
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 27 May 2021
Sec. B Cell Biology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.635326
This article is part of the Research TopicNew Insights into B cell Subsets in Health and DiseaseView all 18 articles
 Zuhui Su1†
Zuhui Su1† Yabin Jin1†
Yabin Jin1† Yu Zhang2†
Yu Zhang2† Zhanwen Guan1
Zhanwen Guan1 Huishi Li2
Huishi Li2 Xiangping Chen1
Xiangping Chen1 Chao Xie2
Chao Xie2 Chuling Zhang1
Chuling Zhang1 Xiaofen Liu2
Xiaofen Liu2 Peixian Li1
Peixian Li1 Peiyi Ye2
Peiyi Ye2 Lifang Zhang1
Lifang Zhang1 Yaozhong Kong2*
Yaozhong Kong2* Wei Luo1*
Wei Luo1*Membranous nephropathy (MN), an autoimmune glomerular disease, is one of the most common causes of nephrotic syndrome in adults. In current clinical practice, the diagnosis is dependent on renal tissue biopsy. A new method for diagnosis and prognosis surveillance is urgently needed for patients. In the present study, we recruited 66 MN patients before any treatment and 11 healthy control (HC) and analyzed multiple aspects of the immunoglobulin heavy chain (IGH) repertoire of these samples using high-throughput sequencing. We found that the abnormalities of CDR-H3 length, hydrophobicity, somatic hypermutation (SHM), and germ line index were progressively more prominent in patients with MN, and the frequency of IGHV3-66 in post-therapy patients was significantly lower than that in pre-therapy patients. Moreover, we found that the IGHV3-38 gene was significantly related to PLA2R, which is the most commonly used biomarker. The most important discovery was that several IGHV, IGHD transcripts, CDR-H3 length, and SHM rate in pre-therapy patients had the potential to predict the therapeutic effect. Our study further demonstrated that the IGH repertoire could be a potential biomarker for prognosis prediction of MN. The landscape of circulating B-lymphocyte repertoires sheds new light on the detection and surveillance of MN.
Membranous nephropathy (MN) is the most common cause of idiopathic nephrotic syndrome in non-diabetic adults worldwide, representing between 20 and 37% in most series and rising to as high as 40% in adults over 60 (1, 2). MN is characterized by thickening of the glomerular basement membrane (GBM) and rigid capillaries (3). It is widely accepted that MN is an autoimmune reaction to inherent podocyte antigens. The antigenic targets of these antibodies are most often PLA2R and THSD7A (4, 5).
Clinically, the diagnosis of MN depends on the histopathologic features of renal biopsy. However, renal biopsy is an invasive procedure, and it could increase the potential for kidney infection. Moreover, during pre- and post-treatments, repeating kidney biopsies to monitor disease progression is inconvenient. Patients badly need a dependable and minimally invasive biomarker to diagnose MN, assess disease stage, and evaluate treatment effect. Thanks to recent advances, we now know that some novel biomarkers have been developed, and the most commonly used biomarker is APLA2R (6–8). APLA2R has shown some potential in MN, but there are still some false positives and false negatives. As a result, more novel biomarkers are needed.
In recent years, arguments about MN have led to the conclusion that B cells play a key role in the disease, and the treatment targeting the B lymphocyte could be more significant. Rituximab (RTX) has been a novel anti-CD20 monoclonal antibody in patients with nephrotic syndrome due to MN (9). After RTX, the use of belimumab is a new step in B-lymphocyte-targeting therapy (10). Belson et al. (11) assessed the effect of belimumab on proteinuria and PLA2R serology in patients with PLA2R-related MN. These treatments targeting the B lymphocyte indicated that the pathogenesis of MN was strongly associated with B-lymphocyte repertoire.
At present, many studies using B-cell receptor repertoire high-throughput sequencing (BCR-HTS) have characterized the B-cell repertoires and demonstrated their diagnostic, therapeutic, and prognostic significance in different immune-related diseases, including autoimmune diseases (12), immunoglobulin A nephropathy (13), and gastric cancer (14). The B-lymphocyte repertoire could reflect the B-cell immune condition in MN, and sequencing the B-cell receptors to investigate the diversity of B lymphocytes provided a new path to understand the pathogenesis of the disease.
Numerous studies have focused mostly on the immune status of immunoglobulin A nephropathy rather than on MN. Chen Huang et al. (13) presented a comprehensive landscape of T-cell receptor beta chain (TCRβ) and IGH in immunoglobulin A nephropathy patients. Dapeng Chen et al. (15) analyzed the CDR-H3 clones of immunoglobulin A nephropathy patients and Minglin Ou et al. (16) also reported B- and T-cell repertoires CDR3 sequences in immunoglobulin A nephropathy patients. Compared to it, there are few studies about IGH genes in MN.
In this study, we analyzed the IGH repertoire of healthy control (HC) and pre- and post-therapy MN patients by BCR-HTS. We found that six IGHC, one IGHD, and 24 IGHV genes were significantly different between MN patients and HC. For the IGHV genes, most of them belong to the IGHV3 and IGHV4 families. The analysis of the CDR-H3 length, hydrophobicity, and somatic hypermutation (SHM) also revealed significant difference between MN patients and HC. Moreover, we found that the frequency of IGHV3-66 in post-therapy patients was significantly lower than that in pre-therapy patients, and IGHV3-38 genes were significantly related to PLA2R, which is the commonly used biomarker. Importantly, we identified 12 IGHV genes, and two IGHD genes were significantly different between complete remission (CR) and non-CR patients. Moreover, the CDR-H3 length and SHM rate in the IgM subtype were observed to be higher in the CR group. All the results demonstrated that the IGH repertoire has the potential to be a novel biomarker for prognosis prediction in MN.
This study was approved by the Ethics Committee of the Affiliated Foshan Hospital of Sun Yat-Sen University. From 2017 to 2018, we recruited 66 patients with MN before any treatment. Clinical characteristics of the MN patients are shown in Tables 1 and S1. The disease diagnosis criterion of MN was established by renal biopsy. The kidney tissue biopsies of 66 patients were examined by light microscopy, electron microscopy, and immunofluorescence. Peripheral blood samples were collected before treatment for the 66 patients. Among them, the peripheral blood samples of 13 patients were collected at 6 months after treatment. Assessment of disease state indicators included 24 h urine protein, serum albumin, PLA2R status, uric acid, and serum creatinine, which were collected in renal biopsy and the follow-up at 6 months after therapy. According to Kidney Disease: Improving Global Outcomes (KDIGO), we evaluated the prognostic condition of the MN patients (17, 18). Without a history of cancer, autoimmune disorder, or surgery, 11 healthy volunteers were recruited as HC, and peripheral blood samples were drawn from each person. Peripheral blood mononuclear cells (PBMCs) were isolated immediately and lysed with TRIzolR reagent (Life, US), then frozen at −80°C.
Following the manufacturer’s instructions, we extracted RNA from PBMC lysates using a total RNA Kit (OMEGA Bio-tek, US). For each sample, we used the SMARTer PCR cDNA synthesis kit (Clontech, US) to reverse transcribe 1 µg total RNA into cDNA. To amplify a fully sequenced IGH strand fragment, the 5′RACE-ready cDNA was used as a template for a 5′RACE-PCR with forward universal primer and reverse mix primers specific to the IGH constant region Cα (IGHA), Cμ (IGHM), Cγ (IGHG), Cδ (IGHD), and Cϵ (IGHE) (The primer list is shown in Table S3). The PCR conditions were as follows: 3 min denaturation at 94°C was followed by 40 cycles of 15 s at 94°C, 30 s at 58°C, and 45 s at 72°C, plus a final extension for 10 min at 72°C. Next, we used the gel extraction kit (QIAGEN, German) to purify the products by 2% agarose gel electrophoresis. Illumina HiSeq sequence adaptors were ligated to construct sequencing libraries, which were then sequenced on an Illumina HiSeq Xten platform. Data from the IGH repertoire library sequencing can be provided on request.
The sequencing data was stored in a FASTQ format. After filtering low-quality sequences, the high-quality sequences were aligned with the BCR reference sequences by BLAST (-stepSize = 5 –minIdentity = 0 –minScore = 0) (19). The reference sequences were downloaded from the IMGT/GENE database (20). If V, J, and C genes in a given sequence were all identified, we further translated them into an amino acid (aa) sequence. The aa sequences without a terminator were selected as the productive BCR sequences. At the V–D–J junctions, the sequences that started with cysteine and ended with the tryptophan were defined as CDR-H3. The source code of our own BCR sequence bioinformatical analysis tool is available on request.
We selected the Shannon index and Simpson index to estimate the diversity in the BCR repertoire. Both of them take into consideration the two components that constitute the concept of diversity, the richness of a population and its homogeneity. The richness of a population is defined by its total number of species, and homogeneity measures the distribution of the species (21–23). The formulas of the Shannon index and Simpson index are as follows:
where i is an index that is chosen between 1 and the number of species s; ni is the number of sequencing reads in species i; and N is the total number of reads.
Kyte–Doolittle index of hydrophobicity in CDR-H3 was calculated based on the normalized Kyte–Doolittle scale which assigned one value to each amino acid (24). SHM introduces additional diversity in the IGH repertoire of mature B cells and allows selection of high-affinity antibodies. SHM rate was calculated by dividing the number of point mutations by the total number of nucleotides in the V gene of our sequences (25). The germline index (GI) can be used to estimate the abundance of palindromic and “N” nucleotides and was calculated by dividing the number of nucleotides in the CDR3 that were encoded by V, D, and J genes by the total number of nucleotides contained in the CDR3, generating a value between 0 and 1 (26).
Comparisons between groups were conducted using the Mann–Whitney U test or the Wilcoxon signed rank test if appropriate, and P <0.05 was considered statistically significant. The Spearman correlation coefficient (r) was used to measure the linear correlation between pairs of variables. These analyses were performed using GraphPad Prism software (version 5.1), SPSS (version 20.0), and R software (version 3.4.1; http://www.Rproject.org).
The demographic and clinical characteristics of the patients are summarized in Tables 1 and S1. With regard to the pre-therapy MN patients, 42 of them were male, and 24 of them were female. The average age of the patients was 51 years old. There we no significant differences in gender and age between the MN patients and HC. The average 24 h proteinuria measurement was 4.15 g for MN patients and varied largely among patients (0.32 g/24 h–15.9 g/24 h). Serum creatinine, serum albumin, uric acid, and PLA2R also varied extensively among MN patients, with an average level of 76.5 µmol/L, 26.8 g/L, 413 µmol/L, and 76.39 Ru/ml, respectively.
A total of 119,300,197 productive aa sequences were obtained from 92 blood samples of the patients and HC, with an average of 1,296,741 productive sequences generated per sample. The average number of productive unique sequences per sample was 270,904. Eighty-six distinct IGHV, 25 distinct IGHD, and 10 distinct IGHC segments were identified, and the usage frequencies of these segments were analyzed in each sample, which are shown in Figure 1 and Table S2.
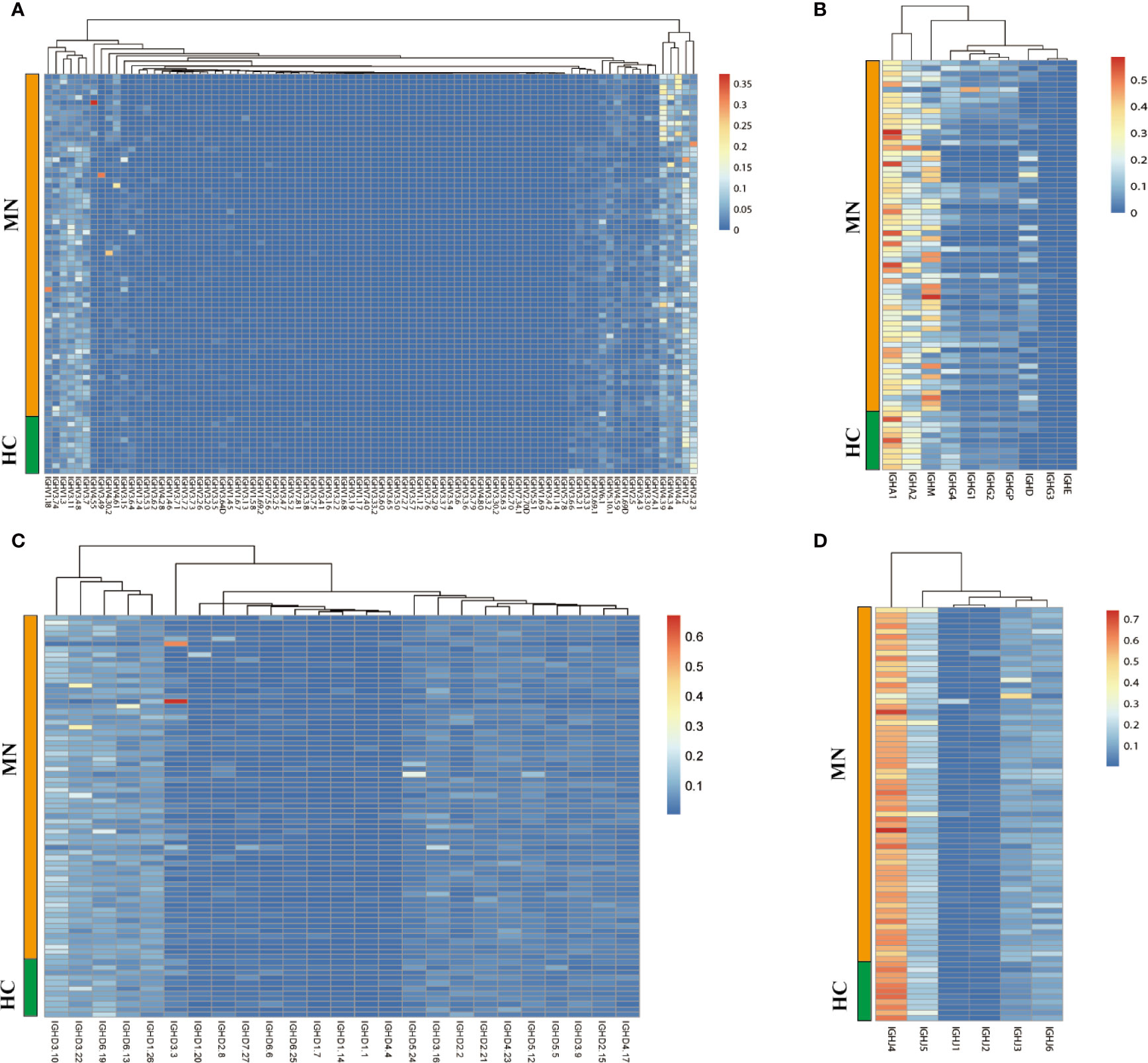
Figure 1 The heat maps of IGH gene usage frequencies in BCR repertoire from the sample of the MN patients and HC. (A): IGHV; (B): IGHC; (C): IGHD; (D): IGHJ.
We compared the IGHC frequency between MN patients and HC. The frequency was delineated in heat maps (Figure 1A). There are six transcripts (IGHA1, IGHA2, IGHM, IGHG4, IGHD, IGHE) in Figure 1 that are observably different between MN patients and HC. IGHA is composed of two subclasses: IGHA1 and IGHA2. A more detailed description is presented in Figure 2. We found that the frequency of IGHM, IGHD, and IGHE in PBMCs was significantly higher than those in MN patients. In contrast, the frequencies of IGHA and IGHG4 were significantly lower in MN patients than in the HC. It showed that, compared with HC, the proportion of IG in the BCR repertoire of MN patients has changed. This result is consistent with previously reported results (27).
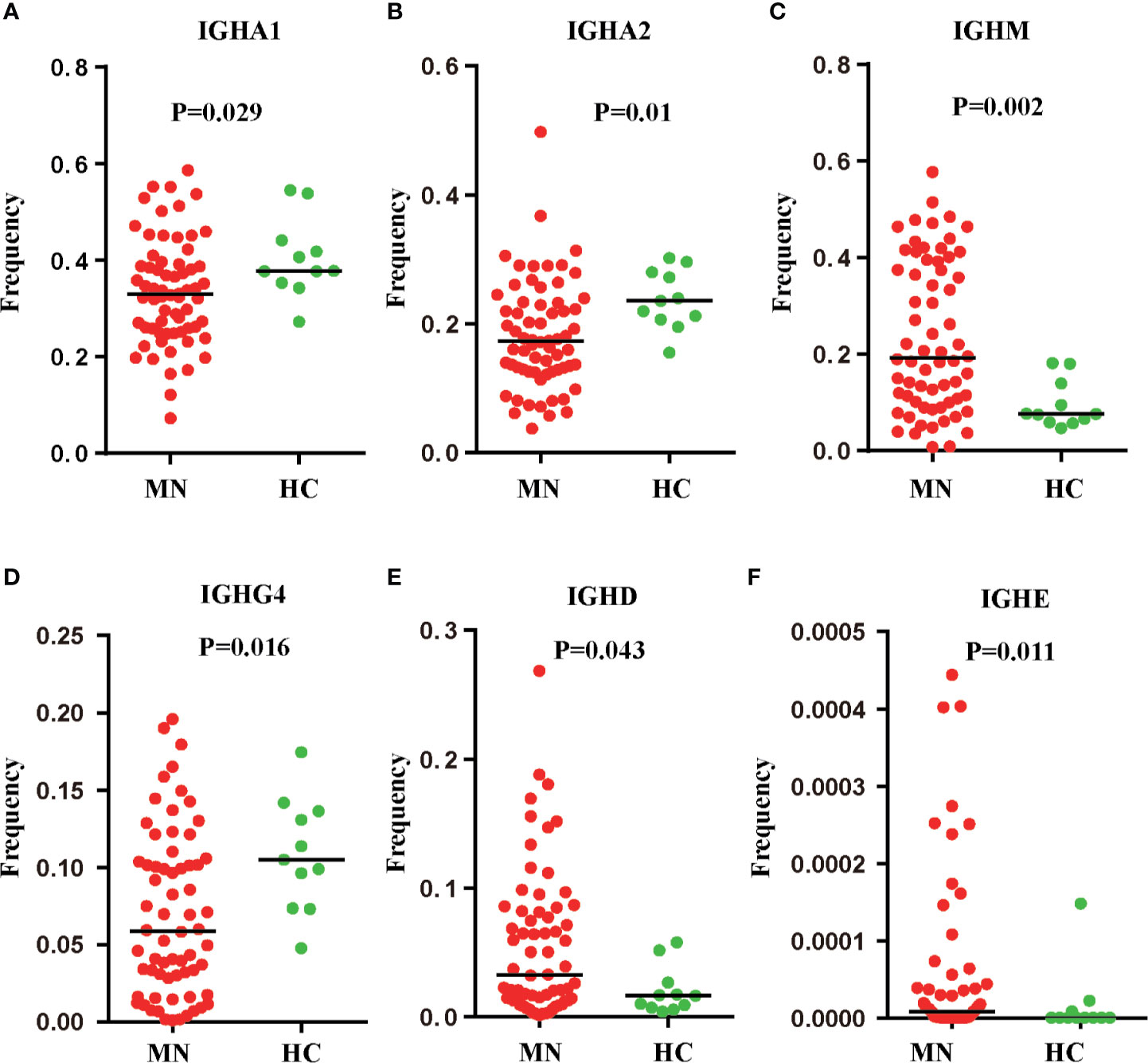
Figure 2 Six IGH transcripts were significantly different between MN patients and HC. IGHA1 (A), IGHA2 (B), IGHM (C), IGHG4 (D), IGHD (E), IGHE (F) frequency distribution in PBMC of MN patients (n = 66) and HC (n = 11) with line at the median.
We also compared the IGHV, IGHD, and IGHJ gene usage between MN patients and HC. There were 24 IGHV genes that were significantly different between the two groups (P < 0.05; Figure 3A; IGHV3-72, IGHV1-2, IGHV3-23, IGHV3-64D, IGHV3-36, IGHV3-33, IGHV3-35, IGHV3-69-1, IGHV5-78, IGHV3-16, IGHV3-15, IGHV3-7, IGHV3-48, IGHV3-64, IGHV3-13, IGHV1-58, IGHV4-39, IGHV3-73, IGHV4-34, IGHV1-69-2, IGHV2-5, IGHV4-4, IGHV4-30-2, IGHV4-59, IGHV3-49). We investigated the frequency of the IGHV3 and IGHV4 families and found that the frequency of the IGHV3 family was lower in MN patients, but the frequency of the IGHV4 family was higher than that in HC, with significant difference (Figure 3B). Moreover, we found the frequency of the IGHD2-2 was significantly lower in MN patients (P = 0.021; Figure 3C). These results demonstrated that usage patterns of IGHV and IGHD genes were skewed in patients, and the B-cell immune status in MN patients had been altered.
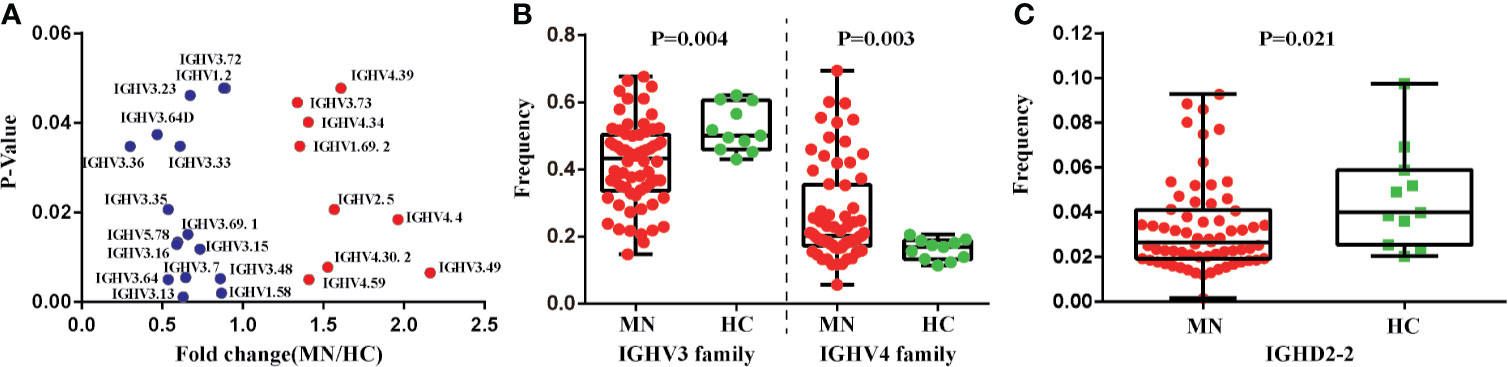
Figure 3 The IGHV and IGHD gene usage between MN patients and the HC. (A) Twenty-four IGHV genes which are significantly different between MN patients and the HC (without correction for multiple comparison). (B) The frequencies of IGHV3 family and IGHV4 family have significant differences in MN patients and the HC. (C) The frequency of IGHD2-2 has significant differences in MN patients and the HC (p = 0.021).
We compared the CDR-H3 length, hydrophobicity, SHM, and GI between MN patients and HC (Figures 4A–F); the CDR-H3 length distribution of IGHA, IGHM, IGHG, and IGHD transcripts was observed to be more prominent in patients with MN (P < 0.05; Figure 4C), and we found abnormalities of the hydrophobicity profile of the IGHM and IGHD in MN patients, as measured by the normalized Kyte–Doolittle index (P < 0.05; Figure 4D). In particular, MN patients showed a higher rate of SHM in IGHA, IGHM, IGHG, and IGHD transcripts compared with HC (P < 0.05; Figure 4E). Overall, these results showed the CDR-H3 length, hydrophobicity, and SHM had a significant difference between the patients with MN and HC. It indicated that CDR-H3 length, hydrophobicity, and SHM of the peripheral BCR repertoire were influenced significantly by MN.
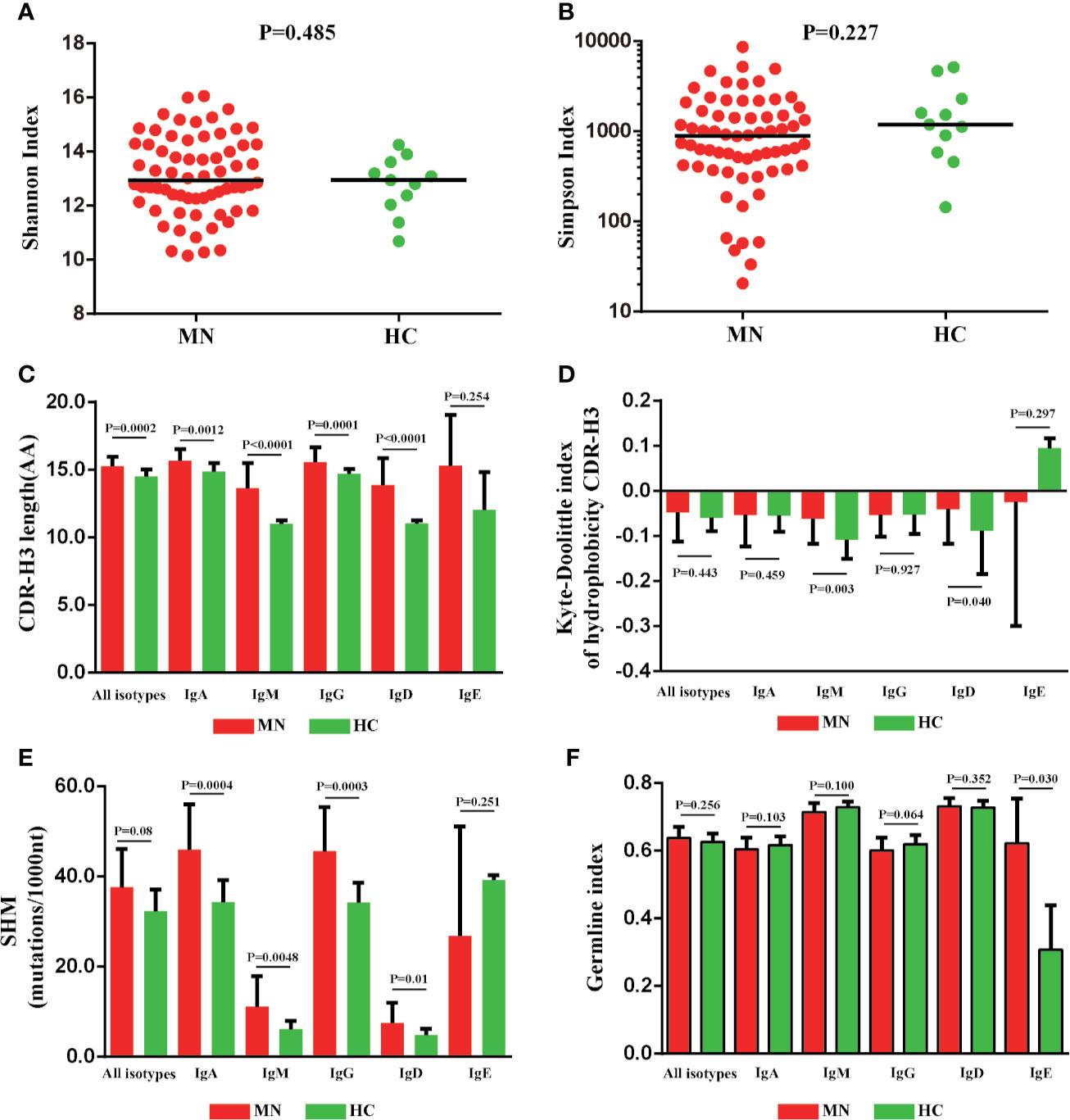
Figure 4 The CDR-H3 length, hydrophobicity somatic hypermutation and GI of MN patients showed significant differences compared with HC. The Shannon index (A), Simpson index (B), CDR-H3 length distribution (C), the Kyte–Doolittle index of hydrophobicity (D), the rate of SHM (E), and the germline index (F) of IGH repertoire in MN patients and HC.
When comparing the BCR repertoire between the pre- and post-therapy patients, no significant difference was observed for the diversity index, CDR-H3 length, SHM, hydrophobicity, GI, IGHD genes, IGHJ genes, and IGHC genes. However, we found that the frequency of IGHV3-66 was significantly lower in post-therapy patients than that in pre-therapy patients (Figure 5).
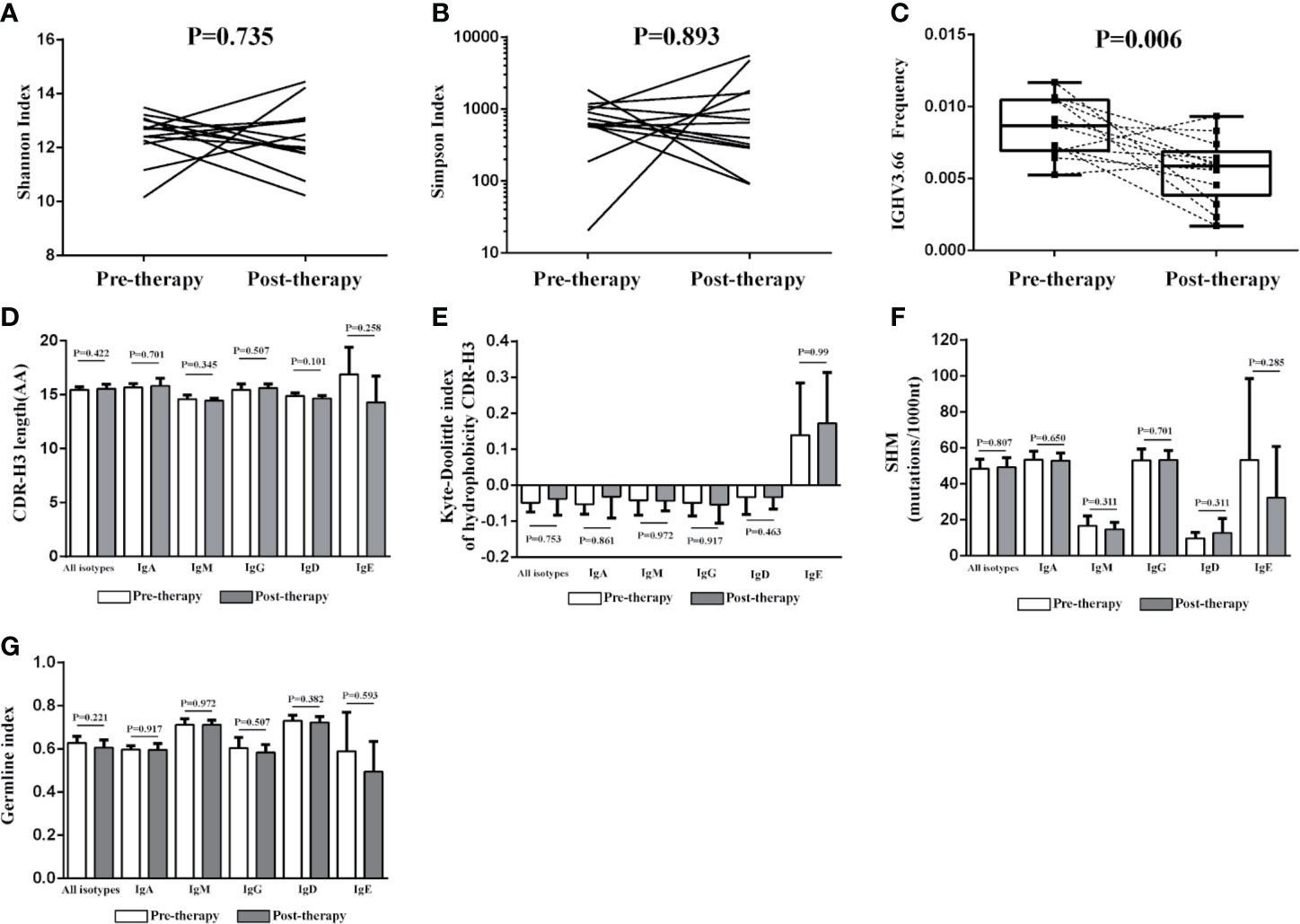
Figure 5 Change of the IGH repertoire between the pre-therapy and post-therapy patients. The Shannon index (A), Simpson index (B), CDR-H3 length distribution (D), the Kyte–Doolittle index of hydrophobicity (E), the rate of SHM (F) and the germline index (G) of IGH repertoire have no significant differences between pre-therapy and post-therapy, but the frequency of IGHV3-66 was significantly lower in the samples of post-therapy patients (n = 13) than paired pre-therapy samples (n = 13) (C).
According to KDIGO, we divided the patients into two groups: CR and non-CR, after 6 months of therapy. We tried to investigate the diversity, hydrophobicity, GI, IGHJ gene, and IGHC gene between the two groups and found that no significant difference was observed (Figures 6E, G). However, we identified three IGHV genes (IGHV4.61, IGHV4.59, V4 family) were significantly increased in the CR group (Figure 6A). Interestingly, two of these genes (red in Figure 6A) in MN patients were significantly higher than those in HC (Figure 3A). Moreover, 11 IGHV and IGHD genes (IGHV1.12, IGHV1.2, IGHV3.21, IGHV3.48, IGHV3.69.1, IGHV3.2, IGHV3.35, IGHV3.64, IGHV3.16, IGHD1.26, IGHD1.14) were significantly lower in the CR group (Figure 6A), and six of these genes (blue in Figure 6A) in MN patients were significantly lower than those in HC (Figure 3A). In addition, APLA2R, which is the most commonly used biomarker, was found to be correlated with the CR group and non-CR group. APLA2R in the non-CR group was significantly higher than that in the CR group (Figure 6B). More importantly, we found the IGHV3-38 gene had a negative correlation with APLA2R (Figure 6C). In addition, we found that CDR-H3 length distribution of all isotypes (P = 0.014; Figure 6D) and SHM rate of IGHM (P = 0.032; Figure 6F) were more prominent in the CR group.
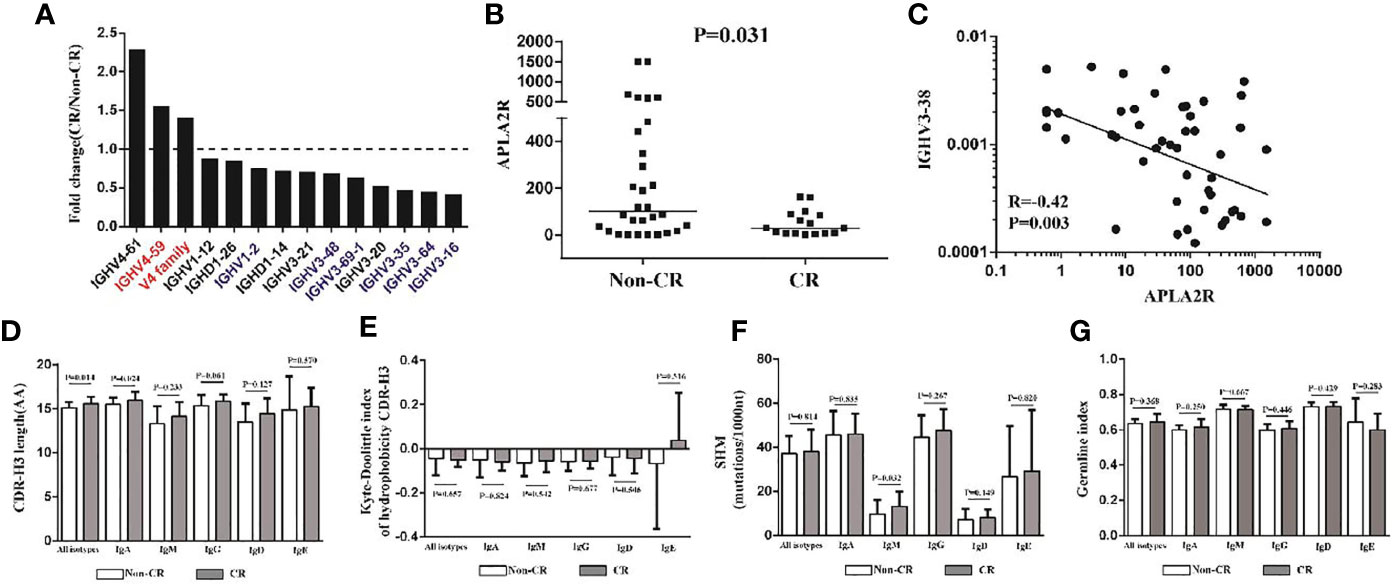
Figure 6 The IGHV genes, CDR-H3 length, SHM rate, and APLA2R between the CR and non-CR patients. (A) Three IGHV genes were significantly increased in the CR group (n = 22), two of them in MN patients are significantly higher than in HC, which is represented by red. Nine IGHV genes and two IGHD genes that were significantly lower than non-CR (n = 43), and six of them in MN patients are significantly higher than in HC, which is represented by blue. (B) The APLA2R in non-CR group and CR group. (C) IGHV3-38 gene has a negative correlation with APLA2R. (D–G) CDR-H3 length, the Kyte–Doolittle index of hydrophobicity, SHM rate and germline index of IGH repertoire in CR and non-CR patients.
B cells and T cells serve important roles in the human immune system, and understanding their receptor characteristics in human autoimmune diseases may help the development of specific immunotherapeutic interventions for diseases in the clinic (28). Each B cell expresses a single BCR (29), and the diverse range of BCRs expressed by the total B-cell population of an individual is termed the ‘BCR repertoire’. Further diversification of BCRs occurs in specialized germinal centers, in which SHM of genes that encode the variable (V) regions of antibodies may enhance BCR affinity and specificity (30). This diversification of B-cell clones after exposure to antigen is tempered by tolerance checkpoints to reduce the risk of autoimmunity (31). The peripheral BCR repertoire is thus a composite of both the naive repertoire and that generated by antigenic encounter. Abnormal use of the IGHV genes can exhibit abnormal isotype-specific clonal expansion or diversity in some immune-mediated diseases like SLE, Crohn’s disease, and EGPA (32). Recently, Meng Yuan et al. analyzed 294 anti-SARS-CoV-2 antibodies and found that IGHV3-53 is the most frequently used IGHV gene for targeting the receptor-binding domain of the spike protein (33). Most cases of MN have circulating IgG4 autoantibody on the podocyte membrane antigen PLA2R (70%), which is characterized by glomerular subepithelial deposits of immune complexes, severe basement membrane thickening, and little or no inflammatory infiltrate (PMN); B-cell dysfunction plays a role in the pathogenesis of MN, and differential skewing of IGHV genes could reflect the B-cell immune status in MN patients.
We previously demonstrated the usage patterns of the TCRβ V gene and J gene were skewed in autoimmune diseases and tumors (34–37). In our previously reported study, we analyzed the TCRβ repertoire of the circulating T lymphocytes of MN patients (pre-therapy and post-therapy) and HC using TCR-HTS, and we found that the diversities of VJ cassette combination in the peripheral blood of MN patients were lower than those of HC. We also found the TCRβ repertoire similarity between pre- and post-therapy could reflect the clinical outcome, and two Vβ genes in pre-therapy also had the potential to predict the therapeutic effect (38). In this work, we analyzed the IGH repertoire of PBMCs of MN patients pre- and post-therapy, as well as the PBMCs of HC, to investigate the B-cell immunity in MN patients. The patient cohort in this study included the patients from the previous reported study (38), and new patients were recruited. Changes of IGH genes in MN patients might reveal the occurrence and development of MN disease and could be used for diagnosis.
Numerous studies focused on IGH CDR-H3 clones of IgA nephropathy, with few studies about IGHV genes in MN. MN is an immune-related disease. If the treatment is effective, the immune status of the human body will be different from that before treatment, and the IGH repertoire will also be changed significantly, which has been reported in numerous studies (39–41). In this study, we found the frequency of IGHV3-66 was significantly lower in the post-therapy patients than in the pre-therapy patients, and some IGHV genes skewed in MN patients were also significantly different between the CR and non-CR groups. All the results demonstrated that the IGHV gene has the potential to assist diagnosis and predict efficacy. However, due to the limited number of samples in our work, we could not confirm the performance of IGHV genes associated with MN disease. In fact, those biomarkers that show some potential for prognosis prediction of MN need more studies to demonstrate their accuracy and reproducibility.
The length and amino acid composition of CDR-H3 region affect recognition of antigens. Progressive reduction of CDR-H3 length and increase of highly hydrophobic and hydrophilic sequences during differentiation from immature to naive and memory B cells are paralleled by a progressive decrease in the proportion of self-reactive B-cell specificities during B-cell ontogeny (42, 43). The increased SHM indicates an enhanced in-vivo activation of B cells, thus suggesting abnormal activation of B cell in MN patients. In this study, these parameters were significantly different between MN and HC, CR and non-CR.
As previously discussed, proteinuria and serum creatinine may not accurately reflect disease activity and do not discriminate between immunologically active disease and irreversible structural glomerular damage (44). In fact, some studies have reported a new individualized serology-based approach that complements and refines the traditional proteinuria-driven approach, which has recently been proposed (45) and has been the subject of recent reviews (46). The hope is that we can find a personalized approach that can improve prognostic accuracy and that can provide individualized treatment for patients with MN while limiting unnecessary exposure to immunosuppressive therapy, which has many adverse effects, such as liver function damage, infection, and myelosuppression. This study indicated that some parameters of IGH repertoire could be a predictive biomarker for the treatment efficacy of MN.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the ethics committee of the Affiliated Foshan Hospital of Sun Yat-Sen University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
YZ, HL, CX, XL, PY, and YK provided patients’ samples and clinical information. ZS, ZG, XC, CZ, PL, and LZ performed the experiments. YJ performed the statistical analysis. YJ and WL analyzed and interpreted the data. YK and WL designed and supervised the study. ZS and YJ wrote the manuscript. WL did an extensive revision. All authors contributed to the article and approved the submitted version.
This work was funded by grants from the National Natural Science Foundation of China (81972335), Foundation and Applied Basic Research Fund of Guangdong Province (2019A1515110676), Science and Technology Innovation Platform in Foshan City (FS0AA-KJ218-1301-0007), and Foshan city climbing peak plan (2019A004, 2019A025), Medical Engineering Technology Research and Development Center of Immune Repertoire in Foshan and Medical Scientific Research Foundation of Guangdong Province of China (A2021493).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the patients and their families for participation in the study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.635326/full#supplementary-material
1. Cattran DC, Brenchley PE. Membranous Nephropathy: Integrating Basic Science Into Improved Clinical Management. Kidney Int (2017) 91(3):566–74. doi: 10.1016/j.kint.2016.09.048
2. Beck LH Jr, Salant DJ. Membranous Nephropathy: From Models to Man. J Clin Invest (2014) 124(6):2307–14. doi: 10.1172/JCI72270
3. Cai Q, Hendricks AR. Membranous Nephropathy: A Ten-Year Journey of Discoveries. Semin Diagn Pathol (2020) 37(3):116–20. doi: 10.1053/j.semdp.2020.01.001
4. Beck LH Jr, Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, et al. M-Type Phospholipase A2 Receptor Astarget Antigen in Idiopathic Membranous Nephropathy. N Engl J Med (2009) 361(1):11–21. doi: 10.1056/NEJMoa0810457
5. Tomas NM, Beck LH Jr, Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, et al. Thrombospondin Type-1 Domaincontaining 7A in Idiopathic Membranous Nephropathy. N Engl J Med (2014) 371(24):2277–87. doi: 10.1056/NEJMoa1409354
6. Beck LH Jr, Fervenza FC, Beck DM, Bonegio RGB, Malik FA, Erickson SB, et al. Rituximab-Induced Depletion of anti-PLA2R Autoantibodies Predicts Response in Membranous Nephropathy. J Am Soc Nephrol (2011) 22(8):1543–50. doi: 10.1681/ASN.2010111125
7. Hoxha E, Harendza S, Pinnschmidt HO, Tomas NM, Helmchen U, Panzer U, et al. Spontaneous Remission of Proteinuria Is a Frequent Event in Phospholipase A2 Receptor Antibody-Negative Patients With Membranous Nephropathy. Nephrol Dial Transplant (2015) 30(11):1862–9. doi: 10.1093/ndt/gfv228
8. Hu SL, Wang D, Gou WJ, Lei Q-F, Ma T-A, Cheng J-Z, et al. Diagnostic Value of Phospholipase A2 Receptor in Idiopathic Membranous Nephropathy: A Systematic Review and Meta-Analysis. J Nephrol (2014) 27(2):111–7. doi: 10.1007/s40620-014-0042-7
9. Esposito P, Grosjean F. Rituximab in Primary Membranous Nephropathy: Beyond a B-Cell-Centered Paradigm? Clin Exp Nephrol (2018) 22(1):208–9. doi: 10.1007/s10157-017-1437-2
10. Dahan K, Boffa J-J. Membranous Glomerulonephritis: A Step Forward in B-cell Targeting Therapy? Nephrol Dial Transplant (2020) 35(4):549–51. doi: 10.1093/ndt/gfz139
11. Barrett C, Willcocks LC, Jones RB, Tarzi RM, Henderson RB, Cai G, et al. Effect of Belimumab on Proteinuria and Anti-Phospholipase A2 Receptor Autoantibody in Primary Membranous Nephropathy. Nephrol Dial Transplant (2020) 35(4):599–606. doi: 10.1093/ndt/gfz086
12. Pollastro S, Klarenbeek PL, Doorenspleet ME, van Schaik BDC, Esveldt REE, Thurlings RM, et al. Non-Response to Rituximab Therapy in Rheumatoidarthritis Is Associated With Incomplete Disruption Ofthe B Cell Receptor Repertoire. Ann Rheum Dis (2019) 0:1–7. doi: 10.1136/annrheumdis-2018-214898
13. Huang C, Li X. The Landscape and Diagnostic Potential of T and Bcell Repertoire in Immunoglobulin A Nephropathy. Front Immunol (2018) 97:2729. doi: 10.3389/fimmu.2018.02729
14. Konishi H, Komura D, Katoh H, Atsumi S, Koda H, Yamamoto A, et al. Capturing the Differences Between Humoral Immunity in the Normal and Tumor Environments From Repertoire-Seq of B-Cell Receptors Using Supervised Machine Learning. BMC Bioinf (2019) 20:267. doi: 10.1186/s12859-019-2853-y
15. Chen D, Zhang Z, Yang Y, Hong Q, Li W, Zhuo L, et al. High-Throughput Sequencing Analysis of Genes Encoding the B-Lymphocyte Receptor Heavy-Chain CDR3 in Renal and Peripheral Blood of IgA Nephropathy. Biosci Rep (2019) 39(10):BSR20190482. doi: 10.1042/BSR20190482
16. Ou M, Zheng F, Zhang X, Liu S, Tang D, Zhu P, et al. Integrated Analysis of B-Cell and T-Cell Receptors by High-Throughput Sequencing Reveals Conserved Repertoires in IgA Nephropathy. Mol Med Rep (2018) 17(15):7027–36. doi: 10.3892/mmr.2018.8793
17. Gordon CE, Balk EM, Francis JM. Summary of the 2018 Kidney Disease Improving Global Outcomes (KDIGO) Guideline on Hepatitis C in Chronic Kidney Disease. Semin Dial (2019) 32(2):187–95. doi: 10.1111/sdi.12768
18. Eckardt KU, Kasiske BL. Kidney Disease: Improving Global Outcomes. Nat Rev Nephrol (2009) 5(11):650–7. doi: 10.1038/nrneph.2009.153
19. Ruggiero E, Nicolay JP, Fronza R, Arens A, Paruzynski A, Nowrouzi A, et al. High-Resolution Analysis of the Human T-Cell Receptor Repertoire. Nat Commun (2015) 6:8081. doi: 10.1038/ncomms9081
20. Giudicelli V, Chaume D, Lefranc MP. Imgt/Gene-DB: A Comprehensive Database for Human and Mouse Immunoglobulin and T Cell Receptor Genes. Nucleic Acids Res (2005) 33(Database issue):D256–61. doi: 10.1093/nar/gki010
21. Shannon CE. The Mathematical Theory of Communication. 1963 MD Comput (1997) 14(4):306–17. doi: 10.2307/3498901
22. Schneider-Hohendorf T, Mohan H, Bien CG, Breuer J, Becker A, Görlich D, et al. CD8(+) T-Cell Pathogenicity in Rasmussen Encephalitis Elucidated by Large-Scale T-Cell Receptor Sequencing. Nat Commun (2016) 7:11153. doi: 10.1038/ncomms11153
23. Sims JS, Grinshpun B, Feng Y, Ung TH, Neira JA, Samanamud JL, et al. Diversity and Divergence of the Glioma-Infiltrating T-cell Receptor Repertoire. Proc Natl Acad Sci USA (2016) 113(25):E3529–37. doi: 10.1073/pnas.1601012113
24. Eisenberg D. Three-Dimensional Structure of Membrane and Surface. proteins Annu Rev Biochem (1984) 53:595–623. doi: 10.1146/annurev.bi.53.070184.003115
25. Rogosch T, Kerzel S, Hoi KH, Zhang Z, Maier RF, Ippolito GC, et al. Immunoglobulin Analysis Tool: A Novel Tool for the Analysis of Human and Mouse Heavy and Light Chain Transcripts. Front Immunol (2012) 3:176. doi: 10.3389/fimmu.2012.00176. eCollection 2012.
26. Yu X, Almeida JR, Darko S, van der Burg M, DeRavin SS, Malech H, et al. Human Syndromes of Immunodeficiency and Dysregulation are Characterized by Distinct Defects in T-Cell Receptor Repertoire Development. Allergy Clin Immunol (2014) 133:1109–15. doi: 10.1016/j.jaci.2013.11.018
27. Zhu H, Han Q, Zhang D, Wang Y, Gao J, Yang X, et al. The Clinicopathological Features of Patients With Membranous Nephropathy. Int J Nephrol Renovasc Dis (2018) 11:33–40. doi: 10.2147/ijnrd.s149029
28. Borroto A, Reyes-Garau D, Jiménez MA, Carrasco E, Moreno B, Martínez-Pasamar S, et al. First-in-Class Inhibitor of the T Cell Receptor for the Treatment of Autoimmune Diseases. Sci Transl Med (2016) 8(370):370ra184. doi: 10.1126/scitranslmed.aaf2140
29. Nossal GJV, Lederberg J. Antibody Production by Single Cells. Nature (1958) 181:1419–20. doi: 10.1038/1821383a0
30. De Silva NS, Klein U. Dynamics of B Cells in Germinal Centres. Nat Rev Immunol (2015) 15:137–48. doi: 10.1038/nri3804
31. Giltiay NV, Chappell CP, Clark EA. B-Cell Selection and the Developmentof Autoantibodies. Arthritis Res Ther (2012) 14:S1. doi: 10.1186/ar3918
32. Bashford-Rogers RJM, Bergamaschi1 L, McKinney EF, Pombal DC, Mescia F, Lee JC, et al. Analysis of the B Cell Receptor Repertoire in Six Immune-Mediated Diseases. Nature (2019) 574(7776):122–6. doi: 10.1038/s41586-019-1595-3
33. Yuan M, Liu H, Wu NC, Lee CD, Zhu X, Zhao F, et al. Structural Basis of a Shared Antibody Response to SARS-Cov-2. Science (2020) 369(6507):1119–23. doi: 10.1126/science.abd2321
34. Jin YB, Luo W, Zhang GY, Lin K-R, Cui J-H, Chen X-P, et al. TCR Repertoire Profiling of Tumors, Adjacent Normal Tissues, and Peripheral Blood Predicts Survival in Nasopharyngeal Carcinoma. Cancer ImImmunother (2018) 67(11):1719–30. doi: 10.1007/s00262-018-2237-6
35. Lin KR, Pang DM, Jin YB, Hu Q, Pan YM, Cui JH, et al. Circulating CD8(+) T-Cell Repertoires Reveal the Biological Characteristics of Tumors and Clinical Responses to Chemotherapy in Breast Cancer Patients. Cancer Im Immunother> (2018) 67(11):1743–52. doi: 10.1007/s00262-018-2213-1
36. Cui JH, Lin KR, Yuan SH, Jin YB, Chen XP, Su XK, et al. Tcr Repertoire as a Novel Indicator for Immune Monitoring and Prognosis Assessment of Patients With Cervical Cancer. Front Immunol (2018) 9:2729. doi: 10.3389/fimmu.2018.02729
37. Fervenza FC, Appel GB, Barbour SJ, Rovin BH, Lafayette RA, Aslam N, et al. Rituximab or Cyclosporine in the Treatment of Membranous Nephropathy. N Engl J Med (2019) 381(1):36–46. doi: 10.1056/NEJMoa1814427
38. Zhang Y, Jin Y, Guan Z, Li H, Su Z, Xie C, et al. The Landscape and Prognosis Potential of the T-Cell Repertoire in Membranous Nephropathy. Front Immunol (2020) 11:387. doi: 10.3389/fimmu.2020.00387
39. Tucci FA, Kitanovski S. Biased IGH VDJ Gene Repertoire and Clonal Expansions in B Cells of Chronically Hepatitis C Virus-Infected Individuals. Blood (2018) 131(5):546–57. doi: 10.1182/blood-2017-09-805762
40. Zhang W, Feng Q, Wang C. Characterization of the B Cell Receptor Repertoire in the Intestinal Mucosa and of Tumor-Infiltrating Lymphocytes in Colorectal Adenoma and Carcinoma. J Immunol (2017) 198(9):3719–28. doi: 10.4049/jimmunol.1602039
41. Wu J, Jia S. Minimal Residual Disease Detection and Evolved IGH Clones Analysis in Acute B Lymphoblastic Leukemia Using Igh Deep Sequencing. Front Immunol (2016) 7:403. doi: 10.3389/fimmu.2016.00403
42. Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant Autoantibody Production by Early Human B Cell Precursors. Science (2003) 301:1374–7. doi: 10.1126/science.1086907
43. Lee YN, Frugoni F, Dobb K, Tirosh I, Du L, Ververs FA, et al. Characterization of T and B Cell Repertoire Diversity in Patients With RAG Deficiency. Sci Immunol (2016) 6:eaah6109. doi: 10.1126/sciimmunol.aah6109
44. Bomback AS, Fervenza FC. Membranous Nephropathy: Approaches to Treatment. Am J Nephrol (2018) 47(Suppl 1):30–42. doi: 10.1159/000481635
45. De Vriese AS, Glassock RJ, Nath KA, Sethi S, Fervenza FC. A Proposal for a Serology-Based Approach to Membranous Nephropathy. J Am Soc Nephrol (2017) 28(2):421–30. doi: 10.1681/ASN.2016070776
Keywords: B-cell receptor repertoire, membranous nephropathy, high-throughput sequencing, immunoglobulin heavy chain, biomarkers
Citation: Su Z, Jin Y, Zhang Y, Guan Z, Li H, Chen X, Xie C, Zhang C, Liu X, Li P, Ye P, Zhang L, Kong Y and Luo W (2021) The Diagnostic and Prognostic Potential of the B-Cell Repertoire in Membranous Nephropathy. Front. Immunol. 12:635326. doi: 10.3389/fimmu.2021.635326
Received: 30 November 2020; Accepted: 07 May 2021;
Published: 27 May 2021.
Edited by:
Mirjam van der Burg, Leiden University Medical Center, NetherlandsCopyright © 2021 Su, Jin, Zhang, Guan, Li, Chen, Xie, Zhang, Liu, Li, Ye, Zhang, Kong and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Luo, bHVvd2VpXzQyMUAxNjMuY29t; Yaozhong Kong, a3l6aG9uZ0Bmc3l5eS5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.