
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 24 May 2021
Sec. Cancer Immunity and Immunotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.679829
This article is part of the Research TopicDeciphering the Microbiome-Immunity-Cancer AxisView all 9 articles
Commensal microbiota has emerged as an essential biomarker and regulator of both tumorigenesis and response to cancer therapy. However, our current knowledge about microbiota in cancer has been largely limited to intestinal microbiota. As a mucosal organ harboring one of the largest surface areas in the body, the lung is exposed to a variety of microbes through inhalation and micro-aspiration, and is colonized by a diverse bacterial community in both physiological and pathological conditions. Importantly, increasing evidence has linked the lung microbiome to cancer development. Studies in lung cancer patients and mouse models have revealed tumor-associated dysregulation of the local microbiome in the lung, which in turn impacts cancer progression by shaping the tumor microenvironment and modulating the activity of tumor-infiltrating immune cells. These findings not only provide novel mechanistic insight into the biology of lung cancer but also shed light on new therapeutic targets and strategies for lung cancer prevention and treatment. The goal of this review is to discuss the key findings, remaining questions, and future directions in this new and exciting field.
Mucosal surfaces exposed to the external environment are colonized by a vast number of microbes consisting of bacteria, viruses, fungi and archaea, which are collectively referred to as the commensal microbiota (1, 2). While oral cavity, skin, respiratory tract, urogenital tract and gastrointestinal tract are major body sites that each harbor unique microbiota, the primary habitat of commensal bacteria is the gastrointestinal tract (3). Numerous studies in the past two decades have explored the interactions between the host and microbiome, and have demonstrated the critical role of microbiome in regulating diverse physiological and pathological processes of the host. For example, commensal microbiota was essential in maintaining host metabolic homeostasis, instructing the development of specific organ structures, and driving the maturation of the host immune system (4). On the other hand, dysregulation of the microbiota with altered bacterial composition, quantity, and diversity has been found to increase host susceptibility to various pathogen infections, exacerbate intestinal inflammation and autoimmunity, induce different forms of metabolic disorders, and promote the development of neurological diseases (5, 6). Importantly, the microbiome is increasingly being recognized for its role in multiple aspects of cancer development and treatment across different types of cancer (7).
Lung cancer is the second most common cancer and is the leading cause of cancer-related deaths in both men and women (8). While small cell lung cancer (SCLC) accounts for 10-15% of lung cancer cases, up to 85% of lung cancers are non-small cell lung cancer (NSCLC), of which the main subtypes are adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. At the molecular level, both the genomic landscape and genetic heterogeneity of lung cancer have been extensively characterized, suggesting that it is a highly heterogeneous set of diseases. Despite the recent development of targeted therapies for some genetic subtypes of human lung cancer, overall survival rates for this disease remain very low. Cigarette smoking is the number one risk factor for lung cancer, while other established risk factors may include second-hand smoke, air pollution, exposure to radon, asbestos, and other occupational carcinogens (9, 10). However, the mechanisms by which these environmental risk factors and other tumor-extrinsic factors control lung carcinogenesis remain poorly understood. As the organ with the largest surface area in the human body and with gas exchange functions, the lung is inevitably exposed to various environmental microorganisms. Therefore, the critical questions are: does the lung have its own indigenous microbiota, and do the lung microbiota change with cancer? How do the microbiota impact lung carcinogenesis? What are the underlying mechanisms and any potential clinical implications? Focusing on these key questions, we hereby summarize the recent advances in this rising area of host-microbiome interaction in lung cancer.
Lung, as an organ for gas exchange, harbors one of the largest interfaces between the human host and the external environment, and is therefore a unique mucosal site for host-microbiota interaction (11). Yet the healthy lung is traditionally believed to be sterile due to the failure to grow bacteria from lower airway samples using traditional culture-based microbiological approaches. However, it is now recognized that only 20~40% of bacteria in human feces can be cultivated (12). Until very recently, a substantial number of studies applying 16S-based culture-independent analysis have identified the presence of microbial communities containing a complex diversity of bacteria in the lung in both health and diseases (13–18). Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria are found to be the major phyla of the lung microbiome, while the genera including Prevotella, Veillonella, Streptococcus, Neisseria, Haemophilus, Fusobacterium, Sphingomonas, Pseudomonas, Acinetobacter, Megasphaera, Staphylococcus, and Corynebacterium have all been described in the airway microbiota (14, 16, 19). About 20% of OTUs observed by sequencing-based technology have been confirmed by culture-based studies (20). In general, the bacterial composition of the lung microbiota is distinct from the gut or skin microbiota, but shares considerable similarity with the upper respiratory tract and oral microbiome (13, 16).
One of the key features of lung microbiota is its low bacterial biomass, especially when compared to the gastrointestinal microbiome (21). Human lungs harbor 2,000 bacterial genomes per cm2 of surface area when estimated based on 16S rRNA analysis of bronchial brushings (16). Experimentally, this introduces technical challenges with sequencing, particularly when using techniques designed for high biomass samples, such as gut microbiome analyses. Low biomass can often result in increased erroneous read frequency as well as background 16s rRNA contamination from the environment or reagents used (22). To combat this, certain strategies with different strengths and weaknesses have been developed, such as the exclusion method and correlation analysis. The abundance and composition of the lung microbiome are mainly regulated by the balance of these three factors: bacterial immigration, elimination, and replication (23, 24). Regarding bacterial immigration, there are two major sources of lung microbiota while inhalation constantly exposes the lung to the air-borne bacteria and microbiota from the upper respiratory tract, micro-aspiration of oral fluids also seeds the lungs with oral bacteria. On the other hand, the known elimination pathways of the pulmonary microbes include cough, mucociliary clearance, and innate and adaptive host defenses (23, 24). Lastly, the bacterial replication rate is mostly determined by the regional growth conditions such as oxygen level, pH, nutrient availability, and anatomical location. Importantly, lung diseases can cause changes in the local environment, favoring the growth of certain bacterial species over others, thereby leading to the development of dysbiosis (24).
Emerging evidence has linked local dysbiosis with both acute and chronic pulmonary diseases (25). For example, although the lung microbiome varies across individuals, no spatial variation was observed within the healthy lung (23). In contrast, in patients with advanced chronic obstructive pulmonary disease (COPD), the marked micro-anatomic differences in bacterial communities within the lungs were identified even when there was no change in bronchiectasis by CT scan (14). Similar spatial heterogeneity was also discovered in the lung with cystic fibrosis (26). Besides microbial distribution, both the biomass and composition of the lung microbiome are altered in pathological conditions. In asthmatic patients, the bacterial burden is much higher than the healthy controls, and the relative abundance of specific bacterial families within the community is closely correlated to the severity of bronchial hyperresponsiveness (18). In patients with COPD, the overgrowth of certain pathogenic microorganisms and the appearance of new commensal strains in the lung are associated with disease exacerbation (27, 28).
Multiple lines of clinical evidence have suggested the link between lung microbiota dysbiosis and cancer. Early epidemiology data showed that bacterial infections are very common in lung cancer patients. Up to 50%-70% of lung cancer cases are complicated by pulmonary infections over the course of the disease (29), and post-obstructive pneumonia can negatively affect the efficacy of lung cancer therapy and overall survival of cancer patients (30). However, the microbiology of these pneumonias is generally poorly understood, making antibiotic therapies hard to direct. More recently, with the advent of high throughput sequencing techniques, an increasing number of studies have revealed the strong association between local dysbiosis and lung cancer. For example, in comparison to that of the non-malignant lung tissue samples, the microbiota of lung tumors was found to exhibit substantially lower taxonomic alpha diversity, while the bacterial compositions were correlated with epidemiologic exposures and cancer stages (31). Regarding the specific bacterial taxon, the genus Thermus was reported to be more abundant in tumor tissues from advanced-stage patients, and Legionella was higher in patients who developed metastases, implicating the role of these bacteria in cancer progression (31). Similarly, various other bacterial taxa have also been associated with lung cancer by a number of studies. Yan et al. showed that the oral microorganisms Veillonella and Capnocytophaga were significantly higher in the saliva samples of lung cancer patients and that they may be used as biomarkers for early detection of both small-cell carcinoma and adenocarcinoma (32); Lee et al. detected increased abundance of Veillonella and Megasphaera in bronchoalveolar lavage samples from lung cancer patients (33); Cameron et al. found that Granulicatella adiacens and six other opportunistic pathogens were more common in spontaneous sputum samples from lung cancer patients (34); Greathouse and colleagues demonstrated a correlation between Acidovorax genus and squamous cell carcinoma, and identified a specific group of taxa enriched in smokers with TP53 mutation (35); Gomes and colleagues described that Brevundimonas, Acinetobacter, and Propionibacterium were more enriched in lung adenocarcinoma while Enterobacter was relatively more enriched in squamous-cell carcinoma (19); it was also found that the lower airway samples from lung cancer individuals were enriched more supraglottic-predominant taxa including Streptococcus, Prevotella, Veillonella, and Rothia (36).
Overall, although the exact bacterial taxa identified in lung cancer may vary from one study to another depending on the sample type, sampling method, and patient cohort, the consistent findings are that lung cancer is associated with a dysregulated local microbiome, featured by increased total bacterial abundance, reduced alpha-diversity, and altered bacterial composition. While the specific consequences of altered bacterial diversity in lung cancer have yet to be elucidated, previous studies have shown that increased alpha diversity tends to be associated with better survival and treatment response outcomes in certain cancers, such as cervical cancer and resected pancreatic adenocarcinoma, potentially by influencing the host immune response (37, 38). Interestingly, there seems to an overall enrichment of oral bacteria in lung tumors as observed by multiple groups, and certain bacterial taxa are particularly over-represented in specific subtypes of lung cancer and are correlated with smoking status, and genetic mutations. Altogether, these recent studies suggest that the microbiome may soon become a critical diagnostic and preventive biomarker for lung cancer status, stage, host genotypes and risk factors (Table 1).
Despite the established link between carcinogenesis and altered lung microbiota, it remains unclear whether the lung microbiome has a causal effect in lung cancer development, and whether it can be a therapeutic target for lung cancer treatment. To address this question, it is key is to elucidate the role of microbiota in lung carcinogenesis and to dissect the mechanisms by which microbiota regulate tumor initiation and progression. Similar to the findings in gut microbiota, it has been suggested that the lung microbiome may contribute to lung tumorigenesis either by directly acting on tumor cells or indirectly via modulation of the tumor-associated immune response (Figure 1).
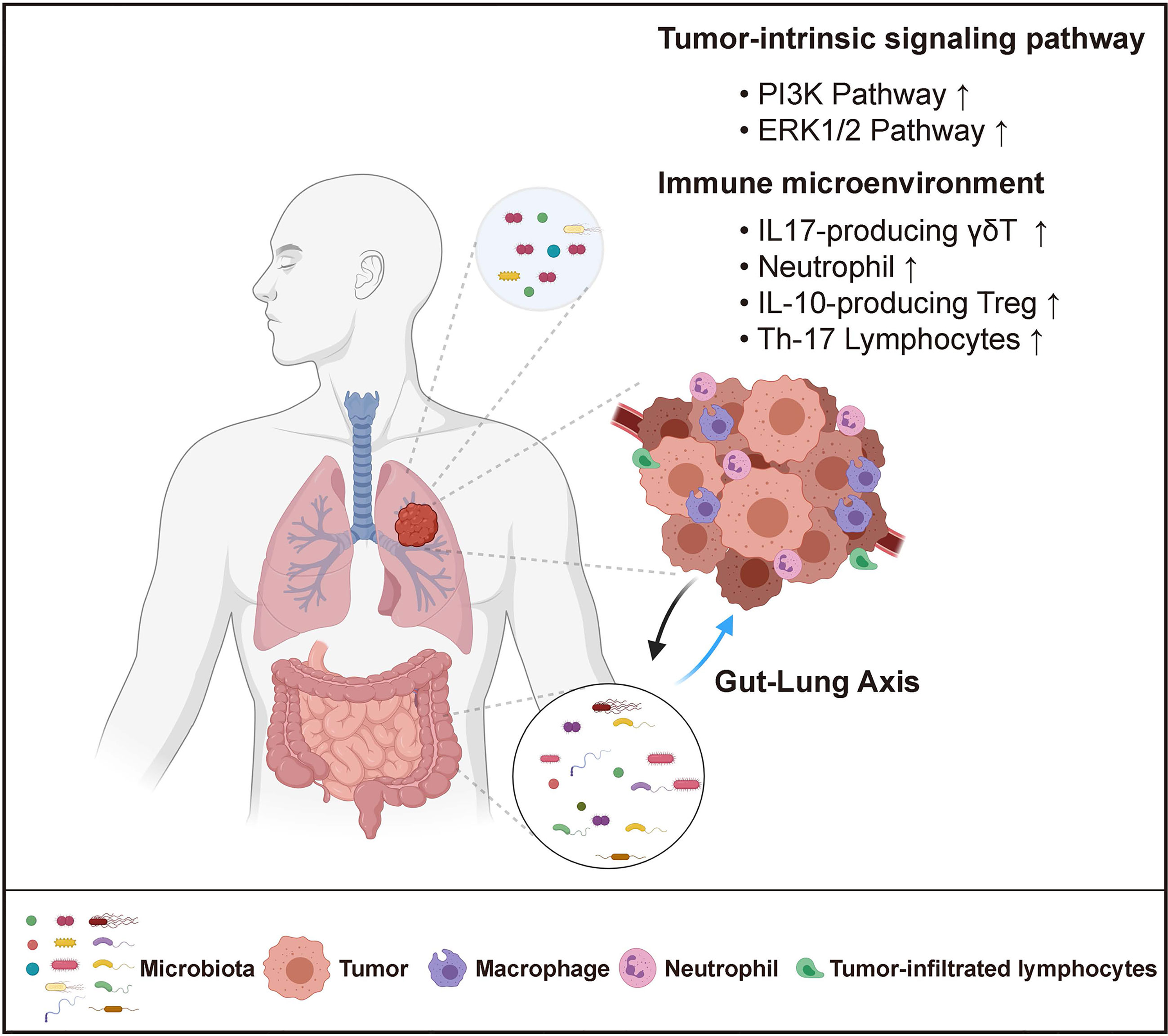
Figure 1 Host-Microbiome Interaction in Lung Cancer. Both local (lung) and distal (gut) microbiota play critical roles in lung cancer. Local microbiota derived metabolites may serve as signaling molecules that directly impact tumor cell intrinsic pathways. They can also reshape the local immune microenvironment by promoting inflammation and inducing immune-suppression. Distal gut microbiota may affect lung tumor development through the regulation of the systemic immune response.
Lung microbiota can regulate specific oncogenic pathways that directly drive carcinogenesis. As a number of bacterial metabolites have been implicated in the regulation of host metabolism and signaling pathways (41), it is possible that the alteration of bacteria-derived molecules resulting from dysbiosis in the tumor microenvironment can affect the metabolism and oncogenic signaling of lung cancer cells. Supporting this notion, it was reported that the local microbiota of lung cancer patients exhibited a decreased abundance of the KEGG modules of energy metabolism and ABC-type transport, with increased modules involved in amino acid metabolism, lipid metabolism, xenobiotic biodegradation and metabolism (31, 42). This altered microbial metabolic profile could further impact the gene expressions in airway epithelia cells. In vitro stimulation of the human lung adenocarcinoma cell line A549 with bacterial products isolated from lung cancer patient enriched bacteria was shown to upregulate the expression of genes involved in the PI3K and ERK1/2 signaling pathway, which was consistent with the transcriptomic changes observed in lung cancer patients compared with healthy individuals (36). Importantly, PI3K pathway upregulation has been considered as an early event in the process of lung tumorigenesis (43), suggesting a direct link between lung microbiota and oncogenesis.
Additionally, it has also been demonstrated that the microbiota shape the immune microenvironment in the lung to promote tumor growth. A network of lung-resident immune cells maintains pulmonary tissue homeostasis in the steady-state and provide immune defense in response to invading pathogens (44). The development of lung cancer is closely associated with chronic inflammation characterized by infiltration of inflammatory cells and accumulation of pro-inflammatory factors including cytokines, chemokines and prostaglandins that stimulate cell proliferation, angiogenesis, tissue remodeling, or metastasis (45). However, the source of inflammation has not been clearly defined, and the contribution of specific cellular and molecular components of the immune system remains to be determined. Using an autochthonous GEMM of lung adenocarcinoma driven by an activating point mutation of Kras and loss of p53, our previous study has demonstrated the critical role of microbiota-immune crosstalk in driving inflammation and lung tumorigenesis (46). Mechanistically, we found that lung tumor growth is associated with increased bacterial load and altered bacterial composition in the airway. This dysregulated local microbiota stimulated Myd88-dependent IL-1β and IL-23 production from myeloid cells, inducing proliferation and activation of lung-resident Vγ6+Vδ1+ γδ T cells. These γδ T cells then produced IL-17 to promote neutrophil infiltration and inflammation in the tumor microenvironment; they also expressed IL-22, amphiregulin, and other effector molecules to directly enhance tumor cell proliferation. Eliminating commensal bacteria by systemic antibiotic treatment or rederiving the mice to the germ-free (GF) condition, as well as blocking the γδ T or downstream IL-17 all significantly suppressed tumor growth in the lung. The tumor-promoting role of microbiota elucidated in our study is consistent with the findings from another study showing that treatment of aerosolized vancomycin and neomycin reduced lung tumor implantation. In this study, aerosolized antibiotic treatment led to decreased IL-10 producing Tregs and increased activation of the anti-tumoral NK and T cell response, thereby alleviating the immune suppression in the tumor microenvironment (9).
It is also becoming clear that the specific bacterial composition of the lung is key to regulating lung inflammation, especially in the context of cancer. Interestingly, de-repression of the anti-tumor immunity was found to be associated with a shift from Firmicutes to Proteobacteria or other Gram-positive microbes rather than specific species after antibiotic treatment (9). Therefore, the balance between different bacterial taxa in the lung seems to be critical for maintaining the local immune homeostasis. Further supporting this, Segal and colleagues found that enrichment of oral bacterial taxa in the lung, such as Prevotella and Veillonella, was associated with an inflammatory phenotype, including an increased level of Th17 lymphocytes, enhanced expression of inflammatory cytokines, and a blunted alveolar macrophage TLR4 response (39). In addition, a previous study showed that immediately following birth, a shift from a predominance of Gammaproteobacteria and Firmicutes towards Bacteroidetes promoted transient PD-L1 expression in lung CD11b+ CD103- dendritic cells, regulating regulatory T cell activation and the general aeroallergen response in a two-week window after birth (47). Although this specific study did not discuss this effect in cancer, it is likely that shifting bacterial diversity may be able to regulate key molecules like PD-L1 as well as various innate and adaptive immune populations. Altogether, these studies not only provide mechanistic insight into the host-microbiome interaction in lung cancer, but also define novel targets for lung cancer prevention and treatment.
The gastrointestinal tract harbors 99% of the commensal bacterial mass of the human microbiome. The bi-directional crosstalk between the gut and the lung has been well established, and the gut microbiota was previously found to regulate the lung immune response in allergic airway diseases and pulmonary viral infection (48–50). Gut microbiota has also emerged as a biomarker and regulator of both tumorigenesis and host response to cancer therapy. Interestingly, while the gut microbiota is found to promote local inflammation and development of gastrointestinal cancers (7, 51), it seems to exert an opposing effect on transplanted tumors at distal sites by priming the host immune system and enhancing the systemic anti-tumor immune response (52). Using a transplanted mouse model, previous studies showed that oral antibiotic treatment could increase the susceptibility to Lewis lung carcinoma (53), and reduced the anti-cancer response of the chemotherapeutic agent cisplatin (54). In a landmark study by Routy et al., it was shown that the patients with advanced non-small cell lung cancer who took ATB orally shortly before or after the first administration of PD-1/PD-L1 mAb have significantly decreased progression-free survival and overall survival (55). A similar conclusion was also drawn by many other studies (56–58). Given that the intestinal microbiome is the main target of oral antibiotic treatment, these findings suggest that distal intestinal microbiota may indirectly regulate lung cancer development and the therapeutic efficacy of chemotherapy or immunotherapy through regulation of the systemic immune response.
However, these studies mainly focused on load and diversity in the intestinal microbiome and did not specifically study changes in the lung microbiome. Recently, new work has come out specifically describing how lower airway dysbiosis leads to worse NSCLC progression and decreased survival. Tsay et al. showed that Veillonella parvula was the most abundant taxon driving this dysbiotic association, leading to upregulation of the IL17, PI3K, MAPK, and ERK pathways in the airway transcriptome. They then used V. parvula in the murine KP lung cancer model, characterizing the mechanistic effects on lower airway CD4+ and CD8+, neutrophils, and Th17 recruitment (59). Also, Peters et al. recently demonstrated through 16s rRNA gene sequencing of lung samples from NSCLC patients that lung tumor tissue was correlated with significantly lower bacterial richness and diversity, and that alterations in the relative abundance of families such as Koribacteraceae, Bacteroidaceae, Lachnospiraceae, and Ruminococcaceae were associated with increased or decreased recurrence-free and disease-free survival (60). It is clear that particularly in lung cancer patients, it may be necessary to conduct deeper analysis specifically into the role of lung bacteria and seek to isolate these effects from the gut.
On the other hand, lung cancer development is also associated with changes in the gut microbiome. A recent study found a significant difference in the beta-diversity but not the alpha diversity of the gut microbiota between lung cancer patients and healthy control individuals, with a particular over-representation of Enterococcus in the fecal material from lung cancer patients (61). Similarly, several other bacteria tax, including Streptococcus, Prevotella, Blautia, Coprococcus, Bifidobacterium, and Lachnospiraceae were found to be enriched in the gut microbiota of lung cancer patients (42). Additionally, Ming et al. showed that the gut microbiome in lung cancer patients have a lower abundance of Prevotella but a higher level of Actinomyces and Streptococcus. Interestingly, the alteration of the gut microbiome in lung cancer patients seems to be dependent on the different lung cancer subtypes: lower alpha diversity of the gut microbiome was only observed in patients with small cell lung cancer but not adenocarcinoma or squamous cell carcinoma; in terms of the taxonomical composition, gut microbiota in adenocarcinoma patients was characterized by the dominant phylotypes including Bifidobacterium, Clostridium, and Prevotella, squamous cell carcinoma patients exhibited a high abundance of Ruminococcus, Lachnospira, and Lactobacillus in their gut, and Streptococcus, Anaerotruncus, and Bacillus were enriched in small cell lung cancer (62). Taken together, these observations indicate that the distal gut microbiome may also serve as a robust biomarker for lung cancer.
Based on our current knowledge about the host-microbiome interaction in lung cancer, the research in this area is progressing towards these two main directions: first is to understand how the microbiota co-evolves with tumors during cancer initiation and progression to identify the changes in bacterial composition, quantity, diversity and metabolic activity that can be used as reliable biomarkers for lung cancer diagnosis; second is to further elucidate the cellular and molecular mediators of host-microbiota interaction that can be therapeutically targeted for lung cancer treatment.
Supporting the idea that specific bacterial species may serve as unique biomarkers for lung cancer subtypes and tumor stages, Greathouse et al. showed the higher abundance of Acidovorax among the subset of squamous cell carcinoma cases with TP53 mutations, but this association was not present in adenocarcinomas. Another study analyzed the lower airway microbiota in patients undergoing diagnostic bronchoscopy for suspected lung cancer and found that malignancy could be determined with an accuracy of 70% by isolating Enterococcus, Capnocytophaga, or Actinomyces, whereas benign diseases can be indicated by Microbispora with a sensitivity of 55%, specificity of 88%, and accuracy of 78% (63). Similarly, enrichment of Capnocytophaga and Veillonella were observed in both small cell lung cancer and lung adenocarcinoma, and the combination of these two bacterial biomarkers succeeded in distinguishing patients with small cell lung cancer or lung adenocarcinoma from healthy individuals (32).
It is worth mentioning that in one of the latest studies, Nejman and colleagues performed a comprehensive analysis of the tumor microbiome across seven cancer types, including lung cancer (64). Importantly, bacterial components were detected within lung tumors as measured by 16S qPCR, IHC for LPS, and in situ hybridization of 16S rRNA. While each tumor type was shown to have distinct microbial compositions, it was found that bacterial taxa that can degrade chemicals in cigarette smoke were significantly enriched in lung tumors compared with other tumor types, suggesting that high levels of these metabolites create a preferred niche for bacteria that can use these molecules. This enrichment was even more apparent in lung tumors from current smokers in comparison to never-smokers. Moreover, bacterial pathways involved in the biosynthesis of plant metabolites present in cigarette tobacco were also enriched in the lung tumors of smokers, further reflecting the impact of environmental exposure on the lung microbiome. Specifically, with regards to smoking, many different studies have observed significant changes to the lung microbiome in different smoking condition. While some have specifically shown a positive correlation between smoking and alpha diversity, others have shown specific enhancement or reduction in key populations such as Pseudomonas, Streptococcus, and Granulicatella (14, 31, 65). This suggests that in addition to the direct effects of smoking and other environmental factors on tumor cells, the smoking-microbiome-immune axis also likely plays an important role in regulating overall tumor progression.
Moving forward, we think the critical questions in this field that require further investigations are (1) How do tumors shape the local microbial community in the lung? (2) What are the specific tumor-promoting component(s) in lung microbiota? To this end, it is critical to interrogate the interactions between the microbiome and host genes, as well as the interactions between the microbiome and environmental exposure such as cigarette smoking. Furthermore, given the potent impact of gut microbiome on the efficacy of various anti-cancer therapies including the lung cancer response to cisplatin and immune checkpoint blockade, it is also essential to further delineate the specific roles of the local lung microbiome vs. the distal gut microbiome in tumor growth and tumor-associated immune responses. It is possible that selectively targeting these two compartments may exert distinct effects on lung cancer progression and treatment, thereby shedding light on new strategies for future lung cancer therapies.
QD, ESC, CZ, and CJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
CJ is supported in part by a R00 award from NIH/NCI (CA226400), an Emerson Collective Cancer Research fund, and a Lung Cancer Research Foundation (LCRF) pilot grant. CZ is supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, SITC-AstraZeneca Immunotherapy in Lung Cancer (Early Stage NSCLC) Clinical Fellowship Award. CJ and CZ are supported by NIH Bench-to-Bedside and Back Program (BtB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The figure was created with BioRender.com.
1. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current Understanding of the Human Microbiome. Nat Med (2018) 24(4):392–400. doi: 10.1038/nm.4517
2. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the Human Microbiome. Nutr Rev (2012) 70(Suppl 1):S38–44. doi: 10.1111/j.1753-4887.2012.00493.x
3. Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature (2012) 486(7402):207–14. doi: 10.1038/nature11234
4. Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell (2014) 157(1):121–41. doi: 10.1016/j.cell.2014.03.011
5. Butler MI, Cryan JF, Dinan TG. Man and the Microbiome: A New Theory of Everything? Annu Rev Clin Psychol (2019) 15:371–98. doi: 10.1146/annurev-clinpsy-050718-095432
6. Wang B, Yao M, Lv L, Ling Z, Li L. The Human Microbiota in Health and Disease. Engineering (2017) 3(1):71–82. doi: 10.1016/j.Eng.2017.01.008
7. Garrett WS. Cancer and the Microbiota. Science (2015) 348(6230):80–6. doi: 10.1126/science.aaa4972
8. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
9. Le Noci V, Guglielmetti S, Arioli S, Camisaschi C, Bianchi F, Sommariva M, et al. Modulation of Pulmonary Microbiota by Antibiotic or Probiotic Aerosol Therapy: A Strategy to Promote Immunosurveillance Against Lung Metastases. Cell Rep (2018) 24(13):3528–38. doi: 10.1016/j.celrep.2018.08.090
10. Kaderbhai C, Richard C, Fumet JD, Aarnink A, Foucher P, Coudert B, et al. Antibiotic Use Does Not Appear to Influence Response to Nivolumab. Anticancer Res (2017) 37(6):3195–200. doi: 10.21873/anticanres.11680
11. Pilette C, Ouadrhiri Y, Godding V, Vaerman JP, Sibille Y. Lung Mucosal Immunity: Immunoglobulin-a Revisited. Eur Respir J (2001) 18(3):571–88. doi: 10.1183/09031936.01.00228801
12. Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct Analysis of Genes Encoding 16S rRNA From Complex Communities Reveals Many Novel Molecular Species Within the Human Gut. Appl Environ Microbiol (1999) 65(11):4799–807. doi: 10.1128/AEM.65.11.4799-4807.1999
13. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical Continuity of Bacterial Populations in the Healthy Human Respiratory Tract. Am J Respir Crit Care Med (2011) 184(8):957–63. doi: 10.1164/rccm.201104-0655OC
14. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the Lung Microbiome in the “Healthy” Smoker and in COPD. PloS One (2011) 6(2):e16384. doi: 10.1371/journal.pone.0016384
15. Harris JK, De Groote MA, Sagel SD, Zemanick ET, Kapsner R, Penvari C, et al. Molecular Identification of Bacteria in Bronchoalveolar Lavage Fluid From Children With Cystic Fibrosis. Proc Natl Acad Sci U S A (2007) 104(51):20529–33. doi: 10.1073/pnas.0709804104
16. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered Microbial Communities in Asthmatic Airways. PloS One (2010) 5(1):e8578. doi: 10.1371/journal.pone.0008578
17. Huang YJ, Kim E, Cox MJ, Brodie EL, Brown R, Wiener-Kronish JP, et al. A Persistent and Diverse Airway Microbiota Present During Chronic Obstructive Pulmonary Disease Exacerbations. OMICS (2010) 14(1):9–59. doi: 10.1089/omi.2009.0100
18. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway Microbiota and Bronchial Hyperresponsiveness in Patients With Suboptimally Controlled Asthma. J Allergy Clin Immunol (2011) 127(2):372–81.e1-3. doi: 10.1016/j.jaci.2010.10.048
19. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of Lung Microbiota Discloses Differences in Adenocarcinoma and Squamous Cell Carcinoma. Sci Rep (2019) 9(1):12838. doi: 10.1038/s41598-019-49195-w
20. Venkataraman A, Bassis CM, Beck JM, Young VB, Curtis JL, Huffnagle GB, et al. Application of a Neutral Community Model to Assess Structuring of the Human Lung Microbiome. mBio (2015) 6(1):e02284-14. doi: 10.1128/mBio.02284-14
21. Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, et al. Paradigms of Lung Microbiota Functions in Health and Disease, Particularly, in Asthma. Front Physiol (2018) 9:1168. doi: 10.3389/fphys.2018.01168
22. Claassen-Weitz S, Gardner-Lubbe S, Mwaikono KS, du Toit E, Zar HJ, Nicol MP. Optimizing 16S rRNA Gene Profile Analysis From Low Biomass Nasopharyngeal and Induced Sputum Specimens. BMC Microbiol (2020) 20(1):113. doi: 10.1186/s12866-020-01795-7
23. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc (2015) 12(6):821–30. doi: 10.1513/AnnalsATS.201501-029OC
24. Dickson RP, Martinez FJ, Huffnagle GB. The Role of the Microbiome in Exacerbations of Chronic Lung Diseases. Lancet (2014) 384(9944):691–702. doi: 10.1016/S0140-6736(14)61136-3
25. O’Dwyer DN, Dickson RP, Moore BB. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J Immunol (2016) 196(12):4839–47. doi: 10.4049/jimmunol.1600279
26. Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, et al. Spatial Distribution of Microbial Communities in the Cystic Fibrosis Lung. ISME J (2012) 6(2):471–4. doi: 10.1038/ismej.2011.104
27. Rosell A, Monso E, Soler N, Torres F, Angrill J, Riise G, et al. Microbiologic Determinants of Exacerbation in Chronic Obstructive Pulmonary Disease. Arch Intern Med (2005) 165(8):891–7. doi: 10.1001/archinte.165.8.891
28. Sethi S, Sethi R, Eschberger K, Lobbins P, Cai X, Grant BJ, et al. Airway Bacterial Concentrations and Exacerbations of Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med (2007) 176(4):356–61. doi: 10.1164/rccm.200703-417OC
29. Akinosoglou KS, Karkoulias K, Marangos M. Infectious Complications in Patients With Lung Cancer. Eur Rev Med Pharmacol Sci (2013) 17(1):8–18.
30. Qiao D, Wang Z, Lu Y, Wen X, Li H, Zhao H. A Retrospective Study of Risk and Prognostic Factors in Relation to Lower Respiratory Tract Infection in Elderly Lung Cancer Patients. Am J Cancer Res (2015) 5(1):423–32.
31. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing Human Lung Tissue Microbiota and its Relationship to Epidemiological and Clinical Features. Genome Biol (2016) 17(1):163. doi: 10.1186/s13059-016-1021-1
32. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and Validation of Potential Bacterial Biomarkers for Lung Cancer. Am J Cancer Res (2015) 5(10):3111–22.
33. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of Microbiome in Bronchoalveolar Lavage Fluid of Patients With Lung Cancer Comparing With Benign Mass Like Lesions. Lung Cancer (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
34. Cameron SJS, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A Pilot Study Using Metagenomic Sequencing of the Sputum Microbiome Suggests Potential Bacterial Biomarkers for Lung Cancer. PloS One (2017) 12(5):e0177062. doi: 10.1371/journal.pone.0177062
35. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction Between the Microbiome and TP53 in Human Lung Cancer. Genome Biol (2018) 19(1):123. doi: 10.1186/s13059-018-1501-6
36. Tsay JJ, Wu BG, Badri MH, Clemente JC, Shen N, Meyn P, et al. Airway Microbiota is Associated With Upregulation of the PI3K Pathway in Lung Cancer. Am J Respir Crit Care Med (2018) 198(9):1188–98. doi: 10.1164/rccm.201710-2118OC
37. Sims TT, El Alam MB, Karpinets TV, Dorta-Estremera S, Hegde VL, Nookala S, et al. Gut Microbiome Diversity is an Independent Predictor of Survival in Cervical Cancer Patients Receiving Chemoradiation. Commun Biol (2021) 4(1):237. doi: 10.1038/s42003-021-01741-x
38. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell (2019) 178(4):795–806 e12. doi: 10.1016/j.cell.2019.07.008
39. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the Lung Microbiome With Oral Taxa is Associated With Lung Inflammation of a Th17 Phenotype. Nat Microbiol (2016) 1:16031. doi: 10.1038/nmicrobiol.2016.31
40. Oriano M, Gramegna A, Terranova L, Sotgiu G, Sulaiman I, Ruggiero L, et al. Sputum Neutrophil Elastase Associates With Microbiota and P. Aeruginosa in Bronchiectasis. Eur Respir J (2020) 56:2000769. doi: 10.1183/13993003.00769-2020
41. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-Gut Microbiota Metabolic Interactions. Science (2012) 336(6086):1262–7. doi: 10.1126/science.1223813
42. Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, et al. Dysbiosis of the Gut Microbiome is Associated With Tumor Biomarkers in Lung Cancer. Int J Biol Sci (2019) 15(11):2381–92. doi: 10.7150/ijbs.35980
43. Gustafson AM, Soldi R, Anderlind C, Scholand MB, Qian J, Zhang X, et al. Airway PI3K Pathway Activation is an Early and Reversible Event in Lung Cancer Development. Sci Trans Med (2010) 2(26):26ra5–ra5. doi: 10.1126/scitranslmed.3000251
44. Lloyd CM, Marsland BJ. Lung Homeostasis: Influence of Age, Microbes, and the Immune System. Immunity (2017) 46(4):549–61. doi: 10.1016/j.immuni.2017.04.005
45. Palucka AK, Coussens LM. The Basis of Oncoimmunology. Cell (2016) 164(6):1233–47. doi: 10.1016/j.cell.2016.01.049
46. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal Microbiota Promote Lung Cancer Development Via Gammadelta T Cells. Cell (2019) 176(5):998–1013.e16. doi: 10.1016/j.cell.2018.12.040
47. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung Microbiota Promotes Tolerance to Allergens in Neonates Via PD-L1. Nat Med (2014) 20(6):642–7. doi: 10.1038/nm.3568
48. Anand S, Mande SS. Diet, Microbiota and Gut-Lung Connection. Front Microbiol (2018) 9:2147. doi: 10.3389/fmicb.2018.02147
49. Dumas A, Bernard L, Poquet Y, Lugo-Villarino G, Neyrolles O. The Role of the Lung Microbiota and the Gut-Lung Axis in Respiratory Infectious Diseases. Cell Microbiol (2018) 20(12):e12966. doi: 10.1111/cmi.12966
50. Vaughan A, Frazer ZA, Hansbro PM, Yang IA. COPD and the Gut-Lung Axis: The Therapeutic Potential of Fibre. J Thorac Dis (2019) 11(Suppl 17):S2173–80. doi: 10.21037/jtd.2019.10.40
51. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-Induced Cancer: Crosstalk Between Tumours, Immune Cells and Microorganisms. Nat Rev Cancer (2013) 13(11):759–71. doi: 10.1038/nrc3611
52. Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, et al. Microbes and Cancer. Annu Rev Immunol (2017) 35:199–228. doi: 10.1146/annurev-immunol-051116-052133
53. Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, et al. Microbiota Modulate Tumoral Immune Surveillance in Lung Through a gammadeltaT17 Immune Cell-Dependent Mechanism. Cancer Res (2014) 74(15):4030–41. doi: 10.1158/0008-5472.CAN-13-2462
54. Gui QF, Lu HF, Zhang CX, Xu ZR, Yang YH. Well-Balanced Commensal Microbiota Contributes to Anti-Cancer Response in a Lung Cancer Mouse Model. Genet Mol Res (2015) 14(2):5642–51. doi: 10.4238/2015.May.25.16
55. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut Microbiome Influences Efficacy of PD-1-based Immunotherapy Against Epithelial Tumors. Science (2018) 359(6371):91–7. doi: 10.1126/science.aan3706
56. Ahmed J, Kumar A, Parikh K, Anwar A, Knoll BM, Puccio C, et al. Use of Broad-Spectrum Antibiotics Impacts Outcome in Patients Treated With Immune Checkpoint Inhibitors. Oncoimmunology (2018) 7(11):e1507670. doi: 10.1080/2162402X.2018.1507670
57. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative Association of Antibiotics on Clinical Activity of Immune Checkpoint Inhibitors in Patients With Advanced Renal Cell and non-Small-Cell Lung Cancer. Ann Oncol (2018) 29(6):1437–44. doi: 10.1093/annonc/mdy103
58. Zhao S, Gao G, Li W, Li X, Zhao C, Jiang T, et al. Antibiotics are Associated With Attenuated Efficacy of anti-PD-1/PD-L1 Therapies in Chinese Patients With Advanced non-Small Cell Lung Cancer. Lung Cancer (2019) 130:10–7. doi: 10.1016/j.lungcan.2019.01.017
59. Tsay JJ, Wu BG, Sulaiman I, Gershner K, Schluger R, Li Y, et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov (2021) 11(2):293–307. doi: 10.1158/2159-8290.CD-20-0263
60. Peters BA, Hayes RB, Goparaju C, Reid C, Pass HI, Ahn J. The Microbiome in Lung Cancer Tissue and Recurrence-Free Survival. Cancer Epidemiol Biomarkers Prev (2019) 28(4):731–40. doi: 10.1158/1055-9965.EPI-18-0966
61. Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, et al. Dysbiosis of the Gut Microbiome in Lung Cancer. Front Cell Infect Microbiol (2019) 9:112. doi: 10.3389/fcimb.2019.00112
62. Ming L, Fang Y, Xiaohui C, Huan Z, Xiaoqing W, Yinhui L, et al. Alteration of Gut Microbiome in Lung Cancer Patients. bioRxiv (2019). doi: 10.1101/640359
63. Sanchez-Hellin V, Galiana A, Zamora-Molina L, Soler-Sempere M, Grau-Delgado J, Barbera VM, et al. Lower Airway Microbiota and Lung Cancer. Microbiol Biotechnol Lett (2019) 47(3):441–8. doi: 10.4014/mbl.1811.11002
64. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The Human Tumor Microbiome is Composed of Tumor Type-Specific Intracellular Bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
Keywords: lung cancer, microbiota, cancer therapy, tumor microenvironment (TME), tumor immunology, Gut-Lung Axis, immunotherapy
Citation: Dong Q, Chen ES, Zhao C and Jin C (2021) Host-Microbiome Interaction in Lung Cancer. Front. Immunol. 12:679829. doi: 10.3389/fimmu.2021.679829
Received: 12 March 2021; Accepted: 07 May 2021;
Published: 24 May 2021.
Edited by:
Peter Brossart, University of Bonn, GermanyReviewed by:
Thomas Clarke, Imperial College London, United KingdomCopyright © 2021 Dong, Chen, Zhao and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Zhao, Y2hlbi56aGFvQG5paC5nb3Y=; Chengcheng Jin, Y2hlbmdjaGVuZy5qaW5AcGVubm1lZGljaW5lLnVwZW5uLmVkdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.