
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 May 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.678699
This article is part of the Research Topic Metal Ions in Innate Immune Antimicrobial Responses: How the Bugs are Fighting Back View all 5 articles
Previously, the dual RNA-seq was carried out in a Pseudomonas plecoglossicida- Epinephelus coioides infection model to investigate the dynamics of pathogen-host interplay in vivo. ZnuC, a member of ZnuCBA Zn importer, was found transcriptionally up-regulated during infection. Thus, this study aimed to assess its role during the trade-off for Zn between host and P. plecoglossicida. ICP-MS analysis and fluorescent staining showed that Zn was withheld from serum and accumulated in the spleen, with increased Zn uptake in the Golgi apparatus of macrophages after infection. Additionally, growth assay, macrophage infection and animal infection after gene knockout / silencing revealed that znuC was necessary for growth in Zn-limiting conditions, colonization, intracellular viability, immune escape and virulence of P. plecoglossicida. Further analysis with dual RNA-seq revealed associations of host’s Zn nutritional immunity genes with bacterial Zn assimilation genes. IL6 and ZIP4 played key roles in this network, and markedly affected znuB expression, intracellular viability and immune escape, as revealed by gene silencing. Moreover, EMSA and GFP reporter gene analysis showed that Fur sensed changes in Fe concentration to regulate znuCBA in P. plecoglossicida. Jointly, these findings suggest a trade-off for Zn between host and P. plecoglossicida, while ZnuC is important for P. plecoglossicida Zn acquisition.
Manganese (Mn), zinc (Zn) and iron (Fe), as well as other transition metals, are vital to living beings, because they play important roles in protein structure and function (1). Pathogenic microorganisms must obtain these micronutrients from the host, while the latter attempts to intercept them through a process called nutritional immunity (2–5).
Fe is a common co-factor, which plays an important role in various physiological processes. Therefore, almost all bacterial pathogens need Fe, whose availability is limited by vertebrates to take advantage of this demand as an effective defense against infection (4, 6). Although the systems by which bacteria obtain Fe and the mechanism of vertebrate host interception of Fe from invading bacteria have been studied longer and more extensively (7–10), a large body of literature exists for other metals.
Zn also plays vital roles in bacteria (5). As the second most abundant transition metal in the majority of organisms, it has both catalytic and structural functions in proteins (11). Indeed, Zn-binding proteins account for about 4 to 8% of all proteins produced by prokaryote organisms (12). In some bacterial pathogens, the lack of a high affinity Zn transport system leads to reduced virulence (4). In the light of these important roles of Zn in bacterial physiology, it is not surprising that Zn sequestration represents an essential innate defense strategy. A typical example of Zn restriction is the staphylococcal abscess, which lacks Zn (13, 14).
Recent studies have elucidated some mechanisms of Zn chelation in the host. The S100 protein family comprises multiple calcium-binding proteins found in vertebrates, some of which are related to resistance to infection. S100A7 (psoriasin), secreted by keratinocytes, exerts antimicrobial effects via Zn chelation (15). S100A12 (calgranulin C) can bind Zn as well (16, 17). Calprotectin, a heterodimer comprising S100A8 and S100A9, accounts for about 40% of neutrophil cytoplasmic proteins. It has strong antimicrobial activity against various bacterial and fungal pathogens (13, 18, 19). Calprotectin achieves its antibacterial activity by chelating Mn and Zn (13). It is involved in host defense against Salmonella typhimurium, Staphylococcus aureus and fungal pathogens such as Candida spp. and Aspergillus spp (18–21). Nevertheless, some pathogenic organisms have evolved ingenious mechanisms for combating or twisting these inhibitory features.
To counteract nutritional immunity, pathogens have evolved ingenious strategies for acquiring Zn from the host (22, 23). Although not completely defined, the transport of Zn through the outer membrane in Gram-negative bacteria may be driven by the TonB-ExbBExbD system. ZnuD, a Zn-modulated TonB-dependent receptor, has been characterized in Neisseria meningitidis and numerous other pathogens (24). ZnuD is similar to HumA, a Morexalla catarrhalis heme transporter. ZnuD expression in Escherichia coli promotes the acquisition of heme (25). The dual regulation of ZnuD by Zur and Fur (Zn and Fe uptake regulators, respectively) further indicates ZnuD may be involved in the acquisition of Zn and heme (25). In addition, the cross regulation of ZnuD may be due to the increased demand for exogenous heme under Zn limitation, because some endogenous heme biosynthesis enzymes need Zn (26). In Gram-positive and Gram-negative bacteria, Zn import through the plasma membrane is mainly promoted by the ABC-family of transporters, such as ZnuABC (5, 27–30). The importance of Zn transporters to overcoming nutritional immunity and calprotectin has been demonstrated for several pathogens. For example, Pseudomonas aeruginosa relys on the znuABC Zn transporter to overcoming calprotectin-mediated growth inhibition (31). Recent studies have shown that the ZnuABC Zn uptake system of S. typhimurium is necessary for resistance to Zn chelation mediated by calprotectin accumulation in the intestine after infection (20). In addition, S. typhimurium uses calprotectin-mediated Zn-chelation to compete with the host microbiota, which is not well adapted to nutritional deficiency (20).
The mechanism of Zn nutritional immunity has not been fully elucidated. In addition, the majority of existing research evaluating nutritional immunity and the competition for micronutrients between pathogen and host have been carried out in mammalian and human pathogenic bacteria. For example, many P. aeruginosa resistance genes were induced during human infection, including those involved in zinc transport (32). At present, studies assessing fish nutritional immunity and its function in pathogen-host interactions are scarce. Through some strategies, it is possible to protect fish from disease by providing balanced food that increase immunity (33). Thus, it is necessary to understand the interaction between the bacterial pathogens and fish host, as well as the most important reasons that can increase or suppress nutritional metal homeostasis.
Pseudomonas plecoglossicida is a marine pathogenic bacterium, which can infect Plecoglossus altivelis (Ayu) (34), Pseudosciaena crocea (large yellow croaker) (35), Epinephelus coioides (orange spotted grouper) (36), and Oncorhynchus mykiss (rainbow trout) (37). Outbreaks of P. plecoglossicida infections in P. crocea and E. coioides are characterized by internal organ granulomas, and cause severe economic losses. Furthermore, as there is no effective prevention and control measures for this fish infectious disease, there is a risk of antibiotic abuse. “No antibiotics” is the future trend of aquaculture, so it is urgent to find a safe, efficient and healthy method to control P. plecoglossicida infections. Our previous dual RNA-seq analysis showed that znuC is significantly up-regulated during infection by Pseudomonas plecoglossicida, which indicated a critical role for znuC during infection (38). In several bacterial pathogens, as a member of the high-affinity Zn transporter ZnuCBA, znuC is necessary for virulence and overcoming Zn nutritional immunity. However, its contribution to infection, Zn import and resistance to nutritional immunity have not been experimentally demonstrated. Thus, this study aimed to assess its role during the trade-off established between the host and P. plecoglossicida in terms of Zn utilization. The research on the molecular mechanism will contribute to the formulation of the disease control strategy on the basis of nutritional immunity and the development of highly effective live attenuated vaccine.
P. plecoglossicida (NZBD9) was obtained from Pseudosciaene crocea spleen after natural infection, and confirmed for pathogenicity by artificial infection (35). Storage was performed in saline containing 10% glycerol at −80°C. NZBD9 was cultured in LB (Luria Bertani) medium (18 or 28°C;, 220 rpm). E. coli DH5α (TransGen Biotech, China) was also cultured in LB medium (37°C; 220 rpm).
Gene knockout was performed according to previous research (39). The pKD46 plasmid was introduced into P. plecoglossicida, and induced by 30 mM L-arabinose (40). The targeted fragments with the termini showing homology to the 50-bp upstream and downstream the flanking regions of znuC and fur, respectively, were constructed by PCR amplification and introduced into P. plecoglossicida via electroporation. Tetracycline (10 μg / mL) was utilized for screening, and clones were verified by PCR. All primers utilized are described in Table S1.
To construct the complements for ΔznuC and Δfur, the znuC and fur genes were amplified by PCR and then cloned into the linearized pBAD33 vector using T4 DNA ligase (New England Biolabs) based on the manufacturer’s recommendations. The vectors were transferred into ΔznuC and Δfur, and Chloramphenicol was used to screen the positive clones.
RNAi was performed based on a previous report (41). Five short hairpin RNAs against znuC and fur were provided by Shanghai Generay Biotech (China), respectively. NsiI and BsrGI (New England Biolabs, USA) were used to linearize the pCM130/tac vector. Then, T4 DNA ligase (New England Biolabs) was used to anneal the oligonucleotides and ligate them to linearized pCM130/tac (42). Through heat shock, recombinant pCM130/tac plasmids were firstly transferred into DH5α. Then, the recombinant pCM130/tac plasmids were purified and introduced into P. plecoglossicida. Positive clone screening was performed with tetracycline (10 μg/mL). Finally, quantitative real time PCR (qRT-PCR) was undertaken for validating znuC and fur expression in RNAi strains.
Healthy E. coioides (47.0 ± 2.0 g) were randomized to multiple groups (n=40/group, triplicate assays were performed). Intraperitoneal injection of P. plecoglossicida was administered to each fish at 103 CFU/g (43). The negative control group received the injection of sterile PBS. Daily observation was carried out.
For dual RNA-Seq, spleen specimens from six weight-matched E. coioides after P. plecoglossicida infection were collected at 48 h post-infection (hpi); two consecutive samples were pooled (44). For bacteria distribution assessment, spleens from 3 E. coioides were obtained at 1, 6, 12, 24, 48, 72 and 96 hpi, respectively (45, 46). Spleens and blood specimens from 3 E. coioides were collected at 48 hpi, for Zn concentration measurements by inductively coupled plasma mass spectrometry (ICP-MS).
Experiments involving animals were performed as recommended by the “Guide for the Care and Use of Laboratory Animals” put forth by the National Institutes of Health. The involved protocols had approval from the Animal Ethics Committee of Jimei University (JMULAC201159).
E. coioides macrophages were isolated as previously described (47). E. coioides were anaesthetised with 4-ethyl-amino-benzocaine, and the head-kidneys were sampled, pushed through a 100 mesh nylon screen and then suspended in L-15 medium (Biological Industries, Israel) supplemented with 100 IU streptomycin/penicillin (S/P)/mL, 10 IU/mL heparin and 2% foetal calf serum (FCS). The cell suspension was layered onto a 34%/51% discontinuous Percoll (Amersham Pharmacia Biotech, UK) density gradient with a syringe and centrifuged at 400×g for 30 min at 4 °C. The band of cells in the layer above the 34%/51% interface was collected, washed twice and re-suspended with L-15 medium with 100 IU S/P/mL, 10 IU heparin/mL and 10% FCS. The cells were then incubated at 28 °C. After 4 h, non-adherent cells were removed by washing with L-15 and monolayers were collected. Then, the cell suspension was adjusted to ~2×106 cells/mL in L-15 medium with 100 IU S/P/mL, 10 IU heparin/mL and 10% FCS and transferred to 6-well plates at 1 mL/well.
Macrophage nucleofection with siRNA was carried out based on a previous report (48). Small interfering RNAs (siRNAs) were manufactured based on target gene sequences by GenePharma (China) (Table S1). Macrophages (107/mL) were cultured in L-15 medium (PAN-Biotech GmbH, Germany) with 10% fetal calf serum. 2 µl of each siRNA and 20 µl per 200,000 cells suspended in Nucleofector™ solution SF were then added to each well of a sterile 96-well plate to be transfected. Then the nucleofection was carried out on an Amaxa nucleofector 96-well shuttle system according to the manufacturer’s recommendations. qRT-PCR and P. plecoglossicida infection were performed at 24 h post-nucleofection.
Macrophages from E. coioides head-kidney were isolated as the method described by Zhang et al. (47) and seeded in a 6-well culture plate which contained 2 mL of L-15 medium supplemented with 10% FCS and 100 IU/mL of S/P in each well. After incubation at 28°C for 4h, the cultures were washed twice and the macrophages (107 cells/mL) were incubated in L-15 medium containing 10% FCS with the wild-type and mutant/knock-down strains of P. plecoglossicida [multiplicity of infection (MOI)]=100 (100 bacteria per macrophages added). After incubation at 18°C for 1 h, the macrophages were washed twice with cold PBS, re-suspended in 2 mL PBS, treated with 250 mg/mL gentamycin for 20 min at 18°C to eliminate extracellular bacteria, and then washed twice with PBS. The supernatant was withdrawn and tested for sterility. The cells were re-suspended in fresh L-15 medium with 10 IU/mL heparin, 100 IU/mL S/P, and 10% FCS, and this time point was denoted as 0 h. Then, the 0 h samples were further incubated for 1 h and 3 h and the samples were denoted as 1 h and 3 h, respectively. After that, the cells from 0 h to 3 h samples were centrifuged for 5 min at 100×g at 18 °C, and 1 mL sterile distilled water was added for 30 min to lyse the cells. The CFU of the precipitate was determined by plating appropriate dilutions on TSA plates. For the 3 h sample, the precipitate of macrophages was removed by centrifugation and the CFU of bacteria in the supernatant was counted on TSA plates. The intracellular survival rate was defined as the number of CFU at 1 h divided by the number of CFU at 0 h. The escape rate was defined as the number of CFU of the supernatant at 3 h divided by the number of CFU at 0 h.
20 mg spleen samples were freeze-dried for 24 h at -80°C and homogenized individually with an agate mortar prior to analysis. Then, acid digestion with HNO3+H2O2 (5:1) by microwave-assisted digestion was carried out (49). 500 μL of serum samples were mixed with 500 μL of H2O2 and 1 mL of 14 M HNO3 in PET bottles and diluted to 30 g with MilliQ deionized water. Zinc content was determined by ICP-MS (Agilent 7700x) and normalized to mass and volume, respectively.
E. coioides macrophages were infected with P. plecoglossicida for 24 h followed by incubation with Golgi and Zn dyes for 30 min (50). Zinquin ethyl ester (MKBio, China) was used for the visualization of labile Zn. BODIPY TR Ceramide (MKBio, China) was used for the staining of Golgi. Images were generated with a LEICA SP8 confocal microscopy.
The Wizard genomic DNA purification kit (Promega, USA) was utilized for genomic DNA extraction (51).
Total RNA extraction used TRIzol reagent (Invitrogen, USA) as we described before (52, 53). The mixed genomic DNA in total RNA was digested with the Turbo DNA-free DNase (Ambion, Austin, TX, USA). The RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The Ribo-Zero rRNA removal kit (Epicentre, USA) was utilized for rRNA removal.
Gene expression was detected on a QuantStudio 6 Flex real-time PCR system (Life Technologies, USA), with gyrB and β-actin used as reference genes for P. plecoglossicida and E. coioides, respectively (54–56). The 2−ΔΔCt method was applied for analysis. The number of gyrB DNA copies/milligram of tissue was utilized for dynamically estimating P. plecoglossicida distribution in host spleen (57). Table S1 lists all primers.
RNA-seq library preparation utilized the TruSeqTM RNA sample preparation kit (Illumina, USA). Then, rRNA-free RNA samples were incubated with fragmentation buffer, and the SuperScript double-stranded cDNA synthesis kit (Invitrogen) was employed for cDNA synthesis. Amplification utilized Phusion DNA polymerase (New England Biolabs), and Illumina HiSeq4000 sequencing was then performed by Majorbio Biotech (China) (58). The PE150 sequencing strategy was selected, with a 250-300 bp insert strand specific library.
Sickle (Version 1.33) and SeqPrep (Version 1.3.2-2) were used for the trimming and quality control of raw reads. With Bowtie2 (2.4.2), clean reads mapping to the genome of P. plecoglossicida strain NZBD9 [NCBI Sequence Read Archive (SRA); accession number SRP062985] was carried out (59). Mapped reads were considered to belong to P. plecoglossicida, and the remaining ones underwent de novo assembly to yield E. coioides unigenes (58). The summary of the number of mapped reads/input reads per replicate and the coverage have been listed in Table S2.
Clean reads not mapped to P. plecoglossicida genome from spleen specimens after infection with wild-type and RNAi strain, respectively, were considered a pool of reads. Then, de novo assembly into unigenes was carried out with Trinity (Version 2.12.0) using a kmer size of 25 (60). The bacterial NCBI non-redundant (NR) protein database was utilized for removing any possible prokaryote contaminants. Clean unigenes were next assessed in various databases such as NCBI NR protein, STRING, SWISS-PROT and Kyoto Encyclopedia of Genes and Genomes (KEGG) with BLASTX for identifying proteins showing highest sequence similarities with the obtained unigenes. Blast2GO was utilized for Gene Ontology (GO) analysis (61), and KEGG was employed for metabolic pathway analysis (62).
Based on NCBI (NZ_ASJX00000000.1) annotations and the abovementioned reference transcriptome annotation, analysis of E. coioides transcriptome was carried out. DEGs with | log2FC | ≥ 1 and a false discovery rate (FDR) below 0.05 were tested by the edgeR package (63).
With the NetGenerator algorithm (64, 65), a gene regulatory network among E. coioides Zn nutritional immunity related DEGs and P. plecoglossicida Zn acquisition related DEGs was predicted. Its robustness was assessed, and edges detected by >50% of all iterations were included in the final network.
Wild type, znuC-50%RNAi, znuC-95%RNAi, ΔznuC and znuC+ strains of P. plecoglossicida were cultured in LB medium supplemented with 2 μM Tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN) overnight (28°C; 220 rpm). The overnight culture was diluted to OD600=0.2, and incubated at 18°C; for 24 h while measuring OD600 at 0, 3, 6, 9, 12, 15, 18, 21, 24 h (66).
GFP reporter gene systems were constructed based on previously reported descriptions (67). Briefly, the pET16b-EGFP plasmid containing the Fur binding site was transformed into wild type strain and fur knockout/knockdown strains, respectively, via electroporation. The primers used here are listed in Table S1.
Recombinant Fur protein was expressed and purified according to previous descriptions (68). Table S1 lists all the primers utilized to amplify the full-length of fur, which was then cloned into the pET-32a (+) expression vector linearized with EcoR I and Xho I digestion. The plasmid was then transformed into E. coli BL21, and the recombinant Fur protein was induced with isopropyl-β-D-thiogalactopyranoside (IPTG) and purified with a nickel-nitrilotriacetic acid (Ni-NTA) column (TaKaRa).
EMSA was performed with mixtures of the Fur protein at different levels (0, 0.5, 1, 1.5, and 2 μM) and znuC promoter DNA fragments (2 μg) conjugated to 6-carboxyfluorescein at the 5’ terminus (GenePharma) according to previous descriptions (69). The reaction mixture (50 μL) contained 200 mM KCl, 50 mM Tris-HCl, 5% v/v glycerol and 0.1 mM EDTA. The mixture was incubated at 25 °C for 2 h and then loaded (15 μL) onto each lane of a 5% native polyacrylamide gel for electrophoresis (Chakraborty et al., 2010).
Based on the genomic DNA sequence of P. plecoglossicida, four pairs of primers were used to determine whether znuCBA genes were transcribed into a single operon as previously reported (69–71). Table S1 lists all the primers utilized.
Data are mean ± SD, and were compared by one-way ANOVA with post-hoc Dunnett’s test. SPSS 13.0 was used for data analysis. P<0.05 indicated statistical significance.
Read data can be retrieved from the SRA database (accession number: PRJNA613574).
To determine whether infection of P. plecoglossicida affects the Zn distribution in E. coioides, serum and spleen Zn concentrations were measured via ICP-MS in E. coioides infected with P. plecoglossicida at 48 h post-infection (hpi) and compared to the healthy E. coioides. As shown in Figure 1A, P. plecoglossicida infection produced hypozincemia in E. coioides. The serum Zn concentration in P. plecoglossicida infected E. coioides decreased ~4-fold compared to the healthy E. coioides, indicating that Zn was withheld from serum upon P. plecoglossicida infection. In the E. coioides spleen, a significant increase of Zn was observed after P. plecoglossicida infection (Figure 1B). The spleen Zn concentration in P. plecoglossicida infected E. coioides increased by ~1.57 μg/g compared to the healthy E. coioides, indicating that Zn was accumulated in spleen upon P. plecoglossicida infection.
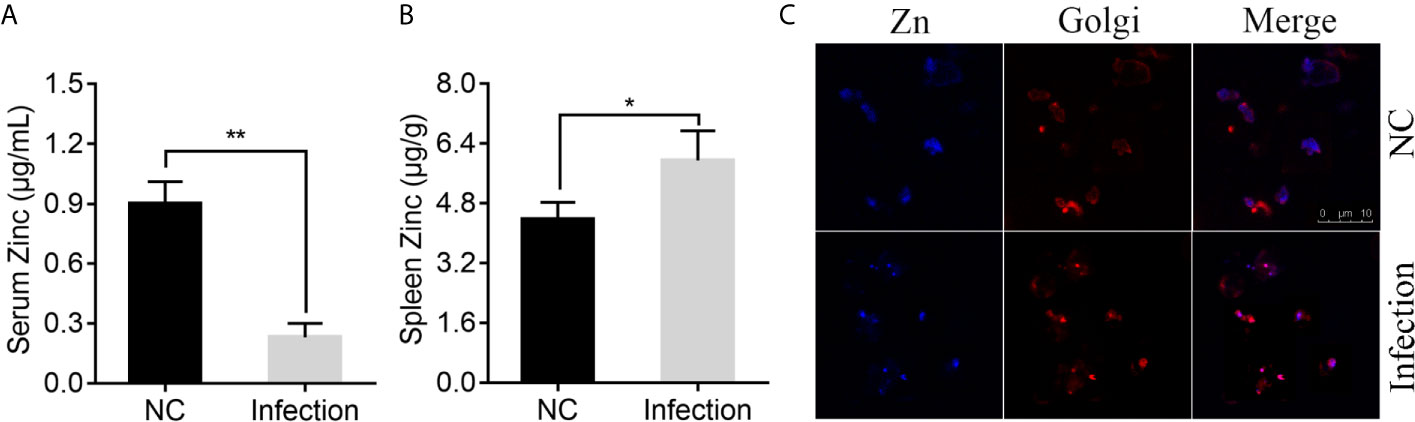
Figure 1 Effect of Pseudomonas plecoglossicida infection on Zinc (Zn) concentration in serum, spleen, and macrophages. (A) Serum Zn concentrations and (B) spleen Zn concentrations were measured via inductively coupled plasma mass spectrometry (ICP-MS) in Epinephelus coioides infected with P. plecoglossicida at 48 hpi and compared to the healthy E. coioides. Values are mean ± SD (n = 3, *P < 0.05, **P < 0.01). (C) Confocal microscopy was used to visualize labile Zn with the fluorescent probe, Zinquin ethyl ester. Staining for Zn (blue) and Golgi (red) in macrophages. Purple shows a co-localization of Golgi and Zn. Scale bars represent 10 μm, 3 independent experiments.
According to previous study, P. plecoglossicida are phagocytosed by macrophages in the spleen of the infected fish, while it is proved to be capable of intracellular survival and replication (72, 73). Macrophages possess numerous mechanisms to combat microbial invasion, including sequestration of essential nutrients, like Zn (50). Therefore, we speculated that Zn nutritional immunity in macrophages may affect the intracellular survival of P. plecoglossicida, and detected the distribution of Zn after P. plecoglossicida infecting macrophages. Confocal microscopy was used to visualize labile Zn with the fluorescent probe, Zinquin ethyl ester. Uninfected resting macrophages showed labile Zn diffuse distribution in the cytoplasm (Figure 1C); however, P. plecoglossicida infection caused focal Zn accumulation into the Golgi apparatus (Figure 1C), suggesting that P. plecoglossicida infection induced Zn redistribution in macrophages, which may limit its accessibility to P. plecoglossicida.
Collectively, these data indicated the existence of Zn nutritional immunity induced by P. plecoglossicida infection.
By dual RNA-seq performed previously (38) and qRT-PCR performed here, we assessed znuC expression at 1~4 days after infection. znuC was significantly induced from 2 to 4 days after infection, while the peak appeared on the third day, indicating the potential role of znuC during P. plecoglossicida infection (Figure 2A).
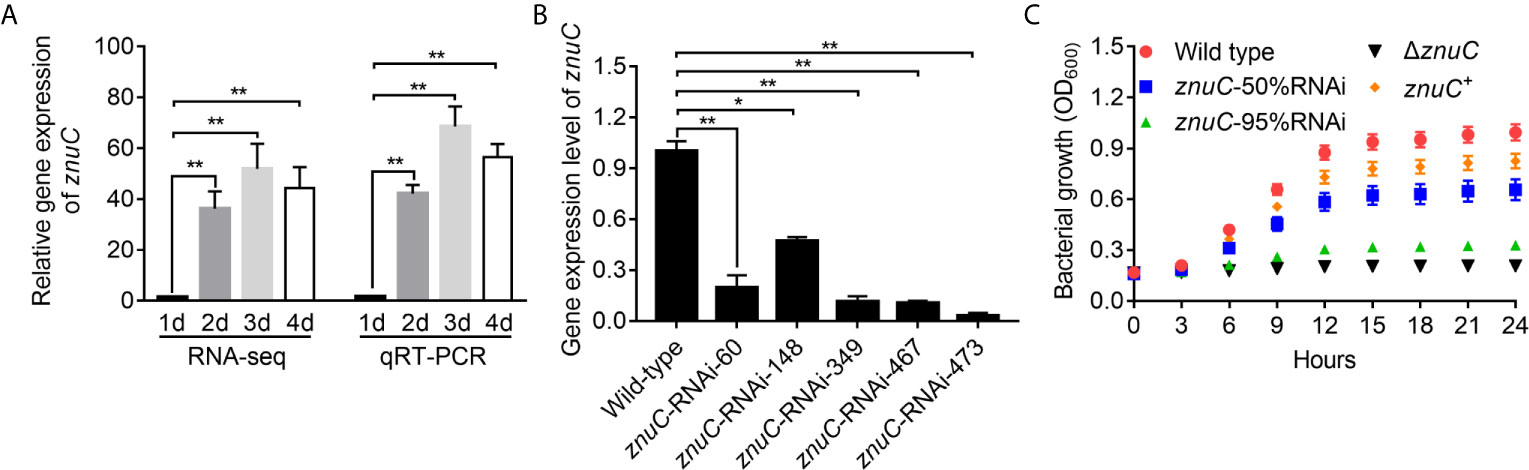
Figure 2 ZnuC is essential for Pseudomonas plecoglossicida growth under Zinc (Zn) limiting conditions. (A) Expression of znuC during infection was determined by dual RNA-seq and quantitative real time PCR (qRT-PCR). (B) To characterize znuC’s function, five znuC-RNAi strains were constructed, and the silencing efficiencies were validated by qRT-PCR. (C) To validate whether the znuC in P. plecoglossicida is important for growth during Zn limitation, growth of wild type, znuC-50%RNAi, znuC-95%RNAi, ΔznuC and znuC+ strains were compared in Zn-limiting condition with 2 μM TPEN. gyrB was used as a reference gene in the qRT-PCR analysis. Values are mean ± SD (n = 3, *P < 0.05, **P < 0.01).
To characterize znuC’s function, five znuC-RNAi strains were constructed, and the silencing efficiencies were validated by qRT-PCR. Among them, znuC-RNAi-148 and znuC-RNAi-473 strains, with about 50% and 95% znuC silencing efficiencies, respectively, were assessed in further studies (Figure 2B). Meanwhile, a znuC null mutant strain ΔznuC, as well as a complemented strain znuC+ were constructed (Figure S1) and also used to characterize the function of znuC.
To validate whether the znuC in P. plecoglossicida is important for growth during Zn limitation, growth of wild type, znuC-50%RNAi, znuC-95%RNAi, ΔznuC and znuC+ strains were compared in Zn-limiting condition with 2 μM Tetrakis-(2-pyridylmethyl)-ethylenediamine (TPEN). The ΔznuC mutant grew to a significantly lower terminal optical density than wild type (Figure 2C), while the growth defect could be complemented by ectopic expression of znuC (Figure 2C). After znuC knockdown, P. plecoglossicida showed significantly decreased growth under Zn limiting culture conditions in a znuC dependent manner (Figure 2C). These results supported a role for znuC of P. plecoglossicida in resisting Zn starvation.
To assess the importance of znuC to P. plecoglossicida pathogenesis, E. coioides were infected with P. plecoglossicida wild type and znuC-95%RNAi. Infection with znuC-95%RNAi resulted in significantly delayed death and lower mortality compared to infection with wild type (Figure 3A). The initial death time was delayed by 3 days, and the cumulative mortality rate was reduced by 80%. The E. coioides infected with P. plecoglossicida were dissected and observed at 96 hpi. It was found showed that the spleen specimens from the wild-type P. plecoglossicida group had characteristic symptoms (spleen blanketed by a large number of white spots), which were almost inexistent in E. coioides administered the znuC-95% RNAi strain (Figure 3B). These findings indicated that znuC was important for P. plecoglossicida virulence.
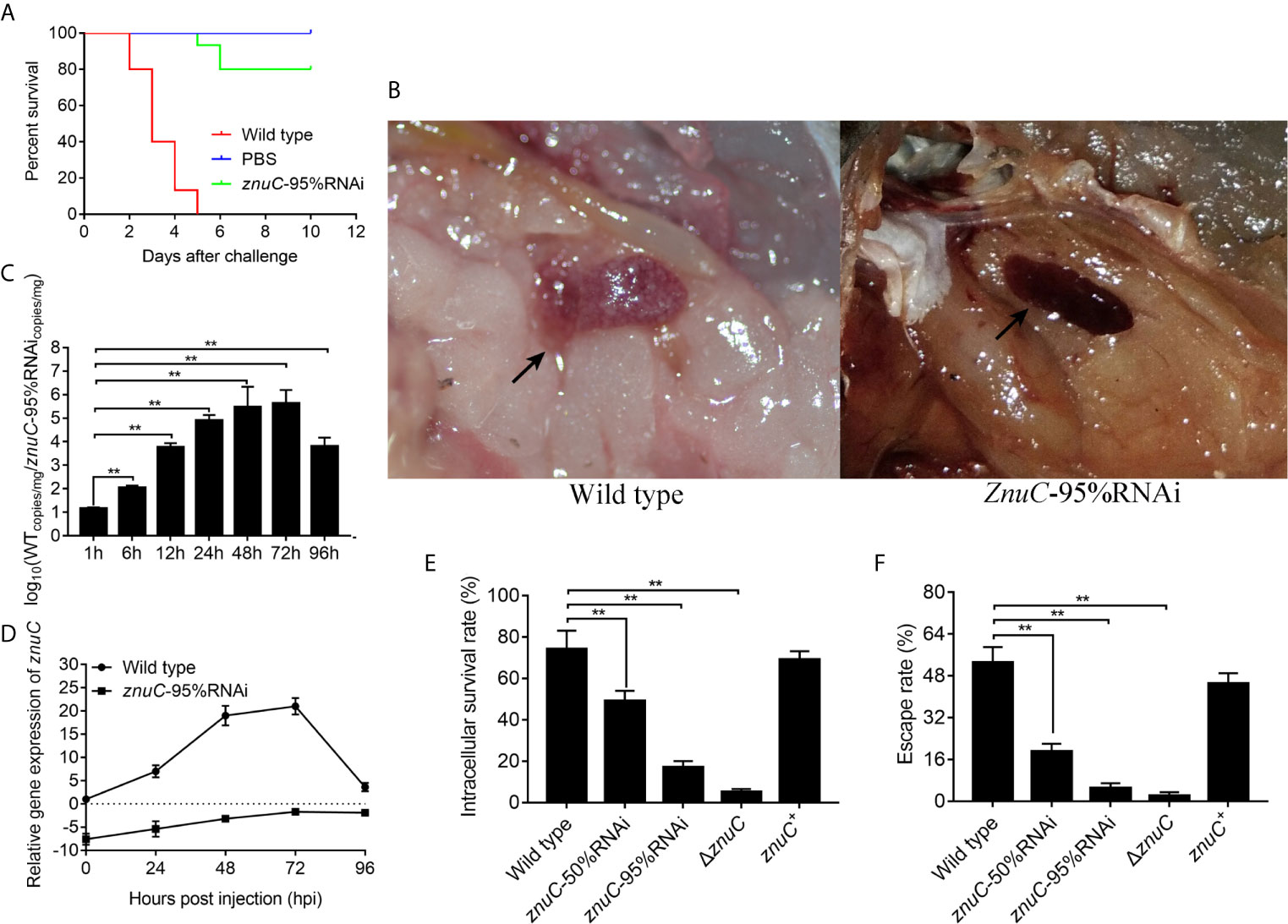
Figure 3 ZnuC is essential for Pseudomonas plecoglossicida virulence. (A) To assess the importance of znuC to P. plecoglossicida pathogenesis, Epinephelus coioides were infected with P. plecoglossicida wild type and znuC-95%RNAi. Amounts of E. coioides that survived after infection with the indicated strains were compared (n=3). (B) The E. coioides infected with P. plecoglossicida were dissected and observed at 96 hpi. Symptoms of E. coioides spleen after infection with P. plecoglossicida were compared. (C) The bacterial burdens of P. plecoglossicida wild type and znuC-95%RNAi in E. coioides spleen were measured by quantitative real time PCR (qRT-PCR) at 1, 6, 12, 24, 48, 72, and 96 hpi. Temporal dynamic distribution of P. plecoglossicida in E. coioides spleen were compared. (D) To determine whether the reduced bacterial burdens of the znuC-95%RNAi in E. coioides spleen was due to the stable low expression of znuC, we compared the expression of znuC in P. plecoglossicida wild type and znuC-95%RNAi during the infection at 0, 24, 48, 72 and 96 hpi. (E, F) Given our observation of increased Zinc (Zn) uptake in the Golgi apparatus of E. coioides macrophages after infection, we sought to validate whether the znuC in P. plecoglossicida is important for intracellular survival. The intracellular viability and immune escape of wild type, znuC-50%RNAi, znuC-95%RNAi, ΔznuC and znuC+ strains were compared in E. coioides macrophages. Values are mean ± SD (n = 3, **P < 0.01).
The bacterial burdens of P. plecoglossicida wild type and znuC-95%RNAi in E. coioides spleen were measured by qRT-PCR at 1, 6, 12, 24, 48, 72, and 96 hpi. Spleen amounts of the znuC-95% RNAi strain were markedly reduced than those of wild-type P. plecoglossicida at most times following infection (Figure 3C). To determine whether the reduced bacterial burdens of the znuC-95%RNAi in E. coioides spleen was due to the stable low expression of znuC, we compared the expression of znuC in P. plecoglossicida wild type and znuC-95%RNAi during the infection at 0, 24, 48, 72 and 96 hpi. Within 96 h after infection, znuC levels in the spleen were starkly reduced in the znuC-95% RNAi strain group in comparison with the wild-type group (Figure 3D). These findings indicated that znuC was important for P. plecoglossicida colonization.
Given our observation of increased Zn uptake in the Golgi apparatus of E. coioides macrophages after infection, we sought to validate whether the znuC in P. plecoglossicida is important for intracellular survival. The intracellular viability and immune escape of wild type, znuC-50%RNAi, znuC-95%RNAi, ΔznuC and znuC+ strains were compared in E. coioides macrophages. The ΔznuC mutant displayed a significantly lower intracellular viability and immune escape than wild type, while the defects could be complemented by ectopic expression of znuC (Figures 3E, F). After znuC knockdown, P. plecoglossicida showed significantly decreased intracellular viability and immune escape in a znuC dependent manner (Figures 3E, F). These results supported a role for znuC of P. plecoglossicida intracellular survival.
Taken together, these findings illuminated an important role for znuC in P. plecoglossicida pathogenesis.
In order to simultaneously detect the gene expression patterns of pathogen and host in vivo, so as to understand the trade-off established between the host and the pathogen in terms of Zn utilization, especially the role of znuC in this process, dual RNA-seq was carried out in E. coioides injected with PBS, wild-type or znuC-95% RNAi P. plecoglossicida.
Dual RNA-seq was carried out to assess spleen specimens from E. coioides upon infection with P. plecoglossicida for 48h. E. coioides transcriptomes after injection of PBS, and wild-type and znuC-95% RNAi P. plecoglossicida were significantly different. Totally 166517 genes were detected in E. coioides spleen. Compared to the wild-type P. plecoglossicida group, spleen specimens from animals infected with the znuC-95%RNAi strain had 23,059 differentially expressed genes (DEGs), including 12,005 downregulated and 11,054 upregulated genes (Figure 4A). These large amounts of differentially expressed genes revealed the important role of znuC in P. plecoglossicida pathogenicity. According to the latest version of the KEGG database, these 23,059 DEGs were clustered in 56 KEGG pathways, including those tightly associated with immunomodulation, e.g., phagosome, cytokine-cytokine receptor interaction, ECM-receptor interaction, antigen processing and presentation, complement and coagulation cascades, intestinal immune network for IgA production, and platelet activation (Figure S2). 108 DEGs, such as stx13, tfr, tap, sec61, and p40phox were enriched in the phagosome pathway, while phagocytosis is a central mechanism in the inflammation and defense against infectious agents. 91 DEGs, such as IL8, IL8RB, CCL4, tbfB2, and tgfBR2 were enriched in the cytokine-cytokine receptor interaction pathway, while cytokines are crucial intercellular regulators in innate and adaptive inflammatory host defenses, cell differentiation, and repair processes aimed at the restoration of homeostasis. 68 DEGs, such as reelin, thbs, tenascin, rhamiv, and syndecan were enriched in the ECM-receptor interaction pathway, while these interactions lead to a direct or indirect control of cellular activities such as adhesion, migration, differentiation, proliferation, and apoptosis. 48 DEGs, such as tnfA, hsp70, tapbp, aep, and slip were enriched in the antigen processing and presentation pathway, whose importance in resisting bacterial infection is self-evident. These findings indicated quite distinct physiological states of the host under infection of P. plecoglossicida wild type and znuC-95%RNAi. It is worth noting that many immune related genes are up-regulated instead of down regulated in the infection with znuC-95%RNAi strain compared to the infection with wild type strain. Thus, we believe that this is not only due to the differential bacterial burdens, which results in less stimulation to the immune system, but also due to the decreased zinc uptake of znuC-RNAi strain, which changes the host’s resistance strategies.
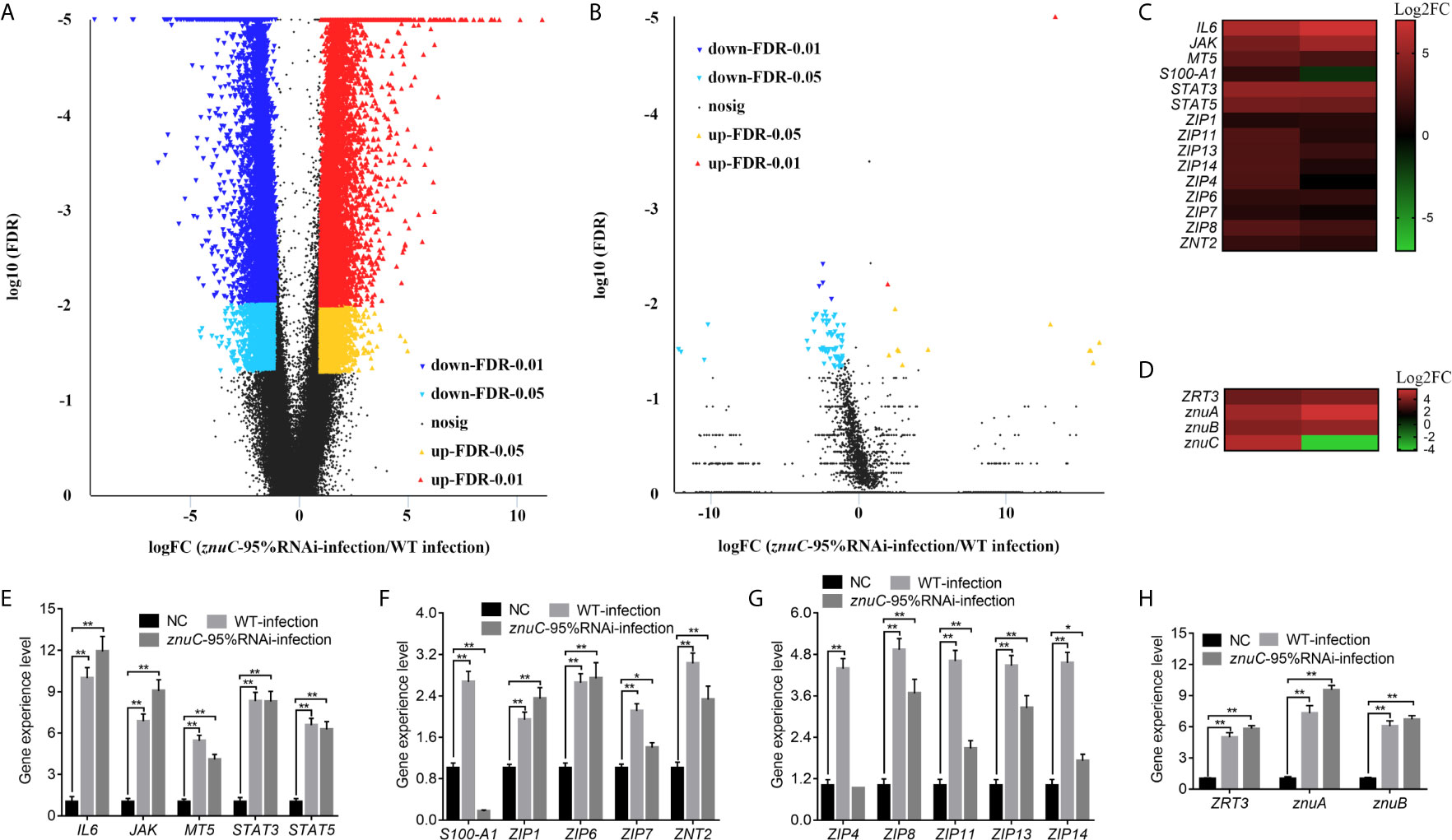
Figure 4 Dual RNA-seq reveals differentially expressed genes (DEGs) associated with the trade-off for Zinc (Zn) between host and pathogen during Pseudomonas plecoglossicida infection. (A, B) Dual RNA-seq was carried out to assess spleen specimens from Epinephelus coioides upon infection with P. plecoglossicida for 48h. Compared to the wild-type P. plecoglossicida group, spleen specimens from animals infected with the znuC-95%RNAi strain had 23,059 DEGs, including 12,005 downregulated and 11,054 upregulated genes. Totally 5,103 genes were detected in the transcriptome of P. plecoglossicida from E. coioides spleen. In comparison to wild-type P. plecoglossicida in E. coioides spleen, the znuC-95%RNAi strain had 88 DEGs, including 74 downregulated and 14 upregulated genes. Volcano plot indicated upregulated and downregulated genes of E. coioides (A) and P. plecoglossicida (B) detected by dual RNA-seq here. (C, D) Among all the DEGs, 15 Zn nutritional immunity related genes were identified in E. coioides, while 4 DEGs related to Zn acquisition were found in P. plecoglossicida. Heat maps indicated E. coioides Zn nutritional immunity related genes and P. plecoglossicida Zn acquisition genes (adjusted FDR < 0.05; | log2FC | ≥ 1; n=3) compared to healthy E. coioides. Upregulated and downregulated genes are colored in red and green, respectively. (E, F) quantitative real time PCR (qRT-PCR) analysis of E. coioides Zn nutritional immunity related genes and P. plecoglossicida Zn acquisition genes in E. coioides injected with PBS, wild type and znuC-RNAi P. plecoglossicida, respectively. Values are mean ± SD (n = 3, *P < 0.05, **P < 0.01).
Compared to wild-type P. plecoglossicida, a large number of genes in znuC-95% RNAi strain changed significantly in vivo. This may be due to the loss of znuC increases the extent of Zn limitation experienced by the bacterium, which suggested the important role of znuC during the pathogenesis. Totally 5,103 genes were detected in the transcriptome of P. plecoglossicida from E. coioides spleen. In comparison to wild-type P. plecoglossicida in E. coioides spleen, the znuC-95%RNAi strain had 88 DEGs, including 74 downregulated and 14 upregulated genes (Figure 4B). These 88 DEGs were clustered in 25 KEGG pathways, including those tightly associated with bacterial virulence regulation, e.g., flagellar assembly, bacterial chemotaxis, ABC transporters, bacterial secretion system, two component system and protein export (Figure S3). 8 DEGs, including flgF, flgI, motB, motA, fliE, FliF, FliG, and FliS were enriched in the flagellar assembly pathway, while the flagellar assembly plays important roles in host-microbial interactions, bacterial colonization and virulence. It is worth noting that all these 8 genes were significantly down-regulated in the znuC-95%RNAi strain compared to the wild type strain. 4 DEGs, including fliG, cheB, motB, and motA were enriched in the bacterial chemotaxis pathway, while bacterial chemotaxis plays a critical role for fitness and virulence during infections. 6 DEGs, including cysP, porH, livM, metQ and metN were enriched in the ABC transporters pathway, which are responsible for the transport of sulfate, putrescine, branched-chain amino acid, and D-methionine, respectively. It is worth noting that all these 6 genes were significantly down-regulated in the znuC-95%RNAi strain compared to the wild type strain, which indicated that there might be a cross-talk between these nutrition transport and the Zn transport. These findings suggested a critical role of znuC in the pathogenesis.
Among all the DEGs, 15 Zn nutritional immunity related genes were identified in E. coioides, including IL6, JAK, MT5, S100-A1, STAT3, STAT5, ZIP1, ZIP4, ZIP6, ZIP7, ZIP8, ZIP11, ZIP13, ZIP14, and ZNT2. Compared with healthy E. coioides, these genes were significantly upregulated in wild-type and znuC-95%RNAi strain infected E. coioides, except for S100-A1 that was downregulated in E. coioides upon infection with the znuC-95%RNAi strain (Figure 4C). Meanwhile, only 4 DEGs related to Zn acquisition were found in P. plecoglossicida, including ZRT3, znuA, znuB and znuC (Figure 4D). The results of transcriptome analysis were confirmed by qRT-PCR, further supporting dual RNA-seq’s reliability (Figures 4E–H). Since the purpose of this research was to better understand the contention between host and P. plecoglossicida for Zn, we selected the above 19 genes to further analyze host-pathogen interactions.
Using dual RNA-seq, researchers could predict essential and also indirect interactions between pathogen and host. These analyses and predictions can provide clues for our research. Thus, 15 Zn nutritional immunity related genes identified in E. coioides (including IL6, JAK, MT5, S100-A1, STAT3, STAT5, ZIP1, ZIP4, ZIP6, ZIP7, ZIP8, ZIP11, ZIP13, ZIP14, and ZNT2) and 4 DEGs related to Zn acquisition found in P. plecoglossicida (including ZRT3, znuA, znuB and znuC) were used to predict a pathogen-host gene regulatory network (Figure 5A). From the perspective of correlations between bacterial Zn acquisition- and host Zn nutritional immunity-related genes, first, infection induced most Zn nutritional immunity- and bacterial Zn acquisition-related genes. Secondly, there were interactions between bacteria and host genes. Zn acquisition related genes such as znuC and znuA could influence host Zn nutritional immunity, thereby activating IL6 expression or repressing ZIP4 and ZIP11 expression. Meanwhile, IL6 and ZIP4 could repress and induce the expression of znuB, respectively. Finally, there was a mutual regulatory relationship between host genes. For instance, IL6 inhibited ZIP1 and ZIP7; ZIP4 activated ZIP1 and ZIP7; ZIP6 induced ZIP11, ZNT2, and S100-A1. In this interaction network, IL6 and ZIP4 were in the center of the battlefield, suggesting that they had critical roles in pathogen-host interactions.
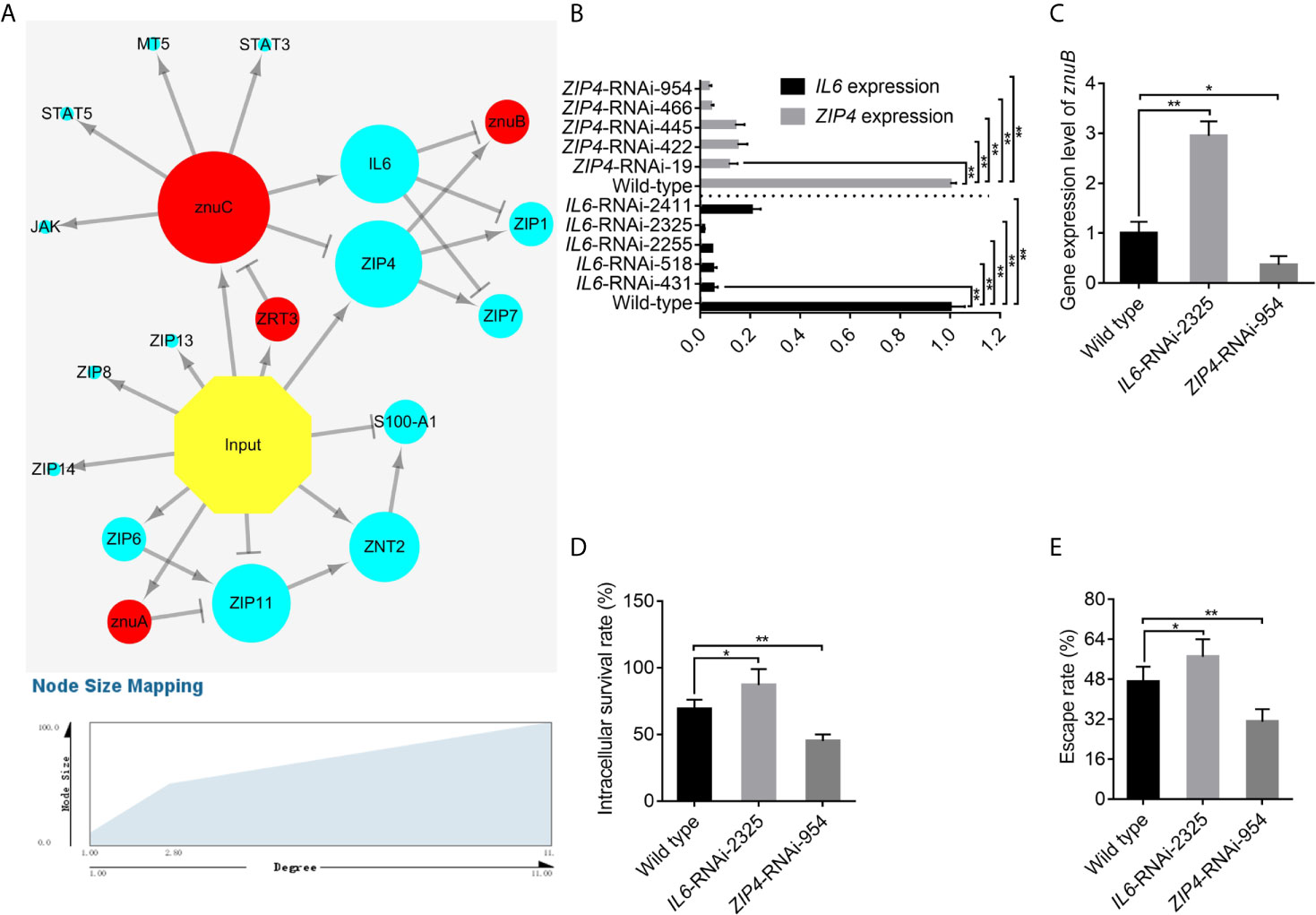
Figure 5 IL6 and ZIP4 play key roles in the trade-off for Zinc (Zn) between Epinephelus coioides and Pseudomonas plecoglossicida. (A) Predicted regulation network among E. coioides Zn nutritional immunity related genes and P. plecoglossicida Zn acquisition genes. Occurrence of infection, E. coioides genes and P. plecoglossicida genes are colored in yellow, blue and red, respectively. Positive and negative correlations are represented by lines. (B) To characterize the importance of IL6 and ZIP4, five IL6-RNAi and ZIP4-RNAi E. coioides macrophages were constructed, and the silencing efficiencies were validated by quantitative real time PCR (qRT-PCR). (C) According to the pathogen-host gene regulatory network, IL6 and ZIP4 were supposed to promote and repress znuB expression, respectively. To validate this, qRT-PCR was carried out on wild type, IL6-RNAi-2325 and ZIP4-RNAi-954 E. coioides macrophages. (D, E) In order to validate whether the IL6 and ZIP4 in E. coioides macrophages is important for intracellular killing of P. plecoglossicida, the intracellular viability and immune escape of wild type P. plecoglossicida was compared in wild type, IL6-RNAi-2325 and ZIP4-RNAi-954 E. coioides macrophages. Values are mean ± SD (n = 3, *P < 0.05, **P < 0.01).
According to previous study, P. plecoglossicida are phagocytosed by macrophages in the spleen of the infected fish, while it is proved to be capable of intracellular survival and replication (72, 73). Macrophages possess numerous mechanisms to combat microbial invasion, including sequestration of essential nutrients, like Zn (50). As we have shown in Figure 1C, increased Zn uptake in the Golgi apparatus of E. coioides macrophages after infection was observed, which indicated the existence of Zn nutritional immunity in macrophages induced by P. plecoglossicida infection. Therefore, we speculated that Zn nutritional immunity in macrophages may affect the intracellular survival of P. plecoglossicida, and further study was carried out on E. coioides macrophages to investigate the host-pathogen interaction during Zn nutritional immunity.
To characterize the importance of IL6 and ZIP4, five IL6-RNAi and ZIP4-RNAi E. coioides macrophages were constructed, and the silencing efficiencies were validated by qRT-PCR. IL6 and ZIP4 levels in E. coioides macrophages treated with RNAi were significantly reduced (Figure 5B). Among them, IL6-RNAi-2325 and ZIP4-RNAi-954 with the best gene silencing efficiencies were selected for further research.
According to the pathogen-host gene regulatory network, IL6 and ZIP4 were supposed to promote and repress znuB expression, respectively. To validate this, qRT-PCR was carried out on wild type, IL6-RNAi-2325 and ZIP4-RNAi-954 E. coioides macrophages. Our results showed that, the silencing of IL6 and ZIP4 significantly increased and decreased znuB expression, respectively, which confirmed the above pathogen-host gene regulatory prediction network (Figure 5C).
Furthermore, in order to validate whether the IL6 and ZIP4 in E. coioides macrophages is important for intracellular killing of P. plecoglossicida, the intracellular viability and immune escape of wild type P. plecoglossicida was compared in wild type, IL6-RNAi-2325 and ZIP4-RNAi-954 E. coioides macrophages. As depicted in Figures 5D, E, compared with WT macrophages, viability and immune escape of P. plecoglossicida in IL6 and ZIP4 silenced macrophages were markedly enhanced and decreased, respectively.
Taken together, these findings illuminated an important role for IL6 and ZIP4 in intracellular killing of P. plecoglossicida. Some physiological property of the phagocytic cell that increases metal availability is changing after the silence of IL6 and ZIP4.
The three genes, znuA, znuB and znuC in E. coli are transcribed into a single mRNA molecule (74). In order to validate this in P. plecoglossicida, we performed RT-PCR to assess RNA isolated from P. plecoglossicida using primers spanning adjacent genes, and znuC, znuB and znuA, flanked by WP_016394241.1 and katE, were transcribed into one operon as znuCBA (Figures 6A, B). This corroborated previously reported studies, and indicated that these three genes may be controlled by the promoter upstream of znuC.
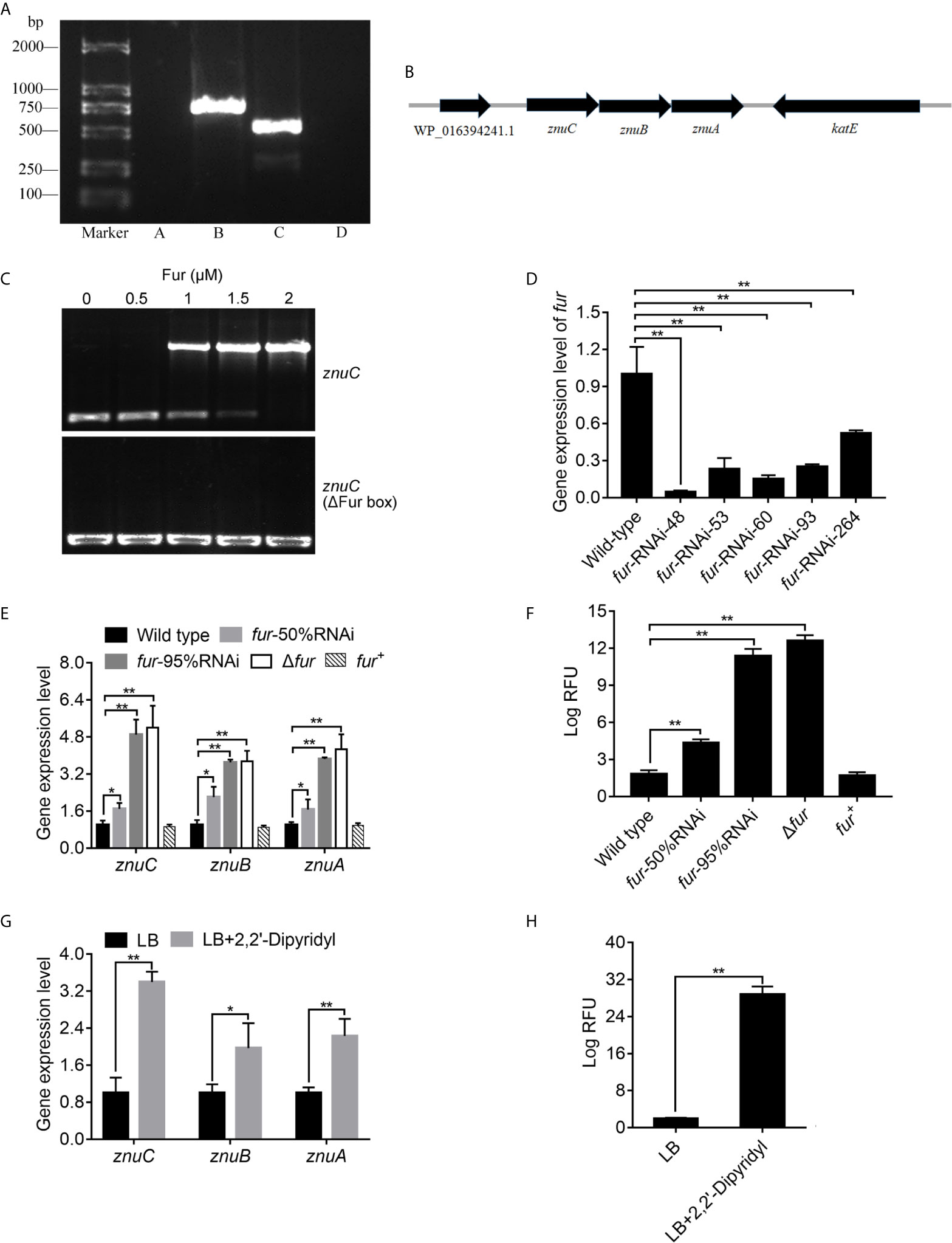
Figure 6 ZnuCBA is negatively regulated by Fur. (A) In order to validate whether znuA, znuB and znuC were transcribed into a single mRNA molecule, reverse transcription PCR (RT-PCR) was carried out to assess RNA isolated from Pseudomonas plecoglossicida using four pairs of primers. Pair A was designed to amplify the region between WP_016394241.1 and znuC, pair B between znuC and znuB, pair C between znuB and znuA, and pair D between znuA and katE. Primer pairs A and D produced no bands, while primer pairs B and C produced bands of correct sizes. (B) Genetic organization of the znuCBA operon. (C) To validate the binding of Fur on the znuC promoter, the electrophoretic mobility shift assay (EMSA) was carried out. 6-carboxyfluorescein-labeled DNA fragment from the promoter region of znuC (upper panel) or DNA fragment with the same boundaries but with Fur box removal (lower panel) was added to the reaction mixture containing different concentrations of the Fur protein. (D) In order to assess the function of Fur, five fur-RNAi strains were constructed, and the silencing efficiencies were validated by quantitative real time PCR (qRT-PCR). (E) To validate whether the Fur in P. plecoglossicida is important for the regulation of znuCBA, qRT-PCR was carried out in wild type, fur-50%RNAi, fur-95%RNAi, Δfur and fur+ strains. (F) For further validating znuCBA downregulation via Fur, a znuCBA-reporter gene fusion controlled by the putative Fur promoter (nt -900 to +150) was constructed. Expression was assessed by measuring fluorescence with fur knocked down or knocked out. (G) To validate the effect of Iron (Fe) limitation on znuCBA expression, expression of znuCBA in wild type were compared in LB and Fe-limiting condition with 2’,2-Dipyridyl. (H) For further validating znuCBA upregulation under Fe limitation, transcription levels of the znuC-GFP reporter gene fusion in P. plecoglossicida under Fe limiting conditions was detected. Values are mean ± SD (n = 3, *P < 0.05, **P < 0.01).
In order to investigate the regulation mechanism of znuCBA in P. plecoglossicida, Virtual Footprint was used to analysis the promoter of znuC. Virtual Footprint indicated a Fur putative binding site (5’-TGAATACT-3’) located within nt -545 to -538 upstream the start codon of znuC. To validate the binding of Fur on the znuC promoter, the EMSA assay was carried out. The EMSA assay (Figure 6C) revealed that Fur was able to strongly interact with a 364-bp DNA fragment (nt -361 to +3; part of the znuC promoter region), but not with another DNA fragment with the same boundaries after Fur binding site (nt -545 and -538) removal.
In order to assess the function of Fur, we constructed a fur null mutant Δfur, as well as a complemented strain fur+, and identified them by PCR (Figure S4). Meanwhile, five fur-RNAi strains were constructed, and the silencing efficiencies were validated by qRT-PCR (Figure 6D). Then, fur-RNAi strains with approximately 50% (fur-RNAi-264) and 95% (fur-RNAi-48) silencing efficiencies for fur were used for further research. To validate whether the Fur in P. plecoglossicida is important for the regulation of znuCBA, qRT-PCR was carried out in wild type, fur-50%RNAi, fur-95%RNAi, Δfur andfur+ strains. The Δfur mutant displayed a significantly higher znuCBA expression than wild type (Figure 6E), while the high expression of znuCBA could be complemented by ectopic expression of fur (Figure 6E). After fur knockdown, P. plecoglossicida showed significantly increased znuCBA expression in a fur-dependent manner (Figure 6E).
For further validating znuCBA downregulation via Fur, a znuCBA-reporter gene fusion controlled by the putative Fur promoter (nt -900 to +150) was constructed. A significant increase of znuCBA-GFP activity was observed in fur knockout and knockdown strains, and complemented by ectopic expression of fur (Figure 6F). Simultaneously, the znuCBA expression (Figure 6G) and znuCBA-GFP activity (Figure 6H) of P. plecoglossicida significantly increased when cultured under Fe limiting conditions.
Taken together, these results suggested that Fur was a transcriptional regulator targeting and regulating the znuCBA promoter.
Micronutrients including Fe, Zn and Mn are universally essential for life. In order to survive and proliferate, pathogenic bacteria must plunder micronutrients from infected host tissues. Taking advantage of pathogens’ requirement for micronutrients, mammals have developed a number of strategies to control bacterial infections via limiting microbial access to these micronutrients by evolving complex isolation mechanisms (75). However, research on fish nutritional immunity and its function in pathogen-host interactions remain scarce. What’s more, the understanding of fish Zn nutritional immunity is almost inexistent. The above results displayed Zn was withheld from serum and accumulated in the spleen after infection, which is probably a strategy that sequestrates Zn from the extracellular environment. Meanwhile, increased Zn uptake in the Golgi apparatus of macrophages after infection was also observed. Furthermore, 15 Zn nutritional immunity related genes were induced in E. coioides after P. plecoglossicida infection, including IL6, JAK, MT5, S100-A1, STAT3, STAT5, ZIP1, ZIP4, ZIP6, ZIP7, ZIP8, ZIP11, ZIP13, ZIP14, and ZNT2. ZIP1, ZIP4, ZIP6, ZIP7, ZIP8, ZIP11, ZIP13, ZIP14, and ZNT2 belong to solute carrier 39A (SLC39A) Zn transporters (ZIPs) and solute carrier 30A (SLC30A) Zn transporters (ZnT), which are responsible for the homeostasis of Zn and other metals in mammalian species (76). Liuzzi et al. (77) demonstrated that the expression of ZIP14 is positively regulated by IL-6, and this Zn transporter is likely to have a critical function in hypozincemia, which leads to acute phase reactions accompanied by infection and inflammation. The other genes are closely related to extracellular and intracellular Zn sequestration (4, 75), which is discussed in detail below. These results indicated the existence of E. coioides Zn nutritional immunity induced by P. plecoglossicida infection (Figure 7).
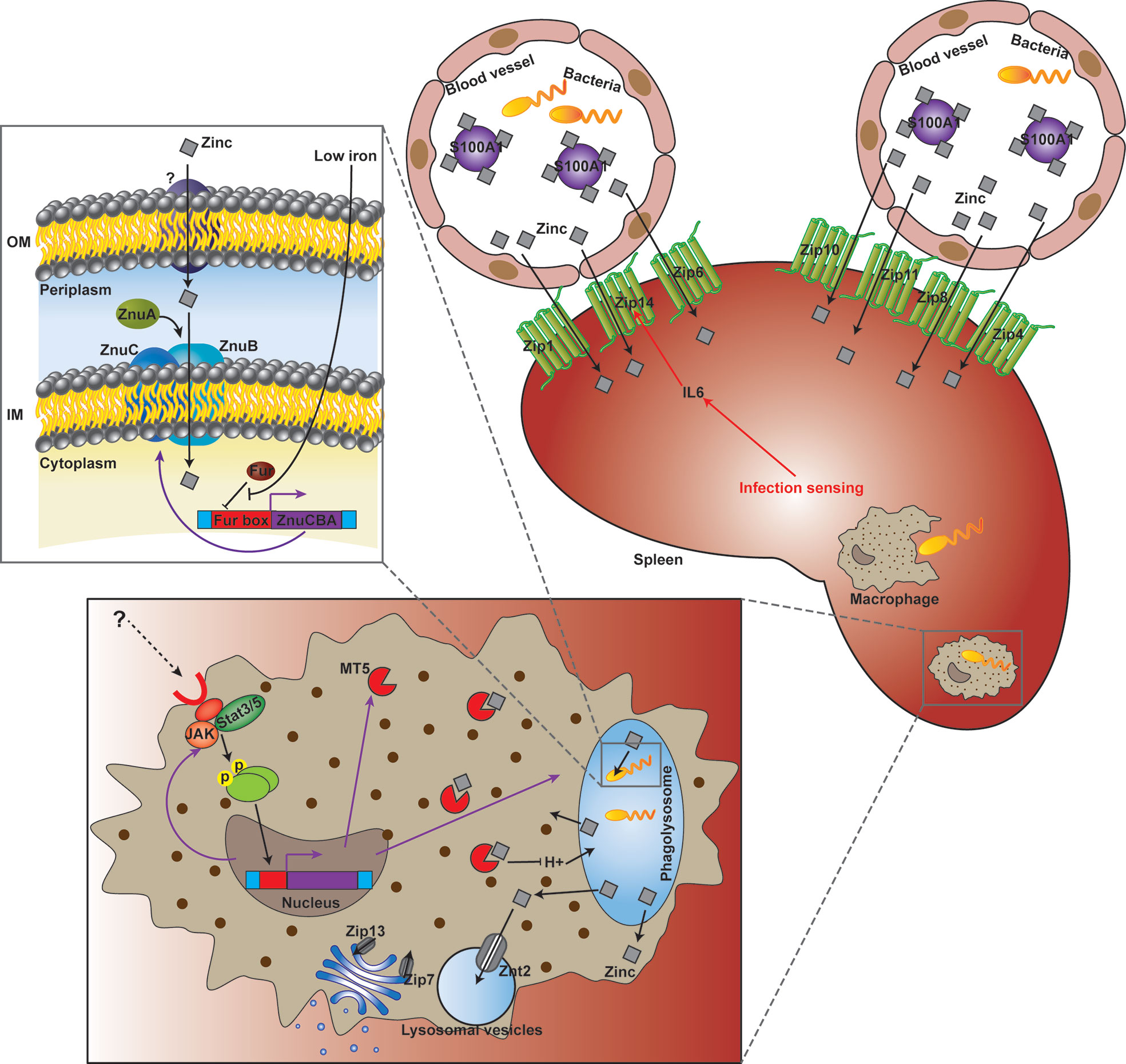
Figure 7 Working model of the struggle for Zinc (Zn) between Epinephelus coioides and Pseudomonas plecoglossicida.
As a transition metal, Zn in living systems ranks second in content and is subject to strict control. Although Zn nutritional immunity remains unclear, in recent years, studies have begun to clarify the mechanism of Zn sequestration in organisms, organs, tissues and cells in mammals during bacterial infection (4, 5). The main mechanism of extracellular Zn restriction is through the heterodimer calprotectin composed of S100A8 and S100A9. Calprotectin limits the utilization of Zn by microorganisms in the local environment through binding of Zn with femtomola affinity (13). S100A7 can control the growth of microorganisms on the body surface, including mucous membranes, by isolating Zn (15). The roles of other members of the S100A family in infection control remain undefined, although they have been reported to perform Zn binding functions. In the present study, S100-A1 was significantly induced in wild type strain-infected E. coioides, but downregulated in the znuC-95%RNAi strain group, which indicated that S100-A1 is not only involved in Zn sequestration, but could also sense and response to the status of Zn acquisition in bacteria infecting fish (Figure 7).
Intracellular Zn nutritional immunity has been also reported in mammalian macrophages (78, 79). Phagolysosomes of macrophages containing pathogens may form a Zn-deficient environment. There is evidence that phagocytosed C. albicans yeasts upregulate ZRT2, a Zn transporter (80). Zn homeostasis in pathogen-containing macrophages is a very dynamic and strongly modulated process. Winters and colleagues (81) were the first to describe the opposite effects of GM-CSF and IL-4 on Zn homeostasis in macrophages with H. capsulatum. Subramanian Vignesh and collaborators (50) demonstrated that after inactivated macrophages undergo infection with H. capsulatum, Zn content in cells is high, indicating that without macrophage stimulation, H. capsulatum may obtain this key micronutrient any time. After treatment with GM-CSF, Zn was removed from phagocytosed yeasts and transported to the Golgi apparatus. This Zn migration in macrophages was accompanied by a decrease in Zn utilization by H. capsulatum. Subramanian Vignesh and co-workers (50) speculated that the regionalization of Zn in the Golgi apparatus might be due to the actions of ZnT4 and ZnT7, which are transcriptionally upregulated in H. capsulatum infected macrophages. These cationic efflux proteins transport Zn from the cytoplasm to subcellular organelles (or pump the metal out of cells), whereas Znt4 and ZnT7 were previously shown to be associated with Zn transport in other cell types (82, 83). In the present study, intra-Golgi compartmentalization of Zn was also observed, while ZnT4 and ZnT7 were not found in E. coioides. Instead, Zip13, another iron efflux protein encoding gene previously implicated in Golgi zinc transport (84), was upregulated in P. plecoglossicida-containing macrophages. Therefore, in E. coioides, Zip3 might be a zinc gatekeeper in the Golgi apparatus, which contributed to Zn sequestration from the cytosol into the Golgi apparatus (Figure 7).
In mammalian macrophages, the availability of Zn might be further constricted by Zn-binding metallothioneins (Mts), because Mt1 and Mt2 amounts STAT3/STAT5-dependently are increased, accompanied by Zn restriction. These phenomena were also related to increased production of ROS in the phagolysosome, which created a “perfect storm” of antibacterial effects. In the present study, STAT3 and STAT5 were both significantly induced, which might contribute to Zn restriction and ROS production in E. coioides macrophages. However, Mt1 and Mt2 were not found in E. coioides. Instead, Mt5, another Zn-binding metallothionein (85), was found to be upregulated as STAT3 and STAT5 in P. plecoglossicida-containing macrophages. Therefore, in E. coioides, MT5 might further limit the availability of Zn in a STAT3/STAT5-dependent manner. In addition, the transcription levels of ZNT2, a Zn transporter sequestering Zn into intracellular vesicles (such as lysosomal vesicles) for protection from Zn cytotoxicity, were increased. Hence, ZNT2 might be also involved in Zn homeostasis during the pathogen-host interaction (Figure 7), which deserves further investigation.
Although the host has Zn nutritional immunity, the pathogen can still reproduce in the infected host, which indicates that the pathogen has evolved an effective Zn scavenging strategy under the limitation of Zn nutritional immunity. For example, in S. cerevisiae, the Zn responsive transcription factor Zap1 addresses a decrease in metal amounts by upregulating the Zn importers Zrt1 and Zrt2 and the vacuolar Zn exporter Zrt3 (86–88). The Zn stored in vacuoles can be quickly transported to the cytosol through Zrt3 (89). Here, ZRT3 was significantly increased in both wild type and znuC-RNAi P. plecoglossicida which infected E. coioides. This indicated that ZRT3 might contribute to counteracting Zn nutritional immunity independent of znuC. Another important “neutralizer” against Zn nutritional immunity is the Zn high-affinity uptake system ZnuABC (28–30), which is responsible for importing Zn through the cytoplasmic membrane of bacteria (5, 27). ZnuABC has demonstrated roles in the pathogeneses of some bacterial infections. For example, ZnuABC has a critical function in S. typhimurium resistance to calprotectin-mediated Zn chelation in the intestine (20). P. aeruginosa relies on the znuABC Zn transporter to overcoming calprotectin-mediated growth inhibition (31). Interestingly, the P. aeruginosa intracellular zinc content is not affected by the dysfunction of the ZnuABC transporter evidently, which suggested a redundant mechanisms for the acquisition of Zn in P. aeruginosa at Zn limiting condition. Moreover, a study showed that ZnuABC is very important for bacteria to infect fish (66). Our previous study identified the ZnuCBA transporter in P. plecoglossicida, with all three components, including ZnuC (ATPase), ZnuB (inner membrane permease) and ZnuA (periplasmic zinc binding protein) (38). According to the current results, works done in other systems and homology, ZnuCBA was necessary for virulence in P. plecoglossicida by promoting Zn uptake. Furthermore, these 3 genes in P. plecoglossicida were transcribed into a single mRNA molecule and negatively regulated by a Fur binding box located upstream of znuC (Figure 7). Fur represents an important regulator of Fe absorption, and is closely related to oxidative stress. It can also regulate the expression of virulence genes by sensing changes in Fe concentration. During infection, vertebrates use the serum protein transferrin to chelate Fe, thereby forming a Fe-deficient environment. Many pathogenic bacteria perceive this Fe depletion as a signal in their vertebrate host and trigger the expression of pathogenic genes, so that they can adapt to the Fe-deficient environment (90). For example, Fur helps Staphylococcus aureus perceive changes in Fe concentration, thereby regulating the expression of many virulence factors, such as cytolysins and immunomodulatory proteins, to resist the attack of neutrophils (91). Fur also modulates the hemolysin system, which is widely distributed in pathogenic Edwardsiella tarda. The regulatory effect of Fur on ZnuCBA indicates a cross-talk between Zn and Fe acquisition against host nutritional immunity. This was not the first clue to such cross-talk. For example, in Neisseria meningitidis, the transport of Zn through the outer membrane is driven by the Zn-modulated TonB-dependent receptor ZnuD (24), which is similar to the M. catarrhalis heme transporter HumA in sequence. ZnuD expression in E. coli promotes the acquisition of heme (25). The dual regulation of ZnuD by Zur and Fur (Zn and Fe uptake regulators, respectively) further indicates ZnuD may be involved in the acquisition of Zn and heme (25). In addition, the cross regulation of ZnuD may be due to the increased demand for exogenous heme under Zn limitation, because some endogenous heme biosynthesis enzymes utilize Zn (26). Hence, the above results revealed a Fur and ZnuCBA dependent cross-talk between Zn and Fe acquisition against host nutritional immunity in P. plecoglossicida. The regulatory effect of Fur on ZnuCBA also indicated that sequestration of Fe in the host might occur before Zn sequestration. Microorganisms have a feature called “predictive adaptation”, which can predict events that will occur in their natural habitat (92). For example, E. coli exposed to lactose expresses genes related to the metabolisms of lactose and maltose simultaneously, predicting that when passing through the mammalian digestive tract, it will be sequentially exposed to both monosaccharides (93). For the same reason, when P. plecoglossicida Fur senses low Fe concentration, it predicts that after the sequestration of Fe, it will undergo Zn starvation, and optimally induces the expression of znuCBA. However, since ZnuD and Zur were not found in P. plecoglossicida genome, this cross-talk was independent of these molecules. In addition, which transporter is used by P. plecoglossicida to transport Zn through the outer membrane in the absence of ZnuD deserves further investigation.
Previous study by Pederick et al. (94) investigated the effect of ΔznuA-induced zinc limitation on the transcriptome of P. aeruginosa. Genes that enable adaptation to zinc limitation were significantly changed. Non-zinc-requiring paralogs of zinc-dependent proteins and a number of novel import pathways associated with zinc acquisition were significantly up-regulated. For example, znuD was dramatically increased by 172.2 folds in P. aeruginosa ΔznuA, which was proved to be involved in the acquisition of Zn and heme (25) but not found in P. plecoglossicida genome as we mentioned above. The Zn2+-independent transcription regulator DksA2 and P-type ATPase importer HmtA was transcriptionally increased by 134.6 and 5.6 folds in P. aeruginosa ΔznuA respectively (94), but not found in P. plecoglossicida genome. The cobaltochelatase subunit CobN and the biopolymer transport protein ExbD was transcriptionally increased by 122.2 and 44.7 folds in ΔznuA respectively (94), but not significantly affected in the present study. There are many other differences between the two studies like these. We also compared Mastropasqua et al. (95) with our study and found many similar differences. For example, rpmE2, zmrABCD and xdhA were significantly up-regulated in P. aeruginosa under zinc deficiency (95), while rpmE2 and zmrABCD were not found in P. plecoglossicida genome, xdhA was down-regulated in P. plecoglossicida. These differences may be one of the reasons why the P. aeruginosa is more common in mammals infection and the P. plecoglossicida is more common in fish infection. On the other hand, these differentially expressed genes found by Pederick et al. (94) are predicted to be under the regulation of Zur, while the P. plecoglossicida do not have Zur, which may be an important reason for such a big difference in gene expression patterns under Zn restriction.
In conclusion, the importance of ZnuC was revealed by investigating how P. plecoglossicida responds to Zn nutritional immunity during infection. While diverse Zn sequestration strategies against P. plecoglossicida have been observed in E. coioides, a common denominator of all these strategies is pushing P. plecoglossicida out of its ideal environment. With the importance of pathogen-host interaction continued to be reported, current and previous studies have revealed the importance of considering the host’s internal environment while evaluating the contribution of virulent genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by The Animal Ethics Committee of Jimei University (JMULAC201159).
All authors contributed to the article and approved the submitted version. QY and LH conceived the experiments. YZ, YQ, LZ and ML conducted the experiments. All authors assisted in the collection and interpretation of data. LH and QY wrote the manuscript.
This work was supported by the Natural Science Foundation of Fujian Province (No. 2019J06020 and 2019J01695) and the “Distinguished Young Scientific Research Talents Plan in Universities of Fujian Province”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.678699/full#supplementary-material
Supplementary Figure 1 | Construction and confirmation of the knockout mutant strain ΔznuC and its complement znuC+. WT, Amplification of wild-type using primers znuCmut-F-R; ΔznuC, Amplification of ΔznuC using primers znuCmut-F-R; znuC+, Amplification of znuC+ using primers znuCmut-F-R.
Supplementary Figure 2 | KEGG pathway analysis of differentially expressed genes (DEGs) between wild-type Pseudomonas plecoglossicida and znuC-95%RNAi strain infected Epinephelus coioides. Spleen samples were assessed.
Supplementary Figure 3 | KEGG pathway analysis of differentially expressed genes (DEGs) between wild-type and znuC-95%RNAi Pseudomonas plecoglossicida in the spleen of Epinephelus coioides.
Supplementary Figure 4 | Construction and confirmation of the knockout mutant strain Δfur and its complement fur+. WT, Amplification of wild-type using primers furmut-F-R; Δfur, Amplification of Δfur using primers furmut-F-R, fur+, Amplification of fur+ using primers furmut-F-R.
Supplementary Table 2 | The summary of the number of mapped reads/input reads per replicate and the coverage.
1. Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal Ions in Biological Catalysis: From Enzyme Databases to General Principles. J Biol Inorg Chem (2008) 13:1205–18. doi: 10.1007/s00775-008-0404-5
2. Besold AN, Gilston BA, Radin JN, Ramsoomair C, Culbertson EM, Li CX, et al. Role of Calprotectin in Withholding Zinc and Copper From Candida Albicans. Infect Immun (2018) 86:e00779–17. doi: 10.1128/IAI.00779-17
3. Cassat JE, Skaar EP. Iron in Infection and Immunity. Cell Host Microbe (2013) 13:509–19. doi: 10.1016/j.chom.2013.04.010
4. Hood MI, Skaar EP. Nutritional Immunity: Transition Metals At the Pathogen–Host Interface. Nat Rev Microbiol (2012) 10:525. doi: 10.1038/nrmicro2836
5. Kehl-Fie TE, Skaar EP. Nutritional Immunity Beyond Iron: a Role for Manganese and Zinc. Curr Opin Chem Biol (2010) 14:218–24. doi: 10.1016/j.cbpa.2009.11.008
6. Weinberg ED. Iron Availability and Infection. Biochim Biophys Acta (2009) 1790:600–5. doi: 10.1016/j.bbagen.2008.07.002
7. Cassat JE, Skaar EP. Metal Ion Acquisition in Staphylococcus Aureus: Overcoming Nutritional Immunity. Semin Immunopathol (2012) 34:215–35. doi: 10.1007/s00281-011-0294-4
8. Haley KP, Skaar EP. A Battle for Iron: Host Sequestration and Staphylococcus Aureus Acquisition. Microbes Infect (2012) 14:217–27. doi: 10.1016/j.micinf.2011.11.001
9. Nobles CL, Maresso AW. The Theft of Host Heme by Gram-Positive Pathogenic Bacteria. Metallomics (2011) 3:788–96. doi: 10.1039/c1mt00047k
10. Braun V, Hantke K. Recent Insights Into Iron Import by Bacteria. Curr Opin Chem Biol (2011) 15:328–34. doi: 10.1016/j.cbpa.2011.01.005
11. Hantke K. Bacterial Zinc Uptake and Regulators. Curr Opin Microbiol (2005) 8:196–202. doi: 10.1016/j.mib.2005.02.001
12. Andreini C, Banci L, Bertini I, Rosato A. Zinc Through the Three Domains of Life. J Proteome Res (2006) 5:3173–8. doi: 10.1021/pr0603699
13. Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal Chelation and Inhibition of Bacterial Growth in Tissue Abscesses. Science (2008) 319:962–5. doi: 10.1126/science.1152449
14. Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, et al. Mntabc and Mnth Contribute to Systemic Staphylococcus Aureus Infection by Competing With Calprotectin for Nutrient Manganese. Infect Immun (2013) 81:3395–405. doi: 10.1128/IAI.00420-13
15. Gläser R, Harder J, Lange H, Bartels J, Christophers E, Schröder JM. Antimicrobial Psoriasin (S100A7) Protects Human Skin From Escherichia Coli Infection. Nat Immunol (2005) 6:57–64. doi: 10.1038/ni1142
16. Moroz OV, Antson AA, Grist SJ, Maitland NJ, Dodson GG, Wilson KS, et al. Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence. Acta Crystallogr D Biol Crystallogr (2003) 59:859–67. doi: 10.1107/S0907444903004700
17. Moroz OV, Burkitt W, Wittkowski H, He W, Ianoul A, Novitskaya V, et al. Both Ca2+ and Zn2+ are Essential for S100A12 Protein Oligomerization and Function. BMC Biochem (2009) 10:11. doi: 10.1186/1471-2091-10-11
18. McCormick A, Heesemann L, Wagener J, Marcos V, Hartl D, Loeffler J, et al. Nets Formed by Human Neutrophils Inhibit Growth of the Pathogenic Mold Aspergillus Fumigatus. Microbes Infect (2010) 12:928–36. doi: 10.1016/j.micinf.2010.06.009
19. Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, et al. Neutrophil Extracellular Traps Contain Calprotectin, a Cytosolic Protein Complex Involved in Host Defense Against Candida Albicans. PLoS Pathog (2009) 5:e1000639. doi: 10.1371/journal.ppat.1000639
20. Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, et al. Zinc Sequestration by the Neutrophil Protein Calprotectin Enhances Salmonella Growth in the Inflamed Gut. Cell Host Microbe (2012) 11:227–39. doi: 10.1016/j.chom.2012.01.017
21. Bianchi M, Niemiec MJ, Siler U, Urban CF, Reichenbach J. Restoration of Anti-Aspergillus Defense by Neutrophil Extracellular Traps in Human Chronic Granulomatous Disease After Gene Therapy is Calprotectin-Dependent. J Allergy Clin Immunol (2011) 127:1243–52. doi: 10.1016/j.jaci.2011.01.021
22. Barnett JP, Millard A, Ksibe AZ, Scanlan DJ, Schmid R, Blindauer CA. Mining Genomes of Marine Cyanobacteria for Elements of Zinc Homeostasis. Front Microbiol (2012) 3:142. doi: 10.3389/fmicb.2012.00142
23. Gonzalez MR, Ducret V, Leoni S, Perron K. Pseudomonas Aeruginosa Zinc Homeostasis: Key Issues for an Opportunistic Pathogen. Biochim Biophys Acta Gene Regul Mech (2019) 1862(7):722–33. doi: 10.1016/j.bbagrm.2018.01.018
24. Stork M, Bos MP, Jongerius I, de Kok N, Schilders I, Weynants VE, et al. An Outer Membrane Receptor of Neisseria Meningitidis Involved in Zinc Acquisition With Vaccine Potential. PLoS Pathog (2010) 6:e1000969. doi: 10.1371/journal.ppat.1000969
25. Kumar P, Sannigrahi S, Tzeng YL. The Neisseria Meningitidis Znud Zinc Receptor Contributes to Interactions With Epithelial Cells and Supports Heme Utilization When Expressed in Escherichia Coli. Infect Immun (2012) 80:657–67. doi: 10.1128/IAI.05208-11
26. Li JM, Russell CS, Cosloy SD. The Structure of the Escherichia Coli Hemb Gene. Gene (1989) 75:177–84. doi: 10.1016/0378-1119(89)90394-6
27. Klein JS, Lewinson O. Bacterial ATP-Driven Transporters of Transition Metals: Physiological Roles, Mechanisms of Action, and Roles in Bacterial Virulence. Metallomics (2011) 3:1098. doi: 10.1039/c1mt00073j
28. Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, et al. High-Affinity Zn2+ Uptake System Znuabc is Required for Bacterial Zinc Homeostasis in Intracellular Environments and Contributes to the Virulence of Salmonella Enterica. Infect Immun (2007) 75:5867–76. doi: 10.1128/IAI.00559-07
29. Campoy S, Jara M, Busquets N, Pérez De Rozas AM, Badiola I, Barbé J. Role of the High-Affinity Zinc Uptake Znuabc System in Salmonella Enterica Serovar Typhimurium Virulence. Infect Immun (2002) 70:4721–5. doi: 10.1128/IAI.70.8.4721-4725.2002
30. Nielubowicz GR, Smith SN, Mobley HLT. Zinc Uptake Contributes to Motility and Provides a Competitive Advantage to Proteus Mirabilis During Experimental Urinary Tract Infection. Infect Immun (2010) 78:2823–33. doi: 10.1128/IAI.01220-09
31. D’Orazio M, Mastropasqua MC, Cerasi M, Pacello F, Consalvo A, Chirullo B, et al. The Capability of Pseudomonas Aeruginosa to Recruit Zinc Under Conditions of Limited Metal Availability is Affected by Inactivation of the Znuabc Transporter. Metallomics (2015) 7(6):1023–35. doi: 10.1039/C5MT00017C
32. Cornforth DM, Dees JL, Ibberson CB, Huse HK, Mathiesen IH, Kirketerp-Møller K, et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A (2018) 115(22):E5125–34. doi: 10.1073/pnas.1717525115
33. Mahmoud AOD. Nutritional Immunity of Fish Intestines: Important Insights for Sustainable Aquaculture. Rev Aquacult (2021) 13:642–63. doi: 10.1111/raq.12492
34. Nishimori E, Kita-Tsukamoto K, Wakabayashi H. Pseudomonas Plecoglossicida Sp. nov., the causative agent of bacterial hemorrhagic ascites of ayu, Plecoglossus altivelis. Int J Syst Evol Microbiol (2000) 50:83–9. doi: 10.1099/00207713-50-1-83
35. Hu J, Zhang F, Xu X, Su Y, Qin Y, Ma Y, et al. Isolation, Identification and Virulence of the Pathogen of White-Spots Disease in Internal Organs of Pseudosciaena Crocea. Oceanol Limnol Sin (2014) 45:409–17.
36. Zhang B, Luo G, Zhao L, Huang L, Qin Y, Su Y, et al. Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish Shellfish Immunol (2018) 81:121–9. doi: 10.1016/j.fsi.2018.06.051
37. Akayli T, Canak O, Basaran B. A New Pseudomonas Species Observed in Cultured Young Rainbow Trout (Oncorhynchus Mykiss Walbaum, 1792): Pseudomonas Plecoglossicida. Res J Biol Sci (2011) 4:107–11.
38. Luo G, Sun Y, Huang L, Su Y, Zhao L, Qin Y, et al. Time-Resolved Dual RNA-Seq of Tissue Uncovers Pseudomonas Plecoglossicida Key Virulence Genes in Host-Pathogen Interaction With Epinephelus Coioides. Environ Microbiol (2019) 22:677–93. doi: 10.1111/1462-2920.14884
39. Wang X, Du Y, Hua Y, Fu M, Niu C, Zhang B, et al. The EspF N-terminal of enterohemorrhagic Escherichia coli O157: H7 EDL933w imparts stronger toxicity effects on HT-29 cells than the C-terminal. Front Cell Infect Microbiol (2017) 7:410. doi: 10.3389/fcimb.2017.00410
40. Huang L, Zhao L, Qi W, Xu X, Zhang J, Zhang J, et al. Temperature-Specific Expression of Cspa1 Contributes to Activation of Sigx During Pathogenesis and Intracellular Survival in Pseudomonas Plecoglossicida. Aquaculture (2020) 518:734861. doi: 10.1016/j.aquaculture.2019.734861
41. Sun Y, Luo G, Zhao L, Huang L, Qin Y, Su Y, et al. Integration of Rnai and RNA-Seq Reveals the Immune Responses of Epinephelus Coioides to Sigx Gene of Pseudomonas Plecoglossicida. Front Immunol (2018) 9:1624. doi: 10.3389/fimmu.2018.01624
42. Huang L, Zhang Y, He R, Zuo Z, Luo Z, Xu W, et al. Phenotypic Characterization, Virulence, and Immunogenicity of Pseudomonas Plecoglossicida Rpoe Knock-Down Strain. Fish Shellfish Immunol (2019) 87:772–7. doi: 10.1016/j.fsi.2019.02.028
43. Luo G, Xu X, Zhao L, Qin Y, Huang L, Su Y, et al. Clpv is a Key Virulence Gene During In Vivo Pseudomonas Plecoglossicida Infection. J Fish Dis (2019) 42(7):991–1000. doi: 10.1111/jfd.13001
44. Sun Y, Nie P, Zhao L, Huang L, Qin Y, Xu X, et al. Dual RNA-Seq Unveils the Role of the Pseudomonas Plecoglossicida Flia Gene in Pathogen-Host Interaction With Larimichthys Crocea. Microorganisms (2019) 7(10):443. doi: 10.3390/microorganisms7100443
45. Qi W, Xu W, Zhao L, Xu X, Luo Z, Huang L, et al. Protection Against Pseudomonas Plecoglossicida in Epinephelus Coioides Immunized With a Cspa1-Knock-Down Live Attenuated Vaccine. Fish Shellfish Immunol (2019) 89:498–504. doi: 10.1016/j.fsi.2019.04.029
46. Tang R, Luo G, Zhao L, Huang L, Qin Y, Xu X, et al. The Effect of a Lysr-Type Transcriptional Regulator Gene of Pseudomonas Plecoglossicida on the Immune Responses of Epinephelus Coioides. Fish Shellfish Immunol (2019) 89:420–7. doi: 10.1016/j.fsi.2019.03.051
47. Zhang M, Yan Q, Mao L, Wang S, Huang L, Xu X, et al. Katg Plays an Important Role in Aeromonas Hydrophila Survival in Fish Macrophages and Escape for Further Infection. Gene (2018) 672:156–64. doi: 10.1016/j.gene.2018.06.029
48. Arras LD, Guthrie BS, Alper S. Using RNA-Interference to Investigate the Innate Immune Response in Mouse Macrophages. J Visual Exp Jove (2014) 93:e51306–6. doi: 10.3791/51306
49. Xu C, Wang Y, Rezeng C, Zhang L, Zhao B, Wang X, et al. Tissue Metabolomics Study to Reveal the Toxicity of a Traditional Tibetan Medicine ‘Renqing Changjue’ in Rats. RSC Adv (2018) 8(66):37652–64. doi: 10.1039/C8RA07058J
50. Subramanian Vignesh K, Landero Figueroa JA, Porollo A, Caruso JA, Deepe GS Jr. Granulocyte Macrophage-Colony Stimulating Factor Induced Zn Sequestration Enhances Macrophage Superoxide and Limits Intracellular Pathogen Survival. Immunity (2013) 39:697–710. doi: 10.1016/j.immuni.2013.09.006
51. Huang L, Xi Z, Wang C, Zhang Y, Yang Z, Zhang S, et al. Phenanthrene Exposure Induces Cardiac Hypertrophy Via Reducing Mir-133a Expression by DNA Methylation. Sci Rep (2016) 6:20105. doi: 10.1038/srep20105
52. Zhang B, Zhuang Z, Wang X, Huang H, Fu Q, Yan Q. Dual RNA-Seq Reveals the Role of a Transcriptional Regulator Gene in Pathogen-Host Interactions Between Pseudomonas Plecoglossicida and Epinephelus Coioides. Fish Shellfish Immunol (2019) 87:778–87. doi: 10.1016/j.fsi.2019.02.025
53. He R, Zhao L, Xu X, Zheng W, Zhang J, Zhang J, et al. Aryl Hydrocarbon Receptor is Required for Immune Response in Epinephelus Coioides and Danio Rerio Infected by Pseudomonas Plecoglossicida. Fish Shellfish Immunol (2020) 97:564–70. doi: 10.1016/j.fsi.2019.12.084
54. Pang H, Zhang X, Wu Z, Jian J, Cai S, Liang J. Identification of Novel Immunogenic Proteins of Vibrio Alginolyticus by Immunoproteomic Methodologies. Aquacult Res (2013) 44(3):472–84. doi: 10.1111/j.1365-2109.2012.03150.x
55. Liu WC, Zhou SH, Balasubramanian B, Zeng FY, Sun CB, Pang HY. Dietary Seaweed (Enteromorpha) Polysaccharides Improves Growth Performance Involved in Regulation of Immune Responses, Intestinal Morphology and Microbial Community in Banana Shrimp Fenneropenaeus Merguiensis. Fish Shellfish Immunol (2020) 104:202–12. doi: 10.1016/j.fsi.2020.05.079
56. Tang Y, Sun Y, Zhao L, Xu X, Huang L, Qin Y, et al. Mechanistic Insight Into the Roles of Pseudomonas Plecoglossicida Clpv Gene in Host-Pathogen Interactions With Larimichthys Crocea by Dual RNA-Seq. Fish Shellfish Immunol (2019) 93:344–53. doi: 10.1016/j.fsi.2019.07.066
57. Wang L, Sun Y, Zhao L, Xu X, Huang L, Qin Y, et al. Dual RNA-Seq Uncovers the Immune Response of Larimichthys Crocea to the Secy Gene of Pseudomonas Plecoglossicida From the Perspective of Host-Pathogen Interactions. Fish Shellfish Immunol (2019) 93:949–57. doi: 10.1016/j.fsi.2019.08.040
58. Luo G, Zhao L, Xu X, Qin Y, Huang L, Su Y, et al. Integrated Dual RNA-Seq and Dual Itraq of Infected Tissue Reveals the Functions of a Diguanylate Cyclase Gene of Pseudomonas Plecoglossicida in Host-Pathogen Interactions With Epinephelus Coioides. Fish Shellfish Immunol (2019) 95:481–90. doi: 10.1016/j.fsi.2019.11.008
59. Langmead B, Salzberg SL. Fast Gapped-Read Alignment With Bowtie 2. Nat Methods (2011) 9:357–9. doi: 10.1038/nmeth.1923
60. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-Length Transcriptome Assembly From RNA-Seq Data Without a Reference Genome. Nat Biotechnol (2011) 29:644. doi: 10.1038/nbt.1883
61. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
62. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res (2000) 28:27–30. doi: 10.1093/nar/28.1.27
63. Robinson MD, McCarthy DJ, Smyth GK. Edger: a Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
64. Schulze S, Henkel SG, Driesch D, Guthke R, Linde J. Computational Prediction of Molecular Pathogen-Host Interactions Based on Dual Transcriptome Data. Front Microbiol (2015) 6:65. doi: 10.3389/fmicb.2015.00065
65. Schulze S, Schleicher J, Guthke R, Linde J. How to Predict Molecular Interactions Between Species. Front Microbiol (2016) 7:442. doi: 10.3389/fmicb.2016.00442
66. Dahiya I, Stevenson RM. The Znuabc Operon is Important for Yersinia Ruckeri Infections of Rainbow Trout, Oncorhynchus Mykiss (Walbaum). J Fish Dis (2010) 33:331–40. doi: 10.1111/j.1365-2761.2009.01125.x
67. Kelliher JL, Radin JN, Grim KP, Párraga Solórzano PK, Degnan PH, Kehl-Fie TE. Acquisition of the Phosphate Transporter Npta Enhances Staphylococcus Aureus Pathogenesis by Improving Phosphate Uptake in Divergent Environments. Infect Immun (2017) 86(1):e00631–17. doi: 10.1128/IAI.00631-17
68. Yu L, Li C, Chen J. A Novel CC Chemokine Ligand 2 Like Gene From Ayu Plecoglossus Altivelis is Involved in the Innate Immune Response Against to Vibrio Anguillarum. Fish Shellfish Immunol (2019) 87:886–96. doi: 10.1016/j.fsi.2019.02.019
69. Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, Mok YK. Temperature and Mg2+ Sensing by a Novel Phop-Phoq Two-Component System for Regulation of Virulence in Edwardsiella Tarda. J Biol Chem (2010) 285:38876–88. doi: 10.1074/jbc.M110.179150
70. Huang L, Zhao L, Liu W, Xu X, Qin Y, Su Y, et al. Dual RNA-Seq Unveils Pseudomonas Plecoglossicida Htpg Gene Functions During Host-Pathogen Interactions With Epinephelus Coioides. Front Immunol (2019) 10:984. doi: 10.3389/fimmu.2019.00984
71. Huang L, Zuo Y, Jiang Q, Su Y, Qin Y, Xu X, et al. A Metabolomic Investigation Into the Temperature-Dependent Virulence of Pseudomonas Plecoglossicida From Large Yellow Croaker (Pseudosciaena Crocea). J Fish Dis (2019) 42(3):431–46. doi: 10.1111/jfd.12957
72. Mao Z, Ye J, Li M, Xu H, Chen J. Vaccination Efficiency of Surface Antigens and Killed Whole Cell of Pseudomonas putida in large yellow croaker (Pseudosciaena crocea). Fish Shellfish Immunol (2013a) 35(2):375–81. doi: 10.1016/j.fsi.2013.04.030
73. Mao Z, Li M, Chen J. Draft Genome Sequence of Pseudomonas plecoglossicida strain NB2011, the causative agent of white nodules in large yellow croaker (Larimichthys crocea). Genome Announce (2013b) 1(4):e00586–13. doi: 10.1128/genomeA.00586-13
74. Patzer SI, Hantke K. The Znuabc High-Affinity Zinc Uptake System and its Regulator Zur in Escherichia Coli. Mol Microbiol (1998) 28(6):1199–210. doi: 10.1046/j.1365-2958.1998.00883.x
75. Crawford A, Wilson D. Essential metals at the host–pathogen interface: nutritional immunity and micronutrient assimilation by human fungal pathogens. FEMS Yeast Res (2015) 15(7):fov071. doi: 10.1093/femsyr/fov071
76. Thokala S, Bodiga VL, Kudle MR, Bodiga S. Comparative Response of Cardiomyocyte Zips and Znts to Extracellular Zinc and TPEN. Biol Trace Element Res (2019) 192:297–307. doi: 10.1007/s12011-019-01671-0
77. Liuzzi JP, Lichten LA, Rivera S, Blanchard RK, Aydemir TB, Knutson MD, et al. Interleukin-6 Regulates the Zinc Transporter Zip14 in Liver and Contributes to the Hypozincemia of the Acute-Phase Response. Proc Natl Acad Sci (2005) 102:6843–8. doi: 10.1073/pnas.0502257102
78. Seider K, Heyken A, Lüttich A, Miramón P, Hube B. Interaction of Pathogenic Yeasts With Phagocytes: Survival, Persistence and Escape. Curr Opin Microbiol (2010) 13:392–400. doi: 10.1016/j.mib.2010.05.001
79. Miramon P, Kasper L, Hube B. Thriving Within the Host: Candida Spp. interactions with phagocytic cells. Med Microbiol Immunol (2013) 202:183–95. doi: 10.1007/s00430-013-0288-z
80. Lorenz MC, Bender JA, Fink GR. Transcriptional Response of Candida Albicans Upon Internalization by Macrophages. Eukaryot Cell (2004) 3:1076–87. doi: 10.1128/EC.3.5.1076-1087.2004
81. Winters MS, Chan Q, Caruso JA, Deepe GS Jr. Metallomic Analysis of Macrophages Infected With Histoplasma Capsulatum Reveals a Fundamental Role for Zinc in Host Defenses. J Infect Dis (2010) 202:1136–45. doi: 10.1086/656191
82. Gao HL, Feng WY, Li XL, Xu H, Huang L, Wang ZY. Golgi Apparatus Localization of ZNT7 in the Mouse Cerebellum. Histol Histopathol (2009) 24:567–72. doi: 10.14670/HH-24.567
83. McCormick NH, Kelleher SL. ZnT4 provides zinc to zinc dependent proteins in the trans-Golgi network critical for cell function and Zn export in mammary epithelial cells. Am J Physiol Cell Ph (2012) 303:C291–7. doi: 10.1152/ajpcell.00443.2011
84. Lee MG, Bin BH. Different Actions of Intracellular Zinc Transporters ZIP7 and ZIP13 are Essential for Dermal Development. Int J Mol Sci (2019) 20:3941. doi: 10.3390/ijms20163941
85. Pei D. Identification and Characterization of the Fifth Membrane-Type Matrix Metalloproteinase MT5-MMP. J Biol Chem (1999) 274:8925–32. doi: 10.1074/jbc.274.13.8925
86. Zhao H, Eide DJ. Zap1p, a Metalloregulatory Protein Involved in Zinc-Responsive Transcriptional Regulation in Saccharomyces Cerevisiae. Mol Cell Biol (1997) 17:5044–52. doi: 10.1128/MCB.17.9.5044
87. Zhao H, Eide D. The Yeast ZRT1 Gene Encodes the Zinc Transporter Protein of a High-Affinity Uptake System Induced by Zinc Limitation. PNAS (1996a) 93:2454–8. doi: 10.1073/pnas.93.6.2454
88. Zhao H, Eide D. The ZRT2 Gene Encodes the Low Affinity Zinc Transporter in Saccharomyces cerevisiae. J Biol Chem (1996b) 271:23203–10. doi: 10.1074/jbc.271.38.23203
89. MacDiarmid CW, Gaither LA, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J (2000) 19:2845–55. doi: 10.1093/emboj/19.12.2845
90. Skaar EP. The Battle for Iron Between Bacterial Pathogens and Their Vertebrate Hosts. PLoS PPathog (2010) 6:e1000949. doi: 10.1371/journal.ppat.1000949
91. Torres VJ, Attia AS, Mason WJ, Hood MI, Corbin BD, Beasley FC, et al. Staphylococcus Aureus Fur Regulates the Expression of Virulence Factors That Contribute to the Pathogenesis of Pneumonia. Infect Immun (2010) 78(4):1618–28. doi: 10.1128/IAI.01423-09
92. Tagkopoulos I, Liu YC, Tavazoie S. Predictive Behavior Within Microbial Genetic Networks. Science (2008) 320:1313–7. doi: 10.1126/science.1154456
93. Mitchell A, Romano GH, Groisman B. Adaptive Prediction of Environmental Changes by Microorganisms. Nature (2009) 460:220–4. doi: 10.1038/nature08112
94. Pederick VG, Eijkelkamp BA, Begg SL, Ween MP, McAllister LJ, Paton JC, et al. Znua and Zinc Homeostasis in Pseudomonas Aeruginosa. Sci Rep (2015) 5:13139. doi: 10.1038/srep13139
Keywords: Pseudomonas plecoglossicida, Epinephelus coioides, dual RNA-seq, znuC, nutritional immunity
Citation: Huang L, Zuo Y, Qin Y, Zhao L, Lin M and Yan Q (2021) The Zinc Nutritional Immunity of Epinephelus coioides Contributes to the Importance of znuC During Pseudomonas plecoglossicida Infection. Front. Immunol. 12:678699. doi: 10.3389/fimmu.2021.678699
Received: 10 March 2021; Accepted: 19 April 2021;
Published: 04 May 2021.
Edited by:
Zhihong Sun, Inner Mongolia Agricultural University, ChinaReviewed by:
Qixiao Zhai, Jiangnan University, ChinaCopyright © 2021 Huang, Zuo, Qin, Zhao, Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingpi Yan, eWFucXBAam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.