
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 22 March 2021
Sec. NK and Innate Lymphoid Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.649311
This article is part of the Research TopicThe Role of Tissue Resident NK Cells During Homeostasis, Primary and Secondary InfectionView all 10 articles
Diverse populations of natural killer (NK) cells have been identified in circulating peripheral blood and a wide variety of different tissues and organs. These tissue-resident NK cell populations are phenotypically distinct from circulating NK cells, however, functional descriptions of their roles within tissues are lacking. Recent advances in single cell RNA sequencing (scRNA-seq) have enabled detailed transcriptional profiling of tissues at the level of single cells and provide the opportunity to explore NK cell diversity within tissues. This review explores potential novel functions of human liver-resident (lr)NK cells identified in human liver scRNA-seq studies. By comparing these datasets we identified up-regulated and down-regulated genes associated with lrNK cells clusters. These genes encode a number of activating and inhibiting receptors, as well as signal transduction molecules, which highlight potential unique pathways that lrNK cells utilize to respond to stimuli within the human liver. This unique receptor repertoire of lrNK cells may confer the ability to regulate a number of immune cell populations, such as circulating monocytes and T cells, while avoiding activation by liver hepatocytes and Kupffer cells. Validating the expression of these receptors on lrNK cells and the proposed cellular interactions within the human liver will expand our understanding of the liver-specific homeostatic roles of this tissue-resident immune cell population.
The phenotype and function of natural killer (NK) cells have been extensively studied in both mice and humans, predominantly utilizing NK cells isolated from circulating peripheral blood (PB). Circulating NK cells account for 5–15% of the total lymphoid population in PB. In humans two functionally distinct subsets of NK cells are recognized, based on their expression of CD56 and CD16: CD56dimCD16+ and CD56brightCD16lo/− (1). The CD56dim NK cells represent the majority of circulating NK cells (~90%) and are more cytotoxic than CD56bright NK cells (2).
Circulating NK cells are equipped to recognize and kill both tumor and virally infected cells as well as possessing the ability to regulate other immune cells (3, 4). To carry out their effector functions NK cells have an extensive repertoire of stimulatory, costimulatory and inhibitory receptors but, unlike T and B cells, the genes encoding these receptors do not undergo somatic gene recombination. These NK cell receptors include the killer immunoglobulin-like receptor (KIR) family, the CD94(KLRD1)/NKG2 family of C-type lectins, the natural cytotoxicity receptors (NCR) and as well as a variety other activating and inhibitory surface receptors, as reviewed previously (5–7). It is the balance of expression of these receptors that is necessary to discriminate between healthy host cells and cells in distress, as well as self and non-self-cells. This balance ensures NK cells remain tolerant of healthy tissue whilst effectively clearing any potential threats, such as viral infection or cancerous cells.
In addition to these circulating NK cell populations it is evident that a variety of tissue-resident NK cell subsets exist (8). Tissue-resident NK cells are enriched in lymphoid tissues, the uterus and the liver, as well as a number of other tissues (9–12). Tissue-resident NK cell populations are predominantly CD56bright in humans and they appear to be phenotypically and functionally distinct from their circulating counterparts (8). In the human liver NK cells constitute up to half of total lymphocytes. The majority of intrahepatic NK cells possess a CD56bright phenotype, however their functional roles within the liver remains unclear (8, 13–16).
The liver has vital metabolic functions, detoxification functions, and immunological functions (17, 18). It is home to a diverse repertoire of resident immune cells including Kupffer cells (KC), NK cells and other innate and adaptive immune cells which are thought to contribute to maintain hepatic tolerance (19–21). The liver receives up to 80% of its blood supply from the gut via the hepatic portal vein, and this blood contains a high concentration of foreign molecules derived from our diet and gut microbiome. Resident immune cells must remain tolerant to this continuous exposure to harmless dietary and commensal bacterial products, while simultaneously being ready to clear any pathogens, malignant cells or toxic products that it may encounter, ensuring organ homeostasis (18).
Intrahepatic NK cells were first described in rat livers as “pit cells” in the 1970s after their cytoplasmic granules which resembled grape pits (22). For many years it was known that NK cells were enriched in the liver, however the functional significance of this was unknown (14, 23). Murine studies investigating memory-like NK cell responses identified a population of intrahepatic NK cells in murine livers defined as DX5−CD49a+CXCR6+TRAIL+ (24– 28). In 2015 an equivalent human intrahepatic NK cell population was described within hepatic sinusoids defined by the expression of CD49a (29). These CD49a+ intrahepatic NK cells display enhanced pro-inflammatory cytokine responses and memory-like recall responses in in vivo murine and ex vivo human studies (25, 29, 30).
In human liver this CD49a+ intrahepatic NK cell population is highly variable between donors and only represents a minority of the total intrahepatic NK cell population. The majority of human intrahepatic NK cells consist of a CD56bright liver-resident (lr)NK cell population, and are defined phenotypically as CD56brightCXCR6+CD69+EomeshiTbetloCD49a−CD49e− (11, 13, 16, 29, 31–33). Due to the species specific differences between mice and humans this review will focus only on human studies, with the term “lrNK” being used to specifically refer to the human CD56brightCXCR6+CD69+EomeshiTbetloCD49a−CD49e− NK cell population.
Using these defined phenotypical markers the effector function of human lrNK cells have been explored in a number of studies (summarized in Table 1). These studies have identified equivalent or reduced cytokine responses and increased degranulation compared PB NK cells and intrahepatic CD56dim populations (Table 1). However, to date these findings are limited to functional assays assessing a limited range of markers, upon stimulation with cytokines or HLA class I deficient immortalized cell lines. In this context, data generated from single cell RNA sequencing (scRNA-seq) studies can provide novel insights into potential roles for human lrNK cells beyond IFN-γ and TNF-α production and cytotoxic responses to cytokine activation or HLA-deficient tumor cells.
Recent advancements in scRNA-seq have provided us with a detailed view of the cellular components that make up the human liver (17, 32, 36–39). A number of studies utilizing scRNA-seq technologies have profiled the human liver (summarized in Table 2) (36–40). These studies have provided a global overview of the unique liver-specific adaptations of both parenchymal and non-parenchymal cells, including tissue-resident immune cells. This data represents an important resource for the international research community.
These scRNA-seq datasets have sampled all tissue-resident immune cell populations (limited only by the number of single cells sequenced in each dataset). However, subsequent validation experiments and functional work in these studies have, out of necessity, focused on only a few selected cell populations present in the human liver. One population currently lacking a detailed interpretation of the transcription profiles generated in these datasets is lrNK cells.
In a number of published scRNA-seq studies of the human liver NK cell clusters have been identified although the composition of these NK cell clusters varies between studies (Table 2). The requirement to define clusters based on genes characteristic of specific cell types represents a limitation of scRNA-seq studies. While phenotypic markers have been well defined, the transcriptional profiles that distinguish between immune cell subpopulations, in particular subpopulations of NK cells, lack extended validation. This issue is compounded by the fact that the optimal number of clusters differs depending on the specific research question. The datasets generated by MacParland et al. (37), Aizarani et al. (38), and Zhang et al. (40), all identified a single cluster of NK cells (Table 2). It is likely this single NK cell cluster contains both lrNK cells and circulating NK cells. As such, further analysis of NK cell subpopulations is not possible without reanalysing the raw sequencing datasets in order to generate additional NK cell clusters.
In contrast both the study by Zhao et al. (36) and the study by Ramachandran et al. (39) identified multiple NK cell clusters (Table 2). The study by Zhao and colleagues identified differentially expressed genes comparing between CXCR6+ lrNK cells and CX3CR1+ circulating (c)NK cells (36). The study by Ramachandran and colleagues identified two lrNK cell clusters and one cNK cell cluster, and included lists of differentially expressed genes comparing each individual cluster to all others (39). This study includes liver samples from both healthy controls and patients with liver cirrhosis, and one of the lrNK cell clusters is largely absent in cirrhotic patients highlighting the possibility that NK cell subpopulations may be altered in the diseased liver (39).
To explore the transcriptional profile unique to lrNK cells our review focuses on genes identified in the Ramachandran et al. and Zhao et al. datasets (36, 39). Comparing the lrNK cell and cNK cell clusters between these 2 datasets identifies 180 up-regulated genes associated with lrNK cells, 146 up-regulated genes associated with cNK cells and 19 up-regulated genes shared between lrNK and cNK clusters (Figure 1 and Supplementary Data). Genes up-regulated in lrNK cell clusters include EOMES and CXCR6, which have been validated at a protein level as phenotypic markers of lrNK cells (11, 13, 16). Genes up-regulated in cNK cell clusters include ITGAM, S1PR1, and SELL, which are markers of immune cell migration that are down-regulated at a protein level in tissue-resident immune cell populations (31, 33). A variety of activating and inhibitory receptors, downstream signaling molecules, and effector molecules are differentially expressed between lrNK cells and cNK cells (Figure 1 and Supplementary Data). These differentially regulated genes provide opportunities to identify novel markers of lrNK cells as well as providing intriguing insights into potential functions of lrNK cells within the human liver, which are distinct from cNK cells and extend beyond the recognition of virally infected cells or tumors.
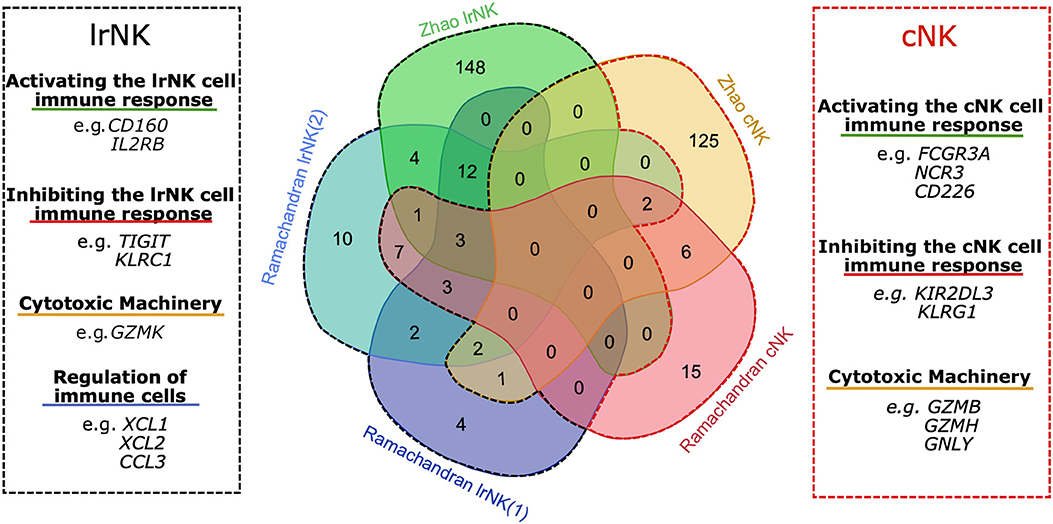
Figure 1. Overlap of up-regulated genes in lrNK cell populations from human liver scRNA-seq datasets. Venn diagram illustrating the overlap of DEG within each NK cell cluster of both the Zhao et al. (36) and Ramachandran et al. (39) datasets. The black dashed line and the red dashed line indicate genes up-regulated in lrNK cell clusters and cNK clusters respectively.
A number of genes involved in the activation of NK cells are differentially regulated in lrNK and cNK cells in these scRNA-seq datasets (Figure 1 and Supplementary Data). The lrNK cell clusters up-regulate activating receptor genes CD160, CD27, CD7, IL2RB, TMIGD2, and TNFSF14 as well as SH2D1A, which is involved in signal transduction. In contrast the cNK cell clusters up-regulate activating receptor genes such as CD226, FCGR3A, IL12RB1, and NCR3.
A number of these genes are involved in NK cell interactions with hepatocytes and other immune cell populations. IL2RB encodes the interleukin-2 receptor β-chain (IL-2Rβ; CD122) and is a major marker of NK-committed cells or NK-progenitors, which allows the cells to respond to IL-15 (41). IL-2Rα forms part of the IL-2 as well as the IL-15 receptor (in complex with IL-15Rα) (42). IL-15 is important for NK cell survival, activation and function (43, 44), is strongly expressed in the liver (45–47), and hepatocyte- and KC-derived IL-15/IL-15Rα directly regulates the homeostasis of liver NK cells via trans-presentation (48). Conversely IL12RB, which encodes IL-12Rβ, is upregulated on cNK cells and induces IFN-γ production when stimulated by IL-12, highlighting that cNK and lrNK cells are regulated by different cytokines in the liver microenvironment (49).
CD27+ NK cells are rare in the PB yet are enriched in tissues and associated with lower cytotoxic abilities with reduced granzyme B and perforin in comparison to their CD27− counterparts (50). Interestingly, upon binding of its ligand CD70, CD27 is downregulated in an IL-15R-dependent manner suggesting that these lrNK cell's cytotoxicity is tightly controlled by their expression of IL-15R and CD27 and the local availability of IL-15 and CD70 (51). In the steady state, CD70 levels are quite low, with tightly regulated expression observed upon activation on T cells, B cells and subsets of DCs (52).
Both CD160 and TNFSF14 (also known as LIGHT) encode proteins capable of binding to herpes virus entry mediator (HVEM; also known as TNFRSF14) (53, 54). HVEM is widely expressed by monocytes, dendritic cells (DC), neutrophils, NK cells, T- and B-cells and downregulated in activated T cells (55, 56). CD160 also binds weakly to classical MHC-I such as HLA-C (57, 58) and the non-classical MHC-I soluble HLA-G (59), while LIGHT also binds to lymphotoxin-receptor (LT-R, also known as TNFRSF3) (60).
Engagement of CD160 or LIGHT on PB CD56dim NK cells activates their cytotoxic effector function enhancing NK-mediated lysis of target cells (54, 58, 61), indicating the possibility that NK cells directly regulate HVEM-expressing immune cells in the liver. Both KCs and hepatocytes express LT-R within the liver. The engagement of LIGHT and LT-R on hepatocytes has also been shown to regulate hepatic lipase expression (62) as well as directly regulating hepatocyte proliferation and liver regeneration in response to partial hepatectomy (63–65).
CD7 encodes an early differentiation marker on common lymphoid progenitors and crosslinking of CD7 induces NK cell activation, proliferation, cytokine production and cytotoxicity (66, 67). Ligands for CD7 include SECTM1 (68) (secreted and transmembrane protein 1) and LGALS1 (galectin-1) (69). SECTM1 is highly expressed by epithelial cells and acts as a chemoattractant for CD7 expressing cells (70, 71). Galectin-1 is also expressed by epithelial cells as well as T regulatory cells. Galectin-1 is capable of suppressing inflammatory immune responses and is essential for regulating inflammation and hepatocyte proliferation during liver regeneration (72).
SH2D1A encodes the signaling lymphocyte activation molecule (SLAM)-associated protein SAP. This adaptor protein mediates downstream signaling from SLAM family receptors on NK cells leading to stable conjugate formation with target cells via LFA-1 (73) and target cell-directed cytotoxic granule polarization and exocytosis (74). SLAM family receptors are immunoglobulins expressed on a variety of immune cell populations that are activated via homophilic binding (except for SLAMF4/2B4 which binds SLAMF2/CD48) (75).
In addition to these up-regulated genes, a number of activating receptors are expressed by cNK cell but not lrNK cells. cNK cells upregulate FCGR3A which encodes the CD16 protein, a key marker of CD56dim cNK cells, which is responsible for antibody-dependent cell-mediated cytotoxicity (ADCC). NCR3 encodes NKp30 and is upregulated on cNK cells equipping them with anti-tumor specific cytotoxic abilities (76). Likewise the CD226 gene is upregulated on cNK cells and encodes the activating receptor DNAM-I. Interestingly this receptor shares ligands, which include polio virus receptor (PVR/CD155) and Nectin-2 (PVRL2/ CD112), with the inhibitory molecule T cell Ig and ITIM domain (TIGIT) that is upregulated in lrNK cells and is discussed in detail in the next section (77). This differential expression emphasizes how certain stimuli can regulate cNK and lrNK cell populations in different directions.
These differentially expressed activating receptor genes highlight the importance of hepatocytes in regulating lrNK cell survival and imply an important role for lrNK cells in regulating infiltrating immune cell populations within the human liver (Figure 2).
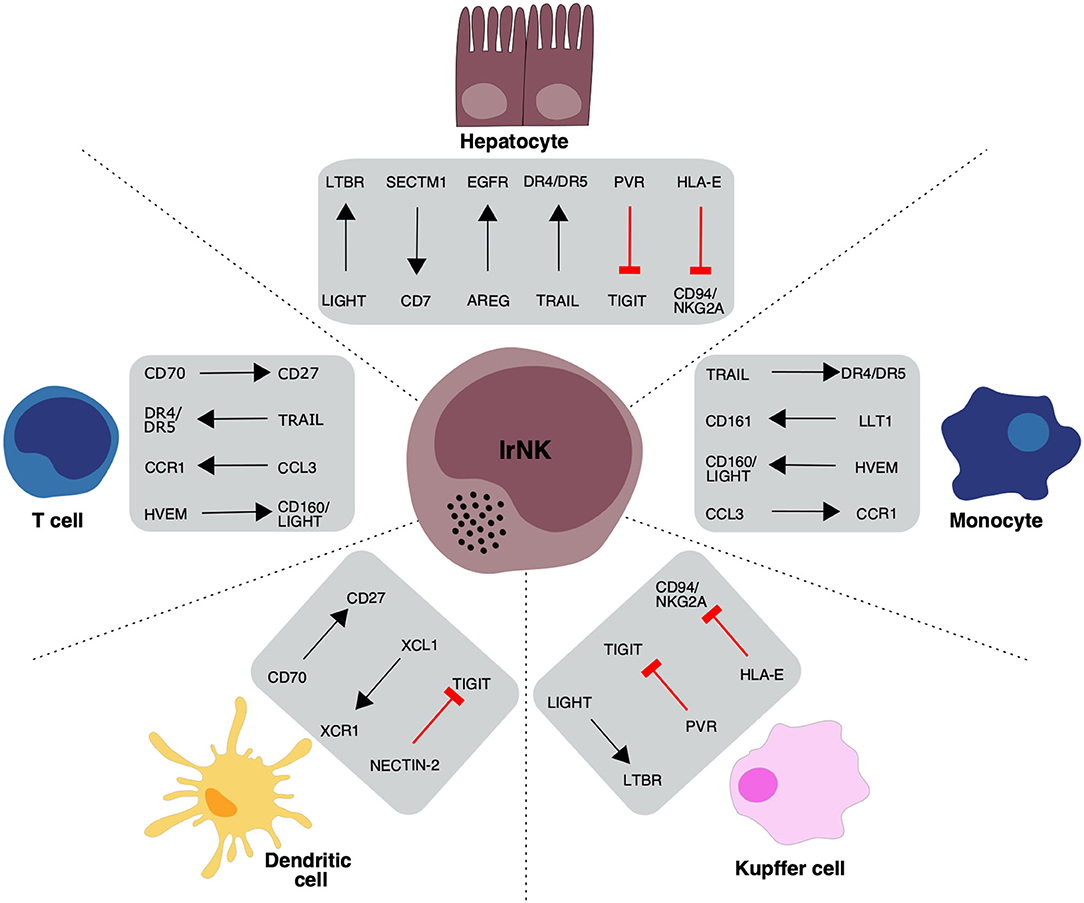
Figure 2. Overview of proposed receptor-ligand interactions between lrNK cells and haemopoietic and non-haemopoietic cell populations in the liver. Black arrows indicate the direction of a stimulatory ligand-receptor interaction and red lines indicate an inhibitory ligand-receptor interaction between interacting cell types.
In addition to activating receptors, a number of inhibitory receptors are differentially regulated in lrNK and cNK cells in these scRNA-seq datasets (Figure 1 and Supplementary Data). The lrNK cell clusters up-regulate genes including KLRB1, KLRC1, and TIGIT. Conversely, the cNK cell clusters up-regulate CLEC2D, KIR2DL3, KIR3DL2, and KLRG1.
TIGIT is an inhibitory receptor expressed by T and NK cells and up-regulated upon activation (78, 79). As mentioned previously TIGIT shares its ligands PVR and Nectin-2 with the activating receptor DNAM-I that is up-regulated in cNK cells (80). TIGIT binds with high affinity to PVR and with lower affinity to Nectin-2, with both interactions resulting in inhibition of NK cell cytotoxicity and IFN-γ production (80–82). Hepatocytes and KCs in the liver express PVR equipping them with the ability to directly inhibit NK cell function (83). Hepatocyte PVR levels are increased during liver regeneration, which is suggested to be a mechanism by which hepatocytes safeguard themselves from TIGIT+ NK cell killing (83, 84). DCs express both PVR and Nectin-2, and engagement of PVR with a TIGIT-Fc fusion protein increased DC IL-10 production, which may further suppress NK cell function in the liver (79). Using ligand-receptor pair databases Zhang and colleagues predicted that LAMP3+ DCs in the liver interact with conventional circulating NK cells via the activating Nectin2-DNAM-I axis and with lrNK cells via inhibitory Nectin-2-TIGIT interactions suggesting that liver DCs may be regulating different NK subsets in opposing directions (85, 86).
KLRD1 (CD94) is up-regulated in both cNK and lrNK cells and can combine with KLRC1 (NKG2A) which is specifically up-regulated in lrNK cells. This heterodimeric complex is a C-type lectin receptor and interacts with the non-classical HLA class I molecule HLA-E on target cells resulting in inhibition of NK cell function. High protein levels of NKG2A have been confirmed on CD49e− intrahepatic NK cells (29, 35), and NKG2A can be induced by the immunosuppressive cytokines TGF-β and IL-10 which are present at high levels in the liver (87, 88). HLA-E is expressed in the liver by both KCs and hepatocytes providing another molecular pathway through which hepatocytes, and KCs, can inhibit lrNK cell activation. In circulating PB NKG2A+ NK cells that lack KIR have been shown to kill autologous immature DCs that have reduced HLA-E expression in vitro (89), and it is possible lrNK cells are likewise capable of effectively targeting immature DCs that express low levels of HLA-E.
An up-regulation of the KLRB1 gene in lrNK cells provides a further mechanism that may lead to inhibition of lrNK cell function. Engagement of KLRB1 (CD161) by its ligand lectin-like transcript 1 (LLT1 also known as CLEC2D) inhibits NK cell cytotoxicity and cytokine production (90, 91). LLT1 is expressed by B cells and monocytes and the level of LLT1 decreases on monocytes upon activation (92) but it also increased in cNK cell clusters in these scRNA-seq datasets suggesting that cNK cells may regulate lrNK cell function. While hepatocytes in healthy livers express relatively low levels of LLT1, it is increased in the context of liver cirrhosis and this results in reduced NK cell function (93).
In contrast to lrNK cells, cNK cells express a distinct profile of inhibitory receptors. The scRNA-seq datasets reveal an up-regulation of inhibitory KIRs in cNK cells (KIR3DL2, KIR2DL3). This low expression or absence of KIRs on lrNK cells has been validated at the protein level (33). KLRG1 is also up-regulated in cNK cells. KLRG1 encodes an inhibitory C-type lectin receptor that recognizes E-cadherin and N-cadherin and inhibits NK cell activation (94).
These differentially expressed inhibitory receptor genes highlight a number of key regulatory mechanisms by which liver hepatocytes and intrahepatic KCs restrain lrNK cell activity and maintain overall organ homeostasis (Figure 2).
Upon activation by a target cell, NK cells can release cytotoxic granules and up-regulate death-inducing ligands, each of which result in target cell apoptosis. These scRNA-seq datasets indicate an up-regulation of GZMK and TNFSF10 in lrNK cells, and an up-regulation of GNLY, GZMB, and GZMH in cNK cells (Figure 1 and Supplementary Data).
TNFSF10 encodes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), a transmembrane protein that mediates a death-receptor mediated pathway of cytotoxicity. It has been described as important for both NK cell anti-viral and anti-tumor activity in the liver as it binds to TRAIL death receptors (DR) including DR4 and DR5 on target cells to induce target cell-apoptosis (95–97). Decoy receptors (DcR) also exist that do not induce apoptosis and include DcR1 and DcR2. DR4 and DR5 are expressed in by hepatocytes (98), monocytes, monocyte derived macrophages and activated T cells (99).
Granule-mediated cytotoxicity involves the targeted release of secretory granules containing granzymes (e.g., granzyme-A, -B, -H, -K, and -M), together with perforin and granulysin (100). LrNK cells have elevated transcription of GZMK which encodes granzyme-K. Granzyme-K has trypsin-like activity and can induce a rapid cell death independently of caspases (100). In contrast cNK cells have elevated expression of GZMB encoding for granzyme-B and GZMH encoding for granzyme-H. Granzyme-B mediates apoptosis of target cells using caspase-dependent mechanisms (101–103), while granzyme-H can induce cell death independently of caspases (104). Granulysin is a saposin-like protein that has pore forming abilities as well as anti-tumor and anti-microbe cytotoxic activity and is selectively up-regulated in cNK cells (105–107).
The elevated expression of GZMK and TNFSF10 indicates lrNK cells can utilize both death-receptor and granule-mediated cytotoxic pathways to kill target cells, however, they use distinct mediators in comparison to cNK cells where granzyme-B and granulysin are key cytotoxic mediators.
In addition to mediating cytotoxic responses, NK cells are also capable of producing a wide variety of immunoregulatory molecules, most notably IFN-γ. Within the scRNA-seq datasets lrNK cells upregulate a variety of immunoregulatory genes including XCL1, XCL2, CCL3, and AREG which may regulate both parenchymal and non-parenchymal cells in the liver and shape the hepatic immune response (Figure 2).
A number of chemokines were identified in lrNK clusters highlighting potential roles in regulating the migration of other immune cell subsets in the liver. XCL1 and XCL2 bind to the XC chemokine receptor 1 (XCR1), which is selectively expressed by cross-presenting CD141+ DC's in circulating PB, commonly referred to as cDC1 (108). These cDC1s are functionally equivalent to CD8+ DC in the mouse, a subset of DC that endocytose stressed cells and dying cells (109, 110) and preferentially present this processed antigen to CD8+ T cells (111, 112). This XCL1-XCR1 axis may provide lrNK cells the ability to regulate DCs within the human liver (38, 39, 113, 114).
The transcripts for CCL3, up-regulated in lrNK cells, and CCL4, up-regulated in both cNK and lrNK cells, encode CCL3 (also known as MIP-1α) and CCL4 (also known as MIP-1β). Both of these chemokines signal via the CCR1 and CCR5 receptors (115) which are expressed by several cell types including monocytes, NK cells and activated CD4+ and CD8+ T cells (116, 117). Expression of CCL3 and CCL4 allows lrNK cells to directly regulate migration of myeloid cell populations and T cells into the liver. HSCs also express CCR5 and CCL3-engagement induced their proliferation in vitro and implicating the CCL3-CCR5 axis in the progression of liver fibrosis (118, 119).
The liver has a remarkable ability to regenerate itself via hepatocyte proliferation. One of the key pathways involved in regulating hepatocyte proliferation is the epidermal growth factor (EGF) receptor (EGFR). A number of EGFR ligands are known, one of which is amphiregulin (AREG) (120–122), which is up-regulated at a transcriptional level in lrNK cells. Interestingly, AREG was only identified in lrNK cell cluster from the Ramachandran et al. dataset. This cluster included lrNK cells from both healthy and cirrhotic liver tissues, suggesting this gene may be up-regulated in lrNK cells in the context of liver disease. AREG is a membrane-bound precursor protein that can be processed and secreted to act in a paracrine or autocrine fashion and is involved in in multiple processes including proliferation, apoptosis and migration of both epithelial and immune cells (123). EGFR is expressed by hepatocytes and HSCs in the liver and the AREG-EGFR axis is known to specifically modulate the hepatic acute phase reaction in the liver regeneration process and is essential for normal hepatocellular survival and proliferation (124, 125). Overall, immune cell derived AREG is suggested to be associated with type 2 immune-mediated resistance and tissue protective mechanisms (123, 126, 127).
The up-regulation of specific signaling molecules and receptors on lrNK cells and their enrichment in the liver suggest an important liver-specific role for these cells. Attempts to understand these NK cell functions have been limited by the use of functional assays that assess a limited range of markers. Here we show that scRNA-seq datasets can provide novel insights into possible functional roles of human lrNK cells (Figure 2) that may support the development of novel hypotheses to be explored in future functional investigations, once expression at the protein level is validated.
The increased expression of cytotoxic machinery components, including perforin and granzyme-K, equip these lrNK cells with the ability to mediate cytotoxic responses. The activation of this cytotoxicity is tightly controlled by the careful balance of activating and inhibitory receptors, of which lrNK cells express a distinct repertoire. This distinct receptor repertoire may confer the ability to regulate a number of infiltrating immune cell populations within the liver. At the same time this distinct receptor repertoire ensures hepatocytes and KCs can restrain lrNK cell activity and promote organ homeostasis.
Detailed investigations into specific intrahepatic cells that express these receptor ligands and the conditions under which they are induced will aid in our understanding of the liver-specific role of lrNK cells. Validation of potential liver-specific functions of lrNK cells requires a comprehensive analysis of human liver parenchymal and non-parenchymal cell populations, together with the development of novel functional assays. It is evident that a wide diversity of resident intrahepatic NK cell populations exist in the human liver and a detailed understanding of their roles in liver homeostasis and liver disease is essential for our understanding of tissue-resident immune responses within the liver.
GJ and MR contributed equally to the conception and design of the article, interpreting the relevant literature, and drafting and reviewing the manuscript. Both authors contributed to the article and approved the submitted version.
This research was funded by the Health Research Board (Grant number EIA-2017-013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Professor Derek Doherty and Professor Cliona O'Farrelly for critical discussions and feedback on this manuscript. Figures 1, 2 were generated using artwork from smart.servier.com.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.649311/full#supplementary-material
1. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. (1975) 5:112–7. doi: 10.1002/eji.1830050208
2. Lanier LL, Le AM, Phillips JH, Warner NL, Babcock GF. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J Immunol. (1983) 131:1789–96. doi: 10.1002/jlb.35.1.11
3. Jost S, Altfeld M. Control of human viral infections by natural killer cells. Annu Rev Immunol. (2013) 31:163–94. doi: 10.1146/annurev-immunol-032712-100001
4. Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. (2012) 481:394–8. doi: 10.1038/nature10624
5. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity. (2017) 47:820–33. doi: 10.1016/j.immuni.2017.10.008
6. Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. (2013) 5:208ra145. doi: 10.1126/scitranslmed.3006702
7. Lanier LL. NK Cell recognition. Annu Rev Immunol. (2005) 23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526
8. Björkström NK, Ljunggren H-GG, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immun. (2016) 16:310–20. doi: 10.1038/nri.2016.34
9. Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. (2003) 198:1201–12. doi: 10.1084/jem.20030305
10. Manaster I, Mizrahi S, Goldman-Wohl D, Sela HY, Stern-Ginossar N, Lankry D, et al. Endometrial NK cells are special immature cells that await pregnancy. J Immunol. (2008) 181:1869–76. doi: 10.4049/jimmunol.181.3.1869
11. Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, et al. CXCR6 marks a novel subset of T-bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. (2016) 6:26157. doi: 10.1038/srep26157
12. Lugthart G, Melsen JE, Vervat C, van Ostaijen-Ten Dam MM, Corver WE, Roelen DL, et al. Human lymphoid tissues harbor a distinct CD69+CXCR6+ NK cell population. J Immunol. (2016) 197:78–84. doi: 10.4049/jimmunol.1502603
13. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver-resident CD56 bright /CD16 neg NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways HHS Public Access. J Autoimmun. (2016) 66:40–50. doi: 10.1016/j.jaut.2015.08.011
14. Norris S, Collins C, Doherty DG, Smith F, McEntee G, Traynor O, et al. Resident human hepatitis lymphocytes are phenotypically different from circulating lymphocytes. J Hepatol. (1998) 28:84–90. doi: 10.1016/S0168-8278(98)80206-7
15. Norris S, Doherty DG, Collins C, McEntee G, Traynor O, Hegarty JE, et al. Natural T cells in the human liver: cytotoxic lymphocytes with dual T cell and natural killer cell phenotype and function are phenotypically heterogenous and include Valpha24-JalphaQ and gammadelta T cell receptor bearing cells. Hum Immunol. (1999) 60:20–31. doi: 10.1016/S0198-8859(98)00098-6
16. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, et al. Tissue-resident Eomes hi T-bet lo CD56 bright NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol. (2016) 46:2111–20. doi: 10.1002/eji.201646559
17. Stamataki Z, Swadling L. The liver as an immunological barrier redefined by single-cell analysis. Immunology. (2020) 160:157–70. doi: 10.1111/imm.13193
18. Robinson MW, Harmon C, O'Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cellul Mol Immunol. (2016) 13: 267–76. doi: 10.1038/cmi.2016.3
19. Crispe IN. Hepatic T cells and liver tolerance. Nat Rev Immunol. (2003) 3:51–62. doi: 10.1038/nri981
20. Doherty DG, O'Farrelly C. Dendritic cells: regulators of hepatic immunity or tolerance? J Hepatol. (2001) 34:156–60. doi: 10.1016/S0168-8278(00)00020-9
21. Harmon C, Sanchez-Fueyo A, O'Farrelly C, Houlihan DD. Natural killer cells and liver transplantation: orchestrators of rejection or tolerance? Am J Transplant. (2016) 16:751–7. doi: 10.1111/ajt.13565
22. Wisse E, van't Noordende JM, van der Meulen J, Daems WT. The pit cell: Description of a new type of cell occurring in rat liver sinusoids and peripheral blood. Cell Tissue Res. (1976) 173:423–35. doi: 10.1007/BF00224305
23. Doherty DG, Suzanne N, Madrigal-Estebas L, McEntee G, Traynor O, Hegarty JE, et al. The human liver contains multiple populations of NK cells, T cells, and CD3+CD56+ natural T cells with distinct cytotoxic activities and Th1, Th2, and Th0 cytokine secretion patterns. J Immunol. (1999) 163:2314–21.
24. Kim S, Iizuka K, Kang HSP, Dokun A, French AR, Greco S, et al. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. (2002) 3:523–8. doi: 10.1038/ni796
25. Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. (2013) 123:1444–56. doi: 10.1172/JCI66381
26. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. (2014) 3:e01659. doi: 10.7554/eLife.01659
27. Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood. (2005) 105:2082–9. doi: 10.1182/blood-2004-08-3262
28. Paust S, Gill HS, Wang B-Z, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. (2010) 11:1127–35. doi: 10.1038/ni.1953
29. Marquardt N, Béziat V, Nyström S, Hengst J, Ivarsson MA, Kekäläinen E, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol. (2015) 194:2467–71. doi: 10.4049/jimmunol.1402756
30. Stary V, Pandey RV, Strobl J, Kleissl L, Starlinger P, Pereyra D, et al. A discrete subset of epigenetically primed human NK cells mediates antigen-specific immune responses. Sci Immunol. (2020) 5:eaba6232. doi: 10.1126/sciimmunol.aba6232
31. Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR, et al. Eomeshi NK Cells in human liver are long-lived and do not recirculate but can be replenished from the circulation. J Immunol. (2016) 197:4283–91. doi: 10.4049/jimmunol.1601424
32. Filipovic I, Sönnerborg I, Strunz B, Friberg D, Cornillet M, Hertwig L, et al. 29-Color flow cytometry: unraveling human liver NK cell repertoire diversity. Front Immunol. (2019) 10:2692. doi: 10.3389/fimmu.2019.02692
33. Aw Yeang HX, Piersma SJ, Lin Y, Yang L, Malkova ON, Miner C, et al. Cutting edge: human CD49e- NK cells are tissue resident in the liver. J Immunol. (2017) 198:1417–22. doi: 10.4049/jimmunol.1601818
34. Sun H, Xu J, Huang Q, Huang M, Li K, Qu K, et al. Reduced CD160 expression contributes to impaired NK-cell function and poor clinical outcomes in patients with HCC. Cancer Res. (2018) 78:6581–93. doi: 10.1158/0008-5472.CAN-18-1049
35. Lunemann S, Langeneckert AE, Martrus G, Hess LU, Salzberger W, Ziegler AE, et al. Human liver-derived CXCR6 + NK cells are predominantly educated through NKG2A and show reduced cytokine production. J Leukoc Biol. (2019) 105:1331–40. doi: 10.1002/JLB.1MA1118-428R
36. Zhao J, Zhang S, Liu Y, He X, Qu M, Xu G, et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. (2020) 6:22. doi: 10.1038/s41421-020-0157-z
37. MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. (2018) 9:4383. doi: 10.1038/s41467-018-06318-7
38. Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. (2019) 572:199–204. doi: 10.1038/s41586-019-1373-2
39. Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. (2019) 575:512–8. doi: 10.1101/766113
40. Zhang M, Yang H, Wan L, Wang Z, Wang H, Ge C, et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. (2020) 73:1118–30. doi: 10.1016/j.jhep.2020.05.039
41. Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. (2005) 22:295–304. doi: 10.1016/j.immuni.2005.01.013
42. Wu Y, Tian Z, Wei H. Developmental and functional control of natural killer cells by cytokines. Front Immunol. (2017) 8:930. doi: 10.3389/fimmu.2017.00930
43. Dunne J, Lynch S, O'farrelly C, Todryk S, Hegarty JE, Feighery C, et al. Selective expansion and partial activation of human NK cells and NK receptor-positive T cells by IL-2 and IL-15. J Immunol. (2001) 167:3129–38. doi: 10.4049/jimmunol.167.6.3129
44. Easom NJW, Stegmann KA, Swadling L, Pallett LJ, Burton AR, Odera D, et al. IL-15 Overcomes hepatocellular carcinoma-induced NK cell dysfunction. Front Immunol. (2018) 9:1009. doi: 10.3389/fimmu.2018.01009
45. Golden-Mason L, Kelly AM, Doherty DG, Traynor O, McEntee G, Kelly J, et al. Hepatic interleukin 15 (IL-15) expression: Implications for local NK/NKT cell homeostasis and development. Clin Exp Immunol. (2004) 138:94–101. doi: 10.1111/j.1365-2249.2004.02586.x
46. Stienstra R, Saudale F, Duval C, Keshtkar S, Groener JEM, Van Rooijen N, et al. Kupffer cells promote hepatic steatosis via interleukin-1β-dependent suppression of peroxisome proliferator-activated receptor α activity. Hepatology. (2010) 51:511–22. doi: 10.1002/hep.23337
47. Cui G, Hara T, Simmons S, Wagatsuma K, Abe A, Miyachi H, et al. Characterization of the IL-15 niche in primary and secondary lymphoid organs in vivo. Proc Natl Acad Sci U S A. (2014) 111:1915–20. doi: 10.1073/pnas.1318281111
48. Cepero-Donates Y, Rakotoarivelo V, Mayhue M, Ma A, Chen YG, Ramanathan S. Homeostasis of IL-15 dependent lymphocyte subsets in the liver. Cytokine. (2016) 82:95–101. doi: 10.1016/j.cyto.2015.12.012
49. Zwirner NW, Ziblat A. Regulation of NK cell activation and effector functions by the IL-12 family of cytokines: The case of IL-27. Front Immunol. (2017) 8:25. doi: 10.3389/fimmu.2017.00025
50. Fu B, Wang F, Sun R, Ling B, Tian Z, Wei H. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology. (2011) 133:350–9. doi: 10.1111/j.1365-2567.2011.03446.x
51. Vossen MTM, Matmati M, Hertoghs KML, Baars PA, Gent M-R, Leclercq G, et al. CD27 defines phenotypically and functionally different human NK cell subsets. J Immunol. (2008) 180:3739–45. doi: 10.4049/jimmunol.180.6.3739
52. Arroyo Hornero R, Georgiadis C, Hua P, Trzupek D, He LZ, Qasim W, et al. CD70 expression determines the therapeutic efficacy of expanded human regulatory T cells. Commun Biol. (2020) 3:1–17. doi: 10.1038/s42003-020-1097-8
53. Mauri DN, Ebner R, Montgomery RI, Kochel KD, Cheung TC, Yu GL, et al. LIGHT, a new member of the TNF superfamily, and lymphotoxin α are ligands for herpesvirus entry mediator. Immunity. (1998) 8:21–30. doi: 10.1016/S1074-7613(00)80455-0
54. Šedý JR, Bjordahl RL, Bekiaris V, Macauley MG, Ware BC, Norris PS, et al. CD160 Activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J Immunol. (2013) 191:828–36. doi: 10.4049/jimmunol.1300894
55. Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. (2008) 29:138–49. doi: 10.1016/j.immuni.2008.05.009
56. Steinberg MW, Cheung TC, Ware CF. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol Rev. (2011) 244:169–87. doi: 10.1111/j.1600-065X.2011.01064.x
57. Le Bouteiller P, Barakonyi A, Giustiniani J, Lenfant F, Marie-Cardine A, Aguerre-Girr M, et al. Engagement of CD160 receptor by HLA-C is a triggering mechanism used by circulating natural killer (NK) cells to mediate cytotoxicity. Proc Natl Acad Sci U S A. (2002) 99:16963–8. doi: 10.1073/pnas.012681099
58. Barakonyi A, Rabot M, Marie-Cardine A, Aguerre-Girr M, Polgar B, Schiavon V, et al. Cutting edge: engagement of CD160 by its HLA-C physiological ligand triggers a unique cytokine profile secretion in the cytotoxic peripheral blood NK cell subset. J Immunol. (2004) 173:5349–54. doi: 10.4049/jimmunol.173.9.5349
59. Fons P, Chabot S, Cartwright JE, Lenfant F, L'Faqihi F, Giustiniani J, et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood. (2006) 108:2608–15. doi: 10.1182/blood-2005-12-019919
60. Rooney IA, Butrovich KD, Glass AA, Borboroglu S, Benedict CA, Whitbeck JC, et al. The lymphotoxin-β receptor is necessary and sufficient for LIGHT- mediated apoptosis of tumor cells. J Biol Chem. (2000) 275:14307–15. doi: 10.1074/jbc.275.19.14307
61. Holmes TD, Wilson EB, Black EVI, Benest AV, Vaz C, Tan B, et al. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc Natl Acad Sci U S A. (2014) 111:E5688–96. doi: 10.1073/pnas.1411072112
62. Lo JC, Wang Y, Tumanov AV, Bamji M, Yao Z, Reardon CA, et al. Lymphotoxin β receptor-dependent control of lipid homeostasis. Science. (2007) 316:285–8. doi: 10.1126/science.1137221
63. Anders RA, Subudhi SK, Wang J, Pfeffer K, Fu Y-X. Contribution of the lymphotoxin β receptor to liver regeneration. J Immunol. (2005) 175:1295–300. doi: 10.4049/jimmunol.175.2.1295
64. Behnke K, Sorg UR, Herebian D, Häussinger D, Keitel V, Pfeffer K. The role of the lymphotoxin-β receptor (LTβR) in hepatocyte-mediated liver regeneration. Eur J Med Res. (2014) 19(Suppl. 1):S3. doi: 10.1186/2047-783X-19-S1-S3
65. Tumanov AV, Koroleva EP, Christiansen PA, Khan MA, Ruddy MJ, Burnette B, et al. T cell-derived lymphotoxin regulates liver regeneration. Gastroenterology. (2009) 136:694–704.e4. doi: 10.1053/j.gastro.2008.09.015
66. Rabinowich H, Lin WC, Herberman RB, Whiteside TL. Signalling via CD7 molecules on human NK cells. Induction of tyrosine phosphorylation and beta 1 integrin-mediated adhesion to fibronectin. J Immunol. (1994) 153:3504–13.
67. Rabinowich H, Pricop L, Herberman RB, Whiteside TL. Expression and function of CD7 molecule on human natural killer cells. J Immunol. (1994) 152:517–26.
68. Slentz-Kesler KA, Hale LP, Kaufman RE. Identification and characterization of K12 (SECTM1), a novel human gene that encodes a Golgi-associated protein with transmembrane and secreted isoforms. Genomics. (1998) 47:327–40. doi: 10.1006/geno.1997.5151
69. Pace KE, Hahn HP, Pang M, Nguyen JT, Baum LG. Cutting edge: CD7 delivers a pro-apoptotic signal during galectin-1-induced T cell death. J Immunol. (2000) 165:2331–4. doi: 10.4049/jimmunol.165.5.2331
70. Wang T, Ge Y, Xiao M, Lopez-Coral A, Li L, Roesch A, et al. SECTM1 produced by tumor cells attracts human monocytes via CD7-mediated activation of the PI3K pathway. J Invest Dermatol. (2014) 134:1108–18. doi: 10.1038/jid.2013.437
71. Lam GK, Liao H-X, Xue Y, Munir Alam S, Scearce RM, Kaufman RE, et al. Expression of the CD7 ligand K-12 in human thymic epithelial cells: regulation by IFN-γ. J Clin Immunol. (2005) 25:41–9. doi: 10.1007/s10875-005-0356-5
72. Potikha T, Ella E, Cerliani JP, Mizrahi L, Pappo O, Rabinovich GA, et al. Galectin-1 is essential for efficient liver regeneration following hepatectomy. Oncotarget. (2016) 7:31738–54. doi: 10.18632/oncotarget.9194
73. Dong Z, Davidson D, Pérez-Quintero LA, Kurosaki T, Swat W, Veillette A. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. (2012) 36:974–85. doi: 10.1016/j.immuni.2012.03.023
74. Pérez-Quintero L-A, Roncagalli R, Guo H, Latour S, Davidson D, Veillette A. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J Exp Med. (2014) 211:727–42. doi: 10.1084/jem.20132038
75. Claus M, Urlaub D, Fasbender F, Watzl C. SLAM family receptors in natural killer cells - mediators of adhesion, activation and inhibition via cis and trans interactions. Clin Immunol. (2019) 204:37–42. doi: 10.1016/j.clim.2018.10.011
76. von Strandmann EP, Shatnyeva O, Hansen HP. NKp30 and its ligands: Emerging players in tumor immune evasion from natural killer cells. Ann Transl Med. (2015). 3:314. doi: 10.3978/j.issn.2305-5839.2015.09.08
77. Bottino C, Castriconi R, Pende D, Rivera P, Nanni M, Carnemolla B, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. (2003) 198:557–67. doi: 10.1084/jem.20030788
78. Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol. (2015) 15:243–54. doi: 10.1038/nri3799
79. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. (2009) 10:48–57. doi: 10.1038/ni.1674
80. Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A. (2009) 106:17858–63. doi: 10.1073/pnas.0903474106
81. Liu S, Zhang H, Li M, Hu D, Li C, Ge B, et al. Recruitment of Grb2 and SHIP1 by the ITT-like motif of TIGIT suppresses granule polarization and cytotoxicity of NK cells. Cell Death Differ. (2013) 20:456–64. doi: 10.1038/cdd.2012.141
82. Li M, Xia P, Du Y, Liu S, Huang G, Chen J, et al. T-cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon-γ production of natural killer cells via β-arrestin 2-mediated negative signaling. J Biol Chem. (2014) 289:17647–57. doi: 10.1074/jbc.M114.572420
83. Bi J, Zheng X, Chen Y, Wei H, Sun R, Tian Z. TIGIT safeguards liver regeneration through regulating natural killer cell-hepatocyte crosstalk. Hepatology. (2014) 60:1389–98. doi: 10.1002/hep.27245
84. Erickson BM, Thompson NL, Hixson DC. Tightly regulated induction of the adhesion molecule Necl-5/CD155 during rat liver regeneration and acute liver injury. Hepatology. (2006) 43:325–34. doi: 10.1002/hep.21021
85. Zhang Q, He Y, Luo N, Peng J, Ren X, Zhang Correspondence Z. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. (2019) 179:829–45. doi: 10.1016/j.cell.2019.10.003
86. de Andrade LF, Smyth MJ, Martinet L, Ferrari De Andrade L, Smyth MJ, Martinet L. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol Cell Biol. (2014) 92:237–44. doi: 10.1038/icb.2013.95
87. Jinushi M, Takehara T, Tatsumi T, Yamaguchi S, Sakamori R, Hiramatsu N, et al. Natural killer cell and hepatic cell interaction via NKG2A leads to dendritic cell-mediated induction of CD4+ CD25+ T cells with PD-1-dependent regulatory activities. Immunology. (2007) 120:73–82. doi: 10.1111/j.1365-2567.2006.02479.x
88. Lassen MG, Lukens JR, Dolina JS, Brown MG, Hahn YS. Intrahepatic IL-10 Maintains NKG2A + Ly49 – liver NK cells in a functionally hyporesponsive state. J Immunol. (2010) 184:2693–701. doi: 10.4049/jimmunol.0901362
89. Della Chiesa M, Vitale M, Carlomagno S, Ferlazzo G, Moretta L, Moretta A, et al. The natural killer cell-mediated killing of autologous dendritic cells is confined to a cell subset expressing CD94/NKG2A, but lacking inhibitory killer Ig-like receptors. Eur J Immunol. (2003) 33:1657–66. doi: 10.1002/eji.200323986
90. Aldemir H, Prod'homme V, Dumaurier M-J, Retiere C, Poupon G, Cazareth J, et al. Cutting Edge: Lectin-Like Transcript 1 Is a Ligand for the CD161 Receptor. J Immunol. (2005) 175:7791–5. doi: 10.4049/jimmunol.175.12.7791
91. Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting Edge: Lectin-Like Transcript-1 Is a Ligand for the Inhibitory Human NKR-P1A Receptor. J Immunol. (2005) 175:7796–9. doi: 10.4049/jimmunol.175.12.7796
92. Llibre A, Garner L, Partridge A, Freeman GJ, Klenerman P, Willberg CB. Expression of lectin-like transcript-1 in human tissues. F1000Res. (2016) 5:2929. doi: 10.12688/f1000research.10009.1
93. Jeffery HC, Braitch MK, Bagnall C, Hodson J, Jeffery LE, Wawman RE, et al. Changes in natural killer cells and exhausted memory regulatory T Cells with corticosteroid therapy in acute autoimmune hepatitis. Hepatol Commun. (2018) 2:421–36. doi: 10.1002/hep4.1163
94. Tessmer MS, Fugere C, Stevenaert F, Naidenko OV, Chong HJ, Leclercq G, et al. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int Immunol. (2007) 19:391–400. doi: 10.1093/intimm/dxm004
95. Smyth MJ, Cretney E, Takeda K, Wiltrout RH, Sedger LM, Kayagaki N, et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon γ-dependent natural killer cell protection from tumor metastasis. J Exp Med. (2001) 193:661–70. doi: 10.1084/jem.193.6.661
96. Takeda K, Hayakawa Y, Smyth MJ, Kayagaki N, Yamaguchi N, Kakuta S, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat Med. (2001) 7:94–100. doi: 10.1038/83416
97. Sato K, Hida S, Takayanagi H, Yokochi T, Kayagaki N, Takeda K, et al. Antiviral response by natural killer cells throughTRAIL gene induction by IFN-α/β. Eur J Immunol. (2001) 31:3138–46. doi: 10.1002/1521-4141(200111)31:11<3138::AID-IMMU3138>3.0.CO;2-B
98. Mobley C, Zarrinpar A. Molecular and cellular basis of liver failure. In: Busuttil RW, Klintmalm GBG, editors. Transplantation of the Liver. 3rd ed. Philadelphia, PA: Elsevier Inc. (2015). p. 40–57. doi: 10.1016/B978-1-4557-0268-8.00003-8
99. Sag D, Ayyildiz ZO, Gunalp S, Wingender G. The role of trail/drs in the modulation of immune cells and responses. Cancers. (2019) 11:1469. doi: 10.3390/cancers11101469
100. Prager I, Watzl C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J Leukoc Biol. (2019) 105:1319–29. doi: 10.1002/JLB.MR0718-269R
101. Wang S, Xia P, Shi L, Fan Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. (2012) 19:605–15. doi: 10.1038/cdd.2011.130
102. Baschuk N, Wang N, Watt SV, Halse H, House C, Bird PI, et al. NK cell intrinsic regulation of MIP-1α by granzyme M. Cell Death Dis. (2014) 5:e1115. doi: 10.1038/cddis.2014.74
103. Ng SS, De Labastida Rivera F, Yan J, Corvino D, Das I, Zhang P, et al. The NK cell granule protein NKG7 regulates cytotoxic granule exocytosis and inflammation. Nat Immunol. (2020) 21:1205–18. doi: 10.1038/s41590-020-0758-6
104. Fellows E, Gil-Parrado S, Jenne DE, Kurschus FC. Natural killer cell-derived human granzyme H induces an alternative, caspase-independent cell-death program. Blood. (2007) 110:544–52. doi: 10.1182/blood-2006-10-051649
105. Anderson DH, Sawaya MR, Cascio D, Ernst W, Modlin R, Krensky A, et al. Granulysin crystal structure and a structure-derived lytic mechanism. J Mol Biol. (2003) 325:355–65. doi: 10.1016/S0022-2836(02)01234-2
106. Lu C-C, Wu T-S, Hsu Y-J, Chang C-J, Lin C-S, Chia J-H, et al. NK cells kill mycobacteria directly by releasing perforin and granulysin. J Leukoc Biol. (2014) 96:1119–29. doi: 10.1189/jlb.4A0713-363RR
107. Al-Wasaby S, de Miguel D, Aporta A, Naval J, Conde B, Martínezlostao L, et al. In vivo potential of recombinant granulysin against human tumors. Oncoimmunology. (2015) 4:1–13. doi: 10.1080/2162402X.2015.1036213
108. Dorner BG, Dorner MB, Zhou X, Opitz C, Mora A, Güttler S, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. (2009) 31:823–33. doi: 10.1016/j.immuni.2009.08.027
109. Iyoda T, Shimoyama S, Liu K, Omatsu Y, Akiyama Y, Maeda Y, et al. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J Exp Med. (2002) 195:1289–302. doi: 10.1084/jem.20020161
110. Schulz O, Reis E, Sousa C. Cross-presentation of cell-associated antigens by CD8α+ dendritic cells is attributable to their ability to internalize dead cells. Immunology. (2002) 107:183–9. doi: 10.1046/j.1365-2567.2002.01513.x
111. Pooley JL, Heath WR, Shortman K. Cutting edge: intravenous soluble antigen is presented to CD4 T cells by CD8 – dendritic cells, but cross-presented to CD8 T cells by CD8 + dendritic cells. J Immunol. (2001) 166:5327–30. doi: 10.4049/jimmunol.166.9.5327
112. Den Haan JMM, Lehar SM, Bevan MJ. CD8+ but not CD8- dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. (2000) 192:1685–95. doi: 10.1084/jem.192.12.1685
113. Hartung E, Becker M, Bachem A, Reeg N, Jäkel A, Hutloff A, et al. Induction of potent CD8 T cell cytotoxicity by specific targeting of antigen to cross-presenting dendritic cells in vivo via murine or human XCR1. J Immunol. (2015) 194:1069–79. doi: 10.4049/jimmunol.1401903
114. Matsuo K, Kitahata K, Kawabata F, Kamei M, Hara Y, Takamura S, et al. A highly active form of XCL1/lymphotactin functions as an effective adjuvant to recruit cross-presenting dendritic cells for induction of effector and memory CD8+ T cells. Front Immunol. (2018) 9:2775. doi: 10.3389/fimmu.2018.02775
115. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. (2014) 32:659–702. doi: 10.1146/annurev-immunol-032713-120145
116. Rucker J, Samson M, Doranz BJ, Libert F, Berson JF, Yi Y, et al. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. (1996) 87:437–46. doi: 10.1016/S0092-8674(00)81364-1
117. Bluman EM, Bartynski KJ, Avalos BR, Caligiuri MA. Human natural killer cells produce abundant macrophage inflammatory protein-1a in response to monocyte-derived cytokines. J Clin Invest. (1996) 97:2722–7. doi: 10.1172/JCI118726
118. Heinrichs D, Berres ML, Nellen A, Fischer P, Scholten D, Trautwein C, et al. The chemokine CCL3 promotes experimental liver fibrosis in Mice. PLoS ONE. (2013) 8:66106. doi: 10.1371/journal.pone.0066106
119. Seki E, De Minicis S, Gwak GY, Kluwe J, Inokuchi S, Bursill CA, et al. CCR1 and CCR5 promote hepatic fibrosis in mice. J Clin Invest. (2009) 119:1858–70. doi: 10.1172/JCI37444
120. Santamaría E, Rodríguez-Ortigosa CM, Uriarte I, Latasa MU, Urtasun R, Alvarez-Sola G, et al. The epidermal growth factor receptor ligand amphiregulin protects from cholestatic liver injury and regulates bile acids synthesis. Hepatology. (2019) 69:1632–47. doi: 10.1002/hep.30348
121. Berasain C, García-Trevijano ER, Castillo J, Erroba E, Lee DC, Prieto J, et al. Amphiregulin: an early trigger of liver regeneration in mice. Gastroenterology. (2005) 128:424–32. doi: 10.1053/j.gastro.2004.11.006
122. Michalopoulos GK, Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology. (2005). 128:503–6. doi: 10.1053/j.gastro.2004.12.039
123. Berasain C, Avila MA. Amphiregulin. Semin Cell Dev Biol. (2014) 28:31–41. doi: 10.1016/j.semcdb.2014.01.005
124. Carver RS, Stevenson MC, Scheving LA, Russell WE. Diverse expression of ErbB receptor proteins during rat liver development and regeneration. Gastroenterology. (2002) 123:2017–27. doi: 10.1053/gast.2002.37060
125. Pardo-Saganta A, Latasa MU, Castillo J, Alvarez-Asiain L, Perugorría MJ, Sarobe P, et al. The epidermal growth factor receptor ligand amphiregulin is a negative regulator of hepatic acute-phase gene expression. J Hepatol. (2009) 51:1010–20. doi: 10.1016/j.jhep.2009.06.030
126. Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. (2015) 42:216–26. doi: 10.1016/j.immuni.2015.01.020
Keywords: liver, RNA-seq, liver-resident, intrahepatic, NK cell
Citation: Jameson G and Robinson MW (2021) Insights Into Human Intrahepatic NK Cell Function From Single Cell RNA Sequencing Datasets. Front. Immunol. 12:649311. doi: 10.3389/fimmu.2021.649311
Received: 04 January 2021; Accepted: 22 February 2021;
Published: 22 March 2021.
Edited by:
Silke Paust, The Scripps Research Institute, United StatesReviewed by:
Carsten Watzl, Leibniz Research Centre for Working Environment and Human Factors (IfADo), GermanyCopyright © 2021 Jameson and Robinson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark W. Robinson, bWFyay5yb2JpbnNvbkBtdS5pZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.