
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 14 June 2021
Sec. Inflammation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.646259
This article is part of the Research Topic Unveiling Immunological Mechanisms of Periodontal Diseases View all 18 articles
 Sejal Patel1‡
Sejal Patel1‡ Derek Howard1‡
Derek Howard1‡ Nityananda Chowdhury2
Nityananda Chowdhury2 Casey Derieux2
Casey Derieux2 Bridgette Wellslager2
Bridgette Wellslager2 Özlem Yilmaz2,3†
Özlem Yilmaz2,3† Leon French1,4,5,6*
Leon French1,4,5,6*Porphyromonas gingivalis, a bacterium associated with periodontal disease, is a suspected cause of Alzheimer’s disease. This bacterium is reliant on gingipain proteases, which cleave host proteins after arginine and lysine residues. To characterize gingipain susceptibility, we performed enrichment analyses of arginine and lysine proportion proteome-wide. Genes differentially expressed in brain samples with detected P. gingivalis reads were also examined. Genes from these analyses were tested for functional enrichment and specific neuroanatomical expression patterns. Proteins in the SRP-dependent cotranslational protein targeting to membrane pathway were enriched for these residues and previously associated with periodontal and Alzheimer’s disease. These ribosomal genes are up-regulated in prefrontal cortex samples with detected P. gingivalis sequences. Other differentially expressed genes have been previously associated with dementia (ITM2B, MAPT, ZNF267, and DHX37). For an anatomical perspective, we characterized the expression of the P. gingivalis associated genes in the mouse and human brain. This analysis highlighted the hypothalamus, cholinergic neurons, and the basal forebrain. Our results suggest markers of neural P. gingivalis infection and link the cholinergic and gingipain hypotheses of Alzheimer’s disease.
Porphyromonas gingivalis (P. gingivalis), a keystone species in the development of periodontal disease is believed to play a pathogenic role in several systemic inflammatory diseases (1). This Gram-negative anaerobe is unique in its ability to secrete gingipain proteases, which are its primary virulence factors and are required for its survival in vivo (2). P. gingivalis is asaccharolytic and uses gingipains to degrade host peptides for nutrition and energy. These gingipain peptidases are cysteine proteases that cleave bonds after arginine (RgpA and RgpB) and lysine (Kgp) (3, 4). These two positively charged amino acid residues facilitate binding of negatively charged nucleic acids (5, 6). This electrostatic relationship suggests gingipains may severely disrupt host cell protein-RNA and protein-DNA interactions.
Positive associations between periodontal disease and orodigestive cancer (7), rheumatoid arthritis (8), heart disease (9), male infertility (10), and Alzheimer’s disease have been reported (11, 12). Several studies have found links that implicate P. gingivalis in these associations. For example, serum P. gingivalis antibody levels are a risk factor for orodigestive cancer death (13), and are more common in rheumatoid arthritis subjects (14). More directly, P. gingivalis expresses enzymes that convert arginine residues to citrulline, which is thought to trigger inflammatory responses in rheumatoid arthritis (15). In the context of heart disease, P. gingivalis DNA has been found in human atherosclerotic plaques (16, 17) and gingipains modify high- and low-density lipoproteins (18). More specifically, arginine‐specific gingipains are able to fragment Apolipoprotein E (ApoE) (18). ApoE is the strongest genetic risk factor for late-onset Alzheimer’s disease, differences between the genetic variants increase the number of arginine residues at two positions. The lowest risk ApoE2 isoform encodes only cysteines at these positions, while ApoE3 contains one arginine and ApoE4, which confers the highest risk contains two (19). This connection to genetic risk is supported by significant evidence of P. gingivalis and specifically gingipains in Alzheimer’s disease pathogenesis (11, 20–23). This evidence linking P. gingivalis to several chronic diseases combined with the direct link between arginine count and genetic risk for Alzheimer’s disease motivated our genome- and brain-wide study of gingipain susceptibility.
In this study, we sought to determine which human proteins are susceptible to gingipain cleavage by characterizing proteins with high proportions of arginine and lysine. Motivated by evidence linking P. gingivalis to Alzheimer’s disease, we tested if the genes encoding these proteins are differentially expressed in brain tissue with detected P. gingivalis RNA. We then performed neuroanatomical enrichment analyses to better understand tissue specific susceptibility. We extend these analyses to a single-cell atlas of the mouse nervous system to identify cell-types that may be specifically susceptible to gingipains.
Translated human protein sequences were obtained from GENCODE version 32 (24). Amino acid proportions were mean averaged across multiple transcripts that were annotated to the same gene symbol. Protein sequences annotated to more than one gene were removed to prevent amplification of single sequences in the following enrichment analyses.
The Gene Ontology (GO) provides gene-level annotations that span specific cellular components, biological processes, and molecular functions (25). These annotations, defined by GO terms, were required to have annotations for 10 to 200 tested genes (6,807 unique GO groups annotating 14,655 unique genes). To test for enrichment, we sorted the genes from the most enriched to the most depleted proportions of arginine and lysine residues. Within this ranking, the area under the receiver operating characteristic curve (AUC) was used to test for gene ontology terms that are enriched in either direction (enriched: AUC > 0.5, depleted: AUC < 0.5). The Mann–Whitney U test was used to determine statistical significance with FDR correction for the GO groups tested. We used GO annotations from the GO.dB and org.Hs.eg.db packages in R, version 3.8.2, which were dated April 24, 2019 (26, 27).
Gingipain activity was tested against the 70S ribosome of Escherichia coli (New England Biolabs, Ipswich, MA). Recombinant Lys-gingipain (rKgp; CUSABIO, Houston, TX) or recombinant Arg-gingipains (rRgpA and rRgpB; CUSABIO) at 120nM concentrations were incubated with 5 µg of ribosome in TC150 buffer (50 mM Tris, pH 8.0; 150 mM NaCl, 5 mM Cysteine, 5 mM CaCl2) (28) for 1, 3, 6, or 12 hours at 30°C. One unit of recombinant human Caspase-3 (Sigma, St. Louis, MO), a cysteine protease, was used as a control favoring aspartic acid rich peptides to lysine- or arginine-rich peptides (29, 30). All the reactions were ceased via boiling with a laemmli sample buffer (Bio-Rad, Hercules, CA) for 5 minutes. The samples were then resolved by SDS PAGE using 4–20% Criterion TGX stain-free gel (Bio-Rad) at 100V for 80 min. Gels were visualized after staining with 0.25% coomassie brilliant blue (in 50% methanol and 7% acetic acid) for 2-4 hours, followed by de-staining (10% methanol and 7% glacial acetic acid) until bands were visible. The assay was also repeated at 37°C and/or using twofold amounts of gingipains (240 nM) for 1 and 3 hours. In all assays, ribosomes without any enzymes or enzymes without any ribosomes were used as reagent controls. To confirm that the gingipains used in this study were biologically active, each of them was tested against recombinant human ApoE4 protein (Novus Biologicals, Centennial, CO), a known substrate of gingipains (18, 20).
Gene expression profiles from a postmortem study of Parkinson’s disease were used to test for gene expression differences in tissue samples with P. gingivalis reads (31). To exclude confounding effects of disease processes, we limited our analyses to the 44 neurologically normal control samples of this study. As detailed by Dumitriu et al., prefrontal cortex samples were profiled with Illumina’s HiSeq 2000 sequencers. All samples were from males of European ancestry. Neuropathology reports were used by Dumitriu et al. to exclude brains with Alzheimer’s disease pathology beyond that observed in normal aging.
We used the Sequence Read Archive analysis tool to identify samples with sequencing reads that align to the P. gingivalis genome (32). To test for differential expression, we obtained the normalized count matrix for GSE68719 (GSE68719_mlpd_PCG_DESeq2_norm_counts.txt.gz). Batch information was obtained from the Gemma bioinformatics system (33). To identify genes differentially expressed in samples with detected P. gingivalis, we modelled log(expression +1) with an ordinary least squares linear model with covariates for age, RIN, PMI, and total bases read. Gene ontology enrichment analyses were performed using the same AUC method described above.
To examine differential expression of cell-type markers, we used the top marker genes obtained from a single cell study of the adult human temporal cortex (34). This study used gene expression profiles to cluster cells into astrocyte, neuron, oligodendrocyte, oligodendrocyte precursor, microglia and endothelial groups. These marker genes were used to calculate AUC values.
The Allen Human Brain Atlas was used to determine regional enrichment of the genes identified in the differential expression analyses throughout the brain (35). Thus, in contrast to the differential analyses that focused on samples from the cerebral cortex, this approach used the whole brain, and expression profiles were summarized to named brain regions. For each donor, samples mapping to the same-named brain region were mean-averaged to create a single expression profile for each region. Values from analogous named brain regions of both hemispheres were pooled because no significant differences in molecular architecture have been detected between the left and right hemispheres (36). Expression levels of the 48,170 probes were then summarized by mean averaging for each of 20,778 gene transcripts. Expression values were next converted to ranks within a named brain region, and then z-score normalized across brain regions. For the analyses that combine across donors, z-scores for each donor were averaged to obtain a single gene-by-region reference matrix of expression values.
The RNA sequencing data that were used to characterize gene expression across the mouse nervous system were obtained from Zeisel et al. (37). This dataset of more than 500,000 isolated cells from 19 defined regions was obtained from male and female mice that ranged in age from 12 to 56 days old. Following quality control, Zeisel et al. identified 265 clusters of cells. These clusters are thought to represent different cell types. Aggregate expression values for the identified transcriptomic cell-type clusters were obtained from the Mousebrain.org downloads portal. These values were log(expression+1) transformed and then z-scored at the gene level. We restricted our analysis to genes with human homologs by filtering mouse gene symbols for those that have human homologs (38). Expression z-scores are then ranked genome-wide within a defined cell-type cluster.
To calculate enrichment for gene sets of interest within a brain region or a cell type, z-scores in the processed expression matrices are first ranked. This provides a genome-wide list for each cell type or brain region with high ranks marking specific and low for depleted expression. We project our genes of interest into these ranked lists to determine enrichment. The AUC statistic was used to quantify if the genes of interest are more specifically expressed (ranked higher) in this sorted list of genes for a specific region or cell cluster. In this context, AUC > 0.5 means the associated genes have specific or enriched expression in a brain region/cell-type cluster. For AUC < 0.5, the reverse is true, with a bias toward lower relative expression. The Mann-Whitney U test was used to test the statistical significance, and the FDR procedure was used to correct for testing of multiple brain regions or cell clusters within a dataset.
The average proportion of arginine or lysine residues was 11.4% across the 20,024 human protein-coding genes in our analysis. A broad range was observed, with 95% of genes having proportions between 5.5% and 19.4% (full listing in Supplement Table 1). The gene with the highest proportion was Ribosomal Protein L41 (RPL41, 68%) and joined three other ribosomal proteins within the top ten list (RPL39, RPS25, and RPL19). This high proportion of arginine and lysine in these top proteins suggests they are highly susceptible to gingipain cleavage.
To characterize the proteins with high arginine and lysine residues genome-wide, we performed a GO enrichment analysis. Of the 6,806 tested GO groups, 881 are enriched for higher proportions of arginine and lysine after multiple test correction (top ten in Table 1, full listing in Supplement Table 2). In agreement with inspection of the top ten proteins, genes encoding parts of the ribosomal subunit are the most strongly enriched with one in five residues being arginine or lysine on average (AUC = 0.87, pFDR < 10-58). The remaining top groups largely contain ribosomal protein genes. Ranked 15th are genes annotated to the nucleosome, which includes many histones (60 genes, 20.9% average proportion, AUC = 0.926, pFDR < 10-29). The high enrichment for this set is partially due to 9 highly similar protein sequences from a histone microcluster that has the same arginine and lysine proportion (22.2%). Overall, this enrichment for positively charged arginine and lysine residues mirrors their known ability to facilitate the binding of DNA and ribosomal RNA (6, 39).
Within the top ten most enriched GO groups, ‘SRP-dependent cotranslational protein targeting to membrane’ appears to be the most specific with the lowest number of annotated genes (Figure 2B). These genes are primarily components of the cytosolic ribosome that facilitate translation into the endoplasmic reticulum (ER) but also include genes encoding the signal recognition particle (SRP) and its receptor. For brevity, we refer to this GO group as the ‘ER translocation’ genes. This specific GO group has been previously associated with disorders that P. gingivalis is believed to play a pathogenic role. Specifically, a spatial transcriptomics study reported higher expression of the ER translocation genes in inflamed areas of periodontitis-affected gingival connective tissue compared to non-inflamed areas (40). A second study that examined peri-implant soft tissue found that ER translocation genes are expressed at higher levels in diseased mucosa samples (41). In the context of Alzheimer’s disease, the ER translocation genes are upregulated across neuroinflammation, in cases from a Caribbean Hispanic postmortem study, amyloid associated dystrophic microglia, and in regions of Alzheimer’s disease-associated hypometabolism (42–44). Given this upregulation in periodontal and Alzheimer’s disease, we pursued further characterization of the ER translocation genes.
Genome-wide, the ER translocation genes have a high proportion of arginine and lysine, but it’s not clear if other residues are enriched. To determine the specificity of this enrichment, we tested proportions of other amino acids. For single residues, lysine (AUC = 0.89) and arginine (AUC = 0.76) are the most enriched in the ER translocation genes, followed by valine and isoleucine (AUC = 0.64 and 0.63, respectively) (Supplement Table 3). For pairs, Figure 1 shows the AUC values of the 190 possible amino acid combinations. Only the lysine and valine proportion have a higher enrichment score, which is slightly higher (AUC = 0.896 compared to 0.894 for arginine + lysine). Still, this combination is less frequent (18.3% of residues compared to 20.4%). Overall, the ER translocation genes are strongly and specifically enriched for arginine and lysine residues, suggesting they are particularly susceptible to cleavage by gingipains.
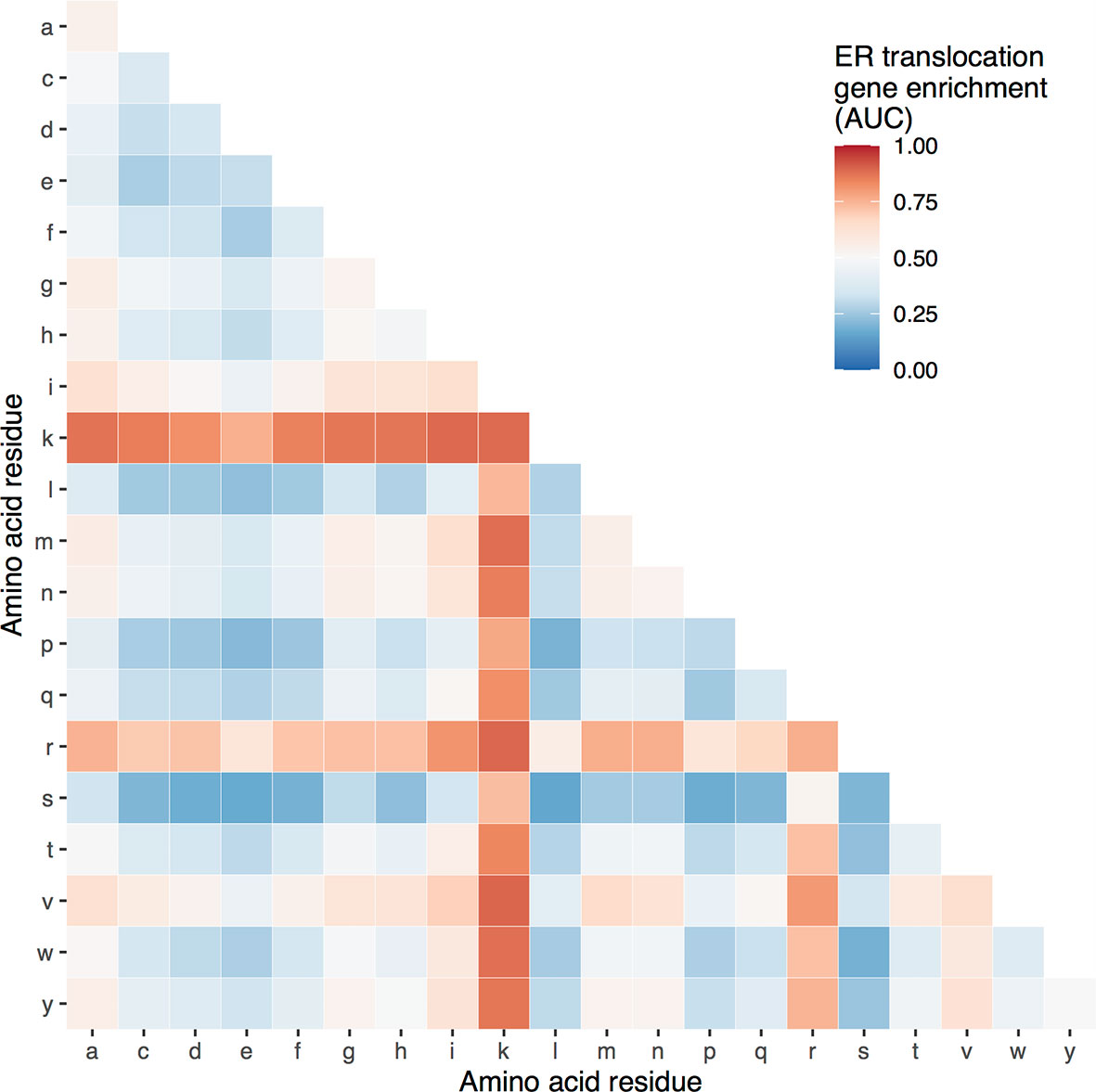
Figure 1 Heatmap of ER translocation gene enrichment for proportions of different amino acid pairs. Relative to all other genes, proportions of specific amino acid pairs for the ER translocation genes range from high (red) to low (blue).
Recombinant lysine and arginine gingipains exhibited weak to no activity against the E. coli 70S ribosome at 30°C for a 1-3 hour incubation period (Supplement Figure 1). Cleavage activity did not change upon increases in gingipains’ concentration, incubation temperature (37°C), or duration (data not shown). Similarly, Caspase-3, which was used as a control cysteine protease did not demonstrate any detectable activity (cleavage) of the ribosome. The recombinant gingipains used were biochemically active as confirmed by the cleavage of recombinant human ApoE4 protein in control assays.
Guided by the findings of the upregulation of ER translocation genes in the context of Alzheimer’s disease, we tested for direct associations with P. gingivalis in postmortem brain tissue. We examined control samples that lacked any neurodegenerative disease pathology to remove any late-stage signals of Parkinson’s or Alzheimer’s disease. In this all-male dataset, age ranged from 46 to 97 years old. Within the 44 prefrontal cortex samples, P. gingivalis sequencing reads were detected in ten. In contrast, reads from the two other bacteria in the red complex, which is associated with severe periodontal disease, were not detected (Tannerella forsythia and Treponema denticola). There was no difference between age, RNA integrity number, and postmortem interval between the samples with and without detected P. gingivalis reads (all p > 0.33). In contrast, there was a significant difference in the number of bases sequenced, with more reads in the samples with detected P. gingivalis. All four of these variables were covariates in our differential expression model. Importantly, batch information is confounded with the detected P. gingivalis reads. Specifically, all 10 ten samples with detected P. gingivalis reads were sequenced on one of the three sequencing machines used for the study (device DGL9ZZQ1). This adds uncertainty to our results from this dataset. We also tested for differential expression in samples from this specific device but note this reduces the sample size to 14 from 44 for this specific analysis.
In total, 2,189 of the 15,936 tested genes were differentially expressed. More genes were down- than up-regulated in samples with detected P. gingivalis reads (1247 versus 942). Arginine and lysine proportion was different between these two sets of genes with an average proportion of 12.8% for the up-regulated genes versus 11.4% for the down-regulated genes (p < 10-16). The top ten most up- and down-regulated genes are provided in Table 2 (full listing in Supplement Table 4). The third most up-regulated gene is signal recognition particle 9 (SRP9, pFDR < 0.0013, Figure 2A). SRP9 is a member of the ER translocation gene set that forms a heterodimer with the next ranked SRP gene (SRP14, pFDR < 0.02) (45). The strongest downregulated genes include putative RNA helicases (DHX30, DHX37) and other genes involved in the regulation of global gene transcription (ZNF696). No genes survive multiple test correction when differential expression is tested in the smaller set samples profiled by device DGL9ZZQ1. However, ZNF696 and DHX37 remain down-regulated in these samples (uncorrected p < 0.05).
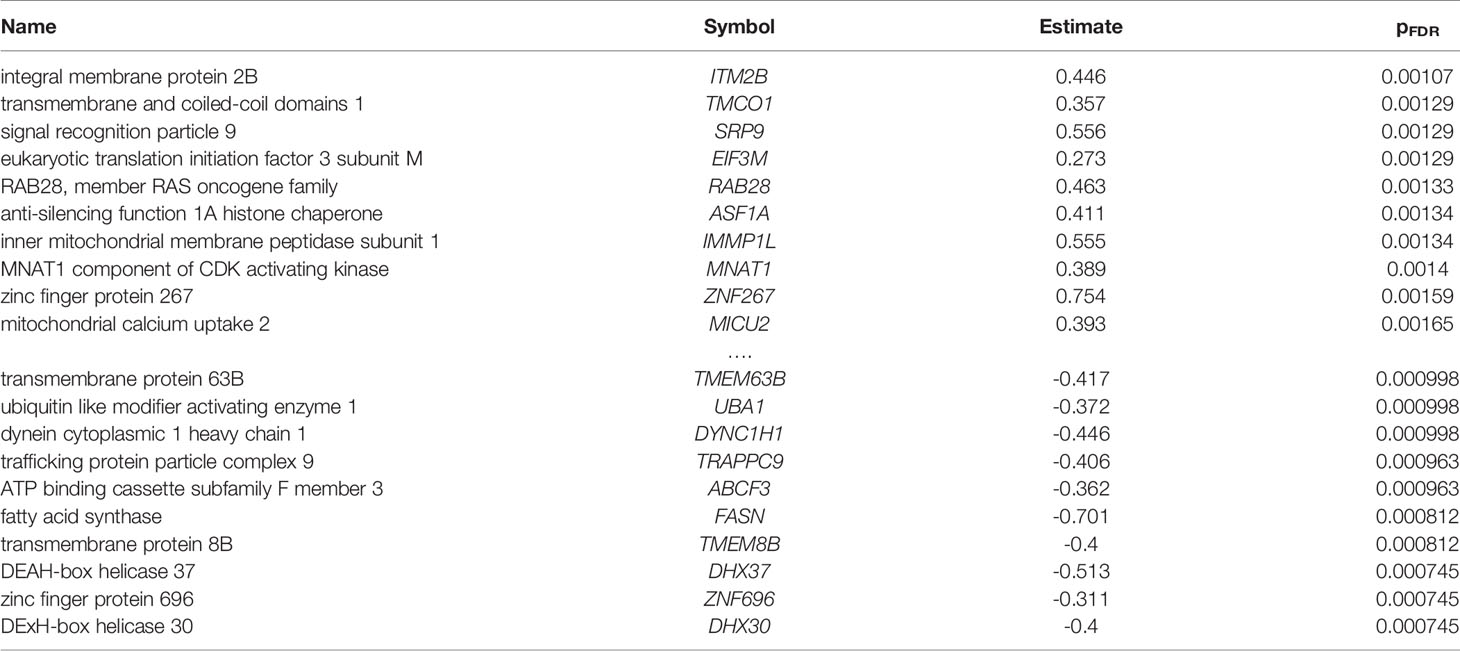
Table 2 Top ten most up- and down-regulated differentially expressed genes in the prefrontal cortex with detected P. gingivalis reads.

Figure 2 Visualization of differential expression results. (A) Dotplot of SRP9 expression is plotted for samples with and without P. gingivalis reads. (B) ROC for the ER translocation genes when all genes are ranked according to direction and significance [signed log(p-value)].
To summarize the hundreds of differentially expressed genes, we again used the AUC metric. Genes were ranked from the most up- to down-regulated in the samples with detected P. gingivalis. Of the 6,538 tested GO groups, 23 were significantly up-regulated, and 49 were down-regulated after multiple test correction (full listing in Supplement Table 5). Across this ranking, ER translocation genes were strongly up-regulated and is the fourth most significant GO group (AUC = 0.698, pFDR < 10-7). Similar GO groups that primarily contain genes encoding ribosomal proteins make up the other top ten up-regulated groups. Of these ten, all are in the top ten list of groups that are enriched for arginine and lysine residues except the ‘protein localization to endoplasmic reticulum’ and ‘protein targeting to membrane’ groups. Within the top ten most down-regulated GO groups, ‘homophilic cell adhesion via plasma membrane adhesion molecules’ ranked first, followed by several synapse associated groups and ‘ATP biosynthetic process’ (all pFDR < 0.005). In the context of amino acid residues, we note that genes annotated to ‘aminoacyl-tRNA ligase activity’ are also down-regulated (AUC = 0.33, pFDR < 0.05). In summary, samples with detected P. gingivalis reads have higher expression of ER translocation genes.
Given the down-regulation of synapse genes, we next tested if cell-type specific markers are differentially expressed. Of the six cell-types, only genes marking endothelial cells (AUC = 0.805, pFDR < 0.00005) and astrocytes (AUC = 0.682, pFDR < 0.02) were enriched, with higher expression in samples with P. gingivalis reads. In contrast, markers of oligodendrocyte precursors, oligodendrocytes, neurons, and microglia were not enriched. Specifically, the 20 neuronal markers are equally split between up- and down-regulated with no genes reaching statistical significance at an alpha of 0.05 (uncorrected). In summary, differential expression of cell-type markers suggests changes in endothelial and astrocyte cell states or proportions but not neurons.
To further characterize the genes associated with P. gingivalis, we determined which brain regions are enriched for their expression. While these genes were identified in samples from the prefrontal cortex, it is believed neocortical degeneration doesn’t occur until later stages of Alzheimer’s disease (46). Testing for neuroanatomical enrichment may reveal other regions of interest. In this analysis, for a given brain region, the genome was ranked from the most specifically expressed gene to the most depleted gene (relative to the rest of the brain). Similar to the preceding analyses, we use the AUC metric to test if genes of interest rank higher in this list to determine region-specific expression. In total, 82 of 232 tested brain regions are enriched for high expression of the genes up-regulated in samples with detected P. gingivalis (Supplement Table 6). The anterior hypothalamic area most specifically expresses these genes (AUC = 0.740, pFDR < 10-120). This result is consistent when tested in individual brain hemispheres (n = 2 brains, left AUC: 0.738; right: 0.698). Within the top ten most enriched regions, two other hypothalamic regions appear (Table 3, AUCs > 0.68, both pFDR < 10-76). Notably, most of the top regions border ventricles (medial habenular nucleus, thalamic paraventricular nuclei, substantia innominata, central gray of the pons, paraventricular nucleus of the hypothalamus, and septal nuclei).
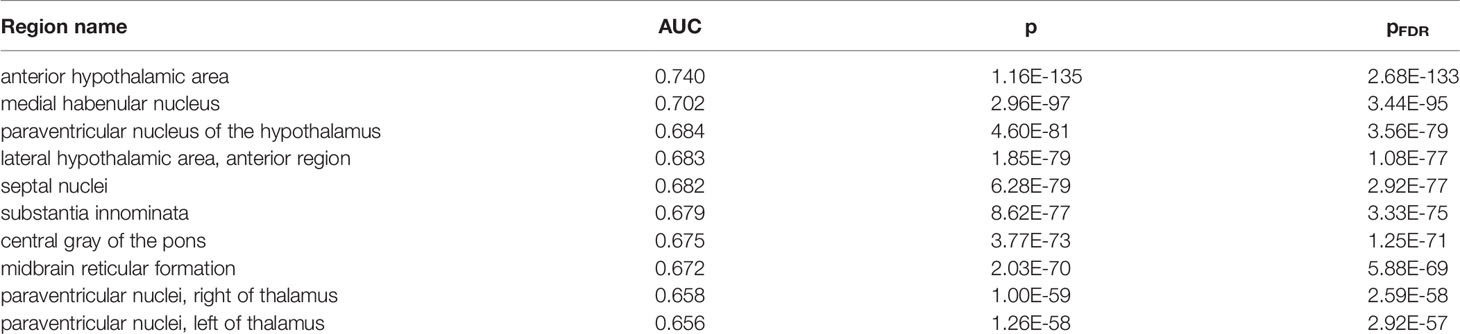
Table 3 Top ten regions enriched for higher expression of genes up-regulated in samples with detected P. gingivalis reads.
We next characterized the expression patterns of the genes up-regulated in samples with detected P. gingivalis at a finer resolution in the mouse nervous system (37). Similar to the regional analyses above, we ranked each gene from the most specific to depleted expression for each transcriptomic cell type cluster. The 873 mouse homologs of the up-regulated genes are enriched for specific expression in 79 of the 265 tested transcriptomic cell type clusters (Supplement Table 7). The top two enriched clusters are nitrergic enteric neurons (both AUC > 0.67, pFDR < 10-65). The next three most enriched are cholinergic neurons with probable locations listed as the sympathetic ganglion or myenteric plexus of the small intestine (all AUC > 0.66, pFDR < 10-56). The top 19 most enriched clusters are located in the peripheral nervous system. Focusing on the central nervous system, the top two clusters described as “Afferent nuclei of cranial nerves VI-XII” and “Cholinergic neurons, septal nucleus, Meynert [sic] and diagonal band” (both AUC < 0.59, pFDR < 10-19). Broadly, cells in the mouse peripheral nervous system and cholinergic neurons strongly express genes that are up-regulated in samples with detected P. gingivalis.
To extend our results beyond protein level susceptibility, we next identified brain regions that express high levels of the ER translocation genes. This provided a neuroanatomical perspective of arginine and lysine proportions. While our preceding analyses used genes identified from brain tissue samples, the ER translocation genes were identified from analyses of protein sequences alone. This provides independence from tissue- and state-specific expression. In a combined analysis of all six brains, 83 of the 232 brain regions showed overexpression of the ER translocation genes. The substantia innominata was top-ranked with the most specific expression of the ER translocation genes (AUC = 0.846, pFDR < 10-28, full listing in Supplement Table 8). This enrichment appears when hemispheres are tested separately (n = 2 brains, AUC left: 0.847; right: 0.816). Of the nine remaining top 10 regions, seven are located near the substantia innominata: internal and external globus pallidus, substantia nigra pars reticulata, the septal nuclei, nucleus accumbens, subcallosal cingulate gyrus, head of the caudate nucleus (AUCs > 0.774, all pFDR < 10-18). Of the 34 assayed cerebellar cortex regions, all are significantly enriched for expression of the ER translocation genes. Within specific brains, the substantia innominata is the 10th ranked brain region (of 194) for selective expression of ER translocation associated genes in donor 10021/H0351.2002, and is ranked 3rd of 182 in donor 9861/H0351.2001. There are no samples from the substantia innominata in the other four donors, but its constituent nuclei are. Testing for anatomical enrichment of the ER translocation associated genes in these nuclei reveals a highly heterogeneous pattern (Figure 3). A key characteristic of the substantia innominata is a high proportion of cholinergic neurons (47). Alzheimer’s disease has been previously associated with cholinergic neuron loss in the basal forebrain and deficits in choline O-acetyltransferase (encoded by CHAT) (48–50). Figure 3 shows the relationships between CHAT gene expression and ER translocation genes across the brain, marking the substantia innominata as having high expression of both CHAT and the ER translocation genes.
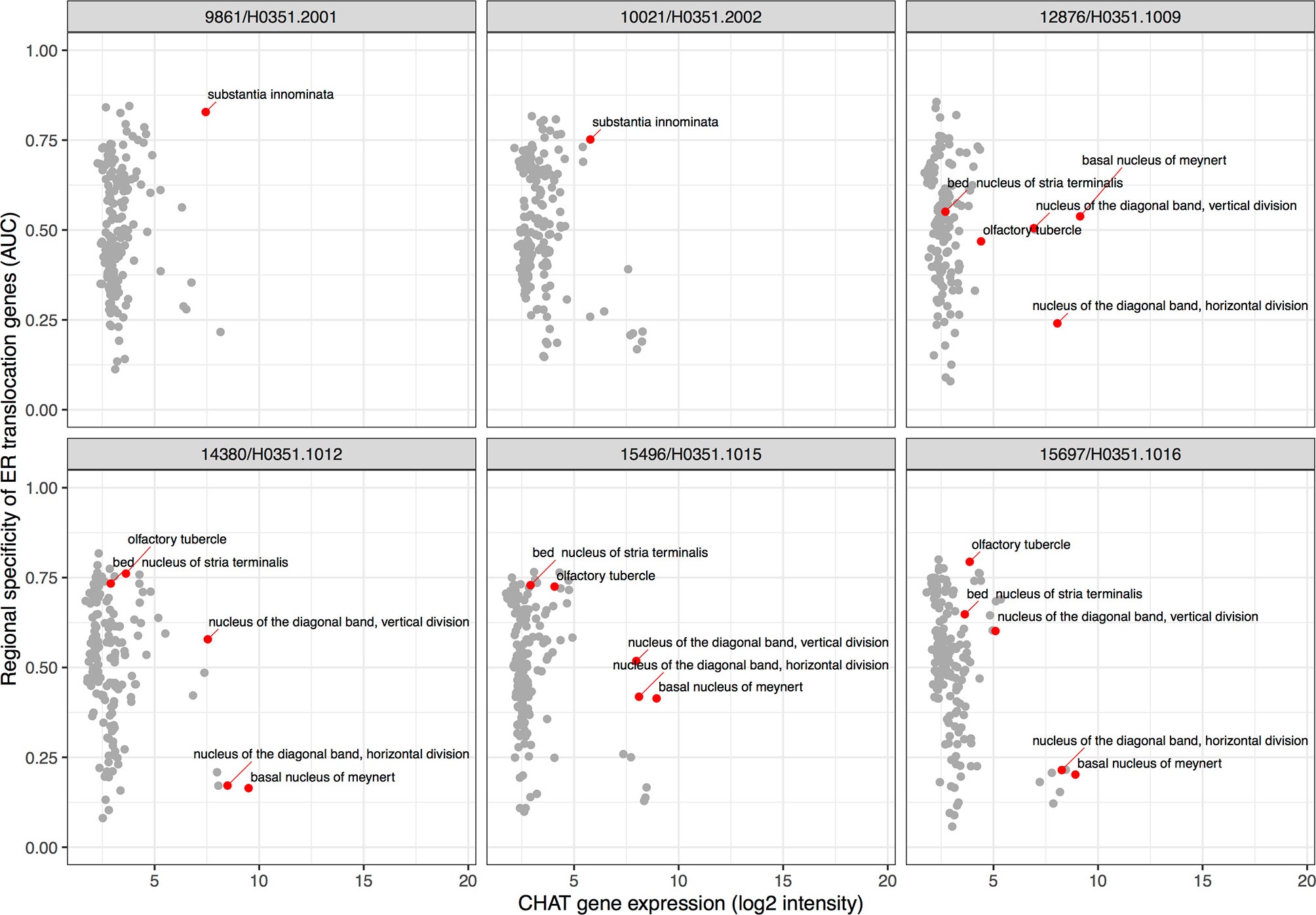
Figure 3 Scatter plots showing specific enrichment for ER translocation genes (y-axis) and Choline O-Acetyltransferase (CHAT) gene expression on the x-axis in each brain. Each point is a brain region with red marking the substantia innominata and its nuclei.
Mirroring our analyses of P. gingivalis associated genes, we next characterized the expression patterns of the ER translocation genes in the mouse nervous system. We observe that the 79 mouse homologs of the ER translocation genes are not evenly expressed across clusters in this single-cell atlas. The top 20 most enriched transcriptomic cell type clusters are provided in Table 4 (full listing in Supplement Table 9). Four cholinergic enteric neuron clusters are within the top ten cell clusters. Of the 14 cholinergic clusters in this atlas, 12 are enriched for ER translocation gene expression (all AUC > 0.5 and pFDR < 0.05). Relative to the 265 tested cell type clusters, the cholinergic groups are enriched for higher AUC values (p < 0.0001, Mann-Whitney U test). The top-ranked cholinergic cell clusters from brain tissue are listed as telencephalon interneurons with an annotated location of striatum and amygdala (TECHO, AUC = 0.822, ranked 21st). In Table 4, it is also clear that hypothalamic cells strongly express the ER translocation genes. Of the 14 total hypothalamic cell clusters, 7 are enriched for ER translocation gene expression (AUC > 0.57, pFDR < 0.05). All of the hypothalamic cell type clusters enriched for ER translocation genes are peptidergic (or produce peptide hormone precursors). Three of the top hypothalamic transcriptomic cell types were annotated with locations that are in or near the basal forebrain. Specifically, the HYPEP5 cluster lists the nucleus of the diagonal band and the HYPEP8 cluster names the medial septal nucleus as probable locations of those clustered cells (37). While substantia innominata is not mentioned in this atlas, both the mouse and human regional analyses highlight the basal forebrain, hypothalamus and cholinergic system.
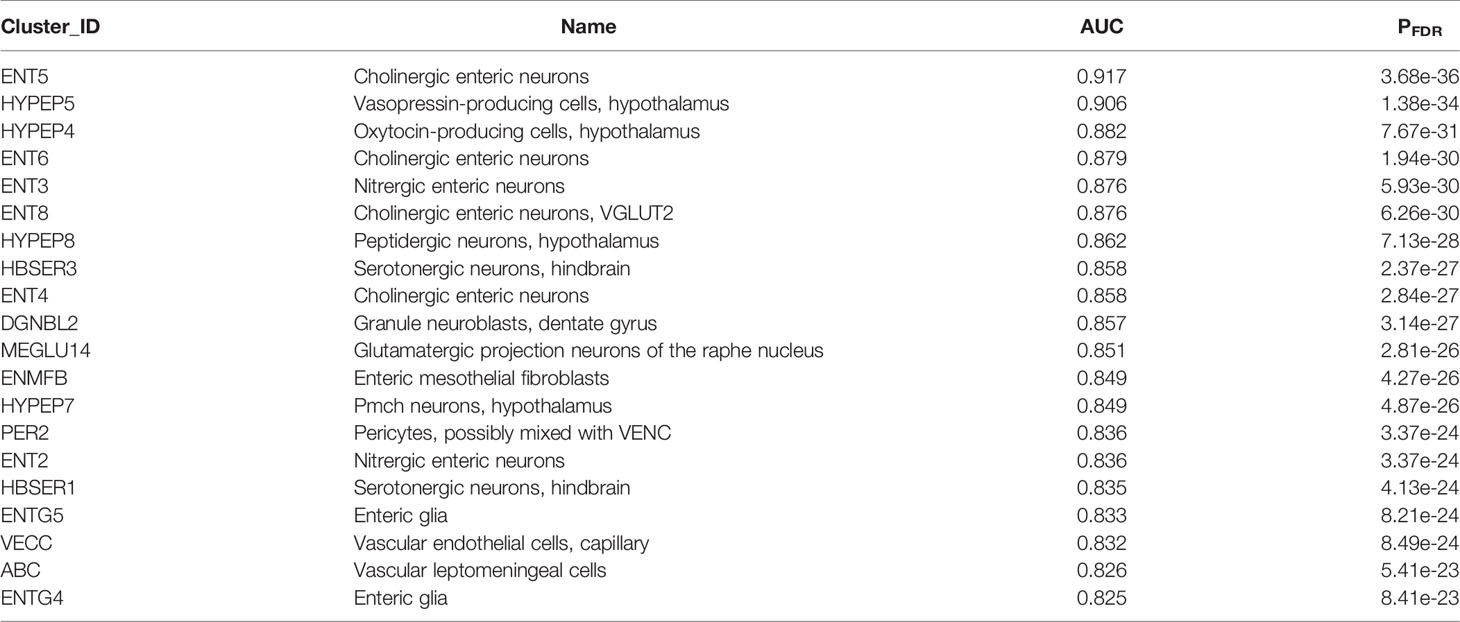
Table 4 Top 20 transcriptomic cell type clusters enriched for higher expression of the ER translocation genes.
We next tested for overlap between the two brain-wide patterns derived from separate sources of P. gingivalis associated genes. Within the top ten regions, the central gray of the pons, septal nuclei, and substantia innominata appear in both enrichment results. More specifically, the anterior hypothalamic area, the most enriched region for the genes associated with P. gingivalis reads, is the top-ranked hypothalamic region for the ER translocation results (ranked 51 of 232 overall, AUC = 0.697, pFDR < 10-10). Conversely, the substantia innominata, which is most enriched for the ER translocation genes, is ranked 4th for genes associated with P. gingivalis reads. This spatial convergence within the top hits extends brain-wide with 48 brain regions overlapping between two lists of significantly enriched structures (hypergeometric test, p < 10-6) and correlation coefficient of AUC values of 0.36 (p < 10-7). This brain-wide spatial agreement is visualized in Figures 4 and 5A. As shown in Figure 5B, high agreement is also observed in the single-cell atlas of the mouse nervous system (r = 0.72, p < 10-43). In particular, enriched expression in cholinergic neurons is evident for both sets of P. gingivalis associated genes. In summary, genes associated with gingipain susceptibility and P. gingivalis presence are highly expressed in hypothalamic, cholinergic neurons, and basal forebrain regions.
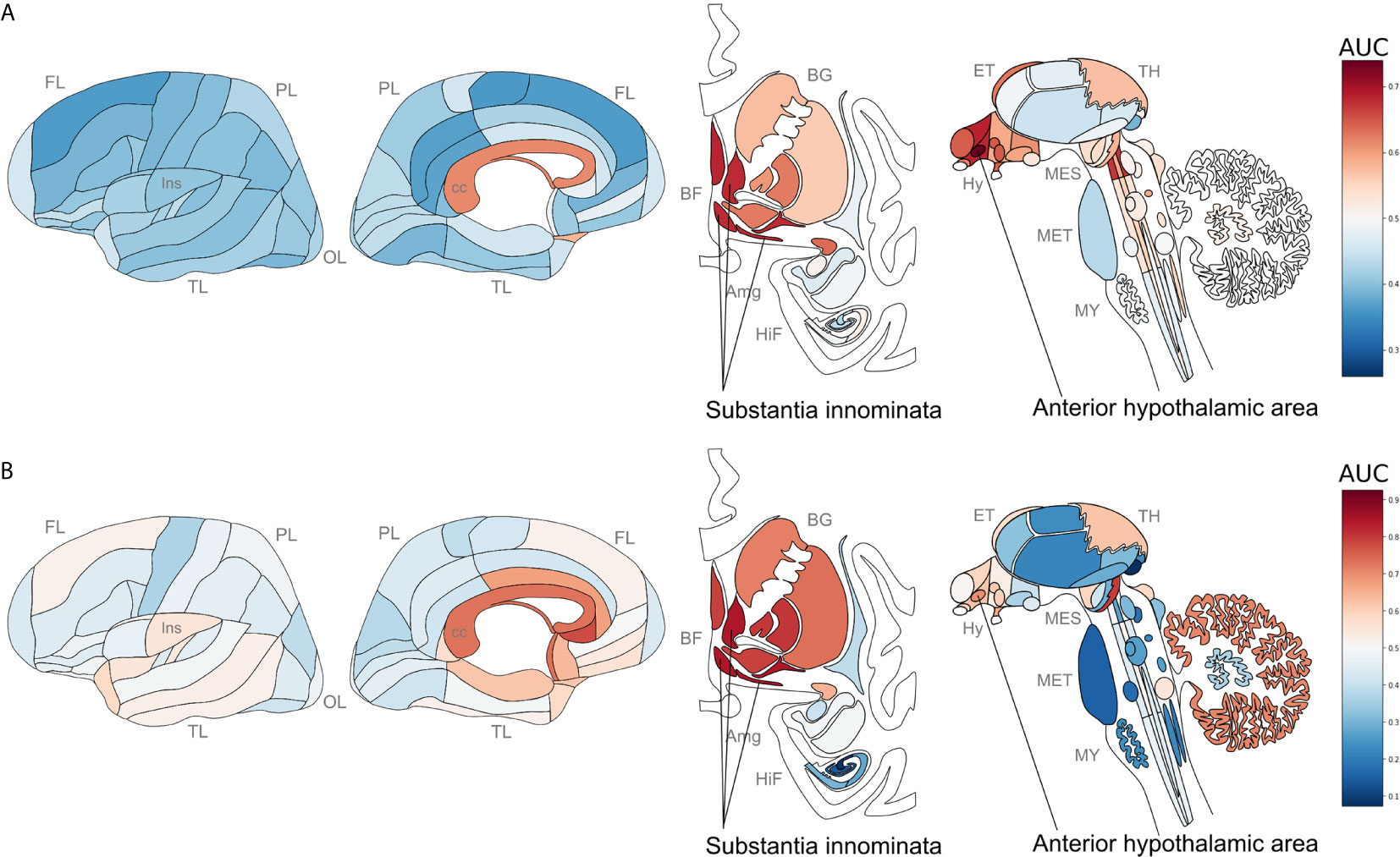
Figure 4 Neuroanatomical heatmaps marking specific expression of the genes up-regulated in samples with P. gingivalis reads (A) and ER translocation genes (B). AUC values range from depleted expression in dark blue to enriched in dark red. Brain region abbreviations: FL, frontal lobe; PL, parietal lobe; TL, temporal lobe; OL, occipital lobe; BF, basal forebrain; BG, basal ganglia; AmG, amygdala; HiF, hippocampal formation; EP, epithalamus; TH, thalamus; Hy, hypothalamus; MES, mesencephalon; MET, metencephalon; and MY, myelencephalon. Anatomical template images are from the Allen Human Brain Reference Atlas (51).
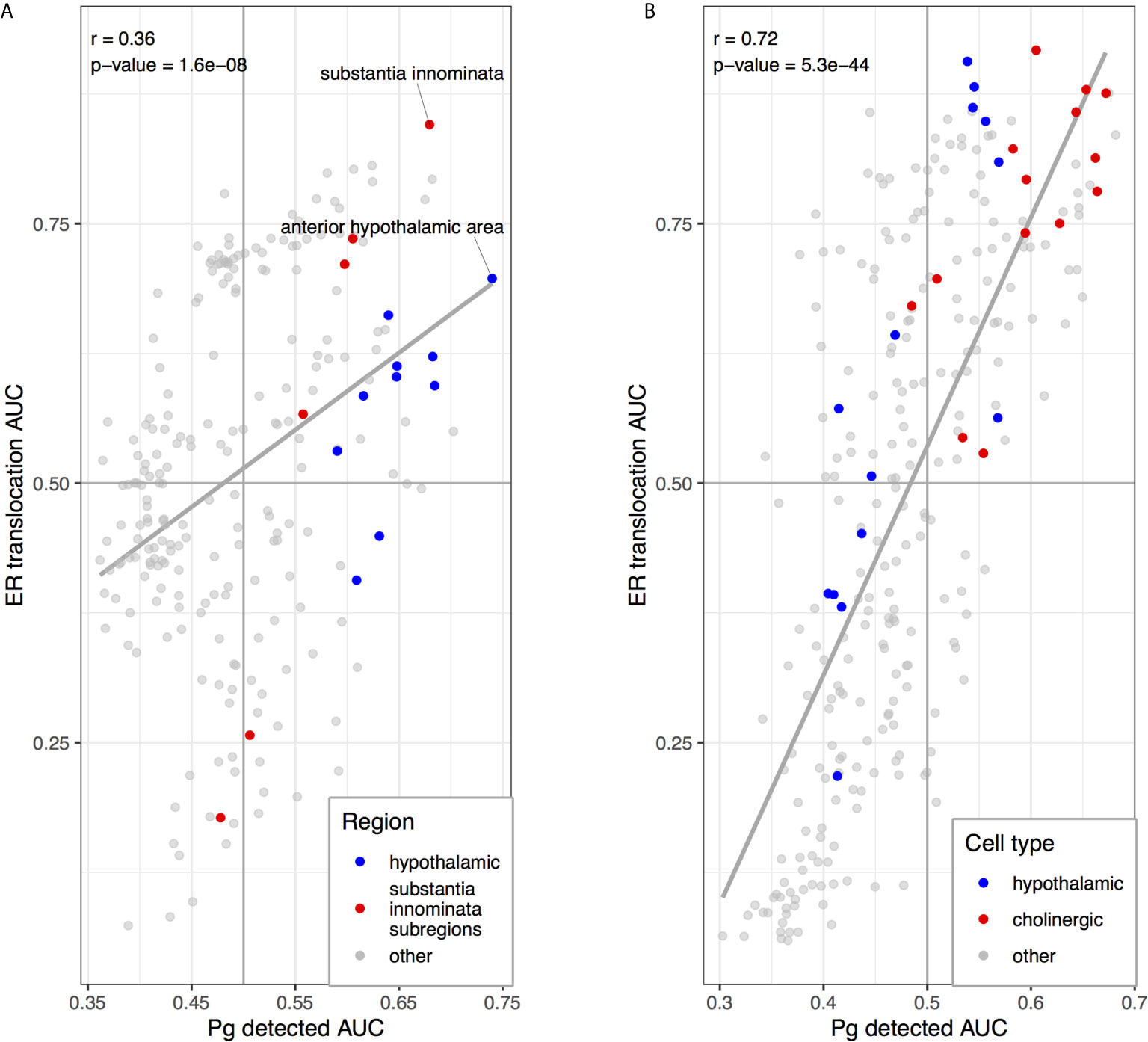
Figure 5 Scatterplots comparing anatomical enrichment of ER translocation genes (y-axis) and up-regulated in samples with P. gingivalis reads (x-axis). AUC values are plotted to show enrichment in the human brain atlas (A) and the mouse nervous system (B). Hypothalamic regions and transcriptomic cell type clusters are marked in blue. Cholinergic clusters, substantia innominata and its subregions are marked in red. Fitted linear regression lines for all points are shown in grey.
In this study, we characterized protein susceptibility to gingipain cleavage and genes differentially expressed in brain tissue with P. gingivalis. As expected, we found that genes with high arginine and lysine proportions are enriched in proteins that bind RNA and DNA. We focused on a specific set of these genes that participate in ER translocation and have previously been shown to be up-regulated in periodontitis, dystrophic microglia, and Alzheimer’s disease (40, 42–44). We directly link these findings to P. gingivalis by showing that these genes are also up-regulated in brain tissue with detected P. gingivalis RNA. This convergence between proteins susceptible to gingipain cleavage and the transcriptomic response to P. gingivalis motivated our neuroanatomical characterization of these genes. In this spatial analysis, we again observe agreement, with enrichment for cholinergic neurons, basal forebrain and hypothalamic regions. Regions near ventricles and peripheral neurons are also enriched, suggesting relevance to P. gingivalis brain entry. While gingipain levels have been shown to correlate with tau and amyloid pathology (20–22), we are the first to associate this virulence factor with the cholinergic hypothesis of Alzheimer’s disease.
In our postmortem brain tissue analyses, several differentially expressed genes are related to amyloid processing and inflammation. For example, ZNF267, the 9th most up-regulated gene, was selected as one of the ten genes used in a blood-based transcriptomic panel for diagnosis AD (52). The most significantly up-regulated gene, integral membrane protein 2B (ITM2B, pFDR < 0.0011), regulates the processing of amyloid-beta and inhibits amyloid aggregation (53). Mutations in this gene that encodes the BRI2 protein cause familial British and Danish dementia (54, 55). We also note that the 12th most downregulated gene encodes Myc associated zinc finger protein (MAZ) and is also known as serum amyloid A-activating factor-1 (SAF-1), due to its role in regulating serum amyloid A in response to inflammation (56). Expression of the mouse homolog of MAZ is increased in an Alzheimer’s disease mouse model (57). Furthermore, murine overexpression increases the risk of severe arthritis (58). Like P. gingivalis, serum amyloid A is suspected to be involved in the pathogenesis of arthritis, atherosclerosis, amyloidosis and Alzheimer’s disease (59–62). Lastly, the 3rd most down-regulated gene, DHX37, harbors a rare frameshift mutation that segregates with Alzheimer’s disease in one family (63). While our differential expression analysis highlighted ER translocation genes, several others link amyloid processing and diseases associated with P. gingivalis. While these differential expression results are uncertain due to a batch confound, the prior associations of these genes suggest follow-up experiments.
Recently, an in situ examination of human periodontitis gingiva samples identified a markedly increased presence of microvasculature with heavily invaded microvessels by live P. gingivalis (64). In support, we observe strong up-regulation of endothelial marker genes in brain samples with detected P. gingivalis reads. This upregulation indicates a higher proportion of endothelial cells. Together, these findings suggest the vasculature as a potential dissemination route for the microorganism from the oral cavity.
Genes that function in cell-cell contact were down-regulated in brain tissue with detected P. gingivalis. Specifically, the top two GO groups enriched for down-regulation were “homophilic cell adhesion via plasma membrane adhesion molecules” and “regulation of synaptic plasticity”. These are independent signals as only two genes are in both of these sets. A study of cell-free mRNA found that “homophilic cell adhesion via plasma membrane adhesion molecules” was the most significantly enriched GO group within genes down-regulated in AD cases (65). In mice, Huang and colleagues found that P. gingivalis infection led to synaptic loss in cerebral cortex neurons. They determined this synaptic failure was driven by Cathepsin B mediated interleukin-1β upregulation in leptomeningeal cells (66). In support, a recent study by Haditsch et al. found that neurons derived from human inducible pluripotent stem cells, when infected with P. gingivalis, had a significant loss of synapse density (67). This loss was more pronounced than neuron cell death. In agreement, our results do not suggest differences in estimated neuron proportions. It is known that synapse loss is an early and significant event in Alzheimer’s disease (68). In addition, in the aging and Alzheimer’s brain, decreases in synapse number and gene expression are observed but not neuron counts (69, 70). Haditsch and colleagues suggest synapse loss is due to microtubule destabilization caused by tau degradation by gingipains. In support, the most differentially expressed gene in the cell adhesion GO group is microtubule-associated protein tau (MAPT, pFDR < 0.005, 198th most down-regulated gene). However, our results suggest that transcriptional responses may partially cause reduced synapse density and cell adhesion. Cell-to-cell contact has been shown to increase P. gingivalis transmission rate (71, 72), suggesting that these transcriptional responses may reduce P. gingivalis persistence.
The ribosome, RNA processing, and protein synthesis have been previously associated with mild cognitive impairment, atherosclerosis, and Alzheimer’s disease (73–76). In agreement with gingipain susceptibility, a proteomic study found a lower abundance of RNA splicing proteins in cerebral atherosclerosis (77). Dysregulated splicing is also demonstrated by increased intron retention in Alzheimer’s disease (78). Similarly, RNA quality, which is measured from ribosomal RNA is lower in brain tissue from dementia cases (79). However, gingipains did not clearly cleave the ribosome in our experiments. We suspect this is due to the stability of the ribosome and inability to detect the cleavage of small ribosomal proteins. Pathological tau, a key marker of Alzheimer’s disease, has been shown to affect translational selectivity and associate with ribosomes on the ER [reviewed in (80)]. Linking arginine, tau was also found to strongly bind transfer RNAs with a preference for tRNAArg (81). Also, major histone acetylation differences have been associated with tau pathology (82). These findings of disruptions in processes involving nucleic acid interactions, match the functions of proteins enriched for arginine and lysine residues.
While proteins that interact with RNA and DNA are broadly enriched for arginine and lysine, protein targeting to the ER is the top GO group enriched for up-regulation in samples with detected P. gingivalis. While speculative, the ER translocation genes point to mechanisms that support the gingipain hypothesis of Alzheimer’s disease. The gingipain hypothesis proposes that P. gingivalis infection of brain tissue causes Alzheimer’s disease (20). In infected human cells, P. gingivalis is found in vacuoles that contain undegraded ribosomes (83). P. gingivalis uses amino acids as an energy source (84). Colocalized ribosomes may provide a particularly digestible source of amino acids because of their enrichment for the positively charged residues that gingipains cleave. Mirroring the movement of the SRP after recognizing a signal peptide, cytosolically free P. gingivalis colocalizes with the rough ER upon cell entry (85). It then forms autophagosome-like vacuoles, which support intracellular persistence and multiplication (85). It is tempting to speculate that sequestration of ribosomes in autophagosome-like vacuoles by P. gingivalis may cause increased transcription of cytosolic ribosomal protein genes because ribosome biogenesis is highly regulated (86).
The anterior hypothalamic area, oxytocin-, orexin- and vasopressin-expressing neurons were strongly enriched for our P. gingivalis associated genes. The hypothalamus releases these neuropeptides and peptide hormones after extensive pre-processing in the ER. Evidence of hypothalamic dysfunction and decreases in hypothalamic volume have been found in Alzheimer’s disease patients [reviewed in (87)]. Functionally, significant deficits in sleep and circadian rhythm have been reported with associations to vasopressin in human studies (88). Orexin, which plays a major role in the sleep-wake cycle, has been associated with amyloid pathology in mouse models (89). In Alzheimer’s patients, orexin levels in cerebrospinal fluid were correlated with amyloid-β42, sleep disruption and fragmentation (90). In addition, galanin, which is strongly expressed in the hypothalamic neurons enriched in our analysis, is associated with sleep fragmentation in Alzheimer’s disease (91). Hypothalamic dysfunction in Alzheimer’s disease and its enrichment for P. gingivalis associated genes suggest this region is relevant to the gingipain hypothesis.
Our findings associate the substantia innominata and cholinergic neurons with P. gingivalis transcriptomic response and gingipain susceptibility. Enrichment in the medial habenula, a region with high expression of nicotinic cholinergic receptors, is also observed (92). While basal forebrain neurons receive inputs from many cells, rodent studies have found 2-11% are from the hypothalamus (93, 94). These circuits that connect regions with the highest P. gingivalis associated enrichment have been explored in the context of cognitive impairment and Alzheimer’s disease. Specifically, studies have highlighted the hypothalamic-pituitary-adrenal axis and upstream modulation of specific cortical function (95, 96). More directly, extensive loss of cholinergic neurons in the substantia innominata has been found in Alzheimer’s patients (97). This finding and many others form the basis of the cholinergic hypothesis of AD, which proposes that degeneration of cholinergic neurons in the basal forebrain substantially contributes to cognitive decline in AD patients (49). This subcortical degeneration is thought to occur early in the disease process (98). In the context of the gingipain hypothesis, we note that reduced basal forebrain volume and cholinergic function follow the removal of teeth in rodents (99–101), and oral acetylcholine levels are correlated with periodontal disease severity (102). High expression of the ER translocation genes in cholinergic neurons may be required to support acetylcholinesterase processing in the ER (103). In the mouse atlas, cholinergic neurons in the enteric nervous system had the highest expression of the ER translocation genes. In the context of AD, loss of enteric cholinergic neurons has been observed in a transgenic mouse model of the disease (104). Taken together, our P. gingivalis associated genes highlight cholinergic neurons and the substantia innominata, providing an anatomical between the gingipain and cholinergic hypotheses.
In conclusion, we show that proteins enriched for arginine and lysine residues, which are potential gingipain cleavage sites, bind RNA and DNA. Neuroanatomical analysis of the P. gingivalis associated genes marked cholinergic neurons, the basal forebrain, and hypothalamic regions. These results link the gingipain and cholinergic hypotheses of AD. Our findings detail P. gingivalis response and susceptibility at the molecular and anatomical levels that suggest new associations relevant to AD pathogenesis.
Scripts, supplementary tables, and data files for reproducing the analyses are publicly available online at https://figshare.com/articles/dataset/Susceptibility_to_gingipains_and_transcriptomic_response_to_P_gingivalis_highlights_the_ribosome_hypothalamus_and_cholinergic_neurons/12782576 and https://github.com/leonfrench/gingipain_release.
SP, DH, and LF designed and performed bioinformatic analyses. NC, CD, BW, and ÖY designed and performed the protease digestion experiments. All authors contributed to the drafting of the manuscript. ÖY and LF supervised the research. All authors contributed to the article and approved the submitted version.
This study was supported by the CAMH Foundation, CAMH Discovery Fund, and a National Science and Engineering Research Council of Canada (NSERC) Discovery Grant to LF. This work was also supported by funding from the NIDCR grants, R56DE016593, R01DE030313, and R01DE030313S1.
LF owns shares in, and has received consulting fees from Cortexyme Inc., a company that is developing gingipain inhibitors to treat neurodegenerative diseases.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Navona Calarco for help with the analysis of the single-cell atlas of the mouse nervous system. We thank the Allen Institute for Brain Science for creating the transcriptomic atlas of the human brain. We thank Ed Lein, Michael Hawrylycz, Jeremy Miller, Taylor Schmitz, Shreejoy Tripathy, Karina Carneiro, and Daniel Felsky for their insightful comments and suggestions.
The Supplementary Material for this article can be found online at: https://figshare.com/articles/dataset/Susceptibility_to_gingipains_and_transcriptomic_response_to_P_gingivalis_highlights_the_ribosome_hypothalamus_and_cholinergic_neurons/12782576
Supplementary Table 1 | Proteome-wide arginine and lysine counts and proportion.
Supplementary Table 2 | Gene Ontology enrichment results for arginine and lysine proportions.
Supplementary Table 3 | SRP-dependent cotranslational protein targeting to membrane amino acid specificity tests.
Supplementary Table 4 | Results of differential expression tests for P. gingivalis detection.
Supplementary Table 5 | Gene Ontology enrichment results for genes associated with P. gingivalis detection.
Supplementary Table 6 | Neuroanatomical enrichment results for genes associated with P. gingivalis detection.
Supplementary Table 7 | Nervous system cell-type enrichment results for genes associated with P. gingivalis detection.
Supplementary Table 8 | Neuroanatomical enrichment results for the SRP-dependent cotranslational protein targeting to membrane genes.
Supplementary Table 9 | Nervous system cell-type enrichment results for the SRP-dependent cotranslational protein targeting to membrane genes.
Supplementary Figure 1 | Gingipain digestion of 70S ribosomes. Fifteen microliter reactions containing 2.5 ug of 70S ribosome and either 120 nM of recombinant Lys-gingipain (rKgp), Arg-gingipains (rRgpA and rRgpB), or about 206 – 412 µM (one unit in a 15 µL reaction volume) of Caspase-3 were incubated at 30°C for 1 h or 3 h. After incubation, results were visualized using SDS PAGE &Coomassie brilliant blue staining. The Precision Plus Protein WesternC Dual Color ladder was used for band sizing. The asterisk (*) to the left of the ribosome only control indicates a faint band present at about 60 kDa. This faint band appeared to be digested by Kgp and RgpB after 1 and 3 hours of incubation, as indicated by carets (^) to the left of the Kgp and RgpB.
AD, Alzheimer’s disease; AmG, amygdala; ApoE, apolipoprotein E; AUC, area under the receiver operating characteristic curve; BF, basal forebrain; BG, basal ganglia; CHAT, choline o-acetyltransferase; EP, epithalamus; ER, endoplasmic reticulum; FL, frontal lobe; GO, Gene Ontology; HiF, hippocampal formation; Hy, hypothalamus; ITM2B, integral membrane protein 2B; Kgp, Lys-gingipain; MAPT, microtubule-associated protein tau; MAZ, Myc associated zinc finger protein; MES, mesencephalon; MET, metencephalon; MY, myelencephalon; NOS, nitric oxide synthase; OL, occipital lobe; P. gingivalis, Porphyromonas gingivalis; PL, parietal lobe; RgpA, gingipain R1; RgpB, gingipain R2; RPL41, ribosomal protein L41; SAF-1, serum amyloid A-activating factor-1; SRP, signal recognition particle; SRP9, signal recognition particle 9; TH, thalamus; TL, temporal lobe.
1. Fiorillo L, Cervino G, Laino L, D’Amico C, Mauceri R, Tozum TF, et al. Porphyromonas Gingivalis, Periodontal and Systemic Implications: A Systematic Review. Dent J (2019) 7:114. doi: 10.3390/dj7040114
2. Guo Y, Nguyen K-A, Potempa J. Dichotomy of Gingipains Action as Virulence Factors: From Cleaving Substrates With the Precision of a Surgeon’s Knife to a Meat Chopper-Like Brutal Degradation of Proteins. Periodontol 2000 (2010) 54:15–44. doi: 10.1111/j.1600-0757.2010.00377.x
3. de Diego I, de Diego I, Veillard F, Sztukowska MN, Guevara T, Potempa B, et al. Structure and Mechanism of Cysteine Peptidase Gingipain K (Kgp), A Major Virulence Factor of Porphyromonas Gingivalisin Periodontitis. J Biol Chem (2014) 289:32291–302. doi: 10.1074/jbc.m114.602052
4. Bostanci N, Belibasakis GN. Porphyromonas Gingivalis: An Invasive and Evasive Opportunistic Oral Pathogen. FEMS Microbiol Lett (2012) 333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x
5. Blanco C, Bayas M, Yan F, Chen IA. Analysis of Evolutionarily Independent Protein-RNA Complexes Yields a Criterion to Evaluate the Relevance of Prebiotic Scenarios. Curr Biol (2018) 28:526–37.e5. doi: 10.1016/j.cub.2018.01.014
6. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc Natl Acad Sci USA (2001) 98:10037–41. doi: 10.1073/pnas.181342398
7. Olsen I, Yilmaz Ö. Possible Role of Porphyromonas Gingivalis in Orodigestive Cancers. J Oral Microbiol (2019) 11:1563410. doi: 10.1080/20002297.2018.1563410
8. Bingham CO3, Moni M. Periodontal Disease and Rheumatoid Arthritis: The Evidence Accumulates for Complex Pathobiologic Interactions. Curr Opin Rheumatol (2013) 25:345–53. doi: 10.1097/BOR.0b013e32835fb8ec
9. Bui FQ, Almeida-da-Silva CLC, Huynh B, Trinh A, Liu J, Woodward J, et al. Association Between Periodontal Pathogens and Systemic Disease. BioMed J (2019) 42:27–35. doi: 10.1016/j.bj.2018.12.001
10. Kellesarian SV, Yunker M, Malmstrom H, Almas K, Romanos GE, Javed F. Male Infertility and Dental Health Status: A Systematic Review. Am J Mens Health (2018) 12:1976–84. doi: 10.1177/1557988316655529
11. Singhrao SK, Olsen I. Assessing the Role of Porphyromonas Gingivalis in Periodontitis to Determine a Causative Relationship With Alzheimer’s Disease. J Oral Microbiol (2019) 11:1563405. doi: 10.1080/20002297.2018.1563405
12. Nadim R, Tang J, Dilmohamed A, Yuan S, Wu C, Bakre AT, et al. Influence of Periodontal Disease on Risk of Dementia: A Systematic Literature Review and a Meta-Analysis. Eur J Epidemiol (2020) 12:1–3. doi: 10.1007/s10654-020-00648-x
13. Ahn J, Segers S, Hayes RB. Periodontal Disease, Porphyromonas Gingivalis Serum Antibody Levels and Orodigestive Cancer Mortality. Carcinogenesis (2012) 33:1055–8. doi: 10.1093/carcin/bgs112
14. Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody Responses to Porphyromonas Gingivalis (P. Gingivalis) in Subjects With Rheumatoid Arthritis and Periodontitis. Int Immunopharmacol (2009) 9:38–42. doi: 10.1016/j.intimp.2008.09.008
15. Klareskog L, Rönnelid J, Lundberg K, Padyukov L, Alfredsson L. Immunity to Citrullinated Proteins in Rheumatoid Arthritis. Annu Rev Immunol (2008) 26:651–75. doi: 10.1146/annurev.immunol.26.021607.090244
16. Nakano K, Inaba H, Nomura R, Nemoto H, Takeda M, Yoshioka H, et al. Detection of Cariogenic Streptococcus Mutans in Extirpated Heart Valve and Atheromatous Plaque Specimens. J Clin Microbiol (2006) 44:3313–7. doi: 10.1128/JCM.00377-06
17. Haraszthy VI, Zambon JJ, Trevisan M, Zeid M, Genco RJ. Identification of Periodontal Pathogens in Atheromatous Plaques. J Periodontol (2000) 71:1554–60. doi: 10.1902/jop.2000.71.10.1554
18. Lönn J, Ljunggren S, Klarström-Engström K, Demirel I, Bengtsson T, Karlsson H. Lipoprotein Modifications by Gingipains of Porphyromonas Gingivalis. J Periodontal Res (2018) 53:403–13. doi: 10.1111/jre.12527
19. Liu C-C, Liu C-C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer Disease: Risk, Mechanisms and Therapy. Nat Rev Neurol (2013) 9:106–18. doi: 10.1038/nrneurol.2012.263
20. Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas Gingivalis in Alzheimer’s Disease Brains: Evidence for Disease Causation and Treatment With Small-Molecule Inhibitors. Sci Adv (2019) 5:eaau3333. doi: 10.1126/sciadv.aau3333
21. Ilievski V, Zuchowska PK, Green SJ, Toth PT, Ragozzino ME, Le K, et al. Chronic Oral Application of a Periodontal Pathogen Results in Brain Inflammation, Neurodegeneration and Amyloid Beta Production in Wild Type Mice. PloS One (2018) 13:e0204941. doi: 10.1371/journal.pone.0204941
22. Poole S, Singhrao SK, Chukkapalli S, Rivera M, Velsko I, Kesavalu L, et al. Active Invasion of Porphyromonas Gingivalis and Infection-Induced Complement Activation in ApoE-/- Mice Brains. J Alzheimers Dis (2015) 43:67–80. doi: 10.3233/JAD-140315
23. Ishida N, Ishihara Y, Ishida K, Tada H, Funaki-Kato Y, Hagiwara M, et al. Periodontitis Induced by Bacterial Infection Exacerbates Features of Alzheimer’s Disease in Transgenic Mice. NPJ Aging Mech Dis (2017) 3:15. doi: 10.1038/s41514-017-0015-x
24. Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, et al. GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res (2019) 47:D766–73. doi: 10.1093/nar/gky955
25. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat Genet (2000) 25:25–9. doi: 10.1038/75556
28. Zhang L, Veith PD, Huq NL, Chen Y-Y, Seers CA, Cross KJ, et al. Porphyromonas Gingivalis Gingipains Display Transpeptidation Activity. J Proteome Res (2018) 17:2803–18. doi: 10.1021/acs.jproteome.8b00286
29. Julien O, Wells JA. Caspases and Their Substrates. Cell Death Differ (2017) 24:1380–9. doi: 10.1038/cdd.2017.44
30. Dhar R, Persaud SD, Mireles JR, Basu A. Proteolytic Cleavage of p70 Ribosomal S6 Kinase by Caspase-3 During DNA Damage-Induced Apoptosis. Biochemistry (2009) 48:1474–80. doi: 10.1021/bi801840s
31. Dumitriu A, Golji J, Labadorf AT, Gao B, Beach TG, Myers RH, et al. Integrative Analyses of Proteomics and RNA Transcriptomics Implicate Mitochondrial Processes, Protein Folding Pathways and GWAS Loci in Parkinson Disease. BMC Med Genomics (2016) 9:5. doi: 10.1186/s12920-016-0164-y
32. Leinonen R, Sugawara H, Shumway M. International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res (2011) 39:D19–21. doi: 10.1093/nar/gkq1019
33. Lim N, Tesar S, Belmadani M, Poirier-Morency G, Mancarci BO, Sicherman J, et al. Curation of Over 10, 000 Transcriptomic Studies to Enable Data Reuse. Database (2021) 2021:baab006. doi: 10.1093/database/baab006
34. Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A Survey of Human Brain Transcriptome Diversity At the Single Cell Level. Proc Natl Acad Sci USA (2015) 112:7285–90. doi: 10.1073/pnas.1507125112
35. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An Anatomically Comprehensive Atlas of the Adult Human Brain Transcriptome. Nature (2012) 489:391–9. doi: 10.1038/nature11405
36. Miller JA, Menon V, Goldy J, Kaykas A, Lee C-K, Smith KA, et al. Improving Reliability and Absolute Quantification of Human Brain Microarray Data by Filtering and Scaling Probes Using RNA-Seq. BMC Genomics (2014) 15:154. doi: 10.1186/1471-2164-15-154
37. Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. Molecular Architecture of the Mouse Nervous System. Cell (2018) 174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021
38. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference Sequence (RefSeq) Database At NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res (2016) 44:D733–45. doi: 10.1093/nar/gkv1189
39. Lott BB, Wang Y, Nakazato T. A Comparative Study of Ribosomal Proteins: Linkage Between Amino Acid Distribution and Ribosomal Assembly. BMC Biophys (2013) 6:13. doi: 10.1186/2046-1682-6-13
40. Lundmark A, Gerasimcik N, Båge T, Jemt A, Mollbrink A, Salmén F, et al. Gene Expression Profiling of Periodontitis-Affected Gingival Tissue by Spatial Transcriptomics. Sci Rep (2018) 8:9370. doi: 10.1038/s41598-018-27627-3
41. Ludden CW. Transcriptomic Evaluation of Peri-Implant Soft Tissue in Health and Disease (2015). Available at: https://etd.ohiolink.edu/!etd.send_file?accession=osu1435233979&disposition=inline.
42. Felsky D, Sariya S, Schneider JA, Bennett DA, Mayeux R, De Jager PL, et al. The Caribbean‐Hispanic Alzheimer’s brain transcriptome reveals ancestry‐specific disease mechanisms: Genetics/omics and systems biology. Alzheimers Dement (2020) 16:e043068.
43. Patel S, Howard D, Man A, Schwartz D, Jee J, Felsky D, et al. Donor Specific Transcriptomic Analysis of Alzheimer’s Disease Associated Hypometabolism Highlights a Unique Donor, Ribosomal Proteins and Microglia. eNeuro (2020) 7:1–11. doi: 10.1523/ENEURO.0255-20.2020
44. Nguyen AT, Wang K, Hu G, Wang X, Miao Z, Azevedo JA, et al. APOE and TREM2 Regulate Amyloid-Responsive Microglia in Alzheimer’s Disease. Acta Neuropathol (2020) 140:477–93. doi: 10.1007/s00401-020-02200-3
45. Birse DE, Kapp U, Strub K, Cusack S, Aberg A. The Crystal Structure of the Signal Recognition Particle Alu RNA Binding Heterodimer, SRP9/14. EMBO J (1997) 16:3757–66. doi: 10.1093/emboj/16.13.3757
46. Braak H, Braak E. Neuropathological Stageing of Alzheimer-Related Changes. Acta Neuropathol (1991) 82:239–59. doi: 10.1007/BF00308809
47. Mesulam MM, Mufson EJ, Wainer BH, Levey AI. Central Cholinergic Pathways in the Rat: An Overview Based on an Alternative Nomenclature (Ch1-Ch6). Neuroscience (1983) 10:1185–201. doi: 10.1016/0306-4522(83)90108-2
48. Sims NR, Bowen DM, Allen SJ, Smith CC, Neary D, Thomas DJ, et al. Presynaptic Cholinergic Dysfunction in Patients With Dementia. J Neurochem (1983) 40:503–9. doi: 10.1111/j.1471-4159.1983.tb11311.x
49. Hampel H, Mesulam M-M, Cuello AC, Khachaturian AS, Vergallo A, Farlow MR, et al. Revisiting the Cholinergic Hypothesis in Alzheimer’s Disease: Emerging Evidence From Translational and Clinical Research. J Prev Alzheimer’s Dis (2019) 6:2–15. doi: 10.14283/jpad.2018.43
50. DeKosky ST, Harbaugh RE, Schmitt FA, Bakay RA, Chui HC, Knopman DS, et al. Cortical Biopsy in Alzheimer’s Disease: Diagnostic Accuracy and Neurochemical, Neuropathological, and Cognitive Correlations. Intraventricular Bethanecol Study Group. Ann Neurol (1992) 32:625–32. doi: 10.1002/ana.410320505
51. Ding S-L, Royall JJ, Sunkin SM, Ng L, Facer BAC, Lesnar P, et al. Comprehensive Cellular-Resolution Atlas of the Adult Human Brain. J Comp Neurol (2016) 524:3127–481. doi: 10.1002/cne.24097
52. Fehlbaum-Beurdeley P, Sol O, Désiré L, Touchon J, Dantoine T, Vercelletto M, et al. Validation of AclarusDxTM, A Blood-Based Transcriptomic Signature for the Diagnosis of Alzheimer’s Disease. J Alzheimers Dis (2012) 32:169–81.
53. Kim J, Miller VM, Levites Y, West KJ, Zwizinski CW, Moore BD, et al. BRI2 (ITM2b) Inhibits Abeta Deposition In Vivo. J Neurosci (2008) 28:6030–6. doi: 10.1523/JNEUROSCI.0891-08.2008
54. Vidal R, Frangione B, Rostagno A, Mead S, Révész T, Plant G, et al. A Stop-Codon Mutation in the BRI Gene Associated With Familial British Dementia. Nature (1999) 399:776–81. doi: 10.1038/21637
55. Vidal R, Revesz T, Rostagno A, Kim E, Holton JL, Bek T, et al. A Decamer Duplication in the 3’ Region of the BRI Gene Originates an Amyloid Peptide That is Associated With Dementia in a Danish Kindred. Proc Natl Acad Sci USA (2000) 97:4920–5. doi: 10.1073/pnas.080076097
56. Ray A, Ray BK. A Novel Cis-Acting Element Is Essential for Cytokine-Mediated Transcriptional Induction of the Serum Amyloid A Gene in Nonhepatic Cells. Mol Cell Biol (1996) 16:1584–94. doi: 10.1128/MCB.16.4.1584
57. Liu L, Lai Y-J, Zhao L-G, Chen G-J. Increased Expression of Myc-Interacting Zinc Finger Protein 1 in APP/PS1 Mice. Exp Ther Med (2017) 14:5751–6. doi: 10.3892/etm.2017.5289
58. Ray A, Kumar D, Shakya A, Brown CR, Cook JL, Ray BK. Serum Amyloid A-activating Factor-1 (SAF-1) Transgenic Mice Are Prone to Develop a Severe Form of Inflammation-Induced Arthritis. J Immunol (2004) 173:4684–91. doi: 10.4049/jimmunol.173.7.4684
59. Kindy MS, Yu J, Guo J-T, Zhu H. Apolipoprotein Serum Amyloid A in Alzheimer’s Disease. J Alzheimers Dis (1999) 1:155–67. doi: 10.3233/JAD-1999-1303
60. Ray A, Shakya A, Kumar D, Benson MD, Ray BK. Inflammation-Responsive Transcription Factor SAF-1 Activity Is Linked to the Development of Amyloid A Amyloidosis. J Immunol (2006) 177:2601–9. doi: 10.4049/jimmunol.177.4.2601
61. Getz GS, Krishack PA, Reardon CA. Serum Amyloid A and Atherosclerosis. Curr Opin Lipidol (2016) 27:531–5. doi: 10.1097/MOL.0000000000000331
62. Targońska-Stępniak B, Majdan M. Serum Amyloid A as a Marker of Persistent Inflammation and an Indicator of Cardiovascular and Renal Involvement in Patients With Rheumatoid Arthritis. Mediators Inflamm (2014) 2014:793628. doi: 10.1155/2014/793628
63. Zhang X, Zhu C, Beecham G, Vardarajan BN, Ma Y, Lancour D, et al. A Rare Missense Variant of CASP7 Is Associated With Familial Late-Onset Alzheimer’s Disease. Alzheimers Dement (2019) 15:441–52. doi: 10.1016/j.jalz.2018.10.005
64. Lee JS, Spooner R, Chowdhury N, Pandey V, Wellslager B, Atanasova KR, et al. In Situ Intraepithelial Localizations of Opportunistic Pathogens, Porphyromonas Gingivalis and Filifactor Alocis, in Human Gingiva. Curr Res Microbial Sci (2020) 1:7–17. doi: 10.1016/j.crmicr.2020.05.001
65. Toden S, Zhuang J, Acosta AD, Karns AP, Salathia NS, Brewer JB, et al. Noninvasive Characterization of Alzheimer’s Disease by Circulating, Cell-Free Messenger RNA Next-Generation Sequencing. Sci Adv (2020) 6:eabb1654. doi: 10.1126/sciadv.abb1654
66. Huang W, Zeng F, Gu Y, Jiang M, Zhang X, Yan X, et al. Leptomeningeal Cells Induce Synaptic Failure Via Cathepsin B-Mediated IL-1β Production After Porphyromonas Gingivalis Infection. Preprint (2020). doi: 10.21203/rs.3.rs-111412/v1
67. Haditsch U, Roth T, Rodriguez L, Hancock S, Cecere T, Nguyen M, et al. Alzheimer’s Disease-Like Neurodegeneration in Porphyromonas Gingivalis Infected Neurons With Persistent Expression of Active Gingipains. J Alzheimer’s Dis (2020) 75:1361–76. doi: 10.3233/jad-200393
68. Scheff SW, Price DA, Ansari MA, Roberts KN, Schmitt FA, Ikonomovic MD, et al. Synaptic Change in the Posterior Cingulate Gyrus in the Progression of Alzheimer’s Disease. J Alzheimers Dis (2015) 43:1073–90. doi: 10.3233/JAD-141518
69. French L, Ma T, Oh H, Tseng GC, Sibille E. Age-Related Gene Expression in the Frontal Cortex Suggests Synaptic Function Changes in Specific Inhibitory Neuron Subtypes. Front Aging Neurosci (2017) 9:162. doi: 10.3389/fnagi.2017.00162
70. Mostany R, Anstey JE, Crump KL, Maco B, Knott G, Portera-Cailliau C. Altered Synaptic Dynamics During Normal Brain Aging. J Neurosci (2013) 33:4094–104. doi: 10.1523/JNEUROSCI.4825-12.2013
71. Li L, Michel R, Cohen J, Decarlo A, Kozarov E. Intracellular Survival and Vascular Cell-to-Cell Transmission of Porphyromonas Gingivalis. BMC Microbiol (2008) 8:26. doi: 10.1186/1471-2180-8-26
72. Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular Spreading of Porphyromonas Gingivalis Infection in Primary Gingival Epithelial Cells. Infect Immun (2006) 74:703–10. doi: 10.1128/IAI.74.1.703-710.2006
73. Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome Dysfunction is an Early Event in Alzheimer’s Disease. J Neurosci (2005) 25:9171–5. doi: 10.1523/JNEUROSCI.3040-05.2005
74. Langstrom NS, Anderson JP, Lindroos HG, Winblad B, Wallace WC. Alzheimer’s Disease-Associated Reduction of Polysomal mRNA Translation. Brain Res Mol Brain Res (1989) 5:259–69. doi: 10.1016/0169-328X(89)90060-0
75. Hernández-Ortega K, Garcia-Esparcia P, Gil L, Lucas JJ, Ferrer I. Altered Machinery of Protein Synthesis in Alzheimer’s: From the Nucleolus to the Ribosome. Brain Pathol (2016) 26:593–605. doi: 10.1111/bpa.12335
76. Nyhus C, Pihl M, Hyttel P, Hall VJ. Evidence for Nucleolar Dysfunction in Alzheimer’s Disease. Rev Neurosci (2019) 30(7):685–700. doi: 10.1515/revneuro-2018-0104
77. Wingo AP, Fan W, Duong DM, Gerasimov ES, Dammer EB, Liu Y, et al. Shared Proteomic Effects of Cerebral Atherosclerosis and Alzheimer’s Disease on the Human Brain. Nat Neurosci (2020) 23:696–700. doi: 10.1038/s41593-020-0635-5
78. Adusumalli S, Ngian Z-K, Lin W-Q, Benoukraf T, Ong C-T. Increased Intron Retention is a Post-Transcriptional Signature Associated With Progressive Aging and Alzheimer’s Disease. Aging Cell (2019) 18:e12928. doi: 10.1111/acel.12928
79. Miller JA, Guillozet-Bongaarts A, Gibbons LE, Postupna N, Renz A, Beller AE, et al. Neuropathological and Transcriptomic Characteristics of the Aged Brain. Elife (2017) 6:e31126. doi: 10.7554/eLife.31126
80. Koren SA, Galvis-Escobar S, Abisambra JF. Tau-Mediated Dysregulation of RNA: Evidence for a Common Molecular Mechanism of Toxicity in Frontotemporal Dementia and Other Tauopathies. Neurobiol Dis (2020) 141:104939. doi: 10.1016/j.nbd.2020.104939
81. Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, Hernandez I, et al. RNA Stores Tau Reversibly in Complex Coacervates. PloS Biol (2017) 15:e2002183. doi: 10.1371/journal.pbio.2002183
82. Klein H-U, McCabe C, Gjoneska E, Sullivan SE, Kaskow BJ, Tang A, et al. Epigenome-Wide Study Uncovers Large-Scale Changes in Histone Acetylation Driven by Tau Pathology in Aging and Alzheimer’s Human Brains. Nat Neurosci (2019) 22:37–46. doi: 10.1038/s41593-018-0291-1
83. Dorn BR, Dunn WA Jr, Progulske-Fox A. Porphyromonas Gingivalis Traffics to Autophagosomes in Human Coronary Artery Endothelial Cells. Infect Immun (2001) 69:5698–708. doi: 10.1128/IAI.69.9.5698-5708.2001
84. Nelson KE, Fleischmann RD, DeBoy RT, Paulsen IT, Fouts DE, Eisen JA, et al. Complete Genome Sequence of the Oral Pathogenic Bacterium Porphyromonas Gingivalis Strain W83. J Bacteriol (2003) 185:5591–601. doi: 10.1128/JB.185.18.5591-5601.2003
85. Lee K, Roberts JS, Choi CH, Atanasova KR, Yilmaz Ö. Porphyromonas Gingivalis Traffics Into Endoplasmic Reticulum-Rich-Autophagosomes for Successful Survival in Human Gingival Epithelial Cells. Virulence (2018) 9:845–59. doi: 10.1080/21505594.2018.1454171
86. Lempiäinen H, Shore D. Growth Control and Ribosome Biogenesis. Curr Opin Cell Biol (2009) 21:855–63. doi: 10.1016/j.ceb.2009.09.002
87. Hiller AJ, Ishii M. Disorders of Body Weight, Sleep and Circadian Rhythm as Manifestations of Hypothalamic Dysfunction in Alzheimer’s Disease. Front Cell Neurosci (2018) 12:471. doi: 10.3389/fncel.2018.00471
88. Hu K, Harper DG, Shea SA, Stopa EG, Scheer FAJL. Noninvasive Fractal Biomarker of Clock Neurotransmitter Disturbance in Humans With Dementia. Sci Rep (2013) 3:2229. doi: 10.1038/srep02229
89. Roh JH, Jiang H, Finn MB, Stewart FR, Mahan TE, Cirrito JR, et al. Potential Role of Orexin and Sleep Modulation in the Pathogenesis of Alzheimer’s Disease. J Exp Med (2014) 211:2487–96. doi: 10.1084/jem.20141788
90. Gabelle A, Jaussent I, Hirtz C, Vialaret J, Navucet S, Grasselli C, et al. Cerebrospinal Fluid Levels of Orexin-a and Histamine, and Sleep Profile Within the Alzheimer Process. Neurobiol Aging (2017) 53:59–66. doi: 10.1016/j.neurobiolaging.2017.01.011
91. Lim ASP, Ellison BA, Wang JL, Yu L, Schneider JA, Buchman AS, et al. Sleep Is Related to Neuron Numbers in the Ventrolateral Preoptic/Intermediate Nucleus in Older Adults With and Without Alzheimer’s Disease. Brain (2014) 137:2847–61. doi: 10.1093/brain/awu222
92. Le Foll B, French L. Transcriptomic Characterization of the Human Habenula Highlights Drug Metabolism and the Neuroimmune System. Front Neurosci (2018) 12:742. doi: 10.3389/fnins.2018.00742
93. Gielow MR, Zaborszky L. The Input-Output Relationship of the Cholinergic Basal Forebrain. Cell Rep (2017) 18:1817–30. doi: 10.1016/j.celrep.2017.01.060
94. Zaborszky L, Gombkoto P. The Cholinergic Multicompartmental Basal Forebrain Microcircuit. In: Shepherd GM, Grillner S (eds). Handbook of Brain Microcircuits. New York, NY, USA: Oxford University Press. (2018). pp. 163–83
95. Paul S, Jeon WK, Bizon JL, Han J-S. Interaction of Basal Forebrain Cholinergic Neurons With the Glucocorticoid System in Stress Regulation and Cognitive Impairment. Front Aging Neurosci (2015) 7:43. doi: 10.3389/fnagi.2015.00043
96. Zaborszky L, van den Pol A, Gyengesi E. The Basal Forebrain Cholinergic Projection System in Mice. In: The Mouse Nervous System. San Diego: Academic Press (2012). p. 684–718.
97. Whitehouse PJ, Price DL, Struble RG, Clark AW, Coyle JT, Delon MR. Alzheimer’s Disease and Senile Dementia: Loss of Neurons in the Basal Forebrain. Science (1982) 215:1237–9. doi: 10.1126/science.7058341
98. Mesulam M, Shaw P, Mash D, Weintraub S. Cholinergic Nucleus Basalis Tauopathy Emerges Early in the Aging-MCI-AD Continuum. Ann Neurol (2004) 55:815–28. doi: 10.1002/ana.20100
99. Kato T, Usami T, Noda Y, Hasegawa M, Ueda M, Nabeshima T. The Effect of the Loss of Molar Teeth on Spatial Memory and Acetylcholine Release From the Parietal Cortex in Aged Rats. Behav Brain Res (1997) 83:239–42. doi: 10.1016/S0166-4328(97)86078-0
100. Onozuka M, Watanabe K, Fujita M, Tomida M, Ozono S. Changes in the Septohippocampal Cholinergic System Following Removal of Molar Teeth in the Aged SAMP8 Mouse. Behav Brain Res (2002) 133:197–204. doi: 10.1016/S0166-4328(02)00006-2
101. Avivi-Arber L, ev ‘Seltzer Z, Friedel M, Lerch JP, Moayedi M, Davis KD, et al. Widespread Volumetric Brain Changes Following Tooth Loss in Female Mice. Front Neuroanat (2016) 10:121. doi: 10.3389/fnana.2016.00121
102. Apatzidou DA, Iskas A, Konstantinidis A, Alghamdi AM, Tumelty M, Lappin DF, et al. Clinical Associations Between Acetylcholine Levels and Cholinesterase Activity in Saliva and Gingival Crevicular Fluid and Periodontal Diseases. J Clin Periodontol (2018) 45:1173–83. doi: 10.1111/jcpe.12989
103. Dobbertin A, Hrabovska A, Dembele K, Camp S, Taylor P, Krejci E, et al. Targeting of Acetylcholinesterase in Neurons In Vivo: A Dual Processing Function for the Proline-Rich Membrane Anchor Subunit and the Attachment Domain on the Catalytic Subunit. J Neurosci (2009) 29:4519–30. doi: 10.1523/JNEUROSCI.3863-08.2009
Keywords: Alzheimer’s disease, cholinergic system, gingipains, transcriptomics, hypothalamus, Porphyromonas gingivalis (P. gingivalis)
Citation: Patel S, Howard D, Chowdhury N, Derieux C, Wellslager B, Yilmaz Ö and French L (2021) Characterization of Human Genes Modulated by Porphyromonas gingivalis Highlights the Ribosome, Hypothalamus, and Cholinergic Neurons. Front. Immunol. 12:646259. doi: 10.3389/fimmu.2021.646259
Received: 25 December 2020; Accepted: 20 May 2021;
Published: 14 June 2021.
Edited by:
Teun J. De Vries, VU University Amsterdam, NetherlandsCopyright © 2021 Patel, Howard, Chowdhury, Derieux, Wellslager, Yilmaz and French. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Leon French, bGVvbmZyZW5jaEBnbWFpbC5jb20=
†ORCID: Özlem Yilmaz, orcid.org/0000-0003-3487-7217
‡These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.