
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 27 May 2021
Sec. Autoimmune and Autoinflammatory Disorders
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.667690
Background: Although more and more evidence has supported psoriasis is prone to atherosclerosis, the common mechanism of its occurrence is still not fully elucidated. The purpose of this study is to further explore the molecular mechanism of the occurrence of this complication.
Methods: The gene expression profiles of psoriasis (GSE30999) and atherosclerosis (GSE28829) were downloaded from the Gene Expression Omnibus (GEO) database. After identifying the common differentially expressed genes (DEGs) of psoriasis and atherosclerosis, three kinds of analyses were performed, namely functional annotation, protein‐protein interaction (PPI) network and module construction, and hub gene identification and co-expression analysis.
Results: A total of 94 common DEGs (24 downregulated genes and 70 upregulated genes) was selected for subsequent analyses. Functional analysis emphasizes the important role of chemokines and cytokines in these two diseases. In addition, lipopolysaccharide-mediated signaling pathway is closely related to both. Finally, 16 important hub genes were identified using cytoHubba, including LYN, CSF2RB, IL1RN, RAC2, CCL5, IRF8, C1QB, MMP9, PLEK, PTPRC, FYB, BCL2A1, LCP2, CD53, NCF2 and TLR2.
Conclusions: Our study reveals the common pathogenesis of psoriasis and atherosclerosis. These common pathways and hub genes may provide new ideas for further mechanism research.
More and more evidence show that psoriasis increases the relative risk of atherosclerosis (about 25%) and is independent of risk factors, such as smoking, obesity, and hyperlipidemia (1). Patients with a course of psoriasis more than 8 years are more likely to develop atherosclerosis than non-psoriatic patients (2). Atherosclerosis is the pathological basis of psoriasis complicated with cardiovascular disease. Atherosclerosis and psoriasis have many overlapping inflammatory environmental factors, including local and systemic immune processes, inflammatory cytokines/chemokines, such as serum TNF-α, vascular endothelial growth factor, IL12, monocyte chemotaxis Protein 1, S100A8/A9 and significantly elevated IL17A, etc. (3).
Although psoriasis is considered as a risk factor for developing atherosclerosis, the exact mechanisms that explain the coexistence of these two disorders remain unclear. The IL23/IL17 axis could be the most fundamental pathway. Psoriasis and atherosclerosis have overlapping mechanisms in pathogenic pathways. In psoriasis, myeloid dendritic cells secrete IL12 and IL23, prompting T cells to differentiate into Th1 and Th17 cell subtypes; Th1 cells secrete TNF-α and IFN-γ, which promote the activation and proliferation of keratinocytes and the expression of adhesion molecules such as intercellular adhesion molecule 1 (4); Th17 cells secrete IL17 and IL22 to promote the proliferation of keratinocytes and angiogenesis in the lesion (5). In atherosclerosis, the activation of endothelial cells in the plaques of neonatal arteries promotes monocyte and lymphocyte overflow, and granulocyte-macrophage colony stimulating factor can promote the synthesis of IL12 and IL23 by macrophages and dendritic cells, reducing BCL2 and activating Th1 and Th17 responses (6). Th1 cells secrete TNF-α and IFN-γ to promote the further growth of atherosclerotic plaques, while Th17 cells secrete IL17 and IL22, and the release of reactive oxygen species increases, which promotes the formation of new blood vessels and intraplaque hemorrhage; Elevated IL17 levels in plaques may further weaken the fibrous caps in atherosclerotic plaques, leading to plaque rupture and myocardial infarction (1, 7).
The common transcription feature may provide new insights into the common pathogenesis of psoriasis and atherosclerosis. The purpose of this study is to identify hub genes related to the pathogenesis of psoriasis complicated with atherosclerosis. We analyzed two gene expression data sets (GSE30999 and GSE28829) downloaded from the GEO database. Comprehensive bioinformatics and enrichment analysis were used to determine the common DEGs and its functions of psoriasis and atherosclerosis. In addition, PPI network was constructed to analyze gene modules and identify hub genes by using STRING database and Cytoscape software. In the end, we identified 16 important hub genes, and we further analyzed the transcription factors of these genes and verified their expression. The hub genes identified here between psoriasis and atherosclerosis are expected to provide new insights into the biological mechanisms of these two diseases.
GEO (http://www.ncbi.nlm.nih.gov/geo) (8) is a public database containing a large number of high-throughput sequencing and microarray data sets submitted by research institutes worldwide. We searched for related gene expression datasets using psoriasis and atherosclerosis as keywords. The inclusion criteria are set as: two independent expression profiles come from the same sequencing platform and contain the largest sample size. In addition, the test specimens included should be from humans. finally, two microarray datasets [GSE30999 (9) and GSE28829 (10)] were downloaded from it (Affymetrix GPL570 platform, Affymetrix Human Genome U133 Plus 2.0 Array). The GSE30999 dataset contains 85 paired psoriasis patients with skin lesions (LS) and adjacent normal tissues (NL). GSE28829 consists of 13 early atherosclerotic plaque samples (EA) and 16 advanced atherosclerotic plaque samples (AA) from the human carotid artery.
GEO2R (www.ncbi.nlm.nih.gov/geo/ge2r) (11) is an online analysis tool developed based on two R packages (GEOquery and Limma). The GEOquery package is used to read data, and the Limma package is used to calculate the differential expression multiple. We used GEO2R to compare gene expression profiles between different groups to determine the DEGs between the diseased group and the control group. Probe sets with no corresponding gene symbols or genes with more than one probe set were removed or averaged, respectively. Only the genes with adjusted P-value < 0.05 and |logFC (fold change) | ≥ 1 were identified as DEGs. The online Venn diagram tool was used to obtain their common DEGs.
Gene ontology (GO) is a database established by the Gene Ontology Federation, which provides simple annotations of gene products from functions, biological pathways involved, and location in cells. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway is a database dedicated to storing information on gene pathways in different species. KEGG Orthology Based Annotation System (KOBAS) (http://kobas.cbi.pku.edu.cn) (12) is a Web server for gene/protein functional annotation and functional enrichment developed by Peking University, which collects 4325 species functional annotation information. The enrichment analysis results of GO and KEGG Pathway were obtained from the KOBAS 3.0 database. Adjusted P-value < 0.05 was considered significant.
Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) (version 11.0) (13) can search for the relationship between proteins of interest, such as direct binding relationships, or coexisting upstream and downstream regulatory pathways, to construct a PPI network with complex regulatory relationships. Interactions with a combined score over 0.4 were considered statistically significant. Cytoscape (http://www.cytoscape.org) (version 3.7.2) (14) was used to visualize this PPI network. Cytoscape’s plug-in molecular complex detection technology (MCODE) was used to analyze key functional modules. Set the selection criteria as: K-core = 2, degree cutoff = 2, max depth = 100, and node score cutoff = 0.2. Then the KEGG and GO analysis of the involved modular genes were performed with KOBAS 3.0.
The hub genes were identified by using the cytoHubba plug-in of Cytoscape. Here, we used seven common algorithms (MCC, MNC, Degree, Closeness, Radiality, Stress, EPC) to evaluate and select hub genes. Subsequently, we constructed a co-expression network of these hub genes via GeneMANIA (http://www.genemania.org/) (15), which is a reliable tool for identifying internal associations in gene sets.
The mRNA expression of identified hub genes was verified in GSE14905 (16) and GSE100927 (17). The GSE14905 dataset contains 33 LS, 28 NL and 21 normal skin (NS). GSE100927 consists of 69 AA and 35 control artery samples (CA) from the human artery. The comparison between the two sets of data was performed with the T-test. P-value < 0.05 was considered significant.
Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) (18) is a database for the prediction of transcriptional regulatory networks, which contains the target genes corresponding to TFs and the regulatory relationships between TFs. TRRUST currently includes two species: human and mouse, which contain 8,444 and 6,552 TFs target regulatory relationships of 800 human TFs and 828 mouse TFs, respectively. TFs that regulate the hub genes were obtained through the TRRUST database, and adjusted P-value < 0.05 was considered significant. Subsequently, we verified the expression levels of these TFs in GSE30999 and GSE28829 with the T-test.
The research flowchart of this research was shown in Figure 1. After standardizing the microarray results, DEGs (3318 in GSE30999 and 338 in GSE28829) were identified (Figures 2A, B). After taking the intersection of the Venn diagram, 105 common DEGs were obtained (Figure 2C). Subsequently, we obtained 94 DEGs (Table S1) after excluding genes with opposite expression trends in GSE30999 and GSE28829.
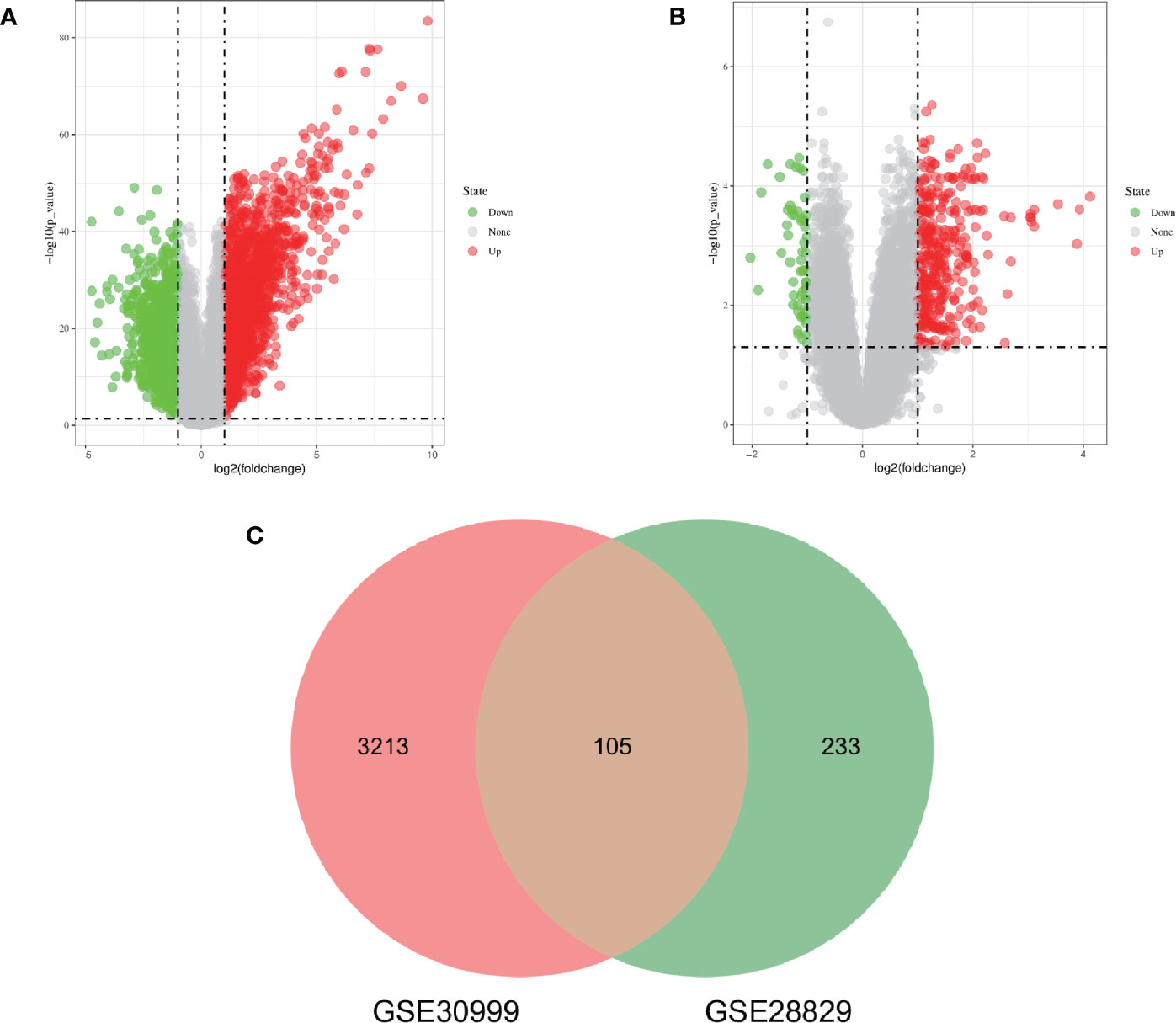
Figure 2 Volcano diagram and Venn diagram. (A) The volcano map of GSE30999. (B) The volcano map of GSE28829. Upregulated genes are marked in light red; downregulated genes are marked in light green. (C) The two datasets showed an overlap of 105 DEGs.
In order to analyze the biological functions and pathways involved in the 94 common DEGs, GO and KEGG Pathway enrichment analysis were performed. GO analysis results show that these genes were mainly enriched in protein binding (P = 7.11E-15), neutrophil degranulation (P = 5.81E-10), immune response (P = 1.55E-07), chemotaxis (P = 3.91E-06) and cytokine-mediated signaling pathway (P = 5.19E-06) (Figure 3B). In terms of KEGG Pathway, the three significant enrichment pathways are chemokine signaling pathway (P = 7.44E-05), cytokine-cytokine receptor interaction (P = 4.68E-04) and leukocyte transendothelial migration (P = 4.68E-04) (Figure 3C). These results strongly indicate that chemokines and cytokines are jointly involved in the occurrence and development of these two inflammatory diseases.
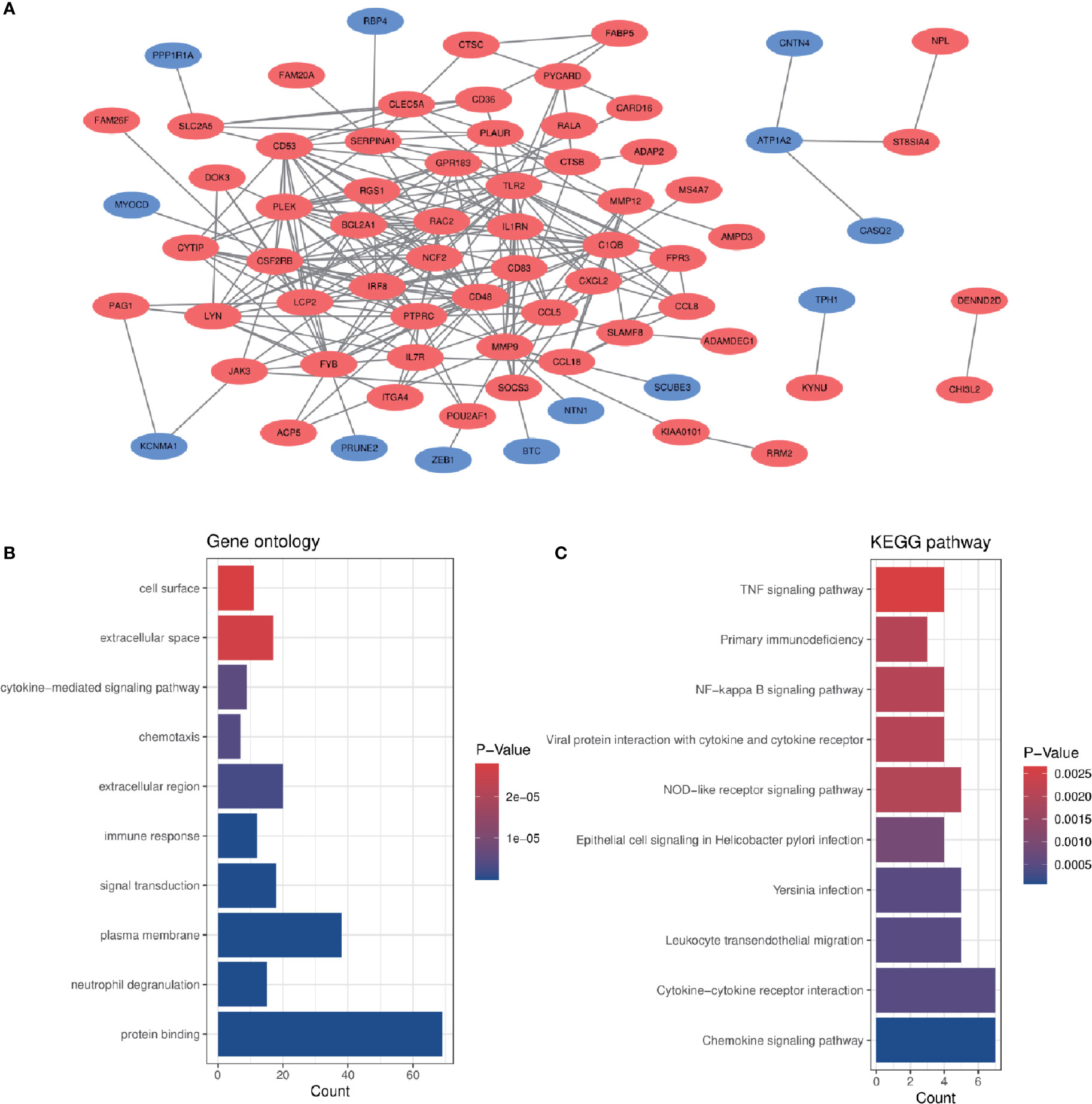
Figure 3 PPI network and common DEGs enrichment analysis results. (A) PPI network diagram. Red indicates up-regulated genes, and blue-violet indicates down-regulated genes. (B, C) The enrichment analysis results of GO and KEGG Pathway. Adjusted P-value < 0.05 was considered significant.
The PPI network of the common DEGs with combined scores greater than 0.4 was constructed using Cytoscape, which contained 72 nodes and 230 interaction pairs (Figure 3A). Three closely connected gene modules were obtained through MCODE plug-in of Cytoscape, including 24 common DEGs and 59 interaction pairs (Figures 4A–C). GO analysis showed that these genes are related to inflammation and immune response (Figure 4D). KEGG Pathway analysis showed that them to be mainly involved in TNF signaling pathway, cytokine-cytokine receptor interaction and leukocyte transendothelial migration (Figure 4E).
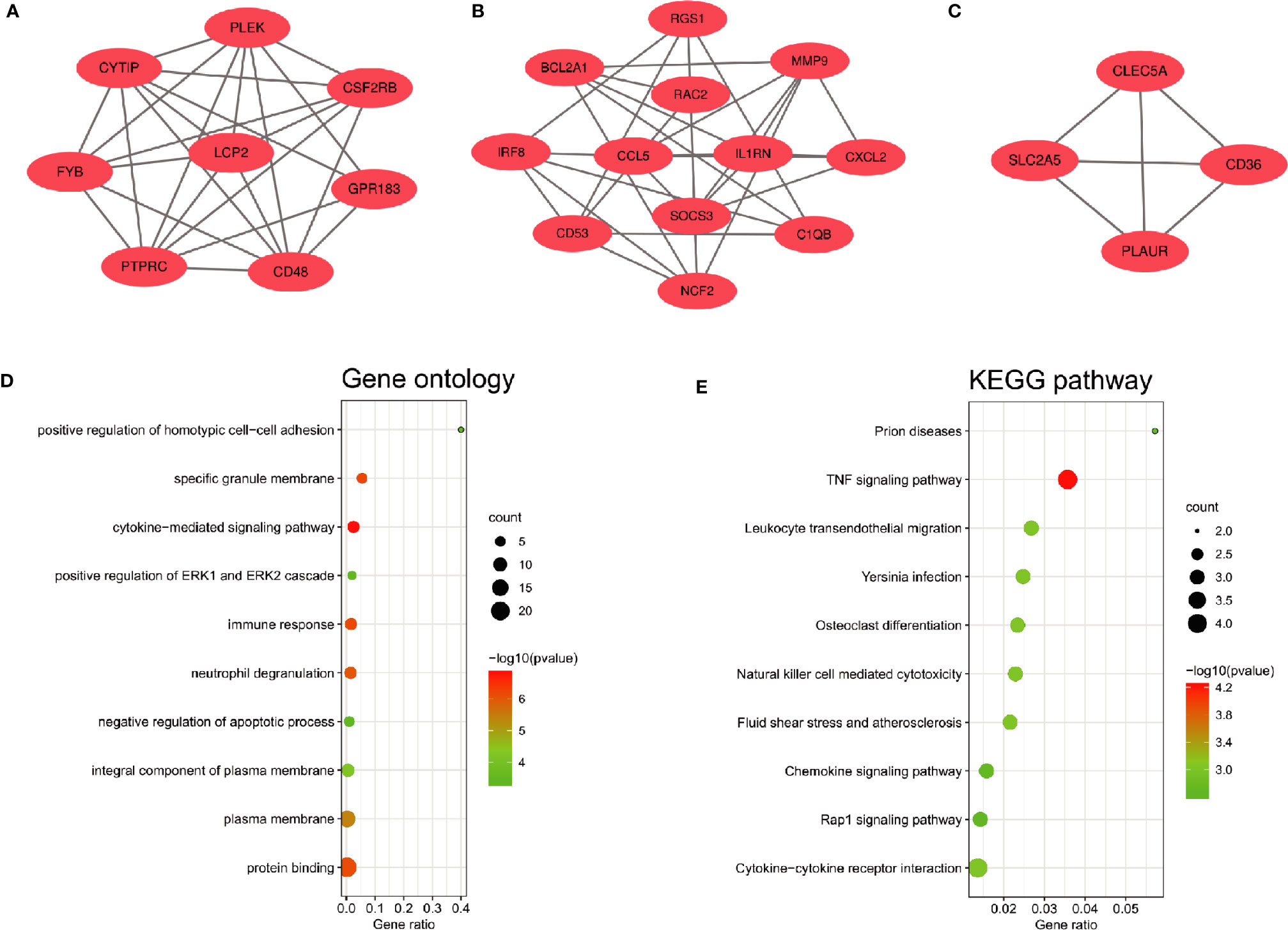
Figure 4 Significant gene module and enrichment analysis of the modular genes (A–C) Three significant gene clustering modules. (D, E) GO and KEGG enrichment analysis of the modular genes. The size of the circle represents the number of genes involved, and the abscissa represents the frequency of the genes involved in the term total genes.
Through the seven algorithms of plug-in cytoHubba, we have calculated the top 20 hub genes (Table 1). After taking the intersection of the Venn diagrams, we found 16 common hub genes, including LYN, CSF2RB, IL1RN, RAC2, CCL5, IRF8, C1QB, MMP9, PLEK, PTPRC, FYB, BCL2A1, LCP2, CD53, NCF2 and TLR2 (Figure 5A). Table 2 shows their full names and related functions. Based on the GeneMANIA database, we analyzed the co-expression network and related functions of these genes. These genes showed the complex PPI network with the co-expression of 80.18%, physical interactions of 11.58%, co-localization of 4.81%, predicted of 2.45% and pathway of 0.99% (Figure 5B). GO analysis showed that these genes are mainly involved in protein binding, lipopolysaccharide-mediated signaling pathway, immune response, cytokine-mediated signaling pathway and cellular response to interferon-gamma (Figure 6A). These results emphasized the important role of lipopolysaccharide and cytokines in these two diseases. In addition, KEGG Pathway analysis showed that they are mainly involved in Fc epsilon RI signaling pathway, Fc gamma R-mediated phagocytosis, leukocyte transendothelial migration and chemokine signaling pathway. Interestingly, three genes (NCF2, RAC2 and MMP9) were also involved in fluid shear stress and atherosclerosis (Figure 6B).
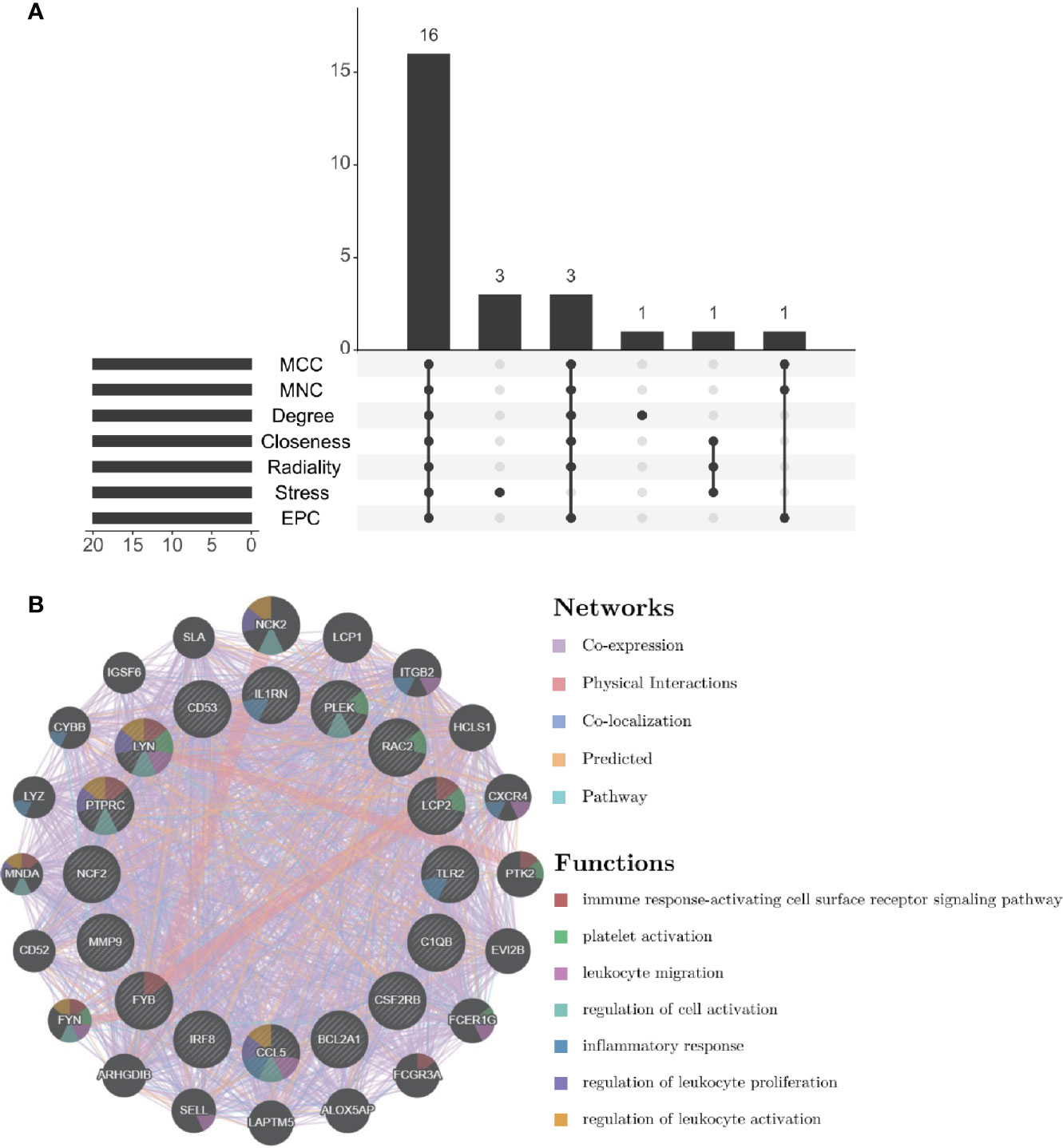
Figure 5 Venn diagram and co-expression network of hub genes. (A) The Venn diagram showed that seven algorithms have screened out 16 overlapping hub genes. (B) Hub genes and their co-expression genes were analyzed via GeneMANIA.
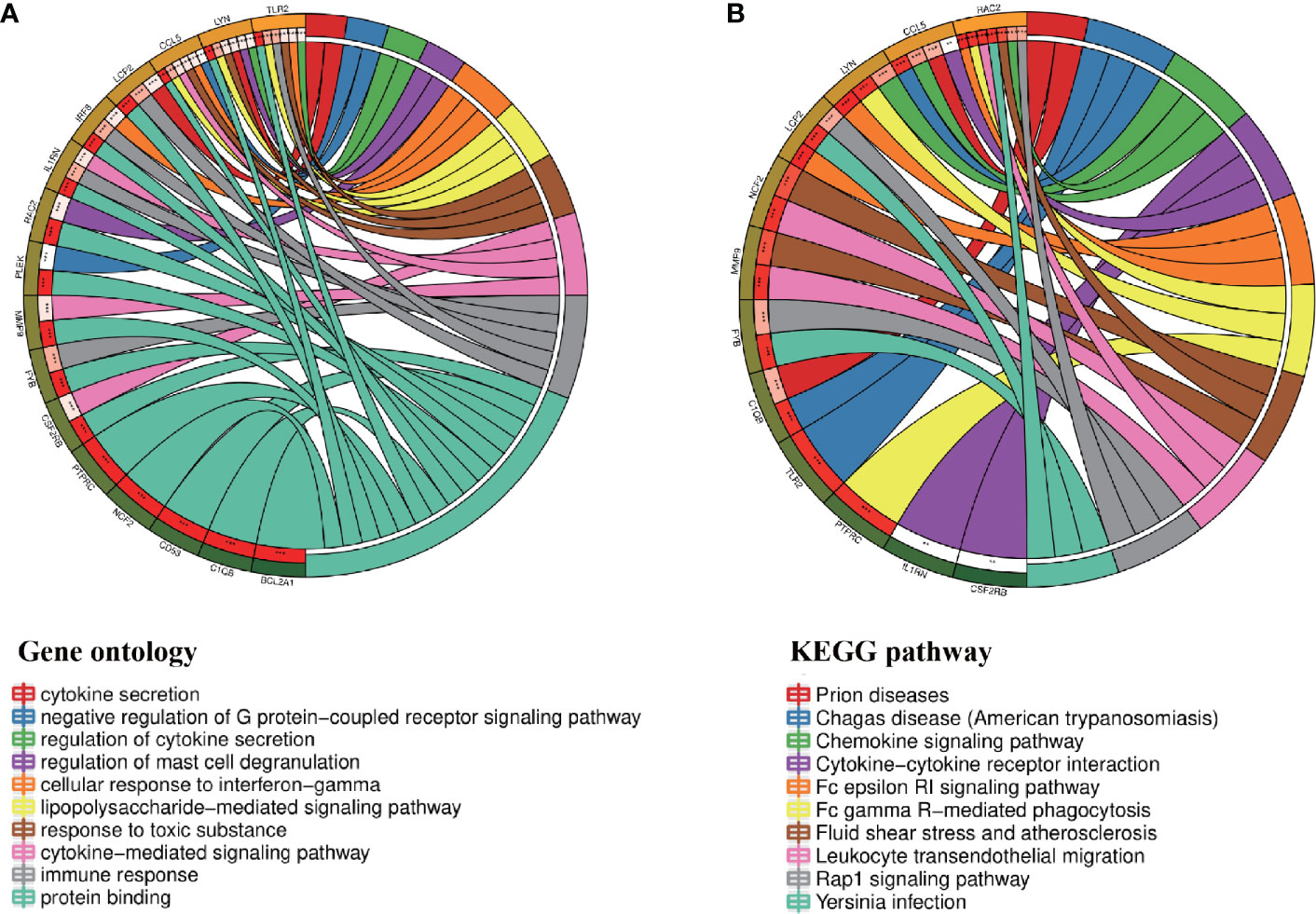
Figure 6 Enrichment analysis of the hub genes (A, B) GO and KEGG enrichment analysis of the hub genes. The outermost circle is term on the right and the inner circle on the left represents the significant P-value of the corresponding pathway of the gene.
In order to verify the reliability of these hub gene expression levels. We selected two other data sets containing psoriasis lesions and atherosclerotic plaques and analyzed the expression levels of these hub genes. The results showed that compared with normal skin and normal tissue adjacent to the skin lesion, all hub genes were significantly up-regulated in psoriatic skin lesions (Figure 7). Similarly, the expression of all genes (the expression value of PLEK missing in GSE100927) in atherosclerotic plaques was also higher than in normal vascular tissues (Figure 8).

Figure 7 The expression level of hub gene in GSE14905. The comparison between the two sets of data uses the mean T test. P-value < 0.05 was considered statistically significant. LS, skin lesions; NL, adjacent normal tissues; NS, normal skin. *p < 0.05; ***p < 0.001; ****p < 0.0001.

Figure 8 The expression level of hub gene in GSE100927. The comparison between the two sets of data uses the mean T test. P-value < 0.05 was considered statistically significant. AA, advanced atherosclerotic plaque; CA, control artery samples. ****p < 0.0001.
Based on the TRRUST database, we found that 11 TFs may regulate the expression of these genes (Figure 9A and Table 3). Further verification, we found that four TFs are highly expressed in psoriatic lesions and atherosclerotic plaques (Figures 9B, C). They coordinately participated in the regulation of four hub genes (CCL5, NCF2, MMP9 and BCL2A1).
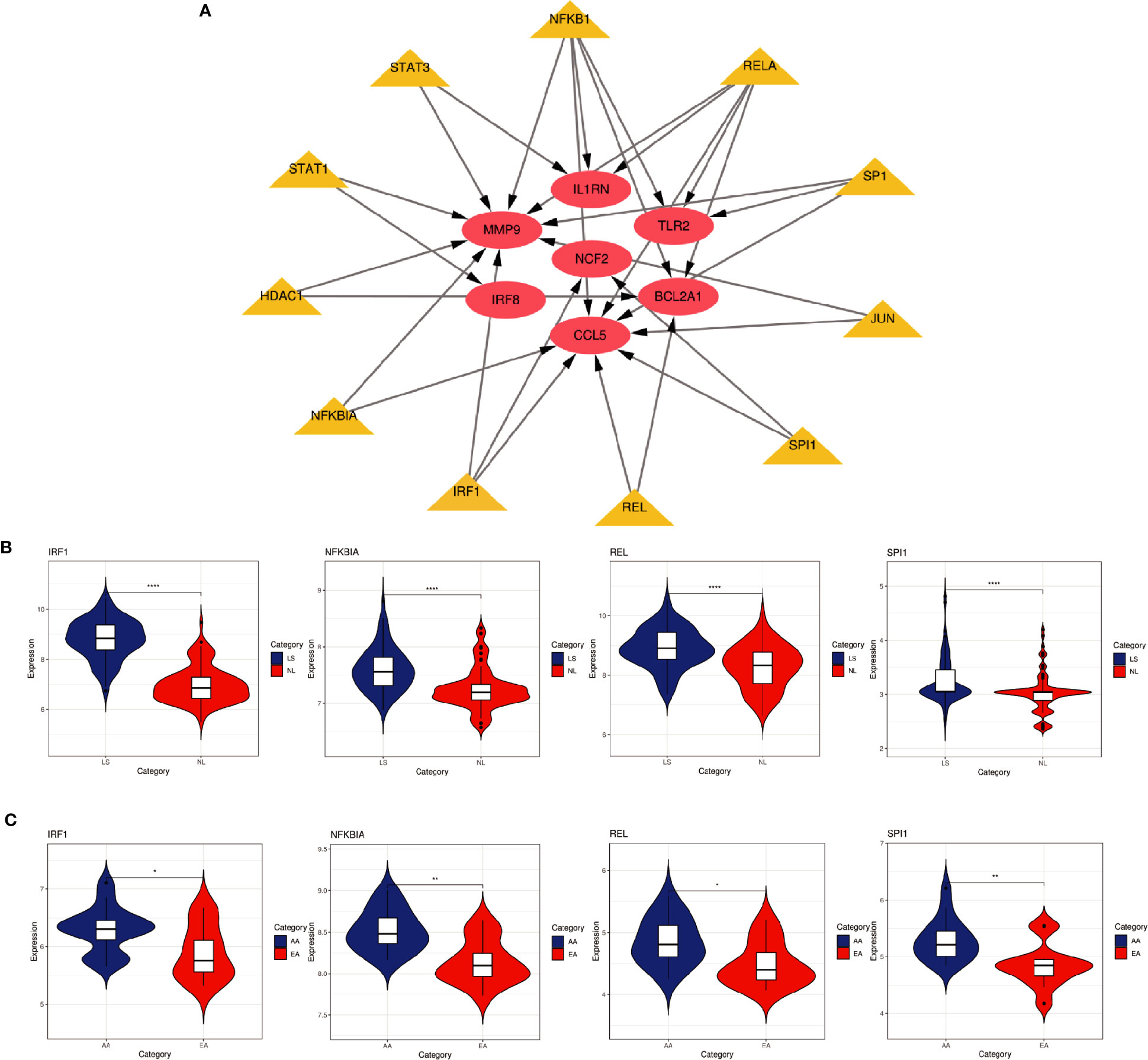
Figure 9 TFs regulatory network and its expression in GSE30999 and GSE28829. (A) TFs regulatory network. TFs were marked in yellow, and the hub genes were marked in red. (B, C) The expression level of TFs in GSE30999 and GSE28829. The comparison between the two sets of data uses the mean T test. P-value < 0.05 was considered statistically significant. LS, skin lesions; NL, adjacent normal tissues; AA, advanced atherosclerotic plaque; EA, early atherosclerotic plaque samples. *p < 0.05; **p < 0.01; ****p < 0.0001.
Insulin resistance induced by the inflammatory response in psoriasis can cause atherosclerosis. Research showed that a variety of inflammatory markers can be detected in the blood of patients with psoriasis, confirming that the body was in a systemic inflammatory state during the onset of psoriasis, which induced insulin resistance and reduced the release of vasodilator factors, then it provided an environmental basis for the formation of atherosclerotic plaques (19). Studies have shown that psoriasis is associated with atherosclerosis through dyslipidemia, including increased levels of total cholesterol, triglycerides, low-density lipoprotein, ultra-low-density lipoprotein and lipoprotein a, and the reduction of high-density lipoprotein and apolipoprotein B levels (20). Adipokines, including adiponectin, leptin, resistin, omentin, visfatin, chemoattractant and retinol binding protein 4, are considered to be risk factors for atherosclerosis. Among them, leptin, resistin, visfatin, chemoattractant and retinol binding protein 4 may be the common pathogenic factors of psoriasis and atherosclerosis. Adiponectin deficiency, leptin, visfatin and chemokine can induce and aggravate psoriasis by activating plasmacytoid dendritic cells and T cells (21, 22). It has been demonstrated that low-density granulocyte (LDG), which is a subset of neutrophils, is elevated in psoriasis and is related to skin disease severity and noncalcified coronary plaque burden. The main mechanism of neutrophil infiltration in patients with psoriasis complicated with atherosclerosis is that psoriasis LDGs may have a cytotoxic effect on endothelial cells, and in an inflammatory environment the adherence of LDG to platelets might be an important link between psoriasis skin disease severity and early atherogenesis (23).
Psoriasis and atherosclerosis might have overlapping pathogenic pathways. Inflammatory and immune regulatory pathways, such as IL23/IL17A pathways are linked to the pathogenesis of these two diseases. IL17A inhibitors are currently commonly used to treat psoriasis. The main purpose of our study is to identify the common DEGs in psoriasis and atherosclerosis. Thereby, revealing potential targets is to predict the therapeutic effect of biologics and treating psoriasis complicated with atherosclerosis.
We must acknowledge the genetic similarity between psoriasis and cardiovascular disease. Study found that single-nucleotide polymorphisms (SNPs) associated with increased risk for dyslipidemia (rs2247056, rs3177928, rs492602, and rs181362), increased blood pressure levels (rs805303, rs653178, and rs3184504), and increased risk for cardiovascular diseases (rs3184504) were associated with increased risk for psoriasis (24). The top three SNPs (rs2247056, rs3177928, and rs805303) were located in the HLA gene region. They also identified four non-HLA SNPs with evidence of shared genetic risk between psoriasis and cardiovascular and metabolic diseases: rs492603 in FUT2, rs181362 in UBE2L3, and rs653178 and rs3184504 near or in SH2B3 (24).
In this study, we identified 94 overlapping DEGs in both, of which 16 were hub genes, including LYN, CSF2RB, IL1RN, RAC2, CCL5, IRF8, C1QB, MMP9, PLEK, PTPRC, FYB, BCL2A1, LCP2, CD53, NCF2 and TLR2. GO and KEGG Pathway enrichment analysis revealed that these genes were significantly enriched in inflammatory and immune pathways. Chemokines and cytokines are jointly involved in the occurrence and development of these two inflammatory diseases, such as serum TNF-α, vascular endothelial growth factor, IL12, monocyte chemotaxis Protein 1, S100A8/A9 and significantly increased IL17A, which play a central role in the development of psoriasis and atherosclerosis. According to GO analysis, the lipopolysaccharide-mediated signaling pathway also play an important role in these two diseases. In psoriasis, lipopolysaccharide mediates nuclear transduction of NF-κB, then proinflammatory factors including TNF-α, IL6, and prostaglandin E (PGE) up- regulate in keratinocytes (25). In atherosclerosis, lipopolysaccharide, a Gram-negative endotoxin and an exogenous ligand for TLR4, can induce the activation of TLR4-MyD88-NF-κB signaling, followed by the release of atherosclerosis-related inflammatory factors such as TNF-α, IL1β, IL6, and MCP-1, leading to the development and progression of atherosclerosis (26). In addition, we found that 11 TFs may regulate the expression of these genes. By further verification, we found that four TFs are highly expressed in psoriatic lesions and atherosclerotic plaques, including IRF1, NFKBIA, REL, SPI1. They coordinately participated in the regulation of four hub genes (MMP9, CCL5, NCF2, and BCL2A1).
MMP9 is a member of MMPs, which endoproteases with collagenase and/or gelatinase activity which exert deleterious effects on the endothelium integrity and collagen fibers, promoting atherosclerotic plaque destabilization and accelerating the process of atherothrombosis (27, 28). Visfatin, which is the common pathogenic factors of psoriasis and atherosclerosis, induces the increased expression of MMP9 in the carotid plaques in an indirect way (29). Studies have confirmed MMP9 is overexpressed in psoriatic neutrophils, and detected to be abundant in psoriatic skin lesions, especially adjacent to endovascular and perivascular tissues, indicating MMP9 may play a role in vascular remodeling in psoriasis. In psoriasis, MMP9 also significantly upregulates the mRNA levels of ICAM-1, IL1β and CXCL1 in vascular endothelial cells (VECs), which enables VECs interaction with more leukocytes (30). CCL5 is expressed and secreted by normal T cell. It is reported that CCL5 production by keratinocytes is increased in psoriatic skin lesions. CCL5 may contribute to the T helper-1-dominant infiltration in psoriatic lesions as well as CXC-chemokines. CCL5 also recruits macrophages. T helper-1 cells or macrophages infiltrating into psoriatic lesions in turn may produce CCL5 and proinflammatory cytokines such as TNF-α, IL1β, or INF-γ, which promote CCL5 production by keratinocytes. Thus, CCL5 production in psoriatic lesions may amplify the inflammation in the lesions (31). In atherosclerosis, macrophages are the major CCL5-expressing cells in the atherosclerotic plaque, CCL5 expression in macrophages is promoted by IFN-γ. CD8α+ dendritic cells aggravate atherosclerosis, likely by inducing Th1 cell response, which promotes CCL5 expression in macrophages and increased infiltration of leukocytes and lesion inflammation (32). BCL2A1 is a member of the BCL2 family. The process of apoptosis is controlled by proteins of the BCL2 family, Increased expression of BCL2A1 protein was associated with psoriatic epidermal hyperplasia (33). It’s also involved in vascular endothelial cell apoptosis and the formation of atherosclerotic plaque. Macrophage resistance to apoptosis was enhanced using macrophage-specific human BCL2 overexpression, have also shown accelerated early atherosclerotic plaque development (34). NCF2 gene encodes neutrophil cytosolic factor 2, the 67-kilodalton cytosolic subunit of the multi-protein complex, which is known as nicotinamide-adenine dinucleotide phosphate (NADPH) oxidase found in neutrophils. This oxidase produces a burst of superoxide which is delivered to the lumen of the neutrophil phagosome (35). As the main source of reactive oxygen species (ROS), the NADPH oxidase family plays an important role in various cellular functions such as antibacterial, anti-inflammatory, and redox signaling (36). ROS are messenger molecules that play a prominent role in cell metabolism-regulating cell proliferation, differentiation and death (37). In psoriasis, it has been reported that an increased intracellular ROS production and an enhanced NADPH oxidase activity in fibroblasts obtained from skin lesions of psoriatic patients (38). NADPH oxidase 4 expressed in dermal fibroblasts is essential for the redox paracrine regulation of epidermal keratinocytes proliferation (39). In atherosclerosis, oxidative stress is a risk factor for it. NADPH seems to mediate its detrimental effects by promoting inflammation in atherosclerosis (40). However, few studies directly analyze the role of NCF2 in psoriasis and atherosclerosis, which emphasizes its importance in future research.
Prior study explored the inflammatory and lipid genes related to atherosclerotic cardiovascular disease in skin and serum from patients with psoriasis (41). It found an increase in skin and serum gene expression for MCP-1 and MDC in psoriasis compared with healthy controls, and lipid metabolism (LXR-α, PPAR-α) genes decreased in psoriasis versus healthy controls (41). Our study also found that new genes like CSF2RB, IL1RN, CCL5, MMP9, CD53, NCF2 and TLR2 associated with Inflammation present high expression both in psoriasis and atherosclerosis. In addition, the lipopolysaccharide-mediated signaling pathway which was found involving many co-expressed genes would induce the release of MCP-1.
Mehta et al. used a bioinformatics approach of lesional psoriasis skin and atherosclerotic plaques, showed that IFN-γ and TNF-α are the dominant pro-inflammatory signals linking atherosclerosis and psoriasis, more importantly, both IFN-γ and TNF-α are increased in the serum of patients with moderate-to-severe psoriasis and their respective receptors in atherosclerotic plaques are increased. Stimulation of primary aortic endothelial cells and ex-vivo atherosclerotic tissue with IFN-γ and TNF-α synergistically increased monocyte and T-cell chemoattractants and adhesion molecules, concomitant with a decrease in endothelial barrier integrity. Their finding suggested IFN-γ/TNF-α synergy may provide a critical pro-inflammatory link between psoriatic skin inflammation and distant vessel atherosclerosis (42). Because of the strong genetic similarity between psoriasis and cardiovascular diseases (24), unlike previous studies, our research pays more attention to the exploration of hub genes and related TFs that are common to psoriasis and atherosclerosis. We have constructed a complex interaction network to identify the key nodes through their common DEGs. This comprehensive bioinformatics method has been proven to be reliable in a variety of diseases (43, 44). In addition, we also analyzed related TFs and verified their expression levels in the original data set. Our research will provide potential directions for the study of the molecular mechanism of psoriasis complicated by atherosclerosis.
Although previous studies have separately explored the hub genes associated with psoriasis (45) and atherosclerosis (46). However, few studies have explored the common molecular mechanism between them by advanced bioinformatics methods. Due to the high comorbidity rate between psoriasis and atherosclerosis, we explored and identified the common DEGs, hub genes and TFs between the two for the first time, which helped to further clarify the mechanism of psoriasis and atherosclerosis. However, our research also has some limitations. First of all, this is a retrospective study that requires external verification to verify our findings; Secondly, the function of the hub gene needs to be further verified in an in vitro model, which will be the focus of our future work.
In summary, we identified the common DEGs of psoriasis and atherosclerosis and performed enrichment and PPI network analysis. We found that psoriasis and atherosclerosis had many common pathogenic mechanisms, which may be mediated by specific hub genes. This study provides new insights for the further study of the molecular mechanism of psoriasis complicated with atherosclerosis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
JJ and YS developed a major research plan. WS and YZ analyze data, draw charts and write manuscripts. XZ and YW helped collect data and references. All authors contributed to the article and approved the submitted version.
This work was supported by Suzhou Minsheng Technology - Medical and Health Application Foundation (SYS2020135) and 2020 Medical Research Project of Jiangsu Provincial Health Commission (Z2020017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.667690/full#supplementary-material
Supplementary Table 1 | The details of the common DEGs.
GEO, Gene Expression Omnibus; DEGs, differentially expressed genes; PPI, protein‐protein interaction; LS: skin lesions; NL, adjacent normal tissues; EA, early atherosclerotic plaque samples; AA, advanced atherosclerotic plaque samples; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; KOBAS, KEGG Orthology Based Annotation System; MCODE, molecular complex detection technology; NS, normal skin; CA, control artery samples; TFs, transcription factors; TRRUST, Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining; LDG, low-density granulocyte; SNP, single-nucleotide polymorphism; PGE, prostaglandin E; VECs, vascular endothelial cells vascular endothelial cells; ROS, reactive oxygen species; NADPH, nicotinamide-adenine dinucleotide phosphate.
1. Lockshin B, Balagula Y, Merola J. Interleukin 17, Inflammation, and Cardiovascular Risk in Patients With Psoriasis. J Am Acad Dermatol (2018) 79(2):345–52. doi: 10.1016/j.jaad.2018.02.040
2. Armstrong A, Harskamp C, Ledo L, Rogers J, Armstrong E. Coronary Artery Disease in Patients With Psoriasis Referred for Coronary Angiography. Am J Cardiol (2012) 109(7):976–80. doi: 10.1016/j.amjcard.2011.11.025
3. Golden J, McCormick T, Ward N. Il-17 in Psoriasis: Implications for Therapy and Cardiovascular Co-Morbidities. Cytokine (2013) 62(2):195–201. doi: 10.1016/j.cyto.2013.03.013
4. Forkel S, Schön M, Hildmann A, Claßen A, John S, Danker K, et al. Inositoylated Platelet-Activating Factor (Ino-C2-PAF) Modulates Dynamic Lymphocyte-Endothelial Cell Interactions and Alleviates Psoriasis-Like Skin Inflammation in Two Complementary Mouse Models. J Invest Dermatol (2014) 134(10):2510–20. doi: 10.1038/jid.2014.170
5. Liu T, Li S, Ying S, Tang S, Ding Y, Li Y, et al. The IL-23/IL-17 Pathway in Inflammatory Skin Diseases: From Bench to Bedside. Front Immunol (2020) 11:594735. doi: 10.3389/fimmu.2020.594735
6. Wolf D, Ley K. Immunity and Inflammation in Atherosclerosis. Circ Res (2019) 124(2):315–27. doi: 10.1161/circresaha.118.313591
7. Taleb S, Tedgui A. Il-17 in Atherosclerosis: The Good and the Bad. Cardiovasc Res (2018) 114(1):7–9. doi: 10.1093/cvr/cvx225
8. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res (2002) 30(1):207–10. doi: 10.1093/nar/30.1.207
9. Suárez-Fariñas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger J. Expanding the Psoriasis Disease Profile: Interrogation of the Skin and Serum of Patients With Moderate-to-Severe Psoriasis. J Invest Dermatol (2012) 132(11):2552–64. doi: 10.1038/jid.2012.184
10. Döring Y, Manthey H, Drechsler M, Lievens D, Megens R, Soehnlein O, et al. Auto-Antigenic Protein-DNA Complexes Stimulate Plasmacytoid Dendritic Cells to Promote Atherosclerosis. Circulation (2012) 125(13):1673–83. doi: 10.1161/circulationaha.111.046755
11. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. Ncbi GEO: Archive for Functional Genomics Data Sets–Update. Nucleic Acids Res (2013) 41(Database issue):D991–5. doi: 10.1093/nar/gks1193
12. Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS Server: A Web-Based Platform for Automated Annotation and Pathway Identification. Nucleic Acids Res (2006) 34(Web Server Issue):W720–4. doi: 10.1093/nar/gkl167
13. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. String v9.1: Protein-Protein Interaction Networks, With Increased Coverage and Integration. Nucleic Acids Res (2013) 41(Database issue):D808–15. doi: 10.1093/nar/gks1094
14. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New Features for Data Integration and Network Visualization. Bioinformatics (2011) 27(3):431–2. doi: 10.1093/bioinformatics/btq675
15. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res (2010) 38(Web Server Issue):W214–20. doi: 10.1093/nar/gkq537
16. Yao Y, Richman L, Morehouse C, de los Reyes M, Higgs B, Boutrin A, et al. Type I Interferon: Potential Therapeutic Target for Psoriasis? PloS One (2008) 3(7):e2737. doi: 10.1371/journal.pone.0002737
17. Steenman M, Espitia O, Maurel B, Guyomarch B, Heymann M, Pistorius M, et al. Identification of Genomic Differences Among Peripheral Arterial Beds in Atherosclerotic and Healthy Arteries. Sci Rep (2018) 8(1):3940. doi: 10.1038/s41598-018-22292-y
18. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. Trrust v2: An Expanded Reference Database of Human and Mouse Transcriptional Regulatory Interactions. Nucleic Acids Res (2018) 46(D1):D380–d386. doi: 10.1093/nar/gkx1013
19. Gonzalez-Cantero A, Gonzalez-Cantero J, Sanchez-Moya A, Perez-Hortet C, Arias-Santiago S, Schoendorff-Ortega C, et al. Subclinical Atherosclerosis in Psoriasis. Usefulness of Femoral Artery Ultrasound for the Diagnosis, and Analysis of Its Relationship With Insulin Resistance. PLoS One (2019) 14(2):e0211808. doi: 10.1371/journal.pone.0211808
20. Praveenkumar U, Ganguly S, Ray L, Nanda S, Kuruvila S. Prevalence of Metabolic Syndrome in Psoriasis Patients and Its Relation to Disease Duration: A Hospital Based Case-Control Study. J Clin Diagn Res JCDR (2016) 10(2):WC01–5. doi: 10.7860/jcdr/2016/17791.7218
21. Shibata S, Tada Y, Hau C, Mitsui A, Kamata M, Asano Y, et al. Adiponectin Regulates Psoriasiform Skin Inflammation by Suppressing IL-17 Production From γδ-T Cells. Nat Commun (2015) 6:7687. doi: 10.1038/ncomms8687
22. Baran A, Flisiak I, Jaroszewicz J, Świderska M. Effect of Psoriasis Activity on Serum Adiponectin and Leptin Levels. Postepy Dermatol i Alergol (2015) 32(2):101–6. doi: 10.5114/pdia.2014.40960
23. Teague H, Varghese N, Tsoi L, Dey A, Garshick M, Silverman J, et al. Neutrophil Subsets, Platelets, and Vascular Disease in Psoriasis. JACC Basic to Trans Sci (2019) 4(1):1–14. doi: 10.1016/j.jacbts.2018.10.008
24. Lu Y, Chen H, Nikamo P, Qi Low H, Helms C, Seielstad M, et al. Association of Cardiovascular and Metabolic Disease Genes With Psoriasis. J Invest Dermatol (2013) 133(3):836–9. doi: 10.1038/jid.2012.366
25. Donnarumma G, Paoletti I, Buommino E, Fusco A, Baudouin C, Msika P, et al. AV119, a Natural Sugar From Avocado Gratissima, Modulates the LPS-Induced Proinflammatory Response in Human Keratinocytes. Inflammation (2011) 34(6):568–75. doi: 10.1007/s10753-010-9264-6
26. Kapelouzou A, Giaglis S, Peroulis M, Katsimpoulas M, Moustardas P, Aravanis C, et al. Overexpression of Toll-Like Receptors 2, 3, 4, and 8 Is Correlated to the Vascular Atherosclerotic Process in the Hyperlipidemic Rabbit Model: The Effect of Statin Treatment. J Vasc Res (2017) 54(3):156–69. doi: 10.1159/000457797
27. Cowan K, Jones P, Rabinovitch M. Elastase and Matrix Metalloproteinase Inhibitors Induce Regression, and Tenascin-C Antisense Prevents Progression, of Vascular Disease. J Clin Invest (2000) 105(1):21–34. doi: 10.1172/jci6539
28. Shi Y, Patel S, Niculescu R, Chung W, Desrochers P, Zalewski A. Role of Matrix Metalloproteinases and Their Tissue Inhibitors in the Regulation of Coronary Cell Migration. Arterioscler Thromb Vasc Biol (1999) 19(5):1150–5. doi: 10.1161/01.atv.19.5.1150
29. Li B, Zhao Y, Liu H, Meng B, Wang J, Qi T, et al. Visfatin Destabilizes Atherosclerotic Plaques in Apolipoprotein E-Deficient Mice. PLoS One (2016) 11(2):e0148273. doi: 10.1371/journal.pone.0148273
30. Chen J, Zhu Z, Li Q, Lin Y, Dang E, Meng H, et al. Neutrophils Enhance Cutaneous Vascular Dilation and Permeability to Aggravate Psoriasis by Releasing Matrix Metallopeptidase 9. J Invest Dermatol (2020) 141(4):787–99. doi: 10.1016/j.jid.2020.07.028
31. Kanda N, Watanabe S. 17beta-Estradiol Inhibits the Production of RANTES in Human Keratinocytes. J Invest Dermatol (2003) 120(3):420–7. doi: 10.1046/j.1523-1747.2003.12067.x
32. Li Y, Liu X, Duan W, Tian H, Zhu G, He H, et al. Batf3-dependent Cd8α Dendritic Cells Aggravates Atherosclerosis Via Th1 Cell Induction and Enhanced CCL5 Expression in Plaque Macrophages. EBioMedicine (2017) 18:188–98. doi: 10.1016/j.ebiom.2017.04.008
33. Batinac T, Zamolo G, Hadzisejdić I, Zauhar G, Brumini G, Ruzić A, et al. Expression of Bcl-2 Family Proteins in Psoriasis. Croatian Med J (2007) 48(3):319–26.
34. Shearn A, Deswaerte V, Gautier E, Saint-Charles F, Pirault J, Bouchareychas L, et al. Bcl-X Inactivation in Macrophages Accelerates Progression of Advanced Atherosclerotic Lesions in Apoe(-/-) Mice. Arterioscler Thromb Vasc Biol (2012) 32(5):1142–9. doi: 10.1161/atvbaha.111.239111
35. Martner A, Aydin E, Hellstrand K. NOX2 in Autoimmunity, Tumor Growth and Metastasis. J Pathol (2019) 247(2):151–4. doi: 10.1002/path.5175
36. Breitenbach M, Rinnerthaler M, Weber M, Breitenbach-Koller H, Karl T, Cullen P, et al. The Defense and Signaling Role of NADPH Oxidases in Eukaryotic Cells: Review. Wiener Med Wochenschr (1946) (2018) 168:286–99. doi: 10.1007/s10354-018-0640-4
37. Lisse T, King B, Rieger S. Comparative Transcriptomic Profiling of Hydrogen Peroxide Signaling Networks in Zebrafish and Human Keratinocytes: Implications Toward Conservation, Migration and Wound Healing. Sci Rep (2016) 6:20328. doi: 10.1038/srep20328
38. Barygina V, Becatti M, Lotti T, Taddei N, Fiorillo C. Low Dose Cytokines Reduce Oxidative Stress in Primary Lesional Fibroblasts Obtained From Psoriatic Patients. J Dermatol Sci (2016) 83(3):242–4. doi: 10.1016/j.jdermsci.2016.06.002
39. Barygina V, Becatti M, Prignano F, Lotti T, Taddei N, Fiorillo C. Fibroblasts to Keratinocytes Redox Signaling: The Possible Role of ROS in Psoriatic Plaque Formation. Antioxid (Basel Switzerland) (2019) 8(11):566. doi: 10.3390/antiox8110566
40. Marqués J, Cortés A, Pejenaute Á, Zalba G. Implications of NADPH Oxidase 5 in Vascular Diseases. Int J Biochem Cell Biol (2020) 128:105851. doi: 10.1016/j.biocel.2020.105851
41. Mehta N, Li K, Szapary P, Krueger J, Brodmerkel C. Modulation of Cardiometabolic Pathways in Skin and Serum From Patients With Psoriasis. J Trans Med (2013) 11:194. doi: 10.1186/1479-5876-11-194
42. Mehta N, Teague H, Swindell W, Baumer Y, Ward N, Xing X, et al. Ifn-γ and TNF-α Synergism may Provide a Link Between Psoriasis and Inflammatory Atherogenesis. Sci Rep (2017) 7(1):13831. doi: 10.1038/s41598-017-14365-1
43. Fang X, Duan S, Gong Y, Wang F, Chen X. Identification of Key Genes Associated With Changes in the Host Response to Severe Burn Shock: A Bioinformatics Analysis With Data From the Gene Expression Omnibus (Geo) Database. J Inflammation Res (2020) 13:1029–41. doi: 10.2147/jir.S282722
44. Yang S, Cao C, Xie Z, Zhou Z. Analysis of Potential Hub Genes Involved in the Pathogenesis of Chinese Type 1 Diabetic Patients. Ann Trans Med (2020) 8(6):295. doi: 10.21037/atm.2020.02.171
45. Gao L, Shen J, Ren Y, Shi J, Wang D, Cao J. Discovering Novel Hub Genes and Pathways Associated With the Pathogenesis of Psoriasis. Dermatol Ther (2020) 33(6):e13993. doi: 10.1111/dth.13993
Keywords: psoriasis, atherosclerosis, bioinformatics, differentially expressed genes, hub genes
Citation: Su W, Zhao Y, Wei Y, Zhang X, Ji J and Yang S (2021) Exploring the Pathogenesis of Psoriasis Complicated With Atherosclerosis via Microarray Data Analysis. Front. Immunol. 12:667690. doi: 10.3389/fimmu.2021.667690
Received: 15 February 2021; Accepted: 12 May 2021;
Published: 27 May 2021.
Edited by:
Trine N. Jorgensen, Case Western Reserve University, United StatesReviewed by:
Wilson Liao, University of California, San Francisco, United StatesCopyright © 2021 Su, Zhao, Wei, Zhang, Ji and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Ji, amlqaWFuZ0BzdWRhLmVkdS5jbg==; Shun Yang, eWFuZ3NodW5zZGZleUAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.