
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 19 March 2021
Sec. Viral Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.654165
In the year since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and with understanding of the etiology of the coronavirus disease 2019 (COVID-19) pandemic, it has become clear that most infected individuals achieve some form of immunity against the virus with relatively few reported reinfections. A number of vaccines have already achieved emergency use authorization based on data from large phase 3 field efficacy clinical trials. However, our knowledge about the extent and durability of this immunity, and the breadth of vaccine coverage against SARS-CoV-2 variants is still evolving. In this narrative review, we summarize the latest and rapidly developing understanding of immunity to SARS-CoV-2 infection, including what we have learned about the key antigens of SARS-CoV-2 (i.e., the spike protein and its receptor-binding domain), their importance in vaccine development, the immediate immune response to SARS-CoV-2, breadth of coverage of emerging SARS-CoV-2 variants, contributions of preexisting immunity to related coronaviruses, and duration of immunity. We also discuss lessons from newer approaches, such as systems serology, that provide insights into molecular and cellular immune responses elicited and how they relate to the trajectory of infection, and potentially inform immune correlates of protection. We also briefly examine the limited research literature on immune responses in special populations, such as pregnant women and children.
In December 2019 in Wuhan, China, a novel coronavirus (CoV) emerged, causing severe acute respiratory syndrome (SARS) in humans (1). The virus, SARS coronavirus 2 (SARS-CoV-2), causes coronavirus disease 2019 (COVID-19), which has subsequently caused a global pandemic (2). As of February 15, 2021, >108 million cases of COVID-19 and almost 2.4 million deaths have been reported worldwide (3).
Seven CoVs are known to infect humans: 4 seasonal CoVs that cause self-limiting upper respiratory tract infections and 3 highly pathogenic CoVs (i.e., SARS coronavirus [SARS-CoV-1], Middle East respiratory syndrome [MERS], and SARS-CoV-2), which emerged in 2003, 2012, and 2019, respectively (4, 5). Knowledge of immunity to all CoVs is sparse (4). Immunity to the seasonal CoVs appears to last ~1 year, whereas for patients with SARS-CoV-1 and MERS, antibody levels are greatly reduced 2–3 years after symptom onset, indicating that patients might be susceptible to reinfection at that time (6–8). Less than a year into the COVID-19 pandemic, our understanding of immunity to SARS-CoV-2 is developing rapidly. A substantial portion of our current knowledge is derived from serosurveillance studies conducted in the very early stages of the pandemic and subsequently from evaluations of B- and T-cell responses among convalescent patients with varying degrees of disease severity. Hospitalized patients who eventually died of COVID-19 have also been analyzed to help correlate the immune response with disease trajectory. In addition, several studies have evaluated sera from the prepandemic period and from longitudinal studies with follow-up from studies on the SARS-CoV-1 pandemic and other CoVs. Inevitably, there are several unanswered questions; however, a remarkable body of knowledge has rapidly accumulated from the scientific enterprise during this pandemic, yielding actionable insights that continue to inform development of vaccines and therapeutics to combat COVID-19. As of this writing, there are ~60 vaccine candidates at various stages of clinical trials and about 172 candidates in preclinical development spanning diverse vaccine platforms, such as inactivated and live virus, protein subunits, viral vector, DNA, and mRNA (9). In a remarkable, unprecedented scientific achievement, after only <10 months in clinical development, 2 vaccine candidates have reported ~95% vaccine efficacy (VE) against COVID-19 from large (>30,000 subjects) prospective placebo-controlled phase 3 clinical trials (10, 11). Before the end of 2020, both vaccines were granted emergency use authorization (EUA) by the US Food and Drug Administration (FDA) (12, 13), and conditional marketing authorization by the European Medicines Agency (14, 15), with vaccinations currently underway in multiple countries. Adenovirus (Ad)-based vaccines in late-stage clinical trials have reported efficacies of 62−92%, depending on the vaccine, dosing regimen, and disease severity (16–18); the ChAdOx-1 nCov-19 vaccine has been authorized for use during the pandemic in the United Kingdom, and in several European and other countries, and vaccinations are in progress (19–22). Interim analysis of a late-stage protein-based vaccine trial reported 89% efficacy against COVID-19 (with lower VE against disease caused by emerging SARS-CoV-2 variants) (23). Finally, multiple whole inactivated SARS-CoV-2 vaccines currently in clinical trials have reported promising safety and immunogenicity findings (24–27). In addition, numerous SARS-CoV-2 neutralizing monoclonal antibodies (mAbs) have been derived from convalescent patient sera and are being evaluated in clinical trials, and 2 mAb therapeutics have been also granted EUA by the FDA (28, 29).
Although it is becoming clear that most infected individuals achieve some form of immunity against COVID-19, with extremely few reported reinfections (30–32), the extent and durability of this immunity, including breadth of coverage of SARS-CoV-2 variants, are among the key research questions. Addressing these queries will guide research into potential vaccines and immunotherapeutics and inform vaccination recommendations from national vaccine technical committees. Therefore, the aim of this narrative review is to summarize the latest understanding of immunity to SARS-CoV-2 infection. From a rapidly advancing body of scientific literature, we examine the key antigens of SARS-CoV-2 and what is known about the immediate immune response to SARS-CoV-2 infection, the contributions of preexisting immunity to related CoVs, the breadth of immune coverage of emerging SARS-CoV-2 variants, and the duration of immunity. We also discuss newer approaches such as systems serology that can provide insights into the molecular and cellular immune responses in relation to the trajectory of infection and potentially inform immune correlates of protection. Finally, we examine the limited research literature on immune responses in special populations, such as pregnant women and children.
A targeted literature search was performed using PubMed databases. We used the search term “coronavirus” and each of the following terms or phrases singly and in combination: “SARS-CoV-2,” “immunity,” “systems serology,” “cellular immunity,” “duration of protection,” “breadth of coverage,” and “vaccination.” We also searched for each of these terms with the search terms “SARS” and “MERS.” References cited within the retrieved publications were used as appropriate. The search was limited to English-language articles published up to December 31, 2020. We included ahead-of-print publications available in the PubMed database. Additionally, references from the authors' personal files were reviewed.
It is now understood that the dominant antigen is the SARS-CoV-2 spike protein, which via its receptor-binding domain (RBD) is responsible for binding the virus to the human angiotensin-converting enzyme 2 (ACE2) receptor and its subsequent cellular uptake (33–35).
One of the earliest studies to characterize the humoral responses to SARS-CoV-2 infection used HIV-1–based virions pseudotyped with SARS-CoV-2 spike protein (SARS-CoV-2 pseudovirus) to evaluate the human antibody response to SARS-CoV-2 in 149 individuals recovering from COVID-19 of varying severity (36). Neutralization of SARS-CoV-2 pseudovirus by plasma from convalescent patients with COVID-19, collected on an average of 39 days after the onset of symptoms, had variable half-maximal neutralizing titers (NT50). In 33% of the samples, NT50 values were <50, and in 79% of the samples, they were <1,000. It is interesting to contemplate the parallels here with data from non-human primate challenge studies, in which low titers (1:20 and above) of circulating neutralizing antibodies were reported in animals protected from COVID-19 (37). Antibody cloning showed expanded clones of RBD-specific memory B cells expressing closely related antibodies in different individuals. Despite low plasma NT50 values (e.g., 297–10,433), antibodies to 3 distinct epitopes on the RBD neutralized the pseudovirus at half-maximal inhibitory concentrations (IC50) ranging from 1.6 to 3.0 ng/mL (36). This study was the earliest report noting no significant cross-reactivity to the RBDs of other key CoVs, such as MERS or human coronaviruses HCoV-OC43,−229E, or NL63 (36). Similar findings regarding neutralizing titers were reported in a cross-sectional study of hospitalized patients acutely infected with COVID-19 (n = 44): RBD-specific immunoglobulin G (IgG) responses and neutralizing titers were detectable in all patients within 6 days after confirmation with polymerase chain reaction (PCR) (38). Additionally, the magnitude of RBD-specific IgG titers was positively correlated with the neutralizing potency (38).
A study that screened and selected convalescent patient sera with high neutralizing titers against SARS-CoV-2 found that most epitopes targeted the spike RBD (39). Pseudovirus-neutralizing titers varied for SARS-CoV-2 and were low to undetectable against SARS-CoV-1, whereas anti-RBD mAbs were of extremely high potency, down to 0.019 μg/mL (39). Similar findings were reported by Piccoli and colleagues in which SARS-CoV-2 RBD accounted for 90% of serum neutralizing the activity in convalescent patient sera (40).
Grifoni and colleagues identified circulating SARS-CoV-2–specific CD8+ and CD4+ T cells in 70 and 100% of patients, respectively (41). Moreover, a robust CD4+ T-cell response to the SARS-CoV-2 spike protein was observed, which correlated with the magnitude of the anti–SARS-CoV-2 IgG and immunoglobulin A (IgA) titers (41).
Antibody responses are triggered by the initial interaction between the SARS-CoV-2 spike protein and the human ACE2 receptor and subsequent cellular uptake of the virus (33–35). The immune response kinetics, magnitude, and relationship to disease severity during this acute-phase response have been defined extensively. SARS-CoV-2 elicits humoral and cellular immune responses; within 7 days of infection, virus-specific memory CD4+ and CD8+ T cells emerge, peaking within 2 weeks but remaining detectable at comparatively lower levels for ≥100 days. Simultaneously, there are strong B-cell responses with immunoglobulin M (IgM) and IgA antibodies detected by days 5–7 and IgG antibodies by days 7–10 (42). The magnitude of both antibody and T-cell responses is not uniform among individuals with COVID-19 and appears to be influenced by disease severity (42–45). Antibody levels and the CD8+ T-cell response subsequently decline after the acute phase of infection (42).
Similar immune response kinetics immediately postvaccination have been observed and noted in data published from human clinical trials of late-stage COVID-19 vaccine candidates using spike protein as antigen: spike-protein–specific neutralizing antibodies were elicited by vaccination, and the titers peaked 7–14 days postcompletion of the vaccination series, and in most cases, these humoral responses were comparable with those observed in the respective panels of convalescent patient sera (46–48). Robust antigen-specific CD4+ and CD8+ T cell responses were observed with the BNT162b2 mRNA vaccine and the mRNA-1273 vaccine (47, 49, 50); notably both vaccines induce strong T-helper type 1 (Th1) CD4+ T cell responses with minimal to no T-helper type 2 (Th2) responses (49, 50). T cell-mediated responses have also been reported for 3 Ad-based vaccines: chimpanzee ChAd-Ox1 n-CoV-19, Ad5-vectored COVID-19 vaccine and recombinant (r) Ad26 and rAd5 vector-based vaccine (48, 51–53).
Like other pathogenic viruses, SARS-CoV-2 continues to accumulate mutations due to natural evolution and immune pressure as it propagates through the human population. CoV RNA replicases have been reported to be 10-fold less error-prone than other RNA viruses (54). This may explain the substantially lower sequence diversity of glycoprotein antigens like the SARS-CoV-2 spike protein compared with influenza A virus antigens hemagglutinin and neuraminidase (437-fold greater diversity), and with viruses that cause chronic infections and are influenced by immune pressure on the virus, such as HIV and hepatitis C virus (54). However, natural viral evolution during 2020 has produced novel SARS-CoV-2 variants that carry several distinct mutations in the spike protein (55), which have materially changed the COVID-19 pandemic. Key variants include the D614G spike protein mutant that has shown a modest ability for faster spread (56), and the mink variant with multiple mutations (e.g., “cluster 5”) that demonstrate spillover transmission across species and highlight the risk of incrementally evolved SARS-CoV-2 viruses with broad host range and/or greater pathogenicity (57–59). Most recently, at least 3 new variants, B.1.1.7 (first observed in the United Kingdom), B.1.351 (first observed in South Africa), and P.1 (first observed in Brazil), have arisen and are spreading across the globe (55, 60).
Questions persist regarding difference in clinical severity of COVID-19 caused by these variants and the breadth of immune coverage against these new SARS-CoV-2 variants. The D614G mutation causes an allosteric change in the spike RBD (61, 62), and exhibits efficient replication in vitro and transmission in animal models (63, 64); however there are no indications thus far of higher clinical severity or mortality (65). Additionally, D614G mutant pseudovirus is neutralized by both convalescent patient sera and vaccine-elicited immune sera (66, 67). Mink-associated variants are not associated with rapid spread or increased disease severity among humans, and preliminary serology assessments show that convalescent sera with varying antibody titers neutralize both wildtype and mink-variant virus suggesting that it is unlikely that these mutations will jeopardize the effects of vaccines or therapeutics (68, 69). The B.1.1.7, variant emerged quite rapidly through December 2020 in the United Kingdom (70, 71). Analysis of 228,361 secondary cases identified via England's National Health Service Test and Trace system between October 5 and December 6, 2020 revealed the index case was infected with B.1.1.7 in 15.1%, and other variants in 9.8% of cases (72), this 30–50% increased transmissibility is consistent with the earlier modeling data (70, 71). A matched cohort study of confirmed wildtype and B.1.1.7 cases in England (1,769 in each group) identified between September 20 and December 15, 2020 demonstrated no statistically significant difference in hospitalization, 28-day case fatality, or likelihood of reinfection (72). However, additional accumulating evidence in January and February 2021 suggests B.1.1.7 infection is associated with increased risk of hospitalization and death, though the absolute mortality risk per infection remains low (73). During this same period, a similar, but not identical, variant named 501Y.V2 (B.1.351) has become prominent in South Africa, wherein 3 of 8 defined mutations in the spike protein – N501Y, E484K and K417N – lie within the receptor-binding motif that forms the interface with the human ACE2 receptor (60, 74).
Studies have shown that certain mutations/variants (E484K mutation in particular) are able to escape neutralization by convalescent plasma and monoclonal antibodies that target single epitopes (75–77). Currently authorized vaccines and almost all late-clinical stage candidates contain the full-length SARS-CoV-2 spike protein as antigen (46–48, 51, 53), thereby targeting epitopes outside the RBD (49). The 2 mRNA vaccines have demonstrated ~95% VE against symptomatic COVID-19 and have been shown to elicit polyclonal antibodies similar to natural infection (10, 11, 78); these newly developed vaccines have demonstrated breadth of neutralizing antibody activity against assorted variants, albeit with small declines. BNT162b2 mRNA vaccine immune sera tested against isogenic wildtype and N501Y SARS-CoV-2 on a strain Wa-1 genetic background found no loss of neutralization titer against the N501Y variant strain (79). Vaccine-elicited sera were able to neutralize recombinant viruses containing key variant mutations in vitro: BNT162b2 induced equivalent geometric mean titers (GMTs) of neutralizing antibodies against the B.1.1.7 variant vs. the reference (Wuhan) strain (80), and showed a 0.81- to 1.46-fold reductions in GMTs against mutations in B.1.1.7 and B.1.351 variants relative to the parental virus (81); mRNA-1273 demonstrated no reduction against the B.1.1.7 variant, and a 6.4-fold reduction in neutralizing titers against the B.1.351 variant (82). Adenoviral vector vaccine ChAdOx1-nCoV-19-elicited sera showed 9-fold lower in vitro neutralization activity against the B.1.1.7 variant vs. a canonical non-B.1.1.7 lineage virus (83). An inactivated whole-virus vaccine, BBV152, elicited sera with comparable in vitro neutralization activity against a B.1.17-like virus of Indian origin vs. the homologous vaccine viral strain (84).
Finally, reduced VE against clinical disease has been observed in late-stage clinical trials in regions with high circulation of emerging variants. Interim analysis in January 2021 for an adenoviral vector vaccine (Ad26.COV2.S) (85) in a single-dose regimen reported 66% VE overall against moderate to severe COVID-19, with 72% VE in the United States and 66% VE in Latin America cohorts, but VE dropped to 57% in the South African cohort, wherein 95% of the accrued cases were caused by the B.1.351 variant (18). Similarly, phase 3 results for the adjuvanted protein subunit vaccine NVX-CoV2372 showed 89.3% VE in the UK cohort, where >50% of accrued cases were caused by the B.1.1.7 variant, while VE in the South African cohort was 49.4%, wherein 93% of the accrued cases were caused by the B.1.351 variant (23). Notably, COVID-19 cases in the placebo group accrued regardless of serologic evidence of prior SARS-CoV-2 infection – attack rate in seropositive 3.9% (26/674 cases) vs. seronegative 3.9% (58/1494 cases) (86), suggesting that B.1.351 variant could escape antibodies elicited by previous natural infection. Novel SARS-CoV-2 variants may thus warrant re-designing vaccines and/or implementing booster vaccines for the global population without immunity arising from natural infection. Regulators are collaborating with manufacturers to develop guidance for the types of data needed to support updated vaccine compositions and streamlined clinical programs that can demonstrate vaccine effectiveness expeditiously (87).
SARS-CoV-2 is a new pathogen, and most of the global population has yet to achieve substantial immunity to it. However, this virus belongs to a known family of CoVs, and it is informative to investigate the contributions of preexisting and cross-reactive immunity, which potentially could contribute to protection against SARS-CoV-2. It is important to understand how that might affect the susceptibility to and the severity of SARS-CoV-2 infection. Although studies of convalescent patient sera showed that natural infection elicited an immune response, the fundamental evidence that natural infection with SARS-CoV-2 confers protective immunity as well as protection against reinfection was obtained from a non-human primate challenge study (88). Inoculation of 9 adult rhesus macaques with a total of 1.1 × 104 to 1.1 × 106 plaque-forming units (PFU, n = 3/group) resulted in high levels of viral RNA in both the upper and lower respiratory tracts as well as symptoms suggestive of mild clinical disease and, most importantly, induction of humoral (anti–SARS-CoV-2 spike protein) and cellular immune responses (88). On rechallenge 35 days after the initial infection, only prechallenged animals exhibited a rapid anamnestic immune response and minimal clinical disease compared with those of naive animals. Minimal to no viral RNA was detected in the respiratory tract of prechallenged animals, although viral RNA concentrations were higher in nasal swabs compared with bronchoalveolar lavage, reminiscent of disease and increased transmissibility in humans (88).
Another study profiled the immune response across multiple human CoV RBDs, respiratory viruses, and SARS-CoV-2 to determine whether heterologous immunity to other CoV-RBDs or other infections influenced the evolution of the SARS-CoV-2 humoral immune response (89). The study found little evidence of correlation between SARS-CoV-2 responses and HKU1, NL63, and respiratory infection (influenza and respiratory syncytial virus) responses, suggesting that common viral infections including common CoV immunity, targeting the RBD involved in viral infection, do not influence the rapid functional evolution of SARS-CoV-2 immunity and thus should not affect diagnostics or vaccine-induced immunity (89).
Another study characterized spike RBD antibody kinetics and isotype profile in COVID-19 cases (n = 343) and prepandemic controls (n = 1,548) evaluating antibody responses to RBDs derived from spike proteins of endemic human CoVs (HKU1, 229E, OC43, and NL63), SARS-CoV-1, and MERS (90). No cross-reactive responses to endemic human CoV RBDs or MERS-CoV RBD were seen in COVID-19 cases, but significant cross-reactivity to SARS-CoV-1 RBD was observed (90).
Individuals with high levels of T cells that are able to recognize SARS-CoV-2 could potentially mount fast and strong immune responses to reinfection, possibly limiting disease severity (91). In a study of 36 individuals recovering from mild to severe COVID-19, all had SARS-CoV-2–specific CD4+ and CD8+ T cells that recognized multiple regions of the SARS-CoV-2 nucleocapsid (N) protein (92). The same study examined T cells in 23 individuals who had recovered from SARS in 2003. Seventeen years after infection, these individuals possessed long-lasting memory T cells that were reactive to the SARS-CoV-1 N protein and also cross-reactive to the N protein of SARS-CoV-2. Similarly, Mathew and colleagues examined blood from 125 hospitalized COVID-19 patients and found that more severe COVID-19 was associated with lower frequencies of both CD4+ and CD8+ T cells (93). However, there is much evidence showing that antibodies to the spike protein rather than the N protein are responsible for neutralizing activity against SARS-CoV-2 (94).
Finally, a retrospective analysis of nearly 16,000 electronic health records from a single academic medical center of patients with and without PCR evidence of recent or ongoing infection with at least 1 of 4 seasonal endemic human CoVs demonstrated a similar propensity to acquire SARS-CoV-2 (95). These data suggested that any immunity elicited by prior CoV exposure was insufficient to prevent subsequent infection by SARS-CoV-2 but may be associated with less severe COVID-19 (95).
The persistence of the immune response to SARS-CoV-2 and how this might contribute to preventing reinfection is not well-understood at this stage of the pandemic. Furthermore, duration of protection is a key question for vaccine development and the need for periodic revaccination to overcome antibody waning, and has substantial implications for achieving and maintaining herd immunity.
Initial evidence for potential duration of protection from novel SARS-CoV-2 infection comes from a long-term epidemiologic study on immunity to seasonal CoVs. Edridge and colleagues followed a cohort of 10 adult men since the 1980's, evaluating immunity to seasonal CoVs—NL63, 229E, OC43, and HKU1—by measuring the frequency of reinfection, sampling every 3–6 months annually for >35 years (8). They concluded that protective immunity against seasonal CoVs is short term (i.e., lasting 6–12 months) because they observed that natural reinfections occurred for all seasonal CoVs at 6 and 9 months but most frequently within 12 months (8).
In a large US study of >30,000 individuals who tested positive for SARS-CoV-2, ~93% had moderate to high titers of antispike antibodies, with >90% of these individuals having detectable neutralizing antibody responses (96). The researchers reported that antibody titers were stable over 3 months with modest declines at 5 months; longer-term studies are needed to help establish a correlate of protection (96).
Persistent immune responses to SARS-CoV-2 have also been assessed by antibody isotype in a cohort of patients with symptomatic SARS-CoV-2 from North America (n = 259); IgM and IgA responses to SARS-CoV-2 RBD were transient, and most patients seroreverted within 2.5 months after symptom onset (90). However, IgG responses were sustained for ≥90 days, and seroreversion was minimal (90). Similarly, in a study of serum (n = 439) and saliva (n = 128) samples from patients with COVID-19, antibody responses to the SARS-CoV-2 spike RBD were detected in both fluids, with peak IgG levels attained by 16–30 days after symptom onset (97). IgA and IgM levels declined, while IgG antibodies remained relatively stable for ≥3 months after symptom onset. Neutralizing antibodies peaked between 31 and 45 days and then declined up to 105 days (97).
A population-based serosurveillance study (n = 30,576) in Iceland, where 15% of the country's citizens had been tested for COVID-19 by quantitative PCR (qPCR), showed that ≥90% (n = 1,107/1,215) of qPCR-positive individuals remained seropositive 120 days after diagnosis, and there were no observed declines in antibody titers (98). When these responses were broken down by isotype and antigen, IgM anti-N antibody levels were transient and no longer detectable within 2 months, whereas IgA anti-S1 antibodies declined 1 month after diagnosis but remained detectable, and IgG anti-N and anti-S1 increased within 6 weeks after diagnosis and persisted but with a slight decrease in levels thereafter (98). A longitudinal cohort study of >1,100 seropositive healthcare workers reported that previous SARS-CoV-2 infection that generated antibody responses offered protection from reinfection for most people in the 6 months after infection (99).
More recently, Dan and colleagues measured circulating SARS-CoV-2 antigen-specific antibodies, memory B cells, and CD8+ and CD4+ T cells for >6 months postinfection in a cohort of patients with COVID-19 (n = 185) across a range of disease including asymptomatic, mild (non-hospitalized), moderate (hospitalized), and severe (hospitalized) (37). Spike RBD IgG titers were almost stable from days 20 to 240 postsymptom onset (half-life [t½] = 140 days), and antigen-specific memory B cells increased over the first ~150 days and then plateaued. Half of the patients were positive for circulating SARS-CoV-2 memory CD8+ T cells at ≥6 months (t½ = 166 days), and 89% of patients were positive for circulating SARS-CoV-2 memory CD4+ T cells at ≥6 months (t½ = 150 days) (37). The authors combined 5 immune components to devise a composite measurement of SARS-CoV-2 immune memory: RBD-specific IgG, IgA, and memory B cells and SARS-CoV-2–specific CD4+ and CD8+ T cells. They found that at 1 to 2 months postinfection, 59% of individuals were positive for 5/5 components, and at ≥5 months postinfection, 40% were positive for 5/5 components; however, 96% of individuals were positive for 3/5 components (37).
For SARS-CoV-2 mRNA vaccines recently authorized for emergency use (12, 13), data on longer-term persistence of vaccine-elicited immune responses will become available as the participants from the phase 3 vaccine clinical studies are followed for 2 years postprimary vaccination (10). An interim readout of immune persistence has been reported for mRNA-1273 suggesting that antibody binding and neutralization titers were maintained for 3 months postprimary vaccination among study participants (100). An Ad vector vaccine against a related CoV, ChAdOx1 MERS, showed that vaccine-elicited neutralizing antibodies waned from peak postprimary immunization levels and remained stable above baseline for ≥1 year (101).
Systems serology is an experimental approach that aims to define comprehensively the varied humoral responses to vaccine antigen(s) by measuring both polyclonal antibody features (antigen-binding portion) and functional properties (Fc portion) and can help elucidate mechanisms of immunity, define multifactorial correlates of protection, and evaluate vaccine candidates (102). We summarize a number of promising system serology investigations that have enhanced our understanding of the distinct early immune response profiles for COVID-19, some of which could serve as biomarkers for predicting COVID-19 trajectory.
Using systems serology, Atyeo and colleagues characterized the antibody features and functions of the SARS-CoV-2–specific humoral immune response in a cohort of 22 SARS-CoV-2–positive patients (recovered, n = 12; deceased, n = 10) (103). They identified distinct humoral profiles comprising 5 antibody features that could differentiate between patients who recovered or eventually died of SARS-CoV-2 disease and could potentially be developed into a biomarker that predicts COVID-19 severity in humans (103). Specifically, convalescent and deceased patients presented distinct humoral profiles: more spike-focused response in convalescent patients and stronger N-specific responses in deceased patients. Convalescent patients showed enriched spike-specific phagocytic and complement activity, whereas those who died of disease had poorly coordinated RBD-specific antibody-dependent complement deposition and natural killer (NK) cell functions as well as a greater N-specific response (103). This early divergence (spike:N ratio) in their humoral immune response was confirmed in a larger validation cohort of acutely infected individuals (recovered, n = 20; died, n = 20) (103). Equivalent titers and functional profiles were observed across both groups, indicating that the magnitude of the responses was comparable and did not contribute to the observed differences in either population (103).
Potent neutralizing antibodies against SARS-CoV-2 appear to increase survival and may protect against reinfection with variants of SARS-CoV-2. In a cohort of 113 patients infected with SARS-CoV-2 stratified by disease severity and outcomes (i.e., non-hospitalized, hospitalized, intubated, immunosuppressed, and deceased), anti-RBD antibody levels, neutralization titers, and neutralization potency indices predicted disease severity and survival (66). Comprehensive immunophenotyping of peripheral blood in 42 individuals with divergent clinical trajectories of SARS-CoV-2 infection and COVID-19 (moderate n = 7, severe n = 28, recovered n = 7) showed perturbations in multiple leukocyte populations, which distinguished cases of severe COVID-19 from healthy donors, cases of moderate COVID-19, and from patients who have recovered from COVID-19 (44). This distinct immune phenotype included activation of CD4+ and CD8+ T-cell memory subsets, substantial oligoclonal expansion of B cells, and dysregulation of innate immune subsets (44). Moreover, these analyses further confirmed the prognostic value of the neutrophil to lymphocyte ratio as a biomarker of disease severity and organ failure. Similarly, longitudinal immune profiling of peripheral blood mononuclear cells and plasma samples from 113 patients with moderate (n = 80) or severe (n = 33) COVID-19 identified 4 distinctive immune response profiles that predicted divergent disease courses within 10 days of infection (104). Patients with severe COVID-19 and poor clinical outcome showed an aberrant immune response that included an early elevation in proinflammatory cytokines. In all patients, an overall increase in innate cell lineages such as monocytes, low-density neutrophils, and eosinophils was seen, with a parallel decrease in CD4+ and CD8+ T-cell levels (104). A study by Mathew and colleagues found 3 distinct immunotypes in hospitalized COVID-19 patients: (1) robust activation of CD4+ T cells and highly activated or exhausted CD8+ T cells, (2) less robust CD4+ T cells and highly functional effector-like CD8+ T cell responses, and (3) a lack of lymphocyte response (93). These major patterns may represent different suboptimal responses to infection (93).
In a study from Spain, investigators observed that plasma SARS-CoV-2 RNAemia and viral RNA load were significantly correlated with severe illness in COVID-19 (105). Among patients with COVID-19, 78% of critically ill patients had a significantly higher frequency of plasma SARS-CoV-2 RNAemia compared with 27% of ward patients and 2% of outpatients (P < 0.001) (105). Viral RNA load in plasma was also associated with a maladaptive immune response profile that included higher levels of chemokines, NK cell activation as well as evidence of a systemic inflammatory response (e.g., IL-6), endothelial dysfunction, coagulation activation, and tissue damage (105). Another study also reported the presence of activated NK cells with upregulated levels of chemokines and effector molecules, nutrient receptors, and activating receptors in persons with moderate to severe COVID-19 (106).
Time to seroconversion may also be predictive of more severe disease. In 159 symptomatic patients with COVID-19 from North America, the median time to seroconversion to SARS-CoV-2 RBD was, on average, 4 days earlier for hospitalized patients compared with non-hospitalized patients over all isotypes (IgG, IgM, and IgA), suggesting an association between antibody kinetics and disease severity (90).
Collectively, the evidence from these studies suggests that uncontrolled viral replication coupled with a reduced innate antiviral defense and a maladaptive proinflammatory response are intrinsic elements to the pathogenesis of COVID-19 (105, 107). Further research is needed to identify which biomarkers may predict those at risk of severe disease ideally to determine a limited number of readily measured markers that are useful in predicting clinical outcomes (45).
At this stage in the pandemic, there is limited evidence regarding the immune response to COVID-19 during pregnancy, postpartum, breastfeeding, and in neonates. None of the completed or ongoing phase 3 clinical studies of vaccines include pregnant women (108), and further research is needed to understand the immune response in these special populations.
A study of immune responses in pregnant and non-pregnant women who either tested positive (n = 22) or negative (n = 11) for SARS-CoV-2 before delivery, found that pregnant and non-pregnant women showed similar IgG responses recognizing the full-length SARS-CoV-2 spike protein, whereas pregnant women had significantly lower anti–spike-RBD IgG titers than did non-pregnant women (108). Pregnant women were also less likely to have detectable neutralizing antibody titers (108). Studies of maternal and cord blood serum found that maternal transfer of neutralizing antibodies may be reduced, but SARS-CoV-2 infection did not generally affect maternal transfer of humoral immunity, as measured by placental Fc receptor and maternal and cord blood serum anti-tetanus IgG titers (108).
A study in the United States found no SARS-CoV-2 RNA detected in breast milk collected from 18 women with mild to moderate COVID-19 (109), however, breast milk did contain anti–SARS-CoV-2 antibodies (IgA and IgG), and concentrations correlated with virus neutralization titers (109). These results are supported by another study in China in 14 women in whom no SARS-CoV-2 RNA was found in breast milk, but in this study, SARS-CoV-2 antibodies (IgM or IgG) were found in only 3 samples (110). Both studies recommended that breastfeeding should continue with care in women infected with SARS-CoV-2 (109, 110). Another study in China investigated the presence of SARS-CoV-2 antibodies in 64 infants, of whom 24 were born to mothers with confirmed COVID-19. Among these 24 infants, 15 had detectable IgG antibodies and 6 had IgM antibodies; none had positive nucleic acid test results (111). IgM and IgG titers gradually declined in all infants; however, IgG titers had a slower decline in infants with positive IgM results (111). Further research is needed to determine a correlation of immunity in infants with antibodies to SARS-CoV-2 (111).
In a study conducted in the United Kingdom, the kinetics of SARS-CoV-2 antibody responses was investigated by measuring seroprevalence in 849 children aged 2–15 years (112). Sixty-five children (7.7%) were positive when first tested, and 45 of those had detectable antibody titers when tested ~2 months later. A systematic review of SARS-CoV-2 in pediatric populations reports that while children have lower case rates compared with adults, they may be just as susceptible to infection but have fewer symptomatic infections (113). While it is not fully understood why children appear to have milder illness compared to adults, it has been suggested that this may be partly due to trained immunity due to routine live childhood vaccinations and frequent viral infections (114). However, additional rigorous studies are essential to better understand the clinical course, transmission, and immune response to COVID-19 in children (113, 114).
Among the 2 mRNA vaccines recently authorized for emergency use (12, 13), BNT162b2 mRNA vaccine is authorized for individuals 16 years and older and is being evaluated in children 12–15 years of age (115). In addition, the mRNA-1273 vaccine is authorized for individuals 18 years and older (13), and is being evaluated among children 12–17 years of age (116).
Unprecedented collaborative research among the global scientific and medical community continues to rapidly advance our understanding of COVID-19 and the associated immune response. This emerging knowledge enabled the development of immunotherapeutics and vaccines. Figure 1 summarizes what we have learned in <1 year about the SARS-CoV-2 antigens and humoral and cellular immunity, and what we have learned from both in terms of predicting COVID-19 trajectory (biomarkers) and vaccine efficacy (immune correlates). Yet, there is still much to understand about this novel virus – SARS-CoV-2.
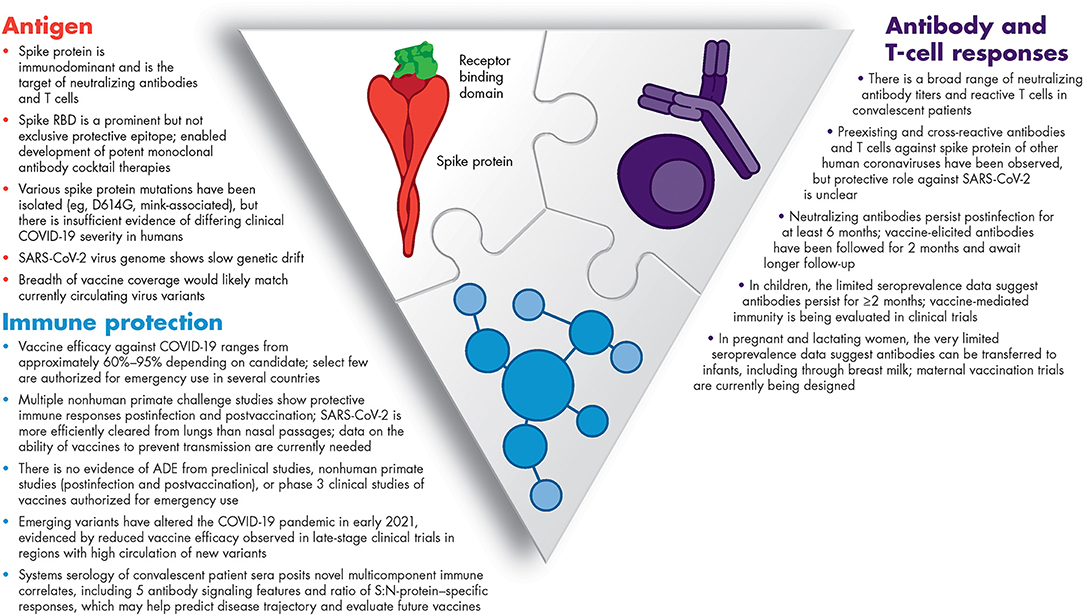
Figure 1. Summary of our current knowledge of immunity to SARS-CoV-2. ADE, antibody-dependent enhancement; COVID-19, coronavirus disease 2019; RBD, receptor-binding domain; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
An effective vaccine against SARS-CoV-2 is thought likely to be the best strategy to control the current COVID-19 pandemic (117), and as of writing, 2 mRNA vaccines have been granted EUA based on ~95% vaccine efficacy against COVID-19 (10, 11) and prioritized individuals are being vaccinated in stages in several countries across the globe, marking a key turning point in this pandemic. Several other candidates—notably viral vector and inactivated virus vaccines—are in late stage clinical trials and are anticipated to become available in the coming months and improve the global vaccine supply (118). In addition to vaccines, neutralizing mAbs to SARS-CoV-2 isolated from convalescent patient sera have been developed, and 2 mAb therapies have been granted EUA (28, 29, 119). These mAbs can be deployed in acute situations to block disease progression and protect those exposed or unvaccinated in high risk settings, or individuals who may be allergic or unlikely to respond to a vaccine (119). In these settings, early and rapid testing of individuals at high risk of infection have been suggested (120). Phase 3 study results report that REGN-COV2, a mAb cocktail, reduces viral load with the most benefit observed in patients with high viral loads at baseline and in those who had not yet initiated an immune response (i.e., those positive for SARS-CoV-2 but serum antibody-negative) (121). In contrast, a study of mAb LY-CoC555 did not show any benefit in patients hospitalized with COVID-19 (122). More research is required to fully realize the potential of mAb therapy in COVID-19.
A major role of neutralizing antibodies is binding of antigens as well as interaction with cells with Fc receptors to induce Fc effector functions (4, 123). There is a potential risk, however, of antibody-dependent enhancement (ADE) of disease mediated by these Fc functional antibodies (4, 123). This phenomenon has been observed with SARS-CoV-1 (123). It is important to understand the balance between Fc signaling that promotes protective immunity and that which promotes inflammatory pathology, and whether systems serology can identify those at risk (124). To date, there has been no evidence of ADE due to SARS-CoV-2 vaccines from preclinical studies or non-human primate studies (125–128). Similarly, no evidence of ADE emerged in a phase 3 clinical study of an mRNA vaccine against SARS-CoV-2 after a median 2 months of follow-up (10). While rare anaphylactic reactions to both mRNA vaccines have been reported (between 2.5–4.7 cases per million doses of vaccine administered) (129), vaccination, along with specified postvaccination observation periods, continues to be recommended except in individuals with history of severe/immediate allergic reaction to previous mRNA vaccines or immediate allergic reaction to any of its components (130).
For many different viral infections, such as influenza, measles, and hepatitis A and B viruses, correlates of protection have been established; these are usually based on the level of antibody that is acquired from vaccination or natural infection that is able to significantly reduce the risk of infection or reinfection (96). However, correlates of protection have yet to be defined for COVID-19 (4, 42, 90). Immunogenicity data from longer term follow-up of vaccine phase 3 trial participants should be helpful to define immune correlates, especially in the case of the vaccines that have already achieved emergency authorization for pandemic use (12, 13). In addition, systems serology studies might yield novel immune correlates based on composite antibody and cellular immune measurements. Regardless, immune correlates will catalyze rapid vaccine development for special populations such as pediatric populations, pregnant and lactating women, and those with immunosuppressive conditions, where traditional field efficacy trials would be unfeasible, and potentially for vaccines with updated compositions to address novel SARS-CoV-2 variants.
All authors contributed to the conception, design of the article, interpreting the relevant literature, drafting of the manuscript, and/or critically revising it for intellectual content.
This work was sponsored by Pfizer Inc.
The authors declare that this study received funding from Pfizer Inc. The funder had the following involvement in the study: Editorial/medical writing support. JF is a speaker for Pfizer and a consultant/advisory board member for Pfizer and GlaxoSmithKline. AS is a Pfizer employee and may hold stock or stock options.
Editorial/medical writing support was provided by Sheena Hunt, Ph.D., and Emily Stackpole, Ph.D., of ICON plc (North Wales, PA) and was funded by Pfizer Inc.
1. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
2. World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 – 11 March 2020. (2020). Available online at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19–11-march-2020 (accessed October 23, 2020).
3. World Health Organization. Coronavirus Disease (COVID-19) Dashboard. (2021). Available online at: https://covid19.who.int/ (accessed February 15, 2021).
4. Poland GA, Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. (2020) 396:1595–606. doi: 10.1016/S0140-6736(20)32137-1
5. Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. (2016) 15:327–47. doi: 10.1038/nrd.2015.37
6. Wu LP, Wang NC, Chang YH, Tian XY, Na DY, Zhang LY, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. (2007) 13:1562–4. doi: 10.3201/eid1310.070576
7. Payne DC, Iblan I, Rha B, Alqasrawi S, Haddadin A, Al Nsour M, et al. Persistence of antibodies against Middle East respiratory syndrome coronavirus. Emerg Infect Dis. (2016) 22:1824–6. doi: 10.3201/eid2210.160706
8. Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. (2020) 26:1691–3. doi: 10.1038/s41591-020-1083-1
9. World Health Organization. Draft landscape of COVID-19 candidate vaccines. (2020). Available online at: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed December 16, 2020).
10. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
11. Mahase E. Covid-19: Moderna applies for US and EU approval as vaccine trial reports 94.1% efficacy. BMJ. (2020) 371:m4709. doi: 10.1136/bmj.m4709
12. US Food and Drug Administration. Pfizer-BioNTech COVID-19 vaccine emergency use authorization review memorandum. (2020). Available online at: https://www.fda.gov/media/144416/download (accessed December 16, 2020).
13. US Food and Drug Administration. Moderna COVID-19 vaccine EUA letter of authorization. (2020). Available online at: https://www.fda.gov/media/144636/download (accessed December 22, 2020).
14. Mahase E. Covid-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. (2020) 371:m4714. doi: 10.1136/bmj.m4714
15. European Medicines Agency. EMA recommends COVID-19 vaccine Moderna for authorisation in the EU. (2021). Available online at: https://www.ema.europa.eu/en/news/ema-recommends-covid-19-vaccine-moderna-authorisation-eu (accessed January 7, 2021).
16. Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. (2021) 397:99–111. doi: 10.1016/S0140-6736(20)32661-1
17. Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. (2021) 397:671–81. doi: 10.1016/S0140-6736(21)00234-8
18. Johnson & Johnson Press Release. Johnson & Johnson Announces Single-Shot Janssen COVID-19 Vaccine Candidate Met Primary Endpoints in Interim Analysis of its Phase 3 ENSEMBLE Trial. (2021). Available online at: https://www.jnj.com/johnson-johnson-announces-single-shot-janssen-covid-19-vaccine-candidate-met-primary-endpoints-in-interim-analysis-of-its-phase-3-ensemble-trial (accessed February 5, 2021).
19. AztraZeneca Press Release. AstraZeneca's COVID-19 vaccine authorised in five other countries. (2021). Available online at: https://www.astrazeneca.com/media-centre/press-releases/2021/serum-institute-of-india-obtains-emergency-use-authorisation-in-india-for-astrazenecas-covid-19-vaccine.html (accessed February 16, 2021).
20. European Medicines Agency. COVID-19 Vaccine AstraZeneca. (2021). Available online at: https://www.ema.europa.eu/en/medicines/human/summaries-opinion/covid-19-vaccine-astrazeneca#opinion-section (accessed February 16, 2021).
21. European Medicines Agency. European Medicines Agency recommends conditional marketing authorisation to ChAdOx1 coronavirus vaccine. (2021). Available online at: https://www.ox.ac.uk/news/2021–01-29-european-medicines-agency-recommends-conditional-marketing-authorisation-chadox1# (accessed February 16, 2021).
22. Medicines & Healthcare Products Regulatory Agency UK. Summary of the Public Assessment Report for AstraZeneca COVID-19 vaccine. (2021). Available online at: https://www.gov.uk/government/publications/regulatory-approval-of-covid-19-vaccine-astrazeneca/summary-of-the-public-assessment-report-for-astrazeneca-covid-19-vaccine (accessed February 16, 2021).
23. Novavax Press Release. Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. (2021). Available online at: https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3 (accessed February 5, 2021).
24. Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. (2021) 21:39–51. doi: 10.1016/S1473-3099(20)30831-8
25. Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis. (2021) 21:181–92. doi: 10.1016/S1473-3099(20)30843-4
26. Ella R, Reddy S, Jogdand H, Sarangi V, Ganneru B, Prasad S, et al. Safety and immunogenicity clinical trial of an inactivated SARS-CoV-2 vaccine, BBV152 (a phase 2, double-blind, randomised controlled trial) and the persistence of immune responses from a phase 1 follow-up report. medRxiv [Preprint]. (2020). doi: 10.1101/2020.12.21.20248643
27. Ella R, Vadrevu KM, Jogdand H, Prasad S, Reddy S, Sarangi V, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: a double-blind, randomised, phase 1 trial. Lancet Infect Dis. (2021). doi: 10.1016/S1473-3099(20)30942-7. [Epub ahead of print].
28. US Food and Drug Administration. Issuance of emergency use authorization, Regeneron Pharmaceuticals. (2020). Available online at: https://www.fda.gov/media/143891/download (accessed December 16, 2020).
29. US Food and Drug Administration. Issuance of emergency use authorization, Eli Lilly. (2020). Available online at: https://www.fda.gov/media/143602/download (accessed December 16, 2020).
30. Centers for Disease Control and Prevention. Reinfection with COVID-19. (2020). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/your-health/reinfection.html (accessed November 17, 2020).
31. Kim AY, Gandhi RT. Re-infection with SARS-CoV-2: what goes around may come back around. Clin Infect Dis. (2020) ciaa1541. doi: 10.1093/cid/ciaa1541
32. Abu-Raddad LJ, Chemaitelly H, Malek JA, Ahmed AA, Mohamoud YA, Younuskunju S, et al. Assessment of the risk of SARS-CoV-2 reinfection in an intense re-exposure setting. medRxiv. (2020). doi: 10.1101/2020.08.24.20179457
33. Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity. (2020) 52:583–9. doi: 10.1016/j.immuni.2020.03.007
34. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. (2020) 581:215–20. doi: 10.1038/s41586-020-2180-5
35. Thanh Le T, Andreadakis Z, Kumar A, Gómez Román R, Tollefsen S, Saville M, et al. The COVID-19 vaccine development landscape. Nat Rev Drug Discov. (2020) 19:305–6. doi: 10.1038/d41573-020-00073-5
36. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. (2020) 584:437–42. doi: 10.1038/s41586-020-2456-9
37. Dan JM, Mateus J, Kato Y, Hastie KM, Faliti CE, Ramirez SI, et al. Immunological memory to SARS-CoV-2 assessed for greater than six months after infection. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.11.15.383323
38. Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. (2020) 1:100040. doi: 10.1101/2020.05.03.20084442
39. Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. (2020) 369:956–63. doi: 10.1126/science.abc7520
40. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. (2020) 183:1024–42.e1021. doi: 10.1016/j.cell.2020.09.037
41. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. (2020) 181:1489–501 e1415. doi: 10.1016/j.cell.2020.05.015
42. Stephens DS, McElrath MJ. COVID-19 and the path to immunity. JAMA. (2020) 324:1279–81. doi: 10.1001/jama.2020.16656
43. Lynch KL, Whitman JD, Lacanienta NP, Beckerdite EW, Kastner SA, Shy BR, et al. Magnitude and kinetics of anti-SARS-CoV-2 antibody responses and their relationship to disease severity. Clin Infect Dis. (2020) 72:301–8. doi: 10.1093/cid/ciaa979
44. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. (2020) 5:eabd7114 doi: 10.1126/sciimmunol.abd7114
45. Perlman S. COVID-19 poses a riddle for the immune system. Nature. (2020) 584:345–6. doi: 10.1038/d41586-020-02379-1
46. Walsh EE, Frenck RW Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. (2020) 383:2439–50. doi: 10.1056/NEJMoa2027906
47. Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N Engl J Med. (2020) 383:2427–38. doi: 10.1056/NEJMoa2028436
48. Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. (2021) 396:1979–93. doi: 10.1016/S0140-6736(20)32466-1
49. Sahin U, Muik A, Vogler I, Derhovanessian E, Kranz LM, Vormehr M, et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. (2020). doi: 10.1101/2020.12.09.20245175
50. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. (2020) 383:1920–31. doi: 10.1056/NEJMoa2022483
51. Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. (2020) 396:467–78. doi: 10.1016/S0140-6736(20)31604-4
52. Zhu FC, Li YH, Guan XH, Hou LH, Wang WJ, Li JX, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. (2020) 395:1845–54. doi: 10.1016/S0140-6736(20)31208-3
53. Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatulin AI, Shcheblyakov DV, Dzharullaeva AS, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. (2020) 396:887–97. doi: 10.1016/S0140-6736(20)31866-3
54. Rausch JW, Capoferri AA, Katusiime MG, Patro SC, Kearney MF. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc Natl Acad Sci USA. (2020) 117:24614–6. doi: 10.1073/pnas.2017726117
55. Centers for Disease Control and Prevention. Emerging SARS-CoV-2 variants. (2021). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html (accessed February 9, 2021).
56. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. (2020) 182:812–27 e819. doi: 10.1016/j.cell.2020.06.043
57. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. (2021) 371:172–7. doi: 10.1126/science.abe5901
58. Zhou P, Shi ZL. SARS-CoV-2 spillover events. Science. (2021) 371:120–2. doi: 10.1126/science.abf6097
59. Lassaunière R, Fonager J, Rasmussen M. Working paper on SARS-CoV-2 spike mutations arising in Danish mink, their 1 spread to humans and neutralization data. (2020). Available online at: https://files.ssi.dk/Mink-cluster-5-short-report_AFO2 (accessed December 16, 2020).
60. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. (2020). doi: 10.1101/2020.12.21.20248640
61. Mansbach RA, Chakraborty S, Nguyen K, Montefiori D, Korber B, Gnanakaran S. The SARS-CoV-2 spike variant D614G favors an open conformational state. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.07.26.219741
62. Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. (2021) 29:23–31.e4. doi: 10.1016/j.chom.2020.11.012
63. Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH 3rd, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. (2020) 370:1464–8. doi: 10.1126/science.abe8499
64. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. (2020) doi: 10.1038/s41586-020-2895-3. [Epub ahead of print].
65. Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. (2020) 184:64–75. doi: 10.1016/j.cell.2020.11.020
66. Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19 neutralizing antibodies predict disease severity and survival. medRxiv. (2020) doi: 10.1101/2020.10.15.20213512
67. Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature. (2020) 586:594–9. doi: 10.1038/s41586-020-2814-7
68. World Health Organization. SARS-CoV-2 mink-associated variant strain – Denmark. (2020). Available online at: https://www.who.int/csr/don/03-december-2020-mink-associated-sars-cov2-denmark/en/ (accessed December 16, 2020).
69. Mallapaty S. COVID mink analysis shows mutations are not dangerous - yet. Nature. (2020) 587:340–1. doi: 10.1038/d41586-020-03218-z
70. Davies N, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. Centre for Mathematical Modelling of Infectious Diseases, London School of Hygiene and Tropical Medicine. (2020). Available online at: https://cmmid.github.io/topics/covid19/uk-novel-variant.html (accessed December 30, 2020).
71. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. MRC Centre for Global Infectious Disease Analysis COVID-19 Report 42 - Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. (2020). Available online at: https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-42-sars-cov-2-variant/ (accessed January 6, 2021).
72. Public Health England. Investigation of novel SARS-CoV-2 variant. Variant of Concern 202012/01 Technical briefing 2. (2020). Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/949639/Technical_Briefing_VOC202012–2_Briefing_2_FINAL.pdf (accessed January 7, 2021).
73. Scientific Advisory Group for Emergencies (SAGE). NERVTAG: Update note on B.1.1.7 severity, 11 February 2021. (2021). Available online at: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961042/S1095_NERVTAG_update_note_on_B.1.1.7_severity_20210211.pdf (accessed February 16, 2021).
74. Centers for Disease Control and Prevention. Emerging SARS-CoV-2 variants. (2020). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html (accessed December 30, 2020).
75. Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.12.28.424451
76. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv [Preprint]. (2021). doi: 10.1101/2020.12.31.425021
77. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Lambson BE, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv [Preprint]. (2021). doi: 10.1101/2021.01.18.427166
78. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv [Preprint]. (2021). doi: 10.1038/s41586-021-03324-6
79. Xie X, Zou J, Fontes-Garfias C, Xia H, Swanson KA, Cutler M, et al. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. bioRxiv [Preprint]. (2021). doi: 10.1101/2021.01.07.425740
80. Muik A, Wallisch AK, Sänger B, Swanson KA, Mühl J, Chen W, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. (2021) 71:1152–1153. doi: 10.1126/science.abg6105
81. Xie X, Liu Y, Liu J, Zhang X, Zou J, Fontes-Garfias CR, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K, and N501Y variants by BNT162b2 vaccine-elicited sera. bioRxiv [Preprint]. (2021). doi: 10.1038/s41591-021-01270-4
82. Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv [Preprint]. (2021). doi: 10.1101/2021.01.25.427948
83. Emary KRW, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 VOC 202012/01 (B.1.1.7). Lancet. (2021). doi: 10.2139/ssrn.3779160. [Epub ahead of print].
84. Sapkal GN, Yadav PD, Ella R, Deshpande GR, Sahay RR, Gupta N, et al. Neutralization of UK-variant VUI-202012/01 with COVAXIN vaccinated human serum. bioRxiv [Preprint]. (2021). doi: 10.1101/2021.01.26.426986
85. Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, et al. Interim results of a phase 1–2a trial of Ad26.COV2.S covid-19 vaccine. N Engl J Med. (2021). doi: 10.1056/NEJMoa2034201
86. Novavax announcement. Efficacy Data Updates from Novavax' Protein-based Vaccine Candidate. (2021). Available online at: https://www.novavax.com/sites/default/files/2021–02/20210202-NYAS-Novavax-Final.pdf (accessed February 16, 2021).
87. Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Continues Important Work to Support Medical Product Development to Address New Virus Variants. (2021). Available online at: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-continues-important-work-support-medical-product-development-address (accessed February 9, 2021).
88. Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. (2020) 369:812–7. doi: 10.1126/science.abc4776
89. Loos C, Atyeo C, Fischinger S, Burke J, Slein MD, Streeck H, et al. Evolution of early SARS-CoV-2 and cross-coronavirus immunity. mSphere. (2020) 5:e00622-20. doi: 10.1128/mSphere.00622-20
90. Iyer AS, Jones FK, Nodoushani A, Kelly M, Becker M, Slater D, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. (2020) 5:eabe0367. doi: 10.1126/sciimmunol.abe0367
91. Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. (2020) 20:457–8. doi: 10.1038/s41577-020-0389-z
92. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. (2020) 584:457–62. doi: 10.1038/s41586-020-2550-z
93. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. (2020) 369:eabc8511. doi: 10.1126/science.abc8511
94. Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. (2020) 21:73–82. doi: 10.1038/s41577-020-00480-0
95. Sagar M, Reifler K, Rossi M, Miller NS, Sinha P, White L, et al. Recent endemic coronavirus infection is associated with less severe COVID-19. J Clin Invest. (2020) 131:e143380. doi: 10.1172/JCI143380
96. Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. (2020) 370:1227–30. doi: 10.1126/science.abd7728
97. Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol. (2020) 5:eabe5511. doi: 10.1126/sciimmunol.abe5511
98. Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med. (2020) 383:1724–34. doi: 10.1056/NEJMoa2026116
99. Lumley SF, O'Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. (2020) 384:533–40. doi: 10.1056/NEJMoa2034545
100. Widge AT, Rouphael NG, Jackson LA, Anderson EJ, Roberts PC, Makhene M, et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med. (2020) 384:80–2. doi: 10.1056/NEJMc2032195
101. Folegatti PM, Bittaye M, Flaxman A, Lopez FR, Bellamy D, Kupke A, et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect Dis. (2020) 20:816–26. doi: 10.1016/S1473-3099(20)30160-2
102. Loos C, Lauffenburger DA, Alter G. Dissecting the antibody-OME: past, present, and future. Curr Opin Immunol. (2020) 65:89–96. doi: 10.1016/j.coi.2020.06.003
103. Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. (2020) 53:524–32 e524. doi: 10.1016/j.immuni.2020.07.020
104. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. (2020) 584:463–9. doi: 10.1038/s41586-020-2588-y
105. Bermejo-Martin JF, Gonzalez-Rivera M, Almansa R, Micheloud D, Tedim AP, Dominguez-Gil M, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit. Care. (2020). doi: 10.1101/2020.08.25.20154252
106. Maucourant C, Filipovic I, Ponzetta A, Aleman S, Cornillet M, Hertwig L, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. (2020) 5:eabd6832. doi: 10.1126/sciimmunol.abd6832
107. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. (2020) 181:1036–45 e1039. doi: 10.1016/j.cell.2020.04.026
108. Sherer ML, Lei J, Creisher P, Jang M, Reddy R, Voegtline K, et al. Dysregulated immunity in SARS-CoV-2 infected pregnant women. medRxiv. (2020). doi: 10.1101/2020.11.13.20231373
109. Pace RM, Williams JE, Jarvinen KM, Belfort MB, Pace CD, Lackey KA, et al. COVID-19 and human milk: SARS-CoV-2, antibodies, and neutralizing capacity. medRxiv. (2020). doi: 10.1101/2020.09.16.20196071
110. Gao X, Wang S, Zeng W, Chen S, Wu J, Lin X, et al. Clinical and immunologic features among COVID-19-affected mother-infant pairs: antibodies to SARS-CoV-2 detected in breast milk. New Microbes New Infect. (2020) 37:100752. doi: 10.1016/j.nmni.2020.100752
111. Gao J, Li W, Hu X, Wei Y, Wu J, Luo X, et al. Disappearance of SARS-CoV-2 antibodies in infants born to women with COVID-19, Wuhan, China. Emerg Infect Dis. (2020) 26:2491–4. doi: 10.3201/eid2610.202328
112. Roarty C, Tonry C, McFetridge L, Mitchell H, Watson C, Waterfield T. Kinetics and seroprevalence of SARS-CoV-2 antibodies in children. Lancet Infect Dis. (2020). doi: 10.1016/S1473-3099(20)30884-7. [Epub ahead of print].
113. Mehta NS, Mytton OT, Mullins EWS, Fowler TA, Falconer CL, Murphy OB, et al. SARS-CoV-2 (COVID-19): what do we know about children? a systematic review. Clin Infect Dis. (2020) 71:2469–79. doi: 10.1093/cid/ciaa556
114. Dhochak N, Singhal T, Kabra SK, Lodha R. Pathophysiology of COVID-19: why children fare better than adults? Indian J Pediatr. (2020) 87:537–46. doi: 10.1007/s12098-020-03322-y
115. ClinicalTrials.gov. Study to describe the safety, tolerability, immunogenicity, and efficacy of RNA vaccine candidates against COVID-19 in healthy individuals. (2020). Available online at: https://clinicaltrials.gov/ct2/show/NCT04368728?term=bnt162&draw=2&rank=6 (accessed December 16, 2020).
116. ClinicalTrials.gov. A study to evaluate the safety, reactogenicity, and effectiveness of mRNA-1273 vaccine in adolescents 12 to<18 years old to prevent COVID-19 (TeenCove). (2020). Available online at: https://www.clinicaltrials.gov/ct2/show/NCT04649151 (accessed December 16, 2020).
117. Koirala A, Joo YJ, Khatami A, Chiu C, Britton PN. Vaccines for COVID-19: the current state of play. Paediatr Respir Rev. (2020) 35:43–9. doi: 10.1016/j.prrv.2020.06.010
118. Li Q, Lu H. Latest updates on COVID-19 vaccines. Biosci Trends. (2020) 14:463–6. doi: 10.5582/bst.2020.03445
119. Marovich M, Mascola JR, Cohen MS. Monoclonal antibodies for prevention and treatment of COVID-19. JAMA. (2020) 324:131–2. doi: 10.1001/jama.2020.10245
120. Cohen MS. Monoclonal antibodies to disrupt progression of early covid-19 infection. N Engl J Med. (2021) 384:289–91. doi: 10.1056/NEJMe2034495
121. Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with covid-19. N Engl J Med. (2021) 384:238–51. doi: 10.1056/NEJMoa2035002
122. Lundgren JD, Grund B, Barkauskas CE, Holland TL, Gottlieb RL, Sandkovsky U, et al. A neutralizing monoclonal antibody for hospitalized patients with covid-19. N Engl J Med. (2021) 384:905–14. doi: 10.1056/NEJMoa2033130
123. Selva KJ, van de Sandt CE, Lemke MM, Lee CY, Shoffner SK, Chua BY, et al. Distinct systems serology features in children, elderly and COVID patients. medRxiv [Preprint]. (2020). doi: 10.1101/2020.05.11.20098459
124. Chakraborty S, Gonzalez J, Edwards K, Mallajosyula V, Buzzanco AS, Sherwood R, et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. (2020) 22:67–73. doi: 10.1038/s41590-020-00828-7
125. Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, et al. A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.09.08.280818
126. Krammer F. SARS-CoV-2 vaccines in development. Nature. (2020) 586:516–27. doi: 10.1038/s41586-020-2798-3
127. Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in non-human primates. N Engl J Med. (2020) 383:1544–55. doi: 10.1056/NEJMoa2024671
128. van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. (2020) 586:578–82. doi: 10.1038/s41586-020-2608-y
129. Shimabukuro TT, Cole M, Su JR. Reports of anaphylaxis after receipt of mRNA COVID-19 vaccines in the US-December 14, 2020-January 18, 2021. JAMA. (2021). doi: 10.1001/jama.2021.1967. [Epub ahead of print].
130. Centers for Disease Control and Prevention. Interim Clinical Considerations for Use of mRNA COVID-19 Vaccines Currently Authorized in the United States. (2021). Available online at: https://www.cdc.gov/vaccines/covid-19/info-by-product/clinical-considerations.html (accessed February 5, 2021).
Keywords: immunity, COVID-19, SARS-CoV-2, duration of protection, receptor-binding domain, spike protein, vaccination, systems serology
Citation: Fergie J and Srivastava A (2021) Immunity to SARS-CoV-2: Lessons Learned. Front. Immunol. 12:654165. doi: 10.3389/fimmu.2021.654165
Received: 15 January 2021; Accepted: 23 February 2021;
Published: 19 March 2021.
Edited by:
Xu Yu, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Ciputra Hartana, Ragon Institute of MGH, MIT and Harvard, United StatesCopyright © 2021 Fergie and Srivastava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Srivastava, YW1pdC5rLnNyaXZhc3RhdmFAcGZpemVyLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.