- 1Department of Laboratory Medicine, Affiliated People's Hospital, Jiangsu University, Zhenjiang, China
- 2Institute of Laboratory Medicine, Jiangsu Key Laboratory for Laboratory Medicine, Jiangsu University School of Medicine, Zhenjiang, China
- 3Department of General Surgery, Affiliated Hospital of Jiangsu University, Zhenjiang, China
Myeloid-derived suppressor cells (MDSCs) are immature heterogeneous cells derived from the bone marrow and they are the major component of the tumor-induced immunosuppressive environment. Tumor necrosis factor receptor-associated factor 6 (TRAF6), an E3 ubiquitin ligase, catalyzes the polyubiquitination of target proteins. TRAF6 plays a critical role in modulating the immune system. However, whether TRAF6 is involved in the regulation of MDSCs has not been thoroughly elucidated to date. In this study, we found that the expression of TRAF6 in MDSCs derived from tumor tissue was significantly upregulated compared with that of MDSCs from spleen of tumor-bearing mice. Knockdown of TRAF6 remarkably attenuated the immunosuppressive effects of MDSCs. Mechanistically, TRAF6 might improve the immunosuppression of MDSCs by mediating K63-linked polyubiquitination and phosphorylation of signal transducer and activator of transcription 3 (STAT3). Additionally, it was discovered that the accumulation of MDSCs was abnormal in peripheral blood of lung cancer patients. TRAF6 and arginase 1 were highly expressed in MDSCs of patients with lung cancer. Taken together, our study demonstrated that TRAF6 participates in promoting the immunosuppressive function of MDSCs and provided a potential target for antitumor immunotherapy.
Introduction
The tumor microenvironment provides a suitable environment for the survival and development of tumor cells. There are a large number of cytokines and chemokines in the tumor microenvironment that recruit immunosuppressive cells to accumulate in local tumors and promote tumor immune escape (1–3). Myeloid-derived suppressor cells (MDSCs) are crucial immunosuppressive cells in the tumor microenvironment, which have powerful suppressive effects (4–6). In mice, MDSCs are immature myeloid cells that co-express CD11b and Gr1, which are further divided into 2 subsets: CD11b+Ly6G+Ly6Clow polymorphonuclear MDSCs (PMN-MDSCs) and CD11b+Ly6G−Ly6Chi mononuclear MDSCs (M-MDSCs) according to the expression of LY6G and LY6C (7–10). However, human MDSCs have a more complex phenotype and are usually characterized by CD11b+CD33+HLA-DRlow/− expression (9, 11). In tumor-bearing mice, the proportion of MDSCs is significantly increased compared with that in wild type mice. MDSCs reach 20 to 40% in the spleen in tumor-bearing mice, and the massive aggregation of MDSCs in tumor tissue is particularly obvious (12). Studies have shown that the immunosuppressive function of MDSCs derived from tumor tissue was stronger than that of spleen-derived MDSCs from tumor-bearing mice (13–15).
Accumulating evidence has indicated that the suppressive function of MDSCs is correlated with the high production of arginase 1 (Arg1), inducible nitric oxide synthase (iNOS) and reactive oxygen species (ROS), which can inhibit T cell proliferation and antitumor responses (16–18). The two subsets of MDSCs exert immunosuppressive effects in different ways. Although both PMN-MDSCs and M-MDSCs highly express Arg1, the expression of iNOS and ROS are notably different (19, 20). Signal transducer and activator of transcription 3 (STAT3) is one of the key transcription factors regulating the expansion, activation and function of MDSCs (21–24). Abnormal activation of STAT3 signaling hinders the normal differentiation of myeloid cells, leading to the expansion and activation of MDSCs (25–27). STAT3 activation is mainly affected by posttranslational modifications. In addition to phosphorylation modification, ubiquitination also plays a critical role in modulating the activity of STAT3 (28, 29).
Tumor necrosis factor receptor-associated factor 6 (TRAF6) is an important member of the TRAF family (30). Unlike other TRAF members, TRAF6 is both an adaptor protein and E3 ubiquitin ligase (31–33). As a non-conventional E3 ubiquitin ligase, TRAF6 catalyzes the K63-linked polyubiquitination of target proteins further affecting functions. Studies have shown that the E3 ubiquitin ligase TRAF6 binds to STAT3 (28, 34). Moreover, TRAF6 promotes STAT3 phosphorylation by mediating the K63-linked polyubiquitination of STAT3 (29). However, whether TRAF6 affects the function of MDSCs by mediating STAT3 activity is unclear.
In this study, we explored the role of TRAF6 in modulating the immunosuppressive function of MDSCs and elaborated the potential molecular mechanism. Our research pointed out that the expression of TRAF6 is significantly increased in MDSCs derived from the tumor tissue of tumor-bearing mice. TRAF6 promoted the immunosuppressive function of MDSCs by mediating the activation of STAT3. These data elucidate a novel mechanism of the regulation of MDSCs in the tumor microenvironment and propose new ideas for therapeutic strategies targeting TRAF6.
Methods
Cell Line, Mice, and Tumor Models
Male C57BL/6 mice, aged 6–8 weeks, were purchased from the Laboratory Animal Center of Jiangsu University (Zhenjiang, China). Murine Lewis lung carcinoma (LLC) cells were obtained from the Cell Bank of Shanghai Institutes for Biological Sciences (Shanghai, China). The cells were cultured in DMEM (Gibco, Carlsbad, CA) with 10% fetal calf serum (Gibco, Carlsbad, CA) at 37°C in a humidified 5% CO2 atmosphere. 1 × 106 LLC cells were injected subcutaneously into the mice to establish tumor-bearing mouse model. All animal experiments were approved by the Committee on the Use of Live Animals in Research and Teaching of Jiangsu University.
Preparation of Single-Cell Suspensions
The murine spleen was ground and treated with ACK buffer. After centrifugation, PBE buffer was added to obtain a spleen cell suspension. In addition, the tumor tissue was stripped and cut into pieces, and then collagenase, hyaluronidase and DNase I (Sigma-Aldrich, St. Louis, MO) were used to digest the tissue in a water bath at 37°C for 2 h. The filtrate was collected through a 70 μm cell strainer. After centrifugation, PBE buffer was added to obtain single-cell suspensions.
Isolation of MDSCs and CD4+ T Cells
Murine Gr1+ CD11b+ MDSCs were isolated using a mouse MDSC kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. To improve the purity of MDSCs isolated from tumor tissues, enriched MDSCs were subsequently isolated using flow cytometry (FCM).
Simultaneously, murine CD4+ T cells were isolated from the spleens of wild-type C57BL/6 mice via using a mouse CD4+ T cell isolation kit (Miltenyi Biotec, Auburn, CA). The purity of MDSCs and CD4+ T cells were determined by FCM.
Flow Cytometry
Single-cell suspensions were stained with relevant fluorochrome-conjugated anti-mouse/human CD11b, anti-mouse Gr-1, Ly6G, and Ly6C antibodies (Biolegend, San Diego, CA) and anti-human-CD33 and HLA-DR antibodies (eBioscience, San Diego, CA). To examine cytotoxic T lymphocytes (CTLs) and T helper 1 (Th1) cells, single-cell suspensions derived from tumor tissues of tumor-bearing mice were treated with 1 μg/mL ionomycin, 2 ng/mL monensin (eBioscience, San Diego, CA), and 50 ng/mL PMA (Sigma-Aldrich, St. Louis, MO) for 5 h. After resuspending in PBS, the cells were stained with anti-mouse CD3e, anti-mouse CD8a or anti-mouse CD4 mAb (eBioscience, San Diego, CA). The cells were incubated at 4°C for 30 min, fixed, permeabilized, and stained with anti-mouse IFN-γ mAbs (BD Pharmingen™) according to the instructions in the intracellular cytokine staining kit (eBioscience, San Diego, CA). Flow cytometry (BD FACSCalibur) was used to determine the proportion of cells.
Quantitative Real-Time PCR (qRT-PCR)
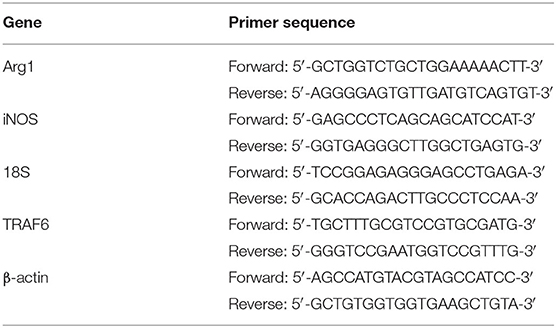
Total RNA was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA) and reverse-transcribed to cDNA with a PrimeScript RT reagent kit (Takara, Osaka, Japan) according to the manufacturer's instructions. Quantitative PCR was performed by using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara, Osaka, Japan). The primer sequences are listed in Table 1.
Immunoprecipitation and Western Blotting
MDSCs were lysed by immunoprecipitation cell lysis buffer. After centrifugation, the supernatant was collected and incubated with anti-STAT3 Abs (Santa Cruz Biotechnology, Santa Cruz, CA) or IgG Abs (Cell Signaling Technology, Beverly, MA) for 30 min at 4°C. Then, Protein A/G plus-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added and blended on a shaker overnight at 4°C. After being washed 3 times, the lysates were boiled in SDS-PAGE protein loading buffer. TRAF6 and STAT3 protein were detected by Western blotting.
Proteins extracted from cells were denatured and subsequently separated by SDS-PAGE. Then, the proteins were transferred to PVDF membranes (Bio-Rad, Hercules, CA), and the membranes were incubated in primary antibodies overnight at 4°C, followed by incubation with HRP-conjugated secondary antibodies. Detection was performed by using LAS4000 chemiluminescence gel imaging and analysis system (Champion Chemical, Whittier, CA). Rabbit anti-TRAF6 mAb, rabbit anti-ubiquitin (linkage-specific K63) mAb, and HRP-conjugated goat anti-rat IgG Ab were purchased from Abcam (Cambridge, UK). Rabbit anti–p-STAT3 (Y705) mAb and rat anti-β-actin mAb were obtained from Cell Signaling Technology (Beverly, MA).
Transfection
Tumor tissue-derived MDSCs were cultured in 24-well plates with RPMI 1640 medium containing 10% FBS. MDSCs were transfected with TRAF6 siRNA or negative control siRNA using Lipofectamine ™ 2,000 transfection reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The siRNA transfection efficiency was determined by qRT-PCR and Western blotting. TRAF6 siRNA and negative control siRNA were purchased from RiboBio (Guangzhou, China).
Assessment of MDSCs Suppressive Activity
In order to determine the immunosuppressive function of MDSCs, the sorted MDSCs were co-cultured with CFSE-labeled splenic CD4+ T cells in the study. Splenic CD4+ T cells were stained with fluorescent dye CFSE (5 μM, Invitrogen) for 10 min at 37°C protected from light. RPMI 1640 medium (Gibco, Carlsbad, CA) containing 10% fetal calf serum (Gibco, Carlsbad, CA) was added to wash the cell pellets for 3 times. Sorted MDSCs were co-cultured with CFSE-stained CD4+ T cells at a ratio of 1:1 in 96-well round-bottomed plates (Costar, Corning, NY) in the presence of anti-CD3 mAbs and anti-CD28 mAbs (Biolegend, San Diego, CA). The cells were incubated in RPMI 1640 medium supplemented with 10% fetal calf serum at 37°C in a humidified 5% CO2 atmosphere for 72 h protected from light. The proliferation of CD4+ T cells was detected by flow cytometry at 488 nm excitation light to determinate the suppressive activity of MDSCs.
Measurement of Arginase 1 Activity and NO Content
MDSCs were lysed with an appropriate amount of RIPA buffer for 30 min, and the lysate supernatant was collected after centrifugation. A QuantiChrom arginase assay kit (BioAssay systems, Hayward, CA) was used to determine the Arg1 activity in the lysate supernatant.
The content of NO was measured by the Griess reagent system kit (Promega, Madison, WI) according to the manufacturer's instructions.
In vivo Experiments
To study the effect of TRAF6 on the immunosuppressive function of MDSCs in vivo, C57BL/6 mice were divided into siRNA control group and siTRAF6-MDSC group. The mice in the siRNA control group were subcutaneously injected with 1 × 106 MDSCs transfected with siNC mixed with 1 × 106 LLC cells, and the mice in the siTRAF6-MDSC group were subcutaneously injected with 1 × 106 MDSCs transfected with siTRAF6 mixed with 1 × 106 LLC cells. We constantly monitored the length and width of the tumor, and the tumor volume was calculated using the formula V = 1/2 × a2 × b (“a” represents the width and “b” represents the length). The mice were sacrificed on the 28th day after inoculation with LLC cells and MDSCs. The proportions of Th1 cells and CTLs from tumor tissue were analyzed by FCM.
Patients and Sampling
Fresh peripheral blood samples from 33 patients with lung cancer and healthy controls were collected at the Affiliated People's Hospital, Jiangsu University. The peripheral blood mononuclear cells (PBMCs) were isolated by density-gradient centrifugation via using Ficoll-Hypaque solution (Haoyang Biological Technology Co.) and subsequently analyzed by flow cytometry. The protocol was approved by the Ethics Committee of the Affiliated People's Hospital of Jiangsu University (Zhenjiang, China). Written informed consent was obtained from all patients before study enrollment.
Statistical Analysis
The experimental data are expressed as the mean ± SD. Student's t-test and ANOVA were used to determine significant differences. Correlations were determined by the Spearman correlation coefficient. Differences were considered significant at a p < 0.05.
Results
TRAF6 Is Highly Expressed in MDSCs Derived From Tumor Tissue of Tumor-Bearing Mice
To investigate whether TRAF6 is involved in regulating MDSCs, we evaluated the expression of TRAF6 in MDSCs. The purity of MDSCs isolated from the spleen or tumor tissue of mice was >90% (Figure 1A). Compared with that of MDSCs from the spleens of wild-type (WT) mice or tumor-bearing (TB) mice, the expression of TRAF6 in MDSCs from tumor tissue was significantly increased (Figures 1B,C). The expression of TRAF6 in PMN-MDSCs and M-MDSCs were determined by qRT-PCR, and the results showed that there was no difference in the mRNA expression of TRAF6 in the two subgroups of MDSCs (Figures 1D,E).
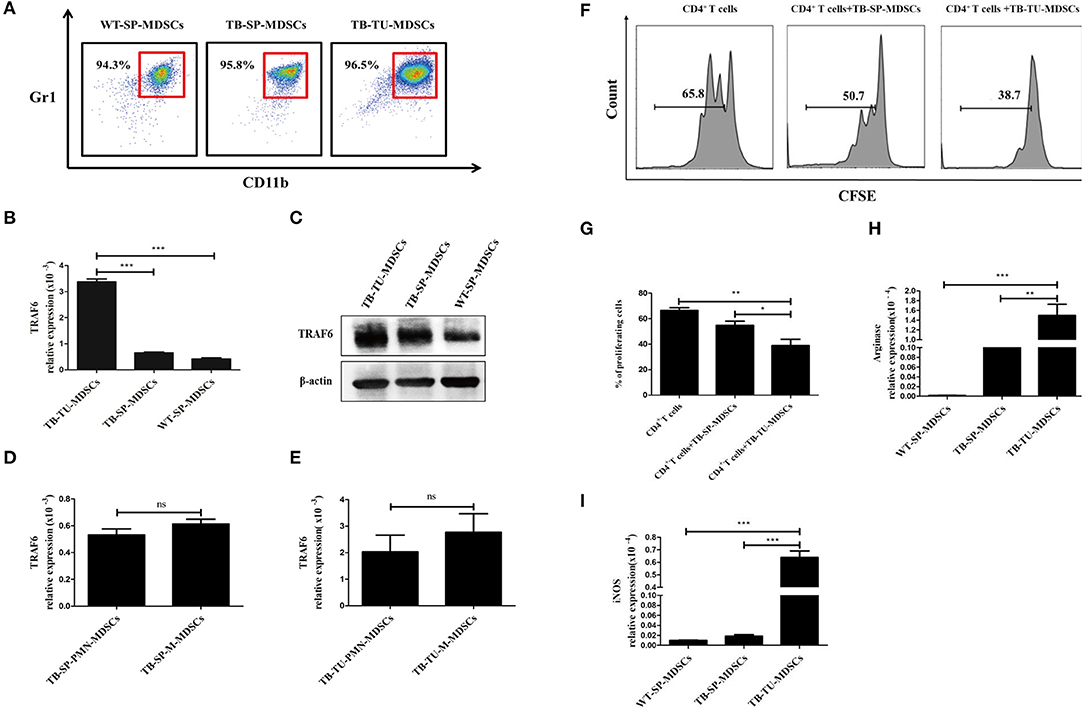
Figure 1. TRAF6 is highly expressed in MDSCs derived from the tumor tissue of tumor-bearing mice. Approximately 1 × 106 LLC cells were s.c. injected in the backs of C57BL/6 mice for 28 d to establish a tumor-bearing (TB) mouse model. MDSCs were isolated by immunomagnetic beads from the spleens of TB mice, the tumor tissue of TB mice or the spleens of wild-type (WT) mice. (A) The purity of the isolated MDSCs was determined using flow cytometry via the detection of the CD11b+Gr1+ phenotype. The expression of TRAF6 in MDSCs derived from different sources was determined by qRT-PCR (B) or Western blotting (C). The mRNA expression of TRAF6 in PMN-MDSCs and M-MDSCs derived from the spleen (D) or tumor tissue (E). (F) CFSE-labeled CD4+ T cells were co-cultured with MDSCs derived from the spleen or tumor tissue in the presence of CD3 and CD28 stimulation. After 72 h, the proliferation of CD4+ T cells was tested via flow cytometry. (G) Statistical analyses of the percentage of proliferating CD4+ T cells co-cultured with MDSCs derived from the spleen or tumor tissue of TB mice. The mRNA expression levels of Arg1 (H) and iNOS (I) in MDSCs were measured by qRT-PCR. ***p < 0.001, **p < 0.01, *p < 0.05; ns, no significance; TB-TU-MDSCs, MDSCs derived from the tumor tissue of tumor-bearing mice; TB-SP-MDSCs, MDSCs derived from the spleens of tumor-bearing mice; WT-SP-MDSCs, MDSCs derived from the spleens of wild-type mice.
In addition, we analyzed the suppressive function of tumor tissue-derived MDSCs. Compared with MDSCs from spleen of TB mice, tumor tissue-derived MDSCs had stronger suppressive effects on the proliferation of CD4+ T cells (Figures 1F,G). Moreover, the expressions of Arg1 and iNOS in tumor tissue-derived MDSCs were higher than that in splenic MDSCs (Figures 1H,I). These results indicated that the immunosuppressive function of MDSCs derived from tumor tissue was stronger than those of splenic MDSCs from TB mice, which was consistent with previous reports (1).
TRAF6 Knockdown Impairs the Immunosuppressive Activity of MDSCs in vitro
To evaluate whether TRAF6 is involved in the suppressive effects of MDSCs, specific siRNA was used to knock down TRAF6 in MDSCs. After treatment with TRAF6-specific siRNA, the expression of TRAF6 in MDSCs from tumor tissue was effectively decreased (Figures 2A,B). Remarkably, the inhibitory effect of MDSCs transfected with siTRAF6 on CD4+ T cell proliferation was significantly attenuated (Figures 2C,D). Consistently, knockdown of TRAF6 distinctly decreased the activity of Arg1 in MDSCs (Figure 2E), although the content of NO did not noticeably altered (Figure 2F). These data indicate that knockdown of TRAF6 impairs the immunosuppressive effects of MDSCs in vitro.
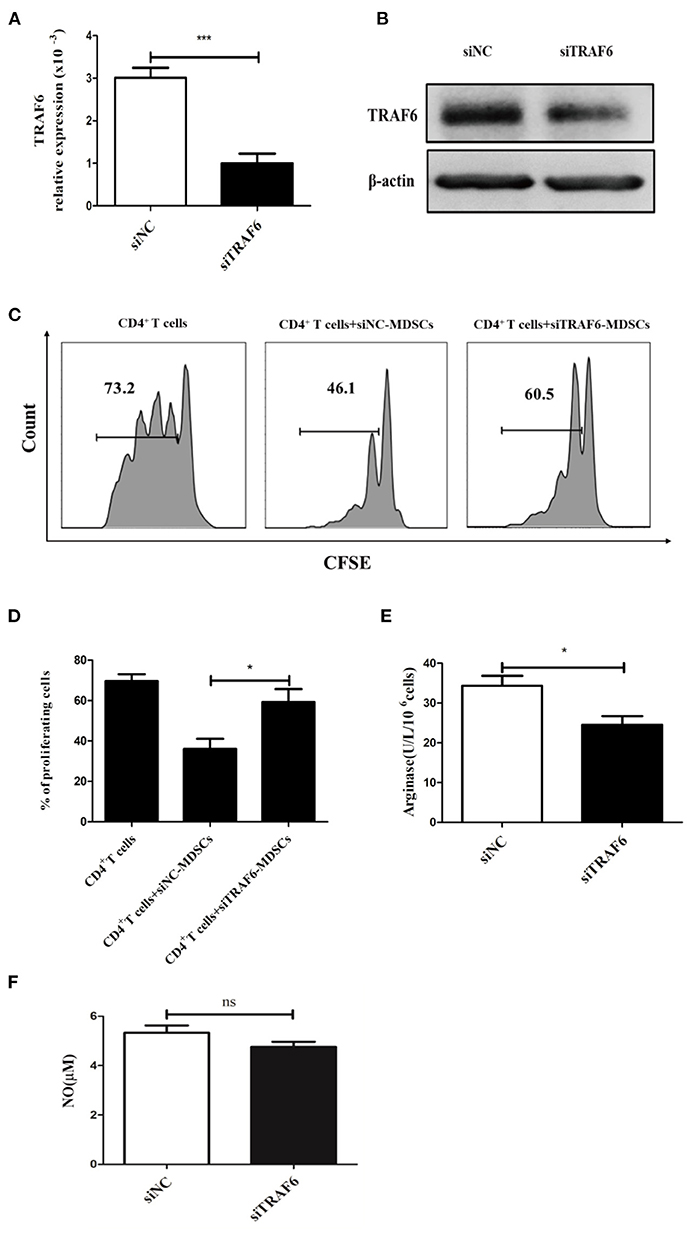
Figure 2. TRAF6 knockdown impairs the immunosuppressive effects of MDSCs in vitro. Specific siRNA (siTRAF6) was used to knockdown the expression of TRAF6 in MDSCs, and the efficiency of siTRAF6 knockdown was validated by qRT-PCR (A) and Western blotting (B). (C) Tumor-derived MDSCs were transfected with siTRAF6 and cocultured with CFSE-labeled CD4+ T cells, and proliferation was measured by flow cytometry after 72 h. (D) Statistical analyses of the percentage of proliferating CD4+ T cells co-cultured with MDSCs transfected with siTRAF6. After TRAF6 knockdown, the activity of Arg1 was measured by a QuantiChrom arginase assay kit (E), and the concentration of NO was determined via a Griess reagent system kit (F). ***p < 0.001, *p < 0.05; ns, no significance.
TRAF6 Alters the Activity of STAT3
Next, we explored the potential molecular mechanism by which TRAF6 regulates MDSC function. It is widely known that the transcription factor STAT3 plays a dominant role in MDSC expansion, activation and function. Recent studies have revealed that the E3 ubiquitin ligase TRAF6 mediates the K63-linked polyubiquitination of STAT3 but has no effect on STAT3 degradation (34). To determine whether TRAF6 impacts the function of MDSCs by STAT3, we conducted the interaction between TRAF6 and STAT3 in MDSCs by co-immunoprecipitation (Co-IP) assays. The results of Co-IP showed that TRAF6 was co-immunoprecipitated with STAT3, suggesting that endogenous TRAF6 binds to endogenous STAT3 in MDSCs (Figure 3A). Furthermore, we examined the regulation of TRAF6 on the posttranslational modification of STAT3. After knockdown of TRAF6 in MDSCs, the K63-linked polyubiquitination of STAT3 was significantly downregulated (Figure 3B). Moreover, silencing of TRAF6 remarkably decreased the levels of phosphorylated STAT3 in MDSCs from tumor tissue of TB mice (Figure 3C).
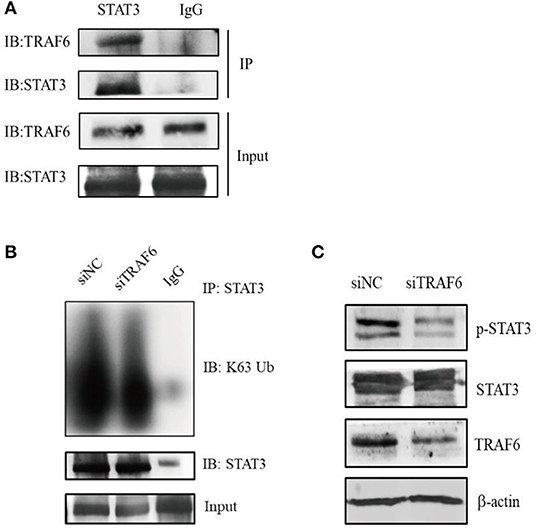
Figure 3. TRAF6 alters the activity of STAT3 by mediating the K63-linked polyubiquitination of STAT3 in MDSCs. (A) The interaction between TRAF6 and STAT3 in tumor-derived MDSCs was determined by co-immunoprecipitation (Co-IP) assays. (B) After knockdown of TRAF6 in MDSCs, the level of STAT3 K63-linked polyubiquitination was measured by Co-IP assays. (C) After knockdown of TRAF6 in MDSCs, the level of STAT3 phosphorylation was assessed by Western blotting.
TRAF6 Knockdown Attenuates the Ability of MDSCs to Accelerate Tumor Progression in Tumor-Bearing Mice
As shown in the in vitro experiments, knockdown of TRAF6 impaired the immunosuppressive activity of MDSCs. Thus, we further investigated the effects of TRAF6 on the suppressive effects of MDSCs in vivo. 1 × 106 Lewis lung carcinoma cells and 1 × 106 MDSCs transfected with TRAF6 siRNA or negative control siRNA were subcutaneously injected into C57BL/6 mice. As shown in Figure 4A, the tumor growth of mice injected with siTRAF6-transfected MDSCs (siTRAF6) group was evidently delayed. Furthermore, the tumor volume and weight were significantly less than those of the control (siNC) group (Figures 4B,C). Given that MDSCs mainly suppress the function of T cells in the tumor microenvironment, we measured the proportion of CD4+ Th1 cells and CD8+ CTLs from tumor tissue of TB mice. Compared with that of the control group, the proportion of Th1 cells was no significant difference in the siTRAF6 group (Figure 4D), while the proportion of CTLs in the siTRAF6 group was significantly increased (Figure 4E). Our data showed that knockdown of TRAF6 attenuated the ability of MDSCs to accelerate tumor progression and partly suppressed the antitumor T cell response.
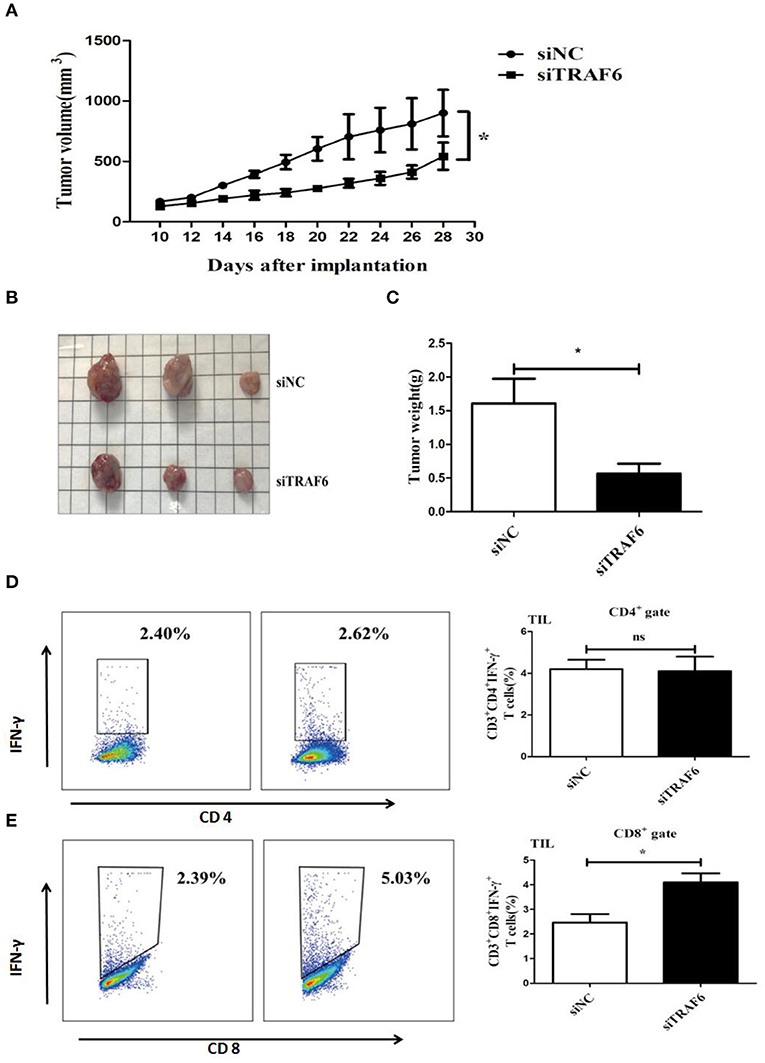
Figure 4. TRAF6 knockdown attenuates the ability of MDSCs to accelerate tumor progression in tumor-bearing mice. To investigate the effects of TRAF6 on the suppressive activity of MDSCs in vivo, 2 groups of wild-type C57BL/6 mice were s.c. injected with 1 × 106 LLC cells and 1 × 106 MDSCs transfected with siTRAF6 (siTRAF6 group) or MDSCs transfected with siNC (control group). (A) Tumor growth was constantly monitored. The width “a” and length “b” were measured, and tumor volume was calculated. (B,C) On the 28th day after the inoculation of LLC cells, the mice were sacrificed, and the tumor image and weights were showed in both groups. (D) The proportion of CD4+IFN-γ+ Th1 cells in the tumor tissue of both groups was analyzed by FCM. (E) The proportion of CD8+IFN-γ+ CTLs in the tumor tissue of both groups was analyzed by FCM. *p < 0.05; ns, no significance.
TRAF6 Expression Was Augmented in MDSCs From the PBMCs of Lung Cancer Patients
Considering that TRAF6 improves the function of MDSCs in mice, we examined whether TRAF6 had similar characteristics in MDSCs from lung cancer patients. We performed flow cytometry to analyze the proportion of CD11b+CD33+HLA-DR− MDSCs in PBMCs from lung cancer patients. The percentage of MDSCs in PBMCs from lung cancer patients was higher than that from healthy controls, which indicated that the abnormal accumulation of MDSCs in peripheral blood was correlated with lung cancer (Figure 5A). We further examined the expression of TRAF6 in human MDSCs. Compared with that of healthy control, the expression of TRAF6 was markedly augmented in MDSCs from PBMCs of lung cancer patients (Figure 5B). In addition, Arg1 is the important immunosuppressive molecule in MDSCs, which was also upregulated in MDSCs from PBMCs of patients (Figure 5C). Furthermore, there was a positive correlation between the expression levels of TRAF6 and Arg1 (Figure 5D), indicating that TRAF6 may be participate in the regulation of Arg1 expression in MDSCs.
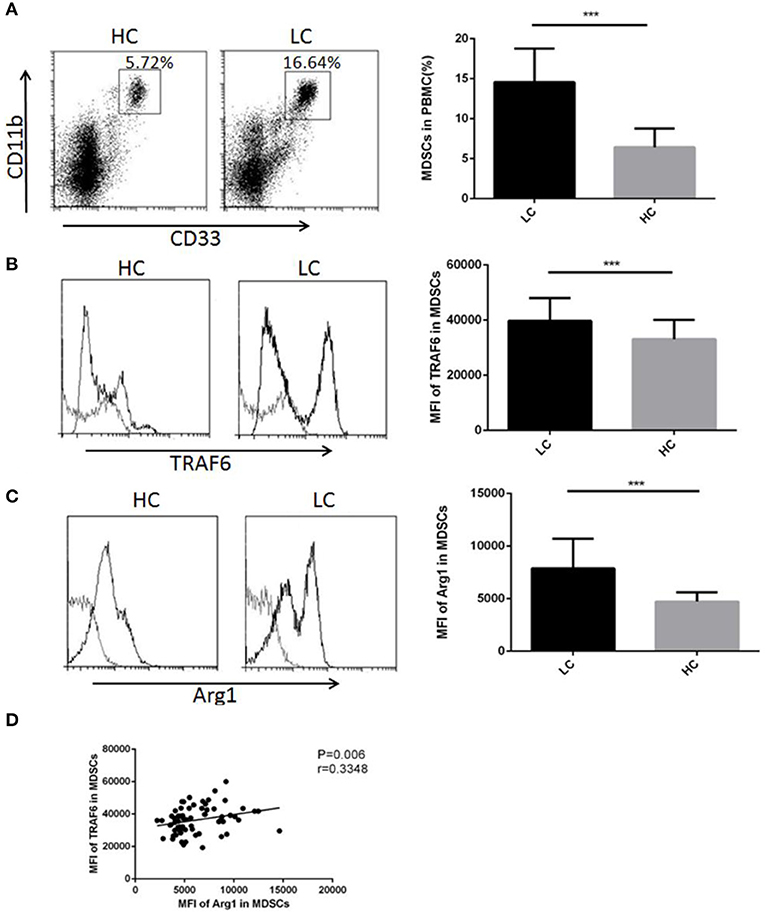
Figure 5. TRAF6 expression was augmented in MDSCs from lung cancer patients. To examine the modulation of TRAF6 in MDSCs from lung cancer patients, the level of TRAF6 in MDSCs was measured in the lung cancer patient group (LC) and healthy control group (HC). (A) The proportions of MDSCs in the PBMCs of lung cancer patients and healthy persons were analyzed by flow cytometry. Representative dot plots of CD11b+CD33+HLA-DR− MDSCs in the blood of patients with LC and healthy controls are shown. (B) The mean fluorescence intensity (MFI) of TRAF6 in MDSCs was determined by flow cytometry. (C) The MFI of arginase-1 in MDSCs was determined by flow cytometry. (D) The correlation between TRAF6 and arginase-1 in MDSCs was analyzed. ***p < 0.001.
Discussion
As an adaptor protein and E3 ubiquitin ligase, TRAF6 is involved in mediating various cellular signaling pathways and regulating a series of physiological process (35–40). In addition, the oncogenic role of TRAF6 in tumors has been widely reported. TRAF6 is abnormally highly expressed in multiple tumor tissues and modulates the malignant behavior of tumor cells (41–46). Furthermore, mounting evidence has revealed that TRAF6 plays a critical role in the development and activation of lymphocytes and myeloid cells (42, 47, 48). However, whether TRAF6 regulates MDSCs which are pivotal immunosuppressive cells has not been reported to date.
To shed light on the potential effect of TRAF6 on MDSC functions, we constructed murine Lewis lung carcinoma models. Our research suggested that the expression of TRAF6 was higher in MDSCs from tumor tissue than that in MDSCs from spleen. Consistent with previous research, we noted that the suppressive function of MDSCs derived from tumor tissue was stronger than that of spleen-derived MDSCs. Thus, we hypothesized that the strong suppressive activity of MDSCs from tumor tissues may be attributed to the high expression of TRAF6. After TRAF6 knockdown, the inhibitory effect of MDSCs on CD4+ T cell proliferation was significantly decreased, with reduced Arg1 activity. Moreover, knockdown of TRAF6 attenuated the ability of MDSCs to accelerate tumor progression in tumor-bearing mice. Many clinical studies have demonstrated that increased levels of both circulating and tumor-infiltrating MDSCs were associated with poor prognosis in cancer patients. We further confirmed that the percentage of MDSCs was markedly increased in lung cancer patients. In addition, the expression of TRAF6 was increased in MDSCs from lung cancer patients, which was positively correlated with the level of Arg1 in MDSCs. These results highlighted the key role of TRAF6 in enhancing the function of MDSCs in vitro and in vivo.
STAT3 is one of the most important transcription factors regulating the expansion, activation and function of MDSCs. STAT3 is abnormally activated in MDSCs in the tumor microenvironment (23, 24, 49, 50). In addition to phosphorylation, ubiquitination is also involved in STAT3 activation. In our study, we demonstrated that TRAF6 binds to STAT3 in MDSCs. TRAF6 knockdown markedly reduced the K63-linked polyubiquitination and phosphorylation of STAT3 in MDSCs, indicating that TRAF6 elevated the suppressive function of MDSCs by interacting with STAT3. Indeed, it has been reported that TRAF6 mediates K63 ubiquitination via the SH2 domain of STAT3, which is an essential step for STAT3 phosphorylation in response to bacterial infections (29). Thus we assumed that TRAF6 fosters the suppressive effect of MDSCs by inducing STAT3 K63 ubiquitination and subsequent STAT3 phosphorylation. However, further studies are required to reveal the detailed molecular mechanism by which TRAF6 affects the function of MDSCs via STAT3.
Conclusions
In summary, we demonstrate that TRAF6 regulates the immunosuppressive activity of MDSCs by modulating the K63-linked polyubiquitination and phosphorylation of STAT3. Targeting of TRAF6 might be a potential clinical therapeutic strategy for enhancing antitumor immune response.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the Affiliated People's Hospital of Jiangsu University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Committee on the Use of Live Animals in Research and Teaching of Jiangsu University. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
GS performed the experiments, analyzed the data, and wrote the paper. YZ analyzed the data, and wrote the paper. JT and JM analyzed the data. KY and HX revised the paper. SW designed the study and wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the Jiangsu Province's Key Medical Talents Program (Grant No. ZDRCB2016018), Research Project of the Jiangsu Commission of Health (Grant No. K2019019), Natural Science Foundation of Jiangsu (Grant No. BK20190242), and Science and Technology Support Program (Social Development) of Zhenjiang (Grant No. SH 2020041).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
Arg1, Arginase 1; CTL, cytotoxic T lymphocyte; FCM, flow cytometry; iNOS, inducible nitric oxide synthase; IP, immunoprecipitation; MDSCs, myeloid-derived suppressor cells; M-MDSCs, monocytic myeloid-derived suppressor cells; NO, nitric oxide; PMN-MDSCs, polymorphonuclear myeloid-derived suppressor cells; ROS, reactive oxygen species; STAT3, signal transducer and activator of transcription 3; TRAF6, tumor necrosis factor receptor-associated factor 6; Ub, ubiquitin.
References
1. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. (2016) 37:208–20. doi: 10.1016/j.it.2016.01.004
2. Li M, Zha X, Wang S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim Biophys Acta Rev Cancer. (2021). doi: 10.1016/j.bbcan.2021.188522. [Epub ahead of print].
3. Xie F, Xu M, Lu J, Mao L, Wang S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer. (2019) 18:146. doi: 10.1186/s12943-019-1074-3
4. Ostrand-Rosenberg S, Fenselau C. Myeloid-Derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J Immunol. (2018) 200:422–31. doi: 10.4049/jimmunol.1701019
5. Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, et al. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. (2019) 18:61. doi: 10.1186/s12943-019-0978-2
6. Tian X, Zheng Y, Yin K, Ma J, Tian J, Zhang Y, et al. LncRNA AK036396 inhibits maturation and accelerates immunosuppression of polymorphonuclear myeloid-derived suppressor cells by enhancing the stability of ficolin B. Cancer Immunol Res. (2020) 8:565–77. doi: 10.1158/2326-6066.CIR-19-0595
7. Bosiljcic M, Cederberg RA, Hamilton MJ, LePard NE, Harbourne BT, Collier JL, et al. Targeting myeloid-derived suppressor cells in combination with primary mammary tumor resection reduces metastatic growth in the lungs. Breast Cancer Res. (2019) 21:103. doi: 10.1186/s13058-019-1189-x
8. Tian J, Rui K, Hong Y, Wang X, Xiao F, Lin X, et al. Increased GITRL impairs the function of myeloid-derived suppressor cells and exacerbates primary sjögren's syndrome. J Immunol. (2019) 202:1693–703. doi: 10.4049/jimmunol.1801051
9. Zhou J, Liu M, Sun H, Feng Y, Xu L, Chan AWH, et al. Hepatoma-intrinsic CCRK inhibition diminishes myeloid-derived suppressor cell immunosuppression and enhances immune-checkpoint blockade efficacy. Gut. (2018) 67:931–44. doi: 10.1136/gutjnl-2017-314032
10. Tam JW, Kullas AL, Mena P, Bliska JB, van der Velden AW. CD11b+ Ly6Chi Ly6G- immature myeloid cells recruited in response to Salmonella enterica Serovar typhimurium infection exhibit protective and immunosuppressive properties. Infect Immun. (2014) 82:2606–14. doi: 10.1128/IAI.01590-13
11. Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, et al. Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol. (2018) 9:473. doi: 10.3389/fimmu.2018.00473
12. Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. (2008) 111:4233–44. doi: 10.1182/blood-2007-07-099226
13. Sumida K, Wakita D, Narita Y, Masuko K, Terada S, Watanabe K, et al. Anti-IL-6 receptor mAb eliminates myeloid-derived suppressor cells and inhibits tumor growth by enhancing T-cell responses. Eur J Immunol. (2012) 42:2060–72. doi: 10.1002/eji.201142335
14. Nefedova Y, Nagaraj S, Rosenbauer A, Muro-Cacho C, Sebti SM, Gabrilovich DI. Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res. (2005) 65:9525–35. doi: 10.1158/0008-5472.CAN-05-0529
15. Noman MZ, Desantis G, Janji B, Hasmim M, Karray S, Dessen P, et al. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J Exp Med. (2014) 211:781–90. doi: 10.1084/jem.20131916
16. Fleet JC, Burcham GN, Calvert RD, Elzey BD, Ratliff TL. 1α, 25 Dihydroxyvitamin D (1,25(OH)(2)D) inhibits the T cell suppressive function of myeloid derived suppressor cells (MDSC). J Steroid Biochem Mol Biol. (2020) 198:105557. doi: 10.1016/j.jsbmb.2019.105557
17. Zhu D, Tian J, Wu X, Li M, Tang X, Rui K, et al. G-MDSC-derived exosomes attenuate collagen-induced arthritis by impairing Th1 and Th17 cell responses. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:165540. doi: 10.1016/j.bbadis.2019.165540
18. Si Y, Merz SF, Jansen P, Wang B, Bruderek K, Altenhoff P, et al. Multidimensional imaging provides evidence for down-regulation of T cell effector function by MDSC in human cancer tissue. Sci Immunol. (2019) 4:eaaw9159. doi: 10.1126/sciimmunol.aaw9159
19. Koehn BH, Apostolova P, Haverkamp JM, Miller JS, McCullar V, Tolar J, et al. GVHD-associated, inflammasome-mediated loss of function in adoptively transferred myeloid-derived suppressor cells. Blood. (2015) 126:1621–28. doi: 10.1182/blood-2015-03-634691
20. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. (2012) 12:253–68. doi: 10.1038/nri3175
21. Liu YF, Zhuang KH, Chen B, Li PW, Zhou X, Jiang H, et al. Expansion and activation of monocytic-myeloid-derived suppressor cell via STAT3/arginase-I signaling in patients with ankylosing spondylitis. Arthritis Res Ther. (2018) 20:168. doi: 10.1186/s13075-018-1654-4
22. Namkoong H, Ishii M, Fujii H, Yagi K, Asami T, Asakura T, et al. Clarithromycin expands CD11b+Gr-1+ cells via the STAT3/Bv8 axis to ameliorate lethal endotoxic shock and post-influenza bacterial pneumonia. PLoS Pathog. (2018) 14:e1006955. doi: 10.1371/journal.ppat.1006955
23. Hellsten R, Lilljebjörn L, Johansson M, Leandersson K, Bjartell A. The STAT3 inhibitor galiellalactone inhibits the generation of MDSC-like monocytes by prostate cancer cells and decreases immunosuppressive and tumorigenic factors. Prostate. (2019) 79:1611–21. doi: 10.1002/pros.23885
24. Trovato R, Fiore A, Sartori S, Canè S, Giugno R, Cascione L, et al. Immunosuppression by monocytic myeloid-derived suppressor cells in patients with pancreatic ductal carcinoma is orchestrated by STAT3. J Immunother Cancer. (2019) 7:255. doi: 10.1186/s40425-019-0734-6
25. Jiang M, Chen J, Zhang W, Zhang R, Ye Y, Liu P, et al. Interleukin-6 trans-signaling pathway promotes immunosuppressive myeloid-derived suppressor cells suppression of suppressor of cytokine signaling 3 in breast cancer. Front Immunol. (2017) 8:1840. doi: 10.3389/fimmu.2017.01840
26. Mao F, Zhao Y, Lv Y, Teng Y, Kong H, Liu Y, et al. CD45CD33CD11b myeloid-derived suppressor cells suppress CD8 T cell activity via the IL-6/IL-8-arginase I axis in human gastric cancer. Cell Death Dis. (2018) 9:763. doi: 10.1038/s41419-018-0803-7
27. Guha P, Gardell J, Darpolor J, Cunetta M, Lima M, Miller G, et al. STAT3 inhibition induces Bax-dependent apoptosis in liver tumor myeloid-derived suppressor cells. Oncogene. (2019) 38:533–48. doi: 10.1038/s41388-018-0449-z
28. Xin J, Zhang Z, Su X, Wang L, Zhang Y, Yang R. Epigenetic component p66a modulates myeloid-derived suppressor cells by modifying STAT3. J Immunol. (2017) 198:2712–20. doi: 10.4049/jimmunol.1601712
29. Ruan H, Zhang Z, Wang S, Nickels LM, Tian L, Qiao J, et al. Tumor necrosis factor receptor-associated factor 6 (TRAF6) mediates ubiquitination-dependent STAT3 activation upon Salmonella enterica Serovar typhimurium infection. Infect Immun. (2017) 85(8). doi: 10.1128/IAI.00081-17
30. Dou Y, Tian X, Zhang J, Wang Z, Chen G. Roles of TRAF6 in central nervous system. Curr Neuropharmacol. (2018) 16:1306–13. doi: 10.2174/1570159X16666180412094655
31. Chung J, Lu M, Yin Q, Lin S, Wu H. Molecular basis for the unique specificity of TRAF6. Adv Exp Med Biol. (2007) 597:122–30. doi: 10.1007/978-0-387-70630-6_10
32. Inoue J, Gohda J, Akiyama T. Characteristics and biological functions of TRAF6. Adv Exp Med Biol. (2007) 597:72–9. doi: 10.1007/978-0-387-70630-6_6
33. Mukhopadhyay D, Sangaré LO, Braun L, Hakimi MA, Saeij JP. Toxoplasma GRA15 limits parasite growth in IFNγ-activated fibroblasts through TRAF ubiquitin ligases. EMBO J. (2020) 39:e103758. doi: 10.15252/embj.2019103758
34. Wei J, Yuan Y, Jin C, Chen H, Leng L, He F, et al. The ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination. PLoS ONE. (2012) 7:e49567. doi: 10.1371/journal.pone.0049567
35. Biswas R, Bagchi A. Structural Characterization of the trimerization of TRAF6 protein through molecular dynamics simulations. Interdiscip Sci. (2019) 11:428–36. doi: 10.1007/s12539-017-0259-y
36. Xie C, Zhang LZ, Chen ZL, Zhong WJ, Fang JH, Zhu Y, et al. A hMTR4-PDIA3P1-miR-125/124-TRAF6 regulatory axis and its function in NF kappa B signaling and chemoresistance. Hepatology. (2020) 71:1660–77. doi: 10.1002/hep.30931
37. Wu H, Lu XX, Wang JR, Yang TY, Li XM, He XS, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. (2019) 15:1506–22. doi: 10.1080/15548627.2019.1586250
38. Khusbu FY, Zhou X, Roy M, Chen FZ, Cao Q, Chen HC. Resveratrol induces depletion of TRAF6 and suppresses prostate cancer cell proliferation and migration. Int J Biochem Cell Biol. (2020) 118:105644. doi: 10.1016/j.biocel.2019.105644
39. Peng SJ, Yao RR, Yu SS, Chen HY, Pang X, Zhang Y, et al. UBL4A augments innate immunity by promoting the K63-linked ubiquitination of TRAF6. J Immunol. (2019) 203:1943–1951. doi: 10.4049/jimmunol.1800750
40. Cao C, An R, Yu Y, Dai H, Qu Z, Gao M, et al. BICP0 negatively regulates TRAF6-mediated NF-κB and interferon activation by promoting K48-linked polyubiquitination of TRAF6. Front Microbiol. (2019) 10:3040. doi: 10.3389/fmicb.2019.03040
41. Wu H, Yang TY, Li Y, Ye WL, Liu F, He XS, et al. Tumor necrosis factor receptor-associated factor 6 promotes hepatocarcinogenesis by interacting with histone deacetylase 3 to enhance c-Myc gene expression and protein stability. Hepatology. (2020) 71:148–63. doi: 10.1002/hep.30801
42. Wu X, Xue R, Peng H, Gan X, Lu X, Yan W, et al. Traf6 inhibitor boosts antitumor immunity by impeding regulatory T cell migration in Hepa1-6 tumor model. Int Immunopharmacol. (2019) 77:105965. doi: 10.1016/j.intimp.2019.105965
43. Zhu G, Cheng Z, Huang Y, Zheng W, Yang S, Lin C, et al. TRAF6 promotes the progression and growth of colorectal cancer through nuclear shuttle regulation NF-kB/c-jun signaling pathway. Life Sci. (2019) 235:116831. doi: 10.1016/j.lfs.2019.116831
44. Park Y, Pang K, Park J, Hong E, Lee J, Ooshima A, et al. Destablilization of TRAF6 by DRAK1 suppresses tumor growth and metastasis in cervical cancer cells. Cancer Res. (2020) 80:2537–49. doi: 10.1158/0008-5472.CAN-19-3428
45. Zhang T, Wang H, Han L. Expression and clinical significance of tumor necrosis factor receptor-associated factor 6 in patients with colon cancer. Iran Red Crescent Med J. (2016) 18:e23931. doi: 10.5812/ircmj.23931
46. Luo Z, Zhang X, Zeng W, Su J, Yang K, Lu L, et al. TRAF6 regulates melanoma invasion and metastasis through ubiquitination of basigin. Oncotarget. (2016) 7:7179–92. doi: 10.18632/oncotarget.6886
47. Zeng XZ, Zhang YY, Yang Q, Wang S, Zou BH, Tan YH, et al. Artesunate attenuates LPS-induced osteoclastogenesis by suppressing TLR4/TRAF6 and PLCγ1-Ca(2+)-NFATc1 signaling pathway. Acta Pharmacol Sin. (2020) 41:229–36. doi: 10.1038/s41401-019-0289-6
48. Ni X, Kou W, Gu J, Wei P, Wu X, Peng H, et al. TRAF6 directs FOXP3 localization and facilitates regulatory T-cell function through K63-linked ubiquitination. EMBO J. (2019) 38:99766. doi: 10.15252/embj.201899766
49. Dysthe M, Parihar R. Myeloid-derived suppressor cells in the tumor microenvironment. Adv Exp Med Biol. (2020) 1224:117–40. doi: 10.1007/978-3-030-35723-8_8
Keywords: myeloid-derived suppressor cells, TRAF6, STAT3, tumor immunology, tumor microenvironment
Citation: Song G, Zhang Y, Tian J, Ma J, Yin K, Xu H and Wang S (2021) TRAF6 Regulates the Immunosuppressive Effects of Myeloid-Derived Suppressor Cells in Tumor-Bearing Host. Front. Immunol. 12:649020. doi: 10.3389/fimmu.2021.649020
Received: 03 January 2021; Accepted: 04 February 2021;
Published: 25 February 2021.
Edited by:
Peng Qu, National Institutes of Health (NIH), United StatesReviewed by:
Yafeng He, National Heart, Lung, and Blood Institute (NHLBI), United StatesHongru Zhang, University of Pennsylvania, United States
Copyright © 2021 Song, Zhang, Tian, Ma, Yin, Xu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengjun Wang, c2p3anNAdWpzLmVkdS5jbg==; Kai Yin, anN5aW5rYWlAMTYzLmNvbQ==; Yue Zhang, MTM5NTI4MTU2MzZAMTYzLmNvbQ==
†Present address: Ge Song, Department of Clinical Laboratory, Haian People's Hospital, Haian, China
 Ge Song
Ge Song Yue Zhang
Yue Zhang Jie Tian
Jie Tian Jie Ma
Jie Ma Kai Yin
Kai Yin Huaxi Xu
Huaxi Xu Shengjun Wang
Shengjun Wang