- 1Department of BioChemistry, West Bengal University of Animal and Fishery Sciences, Kolkata, India
- 2Department of LFC, West Bengal University of Animal and Fishery Sciences, Kolkata, India
- 3Department of Animal Science, Visva Bharati University, Bolpur, India
- 4Department of Computer Science, Indian Institute of Technology, Kharagpur, India
- 5Department of Parasitology, West Bengal University of Animal and Fishery Sciences, Kolkata, India
CD14 (also known as the monocyte differentiation antigen) is an important immune response gene known to be primarily responsible for innate immunity against bacterial pathogens, and as a pattern recognition receptor (PRR), binds with LPS (endotoxin), lipoproteins, and lipotechoic acid of bacteria. So far very limited work has been conducted in parasitic immunology. In the current study, we reported the role of CD14 in parasitic immunology in livestock species (sheep) for the first time. Ovine CD14 is characterized as a horse-shoe shaped bent solenoid with a hydrophobic amino-terminal pocket for CD14 along with domains. High mutation frequency was observed, out of total 41 mutations identified, 23 mutations were observed to be thermodynamically unstable and 11 mutations were deleterious in nature, causing major functional alteration of important domains of CD14, an indication of variations in individual susceptibility for sheep against Haemonchus contortus infestations. In silico studies with molecular docking reveal a role of immune response against Haemonchus contortus in sheep, which is later confirmed with experimental evidence through differential mRNA expression analysis for sheep, which revealed better expression of CD14 in Haemonchus contortus infected sheep compared to that of non-infected sheep. We confirmed the above findings with supportive evidence through haematological and biochemical analyses. Phylogenetic analysis was conducted to assess the evolutionary relationship with respect to humans and it was observed that sheep may well be used as model organisms due to better genetic closeness compared to that of mice.
Introduction
The CD14 molecule is an important pattern recognition receptor for innate immunity reported primarily against bacteria through the process of sensitization of host cells to bacteria. CD14 exists in both membranous (mCD14) and soluble (sCD14) forms, ranging from 1 to 371 (1, 2). mCD14 is GPI-anchored, whereas sCD14 form due to bypassing of GPI addition, cleavage of the GPI anchor by phospholipase D, or direct proteolytic cleavage from the cell surface (3–5). Bacterial lipopolysaccharide or LPS (endotoxin), lipoproteins, lipoteichoic acid, and other acylated microbial products are the site of action for CD14. CD14 acts through various TLR (Toll-like Receptor) signalling complexes causing ligand binding followed by induction of intracellular proinflammatory signalling cascades (6, 7). In Gram-negative bacteria, LPS-binding protein (LBP) aids in the process of binding of LPS to CD14 (8, 9), produced by liver hepatocytes (10–12), which ultimately combines to the TLR4 complex, concentrates low levels of LPS, and leads to the increase of the sensitivity of the system (9, 10, 13, 14). MD-2 is another important co-receptor which is physically associated with TLR4 (15, 16). CD14 is a catalyst in the process of transportation of LPS to the TLR4–MD-2 complex (13, 17–20). CD14 may cause opsonisation of whole bacteria and apoptotic cells (21–25) and interact and bind to certain host phospholipids (26–28), and a wide range of acylated bacterial agonists of TLR2, namely lipoteichoic acid, peptidoglycan, mycobacterial lipoarabinomannan, atypical LPSs, and lipoproteins increasing cellular inflammatory responses (29–39). Soluble CD14 induces B cell growth and differentiation as present in bovine colostrums and milk (40). CD14 is an immune response molecule against both gram negative and gram positive bacteria (such as Mycobacterium sp., Pseudomonas sp., and Staphylococcus aureus). CD14 is an important drug target for the treatment of various diseases, like sepsis (37, 38), mastitis (41), treponemiasis (42), and glomerulonephritis (43), since it has a role in TLR agonist delivery. CD14 is equally important for parasitic diseases (44–49). CD14 as an immune response gene may be utilized in various modes for disease management. These include therapeutic use of recombinant protein, gene therapy with cloned insert, evolvement of transgenic, or gene edited disease resistant animals. Genes for innate resistance may be of practical importance for studying phylogenetic relationships. Some preliminary comparisons between humans and other mammals have already been made (50, 51). Since the CD14 gene shows a considerable degree of variability among the species, it can be used as a marker for phylogenetic analysis.
Cloning and sequencing of the CD14 gene in domestic ruminants like cattle (Bos taurus (52), Buffalo (Bubalus bubalis (53), and goat (Capra hircus (54), have already been conducted earlier in our lab. The present investigation aims towards the cloning and sequencing of the CD14 gene in sheep (Ovis aries), another important domestic ruminant. Currently, only the crystal structure of mouse CD14 (unliganded) purified from SF9 insect cells (39) and human CD14 (55) are available in databases. Both mouse and human CD14 have an N-terminal hydrophobic cavity that provides a putative binding site for LPS and other acylated ligands. So, the present work objective is to computationally model the structures of CD14 in sheep using the existing experimentally determined structures of CD14 of livestock we studied earlier. Additionally, we focus on identifying and comparing the structural and functional domain of CD14 derived peptide using bioinformatics tools, and exploring the domains in 3D structure responsible for pathogen binding. The molecular phylogenetic analysis in domestic ruminants with other domestic and wild mammals and avians with respect to CD14 gene (responsible for innate resistance) and derived peptide, has also been studied which might be helpful to look into evolutionary ecology and pharmacogenomic studies. In silico analysis with molecular docking were conducted to detect the role of CD14 in parasitic immunity. Differential mRNA expression profiling for non-infected and infected (Haemonchus infested) sheep with respect to the CD14 gene was conducted to confirm the above findings.
Materials and Methods
Sample Collection and RNA Isolation
Sheep liver tissue (1g) was collected from a slaughter house located under Kolkata Municipality Corporation. Adult males (n=6) of about 1-1.5 years of age were selected for collection of samples. Liver tissue was immersed in Trizol in vial and transported in ice to the laboratory for RNA isolation. Total RNA was isolated using TRIzol extraction method (Life Technologies, USA), as per the standard procedure and was further utilized for cDNA synthesis (56). Since the samples were collected from the slaughter house, ethical approval was not necessary. cDNA concentration was estimated and samples above 1200 microgram per ml were considered for further study. cDNA was employed for the characterization of ovine CD14 gene.
cDNA Synthesis and PCR Amplification of the CD14 Gene
20μL of reaction mixture was composed of 5μg of total RNA, 0.5μg of oligo dT primer (16–18mer), 40U of Ribonuclease inhibitor, 1000M of dNTP mix, 10mM of DTT, and 5U of MuMLV reverse transcriptase in appropriate buffer. The reaction mixture was mixed thoroughly followed by incubation at 37°C for 1 hour. The reaction was allowed for up to 10 minutes by heating the mixture at 70°C unliganded and then chilled on ice. Afterwards, the integrity of the cDNA was checked by performing PCR. Concentration of cDNA was estimated through Nanodrop. Ovine CD14 primer pair was designed based on the CD14 mRNA sequences of cattle (GenBank Acc No.AF141313) using DNASTAR software (Hitachi Miraibio Inc., USA) to amplify full length open reading frame (ORF) of CD14 gene sequence of sheep. The primers were Forward: CD14-1-F ATGGTCTGCGTGCCCTACCTG and Reverse: CD14-70-1- R GGAGCCCGAGGCTTCGCGTAA. 25μL of reaction mixture was comprised of 80–100ng cDNA, 3.0μL 10X PCR assay buffer, 0.5μL of 10mM dNTP, 1U Taq DNA polymerase, 60ng of each primer, and 2mM MgCl2. PCR-reactions were performed in a thermocycler (PTC-200, MJ Research, USA) with cycling conditions as, initial denaturation at 94°C for 3min, further denaturation at 94°C for 30sec, annealing at 61°C for 35sec, and extension at 72°C for 3min were conducted for 35 cycles followed by final extension at 72°C for 10 min.
cDNA Cloning and Sequencing
Amplified product of ovine CD14 was checked by 1% agarose gel electrophoresis. The products were purified from gel using Gel extraction kit (Qiagen GmbH, Hilden, Germany).pGEM-T easy cloning vector (Promega, Madison, WI, USA) was used for cloning. Then, 10μL of ligated product was mixed thoroughly to 200μL competent cells, and heat shock was given at 42°C for 45sec in a water bath. Subsequently, the cells were immediately transferred on chilled ice for 5 min., and SOC medium was added to it. The bacterial culture was centrifuged to obtain the pellet and plated on an LB agar plate containing Ampicillin (100mg/mL) added to the agar plate @1:1000, IPTG (200mg/mL) and X-Gal (20mg/mL) for blue-white screening. Plasmid isolation from overnight-grown culture was carried out by small-scale alkaline lysis method as described in (54). Recombinant plasmids were characterized by PCR using CD14 primers as reported earlier and restriction enzyme digestion. CD14 gene fragments released by enzyme EcoRI (MBI Fermentas, USA) were inserted in recombinant plasmid which was sequenced by dideoxy chain termination method with T7 and SP6 primers in an automated sequencer (ABI prism, Chromous Biotech, Bangalore).
Sequence Analysis
DNASTAR Version 4.0, Inc., USA software was employed for the nucleotide sequence analysis for protein translation, sequence alignments, and contigs comparisons. Novel sequence was submitted to the NCBI Genbank and accession number KY110723.1 was obtained which is available in the public domain. CD14 sequences have already been analysed earlier in our lab for cattle (GU368102), buffalo (DQ457089), and goat (DQ457090). Supplementary Table 1 lists the source and accession numbers for the CD14 sequences used for the current study.
Study of Predicted Ruminant CD14 Protein Using Bioinformatics Tools
Predicted peptide sequences of CD14 gene of sheep and other ruminants (cattle, buffalo, goat), characterized in our lab earlier, were then aligned with the CD14 peptide of other rodent species and Homo sapiens using MAFFT (57). The analysis was conducted for a sequence-based comparative study among ruminants with rodent and human. Signal peptide is essential to prompt a cell to translocate the protein, usually to the cellular membrane and ultimately signal peptide is cleaved to give mature protein. The prediction of presence and location of signal peptide of CD14 gene was conducted using the software (SignalP 3.0 Sewer-prediction results, Technical University of Denmark). Pattern recognition receptor as CD14 is rich in Leucine, hence it is essential to calculate leucine percentage. Leucine percentage was calculated manually from predicted peptide sequence. Di-sulphide bonds are essential for protein folding and stability, ultimately. It is the 3D structure of protein which is biological active. Di-sulphide bonds were predicted using suitable software (http://bioinformatics.bc.edu/clotelab/DiANNA/) (39).
Protein sequence level analysis was employed (http://www.expasy.org./tools/blast/) for assessment of leucine rich repeats (LRR), leucine zipper, detection of Leucine-rich nuclear export signals (NES), and detection of the position of GPI anchor, N-linked glycosylation sites. Since CD14 is a pattern recognition receptor, these are rich in leucine rich repeat, which are essential for pathogen recognition and binding. Leucine zipper is essential for the dimerization of the CD14 molecule. N-linked glycosylation is important for the molecule to determine its membranous or soluble form. Leucine rich nuclear export signal is essential for export of CD14 from nucleus to cytoplasm, whereas GPI anchor is responsible for anchoring in the case of membranous protein. Prosite was used for LRR site detection.
Leucine-rich nuclear export signals (NES) was analysed with NetNES 1.1 Server, Technical university of Denmark. O-linked glycosylation sites were detected using NetOGlyc 3.1 server (http://www.expassy.org/), whereas N-linked glycosylation site were assessed through NetNGlyc 1.0 software (http://www.expassy.org/). Sites for leucine-zipper were detected through Expassy software, Technical university of Denmark (58). Sites for alpha helix and beta sheet were detected using NetSurfP-Protein Surface Accessibility and Secondary Structure Predictions, Technical University of Denmark (59). Domain linker sites were predicted (60). LPS-binding (61) and LPS-signalling sites (62) were predicted based on homology studies with other species of CD14 polypeptide. These sites are important for pathogen recognition and binding.
Three Dimensional Structure Prediction and Model Quality Assessment
A three dimensional model of ovine CD14 polypeptide was predicted through Swissmodel repository (63). Templates possessing the greatest identity of sequences with our target template were identified with PSI-BLAST (http://blast.ncbi.nlm.nih.gov/Blast (64). PHYRE2 server based on ‘Homology modelling approach’ was used to build a three dimensional model of ovine CD14 (65). The molecular visualization tool PyMOL (http://www.pymol.org/) was employed for model generation and visualization of three dimensional structure of ovine CD14. The structure of ovine CD14 molecule was evaluated and assessed for its stereochemical quality (through SAVES, Structural Analysis and Verification Server, http://nihserver.mbi.ucla.edu/SAVES/); then refined and validated through ProSA (Protein Structure Analysis) web server (https://prosa.services.came.sbg.ac.at/prosa) (66). NetSurfP server (http://www.cbs.dtu.dk/services/NetSurfP/ (59) was used for assessing the surface area of ovine CD14 through relative surface accessibility, Z-fit score, and probability of alpha-Helix, beta-strand, and coil score.
Alignment of 3-D structure of ovine CD14 with other ruminant species as cattle, buffalo, and goat were analysed with RMSD estimation to evaluate the structural differentiation by TM-align software (67). Thermodynamic stability of the protein with mutations and the deleterious nature of the mutant amino acid were analysed through I-mutant 2.0 (68) and Provean analysis (69) respectively.
Protein-Protein Interaction Network Depiction
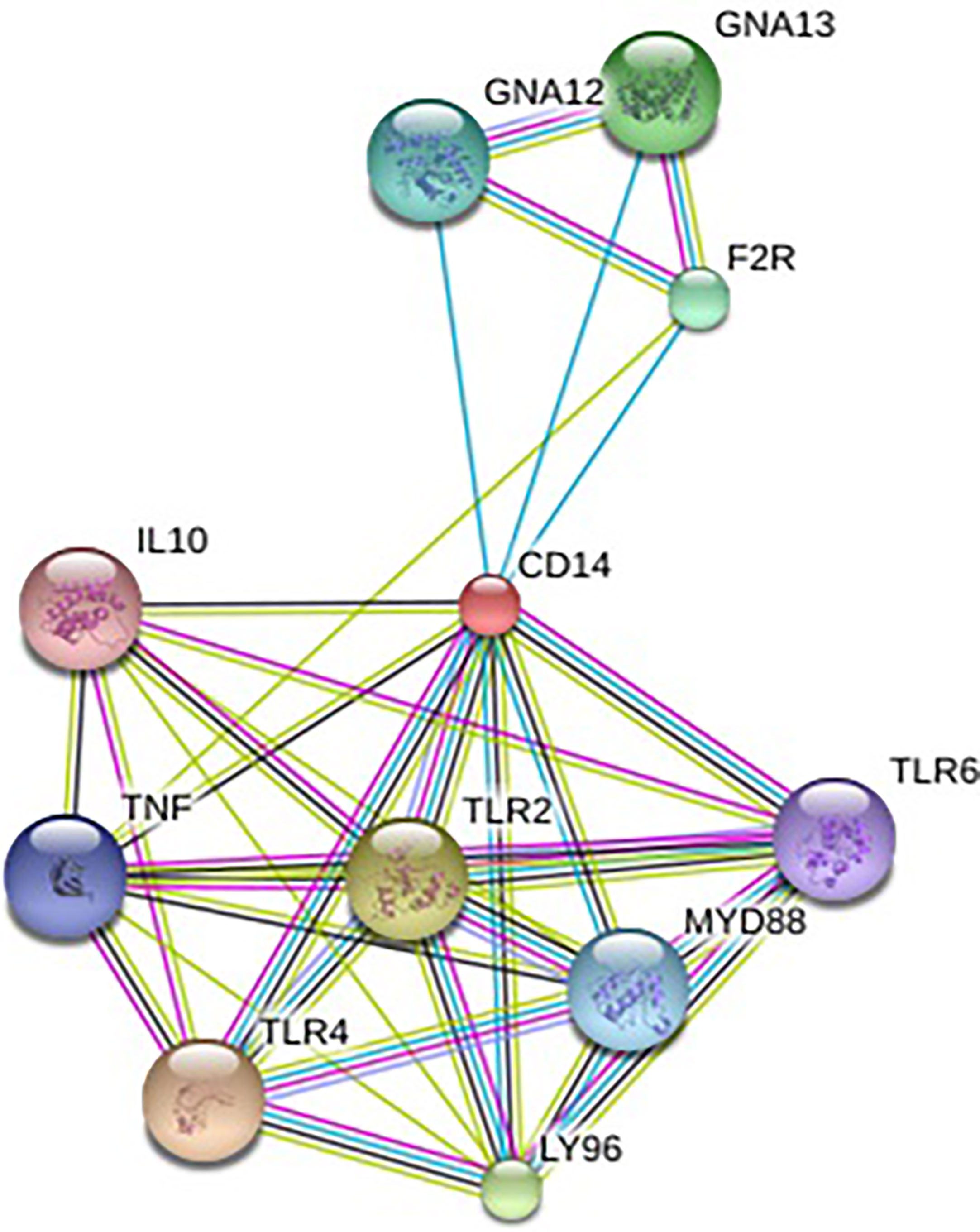
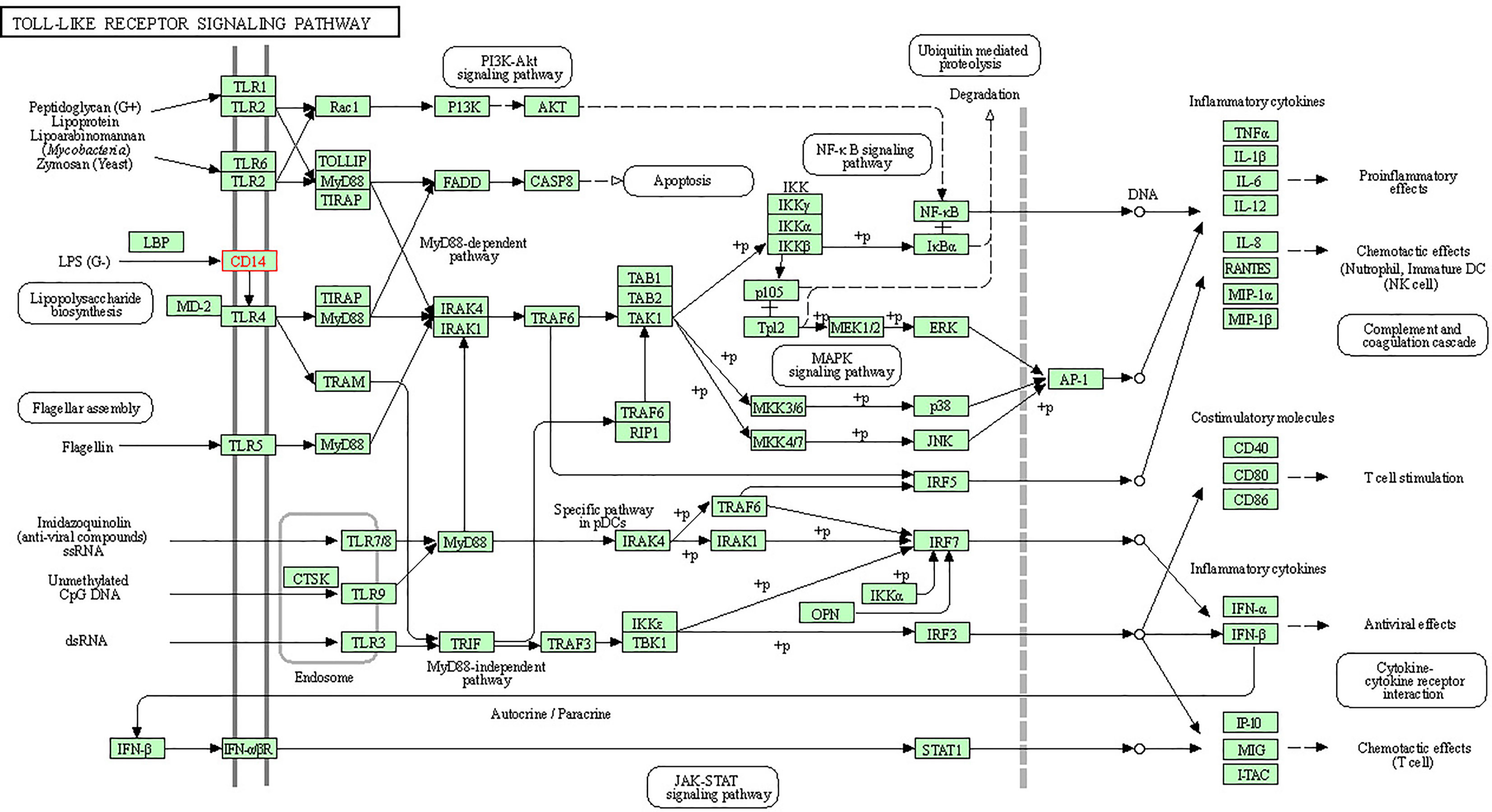
In order to understand the protein interaction network of the CD14 protein, we performed a search in STRING 9.1 database (70). The functional interaction was assessed with confidence scores. Interactions with scores of < 0.3, scores ranging from 0.3 to 0.7, and scores of >0.7 were classified as low, medium, and high confidence respectively. Also, we executed KEGG analysis which depicted the functional association of CD14 with other related proteins.
Phylogenetic Analysis
The nucleotide sequence of Ovine CD14 was aligned with the published nucleotide sequences of other ruminant species we characterized earlier and also with other species obtained from gene bank (http://www.ncbi.nlm.nih.gov/blast) as listed in Supplementary Table 1 for CD14. The ClusterIV method of MegAlign Programme of Lasergene Software (DNASTAR) was employed for the analysis.
Differential mRNA Expression Profiling for the CD14 Gene With Respect to Non-Infected and Infected Sheep
The sheep population under current study was divided in two groups, infected (infected with parasitic infection of Haemonchus contortus) and non-infected (no infection), as assessed through egg per gram count.
Animals, Faecal Sample Collection, and Determination of FEC
We randomly collected 60 sheep samples from LFC farm, WBUAFS, Mohanpur campus. The samples were collected prior to routine deworming procedure, presumed to have pre-existing gastro intestinal parasites since they were not dewormed for the last three months. The animals were presumed to have been exposed to natural infection during grazing.
Faecal samples of the sheep were examined thoroughly by salt floatation method and the Faecal Egg Count were screened for each and every sample. Based on statistical analysis, samples were collected from two groups, designated as non-infected (+SD, where stands for mean and SD means standard deviation) and infected (- SD, where stands for mean and SD means standard deviation). Sheep were regularly sold and slaughtered for the purpose of mutton production unit. Altogether 12 tissue samples were screened, six each with low FEC (grouped as healthy) and six infected samples for further case study. Confirmation of the infected animals were also done through direct visualization of adult parasite in the abomassum and through microscopic examination. Tissue samples were collected for abomassum.
Serum was harvested from blood samples as per the standard procedure. The serum biochemical parameters like glucose, total protein, albumin, globulin, uric acid, creatinine, BUN, triglyceride, cholesterol, HDL, LDL concentrations and serum enzyme variables (AST, ALT, & AP) were estimated by using a semi-auto biochemistry analyser (Span diagnostic Ltd.) with standard kits as per the process described below.
Collection of Blood Samples
Blood sample was collected aseptically from the jugular vein of sheep in the separate sterile vials with anticoagulants in the morning hours between 8 a.m. to 10 a.m. 2-3ml blood was used for haematological parameters, 5ml blood for serum without anticoagulant were collected centrifuged (10 min at 1000 rpm) and preserved at – 20°C until analysis for other biochemical and serological tests.
Haematological Profiles
The haematological parameters like hae moglobin, erythrocyte sedimentation rate (ESR) and packed cell volume (PCV) were estimated in whole blood soon after the collection of blood. Haemoglobin was estimated by acid haematin method (71), E.S.R. and PCV by Wintrobe’s tube (72). The total erythrocyte count (TEC), total leukocyte count (TLC) and Differential leukocyte count (DLC) were studied by standard methods described by Jain (73).
Biochemical Analysis
The serum biochemical parameters estimated in the experiments were total protein, albumin, globulin, albumin: globulin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), Total bilirubin, Indirect bilirubin, direct bilirubin, glucose, uric acid, urea, and BUN by using a semi-auto biochemistry analyser (Span diagnostic Ltd.) with standard kits (Trans Asia Bio-Medicals Ltd., Solan, HP, India). The methodology used for estimation of total protein, albumin, total & direct bilirubin, ALT, ALP, glucose, creatinine urea, and uric acid were biuret method, bromocresol green (BCG) method, 2- 4-DNPH method, modified kind and king’s method, GOD/POD method, modified Jaffe’s Kinetic method. GLDH-urease method, and trinder peroxidise method respectively.
Real Time PCR (qRT-PCR)
An equal quantity of cDNA as quantified through Nanodrop was used in each reaction of 96 well optical plate of ABI 7500 system. Each reaction consisted of 1ng cDNA template, 10 µl of 2X SYBR Green PCR Master Mix, 20pMol each of forward and reverse primers to make up a final volume of up to 20 µl with nuclease free water. Each sample was run in triplicate. Analysis of real-time PCR (qRT-PCR) was performed by delta-delta-Ct (ΔΔCt) method, Ct denotes the threshold value. The list of primers used for QPCR study have been listed below with annealing temperature at 60 C.
CD14 primer: F ACCACCCTCAGTCTCCGTAA,
R- GTGCTTGGGCAATGTTCAG
18S rRNA primer: F-TCCAGCCTTCCTTCCTGGGCAT,
R-GGACAGCACCGTGTTGGCGTAGA.
In Silico Study for the Detection of Binding Site of CD14 With Haemonchus contortus With Molecular Docking
Molecular docking is a bioinformatics tool used for in silico analysis for the prediction of binding mode of a ligand with a protein 3D structure. Patch dock is an algorithm for molecular docking based on shape complementarity principle (74). Patch Dock algorithm was used to predict ligand protein docking for surface antigen for gastrointestinal parasitism with Haemonchus contortus (alpha tubulin and beta tubulin antigen) with ovine CD14.
Result
Cloning and Sequencing of CD14 cDNA of Sheep
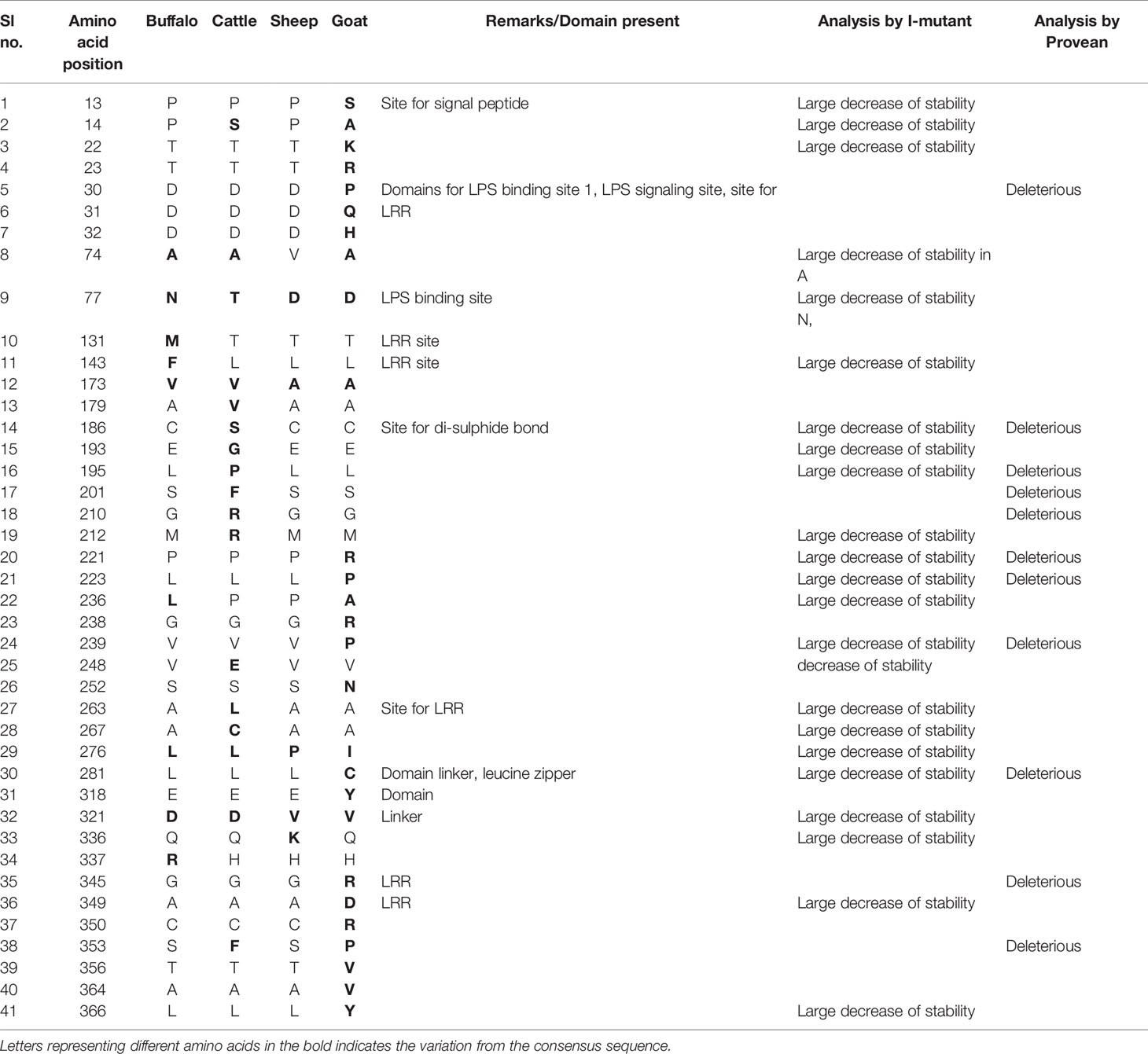
The ovine CD14 was observed to have an open reading frame of 1116 nucleotide, ATG as start codon and TAA as stop codon (KY110723) (Figure 1). Nucleotide sequences as well as derived peptide sequences for Bos taurus (GU368102), Bubalus bubalis (DQ457089), and Capra hircus (DQ457090) had been sequenced earlier in our lab and submitted to the gene bank. Ovine CD14 derived peptide was observed to have 373 amino acids precursor corresponding to the coding sequence of the gene and a signal peptide consisting of 20 amino acids (Figure 2). The molecular weight of sheep CD14 was observed to be 39645 Dalton. Slight differences have been observed in the molecular weight of the CD14 molecule of ruminants. Table 1 lists the amino acid variations in different species of ruminants.
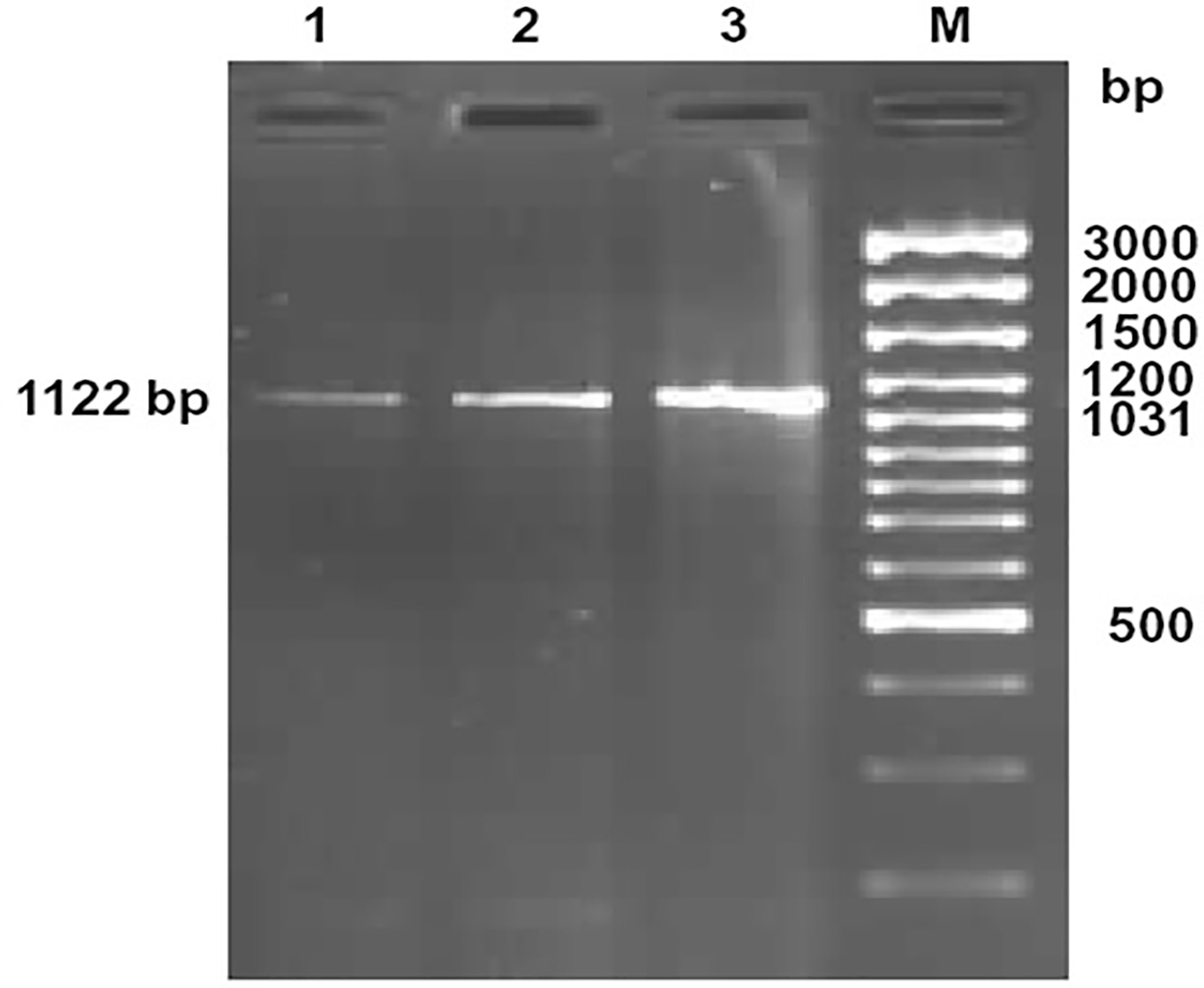
Figure 1 Amplification of 1122bp fragment of CD14 gene of sheep and goat. Lane 1-2: Amplified product from cDNA of CD14 gene of sheep Lane M: 100bp DNA ladder plus.
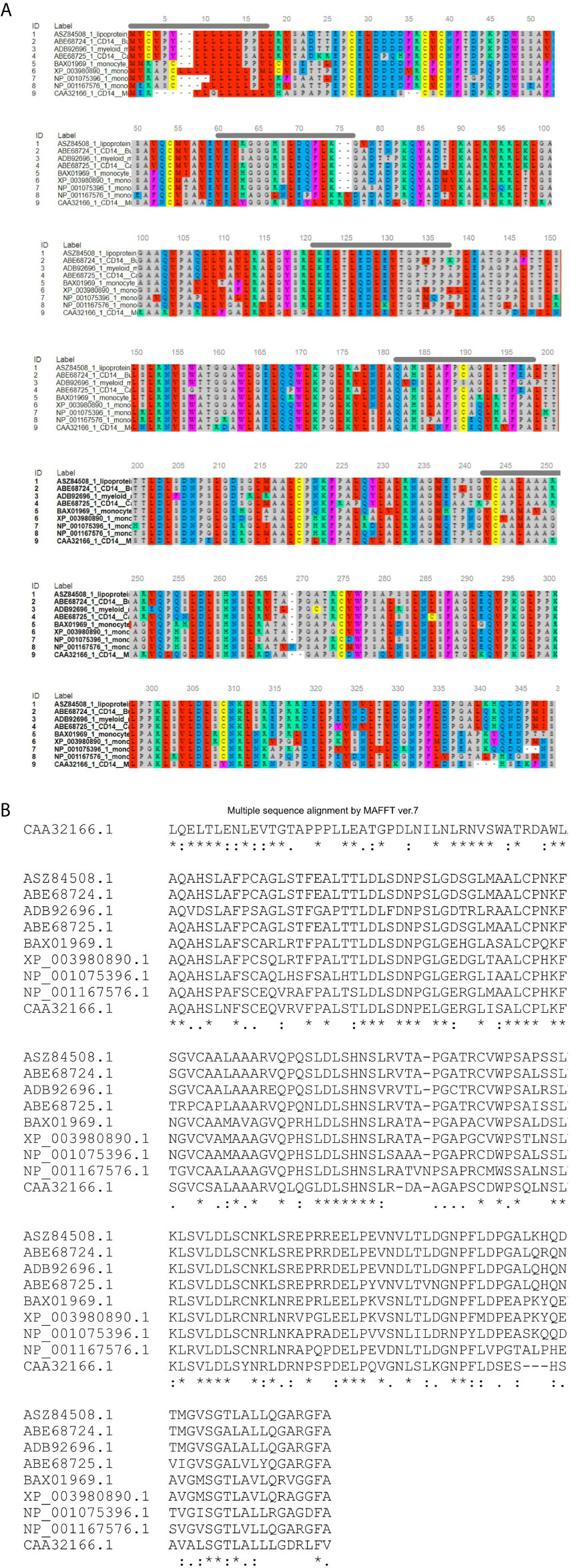
Figure 2 Alignment of ruminant CD14 derived peptide with other species. (A) Alignment through MSA viewer. (B) CLUSTAL format alignment by MAFFT. 1: Sheep (Ovis aries), 2: CB cattle (Bos taurus x Bos indicus), 3: Buffalo (Bubalus bubalis), 4: Goat (Capra hircus), 5: Human (Homo sapiens), 6: Mice (Mus musculus), 7: Dog (Canis lupus familiaris), 8: Cat (Felis catus), 9: Horse (Equus caballus). Green line, Signal peptide; Blue line, LPS binding site; Yellow line, LPS signaling site; Red line, Leucine zipper; Black overline, Leucine rich repeat; Red line, Leucine zipper; Pink line, Domain linker prediction site. * means consensus for single amino acid and it is conserved across the species. ** means two successive amino acids for CD14 correspond to consensus sequence and these amino acid are conserved across the species. Similarly *** corresponds to three amino acids. **** corresponds to four amino acids and so on.
Bioinformatics Analysis for CD14 Gene
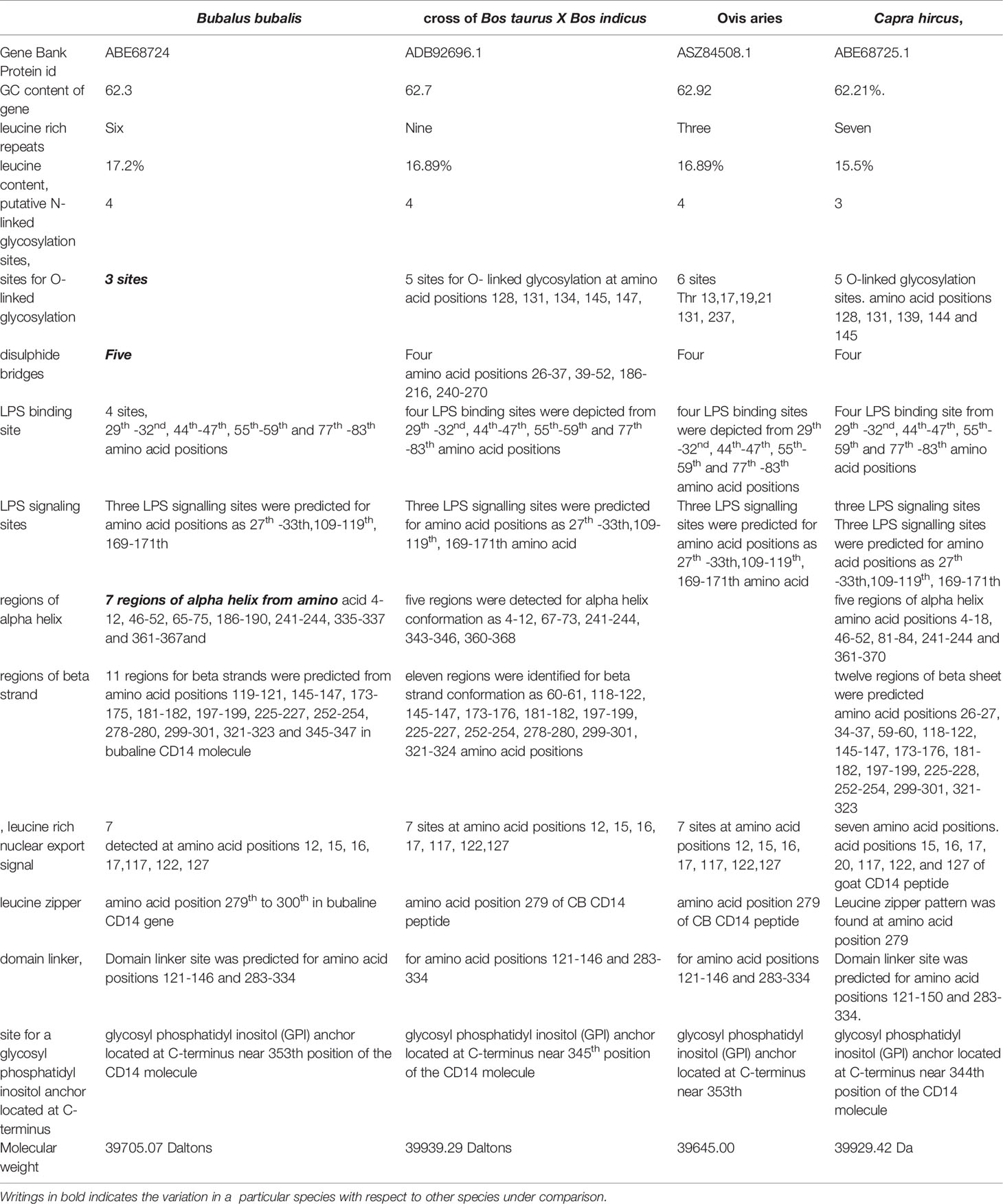
Ovine CD14 was observed to have high GC content. The GC content of the CD14 gene in ruminants was found to be high (62.2 to 62.7%), as depicted in Table 2. Alignment of the derived peptide sequence of ruminants have been depicted in Figure 4. Amino acid cysteine contains sulphur, which is responsible for disulphide bond formation, which in turn are responsible for protein folding. The site for disulphide bond has been predicted in Table 2. LPS binding and LPS signalling sites are very important to determine the potentiality for innate immunity of the CD14 gene. Derived peptide of the ruminant CD14 gene differs in putative N-linked glycosylation sites at amino acid positions, four sites in cattle and buffalo and three sites in goat were predicted (Table 2).
O-linked glycosylation site for ruminant CD14 differs, three in Buffalo and five in both goat and crossbred cattle (Table 2), but absent in human (32). O-linked Glycosylation is needed when hydrophilic clusters of carbohydrates alter the polarity and solubility of protein or protein folding. CD14 and other Toll- like receptors were characterized by presence of LRR and high leucine content. Leucine rich repeats (LRR) predicted peptide sequence of ruminant CD14 cDNA were studied (Table 2 and Figures 2A, B). A protein dynamics study revealed that in ruminants, the position of glycosyl phosphatidyl inositol (GPI) anchor located at C-terminus of the CD14 molecule differs (Table 2). Seven sites for leucine rich nuclear export signal (NES) were detected in the CD14 gene of ruminants (Table 2 and Figures 2A, B). In terms of secondary structure prediction, seven regions of alpha helix and eleven regions of Beta strands with different amino acid position in ruminant CD14 molecule were predicted. (Table 2). A leucine zipper pattern was found at amino acid position 279th in the ruminant CD14 gene (Figures 2A, B). The site for Domain linker for ovine CD14 site was predicted, which is almost similar to amino acid positions in other ruminants (Table 2 and Figures 2A, B).
A 3-D model of the CD14 molecule of sheep was predicted to be a horseshoe shaped structure, with alternating alpha helix and beta chains in a swiss model (Figure 3A), pymol view with LPS binding site (Figure 3B), surface view with LPS binding site (Figure 3C). Figure 3D depicts the groove or pathogen binding site of ovine CD14 surface view. Predicted protein of CD14 molecule revealed a bent solenoid containing a hydrophobic amino terminal pocket. Alignment of 3D structure of ovine CD14 with goat (Figure 4A), cattle (Figure 4B), and buffalo (Figure 4C) have been depicted. The variability in the structure of the pocket of CD14 and alternative rim residues are believed to be important for LPS binding and cell activation, leading to variation in innate immunity across the species. Pockets or grooves (LPS binding site) for TM aligned CD14 sheep and CD14 buffalo have been depicted in Figure 5A. Similarly Figure 5B depicts pathogen binding sites for ovine CD14 with that of goat CD14. The amino terminal pocket containing grooves are involved in ligand binding (Figure 9C). The CD14 receptor exists as a dimer with the help of the leucine zipper in its biologically active form. It had been observed that the amino terminal pocket for ruminant CD14 with grooves is the site for ligand binding, which is the most important factor for CD14. The rim residues of the CD14 hydrophobic pocket are the primary determinants for binding with LPS, or other acylated CD14 ligands. Positively charged residues as K71 and R72 are present on the rim and R80, K87, and R92 are present just outside the rim, whereas hydrophobic residues as W45, F49, V52, F69, Y82, and L89 encircle the rim. These residues were observed to be the primary determinant for ligand binding.
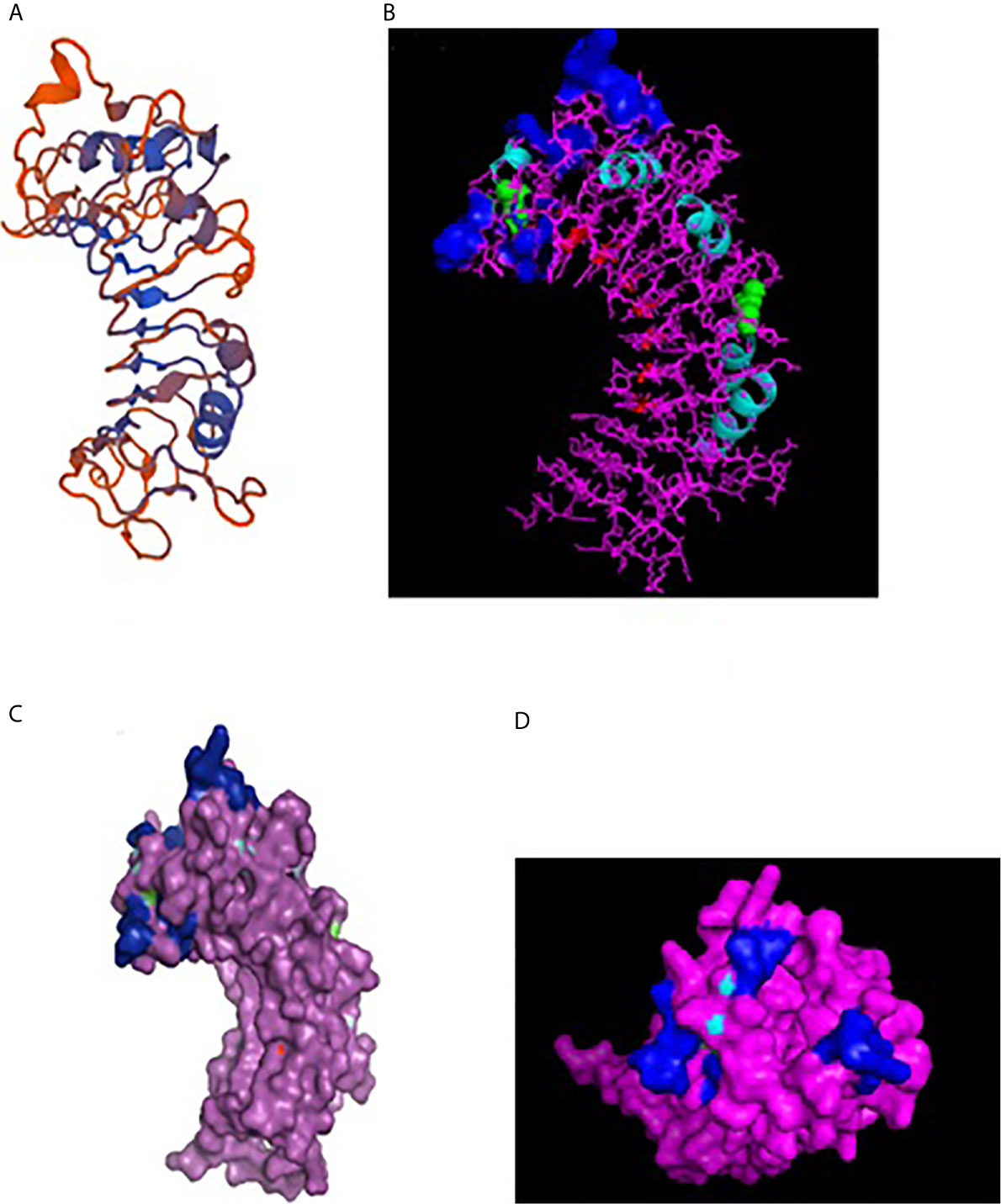
Figure 3 3D structure of sheep CD14. (A) 3D structure of ovine CD14 gene-Swiss prot view. (B) 3D structure of ovine CD14 gene-Pymol view (with LPS binding sites). (C) 3D structure of ovine CD14 gene-Surface view (with LPS binding sites). (D) Sheep CD14- groove (pathogen binding site).
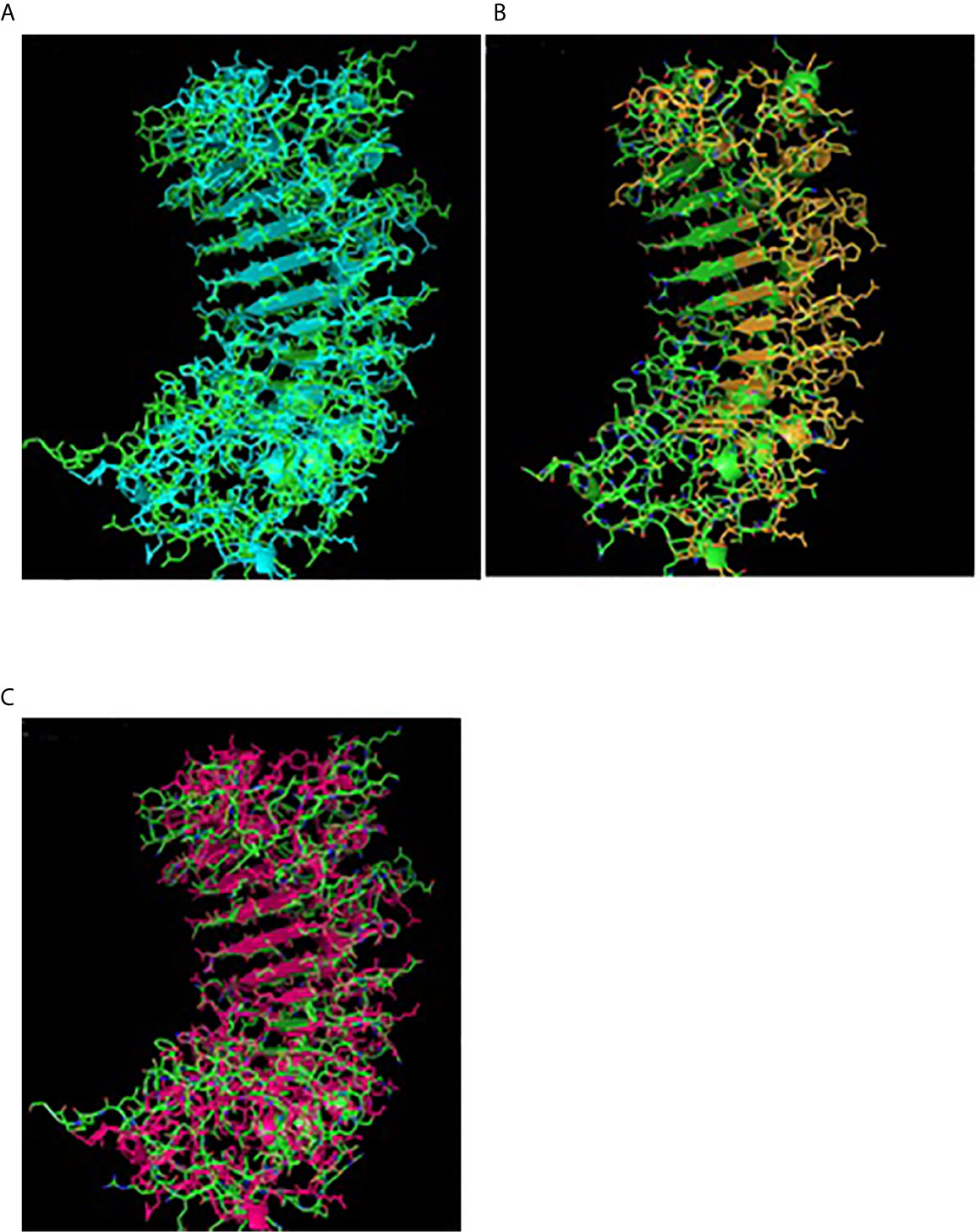
Figure 4 TM alignment of 3D structure of sheep with other ruminants. (A) TM alignment of the 3D structure of sheep and goat CD14 molecules. Green, Sheep CD14 molecule, Cyan, Goat CD14 molecule. (B) TM alignment of the 3D structure of sheep and cattle CD14 molecules. Green, Sheep CD14 molecule, Orange, Cattle CD14 molecule. (C) TM alignment of the 3D structure of sheep and buffalo CD14 molecules; Green, Sheep CD14 molecule; Pink, Buffalo CD14 molecule.
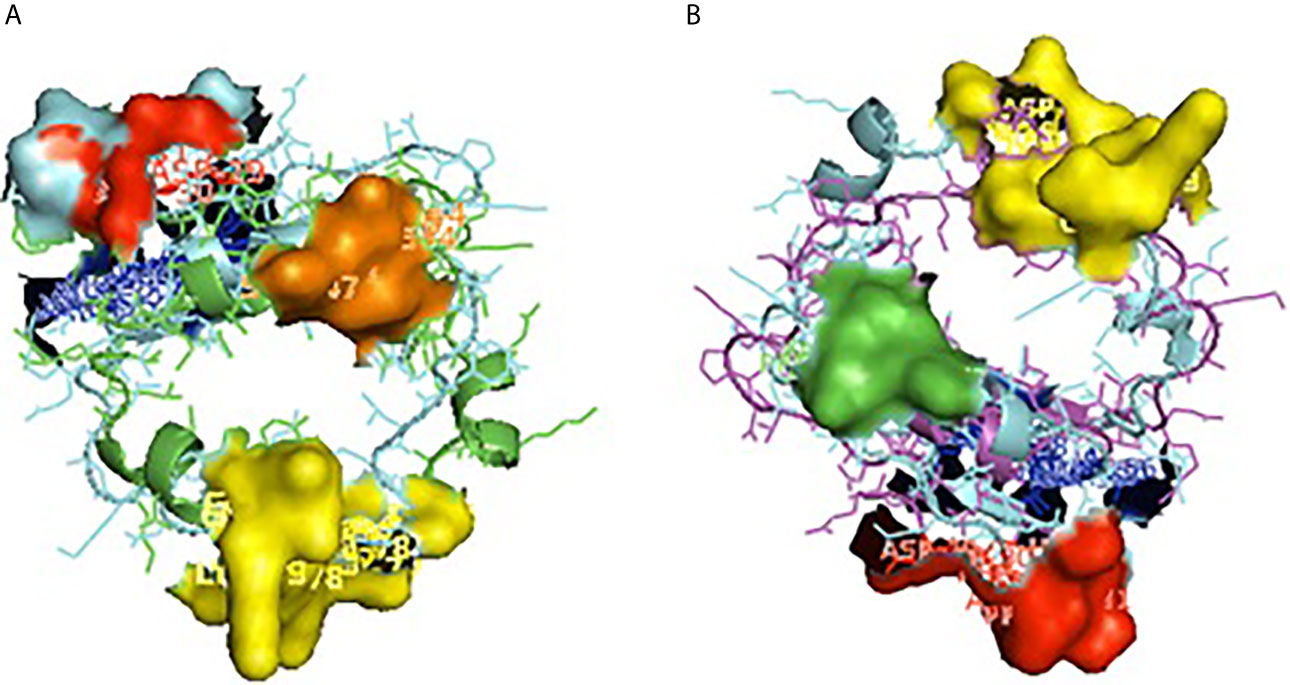
Figure 5 Depiction of receptor binding site for CD14. (A) Pocket or groove (LPS binding site) for TM aligned CD14 sheep and CD14 buffalo. (B) Pocket or groove (LPS binding site) for TM aligned CD14 sheep and CD14 goat.
Comparison of Ovine CD14 With Goat
Small ruminants such as sheep and goats were observed to have significant differences in susceptibility to commonly occurring diseases. Leucine content was observed to be higher in sheep CD14 compared to that of goats. CD14 derived peptide of sheep were observed to have comparatively less GC content, but more leucine content percentage. The number of LRR in buffaloes was observed to be lower in buffalo compared to cattle (Table 2). The GPI anchor was present at 353rd position in buffalo, in contrast to 345th position of cattle. The GPI anchor is important for the membranous form of the CD14 gene. 353rd position implies greater availability of CD14.
Alignment of CD14 (3 D structure) of sheep/goat, sheep/cattle, and sheep/buffalo have been depicted in Figures 4A–C respectively. LPS binding sites were situated at the rim of the groove of the receptor. Four LPS binding sites have been depicted as four sites as 29th -32nd, 44th-47th, 55th-59th and 75th -80th amino acid positions and depicted in red, green, blue, and yellow respectively (Figures 5A, B). A comparison of the CD14 structure of sheep/goats was observed to be highly superimposable with a root mean square deviation of 0.23 A ˚. Observation of the CD14 of goats/buffalo was also observed to be greatly superimposable with RMSD being 1.89. The structural differences arising due to nucleotide differences have been depicted. Comparison of CD14 structure of Sheep/buffalo were observed to be highly super imposable with a root mean square deviation of 0.23. String analysis (Figure 6) and KEGG analysis (Figure 7) depicted that CD14 works in combination with TLR4 and MD2.
Molecular Phylogeny
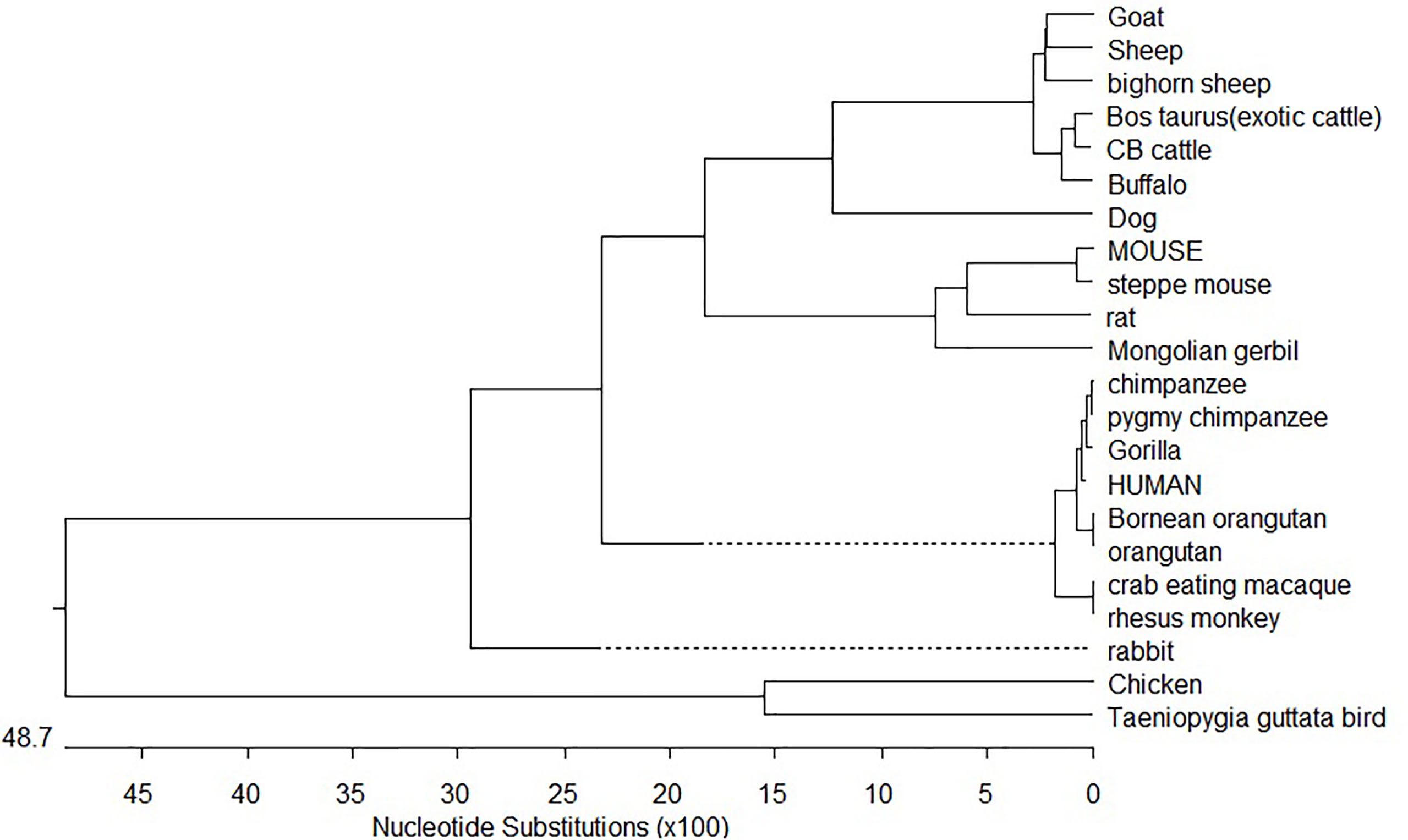
Phylogenetic analysis of CD14 DNA sequences strongly suggests that avians diverged first from mammals with 4870 nucleotide substitutions with respect to the CD14 gene (Figure 8). Rodents were the next to branch off (Figure 8). The phylogenetic tree for ruminants with other mammals and avians of both domestic and wild origin based on the CD14 gene have been depicted in Figure 8. Ruminants were clustered together with about 300 nucleotide substitutions. Small ruminants such as sheep and goat were clustered separately from large ruminants such as cattle and buffalo. The closest ancestors for ruminants were found to be dogs. Primates were genetically distant from ruminants at about 2400 nucleotide substitutions. Based on CD14 gene analysis, avians were observed to be genetically distant to mammals at about 4870 nucleotide substitutions. The genetic similarity has been predicted in Table 1.
Trans-species polymorphism has been detected in ruminants (Supplementary Table 2), where specific alleles were detected for small ruminants (goat and sheep) and large ruminants (cattle and buffalo). Twelve sites were detected (Supplementary Table 2) with five synonymous and seven non-synonymous nucleotide substitutions. Non-synonymous substitution exceeding synonymous indicates positive selection. Comparative sequence analysis of cattle and buffalo CD14 has been depicted in Supplementary Table 2. Twelve non-synonymous nucleotide substitutions have been depicted in cattle and buffalo CD14 gene. Variations in epidemiological data have been observed in large ruminants with reference to common diseases (Supplementary Table 3).
Assessment of Parasitic Infestation in Sheep
Screening of the faecal egg count (FEC) of the sheep (n=60) lead to study the health condition of the sheep which further prevailed us to divide them into two categories of Non-infected and Infected groups. The mean FECs of the animals are given in the Table 3. Direct visualization followed by microscopic examination of the samples (parasites) collected from abomassum reveal the presence of Haemonchus contortus for the slaughtered animals.
Differential mRNA Expression Profile for CD14 Gene With Respect to Non-Infected and Infected Sheep
The current study depicts increased expression profile of CD14 in infected (H. contortus) infected sheep as depicted in Figure 9.
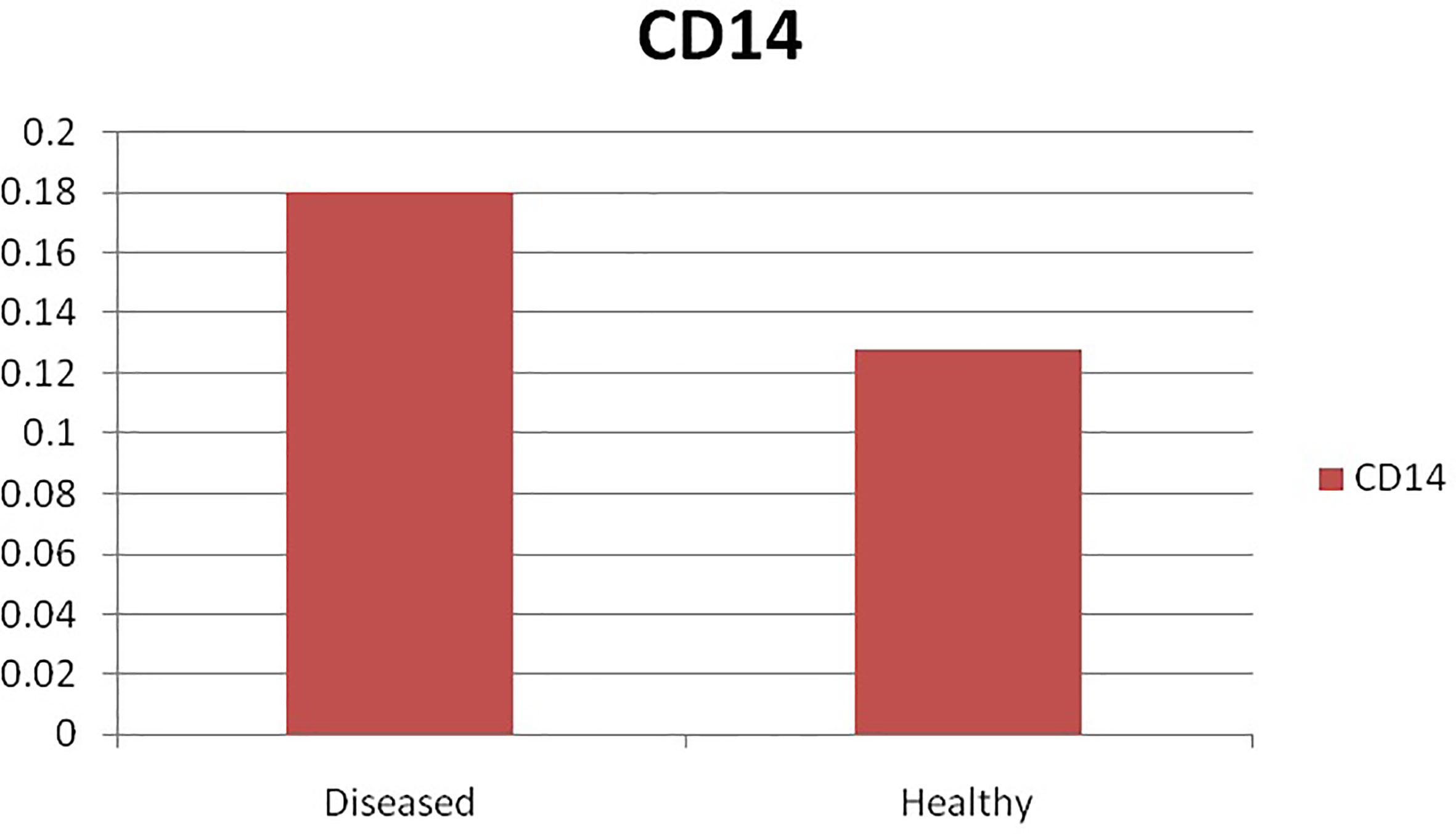
Figure 9 Differential mRNA expression profiling of ovine CD14 with respect to non-infected and infected (Haemonchus infected).
In Silico Study for the Detection of Binding Site of CD14 With Haemonchus contortus
Molecular docking has revealed binding of CD14 with two identified surface molecules of Haemonchus contortus as alpha tubulin (Figures 10A, B) and beta tubulin(Figures 10C, D) with definite binding sites. Figure 10A depicts the structural alignment of alpha tubulin of H. contortus with the CD14 gene, and aligned site being depicted as Glu27 (red sphere) and Ala284 (green sphere). Figure 10B depicts the line diagram of the alignment of alpha tubulin of H. contortus with ovine CD14. Similarly Figure 10C depicts the structural alignment of beta tubulin of H. contortus with the CD14 gene, and the aligned site is depicted as Glu27 (hot pink sphere) and Ala284 (deep teal sphere). Figure 10D depicts the line diagram of the alignment of alpha tubulin of H. contortus with ovine CD14. The study reveals distinct binding sites for CD14 with the two most important structural proteins of Haemonchus contortus, alpha tubulin and beta tubulin. This finding indicates the distinct role of CD14 in parasitic immunity. This is the first novel report of the role of CD14 in parasitic immunity, hence comparison was not possible.
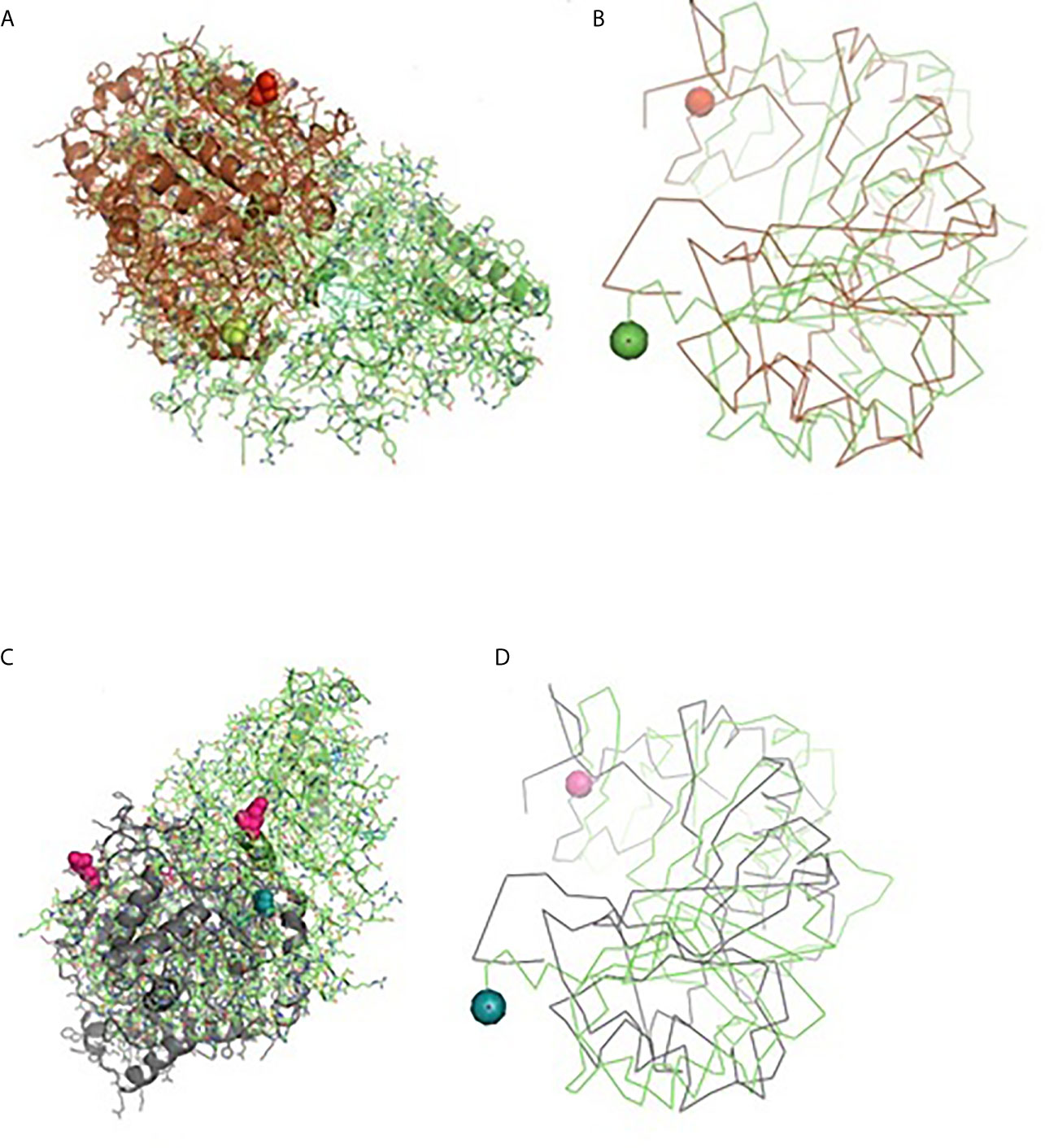
Figure 10 Binding of ovine CD14 with alpha tubulin and beta tubulin of Haemonchus contortus. (A) Alignment of 3D structure of ovine CD14 molecule (green) with alpha tubulin (brown) of Haemonchus contortus. (B) Line drawing of binding site of CD14 molecule with alpha tubulin (brown) of Haemonchus contortus. (C) Alignment of 3D structure of ovine CD14 molecule (green) with beta tubulin (purple) of Haemonchus contortus. (D) Line drawing of binding site of CD14 molecule (green) with beta tubulin (purple) of Haemonchus contortus.
Haematological and Biochemical Parameters With Respect to Non-Infected and Infected Sheep
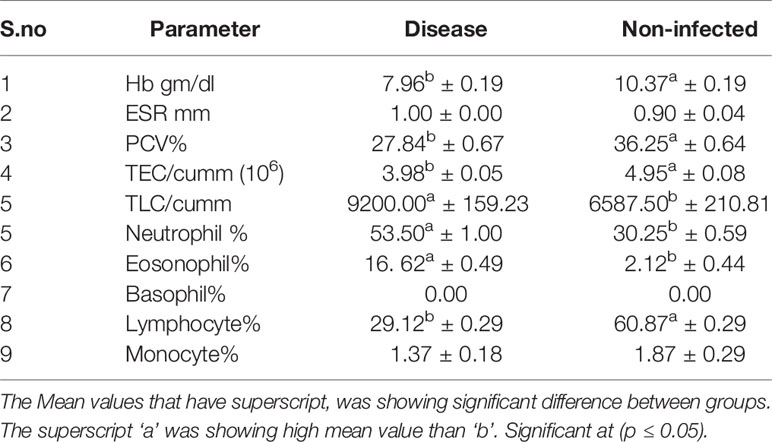
The haematological parameters were assessed with respect to infected (H. contortus infected) vs. non-infected sheep (Table 4). Significant differences were observed in Hb gm/dl, PCV%, TEC/cumm, TLC/cumm, Neutrophil %, Eosonophil%, and Lymphocyte%. Better haemoglobin, PCV, and TEC were observed in non-infected sheep in comparison to that of the infected group. However, total leucocyte count was pronounced in infected sheep. A marked significant increase in eosinophil count was observed in infected sheep. A similarly pronounced increase was observed in neutrophil and lymphocytes, indicative of involvement of immune responsiveness.
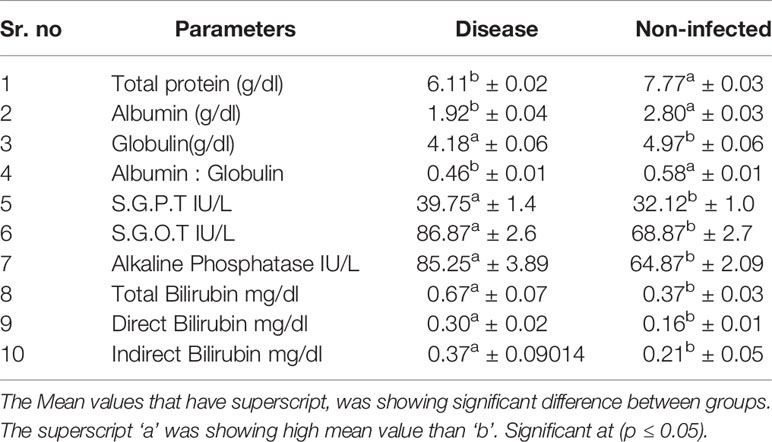
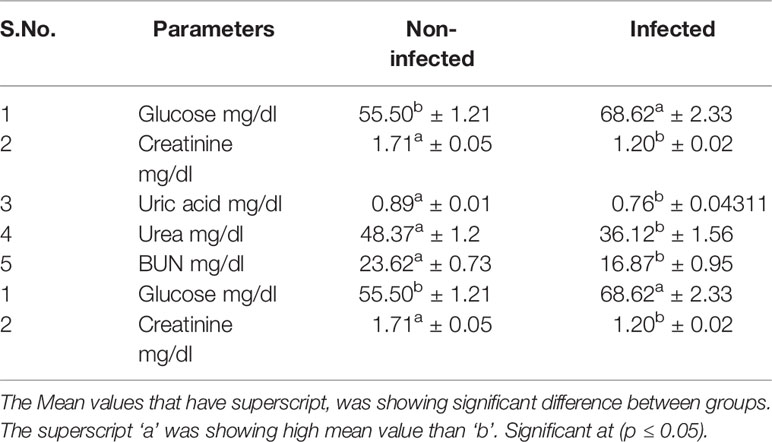
Significant differences were observed for non-infected and infected sheep in terms of liver function test (Table 5) and kidney function test (Table 6). Glucose level was observed to be better in non-infected, while Creatinine, uric acid, urea, and BUN level were better in infected sheep. Total protein, albumin, globulin, and albumin: globulin ratio was observed to be better in non-infected sheep. While other reports indicate more in infected sheep (S.G.P.T. S.G.O.T, Alkaline Phosphatase, Total Bilirubin, Direct Bilirubin, Indirect Bilirubin).
Discussion
Ovine CD14 was characterized and the important domains were identified in comparison to other ruminant species. The GC content of the CD14 gene of humans (62.94%) and dogs (64.07%) was found to be more than ruminants while rodents had less GC content, e.g., rats (56.12%). High GC content was observed to be an inherent characteristic of the CD14 gene. Direct relationship of the length of the coding sequence was observed with the GC content of the gene (75). Species-wise differentiation in GC content may explain the variability in the stability of the CD14 gene among the species. It is known that nucleotide Guanine (G) and cytosine (C) are bounded by triple hydrogen bond in double helix DNA. Thus, it is evident that the GC bond is stronger than the AT bond. Similarly, GC content is reported to have a significant effect on genome functioning and species ecology (75). The importance of GC rich isochores has been predicted. The thermostability, bendability, ability of B-Z transition, and curvature of DNA helix were observed to be correlated positively with GC content (76).
Alignment of the derived peptide sequence of ruminants revealed that leucine residues were mostly conserved across the species (39), except at amino acid position 143. Cysteine residue of CD14 was found to be conserved across ruminant species and other species (Figure 4). The CD14 molecule found in ruminants was predicted to contain the higher percentage of leucine (Table 2), which is similar to mice (17.66%) and humans (15.5%) (77).
Due to protein folding, certain grooves are developed in CD14, which in turn are responsible for the development of a binding site for ligands. Since the models are developed through homology modelling, 5th disulphide is not visualized in the model. Similar five disulphide bonds were also observed in mice (39) and humans (78). Cys287 is tyrosine moiety in the mouse CD14 sequence, hence from the homology modelling, the fifth disulphide could not be detected.
The crystal structure of CD14 was observed to contain five disulphide bonds both in humans (78) and Mus musculus CD14 molecules (39). Biochemical studies have also detected five disulphide bonds in humans (77). The impact for disulphide bonds differ in human CD14 folding. Out of five disulphide bonds, the first and second disulphide bonds were observed to be indispensable. The third and fourth disulphide bonds were reported to be important, while the fifth or the last (Cys287–Cys333) was dispensable (78).
Disulphide bonds have a greater effect on CD14 structure and integrity, as in the case of mice. The first and second disulphide bonds were observed in the β sheets in the inner concave surface (the “core” structure), whereas the third and fourth disulphide bonds were observed in the loops and helices on the outer surface (the “peripheral” structure).The first disulphide bond is responsible for the structural and functional integrity of CD14 molecule. It was inferred that CD14 folding was not determined by third, fourth, and last disulphide bonds. The substitutional mutation had been observed in the third and fourth disulphide bonds. Cysteine was substituted with alanine, but the function of CD14 is not altered (78).
In ruminants, four LPS binding sites and three LPS signalling sites were predicted. The generous size of the pocket of the CD14 receptor may allow structural variation in the hydrophobic portion of the ligand, however, the hydrophobic bottom and walls of the pocket are rigid. The hydrophilic part of the ligand has structural diversity. This is due to the considerable flexibility of the hydrophilic part of the rim leading to the development of multiple grooves for ligand binding. Similar LPS binding sites and LPS signalling sites were reported in mice (39) and humans (77). The LPS signalling site also overlaps with LRR. It has also been observed that the first LRR (Leucine-rich repeat) overlaps with the LPS-binding site region-I. CD14 is a pattern recognition receptor and is rich in leucine-rich repeat, and CD14 are rich in leucine moiety as evident by high leucine percentage. Ligand specificity or flexibility of the rim of CD14 is important for pathogen binding sites for a wide range of pathogens. There was the report of three, five, four, and five glycosylation sites for N-linked glycosylation in Bos taurus (40), Mus musculus (77, 78), Homo sapiens (77), and Rattus norvegicus (79), respectively. N-linked glycosylation is vital for the molecule as it supports the molecule to be present either in membranous or soluble form. O-linked glycosylation site for ruminant CD14 differs, three in Buffalo and five in both goats and crossbred cattle (Table 2), but absent in humans (32). O-linked Glycosylation is needed when hydrophilic clusters of carbohydrates alter the polarity and solubility of protein or protein folding. Homo sapiens, Mus musculus, were reported to have 10 LRR each. A trend was observed for the relationship of number of LRR and disease susceptibility. Pathogen recognition is an important phenomenon in any innate immune response. LRR for CD14 in the extracellular domain has a major role in pathogen recognition.
Similar findings as ovine CD14 for GPI anchor were reported in other species like humans and mice. GPI anchor is an important phenomenon for any membranous protein. GPI anchor for CD14 is important for bridging between CD14 and cell surface. In avians, CD14 was observed to be transmembranous, not GPI anchored (80). Amino acid positions responsible for NES are 12, 15, 16, 17, 117, 122, and 127 in ruminant CD14. Since nuclear export signal (NES) aids in export of the CD14 peptide from the nucleus to the cytoplasm via the nuclear pore complex (81), there is possibility of impairment of this function in goat CD14. In terms of alpha helix and Beta strands of ovine CD14, there are similarities in mice containing seven of alpha helices, while this differs in the Beta helix. Mice CD14 contains 13 Beta helices in contrast to 11 in ruminants. It seems that Beta helices have a relationship with the innate immunity of theCD14 molecule. Nine Beta strands starting from β2, β4, to β11 overlap with leucine rich repeats (LRRs). LRR in turn have roles inn LPS binding and LPS recognition sites, as already discussed, which forms one of the basis for innate immunity.
Since the CD14 molecule forms its functional form by dimerization, the leucine zipper leads to dimerization. Leucine zippers, a common three-dimensional structural motif (66), were observed to be responsible for gene expression, since these were an integral component of the DNA-binding domain in a variety of transcription factors. The presence of the leucine zipper generates adhesion forces in parallel alpha helices (82) and this region was found to be conserved in ruminant CD14.
Domain linker sequences connect two structural domains and in turn act as a scaffold and prevent unfavourable interactions between folding domains (83).
The CD14 molecule was predicted to be mostly expressed on the cell membrane or cell surface for the ruminants under study, which justifies it to be a receptor molecule. Two forms of CD14, membranous and soluble forms, are in existence (84, 85). Cloned ovine CD14 cDNA in the current study was predicted to be existing in membranous form since it contained GPI anchor (84, 85).
The finding is similar to the study conducted by (39), who studied the crystal structure of the Mus musculus CD14 molecule.
Comparison of ovine CD14 with that of goat has revealed certain differences. Epidemiological studies also depict that sheep were better resistant to goats in terms of infectious and parasitic diseases. GC content was lower in sheep. It has been reported that there is preferential fixation of GC alleles leading to biasness of GC nucleotides over AT during DNA repair. GC content is important with respect to the evolving recombination rate, regulated replication or expression timing, bending ability of DNA, and to B-Z transition, transcriptional efficiency as already explained earlier.
It is evident that a higher mutation frequency was detected in goats (0.56), with mutations detected at 25 amino acid positions, out of which thirteen sites were observed to be thermodynamically unstable. Seven sites were observed to be deleterious in nature.
Mutations detected, such as P13S and P14A in goats, are involved in the domain for signal peptides and were observed to have less structural stability. Mutations were observed such as D30P, D31Q, and D32H, which are responsible for important domains as LPS binding site1, LPS signalling site, and LRR site, where D30P was observed to be deleterious. In site 1 of the LPS binding site, there is a drastic shift of acidic amino acid to basic, such as Aspartic acid to proline, aspartic acid to glutamine, and Aspartic acid to histidine. Since there is a change of polarity, the pathogen binding activity is also adversely effected.
P276I was observed to be thermodynamically unstable, but this site constitutes LRR, which is important for LPS signalling. Thus this mutation may cause depressed function of pathogen binding activity of CD14 of goats in contrast to sheep. Another important site for deleterious mutations is L281C, which is responsible for the domain linker, leucine zipper. K336Q was thermodynamically less stable in goat CD14 and is responsible for the site of domain linker.
A cluster of mutations were observed in goat CD14, which are important for the LRR site, ultimately effecting pathogen binding and signalling, which are G345R, A349D, C350R, S353P,T356V, A364V, L366Y.
MD2 is an important component of TLR4 signalling complex. Members of the MD-2–related lipid-recognition protein super- family contains two beta sheets arranged in ab cup fold generating a centralized hydrophobic LPS-binding cavity (86). CD14 sensitizes cells to LPS through the process of delivering this lipid moeity to MD-2. The hydrophobic pocket is occupied mostly by four acyl chains of the ligand buried inside for the complex of both mouse and human CD14 with MD-2 bound to lipid IVa (87, 88). TLR4/MD-2/LPS homodimeric signalling complex depicts the hydrophobic binding pocket sequestering five of the six fatty acid chains. The rest of the unbound acyl chain lies along the surface of MD-2 and, along with the F126 loop of MD-2, generates a new hydrophobic patch promoting homodimer formation through the association of TLR4/MD-2/LPS complex (19).
LPS-binding pocket (hydrophobic) was predicted at the amino terminal end through a homology based study with 3D structure of CD14 of Mus musculus (39) and Homo sapiens (77). CD14 molecule, α-helices were located at the convex surface, and β strands were located at the concave surface of horse-shoe shaped CD14 structure, represented in Figures 3, 5. N terminal hydrophobic pocket revealed a conservation of pocket size and hydrophobicity. Asparagine ladder with conserved asparagines residue had been observed, which is a structural feature for leucine rich repeats. The variable residues in LRR are hydrophilic and exposed to the concave surface of the horseshoe structure. Some of the similar studies had been conducted earlier in ruminants (53, 55).
CD14 has been considered as an important gene to explore a wide range of open questions in evolutionary ecology, since a wide range of nucleotide variability has been observed with the mutational hot spot detected. It was attempted to identify the Trans species polymorphism (TSP).
TSP generally occurs by passage of alleles from ancestral to descendant species. Neutral alleles may be shared by species which are genetically similar. Balancing selection has been regarded as the major evolutionary force responsible for long-lasting TSP. Similar TSP as identified in ruminants with respect to the CD14 gene have also been identified in the MHC Class II gene (89). Speciation is a complex process which involves TSP, divergence of allelic lineages at the coding sites for both synonymous and non-synonymous mutations. These processes took places millions of years ago (90).
Pronounced expression of CD14 was observed in infected sheep compared to that of non-infected sheep. As per our literature search, it is the first report in sheep. A similarly improved expression profile of CD14 was also detected in mice infected with another parasite as Schistosoma mansoni (91).
Some more studies suggested that CD14+ and CD16+ cells help to control the parasitic burden and play important roles in resistance to P. vivax infection in humans (92). However, in our present study increased expression of CD14 gene in infected sheep may be due to its action as a pattern recognition receptor in parasitic infection. It was reported that parasite-derived E/S product was detected by CD14bright/CD16+ intermediate monocytes in case of schistosome-infected patients compared to that of uninfected individuals (93). In addition, CD14 has a significant role in regulating the immunity in S. mansoni-infected mice, through the induction of both TH1 and TH2 through macrophage M1/M2 phenotype. CD14brightCD16+ intermediate monocytes are major producer of IL-10 (94). The Th1 and Th2 cell released different kinds of cytokines such as interleukin (IL-2), interferon-gamma (IFN-γ) and tumour necrosis factor-alpha (TNF-α) from TH1 cells and IL-10 produce from Th2 type cells and these cytokines are responsible to stimulates B cell differentiation, production of different immunoglobulin such as IgE, IgG1, IgG4, and IgA, mastocytosis, and eosinophil activation and function (95).
IL-10 leading to eosinophil mobilization, intestinal mast cell accumulation and production of IgE, mucosal mast cell infiltration, intestinal eosinophilia, elevated serum IgE, increased level of parasite specific IgG-1 which are responsible to eradicate worms (96).
Similar studies that correspond to our finding shows a marked increase in blood eosinophil and abomasal tissue, degranulation of eosinophil and it releases some peroxidases and basic protein and cationic protein which are cytotoxic to helminth (9). Similar studies of caprine CD14 had been conducted by us earlier with focus on evolution and structural analysis (97).
Conclusion
CD 14, an important gene for innate immunity, has revealed immense variability between species, which may lead to variability in resistance to diseases. This variability in disease resistance may be due to variability in ligand binding ability of the CD14 molecule. The greater susceptibility of goat compared to sheep, and greater susceptibility of cattle compared to buffalo revealed mutations in LPS binding site of the goat/cattle CD14 molecule, which causes altered polarity, decreased thermodynamic stability, and the deleterious mutation. Since hypervariable sites or mutational hot spots have been detected in the CD14 gene, due to greater genetic variability, evolutionary study is very promising. Phylogenetic analysis reveals that ruminants were evolutionary closer, but transpecies polymorphism has been detected among the ruminants. The distinct role of CD14 in antiparasitic immunity is reported here for the first time through in silico molecular docking and later confirmed through differential mRNA expression profile with QPCR for non-infected and infected samples.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Ethics committee, West Bengal University of Animal and Fishery Sciences.
Author Contributions
KR has conducted the experimental work, collected samples, performed acquisition of data, drafted the article.*ArP has conceptualized and designed the work, conducted research, analysed and interpreted the data, and drafted the article. SaB has conducted the experimental work. AbP has conducted the bioinformatics analysis and its interpretations.SCM has guided through the experiments on parasitology. SuB has conceptualized the work, guided through the biochemical analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Department of Biotechnology, Ministry of Science and Technology, Govt. of India for providing the financial support for carrying out the research work (Grant No. BT/Bio-CARe/04/10100/2013-14). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors are equally thankful to West Bengal University of Animal and Fishery Sciences and Indian Veterinary Research Institute for carrying out the work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.664877/full#supplementary-material
Supplementary Table 1 | List of mammals & avian under present study analysed for CD14 sequences.
Supplementary Table 2 | Trans species polymorphism detected from molecular evolutionary study of CD14 gene for the ruminants.
Supplementary Table 3 | Epidemiological data (in percentage) for large ruminants with reference to common diseases.
References
1. Engel P, Boumsell L, Balderas R, Bensussan A, Gattei V, Horejsi V, et al. CD Nomenclature 2015: Human Leukocyte Differentiation Antigen Workshops as a Driving Force in Immunology. J Immunol (2015) 195(10):4555–63. doi: 10.4049/jimmunol.1502033
2. Bazil V, Horejsi V, Baudys M, Kristofova H, Strominger JL, Kostka W, et al. Biochemical Characterization of a Soluble Form of the 53-kDa Monocyte Surface Antigen. Eur J Immunol (1986) 16(12):1583–89. doi: 10.1002/eji.1830161218
3. Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The Monocyte Differentiation Antigen, CD14, Is Anchored to the Cell Membrane by a Phosphatidylinositol Linkage. J Immunol (1988) 141(2):547–52.
4. Simmons DL, Tan S, Tenen DG, Nicholson-Weller A, Seed B. Monocyte Antigen CD14 Is a Phospholipid Anchored Membrane Protein. Blood (1989) 73:284–9. doi: 10.1182/blood.V73.1.284.284
5. Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a Receptor for Complexes of Lipopolysaccharide (LPS) and LPS Binding Protein. Science (1990) 249(4975):1431–33. doi: 10.1126/science.1698311
6. Goldsby RA, Kindt TJ, Osborne BA. Kuby Immunology. 4th Edn. New York: W.H. Freeman and Company (2000). 574 p.
7. Rietschel ET, Brade H, Brade L, Kaca W, Kawahara K, Lindner B, et al. Newer Aspects of the Chemical Structure and Biological Activity of Bacterial Endotoxins. Prog Clin Biol Res (1985) 189:31–51.
8. Hailman E, Lichenstein HS, Wurfel MM, Miller DS, Johnson DA, Kelley M, et al. Lipopolysaccharide (LPS)-Binding Protein Accelerates the Binding of LPS to CD14. J Exp Med (1994) 179:269–77. doi: 10.1084/jem.179.1.269
9. Tobias PS, Soldau K, Gegner JA, Mintz D, Ulevitch RJ. Lipopolysaccharide Binding Protein-Mediated Complexation of Lipopolysaccharide With Soluble CD14. J Biol Chem (1995) 270(18):10482–88. doi: 10.1074/jbc.270.18.10482
10. Tobias PS, Soldau KA, Ulevitch RJ. Isolation of a Lipopolysaccharide-Binding Acute Phase Reactant From Rabbit Serum. J Exp Med (1986) 164(3):777–93. doi: 10.1084/jem.164.3.777
11. Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, et al. Structure and Function of Lipopolysaccharide Binding Protein. Science (1990) 249:1429–31. doi: 10.1126/science.2402637
12. Yu B, Wright SD. Catalytic Properties of Lipopolysaccharide (LPS) Binding Protein Transfer of LPS to Soluble CD14. J Biol Chem (1996) 271(8):4100–05. doi: 10.1074/jbc.271.8.4100
13. Goyert SM, Ferrero E, Rettig WJ, Yenamandra AK, Obata F, Le Beau MM. The CD14 Monocyte Differentiation Antigen Maps to a Region Encoding Growth Factors and Receptors. Science (1988) 239(4839):497–500. doi: 10.1126/science.239.4839.497
14. Pugin J, Heumann D, Tomasz A, Kravchenko VV, Akamatsu Y, Nishijima M, et al. CD14 Is a Pattern Recognition Receptor. Immunity (1994) 1(6):509–16. doi: 10.1016/1074-7613(94)90093-0
15. Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, et al. Md-2, a Molecule That Confers Lipopolysaccharide Responsiveness on Toll-like Receptor 4. J Exp Med (1999) 189(11):1777–82. doi: 10.1084/jem.189.11.1777
16. Nunez Miguel R, Wong J, Westoll JF, Brooks HJ, O’Neill LA, Gay NJ, et al. A Dimer of the Toll-Like Receptor 4 Cytoplasmic Domain Provides a Specific Scaffold for the Recruitment of Signaling Adaptor Proteins. PLoS One (2007) 2(8):e788. doi: 10.1371/journal.pone.0000788
17. Gioannini TL, Teghanemt A, Zhang D, Coussens NP, Dockstader W, Ramaswamy S, et al. Isolation of an Endotoxin–MD-2 Complex That Produces Toll-like Receptor 4-Dependent Cell Activation at Picomolar Concentrations. Proc Natl Acad Sci (2004) 101(12):4186–91. doi: 10.1073/pnas.0306906101
18. Prohinar P, Re F, Widstrom R, Zhang D, Teghanemt A, Weiss JP, et al. Specific High Affinity Interactions of Monomeric Endotoxin· Protein Complexes With Toll-like Receptor 4 Ectodomain. J Biol Chem (2007) 282(2):1010–17. doi: 10.1074/jbc.M609400200
19. Teghanemt A, Prohinar P, Gioannini TL, Weiss JP. Transfer of Monomeric Endotoxin From MD-2 to CD14 Characterization AND Functional Consequences. J Biol Chem (2007) 282(50):36250–56. doi: 10.1074/jbc.M705995200
20. Teghanemt A, Widstrom RL, Gioannini TL, Weiss JP. Isolation of Monomeric and Dimeric Secreted MD-2 ENDOTOXIN· Scd14 AND Toll-Like RECEPTOR 4 ECTODOMAIN Selectively REACT With THE Monomeric FORM of SECRETED Md-2. J Biol Chem (2008) 283(32):21881–89. doi: 10.1074/jbc.M800672200
21. Jack RS, Grunwald U, Stelter F, Workalemahu G, Schütt C. Both Membrane-Bound and Soluble Forms of CD14 Bind to Gram-Negative Bacteria. Eur J Immunol (1995) 5):1436–41. doi: 10.1002/eji.1830250545
22. Grunwald U, Fan X, Jack RS, Workalemahu G, Kallies A, Stelter F, et al. Monocytes can Phagocytose Gram-Negative Bacteria by a CD14-Dependent Mechanism. J Immunol (1996) 157(9):4119–25.
23. Fan X, Stelter F, Menzel R, Jack R, Spreitzer I, Hartung T, et al. Structures in Bacillus Subtilis are Recognized by CD14 in a Lipopolysaccharide Binding Protein-Dependent Reaction. Infect Immun (1999) 67(6):2964–68. doi: 10.1128/IAI.67.6.2964-2968.1999
24. Stelter F, Bernheiden M, Menzel R, Jack RS, Witt S, Fan X, et al. Mutation of Amino Acids 39–44 of Human CD14 Abrogates Binding of Lipopolysaccharide and Escherichia Coli. Eur J Biochem (1997) 3(1-2):100–09. doi: 10.1111/j.1432-1033.1997.00100.x
25. Schiff DE, Kline L, Soldau K, Lee JD, Pugin J, Tobias PS, et al. Phagocytosis of Gram-Negative Bacteria by a Unique CD14-dependent Mechanism. J Leukocyte Biol (1997) 62(6):786–94. doi: 10.1002/jlb.62.6.786
26. Wurfel MM, Wright SD. Lipopolysaccharide-Binding Protein and Soluble CD14 Transfer Lipopolysaccharide to Phospholipid Bilayers: Preferential Interaction With Particular Classes of Lipid. J Immunol (1997) 158(8):3925–34.
27. Yu B, Hailman E, Wright SD. Lipopolysaccharide Binding Protein and Soluble CD14 Catalyze Exchange of Phospholipids. J Clin Invest (1997) 99(2):315–24. doi: 10.1172/JCI119160
28. Wang PY, Kitchens RL, Munford RS. Phosphatidylinositides Bind to Plasma Membrane CD14 and can Prevent Monocyte Activation by Bacterial Lipopolysaccharide. J Biol Chem (1998) 273(38):24309–13. doi: 10.1074/jbc.273.38.24309
29. Nakata T, Yasuda M, Fujita M, Kataoka H, Kiura K, Sano H, et al. CD14 Directly Binds to Triacylated Lipopeptides and Facilitates Recognition of the Lipopeptides by the Receptor Complex of Toll-like Receptors 2 and 1 Without Binding to the Complex. Cell Microbiol (2006) 8(12):1899–909. doi: 10.1111/j.1462-5822.2006.00756
30. Dziarski R, Tapping RI, Tobias PS. Binding of Bacterial Peptidoglycan to CD14. J Biol Chem (1998) 273(15):8680–90. doi: 10.1074/jbc.273.15.8680
31. Gupta D, Kirkland TN, Viriyakosol S, Dziarski R. CD14 is a Cell-Activating Receptor for Bacterial Peptidoglycan. J Biol Chem (1996) 271(38):23310–16. doi: 10.1074/jbc.271.38.23310
32. Savedra R Jr, Delude RL, Ingalls RR, Fenton MJ, Golenbock DT. Mycobacterial Lipoarabinomannan Recognition Requires a Receptor That Shares Components of the Endotoxin Signaling System. J Immunol (1996) 157(6):2549–54.
33. Yu W, Soprana E, Cosentino G, Volta M, Lichenstein HS, Viale G, et al. Soluble CD141–152 Confers Responsiveness to Both Lipoarabinomannan and Lipopolysaccharide in a Novel HL-60 Cell Bioassay. J Immunol (1998) 161(8):4244–51.
34. Elass E, Coddeville B, Guérardel Y, Kremer L, Maes E, Mazurier J, et al. Identification by Surface Plasmon Resonance of the Mycobacterial Lipomannan and Lipoarabinomannan Domains Involved in Binding to CD14 and LPS-Binding Protein. FEBS Lett (2007) 581(7):1383–90. doi: 10.1016/j.febslet.2007.02.056
35. Lien E, Sellati TJ, Yoshimura A, Flo TH, Rawadi G, Finberg RW, et al. Toll-Like Receptor 2 Functions as a Pattern Recognition Receptor for Diverse Bacterial Products. J Biol Chem (1999) 274(47):33419–25. doi: 10.1074/jbc.274.47.33419
36. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential Roles of TLR2 and TLR4 in Recognition of Gram-Negative and Gram-Positive Bacterial Cell Wall Components. Immunity (1999) 11(4):443–51. doi: 10.1016/S1074-7613(00)80119-3
37. Salomao R, Brunialti MK, Rapozo MM, Baggio-Zappia GL, Galanos C, Freudenberg M. Bacterial Sensing, Cell Signaling, and Modulation of the Immune Response During Sepsis. Shock (2012) 38(3):227–42. doi: 10.1097/SHK.0b013e318262c4b0
38. Opal SM, Cohen J. Clinical Gram-Positive Sepsis: Does it Fundamentally Differ From Gram-Negative Bacterial Sepsis? Crit Care Med (1999) 27(8):1608–16. doi: 10.1097/00003246-199908000-00039
39. Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, et al. Crystal Structure of CD14 and Its Implications for Lipopolysaccharide Signaling. J Biol Chem (2005) 280(12):11347–51. doi: 10.1074/jbc.M414607200
40. Filipp D, Alizadeh-Khiavi K, Richardson C, Palma A, Paredes N, Takeuchi O, et al. Soluble CD14 Enriched in Colostrum and Milk Induces B Cell Growth and Differentiation. Proc Natl Acad Sci (2001) 98(2):603–08. doi: 10.1073/pnas.98.2.603
41. Lee JW, Paape MJ, Elsasser TH, Zhao X. Recombinant Soluble CD14 Reduces Severity of Intramammary Infection by Escherichia Coli. Infect Immun (2003) 71(7):4034–39. doi: 10.1128/IAI.71.7.4034-4039.2003
42. Schroder NW, Opitz B, Lamping N, Michelsen KS, Zahringer U, Gobel UB, et al. Involvement of Lipopolysaccharide Binding Protein, CD14, and Toll-like Receptors in the Initiation of Innate Immune Responses by Treponema Glycolipids. J Immunol (2000) 165(5):2683–93. doi: 10.4049/jimmunol.165.5.2683
43. Yoon HJ, Shin JH, Yang SH, Chae DW, Kim H, Lee DS, et al. Association of the CD14 Gene–159C Polymorphism With Progression of IgA Nephropathy. J Med Genet (2003) 40(2):104–08. doi: 10.1136/jmg.40.2.104
44. Passos LSA, Gazzinelli-Guimaraes PH, Mendes TAO, Guimaraes ACG, Lemos DS, Ricci ND, et al. Regulatory Monocytes in Helminth Infections: Insights From the Modulation During Human Hookworm Infection. BMC Infect Dis (2017) 17:253. doi: 10.1186/s12879-017-2366-0
45. Parija SC. Intestinal Parasitic Infestation in Children and Other Related Parasitic Infections. Trop Parasitol (2011) 1(2):49. doi: 10.4103/2229-5070.86918
46. Maizels RM. Parasite Immunomodulation and Polymorphisms of the Immune System. J Biol (2009) 8(7):1–4. doi: 10.1186/jbiol166
47. Das M, Laha R. Parasitic Infections of Cattle in North Eastern Region of India–An Overview. Arch Parasitol (2017) 1(1):107.
48. Oakley MS, Majam V, Mahajan B, Gerald N, Anantharaman V, Ward JM, et al. Pathogenic Roles of CD14, Galectin-3, and OX40 During Experimental Cerebral Malaria in Mice. PLoS One (2009) 4(8):e6793. doi: 10.1371/journal.pone.0006793
49. Hirako IC, Gallego-Marin C, Ataide MA, Andrade WA, Gravina H, Rocha BC, et al. Dna-Containing Immunocomplexes Promote Inflammasome Assembly and Release of Pyrogenic Cytokines by CD14+ Cd16+ CD64high Cd32low Inflammatory Monocytes From Malaria Patients. mBio (2015) 6(6):e01605–15. doi: 10.1128/mBio.01605-15
50. Hixson JE, Brown WM. A Comparison of the Small Ribosomal RNA Genes From the Mitochondrial DNA of the Great Apes and Humans: Sequence, Structure, Evolution, and Phylogenetic Implications. Mol Biol Evol (1986) 3(1):1–18. doi: 10.1093/oxfordjournals.molbev.a040379
51. Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC. African Populations and the Evolution of Human Mitochondrial DNA. Science (1991) 253(5027):1503–07. doi: 10.1126/science.1840702
52. Pal A, Sharma A, Bhattacharya TK, Chatterjee PN, Chakravarty AK. Molecular Characterization and SNP Detection of CD14 Gene in Crossbred Cattle. Mol Biol Int (2011) 2011:13. doi: 10.4061/2011/507346
53. Pal A, Chatterjee PN, Sharma A. Sequence Characterization and Polymorphism Detection in Bubaline CD14 Gene. Buffalo Bull (2014) 32(2):138–56.
54. Pal A, Chatterjee PN. Molecular Cloning and Characterization of CD14 Gene in Goat. Small Ruminant Res (2009) 82(2-3):84–7. doi: 10.1016/j.smallrumres.2008.11.016
55. Kelley SL, Lukk T, Nair SK, Tapping RI. The Crystal Structure of Human Soluble CD14 Reveals a Bent Solenoid With a Hydrophobic Amino-Terminal Pocket. J Immunol (2013) 190(3):1304–11. doi: 10.4049/jimmunol.1202446
56. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed Vol. Vol.1. New York, USA: Cold Spring Harbor Laboratory Press (2001). 557 p.
57. Katoh K, Standley DM. A Simple Method to Control Over-Alignment in the MAFFT Multiple Sequence Alignment Program. Bioinformatics (2016) 32(13):1933–42. doi: 10.1093/bioinformatics
58. Glick DM. Glossary of Biochemistry and Molecular Biology. Scissors L, editor. London, UK: Portland Press (1977). 324 p. Revised edition.
59. Petersen B, Petersen TN, Andersen P, Nielsen M Lundegaard C. A Generic Method for Assignment of Reliability Scores Applied to Solvent Accessibility Predictions. BMC Struct Biol (2009) 9:51. doi: 10.1186/1472-6807-9-51
60. Ebina T, Toh H, Kuroda Y. Loop-Length-Dependent SVM Prediction of Domain Linkers for High-Throughput Structural Proteomics. Pept Sci (2009) 92(1):1–8. doi: 10.1002/bip.21105
61. Cunningham MD, Shapiro RA, Seachord C, Ratcliffe K, Cassiano L, Darveau RP. CD14 Employs Hydrophilic Regions to “Capture” Lipopolysaccharides. J Immunol (2000) 164(6):3255–63. doi: 10.4049/jimmunol.164.6.3255
62. Muroi M, Ohnishi T, Tanamoto KI. Regions of the Mouse CD14 Molecule Required for Toll-Like Receptor 2-and 4-Mediated Activation of NF-κb. J Biol Chem (2002) 277(44):42372–9. doi: 10.1074/jbc.M205966200
63. Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISSMODEL Repository and Associated Resources. Nucleic Acids Res (2009) 37:387–92. doi: 10.1093/nar/gkn750
64. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res (1997) 25:3389–402. doi: 10.1093/nar/25.17.3389
65. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat Protoc (2015) 10(6):845–58. doi: 10.1038/nprot.2015.053
66. Wiederstein M, Sippl MJ. ProSA-web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res (2007) 35(suppl_2):W407–10. doi: 10.1093/nar/gkm290
67. Zhang Y, Skolnick J. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res (2005) 33(7):2302–09. doi: 10.1093/nar/gki524
68. Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: Predicting Stability Changes Upon Mutation From the Protein Sequence or Structure. Nucleic Acids Res (2005) 33(2):306–10. doi: 10.1093/nar/gki375
69. Choi Y, Chan AP. PROVEAN Web Server: A Tool to Predict the Functional Effect of Amino Acid Substitutions and Indels. Bioinformatics (2015) 31(16):2745–47. doi: 10.1093/bioinformatics/btv195
70. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. String v9. 1: Protein-Protein Interaction Networks, With Increased Coverage and Integration. Nucleic Acids Res (2015) 41(D1):D808–15. doi: 10.1093/nar/gks1094
71. Benjamin MM. In: Outline of Veterinary Clinical Pathology. 3rd Edn. New Delhi: Kalyani Pub. (1985) p. p 233–54.
72. Hawk PB. In: Hawak’s Physiological Chemistry. 14th Edn. London: McGraw Hill Book Co (1965). 218 p.
73. Jain NC. In: Essentials of Veterinary Haematology. Pennsylvania: Lea and Febiger Publishers Malvern (1986) p. 57–62.
74. Duhovny DS, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: Servers for Rigid and Symmetric Docking. Nucleic Acids Res (2005) 33:363–67. doi: 10.1093/nar/gki481
75. Smarda P, Bures P, Horova L, Leitch IJ, Mucina L, Pacini E, et al. Ecological and Evolutionary Significance of Genomic GC Content Diversity in Monocots. Proc Natl Acad Sci (2014) 111(39):E4096–102. doi: 10.1073/pnas.1321152111
76. Vinogradov AE. DNA Helix: The Importance of Being GC-Rich. Nucleic Acids Res (2003) 31(7):1838–44. doi: 10.1093/nar/gkg296
77. Setoguchi M, Nasu N, Yoshida S, Higuchi Y, Akizuki SI, Yamamoto S. Mouse and Human CD14 (Myeloid Cell-Specific Leucine-Rich Glycoprotein) Primary Structure Deduced From cDNA Clones. Biochim Biophys Acta (BBA)-Gene Structure Expression (1989) 1008(2):213–22. doi: 10.1016/0167-4781(80)90012-3
78. Meng J, Parroche P, Golenbock DT, McKnight CJ. The Differential Impact of Disulfide Bonds and N-linked Glycosylation on the Stability and Function of CD14. J Biol Chem (2008) 283(6):3376–84. doi: 10.1074/jbc.M707640200
79. Takai N, Kataoka M, Higuchi Y, Matsuura K, Yamamoto S. Primary Structure of Rat CD14 and Characteristics of Rat CD14, Cytokine, and NO Synthase mRNA Expression in Mononuclear Phagocyte System Cells in Response to LPS. J Leukocyte Biol (1997) 1(6):736–44. doi: 10.1002/jlb.61.6.736
80. Wu Z, Rothwell L, Hu T, Kaiser P. Chicken CD14, Unlike Mammalian CD14, is Trans-Membrane Rather Than GPI-Anchored. Dev Comp Immunol (2009) 33(1):97–104. doi: 10.1016/j.dci.2008.07.008
81. La Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and Prediction of Leucine-Rich Nuclear Export Signals. Protein Eng Design Selection (2004) 17(6):527–36. doi: 10.1093/protein/gzh062
82. Landschulz WH, Johnson PF, McKnight SL. The Leucine Zipper: A Hypothetical Structure Common to a New Class of DNA Binding Proteins. Science (1988) 240(4860):1759–64. doi: 10.1126/science.3289117
83. George RA, Heringa J. An Analysis of Protein Domain Linkers: Their Classification and Role in Protein Folding. Protein Eng Design Selection (2002) 15(11):871–79. doi: 10.1093/protein/15.11.871
84. Paape MJ, Lilius EM, Wiitanen PA, Kontio MP, Miller RH. Intramammary Defense Against Infections Induced by Escherichia Coli in Cows. Am J Vet Res (1996) 57(4):477–82.
85. Wright SD, Ramos RA, Hermanowski-Vosatka A, Rockwell P, Detmers PA. Activation of the Adhesive Capacity of CR3 on Neutrophils by Endotoxin: Dependence on Lipopolysaccharide Binding Protein and CD14. J Exp Med (1991) 173(5):1281–86. doi: 10.1084/jem.173.5.1281
86. Inohara N, Nunez G. ML—a Conserved Domain Involved in Innate Immunity and Lipid Metabolism. Trends Biochem Sci (2002) 27(5):219–21. doi: 10.1016/S0968-0004(02)02084-4
87. Ohto U, Fukase K, Miyake K, Satow Y. Crystal Structures of Human MD-2 and its Complex With Antiendotoxic Lipid Iva. Science (2007) 316(5831):1632–34. doi: 10.1126/science.1139111
88. Ohto U, Fukase K, Miyake K, Shimizu T. Structural Basis of Species-Specific Endotoxin Sensing by Innate Immune Receptor TLR4/MD-2. Proc Natl Acad Sci (2012) 109(19):7421–6. doi: 10.1073/pnas.1201193109
89. Li L, Zhou X, Chen X. Characterization and Evolution of MHC Class II B Genes in Ardeid Birds. J Mol Evol (2011) 72(5-6):474–83. doi: 10.1007/s00239-011-9446-3
90. Klein J, Sato A, Nagl S, O’hUigín C. Molecular Trans-Species Polymorphism. Annu Rev Ecol Syst (1998) 29(1):1–21. doi: 10.1146/annurev.ecolsys.29.1.1
91. Tundup S, Srivastava L, Nagy T, Harn D. CD14 Influences Host Immune Responses and Alternative Activation of Macrophages During Schistosoma Mansoni Infection. Infect Immun (2014) 82(8):3240–51. doi: 10.1128/IAI.01780-14
92. Antonelli LR, Leoratti FM, Costa PA, Rocha BC, Diniz SQ, Tada MS, et al. The CD14+ CD16+ Inflammatory Monocyte Subset Displays Increased Mitochondrial Activity and Effector Function During Acute Plasmodium Vivax Malaria. PLoS Pathog (2014) 10(9):e1004393. doi: 10.1371/journal.ppat.1004393
93. Turner JD, Bourke CD, Meurs L, Mbow M, Dieye TN, Mboup S, et al. Circulating CD14 Bright CD16+’intermediate’monocytes Exhibit Enhanced Parasite Pattern Recognition in Human Helminth Infection. PLoS Negl Trop Dis (2014) 8(4):e2817. doi: 10.1371/journal.pntd.0002817
94. Skrzeczynska-Moncznik J, Bzowska M, Lo seke S, Grage-Griebenow E, Zembala M, Pryjma J. Peripheral Blood CD14 High CD16+ Monocytes are Main Producers of IL-10. Scand J Immunol (2008) 67(2):152–9. doi: 10.1111/j.1365-3083.2007.02051.x
95. Janeway CA, Travers P, Walport M, Sclomchik M. Immunobiology: The Immune System in Health and Disease. Sixth Edn. Garland Publishe (2004). 432 p.
Keywords: CD14 molecule, in silico analysis, Haemonchus contortus, sheep, differential mRNA expression analysis, molecular docking, molecular phylogeny
Citation: Rawat K, Pal A, Banerjee S, Pal A, Mandal SC and Batabyal S (2021) Ovine CD14- an Immune Response Gene Has a Role Against Gastrointestinal Nematode Haemonchus contortus—A Novel Report. Front. Immunol. 12:664877. doi: 10.3389/fimmu.2021.664877
Received: 06 February 2021; Accepted: 28 April 2021;
Published: 16 July 2021.
Edited by:
Wanderley De Souza, Federal University of Rio de Janeiro, BrazilReviewed by:
Eduardo José Lopes Torres, Rio de Janeiro State University, BrazilYu Zhou, Codiak BioSciences, United States
Copyright © 2021 Rawat, Pal, Banerjee, Pal, Mandal and Batabyal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aruna Pal, YXJ1bmFjaGF0dGVyamVlQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Kavita Rawat
Kavita Rawat Aruna Pal
Aruna Pal Samiddha Banerjee
Samiddha Banerjee Abantika Pal
Abantika Pal Subhas Chandra Mandal5
Subhas Chandra Mandal5 Subhasis Batabyal
Subhasis Batabyal