- 1Department of Blood Transfusion, Huashan Hospital, Fudan University, Shanghai, China
- 2Blood Engineering Laboratory, Shanghai Blood Center, Shanghai, China
- 3Shanghai Key Laboratory of Data Science, School of Computer Science, Fudan University, Shanghai, China
Sepsis is a heterogeneous syndrome caused by a dysregulated host response during the process of infection. Neutrophils are involved in the development of sepsis due to their essential role in host defense. COVID-19 is a viral sepsis. Disfunction of neutrophils in sepsis has been described in previous studies, however, little is known about the role of microRNA-let-7b (miR-let-7b), toll-like receptor 4 (TLR4), and nuclear factor kappa B (NF-κB) activity in neutrophils and how they participate in the development of sepsis. In this study, we investigated the regulatory pathway of miR-let-7b/TLR4/NF-κB in neutrophils. We also explored the downstream cytokines released by neutrophils following miR-let-7b treatment and its therapeutic effects in cecal ligation and puncture (CLP)-induced septic mice. Six-to-eight-week-old male C57BL/6 mice underwent CLP following treatment with miR-let-7b agomir. Survival (n=10), changes in liver and lungs histopathology (n=4), circulating neutrophil counts (n=4), the liver-body weight ratio (n=4–7), and the lung wet-to-dry ratio (n=5–6) were recorded. We found that overexpression of miR-let-7b could significantly down-regulate the expression of human-derived neutrophilic TLR4 at a post-transcriptional level, a decreased level of proinflammatory factors including interleukin-6 (IL-6), IL-8, tumor necrosis factor α (TNF-α), and an upregulation of anti-inflammatory factor IL-10 in vitro. After miR-let-7b agomir treatment in vivo, neutrophil recruitment was inhibited and thus the injuries of liver and lungs in CLP-induced septic mice were alleviated (p=0.01 and p=0.04, respectively), less weight loss was reduced, and survival in septic mice was also significantly improved (p=0.013). Our study suggested that miR-let-7b could be a potential target of sepsis.
Introduction
In 2020, the world was challenged by the COVID-19 pandemic. Despite therapeutic innovations, the novel coronavirus has remained a crude contributor to the mortality rate and has severely endangered public health worldwide (1). Sepsis is defined as a heterogeneous syndrome associated with organ dysfunction (2). The pathogenesis and mechanisms involved in COVID-19 and sepsis both converge on a pivotal role played by the host inflammatory response. Thus, findings concerning sepsis may be useful for anti-inflammatory therapy for patients with COVID-19. Unfortunately, current therapeutic choices are limited and fail to reduce the mortality rate associated with sepsis (3); therefore, a better understanding of the mechanisms of sepsis and providing new targets are critical to improve public health.
Neutrophils are essential for innate immunity and inflammation (4–6). They are primary effectors in immune responses to resist pathogen invasion and are also central contributors of inflammatory reactions (7, 8). They have emerged as important early mediators of inflammatory injury in various tissues, including the skin, heart, joints, and liver (9–13). However, the mechanism by which neutrophils are involved in the regression of inflammation remains unclear and requires further exploration.
There are estimates that 30%–80% of human protein-coding genes are under the control of microRNAs (miRNAs) (14). miRNAs are non-coding small RNA molecules that typically suppress the translation of specific target mRNAs through partial complementarity (15, 16). miRNAs have been identified to be related to inflammation (17). They involve the fine-tuning and moderate suppression of target gene expression, yet master regulators that inhibit various genes in the same pathways (18, 19). MiR-let-7b is a cross-species miRNA, present in multiple genomic locations, including in 10 mature let-7 subtypes and 13 precursor sequences with the same seed sequence (20). MiR-let-7 levels have shown changes in some tumors (21). Gao et al. reported that miR-let-7b targeted TRAF6 in the inflammation response of kidney diseases (22). Reithmair et al. revealed that cellular miR-let-7b was down-regulated in septic shock patients (23). Moreover, restoring miR-let-7b levels reduced the production of IL-6 and TNF-α in neonatal monocytes on LPS stimulation (24). These features suggest that miR-let-7b may be an appropriate regulator for the duration and the magnitude of inflammation.
Toll-like receptors (TLRs) contribute to innate immune recognition of pathogens (25). TLR4 can be easily activated by lipopolysaccharide (LPS), and then triggers the production of proinflammatory mediators. TLRs are a typical innate immune receptor and neutrophils are typical innate immune cells, so the role of TLR4 in neutrophil function has been an area of growing interest (26, 27). Activation of TLR4 by neutrophils can cause the shedding of L-selectin, enhance phagocytosis, reduce chemotaxis, and initiate superoxide generation, and the production of many cytokines (28). Nuclear factor-kappa B (NF-κB) is also a transcriptional factor that regulates a series of genes essential for innate and adaptive immunity and inflammation (29, 30). P65 is a key activation subunit of NF-kB. The canonical NF-kB activation is controlled by its inhibitor, IkB, which blocks the translocation of NF-kB p65 subunit to the nucleus (31, 32). Previous studies have shown that the restoration of the miR-let-7 level can inhibit vascular inflammation mediators, including NF-κB (33). However, the regulatory mechanisms of miR-let-7b on TLR4/NF-κB signaling in neutrophils have not been clearly defined.
Our results suggest that miR-let-7b could regulate immunosuppression by targeting the neutrophilic TLR4/NF-κB signal during CLP-induced sepsis. These results reveal novel mechanisms of the involvement of miR-let-7b in neutrophilic inflammatory activity and provide valuable therapeutic targets for severe inflammation-driven diseases, including sepsis and the current COVID-19.
Materials and Methods
miRNA-Target Prediction and Pathway Analysis
TargetScan (http://www.targetscan.org/vert_71/) was used to predict interactions between miRNA and mRNA using bioinformatics. Supplementary Table 1 outlines the direct targets of hsa-let-7b-5p miRNA. DIANA-mirPath v.3.5 (http://snf-515788.vm.okeanos.grnet.gr/) was used to analyze gene pathways (34) (Supplementary Table 2).
Human Neutrophil Isolation and Coculture
Whole blood was collected from healthy donors from the Shanghai Blood Center, China. Informed consent was provided by donors according to the institutional ethical criteria. Neutrophils were purified from peripheral blood using the MACSxpress® Neutrophil Isolation Kit (Miltenyi Biotec) according to the manufacturer’s guidelines. Red blood cells were lysed with water. Purity was more than 95% using anti-CD15(+) and anti-CD16(+) by flow cytometry carried out on a FACS Calibur cell analyzer (BD Biosciences). The recovery rate was 3 to 5 million neutrophils per milliliter of blood. The cell morphology was examined under a microscope and the cell preparations activated during the separation process were excluded. Cells were then cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS, Sigma, USA) in an incubator with 5% CO2 at 37°C. MiR-let-7b mimics (100 nM), miR-let-7b inhibitor (100 nM), and miR-let-7b mimics negative control (NC, 100 nM) were introduced in cultures with human-derived neutrophils four hours after LPS (100 ng/mL) preconditioning, respectively. MiRNA inhibitor is a single-stranded RNA molecule that can specifically inhibit endogenous miRNA function after binding to mature miRNA. Cells were harvested for subsequent real-time quantitative polymerase chain reaction (RT-qPCR) assay and western blotting analysis after four hours of treatment, and cell culture supernatants were harvested at 10 hours for ELISA detection.
Western Blotting
The lysates were obtained using RIPA buffer (Sigma, USA) premixed with a protease inhibitor cocktail kit (Thermo, USA). Nuclear proteins were extracted using a nuclear and cytoplasmic protein extraction kit (Sangon, China, #C510001). Cells were lysed for 30 minutes and then centrifuged for five minutes at 12,000 ×g at 4°C. Next, the supernatant containing protein was resolved on a 10% acrylamide‐bisacrylamide gel (EpiZyme, China, #PG112), and the proteins were transferred onto a 0.45‐μm PVDF membrane (Millipore, Germany, #IPVH00010). Membranes were blocked using protein-free rapid blocking buffer (EpiZyme, #PS108) and then were incubated with primary antibodies at 4°C overnight, followed by a 1-hour incubation with horseradish-peroxidase (HRP)-conjugated secondary antibodies at room temperature. The primary antibodies used were as follows: anti-TLR4 (1:2000, ABclonal, China, #A17436), anti-NF‐κB p65 (1:3000, Cell Signaling, USA, #8242S), anti-IkBα (1:2000, Proteintech, China, #10268-1-AP), anti-β-actin (1:1000, Beyotime, China, #AF0003), and anti-GAPDH (1:2000, Proteintech, #10494-1-AP). All experiments were performed in triplicate.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from treated neutrophils using Trizol reagent (Invitrogen, USA) and miRNA was isolated with the mir-Vana™ miRNA Isolation Kit (Ambion). Total RNA was reverse transcribed into cDNA using a cDNA Synthesis Kit (Takara) according to the manufacturer’s instructions. The levels of miRNA were determined using Bulge-LoopTM miRNA RT-qPCR system (Ribobio, Guangzhou, China). Next, RT-qPCR was performed using targeted gene primers (BioTNT) following the manufacturer’s cycling parameters and run on an ABI Prism 7500 Sequence Detection System (Applied Biosystems) using TB Green MasterMix (Takara). The primer sequences used were GAPDH forward 5’- GGG AAG GTG AAG GTC GGA GT -3’, GAPDH reverse 5’-GGG GTC ATT GAT GGC AAC A -3’; TLR4 forward 5’-GCA CAT CTT CTG GAG ACG ACT -3’, TLR4 reverse 5’-CAT CCT GTA CCC ACT GTT CCT -3’; IL-6 forward 5’- CAC TGG TCT TTT GGA GTT TGA G -3’, IL-6 reverse 5’- GGA CTT TTG TAC TCA TCT GCA C -3’; IL-8 forward 5’- AAC TGA GAG TGA TTG AGA GTG G -3’, IL-8 reverse 5’- ATG AAT TCT CAG CCC TCT TCA A -3’; TNF-α forward 5’- TGG CGT GGA GCT GAG AGA TAA CC -3’, TNF-α reverse 5’- CGA TGC GGC TGA TGG TGT GG -3’;IL-10 forward 5’- GTT GTT AAA GGA GTC CTT GCT G -3’, and IL-10 reverse 5’- TTC ACA GGG AAG AAA TCG ATG A -3’. Relative fold changes in expression were calculated by normalizing to a housekeeping gene (GADPH) to adjust for loading variation. Primers of miRNAs (miR-let-7b-5p and U6) were designed by RIBOBIO Corporation (Guangzhou, China). U6 was used as the internal control. The miRNAs sequences are covered by a patent.
Flow Cytometry
Human neutrophil purity, mouse neutrophil counts, and human TLR4 expression were both measured by flow cytometry. Cells were collected and centrifuged at 250 ×g for five minutes. After being resuspended in 100 μL binding buffer for cell purity and counts, human neutrophils were marked with 10 μL PE-conjugated CD15 (BD Biosciences, CA) and 10 μL APC-conjugated CD16 (BD Biosciences), and neutrophil counts were marked with FITC-conjugated CD11b, APC-conjugated Ly-6G and PE-conjugated Ly-6C (BioLegend, CA) for 20 minutes at room temperature. Then, 200 μL binding buffer was added and cells were evaluated by a fluorescence-activated cell sorting (FACS) Calibur device (BD Biosciences). Expression of TLR4 was evaluated by flow cytometry using Goat anti-TLR4 primary antibodies (R&D Systems, USA, #AF1478). Donkey anti-Goat IgG-AlexaFluor 647 (1:500, Absin, China, #abs20027) were used as secondary antibodies. Cells were measured by a FACS Calibur device (BD Biosciences). Cell populations were analyzed using the FlowJo software, v10.4.
Enzyme-Linked Immunosorbent Assay
Human IL-6, IL-8, IL-10, and TNF-α levels in the culture supernatants were measured with enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s protocols (R&D Systems, USA). Mouse IL-6 and CXCL1 levels in the serum were measured with ELISA kits following the manufacturer’s instructions (Novus, USA).
Immunofluorescence
Neutrophils were cocultured with FITC-conjugated miR-let-7b mimics in a six-well plate for 30 minutes and were then centrifuged at 250 ×g for five minutes. Cells were then fixed with 4% formaldehyde for 20 minutes and resuspended in 200 μL deionized H2O. A volume of five μL cell suspension was added to the gelatin-coated slide and smeared with a pipette tip. Samples were surrounded with a hydrophobic barrier using a Super Pap Pen (XLPCC, Japan, XL2001). Samples were blocked in a blocking buffer (Beyotime, China, #P0102) for 45 minutes. Slides were then incubated with 10 μg/mL of TLR4 primary antibodies (R&D Systems, USA, #AF1478) and normal goat IgG control (R&D systems, #AB-108-C) overnight at 2–8°C. Slides were washed two times using 1% PBS and then incubated with secondary antibodies (1:500, Absin, China, #abs20027) for one hour. Slides were then washed as described previously. DAPI counterstain was added, incubated two–five minutes at room temperature and coverslips were mounted. The TLR4-miR-let-7b localization was visualized using the inverted FV1000-IX81 microscope (Olympus, Japan). Images were captured at 100× and 180× objectives using the FV10-ASW software, v01.01.
Cecal Ligation and Puncture (CLP) Mice Model
Six-to-eight-week-old male C57BL/6 mice were purchased from the Shanghai Jie Si Jie Laboratory Animal Ltd. Animal experiments were approved by the Institutional Animal Care and Use Committee of Huashan Hospital, Fudan University. Animal models of sepsis induced by cecal ligation and puncture (CLP) were performed on C57BL/6 mice according to Rittirsch et al. (35). Mice were randomly divided into four groups: (1) sham group, sham operation without treatment; (2) CLP group; (3) CLP + agomiR-Let-7b-5p group; (4) CLP + agomiR-NC group. Agomir NC and agomiR-Let-7b-5p (Ribobio, Guangzhou, China) were directly injected into the tail vein at the dose of 10 nmol per mouse suspended in 200 μL of saline, respectively. All mice in groups three and four were injected one hour before surgery, and all four groups of mice were sacrificed after 48 hours. Blood was collected 48 hours after surgery by retro-orbital bleeding. After euthanasia, the lower two-thirds of the right lung was ligated and the remaining parts were lavaged twice with one mL cold sterile PBS to harvest the bronchoalveolar lavage fluid (BALF).
The white blood cell (WBC) and polymorphonuclear (PMN) counts were performed using the auto hematology analyzer (Mindray, China). The pooled BALF was analyzed for pulmonary PMN counts by flow cytometry (C6 Accuri, BD Sciences). The ligated two-thirds of the right lobe of the lung were dissected, one-third was used for analyzing the lung wet-to-dry weight ratios (the wet weight of lung was measured using an electronic scale and was then dried in the oven at 70°C for 24 hours to determine the dry weight (36)) and for macroscopic observation, the other third was fixed in 4% paraformaldehyde for tissue histology. The liver was weighed to calculate the liver-to-body ratio (liver-to-body ratio=liver mass/body mass). To evaluate survival rates, mice were monitored every 12 hours. Survival was estimated from the time of CLP surgery.
Histology and Immunohistochemistry
After harvesting of BALF, one-third of the right lobe of the lung and the whole liver of each mouse were both dissected and fixed in 4% paraformaldehyde for 24 h. Tissues were embedded in paraffin and 4-μm sections were cut. Hematoxylin and eosin (H&E) and immunohistochemistry (IHC)-stained with anti-CD11b rabbit polyclonal (1:500, Servicebio, China) sections were prepared using standard techniques. CD11b positive cells were quantified in five random fields (400× magnification) and imaged using a slide scanner microscope (Nikon 80i, Germany). The area occupied was analyzed by selecting brown areas using Image J software (NIH, USA).
Statistical Analysis
Experiments were performed at least three times with consistent results. R (version 4.0.3) were applied to statistical analyses. The correlation analysis was performed using the “glm” function in R. The prognosis analyses were performed using the R package “survival” to generate the Kaplan–Meier (K-M) curves and to calculate the log-rank p-value. Error bars are represented as means ± standard deviations (SD). Significances of two-group comparisons were determined using a two-tailed Student’s t-test. Comparisons of significant differences of more than two groups were analyzed by one-way ANOVA. Results were considered statistically significant at p-value <0.05.
Results
TLR4 Was Identified as a Potential Target of MiR-let-7b in Neutrophils
Using the data provided in the Human Protein Atlas (https://www.proteinatlas.org), a genome-wide transcriptomic depository of protein-coding genes in human blood cells (37), we first evaluated the presence of TLR4 expression (Figure 1) and the associated functional data of human neutrophils. Prior studies have shown that neutrophils could express TLRs 1, 2, 4, 5, 6, 7, 8, 9, and 10, which are essentially all the TLRs except for TLR3 (28). According to the analysis of the transcriptome, we determined the fraction of transcripts corresponding to different genes in each analyzed cell type and tissue. Thus, we reported the transcriptome usage for each representative blood cell types based on within-sample normalized pTPM values (Figure 1A) and between-sample normalized expression (NX) values (Figure 1B). The pTPM value was calculated by zooming in to a sum of 1 million TPMs of each sample to compensate for the previously deleted non-coding transcripts. The resulting transcriptional expression values, representing NX, were generated using 18 blood cell types and internal normalized channels of total PBMC. In silico analyses of the TLR and NF-κB signaling pathways indicated that miR-let-7b was among the top-ranked miRNA regulators of these two key inflammatory pathways (Supplementary Figure 1 and Supplementary Table 1). The analysis showed that human miR-let-7b targets 15 and 14 genes within the TLR and NF-κB pathways, respectively. We found that TLR4 ranked highly in both pathways. Bioinformatics analysis identified that miR-let-7b could target TLR4 transcripts among the most significant regulators of the TLR signaling pathway (Supplementary Figure 1 and Supplementary Table 2). The potential binding sites are shown in Figure 1C. Neutrophil extracellular traps (NETs) could be easily activated in sepsis or ALI (38, 39), thus we hypothesized that miRNA mimics, as is the case of bacteria, could target neutrophils in NETs-mediated sterile inflammation. After stimulation, resting neutrophils changed from sphere to oblate. Immunofluorescence colocalization analysis in neutrophils was used to examine the location and binding of TLR4 protein and FITC-conjugated miR-let-7b mimics. We found that miR-let-7b-FITC mimics were detectable within the neutrophil cytoplasm (nuclei stained blue DAPI) after 30 minutes of treatment (data showed in Figure 1D) without transfection. Consistent with the data analysis in Figures 1A, B, specific binding of AF647-conjugated TLR4 confirmed its high expression on the neutrophil cell membrane. Taken together, these data identified TLR4 as a direct target of miR-let-7b and miR-let-7b as an important regulator of inflammation-related activity.
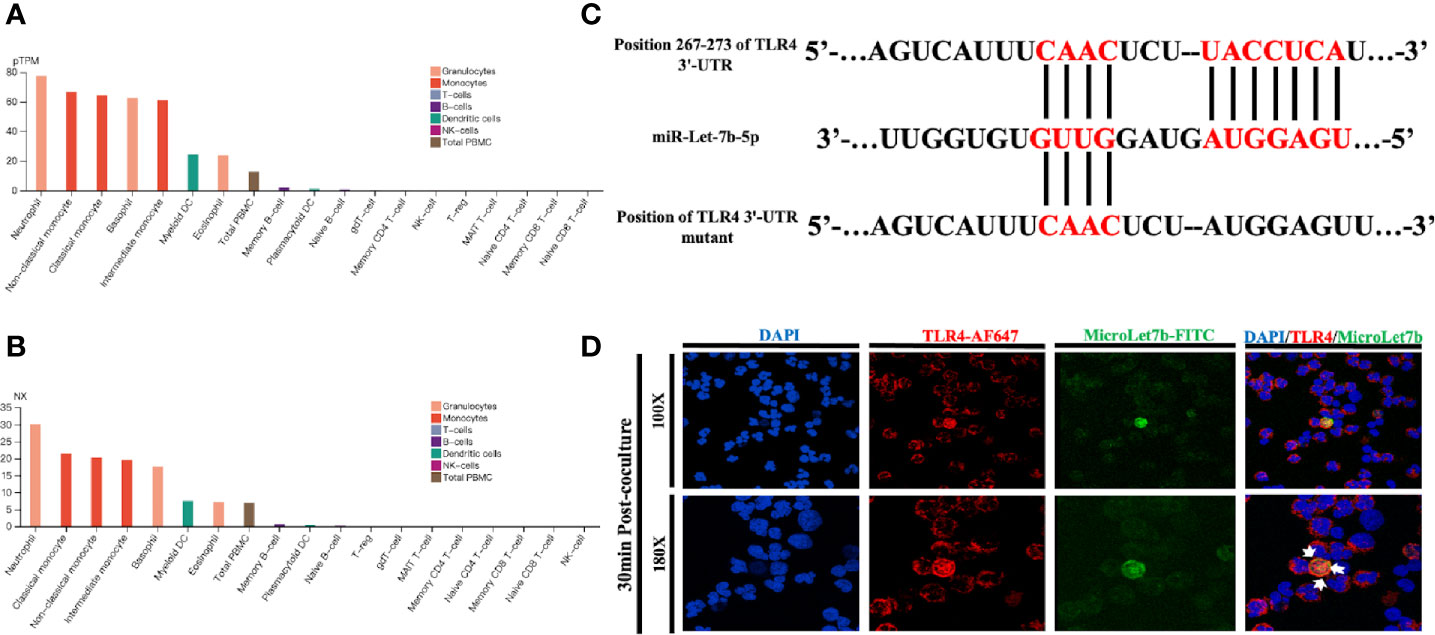
Figure 1 miR-let-7b directly targets TLR4 in neutrophils. (A) Expression profiles based on within-sample normalized pTPM for TLR4 gene enriched in 18 blood cell types and total PBMC ranked according to protein transcript expression values (see www.proteinatlas.org for details). (B) Same as (A), but based on the between-sample NX values. (C) The sequence of human TLR4 3’-UTR (wild type and mutant) is predicted to be targeted by the mature sequence of miR-let-7b. Targeted nucleotides are labeled in red. (D) Confocal microscopy was performed on FITC-labeled miR-let-7b (green fluorescence) and cocultured for 30 minutes with AF647-labeled neutrophil TLR4 (red fluorescence) to demonstrate miR-let-7b internalization. Blue nuclear staining was performed with 4,6-diamidino-2-phenylindole (DAPI). One representative image of the immunostainings is shown. Scale bar denotes 100x and 180x objectives.
MiR-let-7b Activated the TLR4/NF-κB Pathway and Inhibited TLR4 Expression at the Post-transcriptional Level in Neutrophils
To understand the mechanism responsible for miR-let-7b and TLR4-related inflammatory activity, we investigated the influence of miR-let-7b mimics (100 nM), miR-let-7b inhibitor (100 nM), and its negative control (NC, 100 nM), on TLR4 expression in neutrophils. IkB is a regulator of NF-kB upstream. It can bind to NF-kB and be degraded when the NF-kB signaling is activated. The expression level of IkB and nuclear NF-κB p65 can indirectly reflect NF-κB activation. As shown in Figure 2A, culture with miR-let-7b mimics resulted in a reduction in TLR4 and nuclear NF-κB p65 but increasing cytosolic IkBα protein expression (p<0.05, p<0.01, p<0.05, respectively), while treatment with the miR-let-7b inhibitor showed significant upregulation in TLR4 and nuclear NF-κB p65 but reduction in IkBα protein level (p<0.01, p<0.05, p<0.05, respectively). Similar results were also obtained using flow cytometry to detect cell surface expression of TLR4+ (Figure 2B). A lower percentage of TLR4+ (1.26%) was detected on the membrane of human neutrophils cultured with miR-let-7b mimics, whereas a higher percentage (9.41%) of TLR4+ cells on the membrane of human neutrophils cultured with miR-let-7b inhibitor were observed compared with control neutrophils (6.28%). In contrast, the miR-let-7b inhibitor increased TLR4 protein expression in a dose-dependent manner (Figure 2C). These results led us to ask whether miRNAs, which would influence both the translation and stability of mRNA, were involved in this process. But RT-qPCR showed that exposure to miR-let-7b exerted no significant impact on TLR4 mRNA levels (Figure 2D). These findings indicated that miR-let-7b could activate the TLR4/NF-κB pathway and inhibited TLR4 expression via post-transcriptional regulation in neutrophils.
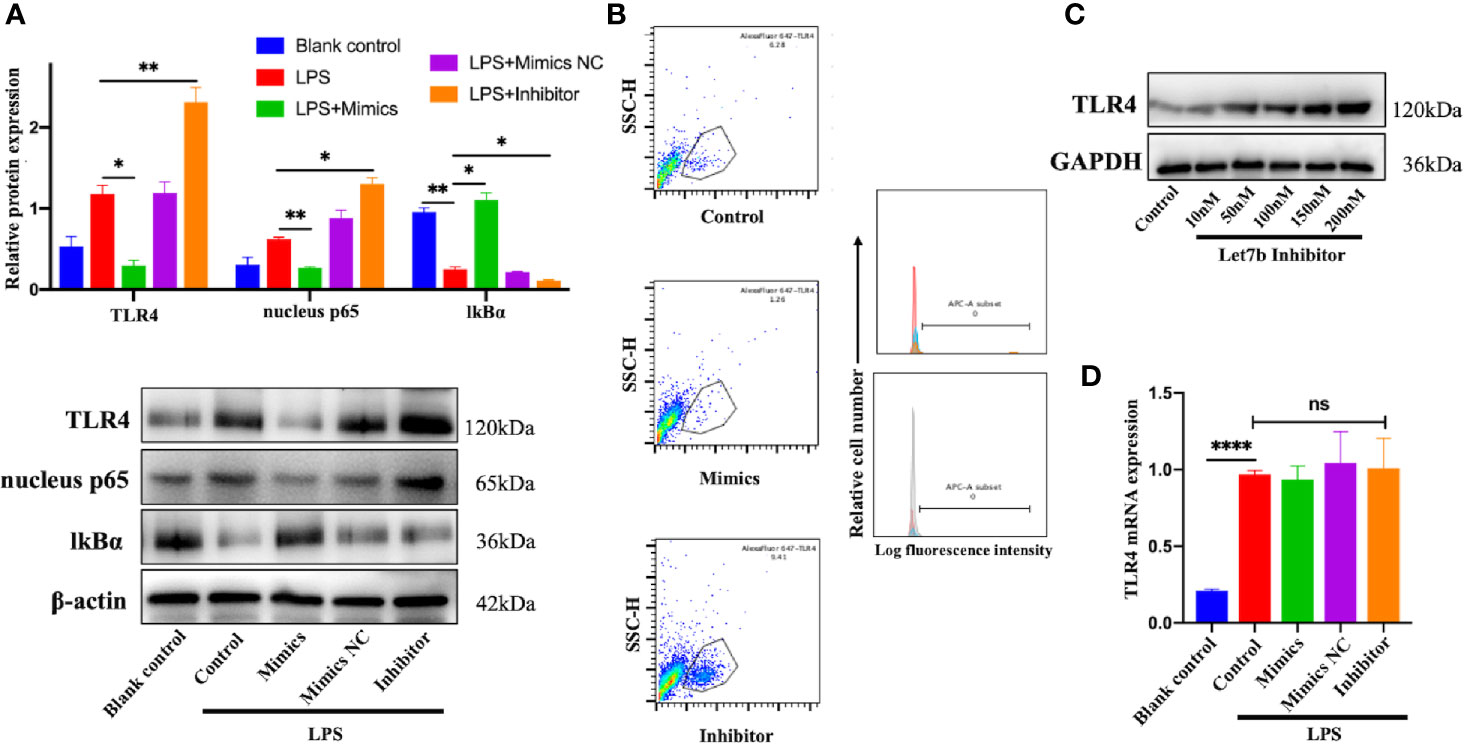
Figure 2 miR-let-7b activates the TLR4/NF-κB signaling and inhibits TLR4 expression via post-transcriptional regulation. (A) Neutrophils treated with 100 ng/mL LPS were then co-incubated with miR-let-7b mimics, miR-let-7b inhibitor, and miR-let-7b mimics NC (100 nM, respectively) for four hours followed by Western blotting to detect TLR4, nucleus p65, and cytosolic IkBα. *P < 0.05, **P < 0.01. (B) Isolated neutrophils were incubated with LPS alone or with indicated concentrations of miR-let-7b mimics and inhibitor (100 nM, each) for four hours followed by measurement of TLR4 binding (staining with AF647) by flow cytometry. The blue line is AF647-stained neutrophils cultured for four hours only (Blank), the red line is AF647-stained neutrophils pretreated with LPS and then cultured for four hours (Control), the brown line is AF647-stained neutrophils pretreated with LPS and then cocultured with miR-let-7b mimics for four hours (Let-7b mimics), and the gray line is AF647-stained neutrophils pretreated with LPS and then cocultured with miR-let-7b inhibitor for four hours (let-7b inhibitor). (C) A dose-dependent increase of TLR4 protein was determined with miR-let-7b inhibitor treatment. (D) RT-qPCR of miR-let-7b expression in human neutrophils. ****P < 0.0001, not significant (ns). All traces are representative of ≥3 independent experiments. Data represent mean ± SD (n=3).
MiR-let-7b Mediated the Differential Effects on Secretion of IL-6, IL-8, TNF-α and IL-10 in Neutrophils
Infection can stimulate the expression of pro-inflammatory genes, such as IL-6, TNF-α, etc., which can effectively eliminate microorganisms, accelerate tissue repair, and secret IL-10 and other anti-inflammatory cytokines to alleviate the inflammatory reaction (40). If the pro-inflammatory cytokine levels are balanced with those of anti-inflammatory cytokines, then the internal microenvironment and homeostasis are maintained; otherwise, a systemic inflammatory response or anti-inflammatory syndrome may result (41, 42), which from a physiological standpoint, would be caused by excessive activation of the body’s preventative mechanisms, rather than the outcome of viral or bacterial infection. To directly assess the effects of miR-let-7b on the secretion of inflammatory-related cytokines, miR-let-7b mimics, inhibitor, and mimics NC were cocultured with human-derived neutrophils four hours after LPS preconditioning, respectively (Figures 3A–H). The neutrophils could be activated by LPS into a pro-inflammatory status. The RT-qPCR and ELISA results showed that after pre-treatment with LPS and incubation with miR-let-7b mimics, neutrophils dramatically produced significantly fewer pro-inflammatory cytokines, including IL-6, IL-8 and TNF-α and more anti-inflammatory cytokine, IL-10 at 10 hours. Conversely, the miR-let-7b inhibitor promoted IL-8 and TNF-α production in neutrophils, but only decreased IL-10 secretion. Taken together, we made the preliminary conclusion that miR-let-7b is a homeostatic regulator of anti-inflammatory activity.
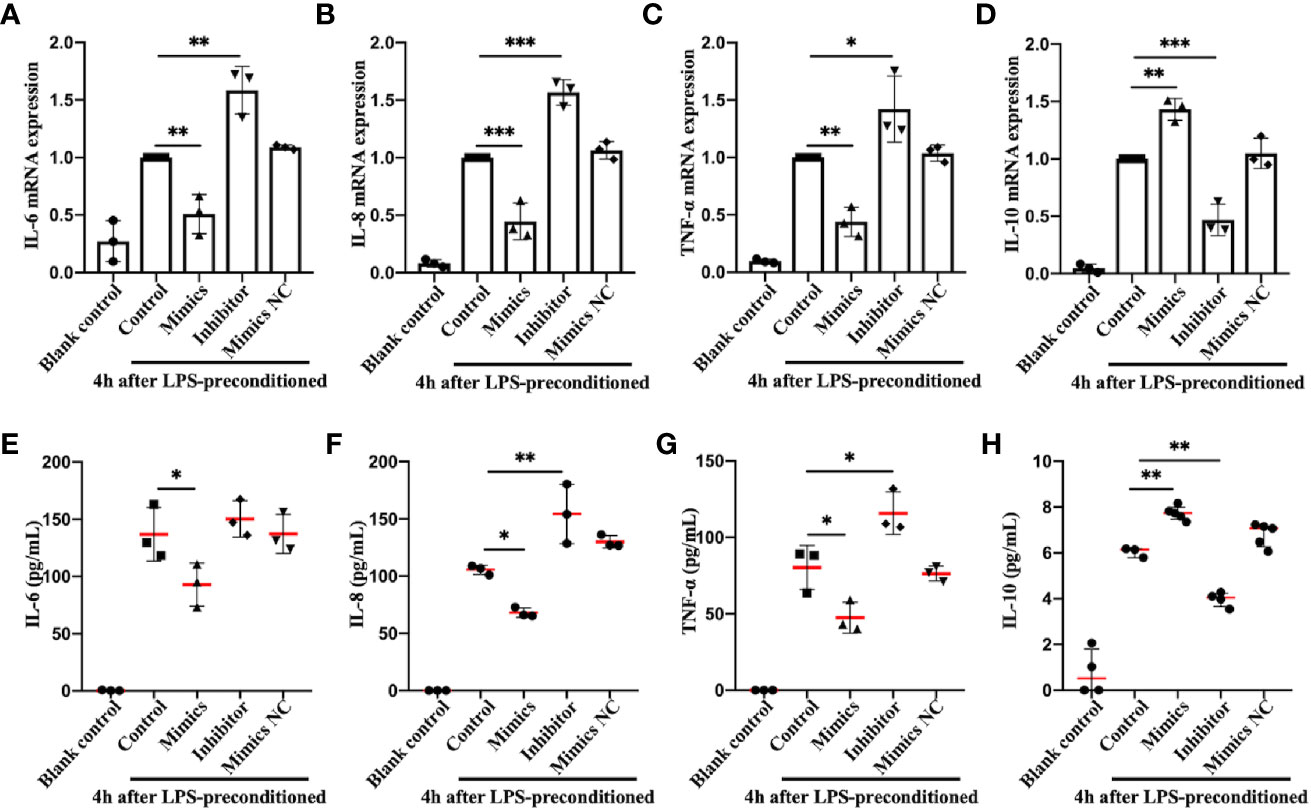
Figure 3 Identification and expression of inflammation-related cytokines in neutrophils in vitro. RT-qPCR (A–D) and ELISA (E–H) detection in the supernatant of IL-6, IL-8, TNF-α, and IL-10 in human freshly isolated neutrophils. Cells were incubated with miR-let-7b mimics, miR-let-7b inhibitor, and miR-let-7b mimics NC (100 nM, respectively), and the supernatant was collected at 10 hours after stimulation with 100 ng/mL LPS for 4 h. Data represent mean ± SD (n=3–5). Tukey’s multiple comparison tests were used to generate the P-values indicated in the figure. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-let-7b Exerted Anti-Inflammatory Effects Through Activating TLR4/NF-κB Pathway in CLP-Induced Septic Mice
To investigate the role of miR-let-7b in inflammation in vivo, we performed the CLP surgery to establish a polymicrobial sepsis model. The expression of miR-let-7b was first detected in blood samples from sham and CLP-induced septic mice. We found that miR-let-7b expression was significantly decreased in septic mice compared with sham treated mice (p<0.0001, Figure 4A). Sepsis is a kind of severe inflammatory host response diagnosed with inflammatory variables like leukocytosis (WBC counts above 12,000/μL) or leukopenia (WBC counts below 4,000/μL) (43–45). The more severe the septic inflammation, the closer the disease to sepsis shock, the lower the WBC level. We, therefore, detected the circulating WBC and found that CLP-induced septic mice had leukopenia and WBC levels that were lower compared to the sham group (p<0.001, Figure 4B). Correlational analysis in Figure 4C showed that miR-let-7b negatively correlated with the degree of septic inflammation (r = -0.8389, p = 0.0024).
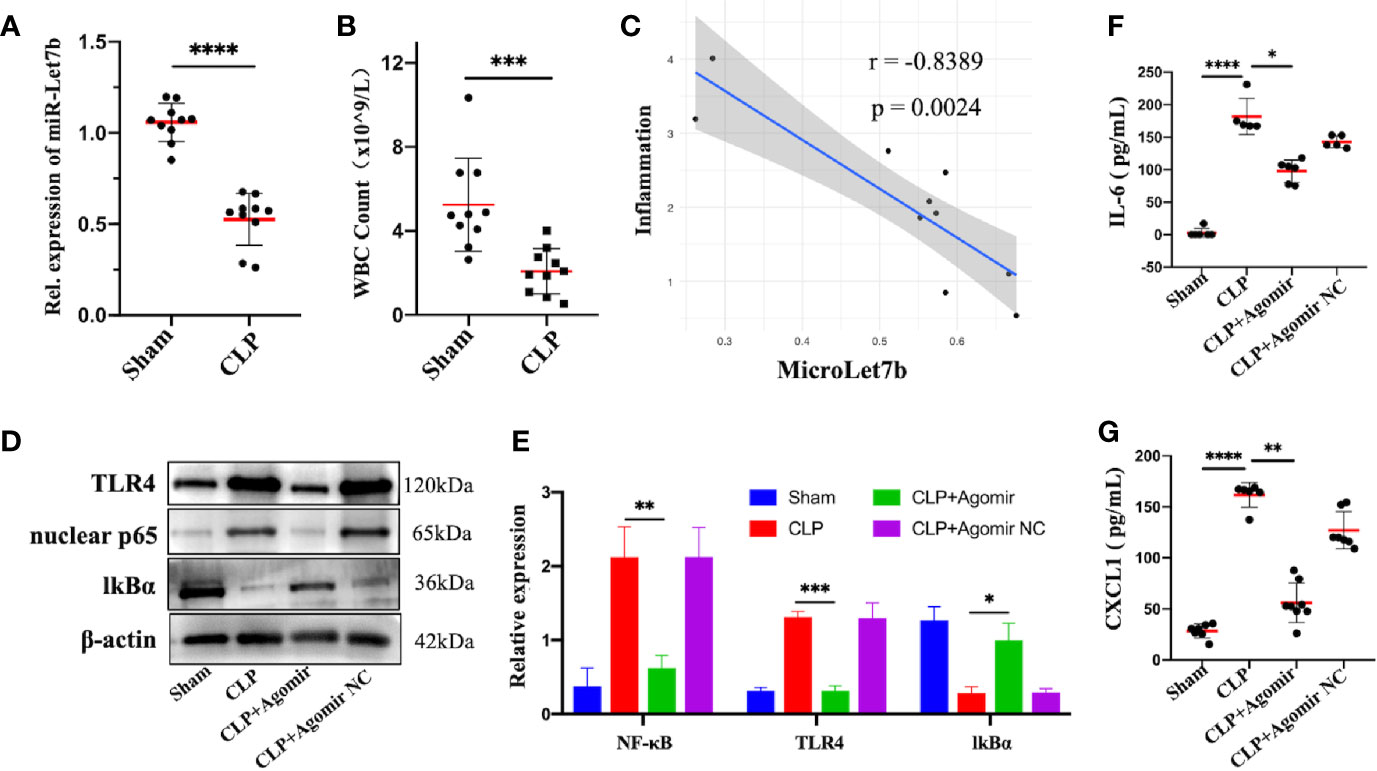
Figure 4 Overexpression of miR-let-7b suppresses neutrophilic inflammation in vivo. (A) Expression of miR-let-7b in sham and CLP-induced septic mice from the peripheral blood using RT-qPCR (n=10, ****P < 0.0001). (B) Total WBC count in sham and CLP-induced septic mice from the peripheral blood (***P < 0.001). (C) Correlation analysis between the level of miR-let-7b and the degree of septic inflammation (r=-0.8389, p=0.0024). (D, E) Effects of miR-let-7b agomir and its NC on neutrophilic TLR4, nuclear NF-κB p65 and IkB expression in CLP-induced septic mice were assessed by western blotting. *P < 0.05, **P < 0.01, ***P < 0.001. (F, G) Serum levels of IL-6 (F) and CXCL1 (G) were measured by ELISA in mice (n = 5-8 mice per group, *P < 0.05, **P < 0.01, ****P < 0.0001).
The critical role of miR-let-7b observed in vitro led us to speculate whether it is also involved in anti-inflammatory activity in vivo. As expected in western blotting analysis, TLR4 and nuclear NF-κB p65 expression increased while cytosolic IkBα decreased in CLP-induced septic mice, similar to that observed following exposure to miR-let-7b agomir NC, while overexpression of miR-let-7b by treatment with its agomir significantly decreased TLR4 and nuclear NF-κB p65 and increased IkBα protein levels in neutrophils (Figures 4D, E). Additionally, treatment with miR-let-7b agomir significantly decreased IL-6 and CXCL1 levels in mice serum relative to the CLP-treated group (Figures 4F, G). Thus, the inhibitory activity of miR-let-7b on CLP-induced pro-inflammatory cytokines secretion was attenuated with TLR4 and nuclear NF-κB p65 co-overexpression. Overall, these results demonstrated that miR-let-7b inhibited CLP-induced inflammation partly through the miR-let-7b/TLR4/NF-κB axis in neutrophils.
Targeting MiR-let-7b Can Ameliorate Lungs and Liver Inflammation and Improve Survival in Sepsis Mice
Pulmonary swelling and liver hyperemia are well-established features of the CLP-induced inflammatory model (35). On the basis of this model, we found that more than 50% of CLP-treated mice had simultaneously exhibited visibly evident liver hyperemia and pulmonary edema compared to the sham group (Figures 5A, B). Our present data revealed miR-let-7b as a potential molecule involved in the inflammatory responses. To confirm that miR-let-7b exerts important anti-inflammatory activity, we observed both inflamed organs in the CLP-induced polymicrobial sepsis. Tissue damage and inflammation in the liver (Figure 5C) and lungs (Figure 5H) were assessed by H&E and IHC. IHC analysis was performed for CD11b, a neutrophil/macrophage marker, in representative cases on the same section exposed to H&E staining. As presented in Figures 5C–E, in both the CLP and agomir NC groups, prominent hepatic inflammation in the form of hepatic sinusoid adherence, shrinkage cracking, hepatic steatosis, vacuoles, and higher liver/body weight ratio (Figure 5E) was observed. A hallmark feature of inflammation is neutrophil recruitment from the blood to the inflammatory tissue (46, 47). The total neutrophil counts in Figure 5F showed that neutrophils in peripheral blood were significantly reduced upon CLP treatment. Results in Figures 5D, F thus mutually corroborated each other. MiR-let-7b agomir treatment produced a significant improvement in these liver injury parameters.
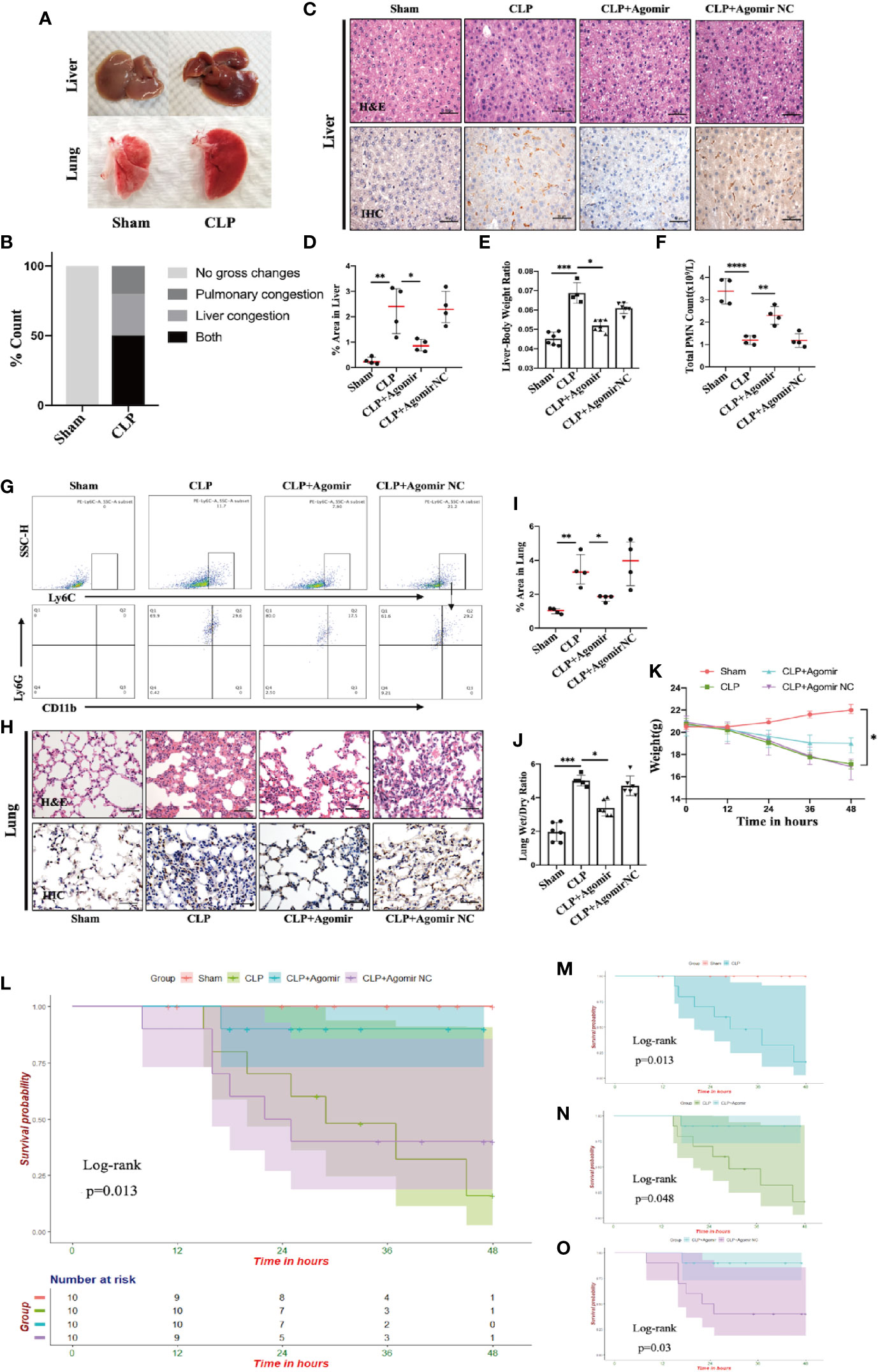
Figure 5 Targeting miR-let-7b alleviates tissue injury and improves overall survival in CLP murine models. (A, B) Gross pathologic changes in mouse livers and lungs showing pulmonary/liver congestion in sham and CLP-treated mice groups. (C) Histological assessment of livers harvested at 48 hours. Shown are representative H&E stain (upper panel) and IHC images (lower panel). Infiltrating neutrophils were identified by CD11b (brown), Scale bars, 50 μm. (D) Area of IHC positive for CD11b expressed as a percentage and assessed in five aleatory images. *P < 0.05; **P < 0.01, one-way ANOVA, n= 4/group. (E) The whole liver was dissected and weighed for determination of liver-body weight ratio (liver to body ratio=liver mass/body mass). n=4–7/group. (F) Total neutrophil counts in the blood. **P < 0.01, ****P < 0.0001, one-way ANOVA, n= 4/group. (G) Percentage of neutrophil infiltration in mice lung was detected in BALF. Representative flow cytometry plots of CD11b+ and Ly6G+ frequency (bottom row) among Ly6Cint cells (top rows). At the end of the experiment (48 h), the lower two-thirds of the right lung were dissected, of which one-third was for lung histology (H, I, same as liver) and the other one-third was weighed for the lung wet-to-dry weight ratio analyses (J) (n=5–6, one-way ANOVA, *P < 0.05, **P < 0.01). (K) The weight of C57BL/6 mice was monitored every 12 hours (*P < 0.05). (L) Survival rate was performed by Kaplan-Meier analysis among four groups. p = 0.013. Survival rates were also performed between sham and CLP group (M), CLP and CLP + miR-let-7b agomir group (N), and CLP + miR-let-7b agomir and CLP + miR-let-7b agomir NC group (O). n=10 mice/group, Log-rank test, p = 0.013, p = 0.048 and p = 0.03, respectively.
Sepsis is an infection-induced systemic inflammatory response syndrome (SIRS) with high morbidity and mortality (48). Sepsis patients are often characterized by lung function impairment (49). We next explored whether miR-let-7b could inhibit CLP-induced lung inflammation. In the BALF of CLP-induced septic mice, neutrophils were labeled with anti-Ly6G, anti-CD11b, and anti-Ly6C markers for flow cytometry analysis. As shown in Figure 5G, pre-treatment with miR-let-7b agomir attenuated lung neutrophilic inflammation, characterized by the reduced recruitment of neutrophils (stained with CD11b+, Ly6G+, and Ly6C int). An increased percentage (29.6%) of neutrophils (+) was observed in the BALF from lungs of CLP mice, whereas lower percentages (17.5%) of neutrophil (+) cells exposed to miR-let-7b agomir were observed compared with of baseline levels of miR-let-7b agomir-NC pre-injection (29.2%). Similarly, the CLP-induced mouse presented alveolar wall congestion, perivascular tissue edema, structural disorder, thickening of the alveolar septum, alveolar cavity narrowing, neutrophils infiltration, and higher lung wet/dry ratio (Figures 5H, J). In comparison, miR-let-7b agomir–treated mice significantly reduced these lung inflammatory responses, although it did not completely rectify them. In Figures 5D, I, CLP-induced septic mice exhibited an augmented positive signals indicative of CD11b expression (versus sham group), while miR-let-7b agomir treatment reduced this effect in both the liver and lungs (p=0.01 and p=0.04, respectively), further supporting the pivotal role of miR-let-7b during sepsis. It was worth mentioning that the total neutrophil counts in Figure 5F corroborated with these two results. During the modeling period, the body weights and survival status of all mice were measured every 12 hours. MiR-let-7b agomir treatment for 48 hours also resulted in reduced weight loss compared to both the CLP and CLP + agomir NC treatment (Figure 5K). Importantly, treatment with miR-let-7b agomir improved the survival rate (p=0.013) as evaluated using R software (survival package) (Figure 5L). CLP group mice showed worse survival compared to the sham group (Figure 5M). Significant improvements in overall survival were then observed in CLP and CLP + miR-let-7b agomir group (Figure 5N), and CLP + miR-let-7b agomir and CLP + miR-let-7b agomir NC group (Figure 5O). Altogether, these data pointed to a protective effect of miR-let-7b on CLP-mediated sepsis in vivo. A possible mechanism of miR-let-7b action in vitro and in vivo is indicated and outlined in Figure 6.
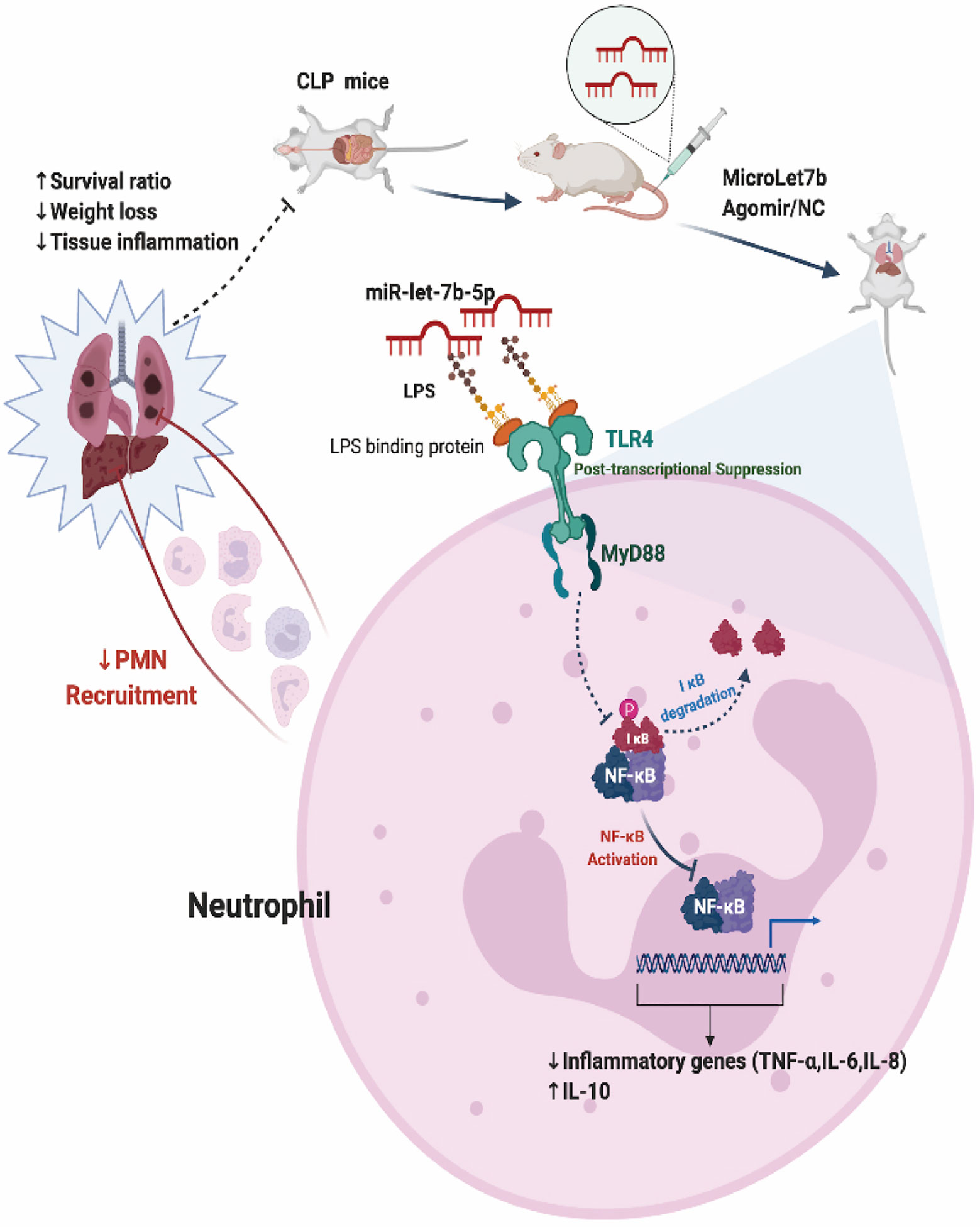
Figure 6 Proposed model for miR-let-7b-mediated regulation of neutrophilic functions that target TLR4/NF-κB axis in vitro and in vivo.
Discussion
In the present study, we demonstrated that miR-let-7b could alter neutrophil function by suppressing the TLR4/NF-κB signaling pathway. Restoring miR-let-7b levels could effectively decrease levels of IL-6, IL-8, and TNF-α, while it increased IL-10 levels in freshly isolated human neutrophils. Treatment with miR-let-7b agomir could effectively protect mice from septic shock by reducing neutrophil recruitment into the liver and lungs. Here, we demonstrated that miR-let-7b expression was highly induced in an experimental murine model of sepsis. Given our findings, miR-let-7b is an attractive candidate for therapeutic strategies for severe inflammatory diseases.
Neutrophils are usually the first cells to reach the injured tissue, and thus may play an important role in the formation of the tissue inflammatory environment and exert profound effects on other somatic and immune cells. The migration of neutrophils into tissues is promoted by infection or sterile inflammation. After stimulation, circulating neutrophils leave the blood flow and accumulate at the site of infection or aseptic inflammation, where they release large numbers of highly toxic chemicals to engulf and kill pathogens. The classical recruitment of neutrophils from the blood into inflamed tissues is mediated by selectins and counter-receptors expressed on neutrophils (46). However, as Choudhury et al. reported in 2019 (50), neutrophil recruitment to the liver and lungs does not follow the classical cascade but require specific molecules.
Individuals with impaired neutrophils experience severe immunodeficiency both in quantity and effector functions (18). Immune cells can release a large number of proinflammatory cytokines, such as IL and TNF, to resist invasion or eliminate the pathogen. These cytokines will recruit additional immune cells in this battle until the immune system wins and achieves stability in the microenvironment. However, when the immune system is over-activated, the immune cells will overexpress cytokines, leading to the occurrence of a cytokine storm. Uncontrolled cytokine release will finally result in systemic inflammation, organ failure, and death. The cytokine storm may eventually lead to severe illness or death, in both sepsis and as observed in the current COVID-19 pandemic (51, 52). Furthermore, levels of IL-6 in severely ill patients were found to have increased significantly and were associated with a poor prognosis (53, 54). Corticosteroids and non-steroidal anti-inflammatory drugs are commonly used to treat inflammation. However, they do not alter the function of neutrophils or even enhance their destructive capacity (46). Excessive release of inflammatory cytokines leads to hyaline membrane formation, diffuse alveolar injury, and protein exudation to the lungs in COVID-19 patients, consistent with the pathological changes we observed in the lung of septic mice.
We have previously reported the therapeutic agents used for treatment of COVID-19 in China and found that the cytokine storm was not affected by neutralizing antibodies in convalescent plasma (55). Currently, different approaches to induce an anti-inflammatory response are being attempted to inhibit immune cell recruitment (allosteric antagonists to CXCR1/2 (56, 57)), release of pro-inflammatory factors (recombinant proteins or antibodies targeting IL-6, IL1-beta, IL-8, or IL-1 receptors), or inducing neutrophil degranulation (phosphodiesterase inhibitors) that have successfully reduced neutrophil recruitment in mice models. However, these findings relative to sepsis have not been translated into the clinic. No anti-inflammatory therapy has been effective in sepsis clinical trials and little is known about the role of miRNAs in regulating neutrophil function. An in-depth understanding of the miRNA function within neutrophils will help to identify potential clinical applications of miRNA as therapeutic agents. We thus developed various approaches including in vitro and in vivo experiments to determine whether let-7b levels could alter inflammation. Herein, we tested the therapeutic potential of restoring let-7b levels in vitro in human-derived neutrophils using let-7b mimics and in vivo via Let-7b agomir tail-vein injection in a CLP murine model.
Since primary neutrophils are granulocytes with a short lifespan, it is difficult to transfect them in vitro. Using a nucleofection plasmid transfection protocol, the transfection efficiency in neutrophils is low (~5%), when evaluated two hours post-transfection (58). Researchers have attempted to overcome this limitation by introducing granulocyte-macrophage colony-stimulating factor (GM-CSF) in the culture medium (59), while use of the leukemia cell line HL-60 as an alternative model (60) or direct electroporation of neutrophils (61) represent additional approaches to overcome this problem. The limitation for HL-60 cells is that they cannot mimic all aspects of neutrophil biology, and the conclusion from our in vitro experiment is that HL-60 cells cannot fully mimic the in vivo environments. As a result, we chose a mouse model to investigate the molecular mechanisms involved in regulating neutrophil function. Further, the extension of neutrophil lifespan by GM-CSF may alter its function and electroporation may also contribute to functional impairment. Since NETs may be easily induced when neutrophils are stimulated with LPS, likely through the activation of TLRs (62), and encouraged by experiments involving coculture of miRNA in activated platelet models (63, 64), we assumed that when neutrophils are cocultured with LPS and miRNA mimics, the former could induce NETs to capture the latter. We finally demonstrated that miR-let-7b mimics are introduced into the neutrophil cytoplasm after a 30-minute coculture following LPS stimulation (data shown in Figure 1D) without the need for transfection. To the best of our knowledge, this is an important technical achievement for introducing miRNA into neutrophils.
Previous studies have shown that a concerted action between different cells and the miR-let-7 family in the microenvironment is crucial for the outcome of the inflammatory response (20). For instance, exosome-derived let-7 can strongly suppress atherosclerotic inflammation (65). Let-7a can reduce the inflammatory response in microglia (66). Let-7g* can attenuate neuroinflammation by reducing microglia activation (67), and let-7d can suppress the atherosclerotic process in modulating PDGF and TNF-α signaling (33). However, not all miRNAs from the let-7 family benefit the vascular space. For instance, let-7a, b, e, and f aggravate neuronal damage following inflammation through TLR7 signaling (68). While, in neutrophils, the role of Let-7b is obscure. Thus, our research adds to the emerging body of evidence about the diverse impacts of inflammation-related members of the let-7 family.
Undeniably, our present study has several limitations to be considered that are mainly associated with its experimental design. Neutrophils are often described as “short-lived cells” with a life span of between 1.5 and 10 hours in mice and humans, which makes antimicrobial function studies and translation to clinical use a major challenge (46). Accordingly, we collected cell culture supernatants to be subjected to ELISA and cells for protein and mRNA detection within 10 hours of treatment. Further, our experiments could be performed under optimal conditions for neutrophil viability for up to 20 hours at 37°C in an anoxic culture medium supplemented with glucose and dimethyloxalylglycine (DMOG) (69) to validate the models in vitro. Moreover, our research lacks the recruitment of sepsis patients from clinical practice. Septic patients are usually treated with a combination of therapies at the time their samples and clinical details are collected. Thus, the effects of specific treatments concerning the expression levels of let-7b in sepsis patients could not be analyzed. Next, we only tested cytokines expression of IL-6 and CXCL1 in mouse serum samples as levels of TNF-α and IL-10 would be too low to be detectable by commercially available ELISA kits (data not shown). Another related problem is represented by our inability to present data relative to intravital imaging of neutrophils, which would provide more detailed behavior tracking of neutrophils under inflammatory condition, as this method is technically challenging. Future studies should be well designed and take these limitations into consideration.
Among miRNAs with altered expression in septic neutrophils, we demonstrate that miR-let-7b is involved in the regulation of genes related to inflammatory processes. Finally, biological therapies that improve miRNA processing should involve those that upregulate the levels of targeted miRNAs, which might contribute to suppress the activation of neutrophils. Thus, we strongly believe that in combination with the advances in miRNA delivery techniques, miR-let-7b will very soon be used to treat perilous human sepsis as well as the complications presented by the COVID-19 pandemic by regulating innate neutrophil function through modulation of the TLR4/NF-κB axis identified in this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
This animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Huashan Hospital, Fudan University.
Author Contributions
BC, JH, and QZ designed the study. BC performed the experiments and wrote the manuscript. SC, RFX, and JY contributed analytic tools. BC and TZ analyzed the results. RX revised the manuscript and acquired the research funding. All authors approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chinese National Natural Science Foundation under Grant No. 82070197.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.653344/full#supplementary-material
Abbreviations
BALF, Bronchoalveolar Lavage Fluid; CLP, Cecal Ligation and Puncture; COVID-19, Corona Virus Disease 2019; LPS, Lipopolysaccharide; IL-6, Interleukin-6; IL-8, Interleukin-8; IL-10, Interleukin-10; miRNA, microRNA; NC, Negative Control; NETs, Neutrophil Extracellular Traps; NF-κB, Nuclear Factor Kappa-B; SIRS, Systemic Inflammatory Response Syndrome; TLR4, Toll-Like Receptor 4; TNF-α, Tumor Necrosis Factor-α.
References
1. Wang YX, Wang YY, Chen Y, Qin QS. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol (2020) 92:568–76. doi: 10.1002/jmv.25748
2. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA (2016) 315:801–10. doi: 10.1001/jama.2016.0287
3. Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med (2017) 43:304–77. doi: 10.1007/s00134-017-4683-6
4. Fan Z, McArdle S, Marki A, Mikulski Z, Gutierrez E, Engelhardt B, et al. Neutrophil recruitment limited by high-affinity bent beta2 integrin binding ligand in cis. Nat Commun (2016) 7:12658. doi: 10.1038/ncomms12658
5. Campbell EL, Bruyninckx WJ, Kelly CJ, Glover LE, McNamee EN, Bowers BE, et al. Transmigrating Neutrophils Shape the Mucosal Microenvironment through Localized Oxygen Depletion to Influence Resolution of Inflammation. Immunity (2014) 40:66–77. doi: 10.1016/j.immuni.2013.11.020
6. Walmsley SR, Chilvers ER, Thompson AA, Vaughan K, Marriott HM, Parker LC, et al. Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J Clin Invest (2011) 121:1053–63. doi: 10.1172/Jci43273
7. Phillipson M, Kubes P. The neutrophil in vascular inflammation. Nat Med (2011) 17:1381–90. doi: 10.1038/nm.2514
8. Van Ziffle JA, Lowell CA. Neutrophil-specific deletion of Syk kinase results in reduced host defense to bacterial infection. Blood (2009) 114:4871–82. doi: 10.1182/blood-2009-05-220806
9. Horckmans M, Ring L, Duchene J, Santovito D, Schloss MJ, Drechsler M, et al. Neutrophils orchestrate post-myocardial infarction healing by polarizing macrophages towards a reparative phenotype. Eur Heart J (2017) 38:187–97. doi: 10.1093/eurheartj/ehw002
10. Cumpelik A, Ankli B, Zecher D, Schifferli JA. Neutrophil microvesicles resolve gout by inhibiting C5a-mediated priming of the inflammasome. Ann Rheum Dis (2016) 75:1236–45. doi: 10.1136/annrheumdis-2015-207338
11. Headland SE, Jones HR, Norling LV, Kim A, Souza PR, Corsiero E, et al. Neutrophil-derived microvesicles enter cartilage and protect the joint in inflammatory arthritis. Sci Transl Med (2015) 7:315ra190. doi: 10.1126/scitranslmed.aac5608
12. Nishio N, Okawa Y, Sakurai H, Isobe K. Neutrophil depletion delays wound repair in aged mice. Age (Dordr) (2008) 30:11–9. doi: 10.1007/s11357-007-9043-y
13. Calvente CJ, Tameda M, Johnson CD, Del Pilar H, Lin YC, Adronikou N, et al. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J Clin Invest (2019) 129:4091–109. doi: 10.1172/JCI122258
14. Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res (2012) 22:1243–54. doi: 10.1101/gr.132514.111
15. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol (2014) 15:509–24. doi: 10.1038/nrm3838
16. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet (2015) 16:421–33. doi: 10.1038/nrg3965
17. Saravanan S, Thirugnanasambantham K, Hanieh H, Karikalan K, Sekar D, Rajagopalan R, et al. miRNA-24 and miRNA-466i-5p controls inflammation in rat hepatocytes. Cell Mol Immunol (2015) 12:113–5. doi: 10.1038/cmi.2014.67
18. Gurol T, Zhou W, Deng Q. MicroRNAs in neutrophils: potential next generation therapeutics for inflammatory ailments. Immunol Rev (2016) 273:29–47. doi: 10.1111/imr.12450
19. Orellana EA, Kasinski AL. MicroRNAs in Cancer: A Historical Perspective on the Path from Discovery to Therapy. Cancers (Basel) (2015) 7:1388–405. doi: 10.3390/cancers7030842
20. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol (2008) 18:505–16. doi: 10.1016/j.tcb.2008.07.007
21. Jiang R, Li Y, Zhang A, Wang B, Xu Y, Xu W, et al. The acquisition of cancer stem cell-like properties and neoplastic transformation of human keratinocytes induced by arsenite involves epigenetic silencing of let-7c via Ras/NF-kappaB. Toxicol Lett (2014) 227:91–8. doi: 10.1016/j.toxlet.2014.03.020
22. Gao C, Zou X, Chen H, Shang R, Wang B. Long Non-Coding RNA Nuclear Paraspeckle Assembly Transcript 1 (NEAT1)Relieves Sepsis-Induced Kidney Injury and Lipopolysaccharide (LPS)-Induced Inflammation in HK-2 Cells. Med Sci Monit (2020) 26:e921906. doi: 10.12659/MSM.921906
23. Reithmair M, Buschmann D, Marte M, Kirchner B, Hagl D, Kaufmann I, et al. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J Cell Mol Med (2017) 21:2403–11. doi: 10.1111/jcmm.13162
24. Yu HR, Hsu TY, Huang HC, Kuo HC, Li SC, Yang KD, et al. Comparison of the Functional microRNA Expression in Immune Cell Subsets of Neonates and Adults. Front Immunol (2016) 7:615:615. doi: 10.3389/fimmu.2016.00615
25. Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol (2002) 14:103–10. doi: 10.1016/s0952-7915(01)00304-1
26. Fan J, Malik AB. Toll-like receptor-4 (TLR4) signaling augments chemokine-induced neutrophil migration by modulating cell surface expression of chemokine receptors. Nat Med (2003) 9:315–21. doi: 10.1038/nm832
27. Remer KA, Brcic M, Jungi TW. Toll-like receptor-4 is involved in eliciting an LPS-induced oxidative burst in neutrophils. Immunol Lett (2003) 85:75–80. doi: 10.1016/S0165-2478(02)00210-9
28. Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood (2003) 102:2660–9. doi: 10.1182/blood-2003-04-1078
29. Chen N, Debnath J. IkappaB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy (2013) 9:1214–27. doi: 10.4161/auto.24870
30. Zhou W, Pal AS, Hsu AY, Gurol T, Zhu X, Wirbisky-Hershberger SE, et al. MicroRNA-223 Suppresses the Canonical NF-kappaB Pathway in Basal Keratinocytes to Dampen Neutrophilic Inflammation. Cell Rep (2018) 22:1810–23. doi: 10.1016/j.celrep.2018.01.058
31. Sen R. Control of B lymphocyte apoptosis by the transcription factor NF-kappaB. Immunity (2006) 25:871–83. doi: 10.1016/j.immuni.2006.12.003
32. Zhou HJ, Zhao K, Li W, Yang N, Liu Y, Chen CY, et al. The interactions between pristine graphene and macrophages and the production of cytokines/chemokines via TLR- and NF-kappa B-related signaling pathways. Biomaterials (2012) 33:6933–42. doi: 10.1016/j.biomaterials.2012.06.064
33. Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, et al. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes (2017) 66:2266–77. doi: 10.2337/db16-1405
34. Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res (2015) 43:W460–6. doi: 10.1093/nar/gkv403
35. Rittirsch D, Huber-Lang MS, Flierl MA, Ward PA. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc (2009) 4:31–6. doi: 10.1038/nprot.2008.214
36. Pittet JF, Wiener-Kronish JP, McElroy MC, Folkesson HG, Matthay MA. Stimulation of lung epithelial liquid clearance by endogenous release of catecholamines in septic shock in anesthetized rats. J Clin Invest (1994) 94:663–71. doi: 10.1172/JCI117383
37. Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science (2019) 366. doi: 10.1126/science.aax9198
38. Caudrillier A, Kessenbrock K, Gilliss BM, Nguyen JX, Marques MB, Monestier M, et al. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J Clin Invest (2012) 122:2661–71. doi: 10.1172/JCI61303
39. Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, Kelly MM, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med (2007) 13:463–9. doi: 10.1038/nm1565
40. Sun Y, Li Q, Gui H, Xu DP, Yang YL, Su DF, et al. MicroRNA-124 mediates the cholinergic anti-inflammatory action through inhibiting the production of pro-inflammatory cytokines. Cell Res (2013) 23:1270–83. doi: 10.1038/cr.2013.116
41. Kapur R, Kim M, Aslam R, McVey MJ, Tabuchi A, Luo A, et al. T regulatory cells and dendritic cells protect against transfusion-related acute lung injury via IL-10. Blood (2017) 129:2557–69. doi: 10.1182/blood-2016-12-758185
42. Plociennikowska A, Hromada-Judycka A, Borzecka K, Kwiatkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci (2015) 72:557–81. doi: 10.1007/s00018-014-1762-5
43. Reinhart K, Brunkhorst FM, Bone HG, Gerlach H, Grundling M, Kreymann G, et al. [Diagnosis and therapy of sepsis]. Clin Res Cardiol (2006) 95:429–54. doi: 10.1007/s00392-006-0414-7
44. Rey C, Los Arcos M, Concha A, Medina A, Prieto S, Martinez P, et al. Procalcitonin and C-reactive protein as markers of systemic inflammatory response syndrome severity in critically ill children. Intens Care Med (2007) 33:477–84. doi: 10.1007/s00134-006-0509-7
45. Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, et al. Surviving Sepsis Campaign: International Guidelines for Management of Severe Sepsis and Septic Shock: 2012. Crit Care Med (2013) 41:580–637. doi: 10.1097/CCM.0b013e31827e83af
46. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol (2013) 13:159–75. doi: 10.1038/nri3399
47. Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol (2007) 7:678–89. doi: 10.1038/nri2156
48. Csoka B, Nemeth ZH, Toro G, Idzko M, Zech A, Koscso B, et al. Extracellular ATP protects against sepsis through macrophage P2X7 purinergic receptors by enhancing intracellular bacterial killing. FASEB J (2015) 29:3626–37. doi: 10.1096/fj.15-272450
49. Genga KR, Russell JA. Update of Sepsis in the Intensive Care Unit. J Innate Immun (2017) 9:441–55. doi: 10.1159/000477419
50. Choudhury SR, Babes L, Rahn JJ, Ahn BY, Goring KAR, King JC, et al. Dipeptidase-1 Is an Adhesion Receptor for Neutrophil Recruitment in Lungs and Liver. Cell (2019) 178:1205–+. doi: 10.1016/j.cell.2019.07.017
51. Cabler SS, French AR, Orvedahl A. A Cytokine Circus with a Viral Ringleader: SARS-CoV-2-Associated Cytokine Storm Syndromes. Trends Mol Med (2020) 26:1078–85. doi: 10.1016/j.molmed.2020.09.012
52. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med (2020) 383:2255–73. doi: 10.1056/NEJMra2026131
53. Gong J, Dong H, Xia Q-S, Huang Z-Y, Wang D-K, Zhao Y, et al. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19: a retrospective study. BMC Infect Dis (2020) 20:963. doi: 10.1186/s12879-020-05681-5
54. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet (2020) 395:507–13. doi: 10.1016/S0140-6736(20)30211-7
55. Chen BZ, Xia R. Early experience with convalescent plasma as immunotherapy for COVID-19 in China: Knowns and unknowns. Vox Sanguinis (2020) 115:507–14. doi: 10.1111/vox.12968
56. Cunha TM, Barsante MM, Guerrero AT, Verri WA, Jr., Ferreira SH, Coelho FM, et al. Treatment with DF 2162, a non-competitive allosteric inhibitor of CXCR1/2, diminishes neutrophil influx and inflammatory hypernociception in mice. Br J Pharmacol (2008) 154:460–70. doi: 10.1038/bjp.2008.94
57. Coelho FM, Pinho V, Amaral FA, Sachs D, Costa VV, Rodrigues DH, et al. The chemokine receptors CXCR1/CXCR2 modulate antigen-induced arthritis by regulating adhesion of neutrophils to the synovial microvasculature. Arthritis Rheumatol (2008) 58:2329–37. doi: 10.1002/art.23622
58. Johnson JL, Ellis BA, Munafo DB, Brzezinska AA, Catz SD. Gene transfer and expression in human neutrophils. The phox homology domain of p47phox translocates to the plasma membrane but not to the membrane of mature phagosomes. Bmc Immunol. (2006) 7:28. doi: 10.1186/1471-2172-7-28
59. Podaza E, Risnik D, Colado A, Elias E, Almejun MB, Grecco HF, et al. Chronic lymphocytic leukemia cells increase neutrophils survival and promote their differentiation into CD16(high)CD62L(dim) immunosuppressive subset. Int J Cancer (2019) 144:1128–34. doi: 10.1002/ijc.31762
60. De la Rosa IA, Perez-Sanchez C, Ruiz-Limon P, Patino-Trives A, Torres-Granados C, Jimenez-Gomez Y, et al. Impaired microRNA processing in neutrophils from rheumatoid arthritis patients confers their pathogenic profile. Modulation by biological therapies. Haematologica. (2020) 105:2250–61. doi: 10.3324/haematol.2018.205047
61. Chen ZQ, Akenhead MA, Sun XH, Sapper H, Shin HY, Hinds BJ. Flow-Through Electroporation of HL-60 White Blood Cell Suspensions using Nanoporous Membrane Electrodes. Adv Healthc Mater (2016) 5:2105–12. doi: 10.1002/adhm.201600204
62. Delgado-Rizo V, Martinez-Guzman MA, Iniguez-Gutierrez L, Garcia-Orozco A, Alvarado-Navarro A, Fafutis-Morris M. Neutrophil extracellular Traps and its implications in inflammation: An Overview. Front Immunol (2017) 8:81. doi: 10.3389/fimmu.2017.00081
63. Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2.microRNA complexes to endothelial cells via microparticles. Blood (2013) 122:253–61. doi: 10.1182/blood-2013-03-492801
64. Risitano A, Beaulieu LM, Vitseva O, Freedman JE. Platelets and platelet-like particles mediate intercellular RNA transfer. Blood (2012) 119:6288–95. doi: 10.1182/blood-2011-12-396440
65. Li SH, Chen L, Zhou X, Li JP, Liu JL. miRNA-223-3p and let-7b-3p as potential blood biomarkers associated with the ischemic penumbra in rats. Acta Neurobiol Exp (2019) 79:205–16. doi: 10.21307/ane-2019-018
66. Cho KJ, Song J, Oh Y, Lee JE. MicroRNA-Let-7a regulates the function of microglia in inflammation. Mol Cell Neurosci (2015) 68:167–76. doi: 10.1016/j.mcn.2015.07.004
67. Bernstein DL, Gajghate S, Reichenbach NL, Winfield M, Persidsky Y, Heldt NA, et al. let-7g counteracts endothelial dysfunction and ameliorating neurological functions in mouse ischemia/reperfusion stroke model. Brain Behav Immun (2020) 87:543–55. doi: 10.1016/j.bbi.2020.01.026
68. Mueller M, Zhou J, Yang L, Gao Y, Wu F, Schoeberlein A, et al. PreImplantation factor promotes neuroprotection by targeting microRNA let-7. Proc Natl Acad Sci U S A (2014) 111:13882–7. doi: 10.1073/pnas.1411674111
Keywords: MicroLet-7b, neutrophil, inflammation, TLR4/NF-κB, sepsis, COVID-19
Citation: Chen B, Han J, Chen S, Xie R, Yang J, Zhou T, Zhang Q and Xia R (2021) MicroLet-7b Regulates Neutrophil Function and Dampens Neutrophilic Inflammation by Suppressing the Canonical TLR4/NF-κB Pathway. Front. Immunol. 12:653344. doi: 10.3389/fimmu.2021.653344
Received: 14 January 2021; Accepted: 04 March 2021;
Published: 29 March 2021.
Edited by:
Felix Ellett, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Stamatia Papoutsopoulou, University of Liverpool, United KingdomEdward Sherwood, Vanderbilt University Medical Center, United States
Copyright © 2021 Chen, Han, Chen, Xie, Yang, Zhou, Zhang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Xia, eGlhcm9uZ2NuQDEyNi5jb20=
†ORCiD: Binzhen Chen, orcid.org/0000-0003-3512-1526
 Binzhen Chen
Binzhen Chen Jia Han
Jia Han Shaoheng Chen
Shaoheng Chen Rufeng Xie
Rufeng Xie Jie Yang
Jie Yang Tongming Zhou
Tongming Zhou Qi Zhang1
Qi Zhang1