacent TADs. TAD borders are frequently demarcated by the binding of CTCF, a TF functioning as a chromatin organizer (68). TAD anchors are frequently bound by the CTCF-cohesin complex usually in a convergent motif orientation (66) through a loop extrusion process mediated by the cohesin ring until the process is blocked at CTCF sites arranged in a convergent orientation (69). Mutations at the TAD boundaries can lead to chromatin reconfiguration at the TAD borders causing enhancers present in one TAD to activate transcription of genes present in the neighboring TAD (70). More specifically, rearrangement of TAD boundaries at the WNT6/IHH/EPHA4/PAX3 locus caused an extensive rewiring of the interactions between enhancers and promoters leading to limb malformations (70). The functional insulatory role of CTCF in gene regulation was further investigated in a study where the disruption of CTCF binding at the TAD boundaries via CRISPR led to the aberrant interaction of a constitutive enhancer with the PDGFRA oncogene in glioma (71). Interestingly, removal of CTCF binding sites both at the boundary and within TADs at the Sox9-Kcnj2 locus led to fusion of neighboring TADs, which, however, did not result in major effects on developmental gene regulation (72). In contrast, however, inversion of CTCF binding sites within the protocadherin enhancer rewires the enhancer-promoter interactions and alters gene expression patterns (73). Taken together, the data presented above indicate that the precise role of chromatin topology in enhancer/promoter function is determined by the specific genomic context.
An important issue regarding the multiplicity of enhancer-promoter network of interactions relates to the mechanisms by which an enhancer selects the correct promoter for activation. While within TADs, the number of candidate promoter targets for a given enhancer is relatively limited, in many cases enhancers and their target promoters are not adjacent in the primary DNA sequence, but interrupted by one or more genes that are not co-expressed or co-regulated with the enhancer’s target gene. For example, expression of the Sex comps reduced (Scr) gene during Drosophila embryogenesis is controlled by an enhancer which is located far upstream. Importantly, the continuity of the Scr gene and its enhancer is interrupted by the localization of the fushi tarazu (ftz) gene, which is regulated by an enhancer positioned at close distance from the Scr gene, whereas the Scr enhancer is present at the 3’ of the ftz gene. In other words, the Scr enhancer is closer to ftz than to Scr promoter, and the ftz enhancer is flanked by the Scr and ftz promoters. However, both genes are expressed at different times during embryogenesis, and their expression is controlled by their individual enhancers. The selectivity of the Scr enhancer for activating transcription from the Scr promoter and not from the closest ftzp promoter depends on promoter proximal DNA elements called tethering elements, which when bound by specific transcription factors might mediate highly specific enhancer-promoter interactions between the Scr enhancer and promoter (19, 74). These data are compatible with the looping and relocation models for enhancer-promoter communication, because they can explain how an enhancer can skip more proximal promoters (75).
The structural chromatin loops responsible for forming TADs are dependent on ubiquitously expressed CTCF and cohesin proteins and therefore are distinct from those assembled by the interactions of regulatory sequences. The latter are generally mediated by enhancer- and promoter-bound transcription factors and cofactors and therefore their formation is dynamic and developmentally regulated (30). Our current understanding is that TADs define regions of the genome where enhancer-promoter interactions are allowed to occur to ensure robust and reliable transcriptional activation, thus precluding inappropriate regulatory interactions between enhancers and promoters located in different TADs (76). Despite the established roles of the architectural protein CTCF in supporting regulatory enhancer-promoter interactions (66, 73), its depletion although leads to a significant reduction of TAD structures results in moderate rather than dramatic changes in global gene expression (77, 78). An emerging idea derived from single-cell 3C and advanced imaging experiments (79, 80) to explain this unexpected finding is that the cell-to-cell variability in the overall TAD structure indicates that the existence of a highly heterogeneous structure of the chromatin could provide a reasonable explanation for the relatively modest effects in transcription upon TAD disruption. Thus, it is not yet fully understood how TADs and TAD boundaries affect the regulation of gene expression, and the regulatory logic that rules their involvement.
The spatial arrangement of the regulatory genome in 3D has been mainly studied by Chromatin Conformation Capture (3C) methodologies and its ChIP or probe-based derivatives and advanced high-resolution microscopy techniques (62, 66, 81–85). Notably, some of the first examples of well-characterized mechanisms of gene regulation by DNA-DNA contacts in mammals, were derived from studies in hematopoietic and immune cells regarding the 3D organization of the β-globin gene locus (86) and the interchromosomal association between the IFN-gamma gene locus and regulatory regions of the T(H)2 cytokine locus in a dynamic and cell-type-specific fashion (87). Additional studies have provided evidence that transcription factors bound to enhancers and promoters mediate this type of DNA-DNA communication by acting as the components exposing complementary interacting protein surfaces required for the establishment and the specificity for the formation of looped structures (20). A well-characterized example of a looping factor essential for chromatin interactions at the β-globin locus is the cofactor Ldb1 (58). Ldb1 together with GATA1 acts as a molecular bridge, which facilitates the communication between the Locus Control Region (LCR) and the β-globin promoter.
In the case of the virus inducible human IFNβ stochastic gene expression, the IFNβ enhancer contributes to the formation of highly specialized 3D associations required for singular and stochastic expression of the IFNβ gene in a subset of infected cells (35, 88, 89). Previous work has indicated that virus infection induces the coordinated activation of three distinct sets of transcription factors, NF-κB, IRF3 and 7, and ATF2/cJun, which together with the architectural protein HMG I(Y) bind cooperatively to the nucleosome-free enhancer/promoter of the IFNβ gene and form a nucleoprotein structure known as the enhanceosome (24, 90–95) (Figure 1). Following its assembly, the IFNβ enhanceosome exposes a continuous recognition surface used to recruit the transcriptional coactivator CBP (93). Enhanceosomes with similar properties have now been identified and characterized on several model enhancers in both Drosophila and mammalian systems. For example, cell type-specific and/or inducible expression of the MHC class I and MHC class II expression, which is crucial for the initiation and regulation of adaptive immunity, requires the assembly of enhanceosomes which activate transcription by recruiting the coactivators NLRC5- and CIITA, respectively (96). As it is the case for the IFNβ enhanceosome, the NLRC5 and CIITA coactivators are recruited to their target promoters by interacting simultaneously with a continuous surface of the MHC Class I and Class II enhanceosome components, respectively.
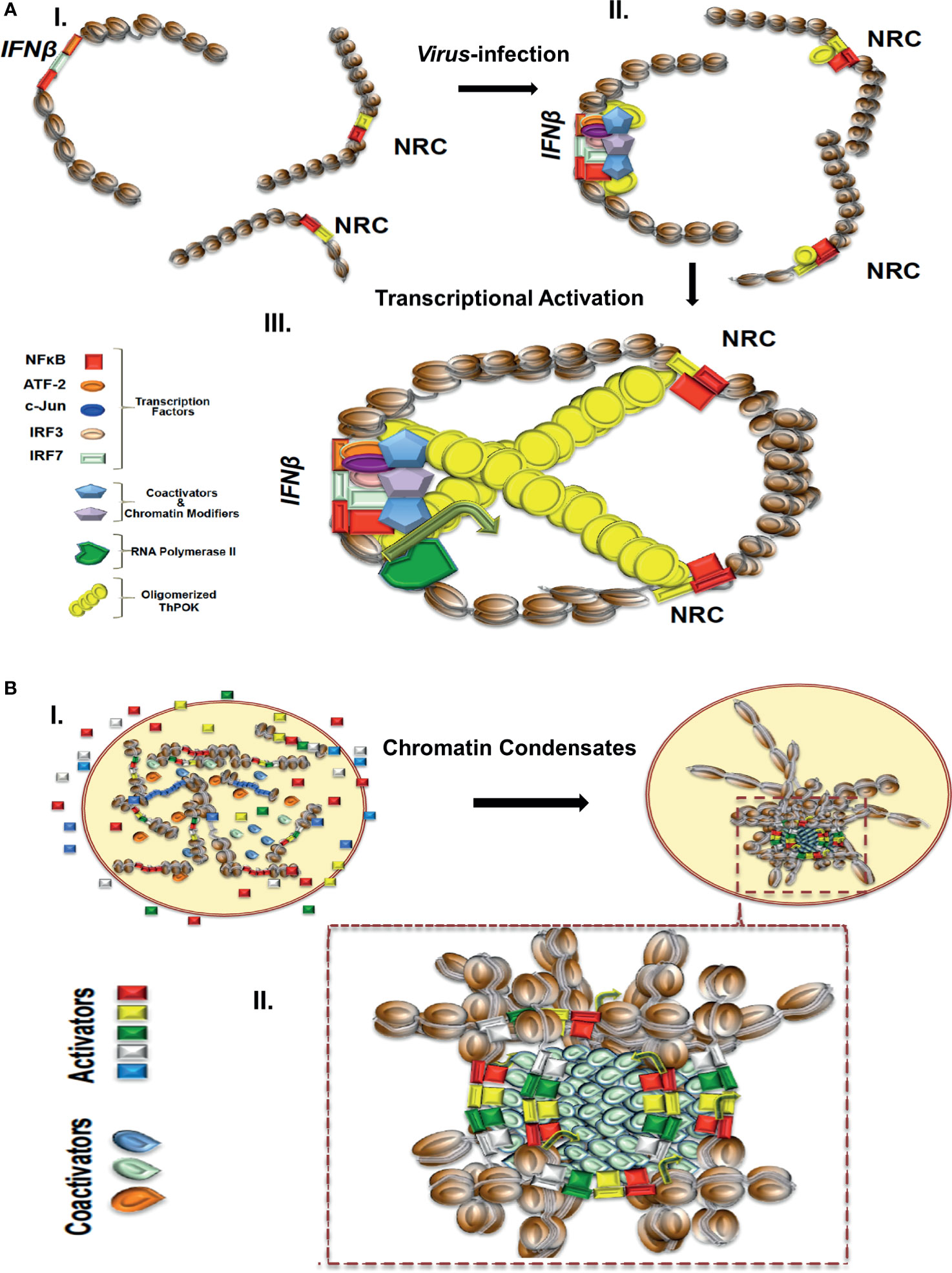
Figure 1 (A) A model depicting the virus infection-induced NRC-IFNβ interchromosomal interactions. In uninfected cells, the IFNB and NRC loci are well separated. Virus infection induces the nuclear localization of NF-κB, which binds cooperatively with ThPOK to appropriately spaced binding sites at NRCs (right part). While bound to NRCs, ThPOK oligomerizes to reach to a single IFNβ allele resulting in physical proximity of the NRCs-bound NF-κB with the IFNβ enhancer triggering enhanceosome assembly and subsequently activation of the IFNβ gene. (B) A model depicting the assembly of enhancer hubs and transcriptional condensates. In resting cells enhancers and transcriptional regulators are diffused in the nucleus. Upon cell activation transcriptional condensates bearing enhancers, promoters and regulators are formed as a result of liquid-liquid phase separation. Components are highly concentrated within the condensates.
The human IFNβ promoter contains one well-positioned nucleosome physically obscuring the TATA box and the start site of transcription. Once assembled, the IFNβ enhanceosome dynamically recruits histone acetyltransferases and chromatin remodelers causing sliding of the nucleosome masking the TATA to a new position 36bp downstream, thus unmasking the TATA box and the start site of transcription (91, 97, 98). The gene is initially expressed from a single allele, but its expression is subsequently becoming biallelic (88, 99). The discovery of the early monoallelic IFNβ expression suggested that gene activation itself is successfully completed only with a certain probability in a given cell. A similar mode of monoallelic stochastic activation has also been described for the IFNα family of genes and it is a typical and general feature of cytokine expression during immune responses (100, 101).
Given the pivotal role of IFNs in antiviral host responses, it has come as a surprise that, at the single-cell level, IFNs are expressed only by a fraction of virus-infected cells. Indeed, approximately ~20% of the virus-infected cells are expressing IFNβ. Moreover, the induction of Interferon Stimulated Genes (ISGs) after acute viral infections has also been found to be highly heterogeneous at the single-cell level (99), with only a fraction of cells inducing an antiviral gene expression program at physiological concentrations of IFNs.
Recent findings have indicated that IFNβ expression heterogeneity is due, at least in part, to stochastic interchromosomal interactions driving the variable patterns of IFNβ gene expression in response to virus infection (35, 88, 89). The important finding was that stochasticity in expression of human IFNβ is due to low intra-cellular concentrations of the transcription factor NF-κB, which is captured onto specialized DNA sites called NRCs (NF-κB Reception Centers) belonging to Alu-like repetitive elements and then it is delivered to the IFNβ enhancer, via stochastic interchromosomal interactions. It was shown that the transcription factor ThPOK (89), a GAGA binding factor, binds cooperatively with NF-κB to NRCs and mediates their physical proximity with the IFNβ gene and other co-regulated genes via its ability to oligomerize when bound to DNA, as it was shown by in vitro experiments. Furthermore, ThPOK knockdown experiments significantly decreased the frequency of interchromosomal interactions, NF-κB DNA binding to the IFNβ enhancer, and virus-induced IFNβ gene activation. Cooperative DNA binding between ThPOK and NF-κB, in vitro and in vivo, on the same face of the double DNA helix, is required for interchromosomal interactions and this distinguishes NRCs from various other Alu elements bearing interspersed NF-κB and ThPOK sites (89) (Figure 1A). These studies showed how DNA binding cooperativity of stereospecifically aligned transcription factors provides the necessary ultrasensitivity for the all-or-none stochastic cell responses to virus infection. The above mechanistic insights strongly suggest that the primary DNA sequence of the interacting DNA elements acts as a blueprint, which contains a specific linear genomic code that is interpreted via the construction of highly specialized high-order nucleoprotein complexes. Additional studies have also shown that stochastic IFNβ expression in response to virus infection is due to cell-to-cell variability due to limiting quantities of components ranging from the recognition of viral RNA by host factors and the activation of signaling pathways, to the exact levels of activated transcription factors (102).
Additional examples of architectural proteins acting as organizers of 3D genome architecture include YY1 (103), ZNF143 (104), and SATB1 (105). In contrast to CTCF, YY1 preferentially binds to active enhancers and promoters mediating their 3D interaction via its ability to dimerize (103). ZNF143 binds to the anchors of chromatin interactions and facilitates the formation of secure connections between promoters and distal regulatory elements (104). Finally, SATB1 can act as a tissue-specific organizer of gene expression with well-established roles in the regulation of TH2 cytokine gene locus (105, 106) by cooperative binding to multiple closely spaced consensus sites in nucleosomes (107). In summary, the examples described above demonstrate that the formation of 3D DNA-DNA nuclear contacts is not a purely random phenomenon but it depends on sequences hosting chromatin architectural proteins, which in many cases are in direct interaction with TFs.
The molecular mechanisms controlling olfactory receptor (OR) gene transcription (108) inducing the monoallelic expression of one out of more than 1,000 genes in each cell, share many common features with the cellular responses leading to virus-induced stochastic monoallelic IFNβ expression. The singular and stochastic expression of OR genes is controlled by a dense network of 3D DNA interactions occurring exclusively in olfactory sensory neurons (109–111). The OR genes, which are interspersed in various chromosomes, aggregate in one nuclear compartment, where they are repressed by acquiring chromatin marks characteristic of heterochromatin. However, this aggregation brings in close proximity many OR enhancers located adjacent to the OR genes via multiple interchromosomal interactions forming a multi-enhancer hub. Monoallelic OR transcription is activated by the multi-enhancer hub, which associates stochastically with the corresponding gene chosen for expression. The TFs Lhx2 and Ebf bind cooperatively in a stereospecific manner on OR enhancers followed by the recruitment of the LIM domain protein Ldb1, which facilitates these interchromosomal interactions. Thus, DNA binding cooperativity between transcription factors, as it is also the case for IFNβ expression, provides the necessary specificity to drive spatiotemporal alterations of the 3D chromatin for the cell-type specific expression of OR genes and cellular responses upon viral infections, respectively.
Many mechanistic similarities to the stochastic monoallelic OR and IFNβ expression are also shared by the NF-κB-dependent TNFα-induced hierarchical co-expression of SAMD4A, TNFAIP2, and SLC6A5 genes (112, 113), which also form a tight signal-dependent multigene complex through intra- and interchromosomal interactions. TNFα induction causes the hierarchical co-transcription of these genes in a small fraction of cells containing tightly associated genes in a multigene complex.
Photoreceptor specification in the Drosophila eye depends on the stochastic and cell-autonomous expression of the transcription factor Spineless (Ss) (114–116). The stochastic Ss expression in individual cells is regulated by a 60 kb region harboring the R7/8 enhancer and the silencer elements 1 and 2 (115, 116). It was shown that the stochastic Ss expression pattern depends on interchromosomal communication and on inter-allelic crosstalk, which averages the frequency of expression of each allele (116). Thus, although each allele follows an independent-stochastic choice of expression, 3D interchromosomal communication coordinates the expression status between alleles, thus ensuring for the proper expression of both alleles within the same randomly selected subset of photoreceptor cells. Conclusively, both the 3D interchromosomal interactions and the action of enhancers from a distance determine stochastic photoreceptor specification.
In summary, the examples described above when taken together with many other similar studies, suggest a general model according to which cell activation by environmental signals or during development and differentiation leads to a cascade of multiprotein and multigene high order dynamic assemblies, ranging from signalsomes to enhanceosomes and chromatin-modifying machines, thus providing local regulatory input from different genomic elements. This mode of information flow ensures that stochastic molecular interactions are canalized to produce specific, robust and reliable outputs. Furthermore, these observations strongly support the view that transcription factors are capable of inducing extensive chromatin rewiring, in many cases stochastic, at specific genomic loci, thus modifying gene expression patterns in a cell-specific manner.
Enhancer hubs containing highly interacting regulatory elements represent a remarkably dynamic and heterogeneous network of multivalent macromolecular interactions. Most of these interactions could be established in a stochastic manner and individual cells within a homogeneous population may contain unique combinations of two or more interacting partners. The higher the number of interacting partners the lower the probability of a cell in the population to contain multi-partner hubs. We speculate that enhancer hubs by containing multiple enhancers targeting a single promoter or many promoters targeted by a single enhancer or many interconnecting enhancers targeting many different promoters, could aggregate into a single nuclear position. A functional consequence of such complex interaction networks could be that different enhancers function synergistically to ensure coordination, tight control and robustness in regulation of expression of spatially connected genes. Thus, enhancer-promoter hubs ensure that all target genes are “on” and are appropriately regulated in a spatiotemporal manner, with stable relative levels of expression. Thus, each gene is subject to the same intrinsic and extrinsic noise e.g. fluctuations in the concentration of transcriptional regulators etc. The high density of transcription factors bound to enhancers and promoters in the context of enhancer hubs or super-enhancers leads to the cooperative and synergistic recruitment of high concentration of coactivators, components of the transcriptional machinery and other cofactors (117). Conclusively, an important implication derived from the assembly of enhancer hubs is the accumulation of transcriptional regulators at high local concentrations, which then are equally available for binding to the enhancer and promoter elements of the associated genes in a locus-specific manner.
Since many transcriptional regulators typically contain large low complexity and/or intrinsically disordered regions (IDRs), like the activation domains of many TFs, the probability to undergo hetero- or homo-typic multivalent interaction or oligomerization is high (118). IDRs have been described as functional protein segments that are not likely to form a defined 3D structure (119) and have been classified mainly by their amino acid profile or by their hypothetical shape (120). High densities of proteins bearing IDRs and nucleic acids have been implicated in the formation of membraneless compartments through the well-understood mechanism of liquid-liquid phase separation (LLPS) (121–125). The term phase separation describes a process by which distinct liquid droplets bearing high concentration of molecules are formed. The formation of such nuclear compartments increases the likelihood for certain biochemical reactions to occur, such as transcription, thus bypassing partially or entirely the necessity for direct physical contacts between individual enhancers and promoters (Figure 1B). Theoretically, an additional advantage of regulating coordination of transcription via a phase separation model is the plasticity and reversibility by which LLPS formations are driven, and thus provide a reliable mechanism of transcriptional initiation and termination of many coregulated genes at once. For example, clusters of enhancer elements have been proposed to regulate biological processes implicated in development and disease (47), as well as regulating mammalian cell identity (45). We speculate that in the case of the SEs assembling upon TNFα induction of endothelial cells (see above), the pronounced recruitment of cofactors, like BRD4, etc. could lead to the additional recruitment of RNA Pol II and cofactors via an extensive network of intermolecular interactions between specific regions of intrinsically disordered hydrophilic activation domains (IDRs), thus leading to the formation of phase-separated nuclear liquid droplets (126) (Figure 1B).
The proposed model describes that the highly condensed concentration of biochemical interactions between TFs and coactivators, occurring on TFBSs-enriched extended 3D chromatin configurations, promote the colocalization/co-recruitment of diverse SEs at specific nuclear sites and subsequently the formation of 3D chromatin condensates (Figure 1B).
The ability of eukaryotic cells to assemble high-order transcription condensates ensures that enhancers bearing low-affinity binding sites can efficiently recruit RNA Pol II molecules to target genes to support robust patterns of gene expression (27, 125, 126). It is reasonable to assume that the individual functions of SEs could become modified upon their recruitment to the 3D chromatin condensates, where they could acquire novel regulatory characteristics hardwired in dense networks of macromolecular interactions.
All authors contributed to the article and approved the submitted version.
DT was supported from the Greek Secretariat for Research and Technology (Cooperative Grants SYNERGASIA I #969, Excellence Award ARISTEIA I #1567, European Committee (FP7 projects INTEGER, NANOMA, PREDICTA and BIOFOS), from the European Economic Area (EL0084), BIO-IMAGING GR (MIS-5002755), and from the KMW offsets program. MA was supported from research grants from the Bodossaki Foundation, Fondation Santé, Gilead Hellas, and Antonios and Ioannis Angelicoussis Foundation. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dimitris Valakos and Marianna Koutsi and members of our lab for helpful discussions. We apologize to the authors of many relevant and important studies for not citing their work due to space limitations.
1. Sung MH. Transcriptional Reprogramming. In: Dubitzky W., Wolkenhauer O., Cho KH., Yokota H. (eds) Encyclopedia of Systems Biology. New York, NY: Springer (2013). pp. 2258–9. doi: 10.1007/978-1-4419-9863-7_756
2. Pope SD, Medzhitov R. Emerging Principles of Gene Expression Programs and Their Regulation. Mol Cell (2018) 71(3):389–97. doi: 10.1016/j.molcel.2018.07.017
3. Weake VM, Workman JL. Inducible Gene Expression: Diverse Regulatory Mechanisms. Nat Rev Genet (2010) 11(6):426–37. doi: 10.1038/nrg2781
4. Smale ST, Natoli G. Transcriptional Control of Inflammatory Responses. Cold Spring Harb Perspect Biol (2014) 6(11):a016261. doi: 10.1101/cshperspect.a016261
5. Furlong EEM, Levine M. Developmental Enhancers and Chromosome Topology. Science (2018) 361(6409):1341–5. doi: 10.1126/science.aau0320
6. Voss TC, Hager GL. Dynamic Regulation of Transcriptional States by Chromatin and Transcription Factors. Nat Rev Genet (2014) 15:69–81. doi: 10.1038/nrg3623
7. Lee TI, Young RA. Transcriptional Regulation and its Misregulation in Disease. Cell (2013) 152(6):1237–51. doi: 10.1016/j.cell.2013.02.014
8. Richels R, Shilatifard A. Enhancer Logic and Mechanics in Development and Disease. Trends Cell Bio (2018) 28(8):608–30. doi: 10.1016/j.tcb.2018.04.003
9. Spitz F, Furlong EE. Transcription Factors: From Enhancer Binding to Developmental Control. Nat Rev Genet (2012) 13(9):613–26. doi: 10.1038/nrg3207
10. Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: Five Essential Questions. Nat Rev Genet (2013) 14(4):288–95. doi: 10.1038/nrg3458
11. Calo E, Wysocka J. Modification of Enhancer Chromatin: What, How, and Why? Mol Cell (2013) 49(5):825–37. doi: 10.1016/j.molcel.2013.01.038
12. Heinz S, Romanoski CE, Benner C, Glass CK. The Selection and Function of Cell Type-Specific Enhancers. Nat Rev Mol Cell Biol (2015) 16(3):144–54. doi: 10.1038/nrm3949
13. Reiter F, Wienerroither S, Stark A. Combinatorial Function of Transcription Factors and Cofactors. Curr Opin Genet Dev (2017) 43:73–81. doi: 10.1016/j.gde.2016.12.007
14. Catarino RR, Stark A. Assessing Sufficiency and Necessity of Enhancer Activities for Gene Expression and the Mechanisms of Transcription Activation. Genes Dev (2018) 32(3-4):202–23. doi: 10.1101/gad.310367.117
15. Oudelaar AM, Davies JOJ, Hanssen LLP, Telenius JM, Schwessinger R, Liu Y, et al. Single-Allele Chromatin Interactions Identify Regulatory Hubs in Dynamic Compartmentalized Domains. Nat Genet (2018) 50:1744–51. doi: 10.1038/s41588-018-0253-2
16. Bhuiyan T, Timmers HTM. Promoter Recognition: Putting TFIID on the Spot. Trends Cell Biol (2019) 29(9):752–63. doi: 10.1016/j.tcb.2019.06.004
17. McKnight SL, Kingsbury R. Transcriptional Control Signals of a Eukaryotic Protein-Coding Gene. Science (1982) 217(4557):316–24. doi: 10.1126/science.6283634
18. Myers RM, Tilly K, Maniatis T. Fine Structure Genetic Analysis of a Beta-Globin Promoter. Science (1986) 232(4750):613–8. doi: 10.1126/science.3457470
19. Calhoun VC, Stathopoulos A, Levine M. Promoter-Proximal Tethering Elements Regulate Enhancer-Promoter Specificity in the Drosophila Antennapedia Complex. Proc Natl Acad Sci U S A (2002) 99(14):9243–7. doi: 10.1073/pnas.142291299
20. Nolis IK, McKay DJ, Mantouvalou E, Lomvardas S, Merika M, Thanos D. Transcription Factors Mediate Long-Range Enhancer-Promoter Interactions. Proc Natl Acad Sci U S A (2009) 106(48):20222–7. doi: 10.1073/pnas.0902454106
21. Visel A, Rubin EM, Pennacchio LA. Genomic Views of Distant-Acting Enhancers. Nature (2009) 461(7261):199–205. doi: 10.1038/nature08451
22. Schoenfelder S, Fraser P. Long-Range Enhancer-Promoter Contacts in Gene Expression Control. Nat Rev Genet (2019) 20(8):437–55. doi: 10.1038/s41576-019-0128-0
23. Guerrero L, Marco-Ferreres R, Serrano AL, Arredondo JJ, Cervera M. Secondary Enhancers Synergise With Primary Enhancers to Guarantee Fine-Tuned Muscle Gene Expression. Dev Biol (2010) 337(1):16–28. doi: 10.1016/j.ydbio.2009.10.006
24. Thanos D, Maniatis T. Virus Induction of Human IFN Beta Gene Expression Requires the Assembly of an Enhanceosome. Cell (1995) 83(7):1091–100. doi: 10.1016/0092-8674(95)90136-1
25. Agelopoulos M, Thanos D. Epigenetic Determination of a Cell-Specific Gene Expression Program by ATF-2 and the Histone Variant Macroh2a. EMBO J (2006) 25(20):4843–53. doi: 10.1038/sj.emboj.7601364
26. Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, et al. Cofactor Binding Evokes Latent Differences in DNA Binding Specificity Between Hox Proteins. Cell (2011) 147(6):1270–82. doi: 10.1016/j.cell.2011.10.053
27. Farley EK, Olson KM, Zhang W, Rokhsar DS, Levine MS. Syntax Compensates for Poor Binding Sites to Encode Tissue Specificity of Developmental Enhancers. Proc Natl Acad Sci U S A (2016) 113(23):6508–13. doi: 10.1073/pnas.1605085113
28. Lavigne MD, Vatsellas G, Polyzos A, Mantouvalou E, Sianidis G, Maraziotis I, et al. Composite Macroh2a/NRF-1 Nucleosomes Suppress Noise and Generate Robustness in Gene Expression. Cell Rep (2015) 11(7):1090–101. doi: 10.1016/j.celrep.2015.04.022
29. Inukai S, Kock KH, Bulyk ML. Transcription factor-DNA Binding: Beyond Binding Site Motifs. Curr Opin Genet Dev (2017) 43:110–9. doi: 10.1016/j.gde.2017.02.007
30. Stadhouders R, Fillion GJ, Graf T. Transcription Factors and 3D Genome Conformation in Cell-Fate Decisions. Nature (2019) 569:345–54. doi: 10.1038/s41586-019-1182-7
31. Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A Census of Human Transcription Factors: Function, Expression and Evolution. Nat Rev Genet (2009) 10(4):252–63. doi: 10.1038/nrg2538
32. Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The Human Transcription Factors. Cell (2018) 172(4):650–65. doi: 10.1016/j.cell.2018.01.029
33. Kribelbauer JF, Rastogi C, Bussemaker HJ, Mann RS. Low-Affinity Binding Sites and the Transcription Factor Specificity Paradox in Eukaryotes. Annu Rev Cell Dev Biol (2019) 35:357–79. doi: 10.1146/annurev-cellbio-100617-062719
34. Hottiger MO, Felzien LK, Nabel GJ. Modulation of Cytokine-Induced HIV Gene Expression by Competitive Binding of Transcription Factors to the Coactivator P300. EMBO J (1998) 17(11):3124–34. doi: 10.1093/emboj/17.11.3124
35. Antonaki A, Demetriades C, Polyzos A, Banos A, Vatsellas G, Lavigne MD, et al. Genomic Analysis Reveals a Novel Nuclear Factor-κb (Nf-κb)-Binding Site in Alu-repetitive Elements. J Biol Chem (2011) 286(44):38768–82. doi: 10.1074/jbc.M111.234161
36. Wong D, Teixeira A, Oikonomopoulos S, Humburg P, Lone IN, Saliba D, et al. Extensive Characterization of NF-κb Binding Uncovers non-Canonical Motifs and Advances the Interpretation of Genetic Functional Traits. Genome Biol (2011) 12(7):R70. doi: 10.1186/gb-2011-12-7-r70
37. Simicevic J, Schmid AW, Gilardoni PA, Zoller B, Raghav SK, Krier I, et al. Absolute Quantification of Transcription Factors During Cellular Differentiation Using Multiplexed Targeted Proteomics. Nat Methods (2013) 10(6):570–6. doi: 10.1038/nmeth.2441
38. Gillespie MA, Palii CG, Sanchez-Taltavull D, Shannon P, Longabaugh WJR, Downes DJ, et al. Absolute Quantification of Transcription Factors Reveals Principles of Gene Regulation in Erythropoiesis. Mol Cell (2020) 78(5):960–74.e11. doi: 10.1016/j.molcel.2020.03.031
39. Vockley CM, D’Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, et al. Direct GR Binding Sites Potentiate Clusters of TF Binding Across the Human Genome. Cell (2016) 166(5):1269–81.e19. doi: 10.1016/j.cell.2016.07.049
40. Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, et al. Genomic Determination of the Glucocorticoid Response Reveals Unexpected Mechanisms of Gene Regulation. Genome Res (2009) 19(12):2163–71. doi: 10.1101/gr.097022.109
41. Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive Cooperation of Immune Master Regulators IRF3 and Nfκb in RNA Pol II Recruitment and Pause Release in Human Innate Antiviral Transcription. Cell Rep (2013) 4(5):959–73. doi: 10.1016/j.celrep.2013.07.043
42. Barakat TS, Halbritter F, Zhang M, Rendeiro AF, Perenthaler E, Bock C, et al. Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell (2018) 23(2):276–288.e8. doi: 10.1016/j.stem.2018.06.014
43. Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, et al. Transcription Factor Binding in Human Cells Occurs in Dense Clusters Formed Around Cohesin Anchor Sites. Cell (2013) 154(4):801–13. doi: 10.1016/j.cell.2013.07.034
44. Partridge EC, Chhetri SB, Prokop JW, Ramaker RC, Jansen CS, Goh ST, et al. Occupancy Maps of 208 Chromatin-Associated Proteins in One Human Cell Type. Nature (2020) 583(7818):720–8. doi: 10.1038/s41586-020-2023-4
45. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master Transcription Factors and Mediator Establish Super-Enhancers At Key Cell Identity Genes. Cell (2013) 153(2):307–19. doi: 10.1016/j.cell.2013.03.035
46. Shin HY, Willi M, HyunYoo K, Zeng X, Wang C, Metser G, et al. Hierarchy Within the Mammary STAT5-driven Wap Super-Enhancer. Nat Genet (2016) 48(8):904–11. doi: 10.1038/ng.3606
47. Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen L, et al. Genetic Dissection of the α-Globin Super-Enhancer In Vivo. Nat Genet (2016) 48(8):895–903. doi: 10.1038/ng.3605
48. Dukler N, Gulko B, Huang YF, Siepel A. Is a Super-Enhancer Greater Than the Sum of its Parts? Nat Genet (2016) 49(1):2–3. doi: 10.1038/ng.3759
49. Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, et al. Spatial Enhancer Clustering and Regulation of Enhancer-Proximal Genes by Cohesion. Genome Res (2015) 25:504–13. doi: 10.1101/gr.184986.114
50. Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, et al. Nf-κb Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol Cell (2014) 56(2):219–31. doi: 10.1016/j.molcel.2014.08.024
51. Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, et al. A Myc Enhancer Cluster Regulates Normal and Leukaemic Haematopoietic Stem Cell Hierarchies. Nature (2018) 553(7689):515–20. doi: 10.1038/nature25193
52. ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
53. ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, et al. Expanded Encyclopaedias of DNA Elements in the Human and Mouse Genomes. Nature (2020) 583(7818):699–710. doi: 10.1038/s41586-020-2493-4
54. Agelopoulos M, McKay DJ, Mann RS. Developmental Regulation of Chromatin Conformation by Hox Proteins in Drosophila. Cell Rep (2012) 1(4):350–9. doi: 10.1016/j.celrep.2012.03.003
55. McKay DJ, Estella C, Mann RS. The Origins of the Drosophila Leg Revealed by the Cis-Regulatory Architecture of the Distalless Gene. Development (2009) 136(1):61–71. doi: 10.1242/dev.029975
56. Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, et al. Deletion of a Coordinate Regulator of Type 2 Cytokine Expression in Mice. Nat Immunol (2001) 2(9):842–7. doi: 10.1038/ni0901-842
57. Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, et al. Enhancer Loops Appear Stable During Development and are Associated With Paused Polymerase. Nature (2014) 512(7512):96–100. doi: 10.1038/nature13417
58. Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, et al. Controlling Long-Range Genomic Interactions At a Native Locus by Targeted Tethering of a Looping Factor. Cell (2012) 149(6):1233–44. doi: 10.1016/j.cell.2012.03.051
59. Dall’Agnese A, Caputo L, Nicoletti C, di Iulio J, Schmitt A, Gatto S, et al. Transcription Factor-Directed Re-Wiring of Chromatin Architecture for Somatic Cell Nuclear Reprogramming Toward Trans-Differentiation. Mol Cell (2019) 76(3):453–472.e8. doi: 10.1016/j.molcel.2019.07.036
60. Di Giammartino DC, Kloetgen A, Polyzos A, Liu Y, Kim D, Murphy D, et al. KLF4 is Involved in the Organization and Regulation of Pluripotency-Associated Three-Dimensional Enhancer Networks. Nat Cell Biol (2019) 21(10):1179–90. doi: 10.1038/s41556-019-0390-6
61. Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell (2019) 76(3):473–84.e7. doi: 10.1016/j.molcel.2019.07.038
62. Williamson I, Lettice LA, Hill RE, Bickmore WA. Shh and ZRS Enhancer Colocalisation is Specific to the Zone of Polarising Activity. Development (2016) 143(16):2994–3001. doi: 10.1242/dev.139188
63. Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, et al. Live-Cell Imaging Reveals Enhancer-Dependent Sox2 Transcription in the Absence of Enhancer Proximity. Elife (2019) 8:e41769. doi: 10.7554/eLife.41769
64. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science (2009) 326(5950):289–93. doi: 10.1126/science.1181369
65. Rowley MJ, Corces VG. Organizational Principles of 3D Genome Architecture. Nat Rev Genet (2018) 19(12):789–800. doi: 10.1038/s41576-018-0060-8
66. Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D Map of the Human Genome At Kilobase Resolution Reveals Principles of Chromatin Looping. Cell (2014) 159(7):1665–80. doi: 10.1016/j.cell.2014.11.021
67. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature (2012) 485(7398):376–80. doi: 10.1038/nature11082
68. Ghirlando R, Felsenfeld G. CTCF: Making the Right Connections. Genes Dev (2016) 30(8):881–91. doi: 10.1101/gad.277863.116
69. Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep (2016) 15(9):2038–49. doi: 10.1016/j.celrep.2016.04.085
70. Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell (2015) 161(5):1012–25. doi: 10.1016/j.cell.2015.04.004
71. Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator Dysfunction and Oncogene Activation in IDH Mutant Gliomas. Nature (2016) 529(7584):110–4. doi: 10.1038/nature16490
72. Despang A, Schöpflin R, Franke M, Ali S, Jerković I, Paliou C, et al. Functional Dissection of the Sox9-Kcnj2 Locus Identifies Nonessential and Instructive Roles of TAD Architecture. Nat Genet (2019) 51(8):1263–71. doi: 10.1038/s41588-019-0466-z
73. Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. Crispr Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell (2015) 162(4):900–10. doi: 10.1016/j.cell.2015.07.038
74. Zhou J, Levine M. A Novel Cis-Regulatory Element, the PTS, Mediates an Anti-Insulator Activity in the Drosophila Embryo. Cell (1999) 99(6):567–75. doi: 10.1016/s0092-8674(00)81546-9
75. Grosveld F, van Staalduinen J, Stadhouders R. Transcriptional Regulation by (Super)Enhancers: From Discovery to Mechanisms. Annu Rev Genomics Hum Genet (2021) 22:13.1–20. doi: 10.1146/annurev-genom-122220-093818
76. Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, et al. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev Cell (2016) 39(5):529–43. doi: 10.1016/j.devcel.2016.10.015
77. Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains From Genomic Compartmentalization. Cell (2017) 169(5):930–44.e22. doi: 10.1016/j.cell.2017.05.004
78. Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, et al. Cohesin Loss Eliminates All Loop Domains. Cell (2017) 171(2):305–320.e24. doi: 10.1016/j.cell.2017.09.026
79. Cattoni DI, Cardozo Gizzi AM, Georgieva M, Di Stefano M, Valeri A, Chamousset D, et al. Single-Cell Absolute Contact Probability Detection Reveals Chromosomes are Organized by Multiple Low-Frequency Yet Specific Interactions. Nat Commun (2017) 8(1):1753. doi: 10.1038/s41467-017-01962-x
80. Finn EH, Pegoraro G, Brandão HB, Valton AL, Oomen ME, Dekker J, et al. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell (2019) 176(6):1502–1515.e10. doi: 10.1016/j.cell.2019.01.020
81. Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science (2002) 295(5558):1306–11. doi: 10.1126/science.1067799
82. Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An Oestrogen-Receptor-Alpha-Bound Human Chromatin Interactome. Nature (2009) 462(7269):58–64. doi: 10.1038/nature08497
83. Hughes JR, Roberts N, McGowan S, Hay D, Giannoulatou E, Lynch M, et al. Analysis of Hundreds of Cis-Regulatory Landscapes At High Resolution in a Single, High-Throughput Experiment. Nat Genet (2014) 46(2):205–12. doi: 10.1038/ng.2871
84. Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, et al. HiChIP: Efficient and Sensitive Analysis of Protein-Directed Genome Architecture. Nat Methods (2016) 13(11):919–22. doi: 10.1038/nmeth.3999
85. Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. Dynamic Interplay Between Enhancer-Promoter Topology and Gene Activity. Nat Genet (2018) 50(9):1296–303. doi: 10.1038/s41588-018-0175-z
86. Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction Between Hypersensitive Sites in the Active Beta-Globin Locus. Mol Cell (2002) 10(6):1453–65. doi: 10.1016/s1097-2765(02)00781-5
87. Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal Associations Between Alternatively Expressed Loci. Nature (2005) 435(7042):637–45. doi: 10.1038/nature03574
88. Apostolou E, Thanos D. Virus Infection Induces NF-kappaB-dependent Interchromosomal Associations Mediating Monoallelic IFN-beta Gene Expression. Cell (2008) 134(1):85–96. doi: 10.1016/j.cell.2008.05.052
89. Nikopoulou C, Panagopoulos G, Sianidis G, Psarra E, Ford E, Thanos D. The Transcription Factor ThPOK Orchestrates Stochastic Interchromosomal Interactions Required for IFNB1 Virus-Inducible Gene Expression. Mol Cell (2018) 71(2):352–61.e5. doi: 10.1016/j.molcel.2018.06.019
90. Thanos D, Maniatis T. The High Mobility Group Protein HMG I(Y) is Required for NF-kappa B-Dependent Virus Induction of the Human IFN-beta Gene. Cell (1992) 71(5):777–89. doi: 10.1016/0092-8674(92)90554-p
91. Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered Recruitment of Chromatin Modifying and General Transcription Factors to the IFN-beta Promoter. Cell (2000) 103(4):667–78. doi: 10.1016/s0092-8674(00)00169-0
92. Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a Transcriptional Switch by HMGI(Y) Acetylation. Science (2001) 293(5532):1133–6. doi: 10.1126/science.293.5532.1133
93. Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN Beta Enhanceosome is Required for Synergistic Activation of Transcription. Mol Cell (1998) 1(2):277–87. doi: 10.1016/s1097-2765(00)80028-3
94. Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev (2001) 11(2):205–8. doi: 10.1016/s0959-437x(00)00180-5
95. Panne D, Maniatis T, Harrison SC. An Atomic Model of the Interferon-Beta Enhanceosome. Cell (2007) 129(6):1111–23. doi: 10.1016/j.cell.2007.05.019
96. Kobayashi KS, van den Elsen PJ. NLRC5: A Key Regulator of MHC Class I-dependent Immune Responses. Nat Rev Immunol (2012) 12(12):813–20. doi: 10.1038/nri3339
97. Lomvardas S, Thanos D. Nucleosome Sliding Via TBP DNA Binding In Vivo. Cell (2001) 106(6):685–96. doi: 10.1016/s0092-8674(01)00490-1
98. Lomvardas S, Thanos D. Modifying Gene Expression Programs by Altering Core Promoter Chromatin Architecture. Cell (2002) 110(2):261–71. doi: 10.1016/s0092-8674(02)00822-x
99. Rand U, Rinas M, Schwerk J, Nöhren G, Linnes M, Kröger A, et al. Multi-Layered Stochasticity and Paracrine Signal Propagation Shape the Type-I Interferon Response. Mol Syst Biol (2012) 8:584. doi: 10.1038/msb.2012.17
100. Wimmers F, Subedi N, van Buuringen N, Heister D, Vivié J, Beeren-Reinieren I, et al. Single-Cell Analysis Reveals That Stochasticity and Paracrine Signaling Control Interferon-Alpha Production by Plasmacytoid Dendritic Cells. Nat Commun (2018) 9(1):3317. doi: 10.1038/s41467-018-05784-3
101. Fang M, Xie H, Dougan SK, Ploegh H, van Oudenaarden A. Stochastic Cytokine Expression Induces Mixed T Helper Cell States. PloS Biol (2013) 11(7):e1001618. doi: 10.1371/journal.pbio.1001618
102. Zhao M, Zhang J, Phatnani H, Scheu S, Maniatis T. Stochastic Expression of the Interferon-β Gene. PloS Biol (2012) 10(1):e1001249. doi: 10.1371/journal.pbio.1001249
103. Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, et al. Yy1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell (2017) 171(7):1573–88.e28. doi: 10.1016/j.cell.2017.11.008
104. Bailey SD, Zhang X, Desai K, Aid M, Corradin O, Cowper-Sal Lari R, et al. ZNF143 Provides Sequence Specificity to Secure Chromatin Interactions At Gene Promoters. Nat Commun (2015) 2:6186. doi: 10.1038/ncomms7186
105. Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-Specific Nuclear Architecture and Gene Expression Regulated by SATB1. Nat Genet (2003) 34(1):42–51. doi: 10.1038/ng1146
106. Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 Packages Densely Looped, Transcriptionally Active Chromatin for Coordinated Expression of Cytokine Genes. Nat Genet (2006) 38(11):1278–88. doi: 10.1038/ng1913
107. Ghosh RP, Shi Q, Yang L, Reddick MP, Nikitina T, Zhurkin VB, et al. Satb1 Integrates DNA Binding Site Geometry and Torsional Stress to Differentially Target Nucleosome-Dense Regions. Nat Commun (2019) 10(1):3221. doi: 10.1038/s41467-019-11118-8
108. Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal Interactions and Olfactory Receptor Choice. Cell (2006) 126(2):403–13. doi: 10.1016/j.cell.2006.06.035
109. Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, Goh T, Murphy KK, Monahan K, et al. Enhancer Interaction Networks as a Means for Singular Olfactory Receptor Expression. Cell (2014) 159(3):543–57. doi: 10.1016/j.cell.2014.09.033
110. Monahan K, Schieren I, Cheung J, Mumbey-Wafula A, Monuki ES, Lomvardas S. Cooperative Interactions Enable Singular Olfactory Receptor Expression in Mouse Olfactory Neurons. Elife (2017) 6:e28620. doi: 10.7554/eLife.28620
111. Monahan K, Horta A, Lomvardas S. LHX2- and LDB1-mediated Trans Interactions Regulate Olfactory Receptor Choice. Nature (2019) 565(7740):448–53. doi: 10.1038/s41586-018-0845-0
112. Fanucchi S, Shibayama Y, Burd S, Weinberg MS, Mhlanga MM. Chromosomal Contact Permits Transcription Between Coregulated Genes. Cell (2013) 155(3):606–20. doi: 10.1016/j.cell.2013.09.051
113. Banos A, Agelopoulos M, Thanos D. Stochastic Responses are Not Left to Pure “Chance”. Cell (2013) 155(3):499–502. doi: 10.1016/j.cell.2013.10.002
114. Wernet MF, Mazzoni EO, Çelik A, Duncan DM, Duncan I, Desplan C. Stochastic Spineless Expression Creates the Retinal Mosaic for Colour Vision. Nature (2006) 440(7081):174–80. doi: 10.1038/nature04615
115. Anderson C, Reiss I, Zhou C, Cho A, Siddiqi H, Mormann B, et al. Natural Variation in Stochastic Photoreceptor Specification and Color Preference in Drosophila. Elife (2017) 6:e29593. doi: 10.7554/eLife.29593
116. Johnston RJ Jr, Desplan C. Interchromosomal Communication Coordinates Intrinsically Stochastic Expression Between Alleles. Science (2014) 343(6171):661–5. doi: 10.1126/science.1243039
117. Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, et al. Enhancer Features That Drive Formation of Transcriptional Condensates. Mol Cell (2019) 75(3):549–61.e7. doi: 10.1016/j.molcel.2019.07.009
118. Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA Polymerase II Clusters Associate in Transcription-Dependent Condensates. Science (2018) 361, 6400:412–5. doi: 10.1126/science.aar4199
119. van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, et al. Classification of Intrinsically Disordered Regions and Proteins. Chem Rev (2014) 114(13):6589–631. doi: 10.1021/cr400525m
120. Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription Factors Activate Genes Through the Phase-Separation Capacity of Their Activation Domains. Cell (2018) 175(7):1842–55. doi: 10.1016/j.cell.2018.10.042
121. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline P Granules are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science (2009) 324(5935):1729–32. doi: 10.1126/science.1172046
122. Kloetgen A, Thandapani P, Tsirigos A, Aifantis I. 3d Chromosomal Landscapes in Hematopoiesis and Immunity. Trends Immunol (2019) 40(9):809–24. doi: 10.1016/j.it.2019.07.003
123. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell (2017) 169(1):13–23. doi: 10.1016/j.cell.2017.02.007
124. Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, et al. Phase Separation of Ligand-Activated Enhancers Licenses Cooperative Chromosomal Enhancer Assembly. Nat Struct Mol Biol (2019) 26(3):193–203. doi: 10.1038/s41594-019-0190-5
125. Zamudio AV, Dall’Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, et al. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell (2019) 76(5):753–66.e6. doi: 10.1016/j.molcel.2019.08.016
Keywords: regulation of transcription, enhancers, transcription factors, chromatin, enhancer hubs, stochastic expression
Citation: Agelopoulos M, Foutadakis S and Thanos D (2021) The Causes and Consequences of Spatial Organization of the Genome in Regulation of Gene Expression. Front. Immunol. 12:682397. doi: 10.3389/fimmu.2021.682397
Received: 18 March 2021; Accepted: 18 May 2021;
Published: 04 June 2021.
Edited by:
Ageliki Tsagaratou, University of North Carolina at Chapel Hill, United StatesReviewed by:
Chris Benner, University of California, San Diego, United StatesCopyright © 2021 Agelopoulos, Foutadakis and Thanos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dimitris Thanos, dGhhbm9zQGJpb2FjYWRlbXkuZ3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.