- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Pharmacognosy Department, College of Pharmacy, Hawler Medical University, Erbil, Iraq
- 3Department of Electrical Engineering, Sharif University of Technology, Tehran, Iran
- 4Skull Base Research Center, Loghman Hakim Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Protein inhibitors of activated STAT (PIAS) are involved in the regulation of the JAK/STAT signaling pathway and have interactions with NF-κB, p73 and p53. These proteins regulate immune responses; therefore dysregulation in their expression leads to several immune-mediated disorders. In the present study, we examined expression of PIAS1-4 in peripheral blood of patients with acute/chronic inflammatory demyelinating polyradiculoneuropathy (AIDP/CIDP) compared with healthy subjects. We demonstrated down-regulation of all PIAS genes in both AIDP and CIDP cases compared with controls. Similarly, comparisons in gender-based groups revealed down-regulation of these gene0s in patients of each gender compared with gender-matched controls. There was no significant difference in expression of PIAS genes between AIDP and CIDP cases. Based on the area under the receiver operating characteristic curves, PIAS1-4 genes could distinguish between inflammatory demyelinating polyradiculoneuropathy and healthy status with accuracy values of 0.87, 0.87, 0.79 and 0.80, respectively. In differentiation between AIDP cases and healthy controls, these values were 0.92, 0.92, 0.83 and 0.86, respectively. Finally, PIAS1-4 genes could discriminate CIDP from healthy status with accuracy values of 0.82, 0.83, 0.75 and 0.75, respectively. The current study underscores the role of PIAS genes in the pathogenesis of inflammatory demyelinating polyradiculoneuropathy and their potential usage as biomarkers.
Introduction
Being firstly recognized as the inhibitors of signal transducer and activator of transcription (STAT) proteins, the protein inhibitors of activated STAT (PIAS) are involved in the regulation of the JAK/STAT signaling pathway (1, 2). PIAS1, PIAS2 (PIASxα, PIASxβ), PIAS3 and PIAS4 (PIASy) are the main members of this protein family (3). In addition to the STAT family of transcription factors (TFs), PIAS proteins have functional interactions with NF-κB, p73 and p53 (4). PIAS proteins have crucial roles in the regulation of the immune responses particularly through modulation of a number of cytokine-associated genes (4). Among PIAS proteins, PIAS1 has a prominent role in the regulation of innate immune responses as it specifically suppresses expression of IFN-inducible genes. These speculations are based on the observed enhancement of the antiviral function of IFN-β and IFN-γ in Pias1−/− cells (5). PIAS2 has been shown to suppress the IL-12-associated STAT4-dependent gene induction (6). Similarly, PIAS3 has a specific inhibitory effect on STAT3 signaling (7). STAT3 has been primarily identified as a TF induced by the IL-6 family of cytokines (8). Subsequent studies revealed its essential roles in the development of Th17 cells (9), regulation of humoral immune responses (10) and modulation of innate immunity (11). PIAS4 has prominent roles in suppression of STAT1, and inhibition of LEF1 and SMAD3 signaling pathways (12). Consistent with the critical functions of PIAS proteins in the regulation of immune responses, they have been shown to be involved in the pathogenesis of multiple sclerosis (MS) (13). However, their expression pattern and functional relevance with inflammatory demyelinating polyradiculoneuropathies are largely unknown. In the current study, we examined expression of PIAS1-4 in patients with acute/chronic inflammatory demyelinating polyradiculoneuropathy (AIDP/CIDP) compared with healthy subjects. Autoimmune inflammatory responses are considered as the principal mechanisms for development of these neurologic conditions (14, 15). In addition to the pathophysiological events, these two conditions share several features in terms of clinical manifestations and treatment modalities (14). Therefore, identification of the role of PIAS genes in the pathogenesis of AIDP/CIDP might facilitate the process of design of therapeutic options and prediction of disease course in these conditions.
Materials and Methods
Enrollment of AIDP/CIDP Patients and Healthy Subjects
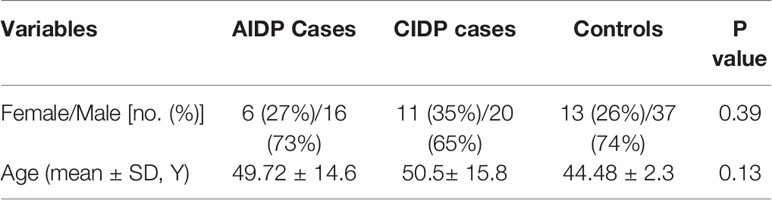
In total, 22 AIDP cases, 31 CIDP cases and 50 healthy individuals with no sign of inflammatory conditions were enrolled in the current study. Diagnostic evaluations were performed using the criteria offered by the American Academy of Neurology (16) and National Institute of Neurological Disorders and Stroke (17). AIDP/CIDP cases were in remission at the time of sampling. Diagnosis was based on clinical manifestations as well as electrophysiological and biochemical tests. Those with recent or chronic infection, cancer, or any systemic diseases were exempted from participation in the study. Persons enlisted in the control group were healthy individuals with no history of recent or chronic infection, malignancy, or systemic diseases. The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1398.853). All AIDP/CIDP cases and controls signed the informed consent forms.
Expression Assay
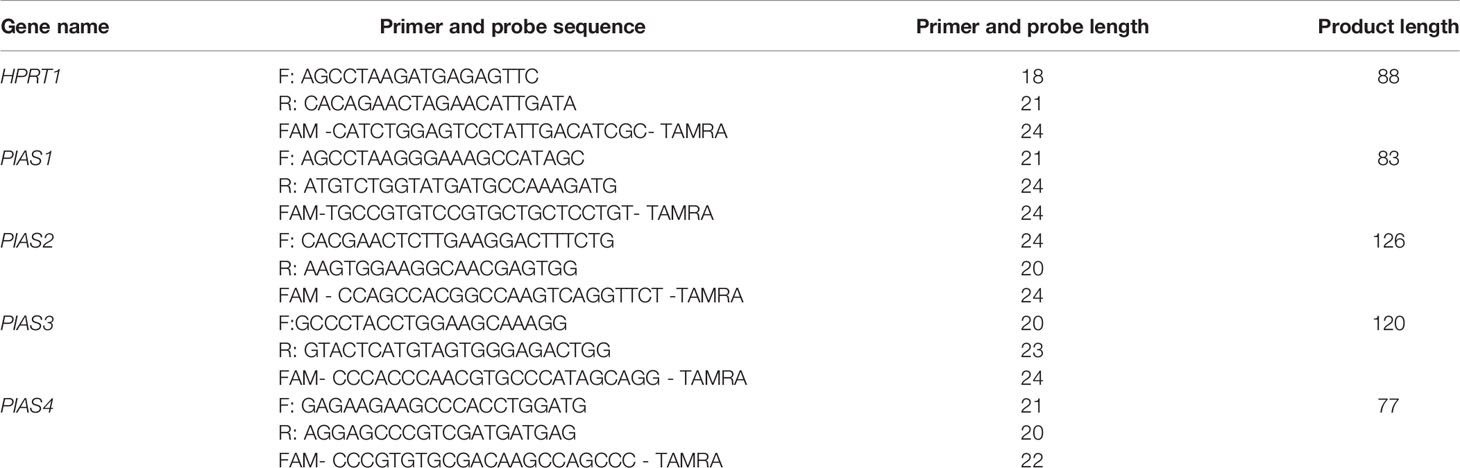
Peripheral blood samples were obtained from controls and AIDP/CIDP patients at the time of remission. Samples were collected in EDTA-containing tubes. RNA was isolated from blood samples using the RNA extraction kit delivered by the GeneAll Company (Seoul, Korea). Then, RNA was converted to cDNA by using the Thermo Fisher Scientific kit (Brussels, Belgium). Expression levels of PIAS genes were measured in AIDP/CIDP cases and control subjects using the master mix supplied by the Ampliqon Company (Odense, Denmark). Expression of PIAS genes were measured using the Step One Plus™ Real-Time PCR system (Applied Biosystems, Foster city, CA, USA). Details of expression analysis of PIAS genes were explained formerly (13, 18). Table 1 shows the primers and probes.
Statistical Methods
R programming language was used for statistical analysis. Transcript quantities of PIAS1, PIAS2, PIAS3, and PIAS4 in relation to the HPRT reference gene was calculated from CT values using the equation:. Then, the values were log2 transformed and used for subsequent analysis. Four comparisons between CIDP/AIDP/All patients and healthy individuals and between CIDP and AIDP patients were done and the significant difference between means was computed using the t-test. Correlations between expressions were evaluated through the calculation of Spearman correlation coefficients. Three predictive machine learning methods namely Bayesian Generalized Linear Model, Generalized Linear Model, and Linear Discriminant Analysis with 10-fold cross validation were used to compute the sensitivity and specificity of each model. The receiver operating characteristic curve was plotted. The Linear Discriminant Analysis Model (LDA) provided the most efficient estimates and in the best setting, the AUC was 0.94. Youden’s J statistic was employed to find the optimum threshold. LDA was then selected based on previous results to investigate efficiency of each gene for separating groups.
Results
General Demographic and Clinical Information of Cases and Controls
The current project enrolled 17 female and 36 male patients along with 13 female and 37 male subjects who enlisted as controls. Cases and controls were matched in the term of age parameter. Demographic information of patients and controls are demonstrated in Table 2.
Expression Assays
Figure 1 shows the relative expression amounts of PIAS genes in AIDP/CIDP patients and healthy subjects. Expression levels of all PIAS genes were significantly decreased in both CIDP cases compared with controls. For PIAS1, ratio of mean expressions (RME) in cases versus controls was 1.12E-03 (P value=8.2E-14). RME (P values) for PIAS2, PIAS3 and PIAS4 genes in CIDP cases versus controls were 2.22E-03 (4.5E-13), 9.02E-03 (1.7E-08) and 1.05E-02 (3.8E-09), respectively. Expression of all genes was also decreased in AIDP cases compared with controls. The corresponding RME and P values for PIAS1-PIAS4 genes in the AIDP cases versus controls were 1.52E-03 (3.3E-07), 2.60E-03 (5.9E-07), 1.47E-02 (5.9E-05) and 1.21E-02 (2.6E-05), respectively.
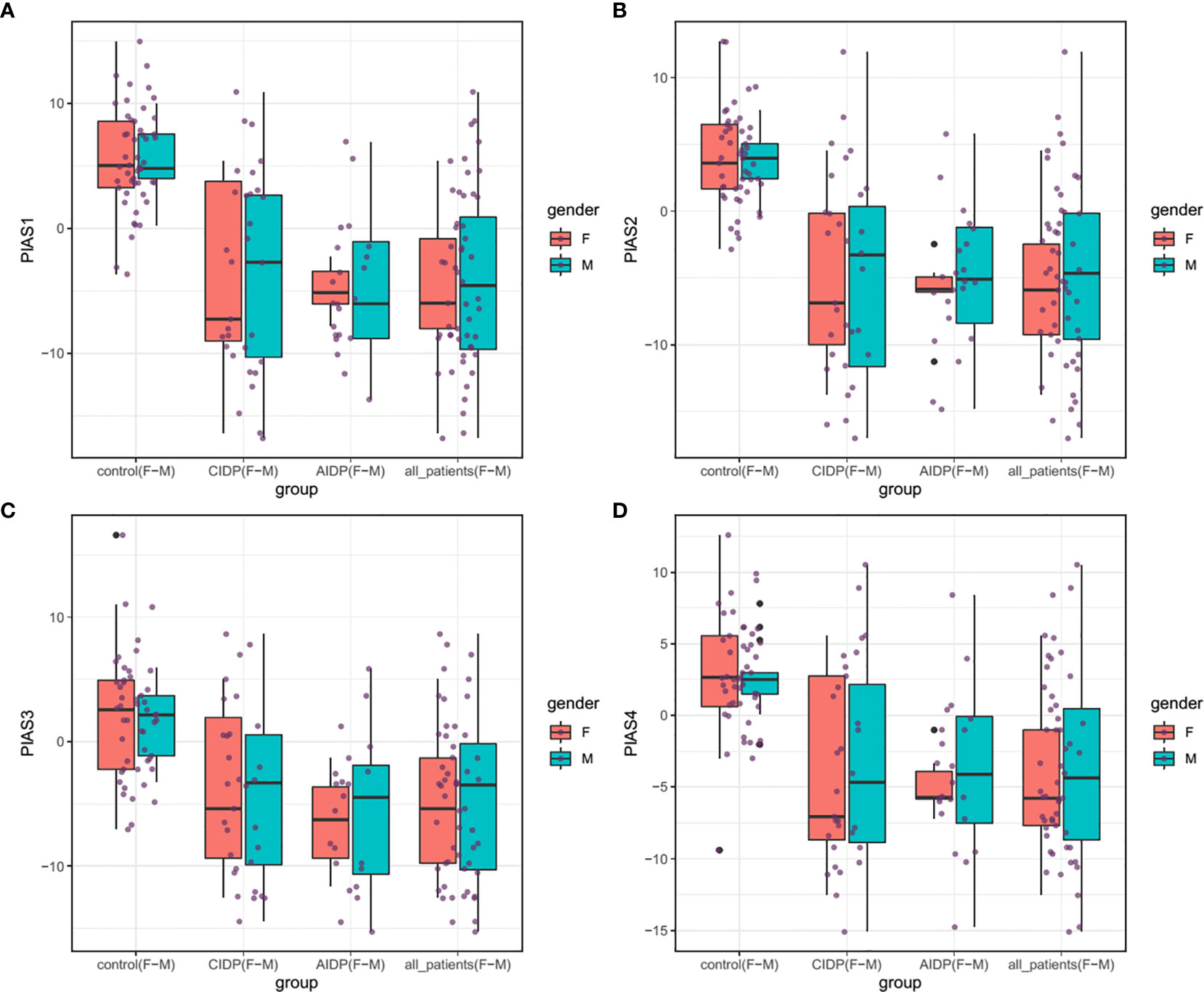
Figure 1 Relative expressions of PIAS genes in AIDP/CIDP patients and healthy subjects. Depicted box plots and whiskers show all data points from maximum to minimum. Boxes are depicted from Q1 to Q3. The horizontal lines in the middle of boxes show the median values. Each black dot shows expression in a certain sample. Mean values and interquartile range are displayed.
Similarly, comparisons in gender-based groups revealed down-regulation of these genes in patients of each gender compared with gender-matched controls. There was no significant difference in expression of PIAS genes between AIDP and CIDP cases. When assessing expression of PIAS genes in total CIDP and AIDP cases versus total controls, all genes were found to be down-regulated in affected individuals which is consistent with their expression pattern in related patients’ subgroups. Table 3 shows the detailed statistics of expression analysis of PIAS genes among study groups.
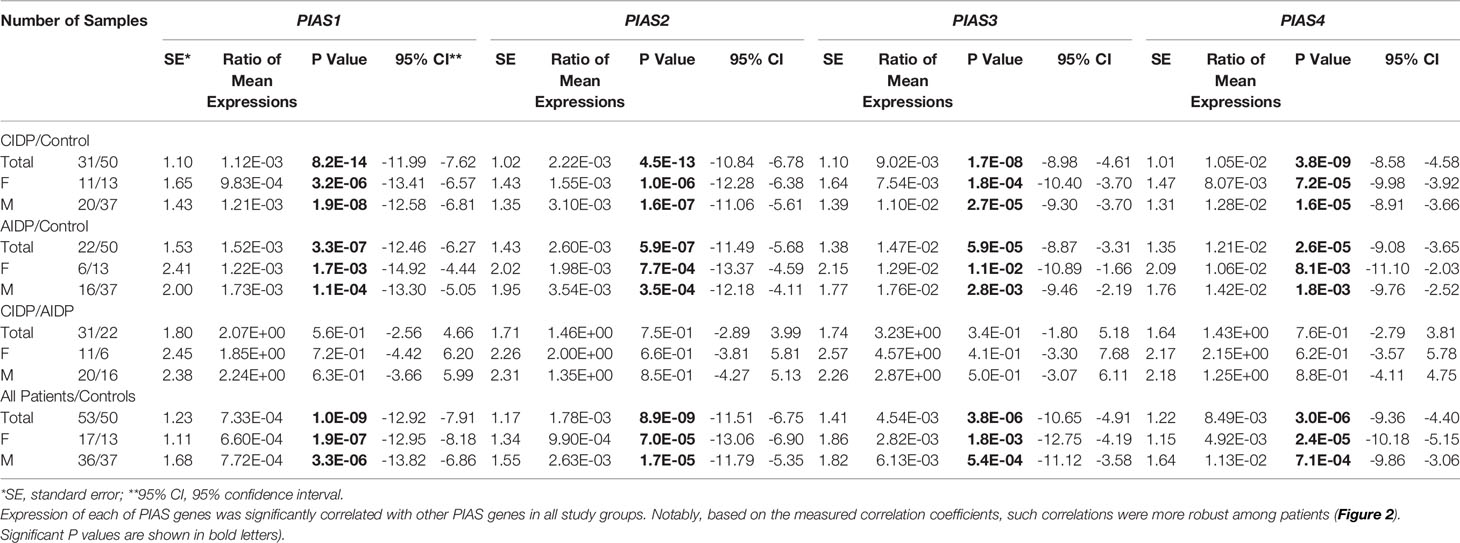
Table 3 The results of Bayesian Regression model for comparison of expression of PIAS genes in AIDP/CIDP patients and healthy persons (Expressions of PIAS genes have been compared between CIDP cases and controls, AIDP cases and controls, CIDP cases and AIDP cases as well as total patients and total control.
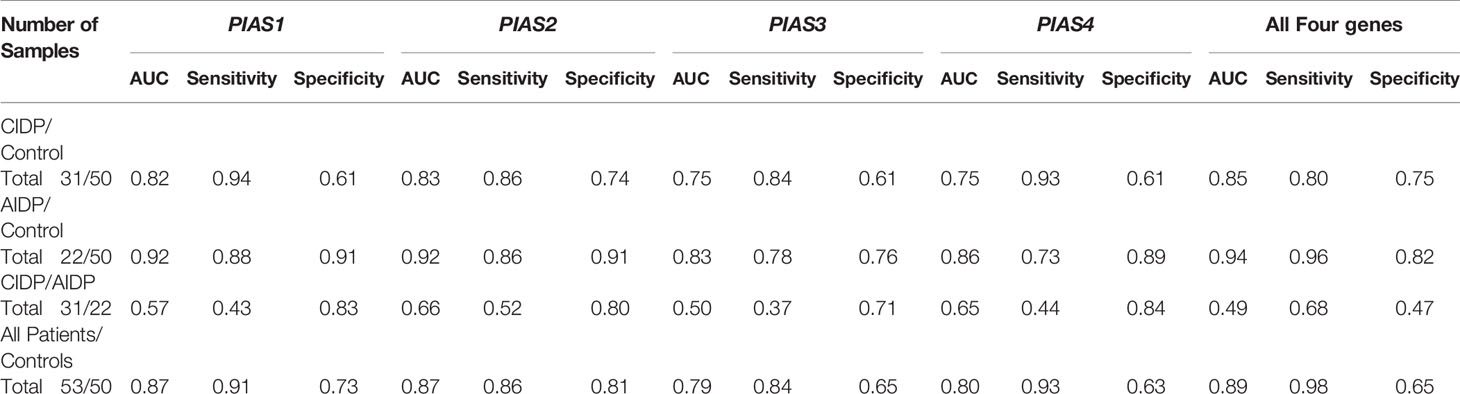
Afterwards, we appraised whether expression of PIAS transcripts can separate AIDP/CIDP patients from controls (Figure 2). Based on the AUC values, PIAS1-4 genes could distinguish between inflammatory demyelinating polyradiculoneuropathy and healthy status with accuracy values of 0.87, 0.87, 0.79 and 0.80, respectively. In differentiation between AIDP cases and healthy controls, these values were 0.92, 0.92, 0.83 and 0.86, respectively. Finally, PIAS1-4 genes could discriminate CIDP from healthy status with accuracy values of 0.82, 0.83, 0.75 and 0.75, respectively. Figure 3 and Table 4 show details of ROC curve analysis.
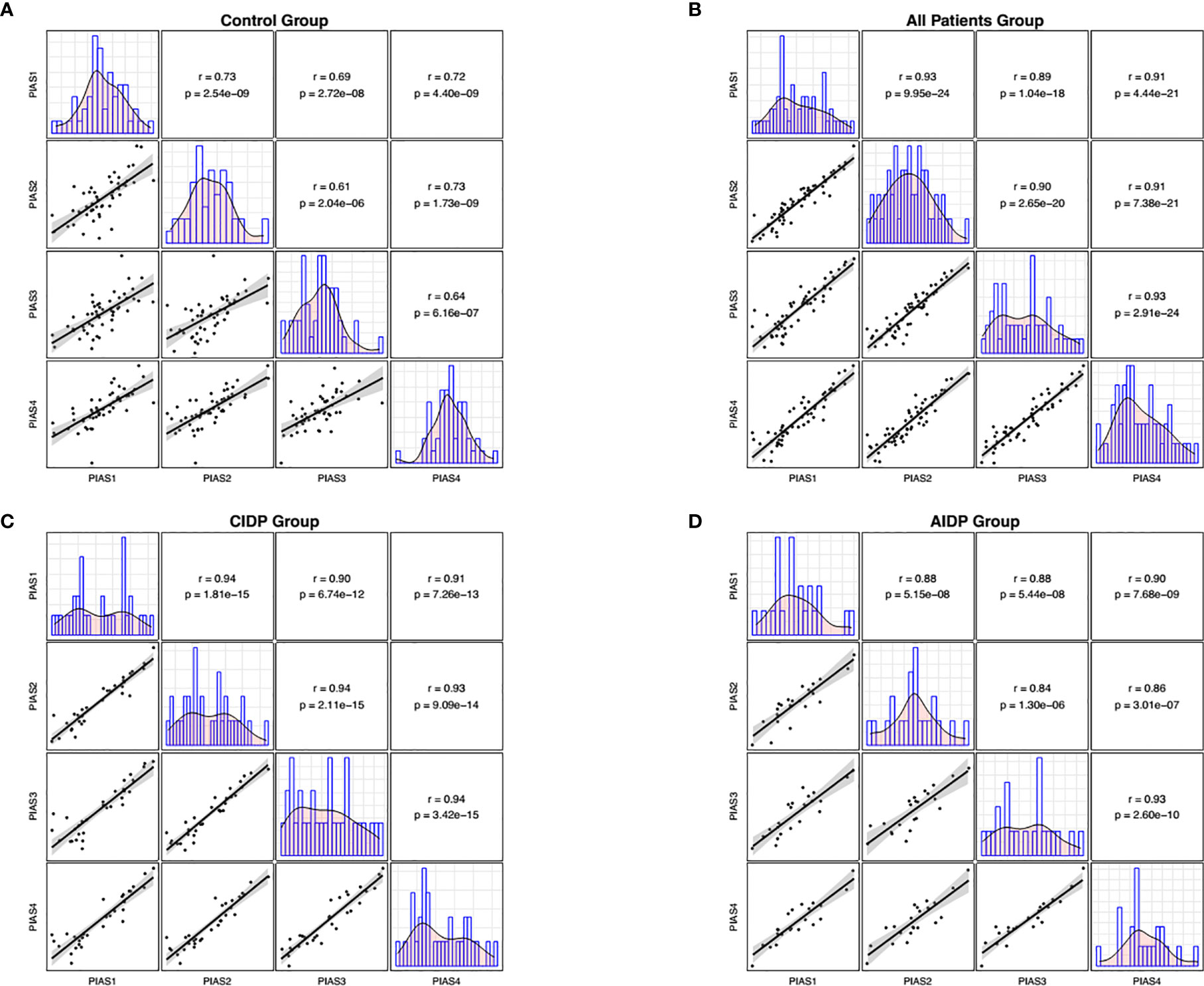
Figure 2 Correlations between transcript quantities of PIAS genes. The distributions of parameters are represented on the diagonals. The bivariate scatter plots with a fitted line are shown on the lower parts of the diagonals. Correlation coefficients and p values of the correlations are displayed on the upper sections of the diagonal.
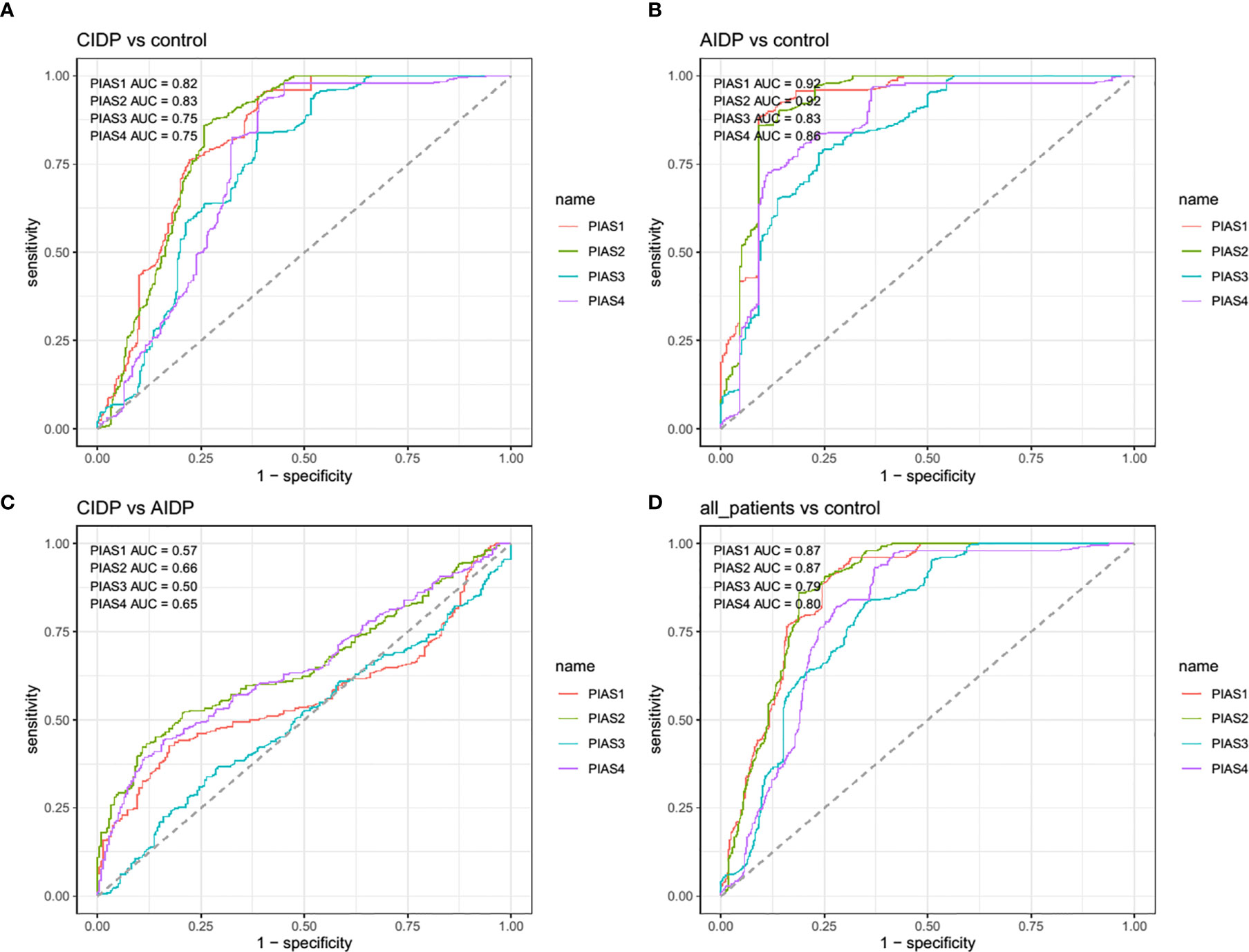
Figure 3 The receiver operating characteristic (ROC) curve of PIAS1-4 in diagnosis of inflammatory demyelinating polyradiculoneuropathy.
Discussion
PIAS proteins have been shown to modulate the function of many TFs such as STATs, NF-κB and SMADs. The Jak-STAT cascade as a main target of PIAS proteins is the main intracellular cascade induced by class I cytokine receptor proteins. Through this signaling pathway, the activated kinase phosphorylates tyrosine amino acids in the intracellular domain of cytokine receptors (19). STATs have important roles in STATs in the development of neutrophils and regulation of their function, polarization of macrophages, and activity of dendritic cells (19). NF-κB+ signaling has also been shown to regulate expression of cytokines and antimicrobial molecules. Moreover, this signaling pathway controls differentiation, subsistence and proliferation of cells that are involved in innate and adaptive immune reactions (20). The SMAD pathway controls IgA secretion by B cells and enhances differentiation of CD4+ T cells into T17 cells and regulatory T cells (21). Therefore, PIAS proteins can affect immune responses via various routes. These proteins exert their regulatory roles via different routes such as inhibition of the DNA-binding activity of TFs and functional interaction with transcriptional corepressors or co-activators. They also have prominent roles in the regulation of innate immune reactions (4). In the current project, we demonstrated down-regulation of PIAS1-4 transcripts in the peripheral blood of AIDP and CIDP patients, with no significant difference between these two groups of patients. We have recently examined expression levels of STAT genes in the same cohort of CIDP and AIDP patients and reported over-expression of STAT1 in female patients compared with sex-matched controls (22). It is worth mentioning that since PIAS1 was similarly down-regulated in males and females in the current study, this system may not be the only one responsible for elevated STAT1 levels. STAT1 has a crucial role in induction of gene expression in response to IFN stimulation. PIAS1 is the only member of the PIAS family of proteins that can block the DNA binding function of STAT1 and suppress STAT1-associated stimulation of gene expression in response to IFN (2). Consistent with the anti-inflammatory role of PIAS1, down-regulation of this transcript has been associated with allograft rejection (23). PIAS proteins have also prominent roles in decreasing the effects of pro-inflammatory cytokines (24). Moreover, PIAS agonists and inducers have immunosuppressive effects (24). Therefore, the observed down-regulation of PIAS transcripts in AIDP and CIDP cases might exacerbate the autoimmune responses in these patients, thus contributing in the pathogenesis of these conditions. In line with the inhibitory role of PIAS proteins on the activity of NF-κB and the observed down-regulation of PIAS transcripts in AIDP/CIDP patients, Andorfer et al. have shown a remarkable increase in NF-κB levels in Guillain-Barré syndrome and CIDP cases compared to controls. They also suggested a critical role for NF-κB in the pathogenesis of these inflammatory conditions (25). Although we did not perform functional studies to assess whether down-regulation of PIAS transcripts is the cause or effect of CIDP/AIDP, based on the role of these transcripts in suppression of STAT signaling we hypothesize that this dysregulation is the cause of this autoimmune condition. Abnormal levels of PIAS-targeting miRNAs might be a possible mechanism of down-regulation of PIAS transcripts. Several miRNAs including miR-146a have been shown to affect expression of PIAS, STAT and other components of this pathway (26). Meanwhile, expression of a number of these miRNAs including miR-146a has been found to be dysregulated in GBS patients (27). Besides, miR-18a has been found to negatively regulate PIAS3 expression and therefore influencing expression of STAT3 target genes (28). Notably, miR-18a has a distinctive inhibitory effects on differentiation of T17 cells (29). Therefore, abnormal functional network between PIAS, STAT and miRNAs might affect expression and function of these pathways.
We also demonstrated significant correlations between PIAS1-4 transcript levels particularly among patients. Such finding implies the presence of a solitary regulatory mechanism for these genes and the robustness of this mechanism in the context of AIDP/CIDP. Finally, we appraised the diagnostic power of PIAS1-4 genes in distinguishing between disease and healthy conditions. The best values were detected for PIAS1 and PIAS2 for differentiation of AIDP cases from healthy condition.
Taken together, the observed down-regulation of PIAS genes in AIDP/CIDP cases implies their possible contribution in the pathogenesis of these conditions and their potential usage as biomarkers. Therapeutic options that alter expression of these genes might improve the response of patients to available therapeutic options and attenuate the course of these disorders. These speculations should be appraised in animal models of inflammatory demyelinating polyradiculoneuropathy. A limitation of this study is lack of assessment of expression levels of PIAS transcripts in different time points during the course of disorder to find whether their expression is changed during the time.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The study protocol was approved by the ethical committee of Shahid Beheshti University of Medical Sciences (IR.SBMU.MSP.REC.1398.853). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
MT and FN performed the experiment. AS and NN collected the data and analyzed it. SG-F wrote the draft and revised it. All authors contributed to the article and approved the submitted version.
Funding
The current study was supported by a grant from Shahid Beheshti University of Medical Sciences (Grant number 18934).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Schmidt D, Müller S. Members of the PIAS Family Act as SUMO Ligases for c-Jun and p53 and Repress p53 Activity. Proc Natl Acad Sci (2002) 99(5):2872–7. doi: 10.1073/pnas.052559499
2. Liu B, Liao J, Rao X, Kushner SA, Chung CD, Chang DD, et al. Inhibition of Stat1-mediated Gene Activation by PIAS1. Proc Natl Acad Sci (1998) 95(18):10626–31. doi: 10.1073/pnas.95.18.10626
3. Niu G-J, Xu J-D, Yuan W-J, Sun J-J, Yang M-C, He Z-H, et al. Protein Inhibitor of Activated STAT (PIAS) Negatively Regulates the JAK/STAT Pathway by Inhibiting STAT Phosphorylation and Translocation. Front Immunol (2018) 9:2392. doi: 10.3389/fimmu.2018.02392
4. Shuai K, Liu B. Regulation of Gene-Activation Pathways by PIAS Proteins in the Immune System. Nat Rev Immunol (2005) 5(8):593–605. doi: 10.1038/nri1667
5. Liu B, Mink S, Wong KA, Stein N, Getman C, Dempsey PW, et al. PIAS1 Selectively Inhibits Interferon-Inducible Genes and is Important in Innate Immunity. Nat Immunol (2004) 5(9):891–8. doi: 10.1038/ni1104
6. Arora T, Liu B, He H, Kim J, Murphy TL, Murphy KM, et al. Piasx is a Transcriptional Co-Repressor of Signal Transducer and Activator of Transcription 4. J Biol Chem (2003) 278(24):21327–30. doi: 10.1074/jbc.C300119200
7. Chung CD, Liao J, Liu B, Rao X, Jay P, Berta P, et al. Specific Inhibition of Stat3 Signal Transduction by PIAS3. Science (1997) 278(5344):1803–5. doi: 10.1126/science.278.5344.1803
8. Zhong Z, Wen Z, Darnell JE. Stat3: A STAT Family Member Activated by Tyrosine Phosphorylation in Response to Epidermal Growth Factor and Interleukin-6. Science (1994) 264(5155):95–8. doi: 10.1126/science.8140422
9. Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 Regulates Cytokine-Mediated Generation of Inflammatory Helper T Cells. J Biol Chem (2007) 282(13):9358–63. doi: 10.1074/jbc.C600321200
10. Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. Critical Role for Stat3 in T-dependent Terminal Differentiation of Igg B Cells. Blood (2006) 107(3):1085–91. doi: 10.1182/blood-2005-07-2871
11. Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z, et al. STAT3 Deletion During Hematopoiesis Causes Crohn’s Disease-Like Pathogenesis and Lethality: A Critical Role of STAT3 in Innate Immunity. Proc Natl Acad Sci (2003) 100(4):1879–84. doi: 10.1073/pnas.0237137100
12. Yagil Z, Nechushtan H, Kay G, Yang CM, Kemeny DM, Razin E. The Enigma of the Role of Protein Inhibitor of Activated STAT3 (PIAS3) in the Immune Response. Trends Immunol (2010) 31(5):199–204. doi: 10.1016/j.it.2010.01.005
13. Taheri M, Azimi G, Sayad A, Mazdeh M, Arsang-Jang S, Omrani MD, et al. Expression Analysis of Protein Inhibitor of Activated STAT (PIAS) Genes in Ifnβ-Treated Multiple Sclerosis Patients. J Inflammation Res (2018) 11:457–63. doi: 10.2147/JIR.S187414
14. Peltier AC, Donofrio PD. Chronic Inflammatory Demyelinating Polyradiculoneuropathy: From Bench to Bedside. Semin Neurol (2012) 32(3):187–95. doi: 10.1055/s-0032-1329194
15. Ubogu EE. Inflammatory Neuropathies: Pathology, Molecular Markers and Targets for Specific Therapeutic Intervention. Acta Neuropathol (2015) 130(4):445–68. doi: 10.1007/s00401-015-1466-4
16. Neurology AAo. Research Criteria for Diagnosis of Chronic Inflammatory Demyelinating Polyneuropathy (CIDP): Report From an Ad Hoc Subcommittee of the American Academy of Neurology Aids Task Force. Neurology (1991) 41:617–8. doi: 10.1212/WNL.41.5.617
17. Van der Meché F, Van Doorn P, Meulstee J, Jennekens F. Diagnostic and Classification Criteria for the Guillain-Barré Syndrome. Eur Neurol (2001) 45(3):133–9. doi: 10.1159/000052111
18. Sayad A, Taheri M, Azari I, Oskoei VK, Ghafouri-Fard S. PIAS Genes as Disease Markers in Bipolar Disorder. J Cell Biochem (2019) 120(8):12937–42. doi: 10.1002/jcb.28564
19. Li HS, Watowich SS. Innate Immune Regulation by STAT-mediated Transcriptional Mechanisms. Immunol Rev (2014) 261(1):84–101. doi: 10.1111/imr.12198
20. Hayden MS, West AP, Ghosh S. NF-Kappab and the Immune Response. Oncogene (2006) 25(51):6758–80. doi: 10.1038/sj.onc.1209943
21. Malhotra N, Kang J. SMAD Regulatory Networks Construct a Balanced Immune System. Immunology (2013) 139(1):1–10. doi: 10.1111/imm.12076
22. Ali ZPM, Taheri M, Sangsefidi S, Arsang-Jang S, Mazdeh M, Zamani A, et al. Evaluation of Expression of STAT Genes in Immune-Mediated Polyneuropathies. J Mol Neurosci (2020) 70:945–52. doi: 10.1007/s12031-020-01494-y
23. Nafar M, Kalantari S, Samavat S, Omrani MD, Arsang-Jang S, Taheri M, et al. Downregulation of Protein Inhibitor of Activated Stat (Pias) 1 Is Possibly Involved in the Process of Allograft Rejection. Transplant Proc (2020) 52(1):414–8. doi: 10.1016/j.transproceed.2019.10.006
24. O’shea J. Targeting the Jak/STAT Pathway for Immunosuppression. Ann Rheumatic Dis (2004) 63(suppl 2):ii67–71. doi: 10.1136/ard.2004.028290
25. Andorfer B, Kieseier BC, Mathey E, Armati P, Pollard J, Oka N, et al. Expression and Distribution of Transcription Factor NF-kappaB and Inhibitor IkappaB in the Inflamed Peripheral Nervous System. J Neuroimmunol (2001) 116(2):226–32. doi: 10.1016/S0165-5728(01)00306-X
26. Zhang L, Li J, Wang Q, Meng G, Lv X, Zhou H, et al. The Relationship Between microRNAs and the STAT3-related Signaling Pathway in Cancer. Tumor Biol (2017) 39(7):1010428317719869. doi: 10.1177/1010428317719869
27. Huang P, Xu M, He X-Y. Correlations Between microRNA-146a and Immunoglobulin and Inflammatory Factors in Guillain-Barré Syndrome. J Int Med Res (2020) 48(3):300060520904842–. doi: 10.1177/0300060520904842
28. Nomair AM, Ahmed SS, Nomeir HM, El Mansy H, Mohammed AF. The Role of Protein Inhibitor of Activated STAT3 and miRNA-18a Expressions in Breast Cancer. Egypt J Med Hum Genet (2019) 20(1):1–9. doi: 10.1186/s43042-019-0021-6
Keywords: protein inhibitor of activated STAT, PIAS, inflammatory demyelinating polyradiculoneuropathy, AIDP, CIDP
Citation: Ghafouri-Fard S, Hussen BM, Nicknafs F, Nazer N, Sayad A and Taheri M (2021) Expression Analysis of Protein Inhibitor of Activated STAT in Inflammatory Demyelinating Polyradiculoneuropathy. Front. Immunol. 12:659038. doi: 10.3389/fimmu.2021.659038
Received: 10 February 2021; Accepted: 21 April 2021;
Published: 12 May 2021.
Edited by:
Poornima Paramasivan, Abertay University, United KingdomReviewed by:
Rezvan Noroozi, Jagiellonian University, PolandAmin Safa, Complutense University of Madrid, Spain
Copyright © 2021 Ghafouri-Fard, Hussen, Nicknafs, Nazer, Sayad and Taheri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arezou Sayad, YXIuc2F5YWRAeWFob28uY29t; Mohammad Taheri, TW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==
 Soudeh Ghafouri-Fard
Soudeh Ghafouri-Fard Bashdar Mahmud Hussen
Bashdar Mahmud Hussen Fwad Nicknafs1
Fwad Nicknafs1 Arezou Sayad
Arezou Sayad Mohammad Taheri
Mohammad Taheri