
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 09 June 2021
Sec. Inflammation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.679482
 Hua Geng1,2†
Hua Geng1,2† Saravanan Subramanian1,2
Saravanan Subramanian1,2 Longtao Wu3
Longtao Wu3 Heng-Fu Bu1,2
Heng-Fu Bu1,2 Xiao Wang1,2
Xiao Wang1,2 Chao Du1,2
Chao Du1,2 Isabelle G. De Plaen2,4
Isabelle G. De Plaen2,4 Xiao-Di Tan1,2,5,6*†
Xiao-Di Tan1,2,5,6*†Infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes COVID-19, a disease that involves significant lung tissue damage. How SARS-CoV-2 infection leads to lung injury remains elusive. The open reading frame 8 (ORF8) protein of SARS-CoV-2 (ORF8SARS-CoV-2) is a unique accessory protein, yet little is known about its cellular function. We examined the cellular distribution of ORF8SARS-CoV-2 and its role in the regulation of human lung epithelial cell proliferation and antiviral immunity. Using live imaging and immunofluorescent staining analyses, we found that ectopically expressed ORF8SARS-CoV-2 forms aggregates in the cytosol and nuclear compartments of lung epithelial cells. Using in silico bioinformatic analysis, we found that ORF8SARS-CoV-2 possesses an intrinsic aggregation characteristic at its N-terminal residues 1-18. Cell culture did not reveal any effects of ORF8SARS-CoV-2 expression on lung epithelial cell proliferation and cell cycle progression, suggesting that ORF8SARS-CoV-2 aggregates do not affect these cellular processes. Interestingly, ectopic expression of ORF8SARS-CoV-2 in lung epithelial cells suppressed basal expression of several antiviral molecules, including DHX58, ZBP1, MX1, and MX2. In addition, expression of ORF8SARS-CoV-2 attenuated the induction of antiviral molecules by IFNγ but not by IFNβ in lung epithelial cells. Taken together, ORF8SARS-CoV-2 is a unique viral accessory protein that forms aggregates when expressing in lung epithelial cells. It potently inhibits the expression of lung cellular anti-viral proteins at baseline and in response to IFNγ in lung epithelial cells, which may facilitate SARS-CoV-2 escape from the host antiviral innate immune response during early viral infection. In addition, it seems that formation of ORF8SARS-CoV-2 aggregate is independent from the viral infection. Thus, it would be interesting to examine whether any COVID-19 patients exhibit persistent ORF8 SARS-CoV-2 expression after recovering from SARS-CoV-2 infection. If so, the pathogenic effect of prolonged ORF8SARS-CoV-2 expression and its association with post-COVID symptoms warrant investigation in the future.
The global coronavirus disease 2019 (COVID-19) pandemic is caused by a novel beta coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This new pathogenic virus infects epithelial cells and macrophages in the lungs, which leads to severe inflammation in the respiratory system and diffuse alveolar damage (1–3). However, the key molecular mechanisms leading to lung injury after SARS-CoV-2 infection are not clear. Other pathogenic beta CoVs, including SARS-CoV and MERS-CoV, caused large-scale outbreaks in recent decades (4, 5). Previous studies demonstrated that the genome of CoVs encodes not only a set of viral structural proteins and a replicase complex but also a group of accessory proteins (6) that disrupt host signaling pathways and cell function (7). For example, previous studies found that the SARS-CoV ORF6 protein is present in the endoplasmic reticulum (ER)/Golgi membrane of infected cells (8), where it sequesters nuclear import factors on the rough ER/Golgi membrane and antagonizes STAT1-mediated antiviral signaling (9). Several SARS-CoV accessory proteins, including ORF3a, ORF8a, and ORF8b, have been shown to induce epithelial cell injury (10–13). SARS-CoV ORF8b can activate NLRP3-mediated inflammasome signaling (12). Similarly, MERS-CoV ORF4b binds to regulatory molecules including TBK1 and IKKϵ in cells and modulates interferon (IFN)-β production through inhibition of IRF3 phosphorylation (14). Furthermore, MERS-CoV ORF4b has 2’-5’-phosphodiesterase activity and antagonizes the host anti-viral OAS-RNase L pathway (15). Like other CoVs, the SARS-CoV-2 genome encodes various accessory proteins; however, the pathogenic roles of these proteins are largely unknown.
It has been reported that SARS-CoV and SARS-CoV-2 share a large amount of sequence identity (1). Interestingly, SARS-CoV-2 accessory protein ORF8 (ORF8SARS-CoV-2) has a low sequence similarity to SARS-CoV ORF8a, 8b, or 8ab. ORF8SARS-CoV-2 protein is highly expressed in patients with SARS-CoV-2 infection (16, 17). Recently, Young et al. (18) observed an association between ORF8SARS-CoV-2 and the severity of COVID-19 disease, suggesting that ORF8SARS-CoV-2 may play an important role in the pathogenesis of COVID-19. However, little is known about whether ORF8SARS-CoV-2 affects the host cell immunological response. Therefore, we investigated the fundamental question of whether and how lung epithelial cells respond to ORF8SARS-CoV-2 expression.
Human lung epithelial cell line A549 (derived from human pulmonary adenocarcinoma) and human embryonic kidney epithelial cell line HEK293 were obtained from American Type Culture Collection (ATCC, Manassas, VA). A549 and HEK cells were respectively cultured in ATCC-formulated F-12K and DMEM medium supplemented with 10% (v/v) fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in a humidified incubator with 5% CO2.
The gBlocks gene fragment of ORF8SARS-CoV-2 (RefSeq MN908947 27894 to 28256) was synthesized by Integrated DNA Technologies (Coralville, IA) and cloned into a pEGFP-N1 expression vector with C-terminal eGFP fusion, resulting in a new plasmid construct, pEGFP-N1-ORF8. The identity of pEGFP-N1-ORF8 was confirmed by Sanger sequencing. To generate a plasmid construct of ORF8SARS-CoV-2 with C-terminal Flag tag, pEGFP-N1-ORF8 was used as a template for PCR amplification and subcloned into pcDNA3.1 BamHI and EcoRI sites using In-Fusion HD Cloning kit (Clontech). The identity of pcDNA3.1-ORF8-Flag construct was confirmed by Sanger sequencing. A549 or HEK293 cells were seeded onto 6-well plates at a density of 3.5 × 105 cells/well and cultured at 37°C for 24 hours before transfection. Cells were then transfected with empty vector pEGFP-N1 or pEGFP-N1-ORF8 plasmids (2.5 µg/well), respectively, using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. In some experiments, cells were co-transfected with a mixture of plasmids (2.5 µg/well) and Poly(I:C) (25 ng/well, InvivoGen, San Diego, CA). Culture medium was refreshed after 4 hours, and cells were processed for analysis 24-48 hours post-transfection. Some cells were treated with IFNβ- or IFNγ-containing medium (100 ng/mL, PeproTech, Rocky Hill, NJ) after transfection and then processed for analysis.
Total protein was isolated from cells using RIPA lysis buffer (Thermo Fisher Scientific) and Halt protease inhibitor cocktail (Thermo Fisher Scientific). Protein concentration was measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific) and cell lysates containing 20 µg of total protein were loaded and separated in 4-20% TGX precast SDS-PAGE gels (Bio-Rad, Hercules, CA) followed by transfer onto PVDF membranes (Bio-Rad). Immunoblot analyses were carried out as previously described (19). Mouse monoclonal antibody against GFP (1:1000, Santa Cruz, Dallas, Texas) and HRP-conjugated mouse monoclonal antibody against β-actin (1:50,000, Sigma-Aldrich, St. Louis, MO) were used for the assay.
Cells were seeded onto a Lab-Tek chambered coverslip (Sigma-Aldrich) and transfected with empty vector pEGFP-N1 or pEGFP-N1-ORF8 plasmids. GFP-live fluorescent images were captured and merged with differential interference contrast (DIC) bright-field images using a Leica Thunder microscope system (Wetzlar, Germany). For immunofluorescent staining, cells seeded onto Lab-Tek removable chamber slides were transfected with plasmids and Poly(I:C) as described above, then fixed using 4% paraformaldehyde (Thermo Fisher Scientific), permeabilized with 0.1% Triton X-100, and blocked with PBS buffer containing 2% normal goat serum (Vector Labs, Burlingame, CA) and 1% bovine serum albumin (Sigma-Aldrich). The slides were incubated at 4°C overnight with chicken anti-GFP primary antibody (1:500; Aves Lab, Tigard, OR). After incubation, slides were washed with PBS and incubated for 1 hour at room temperature in the dark with goat anti-chicken IgY (H+L) antibody labeled with Alexa Fluor 488 (1:250; Thermo Fisher Scientific). Finally, slides were washed with PBS and mounted with 4’,6-diamidino-2-phenylindole (DAPI)-contained mounting solution (Vector Labs). Slides were reviewed under a Leica Thunder microscope system and further processed by Adobe Photoshop software v21 (Adobe Systems Inc., San Jose, CA). For image quantification, 8-14 random images were acquired, and approximately 100-200 GFP-positive cells were counted from each group. To further confirm the subcellular distribution of ORF8SARS-CoV-2, cells were transfected with pcDNA3.1-ORF8-Flag construct and the transfected cells were processed for immunofluorescent staining with mouse anti-Flag M2 antibody (1:500; Sigma).
Cells were plated in 24-well plates and co-transfected with plasmids with or without Poly(I:C) as described above. Cell proliferation was assessed 24 hours after transfection. In brief, cells were trypsinized and mixed with an equal volume of 0.4% trypan blue stain, then subjected to cell counting using a Countess 3 automated cell counter (Thermo Fisher Scientific). All experiments were conducted in triplicate.
To assess transfection efficiency, cells were plated in 6-well plates and transfected with plasmids. Cells were collected 24 hours post-transfection and stained with cell viability dye FVD-eFluor-506 (Thermo Fisher Scientific) at 4°C for 30 min, then subjected to flow cytometry analysis using a BD FACSymphony A5 Cell Analyzer (Indianapolis, IN). The GFP-positive live cell population was calculated by FlowJo (FlowJo LLC, Ashland, OR). For cell cycle analysis, cells were collected and washed twice with PBS, then resuspended in 1 mL PBS and fixed by adding 4 mL ice-cold absolute ethanol at -20°C overnight. After fixation, cells were washed and counter-stained with a PBS-buffered solution containing 0.1% Triton X-100, 3 µM propidium iodide (PI, Thermo Fisher Scientific), and 0.5 mg/mL RNase A (Sigma Aldrich) at 4°C for 30 min. Cell cycle distribution was examined using a BD FACSymphony A5 Cell Analyzer, and the proportion of cells in the G0−G1, S, and G2−M phases was determined using FlowJo software. All experiments were performed in triplicate.
Total RNA was extracted from cells using Trizol (Thermo Fisher Scientific) and purified by Zymo RNA Clean & Concentrator kit (Zymo, Irvine, CA). RNA was quantified with the Nanodrop (Agilent Technologies, Santa Clara, CA) and single-strand cDNA was generated using the iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was performed using SYBR Green PowerUp PCR Universal Mastermix (Thermo Fisher Scientific) and the QuantStudio™ 6 real-time PCR system (Thermo Fisher Scientific), all according to the manufacturer’ manuals. The fold change in expression levels of target genes was calculated using the 2−ΔΔCT method, with GAPDH as the endogenous reference. PCR reactions were run in duplicate for each sample. Primers used for RT-qPCR were synthesized by Integrated DNA Technologies and are listed in Supplementary Table 1. All experiments were repeated at least three times.
The ORF8SARS-CoV-2 sequence was retrieved from NCBI RefSeq MN908947 and submitted to online bioinformatics prediction tool PASTA2 (http://protein.bio.unipd.it/pasta2/). The propensity of ORF8 to form aggregates was assessed based on parallel aggregation probability and free energy.
All experiments were performed at least twice with triplicate samples. Statistical analysis was performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA). Data are presented as mean ± s.d. and the statistical significance was assessed with either student’s t-test or one-way analysis of variance (ANOVA) followed by Fisher’s least significant difference post-hoc test. p<0.05 was considered statistically significant.
ORF8SARS-CoV-2 is an accessory protein encoded by the SARS-CoV-2 genome. This protein has been detected in sera of COVID-19 patients (16, 17), suggesting that ORF8SARS-CoV-2 is expressed in SARS-CoV-2 virus-infected cells. However, the function and pathobiological features of ORF8SARS-CoV-2 are largely unknown. To study ORF8SARS-CoV-2 biology, we prepared a plasmid, pEGFP-ORF8SARS-CoV-2, by inserting the full-length sequence of ORF8SARS-CoV-2 at the N-terminus of the enhanced green fluorescent protein (eGFP) of the pEGFP-N1 mammalian expression vector (Figure 1A). After transfecting the pEGFP-ORF8SARS-CoV-2 plasmid into A549 cells, expression of eGFP-tagged ORF8SARS-CoV-2 protein (42 kDa) was confirmed by immunoblot with anti-GFP. eGFP (27 kDa) was expressed in the A549 cells transfected with pEGFP-N1 empty vector (EV) (Figure 1B). When examined by flow cytometry analysis, approximately 60% of A549 cells were GFP-positive 24 h after transfection with either pEGFP-N1 or pEGFP-ORF8SARS-CoV-2 constructs (Figure 1C), suggesting significant uptake of plasmids by A549 cells. This in vitro system was used to study the function of ORF8SARS-CoV-2 in human lung epithelial cells.
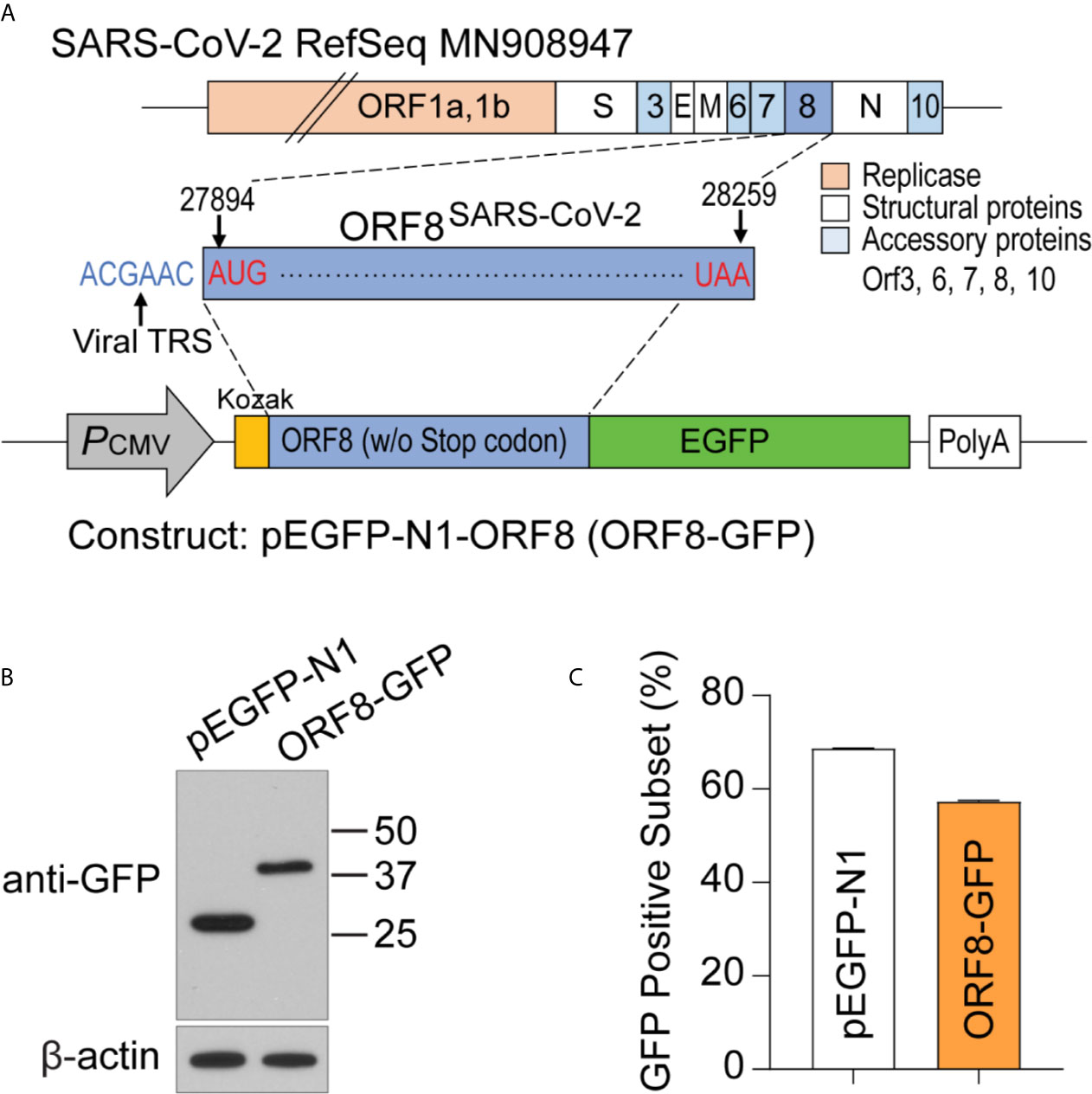
Figure 1 Generating a plasmid construct to study the function of ORF8SARS-CoV-2 in human A549 lung epithelial cells. (A) Cloning strategy to generate the pEGFP-ORF8SARS-CoV-2 (ORF8-GFP) construct. (B) Verification of ORF8-GFP expression in A549 lung epithelial cells transfected with pEGFP-ORF8SARS-CoV-2 by immunoblot analysis using anti-GFP antibody. Whole cell lysates were collected from cells transfected with empty vector (pEGFP-N1; EV) or ORF8-GFP at 24 hours post-transfection. (C) Characterization of the transfection efficiency of ORF8-GFP in A549 lung epithelial cells using FACS analysis. Cells were subjected to flow cytometry 24 hours after transfection with pEGFP-N1 (EV) or ORF8-GFP. The experiments were repeated twice with triplicate samples.
Because SARS-CoV-2 targets human lung epithelial cells (20, 21), we first visualized the distribution of the SARS-CoV-2 ORF8 accessory protein in A549 cells using the pEGFP-ORF8SARS-CoV-2 plasmid construct. Using live fluorescent imaging, we found that pEGFP-N1-transfected A549 cells exhibited homogeneous GFP fluorescence (Figure 2A). In contrast, distinctive fluorescent particles were seen in A549 cells transfected with the pEGFP-ORF8SARS-CoV-2 construct (Figure 2A), suggesting that ORF8SARS-CoV-2 forms aggregates in lung epithelial cells. In addition, immunofluorescence staining revealed that ectopically expressed ORF8SARS-CoV-2-GFP protein was either homogenously distributed in A549 cells or formed aggregates in cytoplasmic and nuclear compartments of the cells (Figure 2B). Image quantification revealed a higher ratio of intracellular aggregates in pEGFP-ORF8SARS-CoV-2-transfected cells compared to cells transfected with pEGFP-N1 empty vector (Figure 2C). To exclude the possibility that eGFP fusion to C-terminal of ORF8SARS-CoV-2 could affect the subcellular distribution of ORF8SARS-CoV-2, cells were transfected with pcDNA3.1-ORF8-Flag construct followed by immunofluorescent staining with anti-Flag antibody. Using this approach, we confirmed a similar subcellular expression pattern of ORF8SARS-CoV-2 (Supplementary Figure 1), suggesting that ectopic expression of ORF8SARS-CoV-2 forms cytoplasmic and nuclear aggregates in the lung epithelial cells without contribution of Tag proteins. Using in silico prediction (http://protein.bio.unipd.it/pasta2/) (22), we found that ORF8SARS-CoV-2 possesses an intrinsic aggregation characteristic at its N-terminal residues 1-18 (Figures 2D, E), with a total of 20 potential aggregations predicted with the best energy -10.07 (Supplementary Table 2).
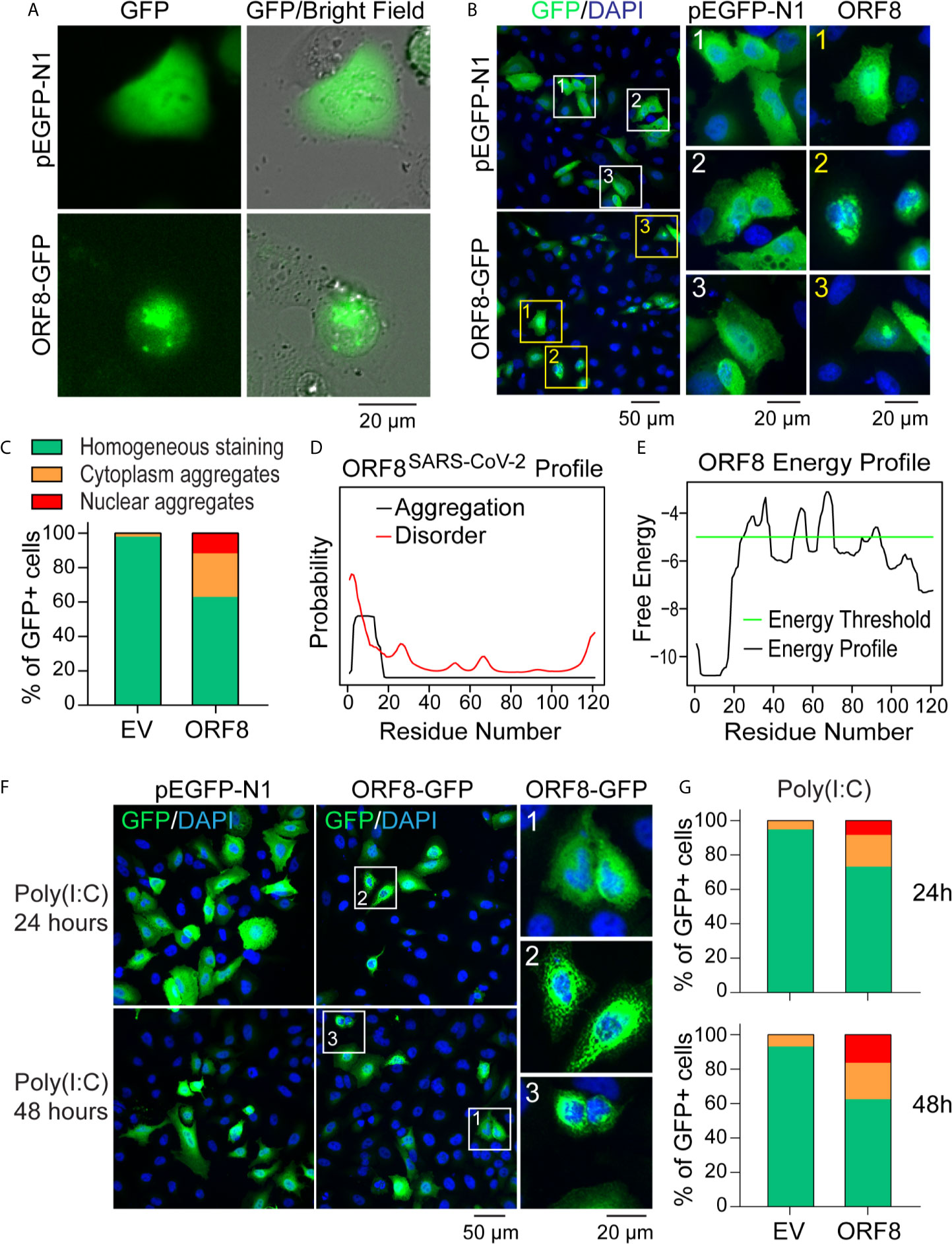
Figure 2 ORF8SARS-CoV-2 forms intracellular aggregates in human lung epithelial cells. (A-C) Aggregation of ORF8SARS-CoV-2 protein in A549 cells viewed by fluorescent microscopy. Human lung epithelial A549 cells were transfected with pEGFP-N1 (empty vector; EV) or pEGFP-ORF8SARS-CoV-2. After 24 h, fluorescent microscopy was performed on either live cells (A) or paraformaldehyde-fixed cells that were stained with anti-GFP antibody (B). In the anti-GFP antibody-stained cells, quantitative analysis of nuclear-cytoplasmic distribution pattern of aggregated ORF8SARS-CoV-2 was performed (C). (D, E) Prediction of aggregation based on the ORF8SARS-CoV-2 protein primary structure using the PASTA 2.0 algorithm (http://protein.bio.unipd.it/pasta2/). Aggregation disorder profile (D) and aggregation-free energy profile (E) showing that N-terminal residues 1-18 have the lowest aggregation free energy and thus is the most aggregation-stabilizing region underlying the propensity for ORF8SARS-CoV-2 aggregates formation. (F, G) The intracellular distribution pattern of ORF8SARS-CoV-2 aggregates is not affected by a poly (I:C)-induced inflammatory response. A549 cells were treated with poly (I:C) during transfection with pEGFP-N1 (EV) or pEGFP-ORF8SARS-CoV-2. After the indicated times, cells were fixed with paraformaldehyde followed by immunofluorescent staining with anti-GFP antibody. The stained cells were viewed by fluorescent microscopy (F) and processed for analysis of nuclear-cytoplasmic distribution pattern of ORF8SARS-CoV-2 aggregates (G). 1 indicates homogenous distribution; 2, cytoplasmic aggregates; and 3, nuclear aggregates.
We next examined whether activation of antiviral immune response affects the formation of ORF8SARS-CoV-2 aggregates in lung epithelial cells. A549 cells were stimulated with Poly(I:C) to mimic the activation of cells by viral infection. As expected, treatment with Poly(I:C) profoundly induced the expression of various antiviral genes in A549 cells (Supplementary Figure 2), indicating activation of the antiviral immune response. Next, cells were co-transfected with a mixture of plasmids and Poly(I:C) to examine the formation of ORF8SARS-CoV-2 aggregates in lung epithelial cells under a Poly(I:C)-mimicked antiviral microenvironment. Immunostaining analysis did not reveal any change in the distribution pattern of ORF8SARS-CoV-2 aggregates in poly(I:C)-treated cells (Figures 2F, G). These data suggest that ORF8SARS-CoV-2 forms aggregates in both the cytosol and nucleus of lung epithelial cells, independent from inflammatory events activated by viral infection.
Intracellular protein aggregates can be toxic, causing injury or death to cells (23, 24). Thus, we next examined whether lung epithelial cell growth is affected by ectopic ORF8SARS-CoV-2 expression. Cell numbers of pEGFP-N1-transfected and pEGFP-ORF8SARS-CoV-2-transfected A549 cells were compared in the presence or absence of poly(I:C). Cells transfected with pEGFP-ORF8SARS-CoV-2 exhibited a similar growth rate to empty vector-transfected cells when cultured with or without poly(I:C) (Figure 3A).
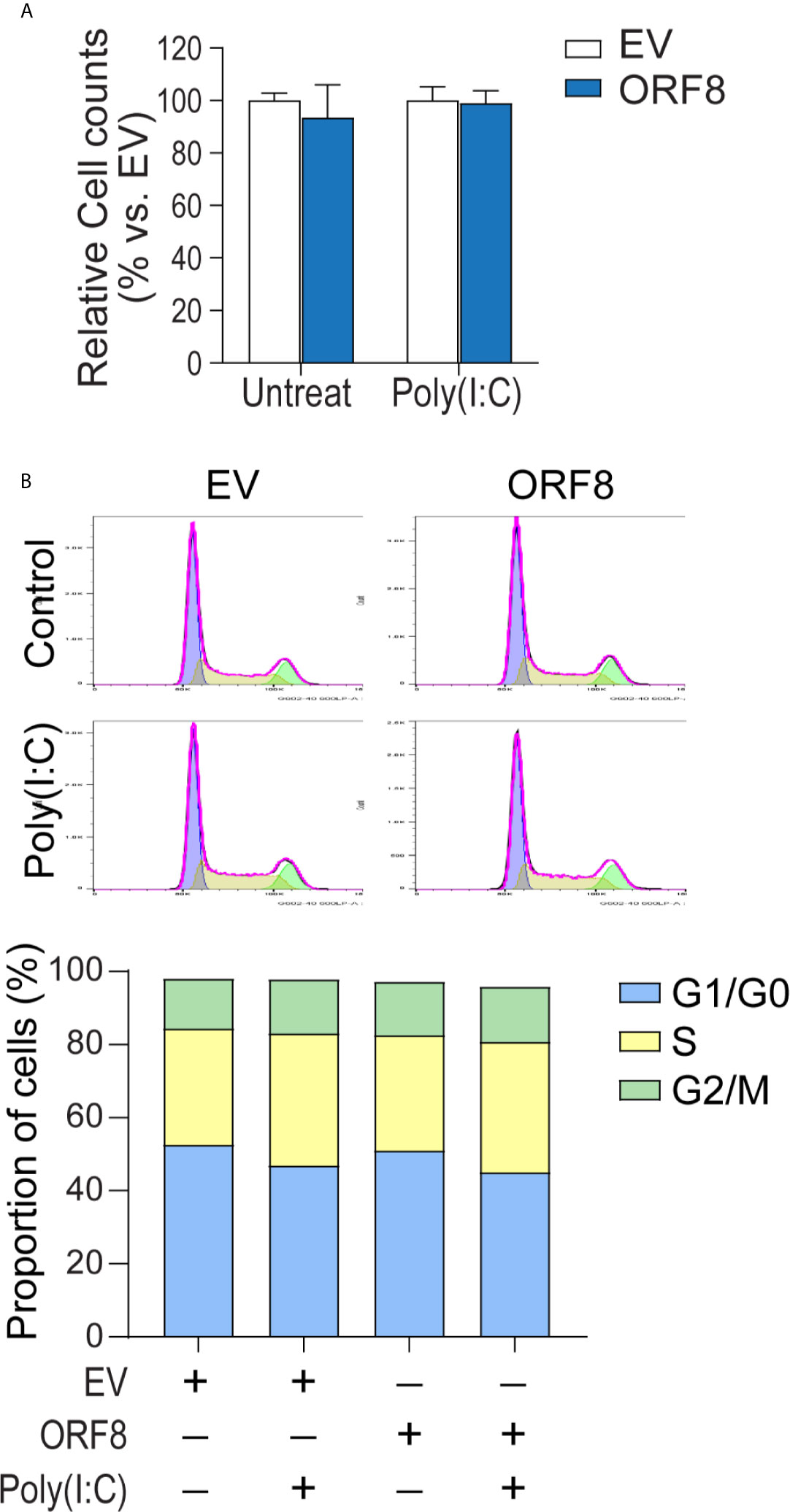
Figure 3 Expression of ORF8SARS-CoV-2 does not affect lung epithelial cell proliferation or cell cycle progression. Human A549 lung epithelial cells were transfected with pEGFP-N1 (empty vector, EV) or pEGFP-ORF8SARS-CoV-2 and treated with or without poly (I:C). At 24 hours post-transfection, cells were processed for in vitro proliferation assay (A) and flow cytometry-based cell cycle analysis (B). n = 3 for each group.
It has been shown that the interference with cell cycle progression is a common host response upon viral infection (25, 26). Thus, we investigated the influence of ORF8SARS-CoV-2 expression on cell cycle progression in A549 cells using flow cytometry analysis. We noted that cell cycle phases (G1/G0, S, G2/M) in pEGFP-ORF8SARS-CoV-2-transfected cells were similar to those in pEGFP-N1-transfected cells, regardless of poly(I:C) stimulation (Figure 3B), suggesting that expression of ORF8SARS-CoV-2 does not change cell cycle status. Therefore, neither expression of ORF8SARS-CoV-2 nor formation of ORF8SARS-CoV-2 protein aggregates influenced lung epithelial cell proliferation and cell cycle progression.
To further determine the effect of ORF8SARS-CoV-2 on the antiviral immune response, we examined whether ORF8SARS-CoV-2 affects expression of viral infection-associated innate immune molecules (Table 1) in A549 cells. Using RT-qPCR, we found that ectopic expression of ORF8SARS-CoV-2 significantly inhibited expression of some members of the IFN-stimulated gene (ISG) family including ZBP1 (an innate sensor of viral infection), MX1 and MX2 (host restriction factors of viral replication), and DHX58 (a critical molecule in the RIG-I cytosolic pattern recognition receptor pathway) in A549 cells (Figure 4A). However, expression of ORF8SARS-CoV-2 did not influence the expression of IFNβ and several other IFN signature genes including IFIH1, DDX60, OAS3, and IFITM1 (Figure 4B). To determine whether ORF8SARS-CoV-2 could affect the basal-level expression of these ISG-associated genes in other type of cells, we further transfected HEK293 embryonic kidney epithelial cells with pEGFP-ORF8SARS-CoV-2. We revealed significant decrease in expression of OAS3 and IFITM1 in ORF8SARS-CoV-2-transfected HEK293 cells comparing to the control cells (Supplementary Figure 3). Interestingly, the expression of these two genes in A549 cells was not affected by ectopic ORF8SARS-CoV-2 (Figure 4B). These data suggest that ORF8SARS-CoV-2 selectively disrupts expression of several innate immunity-associated molecules without affecting IFN signaling in a cell type specific manner.
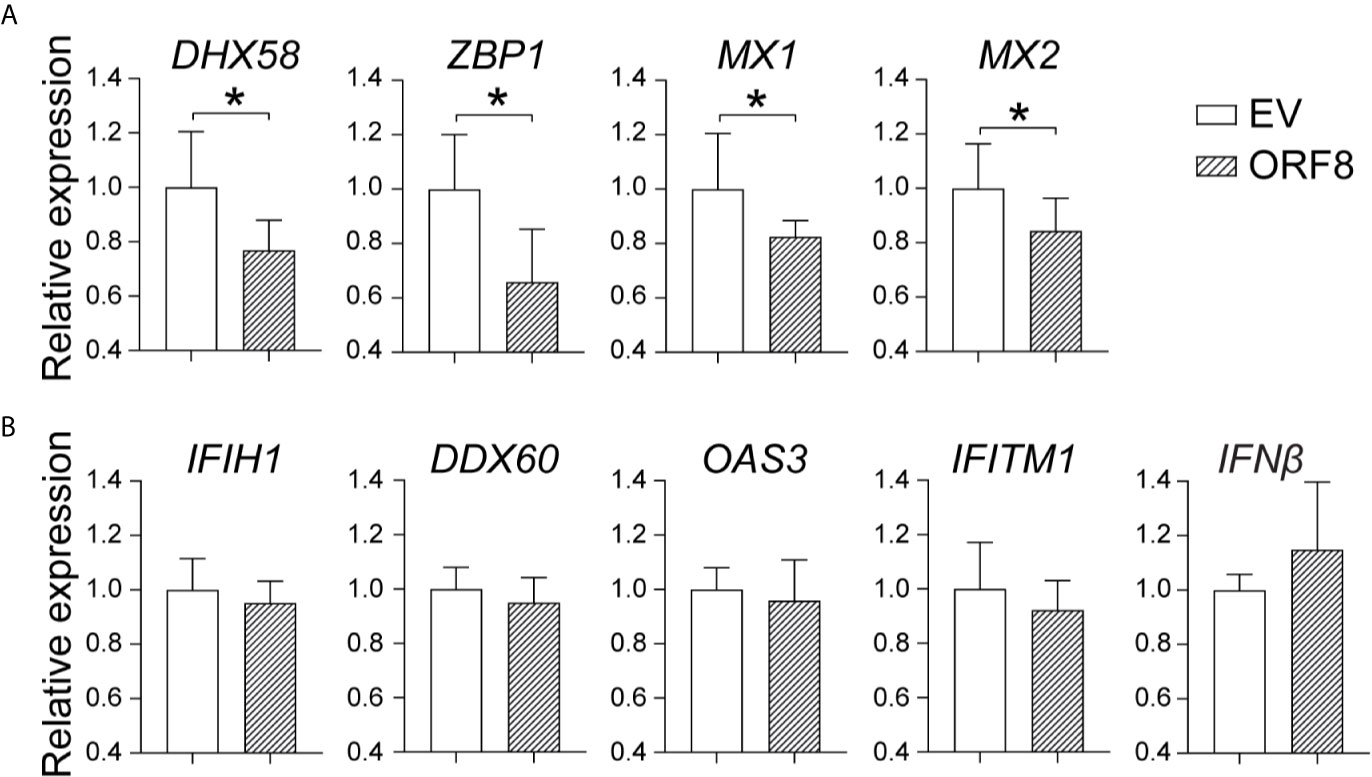
Figure 4 Characterization of the role of ORF8SARS-CoV-2 on expression of antiviral immunity-associated genes in lung epithelial cells. A549 cells were transfected with pEGFP-N1 (empty vector, EV) or pEGFP-ORF8SARS-CoV-2. At 24 hours post-transfection, cells were processed for RNA extraction and RT-qPCR to determine the expression levels of indicated immune response genes. (A) ORF8-inhibited genes. (B) Non-responsive genes. n = 3-5, *p < 0.05 was considered as statistically significant.
IFNγ and IFNβ play critical roles in the regulation of the antiviral immune response (27, 28). Thus, we examined whether ORF8SARS-CoV-2 affects IFNγ and/or IFNβ-induced ISG expression in A549 cells. First, we found that both IFNγ and IFNβ markedly induced expression of several antiviral molecules (Supplementary Figures 4A, B). Expression of ORF8 did not change IFNβ-induced expression of most antiviral molecules assessed, but did increase IFNβ-induced endogenous IFNβ expression in lung epithelial cells (Figure 5A). In contrast, expression of ORF8 in lung epithelial cells significantly decreased IFNγ-induced antiviral molecules (Figure 5B), suggesting that expression of ORF8SARS-CoV-2 inhibits IFNγ-mediated antiviral immunity. Together, it appears that ORF8SARS-CoV-2 affects the IFNγ- but not IFNβ-induced host antiviral response in lung epithelial cells.
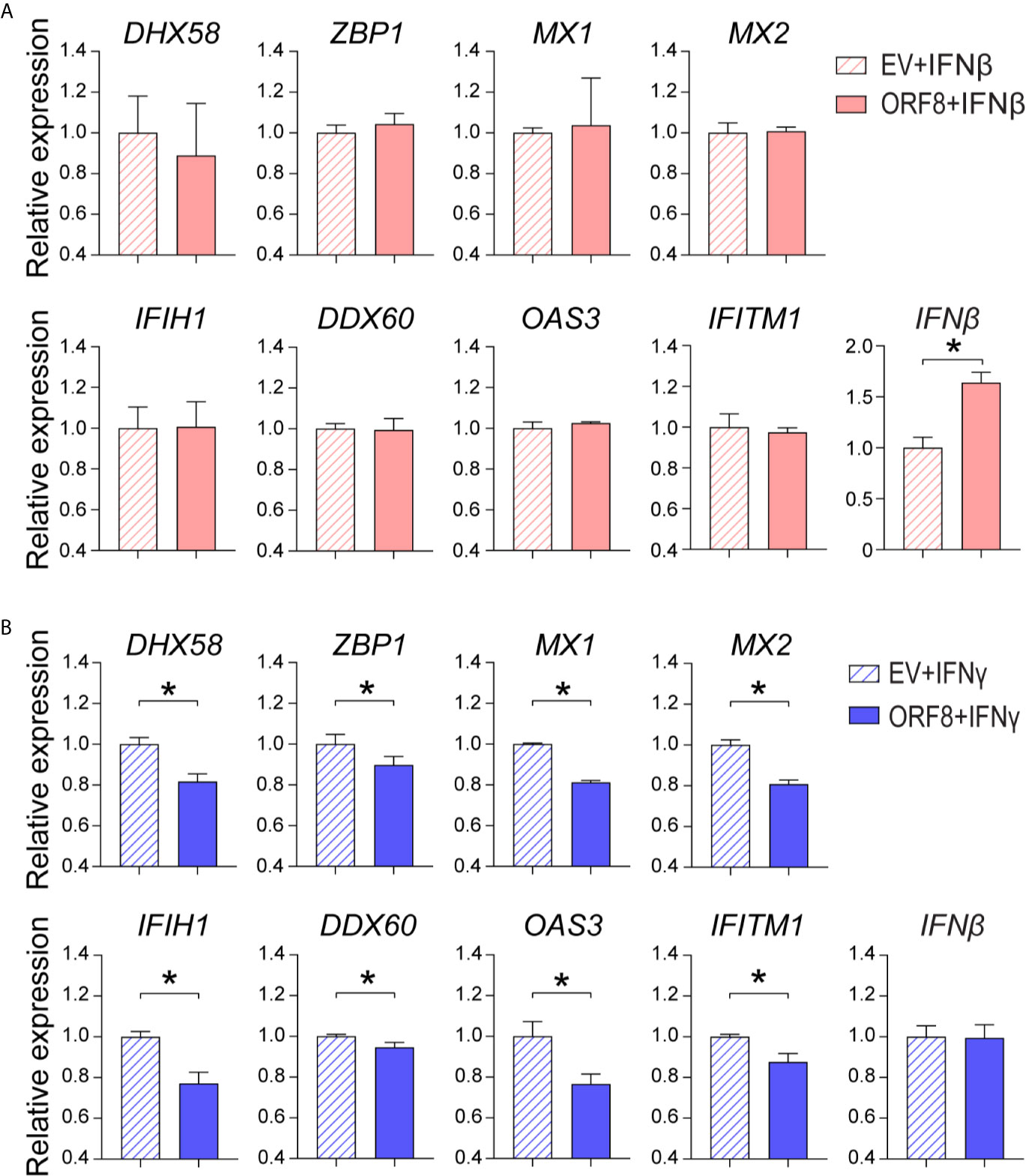
Figure 5 Characterization of the role of ORF8SARS-CoV-2 on regulation of type I and II interferon (IFN)-induced expression of antiviral molecules in human lung epithelial cells. Human A549 lung epithelial cells were transfected with pEGFP-N1 (empty vector, EV) or pEGFP-ORF8SARS-CoV-2. The transfected cells were treated with medium containing IFNβ (100 ng/mL) (A) or IFNγ (100 ng/mL) (B) for 24 h followed by RNA extraction and RT-qPCR analysis of expression of the indicated genes. n = 3-5. *p < 0.05 was considered as statistically significant.
In this study, we characterized the biological and immunobiological effects of the SARS-CoV-2 ORF8 accessory protein in lung epithelial cells. Specifically, we found that the N-terminal sequence of ORF8SARS-CoV-2 is composed of features that favor the formation of protein aggregates. We showed that ectopic expression of ORF8SARS-CoV-2 results in formation of cytoplasmic and nuclear ORF8 protein aggregates in lung epithelial cells, forming structures that can profoundly influence cell processes (12, 23). Interestingly, expression of ORF8SARS-CoV-2 did not affect lung epithelial cell proliferation and cell cycle progression regardless the activation of antiviral responses in the cells. Rather, ORF8SARS-CoV-2 expression was found to attenuate the IFNγ- but not IFNβ-mediated antiviral gene expression in lung epithelial cells. Together, our study suggests that ORF8SARS-CoV-2 forms aggregates in lung cells that then impair the molecular machinery of the antiviral immune response.
Evidence suggests that host cells form insoluble aggregates/inclusions during viral infections (23), which then have various functions in cells. For example, viral infection-associated protein aggregates can form protective structures that facilitate viral escape from cellular degradation machinery (23, 45). Viruses can also utilize aggregates to promote viral replication, gene translation, and intra- and intercellular transportation (23, 45). In addition, protein aggregates and misfolded proteins themselves can cause host cell death by inducing ER stress and the unfolded protein response (UPR) (12). Previously, SARS-CoV ORF8b was found to form insoluble intracellular aggregates through its C-terminus; aggregated ORF8b caused the death of epithelial cells through induction of ER stress and subsequent activation of the autophagy-lysosome pathway (12). In the present study, we detected the intracellular aggregation of ORF8SARS-CoV-2 in lung epithelial cells using a nonvirus-infection approach, but we did not observe cytotoxic effects of ORF8SARS-CoV-2 aggregates in the cells. Thus, we speculate that ORF8SARS-CoV-2 aggregates might not overwhelm cellular homeostatic mechanisms in our experimental settings in vitro.
The mechanism underlying formation of ORF8SARS-CoV-2 aggregates is not clear. Evidence shows that typical residues and regions in ORF proteins of coronavirus play an important role in formation of protein aggregates. For instance, Ng and Liu previously revealed that coronavirus IBV ORF 1a forms aggregates, which seems to be mediated the presence of a hydrophobic domain downstream of the 3C-like proteinase-encoding region in this ORF protein (46). Shi et al. recently reported that ORF8b of SARS-CoV forms intracellular aggregates dependent on a valine at residue 77 of this ORF protein (12). Using a bioinformatic approach, we noted that N-terminal residues 1-18 of ORF8SARS-CoV-2 may affect the aggregation propensity of this protein. Further experimental analysis is required to clarify exact roles of these residues in formation of intracellular aggregates in ORF8SARS-CoV-2 expressing cells.
Our findings are consistent with a growing body of evidence showing that accessory proteins of pathogenic CoVs play a role in disrupting antiviral immune responses (6, 7, 47). For instance, SARS-CoV ORF3a was found to promote ubiquitination and lysosomal pathway-dependent degradation of IFNAR1, which results in attenuation of the type I IFN response (48). SARS-CoV ORF8b and 8ab were reported to act as IFN antagonists through mediating ubiquitin-dependent rapid degradation of IRF3 (49). Recently, two independent studies demonstrated that SARS-CoV-2 ORF6 is a potent IFN antagonist that inhibits the IFNβ promoter, ISRE, and NF-κB element in IFN-stimulated genes (ISGs) (47, 50). Furthermore, Miorin et al. (51) reported that SARS-CoV-2 ORF6 inhibits IFN signaling through blocking STAT nuclear import. Collectively, it appears that accessory proteins encoded by pathogenic CoV genomes influence immunological activities in the host cells.
SARS-CoV-2 infects human lung epithelial cells which are increasingly recognized as an important immunological barrier. In the present study, we found that ORF8SARS-CoV-2 selectively attenuates the basal expression of several ISGs including ZBP1, MX1 and MX2, and DHX58 in the lung epithelial cells. Among these genes, ZBP1 and DHX58 are reported to play an important role in sensing viral pathogens (32–34, 37, 52), whereas MX1 and MX2 are critical antiviral gatekeepers in cells (40, 53). Interestingly, IFN production is impaired in COVID-19 patients (54). Thus, our findings suggest that ORF8SARS-CoV-2 attenuates basal-level expression of diverse viral-sensing molecules; the lack of IFN signaling subsequently facilitates replication of SARS-CoV-2 in the host cells.
Evidence shows that signaling cascades activated by IFNα/β are of particular importance in the host response to viral pathogens (27–29). Recently, several investigators studied the role of ORF8SARS-CoV-2 in the regulation of the type I IFN-associated antiviral immune responses, though the conclusions drawn from these studies remain controversial. For example, Li et al. (47) reported that ORF8SARS-CoV-2 is a potent IFN antagonist that delays the release of IFNs through inhibition of IFNβ promoter activity in HEK 293T cells. Yuen et al. (50) found that ORF8SARS-CoV-2 does not suppress primary IFN production and signaling in 293FT cells. Here, we showed that expression of ORF8SARS-CoV-2 promotes the autocrine effect of IFNβ on its own gene expression but does affect IFNβ-mediated induction of antiviral ISGs family genes in lung epithelial cells. Taken together, our data, combined with findings from other investigators, suggest that ORF8SARS-CoV-2 affects type I IFN signaling in a cell-type dependent manner via a complex mechanism.
In addition to IFNα/β, emerging evidence shows that IFNγ carries out unique functions in antiviral immunity (55). For example, NK cells or innate lymphoid type I cells produce IFNγ to modulate the function of surrounding innate immune cells, including macrophages and dendritic cells, which subsequently regulate the antiviral immune response (27, 55). In the present study, we showed that IFNγ profoundly induces the expression of various antiviral molecules in lung epithelial cells, suggesting that IFNγ enhances the innate immune function in the lung epithelial barrier. Furthermore, we found for the first time that expression of ORF8SARS-CoV-2 significantly inhibits IFNγ-mediated upregulation of antiviral gene expression, suggesting that ORF8SARS-CoV-2 hinders IFNγ-regulated antiviral immunity. The mechanism underlying the blockage of IFNγ activity by ORF8SARS-CoV-2 needs to be studied in the future.
Upon virus infection, the host cells initiate and launch innate antiviral immune responses. In present study, we found ORF8SARS-CoV-2 attenuates basal-level expression of diverse viral-sensing molecules and suppresses IFNγ-regulated antiviral immunity in cultured A549 human lung epithelial cells. However, it remains unclear whether ORF8SARS-CoV-2 hinders host cells antiviral immunity in SARS-CoV-2 infected cells. To firmly elucidate the role of ORF8SARS-CoV-2 in coronavirus pathogenesis by targeting ISGs of lung epithelial cells, it would be of interest to further study cell biological and immunological functions of ORF8SARS-CoV-2 in SARS-CoV-2 infected cells using mutagenesis and RNAseq approaches in the future. In addition, our data suggest that formation of ORF8SARS-CoV-2 aggregates in cells takes place without active SARS-CoV-2 infection, which may affect cell functions. Thus, future studies are needed to determine whether persistent ORF8SARS-CoV-2 expression occurs in COVID-19 patients after recovering from SARS-CoV-2 infection. If so, it is possible that prolonged ORF8SARS-CoV-2 expression may play a pathogenic role in long-haul COVID-19 symptoms.
Collectively, our study suggests that ORF8SARS-CoV-2 displays several potent pathogenic activities in human lung epithelial cells including formation of intracellular aggregates, disruption of basal expression of multiple types of viral sensing molecules, and inhibition of IFNγ-induced antiviral gene expression. However, the link among these ORF8SARS-CoV-2-associated cell biological and immunological events is not clear. Furthermore, molecular mechanisms underlying these processes are of interest to study in the future, which will advance our knowledge in regarding the role of ORF8SARS-CoV-2 in pathogenesis of COVID-19 and development of novel therapeutic approaches for maintaining homeostasis of antiviral immunity in patients with COVID-19.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
HG and X-DT were involved in the overall design of experiments and interpretation of results. HG, SS, LW, and H-FB performed all experiments. HG and LW prepared figures. HG, SS, and X-DT wrote manuscript with input from all authors. XW, CD, and IP critically reviewed and commented on the work. HG and X-DT conceived and orchestrated the project. All authors contributed to the article and approved the submitted version.
HG is a recipient of the COVID-19 Exploratory Springboard Award from the Stanley Manne Children’s Research Institute, the Ann & Robert H. Lurie Children’s Hospital of Chicago. X-DT is funded by US National Institutes of Health (NIH) grants (R01GM117628, R01GM122406, and R01DK123826) and US Department of Veterans Affairs Merit Review Award (I01BX001690). IP is funded by NIH grant R01DK116568. The funders had no role in study design, data collection and analysis, interpretation of data, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.679482/full#supplementary-material
SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID19, coronavirus disease 2019; ORF8, open reading frame 8; ISG, interferon-stimulated gene; IFNβ, interferon beta; IFNγ, interferon gamma; IFIH1, interferon induced with helicase C domain 1; DHX58, DExH-box helicase 58; DDX60, DExD/H-box helicase 60; ZBP1, Z-DNA binding protein 1; OAS3, 2’-5’-oligoadenylate synthetase 3; MX1, MX dynamin like GTPase 1; MX2, MX dynamin like GTPase 2; IFITM1, interferon-induced transmembrane protein 1.
1. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet (2020) 395(10224):565–74. doi: 10.1016/S0140-6736(20)30251-8
2. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A Pneumonia Outbreak Associated With a New Coronavirus of Probable Bat Origin. Nature (2020) 579(7798):270–3. doi: 10.1038/s41586-020-2012-7
3. Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, et al. Circuits Between Infected Macrophages and T Cells in SARS-CoV-2 Pneumonia. Nature (2021) 590(7847):635–41. doi: 10.1038/s41586-020-03148-w
4. Breban R, Riou J, Fontanet A. Interhuman Transmissibility of Middle East Respiratory Syndrome Coronavirus: Estimation of Pandemic Risk. Lancet (2013) 382(9893):694–9. doi: 10.1016/S0140-6736(13)61492-0
5. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A Novel Coronavirus Associated With Severe Acute Respiratory Syndrome. N Engl J Med (2003) 348(20):1953–66. doi: 10.1056/NEJMoa030781
6. Satija N, Lal SK. The Molecular Biology of SARS Coronavirus. Ann N Y Acad Sci (2007) 1102:26–38. doi: 10.1196/annals.1408.002
7. McBride R, Fielding BC. The Role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus Accessory Proteins in Virus Pathogenesis. Viruses (2012) 4(11):2902–23. doi: 10.3390/v4112902
8. Geng H, Liu YM, Chan WS, Lo AW, Au DM, Waye MM, et al. The Putative Protein 6 of the Severe Acute Respiratory Syndrome-Associated Coronavirus: Expression and Functional Characterization. FEBS Lett (2005) 579(30):6763–8. doi: 10.1016/j.febslet.2005.11.007
9. Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J Virol (2007) 81(18):9812–24. doi: 10.1128/JVI.01012-07
10. Yue Y, Nabar NR, Shi CS, Kamenyeva O, Xiao X, Hwang IY, et al. Sars-Coronavirus Open Reading Frame-3a Drives Multimodal Necrotic Cell Death. Cell Death Dis (2018) 9(9):904. doi: 10.1038/s41419-018-0917-y
11. Chen CY, Ping YH, Lee HC, Chen KH, Lee YM, Chan YJ, et al. Open Reading Frame 8a of the Human Severe Acute Respiratory Syndrome Coronavirus Not Only Promotes Viral Replication But Also Induces Apoptosis. J Infect Dis (2007) 196(3):405–15. doi: 10.1086/519166
12. Shi CS, Nabar NR, Huang NN, Kehrl JH. Sars-Coronavirus Open Reading Frame-8b Triggers Intracellular Stress Pathways and Activates NLRP3 Inflammasomes. Cell Death Discov (2019) 5:101. doi: 10.1038/s41420-019-0181-7
13. Law PY, Liu YM, Geng H, Kwan KH, Waye MM, Ho YY. Expression and Functional Characterization of the Putative Protein 8b of the Severe Acute Respiratory Syndrome-Associated Coronavirus. FEBS Lett (2006) 580(15):3643–8. doi: 10.1016/j.febslet.2006.05.051
14. Yang Y, Ye F, Zhu N, Wang W, Deng Y, Zhao Z, et al. Middle East Respiratory Syndrome Coronavirus ORF4b Protein Inhibits Type I Interferon Production Through Both Cytoplasmic and Nuclear Targets. Sci Rep (2015) 5:17554. doi: 10.1038/srep17554
15. Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, et al. Middle East Respiratory Syndrome Coronavirus NS4b Protein Inhibits Host Rnase L Activation. mBio (2016) 7(2):e00258. doi: 10.1128/mBio.00258-16
16. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. ORF8 and ORF3b Antibodies are Accurate Serological Markers of Early and Late SARS-CoV-2 Infection. Nat Immunol (2020) 21(10):1293–301. doi: 10.1038/s41590-020-0773-7
17. Wang X, Lam JY, Wong WM, Yuen CK, Cai JP, Au SW, et al. Accurate Diagnosis of COVID-19 by a Novel Immunogenic Secreted Sars-CoV-2 Orf8 Protein. mBio (2020) 11(5):e02431–20. doi: 10.1128/mBio.02431-20
18. Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a Major Deletion in the SARS-CoV-2 Genome on the Severity of Infection and the Inflammatory Response: An Observational Cohort Study. Lancet (2020) 396(10251):603–11. doi: 10.1016/S0140-6736(20)31757-8
19. Geng H, Bu HF, Liu F, Wu L, Pfeifer K, Chou PM, et al. In Inflamed Intestinal Tissues and Epithelial Cells, Interleukin 22 Signaling Increases Expression of H19 Long Noncoding Rna, Which Promotes Mucosal Regeneration. Gastroenterology (2018) 155(1):144–55. doi: 10.1053/j.gastro.2018.03.058
20. Mason RJ. Pathogenesis of COVID-19 From a Cell Biology Perspective. Eur Respir J (2020) 55(4):2000607. doi: 10.1183/13993003.00607-2020
21. Valyaeva AA, Zharikova AA, Kasianov AS, Vassetzky YS, Sheval EV. Expression of SARS-CoV-2 Entry Factors in Lung Epithelial Stem Cells and its Potential Implications for COVID-19. Sci Rep (2020) 10(1):17772. doi: 10.1038/s41598-020-74598-5
22. Walsh I, Seno F, Tosatto SC, Trovato A. Pasta 2.0: An Improved Server for Protein Aggregation Prediction. Nucleic Acids Res (2014) 42(Web Server issue):W301–7. doi: 10.1093/nar/gku399
23. Moshe A, Gorovits R. Virus-Induced Aggregates in Infected Cells. Viruses (2012) 4(10):2218–32. doi: 10.3390/v4102218
24. Ogen-Shtern N, Ben David T, Lederkremer GZ. Protein Aggregation and ER Stress. Brain Res (2016) 1648(Pt B):658–66. doi: 10.1016/j.brainres.2016.03.044
25. Bagga S, Bouchard MJ. Cell Cycle Regulation During Viral Infection. Methods Mol Biol (2014) 1170:165–227. doi: 10.1007/978-1-4939-0888-2_10
26. Dove B, Brooks G, Bicknell K, Wurm T, Hiscox JA. Cell Cycle Perturbations Induced by Infection With the Coronavirus Infectious Bronchitis Virus and Their Effect on Virus Replication. J Virol (2006) 80(8):4147–56. doi: 10.1128/JVI.80.8.4147-4156.2006
27. Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons At Age 50: Past, Current and Future Impact on Biomedicine. Nat Rev Drug Discovery (2007) 6(12):975–90. doi: 10.1038/nrd2422
28. Samuel CE. Antiviral Actions of Interferons. Clin Microbiol Rev (2001) 14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001
29. Stetson DB, Medzhitov R. Type I Interferons in Host Defense. Immunity (2006) 25(3):373–81. doi: 10.1016/j.immuni.2006.08.007
30. Dias Junior AG, Sampaio NG, Rehwinkel J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol (2019) 27(1):75–85. doi: 10.1016/j.tim.2018.08.007
31. Streicher F, Jouvenet N. Stimulation of Innate Immunity by Host and Viral Rnas. Trends Immunol (2019) 40(12):1134–48. doi: 10.1016/j.it.2019.10.009
32. Bruns AM, Leser GP, Lamb RA, Horvath CM. The Innate Immune Sensor LGP2 Activates Antiviral Signaling by Regulating MDA5-RNA Interaction and Filament Assembly. Mol Cell (2014) 55(5):771–81. doi: 10.1016/j.molcel.2014.07.003
33. Moresco EM, Beutler B. LGP2: Positive About Viral Sensing. Proc Natl Acad Sci U S A (2010) 107(4):1261–2. doi: 10.1073/pnas.0914011107
34. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a Positive Regulator of RIG-I- and MDA5-mediated Antiviral Responses. Proc Natl Acad Sci USA (2010) 107(4):1512–7. doi: 10.1073/pnas.0912986107
35. Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H Box Helicase, is a Novel Antiviral Factor Promoting RIG-I-Like Receptor-Mediated Signaling. Mol Cell Biol (2011) 31(18):3802–19. doi: 10.1128/MCB.01368-10
36. Oshiumi H, Miyashita M, Okamoto M, Morioka Y, Okabe M, Matsumoto M, et al. Ddx60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced Egfr Activation. Cell Rep (2015) 11(8):1193–207. doi: 10.1016/j.celrep.2015.04.047
37. Kuriakose T, Kanneganti TD. Zbp1: Innate Sensor Regulating Cell Death and Inflammation. Trends Immunol (2018) 39(2):123–34. doi: 10.1016/j.it.2017.11.002
38. Ibsen MS, Gad HH, Thavachelvam K, Boesen T, Despres P, Hartmann R. The 2’-5’-Oligoadenylate Synthetase 3 Enzyme Potently Synthesizes the 2’-5’-Oligoadenylates Required for Rnase L Activation. J Virol (2014) 88(24):14222–31. doi: 10.1128/JVI.01763-14
39. Li Y, Banerjee S, Wang Y, Goldstein SA, Dong B, Gaughan C, et al. Activation of Rnase L is Dependent on OAS3 Expression During Infection With Diverse Human Viruses. Proc Natl Acad Sci USA (2016) 113(8):2241–6. doi: 10.1073/pnas.1519657113
40. Verhelst J, Hulpiau P, Saelens X. Mx Proteins: Antiviral Gatekeepers That Restrain the Uninvited. Microbiol Mol Biol Rev (2013) 77(4):551–66. doi: 10.1128/MMBR.00024-13
41. Busnadiego I, Kane M, Rihn SJ, Preugschas HF, Hughes J, Blanco-Melo D, et al. Host and Viral Determinants of Mx2 Antiretroviral Activity. J Virol (2014) 88(14):7738–52. doi: 10.1128/JVI.00214-14
42. Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. MX2 is an Interferon-Induced Inhibitor of HIV-1 Infection. Nature (2013) 502(7472):563–6. doi: 10.1038/nature12653
43. Bailey CC, Zhong G, Huang IC, Farzan M. Ifitm-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu Rev Virol (2014) 1:261–83. doi: 10.1146/annurev-virology-031413-085537
44. Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, et al. Interferon Induction of IFITM Proteins Promotes Infection by Human Coronavirus OC43. Proc Natl Acad Sci USA (2014) 111(18):6756–61. doi: 10.1073/pnas.1320856111
45. Mohammad S, Bouchama A, Mohammad Alharbi B, Rashid M, Saleem Khatlani T, Gaber NS, et al. SARS-Cov-2 ORF8 and SARS-CoV Orf8ab: Genomic Divergence and Functional Convergence. Pathogens (2020) 9(9):677. doi: 10.3390/pathogens9090677
46. Ng LF, Liu DX. Further Characterisation of the Coronavirus IBV ORF 1a Products Encoded by the 3C-Like Proteinase Domain and the Flanking Regions. Adv Exp Med Biol (1998) 440:161–71. doi: 10.1007/978-1-4615-5331-1_21
47. Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, et al. The ORF6, ORF8 and Nucleocapsid Proteins of SARS-CoV-2 Inhibit Type I Interferon Signaling Pathway. Virus Res (2020) 286:198074. doi: 10.1016/j.virusres.2020.198074
48. Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PloS One (2009) 4(12):e8342. doi: 10.1371/journal.pone.0008342
49. Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. Accessory Proteins 8b and 8ab of Severe Acute Respiratory Syndrome Coronavirus Suppress the Interferon Signaling Pathway by Mediating Ubiquitin-Dependent Rapid Degradation of Interferon Regulatory Factor 3. Virology (2018) 515:165–75. doi: 10.1016/j.virol.2017.12.028
50. Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, et al. Sars-CoV-2 nsp13, nsp14, nsp15 and Orf6 Function as Potent Interferon Antagonists. Emerg Microbes Infect (2020) 9(1):1418–28. doi: 10.1080/22221751.2020.1780953
51. Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, et al. SARS-Cov-2 Orf6 Hijacks Nup98 to Block STAT Nuclear Import and Antagonize Interferon Signaling. Proc Natl Acad Sci USA (2020) 117(45):28344–54. doi: 10.1073/pnas.2016650117
52. Rodriguez KR, Bruns AM, Horvath CM. MDA5 and LGP2: Accomplices and Antagonists of Antiviral Signal Transduction. J Virol (2014) 88(15):8194–200. doi: 10.1128/JVI.00640-14
53. Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. Dynamin-Like MxA Gtpase: Structural Insights Into Oligomerization and Implications for Antiviral Activity. J Biol Chem (2010) 285(37):28419–24. doi: 10.1074/jbc.R110.145839
54. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5
Keywords: SARS-CoV-2 accessory protein, ORF8, lung epithelial cells, interferon signaling, inflammation
Citation: Geng H, Subramanian S, Wu L, Bu H-F, Wang X, Du C, De Plaen IG and Tan X-D (2021) SARS-CoV-2 ORF8 Forms Intracellular Aggregates and Inhibits IFNγ-Induced Antiviral Gene Expression in Human Lung Epithelial Cells. Front. Immunol. 12:679482. doi: 10.3389/fimmu.2021.679482
Received: 11 March 2021; Accepted: 17 May 2021;
Published: 09 June 2021.
Edited by:
Haichao Wang, Feinstein Institute for Medical Research, United StatesReviewed by:
Xuping Xie, University of Texas Medical Branch at Galveston, United StatesCopyright © 2021 Geng, Subramanian, Wu, Bu, Wang, Du, De Plaen and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Di Tan, eHRhbkBub3J0aHdlc3Rlcm4uZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.