- 1Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of Applied Pharmaceutical Sciences and Clinical Pharmacy, Faculty of Pharmacy, Isra University, Amman, Jordan
- 3Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia
- 4Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Umm Al-Qura University, Makkah, Saudi Arabia
- 5Vaccines and Immunotherapy Unit, King Fahad Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia
Background: SARS, MERS, and COVID-19 share similar characteristics. For instance, the genetic homology of SARS-CoV-2 compared to SARS-CoV and MERS-CoV is 80% and 50%, respectively, which may cause similar clinical features. Moreover, uncontrolled release of proinflammatory mediators (also called a cytokine storm) by activated immune cells in SARS, MERS, and COVID-19 patients leads to severe phenotype development.
Aim: This systematic review and meta-analysis aimed to evaluate the inflammatory cytokine profile associated with three strains of severe human coronavirus diseases (MERS-CoV, SARS-CoV, and SARS-CoV-2).
Method: The PubMed, Embase, and Cochrane Library databases were searched for studies published until July 2020. Randomized and observational studies reporting the inflammatory cytokines associated with severe and non-severe human coronavirus diseases, including MERS-CoV, SARS-CoV, and SARS-CoV-2, were included. Two reviewers independently screened articles, extracted data, and assessed the quality of the included studies. Meta-analysis was performed using a random-effects model with a 95% confidence interval to estimate the pooled mean of inflammatory biomarkers.
Results: A high level of circulating IL-6 could be associated with the severity of infection of the three coronavirus strains. TNF, IL-10, and IL-8 are associated with the severity of COVID-19. Increased circulating levels of CXCL10/IP10 and CCL2/MCP-1 might also be related to the severity of MERS.
Conclusion: This study suggests that the immune response and immunopathology in the three severe human coronavirus strains are somewhat similar. The findings highlight that nearly all studies reporting severe cases of SARS, MERS, and COVID-19 have been associated with elevated levels of IL-6. This could be used as a potential therapeutic target to improve patients’ outcomes in severe cases.
Systematic Review Registration: PROSPERO registration 94 number: CRD42020209931.
Introduction
Coronavirus disease 2019 (COVID-19) is the third human coronavirus (hCoV) outbreak in only two decades (1). Coronaviruses (CoVs) are enveloped, single-stranded RNA viruses that infect the lower respiratory tract (2). Severe acute respiratory syndrome (SARS), caused by SARS-CoV, was the first global epidemic of the twenty-first century (3, 4). In March 2003, the disease broke out in Hong Kong, quickly spreading throughout the world, including Asia, Europe, and the United States (5). Nearly a decade following the SARS epidemic, in June 2012, Middle East respiratory syndrome (MERS), caused by MERS-CoV, emerged in Saudi Arabia and rapidly spread to more than 20 countries (6). Bats are known to be the natural reservoir of SARS and MERS. Dromedary camels have also been found to be a zoonotic reservoir for transmission of MERS to humans (7).
COVID-19 is the newest emerging infectious disease (8). The infection began in Wuhan, China, in December 2019 and has infected over 100 million people, with over 2 million deaths worldwide as of April, 2021 (9). This version of human coronavirus is a more serious global threat than either of its two predecessors.
The three human coronaviruses share similar characteristics; the genetic homology of SARS-CoV-2 to SARS-CoV and MERS-CoV is 80% and 50%, respectively (10). All three respiratory infectious diseases are also associated with significant morbidity and mortality, as they are highly transmissible, primarily through respiratory droplets and close contact (11). However, the transmission of MERS to humans is relatively inefficient compared to SARS and COVID-19 (12, 13). Conversely, SARS and COVID-19 have a relatively lower mortality rates than MERS (14). This is likely related to the viral kinetics, age and comorbid illnesses (14).
The three respiratory infectious diseases (SARS, MERS, and COVID-19) have similar clinical features that range from a lack of symptoms to severe illness that requires intubation and intensive care management (15, 16). The severity of these diseases is linked to age and chronic illness, including diabetes, hypertension, cardiovascular disease, chronic respiratory disease, and cancer (17). Viral pneumonia, acute respiratory distress syndrome, and multiple organ failure are common in these diseases’ later stages (18–20). Non-specific laboratory tests, including those for leukopenia, lymphopenia, thrombocytopenia, and elevated serum amino transaminases, are often positive in SARS, MERS, and COVID-19 infections (21).
A cytokine release storm (CRS) is the uncontrolled release of several proinflammatory cytokines due to an exuberant host immune response (22–24). In SARS (25), MERS (26), and COVID-19 (27, 28) cases, CRS has been reported to lead to the development of severe phenotypes (Figure 1). Given the potential overlap in presentation and manifestation among severe hCoV infections, and the absence of effective treatment, it is essential to understand hCoVs’ immunopathology, including hCoV-induced CRS. Therefore, this systematic review and meta-analysis aimed to compare the inflammatory biomarkers associated with three significant strains of severe human coronavirus disease (MERS-CoV, SARS-CoV, and SARS-CoV-2) that may have a distinct inflammatory profile. Such results could help to identify potential treatment options for severe patients.
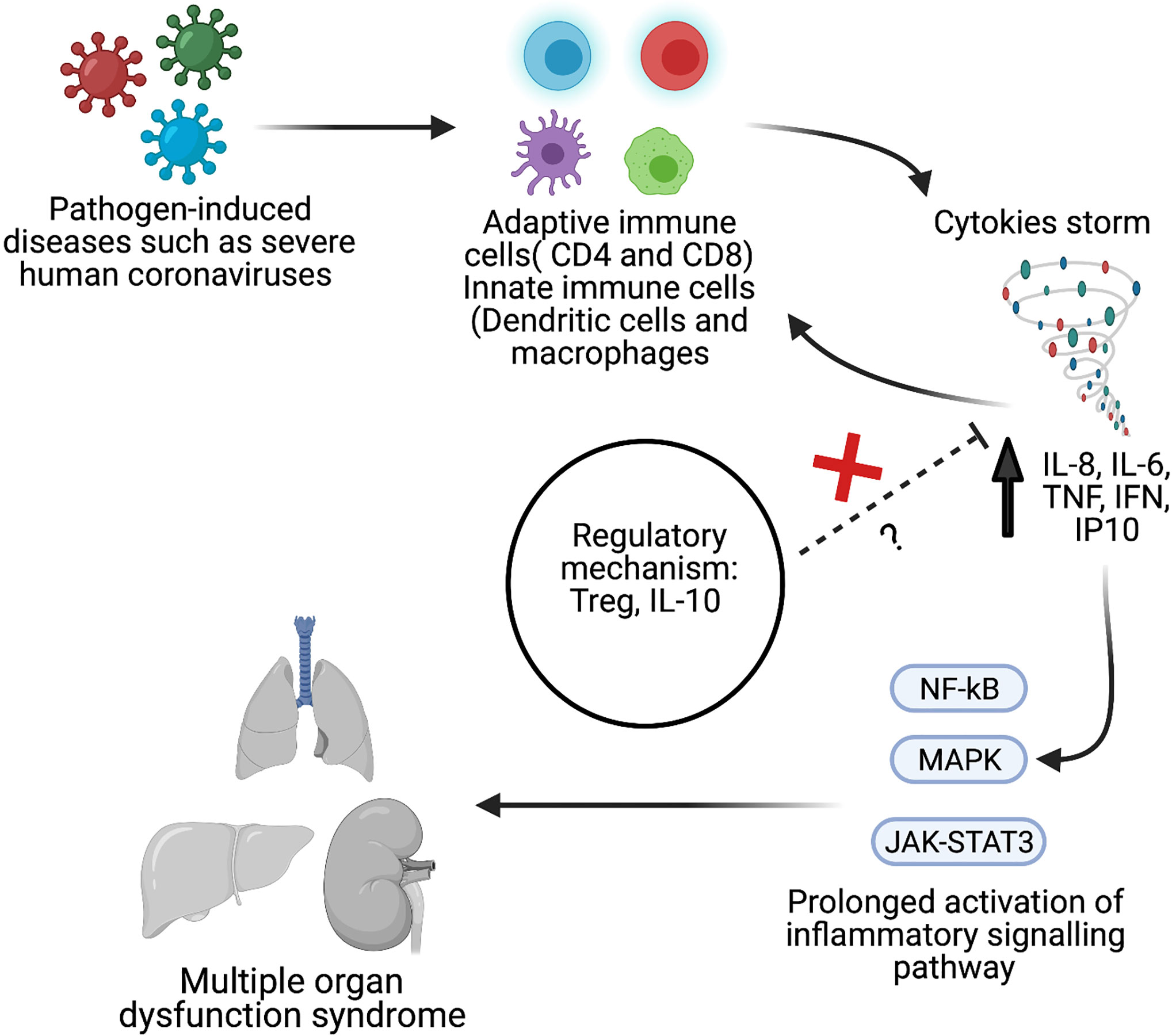
Figure 1 Schematic overview of pathological hallmark of cytokines storm during viral infection such as severe human coronavirus (hCoV). Created with BioRender.com.
Methods
The systematic review and meta-analysis were carried out following the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines (29) and reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (30). The protocol of the study was registered with PROSPERO (provisional registration number CRD42020209931).
Databases and Search Strategy
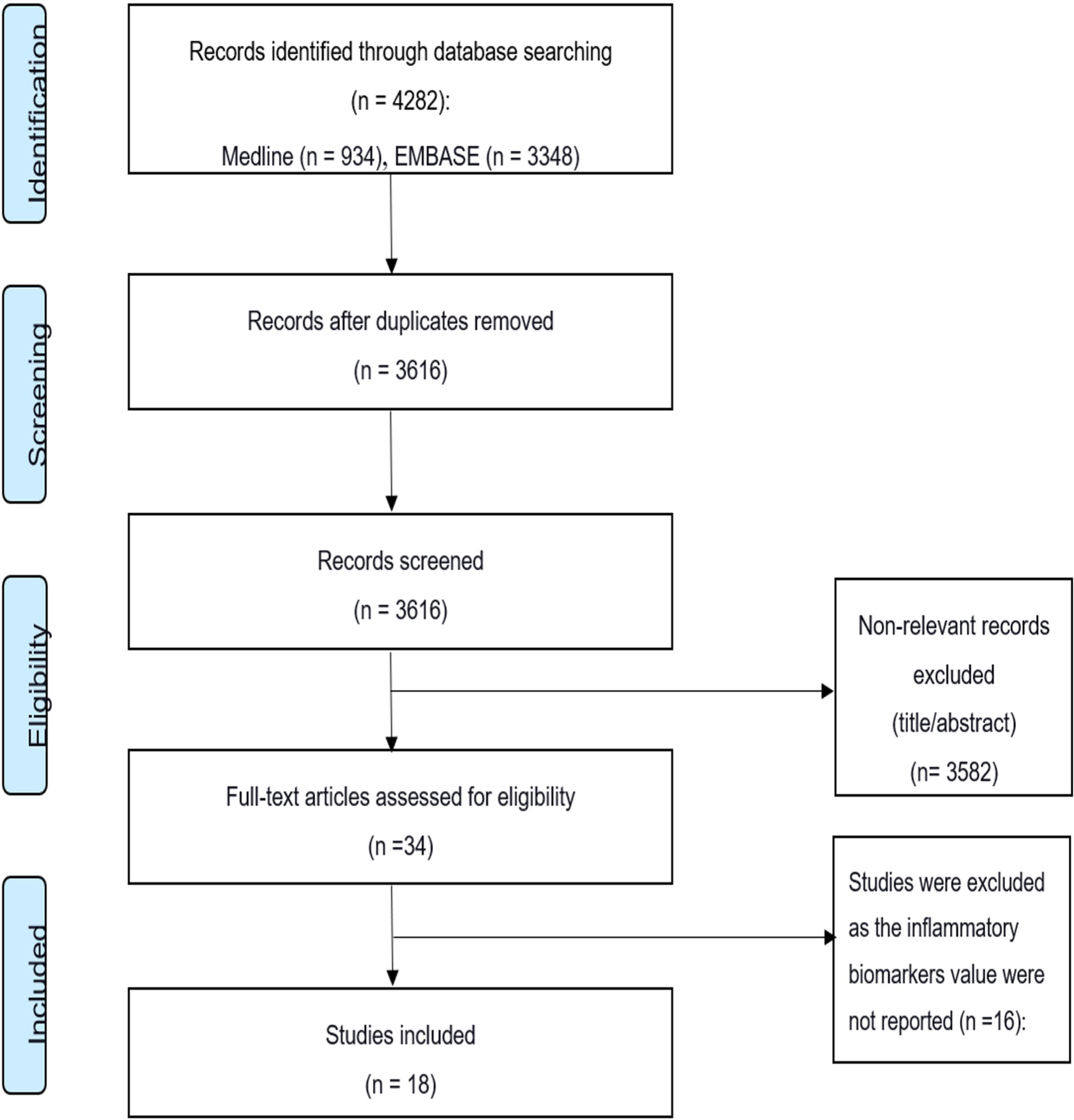
An extensive search strategy (summarized in Figure 2) was developed to identify relevant studies. A detailed electronic search on bibliographic databases, including Medline, Embase, and Cochrane Library, was performed from inception to July 2020. Keywords, Emtree, and MeSH terms were used with both English and American spellings. The search strategy covered the following keywords: cytokines, inflammatory biomarkers, SARS-CoV, SARS-CoV-2, severe acute respiratory syndrome, Middle East respiratory syndrome coronavirus, and COVID-19.
Eligibility Criteria
The selection of included studies was based on inclusion/exclusion criteria. Inclusion criteria include any randomized or observational studies that assessed the concentration of cytokines and chemokines in the peripheral blood of patients with severe SARS, MERS, or COVID-19 infections. Severe or critical patients were defined as patients who had a respiratory failure, required mechanical ventilation, experienced shock, experienced other organ failures, or required a stay in the intensive care unit (ICU). Conference proceedings, reviews, studies not published in English, animal studies, and in vitro studies of cytokine production in stimulated cells were excluded. Studies that did not clearly differentiate between levels of severity or did not report the mean and standard deviation or median of cytokine parameter levels were also excluded.
Data Extraction
Initially, all the researchers (A.Z., A.N., H.A., and F.M.) independently screened the title and abstract of each study to assess its eligibility based on the inclusion/exclusion criteria. Subsequently, all the researchers (A.Z., A.N., H.A., and F.M.) independently conducted a two-step full-text literature search to further assess eligibility. Data extraction from the relevant studies was performed independently by all researchers using a form created for this study. The following demographic details were extracted from each study: study type, data source, year of publication, sample size, age, and sex. The mean, median, standard error, IQR, and range of certain cytokines and chemokines (IL-1, IL-2, IL-4, IL-6, IL-8, IL-10, IL-17, TNF, IFN-γ, RANTS, CCL2/MCP-1, CXCL10/IP10, and TGFß) were also extracted.
Quality Assessment
The methodological quality and risk of bias were assessed using the Newcastle Ottawa Scale for Observational Studies (31), which was modified to meet the requirements of this study (32). A total of six criteria were evaluated: representativeness of the population, sample size, statistical analysis, missing data, methodology for reporting the outcome of interest, and methods to detect or report the outcome of interest. Each criterion was rated on a scale ranging from 0 to 3, where 3 represented the highest quality. The highest possible total score was 18. In addition, we categorized the score into three categories: good quality (>12), moderate quality (>6), and low quality (≤6).
Data Synthesis and Analysis
Descriptive statistics were used to describe the demographic characteristics and quantitative mean concentration of the inflammatory biomarkers reported in each study. To standardize the data for meta-analysoo, we estimated the mean cytokine concentrations from studies that reported the median using the following formula: if the sample size was ≤25, , and if the sample size was >25, (). We calculated the variance as follows: if the sample size was ≤15, ; if the sample size was 15–70, ; and if the sample size was 70, , where m is the median, a is a minimum, b is a maximum, X is a mean, S is variance and R is a range (b-a) (33). A random-effects model was used to estimate the mean concentration of biomarkers (34). Heterogeneity among the studies included in the meta-analysis was assessed using the standard χ2 tests and the I2 statistic. If high heterogeneity was indicated (I2 ≥ 75%), systematic narrative synthesis was provided. All analysis was performed using STATA 15.0.
Results
Study Selection, Characteristics, and Quality Assessment
A total of 4,282 studies were identified through the initial online search of the databases (Figure 2). Of these, 666 studies were duplicates. Title and abstract screening identified 34 studies for final full-text review using the inclusion/exclusion criteria. Finally, the search yielded 18 studies that met the inclusion criteria. Of these, 15 studies were conducted on COVID-19, 2 studies were on MERS, and 1 study was on SARS. The 15 COVID-19 studies included 538 severe patients (67% male, 33% female) in the final analysis. The one SARS (35) and two MERS studies (36, 37) had fewer patients. The SARS study included 30 severe patients (70% male, 30% female). The two MERS studies (36, 37) had 15 severe patients (80% male, 20% female). The 18 studies also included 1,922 non-severe individuals as controls. Thirteen of the 18 studies were conducted in China, 3 were conducted in South Korea, 1 was conducted in Germany, and 1 was conducted in Ireland. Most of the laboratory tests were conducted after admission.
The characteristics of the included studies are summarized in Table 1. Overall, the quality of the included studies was good. The quality assessment score for the included studies ranged from 12 to 18. Most of the studies (94.4%) were of good quality and scored >12 (n = 17), but one study was of moderate quality (12 out of 18). Details regarding the quality assessments are presented in Table 1 and Supplementary Table 1.
The Level of Circulating Cytokines in COVID-19, MERS, and SARS Patients
The dynamic changes of circulating cytokines (IL-1, IL-2, IL-6, TNF, and IFN-γ, IL-4, IL-10, and IL-17) were analyzed in the 18 included studies (Table 2). All the studies reported IL-6 concentrations. Remarkably, all but one study showed a significant elevation in the level of IL-6 in severe COVID-19 patients compared to non-severe groups. The level of circulating IL-6 was also significantly high in severe SARS patients (517 ± 769 pg/ml) compared to non-severe groups (163 ± 796 pg/ml) (35). Furthermore, the two studies (36, 37) that measured the level of circulating cytokines in severe MERS patients showed an elevation in the level of IL-6 in severe MERS patients compared to non-severe groups.
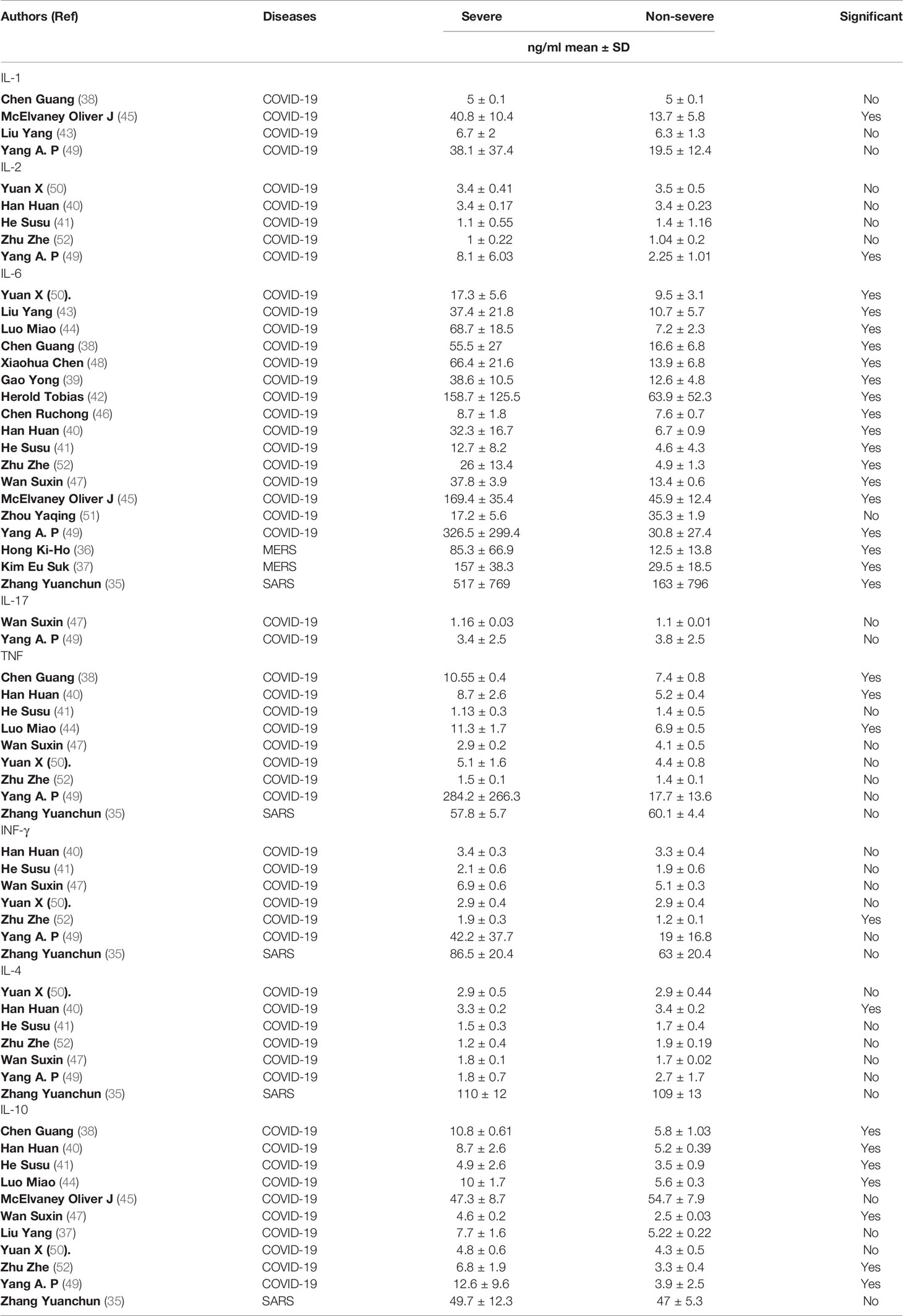
Table 2 Details on the proinflammatory and anti-inflammatory cytokines profile among all study participants.
Other cytokines were substantially elevated in patients with severe COVID-19, but not in patients with severe SARS and MERS. For example, three studies (38, 40, 44) out of eight showed a significant elevation on TNF level in severe COVID-19 patients compared to non-severe groups. The concentration level of IL-10 was measured in eleven studies (39–41, 47, 49, 52), of which seven studies reported a significant difference between severe COVID-19 patients compared to non-severe groups. In contrast (35), and (36, 37) showed that the level of TNF and IL-10 cytokines were not significantly elevated in patients with severe SARS and MERS, respectively, compared to a non-severe group.
Most of the other cytokines were not comparatively high in severe patients with COVID-19, SARS, and MERS. One study (52) out of six showed that the level of IFN-γ was significantly higher in patients with severe COVID-19 (1.9 ± 0.3 pg/ml) compared to non-severe groups (1.2 ± 0.1 pg/ml). However, another study showed that IFN-γ concentrations were not elevated in patients with severe SARS compared to the non-severe group (35).
Similar results were seen for IL-4. One study (40) out of six showed that the level of IL-4 in severe COVID-19 patients was significantly higher (3.3 ± 0.2 pg/ml) than in non-severe groups (3.4 ± 0.2 pg/ml). However (35), reported that the IL-4 concentration in patients with severe SARS was (110 ± 12 pg/ml), similar to the concentration for a non-severe group (109 ± 13 pg/ml). The levels of IFN-γ and IL-4 were not available in any MERS studies.
The concentrations of IL-1, IL-2, and IL-17 were reported in only four COVID-19 studies (38, 43, 45, 49). Of these, one study (45) showed that the level of IL-1 was significantly higher in patients with severe COVID-19 (40.8 ± 10.4 pg/ml) compared to non-severe groups (13.7 ± 5.8 pg/ml). Similar results were seen for IL-2. One study (49) out of five reported a significant difference between severe COVID-19 patients (8.1 ± 6.03 pg/ml) and non-severe groups (2.25 ± 1.01 pg/ml). The only two studies (47, 49) that measured the level of IL-17 in severe COVID-19 patients showed no significant difference between severe COVID-19 patients and non-severe groups. Altogether, our data suggest that high levels of circulating IL-6, TNF, and IL-10 might be associated with the severity of COVID-19, while only an increased level of IL-6 might be related to the severity of SARS and MERS.
The Level of Circulating Chemokines in COVID-19, MERS, and SARS Patients
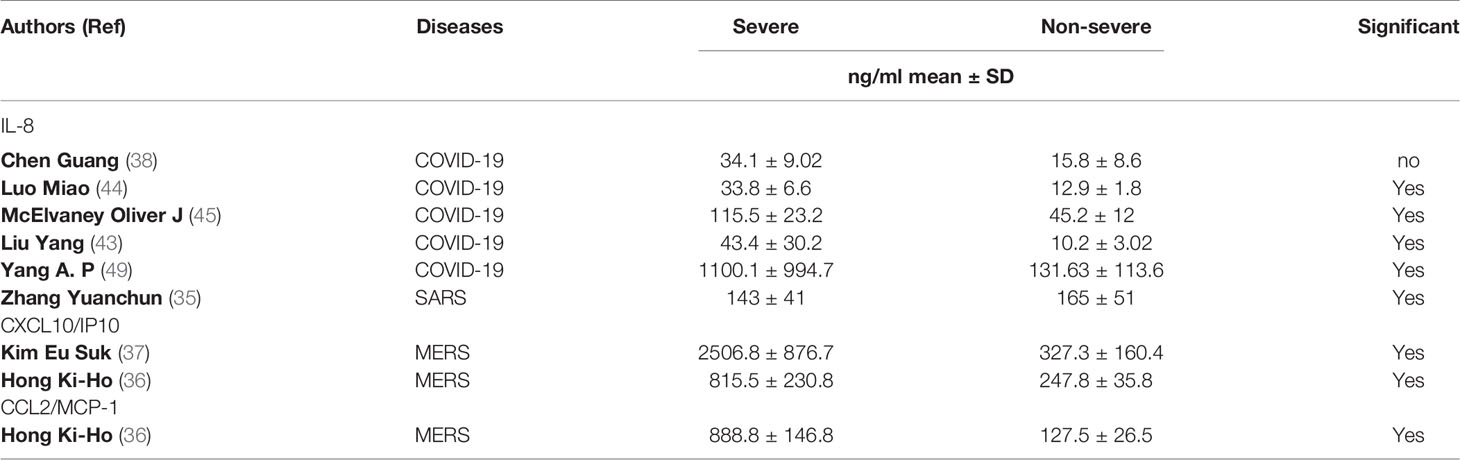
The concentration of the circulating inflammatory chemokine IL-8 were reported in five (35, 38, 43–45, 49) COVID-19 studies and one (35) SARS study. Remarkably, four (38, 43–45, 49) of the five COVID-19 studies reported a significant elevation in the level of IL-8 in severe COVID-19 patients compared to non-severe groups. In contrast, the one study (35) conducted in SARS patients reported a significant reduction in the level of IL-8 in severe SARS patients (143 ± 41 pg/ml) compared to the non-severe group (165 ± 51 pg/ml).
The concentration level of C-X-C motif chemokine 10/interferon-gamma-induced protein 10 (CXCL10/IP10) and chemokine ligand 2/monocyte chemoattractant protein-1 (CCL2/MCP-1) were only reported in two MERS studies (36, 37). The concentrations of CXCL10/IP10 and CCL2/MCP-1 were significantly higher in patients with severe MERS than in non-severe groups.
Although the number of studies measuring the level of chemokines in patients with severe COVID-19, SARS, and/or MERS is limited, the data suggest that circulating IL-8 might be associated with the severity of COVID-19. Moreover, the levels of circulating CXCL10/IP10 and CCL2/MCP-1 might be related to the severity of MERS (Table 3).
The 18 studies included in the meta-analysis had a high level of heterogeneity (I2 = 99.9%). There was considerable variation between the studies in terms of geographic location, study design, time of blood collection, variable assay used to measure the level of cytokines, and data source for both severe and non-severe cases (Supplementary Figure 1).
Discussion
An emerging body of research is focusing on selective targeting of elevated inflammatory cytokines to treat CRS in hCoV cases (53). However, the mutual cytokine profile of hCoV strains is undetermined. Knowledge of this profile could improve the hyperinflammatory state in critically ill hCoV patients. In addition, there remain unanswered questions about the mechanistic role of the cytokine storm caused by the three hCoVs. Therefore, this systemic review and meta-analysis were performed to analyze the circulatory cytokines and chemokines profiles in patients with severe hCoV infections.
Among the inflammatory parameters, our systematic review and analysis demonstrated a marked elevation in the level of circulating IL-6 and TNF in severe COVID-19 and MERS patients in comparison to non-severe groups. Most of the included studies also showed a significant elevation of circulating IL-10 in severe COVID-19 patients compared to non-severe groups. However, due to the limited number of studies conducted on SARS, we could not find a strong association between high levels of circulating IL-6 and TNF and IL-10 and the severity of disease. This could be the topic of further investigation.
Although clinical data regarding CRS in SARS and MERS infections are limited, an animal study of SARS-CoV has shown that SARS spike protein induced the secretion of high levels of IL-6 and TNF through the NF-kB pathway (54). Others indicated that MERS-infected human monocyte derived-macrophages induce the secretion of high levels of TNF and IL-6 (55). It has also been shown that the coronavirus envelope (E) proteins of SARS-CoV-2 and SARS-CoV share 95% homology (53). It is therefore reasonable to speculate that hCoVs induce similar immune responses.
Among the excessively produced cytokines, IL-6 is considered a key cytokine and an early marker of morbidity and mortality in lung diseases (35, 53, 56)56). For example, a recent study reported the significant role of IL-6 as an independent COVID-19 risk factor using multivariate logistic regression analyses (52). Several large retrospective cohort studies also reported that the level of blood IL-6 correlates with severity and mortality in patients with COVID-19 (57, 58). Evidence suggests that the considerable elevation of IL-6 cytokines in severe COVID-19 patients is linked to massive mucus production by stimulating the expression of the two predominant mucin genes (MUC5AC and MUC5B) in tracheobronchial epithelial cells (59, 60). Recent studies have also revealed that excessive IL-6 signaling leads to hypercoagulating, megakaryocyte activation, precipitation of pulmonary immune-mediated thrombosis (61–63), induction of macrophage activation syndrome, and reduction of myocardium contractility, which might contribute to organ damage (57, 64). Given that IL-6 has a key role in CRS, the recombinant human IL-6 monoclonal antibody tocilizumab is currently the subject of clinical trials, and it shows promising efficacy for the treatment of severe COVID-19 (65–67).
IL-6 has also been found to be significantly correlated with the anti-inflammatory cytokine IL-10, which may reflect self-protection (68, 69). IL-10 is a pleiotropic cytokine primarily responsible for an anti-inflammatory effect (70). It has been shown to inhibit the synthesis of proinflammatory cytokines, such as TNF and IL-6 (71). Unlike in SARS and MERS patients, high levels of IL-10 have been demonstrated in severe and critically ill COVID-19 patients (72). However, it is unclear how IL-10 could be correlated to the severity of COVID-19. Moreover, prolonged production of endogenous IL-10 following the onset of a cytokine storm could be detrimental for the host during COVID-19 infection. For instance, IL-10 has been known to introduce anergy T cells during viral infection (73). Exhausted T cells (PD-1+TIM3+CD8+) have been found in severe cases of patients infected with SARS-CoV-2 and have been reported to be correlated with high serum IL-10 (74). These results suggest that blocking IL-10 or IL-10R might be beneficial for T cell recovery and the prevention of viral persistence. However, several studies have indicated that a lack of IL-10 leads to better bacterial clearance, a higher survival rate, and limited sepsis-induced immunosuppression (75, 76). Therefore, the use of IL-10 antagonists should be carefully studied in the context of severe hCoV infections.
In the past decade, anti-cytokine therapy has made great strides in managing CRS-related diseases, including acute respiratory distress syndrome (ARDS), autoimmunity, and sepsis (53). Therefore, treating severe COVID-19 cases with anti-cytokine treatments has attracted substantial attention as a beneficial addition to antiviral therapy (63, 77). At least 20 registered clinical trials have examined the ability of various IL-6, IL-1, and IFN-1 antagonists to treat COVID-induced CRS (62). We believe that the use of tocilizumab, an anti-IL‐6R antibody (78), is a promising strategy for treating severe hCoV-induced CRS (79), as it showed promising results in clinical trials (80). However, the focus on anti-cytokine therapy might have led to the opportunity to explore other potential therapies that target the host’s immune system, such as stem cell therapy, transfusion of convalescent plasma, targeting of the microRNA network, and immune-supportive therapy (53, 81–84). It is therefore recommended to combine immunotherapy with antiviral treatment. Early diagnosis of CRS in hCoV patients is also recommended to guide therapeutic decisions (53).
Less is known about the chemokine spectrum in human coronaviruses compared to cytokine production. Thus, it is difficult to determine the importance of chemokines in the disease progression for hCoV diseases. However, upon comparing the chemokine profile of the three hCoVs, it can be concluded that CCL2/MCP-1, CXCL8, and CXCL10/IP10 have vital roles in the pathogenesis of infection and serve as prognostic markers for hCoVs’ severity (55, 85–89). Many cell types secrete CXCL8, also known as IL-8 and CCL2/MCP-1, in response to IL-6 and TNF-mediated cytokines (90, 91). CXCL10/IP10 is also secreted by many cells, including monocytes, endothelial cells, and fibroblasts, in response to IFN-γ (92, 93). These chemokines and others play crucial roles in the pathogenesis of diseases characterized by thrombosis and the recruiting of leukocytes to inflamed tissues (94, 95). It is worth noting that CXCL10/IP10 is absent in healthy individuals. However, it is detected in asymptomatic patients and is directly linked to disease severity (96, 97), suggesting that early detection of chemokines can help control new outbreaks (98). Several clinical investigations involving autopsies have also provided evidence that thrombosis is an important consequence of hCoV diseases (99). These findings indicate that the thrombosis-related indicators (CXCL10/IP10, MCP-1) could be directly involved in different stages of hCoV infection (98) and could be used as potential biomarkers to predict the risk of death in hCoV patients (89, 100). Moreover, antagonists targeting chemokines, or their receptors provided a therapeutic benefit against various diseases. For instance, CCR5 and CXCL10/IP-10 antagonists are currently being validated in clinical trials to treat HIV and cardiovascular diseases, respectively (101–103). Thus, neutralization of certain chemokines is a promising potential therapeutic approach to fight hCoVs, especially in patients with thrombotic events (104, 105).
Our demographic data demonstrated that most severe hCoV cases are among older males, in line with previous studies. It has been previously reported that older people with comorbidities are more vulnerable than young people to developing more advanced and severe human coronavirus-related diseases (106). It is believed that dysregulated innate immunity and deficiency in the adaptive immune response of older people, mostly males, could play an essential role in this phenomenon (106). For instance, a previous study demonstrated that peripheral blood mononuclear cells from males stimulated with influenza and herpes-simplex-1 viruses induced the production of higher levels of IL-10 compared to females (107). Other studies showed that males had higher levels of proinflammatory cytokines (e.g., TNF) and chemokines (e.g., CXCL10/IP-10) following lipopolysaccharide stimulation (108). In contrast, females had a higher number of proliferating T-cells and B-cells in vitro and antibody response to influenza vaccination than males (109). Patients with comorbidities, such as diabetes, asthma, and hypertension, are at higher risk of mortality because of the excessive production of inflammatory cytokines, including IL-2R, IL-10, and TNF (110–112). These studies and others highlighted that gender and age-specific innate and adaptive immunity and comorbidity might affect hCoV patients’ inflammation reaction, immunotherapy, and immune response to hCoV vaccination.
Limitations of the Study
Our study has several limitations. The most important is the observational nature of studies and the significant heterogeneity of the study results. However, high statistical heterogeneity is more frequent in meta-analyses of prevalence and descriptive studies (113). This can be explained by differences in the patient populations, underlying comorbidities and coinfections, variant treatments, follow-up, time of blood collection, control donors, and cytokine detection assays. Another significant limitation is the variability in laboratory assays used to measure the level of serum cytokines, as local laboratories have different normal ranges based on local data. This confounding variable may somewhat undermine our results. Thus, our data should be interpreted accounting for this important limitation.
The number of included studies on SARS and MERS was low compared to the number of included studies on COVID-19. However, this can be explained by the fact that COVID-19 infection constitutes a larger threat to the global population compared to SARS and MERS.
Despite these limitations, our meta-analysis remained consistent with observational data analysis, demonstrating the importance of CRS for worsening outcomes for the three hCoV diseases. It is also worth considering that less noticeable cytokine elevations associated with the three hCoV infections might suggest a regulated or insufficient inflammatory response to overwhelming viral infections, such as IFN-γ (81).
Conclusions and Future Directions
The immune response and immunopathology of the newly emerging hCoV COVID-19 are somewhat similar to those of SARS and MERS. The three zoonotic viral diseases share clinical outcomes characterized by CRS, for which there is still no specific or effective treatment.
Therefore, there is an urgent need to better understand the role of CRS in the disease progression of hCoVs, as this will provide important insights into the type of immune response or pathophysiological events involved in these diseases, which can help to manage the diseases and design new therapies. We recommend using a combination of existing approved therapies with proven safety profiles, such as IL-6 blockade signaling (tocilizumab), for the treatment of hyperinflammation in severe SARS, MERS, and COVID-19 infections. It would also be helpful to measure the inflammatory biomarkers in routine clinical tests to provide an appropriate early risk-based assessment of hCoV patients and thereby reduce the risk of death from hCoV infections. It would also be relevant to determine whether CRS, direct virus-induced tissue damage, or the synergistic effect of both is associated with severe hCoV complications, such as multiple organ dysfunction.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author Contributions
Conception and design: FM. Acquisition of the data: AZ, AN, HA, and FM. Analysis and interpretation of the data: AZ, AN, HA, and FM. Drafting and revising the article: AZ, AN, HA, and FM. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.666223/full#supplementary-material
References
1. Guarner J. Three Emerging Coronaviruses in Two Decades: The Story of SARS, MERS, and Now COVID-19. Oxford University Press US (2020).
2. Egloff M-P, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, et al. The Severe Acute Respiratory Syndrome-Coronavirus Replicative Protein Nsp9 is a Single-Stranded RNA-binding Subunit Unique in the RNA Virus World. Proc Natl Acad Sci (2004) 101:3792–6. doi: 10.1073/pnas.0307877101
3. Cherry JD, Krogstad P. SARS: The First Pandemic of the 21st Century. Pediatr Res (2004) 56:1–5. doi: 10.1203/01.PDR.0000129184.87042.FC
4. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A Novel Coronavirus Associated With Severe Acute Respiratory Syndrome. N Engl J Med (2003) 348:1953–66. doi: 10.1056/NEJMoa030781
5. Skowronski DM, Astell C, Brunham RC, Low DE, Petric M, Roper RL, et al. Severe Acute Respiratory Syndrome (SARS): A Year in Review. Annu Rev Med (2005) 56:357–81. doi: 10.1146/annurev.med.56.091103.134135
6. Raj VS, Osterhaus AD, Fouchier RA, Haagmans BL. MERS: Emergence of a Novel Human Coronavirus. Curr Opin Virol (2014) 5:58–62. doi: 10.1016/j.coviro.2014.01.010
7. Nowotny N, Kolodziejek J. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Dromedary Camels, Oman, 2013. Euro Surveill (2014) 1920781:20781. doi: 10.2807/1560-7917.ES2014.19.16.20781
8. Spinelli A, Pellino G. Covid-19 Pandemic: Perspectives on an Unfolding Crisis. Br J Surg (2020) 107:785–7. doi: 10.1002/bjs.11627
9. Cucinotta D, Vanelli M. Who Declares Covid-19 a Pandemic. Acta BioMed (2020) 91:157–60. doi: 10.23750/abm.v91i1.9397
10. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
11. Cunha CB, Opal SM. Middle East Respiratory Syndrome (MERS): A New Zoonotic Viral Pneumonia. Virulence (2014) 5:650–4. doi: 10.4161/viru.32077
12. Zumla AI, Memish ZA. Middle East Respiratory Syndrome Coronavirus: Epidemic Potential or a Storm in a Teacup? Eur Respir J (2014) 43:1243–8. doi: 10.1183/09031936.00227213
13. Chen J. Pathogenicity and Transmissibility of 2019-nCoV-A Quick Overview and Comparison With Other Emerging Viruses. Microbes Infect (2020) 22:69–71. doi: 10.1016/j.micinf.2020.01.004
14. Hui DS, Memish ZA, Zumla A. Severe Acute Respiratory Syndrome vs. The Middle East Respiratory Syndrome. Current Opinion in Pulmonary Medicine. Curr Opin Pulm Med (2014) 20:233–41. doi: 10.1097/MCP.0000000000000046
15. Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y. From SARS and MERS to COVID-19: A Brief Summary and Comparison of Severe Acute Respiratory Infections Caused by Three Highly Pathogenic Human Coronaviruses. Respir Res (2020) 21:1–14. doi: 10.1186/s12931-020-01479-w
16. Channappanavar R, Perlman S. Pathogenic Human Coronavirus Infections: Causes and Consequences of Cytokine Storm and Immunopathology. Semin In Immunopathol (2017) 39:529–39. doi: 10.1007/s00281-017-0629-x
17. Lei S, Jiang F, Su W, Chen C, Chen J, Mei W, et al. Clinical Characteristics and Outcomes of Patients Undergoing Surgeries During the Incubation Period of COVID-19 Infection. EClinicalMedicine (2020) 21:100331. doi: 10.1016/j.eclinm.2020.100331
18. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
19. Guery B, Poissy J, el Mansouf L, Sejourne C, Ettahar N, Lemaire X, et al. Clinical Features and Viral Diagnosis of Two Cases of Infection With Middle East Respiratory Syndrome Coronavirus: A Report of Nosocomial Transmission. Lancet (2013) 381:2265–72. doi: 10.1016/S0140-6736(13)60982-4
20. Booth CM, Matukas LM, Tomlinson GA, Rachlis AR, Rose DB, Dwosh HA, et al. Clinical Features and Short-Term Outcomes of 144 Patients With SARS in the Greater Toronto Area. JAMA (2003) 289:2801–9. doi: 10.1001/jama.289.21.JOC30885
21. Petrosillo N, Viceconte G, Ergonul O, Ippolito G, Petersen E. Covid-19, SARS and MERS: Are They Closely Related? Clin Microbiol Infect (2020) 26:729–34. doi: 10.1016/j.cmi.2020.03.026
22. Shimabukuro-Vornhagen A, Godel P, Subklewe M, Stemmler HJ, Schlosser HA, Schlaak M, et al. Cytokine Release Syndrome. J Immunother Cancer (2018) 6:56. doi: 10.1186/s40425-018-0343-9
23. Teijaro JR. Cytokine Storms in Infectious Diseases. Semin Immunopathol (2017) 39:501–3. doi: 10.1007/s00281-017-0640-2
24. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med (2020) 383:2255–73. doi: 10.1056/NEJMra2026131
25. Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human Immunopathogenesis of Severe Acute Respiratory Syndrome (SARS). Virus Res (2008) 133:13–9. doi: 10.1016/j.virusres.2007.02.014
26. Lau SKP, Lau CCY, Chan KH, Li CPY, Chen H, Jin DY, et al. Delayed Induction of Proinflammatory Cytokines and Suppression of Innate Antiviral Response by the Novel Middle East Respiratory Syndrome Coronavirus: Implications for Pathogenesis and Treatment. J Gen Virol (2013) 94:2679–90. doi: 10.1099/vir.0.055533-0
27. Mangalmurti N, Hunter CA. Cytokine Storms: Understanding Covid-19. Immunity (2020) 53:19–25. doi: 10.1016/j.immuni.2020.06.017
28. Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe (2020) 27883-890:e2. doi: 10.1016/j.chom.2020.04.017
29. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-Analysis of Observational Studies in Epidemiology: A Proposal for Reporting. Jama (2000) 283:2008–12. doi: 10.1001/jama.283.15.2008
30. Picot J, Hartwell D, Harris P, Mendes D, Clegg A, Takeda A. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses Checklist, The Effectiveness of Interventions to Treat Severe Acute Malnutrition in Young Children: A Systematic Review. NIHR J Lib (2012) 12(19):117–8. doi: 10.3310/hta16190
31. Wells GA, Shea B, O’Connell DA, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Oxford (2000). doi: 10.1186/1471-2288-5-13
32. Park JY, Howren AM, Zusman EZ, Esdaile JM, De Vera MA. The Incidence of Depression and Anxiety in Patients With Ankylosing Spondylitis: A Systematic Review and Meta-Analysis. BMC Rheumatol (2020) 4:1–8. doi: 10.1186/s41927-019-0111-6
33. Hozo SP, Djulbegovic B, Hozo I. Estimating the Mean and Variance From the Median, Range, and the Size of a Sample. BMC Med Res Method (2005) 5:13. doi: 10.1186/1471-2288-5-13
34. Deeks JJ, Higgins JP, Altman DG, Group CSM. Analysing Data and Undertaking Meta-Analyses. In: Cochrane Handbook for Systematic Reviews of Interventions. The Cochrane Library, Chichester, UK: John Wiley & Sons Ltd. (2019) p. 241–84. doi: 10.1002/9781119536604.ch10
35. Zhang Y, Li J, Zhan Y, Wu L, Yu X, Zhang W, et al. Analysis of Serum Cytokines in Patients With Severe Acute Respiratory Syndrome. Infect Immun (2004) 72:4410–5. doi: 10.1128/IAI.72.8.4410-4415.2004
36. Hong KH, Choi JP, Hong SH, Lee J, Kwon JS, Kim SM, et al. Predictors of Mortality in Middle East Respiratory Syndrome (MERS). Thorax (2018) 73:286–9. doi: 10.1136/thoraxjnl-2016-209313
37. Kim ES, Choe PG, Park WB, Oh HS, Kim EJ, Nam EY, et al. Clinical Progression and Cytokine Profiles of Middle East Respiratory Syndrome Coronavirus Infection. J Kor Med Sci (2016) 31:1717–25. doi: 10.3346/jkms.2016.31.11.1717
38. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J Clin Invest (2020) 130:2620–9. doi: 10.1172/JCI137244
39. Gao Y, Li T, Han M, Li X, Wu D, Xu Y, et al. Diagnostic Utility of Clinical Laboratory Data Determinations for Patients With the Severe COVID-19. J Med Virol (2020) 92:791–6. doi: 10.1002/jmv.25770
40. Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, et al. Profiling Serum Cytokines in COVID-19 Patients Reveals IL-6 and IL-10 are Disease Severity Predictors. Emerg Microbes Infect (2020) 9:1123–30. doi: 10.1080/22221751.2020.1770129
41. He S, Zhou C, Lu D, Yang H, Xu H, Wu G, et al. Relationship Between Chest CT Manifestations and Immune Response in COVID-19 Patients. Int J Infect Dis (2020) 20:125–9. doi: 10.1016/j.ijid.2020.06.059
42. Herold T, Jurinovic V, Arnreich C, Lipworth BJ, Hellmuth JC, Bergwelt-Baildon MV, et al. Elevated Levels of IL-6 and CRP Predict the Need for Mechanical Ventilation in COVID-19. J Allergy Clin Immunol (2020) 146(1):128–36. doi: 10.1016/j.jaci.2020.05.008
43. Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation Between Relative Nasopharyngeal Virus RNA Load and Lymphocyte Count Disease Severity in Patients With COVID-19. Viral Immunol (2020) 10:125–9. doi: 10.1089/vim.2020.0062
44. Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S. IL-6 and CD8+ T Cell Counts Combined are an Early Predictor of in-Hospital Mortality of Patients With COVID-19. JCI Insight (2020) 5:09. doi: 10.1172/jci.insight.139024
45. McElvaney OJ, McEvoy N, McElvaney OF, Carroll TP, Murphy MP, Dunlea DM, et al. Characterization of the Inflammatory Response to Severe Covid-19 Illness. Am J Respir Crit Care Med (2020) 25:812–21. doi: 10.1164/rccm.202005-1583OC
46. Chen R, Sang L, Jiang M, Yang Z, Jia N, Fu W, et al. Longitudinal Hematologic and Immunologic Variations Associated With the Progression of COVID-19 Patients in China. J Allergy Clin Immunol (2020) 146(1):89–100. doi: 10.1016/j.jaci.2020.05.003
47. Wan S, Yi Q, Fan S, Lv J, Zhang X, Guo L, et al. Relationships Among Lymphocyte Subsets, Cytokines, and the Pulmonary Inflammation Index in Coronavirus (COVID-19) Infected Patients. Br J Haematol (2020) 189:428–37. doi: 10.1111/bjh.16659
48. Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable Serum SARS-CoV-2 Viral Load (Rnaaemia) is Closely Correlated With Drastically Elevated Interleukin 6 (IL-6) Level in Critically Ill COVID-19 Patients. Clin Infect Dis (2020) 17:1937–42. doi: 10.1101/2020.02.29.20029520
49. Yang AP, Li HM, Tao WQ, Yang XJ, Wang M, Yang WJ, et al. Infection With SARS-CoV-2 Causes Abnormal Laboratory Results of Multiple Organs in Patients. Aging (2020) 12:10059–69. doi: 10.18632/aging.103255
50. Yuan X, Huang W, Ye B, Chen C, Huang R, Wu F, et al. Changes of Hematological and Immunological Parameters in COVID-19 Patients. Int J Hematol (2020) 112(4):553–9. doi: 10.1007/s12185-020-02930-w
51. Zhou Y, Han T, Chen J, Hou C, Hua L, He S, et al. Clinical and Autoimmune Characteristics of Severe and Critical Cases With COVID-19. Clin Trans Sci (2020) 62(5):388–93. doi: 10.1111/cts.12805
52. Zhu Z, Cai T, Fan L, Lou K, Hua X, Huang Z, et al. Clinical Value of Immune-Inflammatory Parameters to Assess the Severity of Coronavirus Disease 2019. Int J Infect Dis (2020) 95:332–9. doi: 10.1016/j.ijid.2020.04.041
53. Liu B, Li M, Zhou Z, Guan X, Xiang Y. Can We Use Interleukin-6 (IL-6) Blockade for Coronavirus Disease 2019 (COVID-19)-induced Cytokine Release Syndrome (CRS)? J Autoimmun (2020) 111:102452. doi: 10.1016/j.jaut.2020.102452
54. Wang W, Ye L, Ye L, Li B, Gao B, Zeng Y, et al. Up-Regulation of IL-6 and TNF-alpha Induced by SARS-coronavirus Spike Protein in Murine Macrophages Via NF-kappaB Pathway. Virus Res (2007) 128:1–8. doi: 10.1016/j.virusres.2007.02.007
55. Zhou J, Chu H, Li C, Wong BHY, Cheng ZS, Poon VKM, et al. Active Replication of Middle East Respiratory Syndrome Coronavirus and Aberrant Induction of Inflammatory Cytokines and Chemokines in Human Macrophages: Implications for Pathogenesis. J Infect Dis (2014) 209:1331–42. doi: 10.1093/infdis/jit504
56. Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine Release Syndrome in Severe COVID-19: Interleukin-6 Receptor Antagonist Tocilizumab may be the Key to Reduce Mortality. Int J Antimicrob Agents (2020) 55(5):105954–60. doi: 10.1016/j.ijantimicag.2020.105954
57. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
58. Sun H, Guo P, Zhang L, Wang F. Serum Interleukin-6 Concentrations and the Severity of COVID-19 Pneumonia: A Retrospective Study At a Single Center in Bengbu City, Anhui Province, China, in January and February 2020. Med Sci Monit (2020) 26:e926941. doi: 10.12659/MSM.926941
59. Wang C, Xie J, Zhao L, Fei X, Zhang H, Tan Y, et al. Aveolar macrophage activation and cytokine storm in the pathogenesis of severe COVID-19. [Preprint] (2020). doi: 10.21203/rs.3.rs-19346/v1
60. Chen Y, Thai P, Zhao Y-H, Ho Y-S, DeSouza MM, Wu R. Stimulation of Airway Mucin Gene Expression by Interleukin (IL)-17 Through IL-6 Paracrine/Autocrine Loop. J Biol Chem (2003) 278:17036–43. doi: 10.1074/jbc.M210429200
61. Hadid T, Kafri Z, Al-Katib A. Coagulation and Anticoagulation in COVID-19. Blood Rev (2020). 100761 doi: 10.1016/j.blre.2020.100761
62. Rapkiewicz AV, Mai X, Carsons SE, Pittaluga S, Kleiner DE, Berger JS, et al. Megakaryocytes and Platelet-Fibrin Thrombi Characterize Multi-Organ Thrombosis At Autopsy in COVID-19: A Case Series. EClinicalMedicine (2020) 24:100434. doi: 10.1016/j.eclinm.2020.100434
63. McGonagle D, O’Donnell JS, Sharif K, Emery P, Bridgewood C. Immune Mechanisms of Pulmonary Intravascular Coagulopathy in COVID-19 Pneumonia. Lancet Rheumatol (2020) 2:e437–45. doi: 10.1016/S2665-9913(20)30121-1
64. Tanaka T, Narazaki M, Kishimoto T. Immunotherapeutic Implications of IL-6 Blockade for Cytokine Storm. Immunotherapy (2016) 8:959–70. doi: 10.2217/imt-2016-0020
65. Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, et al. The Use of Anti-Inflammatory Drugs in the Treatment of People With Severe Coronavirus Disease 2019 (COVID-19): The Perspectives of Clinical Immunologists From China. Clin Immunol (2020) 214:108393. doi: 10.1016/j.clim.2020.108393
66. Toniati P, Piva S, Cattalini M, Garrafa E, Regola F, Castelli F, et al. Tocilizumab for the Treatment of Severe COVID-19 Pneumonia With Hyperinflammatory Syndrome and Acute Respiratory Failure: A Single Center Study of 100 Patients in Brescia, Italy. Autoimmun Rev (2020) 19(7):102568. doi: 10.1016/j.autrev.2020.102568
67. Hermine O, Mariette X, Tharaux PL, Resche-Rigon M, Porcher R, Ravaud P, et al. Effect of Tocilizumab vs Usual Care in Adults Hospitalized With Covid-19 and Moderate or Severe Pneumonia: A Randomized Clinical Trial. JAMA Intern Med (2021) 181:32–40. doi: 10.1001/jamainternmed.2020.6820
68. Steensberg A, Fischer CP, Keller C, Moller K, Pedersen BK. IL-6 Enhances Plasma IL-1ra, Il-10, and Cortisol in Humans. Am J Physiol Endocrinol Metab (2003) 285:E433–7. doi: 10.1152/ajpendo.00074.2003
69. Dhar SK, Vishnupriyan K, Damodar S, Gujar S, Das M. IL-6 and IL-10 as Predictors of Disease Severity in COVID-19 Patients: Results From Meta-Analysis and Regression. Heliyon (2021) 7:e06155. doi: 10.1016/j.heliyon.2021.e06155
70. Moore KW, O’Garra A, de Waal Malefyt R, Vieira P, Mosmann TR. Interleukin-10. Annu Rev Immunol (1993) 11:165–90. doi: 10.1146/annurev.iy.11.040193.001121
71. Clarke CJ, Hales A, Hunt A, Foxwell BM. Il-10-mediated Suppression of TNF-alpha Production is Independent of its Ability to Inhibit NF Kappa B Activity. Eur J Immunol (1998) 28:1719–26. doi: 10.1002/(SICI)1521-4141(199805)28:05<1719::AID-IMMU1719>3.0.CO;2-Q
72. Li J, Rong L, Cui R, Feng J, Jin Y, Yu Y, et al. Dynamic Changes in Serum IL-6, Il-8, and IL-10 are Associated With the Outcome of Patients With Severe COVID-19 in ICU. [Preprint] (2020). doi: 10.21203/rs.3.rs-83336/v1
73. Maris CH, Chappell CP, Jacob J. Interleukin-10 Plays an Early Role in Generating Virus-Specific T Cell Anergy. BMC Immunol (2007) 8:1–9. doi: 10.1186/1471-2172-8-8
74. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
75. Greenberger MJ, Strieter RM, Kunkel SL, Danforth JM, Goodman RE, Standiford TJ. Neutralization of IL-10 Increases Survival in a Murine Model of Klebsiella Pneumonia. J Immunol (1995) 155:722–9.
76. Hotchkiss RS, Monneret G, Payen D. Sepsis-Induced Immunosuppression: From Cellular Dysfunctions to Immunotherapy. Nat Rev Immunol (2013) 13:862–74. doi: 10.1038/nri3552
77. Chen C, Zhang XR, Ju ZY, He WF. [Advances in the Research of Mechanism and Related Immunotherapy on the Cytokine Storm Induced by Coronavirus Disease 2019]. Zhonghua Shao Shang Za Zhi (2020) 36:471–5. doi: 10.376/cma.j.cn501120-20200224-00088
78. Le RQ, Li L, Yuan W, Shord SS, Nie L, Habtemariam BA, et al. Fda Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist (2018) 23:943–7. doi: 10.1634/theoncologist.2018-0028
79. Corominas H, Castellvi I, Pomar V, Antonijoan R, Mur I, Matas L, et al. Effectiveness and Safety of Intravenous Tocilizumab to Treat COVID-19-associated Hyperinflammatory Syndrome: Covizumab-6 Observational Cohort. Clin Immunol (2021) 223:108631. doi: 10.1016/j.clim.2020.108631
80. Rosas IO, Brau N, Waters M, Go RC, Hunter BD, Bhagani S, et al. Tocilizumab in Hospitalized Patients With Severe Covid-19 Pneumonia. N Engl J Med (2021). doi: 10.1056/NEJMoa2028700
81. Leisman DE, Ronner L, Pinotti R, Taylor MD, Sinha P, Calfee CS, et al. Cytokine Elevation in Severe and Critical COVID-19: A Rapid Systematic Review, Meta-Analysis, and Comparison With Other Inflammatory Syndromes. Lancet Respir Med (2020) 8:1233–44. doi: 10.1016/S2213-2600(20)30404-5
82. Wang J, Wang BJ, Yang JC, Wang MY, Chen C, Luo GX, et al. [Advances in the Research of Mechanism of Pulmonary Fibrosis Induced by Corona Virus Disease 2019 and the Corresponding Therapeutic Measures]. Zhonghua Shao Shang Za Zhi (2020) 36:E006. doi: 10.376/cma.j.cn501120-20200307-00132
83. Gasparello J, Finotti A, Gambari R. Tackling the COVID-19 “Cytokine Storm” With microRNA Mimics Directly Targeting the 3’UTR of Pro-Inflammatory Mrnas. Med Hypotheses (2021) 146:110415. doi: 10.1016/j.mehy.2020.110415
84. Giuliani C, Bucci I, Napolitano G. Phenylmethimazole is a Candidate Drug for the Treatment of Severe Forms of Coronavirus Disease 2019 (COVID-19) as Well as Other Virus-Induced “Cytokines Storm”. Med Hypotheses (2021) 146:110473. doi: 10.1016/j.mehy.2020.110473
85. Alosaimi B, Hamed ME, Naeem A, Alsharef AA, AlQahtani SY, AlDosari KM, et al. Mers-CoV Infection is Associated With Downregulation of Genes Encoding Th1 and Th2 Cytokines/Chemokines and Elevated Inflammatory Innate Immune Response in the Lower Respiratory Tract. Cytokine (2020) 126:154895. doi: 10.1016/j.cyto.2019.154895
86. Yoshikawa T, Hill T, Li K, Peters CJ, Tseng CTK. Severe Acute Respiratory Syndrome (SARS) Coronavirus-Induced Lung Epithelial Cytokines Exacerbate SARS Pathogenesis by Modulating Intrinsic Functions of Monocyte-Derived Macrophages and Dendritic Cells. J Virol (2009) 83:3039–48. doi: 10.1128/JVI.01792-08
87. Cheung CY, Poon LL, Ng IH, Luk W, Sia S-F, Wu MH, et al. Cytokine Responses in Severe Acute Respiratory Syndrome Coronavirus-Infected Macrophages In Vitro: Possible Relevance to Pathogenesis. J Virol (2005) 79:7819–26. doi: 10.1128/JVI.79.12.7819-7826.2005
88. Tincati C, Cannizzo ES, Giacomelli M, Badolato R, Monforte ADA, Marchetti G. Heightened Circulating Interferon-Inducible Chemokines, and Activated Pro-Cytolytic Th1-Cell Phenotype Features Covid-19 Aggravation in the Second Week of Illness. Front Immunol (2020) 11:580987. doi: 10.3389/fimmu.2020.580987
89. Chen Y, Wang J, Liu C, Su L, Zhang D, Fan J, et al. Ip-10 and MCP-1 as Biomarkers Associated With Disease Severity of COVID-19. Mol Med (2020) 26:1–12. doi: 10.1186/s10020-020-00230-x
90. Rollins BJ, Yoshimura T, Leonard EJ, Pober JS. Cytokine-Activated Human Endothelial Cells Synthesize and Secrete a Monocyte Chemoattractant, MCP-1/JE. Am J Pathol (1990) 136:1229–33.
91. Romano M, Sironi M, Toniatti C, Polentarutti N, Fruscella P, Ghezzi P, et al. Role of IL-6 and its Soluble Receptor in Induction of Chemokines and Leukocyte Recruitment. Immunity (1997) 6:315–25. doi: 10.1016/S1074-7613(00)80334-9
92. Bodnar RJ, Yates CC, Wells A. Ip-10 Blocks Vascular Endothelial Growth Factor-Induced Endothelial Cell Motility and Tube Formation Via Inhibition of Calpain. Circ Res (2006) 98:617–25. doi: 10.1161/01.RES.0000209968.66606.10
93. van den Borne P, Quax PH, Hoefer IE, Pasterkamp G. The Multifaceted Functions of CXCL10 in Cardiovascular Disease. BioMed Res Int (2014) 2014:893106. doi: 10.1155/2014/893106
94. Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte Chemoattractant Protein-1 (MCP-1): An Overview. J Interferon Cytokine Res (2009) 29:313–26. doi: 10.1089/jir.2008.0027
95. Xu ZS, Shu T, Kang L, Wu D, Zhou X, Liao BW, et al. Temporal Profiling of Plasma Cytokines, Chemokines and Growth Factors From Mild, Severe and Fatal COVID-19 Patients. Signal Transduct Target Ther (2020) 5:100. doi: 10.1038/s41392-020-0211-1
96. Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, et al. Serum Cytokine and Chemokine Profile in Relation to the Severity of Coronavirus Disease 2019 (COVID-19) in China. J Infect Dis (2020) 21:21. doi: 10.1093/infdis/jiaa363
97. Xi X, Guo Y, Zhu M, Wei Y, Li G, Du B, et al. Higher Expression of Monocyte Chemotactic Protein 1 in Mild COVID-19 Patients Might be Correlated With Inhibition of Type I IFN Signaling. Virol J (2021) 18:1–7. doi: 10.1186/s12985-020-01478-9
98. Khalil BA, Elemam NM, Maghazachi AA. Chemokines and Chemokine Receptors During COVID-19 Infection. Comput Struct Biotechnol J (2021) 19:976–88. doi: 10.1016/j.csbj.2021.01.034
99. Wichmann D, Sperhake JP, Lutgehetmann M, Steurer S, Edler C, Heinemann A, et al. Autopsy Findings and Venous Thromboembolism in Patients With Covid-19: A Prospective Cohort Study. Ann Intern Med (2020) 173:268–77. doi: 10.7326/L20-1206
100. Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, et al. A Dynamic COVID-19 Immune Signature Includes Associations With Poor Prognosis. Nat Med (2020) 26:1623–35. doi: 10.1038/s41591-020-1038-6
101. Zhang H, Kang D, Huang B, Liu N, Zhao F, Zhan P, et al. Discovery of non-Peptide Small Molecular CXCR4 Antagonists as anti-HIV Agents: Recent Advances and Future Opportunities. Eur J Med Chem (2016) 114:65–78. doi: 10.1016/j.ejmech.2016.02.051
102. Kim MB, Giesler KE, Tahirovic YA, Truax VM, Liotta DC, Wilson LJ. CCR5 Receptor Antagonists in Preclinical to Phase II Clinical Development for Treatment of HIV. Expert Opin Investig Drugs (2016) 25:1377–92. doi: 10.1080/13543784.2016.1254615
103. Lupieri A, Smirnova NF, Solinhac R, Malet N, Benamar M, Saoudi A, et al. Smooth Muscle Cells-Derived CXCL10 Prevents Endothelial Healing Through PI3Kγ-Dependent T Cells Response. Cardiovasc Res (2020) 116:438–49. doi: 10.1093/cvr/cvz122
104. Park JH, Lee HK. Re-Analysis of Single Cell Transcriptome Reveals That the NR3C1-CXCL8-neutrophil Axis Determines the Severity of COVID-19. Front Immunol (2020) 11:2145. doi: 10.3389/fimmu.2020.02145
105. Pum A, Ennemoser M, Adage T, Kungl AJ. Cytokines and Chemokines in SARS-CoV-2 Infections—Therapeutic Strategies Targeting Cytokine Storm. Biomolecules (2021) 11:91. doi: 10.3390/biom11010091
106. Qin L, Li X, Shi J, Yu M, Wang K, Tao Y, et al. Gendered Effects on Inflammation Reaction and Outcome of COVID-19 Patients in Wuhan. J Med Virol (2020) 92(11):2684–92. doi: 10.1002/jmv.26137
107. Torcia MG, Nencioni L, Clemente AM, Civitelli L, Celestino I, Limongi D, et al. Sex Differences in the Response to Viral Infections: TLR8 and TLR9 Ligand Stimulation Induce Higher IL10 Production in Males. PLoS One (2012) 7:e39853. doi: 10.1371/journal.pone.0039853
108. Sankaran-Walters S, Macal M, Grishina I, Nagy L, Goulart L, Coolidge K, et al. Sex Differences Matter in the Gut: Effect on Mucosal Immune Activation and Inflammation. Biol Sex Dif (2013) 4:1–12. doi: 10.1186/2042-6410-4-10
109. Abdullah M, Chai P-S, Chong M-Y, Tohit ERM, Ramasamy R, Pei CP, et al. Gender Effect on In Vitro Lymphocyte Subset Levels of Healthy Individuals. Cell Immunol (2012) 272:214–9. doi: 10.1016/j.cellimm.2011.10.009
110. Huang H, Luo Y, Liang Y, Long X, Peng Y, Liu Z, et al. Cd4+ Cd25+ T Cells in Primary Malignant Hypertension Related Kidney Injury. Sci Rep (2016) 6:1–9. doi: 10.1038/srep27659
111. Rucker AJ, Crowley SD. The Role of Macrophages in Hypertension and its Complications. Pflügers Archiv-Eur J Physiol (2017) 469:419–30. doi: 10.1007/s00424-017-1950-x
112. Silawal S, Willauschus M, Schulze-Tanzil G, Gögele C, Geßlein M, Schwarz S. Il-10 Could Play a Role in the Interrelation Between Diabetes Mellitus and Osteoarthritis. Int J Mol Sci (2019) 20:768. doi: 10.3390/ijms20030768
Keywords: cytokine storm, inflammatory cytokines, COVID-19, MERS, SARS, SARS-CoV-2
Citation: Zawawi A, Naser AY, Alwafi H and Minshawi F (2021) Profile of Circulatory Cytokines and Chemokines in Human Coronaviruses: A Systematic Review and Meta-Analysis. Front. Immunol. 12:666223. doi: 10.3389/fimmu.2021.666223
Received: 09 February 2021; Accepted: 09 April 2021;
Published: 05 May 2021.
Edited by:
Aldo Tagliabue, Italian National Research Council, ItalyReviewed by:
Franco Pandolfi, Catholic University of the Sacred Heart, Rome, ItalyCecilia Garlanda, Humanitas University, Italy
Copyright © 2021 Zawawi, Naser, Alwafi and Minshawi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faisal Minshawi, Zm9taW5zaGF3aUB1cXUuZWR1LnNh
 Ayat Zawawi
Ayat Zawawi Abdallah Y. Naser
Abdallah Y. Naser Hassan Alwafi
Hassan Alwafi Faisal Minshawi
Faisal Minshawi