- 1Department of Pathology, University of Texas Medical Branch, Galveston, TX, United States
- 2Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, TX, United States
- 3Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, United States
Microbiome composition and function have been implicated as contributing factors in the pathogenesis of autoimmune diseases (ADs), including systemic lupus erythematosus (SLE), rheumatoid arthritis and autoimmune hepatitis (AIH). Furthermore, dysbiosis of gut microbiome is associated with impaired barrier function and mucosal immune dysregulation. However, mechanisms by which gut microbiome contributes to the ADs and whether antioxidant treatment can restore gut homeostasis and ameliorate the disease outcome are not known. This study was, therefore, focused on examining the involvement of gut microbiome and host responses in the pathogenesis of SLE using unique female mouse models (C57BL/6, MRL+/+ and MRL/lpr) of 6 and 18 weeks with varying degrees of disease progression. Fecal microbiome diversity and composition, gut oxidative stress (OS), barrier function and inflammation, as well as systemic autoimmunity were determined. Interestingly, each mouse strain had distinct bacterial community as revealed by β-diversity. A lower Firmicutes/Bacteroidetes ratio in 6-week-old MRL/lpr mice was observed, evidenced by decrease in Peptostreptococcaceae under Firmicutes phylum along with enrichment of Rikenellaceae under Bacteroidetes phylum. Additionally, we observed increases in colonic OS [4-hydroxynonenal (HNE)-adducts and HNE-specific immune complexes], permeability changes (lower tight junction protein ZO-2; increased fecal albumin and IgA levels) and inflammatory responses (increased phos-NF-κB, IL-6 and IgG levels) in 18-week-old MRL/lpr mice. These changes were associated with markedly elevated AD markers (antinuclear and anti-smooth muscle antibodies) along with hepatic portal inflammation and severe glomerulonephritis. Notably, antioxidant N-acetylcysteine treatment influenced the microbial composition (decreased Rikenellaceae; increased Akkeransiaceae, Erysipelotrichaceae and Muribaculaceae) and attenuated the systemic autoimmunity in MRL/lpr mice. Our data thus show that gut microbiome dysbiosis is associated with increased colonic OS, barrier dysfunction, inflammatory responses and systemic autoimmunity markers. These findings apart from delineating a role for gut microbiome dysbiosis, also support the contribution of gut OS, permeability changes and inflammatory responses in the pathogenesis of ADs.
Introduction
Autoimmune diseases (ADs), like various complex systemic inflammatory disorders, are driven by combination of genetic, hormonal and environmental factors. Emerging evidence suggest that imbalance in the gut microbiome composition (dysbiosis), an important environmental factor, is associated with multiple ADs, including systemic lupus erythematosus (SLE) and autoimmune hepatitis (AIH) (1–4). Based on accumulating evidence, SLE/AIH patients with active disease have lower bacterial diversity and altered bacterial community composition in the gut (5, 6). Enrichment in Veillonella genus, for example, correlates to AIH disease activity, whereas increases in Ruminococcus gnavus of the Lachnospiraceae family is associated with the SLE disease indices (1, 4, 5). However, the contribution and mechanistic link between microbiome dysbiosis and ADs remain unclear, especially bacterial-host interactions.
Gut barrier function involves the crosstalk and interplay of the gut epithelial and mucosal layers, tight junctional proteins, and the immune system (7, 8). Impaired epithelial integrity and barrier function could be central predisposing factors in a number of ADs (9). Loss of barrier integrity may promote intestinal inflammation and translocation of bacterial components into the circulatory or lymphatic systems, resulting in systemic immune responses, and ultimately leading to SLE/AIH (1, 3). In fact, observed translocation of Enterococcus gallinarum and Lactobacillus reuteri into non-intestinal tissues suggests that leaky gut could be critical in promoting systemic autoimmune responses (10). Furthermore, mechanisms delineating tight junction regulation of gut permeability and inflammation suggest that posttranslational oxidative modification of proteins could contribute to gut leakiness and eventually disease activity (11–13). Our previous studies using MRL/lpr and MRL+/+ mice have shown that increased oxidative stress along with induction of lipid peroxidation-derived reactive aldehydes (LDRAs) are closely associated with the severity of autoimmune response/disease (14). Extensive ROS can enhance proinflammatory cytokine production and reduce mucus production in the intestinal epithelium (15–17). However, contribution of LDRAs and oxidative modification of proteins in microbiome dysbiosis-mediated mucosal and systemic immune responses remain unclear.
Intestinal microbiome dysbiosis is associated with chronic low-grade inflammation, and also triggers autoreactive T cells (CD4+ and CD8+ T cells) to drive spontaneous autoimmunity in target organs (18–20). The intestinal commensals interact with the host mucosal immune system leading to an imbalance in Treg and Th17 cells (21). Moreover, recent evidence suggests that fecal microbiome transplantation or probiotic supplementation could have beneficial effects in ADs based on evidence from both animal models and human studies (22–28). Furthermore, dietary intervention with retinoic acid or probiotics improves the disease manifestations by modulating microbiome composition, restoring gut barrier function and mucosal Treg-Th17 balance (6, 29). These studies thus suggest that in addition to disturbances in gut microbiome composition (dysbiosis), the interaction between the gut microbiome and host immune system could contribute to the pathogenesis of ADs. Our studies were, thus, designed to test the hypothesis that gut microbiome dysbiosis, colonic oxidative stress and impaired barrier function contribute to leaky gut and systemic inflammatory response, leading to ADs.
Here, we present data showing differential gut microbiome composition in lupus-resistant and lupus-prone mice at different ages, especially evidenced by altered Firmicutes to Bacteroidetes ratio (F/B ratio) and β-diversity. More importantly, changes in the microbiome composition are associated with compromised gut barrier function, serum AD markers, circulating cytokines and infiltration of immune cells in the liver and kidney. Overall, these observations support that dynamic interaction among intestinal microbiota composition, immune cell responses and autoantibody production are highly linked to renal and hepatic inflammation, eventually leading to corresponding ADs, i.e., SLE and AIH.
Materials and Methods
Animals and Treatments
Four-week-old female C57BL/6, MRL+/+ and MRL/lpr mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were maintained under sterile animal facility. All experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of Texas Medical Branch. In the first set of experiments, 6 and 18 weeks old C57BL/6, MRL+/+ and MRL/lpr mice (n=5 per strain, per time point) were sacrificed and fecal samples were collected from the colon for determining differential microbiome responses. Secondly, to establish the contributory role of oxidative stress in the disease progression, 5-week-old MRL/lpr mice were treated with antioxidant N-acetylcysteine (NAC, 250 mg/kg/day via drinking water) (30, 31) or drinking water only for 7 weeks and designated as NAC and control (CON) groups, respectively (n=6 per group). Mice were euthanized and major organs were weighed, frozen in liquid nitrogen, and stored at −80°C for further analyses. Sera obtained from blood samples were stored in small aliquots at −80°C until further analysis.
Enzyme-Linked Immunosorbent Assays (ELISAs) and Bio-Plex Assay
Autoantibodies [antinuclear antibodies (ANA); anti-smooth muscle antibodies (ASMA)] in the sera were determined using mouse-specific ANA ELISA kit (Alpha Diagnostic Int’l, San Antonio, TX) and mouse ASMA ELISA kit (Cusabio LLC, CA) by following the manufacturer’s instructions. HNE-protein adducts and its specific circulating immune complexes (CICs) in the colon tissues were analyzed according to our earlier published methods (14, 32).
Serum cytokines were determined using Cytokine 17-Plex Mouse ProcartaPlex Panel (Invitrogen, Carlsbad, CA). Samples were assayed and data were collected by Bio-Rad Bio-Plex 200 System. Data were measured as relative fluorescence intensity and then converted to the concentration using the standard curve.
16s rDNA Sequencing in Fecal Samples
Fecal samples were collected and stored at -80°C. DNA isolation and 16S rDNA sequencing was performed at the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine. Briefly, total genomic DNA was extracted using the MagAttract PowerSoil Kit (Qiagen, Redwood City, CA). The 16Sv4 region was amplified by PCR and sequenced on the MiSeq platform (Illumina, San Diego, CA) using a 2x250 bp paired-end protocol, yielding paired-end reads that overlap almost completely. For microbiome data analysis, Agile Toolkit for Incisive Microbial Analyses (ATIMA) was used. ATIMA is a stand-alone tool for analyzing and visualizing trends in taxa abundance, alpha diversity, and beta diversity as they relate to sample metadata (33, 34). Quantitative PCR was performed to further verify certain bacterial changes (10, 35).
Isolation of Lymphocytes From Liver Tissues
Intrahepatic lymphocytes (IHLs) were isolated as previously described (30). In brief, liver was perfused with 10 ml PBS, minced and digested with RPMI 1640 containing 0.05% collagenase IV (Roche, Indianapolis, IN) at 37 °C for 30 min. After digestion, cell suspensions were passed through 70 μm cell strainers, followed by a centrifugation over a 30/70% discontinuous Percoll density gradient (Sigma, St. Louis, MO) at 400 g at room temperature for 30 min. The cells were collected from the interphase, washed, resuspended in complete RPMI 1640, and the total number of IHLs per liver was counted. The relative percentages of immune cell populations were then analyzed by flow cytometry, and the absolute numbers of these lymphocyte subpopulations per liver were calculated according to their percentages and total IHL numbers in each liver.
Flow Cytometry
The specific antibodies and their corresponding isotype controls were purchased from Biolegend (San Diego, CA) and eBioscience (Waltham, MA). Cells were first incubated with FcγR blocker (CD16/32), followed by fluorochrome-labeled antibodies (Abs). The following Abs were used in combinations: PE-Cy7 anti-mouse CD3, Pacific Blue anti-mouse CD4, APC-Cy7 anti-mouse CD8, APC anti-mouse CD11b, APC-Cy7 anti-mouse CD11c, AF700 anti-mouse CD19 and Percp-cy5.5 anti-mouse CD45R/B220. Flow cytometric analysis was done using an LSRII Fortessa (BD Bioscience, San Jose, CA) and analyzed using FlowJo software 10.0 (TreeStar, Ashland, OR).
Quantitative Reverse Transcriptase PCR (qRT-PCR) Analysis
Total RNA was extracted from liver tissues using Trizol reagent (Sigma) followed by treatment with Qiagen DNase I (Qiagen). cDNA was synthesized with the iScript reverse transcription supermix (Bio-Rad, Hercules, CA). qRT-PCR was performed using iTaq universal SYBR green supermix kit (Bio-Rad, Hercules, CA) on a Bio-Rad CFX96 real time PCR machine. The mRNA expression of selected genes related to inflammation (TLR4, CD14, MCP1, IL-6, IL-4, IL-10, CXCR9, CXCR10 and TNF-α), inflammasome (NLRP3, caspase1, ASC, IL-1β and IL-18) and oxidative stress (SOD, catalase, OGG1 and iNOS) was determined. Mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the housekeeping gene. The primer sequences for the genes analyzed were according to our previous publication (30, 36). For the quantification of bacterial translocation, DNA was extracted from liver tissues and 100 ng DNA was used for qPCR analysis. The primer sequences for eubacteria were according to previous publication (35).
Western Blot Analysis
Total colon protein was homogenized in T-PER buffer including 1% protease inhibitor cocktail (Sigma) and protein concentration in the lysates was determined by Pierce™ BCA Protein Assay Kit. Western blot analysis was done for tight junction proteins. Ten µg protein per lane was loaded onto 4-20% Tris-Glycine gel (ThermoFisher scientific, Grand Island, NY) and transferred to PVDF membrane. The membrane was blotted with primary antibodies at 4°C overnight. Antibody detection was accomplished using horseradish peroxidase conjugated secondary antibodies and visualized with ECL. The signal intensity was quantified with Image Studio Lite Ver 5.2 (LI-COR).
Histological Examination
Liver and kidney tissues were fixed in 10% neutral formalin, dehydrated and paraffin embedded. Paraffin sections (5 µM thick) were stained with hematoxylin-eosin (H&E), and then blindly evaluated for cellular and structural changes in the liver and kidney.
Fecal IgA and Albumin Assays
Fecal samples were weighed and homogenized in cold PBS (1:10), centrifuged at 10,000 g at 4°C for 10 minutes. The supernatant was used for ELISA analysis of IgA (Invitrogen) and albumin (Bethyl Laboratories, Montgomery, TX) levels according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software 7.0 (GraphPad, La Jolla, CA). One-way analysis of variance (ANOVA) followed by Tukey-Kramer multiple comparison test was done to determine statistical significance between any two groups. The p values <0.05 were considered to be statistically significant. *p < 0.05; **p < 0.01. For microbiome data processed via ATIMA website, the statistical tests used are Kruskall-Wallis and Mann-Whitney for alpha diversity and taxa abundance, and the PERMANOVA test for beta diversity.
Results
Differential Autoimmune Responses in Lupus-Resistant/-Prone Mice
MRL+/+ mice spontaneously but very slowly develop autoantibodies several months after birth and lupus-like glomerulonephritis (SLE) late in the second year of life (37–39). In contrast, the MRL/lpr mouse strain is a model for more rapid and aggressive onset of SLE due to Fas (CD95) mutation, with the generation of autoantibodies as early as 6 weeks and advanced renal disease (glomerulonephritis) around 16 weeks (40). Female mice were used in this study due to greater propensity to develop ADs in women (41–43). To demonstrate differential autoimmune responses in normal and genetically susceptible mouse strains, we measured serum levels of ANA (a marker for SLE) and ASMA (a marker for AIH) in age-matched female C57BL6, MRL+/+ and MRL/lpr mice. As evidenced from Figures 1A, B, ANA and ASMA levels were very low at 6 weeks of age in both MRL+/+ and MRL/lpr mice, but significantly increased in MRL/lpr mice at 18 weeks compared to other two strains. In addition, the increases in the autoantibodies were associated with severe glomerulonephritis and infiltration of various immune cells in the hepatic portal area of 18-week-old MRL/lpr mice (Figure 1C). These data indicate systemic autoimmune responses are accompanied by immune cell infiltration in key organs associated with SLE and AIH.
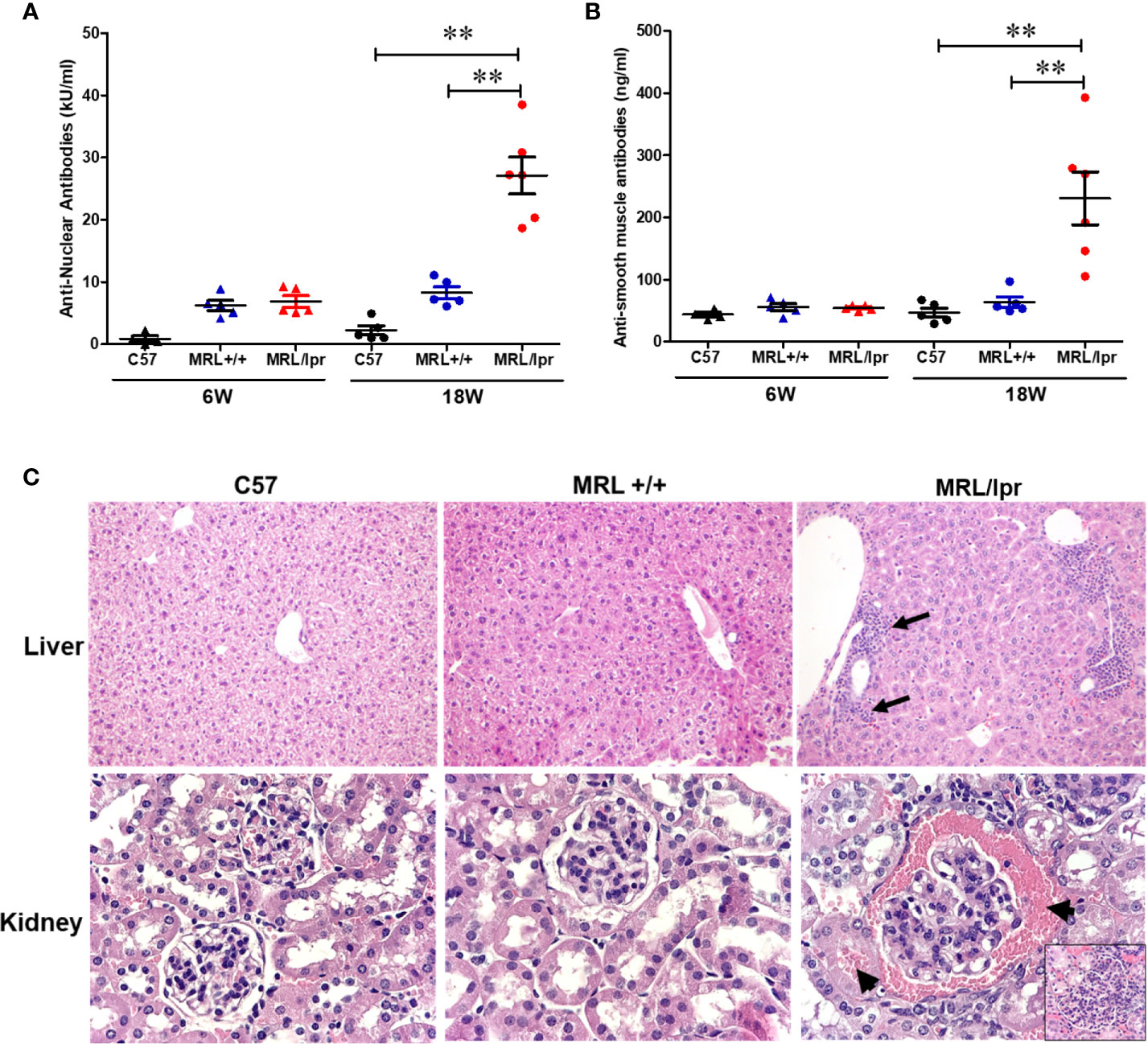
Figure 1 Spontaneous development of SLE- and AIH-like diseases in MRL/lpr mice. Serum ANA levels (A) and serum ASMA levels (B) in C57BL/6 (C57), MRL+/+ and MRL/lpr mice. Histopathology of liver (C) shows marked various peri-portal infiltrates (arrows) in MRL/lpr mice. Renal histopathology (lower panel) shows glomerulus with hypercellularity and red blood cells in Bowman’s spod and adjacent tubule (arrows) in MRL/lpr mice, indicative of acute glomerulonephritis (H&E). n = 5, **p < 0.01.
Hepatic Immune Cell and Pro-Inflammatory Responses in Normal and Lupus-Prone Mice
To address potential cellular involvement in the development of autoimmunity, we measured immune cell populations in the livers of the three mouse strains by FACS. We observed a significant increase of the total hepatic lymphocytes in 18-week-old MRL/lpr mice. Differential analysis showed increased B cells, CD4+ and CD8+ T cells, macrophages as well as dendritic cells in the livers of 18-week-old MRL/lpr mice compared to age-matched MRL+/+ and C57BL6 mice. In contrast, only hepatic CD8+ T cells were elevated in MRL+/+ compared with C57BL6 mice (Figure 2). Significantly higher levels of inflammatory markers TLR4 and CD14 were also found in the livers of 18-week-old MRL/lpr mice (Figure 3A). Furthermore, hepatic expression of MCP-1 and TNF-α was also increased in 18-week-old MRL/lpr mice (Figure 3A) when the disease markers were also highly prevalent. Importantly, inflammasome activation, which is implicated in SLE (30), showed a robust increase in its activation markers (NLRP3, caspase1 and IL-1β) in the livers of 18-week-old MRL/lpr mice (Figure 3B). These findings suggest that infiltration of multiple immune cells and increased pro-inflammatory responses may contribute to breakdown of immune tolerance, leading to autoimmune responses.
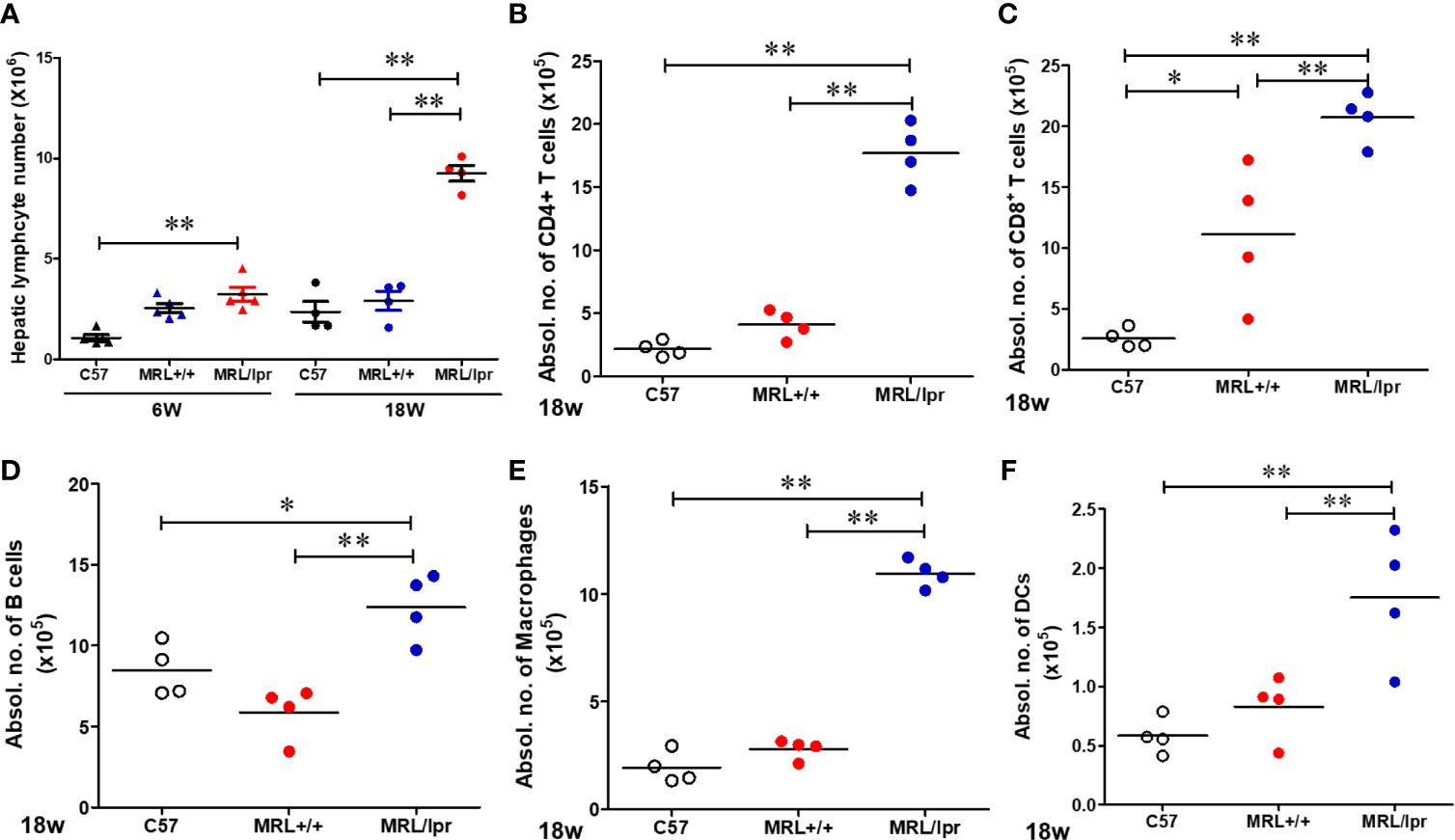
Figure 2 Flow cytometric analysis of hepatic immune cell populations in C57BL/6, MRL+/+ and MRL/lpr mice. (A) Hepatic lymphocytes numbers in 6-week and 18-week old mice. (B–F) Absolute numbers of CD4+ T cells (CD3+ CD4+ CD8−), CD8+ T cells (CD3+ CD4− CD8+), B cells (CD3− B220+), macrophages (CD11b+ F4/80+) and DCs (CD11B+CD11c+) in the livers of 18-week-old mice. Each symbol represents an individual mouse, and horizontal lines indicate the mean. n = 4 *p < 0.05; **p < 0.01.
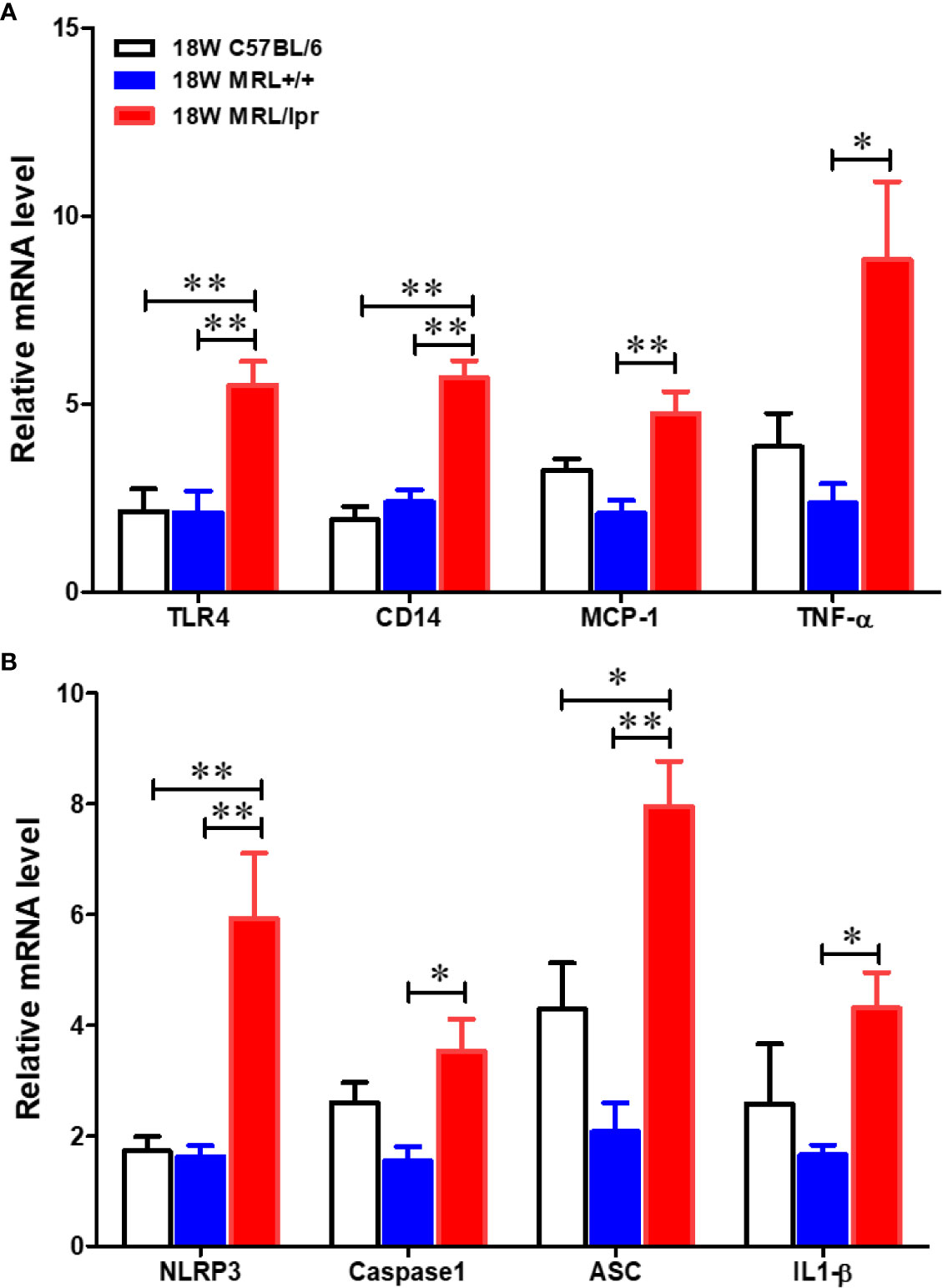
Figure 3 Up-regulation of inflammatory genes in the livers of C57BL/6, MRL+/+ and MRL/lpr mice. Total RNA was extracted from liver tissues and relative mRNA expressions of (A) inflammatory markers [Toll like receptor 4 (TLR4), CD14, MCP-1 and TNF-α] and (B) inflammasome activation markers (NLRP3, caspase-1, ASC and IL-1β) were determined by RT-PCR. Results are Mean ± SEM. n = 5 *p < 0.05; **p < 0.01.
Gut Microbiome Compositional Changes in Lupus-Resistant/-Prone Mice at Different Ages
Gut microbiome composition could have a pivotal role in triggering autoimmunity in susceptible individuals (2). To investigate the involvement of the gut microbiome in SLE, we compared the microbiome composition in C57BL6, MRL+/+ and MRL/lpr mice at different stages in disease development. Fecal samples, collected before disease onset (6 weeks) and at the peak of the disease process in MRL/lpr mice (18 weeks) were subjected to 16S rDNA sequencing for microbiome analysis. Compared to the other two strains, six-week-old MRL/lpr mice had significantly lower alpha diversity as measured by Shannon index but not observed OTU (Figure 4A). However, these observations were no longer evident by 18 weeks. These data suggest early differences in bacterial distribution but not overall community richness. Principal coordinates analysis of beta diversity further revealed differential bacterial community composition in each mouse strain at both 6 and 18 weeks derived from weighted UniFrac (Figure 4B). There was no significant difference between C57BL/6 and MRL+/+ mice for α-diversity at either time point, and no autoantibodies developed in C57BL/6 mice. Therefore, subsequent comparisons were mainly done between MRL+/+ and MRL/lpr mice at both time points. Dynamic changes in the fecal microbiome composition were observed between 6- and 18-week MRL+/+ mice, evidenced by increased levels of Bacteroidetes and Verrucomicrobia phyla and concomitant reductions in Firmicutes and Tenericutes (Figure 4C) at the latter time point. These shifts translated to the family level whereby increases in Akkermansiaceae and Rikenellaceae accompanied by relative losses of Anaeroplasmataceae and Peptostreptococcaceae in MRL+/+ mice over time (Figure 4E). At the lower taxonomic genus level, enrichment of Akkermansia, Alistipes, Blautia and Ruminiclostridium was observed in 18-week compared to 6-week MRL+/+ mice together with reduction in Anaeroplasma and Romboutsia (Figure 4F). Furthermore, the abundance of specific intestinal bacterial strain was also measured by RT-PCR. Increase in Akkermansia muciniphila under Verrucomicrobia, while reduction in Clostridium clostridiiforme and Faecalibacterium prausnitzii under Firmicutes were observed in fecal samples of 18-week-old MRL+/+ mice (Supplementary Figure 1). Collectively, we observed dramatic age-related changes in the gut microbiome of MRL+/+ mice, suggesting dynamic microbiome shift happens before the disease progression which could be a potential early event contributing to autoimmune responses. A lower F/B ratio was observed in the 6-week-old MRL/lpr mice (Figure 4D), evidenced by decreased abundance in Peptostreptococcaceae and Lactobacillaceae under Firmicutes phylum (Figures 5A, B) and significant increase in abundance of Rikenellaceae under Bacteroidetes phylum (Figure 5E) as compared to 6-week-old MRL+/+ mice. Furthermore, lower abundance in Muribaculaceae and Anaeroplasmataceae in 6-week-old MRL/lpr mice than age-matched MRL+/+ mice (Figures 5F, G), and an enrichment of Akkermansiaceae in 18-week-old MRL+/+ mice were observed (Figures 5H). No difference was observed for Lachnospiraceae and Ruminococcaceae (Figures 5C, D) between these two strains at both time points. Taken together, these data provide evidence for an age-related difference in microbiome composition of the lupus-prone mice that might contribute to aberrant gut permeability and also mucosal immune activation.
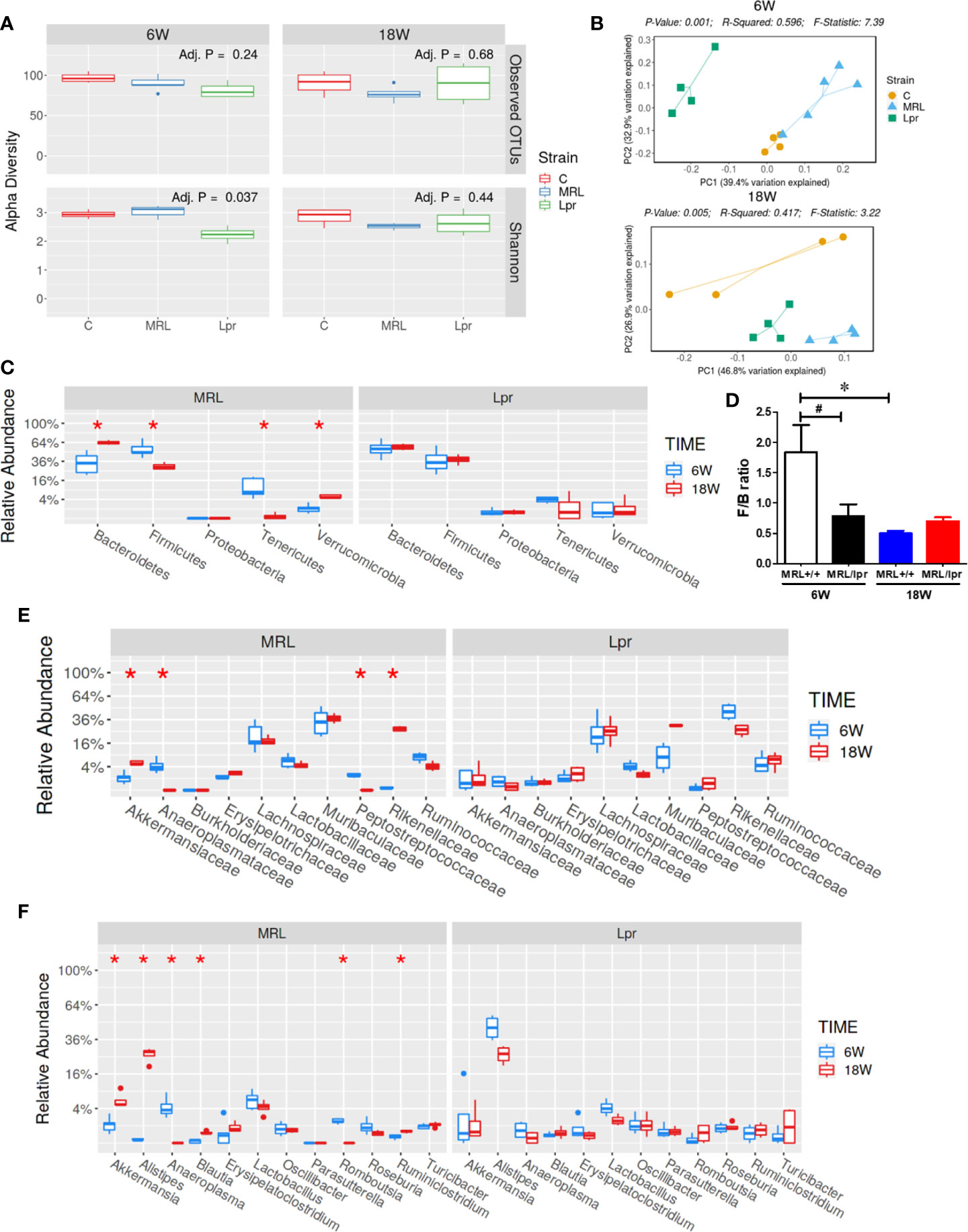
Figure 4 SLE-associated gut microbiome diversity and composition. Fecal pellets from C57BL/6 (C), MRL+/+ (MRL) and MRL/lpr (Lpr) mice were collected and bacterial DNA was isolated and subjected to 16s rDNA sequencing to evaluate the composition of microbiome in each mice. (A) The variety of organisms in a community, termed as alpha diversity. (B) Different mouse strains showed distinct microbiome pattern at 6 week and 18 weeks of age. (C) Age-associated differences in fecal microbiome from MRL+/+ and MRL/lpr mice at phylum level. (D) F/B ratio in MRL+/+ and MRL/lpr mice. (E) Taxa relative abundance at family level in MRL+/+ and MRL/lpr mice. (F) Taxa relative abundance at genus level in MRL+/+ and MRL/lpr mice. Results are mean ± SEM. n = 4 *p < 0.05; #p < 0.1.
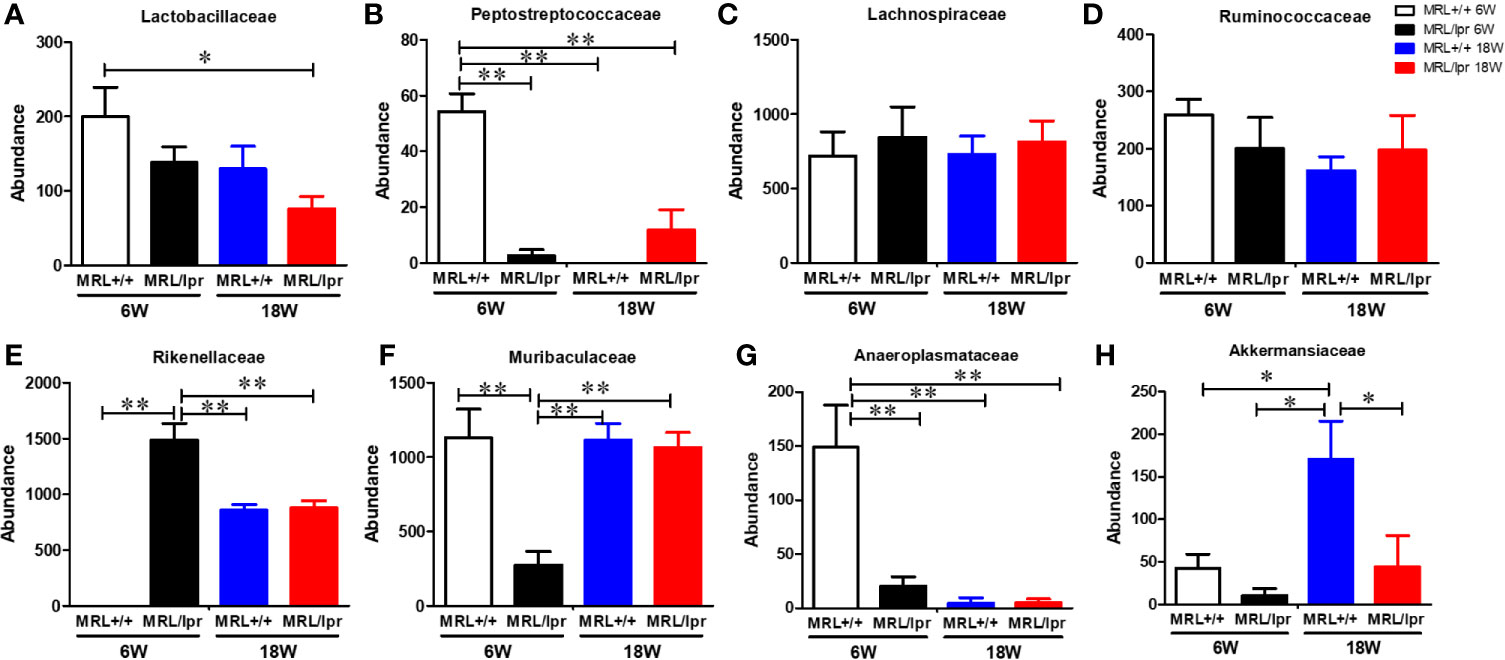
Figure 5 Differential abundance of bacterial families in MRL+/+ and MRL/lpr mice at 6 and 18 weeks of age. (A) Abundance of Lactobacillaceae under Firmicutes. (B) Abundance of Peptostreptococcaceae under Firmicutes. (C) Abundance of Lachnospiraceae under Firmicutes. (D) Abundance of Ruminococcaceae under Firmicutes. (E) Abundance of Rikenellaceae under Bacteroidetes (F) Abundance of Muribaculaceae under Bacteroidetes (G) Abundance of Anaeroplasmataceae under Tenericutes. (H) Abundance of Akkermansiaceae under Verrucomicrobia. Results are mean ± SEM. n = 4 *p < 0.05; **p < 0.01.
Impaired Gut Barrier Function in 18-Week Old Lupus-Prone MRL/lpr Mice
To test whether gut microbiome dysbiosis leads to intestinal inflammation and leakiness, which further contribute to the systemic autoimmune responses, inflammatory markers and tight junction proteins were measured in 18-week samples, when MRL/lpr mice exhibit advanced SLE disease manifestations. Additionally, we also determined the fecal albumin and IgA levels as markers of gut leakage and inflammation. In MRL/lpr mice, altered microbiome composition was associated with increased intestinal permeability, evidenced by increased fecal albumin and IgA levels (Figures 6A, B) and decreased gut tight junction protein ZO-2 (Figure 6D). In addition, intestinal inflammation was observed with increased colonic phos-NF-κB, IL-6 and IgG levels in 18-week-old MRL/lpr mice (Figure 6E). Based on our histological findings and flow cytometry data showing increased hepatic immune cell infiltration and activation, we also determined the bacterial translocation by amplifying the universal eubacteria in the liver tissues. Interestingly, we observed significantly increased eubacteria in the livers of MRL/lpr mice (Figure 6C). Considering that protein oxidation or lipid peroxidation has the potential to affect intestinal permeability in vitro and in vivo, we analyzed HNE-protein adducts and HNE-specific immune complexes (ICs) in the colon. Interestingly, significantly increased levels of both HNE-protein adducts and HNE-specific ICs were observed in the colon of 18-week-old MRL/lpr mice compared to age-matched C57BL/6 or MRL+/+ mice. Moreover, these oxidative stress (OS) markers were markedly increased in MRL+/+ mice compared to C57BL/6 mice (Figure 7). Collectively, these data support an association between gut microbiome dysbiosis-associated OS, intestinal permeability and inflammation in the colon of lupus-prone MRL/lpr mice.
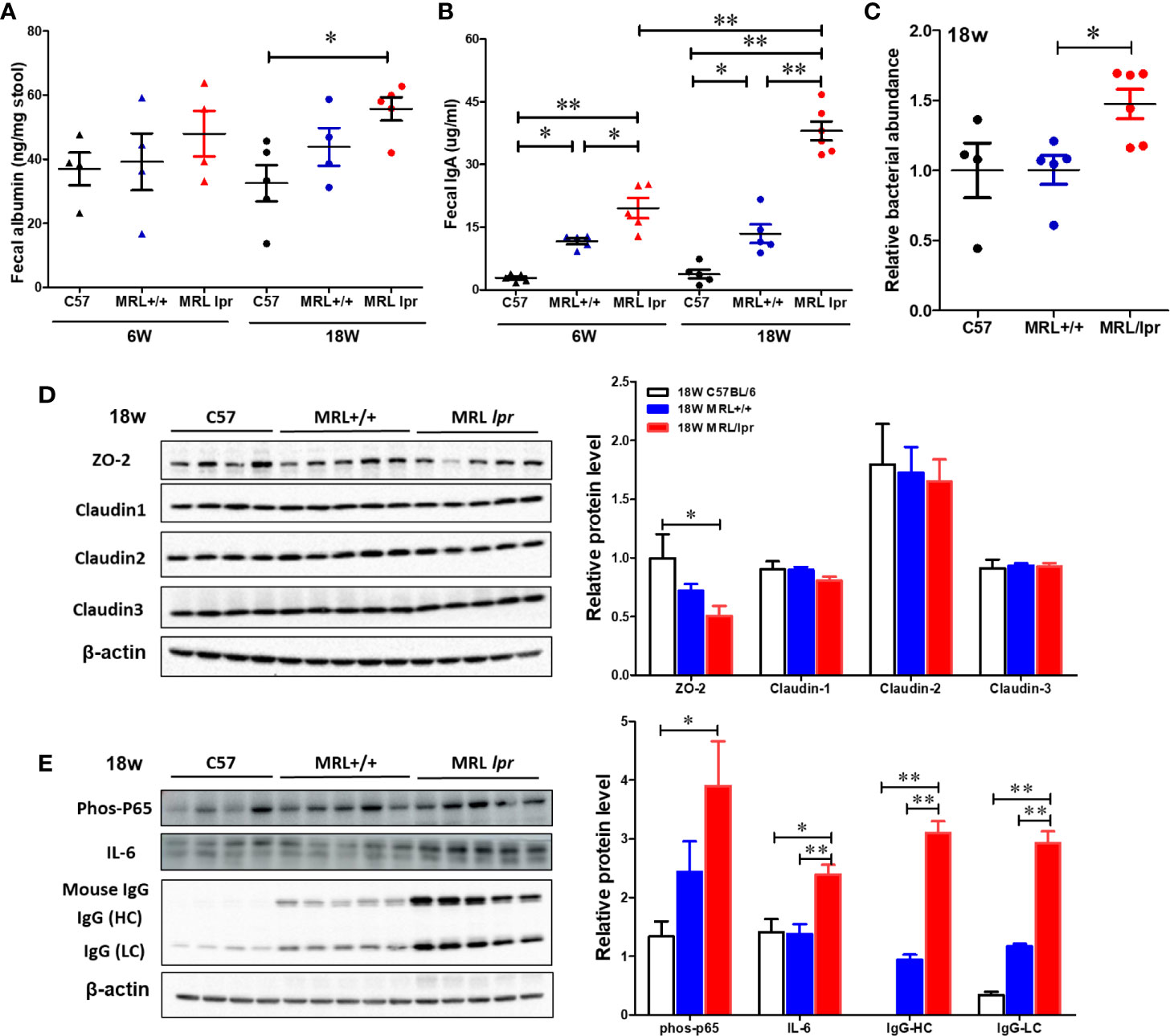
Figure 6 Impaired gut barrier in lupus-prone mice. Fecal albumin and IgA levels were determined by ELISA. Liver DNA was extracted and RT-PCR was performed with Eubacteria primer to detect the bacterial translocation. Colon protein samples were subjected to SDS-PAGE and blotted with specific antibodies. (A) Fecal albumin levels. (B) Fecal IgA levels (C) Bacterial translocation to liver (D, E) Western blot for tight junction proteins (ZO-2, Claudin 1-3) and inflammatory markers. Results are mean ± SEM. n = 4 *p < 0.05; **p < 0.01.
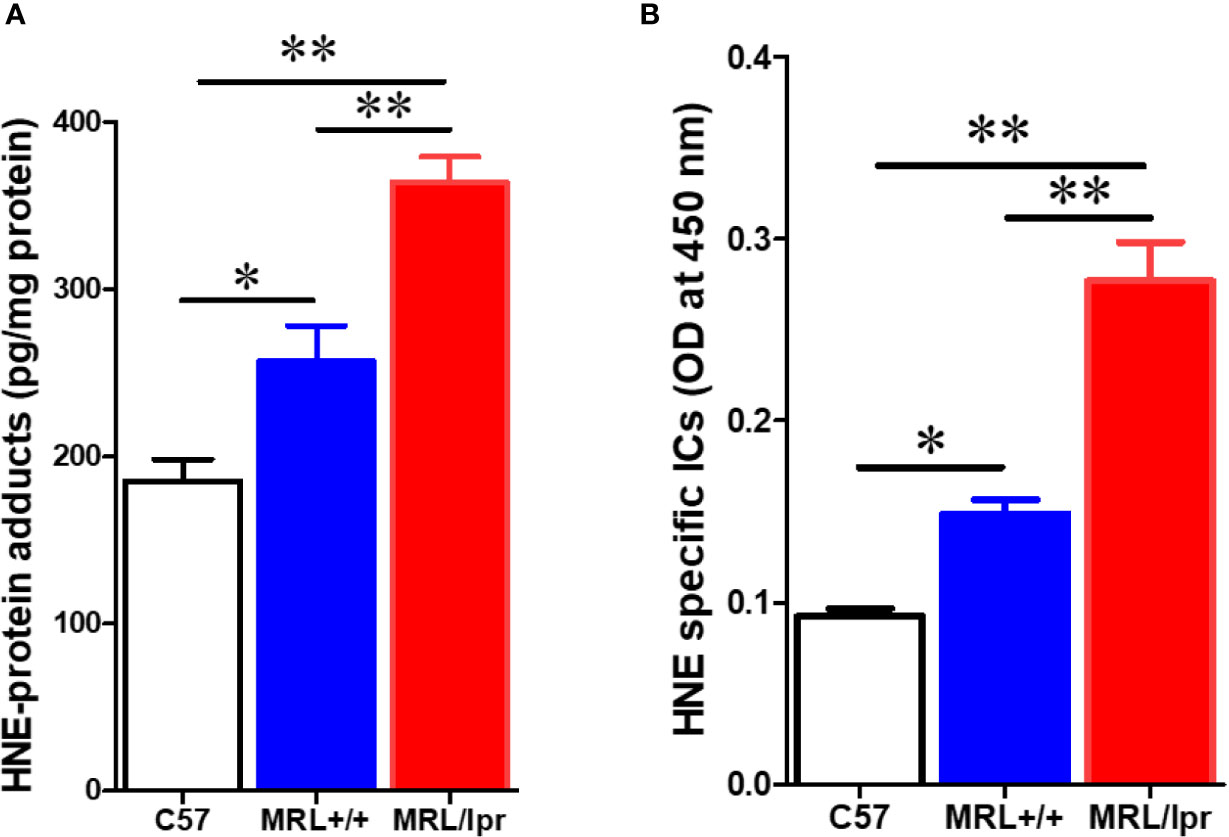
Figure 7 Elevated markers of oxidative stress in the colon of lupus-prone mice. Colon protein samples were subjected to specific ELISA for HNE-protein adducts and HNE-specific ICs. (A) HNE-protein adducts. (B) HNE-specific ICs. Results are mean ± SEM. n = 4 *p < 0.05; **p < 0.01.
NAC Supplementation Modulates the Gut Microbiome and Lupus Pathogenesis
To investigate whether an antioxidant could ameliorate the gut microbiome and pathological changes as well as SLE disease activity, MRL/lpr mice were treated with N-acetylcysteine (NAC), an antioxidant which is a precursor of GSH and detoxifies ROS (31, 44). Although NAC treatment did not alter α-diversity (richness and evenness) (Figure 8A), it did lead to distinct gut microbial community changes revealed by β-diversity and bacterial taxonomy at the family level (Figures 8B, C) evident from relative increases in bacterial families Akkermansiaceae, Erysipelotrichaceae and Muribaculaceae, and decreased Rikenellaceae (Figure 8D). At genus level, NAC supplementation led to enrichment in Akkermansia and Turicibacter, while reduction in Alistipes (Figure 8E). Even though NAC treatment resulted in a decreasing pattern for both ANA and anti-dsDNA levels in the serum, the differences did not reach statistical significance (Figures 9A, B). We also observed significant reduction in hepatic lymphocyte number (Figure 9C) and multiple serum cytokines (GM-CSF, IL-1β, IL-18, IL-5, IL-22, IL-23 and IL-27) following NAC treatment (Figure 9D). Furthermore, levels of hepatic IL-6, iNOS, CXCR9 and CXCR10 were also significantly attenuated following NAC supplementation in MRL/lpr mice (Figure 9E). Overall, our data suggest a beneficial role of NAC for SLE disease attenuation and also provide mechanistic evidence for the involvement of OS in the regulation of mucosal and systemic immune responses.
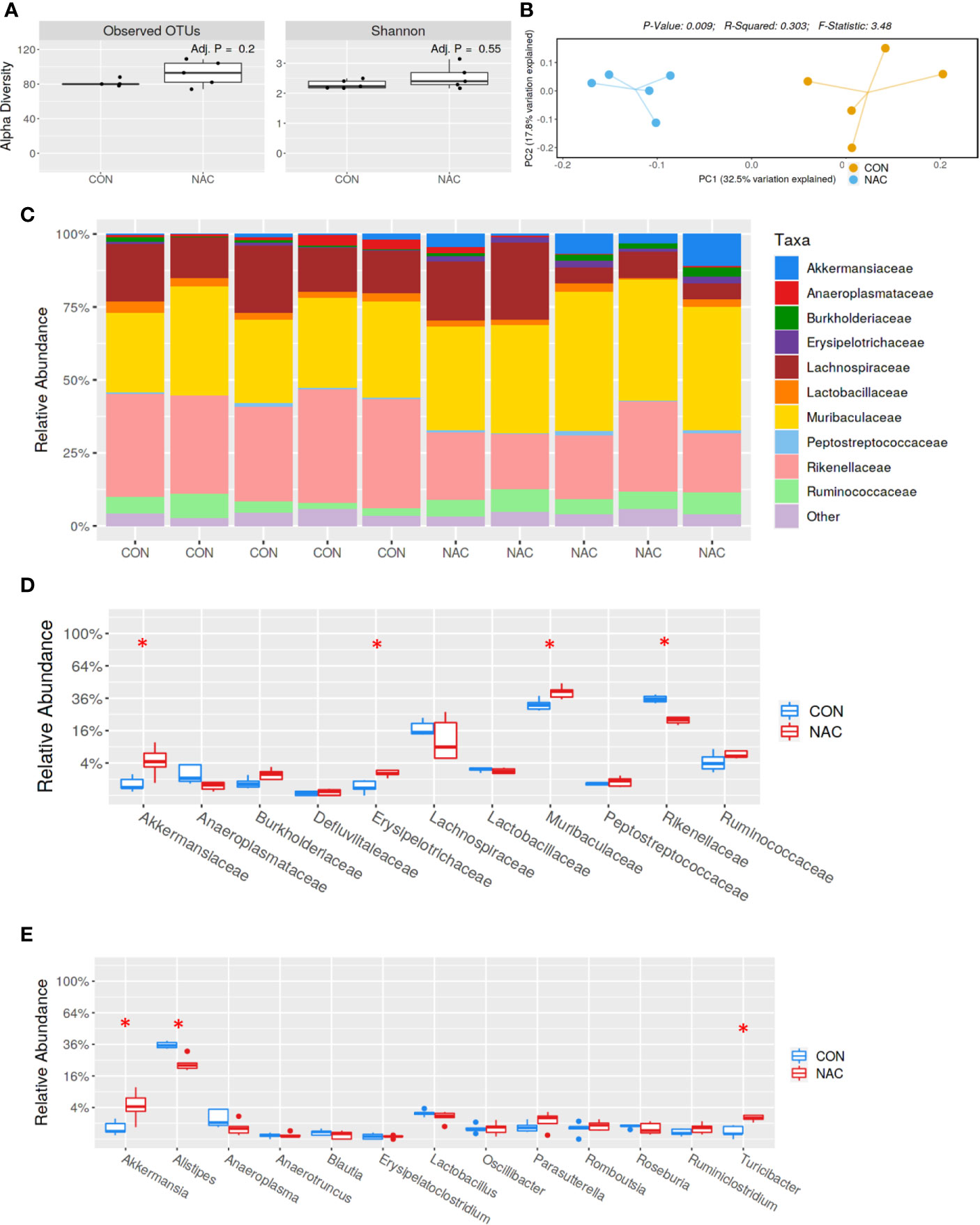
Figure 8 Gut microbiome changes after antioxidant NAC treatment in MRL/lpr mice. Bacterial DNA was isolated from fecal pellets of CON and NAC-treated MRL/lpr mice, and 16s rDNA sequencing was performed to evaluate the microbiome composition in each mice. (A) The variety of organisms in a community termed as alpha diversity. (B) Different mouse strains showed distinct microbiome pattern. (C) Stacked bar plot for composition of common bacterial taxa (>0.1% abundance). (D) Taxa relative abundance at family level. (E) Taxa relative abundance at genus level. Results are mean ± SEM. n = 5 *p < 0.05.
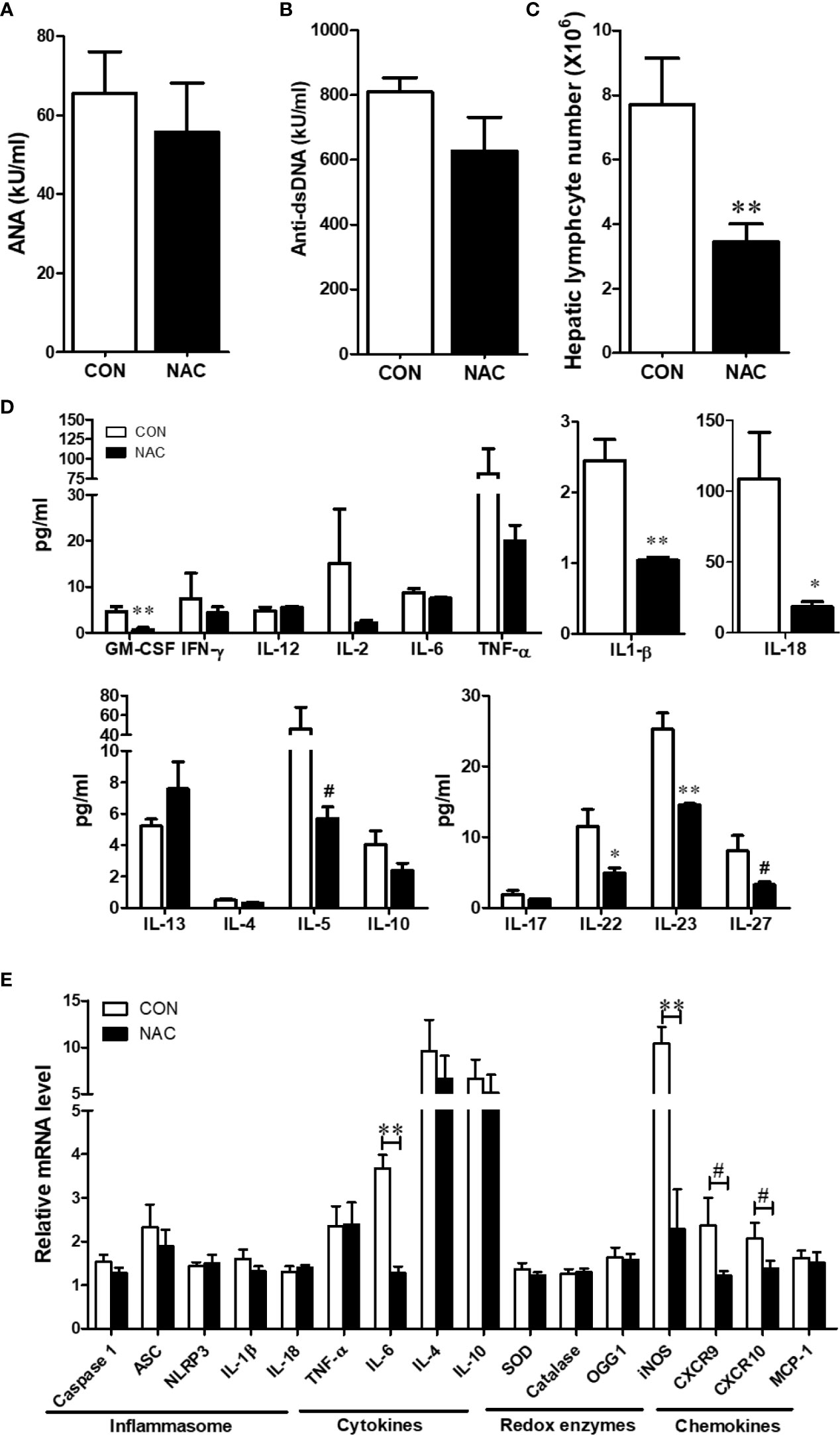
Figure 9 NAC treatment attenuated the auto-inflammatory responses in MRL/lpr mice. Serum ANA (A) and anti-dsDNA (B) levels in the control and NAC-treated mice. (C) Diminution of hepatic lymphocytes following NAC treatment. (D) Serum cytokines, measured using a multiplex assay in the control and NAC-treated mice. (E) RT-PCR for markers of inflammasome activation, cytokines, redox-related enzymes and chemokines in the control and NAC-treated mice. Results are mean ± SEM. n = 5 *p < 0.05; **p < 0.01, #p < 0.1.
Discussion
SLE and AIH are chronic ADs that occur worldwide with a rising prevalence rate (45). These ADs are characterized by loss of tolerance to self-antigens and aberrant immune activation. Recent studies suggest association of gut microbiome dysbiosis with ADs, although it remains to be established how the microbiotia contribute to SLE/AIH pathogenesis. Based on recent evidence, some key perturbations in the gut, including increased OS, changes in permeability (leaky gut) and activation of the mucosal immune system are suggested as few mechanisms that could potentially contribute to the pathogenesis of ADs (2). To better define the role of the gut microbiota and mechanisms involved in disease pathogenesis, in this study, we characterized the gut microbiome and mucosal pathophysiological changes in the progression of systemic autoimmunity using three strains of mice with different manifestations of SLE disease activities.
MRL+/+ mice exhibit a similar autoantibody profile and glomerulonephritis as seen in lupus patients (38, 39). It is well-established that MRL+/+ mice would also develop AIH features, including hepatic immune cell infiltration and cytokine production as seen in response to trichloroethene exposure (30, 46, 47). On the other hand, MRL/lpr mice spontaneously develop accelerated autoimmune responses due to Fas (CD95) mutation (37). Here, we observed that 18-week-old MRL/lpr mice had increased ANA levels and severe glomerulonephritis representing typical SLE disease manifestations, as well as lymphocytic infiltration in the hepatic portal area and elevated ASMA levels indicating prevalence of AIH in these mice. Furthermore, analysis of hepatic immune cell populations revealed significant increases in the absolute numbers for T and B cells in 18-week-old MRL/lpr mice compared to C57BL/6 and MRL+/+ mice. Our data apart from firmly establishing that aged MRL/lpr mice manifest SLE and AIH features, also provide solid support to utilizing our approach of using age-matched different strains of mice with distinct disease activities in exploring the potential contributing factors and mechanisms leading to SLE and AIH.
Based on recent studies, it is becoming clear that altered gut microbiome composition and impaired barrier function are associated with multiple ADs, including SLE/AIH both in patients and murine models (2–4, 48). In fact, evaluation of feces from SLE patients has provided evidence for a lower F/B ratio (49). Here, we also observed a lower F/B ratio in 6-week old MRL/lpr mice compared to age-matched MRL+/+ mice. Interestingly, age-related decrease in F/B ratio was also observed in 18-week-old MRL+/+ mice. Although decreased F/B ratio is not correlated with highly advanced disease activity in 18-week MRL/lpr mice, a lower F/B ratio at 6 weeks could be an important and highly significant contributing factor in promoting early disease onset. Lactobacillus, a known probiotic, exhibits its anti-inflammatory effect in ADs by inducing T regulatory (Treg) cells (50). Lactobacillus colonization attenuates SLE by modulating Treg-Th17 balance and gut barrier function (29). An earlier study showed depletion of Lactobacilli and increases in Lachnospiraceae in 5-week-old young MRL/lpr mice compared to MRL+/+ controls (6). Here, we observed decreasing trend of Lactobacillaceae in 18-week-old MRL/lpr mice compared with 6-week-old counterparts, suggesting a prolonged deficiency of Lactobacillaceae may consequently result in the generation of mucosal inflammatory responses. It has been reported that Akkermansia muciniphila is increased in multiple sclerosis patients and can induce pro-inflammatory responses in vitro and in vivo (51, 52). Interestingly, in this study, Akkermansiaceae was significantly increased in 18-week old MRL+/+ mice compared with 6-week old counterparts. However, MRL/lpr mice had lower level of Akkermansiaceae compared to age-matched MRL+/+ mice. The differential responses in the two strains (MRL+/+ vs MRL/lpr) provide a unique opportunity to explain their role in slow vs. aggressive disease development and thus warrant a detailed investigation of their contribution in SLE pathogenesis.
Gut microbial communities can influence host immune responses and epithelium integrity (7, 8). Therefore, we determined the gut barrier integrity and function, as well as inflammatory markers. Our data show that 18-week-old MRL/lpr mice had significantly higher fecal albumin and IgA levels and reduced tight junction protein ZO-2, which can contribute to gut barrier dysfunction. In fact, these observations are also supported by findings in SLE patients. Azzouz et al. (1) reported an outgrowth of Ruminococcus gnavus under the Lachnospiraceae family, and impaired barrier function evidenced by increased fecal IgA, IgM, IgG and calprotectin levels in SLE active patients. Mucosal IgA level is highly associated with commensal exposure and increased fecal IgA in SLE patients suggests impaired intestinal barrier functions (1, 53). One consequence of impaired intestinal barrier function could be systemic translocation of bacteria or bacterial products due to gut leakiness, and endotoxemia has been reported in chronic inflammatory diseases, including SLE (29, 54). This is further substantiated by findings on the translocation of Enterococcus gallinarum from intestine to systemic tissues (10), which can trigger inflammatory response, autoantibody production and autoimmune disease manifestations in genetically predisposed individuals. The fact that we observed significantly increased eubacteria levels in the liver tissues of 18-week MRL/lpr mice suggests that bacteria or its genetic material (DNA) may translocate to the liver, generating inflammatory response and eventually leading to hepatic autoimmune responses. Further support to this was evidenced by our observation of significantly increased level of inflammatory cytokines in the serum of 18-week MRL/lpr mice, indicating compromised barrier function and likely activation of mucosal immune system resulting in systemic inflammation. Our data thus provide evidence that gut microbiome dysbiosis could be a potential mechanism leading to autoimmunity via epithelial barrier disruption, release of bacteria/bacterial products and consequent systemic immune dysregulation.
Oxidative stress is considered a contributing factor in initiating apoptosis and inflammation leading to ADs (30, 55–57). Our previous studies have shown that OS, including increased MDA-/HNE-protein adducts in the liver and kidney of MRL/lpr mice, is associated with autoimmune responses (14). Here, we provide evidence for significantly increased HNE-protein adducts and HNE-specific ICs in the colon tissue, suggesting that gut microbiome dysbiosis and subsequent interaction with host tissue can change the gut redox potential, which can result in altered gut immune response and barrier function. Loss in intestinal barrier function can facilitate translocation of microbial components and inflammatory mediators, thus contribute to systemic ADs. Since antioxidants can restore normal gut redox status and extra-intestinal redox potential (58–60), we treated MRL/lpr mice with NAC, a ROS scavenger. Ironically, we observed an ameliorated autoimmune response, as evident from decreased pattern for both ANA and anti-dsDNA autoantibodies and suppression of hepatic inflammation after NAC supplementation in MRL/lpr mice. Besides downregulation of hepatic inflammasome activation markers, NAC also reduced the levels of cytokines in the circulation, suggesting the potential of NAC to serve not only as an antioxidant, but also in generating anti-inflammatory response. More importantly, NAC supplementation altered the gut microbiome composition, including increases in Akkermansiaceae. Previous work has established that A. muciniphila is diminished in lupus-prone NZB/W F1 mice as disease progressed, and expansion of A. muciniphila exerts beneficial impact on host metabolism and suppresses autoimmunity (4, 61, 62). In addition, we also observed a reduction of Rikenellaceae family after NAC treatment, which is consistent with higher abundance of Rikenellaceae family associated with more severe SLE-like disease in SNF1 mice (63). The modification of the gut microbiota by NAC supplementation could be an important mechanistic factor for the beneficial role of antioxidants in regulating gut microenvironment and thereby preventing autoimmunity. Whether NAC exerts anti-inflammatory effects through microbial modulation or by acting directly on the mucosal lymphocytes is an interesting and challenging proposition, and deserves an extensive evaluation to clarify the actual role of OS on gut microbiome and/or mucosal immunity.
In summary, the data in this study suggest that microbiome-host interactions can contribute to autoimmune responses. Moreover, our findings on OS appear to be an important and mechanistically critical event associated with microbiome dysbiosis and barrier dysfunction in ADs. Delineating how intestinal microbial dysbiosis and host dysregulation happen will benefit the understanding of the pathophysiology of ADs. Our study addresses this knowledge gap and reveals that manipulation of intestinal microenvironment redox status could lead to devising therapeutic strategies for ADs. Our studies provide solid platform to further explore and exploit the dynamics of microbiome changes and gut barrier dysfunction in causing mucosal immune dysregulation, especially imbalance between Treg and Th17 cells, and ultimately their contribution to systemic immune response leading to devastating diseases like SLE and AIH.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI repository, PRJNA706579.
Ethics Statement
The animal study was reviewed and approved by UTMB IACUC.
Author Contributions
HW, NB, GW, YL, and XD performed the experiments. MK and HW designed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by RO1 grants [ES016302 and ES026887] from the National Institute of Environmental Health Sciences (NIEHS), NIH, and UTMB Institute for Human Infections and Immunity (IHII). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.651191/full#supplementary-material
Supplementary Figure 1 | Dynamic changes in gut bacterial compositions in MRL+/+ and MRL/lpr mice at 6 and 18 weeks of age. Real-time quantitative PCR analysis was performed by amplifying fecal DNA with primers specific for (A) Akkermansia muciniphila, (B) Lactobacillus acidophilus, (C) Faecalibacterium prausnitzii, (D) Clostridium butyricum and (E) Clostridium clostridiiforme. Results are mean ± SEM. n = 5 *p < 0.05; **p < 0.01
References
1. Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis (2019) 78(7):947–56. doi: 10.1136/annrheumdis-2018-214856
2. Khan MF, Wang H. Environmental Exposures and Autoimmune Diseases: Contribution of Gut Microbiome. Front Immunol (2019) 10:3094. doi: 10.3389/fimmu.2019.03094
3. Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol (2015) 8(5):5153–60.
4. Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, et al. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl Environ Microbiol (2018) 84(4):e02288–17. doi: 10.1128/AEM.02288-17
5. Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut (2020) 69(3):569–77. doi: 10.1136/gutjnl-2018-317836
6. Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol (2014) 80(24):7551–60. doi: 10.1128/AEM.02676-14
7. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol (2009) 9(11):799–809. doi: 10.1038/nri2653
8. Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol (2017) 11(9):821–34. doi: 10.1080/17474124.2017.1343143
9. Mu Q, Kirby J, Reilly CM, Luo XM. Leaky Gut As a Danger Signal for Autoimmune Diseases. Front Immunol (2017) 8:598. doi: 10.3389/fimmu.2017.00598
10. Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science (2018) 359(6380):1156–61. doi: 10.1126/science.aar7201
11. Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med (2018) 50(8):103. doi: 10.1038/s12276-018-0126-x
12. Chen Z, Kondrashina A, Greco I, Gamon LF, Lund MN, Giblin L, et al. Effects of Protein-Derived Amino Acid Modification Products Present in Infant Formula on Metabolic Function, Oxidative Stress, and Intestinal Permeability in Cell Models. J Agric Food Chem (2019) 67(19):5634–46. doi: 10.1021/acs.jafc.9b01324
13. Rao R. Oxidative stress-induced disruption of epithelial and endothelial tight junctions. Front Biosci (2008) 13:7210–26. doi: 10.2741/3223
14. Wang G, Li H, Khan MF. Differential oxidative modification of proteins in MRL+/+ and MRL/lpr mice: Increased formation of lipid peroxidation-derived aldehyde-protein adducts may contribute to accelerated onset of autoimmune response. Free Radic Res (2012) 46(12):1472–81. doi: 10.3109/10715762.2012.727209
15. Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev (2014) 94(2):329–54. doi: 10.1152/physrev.00040.2012
16. Jones RM, Mercante JW, Neish AS. Reactive oxygen production induced by the gut microbiota: pharmacotherapeutic implications. Curr Med Chem (2012) 19(10):1519–29. doi: 10.2174/092986712799828283
17. Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal (2014) 20(7):1126–67. doi: 10.1089/ars.2012.5149
18. Horai R, Zarate-Blades CR, Dillenburg-Pilla P, Chen J, Kielczewski JL, Silver PB, et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity (2015) 43(2):343–53. doi: 10.1016/j.immuni.2015.07.014
19. Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. and B Cells Cross-React with Mimotopes Expressed by a Common Human Gut Commensal to Trigger Autoimmunity. Cell Host Microbe (2019) 26(1):100–13 e8. doi: 10.1016/j.chom.2019.05.003
20. Greiling TM, Dehner C, Chen X, Hughes K, Iniguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med (2018) 10(434):eaan2306. doi: 10.1126/scitranslmed.aan2306
21. Khorasani S, Mahmoudi M, Kalantari MR, Lavi Arab F, Esmaeili SA, Mardani F, et al. Amelioration of regulatory T cells by Lactobacillus delbrueckii and Lactobacillus rhamnosus in pristane-induced lupus mice model. J Cell Physiol (2019) 234(6):9778–86. doi: 10.1002/jcp.27663
22. Borody T, Leis S, Campbell J, Torres M, Nowak A. Fecal microbiota transplantation (FMT) in multiple sclerosis (MS). Am J Gastroenterol (2011) 106:S352. doi: 10.14309/00000434-201110002-00942
23. Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology (2013) 145(5):946–53. doi: 10.1053/j.gastro.2013.08.058
24. Makkawi S, Camara-Lemarroy C, Metz L. Fecal microbiota transplantation associated with 10 years of stability in a patient with SPMS. Neurol Neuroimmunol Neuroinflamm (2018) 5(4):e459. doi: 10.1212/NXI.0000000000000459
25. Cani PD. Human gut microbiome: hopes, threats and promises. Gut (2018) 67(9):1716–25. doi: 10.1136/gutjnl-2018-316723
26. Zeng J, Peng L, Zheng W, Huang F, Zhang N, Wu D, et al. Fecal microbiota transplantation for rheumatoid arthritis: A case report. Clin Case Rep (2021) 9(2):906–9. doi: 10.1002/ccr3.3677
27. Lleal M, Sarrabayrouse G, Willamil J, Santiago A, Pozuelo M, Manichanh C. A single faecal microbiota transplantation modulates the microbiome and improves clinical manifestations in a rat model of colitis. EBioMedicine (2019) 48:630–41. doi: 10.1016/j.ebiom.2019.10.002
28. He B, Hoang TK, Tian X, Taylor CM, Blanchard E, Luo M, et al. Lactobacillus reuteri Reduces the Severity of Experimental Autoimmune Encephalomyelitis in Mice by Modulating Gut Microbiota. Front Immunol (2019) 10:385. doi: 10.3389/fimmu.2019.00385
29. Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome (2017) 5(1):73. doi: 10.1186/s40168-017-0300-8
30. Wang H, Wang G, Liang Y, Du X, Boor PJ, Sun J, et al. Redox regulation of hepatic NLRP3 inflammasome activation and immune dysregulation in trichloroethene-mediated autoimmunity. Free Radic Biol Med (2019) 143:223–31. doi: 10.1016/j.freeradbiomed.2019.08.014
31. Wang G, Wang J, Ma H, Ansari GA, Khan MF. N-Acetylcysteine protects against trichloroethene-mediated autoimmunity by attenuating oxidative stress. Toxicol Appl Pharmacol (2013) 273(1):189–95. doi: 10.1016/j.taap.2013.08.020
32. Wang G, Pierangeli SS, Willis R, Gonzalez EB, Petri M, Khan MF. Significance of Lipid-Derived Reactive Aldehyde-Specific Immune Complexes in Systemic Lupus Erythematosus. PloS One (2016) 11(10):e0164739. doi: 10.1371/journal.pone.0164739
33. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods (2016) 13(7):581–3. doi: 10.1038/nmeth.3869
34. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res (2013) 41(Database issue):D590–6. doi: 10.1093/nar/gks1219
35. Wang H, Zhao JX, Hu N, Ren J, Du M, Zhu MJ. Side-stream smoking reduces intestinal inflammation and increases expression of tight junction proteins. World J Gastroenterol (2012) 18(18):2180–7. doi: 10.3748/wjg.v18.i18.2180
36. Soong L, Wang H, Shelite TR, Liang Y, Mendell NL, Sun J, et al. Strong type 1, but impaired type 2, immune responses contribute to Orientia tsutsugamushi-induced pathology in mice. PloS Negl Trop Dis (2014) 8(9):e3191. doi: 10.1371/journal.pntd.0003191
37. Liu J, Karypis G, Hippen KL, Vegoe AL, Ruiz P, Gilkeson GS, et al. Genomic view of systemic autoimmunity in MRLlpr mice. Genes Immun (2006) 7(2):156–68. doi: 10.1038/sj.gene.6364286
38. Heydemann A. The super super-healing MRL mouse strain. Front Biol (Beijing) (2012) 7(6):522–38. doi: 10.1007/s11515-012-1192-4
39. Perry D, Sang A, Yin Y, Zheng YY, Morel L. Murine models of systemic lupus erythematosus. J BioMed Biotechnol (2011) 2011:271694. doi: 10.1155/2011/271694
40. Chu JL, Drappa J, Parnassa A, Elkon KB. The defect in Fas mRNA expression in MRL/lpr mice is associated with insertion of the retrotransposon, ETn. J Exp Med (1993) 178(2):723–30. doi: 10.1084/jem.178.2.723
41. Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: clues from genetic and cytokine studies. Clin Rev Allergy Immunol (2011) 40(1):42–9. doi: 10.1007/s12016-009-8192-4
42. Nusbaum JS, Mirza I, Shum J, Freilich RW, Cohen RE, Pillinger MH, et al. Sex Differences in Systemic Lupus Erythematosus: Epidemiology, Clinical Considerations, and Disease Pathogenesis. Mayo Clin Proc (2020) 95(2):384–94. doi: 10.1016/j.mayocp.2019.09.012
43. Petri M. Epidemiology of systemic lupus erythematosus. Best Pract Res Clin Rheumatol (2002) 16(5):847–58. doi: 10.1053/berh.2002.0259
44. Kerksick C, Willoughby D. The antioxidant role of glutathione and N-acetyl-cysteine supplements and exercise-induced oxidative stress. J Int Soc Sports Nutr (2005) 2:38–44. doi: 10.1186/1550-2783-2-2-38
45. Schramm C, Lohse AW. Autoimmune hepatitis on the rise. J Hepatol (2014) 60(3):478–9. doi: 10.1016/j.jhep.2013.11.025
46. Gilbert KM, Przybyla B, Pumford NR, Han T, Fuscoe J, Schnackenberg LK, et al. Delineating liver events in trichloroethylene-induced autoimmune hepatitis. Chem Res Toxicol (2009) 22(4):626–32. doi: 10.1021/tx800409r
47. Gilbert KM, Reisfeld B, Zurlinden TJ, Kreps MN, Erickson SW, Blossom SJ. Modeling toxicodynamic effects of trichloroethylene on liver in mouse model of autoimmune hepatitis. Toxicol Appl Pharmacol (2014) 279(3):284–93. doi: 10.1016/j.taap.2014.07.003
48. Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol Med (2019) 25(1):35. doi: 10.1186/s10020-019-0102-5
49. Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio (2014) 5(5):e01548–14. doi: 10.1128/mBio.01548-14
50. Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA (2010) 107(5):2159–64. doi: 10.1073/pnas.0904055107
51. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA (2017) 114(40):10713–8. doi: 10.1073/pnas.1711235114
52. Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun (2016) 7:12015. doi: 10.1038/ncomms12015
53. Li H, Limenitakis JP, Greiff V, Yilmaz B, Schären O, Urbaniak C, et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature (2020) 584:274–8. doi: 10.1038/s41586-020-2564-6
54. Shi L, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PloS One (2014) 9(5):e93846. doi: 10.1371/journal.pone.0093846
55. Smallwood MJ, Nissim A, Knight AR, Whiteman M, Haigh R, Winyard PG. Oxidative stress in autoimmune rheumatic diseases. Free Radic Biol Med (2018) 125:3–14. doi: 10.1016/j.freeradbiomed.2018.05.086
56. Wang G, Ma H, Wang J, Khan MF. Contribution of poly(ADP-ribose)polymerase-1 activation and apoptosis in trichloroethene-mediated autoimmunity. Toxicol Appl Pharmacol (2019) 362:28–34. doi: 10.1016/j.taap.2018.10.012
57. Banerjee N, Wang H, Wang G, Khan MF. Enhancing the Nrf2 antioxidant signaling provides protection against trichloroethene-mediated inflammation and autoimmune response. Toxicol Sci (2020) 175(1):64–74. doi: 10.1093/toxsci/kfaa022
58. He L, He T, Farrar S, Ji L, Liu T, Ma X. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell Physiol Biochem (2017) 44(2):532–53. doi: 10.1159/000485089
59. Circu ML, Aw TY. Intestinal redox biology and oxidative stress. Semin Cell Dev Biol (2012) 23(7):729–37. doi: 10.1016/j.semcdb.2012.03.014
60. Circu ML, Aw TY. Redox biology of the intestine. Free Radic Res (2011) 45(11-12):1245–66. doi: 10.3109/10715762.2011.611509
61. Liu S, Rezende RM, Moreira TG, Tankou SK, Cox LM, Wu M, et al. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe (2019) 26(6):779–94 e8. doi: 10.1016/j.chom.2019.10.008
62. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med (2019) 25(7):1096–103. doi: 10.1038/s41591-019-0495-2
Keywords: microbiome, oxidative stress, permeability, inflammation, autoimmunity
Citation: Wang H, Wang G, Banerjee N, Liang Y, Du X, Boor PJ, Hoffman KL and Khan MF (2021) Aberrant Gut Microbiome Contributes to Intestinal Oxidative Stress, Barrier Dysfunction, Inflammation and Systemic Autoimmune Responses in MRL/lpr Mice. Front. Immunol. 12:651191. doi: 10.3389/fimmu.2021.651191
Received: 08 January 2021; Accepted: 11 March 2021;
Published: 12 April 2021.
Edited by:
Giacomo Emmi, Università degli Studi di Firenze, ItalyReviewed by:
Alessandra Bettiol, University of Florence, ItalyNarendra Prasad Singh, University of South Carolina, United States
Copyright © 2021 Wang, Wang, Banerjee, Liang, Du, Boor, Hoffman and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Firoze Khan, bWZraGFuQHV0bWIuZWR1
 Hui Wang
Hui Wang Gangduo Wang1
Gangduo Wang1 Nivedita Banerjee
Nivedita Banerjee Yuejin Liang
Yuejin Liang M. Firoze Khan
M. Firoze Khan