
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 14 May 2021
Sec. T Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.642856
Invariant NKT (iNKT) cells are thymus-generated innate-like T cells, comprised of three distinct subsets with divergent effector functions. The molecular mechanism that drives the lineage trifurcation of immature iNKT cells into the NKT1, NKT2, and NKT17 subsets remains a controversial issue that remains to be resolved. Because cytokine receptor signaling is necessary for iNKT cell generation, cytokines are proposed to contribute to iNKT subset differentiation also. However, the precise roles and requirements of cytokines in these processes are not fully understood. Here, we show that IL-2Rβ, a nonredundant component of the IL-15 receptor complex, plays a critical role in both the development and differentiation of thymic iNKT cells. While the induction of IL-2Rβ expression on postselection thymocytes is necessary to drive the generation of iNKT cells, surprisingly, premature IL-2Rβ expression on immature iNKT cells was detrimental to their development. Moreover, while IL-2Rβ is necessary for NKT1 generation, paradoxically, we found that the increased abundance of IL-2Rβ suppressed NKT1 generation without affecting NKT2 and NKT17 cell differentiation. Thus, the timing and abundance of IL-2Rβ expression control iNKT lineage fate and development, thereby establishing cytokine receptor expression as a critical regulator of thymic iNKT cell differentiation.
Cytokines of the common γ-chain (γc) family play critical roles in the generation and differentiation of T cells in the thymus (1–3). Importantly, γc cytokines not only provide prosurvival signals and metabolic cues, but they also specify the lineage commitment of thymocytes and equip developing T cells with effector function (1). Along these lines, a requirement for intrathymic interleukin-7 (IL-7) in T cell development is well established, as documented in its role to control the expansion of immature thymocytes and to determine CD8 cytotoxic T cell lineage fate (2, 4). IL-4, on the other hand, drives the differentiation of innate-like CD8 T cells (5), whereas IL-2 signaling is critical for the generation of Foxp3+ T regulatory (Treg) cells (6). Collectively, γc cytokine signaling is necessary for the development of αβ T cells; however, the identity of the required cytokine differs depending on the individual lineages or subsets of T cells that are generated.
Cytokine signaling is also important for the generation of invariant NKT (iNKT) cells, a population of innate-like αβ T cells that are produced in the thymus (7, 8). iNKT cells are potent immunoregulatory cells that produce pro-inflammatory cytokines, but they differ from conventional αβ T cells because they acquire effector function before antigen encounter and activation (7, 8). Depending on the cytokines they produce and the transcription factors they express, iNKT cells can be categorized into three distinct subsets (9, 10). iNKT cells that are T-bet+ and produce IFNγ correspond to NKT1 cells. IL-17-producing RORγt+ iNKT cells, on the other hand, are NKT17 cells, and IL-4-producing PLZFhi iNKT cells are NKT2 cells (9, 10). The molecular mechanisms that drive iNKT subset differentiation are not fully mapped. However, cytokines have been implicated in determining iNKT subset specification. Thus, IL-15 promotes NKT1 cell development (11), TGF-β induces NKT17 cell generation (12), and IL-25 produced by thymic tuft cells possibly drives the differentiation of NKT2 cells (13). Consequently, cytokines can specify the differentiation of iNKT cells in the thymus. However, it remains unclear why some iNKT precursors would respond to IL-15 and become NKT1 cells, whereas other precursors are refractory to IL-15 and choose other lineage fates. This problem is further compounded by all iNKT cells expressing the same semi-invariant TCR so that differences in the TCR repertoire are unlikely to play a significant role in subset differentiation. Instead, we consider it likely that differences in the timing or abundance of cytokine receptors could determine the responsiveness to subset-specifying cytokines.
Among iNKT cells, the NKT1 subset is of particular interest because it comprises most of the thymic iNKT cells in C57BL/6 mice. NKT1 cells are also important because they produce copious amounts of IFNγ, which affects T cell lineage differentiation and promotes the terminal maturation of thymic epithelial cells (14, 15). NKT1 cells require IL-15 for their generation, whereby intrathymic IL-15 is primarily provided by medullary thymic epithelial cells (16, 17). IL-15 is a γc family cytokine that signals through γc and IL-2Rβ (3). Consistent with its requirement for IL-15 signaling, IL-2Rβ deficiency was previously shown to dramatically impair the generation of iNKT cells in the thymus (18, 19). However, it remains unclear whether the lack of IL-2Rβ would specifically impede the generation of NKT1 cells or the differentiation of all thymic iNKT subsets. Moreover, if IL-2Rβ is necessary for NKT1 cell differentiation, we wished to assess whether IL-2Rβ would be sufficient to drive NKT1 cell generation. If such were the case, we expected that the forced expression of IL-2Rβ would impose the NKT1 subset fate onto all developing iNKT cells, resulting in a heavily skewed iNKT subset composition where NKT1 cells would be overrepresented and NKT2 and NKT17 cells would be underrepresented. To this end, we examined the generation and differentiation of iNKT cells in IL-2RβTg mice in both the C57BL/6 and BALB/c backgrounds, and further generated mice with conditional deletion of IL-2Rβ upon iNKT lineage commitment. Here, we show that not only the expression itself but also the carefully curated timing of IL-2Rβ induction is important for the generation and subset differentiation of iNKT cells. Collectively, these results unveil previously unappreciated roles and requirements of the cytokine receptor IL-2Rβ in controlling thymic iNKT cell development.
C57BL/6NCrl (C57BL/6) and BALB/cAnNCrl (BALB/c) mice were obtained from the Charles River Laboratories (Frederick, MD). IL-2RβTg mice were generated in house as previously described (20). IL-2Rβ-deficient mice (Il2rb–/–) and IL-2Rβ-floxed mice (Il2rbfl/fl) were obtained from the Jackson Laboratories (21, 22). PLZFCre transgenic mice were kindly provided by Dr. D. Sant’Angelo (Rutgers University, NJ) (23). CA-STAT5Tg mice were a kind gift from by Dr. M. A. Farrar (U. Minnesota, MN) (24). T-bet-ZsGreen reporter transgenic mice (TBGRTg) on C57BL/6 background were a kind gift from Dr. Jinfang Zhu (NIAID, NIH) (25), and these mice were backcrossed to BALB/cAnNCrl for at least 6 generations before analysis. All experimental mice were analyzed between 6 and 12 weeks of age, except for Il2rb–/– and their littermate controls, which were analyzed between 4 and 6 weeks of age. All mice were cared for in accordance with NIH guidelines. Animal experiments were approved by the NCI Animal Care and Use Committee.
Single-cell suspensions were prepared from the indicated tissues and stained with fluorescence-conjugated antibodies as previously described (26). The data were acquired using LSR Fortessa or LSRII flow cytometers (BD Biosciences) and were analyzed using software platforms developed by the EIB Flow Cytometry Facility, CCR, NCI, NIH. Live cells were gated by forward scatter exclusion of dead cells stained with propidium iodide. The following antibodies were used for staining: HSA (M1/69), T-bet (4B10), Foxp3 (FJK-16s), RORγt (AKFJS-9) and isotype control antibodies, all from eBioscience; CD4 (GK1.5 and RM4.5), CD8α (53–6–7), CD69 (H1.2F3), TCRβ (H57-597) and IL2Rβ (TM-β1) from BD Biosciences; CD44 (IM7), NK1.1 (PK136), IL2Rα (PC61), CCR7 (4B12) and PLZF (9E12) from BioLegend. Fluorochrome-conjugated CD1d tetramers loaded with PBS-57 (CD1dTet) and unloaded controls were obtained from the NIH tetramer facility (Emory University, Atlanta, GA). Intracellular Foxp3, PLZF, RORγt, and T-bet proteins were detected using a Foxp3 staining kit according to the manufacturer’s instructions (eBioscience Thermo Fisher).
To identify the subset composition of iNKT cells, total thymocytes were first stained with PBS-57-loaded mouse CD1d tetramers, fixed, permeabilized and then stained for nuclear transcription factors. Briefly, thymocytes were stained with fluorochrome-conjugated CD1d tetramers in FACS buffer (0.5% BSA, 0.1% sodium azide in Ca2+ and Mg2+-free HBSS) for 20 minutes at 4°C. Without removing the tetramer reagents, the cells were incubated for an additional 30 min at 4°C with antibodies for surface markers. Excess reagents were then washed out with FACS buffer, and the cells were resuspended in a 1:3 mixture of concentrate/diluent working solution of the Foxp3 Transcription Factor Staining Buffer kit (eBioscience Thermo Fisher), followed by incubation at room temperature for 20 minutes. Next, the cells were washed twice with 1× permeabilization buffer (eBioscience Thermo Fisher) before adding antibodies for transcription factors, specifically antibodies against PLZF, RORγt and T-bet. The cells were incubated at room temperature for 1 hour before washing out excess reagents with FACS buffer and analysis by flow cytometry.
Recombinant mouse IL-15 (Peprotech) dissolved in PBS was administered into IL-2RβTg BALB/c mice using Alzet osmotic pumps (Durect) following the manufacturer’s instruction. The pumps (model 1002) were set to release IL-15 at a rate of 3 µg of recombinant IL-15 per 24 hours.
The data were shown as the means ± SEM. Statistical significance was determined by unpaired two-tailed Student’s t-test. P values of less than 0.05 were considered significant. *P < 0.05, **P < 0.01, ***P <0.001. All statistical analysis was performed using GraphPad Prism 7 software (GraphPad software).
To understand the role of IL-2Rβ in thymic iNKT cell generation, we first examined IL-2Rβ expression on iNKT cells in the context of αβ T cell development in the thymus. The process of thymocyte positive selection and maturation can be visualized using the combination of the two surface markers CD69 and CCR7 which identifies 5 distinct developmental stages, progressing from stage I to stage V (Figure 1A, top) (27). TCR engagement of immature thymocytes (stage I) induces the expression CD69 (stage II) followed by the upregulation of the chemokine receptor CCR7, so that CD69+CCR7int cells (stage III) correspond to thymocytes undergoing positive selection and lineage commitment. Stage IV and V cells, on the other hand, correspond to postselection thymocytes. We found that iNKT cells, as identified by PBS-57-loaded CD1d tetramers (CD1dTet+), arise as early as in stage I, and that they were then prominent in stage II (Figure 1A, bottom). Importantly, stage I iNKT cells expressed both the cytokine receptor γc and IL-2Rβ (Figure 1B and Supplementary Figure 1), indicating that the induction of IL-2Rβ is one of the earliest events in iNKT cell development. Indeed, such kinetics of IL-2Rβ expression differed substantially from that of CD1dTet-negative conventional αβ T cells, which remained mostly absent for IL-2Rβ expression throughout their development (Figure 1B and Supplementary Figure 2).
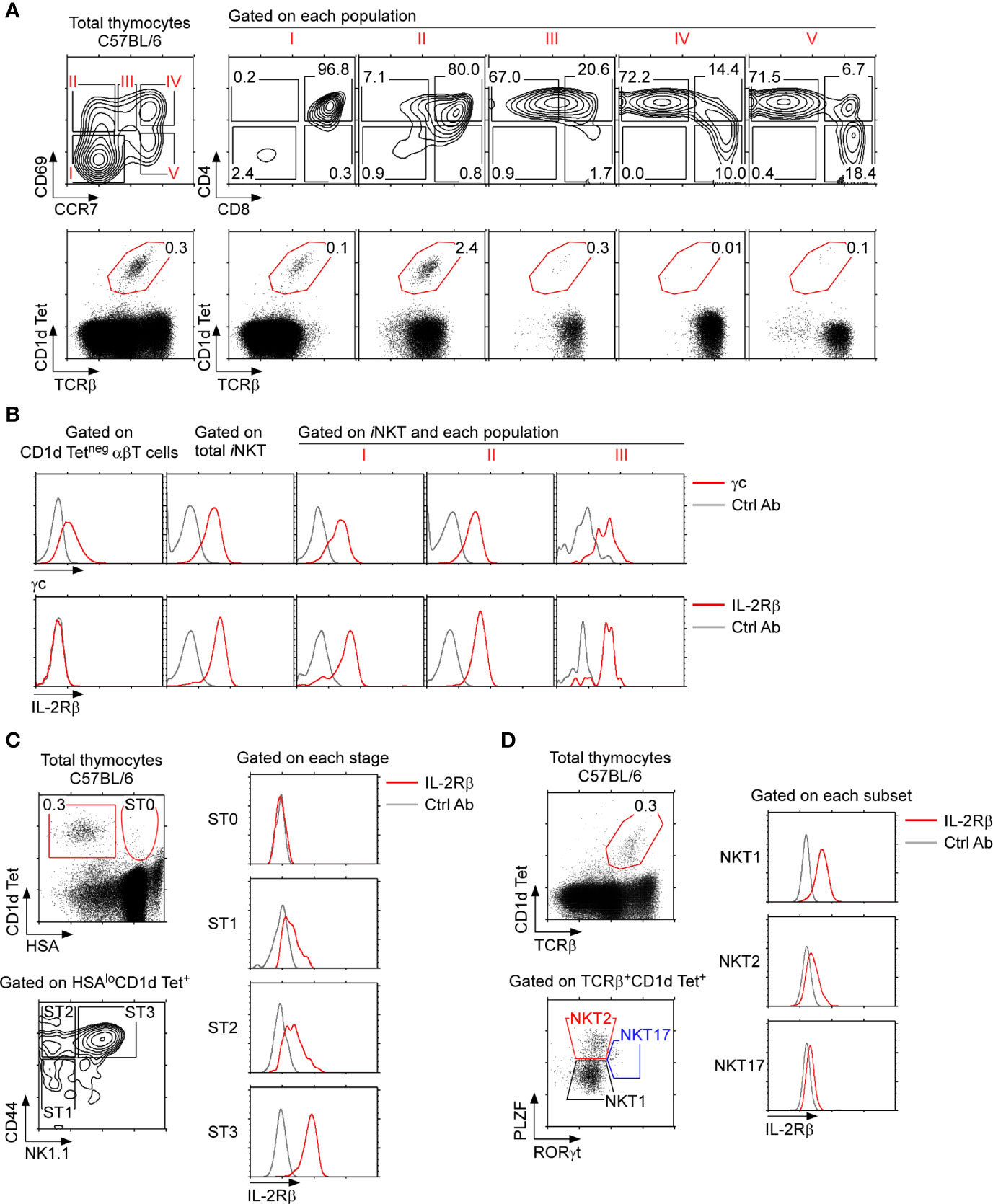
Figure 1 IL-2Rβ expression on thymic iNKT cells (A) C57BL/6 thymocytes were assessed for surface CD69 and CCR7 staining, which visualizes 5 distinct stages of differentiation (i.e., stages I–V), whereby thymocytes undergoing positive selection correspond to population III (top). iNKT cells were identified by PBS57-loaded CD1d tetramer (CD1dTet) staining among the 5 stages defined by CD69 and CCR7 (bottom). The results are representative of 5 independent experiments with a total of 5 WT C57BL/6 mice. (B) Surface cytokine receptor expression on thymic iNKT cell populations. iNKT cells in populations I, II, and III were assessed for γc and IL-2Rβ expression (red lines). Isotype control antibody staining are shown as gray lines. The results are representative of 3 independent experiments. (C) IL-2Rβ expression during iNKT cell differentiation. Thymic iNKT cells were divided into immature ST0 and mature stage 1–3 cells based on HSA expression (left, top). The abundance of IL-2Rβ was then assessed on ST0 (HSAhiCD44–), ST1 (CD44–NK1.1–), ST2 (CD44+NK1.1–), and ST3 (CD44+NK1.1+) iNKT cells (left, bottom). Histograms show surface IL-2Rβ expression for each subset (red line). Isotype control antibody staining are shown as gray lines. The results are representative of 5 independent experiments. (D) IL-2Rβ expression in thymic iNKT subsets. iNKT cells were assessed for intracellular PLZF and RORγt expression to identify NKT1, NKT2, and NK17 cells (left). Each iNKT subset was assessed for IL-2Rβ expression (right, red lines). Isotype control antibody staining are shown as gray lines. The results are representative of 3 independent experiments.
To further align IL-2Rβ expression with the selection and maturation of iNKT cells, we next assessed surface IL-2Rβ expression in developmental stages of thymic iNKT cells. CD44-negative HSAhi thymocytes that bind PBS-57-loaded CD1d tetramers correspond to preselection iNKT cells, and they are usually referred to as stage 0 (ST0) iNKT cells (28). IL-2Rβ was mostly absent on ST0 iNKT cells but was induced following differentiation into HSAlo mature iNKT cells (Figure 1C and Supplementary Figure 1B). Thymic differentiation of HSAlo iNKT cells proceeds along a well-characterized pathway that is marked by the expression of CD44 and NK1.1, whereby CD44–NK1.1– cells correspond to stage 1 (ST1), followed by CD44+NK1.1– cells that are stage 2 (ST2), and terminally differentiate into CD44+NK1.1+ stage 3 cells (ST3) (28). Interestingly, IL-2Rβ was expressed at low levels on ST1 and ST2 cells but highly expressed on ST3 iNKT cells (Figure 1C and Supplementary Figure 1B). Thus, all mature thymic iNKT cells express IL-2Rβ to a certain degree, but the amount of surface IL-2Rβ proteins substantially increases with maturation.
Mature iNKT cells are also categorized into 3 distinct subsets, namely NKT1, NKT2, and NKT17 cells, based on their transcription factor expression profiles (10). Differential expression of the transcription factors PLZF versus RORγt can visualize these three subsets, which we utilized to assess IL-2Rβ expression on individual thymic iNKT subsets (Figure 1D). Consistent with previous observations (10), IL-2Rβ was highly induced on NKT1 cells and minimally expressed on PLZFhiRORγtneg NKT2 and PLZFintRORγt+ NKT17 cells (Figure 1D). Altogether, these results demonstrated that IL-2Rβ is highly upregulated on terminally differentiated ST3 iNKT cells and that such IL-2Rβ+ iNKT cells correspond to NKT1 cells.
Having established that IL-2Rβ expression is associated with iNKT cell maturation, we next wished to examine whether IL-2Rβ would be also required for the development and differentiation of iNKT cells. To this end, we aimed to set up an experimental system where IL-2Rβ would be selectively deleted in iNKT cells. The zinc finger transcription factor PLZF is expressed in iNKT cells but absent in conventional T cells (29, 30). Thus, we bred PLZF-Cre transgenic mice (PLZFCre) with Il2rbfl/fl mice (21, 23) to generate Il2rbfl/flPLZFCre mice and to specifically delete IL-2Rβ in iNKT cells. In such Il2rbfl/flPLZFCre mice, the thymic development of conventional T cells was unaffected (Supplementary Figure 3A). Both TCRβ expression and the CD4 versus CD8 profiles of Il2rbfl/flPLZFCre thymocytes remained comparable to those of littermate Il2rbfl/flPLZFWT thymocytes (Supplementary Figure 3A). Contrary to our expectation, however, the frequency and number of thymic iNKT cells also remained unaltered in Il2rbfl/flPLZFCre mice (Figure 2A).
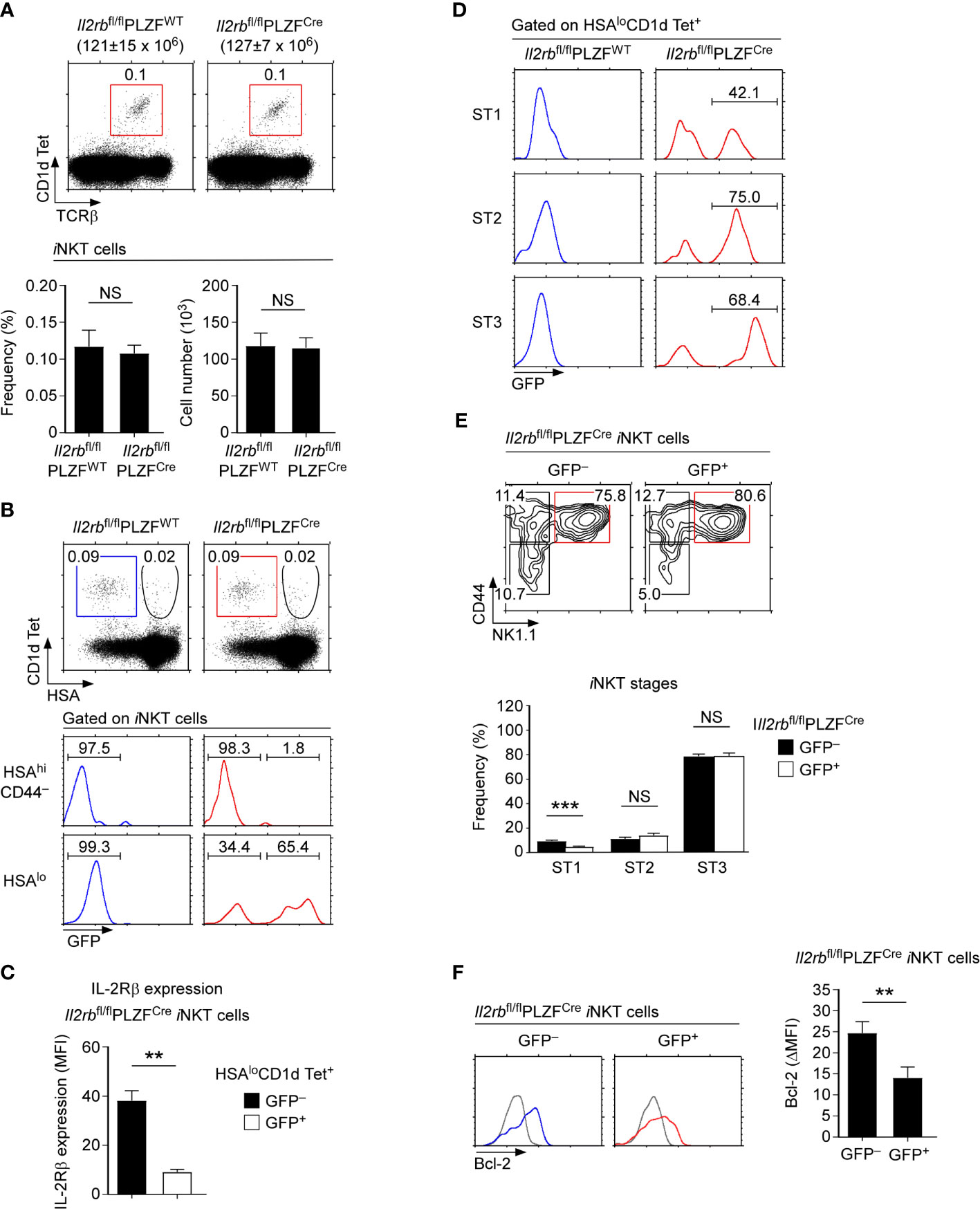
Figure 2 iNKT cell generation and differentiation in IL-2Rβ-floxed PLZF-Cre mice (A) Frequency and number of thymic iNKT cells in Il2rbfl/flPLZFCre and Il2rbfl/fl littermate control mice. The dot plots show thymic iNKT cells as identified by CD1dTet versus TCRβ staining (top). The bar graphs show the frequencies and numbers of iNKT cells from the indicated mice (bottom). The dot plot is representative, and the bar graphs are a summary of 4 independent experiments with a total of 11 Il2rbfl/flPLZFCre and 5 Il2rbfl/fl littermate control mice. (B) IL-2Rβ-deletion efficiency in Il2rbfl/flPLZFCre iNKT cells. The dot plots show thymic iNKT cells of the indicated mice CD1dTet versus HSA staining. HSAhiCD44– cells correspond to ST0 immature iNKT cells while HSAlo cells are mature ST1-3 iNKT cells. The histograms show GFP expression among the indicated population of thymic iNKT cells. (C) IL-2Rβ expression in GFP+ and GFP-negative mature iNKT cells of Il2rbfl/flPLZFCre iNKT mice. Bar graphs show the summary of analyzing 6 Il2rbfl/flPLZFCre mice. (D) GFP expression in ST1, ST2, and ST3 iNKT cells of Il2rbfl/flPLZFCre and littermate control thymocytes. The data are representative of 4 independent experiments. (E) Thymic iNKT cell differentiation in Il2rbfl/flPLZFCre mice. iNKT cells were stained for CD44 and NK1.1 to identify ST1, ST2, and ST3 cells as shown in the contour plots (top). The bar graph shows the frequencies of ST1, ST2, and ST3 cells in GFP+ and GFP– thymic iNKT cells of Il2rbfl/flPLZFCre mice (bottom). Results show the summary of 6 independent experiments with a total of 14 Il2rbfl/flPLZFCre mice. (F) Bcl-2 expression in GFP+ and GFP– iNKT cells of Il2rbfl/flPLZFCre thymocytes. Histograms are representative (left), and the bar graph (right) shows the summary of 3 independent experiments with a total of 8 Il2rbfl/flPLZFCre mice. **P < 0.01; ***P < 0.001. NS, Not Significant.
To understand these results, we considered two possibilities. Firstly, IL-2Rβ expression in the thymus could be dispensable for iNKT cell generation. Alternatively, the PLZF-Cre-mediated deletion of IL-2Rβ could be incomplete. In the latter case, the variegated expression of the PLZF-Cre or inefficient excision of the floxed allele could result in the retention of IL-2Rβ expression. To discriminate these possibilities, it would be necessary to monitor the deletion efficiency of IL-2Rβ in Il2rbfl/flPLZFCre mice. Notably, the Il2rb-floxed allele (Il2rbfl) is engineered to induce the expression of Green Fluorescent Proteins (GFP) upon IL-2Rβ deletion (21), permitting the identification of iNKT cells that have deleted IL-2Rβ. Employing this feature, we next analyzed the efficiency of IL-2Rβ deletion by assessing GFP expression. Among HSAhi immature iNKT cells (ST0), GFP was only found in a very small fraction of iNKT cells (Figure 2B, top histogram). Among HSAlo iNKT cells, however, a large fraction of cells turned out to be GFP+, reporting the successful deletion of the Il2rbfl allele in mature iNKT cells (Figure 2B, bottom histogram, and Supplementary Figure 3B). Assessing the surface IL-2Rβ expression on GFP+ and GFP-negative mature iNKT cells showed a substantial loss of surface IL-2Rβ on GFP+ cells (Figure 2C and Supplementary Figure 3C). Thus, GFP+ cells correspond to iNKT cells that have terminated IL-2Rβ expression. Importantly, the frequencies of such GFP+ iNKT cells increased upon their further differentiation into ST1, ST2, and ST3 iNKT cells (Figure 2D). These results suggested that PLZFCre- mediated deletion of IL-2Rβ is initiated in ST0 cells but not accomplished until later stages of iNKT cell development. Because of this delayed deletion, significant amounts of surface IL-2Rβ remained on GFP+ iNKT cells which could provide residual IL-2R signaling (Figure 2C). Along these lines, we noted that a significant fraction of HSAlo mature iNKT cells remained GFP-negative and fully expressed IL-2Rβ (Figure 2B bottom). Such inefficient deletion could explain why we did not find significant differences in the frequency and number of thymic iNKT cells between Il2rbfl/flPLZFCre and littermate control mice (Figure 2A). We also did not find any significant changes in the ST1-ST3 distribution of GFP+ iNKT cells which expressed dramatically lower amounts of IL-2Rβ compared to GFP-negative iNKT cells (Figure 2E). We also did not find differences in the ST1-3 distribution of Il2rbfl/flPLZFCre mice compared to Il2rbfl/flPLZFWT littermates that express normal amounts of IL-2Rβ (Supplementary Figure 3D). Along these lines, the thymic iNKT subset composition and the frequency of T-bet+ iNKT cells which correspond to ST3 cells were virtually unaltered in Il2rbfl/flPLZFCre mice (Supplementary Figures 4A, B). On the other hand, the protein abundance of Bcl-2, a direct downstream molecule of IL-2Rβ signaling (31), was markedly diminished (Figure 2F), indicating that the lack of IL-2Rβ had physiological consequences for iNKT cells. Nonetheless, the survival of GFP+ thymic iNKT cells remained unaffected, because Bcl-xL and not Bcl-2 provides pro-survival signals for iNKT cells (11). In agreement the frequency and number as well as the subset composition of iNKT cells in the spleen did not differ between Il2rbfl/flPLZFCre and Il2rbfl/flPLZFWT mice (Supplementary Figure 4C). Collectively, the undisturbed differentiation of GFP+ iNKT cells in Il2rbfl/flPLZFCre mice suggested that the late-stage deletion of IL-2Rβ is not detrimental for the ST1 to ST3 iNKT cell maturation.
To further delineate the role of IL-2Rβ in the generation and differentiation of iNKT cells, we next analyzed the development and subset composition of thymic iNKT cells in germline IL-2Rβ-deficient (Il2rb–/–) mice (22). In Il2rb–/– mice, all thymocytes - including ST0 immature iNKT cells - are devoid of IL-2Rβ. Thus, an IL-2Rβ requirement for iNKT cells can be assessed starting at the earliest precursor stages of iNKT cell development. Notably, Il2rb–/– mice are autoimmune because of the impaired generation of immunosuppressive Foxp3+ Treg cells that require IL-2 signaling for their maturation (32, 33). Consequently, we analyzed Il2rb–/– mice before 6 weeks of age to avoid the potential skewing of iNKT cell differentiation due to autoimmunity. We confirmed that the thymus of Il2rb–/– mice at that age did not show an overtly activated phenotype, and that T cell development remained comparable to that of WT littermate mice (Supplementary Figure 5A). We further confirmed the lack of in vivo IL-2Rβ signaling in Il2rb–/– mice by their absence of mature CD25+Foxp3+ Treg cells which require IL-2R signaling for their generation (Supplementary Figure 5B) (32, 33). Consistent with previous reports (18, 19), we confirmed a dramatic reduction in the frequency and number of thymic iNKT cells of Il2rb–/– mice compared with those of WT littermate mice (Figure 3A). Notably, the loss was specific for iNKT cells and did not affect conventional αβ T cells, resulting in a dramatically decreased ratio of iNKT cells over αβ T cells (Supplementary Figure 5C).
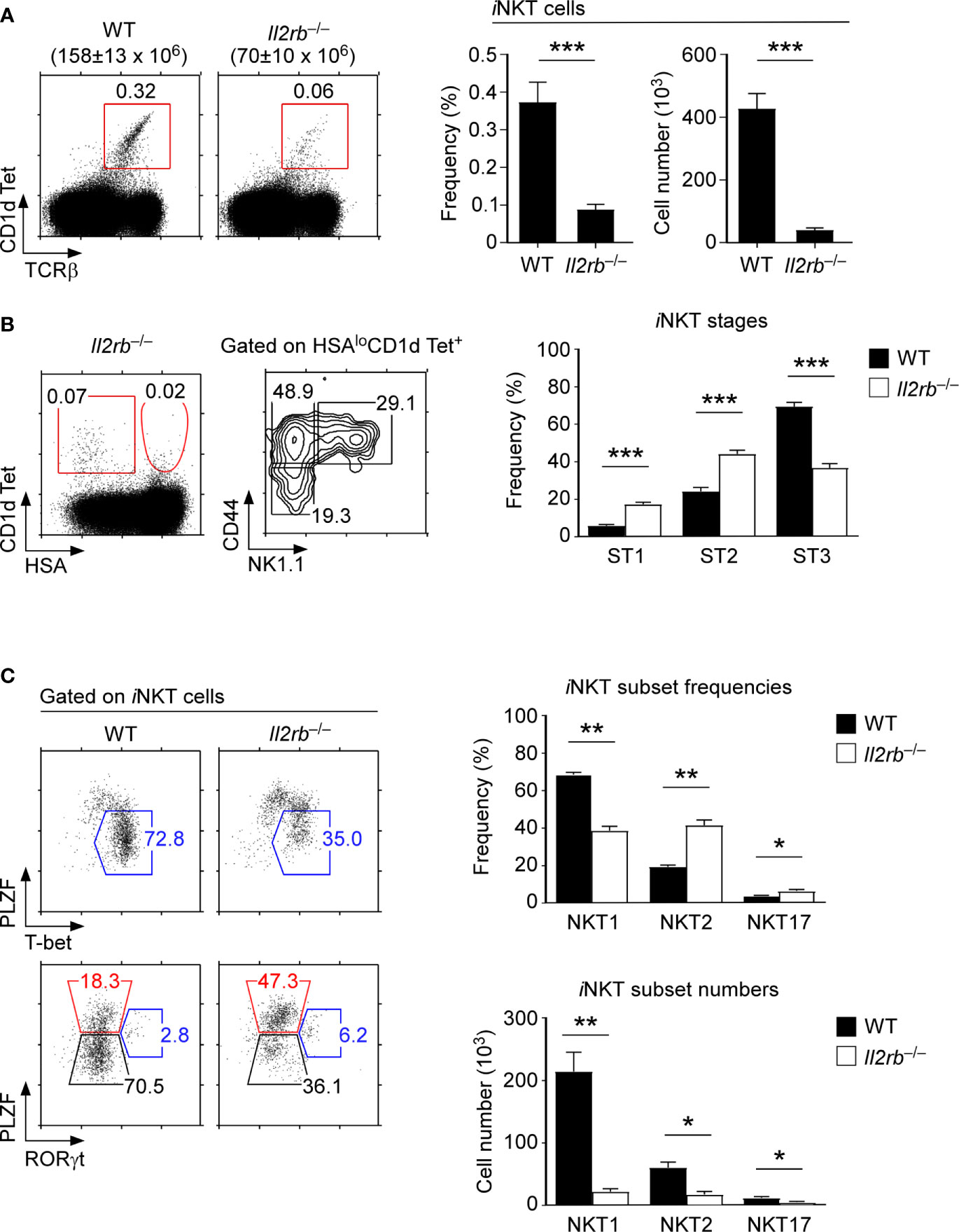
Figure 3 An IL-2Rβ requirement for thymic iNKT cell differentiation (A) Frequency and number of thymic iNKT cells in Il2rb–/– and WT littermate mice. Dot plots show thymic iNKT cells as identified by CD1dTet and TCRβ staining. Total thymocyte numbers are shown above the dot plots as the means ± SEM (left). Bar graphs show the frequencies and numbers of iNKT cells from the indicated mice (right). The dot plot is representative, and the bar graphs are a summary of 4 independent experiments with a total of 13 Il2rb–/– and 7 WT littermate mice. (B) Thymic iNKT cell differentiation in Il2rb–/– mice. HSAlo mature iNKT cells were stained for CD44 and NK1.1 to identify ST1, ST2, and ST3 cells in the indicated mice (left). The bar graph shows the frequencies of ST1, ST2, and ST3 cells in Il2rb–/– and WT littermate mice (right). The dot and contour plots are representative, and the bar graph shows the summary of 4 independent experiments. (C) Dot plots show the thymic iNKT subset composition in Il2rb–/– and WT littermate mice (left). The bar graph on top shows the frequency of thymic iNKT subsets as summary of 5 independent experiments with a total of 14 Il2rb–/– and 8 WT littermate mice. The bottom bar graph shows the cell numbers of thymic iNKT subsets as the summary of 4 independent experiments with a total of 9 Il2rb–/– and 4 WT littermate control mice. *P < 0.05; **P < 0.01; ***P < 0.001.
Most thymic iNKT cells in C57BL/6 background mice correspond to ST3 cells. Therefore, we expected that IL-2Rβ-deficiency would selectively affect the differentiation of ST3 iNKT cells. Indeed, Il2rb–/– thymic iNKT cells were developmentally arrested, resulting in the accumulation of ST1/ST2 iNKT cells and in a substantial decrease in ST3 iNKT cells (Figure 3B). We further found that the fraction of NKT1 cells was significantly diminished among Il2rb–/– iNKT cells, which in turn resulted in the relative overrepresentation of NKT2 and NKT17 cells (Figure 3C). The iNKT cell number for each subset, however, were significantly reduced (Figure 3C), which was consistent with the dramatic decrease in total iNKT cell numbers in Il2rb–/– mice (Figure 3A). Collectively, these results from Il2rb–/– mice documented a major role for the cytokine receptor IL-2Rβ in the generation, but also in the subset differentiation of thymic iNKT cells.
Because IL-2Rβ is important for NKT1 cell generation (Figure 3C), we next asked whether IL-2Rβ would be also sufficient to impose NKT1 lineage fate during iNKT cell development. To this end, we assessed iNKT cell development in IL-2Rβ-transgenic mice (IL-2RβTg) that overexpress mouse IL-2Rβ under the control of human CD2 promoter/enhancer elements (20, 34). In these animals, the transgenic IL-2Rβ is prematurely expressed on preselection thymocytes (Supplementary Figure 6A) and further overexpressed on postselection mature T cells (Supplementary Figure 6A). Consequently, the IL-2Rβ is abundantly expressed on both immature and mature thymocytes of IL-2RβTg mice. Among iNKT cells, immature HSAhiCD44– iNKT cells (ST0) normally do not express IL-2Rβ (Supplementary Figure 6B). However, the ST0 iNKT cells in IL-2RβTg mice displayed substantially increased abundance of IL-2Rβ (Supplementary Figure 6B). Importantly, such premature expression of IL-2Rβ dramatically impaired the generation of iNKT cells, so that the frequency and number of thymic iNKT cells in IL-2RβTg mice were markedly reduced (Figure 4A). These results suggested that IL-2 receptor signaling in immature iNKT is detrimental to the generation of thymic iNKT cells. These findings also indicated that the timing of IL-2Rβ expression during iNKT cell development needs to be carefully controlled, presumably to protect immature iNKT cells from premature IL-2 or IL-15 signaling.
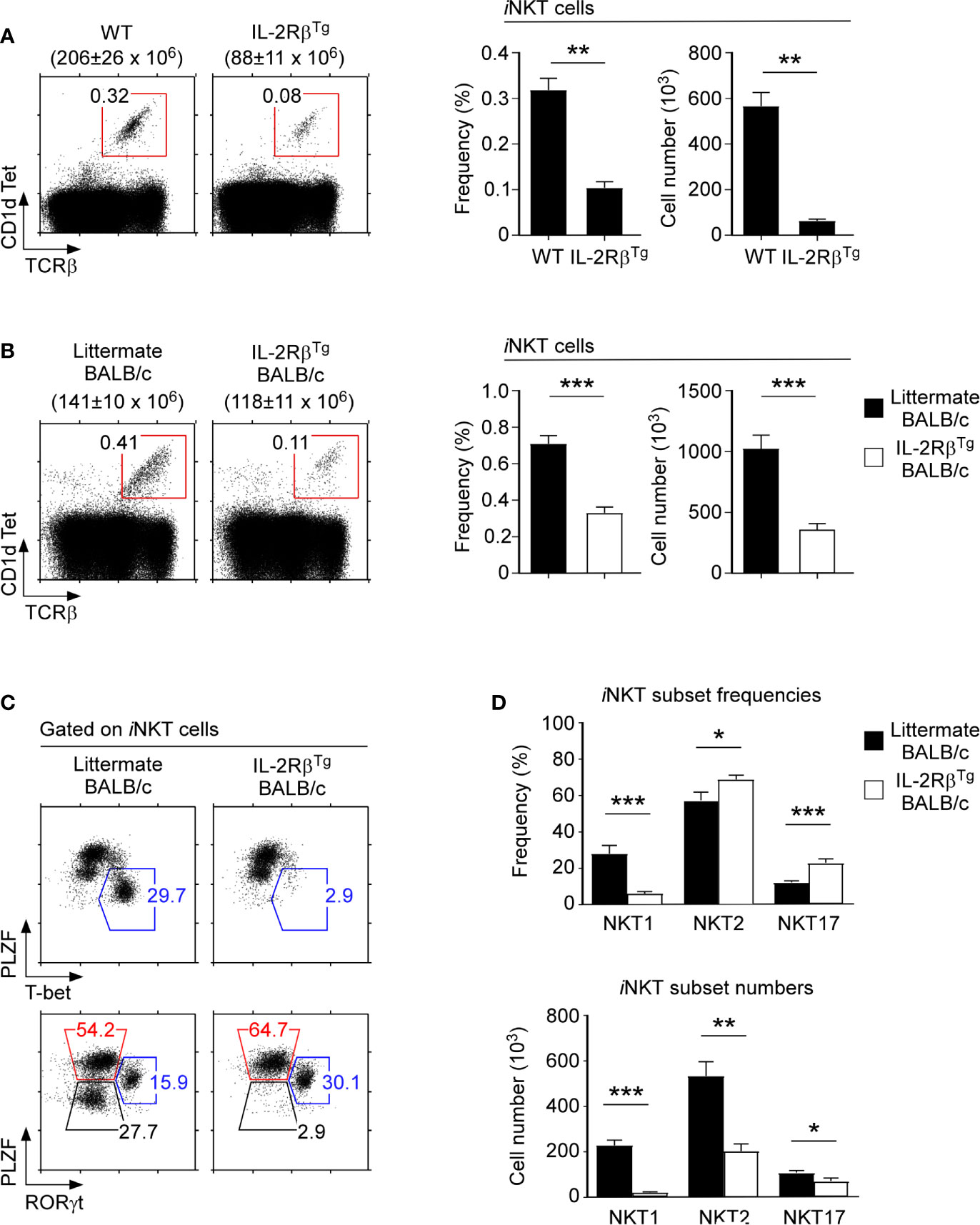
Figure 4 iNKT cell differentiation in IL-2Rβ transgenic mice (A) Frequency and number of thymic iNKT cells in IL-2RβTg and littermate C57BL/6 mice. The dot plots show thymic iNKT cells as identified by CD1dTet versus TCRβ staining (left). The total thymocyte numbers are shown above the dot plots as the means ± SEM. The bar graphs show the frequencies and numbers of iNKT cells from the indicated mice (right). The dot plot is representative, and the bar graphs are a summary of 4 independent experiments with a total of 7 IL-2RβTg and 6 littermate C57BL/6 mice. (B) Frequency and number of thymic iNKT cells in IL-2RβTg BALB/c and littermate WT mice. The dot plots show thymic iNKT cells as identified by CD1dTet versus TCRβ staining (left). The total thymocyte numbers are shown above the dot plots as the means ± SEM. The bar graphs show the frequencies and numbers of iNKT cells from the indicated mice (right). The dot plot is representative, and the bar graphs are a summary of 3 independent experiments with a total of 7 IL-2RβTg and 8 littermate WT mice. (C) The dot plots show the thymic iNKT subset composition in IL-2RβTg BALB/c and WT littermate mice based on PLZF versus T-bet (top) and PLZF versus RORγt analysis (bottom). Results are representative of 4 independent experiments. (D) The bar graphs show the summary of thymic iNKT subset distribution for the indicated mice. Subset frequencies are the summary of 5 independent experiments with a total of 10 IL-2RβTg and 10 littermate WT BALB/c mice (top). Cell numbers of each iNKT subset are the summary of 4 independent experiments with a total of 8 IL-2RβTg and 7 littermate WT BALB/c control mice (bottom). *P < 0.05; **P < 0.01; ***P < 0.001.
Because IL-2Rβ is associated with NKT1 cell differentiation, we next aimed to interrogate if premature IL-2Rβ expression in immature ST0 iNKT cells would affect the development of other iNKT subsets. In C57BL/6 mice, most thymic iNKT cells correspond to NKT1 cells (10), making it difficult to discern how the IL-2RβTg would affect NKT2 and NKT17 cells which are underrepresented in this mouse strain. BALB/c mice differ from C57BL/6 mice in their thymic iNKT subset composition as it is enriched in NKT2 and NKT17 cells and reduced in NKT1 cells compared with that in C57BL/6 mice (10). Thus, we considered BALB/c mice as an appropriate model to assess the effects of premature IL-2Rβ expression on NKT2 and NKT17 cell generation. To this end, we backcrossed the IL-2RβTg onto the BALB/c background and analyzed the development and differentiation of thymic iNKT cells in these mice.
As expected, the thymocytes of IL-2RβTg BALB/c mice expressed substantially increased amounts of surface IL-2Rβ compared with the thymocytes of littermate mice (Supplementary Figure 6C). Such increase in the IL-2Rβ abundance was also observed in HSAhiCD44– (ST0) immature iNKT cells (Supplementary Figure 6D), but curiously not in HSAlo mature iNKT cells (Supplementary Figure 6D). These results suggested that the expression of the IL-2RβTg is either silenced in HSAlo iNKT cells or that the premature expression of IL-2Rβ in HSAhiCD44– (ST0) immature iNKT cells is detrimental for iNKT cell generation so that only iNKT cells that have escaped the transgene expression can mature. In fact, we found that the frequency and number of thymic iNKT cells were indeed dramatically reduced in IL-2RβTg BALB/c mice (Figure 4B). Thus, consistent with our findings in IL-2RβTg C57BL/6 mice (Figure 4A), the forced expression of IL-2Rβ suppresses the generation of thymic iNKT cells in BALB/c mice also (Figure 4B).
To understand the effects of IL-2Rβ on iNKT cell differentiation, we next examined the iNKT subset composition in IL-2RβTg BALB/c mice. Surprisingly, NKT1 cells, as identified by T-bet expression, were virtually absent in IL-2RβTg BALB/c thymocytes. Both the frequency and number of T-bet+ NKT1 cells were dramatically decreased in these mice (Figures 4C, D). Conversely, NKT2 and NKT17 cell frequencies were significantly increased in IL-2RβTg BALB/c mice compared with those of WT littermate controls (Figures 4C, D). Thus, transgenic IL-2Rβ selectively inhibited the development of NKT1 cells without suppressing the generation of NKT2 and NKT17 cells.
To further examine how IL-2RβTg would interfere with the induction of T-bet and impair NKT1 cell generation, we employed the T-bet-ZsGreen reporter mouse (TBGRTg) to monitor T-bet transcription in IL-2RβTg and WT BALB/c mice (Figure 5A) (25). The TBGRTg is designed to express ZsGreen reporter proteins from the Tbx21 gene locus which encodes for T-bet so that ZsGreen expression reflects Tbx21 transcription (25). Indeed, we identified a substantial population of ZsGreen+ cells among thymic iNKT cells of TBGRTg BALB/c mice (Figure 5A), and these ZsGreen+ iNKT cells corresponded to NKT1 cells based on their intracellular PLZF and T-bet protein expression (Figure 5B, left). In contrast, TBGRTg IL-2RβTg BALB/c failed to generate ZsGreen+T-bet+ iNKT cells (Figure 5B, right), affirming the detrimental effect of IL-2RβTg on NKT1 cell generation.
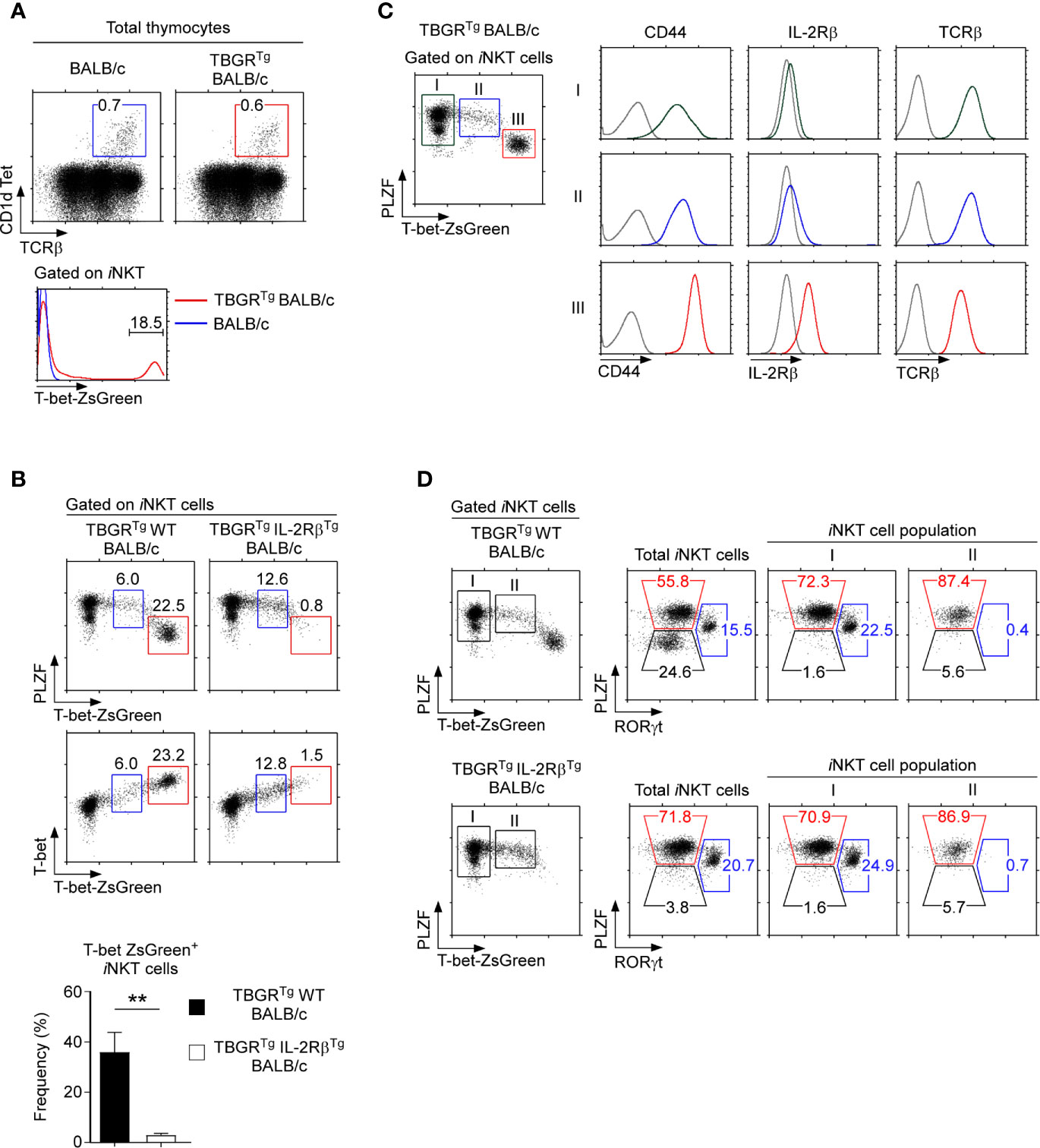
Figure 5 T-bet-ZsGreen reporter expression in iNKT cells of IL-2RβTg BALB/c mice (A) Thymic iNKT cells were identified in BALB/c and TBGRTg BALB/c mice (top) and assessed for T-bet-ZsGreen reporter expression (bottom). Results are representative of 5 independent experiments. (B) T-bet-ZsGreen reporter expression in thymic iNKT cells of TBGRTgIL-2RβTg BALB/c and TBGRTg littermate control mice. Dot plots show either PLZF versus T-bet-ZsGreen reporter expression (top) or T-bet protein versus T-bet-ZsGreen reporter expression (bottom) in HSAlo thymic iNKT cells of TBGRTg IL-2RβTg BALB/c and TBGRTg WT littermate mice. Bar graph shows the frequency of T-bet-ZsGreen+ iNKT cells (red box) of the indicated mice. Results are representative 3 independent experiments with 8 TBGRTgIL-2RβTg BALB/c and 5 TBGRTg littermate WT BALB/c mice. (C) T-bet-ZsGreen reporter expression upon NKT1 cell differentiation. Thymic iNKT cells of TBGRTg BALB/c mice were divided into three distinct populations, i.e. I, II, and III, based on the amount of T-bet-ZsGreen expression (dot plot, left). Surface expression of CD44, IL-2Rβ, and TCRβ was then assessed for each iNKT population (histograms, right). Results are representative of 2 independent experiments with a total of 6 TBGRTg BALB/c mice. (D) Subset compositions of thymic iNKT cells in TBGRTg BALB/c mice based on ZsGreen-reporter expression. Total iNKT cells, T-bet-ZsGreen-negative (population I), and T-bet-ZsGreen-intermediate (population II) iNKT cells were identified in TBGRTg WT BALB/c (top) and TBGRTgIL-2RβTg BALB/c thymocytes (bottom) and examined for their subset composition based on PLZF versus RORγt staining. Results are representative of 3 independent experiments with 8 TBGRTgIL-2RβTg BALB/c and 5 TBGRTg littermate WT BALB/c mice. **P < 0.01.
Of note, we found in both WT and IL-2RβTg thymocytes a distinct population of iNKT cells with intermediate amounts of ZsGreen and T-bet expression, i.e. ZsGreenintT-betint (Figure 5B, cells gated in blue), whose developmental status was unclear to us. To further characterize these ZsGreenintT-betint iNKT cells, we divided TBGRTg BALB/c iNKT cells into 3 populations, i.e. I, II, and III, based on their progressive increase in ZsGreen reporter expression. Accordingly, the ZsGreenintT-betint cells corresponded to population II, while ZsGreenhiT-bethi cells were defined as population III (Figure 5C). Thymic iNKT cells that did not express ZsGreen were referred to as population I. Terminally differentiated NKT1 cells, as identified as population III, expressed large amounts of T-bet and CD44 (Figure 5C and Supplementary Figure 6E) (11). ZsGreenintT-betint cells (population II), on the other hand, expressed significantly smaller amounts of T-bet and CD44 but showed greater abundance of TCRβ. Importantly, ZsGreenintT-betint cells had not yet induced CD122 expression, suggesting that they are NKT1-lineage committed but not fully differentiated NKT1 cells (Figures 5C, D and Supplementary Figure 6E) (35). Along these lines, the ZsGreenintT-betint population did not contain RORγt+ iNKT cells (Figure 5D), indicating that NKT17 cell differentiation is branched off before the appearance of ZsGreenintT-betint population II iNKT cells. Thus, we identified a new subpopulation of thymic iNKT cells, i.e. ZsGreenintT-betint iNKT cells, that are NKT1-lineage committed but are not fully differentiated into mature NKT1 cells. Because ZsGreen+T-bet+ iNKT cells were missing in IL-2RβTg thymocytes, these results further suggest that the forced and premature expression of IL-2Rβ interferes with the terminal differentiation of T-bet+ NKT1 cells but did not inhibit the induction of T-bet expression itself. Collectively, these results unveil an unexpected negative effect of IL-2Rβ on iNKT subset differentiation.
To understand how IL-2Rβ overexpression would affect thymic iNKT cell generation, we considered two alternative but not mutually exclusive hypotheses. First, the increased abundance of IL-2Rβ proteins in IL-2RβTg mice could increase the binding and consumption of IL-15, thus diminishing the availability of IL-15 for iNKT cell development. IL-15 is critical for the generation of iNKT cells in general and specifically for NKT1 cells (11, 36). Consequently, the detrimental effect of IL-2RβTg on iNKT cells could have been due to insufficient IL-15 availability, partly phenocopying the effect of IL-15-deficiency (37). In a second scenario, we focused on the timing of IL-2Rβ expression. IL-2Rβ is normally not induced in immature iNKT cells but forcibly expressed in IL-2RβTg mice. Premature IL-2Rβ signaling could interfere with iNKT cell generation and specifically with NKT1 cell differentiation, albeit the molecular mechanism would be unclear. To discriminate between these possibilities, we implanted IL-2RβTg mice with osmotic pumps that release recombinant IL-15 to supply excess amounts of IL-15 in vivo (Figure 6A). After 10 days of IL-15 pump installation in IL-2RβTg mice, we found a dramatic increase in spleen size and increased frequencies of NK cells whose maintenance depends on IL-15 (38) (Figure 6A and Supplementary Figure 7A). Moreover, the frequency of CD8 T cells and specifically the number of CD44hi CXCR3+ memory-phenotype CD8 T cells were substantially increased in mice that were implanted with IL-15- but not with PBS-releasing pumps (Figure 6A and Supplementary Figure 7B). These results were consistent with the effects of increased IL-15 availability in vivo (39). The generation of thymic iNKT cells in IL-2RβTg BALB/c mice, however, remained unaffected by the IL-15 infusion (Figure 6B), and the iNKT subset composition also remained unaltered (Figures 6C, D). Thus, despite supplemented with excess amounts of IL-15, the IL-2RβTg mice were still impaired in thymic iNKT cell differentiation, with both dramatically reduced frequencies and numbers of NKT1 cells (Figures 6C, D). Along these lines, the thymic generation of Foxp3+ Treg cells which depend on IL-2 and IL-2 receptor signaling (33) was unaffected in IL-2RβTg mice (Supplementary Figure 7C), which further supports that the availability of intrathymic cytokines remains unaltered by IL-2Rβ overexpression. Altogether, these results suggested that it is unlikely that increased consumption and reduced availability of in vivo IL-15 would account for the lack of NKT1 cells in IL-2RβTg mice.
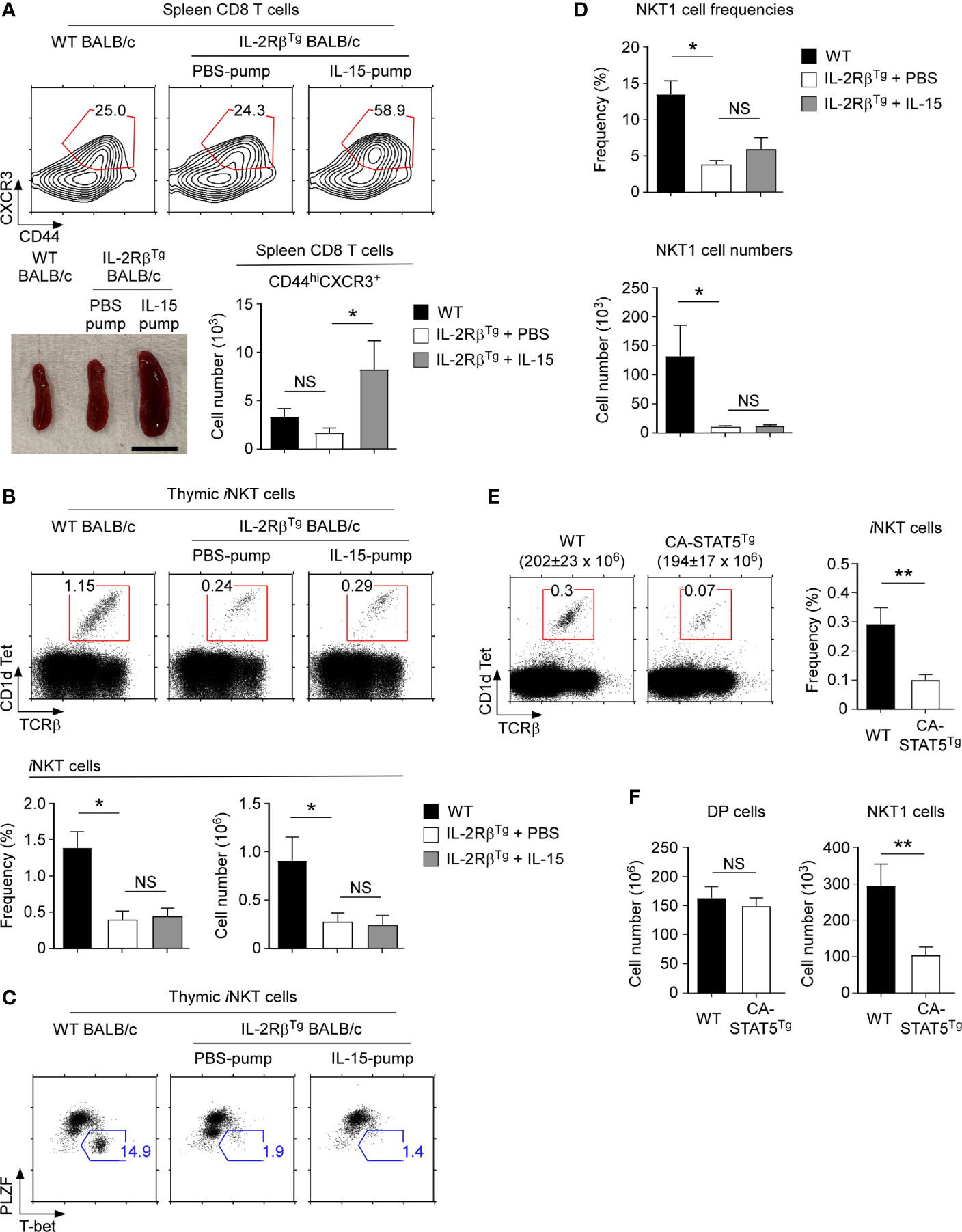
Figure 6 Thymic iNKT cell differentiation in IL-15-infused IL-2RβTg BALB/c mice (A) Effects of recombinant IL-15- or PBS-releasing Alzet osmotic pump installation on IL-2RβTg BALB/c mice. Spleen CD8 T cells of IL-2RβTg BALB/c mice that were infused with IL-15 or PBS for 10 days were assessed for the accumulation of memory-phenotype CD8 T cells by CD44 versus CXCR3 staining. Frequency and numbers of CD44hiCXCR3+ CD8 T cells are representative of 2 independent experiments with 4 IL-15 pump and 3 PBS pump implanted IL-2RβTg BALB/c mice (top). Picture shows spleens of IL-2RβTg BALB/c mice that were implanted with PBS or IL-15 Alzet pumps (bottom). Scale bar = 1 cm. (B) Frequencies and numbers of thymic iNKT cells of IL-2RβTg BALB/c mice implanted with IL-15- or PBS-releasing Alzet osmotic pumps. Dot plots are representative (top) and graphs are summary (bottom) of 3 independent experiments with a total of 5 IL-15 pump and 4 PBS pump implanted IL-2RβTg BALB/c mice. (C) Thymic iNKT subset composition IL-2RβTg BALB/c implanted with IL-15 or PBS-releasing Alzet osmotic pumps. NKT1 cells were identified by PLZF versus T-bet of HSAlo mature thymic iNKT cells. Results are representative of 3 independent experiments with 5 IL-15 pump and 4 PBS pump implanted IL-2RβTg BALB/c mice. (D) Frequencies and numbers of NKT1 cells in IL-2RβTg BALB/c implanted with IL-15 or PBS-releasing Alzet osmotic pumps. Results are summary of 3 independent experiments with 5 IL-15 pump and 4 PBS pump implanted IL-2RβTg BALB/c mice. (E) Frequency of thymic iNKT cells in CA-STAT5Tg and littermate C57BL/6 mice. The dot plots show thymic iNKT cells as identified by CD1dTet versus TCRβ staining (left). The total thymocyte numbers are shown above the dot plots as the means ± SEM (left). The bar graph shows the frequency of iNKT cells from the indicated mice (right). The dot plot is representative, and the bar graphs are a summary of 4 independent experiments with a total of 7 CA-STAT5Tg and 8 littermate WT mice (F) Cell numbers of DP thymocytes and thymic NKT1 cells from CA-STAT5Tg and littermate WT C57BL/6 mice. Bar graph shows the summary from a total of 7 CA-STAT5Tg and 5 littermate WT mice. *P < 0.05; **P < 0.01. NS, Not Significant.
To gain further mechanistic insights into whether and how premature IL-2Rβ expression would impair iNKT cell generation, we considered that STAT5 phosphorylation is a major downstream event of IL-2Rβ signaling (40). The transcription factor STAT5 plays a critical role in T cell development as it induces the expression of prosurvival molecules, such as Bcl-2, and controls the activity of several key transcription factors in T cell differentiation (40, 41). Because immature ST0 iNKT cells lack IL-2Rβ (Figure 1C), we postulated that ST0 iNKT cells of WT mice would not induce phospho-STAT5 (pSTAT5) upon IL-15 stimulation. ST0 iNKT cells in IL-2RβTg mice, on the other hand, would have induced substantial amounts of pSTAT5 due the premature expression and signaling of IL-2Rβ.
If it would be the premature STAT5 activation which suppresses iNKT cell generation, we hypothesized that a constitutively active STAT5b transgene (CA-STAT5Tg) would also impair iNKT cell generation (24). Indeed, we found that both the frequency and number of thymic iNKT cells were dramatically reduced in CA-STAT5Tg mice compared to those in WT littermate controls (Figure 6E). Moreover, assessing the composition of thymic iNKT subsets in CA-STAT5Tg mice (Supplementary Figure 8A) demonstrated that the generation of NKT1 cells was impaired by ectopic STAT5 activation (Figure 6F). T cell development in general, however, was not negatively affected by the constitutive activation of STAT5 (Supplementary Figure 8B). As such, the cell numbers of preselection DP thymocytes remained unaffected (Figure 6F), and the generation of memory-phenotype CD8 T cells was rather increased in CA-STAT5Tg thymocytes (Supplementary Figure 8C). Thus, the detrimental effect of STAT5 activation was limited to the generation of iNKT cells. Collectively, these results identify and demonstrate STAT5 as a major player acting downstream of IL-2Rβ to control the development of thymic iNKT cells.
iNKT cells comprise three major subsets that are characterized by distinct transcription factor and cytokine expression. Although TCR signaling is considered to play a decisive role in the generation and subset differentiation of iNKT cells (42, 43), intrathymic cytokines also play important roles in these processes (44). As such, the γc family cytokine IL-15 is a critical factor in thymic iNKT cell generation that also contributes to the subset specification of iNKT cells (11). Consistent with an IL-15 requirement for NKT1 cell differentiation, the signaling-competent IL-15 receptor heterodimer—i.e., IL-2Rβ/γc—is mostly expressed on NKT1 cells, but not on NKT2 and NKT17 cells (10). This raises the question whether IL-2Rβ expression is specifically induced on NKT1 cells or whether IL-2Rβ could be induced on all iNKT cells but is then terminated upon differentiation into NKT2 and NKT17 cells. Furthermore, it was unclear whether IL-2Rβ expression would be sufficient to impose NKT1 subset fate so that forced expression of IL-2Rβ would direct iNKT cells into NKT1 cells. Here, we addressed these questions using a series of genetically engineered mouse models, and we identified IL-2Rβ as a critical regulator of thymic iNKT cell generation whose abundance and timing of expression dramatically affected the generation of iNKT cells. These findings reveal previously unappreciated aspects of cytokine receptor expression in controlling the development of iNKT cells, and they propose a model of iNKT differentiation that is regulated by the timing and abundance of cytokine receptor expression.
The generation of thymic iNKT cells is currently explained by two competing but not mutually exclusive models of differentiation (7, 45). In the conventional or “linear maturation” model, preselection thymocytes that have recombined a functional Vα14-Jα18 TCRα chain are positively selected to become immature ST0 iNKT cells. ST0 iNKT cells then mature along a developmental pathway defined by CD44 and NK1.1 expression, culminating in end differentiated CD44+NK1.1+ ST3 iNKT effector cells. Under this scenario, CD44+NK1.1– ST2 iNKT cells, which are IL-2Rβ-negative, would give rise to ST3 iNKT cells that are IL-2Rβ-positive. However, the cellular signals that would induce the initial upregulation of IL-2Rβ remain unknown. Notably, such a requirement for IL-2Rβ expression that precedes the IL-15-mediated generation of NKT1 cells is also found in the alternative “lineage differentiation” model (7, 45). While NKT1 cell differentiation depends on IL-15, the IL-2Rβ, which is necessary for IL-15 signaling, is not expressed on immature iNKT cells. Thus, IL-2Rβ expression is carefully timed so that preselection thymocytes lack IL-2Rβ but is then specifically induced on NKT1-committed postselection immature iNKT cells to drive their maturation into NKT1 subset iNKT cells.
The molecular pathway that upregulates IL-2Rβ and drives NKT1 cell fate is not fully understood. However, it likely involves molecules downstream of TCR signaling because differences in TCR signaling strength are acknowledged to drive iNKT subset fate decisions (42). In this regard, the transcription factor Egr2 is of particular interest (46), because its relative abundance is associated with iNKT subset differentiation (42). Egr2 expression is upregulated by TCR-induced calcium signaling and correlates with the strength of TCR signaling (42, 47). Notably, NKT2 cell generation is thought to be driven by strong TCR signaling whereas NKT1 cell differentiation is proposed to be mediated by weak TCR signaling (42). Incidentally, NKT2 cells contain large amounts of Egr2 whereas NKT1 cells express small amounts of Egr2. Such distinct amounts of Egr2 expression presumably results in the graded expression of PLZF whose abundance correlates with iNKT subset specification (48). Thus, the TCR-centric view proposes that distinct amounts of Egr2 would induce different levels of PLZF, which would then determine iNKT subset identity. Interestingly, however, Egr2 was also found to bind the IL-2Rβ promoter and thus to induce IL-2Rβ expression (47). Consequently, we hypothesized that Egr2 could induce the expression of IL-2Rβ in immature iNKT cells to permit their differentiation into NKT1 cells. According to this scenario, TCR signaling would be required to specify NKT1 fate by upregulating IL-2Rβ expression, and cytokine receptor signaling would be then required to drive NKT1 subset differentiation by IL-15 signaling. Altogether, such model reconciles the two concurrent perspectives of iNKT cell development so that TCR and cytokine signals play distinct but nonredundant roles in the subset differentiation of iNKT cells.
Because IL-2Rβ plays such a critical role in NKT1 subset commitment, we were surprised to find that the increased abundance of IL-2Rβ in IL-2RβTg mice did not promote but rather suppressed the generation of thymic iNKT cells, specifically of the NKT1 subset. A detrimental effect of forced IL-2Rβ expression had been previously reported for NK cells where a hematopoietic lineage-specific IL-2Rβ transgene was found to suppress the generation of NK1.1+ cells (49). Whether this NK1.1+ population also comprise iNKT cells, including stage 3 NKT1 cells, is unclear from that study. If such would be the case, however, these results would further bolster a detrimental effect of IL-2Rβ on iNKT cell generation whose molecular basis remains currently unclear to us.
Nonetheless, we considered two important findings regarding the generation of iNKT cells in IL-2RβTg mice. First, unlike NKT1 cells, NKT2 and NKT17 cells in WT mice do not express IL-2Rβ (10) Surprisingly, we found that the few NKT2 and NKT17 cells that arise in IL-2RβTg mice also did not express IL-2Rβ proteins. Therefore, the NKT2 and NKT17 cells in IL-2RβTg mice could be either transgene escapees or they could have downregulated the expression of transgenic IL-2Rβ. We favor the latter case because the IL-2RβTg is driven by the human CD2 mini-cassette, whose activity is downregulated upon positive selection, so that transgene expression substantially diminishes in postselection thymocytes (50). Regardless, we consider the IL-2RβTg model not adequate to assess the effect of forced IL-2Rβ expression on NKT2 and NKT17 cells, simply because the IL-2RβTg fails to be expressed in these iNKT subsets. Second, we wish to underscore that the IL-2RβTg is prematurely expressed on immature thymocytes, including the immature ST0 iNKT cells. IL-2Rβ is normally absent in the ST0 iNKT precursors of WT mice, an observation contrasting the IL-2Rβ expression in IL-2RβTg ST0 iNKT cells which are highly abundant in IL-2Rβ. Consequently, ST0 iNKT cells in IL-2RβTg mice can be signaled by intrathymic IL-15 whereas WT ST0 iNKT cells cannot respond to IL-15. Based on these results, we hypothesize that premature and possibly sustained IL-15 signaling in immature ST0 iNKT cells would be detrimental for NKT1 cell generation.
Additionally, we appreciate the need to determine why premature IL-15 signaling in ST0 iNKT cells would negatively affect thymic iNKT cell generation. IL-15-induced STAT5 phosphorylation is usually considered beneficial for T cells because it induces the expression of anti-apoptotic Bcl-2 and upregulates metabolic activities. Thus, IL-15 signaling should promote and not inhibit iNKT cell generation. However, STAT5 activation also suppresses the induction of Bcl6, which is a critical transcription factor for iNKT cell generation (51). Bcl-6 is selectively and highly expressed on ST0 iNKT cells before they proliferate and differentiate into mature iNKT cells. Bcl-6 is also developmentally necessary because Bcl-6-deficiency is associated with impaired thymic iNKT cell generation (51). Importantly, IL-2 receptor-induced pSTAT5 potently suppresses Bcl-6 expression (52–54). Therefore, STAT5 activation in ST0 iNKT cells could suppress the expression of Bcl-6, which, in turn, could impair the generation of thymic iNKT cells. These results lead to a model of iNKT cell development where IL-2Rβ expression must be precisely timed so that ST0 iNKT cells suppress IL-2Rβ expression to prevent premature IL-15 signaling, but then a subset of post-ST0 iNKT cells would rapidly induce IL-2Rβ to initiate IL-15-induced NKT1 lineage differentiation.
Lastly, we wish to point out that the T-bet-ZsGreen reporter mouse revealed a new developmentally intermediate stage in NKT1 cell development that can be identified by intermediate level of T-bet protein expression and T-bet mRNA transcription (as demonstrated by T-bet-ZsGreen reporter expression). Such ZsGreenintT-betint iNKT cells were NKT1 lineage committed but not fully differentiated NKT1 cells, as they lacked RORγt expression but had not fully upregulated T-bet protein expression. The ZsGreenintT-betint iNKT cells were further marked by their intermediate level of PLZF protein expression which was not downregulated to the amounts found in mature NKT1 cells (48). Thus, ZsGreenintT-betint iNKT cells are NKT1 lineage committed but not fully mature NKT1 cells. Because ZsGreenintT-betint iNKT cells were present in both WT and IL-2RβTg mice, these data indicate that the forced IL-2Rβ expression did not interfere with NKT1 lineage commitment but rather with the maturation of NKT1 cells.
Collectively, here, we demonstrated the importance of cytokine signaling in NKT1 lineage differentiation by altering the timing and abundance of IL-2Rβ expression, independently of TCR expression or signaling. These results document the significance of cytokines and their receptor expression in determining iNKT lineage fate, and further suggest a role for cytokines in parallel or in association with TCR signaling to shape the effector function of T cells.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by National Cancer Institute Animal Care and Use Committee.
HW designed and performed the experiments, analyzed the data, and contributed to the writing of the manuscript. HK, AC, PA, and RG provided expertise, analyzed the data, and commented on the manuscript. J-HP conceived the project, analyzed the data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Drs. Joo-Young Park (Seoul National University) and Damian Kovalovsky (National Cancer Institute, NIH) for critical review and discussion of this manuscript. This study has been supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.642856/full#supplementary-material
1. Etzensperger R, Kadakia T, Tai X, Alag A, Guinter TI, Egawa T, et al. Identification of Lineage-Specifying Cytokines That Signal All CD8(+)-Cytotoxic-Lineage-Fate ‘Decisions’ in the Thymus. Nat Immunol (2017) 18:1218–27. doi: 10.1038/ni.3847
2. Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, et al. Signaling by Intrathymic Cytokines, Not T Cell Antigen Receptors, Specifies CD8 Lineage Choice and Promotes the Differentiation of Cytotoxic-Lineage T Cells. Nat Immunol (2010) 11:257–64. doi: 10.1038/ni.1840
3. Lin JX, Leonard WJ. The Common Cytokine Receptor Gamma Chain Family of Cytokines. Cold Spring Harb Perspect Biol (2018) 10:a028449. doi: 10.1101/cshperspect.a028449
4. Hong C, Luckey MA, Park JH. Intrathymic IL-7: The Where, When, and Why of IL-7 Signaling During T Cell Development. Semin Immunol (2012) 24:151–8. doi: 10.1016/j.smim.2012.02.002
5. Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T Cells Expressing the Transcription Factor PLZF Regulate the Development of Memory-Like CD8+ T Cells. Nat Immunol (2010) 11:709–16. doi: 10.1038/ni.1898
6. Hemmers S, Schizas M, Azizi E, Dikiy S, Zhong Y, Feng Y, et al. IL-2 Production by Self-Reactive CD4 Thymocytes Scales Regulatory T Cell Generation in the Thymus. J Exp Med (2019) 216:2466–78. doi: 10.1084/jem.20190993
7. Hogquist K, Georgiev H. Recent Advances in iNKT Cell Development. F1000Res (2020) 9. doi: 10.12688/f1000research.21378.1
8. Gapin L. Development of Invariant Natural Killer T Cells. Curr Opin Immunol (2016) 39:68–74. doi: 10.1016/j.coi.2016.01.001
9. Krovi SH, Gapin L. Invariant Natural Killer T Cell Subsets-More Than Just Developmental Intermediates. Front Immunol (2018) 9:1393. doi: 10.3389/fimmu.2018.01393
10. Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-State Production of IL-4 Modulates Immunity in Mouse Strains and is Determined by Lineage Diversity of Inkt Cells. Nat Immunol (2013) 14:1146–54. doi: 10.1038/ni.2731
11. Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, et al. IL-15 Regulates Homeostasis and Terminal Maturation of NKT Cells. J Immunol (2011) 187:6335–45. doi: 10.4049/jimmunol.1003965
12. Monteiro M, Almeida CF, Agua-Doce A, Graca L. Induced IL-17-Producing Invariant NKT Cells Require Activation in Presence of TGF-Beta and IL-1beta. J Immunol (2013) 190:805–11. doi: 10.4049/jimmunol.1201010
13. Lucas B, White AJ, Cosway EJ, Parnell SM, James KD, Jones ND, et al. Diversity in Medullary Thymic Epithelial Cells Controls the Activity and Availability of iNKT Cells. Nat Commun (2020) 11:2198. doi: 10.1038/s41467-020-16041-x
14. Catlett IM, Hedrick SM. Suppressor of Cytokine Signaling 1 is Required for the Differentiation of CD4+ T Cells. Nat Immunol (2005) 6:715–21. doi: 10.1038/ni1211
15. Vidalain PO, Laine D, Zaffran Y, Azocar O, Servet-Delprat C, Wild TF, et al. Interferons Mediate Terminal Differentiation of Human Cortical Thymic Epithelial Cells. J Virol (2002) 76:6415–24. doi: 10.1128/jvi.76.13.6415-6424.2002
16. Park JY, Ligons DL, Park JH. Out-Sourcing for Trans-Presentation: Assessing T Cell Intrinsic and Extrinsic IL-15 Expression With Il15 Gene Reporter Mice. Immune Netw (2018) 18:e13. doi: 10.4110/in.2018.18.e13
17. White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, et al. An Essential Role for Medullary Thymic Epithelial Cells During the Intrathymic Development of Invariant NKT Cells. J Immunol (2014) 192:2659–66. doi: 10.4049/jimmunol.1303057
18. Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 Receptor Beta-Chain in Natural Killer 1.1+ T Cell Receptor-Alpha Beta+ Cell Development. J Immunol (1997) 159:5931–5.
19. Minagawa M, Watanabe H, Miyaji C, Tomiyama K, Shimura H, Ito A, et al. Enforced Expression of Bcl-2 Restores the Number of NK Cells, But Does Not Rescue the Impaired Development of NKT Cells or Intraepithelial Lymphocytes, in IL-2/IL-15 Receptor Beta-Chain-Deficient Mice. J Immunol (2002) 169:4153–60. doi: 10.4049/jimmunol.169.8.4153
20. Waickman AT, Ligons DL, Hwang S, Park JY, Lazarevic V, Sato N, et al. CD4 Effector T Cell Differentiation is Controlled by IL-15 That is Expressed and Presented in Trans. Cytokine (2017) 99:266–74. doi: 10.1016/j.cyto.2017.08.004
21. Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, et al. An Essential Role for the IL-2 Receptor in Treg Cell Function. Nat Immunol (2016) 17:1322–33. doi: 10.1038/ni.3540
22. Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, et al. Deregulated T Cell Activation and Autoimmunity in Mice Lacking Interleukin-2 Receptor Beta. Science (1995) 268:1472–6. doi: 10.1126/science.7770771
23. Zhang S, Laouar A, Denzin LK, Sant’Angelo DB. Zbtb16 (PLZF) is Stably Suppressed and Not Inducible in Non-Innate T Cells Via T Cell Receptor-Mediated Signaling. Sci Rep (2015) 5:12113. doi: 10.1038/srep12113
24. Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, et al. Distinct Effects of STAT5 Activation on CD4+ and CD8+ T Cell Homeostasis: Development of CD4+CD25+ Regulatory T Cells Versus CD8+ Memory T Cells. J Immunol (2003) 171:5853–64. doi: 10.4049/jimmunol.171.11.5853
25. Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, et al. The Transcription Factor T-Bet is Induced by Multiple Pathways and Prevents an Endogenous Th2 Cell Program During Th1 Cell Responses. Immunity (2012) 37:660–73. doi: 10.1016/j.immuni.2012.09.007
26. Park JY, Kwon J, Kim EY, Fink J, Kim HK, Park JH. CD24(+) Cell Depletion Permits Effective Enrichment of Thymic iNKT Cells While Preserving Their Subset Composition. Immune Netw (2019) 19:e14. doi: 10.4110/in.2019.19.e14
27. Kadakia T, Tai X, Kruhlak M, Wisniewski J, Hwang IY, Roy S, et al. E-Protein-Regulated Expression of CXCR4 Adheres Preselection Thymocytes to the Thymic Cortex. J Exp Med (2019) 216:1749–61. doi: 10.1084/jem.20182285
28. Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A Thymic Precursor to the NK T Cell Lineage. Science (2002) 296:553–5. doi: 10.1126/science.1069017
29. Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, et al. The Transcriptional Regulator PLZF Induces the Development of CD44 High Memory Phenotype T Cells. Proc Natl Acad Sci USA (2008) 105:17919–24. doi: 10.1073/pnas.0805733105
30. Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant’Angelo DB. PLZF Induces the Spontaneous Acquisition of Memory/Effector Functions in T Cells Independently of NKT Cell-Related Signals. J Immunol (2010) 184:6746–55. doi: 10.4049/jimmunol.1000776
31. Lord JD, McIntosh BC, Greenberg PD, Nelson BH. The IL-2 Receptor Promotes Lymphocyte Proliferation and Induction of the C-Myc, Bcl-2, and Bcl-X Genes Through the Trans-Activation Domain of Stat5. J Immunol (2000) 164:2533–41. doi: 10.4049/jimmunol.164.5.2533
32. Cheng G, Yu A, Dee MJ, Malek TR. IL-2R Signaling is Essential for Functional Maturation of Regulatory T Cells During Thymic Development. J Immunol (2013) 190:1567–75. doi: 10.4049/jimmunol.1201218
33. Soper DM, Kasprowicz DJ, Ziegler SF. IL-2Rbeta Links IL-2R Signaling With Foxp3 Expression. Eur J Immunol (2007) 37:1817–26. doi: 10.1002/eji.200737101
34. Keller HR, Kim HK, Jo Y, Gress RE, Hong C, Park JH. The Abundance and Availability of Cytokine Receptor IL-2Rbeta (CD122) Constrain the Lymphopenia-Induced Homeostatic Proliferation of Naive CD4 T Cells. J Immunol (2020) 204:3227–35. doi: 10.4049/jimmunol.1901276
35. Dickgreber N, Farrand KJ, van Panhuys N, Knight DA, McKee SJ, Chong ML, et al. Immature Murine NKT Cells Pass Through a Stage of Developmentally Programmed Innate IL-4 Secretion. J Leukoc Biol (2012) 92:999–1009. doi: 10.1189/jlb.0512242
36. Park JY, Jo Y, Ko E, Luckey MA, Park YK, Park SH, et al. Soluble Gamma c Cytokine Receptor Suppresses IL-15 Signaling and Impairs iNKT Cell Development in the Thymus. Sci Rep (2016) 6:36962. doi: 10.1038/srep36962
37. Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, et al. Reversible Defects in Natural Killer and Memory CD8 T Cell Lineages in Interleukin 15-Deficient Mice. J Exp Med (2000) 191:771–80. doi: 10.1084/jem.191.5.771
38. Ranson T, Vosshenrich CA, Corcuff E, Richard O, Muller W, Di Santo JP. IL-15 is an Essential Mediator of Peripheral NK-Cell Homeostasis. Blood (2003) 101:4887–93. doi: 10.1182/blood-2002-11-3392
39. Fehniger TA, Suzuki K, Ponnappan A, VanDeusen JB, Cooper MA, Florea SM, et al. Fatal Leukemia in Interleukin 15 Transgenic Mice Follows Early Expansions in Natural Killer and Memory Phenotype CD8+ T Cells. J Exp Med (2001) 193:219–31. doi: 10.1084/jem.193.2.219
40. Spolski R, Li P, Leonard WJ. Biology and Regulation of IL-2: From Molecular Mechanisms to Human Therapy. Nat Rev Immunol (2018) 18:648–59. doi: 10.1038/s41577-018-0046-y
41. Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, et al. Stat5a/B are Essential for Normal Lymphoid Development and Differentiation. Proc Natl Acad Sci USA (2006) 103:1000–5. doi: 10.1073/pnas.0507350103
42. Tuttle KD, Krovi SH, Zhang J, Bedel R, Harmacek L, Peterson LK, et al. TCR Signal Strength Controls Thymic Differentiation of iNKT Cell Subsets. Nat Commun (2018) 9:2650. doi: 10.1038/s41467-018-05026-6
43. Dashtsoodol N, Bortoluzzi S. and Schmidt-Supprian M. T Cell Receptor Expression Timing and Signal Strength in the Functional Differentiation of Invariant Natural Killer T Cells. Front Immunol (2019) 10:841. doi: 10.3389/fimmu.2019.00841
44. Wang H, Hogquist KA. How Lipid-Specific T Cells Become Effectors: The Differentiation of iNKT Subsets. Front Immunol (2018) 9:1450. doi: 10.3389/fimmu.2018.01450
45. Kwon DI, Lee YJ. Lineage Differentiation Program of Invariant Natural Killer T Cells. Immune Netw (2017) 17:365–77. doi: 10.4110/in.2017.17.6.365
46. Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, et al. The Gene Encoding Early Growth Response 2, a Target of the Transcription Factor NFAT, is Required for the Development and Maturation of Natural Killer T Cells. Nat Immunol (2009) 10:306–13. doi: 10.1038/ni.1696
47. Seiler MP, Mathew R, Liszewski MK, Spooner CJ, Barr K, Meng F, et al. Elevated and Sustained Expression of the Transcription Factors Egr1 and Egr2 Controls NKT Lineage Differentiation in Response to TCR Signaling. Nat Immunol (2012) 13:264–71. doi: 10.1038/ni.2230
48. Park JY, DiPalma DT, Kwon J, Fink J, Park JH. Quantitative Difference in PLZF Protein Expression Determines iNKT Lineage Fate and Controls Innate CD8 T Cell Generation. Cell Rep (2019) 27:2548–57.e4. doi: 10.1016/j.celrep.2019.05.012
49. Suwa H, Tanaka T, Kitamura F, Shiohara T, Kuida K, Miyasaka M. Dysregulated Expression of the IL-2 Receptor Beta-Chain Abrogates Development of NK Cells and Thy-1+ Dendritic Epidermal Cells in Transgenic Mice. Int Immunol (1995) 7:1441–9. doi: 10.1093/intimm/7.9.1441
50. Sarafova SD, Van Laethem F, Adoro S, Guinter T, Sharrow SO, Feigenbaum L, et al. Upregulation of CD4 Expression During MHC Class II-Specific Positive Selection is Essential for Error-Free Lineage Choice. Immunity (2009) 31:480–90. doi: 10.1016/j.immuni.2009.07.006
51. Gioulbasani M, Galaras A, Grammenoudi S, Moulos P, Dent AL, Sigvardsson M, et al. The Transcription Factor BCL-6 Controls Early Development of Innate-Like T Cells. Nat Immunol (2020) 21:1058–69. doi: 10.1038/s41590-020-0737-y
52. Walker SR, Nelson EA, Frank DA. STAT5 Represses BCL6 Expression by Binding to a Regulatory Region Frequently Mutated in Lymphomas. Oncogene (2007) 26:224–33. doi: 10.1038/sj.onc.1209775
53. Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a Potent Negative Regulator of TFH Cell Differentiation. J Exp Med (2012) 209:243–50. doi: 10.1084/jem.20111174
Keywords: cytokines, IL-2 (interleukin-2), IL-15, Tbet, thymocytes
Citation: Won HY, Kim HK, Crossman A, Awasthi P, Gress RE and Park J-H (2021) The Timing and Abundance of IL-2Rβ (CD122) Expression Control Thymic iNKT Cell Generation and NKT1 Subset Differentiation. Front. Immunol. 12:642856. doi: 10.3389/fimmu.2021.642856
Received: 16 December 2020; Accepted: 27 April 2021;
Published: 14 May 2021.
Edited by:
Hongbo Chi, St. Jude Children’s Research Hospital, United StatesReviewed by:
Xiao-Ping Zhong, Duke University Medical Center, United StatesCopyright © 2021 Won, Kim, Crossman, Awasthi, Gress and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung-Hyun Park, cGFya2h5QG1haWwubmloLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.