
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 24 May 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.660384
 Guiyi Ji1,2†
Guiyi Ji1,2† Miaomiao Zhang1†
Miaomiao Zhang1† Qianqian Liu3†
Qianqian Liu3† Shouquan Wu1
Shouquan Wu1 Yu Wang1
Yu Wang1 Guo Chen4
Guo Chen4 Andrew J. Sandford5
Andrew J. Sandford5 Jian-Qing He1*
Jian-Qing He1*Background: Nuclear transcription factor erythroid 2 p45-related factor 2 (Nrf2), encoded by NFE2L2, functions as a key transcription factor and regulates expression of antioxidant genes. Our study aimed to investigate the association of single nucleotide polymorphisms of NFE2L2 with tuberculosis (TB) and latent tuberculosis infection (LTBI) and the underlying causal mechanisms.
Methods: 1950 unrelated Chinese Han participants were included in our two independent study groups. Five tag polymorphisms were selected and genotyped. The functional effects of the rs13005431 polymorphism were confirmed by dual-luciferase reporter assays and mRNA level comparisons.
Results: Rs13005431_C and rs2364723_G were associated with increased TB susceptibility (P = 0.010 and P = 0.041) after adjustment for confounding factors. rs6726395_A was associated with increased risk of active TB (P=0.035) in a comparison with the LTBI group. The frequency of haplotype rs1049751- rs13005431 AC was higher in the TB group (P =0.013), while frequency of haplotype AT was higher in the healthy control group (P =0.025). The luciferase activity of a plasmid with the rs13005431C-promoter was significantly lower than that of the rs13005431T-promoter. In addition, neutrophils with the CC/TC genotypes which were activated by GM-CSF showed a decreased level of NFE2L2 mRNA when compared with the rs13005431 TT genotype.
Conclusions: Our study suggests that allele C of rs13005431 might increase the susceptibility to TB by down-regulating the transcriptional activity of NFE2L2.
Tuberculosis (TB) is a disease caused by Mycobacterium tuberculosis (M.TB), which infects approximately one-third of the population worldwide, and remains one of the most important public health problems (1). In 2018, there were an estimated 10 million cases and 1.3 million deaths from tuberculosis worldwide (2). The aim of the End TB Strategy is to achieve a 95% reduction in TB deaths and a 90% reduction in the TB incidence rate by 2035 (2). Thus, great efforts are urgently needed to strengthen the prevention, diagnosis and treatment of TB.
As is well known, encountering M.TB leads to several possible outcomes including M. TB clearance, primary TB, latent TB infection (LTBI) and active TB. It has been suggested that 5-10% of LTBI individuals will progress to active TB during their lifetime (3). Management of LTBI will be required to implement the End TB Strategy (4). Screening and preventive treatment of LTBI have been recommended in populations with high risk (5). There are two available methods for LTBI diagnosis: tuberculin skin test (TST) and Interferon-gamma release assays (IGRAs) (including the Quantiferon GIT Assay and T-SPOT.TB). Although guidelines suggest that either the TST or IGRAs may be used to diagnose LTBI (5), IGRAs have been shown to have higher specificity and sensitivity (6) with no bacille Calmette–Guérin (BCG) vaccination interference and much less environmental mycobacteria interference than that of TST.
Previous studies have suggested that TB is associated with oxidative and antioxidant responses (7, 8). Nuclear factor-erythroid 2 (NF-E2)-related factor 2 (Nrf2), encoded by the NFE2L2 gene, functions as a critical transcription factor in the anti-oxidation process and usually is repressed by Keap1. Nrf2 enters the nucleus upon activation by oxidative stress (9, 10), binds to the GCTGAGTCA site of the antioxidant response element in the promoter of antioxidant phase II genes and promotes the expression of these genes (11, 12). Several studies have revealed a relationship between Nrf2 and M.TB infection. It was reported that Nrf2 participated in autophagy (13), the antioxidant response in M. TB-infected guinea pigs (14), and the reduction of granulomas in Nrf2-deficient mice when infected with M.TB (15).
Recently, oxidant/antioxidant related genes such as FMO2 and CYBA have been associated with TB susceptibility (16, 17). However, there have been no reports of an association between NFE2L2 polymorphisms and risk of LTBI and TB. Therefore, we carried out a discovery study and a replication study to determine the relationship of NFE2L2 variants with susceptibility to TB and LTBI in the Chinese Han population. We also performed dual-luciferase reporter assays and compared NFE2L2 mRNA levels of neutrophils with different genotypes to validate the association findings.
In total, 1950 unrelated Chinese Han participants, consisting of 636 TB patients and 608 healthy controls in the discovery study as well as 301 TB patients, 201 LTBI subjects and 204 uninfected healthy controls (HC) in the replication study, were consecutively recruited from the West China Hospital between July 2013 and August 2017. For the discovery study, the diagnosis of TB was based upon the following criteria: histopathological evidence of TB disease and/or culture positivity for MTB and/or smear positivity for MTB in at least two separate specimens and/or radiological and clinical findings consistent with TB, with positive clinical response to anti-TB therapy (18). TB cases were divided into two subgroups: (1) pulmonary TB patients (PTB, pathological changes limited to the lung) and (2) extra-pulmonary patients (EPTB, pathological changes involving other tissues or organs merely or in combination with the lungs). The healthy controls who did not have a history or evidence of TB on the basis of their syndromes and radiographic examination results were enrolled during their routine health examination in the West China Hospital. For the replication study, both the uninfected HC group and LTBI participants were close contacts of PTB patients. LTBI was defined as adults (age > 18) with a documented positive result of IGRAs and negative results of radiological and clinical manifestations. HC individuals were defined as adults with negative results of IGRAs, radiological and clinical findings. The inclusion criteria of TB patients enrolled in the replication study were the same as the discovery study. Another 60 healthy volunteers were recruited for comparison of mRNA levels of neutrophils. All participants diagnosed with HIV infection, diabetes mellitus, autoimmune disorders, tumors or treated with immunosuppressive drugs were excluded.
NFE2L2 is located on chromosome 2 (2q31) and has five exons and four introns. TagSNPs were selected from the region 3,000 base pairs upstream to 300 base pairs downstream of the NFE2L2 gene based on the Chinese Han Beijing data of the HapMap database (http://hapmap.ncbi.nlm.nih.gov, HapMap Data Rel 27 Phase II + III, Feb09, on NCBI B36 assembly, dbSNP b126) by Haploview software 4.2. Using criteria of minor allele frequency (MAF) ≥ 5% and the Tagger pairwise method (r² ≥0.8), five tagSNPs of NFE2L2 (rs10497511, rs2364723, rs13005431, rs6726395 and rs1962142), representing eleven SNPs with MAF ≥ 0.05 in the covered gene region, were selected for genotyping.
A total of 5 ml peripheral venous blood was collected from each subject and genomic DNA was extracted using the AxyPrep genomic DNA Mini kits (Axygen, USA) following the manufacturer’s protocol. SNPs were genotyped on the MassArray Analyzer 4 system (Sequenom,USA). All probes and primers are available upon request. 5% samples were genotyped twice for quality control. The genotyping related primers were listed in Supplementary Table 1.
Polymerase chain reaction (PCR) was performed to amplify a 651bp sequence surrounding rs13005431 from genotyped genomic DNA with the TT genotype of rs13005431. Primers were designed using Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) and sequences for restriction sites NheI and XhoI were introduced (Supplementary Table 2). Both gel purified PCR product and pGL3-promoter reporter vector (Promega, Madison, WI, USA) were digested by NheI and XhoI restriction enzymes (Takara Bio Inc., Kusatsu, Japan) at 37°C for 4 hours and then ligated by T4 DNA ligase (TaKara Bio Inc.) at 16°C for 8 hours. After transforming into competent E. coli DH5α cells and extraction by plasmid miniprep kit (TianGen, Beijing, China), the recombinant plasmid vector was verified by direct sequencing and named PGL3-rs13005431T-promoter (Supplementary Figure 1). A site-directed mutagenesis strategy was applied to acquire another recombinant plasmid vector of PGL3-rs13005431C-promoter (see Supplementary Table 2 for primer sequences) which was confirmed by DNA sequencing.
HEK293T cells were plated into 96-well culture plates at a density of 3.2×104 cells/well and cultured for 48 hours to acquire 90% confluence at the time of transfection. The PGL3-promoter, PGL3-rs13005431T-promoter and PGL3-rs13005431C-promoter were transfected into the HEK293T cells together with PRL-CMV plasmid vectors which acted as an internal reference using Lipofectamine® 2000 Reagent (Invitrogen (Thermo Fisher Scientific), Waltham, MA USA). Dual-luciferase reporter assays (Promega, USA) were carried out after 30h transfection on a GloMax™ 96 microplate luminometer. The transfection experiment was performed in triplicate. Results are presented as relative luciferase activity by dividing the firefly luciferase activity of each well by the Renilla luciferase activity. The normalized luciferase activity of recombinant plasmids was further normalized to the PGL3-promoter group.
10 ml of peripheral venous blood in a heparin anticoagulant tube was obtained from 60 healthy volunteers. Neutrophils were isolated using Ficoll gradient density centrifugation and confirmed by Wright staining and flow cytometry (CD11b, CD16). The cells were then cultured in 24-well culture plates at a density of 2×106 cells/well for 3 hours, activated or not by GM-CSF (in a final concentration of 1 ng/ml). The Trizol method was then used to extract the total RNA of the neutrophils. Reverse transcription was conducted using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio Inc.). Real-time PCR for NFE2L2 was then performed on an ABI7300 Sequence Detection System (Applied Biosystems (Thermo Fisher Scientific), Waltham, MA USA) using SYBR Premix Ex Taq II (Takara Bio Inc.), with B2M as the house-keeping gene (19). Primers for real-time PCR are shown in the Supplementary Table 2.
Experimental data were analyzed using the Statistical Package for Social Sciences version 17.0 (SPSS, Chicago, IL, USA). Distributions of clinical characteristics between the control group and TB patients were evaluated by the chi-squared test and Fisher exact probability for categorical variables and the student’s t-test for continuous variables. P values, Odds Ratios (OR) and 95% confidence intervals (95%CI) of association between SNPs and TB susceptibility were calculated by binary or multinomial logistic regression under four genetic models (allelic, additive, dominant and recessive). Linkage disequilibrium (LD) analysis and haplotype blocks were created by Haploview software 4.2 (20), while p values, ORs, 95%CIs and the global test of haplotype analysis were calculated using the SHEsis program (21). Genotype and allele frequencies of SNPs in the control group were assessed to determine whether they conformed to Hardy-Weinberg equilibrium (HWE). Gene-environment interaction was detected using the multifactor dimensionality reduction (MDR) constructive induction algorithm. SNP-by-sex additive interactions by the method of Andersson and multiplicative interactions by logistic regression in the discovery study (22). Normalized luciferase activity and relative mRNA levels were evaluated by the student’s t-test. A statistically significant difference was indicated by a two-sided P-value < 0.05.
Table 1 shows the characteristics of all the study subjects. In the discovery study, there was no significant difference in the distribution of gender and age between the case and control groups. However, smoking status was statistically different (P = 0.003). In the replication study, although gender distribution was similar among the three groups, age significantly differed between groups. Smoking status and TB types of the participants in the replication study were not available.
The locations of the five tagSNPs genotyped are presented in Figure 1. The genotype call rates varied from 99.7% to 100% and the genotype reproducibility was 100%. All SNPs in the control group of both studies were in HWE.
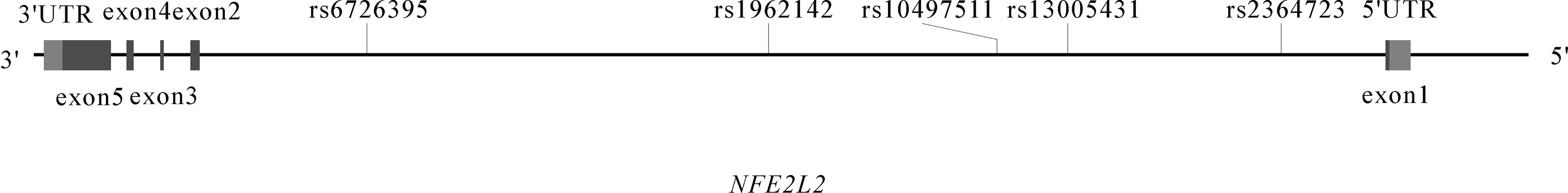
Figure 1 NFE2L2 gene structure and location of five tagSNPs (rs10497511, rs2364723, rs13005431, rs6726395 and rs1962142).
In the discovery study, we analyzed the data between TB patients and healthy controls. As shown in Table 2, allele C of rs13005431 and allele G of rs2364723 were associated with increased TB risk (P = 0.010 and P = 0.041). In stratified analyses, the deleterious effect of rs13005431 C was still seen in the non-smoker, EPTB and female subgroups (P = 0.003; P = 0.015 and P<0.001, respectively). In addition, rs2364723 G still demonstrated a relationship with increased susceptibility to TB in the non-smoker and female subgroups (P = 0.022 and P = 0.004). In addition, allele A of rs6726395 was demonstrated to be a risk factor associated with TB risk in the female subgroup (P = 0.003). There was no significant association observed between the other two tagSNPs (rs10497511 and rs1962142) and TB susceptibility (Table 2).
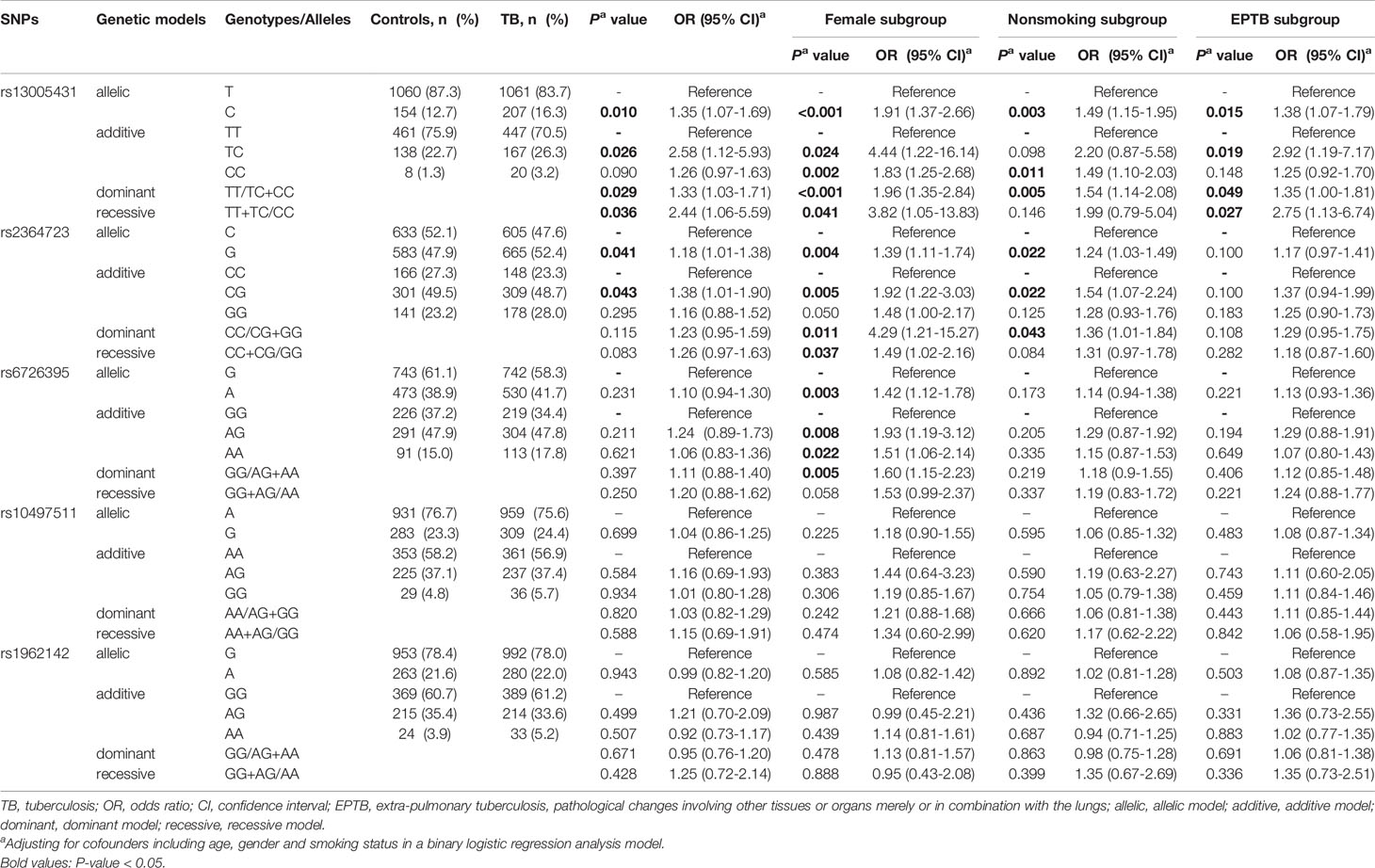
Table 2 Significant associations between five tagSNPs (rs13005431, rs2364723, rs6726395, rs10497511 and rs1962142) and tuberculosis risk in the discovery study and associations stratified by gender, smoking status and TB types.
In the replication study, we analyzed the data among TB patients, LTBI subjects and uninfected healthy controls using a multinomial logistic regression analysis model. When comparing the LTBI group with TB patients, only allele A of rs6726395 was associated with increased TB risk (P = 0.035) (Table 3). When the HC group was compared with the LTBI group, none of the tagSNPs was associated with susceptibility to TB infection. In addition, we combined LTBI subjects and uninfected healthy controls as a healthy control group. When comparing the healthy control group with TB patients, we still seen that rs6726395 A was associated with increased TB risk (P = 0.028) (Table 4).
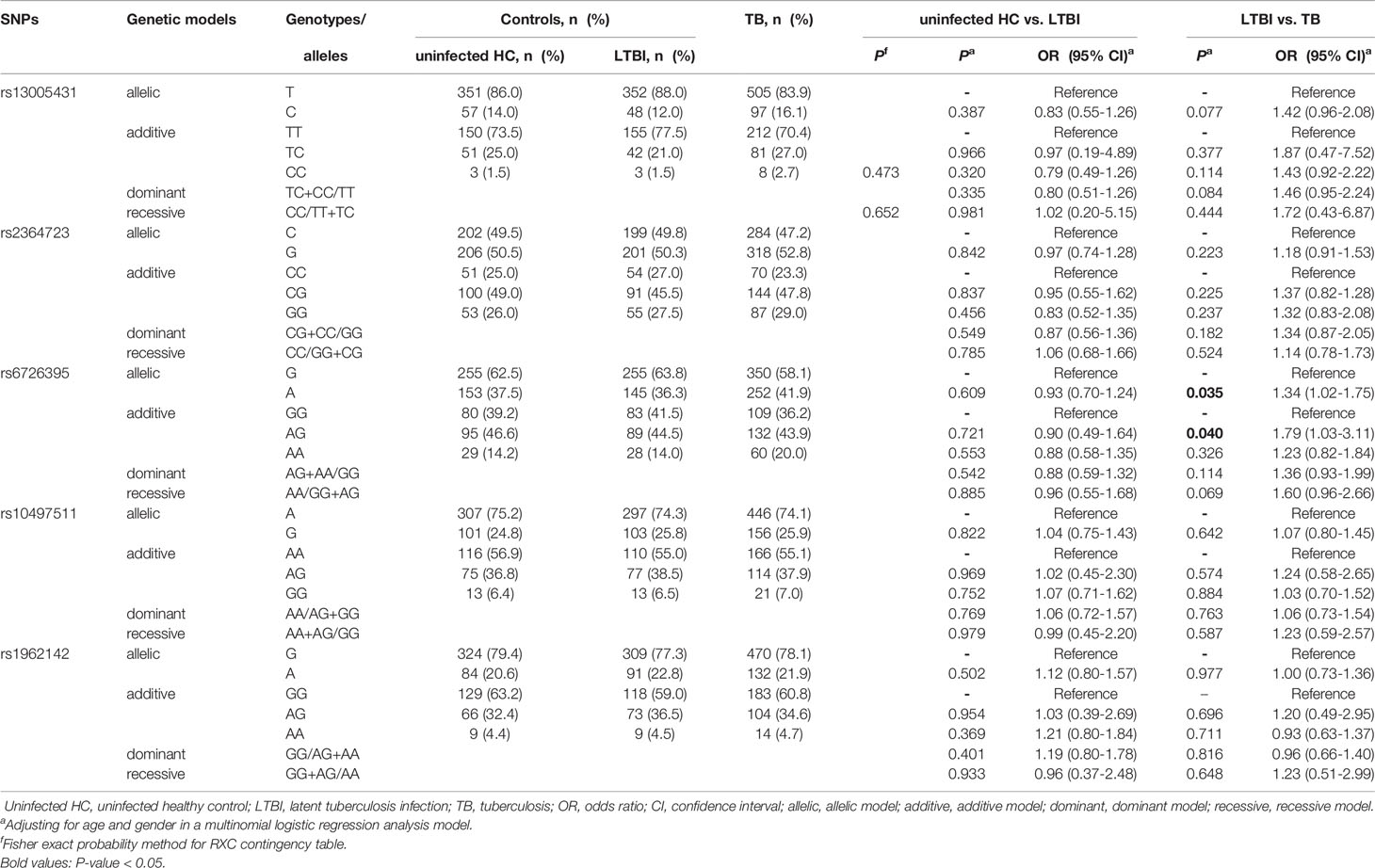
Table 3 Associations between five tagSNPs (rs13005431, rs2364723, rs6726395, rs10497511 and rs1962142) and risk of LTBI and TB in the replication study.
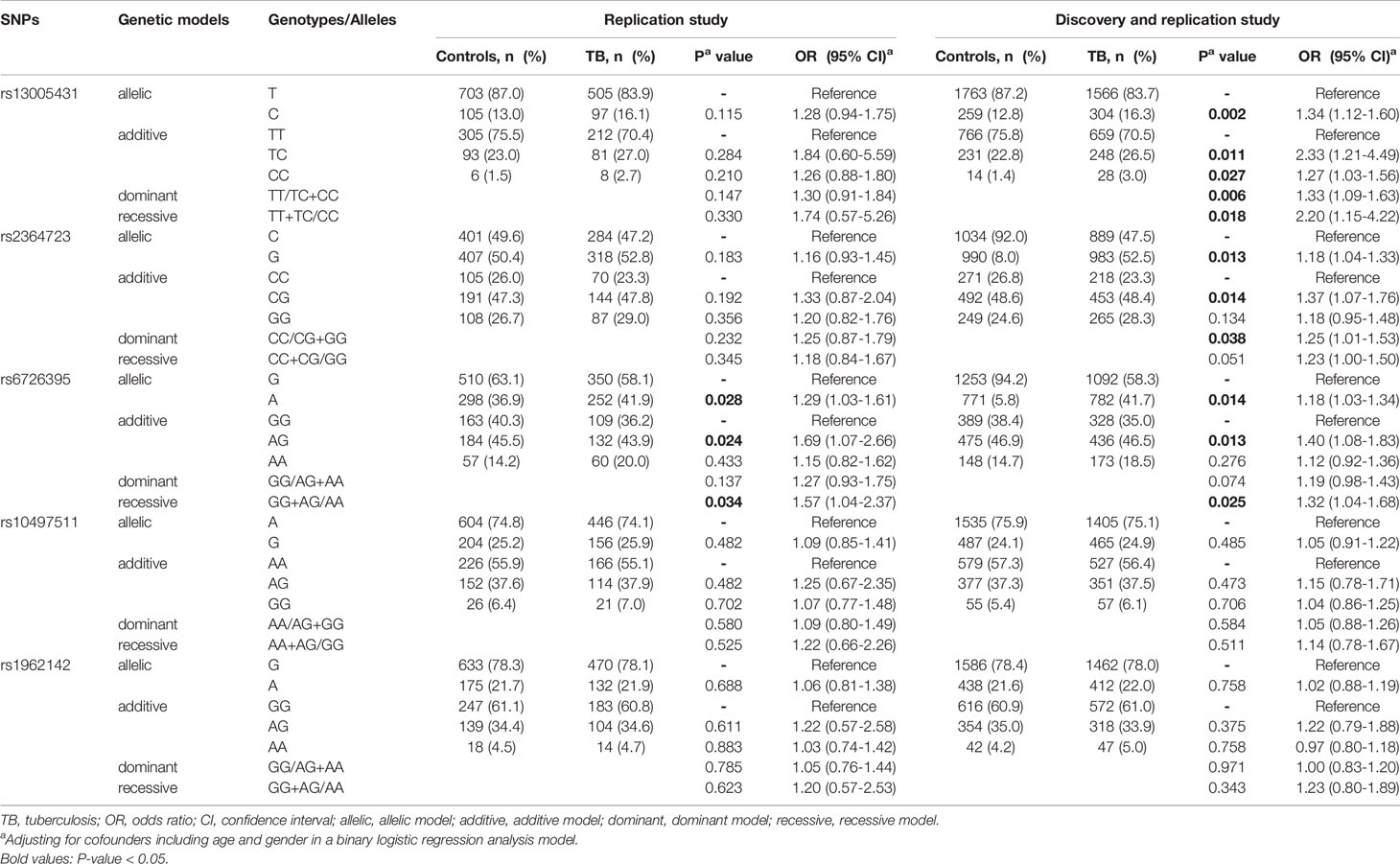
Table 4 Significant associations between five tagSNPs (rs13005431, rs2364723, rs6726395, rs10497511 and rs1962142) and tuberculosis risk in the replication study and the whole study including discovery study and replication study.
Furthermore, we combined the data of the two studies including the discovery study and the replication study. As shown in Table 4, allele C of rs13005431, allele G of rs2364723, and allele A of rs6726395 were associated with increased TB risk (P = 0.002; P = 0.013 and P = 0.014, respectively). There was no significant association observed between the other two tagSNPs (rs10497511 and rs1962142) and TB susceptibility.
The LD analysis (Supplementary Figure 2) demonstrated mild-to-moderate levels of LD between the five tagSNPs of NFE2L2. For the discovery study, haplotype analyses of the two blocks are shown in Supplementary Table 3. The frequency of haplotype rs1049751-rs13005431 AC was significantly higher in the TB group (P = 0.013), while the frequency of haplotype AT was higher in the control group (P = 0.025) and there was a significant global result (P = 0.025). For the replication study, no significant haplotype effect was observed.
The results of gene-environment interactions after MDR analysis are shown in Supplementary Table 4. In the discovery study, smoking-rs13005431 formed the best interaction model with 55.69% testing balanced accuracy and 10/10 cross-validation consistency. Smoking-sex-rs13005431 formed an interaction model with 55.03% testing balanced accuracy and 8/10 cross-validation consistency. After 1000-fold permutation testing, both models were found to be significant (P = 0.001-0.002 and P = 0.023, respectively). As shown in Supplementary Figure 3, smoking status formed the high-risk models regardless the genotypes of rs13005431, non-smoking status and rs13005431 TT formed the low-risk model, while non-smoking status and rs13005431 CC/TC formed the high-risk models. As shown in Supplementary Table 5, the only positive additive interaction was observed between female sex and rs13005431 TC+CC/C genotypes. In addition, there were suggestive positive multiplicative interactions of three SNPs (rs13005431, rs2364723 and rs6726395) with female sex under two genetic models.
We predicted the potential functional significance of SNPs from three aspects including protein coding, splicing regulation and transcription regulation (Supplementary Table 6). The influence factors of transcription regulation can be judged by three terms as following: 1) prediction of transcription factor binding sites altering using TRANSFAC or JASPAR database; 2) other factors affecting transcription regulation, such as high sensitive site of DNase I, histone modification, according to the comprehensive evaluation by GLODEN path; 3) judgment of evolutionary conservatism by PhastCons. According to the predicted results, rs13005431 was identified as a candidate SNP for further functional identification.
rs13005431, located in the first intron of NFE2L2, is outside of the promoter region. However, this SNP might disrupt an enhancer or silencer sequence and thus influence promoter activity indirectly. As previously reported, variants in the first intron might influence gene activity (23). Moreover, rs13005431 was reported to be a cis-eQTL (expression quantitative trait locus) of NFE2L2 in whole blood (P = 1.978×10-4) (24). Therefore, the dual-luciferase reporter assay was used to investigate the regulatory role of rs13005431. As shown in Figure 2, the normalized luciferase activity of PGL3-rs13005431C-promoter was significantly lower than the PGL3-rs13005431T-promoter and PGL3-promoter constructs (P <0.001 and P <0.001, respectively), indicating that the mutant allele C of rs13005431 may decrease promoter activity. In addition, no significant difference was observed between the PGL3-rs13005431T-promoter group and the PGL3-promoter group (P =0.077), suggesting that the wild type allele T of rs13005431 had no significant effect on promoter activity.
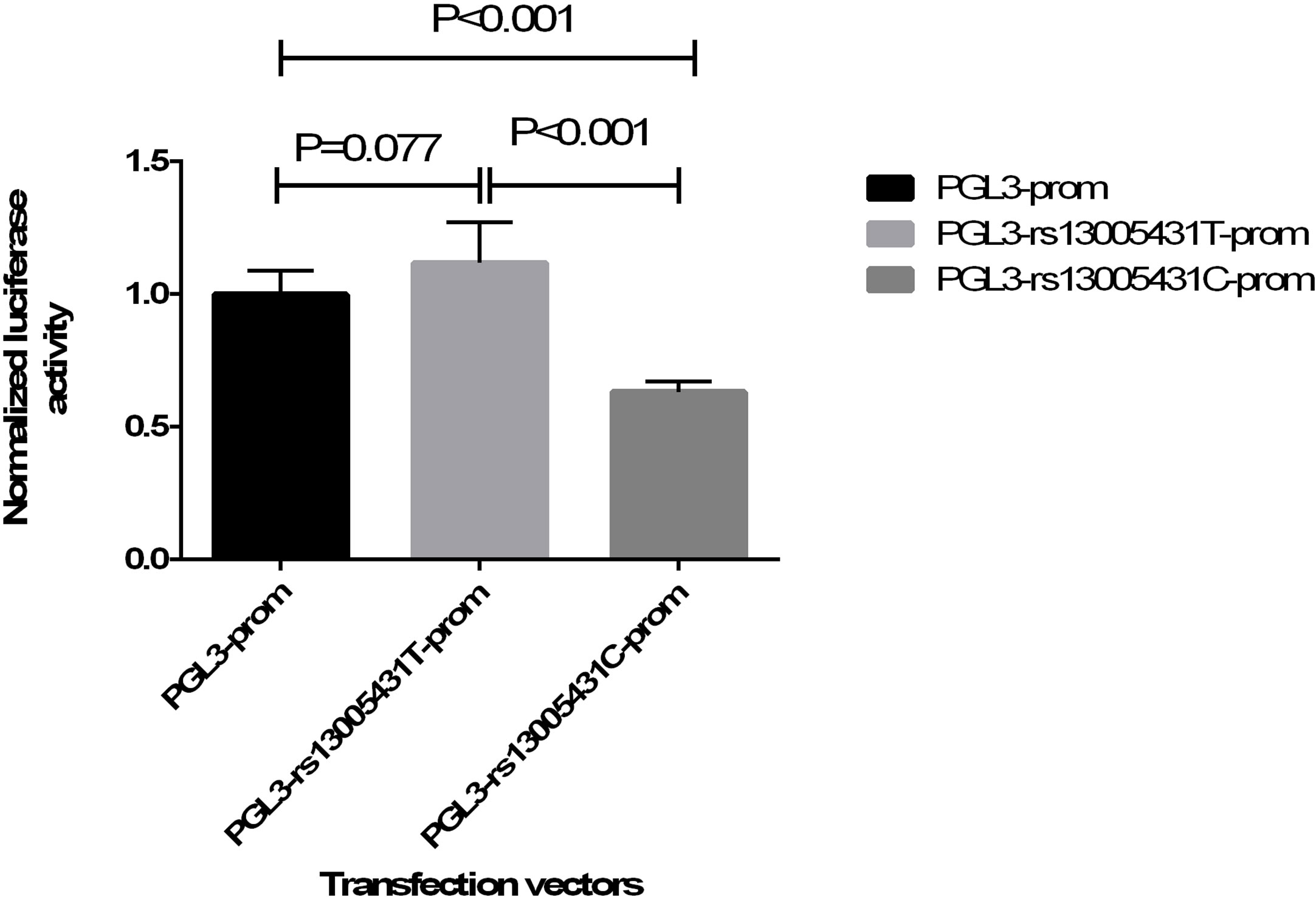
Figure 2 Normalized luciferase activity of three transfection vectors. The relative luciferase activity is presented as the firefly luciferase activity of each well divided by the Renilla luciferase activity. The normalized luciferase activity was expressed as a ratio to the mean relative luciferase activity of the PGL3-promoter group by each relative luciferase activity.
To further confirm whether allele C of rs13005431 influences the promoter activity of the NFE2L2 gene, 60 healthy subjects were enrolled. The relative expression levels of NFE2L2 in neutrophils were analyzed according to different genotypes under basal and GM-CSF activated conditions. As shown in Figure 3A, the mRNA expression levels of NFE2L2 in GM-CSF activated neutrophils were significantly increased compared with the unstimulated group (P <0.001). As shown in Figure 3B, subjects with genotype TC/CC had lower relative expression level than subjects with genotype TT in the GM-CSF activated group (P =0.017) while no difference was found when the neutrophils were unstimulated (P =0.123).
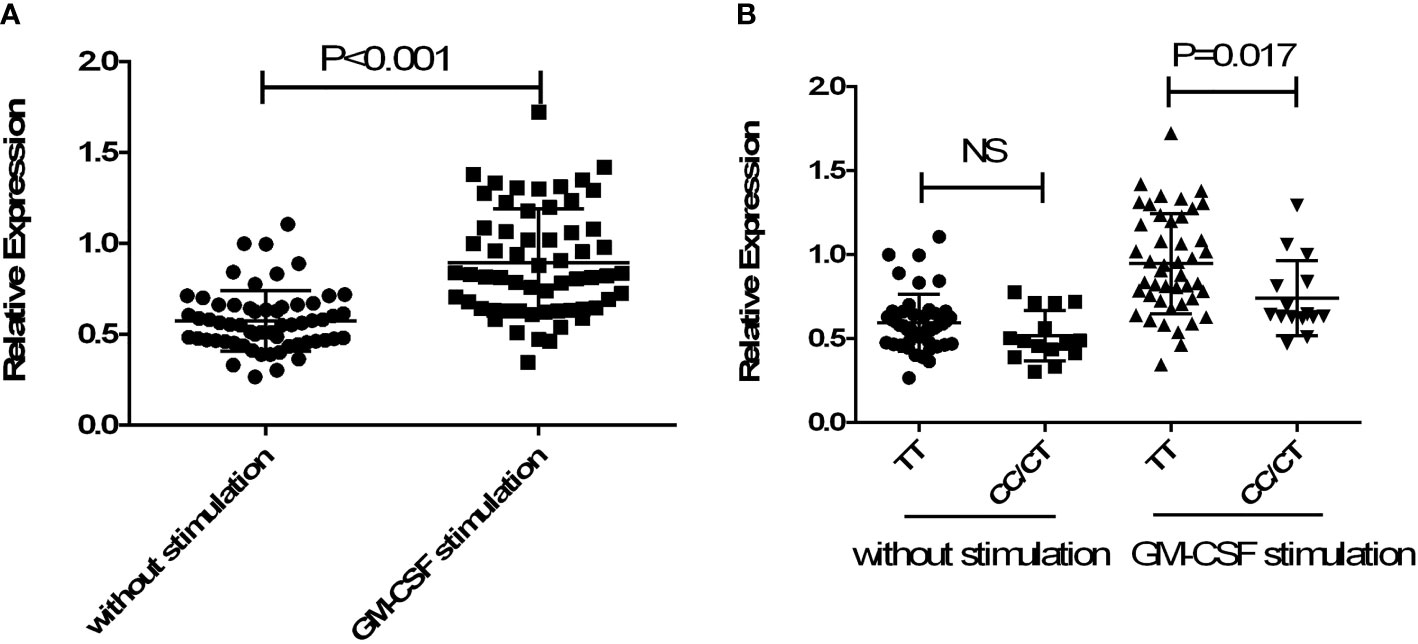
Figure 3 Gene expression analysis of NFE2L2 in neutrophils. (A) Comparison of the mRNA expression level of NFE2L2 between GM-CSF stimulated and without stimulated human neutrophils. (B) Comparison of the effects of different genotypes of rs13005431 on the mRNA expression level of NFE2L2 in the GM-CSF stimulated group and unstimulated group. NS, No statistical difference.
TB is a complex inflammatory process caused by Mycobacterium tuberculosis (M. TB). Active TB in adults is largely due to reactivation of primary infection (25), thus management of LTBI has been promoted for achieving the goals of the End-TB strategy (26). Several gene polymorphisms have shown significant associations with TB and LTBI susceptibility (27), which may help to reveal new aspects of the pathogenesis of TB and discover potential diagnostic genetic markers. It has been suggested that oxidant/antioxidant imbalance is related to TB (7, 8). Nrf2, a critical regulator of antioxidant defenses, has been shown to play a protective role in many lung diseases such as COPD, bleomycin-induced pulmonary fibrosis and so on (28, 29). Palanisamy et al. have shown that deficiency of Nrf2 was associated with progressive oxidative stress in guinea pigs infected with TB and antioxidant drugs could be a beneficial adjunct to anti-TB treatment (14). One study suggested that an Nrf2-mediated 17-gene signature can be used to distinguish TB patients from healthy controls, LTBI, pneumonia, or lung cancer, as well be used as an indicator of the anti-TB response (30). Here, we performed the first study of the relationship of NFE2L2 SNPs with TB and LTBI susceptibility in the Chinese Han population.
Our results showed that tagSNPs of NFE2L2 affect the susceptibility to TB and LTBI. The rs13005431_C and rs2364723_G alleles of NFE2L2 were both risk factors for TB. The haplotype analysis results also showed the association between rs13005431 and TB. The replication study revealed that rs6726395_A was associated with increased risk of active TB when comparing with the LTBI group, while none of the SNPs were associated with TB infection. Different genetic associations between TB and LTBI have been observed for SNPs of genes such as IRGM (27), FOXO3 (31), and TLR9 (32). These data indicate that the underlying genetic mechanisms might differ between susceptibility to LTBI and active TB. It is interesting that NFE2L2 SNPs are also associated with diabetes mellitus (33, 34), which is one of most common comorbidities of TB. Therefore, NFE2L2 dysfunction might serve as the common mechanism leading to TB and other diseases such as diabetes mellitus.
Additionally, we performed subgroup analysis according to TB location, gender and smoking status. Our results revealed that rs13005431_C was associated with increased risk of TB in the EPTB subgroup rather than in PTB subgroup. It has been reported that genetic susceptibility to TB differs between PTB and EPTB (31). Moreover, different mechanisms of immunity in different locations were found in mouse models (35). Our stratified results strengthen the hypothesis that polymorphisms involved in immunological mechanisms and the pathology of different TB types are different. Complicating factors such as gender, smoking and years after primary infection might influence the reactivation site (36). In another subgroup analysis by gender, rs13005431_C, rs2364723_G and rs6726395_A were found to be significant risk alleles in females but not in males. Multiplicative and additive interactions between female sex and rs13005431 genotypes were also observed. Previous studies have revealed that there were gender-specific associations between NFE2L2 gene variants and several diseases (37, 38). Socio-behavioral, cultural factors and gonadal steroids may relate to the gender differences in TB (39, 40).
In subgroup analysis by smoking status, the rs13005431_C and rs2364723_G alleles showed relationships with increased TB risk in the non-smoking subgroup. It is well known that smoking confers a higher risk of M. TB tuberculosis infection, TB development and progression (41–43). The MDR results also revealed smoking-rs13005431 interactions influence TB susceptibility, with smoking contributing to high risk of TB. Animal models and human studies have shown that Nrf2 and downstream activated genes play a critical role in defending against the oxidative stress of cigarette smoke (44, 45). Nrf2 was activated and numerous antioxidant enzymes were increased in healthy smokers (44, 46, 47). The relatively lower antioxidant responses of non-smoking individuals may account for susceptibility to TB in the non-smoking subgroup.
Introns have been shown to influence gene transcription in some reports (23, 48). Intronic polymorphisms may regulate the transcriptional activity or mRNA splicing after transcription. Based on our association results, rs13005431 was investigated using dual-luciferase reporter assays to determine the influence of the SNP on transcriptional activity. The normalized luciferase activity of plasmid PGL3-rs13005431C-promoter was lower than that of PGL3-rs13005431T-promoter, suggesting that the C allele of rs13005431 decreases promoter activity. Neutrophils are one of the main phagocytes involved in M. TB infection. In the sputum and bronchoalveolar lavage fluid of active TB patients, neutrophils are the most prevalent cell type. Therefore, neutrophils are representative of immune response cells associated with M. TB infection. A previous study showed that in a chronic infectious granulomatous disease similar to the pathology of TB, the expression and nuclear translocation of NFE2L2 in neutrophils increased in early granulomatous lesions, but this phenomenon was not found in macrophages (49). Therefore, Nrf2 may be involved in protecting neutrophils from oxidative stress and controlling inflammation to a certain extent (49). Since neutrophils play an important role in the oxidative stress of the microbicidal response to TB, we speculate that the role of Nrf2 in M. TB infection may be mainly reflected in protecting neutrophils from oxidative stress in TB granuloma (50). Therefore, we compared the mRNA level of NFE2L2 in neutrophils with different genotypes to confirm the effect of rs13005431 on NFE2L2 transcriptional activity. After stimulation by GM-CSF, neutrophils with the TC/CC genotype of rs13005431 expressed reduced NFE2L2 mRNA level compared with neutrophils with the TT genotype. From all of the above results, we can speculate that the C allele of rs13005431 may increase the risk of TB by lowering the promoter activity of NFE2L2 and reducing its mRNA level.
The potential weaknesses of our study should be listed. Firstly, we did not further investigate the specific transcription factors binding to the sequence located around rs13005431 and thus clarify the underlying mechanism for the increased TB susceptibility. Secondly, we did not directly provide evidence that during M.TB infection, rs13005431 would be an important player or even an expression quantitative trait locus (eQTL). Finally, since our research was limited to the Chinese Han population, follow up studies in different populations are needed to validate our results.
In conclusion, we have observed significant associations between NFE2L2 variants and TB susceptibility. Further experiments suggested the potential mechanism: allele C of rs13005431 decreased the transcriptional level of NFE2L2. This study may thus pave the way for new treatment modalities targeting anti-oxidative mechanisms in TB.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher. All data excel files are available from the Figshare database (https://figshare.com/s/917cdb79772f89aee2f9).
The studies involving human participants were reviewed and approved by the ethics committee of the West China Hospital of Sichuan University in China. The patients/participants provided their written informed consent to participate in this study.
Conceived and designed the study: GJ and J-QH. Analyzed the data: QL, MZ, YW, SW, GC, and J-QH. Wrote the manuscript: GJ, MZ, QL, AS, and J-QH. All authors contributed to the article and approved the submitted version.
This work was supported by the Research Fund from the National Scientific and Technological Major Project of China (Beijing, China) Grant 2012ZX10004-901, the National Natural Science Foundation of China (Beijing, China) Grants 81072432, 81170042, and 81370121, Science & Technology Department of Sichuan Province (Chengdu, Sichuan, China) Grant 2012SZ0126, Post-Doctor Research Project, West China Hospital, Sichuan University (2019HXBH085), and Investigator-Initiated Clinicaltrial, West China Hospital, Sichuan University (HXCR20001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank all study participants for their cooperation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.660384/full#supplementary-material
M.TB, Mycobacterium tuberculosis; TB, tuberculosis; LTBI, latent tuberculosis infection; TST, tuberculin skin test; IGRAs, Interferon-gamma release assays; BCG, bacille Calmette–Guérin; Nrf2, Nuclear factor-erythroid 2 (NF-E2)-related factor 2; HC, healthy controls; PTB, pulmonary TB patients; EPTB, extra-pulmonary patients; PCR, Polymerase chain reaction; OR, Odds Ratios; 95%CI, 95% confidence intervals; LD, Linkage disequilibrium; HWE, Hardy-Weinberg equilibrium; MDR, multifactor dimensionality reduction; RERI, relative excess risk; AP, attributable proportion; S, synergy index; eQTL, Expression quantitative trait loci.
1. Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. Consensus Statement. Global Burden of Tuberculosis: Estimated Incidence, Prevalence, and Mortality by Country. WHO Global Surveillance and Monitoring Project. JAMA (1999) 282(7):677–86. doi: 10.1001/jama.282.7.677
2. World Health Organization. Global Tuberculosis Report 2019. Who/Htm/Tb/2019.13. Geneva: World Health Organization (2019).
3. Comstock GW, Livesay VT, Woolpert SF. The Prognosis of a Positive Tuberculin Reaction in Childhood and Adolescence. Am J Epidemiol (1974) 99(2):131–8. doi: 10.1093/oxfordjournals.aje.a121593
4. Organization WH. Who Guidelines Approved by the Guidelines Review Committee. In: Guidelines on the Management of Latent Tuberculosis Infection. Geneva: World Health Organization Copyright © World Health Organization (2015) p. 2015.
5. Getahun H, Matteelli A, Abubakar I, Aziz MA, Baddeley A, Barreira D, et al. Management of Latent Mycobacterium Tuberculosis Infection: WHO Guidelines for Low Tuberculosis Burden Countries. Eur Respir J (2015) 46(6):1563–76. doi: 10.1183/13993003.20154606.E60
6. Kahwati LC, Feltner C, Halpern M, Woodell CL, Boland E, Amick HR, et al. Screening for Latent Tuberculosis Infection in Adults: An Evidence Review for the US Preventive Services Task Force. Rockville (MD: Agency for Healthcare Research and Quality (US (2016).
7. Selek S, Aslan M, Horoz M, Celik H, Cosar N, Gunak F, et al. Peripheral DNA Damage in Active Pulmonary Tuberculosis. Environ Toxicol (2012) 27(6):380–4. doi: 10.1002/tox.20674
8. Torun E, Gedik AH, Cakir E, Umutoglu T, Gok O, Kilic U. Serum Paraoxonase 1 Activity and Oxidative Stress in Pediatric Patients With Pulmonary Tuberculosis. Med Princ Pract (2014) 23(5):426–31. doi: 10.1159/000363700
9. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi T, Engel JD, et al. Keap1 Represses Nuclear Activation of Antioxidant Responsive Elements by Nrf2 Through Binding to the Amino-Terminal Neh2 Domain. Genes Dev (1999) 13(1):76–86. doi: 10.1101/gad.13.1.76
10. Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, et al. Oxidative and Electrophilic Stresses Activate Nrf2 Through Inhibition of Ubiquitination Activity of Keap1. Mol Cell Biol (2006) 26(1):221–9. doi: 10.1128/MCB.26.1.221-229.2006
11. Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes Through Antioxidant Response Elements. Biochem Biophys Res Com (1997) 236(2):313–22. doi: 10.1006/bbrc.1997.6943
12. Kensler TW, Wakabayashi N, Biswal S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annu Rev Pharmacol Toxicol (2007) 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046
13. Bonilla DL, Bhattacharya A, Sha Y, Xu Y, Xiang Q, Kan A, et al. Autophagy Regulates Phagocytosis by Modulating the Expression of Scavenger Receptors. Immunity (2013) 39(3):537–47. doi: 10.1016/j.immuni.2013.08.026
14. Palanisamy GS, Kirk NM, Ackart DF, Shanley CA, Orme IM, Basaraba RJ. Evidence for Oxidative Stress and Defective Antioxidant Response in Guinea Pigs With Tuberculosis. PloS One (2011) 6(10):e26254. doi: 10.1371/journal.pone.0026254
15. Mizuno S, Yamamoto M, Sugawara I. Significant Reduction of Granulomas in Nrf2-deficient Mice Infected With Mycobacterium Tuberculosis. Indian J Tuberc (2010) 57(2):108–13.
16. Liu QQ, Wu SQ, Xue M, Sandford AJ, Wu JC, Wang Y, et al. Heterozygote Advantage of the Rs3794624 Polymorphism in CYBA for Resistance to Tuberculosis in Two Chinese Populations. Sci Rep (2016) 6:38213. doi: 10.1038/srep38213
17. Mekonnen E, Bekele E. An Ancestral Human Genetic Variant Linked to an Ancient Disease: A Novel Association of FMO2 Polymorphisms With Tuberculosis (TB) in Ethiopian Populations Provides New Insight Into the Differential Ethno-Geographic Distribution of FMO2*1. PloS One (2017) 12(10):e0184931. doi: 10.1371/journal.pone.0184931
18. Liu QQ, Wang J, Sandford AJ, Wu JC, Wang Y, Wu SQ, et al. Association of CYBB Polymorphisms With Tuberculosis Susceptibility in the Chinese Han Population. Infect Genet Evol (2015) 33:169–75. doi: 10.1016/j.meegid.2015.04.026
19. Zhang X, Ding L, Sandford AJ. Selection of Reference Genes for Gene Expression Studies in Human Neutrophils by Real-Time PCR. BMC Mol Biol (2005) 6:4. doi: 10.1186/1471-2199-6-4
20. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics (2005) 21(2):263–5. doi: 10.1093/bioinformatics/bth457
21. Shi YY, He L. Shesis, a Powerful Software Platform for Analyses of Linkage Disequilibrium, Haplotype Construction, and Genetic Association At Polymorphism Loci. Cell Res (2005) 15(2):97–8. doi: 10.1038/sj.cr.7290272
22. Andersson T, Alfredsson L, Kallberg H, Zdravkovic S, Ahlbom A. Calculating Measures of Biological Interaction. Eur J Epidemiol (2005) 20(7):575–9. doi: 10.1007/s10654-005-7835-x
23. Zhang DM, Chen Y, Chen L, Yang RY, Wang L, Liu WY, et al. Ultraviolet Irradiation Promotes FOXP3 Transcription Via p53 in Psoriasis. Exp Dermatol (2016) 25(7):513–8. doi: 10.1111/exd.12942
24. Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic Identification of Trans eQTLs as Putative Drivers of Known Disease Associations. Nat Genet (2013) 45(10):1238–43. doi: 10.1038/ng.2756
25. Stead WW. Pathogenesis of the Sporadic Case of Tuberculosis. N Engl J Med (1967) 277(19):1008–12. doi: 10.1056/NEJM196711092771906
26. Uplekar M, Weil D, Lonnroth K, Jaramillo E, Lienhardt C, Dias HM, et al. Who’ s New End TB Strategy. Lancet (2015) 385(9979):1799–801. doi: 10.1016/S0140-6736(15)60570-0
27. Lu YJ, Li Q, Peng J, Zhu YW, Wang F, Wang CY, et al. Association of Autophagy-Related IRGM Polymorphisms With Latent Versus Active Tuberculosis Infection in a Chinese Population. Tuberculosis (Edinburgh Scotland) (2016) 97:47–51. doi: 10.1016/j.tube.2016.01.001
28. Kikuchi N, Ishii Y, Morishima Y, Yageta Y, Haraguchi N, Itoh K, et al. Nrf2 Protects Against Pulmonary Fibrosis by Regulating the Lung Oxidant Level and Th1/Th2 Balance. Respir Res (2010) 11:31. doi: 10.1186/1465-9921-11-31
29. Yamada K, Asai K, Nagayasu F, Sato K, Ijiri N, Yoshii N, et al. Impaired Nuclear Factor Erythroid 2-Related Factor 2 Expression Increases Apoptosis of Airway Epithelial Cells in Patients With Chronic Obstructive Pulmonary Disease Due to Cigarette Smoking. BMC Pulm Med (2016) 16:27. doi: 10.1186/s12890-016-0189-1
30. Qian ZQ, Lv JZ, Kelly GT, Wang HT, Zhang XJ, Gu WJ, et al. Expression of Nuclear Factor, Erythroid 2-Like 2-Mediated Genes Differentiates Tuberculosis. Tuberculosis (Edinburgh Scotland) (2016) 99:56–62. doi: 10.1016/j.tube.2016.04.008
31. Lu Y, Zhu Y, Wang X, Wang F, Peng J, Hou HY, et al. FOXO3 Rs12212067: T > G Association With Active Tuberculosis in Han Chinese Population. Inflammation (2016) 39(1):10–5. doi: 10.1007/s10753-015-0217-y
32. Yang Y, Li XW, Cui W, Guan L, Shen F, Xu JS, et al. Potential Association of Pulmonary Tuberculosis With Genetic Polymorphisms of Toll-Like Receptor 9 and Interferon-Gamma in a Chinese Population. BMC Infect Dis (2013) 13:511. doi: 10.1186/1471-2334-13-511
33. Uruno A, Yagishita Y, Yamamoto M. The Keap1-Nrf2 System and Diabetes Mellitus. Arch Biochem Biophys (2015) 566:76–84. doi: 10.1016/j.abb.2014.12.012
34. Wang X, Chen HX, Liu J, Ouyang YY, Wang D, Bao W, et al. Association Between the NF-E2 Related Factor 2 Gene Polymorphism and Oxidative Stress, Anti-Oxidative Status, and Newly-Diagnosed Type 2 Diabetes Mellitus in a Chinese Population. Int J Mol Sci (2015) 16(7):16483–96. doi: 10.3390/ijms160716483
35. Majorov KB, Lyadova IV, Kondratieva TK, Eruslanov EB, Rubakova EI, Orlova MO, et al. Different Innate Ability of I/St and A/Sn Mice to Combat Virulent Mycobacterium Tuberculosis: Phenotypes Expressed in Lung and Extrapulmonary Macrophages. Infect Immun (2003) 71(2):697–707. doi: 10.1128/IAI.71.2.697-707.2003
36. Musellim B, Erturan S, Sonmez Duman E, Ongen G. Comparison of Extra-Pulmonary and Pulmonary Tuberculosis Cases: Factors Influencing the Site of Reactivation. Int J Tuberc Lung Dis (2005) 9(11):1220–3.
37. Okano Y, Nezu U, Enokida Y, Lee MTM, Kinoshita H, Lezhava A, et al. Snp (-617C>A) in ARE-like Loci of the NRF2 Gene: A New Biomarker for Prognosis of Lung Adenocarcinoma in Japanese non-Smoking Women. PloS One (2013) 8(9):e73794. doi: 10.1371/journal.pone.0073794
38. Song P, Li K, Liu L, Wang XW, Jian Z, Zhang WG, et al. Genetic Polymorphism of the Nrf2 Promoter Region is Associated With Vitiligo Risk in Han Chinese Populations. J Cell Mol Med (2016) 20(10):1840–50. doi: 10.1111/jcmm.12874
39. Hudelson P. Gender Differentials in Tuberculosis: The Role of Socio-Economic and Cultural Factors. Tuber Lung Dis (1996) 77(5):391–400. doi: 10.1016/S0962-8479(96)90110-0
40. Neyrolles O, Quintana-Murci L. Sexual Inequality in Tuberculosis. PloS Med (2009) 6(12):e1000199. doi: 10.1371/journal.pmed.1000199
41. Leung CC, Yew WW, Chan CK, Chang KC, Law WS, Lee SN, et al. Smoking Adversely Affects Treatment Response, Outcome and Relapse in Tuberculosis. Eur Respir J (2015) 45(3):738–45. doi: 10.1183/09031936.00114214
42. Lindsay RP, Shin SS, Garfein RS, Rusch MLA, Novotny TE. The Association Between Active and Passive Smoking and Latent Tuberculosis Infection in Adults and Children in the United States: Results From NHANES. PloS One (2014) 9(3):e93137. doi: 10.1371/journal.pone.0093137
43. Maciel EL, Brioschi AP, Peres RL, Guidoni LM, Ribeiro FK, Hadad DJ, et al. Smoking and 2-Month Culture Conversion During Anti-Tuberculosis Treatment. Int J Tuberc Lung Dis (2013) 17(2):225–8. doi: 10.5588/ijtld.12.0426
44. Hubner RH, Schwartz JD, De Bishnu P, Ferris B, Omberg L, Mezey JG, et al. Coordinate Control of Expression of Nrf2-modulated Genes in the Human Small Airway Epithelium is Highly Responsive to Cigarette Smoking. Mol Med (2009) 15(7-8):203–19. doi: 10.2119/molmed.2008.00130
45. Iizuka T, Ishii Y, Itoh K, Kiwamoto T, Kimura T, Matsuno Y, et al. Nrf2-deficient Mice are Highly Susceptible to Cigarette Smoke-Induced Emphysema. Genes Cells (2005) 10(12):1113–25. doi: 10.1111/j.1365-2443.2005.00905.x
46. Cantin AM. Cellular Response to Cigarette Smoke and Oxidants: Adapting to Survive. Proc Am Thorac Soc (2010) 7(6):368–75. doi: 10.1513/pats.201001-014AW
47. Cantin AM, Richter MV. Cigarette Smoke-Induced Proteostasis Imbalance in Obstructive Lung Diseases. Curr Mol Med (2012) 12(7):836–49. doi: 10.2174/156652412801318746
48. Rusin M, Zientek H, Krzesniak M, Małusecka E, Zborek A, Krzyzowska-Gruca S, et al. Intronic Polymorphism (1541-1542delGT) of the Constitutive Heat Shock Protein 70 Gene has Functional Significance and Shows Evidence of Association With Lung Cancer Risk. Mol Carcinog (2004) 39(3):155–63. doi: 10.1002/mc.20009
49. Araujo VC, Demasi AP, Soares AB, Passador-Santos F, Napimoga MH, Martinez EF, et al. Neutrophils in Oral Paracoccidioidomycosis and the Involvement of Nrf2. PloS One (2013) 8(10):e76976. doi: 10.1371/journal.pone.0076976
Keywords: NFE2L2, polymorphism, tuberculosis, latent tuberculosis infection, susceptibility
Citation: Ji G, Zhang M, Liu Q, Wu S, Wang Y, Chen G, Sandford AJ and He J-Q (2021) Functional Polymorphism in the NFE2L2 Gene Associated With Tuberculosis Susceptibility. Front. Immunol. 12:660384. doi: 10.3389/fimmu.2021.660384
Received: 29 January 2021; Accepted: 26 April 2021;
Published: 24 May 2021.
Edited by:
Paul Laszlo Bollyky, Stanford University, United StatesReviewed by:
Guoda Ma, Affiliated Hospital of Guangdong Medical University, ChinaCopyright © 2021 Ji, Zhang, Liu, Wu, Wang, Chen, Sandford and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Qing He, amlhbnFpbmdfaGVAc2N1LmVkdS5jbg==; amlhbnFoZUBnbWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.