
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 16 March 2021
Sec. T Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.659761
This article is part of the Research Topic Multi-Dimensional Genomic Structures in the Immune System View all 12 articles
Cell type-specific gene expression is driven through the interplay between lineage-specific transcription factors (TFs) and the chromatin architecture, such as topologically associating domains (TADs), and enhancer-promoter interactions. To elucidate the molecular mechanisms of the cell fate decisions and cell type-specific functions, it is important to understand the interplay between chromatin architectures and TFs. Among enhancers, super-enhancers (SEs) play key roles in establishing cell identity. Adaptive immunity depends on the RAG-mediated assembly of antigen recognition receptors. Hence, regulation of the Rag1 and Rag2 (Rag1/2) genes is a hallmark of adaptive lymphoid lineage commitment. Here, we review the current knowledge of 3D genome organization, SE formation, and Rag1/2 gene regulation during B cell and T cell differentiation.
Adaptive immunity relies on the assembly of antigen recognition receptor genes (T cell receptor (TCR) and immunoglobulin (Ig) genes) from arrays of variable (V), diversity (D), and joining (J) gene segments that enable the receptors to recognize highly diverse antigens and to invoke antigen-specific immune responses. This process is initiated by a protein complex composed of recombination-activating gene 1 (Rag1) and Rag2, which can recognize and cleave recombination signal sequences (RSSs) flanking the TCR and Ig V, D, J gene segments (1, 2). After T cell lineage commitment is induced by Notch signaling in the thymus, TCRβ V(D)J rearrangement is initiated at the immature CD4–CD8– (double negative, DN) DN2-3 stages. Following the β-selection of the TCR, DN cells differentiate into CD4+CD8+ (double positive, DP) cells, and VJ rearrangement of TCRα is initiated. Similarly, V(D)J recombinations of Ig heavy-chain (Igh) and light-chain occurs in B cell progenitors (pro-B cells) and precursors (pre-B cells), respectively (3, 4). The recombination of TCRγδ occurs concurrently with TCRβ D-J recombination at the DN2 stage, preceding TCRβ V-to-DJ recombination (5). Because RAG-mediated DNA cleavage can lead to oncogenic translocations, Rag1 and Rag2 (Rag1/2) expression is stringently controlled in a highly cell type-specific manner; as are expressed only in T and B cell progenitors and precursors (DN3 and DP thymocytes, pro-B and pre-B cells) (6–9). Therefore, Rag1/2 gene expression typically reflects the cell fate decision toward adaptive immune cells.
Common lymphoid progenitors (CLPs) in the bone marrow (BM) give rise to B cells, T cells, dendritic cells (DCs) and innate lymphoid cells (ILCs), including natural killer (NK) cells. In addition to environmental signals, the adaptive lymphoid cell lineage is specified by the sequential expression of an ensemble of transcription factors (TFs): E2A, Ebf1, Foxo1, and Pax5 for B cell development and E2A/HEB, Gata3, Tcf1, Bcl11b, Runx, Ikaros, and Pu.1 for T cell development (3, 10–12). However, ILCs and T lineage cells express a common set of TFs, such as Gata3, Tcf1, Bcl11b, and Runx, consistent with their similar expression of effector cytokines (13–22). What TFs drive adaptive lymphoid lineages and differences between T cells and ILCs? Namely, E2A and HEB establish T cell identity and suppress the development of thymic ILCs by modulating the repertoires of enhancers, while Pax5 and Ebf1 are required for B cell lineage commitment because they repress genes leading to alternative cell fates for T cells and ILCs (23, 24). Because the biggest difference between T cells and ILCs is RAG-mediated TCR recombination, adaptive lymphoid lineage-specific TFs, which suppress the ILC program, are thought to regulate Rag1/2 genes to make differences between adaptive and innate lymphocytes. Most adaptive lymphocyte development trajectories require regulation by members of the helix-loop-helix families, such as E proteins (E2A, HEB, and E2-2) and Id proteins (Id1-4) (25). The E protein binds to the E-box motif (CANNTG) within enhancer regions of its target genes, the DNA-binding activity of the E protein is antagonized by Id proteins, and Id2 is particularly important for ILC, NK, and LTi cell development (26–28). Therefore, it is proposed that the E-Id protein axis specifies innate and adaptive lymphoid cell fates.
Gene promoters are genetic regulatory elements that provide platforms for TFs to bind and regulate gene expression. However, the regulation of promoter regions is generally insufficient for the cell type-specific regulation of genes that are required for cell functions. Therefore, many genomes contain numerous regulatory elements known as enhancers (29). Lineage-specific TFs alter gene expression patterns by binding to specific DNA sequences within cis-regulatory elements (CREs), including promoters and enhancers. These factors can also change chromatin architecture to determine lineage cell fate and to constrain the development of other lineages. Therefore, the establishment and maintenance of cell type-specific gene expression programs are the result of the interplay between lineage-specific TFs and chromatin architecture, and this interplay can function as a barrier, primer, optimizer, or facilitator to control cell fate. Precisely characterizing this interplay will have profound implications for understanding the development of not only cells but also diseases, such as cancer (30, 31).
Recent studies have identified a novel class of enhancers termed super-enhancers (SEs). An essential feature of SEs is their ability to control genes that have prominent roles in cell type-specific functions, thereby establishing cell identity (32, 33). A SE displays a property of highly cooperative interactions with numerous TFs, mediators and RNA polymerases (34–36). Taking these characteristics into account, many lineage-specific TFs are thought to be associated with each other to form adaptive lymphocyte-specific SEs in Rag1/2 gene loci in developing T and B cells.
Many studies examining the role of chromatin architectures have focused on the recombination of TCR and Ig loci, and many excellent review papers have been published about the importance of three-dimensional (3D) genome organization in antigen receptor loci. Therefore, in this review paper, we focus on adaptive lymphoid cell-specific gene regulation that does not involve TCR or Ig genes. Here, we review the mechanisms of 3D genome organization and SE formation by cell type-specific TFs and explain how cell type-specific expression of the Rag1/2 is mechanistically regulated by CREs, key TFs, and the chromatin architecture. In particular, we focus on the regulation of Rag1/2 genes in developing T and B cells because this locus serves as a template that can be extrapolated to other lineage-specific regulatory events.
The chromatin of the mammalian genome is hierarchically organized into two large compartments, known as transcriptionally permissive (A) and repressive (B) compartments, and smaller domains called topologically associating domains (TADs) (Figures 1A, B) (37–39). TADs contain smaller sub-TADs characterized by higher interaction frequencies. The boundaries of TADs are enriched with insulator binding protein CCCTC-binding factor (CTCF) and highly transcribed housekeeping genes, which play important roles in establishing TAD structure, and the disruption of TAD structures or the dysfunction of these insulators results in the pathogenic reconfiguration of enhancer-promoter interactions (38, 40–42). In the (A) compartment, genes are generally transcribed, and active histone modifications are observed. In contrast, the (B) compartment primarily contains inactive genes and is associated with repressive histone modifications (37). Conspicuously, the Ebf1 gene locus switches from the (B) to the (A) compartment upon B cell lineage commitment (43). Furthermore, chromatin interactions both within and between domains change dramatically, showing extensive (A)/(B) compartment switching during the differentiation of human embryonic stem (ES) cells, while the positioning of TADs remains stable between cell types (44). Interestingly, once a particular cell type is established, extracellular signaling–responsive enhancers are in contact with their target promoter regions even before activation, and looping interactions are largely unchanged after enhancer activation and target gene expression (45).
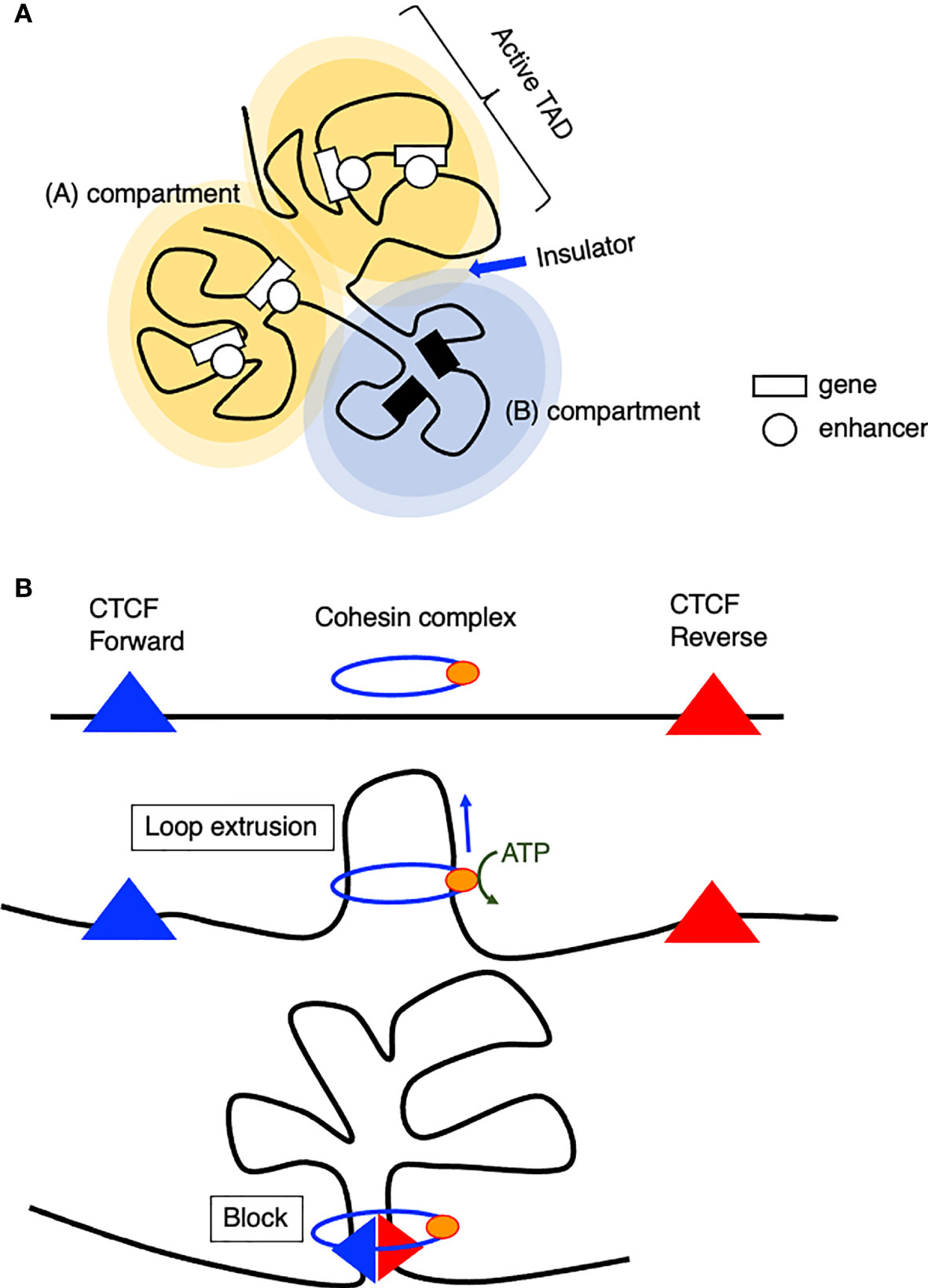
Figure 1 Model of chromatin organization. (A) Spatial chromosome folding in the nucleus separates the genome into (A, B) compartments (A) active compartment, (B): repressive compartment). (A, B) compartments are composed of smaller domains called topological associating domains (TADs), which preferentially interact with themselves rather than with other regions. (B) Model of loop extrusion realized by cohesin ATPase motor activity.
How are TADs organized? The loop extrusion model suggests that the cohesin complex progressively extrudes chromatin until two relevant CTCF molecules encounter and blocks the extruding chromatin at a convergent head-to-head position (46, 47). This chromatin extrusion is mediated by the ATPase activity of the Smc protein, a component of the cohesin or condensin complex (48, 49), extruding tens of kilobase pairs of DNA at a speed of up to 1500 base pairs per second in vitro (50). In addition, this extrusion is asymmetric, which indicates that one site is anchored onto DNA and reels in the chromatin on one side. According to Hi-C data, a loop anchor appears as stripes because it interacts with entire domains, and this stripe- or super-anchor often tethers a SE to a cognate promoter (Figure 1B) (49, 51).
SEs are defined as being highly enriched with transcriptional coactivators and TFs and a high level of histone acetylation (32, 33). SEs exhibit high cooperativity between cell type-specific TFs, mediators, P300, Brd4, and RNA polymerase II, appearing at approximately 10-fold greater density than typical enhancers and contributing to higher transcriptional output (Figure 2A) (36). SEs span large genomic regions, and the clusters of enhancers within SEs are in close physical contact with each other and with promoter regions (Figures 2B, C). SEs are often formed near loci of genes that define cell identity and can be formed as a consequence of the binding of a single TF, a mechanism that makes the SEs vulnerable to perturbation of key components (34–36). On the basis of these features of SEs, a “phase separation model” has been proposed to explain the mechanism by which transcriptional regulators in high concentrations at SEs are associated with higher levels of transcription (transcriptional bursting) and the vulnerability of the SE structure in the absence of its key component (36, 53). TFs control gene expression by binding to DNA in enhancer or promoter regions. While the structure and function of the DNA-binding domains of TFs have been extensively studied, comparatively little is understood about the function of activation domains or other regions of TFs (36). These regions are enriched with intrinsically disordered regions that form liquid-liquid phase-separated droplets in combination with mediator coactivators Med1 and Brd4 at SE regions to compartmentalize and concentrate the transcription apparatus for the induction of robust transcription of cell identity genes (53). In line with this mechanism, the sol-gel phase transition of chromatin enables both rapid and ordered reassembly of the IgH locus for V-D-J recombination in developing B cells, which may be associated with the TF assembly of E2A, PU.1, Foxo1, and Pax5 (54). Therefore, the establishment of cell identity and lineage-specific gene expression programs requires the interplay of lineage-specific TFs and the chromatin architectures, as seen in SE formation.
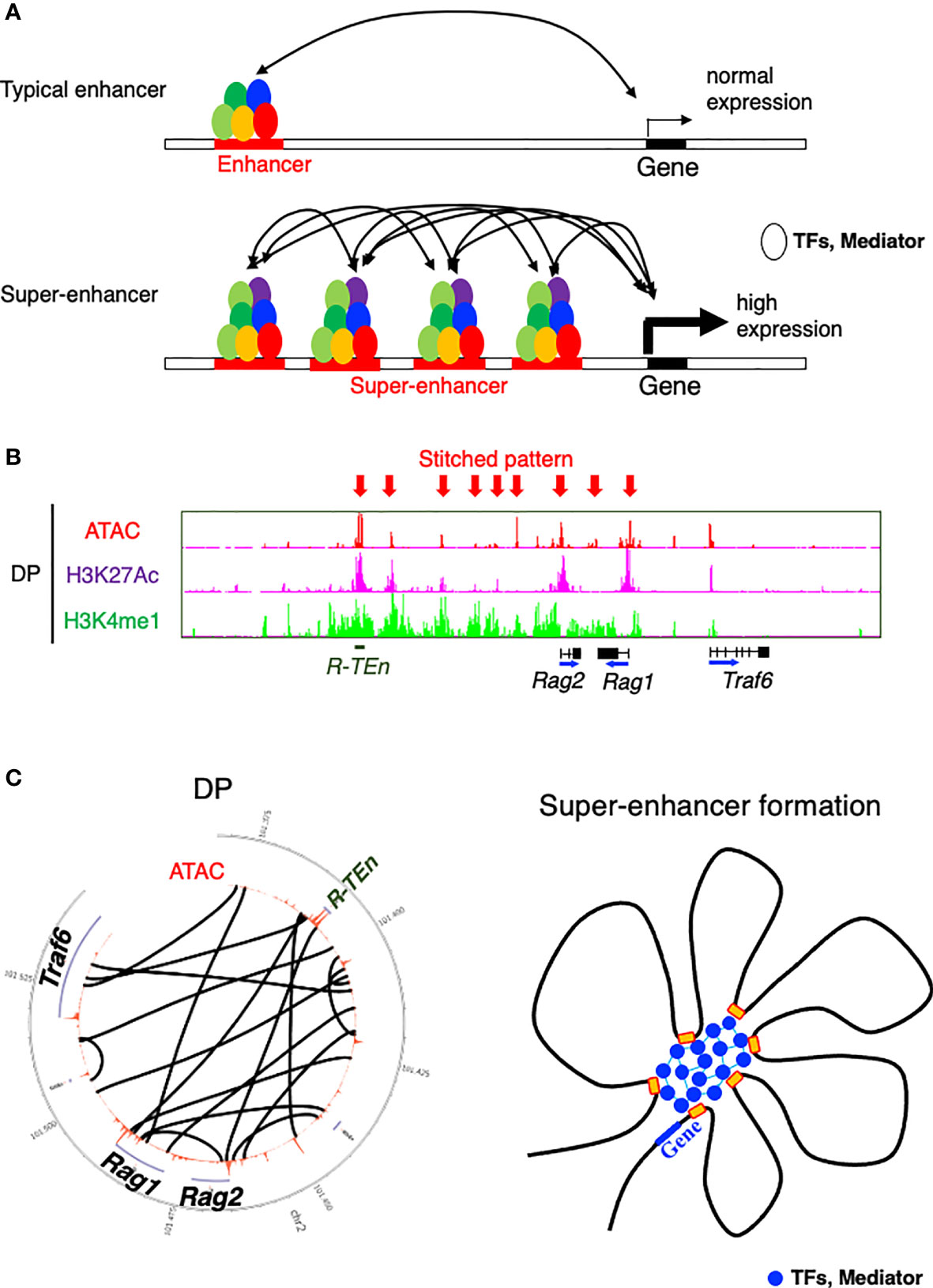
Figure 2 Model of a super-enhancer. (A) Comparison between typical enhancers and super-enhancers (SEs). SEs are occupied by a highly dense population of transcription factors (TFs) and transcriptional mediators. DNA binding regions within SE cooperatively bind to each other, which contributes to the high expression of target genes (36). (B) ChIP-seq [H3K27 acetylation (active region) and H3K4 monomethylation (enhancer)] and the ATAC-seq profile of the Rag gene SE (52). Arrows indicate a stitched pattern of enhancers. (C) Circos diagram representing genomic interactions (black lines) across Rag gene locus in DP cells (left) (52) and model showing TFs and mediators forming phase-separated condensates at SE regions (36).
RAG expression is restricted to developing T and B lymphocytes. In both lineages, there are two waves of RAG expression (8). The first wave of RAG expression is required for the assembly of Ig heavy chain (IgH) and TCRβ genes in pro-B and pro-T cells, respectively. In developing thymocytes, RAG expression also catalyzes TCRγ and TCRδ gene rearrangement to become γδT cells in early DN populations. Rag expression is transiently downregulated in cells that successfully rearrange Igh or TCRβ during the developmental transition from progenitor to precursor (pre-B or pre-T cells). In pre-B and pre-T cells, Rag genes are re-expressed leading to the assembly of Ig light (IgL) chain and TCRα genes, respectively. Following successful recombination of IgL or TCRα and assembly of Ig or TCRαβ, Rag gene expression is suppressed in mature B and T cells. Loss of Rag gene expression results in developmental arrest at the progenitor stages of T cell and B cell development (55, 56), while persistent Rag expression causes aberrant thymic development and profound immunodeficiency (57). Therefore, Rag expression is tightly regulated during adaptive lymphocyte development. Both in vivo and in vitro studies have attempted to identify the CREs for Rag1/2 expression (7). Interestingly, while both B and T cells require Rag1/2 expression for antigen receptor gene recombination, the Rag enhancers in B and T cells differ. In B cells, the deletion of Erag, which is 23 kb upstream of the Rag2 promoter, resulted in a significant reduction in Rag1/2 expression and a partial block at the transition from pro-B to pre-B cells, but did not affect thymocyte development in mice (58). It has been reported that the Erag enhancer is positively regulated by FOXO1 and negatively regulated by Gfi1b, Ebf1, and c-Myb (59–62). In contrast, an anti-silencer element (ASE), which is 8 kb in length and located 73 kb upstream of the Rag2 promoter, is shown to be required for Rag1/2 expression in DN3 and DP cells but not in B cells (63). Since Erag and ASE deletions resulted in the partial loss of RAG activity, additional CREs and their cognate binding factors are likely necessary (7). To my knowledge, there has been no report regarding Rag1/2 expression with respect to TCRγδ gene recombination. Additionally, how enhancers regulates Rag1 and Rag2 in coordinate and how Rag1/2 expression is tightly restrained in innate immune cells will be explored in a later section.
The Krangel group reported that ASE directly interacts with the Rag1 and Rag2 promoters and that the chromatin organizer Satb1 binds to this ASE to promote optimal Rag1/2 gene expression through Rag gene locus organization in DP thymocytes, as proven by chromosome conformation capture (3C) assay (64). In addition, by using the VL3-3M2DP thymocyte cell line, this group demonstrated that the ASE requires Gata3 and E2A, while the Rag1 promoter relies on Runx1 and E2A as critical regulators. Together, the ASE and Rag1 promoter framework functions as a chromatin hub (65). Recently, we have identified cell type-specific CREs for Rag1/2 expression in B and T cells using E2A ChIP-seq and ATAC-seq data, because E2A specifies adaptive lymphocyte and is the driver of differences in enhancer repertoires in T cells and ILCs (24, 52). We mapped E2A binding to one T cell-specific Rag gene enhancer [Rag-T cell enhancer (R-TEn) included in the 8-kb ASE] and to two B cell–specific Rag gene enhancers [Rag B cell enhancer 1/2; R1B (5-kb upstream of the Rag1 promoter) and R2B (partially overlapping with Erag)] (Figure 3A) (52). R-TEn and R1B/R2B loop uniquely to the Rag1/2 promoter regions and form different genome structures. In addition to E2A, T cell- or B cell-specific TFs (T cell: Bcl11b, Tcf1, Runx1, Ikaros, and Gata3; B cell: Pax5, Ets1, Ikaros, and Irf4) bind to these enhancer regions (Figure 3A). Furthermore, the R2B and Rag1 gene promoter region (R1pro) in pre-B cells, and R-TEn along with the Rag1/2 gene cluster in DP cells are SEs according to an analysis of histone acetylation (H3K27Ac) (52). Consistent with this finding, the Rag gene locus and its enhancers show elaborate chromatin interactions and are located in the active developing T cell- and B cell-specific sub-TAD (52). Furthermore, R-TEn deletion in mice specifically blocked thymocyte development during the β-selection of DN3 cells and positive-selection of DP cells, as seen in ASE deletion mice (52, 63). In addition, mice with R1B/R2B double deletions exhibited a severe developmental block at the pro-B stage, whereas the single deletion of either R1B or R2B led to mild or moderate impairments in B cell development. This outcome is comparable to that observed in Erag deficient mice, suggesting enhancer redundancy in R1B and R2B (7, 52, 58). These developmental defects in enhancer-deleted mouse lines were caused by failed Rag1/2 gene expression and TCRαβ or IgH recombination. Interestingly, these cell type-specific enhancers are required not only for enhancer-promoter interactions, but also for other chromatin interactions across the Rag gene locus.
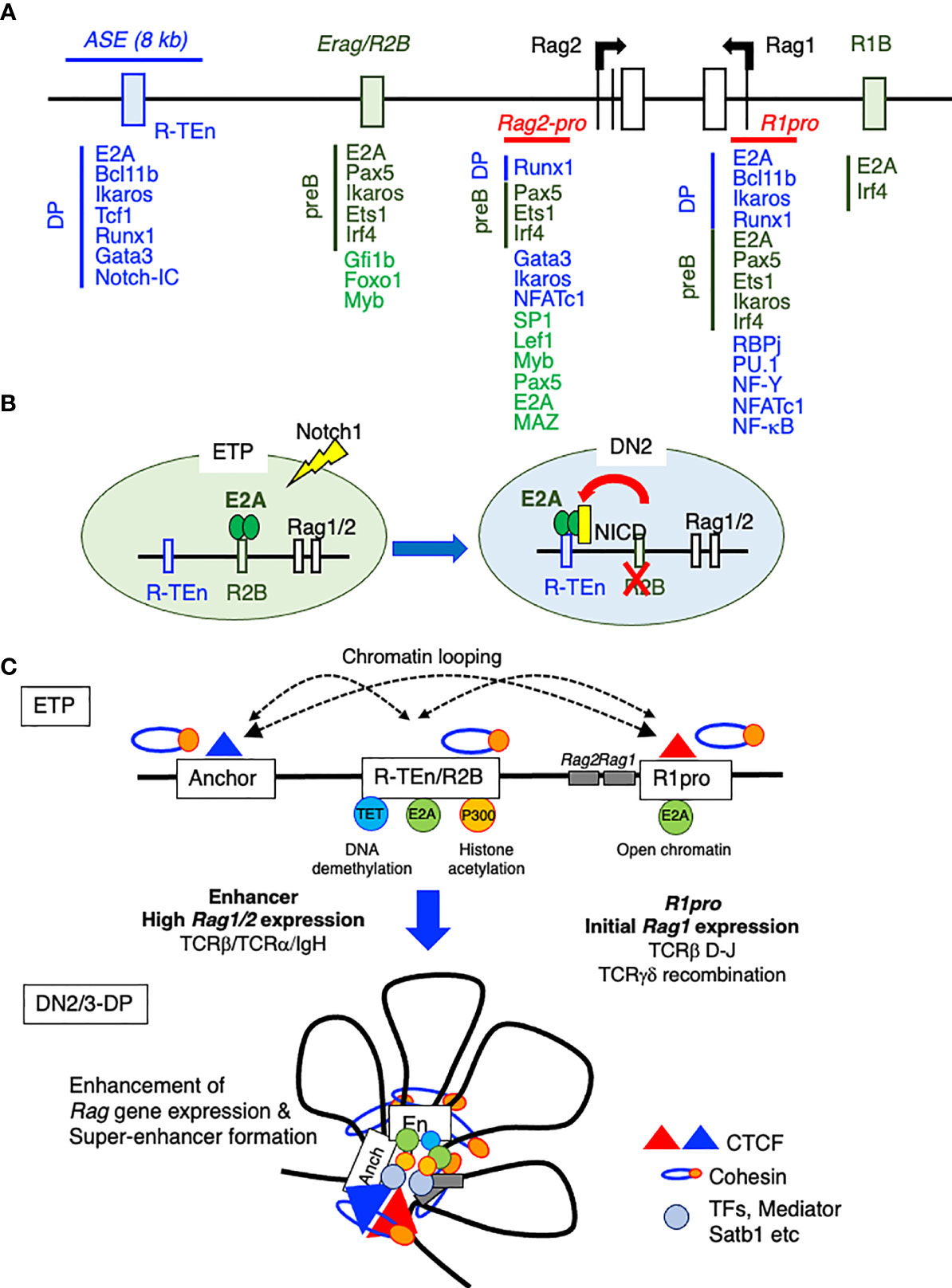
Figure 3 The Rag gene locus and cis-regulatory elements. (A) A schematic diagram of the Rag gene locus and cis-regulatory elements in T and B cell lineages. The green boxes indicate B cell-specific enhancer regions (R1B and Erag/R2B) and transcription factor (TF) binding in proB cells. The blue line and box indicate the T cell-specific enhancer region (ASE and R-TEn) and TF binding in DP cells. The red line indicates the common region between B and T cells (R1pro). (B) The recruitment of E2A to the T cell-specific enhancer region (R-TEn) upon T cell lineage commitment. (C) Model depicting the stepwise establishment of an adaptive lymphocyte-specific super-enhancer for Rag1/2 expression mediated by E2A.
It has been reported that T and B cell-specific TFs bind to Rag1 and Rag2 promoter regions (reviewed in (7)), and we also analyzed the occupancies of TFs in pre-B and pre-T cells (52). In DP cells, Bcl11b, Ikaros, NF-Y, NFATc1, and Runx1 are recruited to the Rag1-promoter region, while Pax5, Ets1, Ikaros, and Irf4 bind to this promoter in pre-B cells (7, 52). Runx1 in DP cells, and Pax5 and Irf4 in pre-B cells, but not other TFs, appear to bind to Rag2 promoter regions in ChIP-seq analysis (7, 52). ChIP-seq data of Myb, Lef1, NFAT1c, and SP1 in pro-T and pro-B and pre-T and B cells are needed for further analysis. Interestingly, we found common E2A-binding sites in the Rag1 gene promoter region (R1pro), but distinct E2A-binding sites in the Rag2 promoter region in pro-T and pro-B cells (52). This finding is consistent with the differing regulation mechanism of the Rag2 promoter in T and B cells (66, 67). To clarify the R1pro activity regulated by E2A, E-box motif mutations in Rag1 promoter region were generated (R1pro-mut) to prevent the loss of other activities of the Rag1 promoter. In addition, by generating E-box mutations in R1pro, the protein level of E2A and the binding of other TFs to this region were not affected. Blocking E protein binding to the R1pro region (R1pro-mut) resulted in the total loss of Rag1 expression without affecting Rag2 expression or enhancer activities in both pro-T cells and pro-B cells, causing developmental arrest at the T cell and B cell progenitor stages (52). This outcome suggests that Rag1 promoter activity is commonly regulated by E protein binding in the adaptive lymphoid lineage. The regulation of Rag2 promoter activity seems to be more complicated. It has been reported that two regions of Rag2 promoter regions (D3 and Ep) are DNaseI hypersensitive sites to which C/EBP and NFKB bind to play important roles in the regulation of Rag2 expression (7, 66–68). In addition, the Pax5/c-Myb/Lef1 protein complex in pre-B and Gata3 in T cells bind to Rag2 promoter region to activate Rag2 expression (69, 70). Although the expression of Rag1, but not Rag2, is severely affected in pro-T and pro-B cells derived from R1pro-mut mice, Rag2 promoter CREs seem to regulate both the Rag1 and Rag2 genes in coordination (66). Therefore, it remains unknown whether Rag2 promoter activity in vivo is independent similar to R1pro.
How do B and T cells utilize the distinct CREs of Rag gene expression? According to a chromatin accessibility analysis, the R2B region first becomes accessible in the CLP stages and stays open until the pre-B cell stage. Interestingly, R2B is accessible at the uncommitted DN1 stage in the thymus but becomes inaccessible at the DN2a commitment stage. At the same time, R-TEn is closed in DN1 cells but open in DN2a cells, indicating mutual exclusivity of enhancer engagement between R2B and R-TEn after adaptive lymphoid lineage commitment. How is this sequential enhancer activity re-established? Because Notch1-Delta-like 4 (DLL4) signaling and E proteins are critically required for T cell lineage commitment and the suppression of ILC development in the thymus (24, 71, 72), Notch-RBP-J machinery possibly alters the binding of E-proteins to both R2B and R-TEn and makes R-TEn accessible (Figure 3B). This speculation is in line with the fact that CpG DNA in R-TEn is hypermethylated in pro-B and pre-B cells and R1B/R2B are hypomethylated in DN3 and DP cells, indicating that R1B/R2B used to be open in the T cell lineage. Notably, R-TEn is never open in the B cell lineage, and the demethylation of CpG islands is correlated with E2A binding. These results are consistent with a previous report showing the functional and physical interaction between E2A and TET proteins in developing B cells (73). We therefore speculate that R1B and R2B are primitive enhancers of Rag1/2 expression in the lymphoid lineage and that R-TEn, a target of Notch signaling, was acquired later in evolution, after thymus anlage development in vertebrates. As Dll4 is a direct target of Foxn1 and Foxn4 in thymic epithelial cells (TECs), it is important for studies of adaptive immunity to understand how the network of Foxn1/2-Dll4 in TECs (Notch ligand expression) and the Notch/E2A-Rag enhancer in progenitor T cells (interaction between Notch signaling and TF) were acquired during thymopoiesis evolution and confer the T cell-specific Rag enhancer activity (74–78).
Notably, Rag1/2 promoter regions are accessible at the progenitor stage (DN3 and pro-B cells) without enhancer activity, while the accessibilities at these promoters depend on the enhancer activity at the precursor stage (DP and pre-B cells), suggesting that enhancer activity maintains the open status of Rag1 and Rag2 promoters through rounds of cell division during the transition from DN to DP stages (52). Consistent with this supposition, the level of E2A occupancy at R-TEn does not decline after pre-TCR signaling, which likely maintains R-TEn activity (79). When DP and pre-B cells differentiate into CD4SP or CD8SP and IgM+ B cells, R1pro, Rag2 promoter, R-TEn, and R2B are immediately close, indicating that TCR and IgH signaling suppress these CRE activities (52). Blocking E2A binding to R-TEn and R1pro CREs is sufficient to disrupt Rag1/2 SE formation and totally eliminate Rag1 promoter activity, respectively (52). These outcomes are in line with the disappearance of chromatin interactions in Rag gene loci in mature T and B cells, which express a low level of E2A and a high level of Id2 and Id3. In fact, Id3 is first induced by pre-TCR signaling and is further upregulated by TCR signaling, and this TCR-Id3 pathway is also important for γδT cell development (79–82). Id2 is upregulated during later positive selection of DP cells, and Id2 and Id3 cooperatively function during positive selection of DP cells (83, 84). Therefore, Id proteins induced by TCR signaling antagonize the E protein to abrogate Rag gene SE and Rag1 expression, inhibiting the subsequent expression of Rag1/2 in mature T and B cells. In line with this process, 3D genome structures in developing T and B cells show changes as they develop. An increasing number of chromatin interaction loops across the Rag gene loci are observed during T and B cell developmental progression from CLPs, and these loops are absent when the Rag1/2 genes are not expressed in mature T and B cells. The timing of loop formation correlates strongly with changes in the PC1 component and TADs and Loop scores during T and B cell development (52).
Interestingly, chromatin accessibility in most hematopoietic cells (granulocytes, erythrocytes, dendritic cells, NK cells, ILC2s and ILC3s), R-TEn and R2B are seldomly accessible in cells other than developing T and B cells (ImmGen data; GSE100738) (Supplementary Figure 1), according to an analysis of published data (68). On the other hand, R1B is slightly open in these cells, which is consistent with the fact that R1B functions as an insulator to sequester Rag1/2 away from active genes (more details are present in the subsequent section). In contrast, we observed clear accessibility at the Nad Wyraz Ciekawy (NWC) locus in most hematopoietic cells, but not T and B cells. NWC is the third evolutionarily conserved gene with unknown function in the Rag gene locus and is located within the Rag2 gene intron (Supplementary Figure 1). NWC seems to be a negative regulator of Rag2 because of the mutual exclusivity of Rag2 and NWC expression; however, it remains unknown whether transcripts from NWC block Rag2 gene expression (7, 85–87). Upon the ILC lineage commitment, the E2A protein is suppressed in the ILC precursor, while Id2 is highly expressed and maintained throughout ILC lineage, suggesting that Id2 inhibits E2A-mediated Rag gene CRE activities in the ILC lineage (24, 88).
Enhancers play important roles in precise gene expression programs during development, and divergence in enhancer sequence and activity is thought to be an important mediators of inter- and intraspecies phenotypic variation (89). Regardless of the level of enhancer sequence conservation, enhancers identified in human ES cells drive cell type- and stage-specific expression when introduced to zebrafish embryos, suggesting the conservation of ancestral functions of TFs (90). Additionally, the evolution of body shape is thought to be tightly coupled to changes in enhancer sequences of vertebrates. For example, a snake-specific sequence alterations within an otherwise highly conserved long-range limb enhancer of Sonic Hedgehog are closely associated with morphological changes of the limb in snakes, demonstrating the critical roles of enhancers in morphological evolution (91). These previous reports suggest that the conservation of DNA sequences within enhancer regions among species reflects the evolution of Rag gene regulation. Rag1/2 genes are conserved among jawed vertebrates, in which they contribute to a diverse repertoire of antibodies and T cell receptors. The generation of this diversity is a pivotal event in the evolution of the adaptive immune system of jawed vertebrates (6). Therefore, to understand the evolution of adaptive immunity, it is also important to investigate the conservation of DNA sequences in T cell- and B cell-specific enhancers. Interestingly, R-TEn and R2B are highly conserved among mammals, as well as most birds and reptiles, but not in amphibians or fish. Furthermore, conserved R-TEn, R1B, and R2B were found to harbor conserved E-boxes (52). These observations reveal the discordance in the evolutionary conservation of Rag genes and their regulatory elements among jawed vertebrates and suggest the possibility that there are various regulatory mechanisms in terrestrial animals, aquatic animals, and amphibians. Here, we propose that adaptive immunity in terrestrial animals is evolutionarily developed via the utilization of E protein activity to increase the expression of Rag genes, which enables receptors to recognize diverse antigens by RAG-mediated TCR and Ig gene recombination. Considering the evolution of the Rag gene, the ancestral Rag transposon (Transib) contained a core region of the Rag1 precursor, and subsequently, the Rag2-like gene was acquired by Transib, leading to the emergence of the Rag transposon that gives rise to the Rag and Rag-like genes found in echinoderms, cephalochordates, and jawed vertebrates (6). Furthermore, recent excellent studies of ProtoRAG transposase illuminate the evolution of RAG transposons and V(D)J recombination for the development of the adaptive immunity system in the human body (92, 93).
Upon the cell lineage decision-making of multipotent cells, abrupt genomewide changes in chromatin accessibility, chromatin interaction and conformation, and transcriptome occur, and these concerted changes in chromatin architecture act as barriers to prevent cells from reversing or being redirected to other lineages (94). As Lin and Murre reported, in B cell development, CTCF occupancies are associated with intradomain interactions, whereas P300, E2A and PU.1 are associated with intra- and interdomain interactions that are developmentally regulated in pro-B cells. During B cell lineage commitment from the pre-pro-B to pro-B stages, Ebf1 is sequestered at the nuclear lamina in the pre-pro-B cells, and following commitment, Ebf1 and other gene loci switch compartments to establish new intra- and interdomain interactions associated with a B cell lineage-specific transcription signature (43, 95). Recently, a prion-like-domain in the C-terminal domain of EBF1 has been shown to have a liquid-liquid phase separation ability, which enables the recruitment of the RNA-binding protein FUS and chromatin remodeler Brg1, which facilitate chromatin opening and acts as pioneer factors to induce the formation of phase-separated SEs for promoting B cell identity gene expression (96). After the global network consisting of E2A, Ebf1, and Foxo1 establishes B cell identity, Ebf1 and Pax5 maintain the B cell signature throughout the changes in the global lineage-specific genome architecture (23, 95, 97).
Upon T lineage commitment, regulome and 3D genome architectures are reorganized by T cell-specific TFs, such as Bcl11b and Tcf1, and noncoding transcripts (ThymoD) at the Bcl11b enhancer (3, 94, 98–100). Likewise Ebf1 locus upon B cell lineage commitment, ThymoD transcripts orchestrate chromatin folding and compartmentalization by activating cohesin-dependent looping to translocate its enhancer from the nuclear lamina to the nuclear interior (98). In each scenario, enhancer activity mediated by lineage-specific TF binding appears to be the first step for chromatin compaction and compartment switching.
Rag1/2 expression and chromatin accessibility of R-TEn were not affected in Tcf1-, Bcl11b-, or ThymoD-deficient thymocytes, as determined by examining in previously published data (52). Furthermore, the expression of Rag1/2 was not reduced in Runx1-deficient pro-B cells (101). Upon the deletion of Satb1 or cohesin (Rad21), histone acetylation (H3K27Ac) levels at the R-TEn and Rag1 promoter regions remained high, although Rag1/2 expression was reduced in Satb1-deficient DP cells (64, 102–104). On the other hand, E-protein deletion (E2A (Tcf3) and HEB (Tcf12)) in DP cells led to the downregulation of Rag1/2 and a reduction in chromatin accessibility at the R-TEn and Rag1/2 promoter regions (52, 105). Mutating the seven E-box motifs in the R-TEn region (R-TEn-E-box-mutant) reduced Rag1/2 expression and blocked thymocyte development. These results are similar to the effects of deleting the entire R-TEn region. Furthermore, the R-TEn-E-box-mutant abolished the chromatin accessibility and SE formation throughout the entire Rag gene locus in DP cells, which was accompanied by a significant reduction in cohesin recruitment. Because the enhancer activity of R-TEn drives active sub-TAD formation, we considered how this active TAD is initiated after the enhancer is activated. For active TAD formation, CTCF and cohesin facilitate loop extrusion by using ATP at the super- or stripe-anchor region, which is closely associated with hypomethylated DNA regulatory regions that tether super-enhancers to cognate promoters (49). Since the recruitment of cohesin, but not CTCF, to the anchor region (200-kb upstream of Rag2 gene) was drastically reduced in R-TEn-E-box-mutant DP cells, E protein binding to the enhancer regions drives cohesin loading for the local compaction of this gene locus (52). In addition to the critical role of CTCF in TCR/IgH recombination (106–111), CTCF stabilizes long-range promoter-enhancer interactions and controls the cell-to-cell variation in gene expression in mammalian cells (112). With respect to the role of cohesin in Rag gene expression, cohesin subunit Rad21-deficient thymocytes, in which cohesin was ablated in resting small DP cells by CD4-Cre, showed the impaired secondary distal Jα recombination (102). However, since Rag gene SE formation is normally formed in Rad21-deficient thymocytes, other molecules may play roles in maintaining Rag gene SE formation and their expression after establishment of the SE structure in DP cells (52). Because Satb1-deficient DP cells showed a significant reduction in the enhancer-promoter interaction of Rag genes, it is reasonable to speculate that cohesin and CTCF drive loop extrusion of the Rag gene locus and that Satb1 may act as a chromatin organizer to maintain the SE structure in DP cells (64, 104). Taken together, the data suggest that E protein binding to CREs facilitates the recruitment of cohesin complexes to enhancer and anchor regions to orchestrate adaptive lymphocyte-specific spatial interactions in the Rag gene locus to induce robust gene expression (Figure 3C). To establish this model, it is necessary to clarify the underlying mechanism of how E proteins recruit the cohesin to enhancer and anchor regions.
Notably, T cell- or B cell-specific enhancers or Rag1 promoter are accessible at the T- or B-progenitor stage without promoter or enhancer activity, even though both activities depend on E protein binding (52). Furthermore, γδT cell development and TCRβ D-J recombination in DN3a cells were less affected in R-TEn deletion mice, while these events were absent in DN3a and pro-B cells obtained from R1pro-E-box-mutant mice. Therefore, upon T lineage commitment, E2A binds to both the Rag enhancer and promoter independently. The low level of Rag1 expression induced by R1pro activity is sufficient to permit TCRβ D-J and TCRγ δ recombination, which is in line with the fact that TCRγ δ recombination occurs concurrently with TCRβ D-J recombination at the DN2 stage before TCRβ V-DJ recombination (Figure 3C) (5, 113). The high level of Rag expression induced by enhancers is required for long-range V-(D)J recombination of TCRβ or TCRα (52). From these data, we propose a two-step Rag gene regulation model. (1) Rag1/2 genes are first initiated by E-protein binding to the CREs near exon 1, which induces D-J and TCRγδ recombination. (2) Rag1/2 gene expression is further enhanced by cell type-specific enhancers to drive their robust expression, which is required for long-range V-(D)J recombination (Figure 3C). Considering a previous report showing that the formation of the RAG recombination center (RC) in T cell- or B cell-progenitors/precursors occurs in a developmental stage-specific manner and that RAG1 preferentially binds to TCRβ D-J regions in pro-T and Igh Jh regions in pro-B, which have a high level of H3K4 trimethylation (2, 114), we speculate that a low level of RAG1 protein can initially bind to the D-J regions but not to V regions, resulting from the insufficient formation of RAG-RC for V-DJ recombination. Further study is needed to clarify the stepwise effect of Rag expression mediated by enhancer-promoter cooperation.
In R-TEn deletion mice, the number of γδT cells was moderately reduced in the adult thymus; however, the percentages of Vγ1.1 and Vγ2 in the γδT cell population were not affected (52). Compared with the recombination of TCRβ V-DJ and TCRα V-J, the assembly of TCRγδ occurs within a short range of TCRγ clusters or local segments of TCRδ in the TCRα locus, depending on their own enhancers. The accessibility and recombination of the TCRγ locus rely on IL-7/IL-15-Stat5 signaling (115, 116). Therefore, we speculate that the low RAG1 and RAG2 protein levels are sufficient for RAG-RC to drive local TCR recombination in cooperation with cytokine signaling.
How do Rag1/2 genes get repressed in innate immune cells? In bone marrow-derived macrophages (BMDMs), a sharp TAD boundary between Rag1 and the neighboring gene Traf6, which is generally expressed, is formed to insulate the entire Rag gene locus from the active compartment and sequester it in the repressive compartment (Figure 4) (52). Tight physical regulation illustrates how harmful genome recombinase enzymes are properly regulated at the chromatin architecture level.
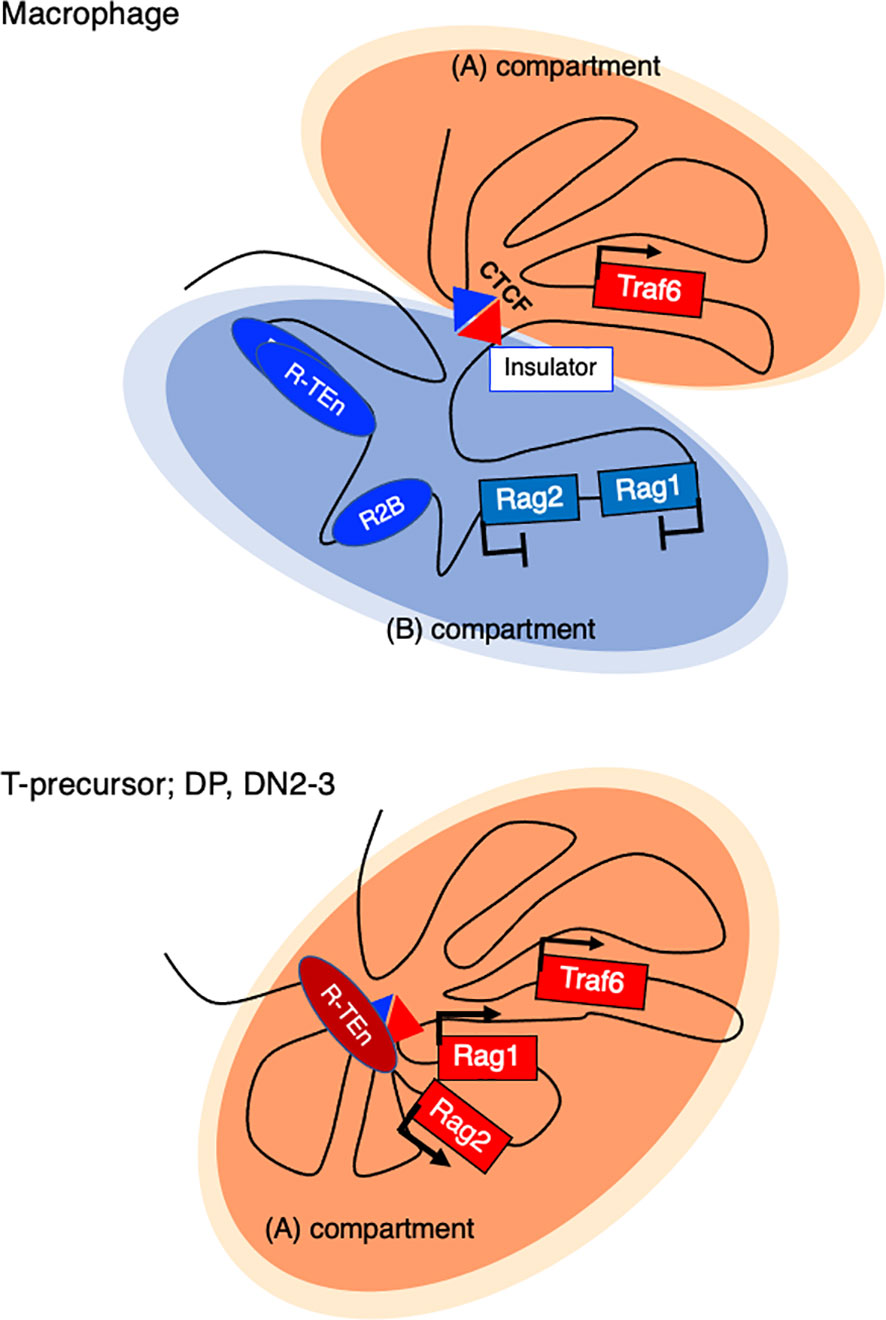
Figure 4 Active and repressive compartmentalization in the Rag gene cluster and neighboring Traf6 gene in a macrophage and T cell precursor. In nonlymphoid cells (Bone marrow derived macrophage (BMDM), a sharp TAD boundary mediated by CTCF forms between Rag1 and Traf6, insulating the entire Rag gene locus from the active (A) compartment and sequestering it in a repressive (B) compartment.
The regulation of Rag1 and Rag2 genes is highly conserved, and the expression of these genes is a hallmark of adaptive immunity. Cell type-specific gene expression is driven by the interplay between E2A/E proteins and chromatin interactions. Future experiments are warranted to explore the role of a potential pioneer TF in regulating enhancer activity with other lineage-specific TFs and to understand how these TFs reorganize the 3D genome architecture through the recruitment of the cohesin complex. These findings may have implications for health and immunological disorders.
KM and MM wrote the manuscript and figures. All authors contributed to the article and approved the submitted version.
This work was funded by the KAKENHI (Grants-in-Aid for Scientific Research) from the MEXT of Japan (19H03487 for MM) and the Mochida Memorial Foundation and the Takeda Science Foundation (MM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank C. Murre and Y.C. Lin for kind suggestions and for reviewing the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.659761/full#supplementary-material
1. Schatz DG, Oettinger MF, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell (1989) 59(6):1035–48. doi: 10.1016/0092-8674(89)90760-5
2. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol (2011) 11(4):251–63. doi: 10.1038/nri2941
3. Rothenberg EV. Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev (2019) 33(17-18):1117–35. doi: 10.1101/gad.327163.119
4. Muljo SA, Schlissel MS. Pre-B and pre-T-cell receptors: conservation of strategies in regulating early lymphocyte development. Immunol Rev (2000) 175:80–93. doi: 10.1111/j.1600-065X.2000.imr017509.x
5. Capone M, Hockett RD Jr, Zlotnik A. Kinetics of T cell receptor beta, gamma, and delta rearrangements during adult thymic development: T cell receptor rearrangements are present in CD44(+)CD25(+) Pro-T thymocytes. Proc Natl Acad Sci U S A (1998) 95(21):12522–7. doi: 10.1073/pnas.95.21.12522
6. Carmona LM, Schatz DG. New insights into the evolutionary origins of the recombination-activating gene proteins and V(D)J recombination. FEBS J (2017) 284(11):1590–605. doi: 10.1111/febs.13990
7. Kuo TC, Schlissel MS. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol (2009) 21(2):173–8. doi: 10.1016/j.coi.2009.03.008
8. Wilson A, Held W, Macdonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med (1994) 179(4):1355–60. doi: 10.1084/jem.179.4.1355
9. Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell (2013) 152(3):417–29. doi: 10.1016/j.cell.2013.01.007
10. Miyazaki K, Miyazaki M, Murre C. The establishment of B versus T cell identity. Trends Immunol (2014) 35(5):205–10. doi: 10.1016/j.it.2014.02.009
11. Mandel EM, Grosshdl R. Transcription control of early B cell differentiation. Curr Opin Immunol (2010) 22(2):161–7. doi: 10.1016/j.coi.2010.01.010
12. Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J Immunol (2014) 192(9):4017–23. doi: 10.4049/jimmunol.1302259
13. Tindemans I, Serafini N, Di Santo JP, Hendriks RW. GATA-3 function in innate and adaptive immunity. Immunity (2014) 41(2):191–206. doi: 10.1016/j.immuni.2014.06.006
14. Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A, et al. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity (2015) 43(2):354–68. doi: 10.1016/j.immuni.2015.07.005
15. Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol (2015) 16(11):1124–33. doi: 10.1038/ni.3272
16. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol (2015) 16(10):1044–50. doi: 10.1038/ni.3248
17. Walker JA, Oliphant CJ, Englezakis A, Yu Y, Clare S, Rodewald HR, et al. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med (2015) 212(6):875–82. doi: 10.1084/jem.20142224
18. Seehus CR, Aliahmad P, De La Torre B, Iliev ID, Spurka L, Funari VA, et al. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol (2015) 16(6):599–608. doi: 10.1038/ni.3168
19. Cortez VS, Robinette ML, Colonna M. Innate lymphoid cells: new insights into function and development. Curr Opin Immunol (2015) 32:71–7. doi: 10.1016/j.coi.2015.01.004
20. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity (2014) 40(3):378–88. doi: 10.1016/j.immuni.2014.01.012
21. Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol (2016) 17(7):765–74. doi: 10.1038/ni.3489
22. Serafini N, Vosshenrich CA, Di Santo JP. Transcriptional regulation of innate lymphoid cell fate. Nat Rev Immunol (2015) 15(7):415–28. doi: 10.1038/nri3855
23. Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol (2013) 14(8):867–75. doi: 10.1038/ni.2641
24. Miyazaki M, Miyazaki, Chen K, Jin Y, Tunner J, Moore AJ, et al. The E-Id Protein Axis Specifies Adaptive Lymphoid Cell Identity and Suppresses Thymic Innate Lymphoid Cell Development. Immunity (2017) 46(5):818–34. doi: 10.1016/j.immuni.2017.04.022
25. Murre C. Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev (2019) 33(1-2):6–25. doi: 10.1101/gad.320663.118
26. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature (2010) 463(7280):540–4. doi: 10.1038/nature08636
27. Klose CS, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell (2014) 157(2):340–56. doi: 10.1016/j.cell.2014.03.030
28. Miyazaki M, Miyazaki K, Chen S, Itoi M, Miller M, Lu LF, et al. Id2 and Id3 maintain the regulatory T cell pool to suppress inflammatory disease. Nat Immunol (2014) 15(8):767–76. doi: 10.1038/ni.2928
29. Johanson TM, Chan WF, Keenan CR, Allan RS. Genome organization in immune cells: unique challenges. Nat Rev Immunol (2019) 19(7):448–56. doi: 10.1038/s41577-019-0155-2
30. Kim S, Shendure J. Mechanisms of Interplay between Transcription Factors and the 3D Genome. Mol Cell (2019) 76(2):306–19. doi: 10.1016/j.molcel.2019.08.010
31. Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature (2019) 569(7756):345–54. doi: 10.1038/s41586-019-1182-7
32. Hnisz, Abraham BJ, TI L, Lau A, Saint-Andre V, Saint-Andre V, et al. - Super-enhancers in the control of cell identity and disease. Cell (2013) 155(4):934–47. doi: 10.1016/j.cell.2013.09.053
33. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell (2013) 153(2):307–19. doi: 10.1016/j.cell.2013.03.035
34. Kreffer-kwon KR, Tand Z, Mathe E, Qian J, Sung MH, Li G, et al. - Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell (2013) 155(7):1507–20. doi: 10.1016/j.cell.2013.11.039
35. Boija A, Klein IA, Sabari BR, Dall!Agnese A, EL C, Zamudio A, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell (2018) 175(7):1842–55. doi: 10.1016/j.cell.2018.10.042
36. Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell (2017) 169(1):13–23. doi: 10.1016/j.cell.2017.02.007
37. Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (2009) 326(5950):289–93. doi: 10.1126/science.1181369
38. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012) 485(7398):376–80. doi: 10.1038/nature11082
39. Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet (2018) 19(12):789–800. doi: 10.1038/s41576-018-0060-8
40. Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell (2014) 159(7):1665–80. doi: 10.1016/j.cell.2014.11.021
41. Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell (2015) 161(5):1012–25. doi: 10.1016/j.cell.2015.04.004
42. Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature (2016) 529(7584):110–4. doi: 10.1038/nature16490
43. Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol (2012) 13(12):1196–204. doi: 10.1038/ni.2432
44. Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature (2015) 518(7539):331–6. doi: 10.1038/nature14222
45. Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature (2013) 503(7475):290–4. doi: 10.1038/nature12644
46. Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A (2015) 112(47):E6456–65. doi: 10.1073/pnas.1518552112
47. Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell (2015) 162(4):900–10. doi: 10.1016/j.cell.2015.07.038
48. Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. The condensin complex is a mechanochemical motor that translocates along DNA. Science (2017) 358(6363):672–76. doi: 10.1126/science.aan6516
49. Vian L, Pekowska A, Rao SSP, Kieffer-Kwon KR, Jung S, Baranello L, et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell (2018) 173(5):1165–78. doi: 10.1016/j.cell.2018.03.072
50. Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, et al. Real-time imaging of DNA loop extrusion by condensin. Science (2018) 360(6384):102–05. doi: 10.1126/science.aar7831
51. Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci U S A (2015) 112(41):12776–81. doi: 10.1073/pnas.1512995112
52. Miyazaki K, Watanabe H, Yoshikawa G, Chen K, Hidaka R, Aitani Y, et al. The transcription factor E2A activates multiple enhancers that drive Rag expression in developing T and B cells. Sci Immunol (2020) 5(51):eabb1455. doi: 10.1126/sciimmunol.abb1455
53. Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science (2018) 361(6400):21. doi: 10.1126/science.aar3958
54. Khanna N, Zhang Y, Lucas JS, Dudko OK, Murre C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun (2019) 10(1):2771. doi: 10.1038/s41467-019-10628-9
55. Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell (1992) 68(5):855–67. doi: 10.1016/0092-8674(92)90029-c
56. Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell (1992) 68(5):869–77. doi: 10.1016/0092-8674(92)90030-g
57. Wayne J, Suh H, Sokol KA, Petrie HT, Witmer-Pack M, Edelhoff S, et al. TCR selection and allelic exclusion in RAG transgenic mice that exhibit abnormal T cell localization in lymph nodes and lymphatics. J Immunol (1994) 153(12):5491–502.
58. Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity (2003) 19(1):105–17. doi: 10.1016/S1074-7613(03)00181-X
59. Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol (2008) 9(6):613–22. doi: 10.1038/ni.1612
60. Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol (2008) 9(12):1388–98. doi: 10.1038/ni.1667
61. Schulz D, Vassen L, Chow KT, Mcwhirter SM, Amin RH, Möröy T, et al. Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J Exp Med (2012) 209(1):187–99. doi: 10.1084/jem.20110645
62. Timblin GA, Xie L, Tjian R, Schlissel MS. Dual Mechanism of Rag Gene Repression by c-Myb during Pre-B Cell Proliferation. Mol Cell Biol (2017) 37(12):e00437–16. doi: 10.1128/mcb.00437-16
63. Yxannoutsos, Barreto V, Misulovin Z, Gazumyan A, Yu W, Rajewsky N, et al. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat Immunol (2004) 5(4):443–50. doi: 10.1038/ni1053
64. Hao B, Naik AK, Watanabe A, Tanaka H, Liang C, Hunter W, et al. An anti-silencer- and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. J Exp Med (2015) 212(5):809–24. doi: 10.1084/jem.20142207
65. Naik AK, Byrd AT, Lucander ACK, Krangel MS. Hierarchical assembly and disassembly of a transcriptionally active RAG locus in CD4(+)CD8(+) thymocytes. J Exp Med (2019) 216(1):231–43. doi: 10.1084/jem.20181402
66. Yu W, Misulovin Z, Suh H, Hardy RR, Jankovic M, Yannoutsos N, et al. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5’ of RAG2. Science (1999) 285(5430):1080–4. doi: 10.1126/science.285.5430.1080
67. Monroe RJ, Chen F, Ferrini R, Davidson L, Alt FW. RAG2 is regulated differentially in B and T cells by elements 5’ of the promoter. Proc Natl Acad Sci U S A (1999) 96(22):12713–8. doi: 10.1073/pnas.96.22.12713
68. Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell (2019) 176(4):897–912.e20. doi: 10.1016/j.cell.2018.12.036
69. Kishi H, Wei XC, Jin ZX, Fujishiro Y, Nagata T, Matsuda T, et al. Lineage-specific regulation of the murine RAG-2 promoter: GATA-3 in T cells and Pax-5 in B cells. Blood (2000) 95(12):3845–52. doi: 10.1182/blood.V95.12.3845
70. Wu CX, Zhao WP, Kishi H, Dokan J, Jin ZX, Wei XC, et al. Activation of mouse RAG-2 promoter by Myc-associated zinc finger protein. Biochem Biophys Res Commun (2004) 317(4):1096–102. doi: 10.1016/j.bbrc.2004.03.159
71. Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, et al. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med (2008) 205(11):2507–13. doi: 10.1084/jem.20080134
72. Koga S, Hozumi K, Hirano KI, Yazawa M, Terooatea T, Minoda A, et al. Peripheral PDGFRα(+)gp38(+) mesenchymal cells support the differentiation of fetal liver-derived ILC2. J Exp Med (2018) 215(6):1609–26. doi: 10.1084/jem.20172310
73. Lio CW, Zhang J, Gonzalez-Avalos E, Hogan PG, Chang X. Rao, Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. Elife (2016) 21(5):18290. doi: 10.7554/eLife.18290
74. Itoi M, Tsukamoto N, Amagai T. Expression of Dll4 and CCL25 in Foxn1-negative epithelial cells in the post-natal thymus. Int Immunol (2007) 19(2):127–32. doi: 10.1093/intimm/dxl129
75. Calderón L, Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell (2012) 149(1):159–72. doi: 10.1016/j.cell.2012.01.049
76. Luo H, Jin K, Xie Z, Qiu F, Li S, Zou M, et al. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc Natl Acad Sci U S A (2012) 109(9):E553–62. doi: 10.1073/pnas.1115767109
77. Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell (2009) 138(1):186–97. doi: 10.1016/j.cell.2009.04.017
78. Trancoso I, Morimoto R, Boehm T. Co-evolution of mutagenic genome editors and vertebrate adaptive immunity. Curr Opin Immunol (2020) 65:32–41. doi: 10.1016/j.coi.2020.03.001
79. Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol (2011) 12(10):992–1001. doi: 10.1038/ni.2086
80. Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity (2000) 12(1):17–26. doi: 10.1016/s1074-7613(00)80155-7
81. Engel I, Murre C. The function of E- and Id proteins in lymphocyte development. Nat Rev Immunol (2001) 1(3):193–9. doi: 10.1038/35105060
82. Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity (2009) 31(4):565–75. doi: 10.1016/j.immuni.2009.07.010
83. Jones-Mason ME, Zhao X, Kappes D, Lasorella A, Iavarone A, Zhuang Y. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity (2012) 36(3):348–61. doi: 10.1016/j.immuni.2012.02.010
84. Miyazaki M, Miyazaki K, Chen S, Chandra V, Wagatsuma K, Agata Y, et al. The E-Id protein axis modulates the activities of the PI3K-AKT-mTORC1-Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis. Genes Dev (2015) 29(4):409–25. doi: 10.1101/gad.255331.114
85. Kisielow P, Miazek A, Cebrat M. NWC, a new gene within RAG locus: could it keep GOD under control? Int J Immunogenet (2008) 35(4-5):395–9. doi: 10.1111/j.1744-313X.2008.00791.x
86. Cebrat M, Miazek A, Kisielow P. Identification of a third evolutionarily conserved gene within the RAG locus and its RAG1-dependent and -independent regulation. Eur J Immunol (2005) 35(7):2230–8. doi: 10.1002/eji.200526225
87. Laszkiewicz A, Bzdzion Ł, Kasztura M, Snieżewski L, Janik S, Kisielow P, et al. Ikaros and RAG-2-mediated antisense transcription are responsible for lymphocyte-specific inactivation of NWC promoter. PloS One (2014) 9(9):e106927. doi: 10.1371/journal.pone.0106927
88. Eberl G, Colonna M, Di Santo JP, Mckenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science (2015) 348(6237):aaa6566. doi: 10.1126/science.aaa6566
89. Long HK, Prescott SL, Wysocka J. Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell (2016) 167(5):1170–87. doi: 10.1016/j.cell.2016.09.018
90. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature (2011) 470(7333):279–83. doi: 10.1038/nature09692
91. Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, et al. Progressive Loss of Function in a Limb Enhancer during Snake Evolution. Cell (2016) 167(3):633–42.e11. doi: 10.1016/j.cell.2016.09.028
92. Huang S, Tao X, Yuan S, Zhang Y, Li P, Beilinson HA, et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell (2016) 166(1):102–14. doi: 10.1016/j.cell.2016.05.032
93. Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, et al. Transposon molecular domestication and the evolution of the RAG recombinase. Nature (2019) 569(7754):79–84. doi: 10.1038/s41586-019-1093-7
94. Hu G, Kairong C, Fang D, Hirose S, Wang X, Wangsa D, et al. Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity (2018) 48(2):227–42. doi: 10.1016/j.immuni.2018.01.013
95. Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol (2010) 11(7):635–43. doi: 10.1038/ni.1891
96. Wang Y, Zolotarev N, Yang CY, Rambold A, Mittler G, Grosschedl R. A Prion-like Domain in Transcription Factor EBF1 Promotes Phase Separation and Enables B Cell Programming of Progenitor Chromatin. Immunity (2020) 53:1–17. doi: 10.1016/j.immuni.2020.10.009
97. Johanson TM, Lun ATL, Coughlan HD, Tan T, Smyth GK, Nutt SL, et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat Immunol (2018) 19(11):1257–64. doi: 10.1038/s41590-018-0234-8
98. Isoda T, Moore A, He Z, Chandra V, Aida M, Denholtz M, et al. Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell (2017) 171(1):103–19. doi: 10.1016/j.cell.2017.09.001
99. Hosokawa H, Romero-Wolf M, Yui MA, Ungerback J, Quiloan ML, Matsumoto M, et al. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol (2018) 19(12):1427–40. doi: 10.1038/s41590-018-0238-4
100. Johnson JL, Georgakilas G, Petrovic J, Kurachi M, Cai S, Harly C, et al. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity (2018) 48(2):243–57.e10. doi: 10.1016/j.immuni.2018.01.012
101. Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med (2012) 209(7):1255–62. doi: 10.1084/jem.20112745
102. Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature (2011) 476(7361):467–71. doi: 10.1038/nature10312
103. Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res (2015) 25(4):504–13. doi: 10.1101/gr.184986.114
104. Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol (2017) 18(2):173–83. doi: 10.1038/ni.3646
105. Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol (2007) 178(9):5717–26. doi: 10.4049/jimmunol.178.9.5717
106. Chen L, Carico Z, Shih HY, Krangel MS. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRδ and TCRα repertoires. Nat Immunol (2015) 16(10):1085–93. doi: 10.1038/ni.3232
107. Chen S, Krangel MS. Diversification of the TCR β Locus V(β) Repertoire by CTCF. Immunohorizons (2018) 2(11):377–83. doi: 10.4049/immunohorizons.1800072
108. Loguercio S, Barajas-Mora EM, Shih HY, Krangel MS, Feeney AJ. Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci. Front Immunol (2018) 9:425. doi: 10.3389/fimmu.2018.00425
109. Jai S, Ba Z, Zhang Y, Dai HQ, Alt FW. CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell (2018) 174(1):102–16. doi: 10.1016/j.cell.2018.04.035
110. Zhang Y, Zhang X, Ba Z, Liang Z, Dring EW, Hu H, et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature (2019) 573(7775):600–04. doi: 10.1038/s41586-019-1547-y
111. Ba Z, Lou J, Ye AY, Dai HQ, Dring EW, Lin SG, et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature (2020) 586(7828):305–10. doi: 10.1038/s41586-020-2578-0
112. Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, et al. CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Mol Cell (2017) 67(6):1049–58.e6. doi: 10.1016/j.molcel.2017.08.026
113. Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol (2009) 21(2):133–9. doi: 10.1016/j.coi.2009.03.009
114. Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell (2010) 141(3):419–31. doi: 10.1016/j.cell.2010.03.010
115. Ye SK, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, et al. The IL-7 receptor controls the accessibility of the TCRgamma locus by Stat5 and histone acetylation. Immunity (2001) 15(5):813–23. doi: 10.1016/s1074-7613(01)00230-8
Keywords: lineage-specific transcription factor, chromatin architecture, enhancer, Rag1 and Rag2 gene, super-enhancer, 3D genome organization
Citation: Miyazaki K and Miyazaki M (2021) The Interplay Between Chromatin Architecture and Lineage-Specific Transcription Factors and the Regulation of Rag Gene Expression. Front. Immunol. 12:659761. doi: 10.3389/fimmu.2021.659761
Received: 28 January 2021; Accepted: 02 March 2021;
Published: 16 March 2021.
Edited by:
Ageliki Tsagaratou, University of North Carolina at Chapel Hill, United StatesReviewed by:
Han-Yu Shih, National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), United StatesCopyright © 2021 Miyazaki and Miyazaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masaki Miyazaki, bW1peWF6YWtpQGluZnJvbnQua3lvdG8tdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.