
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 05 May 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.669241
This article is part of the Research Topic The Role of Epigenetics in Infectious Diseases View all 14 articles
 Ravesh Singh1,2,3†
Ravesh Singh1,2,3† Veron Ramsuran4,5,6,7†
Veron Ramsuran4,5,6,7† Vivek Naranbhai5,6,7
Vivek Naranbhai5,6,7 Nonhlanhla Yende-Zuma7
Nonhlanhla Yende-Zuma7 Nigel Garrett7
Nigel Garrett7 Koleka Mlisana3
Koleka Mlisana3 Krista L. Dong1
Krista L. Dong1 Bruce D. Walker1,2,5
Bruce D. Walker1,2,5 Salim S. Abdool Karim7
Salim S. Abdool Karim7 Mary Carrington5,6
Mary Carrington5,6 Thumbi Ndung’u1,2,5,8,9*
Thumbi Ndung’u1,2,5,8,9*HIV-1 must overcome host antiviral restriction factors for efficient replication. We hypothesized that elevated levels of bone marrow stromal cell antigen 2 (BST-2), a potent host restriction factor that interferes with HIV-1 particle release in some human cells and is antagonized by the viral protein Vpu, may associate with viral control. Using cryopreserved samples, from HIV-1 seronegative and seropositive Black women, we measured in vitro expression levels of BST-2 mRNA using a real-time PCR assay and protein levels were validated by Western blotting. The expression level of BST-2 showed an association with viral control within two independent cohorts of Black HIV infected females (r=-0.53, p=0.015, [n =21]; and r=-0.62, p=0.0006, [n=28]). DNA methylation was identified as a mechanism regulating BST-2 levels, where increased BST-2 methylation results in lower expression levels and associates with worse HIV disease outcome. We further demonstrate the ability to regulate BST-2 levels using a DNA hypomethylation drug. Our results suggest BST-2 as a factor for potential therapeutic intervention against HIV and other diseases known to involve BST-2.
To establish infection and replicate efficiently, HIV-1 must overcome host antiviral restriction factors. Host restriction factors that inhibit HIV-1 replication are an important component of the innate immune system that forms the first line of defense before adaptive immune responses are mobilized and established (1–4). BST-2 (also termed Tetherin/CD317/HM1.24) was discovered as an anti-HIV host factor responsible for the prevention of virus release (5). Subsequently, additional mechanisms of HIV inhibition by BST-2 have been reported (6) and studies have associated BST-2 expression levels with HIV viral control (6–8). Downregulation of BST-2 expression correlated with Vpu expression and elevated BST-2 induced a requirement for Vpu to facilitate HIV particle release in some cells (5, 9). Vpu promotes intracellular down-regulation of BST-2 (10, 11). However, BST-2 is an interferon-induced protein, which gets activated upon HIV infection (7, 12). Factors regulating the expression levels of the BST-2 gene have not been fully resolved.
The “tethering” effect mediated by BST-2 on HIV has subsequently been shown to restrict the replication of a diverse array of other enveloped viruses including other retroviruses, rhabdoviruses, alphaviruses, arenaviruses, filoviruses, herpesviruses, paramyxoviruses, orthomyxoviruses, orthohepadnaviruses and flaviviruses (12–21). In addition, BST-2 expression levels are elevated in several cancers such as head and neck, breast, cervical, lung, endometrial, myelomas, and glioblastomas (22–29) as well as lupus erythematosus (SLE) an autoimmune disease (30), suggesting that BST-2 could be an immunotherapeutic target for several diseases. If BST-2 is directly affecting these conditions, then identifying the factors regulating BST-2 expression could develop strategies against an array of diseases. DNA methylation has been linked with the regulation of BST-2 expression particularly in cancer cells (24), and in lupus (30). A few human genes implicated in HIV control are known to be regulated by DNA methylation. Hypermethylation of FOXP3, EPB41L3, IL-2, CCR5 and HLA-A at gene regulatory sites, are associated with reduced gene expression and worse HIV outcome, whereas reduced methylation corresponds with increased expression of these genes and improved disease outcome (31–36). This highlights the potential importance of this epigenetic mode of gene regulation in HIV disease pathogenesis.
In this study, we found that increased BST-2 levels associated with HIV control. We further show DNA methylation as one of the regulatory mechanisms responsible for BST-2 expression variation within HIV infected individuals. Furthermore, BST-2 methylation levels correlate with HIV outcomes in both ex vivo and in vitro experiments, and experimental manipulation of BST-2 methylation altered its expression levels. Together, these data suggest that manipulation of BST-2 expression levels could be used as a therapeutic target for viral control.
A chronic HIV infection cohort, Sinikithemba (SK; n=21) (37), was compared to the HIV negative arm of the acute infection cohort from the Females Rising through Education, Support, and Health (FRESH; n=65) study (38, 39) in a cross sectional analysis. We further studied a longitudinal HIV acute infection cohort, CAPRISA 002 (n=55) (40, 41), from pre-infection to >36 months of follow-up post HIV infection. All the samples used in this study were from South African females of Black ancestry. The study was approved by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal.
Peripheral blood mononuclear cells (PBMCs) were isolated within 6 hours of blood collection, and frozen in liquid nitrogen until use. Viral load was determined using the automated COBAS AMPLICOR HIV-1 Monitor Test v1.5 (Roche Diagnostics, Mannheim, Germany). CD4+ T cells were enumerated using the Multitest kit (CD4/CD3/CD8/CD45) on a four-parameter FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA).
RNA was extracted from 2 x 106 PBMCs using the TRIzol LS Reagent (Invitrogen, Carlsbad, CA, USA). RNA from each sample was reversed transcribed using the iScript cDNA synthesis kit (Bio-Rad, California, United States of America). PCR primer and cycling conditions for BST-2 and GAPDH (housekeeping gene) are available on request. GAPDH was used as reference gene (42). PCR-product amplification specificity was confirmed via melting curve analysis and agarose gel electrophoresis.
Cell lysates were boiled for 10 minutes in 4X Laemmli sample buffer (Bio-Rad), then separated by SDS-PAGE on 4 to 15% gels (Bio-Rad, California, United States of America) and transferred onto nitrocellulose membrane following standard methods. The membrane was then incubated with the primary antibody (rabbit monoclonal anti-BST-2 [cat. no. ab243229, Abcam, Cambridge, United Kingdom], and mouse polyclonal anti-alpha-tubulin [cat. no. ab7291, Abcam, Cambridge, United Kingdom]), diluted in 5% bovine serum albumin (BSA) (Roche, Basel, Switzerland) in tris-buffered saline and Tween 20 at a 1:100 or 1:5000 dilution overnight, followed by three washes in tris-buffered saline (TBS) and Tween 20 (TBST) for 10 minutes. The membrane was incubated with the secondary antibody (anti-rabbit, or anti-mouse) at a 1:20,000 dilution in 5% BSA in TBST for 1 hour on a rocker, followed by three washes in TBST for 10 minutes. Antibody-antigen complexes were detected via enhanced chemiluminescence reagents (SuperSignal West Dura extended-duration substrate, Thermo Scientific, Pierce Protein Research, United States of America). Proteins were visualized using the ChemiDoc XRS+ system with Image Lab software (Bio-Rad, California, United States of America).
HIV-1 replication in vitro was assessed on PBMCs from 22 donors selected from the 65 healthy HIV uninfected individuals from the FRESH cohort. Individuals with the highest (n =11) and lowest (n=11) BST-2 mRNA levels were included. PBMCs (2×106/mL) were stimulated for 48 hours in R10 buffer [Roswell Park Memorial Institute (RPMI)-1640 medium (Grand Island, NY, USA) supplemented with 10% fetal calf serum (Hyclone Inc., Logan, UT, USA), gentamicin (Gibco-Brl, Gaithersburg, MD, USA) (100 mg/mL)] containing 5 mg/mL phytohemagglutinin (PHA) (Roche, Basel, Switzerland) and 5 mg/mL interleukin-2 (IL-2) (Roche, Basel, Switzerland). Following stimulation with PHA/IL-2, cells were washed with R10 buffer and then infected with HIV IIIB (NIH AIDS Reagent Repository) at 0.1 multiplicity of infection (MOI) by spinoculation (2 hours, 300 x g at 37°C). Infection was performed in a 24-well plate. Virus was subsequently removed by washing the cells, followed by cell culture for 7 days. BST-2 mRNA expression levels and DNA methylation were analyzed by real-time PCR and pyrosequencing. Supernatants from days 2, 4 and 7 were harvested and analyzed by p24 antigen capture enzyme-linked immunosorbent assay (ELISA [Biomérieux, Marcy-l’Étoile, France]).
Primer design for the detection of methylation within the BST-2 promoter region was performed using MethPrimer online software, default settings (43) (Forward meth primer 1 GGTTAGTTTTTGTTGTAGGAGATGG; Reverse meth primer 1 AACTATTACAAAATACCCATAAAAAAC; Forward meth primer 2 TTGATGGTGAAGATAATTAAGGGTATT; Reverse meth primer 2 AAAAACTACTAATCAAAACACTTCCTAAAA). Sodium bisulphite conversion was performed on genomic DNA extracted from PBMCs using the EZ DNA methylation™ kit (Zymo Research, Irvine, USA). Using the BST-2 specific primers on the bisulphite converted DNA, a PCR was run using the following conditions (95°C for 15 minutes, 45 cycles of 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 30 seconds and one cycle of 72°C for 10 minutes). The level of methylation at specific sites within the BST-2 promoter was measured using pyrosequencing (Roche, Basel, Switzerland).
Treatment of cells with the DNA hypomethylation drug, 5’-aza-2-deoxycytidine (5’-Aza-CdR), was performed as previously described (32). Briefly, PBMCs from healthy donors (n=40) were treated with 10 µM 5’-Aza-CdR (Sigma, St. Louis, United States America) or with dimethyl sulfoxide (DMSO; treatment control) for 24 hours at 37°C. BST-2 mRNA levels from 5’-Aza-CdR treated cells were compared to DMSO treated and then plotted against the untreated mRNA levels.
Statistical analyses were conducted using Instat Graphpad Prism V.5 and SAS version 9.4. All expression data was log10 transformed to ensure normality (44, 45). Gene expression levels between HIV negative and HV infected donors were compared using an unpaired t-test. BST-2 mRNA expression levels for HIV positive donors prior to infection and at three months post-infection were compared using paired t-test. Furthermore, we calculated the Pearson correlation coefficient to measure the strength of an association between BST-2 mRNA expression levels and methylation at each time-point.
Univariable linear mixed model with autoregressive order one covariance structure were fitted to determine if there was an association between BST-2 gene expression and viral load. In this model, we included a random effect for the participant or subject.
We investigated the effect of HIV infection on BST-2 mRNA expression levels in PBMCs ex vivo. We found significantly higher levels of BST-2 in HIV negative individuals (n=32, FRESH cohort, black dots) compared to HIV infected late stage antiretroviral (ARV)-naïve individuals (n=21, SK cohort, red dots; p < 0.0001; Figure 1A). To validate these findings for consistency of mRNA expression with protein levels, we randomly selected donors, based on sample availability, from 5 HIV negative donors and 4 HIV infected donors, which formed subsets of the FRESH and SK cohorts, respectively. Western blot assays showed consistent BST-2 protein expression levels relative to mRNA expression levels, with protein expression higher in HIV- compared to HIV+ donors (Figure 1B). We next explored the relationship between BST-2 mRNA expression levels and HIV-1 viral load. A negative correlation was observed in both SK (r=-0.53, p=0.015; Figure 1C), and CAPRISA 002 cohorts (n=28, r=-0.62, p=0.0006; Figure 1D), all individuals analysed cross-sectionally were past 36 months post infection in both cohorts.
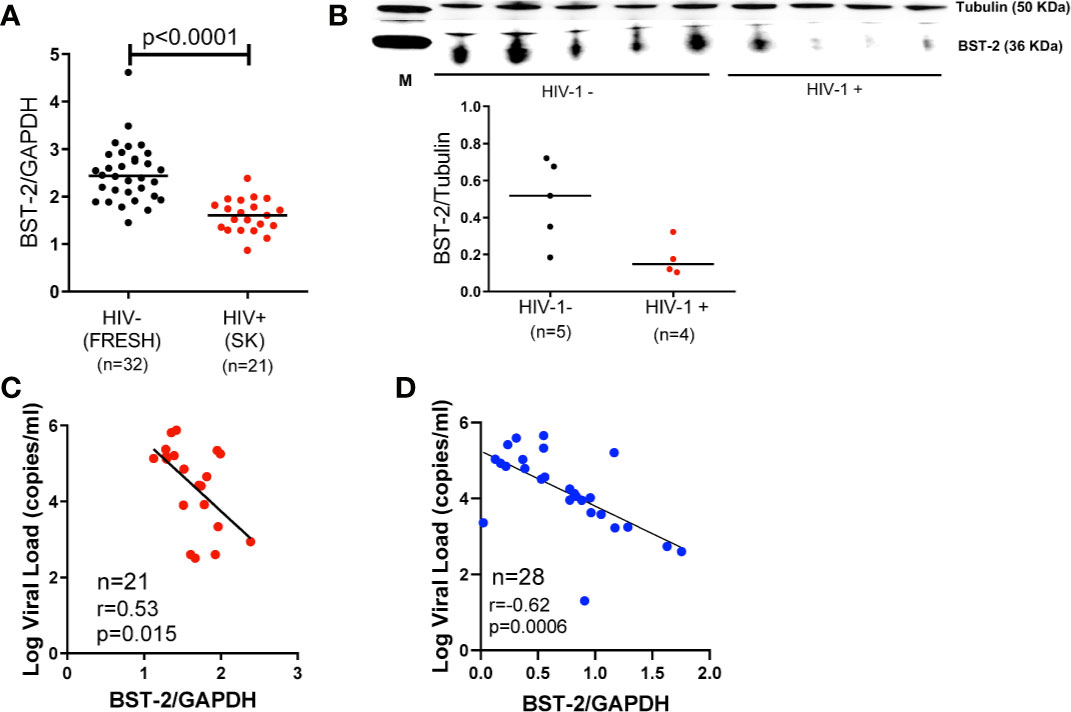
Figure 1 BST-2 mRNA and protein expression levels within HIV-negative and positive individuals. (A) Comparison of BST-2 mRNA expression levels measured in HIV negative and positive donors from the FRESH, (black dots), and SK, (red dots) cohorts, respectively. Significantly elevated BST-2 levels are found within HIV negative donors vs. positives (p<0.0001). These represent unmatched donors from two separate cohorts. The HIV positive donors are ARV naïve chronically infected. (B) Protein levels of BST-2 were measured on 5 HIV negative donors and 4 HIV infected donors from the FRESH and SK cohorts, respectively. BST-2 protein levels were assessed using a Western blot assay. The levels of HIV infected donors are lower than the HIV negative. (C) BST-2 mRNA expression levels were correlated with log viral load within the SK cohort. Higher mRNA levels correlated with lower log viral load levels (r=-0.53, p=0.0150). (D) A negative correlation was also observed when examining the effect of BST-2 mRNA expression levels and viral load using the CAPRISA 002 cohort at the >36 month time point (r=-0.62, p=0.0006).
BST-2 mRNA levels and viral load were also tested longitudinally at three timepoints (3, 12 and >36 months) in the CAPRISA 002 cohort. The results of the generalized estimating equation (GEE) model revealed consistent results to the cross-sectional data where higher mRNA levels associated with decreased viral load (Effect = -0.022; Standard error = 0.009; p=0.0003).
The inverse effect of DNA methylation on BST-2 expression has been shown previously in the context of cancer and autoimmune studies (24, 30). Here, we examined the effect DNA methylation on BST-2 expression levels within an HIV setting. Nine CpG sites located within 200 bp of the transcription start site were evaluated for methylation levels (Figure 2A) in HIV positive and negative individuals (SK vs. FRESH cohort respectively). All sites showed significantly higher methylation levels within the HIV infected group (Figures 2B–J), suggesting that increased BST-2 methylation levels in chronic HIV infection results in decreased expression level of the gene as observed in Figure 1.
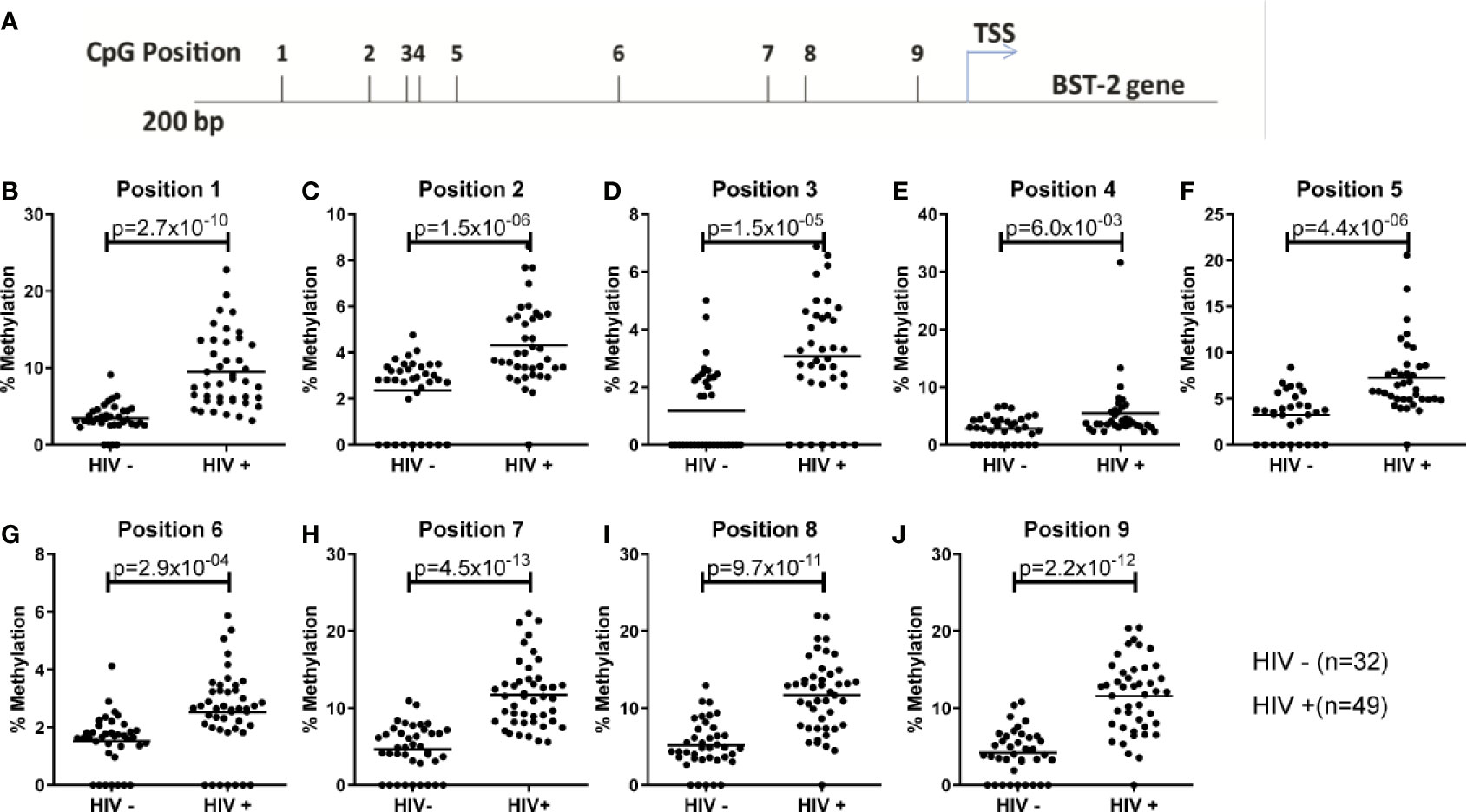
Figure 2 Examining DNA methylation levels across unmatched HIV uninfected and infected donors. (A) Location of nine CpG sites within the BST-2 promoter region 200bp upstream of the TSS. (B-J) Using HIV negative (FRESH) and HIV positive (SK) cohorts the percentage methylation, using pyrosequencing of bisulfite converted DNA, was calculated for each of the nine sites.
The average methylation across the nine CpG sites was compared to BST-2 mRNA expression levels in samples with four different HIV serostatus or disease stages, i.e. pre-infection, 3, 12- and >36-months’ post-infection using n=27 matched ARV-naïve samples, based on sample availability. An inverse correlation was observed at all time points; pre-infection (r=-0.52, p=0.0056; Figure 3A), 3 months (r=-0.50, p=0.0097; Figure 3B), 12 months (r=-0.44, p=0.02; Figure 3C) and >36 months (r=-0.46, p=0.0178; Figure 3D). These data strongly point to methylation as a major contributor in regulation of BST-2 expression levels.
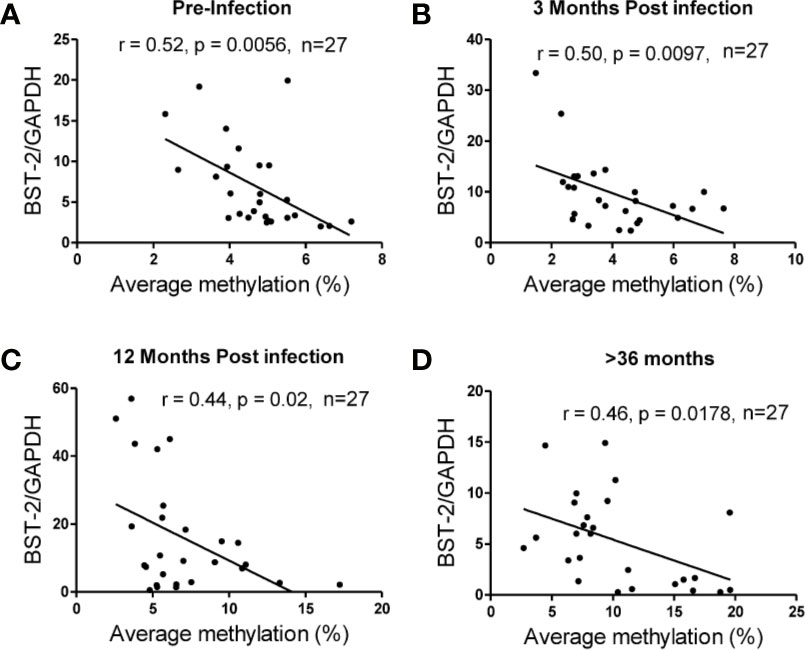
Figure 3 Correlation of DNA methylation and BST-2 mRNA expression levels across HIV disease. Average methylation was calculated as the average methylation level across nine sites within 200bp upstream of the transcription start site. A strong negative correlation was observed at each of the time points examined for a set of n=27 matched samples at varying time points across disease progression. (A) pre-infection (r=-0.52, p=0.0056), (B) 3 months’ post infection (r=-0.50, p=0.0097), (C) 12 months’ post infection (r=-0.44, p=0.02) and (D) >36 months (r=-0.46, p=0.0178).
Comparison of BST-2 DNA methylation pattern with mRNA expression levels indicate distinctions at the four timepoints. At the pre-infection time point, BST-2 expression levels are relatively low, with modest methylation of the gene. Three months after HIV infection, BST-2 expression levels increase with a concomitant decrease in methylation, perhaps as a result of the immune response in acute infection, including IFN-I production, which is known to enhance BST-2 production (3) (Figure 4). Methylation begins to increase at 12 months’ post infection, and by 36 months post infection, the mean expression of BST-2 dips to pre-infection levels while methylation is considerably higher than that at pre-infection timepoint (Figure 4). Overall, these results suggest that DNA methylation is not the sole contributor to BST-2 expression variation.
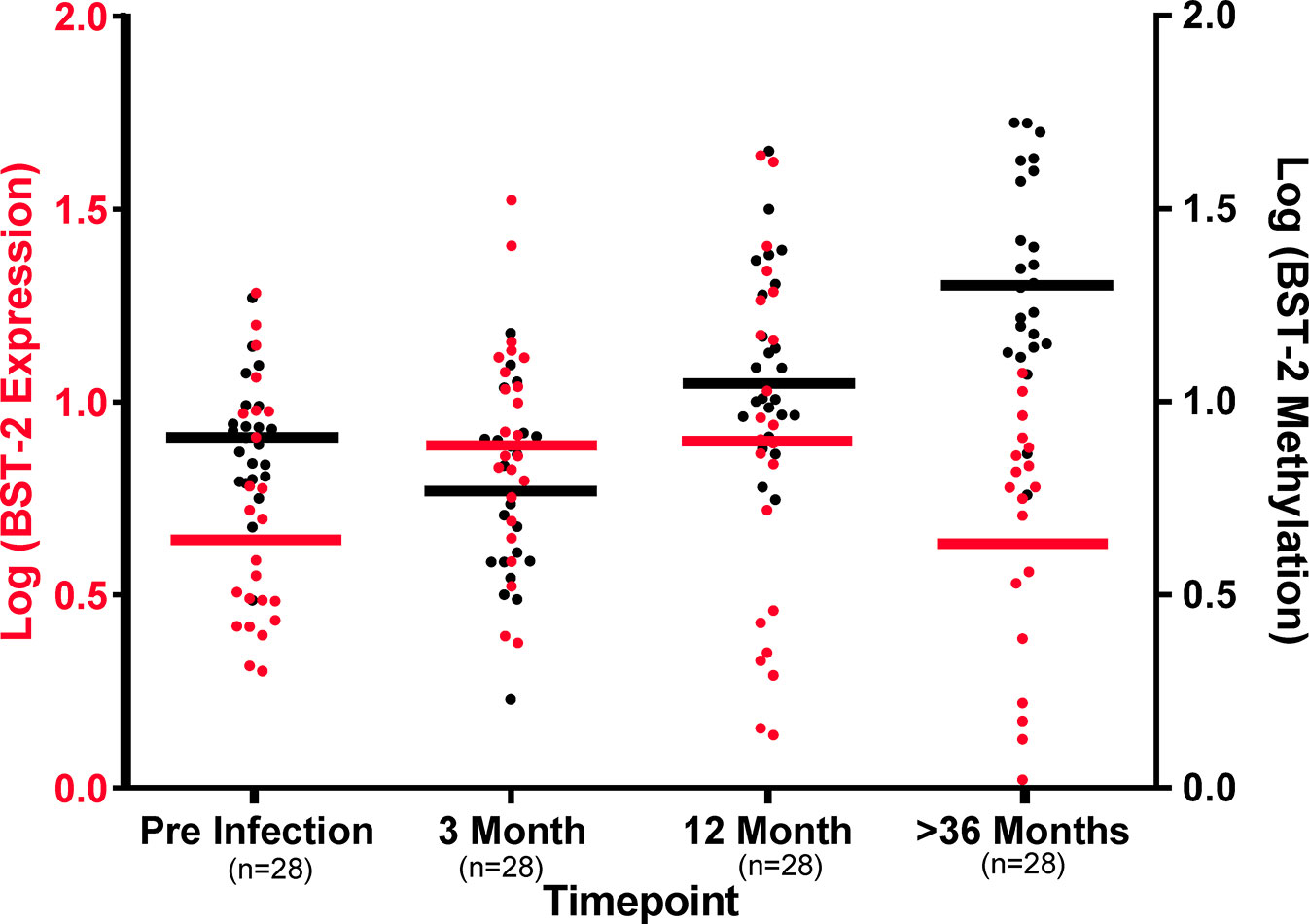
Figure 4 DNA methylation levels dictate BST-2 mRNA levels during HIV disease. Baseline levels of 27 matched samples at varying time points across disease progression show at the pre-infection higher methylation (Black) and low BST-2 expression (Red), while at acute infection (3-month post infection) methylation and expression levels are at similar level, due to IFN induction. The BST-2 expression and methylation levels invert at 12 months’ post infection. The most dramatic difference is observed at the >36 months timepoint, these individuals are at a chronic phase of infection, at this time point we observe the lowest expression and highest methylation.
We next examined the impact of BST-2 mRNA expression levels on HIV replication in vitro. HIV replication was assessed by the amount of p24 released into tissue culture supernatant following infection of PBMCs from HIV negative individuals either having the highest (n=11) or lowest (n=11) BST-2 mRNA expression levels screened from a cohort of 65 HIV negative donors. p24 measurements, taken at days 2, 4 and 7, showed that individuals with higher BST-2 expression (dotted line, Figure 5A) significantly associated with lower viral replication, at days 4 and 7 post infection, compared to lower BST-2 expression individuals (solid line, Figure 5A; ANOVA, p<0.001). Further, a negative correlation between HIV replication and BST-2 mRNA expression levels was observed at day 7 (r=-0.63, p=0.0019, Figure 5B). These data support a model in which higher BST-2 levels diminish HIV replication.
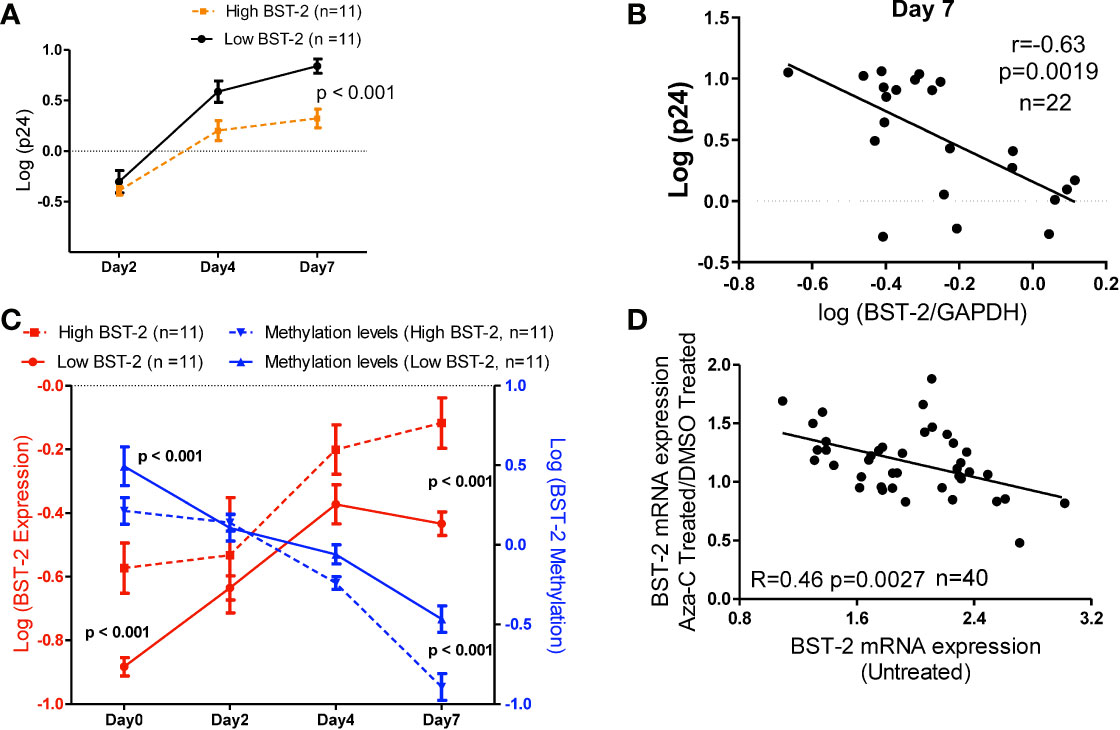
Figure 5 BST-2 mRNA expression and methylation levels correlate in an in vitro viral replication assay, and treatment with a DNA hypo-methylation drug increases BST-2 mRNA expression levels. (A) Individuals were pre-selected based on BST-2 expression levels for a HIV replication assay. PBMCs from HIV negative donors (n=22) were infected with HIV IIIB viral strain, the amount of virus present was determined by measuring the p24 antigen using an ELISA assay. Measurements of p24 for both high and low BST-2 donors were taken at days 2, 4 and 7. Donors with higher BST-2 levels (dotted line) had lower level of p24, while donors with lower BST-2 levels (solid line) had significantly higher p24 levels (p<0.001). (B) A negative correlation was observed when comparing the HIV replication levels against the BST-2 mRNA levels at day 7 from the in vitro HIV infection assay (r=-0.63, p=0.0019). (C) mRNA and DNA were used to measure BST-2 expression (red) and methylation levels (blue), respectively, from high and low BST-2 donors (n=22) at four time points during the viral replication assay, days 0, 2, 4 and 7. Within high BST-2 donors, we find high expression (red dotted line) associated with lower methylation (blue dotted line) and vice versa for low BST-2 levels, where low expression (red solid line) associated with higher methylation (blue solid line). (D) PBMCs from HIV negative donors (n=40) were split into three subsets; the first subset was treated with a DNA methyltransferase inhibitor that causes hypomethylation (5’-Aza-CdR), while the second subset was treated with DMSO. Both subsets were incubated for 24 hours. BST-2 mRNA expression from 5’-Aza-CdR and DMSO treated cells were compared and plotted as a fold change against BST-2 mRNA from an untreated time point (third subset), a significant correlation was observed (R=-0.46, p=0.0027).
Next, we tested whether DNA methylation correlated with BST-2 mRNA expression levels in an in vitro HIV infection assay. Individuals with high BST-2 expression levels (red dotted line, Figure 5C) possessed low methylation levels (blue dotted line) measured at days 0, 2, 4 and 7 days post HIV infection. Conversely, low BST-2 mRNA expression donors (red solid line, Figure 5C) associated with high methylation levels (blue solid line) throughout the time course. Further, the overall difference between the methylation levels within donors either possessing high or low BST-2 expression levels was significant (Figure 5C; ANOVA, p<0.001). Thus, BST-2 mRNA expression levels associate with the level of BST-2 DNA methylation, even within an in vitro time course of HIV infection.
5’-Aza-CdR induces hypomethylation due to its ability to inhibit DNA methyltransferases, the enzymes responsible for methylation. As manipulation of BST-2 expression could be considered as a therapeutic intervention in HIV disease, we tested whether 5’-Aza-CdR enhanced BST-2 expression differentially among donors as a function of the intrinsic expression level of BST-2. BST-2 levels were measured from HIV negative healthy donor PBMCs (n=40) treated with either 5’-Aza-CdR or DMSO (to measure baseline potential for stimulation in each subject). BST-2 ratios of 5’-Aza-CdR/DMSO treated mRNA levels were then plotted against the BST-2 levels measured in corresponding untreated PBMCs (Figure 5D). A negative correlation between levels of BST-2 mRNA expression in untreated and Aza-CdR treated PBMC (R=-0.46, p=0.0027; Figure 5D) was observed. Donors with the lowest intrinsic (i.e. untreated) BST-2 mRNA expression levels had the greatest increase in mRNA expression following 5’-Aza-CdR treatment. These data point directly to DNA methylation as a primary factor in regulating BST-2 gene expression. Increasing BST-2 gene expression by demethylation may therefore enhance resistance to HIV, given the observation that higher BST-2 expression associates with HIV control.
Here we show that expression levels of BST-2, a potent antiviral cellular protein are negatively associated with viral loads in an antiretroviral-naive cohort of women followed longitudinally from acute HIV-1 infection. Similar results were obtained from an ART-naive chronically infected cohort of participants with unknown time of infection. BST-2 levels are lower in chronically infected HIV individuals compared to uninfected persons, however in longitudinally followed matched samples, BST-2 levels first increase significantly over baseline and then decline slowly. We showed BST-2 expression and DNA methylation levels within the gene promoter region are negatively correlated. These findings are consistent in HIV infected subjects in studies performed ex vivo and in vitro. Moreover, we pharmacologically altered BST-2 expression levels by manipulating methylation levels with 5’-Aza-CdR, leading to an increase in BST-2 mRNA expression, especially within cells with lower intrinsic BST-2 levels.
BST-2 levels have been shown to inhibit the production of HIV-1 particles by hindering the release of virion progeny (5, 46). However, HIV-1 has developed the ability to counteract this mechanism through the accessory viral protein, Vpu. BST-2 is trafficked from the viral budding sites on the cell surface by a Vpu-mediated mechanism, which thereafter sequesters the host protein to a perinuclear compartment (47). Vpu-null or defective viruses are most prone to BST-2-mediated inhibition. Previous studies have demonstrated that BST-2 surface levels are elevated during acute infection and then progressively decrease throughout the stages of infection, even after initiation of ART (7, 8). In line with these findings, we observed an elevation in mRNA expression of BST-2 during acute infection both ex vivo and in vitro, with a subsequent decrease observed by 36 months post HIV-infection. The plasticity of BST-2 methylation observed suggests that methylation levels are a strong regulator of BST-2 expression even within a disease setting, although the mechanism regulating the methylation levels requires investigation.
Due to sample availability, we used bulk PBMCs to measure BST-2 mRNA expression, rather than CD4+ T cells specifically. A previous study measuring cell surface BST-2 showed no differences in expression patterns between individual cells types, PBMCs, mononuclear leukocytes, including CD4-positive, CD8-positive T lymphocytes, B cells, across stages of HIV infection (7). Although the level of mRNA expression does not always reflect protein expression levels, our Western blot assay in a small number of participants suggested a fair correlation. Sample limitations prevented us from examining the correlation between BST-2 mRNA levels and cellular surface expression, however, previous studies have demonstrated the correlation between these subsets (24, 48, 49). These studies have shown that BST-2 mRNA and protein levels correlate in mice, monkeys and humans. Furthermore, the studies also show specific tissues and cell types have strong correlations. The effect is observed across diseases (cancer, SIV, and Mouse mammary tumor virus) and healthy human controls (24, 48, 49).
It is plausible that other human HIV restriction factors could be regulated through DNA methylation. Each factor contributing toward the overall HIV disease outcome. Whole genome methylation analysis on a pair of monozygotic twins with discordant HIV status found several distinct differential methylation regions in the HIV infected twins (50, 51). Furthermore genome-wide methylation analysis of 85 unrelated individuals with varying HIV statuses showed differential genome-wide patterns which was associated with their ability to control HIV replication (52). Future studies should focus on larger cohorts of monozygotic twins or consider longitudinal studies such that the changes in DNA methylation profiles may be followed up at the different time points of HIV infection.
DNA methylation is just one of the mechanisms contributing to the variation in BST-2 expression levels. Another mechanism identified is a proposed regulatory variant, rs12609479, located in the BST-2 promoter region, which associated with decreased risk of acquiring HIV-1. The rs12609479-A allele associated with increased BST-2 expression and decreased risk of acquiring HIV-1 (53, 54). The 9 CpG sites that were examined in this study did not contain any polymorphisms and rs12609479 was not located in a CpG site. Despite rs12609479 not being affected by methylation, previous studies have shown diverse changes with respect to minor allele frequency across various ethnic groups (55, 56). Future studies are required to fully understand all the contributing factors responsible for BST-2 expression variation including methylation status across various ethnic groups. Despite these limitations, we found a reproducible association of BST-2 mRNA expression levels with HIV control. Our results were further validated with in vitro data.
In conclusion, we reproducibly demonstrate BST-2 expression levels associate with HIV viral control within a high disease burden setting. DNA methylation was shown to regulate BST-2 levels and observed to associate with HIV disease. The use of the demethylating drug 5’-Aza-CdR in vitro resulted in increased BST-2 expression levels among donors with low baseline expression levels. Thus, HIV control through higher BST-2 expression levels, as determined in part by decreased methylation, may suggest strategic mechanisms for HIV cure therapy.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by Biomedical Research Ethics Committee of the University of KwaZulu-Natal. The patients/participants provided their written informed consent to participate in this study.
RS, VR, MC and TN conceptualized the study. RS, VR, TN, NG, KM, KD, BW, and SK assisted with the cohort setup and proposal design. RS and VR performed the laboratory work. RS, VR, VN, and NY-Z performed the data analysis. RS, VR, MC and TN wrote the paper. All authors contributed to the article and approved the submitted version.
This study was supported by the South African Department of Science and Innovation/National Research Foundation Research Chairs Initiative (SARChI) grant to TN, a grant from the Swiss South Africa Joint Research Programme (SSAJRP) to TN. Partial funding for this work was received from the Gates Foundation, the International AIDS Vaccine Initiative (IAVI) (UKZNRSA1001) and Gilead Sciences (grant ID #00406). RS was supported by the Columbia University-Southern African Fogarty AIDS International Discovery and Research Program (AITRP) through the Fogarty International Center, National Institutes of Health (grant # D43TW000231). VR is a FLAIR Research Fellow (FLAIR Fellowship programme is partnership between the African Academy of Sciences and the Royal Society that is funded by the UK Government as part of the Global Challenge Research Fund [GCRF]), and was supported by the South African Medical Research Council (SAMRC) with funds from the Department of Science and Technology. This work was also supported in part through the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [grant # DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [grant # 107752/Z/15/Z], the Africa Health Research Institute (AHRI) Wellcome Strategic Core award [grant # 201433/Z16/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust or the UK government. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a shared affiliation, though no other collaboration, with one of the authors [MC].
We thank the study participants, CAPRISA and HPP clinical and laboratory staff for providing specimens. Special acknowledgments to the following members of the CAPRISA Acute Infection Study team.
1. Neil S, Bieniasz P. Human Immunodeficiency Virus, Restriction Factors, and Interferon. J Interferon Cytokine Res Off J Int Soc Interferon Cytokine Res (2009) 29:569–80. doi: 10.1089/jir.2009.0077
2. Jolly C. Cell-to-Cell Transmission of Retroviruses: Innate Immunity and Interferon-Induced Restriction Factors. Virology (2011) 411:251–9. doi: 10.1016/j.virol.2010.12.031
3. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I Interferons in Infectious Disease. Nat Rev Immunol (2015) 15:87–103. doi: 10.1038/nri3787
4. Sivro A, Su RC, Plummer FA, Ball TB. Interferon Responses in HIV Infection: From Protection to Disease. AIDS Rev (2014) 16:43–51.
5. Neil SJ, Zang T, Bieniasz PD. Tetherin Inhibits Retrovirus Release and is Antagonized by HIV-1 Vpu. Nature (2008) 451:425–30. doi: 10.1038/nature06553
6. Zhang J, Liang C. BST-2 Diminishes HIV-1 Infectivity. J Virol (2010) 84:12336–43. doi: 10.1128/JVI.01228-10
7. Homann S, Smith D, Little S, Richman D, Guatelli J. Upregulation of BST-2/Tetherin by HIV Infection In Vivo. J Virol (2011) 85:10659–68. doi: 10.1128/JVI.05524-11
8. Mous K, Jennes W, Camara M, Seydi M, Daneau G, Mboup S, et al. Expression Analysis of LEDGF/p75, APOBEC3G, TRIM5alpha, and Tetherin in a Senegalese Cohort of HIV-1-exposed Seronegative Individuals. PloS One (2012) 7:e33934. doi: 10.1371/journal.pone.0033934
9. Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, et al. The Interferon-Induced Protein BST-2 Restricts HIV-1 Release and is Downregulated From the Cell Surface by the Viral Vpu Protein. Cell Host Microbe (2008) 3:245–52. doi: 10.1016/j.chom.2008.03.001
10. Miyagi E, Andrew AJ, Kao S, Strebel K. Vpu Enhances HIV-1 Virus Release in the Absence of Bst-2 Cell Surface Down-Modulation and Intracellular Depletion. Proc Natl Acad Sci USA (2009) 106:2868–73. doi: 10.1073/pnas.0813223106
11. Goffinet C, Allespach I, Homann S, Tervo HM, Habermann A, Rupp D, et al. HIV-1 Antagonism of CD317 is Species Specific and Involves Vpu-Mediated Proteasomal Degradation of the Restriction Factor. Cell Host Microbe (2009) 5:285–97. doi: 10.1016/j.chom.2009.01.009
12. Pan XB, Han JC, Cong X, Wei L. BST2/Tetherin Inhibits Dengue Virus Release From Human Hepatoma Cells. PloS One (2012) 7:e51033. doi: 10.1371/journal.pone.0051033
13. Lopez LA, Yang SJ, Exline CM, Rengarajan S, Haworth KG, Cannon PM. Anti-Tetherin Activities of HIV-1 Vpu and Ebola Virus Glycoprotein do Not Involve Removal of Tetherin From Lipid Rafts. J Virol (2012) 86:5467–80. doi: 10.1128/JVI.06280-11
14. Jones PH, Maric M, Madison MN, Maury W, Roller RJ, Okeoma CM. BST-2/Tetherin-Mediated Restriction of Chikungunya (CHIKV) VLP Budding is Counteracted by CHIKV non-Structural Protein 1 (nsP1). Virology (2013) 438:37–49. doi: 10.1016/j.virol.2013.01.010
15. Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, et al. Ebola Virus Glycoprotein Counteracts BST-2/Tetherin Restriction in a Sequence-Independent Manner That Does Not Require Tetherin Surface Removal. J Virol (2010) 84:7243–55. doi: 10.1128/JVI.02636-09
16. Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, Fruh K. The Great Escape: Viral Strategies to Counter BST-2/Tetherin. PloS Pathog (2010) 6:e1000913. doi: 10.1371/journal.ppat.1000913
17. Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, et al. But Not Filoviruses, is Restricted by BST-2/Tetherin. J Virol (2010) 84:10569–80. doi: 10.1128/JVI.00103-10
18. Yondola MA, Fernandes F, Belicha-Villanueva A, Uccelini M, Gao Q, Carter C, et al. Budding Capability of the Influenza Virus Neuraminidase can be Modulated by Tetherin. J Virol (2011) 85:2480–91. doi: 10.1128/JVI.02188-10
19. Miyakawa K, Matsunaga S, Watashi K, Sugiyama M, Kimura H, Yamamoto N, et al. Molecular Dissection of HBV Evasion From Restriction Factor Tetherin: A New Perspective for Antiviral Cell Therapy. Oncotarget (2015) 6:21840–52. doi: 10.18632/oncotarget.4808
20. Amet T, Byrd D, Hu N, Sun Q, Li F, Zhao Y, et al. BST-2 Expression in Human Hepatocytes is Inducible by All Three Types of Interferons and Restricts Production of Hepatitis C Virus. Curr Mol Med (2014) 14:349–60. doi: 10.2174/1566524013666131118111719
21. Mahauad-Fernandez WD, Okeoma CM. The Role of BST-2/Tetherin in Host Protection and Disease Manifestation. Immunity Inflammation Dis (2016) 4:4–23. doi: 10.1002/iid3.92
22. Fang KH, Kao HK, Chi LM, Liang Y, Liu SC, Hseuh C, et al. Overexpression of BST2 is Associated With Nodal Metastasis and Poorer Prognosis in Oral Cavity Cancer. Laryngoscope (2014) 124:E354–60. doi: 10.1002/lary.24700
23. Wang W, Nishioka Y, Ozaki S, Jalili A, Abe S, Kakiuchi S, et al. Hm1.24 (CD317) is a Novel Target Against Lung Cancer for Immunotherapy Using anti-HM1.24 Antibody. Cancer Immunol Immunother CII (2009) 58:967–76. doi: 10.1007/s00262-008-0612-4
24. Mahauad-Fernandez WD, Borcherding NC, Zhang W, Okeoma CM. Bone Marrow Stromal Antigen 2 (BST-2) DNA is Demethylated in Breast Tumors and Breast Cancer Cells. PloS One (2015) 10:e0123931. doi: 10.1371/journal.pone.0123931
25. Sayeed A, Luciani-Torres G, Meng Z, Bennington JL, Moore DH, Dairkee SH. Aberrant Regulation of the BST2 (Tetherin) Promoter Enhances Cell Proliferation and Apoptosis Evasion in High Grade Breast Cancer Cells. PloS One (2013) 8:e67191. doi: 10.1371/journal.pone.0067191
26. Milutin Gasperov N, Farkas SA, Nilsson TK, Grce M. Epigenetic Activation of Immune Genes in Cervical Cancer. Immunol Lett (2014) 162:256–7. doi: 10.1016/j.imlet.2014.09.019
27. Staudinger M, Glorius P, Burger R, Kellner C, Klausz K, Gunther A, et al. The Novel Immunotoxin HM1.24-ETA’ Induces Apoptosis in Multiple Myeloma Cells. Blood Cancer J (2014) 4:e219. doi: 10.1038/bcj.2014.38
28. Yokoyama T, Enomoto T, Serada S, Morimoto A, Matsuzaki S, Ueda Y, et al. Plasma Membrane Proteomics Identifies Bone Marrow Stromal Antigen 2 as a Potential Therapeutic Target in Endometrial Cancer. Int J Cancer (2013) 132:472–84. doi: 10.1002/ijc.27679
29. Wainwright DA, Balyasnikova IV, Han Y, Lesniak MS. The Expression of BST2 in Human and Experimental Mouse Brain Tumors. Exp Mol Pathol (2011) 91:440–6. doi: 10.1016/j.yexmp.2011.04.012
30. Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-Wide DNA Methylation Patterns in CD4+ T Cells From Patients With Systemic Lupus Erythematosus. Epigenetics (2011) 6:593–601. doi: 10.4161/epi.6.5.15374
31. Gornalusse GG, Mummidi S, Gaitan AA, Jimenez F, Ramsuran V, Picton A, et al. Epigenetic Mechanisms, T-cell Activation, and CCR5 Genetics Interact to Regulate T-cell Expression of CCR5, the Major HIV-1 Coreceptor. Proc Natl Acad Sci USA (2015) 112:E4762–71. doi: 10.1073/pnas.1423228112
32. Ramsuran V, Kulkarni S, O’Huigin C, Yuki Y, Augusto DG, Gao X, et al. Epigenetic Regulation of Differential HLA-A Allelic Expression Levels. Hum Mol Genet (2015) 24:4268–75. doi: 10.1093/hmg/ddv158
33. Pion M, Jaramillo-Ruiz D, Martínez A, Muñoz-Fernández MA, Correa-Rocha R. HIV Infection of Human Regulatory T Cells Downregulates FOXP3 Expression by Increasing DNMT3b Levels and DNA Methylation in the FOXP3 Gene. AIDS (2013) 27:2019–29. doi: 10.1097/QAD.0b013e32836253fd
34. Kelly HA, Chikandiwa A, Warman R, Segondy M, Sawadogo B, Vasiljevic N, et al. Associations of Human Gene EPB41L3 DNA Methylation and Cervical Intraepithelial Neoplasia in Women Living With HIV-1 in Africa. AIDS (London England) (2018) 32:2227–36. doi: 10.1097/QAD.0000000000001932
35. Nakayama-Hosoya K, Ishida T, Youngblood B, Nakamura H, Hosoya N, Koga M, et al. Epigenetic Repression of Interleukin 2 Expression in Senescent CD4+ T Cells During Chronic HIV Type 1 Infection. J Infect Dis (2015) 211:28–39. doi: 10.1093/infdis/jiu376
36. Ramsuran V, Naranbhai V, Horowitz A, Qi Y, Martin MP, Yuki Y, et al. Elevated HLA-A Expression Impairs HIV Control Through Inhibition of NKG2A-expressing Cells. Science (2018) 359:86–90. doi: 10.1126/science.aam8825
37. Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant Influence of HLA-B in Mediating the Potential Co-Evolution of HIV and HLA. Nature (2004) 432:769–75. doi: 10.1038/nature03113
38. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal Bacteria are a Major Modulator of Host Inflammatory Responses in the Female Genital Tract. Immunity (2015) 42:965–76. doi: 10.1016/j.immuni.2015.04.019
39. Dong KL, Moodley A, Kwon DS, Ghebremichael MS, Dong M, Ismail N, et al. Detection and Treatment of Fiebig Stage I HIV-1 Infection in Young At-Risk Women in South Africa: A Prospective Cohort Study. Lancet HIV (2018) 5:e35–44. doi: 10.1016/S2352-3018(17)30146-7
40. van Loggerenberg F, Mlisana K, Williamson C, Auld SC, Morris L, Gray CM, et al. Establishing a Cohort At High Risk of HIV Infection in South Africa: Challenges and Experiences of the CAPRISA 002 Acute Infection Study. PloS One (2008) 3:e1954. doi: 10.1371/journal.pone.0001954
41. Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, Mansoor LE, et al. Effectiveness and Safety of Tenofovir Gel, an Antiretroviral Microbicide, for the Prevention of HIV Infection in Women. Science (2010) 329:1168–74. doi: 10.1126/science.1193748
42. Sewram S, Singh R, Kormuth E, Werner L, Mlisana K, Karim SS, et al. Human TRIM5alpha Expression Levels and Reduced Susceptibility to HIV-1 Infection. J Infect Dis (2009) 199:1657–63. doi: 10.1086/598861
43. Li LC, Dahiya R. MethPrimer: Designing Primers for Methylation Pcrs. Bioinformatics (2002) 18:1427–31. doi: 10.1093/bioinformatics/18.11.1427
44. O’Connor BP. Simple and Flexible SAS and SPSS Programs for Analyzing Lag-Sequential Categorical Data. Behav Res Methods Instruments Comput J Psychonomic Society Inc (1999) 31:718–26. doi: 10.3758/BF03200753
45. Motulsky HJ. Common Misconceptions About Data Analysis and Statistics. Br J Pharmacol (2015) 172:2126–32. doi: 10.1111/bph.12884
46. Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An Interferon-Alpha-Induced Tethering Mechanism Inhibits HIV-1 and Ebola Virus Particle Release but is Counteracted by the HIV-1 Vpu Protein. Cell Host Microbe (2007) 2:193–203. doi: 10.1016/j.chom.2007.08.001
47. Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, et al. Hiv-1 Vpu and HIV-2 Env Counteract BST-2/Tetherin by Sequestration in a Perinuclear Compartment. Retrovirology (2010) 7:51. doi: 10.1186/1742-4690-7-51
48. Jones PH, Mahauad-Fernandez WD, Madison MN, Okeoma CM. Bst-2/tetherin is Overexpressed in Mammary Gland and Tumor Tissues in MMTV-induced Mammary Cancer. Virology (2013) 444:124–39. doi: 10.1016/j.virol.2013.05.042
49. Mussil B, Javed A, Topfer K, Sauermann U, Sopper S. Increased BST2 Expression During Simian Immunodeficiency Virus Infection is Not a Determinant of Disease Progression in Rhesus Monkeys. Retrovirology (2015) 12:92. doi: 10.1186/s12977-015-0219-8
50. Zhang Y, Li S-K, Tsui S K-WJ. Genome-Wide Analysis of DNA Methylation Associated With HIV Infection Based on a Pair of Monozygotic Twins. Genome Data (2015) 6:12–5. doi: 10.1016/j.gdata.2015.07.024
51. Zhang Y, Li S-K, Yang KY, Liu M, Lee N, Tang X, et al. Whole Genome Methylation Array Reveals the Down-Regulation of IGFBP6 and SATB2 by HIV-1. Sci Rep (2015) 5:1–14. doi: 10.1038/srep10806
52. Moron-Lopez S, Urrea V, Dalmau J, Lopez M, Puertas MC, Ouchi D, et al. The Genome-wide Methylation Profile of CD4+ T Cells From Individuals With Human Immunodeficiency Virus (HIV) Identifies Distinct Patterns Associated With Disease Progression. Clin Infect Dis (2020). doi: 10.1093/cid/ciaa1047
53. Hancock DB, Gaddis NC, Levy JL, Bierut LJ, Kral AH, Johnson EO. Associations of Common Variants in the BST2 Region With HIV-1 Acquisition in African American and European American People Who Inject Drugs. AIDS (2015) 29:767–77. doi: 10.1097/QAD.0000000000000604
54. Skelton MM, Kampira EE, Wonkam AA, Mhandire KK, Kumwenda JJ, Duri KK, et al. Frequency Variation Among sub-Saharan Populations in Virus Restriction Gene, BST-2 Proximal Promoter Polymorphisms: Implications for HIV-1 Prevalence Differences Among African Countries. OMICS (2014) 18:461–71. doi: 10.1089/omi.2013.0127
55. Dias BDC, Paximadis M, Martinson N, Chaisson RE, Ebrahim O, Tiemessen CT. The Impact of Bone Marrow Stromal Antigen-2 (BST2) Gene Variants on HIV-1 Control in Black South African Individuals. Infect Genet Evol J Mol Epidemiol Evolutionary Genet Infect Dis (2020) 80:104216. doi: 10.1016/j.meegid.2020.104216
Keywords: BST-2, HIV-1, DNA methylation, epigenetic regulation, expression
Citation: Singh R, Ramsuran V, Naranbhai V, Yende-Zuma N, Garrett N, Mlisana K, Dong KL, Walker BD, Abdool Karim SS, Carrington M and Ndung’u T (2021) Epigenetic Regulation of BST-2 Expression Levels and the Effect on HIV-1 Pathogenesis. Front. Immunol. 12:669241. doi: 10.3389/fimmu.2021.669241
Received: 18 February 2021; Accepted: 20 April 2021;
Published: 05 May 2021.
Edited by:
Ping An, Frederick National Laboratory for Cancer Research (NIH), United StatesReviewed by:
Krishnakumar Devadas, Center for Biologics Evaluation and Research (FDA), United StatesCopyright © 2021 Singh, Ramsuran, Naranbhai, Yende-Zuma, Garrett, Mlisana, Dong, Walker, Abdool Karim, Carrington and Ndung’u. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thumbi Ndung’u, dGh1bWJpLm5kdW5ndUBhaHJpLm9yZw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.