- 1Department of Immunology, Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana
- 2Department of Virology, Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana
- 3Department of Medicine, Washington University School of Medicine in St Louis, St. Louis, MO, United States
- 4Medical and Scientific Research Centre, University of Ghana Medical Centre, University of Ghana, Accra, Ghana
Fc gamma receptors (FcγR) are cell surface glycoproteins which trigger specific effector-cell responses when cross-linked with the Fc portions of immunoglobulin (IgG) antibodies. During HIV-1 infection, the course of disease progression, ART response, and viral reservoir size vary in different individuals. Several factors may account for these differences; however, Fc gamma receptor gene polymorphisms, which influence receptor binding to IgG antibodies, are likely to play a key role. FcγRIIa (CD32) was recently reported as a potential marker for latent HIV reservoir, however, this assertion is still inconclusive. Whether FcγR polymorphisms influence the size of the viral reservoir, remains an important question in HIV cure studies. In addition, potential cure or viral suppression methods such as broadly neutralizing antibody (bNAbs) may depend on FcγRs to control the virus. Here, we discuss the current evidence on the potential role played by FcγR polymorphisms in HIV-1 infection, treatment and vaccine trial outcomes. Importantly, we highlight contrasting findings that may be due to multiple factors and the relatively limited data from African populations. We recommend further studies especially in sub-Saharan Africa to confirm the role of FcγRIIa in the establishment of latent reservoir and to determine their influence in therapies involving bNAbs.
Introduction
An estimated 37.9 million people are infected with the human immunodeficiency virus (HIV). The burden of disease is highest in sub-Saharan Africa, where approximately 25.6 million people live with the virus (1).
For most patients infected with HIV, antibodies elicited by the host immune system have limited potential to neutralize the virus. The result is a gradual decline of host CD4+ T cells leading to full blown acquired immune deficiency syndrome (AIDS) over time. Antiretroviral therapy (ART) is able to suppress the virus and reverse the decline in CD4 count in most patients. However, ART does not provide cure due to a stable latent reservoir established early in the infection process in resting CD4+ T cells, macrophages and other cells.
The course of HIV-1 infection, disease progression, ART response, and reservoir size vary in different individuals. For instance, long term non-progressors can maintain high CD4 count, and control the virus for up to 10 years without ART (2–4), compared to “rapid progressors” who develop full blown AIDS within 3-4 years of infection (5, 6). Then, there is the rare group of HIV-1 infected individuals called “elite controllers”, who can maintain a viral load of less than 50 copies/ml without ART for even longer periods (7, 8). These elite controllers and long term non-progressors, have a smaller viral reservoir (9, 10). Viral factors such as deletions or mutations in key viral genes have been implicated in the differences in natural HIV control. Factors such as source of HIV infection, timing of ART and ethnicity have all been cited as potential determinants of viral reservoir size (11). However, host genetic factors such as human leukocyte antigen (HLA) and polymorphisms in Fc gamma receptor (FCGR) genes which influence the receptor binding to immunoglobulin (Ig) G antibodies are likely to be even more critical. FcγR gene copy number variations (CNVs) and/or single nucleotide polymorphisms (SNPs) could cause differences in Fc gamma receptor (FcγR) expression density on effector cell surface, binding affinity to IgG subclasses and signaling potential which would influence HIV-1 infection risk, disease progression and vaccine efficacy (12, 13). In this review, we summarize current knowledge on the role of FcγR gene polymorphisms and HIV-1 infection, in relation to ART outcomes and control of the viral reservoir. We will explore the idea that FcγR polymorphisms could help explain the differences in HIV-1 infection outcomes, responses to ART and broadly neutralizing antibodies (bNAbs) and influence the size of the viral reservoir.
Overview of FcγRs and Their Role in Host Immunity
FcγRs are cell surface glycoproteins that bind the Fc portions of different IgG subclasses to trigger different cell effector functions (14, 15). FcγRs are expressed on most immune cells including monocytes, natural killer (NK) cells, B cells, eosinophils, basophils, dendritic cells, platelets, macrophages, and some subpopulations of T cells (16–18).
There are three main classes of FcγRs namely FcγRI (CD64), FcγRII (CD32) and FcγRIII (CD16) each with different isoforms encoded by different genes (Figure 1). The FcγRI family of receptors consist of 3 genes (FCGR1A, FCGR1B and FCGR1C) that share about 98% sequence homology and thought to flank the centromere of chromosome 1 at bands 1p12 (FCGR1B) and 1q21 (FCGR1A and FCGR1C) (19). FcγRIA is the only known high affinity FcγR. It is expressed by monocytes, dendritic cells (DC’s), macrophages and neutrophils (20) and plays a role in antibody mediated phagocytosis. The FcγRII family of receptors have low binding affinity for IgG and are encoded by three genes (FCGR2A, FCGR2B and FCGR2C) located on chromosome 1q23.3 (16, 21). They are expressed on neutrophils, DC’s, monocytes, B cells, NK cells, myeloid cells, and platelets. FcγRII family of receptors do not use the common Fcγ-chain for activation (FcγRIIa and FcγRIIc) or inhibitory (FcγRIIb) signaling because their Immunoreceptor Tyrosine-based Activation Motifs (ITAM) or Immunoreceptor Tyrosine-based Inhibitory Motifs (ITIM) are located directly in the intracellular cytoplasmic domain (22). The FcγRIIc gene is expressed on NK cells only with 13Q allele due to the glutamine (Q)/stop (STP) polymorphism at codon 13 located in the first extracellular domain (23, 24).
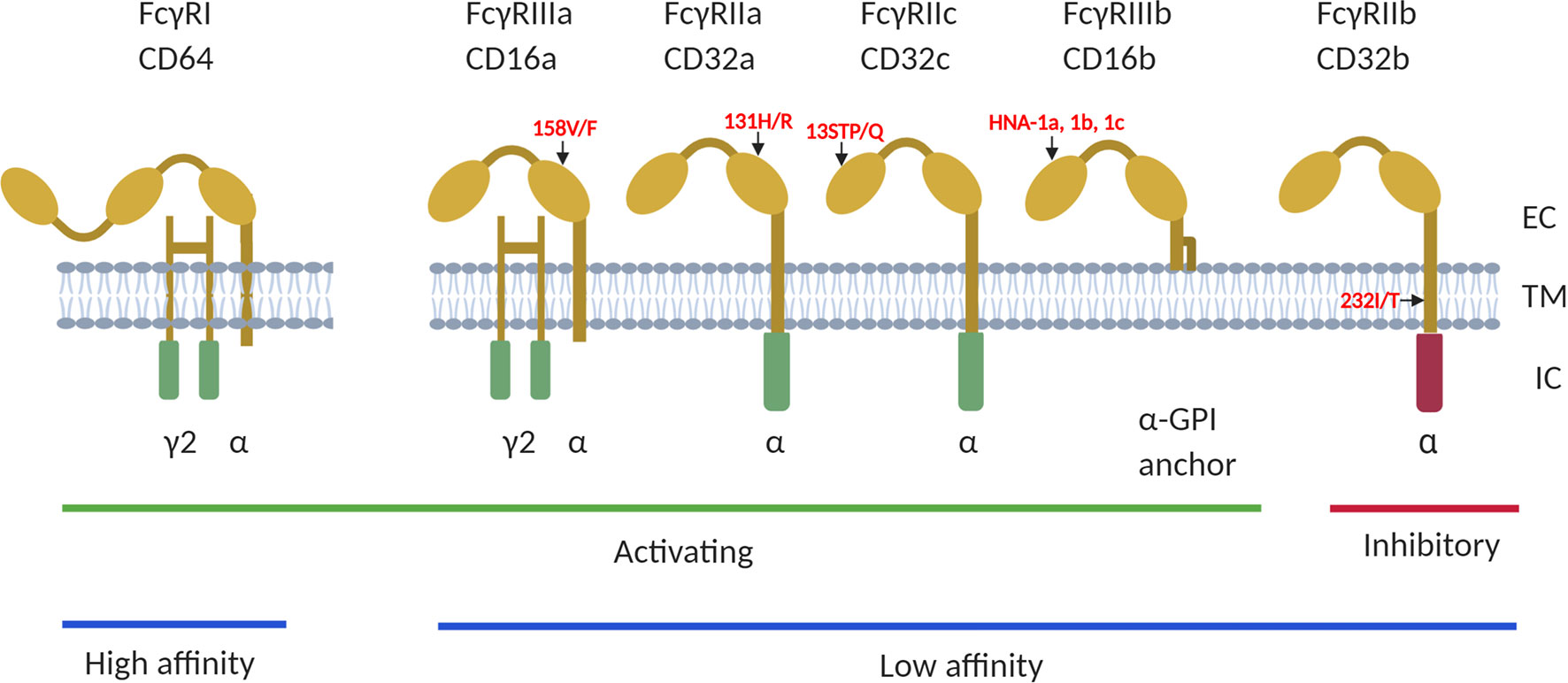
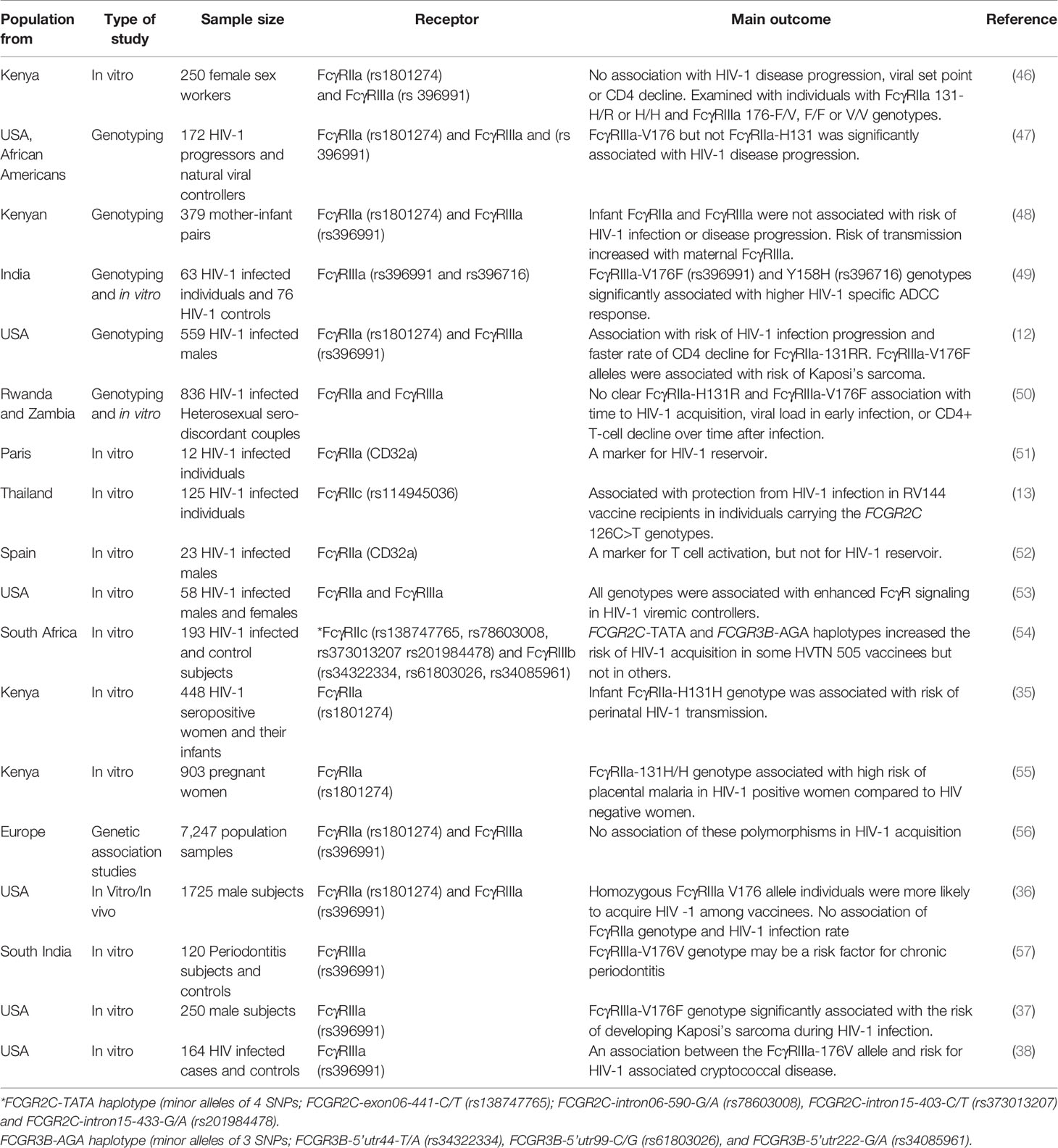
Figure 1 The human FCGRs consists of 3 classes; FcγRI (CD64), FcγRII (CD32) and FcγRIII (CD16), of which FcγRI has high affinity to bind to antibody Fc-fragment. FcγRII and FcγRIII have low affinity for IgG binding. They are also classified as activating or inhibitory due to the signals induced by FcR crosslinking. The FcγRI, FcγRIIa/IIc and FcγRIIIa and FcγRIIIb proteins contain the immunoreceptor tyrosine based activating motifs (ITAM), whiles FcγRIIb is the only receptor that contains the immunoreceptor tyrosine based inhibitory motif (ITIM), in its cytoplasmic domain. Additionally, the FcγRIIIb gene encodes a glycosylphosphatidylinositol (GPI) -anchored receptor, and this encodes human neutrophil specific antigen 1 (HNA1a, b and c). Several functional polymorphisms exist in the FcγR genes, including the FcγRIIa-131H/R of which the 131-H allele has a high binding affinity to IgG2 and FcγRIIIa-176V/F with the 176-V allele being more effective at mediating phagocytosis. The FcγRIIc gene encodes either a glutamine (Q) or a stop codon (STP) at position 13 while the 232-T allele of the FcγRIIb 232I/T polymorphism has a reduced inhibitory signaling capacity. EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain. Summaries of studies that have investigated different interactions between the receptors on different HIV infection outcomes have been provided in Tables 1, 2.
FcγRIII family is encoded by 2 genes (FCGR3A and FCGR3B) for the receptors (FcγRIIIa and FcγRIIIb). Although these receptors are also low affinity binding, they can both bind efficiently to multimeric IgG and immune complexes. FcγRIIIa receptor can also bind with intermediate affinity and expressed on monocytes, tissue specific macrophages, dendritic cells, NK cells and gamma/delta T cells (21). The FCGR3B gene, encodes a glycosylphosphatidylinositol (GPI)-anchored receptor, which is highly expressed on neutrophils (25). These FcγR proteins bind to different IgG subclasses and regulate immunity by causing cell activation or inhibition depending on the receptors engaged (16). The mechanisms for Fc-mediated activities include antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), antibody-dependent cell-mediated virus inhibition (ADCVI), antibody-dependent complement deposition (ADCD), aggregation, and immune activation (26–28). They also induce cytokine production via their immune activating or inhibitory motifs (22). Thus FcγR polymorphisms may influence diverse effector functions such as cytotoxicity, phagocytosis, cytokine production, antigen presentation and degranulation and may contribute to the outcome of infections (29). For instance, in encapsulated bacterial infections, FcγRIIa-131H may be involved in efficient clearance of IgG2-coated particles since it has a higher binding affinity for IgG2 (30–32). Conversely, homozygous FcγRIIa-131R genotype has been associated with severe forms of encapsulated bacterial infection (30, 31, 33, 34).
The allelic differences that affect FcγR function are also important in host immune mechanisms against viral infections. FcγRIIa-131H/H homozygous infants were found to be more susceptible to perinatal HIV transmission (35) but in other studies no associations between FcγRIIa genotypes and HIV infection rates were noted (36), suggesting further investigations are needed. In HIV-1 patients, FcγRIIIa-V176F genotype is associated with the development of Kaposi’s sarcoma and cryptococcal disease (12, 37, 38). Other studies have implicated the FcγRIIa-R131 allele in Dengue (39–41), and other viral infections (34, 42, 43). FcγRIIb inhibits activation signals from activating FcγRs (18) and the FcγRIIb-232T allele elicits reduced inhibitory signaling and has been associated with inflammatory diseases (44), however, it has not been extensively studied in HIV (45). These studies show that different FcγR polymorphisms influence effector functions in diverse ways and subsequently impacts on infection outcomes differently (Table 1).
FcγR Polymorphisms and the Risk of HIV-1 Infection With and Without Vaccines
Studies have not provided conclusive data on the association between FcγR polymorphisms and HIV-1 infection (56). For instance, contrasting results were reported when FcγRIIa and FcγRIIIa were examined with the risk of perinatal HIV-1 infection among infants in Kenya (35, 48). Whereas Brouwer et al., found that infant FcγRIIa-H131H (rs1801274) genotype was associated with susceptibility to perinatal HIV-1 transmission, Milligan and colleagues observed no such association with both FcγRIIa and FcγRIIIa. Rather, they observed that maternal FcγRIIIa-V176F genotypes may lead to higher risk of mother to child transmission compared to the homozygous (V/V or F/F) genotype carriers. The differences in these results could be attributed to differences in the cohort design as well as statistical rigor (48). More work is needed to define the role of these genotypes in mother to child HIV-1 transmission. Such a study may involve using harmonized protocols in a multi-center recruitment of a reasonably large number of HIV-1 infected pregnant women, determining their FcγR genotypes and monitoring their viral load throughout the pregnancy till birth. The HIV-1 infection status and FcγR genotypes of the child could then be determined to assess the genotype association with mother to child HIV-1 transmission.
A study that used samples from the European Multicenter AIDS Cohort Study (MACS), found an association between FcγRIIa-131R/R genotype and a faster rate of CD4+ T cell decline and disease progression compared to individuals with the R131H or H131H genotypes (12). This may be due to the weaker binding of the 131R/R receptor to IgG2 and IgG3 immune complexes. In-vitro experiments have shown that monocytes bearing this receptor do not efficiently internalize HIV-1 complexes, compared to 131H/H receptors (12, 58). Furthermore, the expression of FcγRIIa on immune cells leads to the activation and production of proinflammatory cytokines, an indication that FcγRIIa-mediated T-cell activation would be more efficient in individuals carrying the FcγRIIa-H/H genotype (59, 60). This suggests that FcγRIIa polymorphism may also indirectly influence CD4+ T cell function, and subsequent disease progression through its effect on immune complex internalization by monocytes and dendritic cells leading to their activation (59, 60).
Surprisingly, the 131R/R genotype was found to be associated with decreased AIDS-induced pneumonias compared to the 131H/H genotype in the MACS cohort. Given that FcγRIIa also binds to C-reactive proteins (61), it is possible that during bacterial infections, carriers of the 131R/R genotype may have higher levels of CRP to opsonize IgG2-coated microbes and activate the complement receptors to clear infection (62).
A recent study showed that HIV-1 patients with homozygous 176V for the FcγRIIIa-V176F (rs396991) polymorphisms and/or Y158H (rs396716) genotypes have higher HIV-1 specific ADCC response (49). It was hypothesized that the V176F polymorphism improves the binding capacity between the Fc receptor and anti-HIV-1 antibody, indicating that the FcγRIIIa receptor expressed on NK cells induced strong ADCC response for viral clearance (63, 64). When Geraghty et al. examined the role of FcγRIIa-(rs1801274) and FcγRIIIa-(rs396991) polymorphisms in 6300 HIV+ adults with European ancestry from a previous GWAS (65), they did not find any association between these polymorphisms and HIV acquisition (56). Furthermore, in the largest study of its kind in Africa, Connolly and colleagues also found no association between FcγRIIa-H131R (rs1801274) and FcγRIIIa-V176F (rs396991) variants and HIV infection or CD4+ T-cell decline in two cohorts in Rwanda and Zambia (50).
Accumulating evidence suggest that FcγR polymorphisms may play a key role in HIV-1 acquisition when vaccines are involved (13, 36, 54, 66–68). In the recombinant HIV-1 glycoprotein (gp120) Vax004 vaccine trial in the USA, those homozygous for the FcγRIIIa V176 allele in the lowest behavioral risk group were more likely to acquire HIV compared to individuals carrying the FcγRIIIa 176F or V176F genotypes in the same behavioral risk group (36). However, in that same study, no FcγRIIa-H131R (rs1801274) genotype was associated with increased risk of HIV among vaccine and placebo recipients. Furthermore, in the RV144 trial in Thailand, a SNP in FCGR2C (126C>T, rs114945036) was associated with vaccine efficacy. The study found an 11 to 15% efficacy in CC subjects compared to 64 to 91% in CT/TT subjects with the HIV subtype CRF01_AE 169K HIV-1 (13). On the contrary, in the HVTN 505 vaccine trial, the effect modification of this SNP with respect to vaccine efficacy was not significant. However, other four SNPs; (FCGR2C-exon06-441-C/T, rs138747765, FCGR2C-intron06-590-G/A, rs78603008, FCGR2C-intron15-403-C/T, rs373013207 and FCGR2C-intron15-433-G/A, rs201984478), were associated with increased risk of acquiring HIV in those vaccinated compared to the placebo group (54). These studies provide evidence that FcγR polymorphisms impact HIV-1 infection outcomes differently in vaccine recipients. The affinity of different IgG isotypes and the expression pattern of these FcγR on effector cells could account for these differences. For instance, when neutralizing antibodies induced during HIV-1 vaccination (69, 70) are ineffective against the virus, the antibody-virus complexes formed could lead to antibody dependent enhancement, resulting in virus spread among cells expressing these receptors (71–73).
It is also possible that vaccine types and their modes of delivery could trigger different FcγR responses. For instance, the RV144 vaccine consists of recombinant canarypox vector containing HIV antigens and recombinant gp120 administered in a prime-boosted regimen, while the HVTN 505 vaccine was made of a recombinant adenovirus serotype 5 vector boost (DNA/rAd5). The HVTN 505 vaccination increased the risk of HIV-1 acquisition. In addition, there were additional FcγR SNPs in HVTN 505 (FCGR3B SNPs) that modified vaccine effect in relation to HIV-1 acquisition compared to the RV144 vaccine recipients who had no such SNP associations (54). Furthermore, some minor SNPs identified on FCGR2C intron 6 were found to be in high linkage disequilibrium with others. This SNP may have selectively modulated the expression of FcγR effector functions differently in the two vaccines mentioned above (54). The FCGR2C polymorphisms, which was found in Thai RV144 vaccinees (13), was absent in Africans. This has implications for determining vaccine responses in other populations, if these FCGR2C polymorphisms are used as a proxy for FcγRIIc expression (45). Hence, it is possible that ethnic group, vaccine types and methods for delivering the vaccines may all trigger differences in FcγR dependent responses.
It has been proposed that during vaccination, FcγR polymorphism influence HIV-1 acquisition through their effect on the innate immune response. During HIV-1 infection, plasmacytoid dendritic cells produce high levels of interferon, which restricts the replication of virus (74). However, when viral particles are opsonized, there is suppression of type I and III interferons that are produced via FcγR-mediated mechanisms. This results in the lowering of the interferon levels required to block infection (54, 75).
FcγR polymorphisms also determine the affinity to the Fc region of IgG and alter its functionality after vaccination. For instance, in the HVTN 505 vaccine trial, differences in FcγRIIa binding and gp140-specific ADCP activity among the vaccine and control groups occurred in only one genotype of the FcγRIIa and FcγRIIb SNPs (54). This suggests that FcγR polymorphisms influenced the variation observed in the Fc region of the IgG induced after HVTN 505 vaccination (54). Also, studies have shown variation in Fc glycosylation of HIV-specific antibodies in HIV-1 infected patients and vaccine recipients, an indication that it regulates antibody and FcγR interaction (76, 77). Although these findings suggest possible mechanisms associated with increased risk of HIV-1infection in HVTN-505 vaccine among individuals carrying certain FcγRIIc genotypes, the same effect could not be established among recipients of the RV144 vaccine. These studies underscore the need to consider FcγR polymorphisms in HIV-1 vaccine trials, since they regulate vaccine-induced immunity, which impacts on HIV infection outcomes (36, 54).
FcγR Polymorphisms, HIV Disease Progression and ART Outcomes
Several studies have evaluated FcγR polymorphism and infection progression in European populations, however, studies on FcγR polymorphisms and ART outcomes are lacking. FcγR polymorphisms have been shown to affect the viral reservoir size in acute HIV-1 patients who were put on early ART (78, 79).
In a meta-analysis of several genome-wide association studies comprising 7,266 patients in the International Collaboration for the Genomics of HIV (ICGH), no association between FcγRIIa (rs1801274) and FcγRIIIa (rs396991) polymorphisms and viral set point was found [reviewed in (56)]. Similarly, in a sub-analysis of 467 long-term non-progressors and 517 rapid progressors, there was no association between FcγRIIa polymorphisms and HIV-1 disease progression. Finally, in the same meta-analysis, FcγRIIIa polymorphisms were not associated with HIV-1 disease progression in the Swiss HIV Cohort Study (SHCS) (56). Although another study found homozygous FcγRIIIa-176V/V to be highly prevalent in HIV progressors on ART compared to untreated natural viral suppressors, they could not conclude on this polymorphism’s association with viral set point (47). This is probably because the use of ART for HIV progressors in this study influenced their viral load, hence analysis with this polymorphism with respect to viral load could not be substantiated. Similarly, a study conducted in 253 Kenyan women to evaluate the impact of FcγRIIa-131H/R genotypes and FcγRIIIa-176F/V polymorphisms on HIV-1 disease progression could not find an association between these polymorphisms with viral load set point, decrease in CD4 count or increase in viral load (46).
The studies above agree that FcγRIIa and FcγRIIIa genotypes have no effect on viral load set point, but the results on disease progression differ. The divergent results on disease progression could be due to sample size, different study populations (i.e., men versus women), clinical definitions and the rigorousness of statistical methods employed. Additionally, the differences in these genotypic profiles among the different study populations and association with infection progression may be due to other factors such as viral type, host immunity and genetics. Future studies investigating the role of FcγRs in HIV-1 disease progression should comprehensively address potential population substructure, and longitudinally, pre-existing neutralizing antibodies and genetic variability in the virus to ascertain how these variables influence the outcome. Though haplotype analyses within and between the FcγR genes may offer crucial information on why these differences exist, such studies have been few (11, 80). Differences within the FCGR locus for different populations have been established (81–85). For instance, FcγRIIIa-176V was found to be underrepresented in Kenyan population (23.7%) compared to Europeans and Dutch Caucasians, whiles FcγRIIa-131H and FcγRIIc were highly prevalent in Asians compared with Caucasians (45). In addition, the distribution of FcγRIIIb-HNA1a/HNA1b allotypes were different among different populations, however, the FcγRIIIb-HNA1a and FcγRIIb- 232T variants were highly prevalent in black South Africans compared with Caucasians (45). Different effector functions have been observed in these populations with respect to enhanced cell activation and neutrophil-mediated phagocytosis as a result of these polymorphisms (45). Furthermore, gene copy number variations (CNVs) in FcγRIIc, FcγRIIIa and FcγRIIIb were shown to play a key role in association with HIV-1 infection and ART outcomes (45, 80).
Most of the studies on FcγR polymorphisms were conducted in Europe, USA, Asia and a few in South Africa. Data on the impact of FcγR polymorphisms and CNV on HIV-1 infection are limited in most African populations (12, 13, 45). One study examining the effect of FcγRIIc, FcγRIIIa and FcγRIIIb CNV in Ethiopian and Tanzanian cohorts found no effect on immune reconstitution post ART (80). Thus, supporting the limited role of poorly neutralizing or non-neutralizing antibodies in HIV-1 control (67, 86). This was contrary to the hypotheses that different FcγRs play a key role in mediating a balance between activating and inhibitory functions, IgG binding affinity to receptors and antibody mediated responses in HIV-1 infection progression. These findings in the Tanzanian and Ethiopian cohorts could also be due to epistatic interactions between these FcγR variants and IgG affinity, that will mediate HIV pathogenesis as evidenced in KIR/HLA variants (87).
Importantly, data is lacking on FcγR influence on HIV-1 disease progression and ART responses in African populations, though the continent bears the brunt of the epidemic. Therefore, large studies conducted in populations with African ancestry are needed. This will provide increased power to detect population specific genetic variations in association with disease progression, when combined with data from European populations (11, 88).
FcγR Polymorphisms and Viral Reservoir
Though ART has been used successfully in the management of HIV-1 infection, replication-competent viruses persist as latent reservoirs after long term ART usage (89). Even HIV-1 elite controllers and long term non-progressors, harbor viral reservoirs (9, 10). The extent of viral clearance in HIV-1 infection, will be dependent on both an efficient HIV-1 specific immune response and a very low reservoir size (90). To achieve an HIV cure or remission, the viral reservoir must be eliminated or reduced to a minimum, since the size of the viral reservoir has been shown to be a marker for disease progression and clinical outcomes (91, 92). Therefore, reduction of the reservoir can be used as a criterion for ART interruption in HIV-1 cure studies (92). The impact of FcγRs on ART outcomes needs further evaluation, since it has been hypothesized that some polymorphisms affect the viral reservoir size in patients with acute HIV-1 infection initiated on early ART (78, 79). However, such studies analyzing FcγRs polymorphisms and HIV reservoir size are currently lacking.
There is now increased focus on identifying markers for these latent reservoirs to help in the HIV-1 cure efforts. Receptors expressed on infected cells such as CD30 and CD32 (FcγRIIa) and some immune checkpoint inhibiting molecules on the surfaces of infected cells have been identified as potential markers for latently infected cells (51, 52, 93). Recently, CD32a (FcγRIIa) was identified as a marker of latently infected CD4 T cells. In this study CD4+ T cells expressing CD32a+ molecules were observed to be highly enriched in HIV-1 DNA and contained replication competent proviruses compared to the CD32a- CD4+ T cells (51). However, these findings were not replicated in other studies. Some investigators reported lower enrichment for HIV-1 DNA in cells expressing CD32 in certain individuals (52, 93). Others could not verify that CD32a is expressed on latent reservoirs, nor enriched in CD4+ T cells carrying viral DNA (94–96). A study that used 10 chronic HIV-1 patients on ART also observed no enrichment for HIV-1 DNA in CD32+ CD4 T cells (95). Some investigators proposed that the CD32+ cells may have been derived from adherent non-T cells and other cell conjugates (T-B cell conjugates) expressing this marker (95). Another study proposed that CD32 (FcγRII) is not a specific biomarker for most CD4 T cell populations because they found a greater number of HIV-1 latent reservoirs occurred in CD3+CD4+ CD32- T cells using a quantitative viral outgrowth assay (qVOA) (97).
Some studies have suggested that the inconsistencies in these findings could be attributed to cell sorting techniques that was not able to isolate pure CD4 T cells that express only CD32+ marker, but rather T-B cell doublets (96, 98). In addition, it has been shown that there is a higher proportion of antigen presenting cells (APC) expressing CD32+ cells than what is expressed on CD4+ T cells. Therefore, high number of residual non-T cells found in sorted CD32+ cell population, can bias these findings (93, 95, 99, 100). The cell sorting challenges seem to have been overcome by Darcis and colleagues using an improved isolation and purification technique. They performed two rounds of CD4+ T cell negative selection by magnetic cell sorting before CD32 isolation. Using this method, they observed an increased HIV-1 DNA enrichment in the CD32+ CD4+ T cells (100) as seen in the original study by Descours et al.
Further studies that replicate the sequential cell sorting techniques used by Darcis and colleagues are needed (99) to firmly establish FcγRIIa as bona fide marker of latently infected CD4+ cells. After that, several questions still remain: Is FcγRIIa just a marker of these latently infected cells or does it have functional consequences on the size or reactivation capacity of the reservoir? Do polymorphisms and CNVs in FcγRIIa and other Fc receptors determine the size of the reservoir in individuals treated with ART? Does it matter if the persons are treated early during HIV infection? What about the tissue reservoirs, do they also express FcγRIIa and if so, do polymorphisms and CNV matter in that context? Investigators have started to address some of these important questions. Some have hypothesized that the differences in reservoir size in people who started early on ART may be due to polymorphisms in FcγRIIa (78, 101), since there has been previous association of this gene with HIV-1 disease pathogenesis. However, this hypothesis needs to be tested. Notably, these reservoirs are diverse in nature, occurring in various infected cells and tissues, and thus may enhance the long-term persistence of replication competent viruses (99, 102, 103). Also, since there is some evidence that CD32 (FcγRIIa) marks transcriptionally active HIV-1 infected cells, they could be used to identify persistent HIV-1 infected CD4 T cells that may contribute to viral persistence during antiretroviral treatments (104, 105). The co-expression of CD32a (FcγRIIa) with other markers such as PD-1 in lymph node CD4+ T cells (105) as well as CD32a occurring with CD30 in CD4+ T cell tissues (104), shows these reservoirs are heterogenous in nature.
In a previous study of HIV controllers and non-controllers, epistatic interactions between of genes (GM on chromosome 14) encoding variability in the Fc portion of IgG and FcγR genes (on chromosome 1) were reported to influence the control of HIV-1 viral replication in Caucasian Americans (106). Homozygous FcγRIIa-H individuals who were also GM21 non carriers (homozygous GM5) were more likely to be HIV controllers than GM21 carriers. A similar interaction between GM determinants and FcγRIIIa alleles on HIV control albeit mush weaker compared to that with FcγRIIa. Interestingly, these epistatic interactions between FcγR and GM genes were not observed in the African-American population in the same study highlighting population specific effects of these genes in HIV control (106). In a more recent study, the FcγRIIa AA genotype (rs10800309) was found to increase FcγRIIa expression on myeloid cells and was associated with HIV-1 control independent of HLA-B57 and HLA-B27 (107), which are well established markers involved in controlling viral load and long-term non-progression of HIV infection (108, 109). Taken together, these studies underscore the importance of studies assessing the role of FcγR polymorphisms in HIV to be designed to adequately address inter-ethnic differences in these genes and the potential impact of genetic variability affecting not only the receptor-ligand interphase but also receptor expression levels.
FcγRs, Broadly Neutralizing Antibodies and HIV-1 Remission Attempts
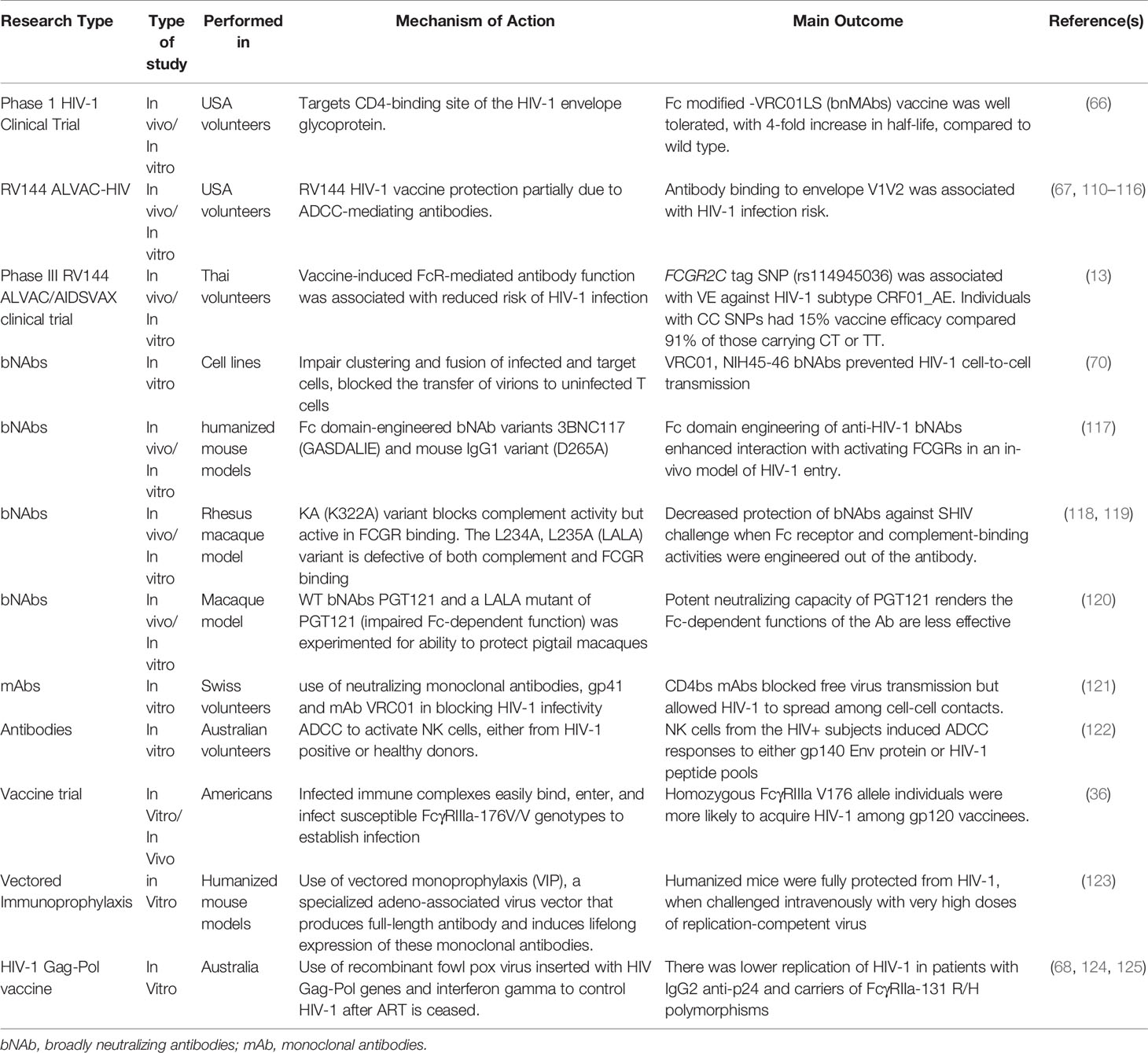
Investigators are exploring different approaches to achieve a remission or cure for HIV (Table 2). First, there are strategies that seek to boost the host immune system using vaccines that produce broadly neutralizing antibodies (bNAbs) to suppress the virus, or chimeric antigen receptors to engage and destroy the latently infected cells. Second, genetic methods like the CRISPR/Cas9 gene editing technology to excise or mutate the integrated virus or render patients resistant to HIV by taking their CD4+ T cells, mutating essential receptors such as CCR5 and reinfusing them back into the patient (126–128). Third, and probably the most studied of the HIV cure methods is the shock and kill approach which seeks to use compounds to reactivate the latent virus with the hope that infected CD+ T cells will die by viral cytopathic effects or immune clearance (129–132). The use of bNAbs, chimeric antigen receptors, and the ‘kill’ part of the shock and kill approach may all involve the engagement of Fc gamma receptors. FcγR binding seems to be a key requirement for bNAbs to efficiently protect from infection, viral suppression, and clearance. Studies conducted in animal models observed a decrease in broadly neutralizing antibody activity from simian-human immunodeficiency virus (SHIV-1) challenge when the FcγR and complement activities were disabled (118). Similarly, bNAbs enhanced binding capacity for activating FcγRs FcγRIIa and FcγRIIIa, which were demonstrated to be protective in humanized murine models (133).
In contrast, though both the WT PGT121 and LALA PGT121 bNAbs administered to macaques, conferred protection against cell associated SHIV SF162P3 challenge, when tested in vitro, the Fc-dependent function was greatly reduced. Although, these two bNAbs have been shown to be associated with reduced plasma viremia both in macaques and humanized mice models (133, 134), indication that the protection conferred by PGT121, may not be dependent on Fc-mediated NK cells responses (120, 135, 136). It has been suggested that the mechanism by which these bNAbs neutralize both cell-free virus and cell-associated virus and confer protection could be through the fragment antigen-binding (Fab) region, as well as epitope specificity of these antibodies (120). Additional studies on other bNAbs and other modified bNAbs in different animal models are needed to validate these results (120). These findings have emphasized the differences in the formulation of bNAbs, and how it affects their function in HIV control. Delineating what influences bNAb function either more through an Fc-mediated mechanism or otherwise would be very important in HIV cure research aimed at using bNAbs as immune-therapeutic tool in effective control and management of HIV.
During HIV-1 infections, broadly neutralizing antibodies (bNAbs) coordinate with Fc receptors to activate effector cells to clear the virus through mechanisms such as ADCC (21, 67, 122). In addition, there is evidence that neutralizing antibodies block cell-free virus through a mechanism that prevents cell-cell transmission. For instance, certain classes of CD4 binding site inhibitors (CD4b) such as mAb VRC01 only block free virus but has low activity during cell to cell transmission (121). Other mAbs such as membrane-proximal external region (MPER), CD4bs and anti-coreceptor agents are involved in blocking free virus transmission (137, 138). Subsequent studies have shown that mAb VRC01 may engage FcγRs and prevent cell to cell HIV-1 transmission and also reduce the half-lives of infected cells and free virions (70, 121). While a potent HIV-1 vaccine must block cell free virus and cell-cell virus transmission (121), using two or more bNAbs that bind different epitopes in combination could enhance efficacy and offer broader protection against HIV-1 infections. This was demonstrated when the combination of CD4bs and VRC07 resulted in the neutralization of about 98% viruses in one study (139). This notwithstanding, the role of antibodies in cell-cell HIV inhibition is divergent, since these findings were based on experiments conducted with several viral strains and antibodies, different donor and target cell types, as well as various HIV-1 transmission models (140). Hence some studies have reported similar HIV-1 inhibition for cell-free virus compared to cell-to-cell transmission (141–143), while others observed a decreased antibody effect in cell-to-cell transmission compared to cell free virus inhibition (70, 121, 144). These controversies appear to have been resolved in comparative studies that obtained similar results when experimental designs were normalized with respect to target cells used, and antibody inhibition activities (141–143, 145). Thus, normalization of these factors must be done in terms of the type and quantity of virus and cell types using standardized assays to establish the role of these antibodies in cell free or cell-to-cell HIV transmission (140).
FcγR differences could also affect the efficacy of bNAbs as HIV-1 prevention agents. In animal studies, bNAbs confer protective immunity against HIV-1 or SHIV-1 challenge in humanized mouse models (Hessell et al., 2007, Balazs et al., 2012, Lu et al., 2016). Recently, Simone and colleagues observed that among a cohort of 23 individuals, 13 developed broadly neutralizing antibodies to HIV-1, which was significantly associated with antibody binding to FcγRIIa. These antibodies were associated with higher Fc polyfunctionality early in the course of infection (146). Furthermore, FcγRIIIa-V176F (rs396991) and FcγRIIIa-Y158H (rs396716) polymorphism are associated with enhanced ADCC in HIV-1 patients (49), giving the indication that the expression of FcγRIIIa receptor on NK cells led to the induction of strong ADCC response for viral clearance (63, 64). These studies suggest that bNAbs have potential as therapeutic or prophylactic treatment in humans. It is becoming clearer that if bNAbs are to become successful in eliminating the reservoir, FcγR-mediated actions will play a pivotal role. It is therefore imperative to determine at this early stage if polymorphisms and CNVs affect how bNAbs can suppress or eliminate HIV-1 in different populations. This is especially crucial since inter-ethnic variations have been reported for polymorphisms in FcγRIIa that influences its affinity for IgG binding (45, 147, 148).
Perspectives and Conclusion
Although several studies have evaluated the impact of FcγR polymorphisms on HIV progression, they do not appear to play a major role in viral load set point or natural control of the virus. However, when it comes to HIV vaccine responses and remission attempts, FcγR polymorphisms and/or CNVs may play a variety of roles. First, if FcγRIIa (CD32) is confirmed as a true marker of the latent reservoir, it will raise many crucial questions as enumerated above. One such critical question is whether FcγR polymorphisms determine the size of the viral reservoir. Second, the use of bNAbs either for reservoir elimination or long-term suppression may depend on engagement with FcγRs. It is important to determine if expression of different variants of FcγRs determine why these antibodies are successful in some persons but not others. Also, it will be significant to determine if FcγRs play a role in the development of resistance or immune tolerance to these antibodies. Third, FcγRs may need to be engaged in cure methods such as the ‘shock and kill’ approach, which seeks to reactivate the reservoir for clearance by the immune system. The ‘kill’ portion of this approach may involve interventions such as bNAbs or chimeric antigen receptors. The role of FcγRs in mediating successful killing of T cells that have been reactivated also need to be investigated especially after ART initiation as well as the synergistic effect between FcγRs and ART outcomes need to be evaluated. Finally, the responses to most HIV-1 vaccines are likely to be dependent on FcγRs as the few unsuccessful vaccine trials have shown. Therefore, investigations into FcγR variations in different populations offer an important area of inquiry in the HIV cure research era.
Author Contributions
HL, BA, and GK designed the review and the concepts. EB assisted in the literature search and manuscript writing. All authors wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project is part of the EDCTP2 program supported by the European Union with grant numbers TMA2017SF-1955 senior fellowship to GK and TMA2018PF-2535 preparatory fellowship to HL. The funder had no role in the design, analysis, or publication of the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ART, Anti-Retroviral Therapy; ADCC, Antibody-Dependent Cellular Cytotoxicity; ADCD, Antibody-Dependent Complement Deposition; ADCP, Antibody-Dependent Cellular Phagocytosis; ADCVI, Antibody-Dependent Cell-mediated Virus Inhibition; bNAbs, Broadly neutralizing antibody; CD4 T cells, Cluster of Differentiation 4 T cells; CNVs, Copy Number Variations; DC’s, Dendritic Cells; EC, Extracellular Domain; FcγR, Fc gamma receptors; FCGR, Fc gamma receptor gene; GPI, Glycosylphosphatidylinositol-anchored; HLA, Human Leukocyte Antigen; HIV, Human Immunodeficiency Virus; ITAM, Immunoreceptor Tyrosine-based Activation Motif; ITIM, Immunoreceptor Tyrosine-based Inhibitory Motif; IgG, Immunoglobulin; IC, Intracellular Domain; NK, Natural Killer Cells; SNPs, Single Nucleotide Polymorphisms; TM, Transmembrane Domain.
References
1. UNAIDS. Global HIV & AIDS Starictics -2019 Fact Sheet (2019). Available at: https://www.unaids.org/en/resources/fact-sheet.
2. Okulicz JF, Marconi VC, Landrum ML, Wegner S, Weintrob A, Ganesan A, et al. Clinical Outcomes of Elite Controllers, Viremic Controllers, and Long-Term Nonprogressors in the US Department of Defense HIV Natural History Study. J Infect Dis (2009) 200(11):1714–23. doi: 10.1086/646609
3. Grabar S, Selinger-Leneman H, Abgrall S, Pialoux G, Weiss L, Costagliola D. Prevalence and Comparative Characteristics of Long-Term Nonprogressors and HIV Controller Patients in the French Hospital Database on HIV. Aids (2009) 23(9):1163–9. doi: 10.1097/QAD.0b013e32832b44c8
4. Diop G, Hirtzig T, Do H, Coulonges C, Vasilescu A, Labib T, et al. Exhaustive Genotyping of the Interferon Alpha Receptor 1 (IFNAR1) Gene and Association of an IFNAR1 Protein Variant With AIDS Progression or Susceptibility to HIV-1 Infection in a French AIDS Cohort. Biomed Pharmacother (2006) 60(9):569–77. doi: 10.1016/j.biopha.2006.08.002
5. Casado C, Colombo S, Rauch A, Martínez R, Günthard HF, Garcia S, et al. Host and Viral Genetic Correlates of Clinical Definitions of HIV-1 Disease Progression. PLoS One (2010) 5(6). doi: 10.1371/journal.pone.0011079
6. Fontaine J, Coutlée F, Tremblay C, Routy J-P, Poudrier J, Infection MPH, et al. HIV Infection Affects Blood Myeloid Dendritic Cells After Successful Therapy and Despite Nonprogressing Clinical Disease. J Infect Dis (2009) 199(7):1007–18. doi: 10.1086/597278
7. Okulicz JF, Lambotte O. Epidemiology and Clinical Characteristics of Elite Controllers. Curr Opin HIV AIDS (2011) 6(3):163–8. doi: 10.1097/COH.0b013e328344f35e
8. Tiemessen CT, Martinson N. Elite Controllers: Understanding Natural Suppressive Control of HIV-1 Infection. Continuing Med Education (2012) 30(8):282–5.
9. Graf EH, Mexas AM, Jianqing JY, Shaheen F, Liszewski MK, Di Mascio M, et al. Elite Suppressors Harbor Low Levels of Integrated HIV DNA and High Levels of 2-LTR Circular HIV DNA Compared to HIV+ Patients on and Off HAART. PLoS Pathogens (2011) 7(2). doi: 10.1371/journal.ppat.1001300
10. Hocqueloux L, Prazuck T, Avettand-Fenoel V, Lafeuillade A, Cardon B, Viard J-P, et al. Long-Term Immunovirologic Control Following Antiretroviral Therapy Interruption in Patients Treated At the Time of Primary HIV-1 Infection. Aids (2010) 24(10):1598–601. doi: 10.1097/QAD.0b013e32833b61ba
11. Lassaunière R, Musekiwa A, Gray GE, Kuhn L, Tiemessen CT. Perinatal HIV-1 Transmission: Fc Gamma Receptor Variability Associates With Maternal Infectiousness and Infant Susceptibility. Retrovirology (2016) 13(1):40. doi: 10.1186/s12977-016-0272-y
12. Forthal DN, Landucci G, Bream J, Jacobson LP, Phan TB, Montoya B. Fcγriia Genotype Predicts Progression of HIV Infection. J Immunol (2007) 179(11):7916–23. doi: 10.4049/jimmunol.179.11.7916
13. Li SS, Gilbert PB, Tomaras GD, Kijak G, Ferrari G, Thomas R, et al. FCGR2C Polymorphisms Associate With HIV-1 Vaccine Protection in RV144 Trial. J Clin Invest (2014) 124(9):3879–90. doi: 10.1172/JCI75539
14. Rosales C, Uribe-Querol E. Antibody-Fc Receptor Interactions in Antimicrobial Functions. Curr Immunol Rev (2013) 9(1):44–55. doi: 10.2174/1573395511309010006
15. Nimmerjahn F, Ravetch JV. Fcγrs in Health and Disease. Curr Topics Microbiol Immunol (2011) 350:105–25. doi: 10.1007/82_2010_86
16. Bournazos S, Ravetch JV. Anti-Retroviral Antibody Fcγr-Mediated Effector Functions. Immunol Rev (2017) 275(1):285–95. doi: 10.1111/imr.12482
17. Boesch AW, Brown EP, Ackerman ME. The Role of Fc Receptors in HIV Prevention and Therapy. Immunol Rev (2015) 268(1):296–310. doi: 10.1111/imr.12339
18. Boross P, van de Poel K, Van de Winkel J, Leusen J. Fc Receptors. In: . Encyclopedia of Life Sciences (Els). Chichester: John Wiley & Sons, Ltd (2008). p. 1–8. doi: 10.1002/9780470015902.a0000916.pub2
19. Maresco DL, Chang E, Theil KS, Francke U, Anderson CL. The Three Genes of the Human FCGR1 Gene Family Encoding Fc Gamma RI Flank the Centromere of Chromosome 1 At 1p12 and 1q21. Cytogen Cell Genet (1996) 73(3):157–63. doi: 10.1159/000134330
20. Hoffmeyer F, Witte K, Schmidt R. The High-Affinity Fcγri on PMN: Regulation of Expression and Signal Transduction. Immunology (1997) 92(4):544–52. doi: 10.1046/j.1365-2567.1997.00381.x
21. Nimmerjahn F, Ravetch JV. Fcγ Receptors as Regulators of Immune Responses. Nat Rev Immunol (2008) 8(1):34–47. doi: 10.1038/nri2206
22. Li X, Ptacek TS, Brown EE, Edberg JC. Fcgamma Receptors: Structure, Function and Role as Genetic Risk Factors in SLE. Genes Immun (2009) 10(5):380–9. doi: 10.1038/gene.2009.35
23. Ernst LK, Metes D, Herberman RB, Morel PA. Allelic Polymorphisms in the Fcγriic Gene can Influence its Function on Normal Human Natural Killer Cells. J Mol Med (2002) 80(4):248–57. doi: 10.1007/s00109-001-0294-2
24. Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, et al. Copy Number Variation of the Activating FCGR2C Gene Predisposes to Idiopathic Thrombocytopenic Purpura. Blood (2008) 111(3):1029–38. doi: 10.1182/blood-2007-03-079913
25. Simmons D, Seed B. The Fcγ Receptor of Natural Killer Cells is a Phospholipid-Linked Membrane Protein. Nature (1988) 333(6173):568–70. doi: 10.1038/333568a0
26. Mayr L, Su B, Moog C. Role of Nonneutralizing Antibodies in Vaccines and/or HIV Infected Individuals. Curr Opin HIV AIDS (2017) 12(3):209–15. doi: 10.1097/COH.0000000000000357
27. Chung AW, Alter G. Systems Serology: Profiling Vaccine Induced Humoral Immunity Against HIV. Retrovirology (2017) 14(1):57. doi: 10.1186/s12977-017-0380-3
28. Forthal DN, Moog C. Fc Receptor-Mediated Antiviral Antibodies. Curr Opin HIV AIDS (2009) 4(5):388. doi: 10.1097/COH.0b013e32832f0a89
29. Li X, Gibson AW, Kimberly RP. Human FcR Polymorphism and Disease. In: Daeron M, Nimmerjahn F, editors. Fc Receptors. Cham: Springer International Publishing (2014). p. 275–302. doi: 10.1007/978-3-319-07911-0_13
30. Endeman H, Cornips MCA, Grutters JC, van den Bosch JM, Ruven HJ, van Velzen-Blad H, et al. The Fcγ Receptor IIA-R/R131 Genotype is Associated With Severe Sepsis in Community-Acquired Pneumonia. Clin Vaccine Immunol (2009) 16(7):1087–90. doi: 10.1128/CVI.00037-09
31. Platonov AE, Shipulin GA, Vershinina IV, Dankert J, van de Winkel JG, Kuijper EJ. Association of Human Fcγriia (CD32) Polymorphism With Susceptibility to and Severity of Meningococcal Disease. Clin Infect Dis (1998) 27(4):746–50. doi: 10.1086/514935
32. Jansen WT, Breukels MA, Snippe H, Sanders LA, Verheul AF, Rijkers GT. Fcγ Receptor Polymorphisms Determine the Magnitude of In Vitro Phagocytosis of Streptococcus Pneumoniae Mediated by Pneumococcal Conjugate Sera. J Infect Dis (1999) 180(3):888–91. doi: 10.1086/314920
33. Yee AM, Phan HM, Zuniga R, Salmon JE, Musher DM. Association Between Fcγriia-R131 Allotype and Bacteremic Pneumococcal Pneumonia. Clin Infect Dis (2000) 30(1):25–8. doi: 10.1086/313588
34. Yuan FF, Tanner J, Chan P, Biffin S, Dyer W, Geczy A, et al. Influence of Fcγriia and MBL Polymorphisms on Severe Acute Respiratory Syndrome. Tissue Antigens (2005) 66(4):291–6. doi: 10.1111/j.1399-0039.2005.00476.x
35. Brouwer KC, Lal RB, Mirel LB, Yang C, Eijk AM, Ayisi J, et al. Polymorphism of Fc Receptor IIa for IgG in Infants is Associated With Susceptibility to Perinatal HIV-1 Infection. Aids (2004) 18(8):1187–94. doi: 10.1097/00002030-200405210-00012
36. Forthal DN, Gabriel EE, Wang A, Landucci G, Phan TB. Association of Fcγ Receptor IIIa Genotype With the Rate of HIV Infection After gp120 Vaccination. Blood J Am Soc Hematol (2012) 120(14):2836–42. doi: 10.1182/blood-2012-05-431361
37. Lehrnbecher TL, Foster CB, Zhu S, Venzon D, Steinberg SM, Wyvill K, et al. Variant Genotypes of FcgammaRIIIA Influence the Development of Kaposi’s Sarcoma in HIV-infected Men. Blood (2000) 95(7):2386–90. doi: 10.1182/blood.V95.7.2386.007k04_2386_2390
38. Rohatgi S, Gohil S, Kuniholm MH, Schultz H, Dufaud C, Armour KL, et al. Fc Gamma Receptor 3A Polymorphism and Risk for HIV-associated Cryptococcal Disease. MBio (2013) 4(5). doi: 10.1128/mBio.00573-13
39. Moi ML, Lim C-K, Takasaki T, Kurane I. Involvement of the Fcγ Receptor IIA Cytoplasmic Domain in Antibody-Dependent Enhancement of Dengue Virus Infection. J Gen Virol (2010) 91(1):103–11. doi: 10.1099/vir.0.014829-0
40. García G, González N, Pérez AB, Sierra B, Aguirre E, Rizo D, et al. Long-Term Persistence of Clinical Symptoms in Dengue-Infected Persons and its Association With Immunological Disorders. Int J Infect Dis (2011) 15(1):e38–43. doi: 10.1016/j.ijid.2010.09.008
41. García G, Sierra B, Pérez AB, Aguirre E, Rosado I, Gonzalez N, et al. Asymptomatic Dengue Infection in a Cuban Population Confirms the Protective Role of the RR Variant of the Fcγriia Polymorphism. Am J Trop Med Hygiene (2010) 82(6):1153–6. doi: 10.4269/ajtmh.2010.09-0353
42. Diamantopoulos PT, Kalotychou V, Polonyfi K, Sofotasiou M, Anastasopoulou A, Galanopoulos A, et al. Correlation of Fc-γ RIIA Polymorphisms With Latent Epstein–Barr Virus Infection and Latent Membrane Protein 1 Expression in Patients With Low Grade B-cell Lymphomas. Leukemia Lymphoma (2013) 54(9):2030–4. doi: 10.3109/10428194.2012.762512
43. Zúñiga J, Buendía-Roldán I, Zhao Y, Jiménez L, Torres D, Romo J, et al. Genetic Variants Associated With Severe Pneumonia in A/H1N1 Influenza Infection. Eur Respir J (2012) 39(3):604–10. doi: 10.1183/09031936.00020611
44. Tarasenko T, Dean JA, Bolland S. FcgammaRIIB as a Modulator of Autoimmune Disease Susceptibility. Autoimmunity (2007) 40(6):409–17. doi: 10.1080/08916930701464665
45. Lassauniere R, Tiemessen C. Variability At the FCGR Locus: Characterization in Black South Africans and Evidence for Ethnic Variation in and Out of Africa. Genes Immun (2016) 17(2):93. doi: 10.1038/gene.2015.60
46. Weis JF, McClelland RS, Jaoko W, Mandaliya KN, Overbaugh J, Graham SM. Fc Gamma Receptors IIa and IIIa Genetic Polymorphisms do Not Predict HIV-1 Disease Progression in Kenyan Women. AIDS Res Hum Retroviruses (2015) 31(3):288–92. doi: 10.1089/aid.2014.0209
47. Poonia B, Kijak GH, Pauza CD. High Affinity Allele for the Gene of FCGR3A is Risk Factor for HIV Infection and Progression. PLoS One (2010) 5(12). doi: 10.1371/journal.pone.0015562
48. Milligan C, Richardson BA, John-Stewart G, Nduati R, Overbaugh J. FCGR2A and FCGR3A Genotypes in Human Immunodeficiency Virus Mother-to-Child Transmission. Open Forum Infect Dis (2015) 2(4). doi: 10.1093/ofid/ofv149
49. Talathi SP, Shaikh NN, Pandey SS, Saxena VA, Mamulwar MS, Thakar MR. Fcγriiia Receptor Polymorphism Influences NK Cell Mediated ADCC Activity Against HIV. BMC Infect Dis (2019) 19(1):1053. doi: 10.1186/s12879-019-4674-z
50. Connolly S, Wall KM, Tang J, Yu T, Kilembe W, Kijak G, et al. Fc-Gamma Receptor IIA and IIIA Variants in Two African Cohorts: Lack of Consistent Impact on Heterosexual HIV Acquisition, Viral Control, and Disease Progression. Virology (2018) 525:132–42. doi: 10.1016/j.virol.2018.09.015
51. Descours B, Petitjean G, López-Zaragoza J-L, Bruel T, Raffel R, Psomas C, et al. CD32a is a Marker of a CD4 T-Cell HIV Reservoir Harbouring Replication-Competent Proviruses. Nature (2017) 543(7646):564–7. doi: 10.1038/nature21710
52. Badia R, Ballana E, Castellví M, García-Vidal E, Pujantell M, Clotet B, et al. CD32 Expression is Associated to T-cell Activation and is Not a Marker of the HIV-1 Reservoir. Nat Commun (2018) 9(1):2739. doi: 10.1038/s41467-018-05157-w
53. Alvarez RA, Maestre AM, Law K, Durham ND, Barria MI, Ishii-Watabe A, et al. Enhanced FCGR2A and FCGR3A Signaling by HIV Viremic Controller Igg. JCI Insight (2017) 2(4):e88226–e. doi: 10.1172/jci.insight.88226
54. Li SS, Gilbert PB, Carpp LN, Pyo C-W, Janes H, Fong Y, et al. Fc Gamma Receptor Polymorphisms Modulated the Vaccine Effect on HIV-1 Risk in the HVTN 505 HIV Vaccine Trial. J Virol (2019) 93(21):e02041–18. doi: 10.1128/JVI.02041-18
55. Brouwer KC, Lal AA, Mirel LB, Otieno J, Ayisi J, Van Eijk AM, et al. Polymorphism of Fc Receptor IIa for Immunoglobulin G is Associated With Placental Malaria in HIV-1-Positive Women in Western Kenya. J Infect Dis (2004) 190(6):1192–8. doi: 10.1086/422850
56. Geraghty DE, Thorball CW, Fellay J, Thomas R. Effect of Fc Receptor Genetic Diversity on HIV-1 Disease Pathogenesis. Front Immunol (2019) 10(970). doi: 10.3389/fimmu.2019.00970
57. Hans VM, Mehta DS, Hans M. Association of Fc Gamma-Receptors IIa, Iiia, and IIIb Genetic Polymorphism With Susceptibility to Chronic Periodontitis in South Indian Population. Contemp Clin Dentistry (2015) 6(Suppl 1):S141–6. doi: 10.4103/0976-237X.166831
58. Parren P, Warmerdam P, Boeije L, Arts J, Westerdaal N, Vlug A, et al. On the Interaction of IgG Subclasses With the Low Affinity Fc Gamma RIIa (CD32) on Human Monocytes, Neutrophils, and Platelets. Analysis of a Functional Polymorphism to Human Igg2. J Clin Invest (1992) 90(4):1537–46. doi: 10.1172/JCI116022
59. Liu Y, Gao X, Masuda E, Redecha PB, Blank MC, Pricop L. Regulated Expression of Fcγr in Human Dendritic Cells Controls Cross-Presentation of Antigen-Antibody Complexes. J Immunol (2006) 177(12):8440–7. doi: 10.4049/jimmunol.177.12.8440
60. Prehn JL, Thomas LS, Landers CJ, Qi TY, Michelsen KS, Targan SR. The T Cell Costimulator TL1A is Induced by Fcγr Signaling in Human Monocytes and Dendritic Cells. J Immunol (2007) 178(7):4033–8. doi: 10.4049/jimmunol.178.7.4033
61. Stein M-P, Edberg JC, Kimberly RP, Mangan EK, Bharadwaj D, Mold C, et al. C-Reactive Protein Binding to Fcγriia on Human Monocytes and Neutrophils is Allele-Specific. J Clin Invest (2000) 105(3):369–76. doi: 10.1172/JCI7817
62. Weiser JN, Pan N, McGowan KL, Musher D, Martin A, Richards J. Phosphorylcholine on the Lipopolysaccharide of Haemophilus Influenzae Contributes to Persistence in the Respiratory Tract and Sensitivity to Serum Killing Mediated by C-reactive Protein. J Exp Med (1998) 187(4):631–40. doi: 10.1084/jem.187.4.631
63. Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased Natural Killer Cell Expression of CD16, Augmented Binding and ADCC Activity to Rituximab Among Individuals Expressing the Fc{gamma}RIIIa-158 V/V and V/F Polymorphism. Blood (2007) 110(7):2561–4. doi: 10.1182/blood-2007-01-070656
64. Oboshi W, Watanabe T, Matsuyama Y, Kobara A, Yukimasa N, Ueno I, et al. The Influence of NK Cell-Mediated ADCC: Structure and Expression of the CD16 Molecule Differ Among Fcγriiia-V158F Genotypes in Healthy Japanese Subjects. Hum Immunol (2016) 77(2):165–71. doi: 10.1016/j.humimm.2015.11.001
65. McLaren PJ, Coulonges C, Ripke S, Van Den Berg L, Buchbinder S, Carrington M, et al. Association Study of Common Genetic Variants and HIV-1 Acquisition in 6,300 Infected Cases and 7,200 Controls. PLoS Pathogens (2013) 9(7). doi: 10.1371/journal.ppat.1003515
66. Gaudinski MR, Coates EE, Houser KV, Chen GL, Yamshchikov G, Saunders JG, et al. Safety and Pharmacokinetics of the Fc-modified HIV-1 Human Monoclonal Antibody VRC01LS: A Phase 1 Open-Label Clinical Trial in Healthy Adults. PLoS Med (2018) 15(1):e1002493. doi: 10.1371/journal.pmed.1002493
67. Bonsignori M, Pollara J, Moody MA, Alpert MD, Chen X, Hwang K-K, et al. Antibody-Dependent Cellular Cytotoxicity-Mediating Antibodies From an HIV-1 Vaccine Efficacy Trial Target Multiple Epitopes and Preferentially Use the VH1 Gene Family. J Virol (2012) 86(21):11521–32. doi: 10.1128/JVI.01023-12
68. French MA, Tanaskovic S, Law MG, Lim A, Fernandez S, Ward LD, et al. Vaccine-Induced IgG2 anti-HIV p24 is Associated With Control of HIV in Patients With a ‘High-Affinity’ FcgammaRIIa Genotype. AIDS (London England) (2010) 24(13):1983–90. doi: 10.1097/QAD.0b013e32833c1ce0
69. Lu C-L, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, et al. Enhanced Clearance of HIV-1-infected Cells by Broadly Neutralizing Antibodies Against HIV-1 In Vivo. Science (New York NY) (2016) 352(6288):1001–4. doi: 10.1126/science.aaf1279
70. Malbec M, Porrot F, Rua R, Horwitz J, Klein F, Halper-Stromberg A, et al. Broadly Neutralizing Antibodies That Inhibit HIV-1 Cell to Cell Transmission. J Exp Med (2013) 210(13):2813–21. doi: 10.1084/jem.20131244
71. Williams WB, Liao H-X, Moody MA, Kepler TB, Alam SM, Gao F, et al. Diversion of HIV-1 Vaccine–Induced Immunity by gp41-microbiota Cross-Reactive Antibodies. Science (2015) 349(6249):aab1253. doi: 10.1126/science.aab1253
72. Takeda A, Tuazon C, Ennis F. Antibody-Enhanced Infection by HIV-1 Via Fc Receptor-Mediated Entry. Science (1988) 242(4878):580–3. doi: 10.1126/science.2972065
73. Homsy J, Meyer M, Tateno M, Clarkson S, Levy J. The Fc and Not CD4 Receptor Mediates Antibody Enhancement of HIV Infection in Human Cells. Science (1989) 244(4910):1357–60. doi: 10.1126/science.2786647
74. Doyle T, Goujon C, Malim MH. HIV-1 and Interferons: Who’s Interfering With Whom? Nat Rev Microbiol (2015) 13(7):403–13. doi: 10.1038/nrmicro3449
75. Newling M, Hoepel W, Vogelpoel LTC, Heineke MH, van Burgsteden JA, Taanman-Kueter EWM, et al. Fc Gamma Receptor IIa Suppresses Type I and III Interferon Production by Human Myeloid Immune Cells. Eur J Immunol (2018) 48(11):1796–809. doi: 10.1002/eji.201847615
76. Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, et al. Natural Variation in Fc Glycosylation of HIV-specific Antibodies Impacts Antiviral Activity. J Clin Invest (2013) 123(5):2183–92. doi: 10.1172/JCI65708
77. Ackerman ME, Dugast A-S, McAndrew EG, Tsoukas S, Licht AF, Irvine DJ, et al. Enhanced Phagocytic Activity of HIV-specific Antibodies Correlates With Natural Production of Immunoglobulins With Skewed Affinity for Fcγr2a and Fcγr2b. J Virol (2013) 87(10):5468–76. doi: 10.1128/JVI.03403-12
78. De Souza MS, Phanuphak N, Pinyakorn S, Trichavaroj R, Pattanachaiwit S, Chomchey N, et al. Impact of Nucleic Acid Testing Relative to Antigen/Antibody Combination Immunoassay on the Detection of Acute HIV Infection. Aids (2015) 29(7):793–800. doi: 10.1097/QAD.0000000000000616
79. Ananworanich J, Fletcher JL, Pinyakorn S, Van Griensven F, Vandergeeten C, Schuetz A, et al. A Novel Acute HIV Infection Staging System Based on 4 Th Generation Immunoassay. Retrovirology (2013) 10(1):56. doi: 10.1186/1742-4690-10-56
80. Machado LR, Bowdrey J, Ngaimisi E, Habtewold A, Minzi O, Makonnen E, et al. Copy Number Variation of Fc Gamma Receptor Genes in HIV-Infected and HIV-Tuberculosis Co-Infected Individuals in Sub-Saharan Africa. PLoS One (2013) 8(11):e78165. doi: 10.1371/journal.pone.0078165
81. Adu B, Dodoo D, Adukpo S, Hedley PL, Arthur F, Gerds TA, et al. Fc Gamma Receptor IIIB (FcgammaRIIIB) Polymorphisms are Associated With Clinical Malaria in Ghanaian Children. PLoS One (2012) 7(9):e46197. doi: 10.1371/journal.pone.0046197
82. Osborne JM, Chacko GW, Brandt JT, Anderson CL. Ethnic Variation in Frequency of an Allelic Polymorphism of Human Fcγ RIIA Determined With Allele Specific Oligonucleotide Probes. J Immunol Methods (1994) 173(2):207–17. doi: 10.1016/0022-1759(94)90299-2
83. van Schie RC, Wilson ME. Evaluation of Human Fcγriia (CD32) and Fcγriiib (CD16) Polymorphisms in Caucasians and African-Americans Using Salivary DNA. Clin Diagn Lab Immunol (2000) 7(4):676–81. doi: 10.1128/CDLI.7.4.676-681.2000
84. Tong Y, Jin J, Yan L, Neppert J, Marget M, Flesch BK. FCGR3B Gene Frequencies and FCGR3 Variants in a Chinese Population From Zhejiang Province. Ann Hematol (2003) 82(9):574–8. doi: 10.1007/s00277-003-0725-y
85. Kaset C, Leetrakool N, Intharanut K, Nathalang O. Frequency of FCGR3B Alleles in Thai Blood Donors. Ann Lab Med (2013) 33(6):426–30. doi: 10.3343/alm.2013.33.6.426
86. Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, et al. Limited or No Protection by Weakly or Nonneutralizing Antibodies Against Vaginal SHIV Challenge of Macaques Compared With a Strongly Neutralizing Antibody. Proc Natl Acad Sci (2011) 108(27):11181–6. doi: 10.1073/pnas.1103012108
87. Bashirova AA, Thomas R, Carrington M. HLA/KIR Restraint of HIV: Surviving the Fittest. Annu Rev Immunol (2011) 29:295–317. doi: 10.1146/annurev-immunol-031210-101332
88. McLaren PJ, Carrington M. The Impact of Host Genetic Variation on Infection With HIV-1. Nat Immunol (2015) 16(6):577. doi: 10.1038/ni.3147
89. Buzon MJ, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, et al. Long-Term Antiretroviral Treatment Initiated At Primary HIV-1 Infection Affects the Size, Composition, and Decay Kinetics of the Reservoir of HIV-1-infected Cd4 T Cells. J Virol (2014) 88(17):10056–65. doi: 10.1128/JVI.01046-14
90. Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, et al. International AIDS Society Global Scientific Strategy: Towards an HIV Cure 2016. Nat Med (2016) 22(8):839. doi: 10.1038/nm.4108
91. Williams JP, Hurst J, Stöhr W, Robinson N, Brown H, Fisher M, et al. Hiv-1 DNA Predicts Disease Progression and Post-Treatment Virological Control. eLife (2014) 3:e03821. doi: 10.7554/eLife.03821
92. Fourati S, Flandre P, Calin R, Carcelain G, Soulie C, Lambert-Niclot S, et al. Factors Associated With a Low HIV Reservoir in Patients With Prolonged Suppressive Antiretroviral Therapy. J Antimicrobial Chemother (2013) 69(3):753–6. doi: 10.1093/jac/dkt428
93. Martin GE, Pace M, Thornhill JP, Phetsouphanh C, Meyerowitz J, Gossez M, et al. Cd32-Expressing Cd4 T Cells Are Phenotypically Diverse and Can Contain Proviral Hiv Dna. Front Immunol (2018) 9(928). doi: 10.3389/fimmu.2018.00928
94. Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, et al. CD32 is Expressed on Cells With Transcriptionally Active HIV But Does Not Enrich for HIV DNA in Resting T Cells. Sci Trans Med (2018) 10(437). doi: 10.1126/scitranslmed.aar6759
95. Pérez L, Anderson J, Chipman J, Thorkelson A, Chun T-W, Moir S, et al. Conflicting Evidence for HIV Enrichment in CD32+ Cd4 T Cells. Nature (2018) 561(7723):E9–16. doi: 10.1038/s41586-018-0493-4
96. Osuna CE, Lim S-Y, Kublin JL, Apps R, Chen E, Mota TM, et al. Evidence That CD32a Does Not Mark the HIV-1 Latent Reservoir. Nature (2018) 561(7723):E20–E8. doi: 10.1038/s41586-018-0495-2
97. Bertagnolli LN, White JA, Simonetti FR, Beg SA, Lai J, Tomescu C, et al. The Role of CD32 During HIV-1 Infection. Nature (2018) 561(7723):E17–9. doi: 10.1038/s41586-018-0494-3
98. Thornhill JP, Pace M, Martin GE, Hoare J, Peake S, Herrera C, et al. CD32 Expressing Doublets in HIV-infected Gut-Associated Lymphoid Tissue are Associated With a T Follicular Helper Cell Phenotype. Mucosal Immunol (2019) 12(5):1212–9. doi: 10.1038/s41385-019-0180-2
99. Darcis G, Binda CS, Klaver B, Herrera-Carrillo E, Berkhout B, Das AT. The Impact of HIV-1 Genetic Diversity on CRISPR-Cas9 Antiviral Activity and Viral Escape. Viruses (2019) 11(3):255. doi: 10.3390/v11030255
100. Darcis G, Kootstra NA, Hooibrink B, van Montfort T, Maurer I, Groen K. CD32+ CD4+ T cells are highly enriched for HIV DNA and can support transcriptional latency. Cell Rep (2020) 30(7):2284–96. doi: 10.1016/j.celrep.2020.01.071
101. Ananworanich J, Chomont N, Eller LA, Kroon E, Tovanabutra S, Bose M, et al. Hiv DNA Set Point is Rapidly Established in Acute HIV Infection and Dramatically Reduced by Early ART. EBioMedicine (2016) 11:68–72. doi: 10.1016/j.ebiom.2016.07.024
102. Pasternak A, Prins J, Berkhout B eds. Cell-Associated HIV-1 Unspliced RNA Level Predicts Both Time to Virological Suppression and Duration of Post-Treatment Virological Control in Patients Treated With Temporary Early ART. Journal OF the INTERNATIONAL Aids Society. GENEVA, 1202, SWITZERLAND: INT AIDS SOCIETY AVENUE DE FRANCE 23 (2016). doi: 10.1016/S2055-6640(20)31009-8
103. Eisele E, Siliciano RF. Redefining the Viral Reservoirs That Prevent HIV-1 Eradication. Immunity (2012) 37(3):377–88. doi: 10.1016/j.immuni.2012.08.010
104. Hogan LE, Vasquez J, Hobbs KS, Hanhauser E, Aguilar-Rodriguez B, Hussien R, et al. Increased HIV-1 Transcriptional Activity and Infectious Burden in Peripheral Blood and Gut-Associated CD4+ T Cells Expressing CD30. PLoS Pathogens (2018) 14(2). doi: 10.1371/journal.ppat.1006856
105. Noto A, Procopio FA, Banga R, Suffiotti M, Corpataux J-M, Cavassini M, et al. CD32+ and PD-1+ Lymph Node CD4 T Cells Support Persistent HIV-1 Transcription in Treated Aviremic Individuals. J Virol (2018) 92(20):e00901–18. doi: 10.1128/JVI.00901-18
106. Deepe RN, Kistner-Griffin E, Martin JN, Deeks SG, Pandey JP. Epistatic Interactions Between Fc (GM) and Fcγr Genes and the Host Control of Human Immunodeficiency Virus Replication. Hum Immunol (2012) 73(3):263–6. doi: 10.1016/j.humimm.2011.12.008
107. Carapito R, Mayr L, Molitor A, Verniquet M, Schmidt S, Tahar O, et al. A FcγRIIa Polymorphism has a HLA-B57 and HLA-B27 Independent Effect on HIV Disease Outcome. Genes Immun (2020) 21(4):263–8. doi: 10.1038/s41435-020-0106-8
108. Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, et al. Influence of HLA-B57 on Clinical Presentation and Viral Control During Acute HIV-1 Infection. Aids (2003) 17(18):2581–91. doi: 10.1097/00002030-200312050-00005
109. Neumann-Haefelin C. Hla-B27-mediated Protection in HIV and Hepatitis C Virus Infection and Pathogenesis in Spondyloarthritis: Two Sides of the Same Coin? Curr Opin Rheumatol (2013) 25(4):426–33. doi: 10.1097/BOR.0b013e328362018f
110. Tomaras GD, Ferrari G, Shen X, Alam SM, Liao H-X, Pollara J, et al. Vaccine-Induced Plasma IgA Specific for the C1 Region of the HIV-1 Envelope Blocks Binding and Effector Function of Igg. Proc Natl Acad Sci U S A (2013) 110(22):9019–24. doi: 10.1073/pnas.1301456110
111. Zolla-Pazner S, deCamp AC, Cardozo T, Karasavvas N, Gottardo R, Williams C, et al. Analysis of V2 Antibody Responses Induced in Vaccinees in the ALVAC/AIDSVAX HIV-1 Vaccine Efficacy Trial. PLoS One (2013) 8(1):e53629. doi: 10.1371/journal.pone.0053629
112. Zolla-Pazner S, DeCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, et al. Vaccine-Induced IgG Antibodies to V1V2 Regions of Multiple HIV-1 Subtypes Correlate With Decreased Risk of HIV-1 Infection. PLoS One (2014) 9(2):e87572. doi: 10.1371/journal.pone.0087572
113. Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, et al. Polyfunctional Fc-Effector Profiles Mediated by IgG Subclass Selection Distinguish RV144 and VAX003 Vaccines. Sci Trans Med (2014) 6(228):228ra38–ra38. doi: 10.1126/scitranslmed.3007736
114. Yates NL, Liao H-X, Fong Y, DeCamp A, Vandergrift NA, Williams WT, et al. Vaccine-Induced Env V1-V2 IgG3 Correlates With Lower HIV-1 Infection Risk and Declines Soon After Vaccination. Sci Trans Med (2014) 6(228):228ra39–ra39. doi: 10.1126/scitranslmed.3007730
115. Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial. N Engl J Med (2012) 366(14):1275–86. doi: 10.1056/NEJMoa1113425
116. Liao H-X, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, et al. Vaccine Induction of Antibodies Against a Structurally Heterogeneous Site of Immune Pressure Within HIV-1 Envelope Protein Variable Regions 1 and 2. Immunity (2013) 38(1):176–86. doi: 10.1016/j.immuni.2012.11.011
117. Bournazos S, DiLillo DJ, Ravetch JV. Humanized Mice to Study FcγR Function. In: Daeron M, Nimmerjahn F, editors. Fc Receptors. Current Topics in Microbiology and Immunology, vol 382. Springer, Cham (2014). doi: 10.1007/978-3-319-07911-0_11
118. Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, et al. Fc Receptor But Not Complement Binding is Important in Antibody Protection Against HIV. Nature (2007) 449(7158):101–4. doi: 10.1038/nature06106
119. Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, et al. Broadly Neutralizing Human anti-HIV Antibody 2G12 is Effective in Protection Against Mucosal SHIV Challenge Even At Low Serum Neutralizing Titers. PLoS Pathog (2009) 5(5):e1000433. doi: 10.1371/journal.ppat.1000433
120. Parsons MS, Lee WS, Kristensen AB, Amarasena T, Khoury G, Wheatley AK, et al. Fc-Dependent Functions are Redundant to Efficacy of anti-HIV Antibody PGT121 in Macaques. J Clin Invest (2019) 129(1):182–91. doi: 10.1172/JCI122466
121. Abela IA, Berlinger L, Schanz M, Reynell L, Günthard HF, Rusert P, et al. Cell-Cell Transmission Enables HIV-1 to Evade Inhibition by Potent CD4bs Directed Antibodies. PLoS Pathogens (2012) 8(4). doi: 10.1371/journal.ppat.1002634
122. Chung AW, Navis M, Isitman G, Wren L, Silvers J, Amin J, et al. Activation of NK Cells by ADCC Antibodies and HIV Disease Progression. J Acquired Immune Deficiency Syndromes (2011) 58(2):127. doi: 10.1097/QAI.0b013e31822c62b9
123. Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-Based Protection Against HIV Infection by Vectored Immunoprophylaxis. Nature (2012) 481(7379):81–4. doi: 10.1038/nature10660
124. Emery S, Kelleher AD, Workman C, Puls RL, Bloch M, Baker D, et al. Influence of IFNgamma Co-Expression on the Safety and Antiviral Efficacy of Recombinant Fowlpox Virus HIV Therapeutic Vaccines Following Interruption of Antiretroviral Therapy. Hum Vaccines (2007) 3(6):260–7. doi: 10.4161/hv.4627
125. Emery S, Workman C, Puls RL, Bloch M, Baker D, Bodsworth N, et al. Randomised, Placebo-Controlled, Phase I/IIa Evaluation of the Safety and Immunogenicity of Fowlpox Virus Expressing HIV Gag-Pol and Interferon-? in HIV-1 Infected Subjects. Hum Vaccines (2005) 1(6):232–8. doi: 10.4161/hv.1.6.2342
127. Battistini A, Sgarbanti M. HIV-1 Latency: An Update of Molecular Mechanisms and Therapeutic Strategies. Viruses (2014) 6(4):1715–58. doi: 10.3390/v6041715
128. Barton KM, Burch BD, Soriano-Sarabia N, Margolis DM. Prospects for Treatment of Latent HIV. Clin Pharmacol Ther (2013) 93(1):46–56. doi: 10.1038/clpt.2012.202
129. Archin NM, Margolis DM. Emerging Strategies to Deplete the HIV Reservoir. Curr Opin Infect Dis (2014) 27(1):29. doi: 10.1097/QCO.0000000000000026
130. Darcis G, Van Driessche B, Van Lint C. Preclinical Shock Strategies to Reactivate Latent HIV-1: An Update. Curr Opin HIV AIDS (2016) 11(4):388–93. doi: 10.1097/COH.0000000000000288
131. Darcis G, Van Driessche B, Van Lint C. HIV Latency: Should We Shock or Lock? Trends Immunol (2017) 38(3):217–28. doi: 10.1016/j.it.2016.12.003
132. Albert BJ, Niu A, Ramani R, Marshall GR, Wender PA, Williams RM, et al. Combinations of Isoform-Targeted Histone Deacetylase Inhibitors and Bryostatin Analogues Display Remarkable Potency to Activate Latent HIV Without Global T-cell Activation. Sci Rep (2017) 7(1):7456. doi: 10.1038/s41598-017-07814-4
133. Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly Neutralizing anti-HIV-1 Antibodies Require Fc Effector Functions for In Vivo Activity. Cell (2014) 158(6):1243–53. doi: 10.1016/j.cell.2014.08.023
134. Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, et al. Therapeutic Efficacy of Potent Neutralizing HIV-1-specific Monoclonal Antibodies in SHIV-infected Rhesus Monkeys. Nature (2013) 503(7475):224–8. doi: 10.1038/nature12744
135. Parsons MS, Lloyd SB, Lee WS, Kristensen AB, Amarasena T, Center RJ, et al. Partial Efficacy of a Broadly Neutralizing Antibody Against Cell-Associated SHIV Infection. Sci Trans Med (2017) 9(402). doi: 10.1126/scitranslmed.aaf1483
136. Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, et al. Highly Potent HIV-specific Antibody Neutralization In Vitro Translates Into Effective Protection Against Mucosal SHIV Challenge In Vivo. Proc Natl Acad Sci (2012) 109(46):18921–5. doi: 10.1073/pnas.1214785109
137. Chen P, Hübner W, Spinelli MA, Chen BK. Predominant Mode of Human Immunodeficiency Virus Transfer Between T Cells is Mediated by Sustained Env-dependent Neutralization-Resistant Virological Synapses. J Virol (2007) 81(22):12582–95. doi: 10.1128/JVI.00381-07
138. Ganesh L, Leung K, Loré K, Levin R, Panet A, Schwartz O, et al. Infection of Specific Dendritic Cells by CCR5-tropic Human Immunodeficiency Virus Type 1 Promotes Cell-Mediated Transmission of Virus Resistant to Broadly Neutralizing Antibodies. J Virol (2004) 78(21):11980–7. doi: 10.1128/JVI.78.21.11980-11987.2004
139. Kong R, Louder MK, Wagh K, Bailer RT, deCamp A, Greene K, et al. Improving Neutralization Potency and Breadth by Combining Broadly Reactive HIV-1 Antibodies Targeting Major Neutralization Epitopes. J Virol (2015) 89(5):2659–71. doi: 10.1128/JVI.03136-14
140. Mayr LM, Su B, Moog C. Non-Neutralizing Antibodies Directed Against HIV and Their Functions. Front Immunol (2017) 8:1590–. doi: 10.3389/fimmu.2017.01590
141. Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. Virological Synapse-Mediated Spread of Human Immunodeficiency Virus Type 1 Between T Cells is Sensitive to Entry Inhibition. J Virol (2010) 84(7):3516–27. doi: 10.1128/JVI.02651-09
142. Su B, Lederle A, Laumond G, Ducloy C, Schmidt S, Decoville T, et al. Broadly Neutralizing Antibody VRC01 Prevents HIV-1 Transmission From Plasmacytoid Dendritic Cells to CD4 T Lymphocytes. J Virol (2014) 88(18):10975–81. doi: 10.1128/JVI.01748-14
143. Su B, Peressin M, Ducloy C, Penichon J, Mayr LM, Laumond G, et al. Short Communication: Exploring Antibody Potential as Prophylactic/Therapeutic Strategies for Prevention of Early Mucosal HIV-1 Infection. AIDS Res Hum Retroviruses (2015) 31(11):1187–91. doi: 10.1089/aid.2015.0041
144. Li H, Zony C, Chen P, Chen BK. Reduced Potency and Incomplete Neutralization of Broadly Neutralizing Antibodies Against Cell-to-Cell Transmission of HIV-1 With Transmitted Founder Envs. J Virol (2017) 91(9):e02425–16. doi: 10.1128/JVI.02425-16
145. Duncan CJ, Russell RA, Sattentau QJ. High Multiplicity HIV-1 Cell-to-Cell Transmission From Macrophages to CD4+ T Cells Limits Antiretroviral Efficacy. Aids (2013) 27(14):2201–6. doi: 10.1097/QAD.0b013e3283632ec4
146. Richardson SI, Chung AW, Natarajan H, Mabvakure B, Mkhize NN, Garrett N, et al. HIV-Specific Fc Effector Function Early in Infection Predicts the Development of Broadly Neutralizing Antibodies. PLoS Pathogens (2018) 14(4):e1006987. doi: 10.1371/journal.ppat.1006987
147. Lehrnbecher T, Foster CB, Zhu S, Leitman SF, Goldin LR, Huppi K, et al. Variant Genotypes of the Low-Affinity Fcγ Receptors in Two Control Populations and a Review of Low-Affinity Fcγ Receptor Polymorphisms in Control and Disease Populations. Blood J Am Soc Hematol (1999) 94(12):4220–32. doi: 10.1182/blood.V94.12.4220.424k08_4220_4232
Keywords: FcγR polymorphisms, HIV-1 latent reservoirs, HIV-1 cure strategies, HIV-1 disease progression, broadly neutralizing antibodies
Citation: Lamptey H, Bonney EY, Adu B and Kyei GB (2021) Are Fc Gamma Receptor Polymorphisms Important in HIV-1 Infection Outcomes and Latent Reservoir Size? Front. Immunol. 12:656894. doi: 10.3389/fimmu.2021.656894
Received: 21 January 2021; Accepted: 06 April 2021;
Published: 04 May 2021.
Edited by:
Ann Jones Hessell, Oregon Health and Science University, United StatesReviewed by:
Michael Anthony Moody, Duke Human Vaccine Institute, United StatesChristiane Moog, Institut National de la Santé et de la Recherche Médicale (INSERM), France
Copyright © 2021 Lamptey, Bonney, Adu and Kyei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bright Adu, QmFkdUBub2d1Y2hpLnVnLmVkdS5naA==; George B. Kyei, Z2t5ZWlAbm9ndWNoaS51Zy5lZHUuZ2g=
†These authors have contributed equally to this work and share last authorship
 Helena Lamptey
Helena Lamptey Evelyn Y. Bonney
Evelyn Y. Bonney Bright Adu
Bright Adu George B. Kyei
George B. Kyei