- Aquaculture Pathology Laboratory, School of Animal & Comparative Biomedical Sciences, The University of Arizona, Tucson, AZ, United States
Acute hepatopancreatic necrosis disease (AHPND) is a lethal disease in marine shrimp that has caused large-scale mortalities in shrimp aquaculture in Asia and the Americas. The etiologic agent is a pathogenic Vibrio sp. carrying binary toxin genes, pirA and pirB in plasmid DNA. Developing AHPND tolerant shrimp lines is one of the prophylactic approaches to combat this disease. A selected genetic line of Penaeus vannamei was found to be tolerant to AHPND during screening for disease resistance. The mRNA expression of twelve immune and metabolic genes known to be involved in bacterial pathogenesis were measured by quantitative RT-PCR in two populations of shrimp, namely P1 that showed susceptibility to AHPND, and P2 that showed tolerance to AHPND. Among these genes, the mRNA expression of chymotrypsin A (ChyA) and serine protease (SP), genes that are involved in metabolism, and crustin-P (CRSTP) and prophenol oxidase activation system 2 (PPAE2), genes involved in bacterial pathogenesis in shrimp, showed differential expression between the two populations. The differential expression of these genes shed light on the mechanism of tolerance against AHPND and these genes can potentially serve as candidate markers for tolerance/susceptibility to AHPND in P. vannamei. This is the first report of a comparison of the mRNA expression profiles of AHPND tolerant and susceptible lines of P. vannamei.
Introduction
Acute hepatopancreatic necrosis disease (AHPND) is a lethal disease of marine shrimp that emerged in China in 2009. Since then it was reported in other countries including Vietnam, Thailand, Mexico, Philippines, Bangladesh, US and South-Korea (1–7). The etiologic agent was initially identified as Vibrio parahaemolyticus carrying plasmid-borne binary toxin genes, pirA and pirB (8, 9). Subsequently other species of Vibrio including V. harveyi and V. owensii have been reported to cause AHPND (10, 11). The clinical signs of AHPND include atrophy and pale discoloration of hepatopancreas, soft shell, gut with discontinuous or no content, and often 100% of mortality occurs in shrimp farms experiencing AHPND outbreaks (12–14). Histopathology of the hepatopancreas tissue from AHPND affected shrimp reveals three different stages of disease development, namely initial, acute/terminal and chronic phases. In the initial phase, elongation of epithelial cells in hepatopancreas is common, whereas during the acute/terminal phase necrosis of tubular epithelial cells and inflammatory responses are observed (13). The chronic phase of AHPND is characterized by the epithelial necrosis and bacterial inflammation in the tubules which is similar to septic hepatopancreatic necrosis (SHPN) (15).
It has widely been accepted that shrimp protect themselves from microbial pathogens by innate immunity that encompass humoral and cellular responses. Recently, it has been reported that the PirAVP- and PirBVP binary toxin encoded by V. parahaemolyticus can be neutralized by either hemocyanin or anti-lipopolysaccharide factor (16). In addition, the interaction between immune and metabolism appears to play a role in AHPND response in hepatopancrease in shrimp (17, 18). Until now, due to the lack of AHPND- tolerant lines of shrimp, no effort could be made to examine the gene expression profiles of AHPND- susceptible and tolerant lines of shrimp.
Recently, a line of P. vannamei shrimp has been identified that displays tolerance to AHPND (15). In this study, we initially compared the mRNA expression of twelve metabolic/immune related genes between AHPND-tolerant and-susceptible lines of P. vannamei by reverse transcriptase quantitative PCR (RT-qPCR). These genes are known to be involved in bacterial pathogenesis (19–24). The expression of seven genes that showed statistically significant differences between the tolerant and susceptible lines were validated. The results showed that there is a significant difference in the expression of these genes between AHPND-tolerant and-susceptible lines of P. vannamei. The potential implications of differential expression of these genes are discussed in the context of immunity and pathogenesis.
Materials and Methods
Bioassay 1- Assessing AHPND Tolerance in P. vannamei
Vibrio parahemolyticus (Strain 13-028A/3) (VpAHPND) was used for the experimental challenges following a previously published protocol (15). Three P. vannamei family lines were obtained from a commercial supplier as a part of an on-going family line screening for AHPND-tolerance. Each line was stocked separately into a total of nine 1000 L tanks in triplicate. From each of these three lines, we screened 56, 57 and 78 organisms from lines one, two and three respectively. A Specific Pathogen Free (SPF) P. vannamei (N=60, average weight 3 g) (AHPND susceptible line, population P1, were obtained from a commercial supplier in the USA and stocked into two 90L tanks and used as positive controls for AHPND challenge. A third tank containing SPF shrimps of the same genetic line (N=64) was used as negative control. An immersion challenge was performed using an inoculum load of 106 cfu/ml (15). The experiment was terminated at 7 days post-inoculation. The mortality in each tank was recorded daily, and a subset of moribund and surviving animals was examined by routine H&E histology.
Histopathology
Moribund P. vannamei were fixed in Davidson’s alcohol-formalin-acetic acid (AFA) fixative. The samples were processed, embedded in paraffin, sectioned (4 μm thick) and analyzed in accordance with standard methods (25).
Bioassay 2- Initial Gene Expression Analyses
To evaluate gene expression in the AHPND-tolerant and-susceptible lines, a second challenge was conducted. A total of 46 animals were utilized from P1 (n= 42, average weight 5g) with 21 animals challenged with VPAHPND, 21 animals utilized as negative controls and 4 animals sampled for gene expression analysis prior to challenge. The tolerant line was named population P2 (n=20, average weight 9g), with 9 animals challenged with VPAHPND, 9 unchallenged as negative controls and 2 animals sampled for gene expression analysis prior to challenge. All shrimp from each family were tagged in the 4th abdominal segment with uniquely colored elastomer tags to allow visual identification. Both families were then held in a single 1000L tank and challenged by immersion, as described above. The negative controls were held in a single 1000L tank but were not challenged.
Twenty-four hours after challenge, animals were collected for gene expression analysis. Limited numbers (N=4) of the susceptible P1 line were available for sampling due to the typically high mortality rates observed in AHPND challenges in the first 24 hours. Two shrimp were collected from the tolerant population P2 at 24 hours post-infection to leave enough animals in the tank for a survival comparison at termination. In order to keep the sample numbers equal, a pool of 10 challenged animals from P2 group in the initial family line challenge were pooled as 2 samples for a total of 4 samples.
Measuring the Expression Levels of Candidate Immune and Metabolic Genes in AHPND Challenged P. vannamei
Four shrimp samples per population were collected from each treatment for RNA extraction using RNAzol following the manufacturer’s protocol (MRC, Ohio, USA). The RNA was treated with DNase 1 (Invitrogen, USA). One µg of DNase treated RNA was used for cDNA synthesis using Superscript IV and following the manufacturer’s recommendation (Invitrogen, USA). After that, cDNA was subjected to gene expression analysis.
The mRNA expression of β-glucan binding protein (BGBP) (AY249858.1), Crustin-P (CRSTP) (AY488497), C-type lectin 1-like (CTL1-like) (DQ858900.1), Extracellular Copper/Zinc Superoxide dismutase (EC-SOD) (HM371157), Kazal protease inhibitor (KPI) (AY544986), lipopolysaccharide and β-1,3-glucan-binding protein (LGBP) (EU102286.1), Penaidin 2 (PEN2) (Y14925), Prophenol oxidase activation system 2 (PPAE2) (AWF98992.1), Serpin8 (SEP8) (KU853046.1), serine protease (SP) (AY368151), Chymotrypsin A (ChyA) (Y10664), Chymotrypsin B (ChyB) (Y10665) and two toxin genes of V. parahaemolyticus (i.e. pirA and pirB toxin genes, KM067908) were measured by quantitative PCR (StepOnePlus Real-time PCR system, Applied biosystems, USA) using PowerUPTM SYBR Green Master Mix (Applied Biosystem, USA). The reaction mixture contained 10 µl of PowerUP™ SYBR Green Master Mix, 0.8 µl (0.4 µM) of each primer, 2.0 µl of cDNA and 6.4 µl of sterile water in a reaction volume of 20 µl. The thermal profile for the reaction was 2 minutes at 50°C, 2 minutes at 95°C followed by 40 cycles of 3 seconds at 95°C and 30 seconds at 60°C. The primer sequence for each gene is given in Supplementary Table S1. In addition, literature search of the responses to pathogens of studied genes was provided in Supplementary Table S4. Each sample was run in duplicate and the mean Ct value was used for gene expression analysis. Expression levels of each gene in various populations are shown relative to the expression in corresponding control treatments according to the formula of Livak and Schmittgen (26). For evaluating the gene expression between challenged animals coming from P1 and P2 population, the expression levels of each gene were shown relative to the expression in the P1 negative control. The expression value was converted to Log based 2 prior to statistical analysis. The genes showing statistical difference in expression level in Bioassay 2 were validated in Bioassay 3.
Bioassay 3- Gene Expression Validation
Two hundred shrimp (average wt. 2 g) with the same genetic characteristics of the AHPND tolerant shrimp line (P2) from Bioassay 2 were stocked in two 1000 L-tanks (100 shrimp/tank) in which one tank was challenged with VPAHPND. The remaining tank was used as negative control. Forty SPF shrimp (AHPND susceptible shrimp) (average wt. ~1.5 g) (P1) were stocked in two 90L-tanks (20 shrimp/tank), one tank was challenged with VPAHPND. The remaining tank was used as negative control. The AHPND challenge protocol was the same as described previously. After 24 hours, nine shrimp from each tank were collected for gene expression analyses.
Statistical Analysis
The statistical significances in the difference in Log based 2 value of gene expression between control and challenged animals in each population and between challenged animals in P1 and P2 populations was determined by using Student’s t-test with P<0.05, SPSS v.16 software. The mortality data was analyzed using Kaplan-Meier method, SPSS v.16 software.
Results
Bioassay 1- Assessing AHPND Tolerance in P. vannamei
In experimental AHPND challenge, the survival rate in the AHPND-tolerant P2 population was over five times higher compared to the AHPND-susceptible P1 population (72% vs. 12.5%). Meanwhile, the survival rate in negative control was 95.45% (Supplementary Figure S1). The histopathology of the P1 and P2 populations are presented in Figure 1. The Davidson-fixed shrimp from the healthy, negative controls from P1 and P2 displayed normal structure of tubules and epithelial cells in the hepatopancreas including high levels of lipid droplets (R-cells), secretory vacuoles (B-cells) and absence of AHPND (Figures 1A, B). In contrast, shrimp from the P1 population challenged with VpAHPND displayed lesions typical of AHPND in the acute phase, including a multifocal necrosis and massive sloughing of HP tubule epithelial cells in the hepatopancreas. At this stage, bacterial colonization was not observed (Figure 1C). Shrimp from the P2 population displayed the typical VPAHPND chronic phase characterized by SHPN-like lesions (Figure 1D).
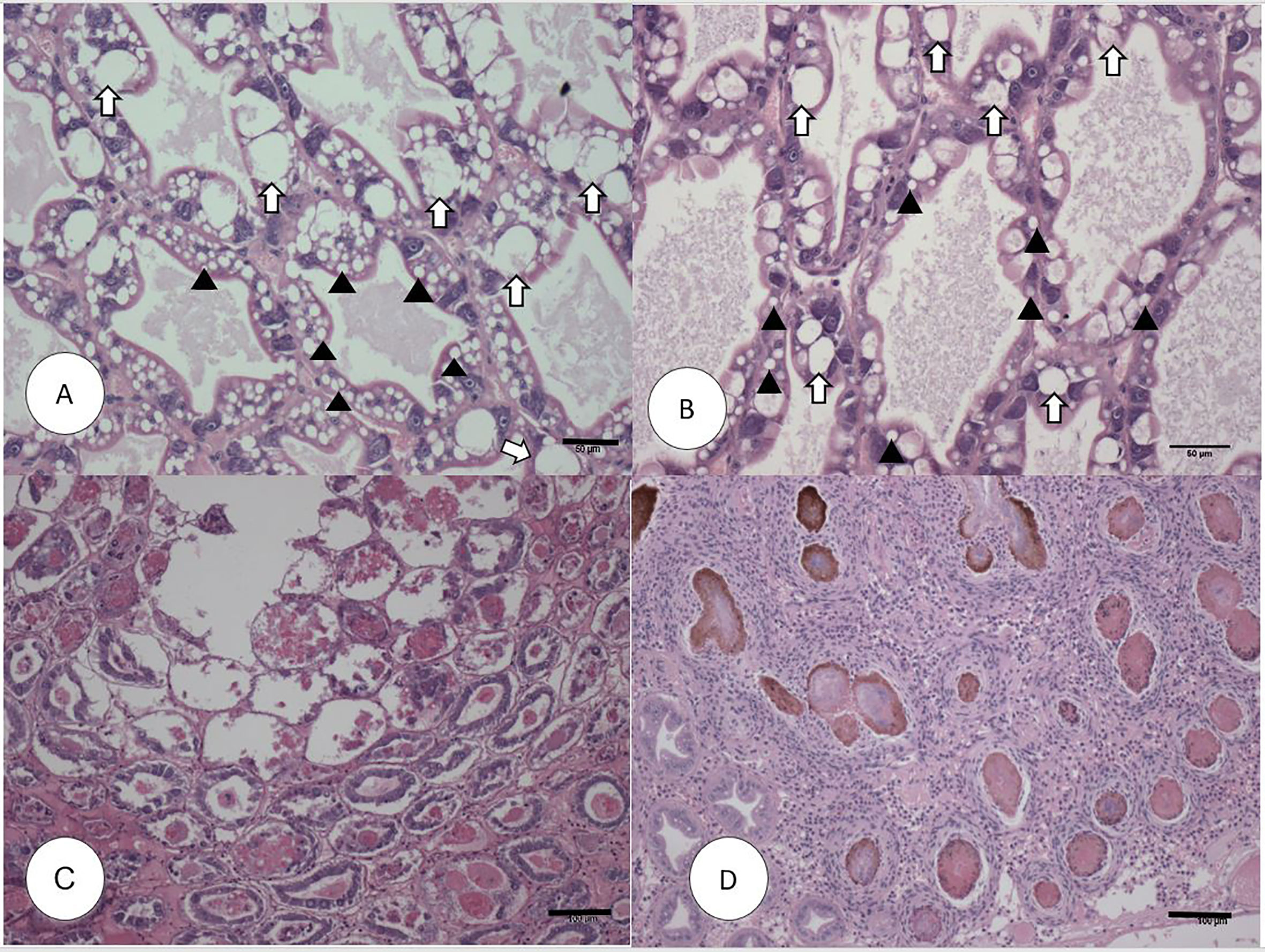
Figure 1 H&E (Mayer–Bennet hematoxylin and eosin-phloxine) histology of Penaeus vannamei from Bioassay 1. Penaeus vannamei from negative control tank from the P1 (A) and P2 populations (B) showing intact tubules and epithelial R-cells (arrowhead) and B-cells (white arrow). Acute phase infection of AHPND in shrimp from P1 population (C) showing a severe sloughing of epithelial tubule cells into the lumen. Terminal phase of AHPND infection in shrimp from P2 population (D) showing a severe intertubular hemocytic infiltration surrounding the affected melanized tubules. Scale bars for (A, B) = 50 μm; (C, D) = 100 μm.
Bioassay 2- Initial Gene Expression Analyses, Measuring the mRNA Expression of Immune and Metabolic Genes in P. vannamei
Interestingly, there was no significant difference in the expression levels of pirA/pirB genes between P1 and P2 population (P>0.05) (Figure 2).
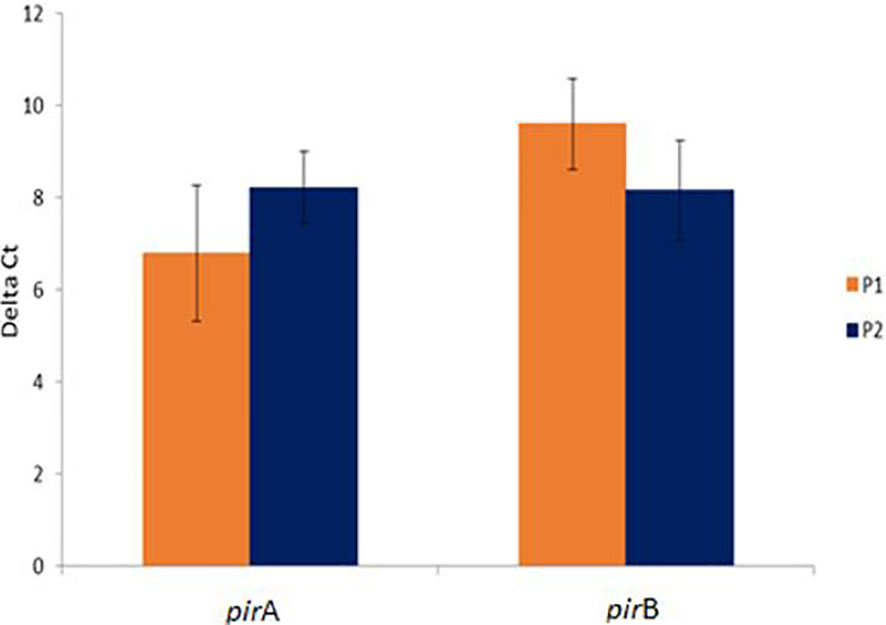
Figure 2 Comparative pirA- and pirB- toxin genes expression in challenged animals from Bioassay 2 in the P1 and P2 populations. The levels of mRNA expression is expressed as a normalized mean Ct value. The data is presented as ΔMean Ct ± SD. Statistical significance between P1 and P2 populations for each of the candidate gene was determined using Student’s t-test.
The mRNA expression of a set of immune and metabolic genes (i.e. SEP8, BGBP, CRSTP, CTL1-like, KPI, LGBP, EC-SOD, PEN2, PPAE2, SP, ChyA and ChyB) were measured by RT-qPCR in control animals and challenged animals in each population. We also compared the levels of gene expression in AHPND susceptible P1 and AHPND tolerant P2 populations.
The V. parahaemolyticus infection led to the significant upregulation of expression of LGBP, PPAE2, and ChyA transcripts in the P1 population (P<0.05) whereas SP mRNA showed significant down regulated expression (P<0.05). The mRNA levels of BGBP, CRSTP, CTL1-like, KPI, PEN2, EC-SOD, SEP8 and ChyB in the challenged and un-challenged groups did not show significant differences (P>0.05) (Figure 3A).
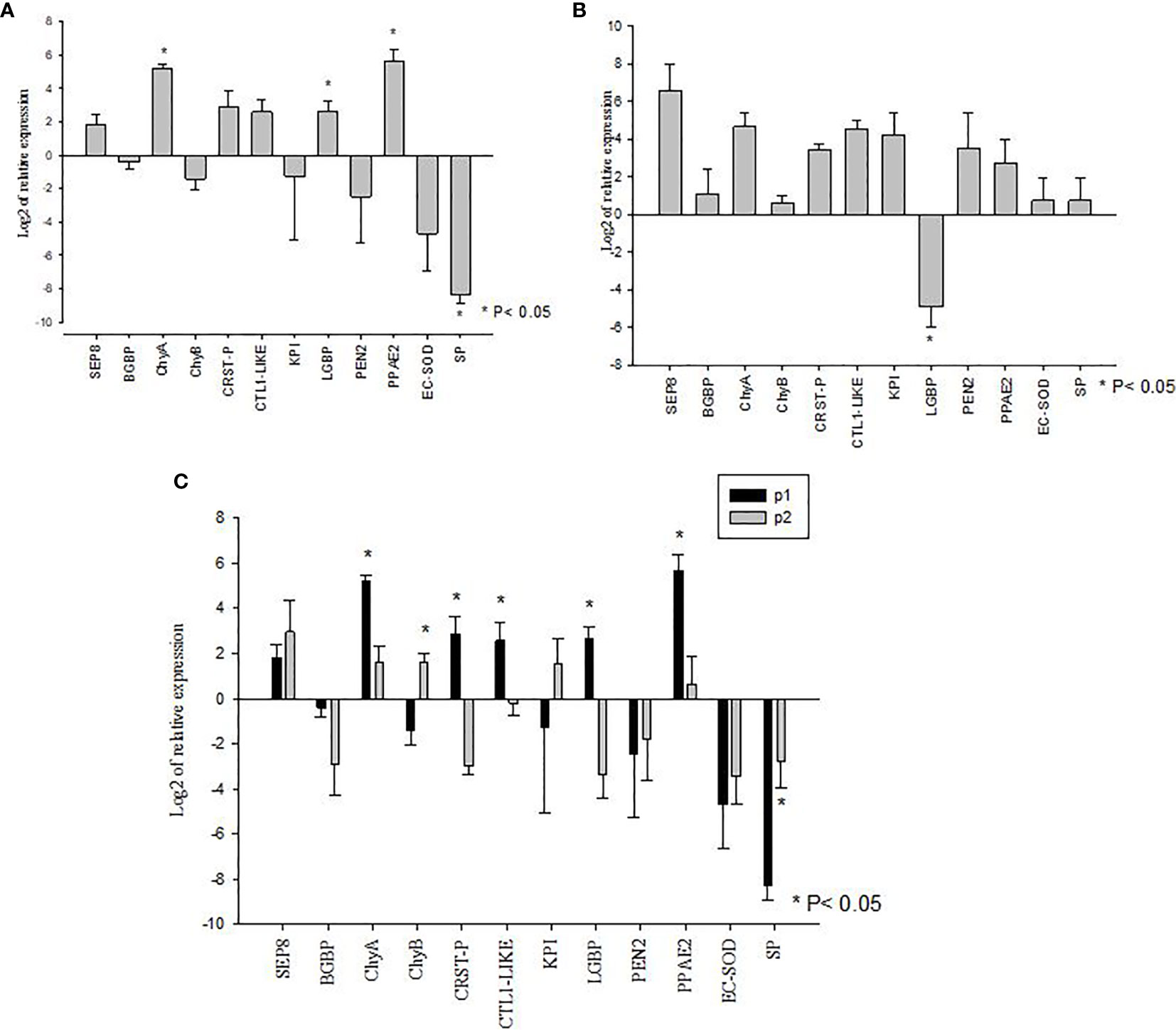
Figure 3 Gene expression profiles of metabolic and immune genes in shrimp Penaeus vannamei from Bioassay 2. (A) The mRNA expression profile in AHPND susceptible (P1) population following experimental challenge. (B) The mRNA expression profile in AHPND tolerant (P2) population following experimental challenge. Expression levels of each gene are shown relative to the expression in correspond control treatment (C) Comparison of gene expression profile from AHPND susceptible (P1) vs. AHPND resistant/tolerant (P2) population. Expression levels of each gene are shown relative to the expression in P1 negative control treatment. The data is presented as log2 of relative expression ± SD. Statistical significance between control and challenged animals for each of the candidate gene was determined using Student’s t-test. *P<0.05. BGBP, β-glucan binding protein; CRST P, Crustin P; CTL, C-type lectin 1-like; ECSOD, Extracellular Copper/Zinc Superoxide dismutase; KPI, Kazal protease inhibitor; LGBP, Lipopolysaccharide and β-1,3-glucan-binding protein; PEN2, Penaidin 2; PPAE2, Prophenol oxidase activation system 2; SEP, Serpin8; SP, Serine protease; ChyA, Chymotrypsin A; ChyB, Chymotrypsin B.
In the P2 population, V. parahaemolyticus infection led to the significant downregulated expression of LGBP mRNA (P<0.05). The expression of BGBP, SEP8, CTL1-like, CRSTP, KPI, EC-SOD, PPAE2, PEN2, SP, ChyA and ChyB showed no significant difference between challenged and un-challenged groups (P>0.05) (Figure 3B).
When the mRNA expression levels were compared between AHPND challenged animals from the P1 and P2 populations, there were significant differences in the expression profiles of some genes. For example, while the susceptible P1 population showed significantly higher levels of expression of ChyA, CRSTP, CTL1-like, LGBP and PPAE2 (P<0.05) (Figure 3C), the AHPND-tolerant population P2 showed higher expression of SP and ChyB compared to the susceptible P1 population (P<0.05) (Figure 3C) (Supplementary Table S2).
Bioassay 3- Gene Expression Validation
The genes showing significant differences in expression levels between susceptible and tolerant shrimp were selected for validations in Bioassay 3. In the P1 population, the expression of CTL1-like, CRSTP, SP and ChyB were significantly down regulated (P<0.05) meanwhile PPAE2 and ChyA expression levels were significantly up regulated (P<0.05) (Figure 4A). LGBP expression level was not significant during the experiment (P>0.05) (Figure 4A).
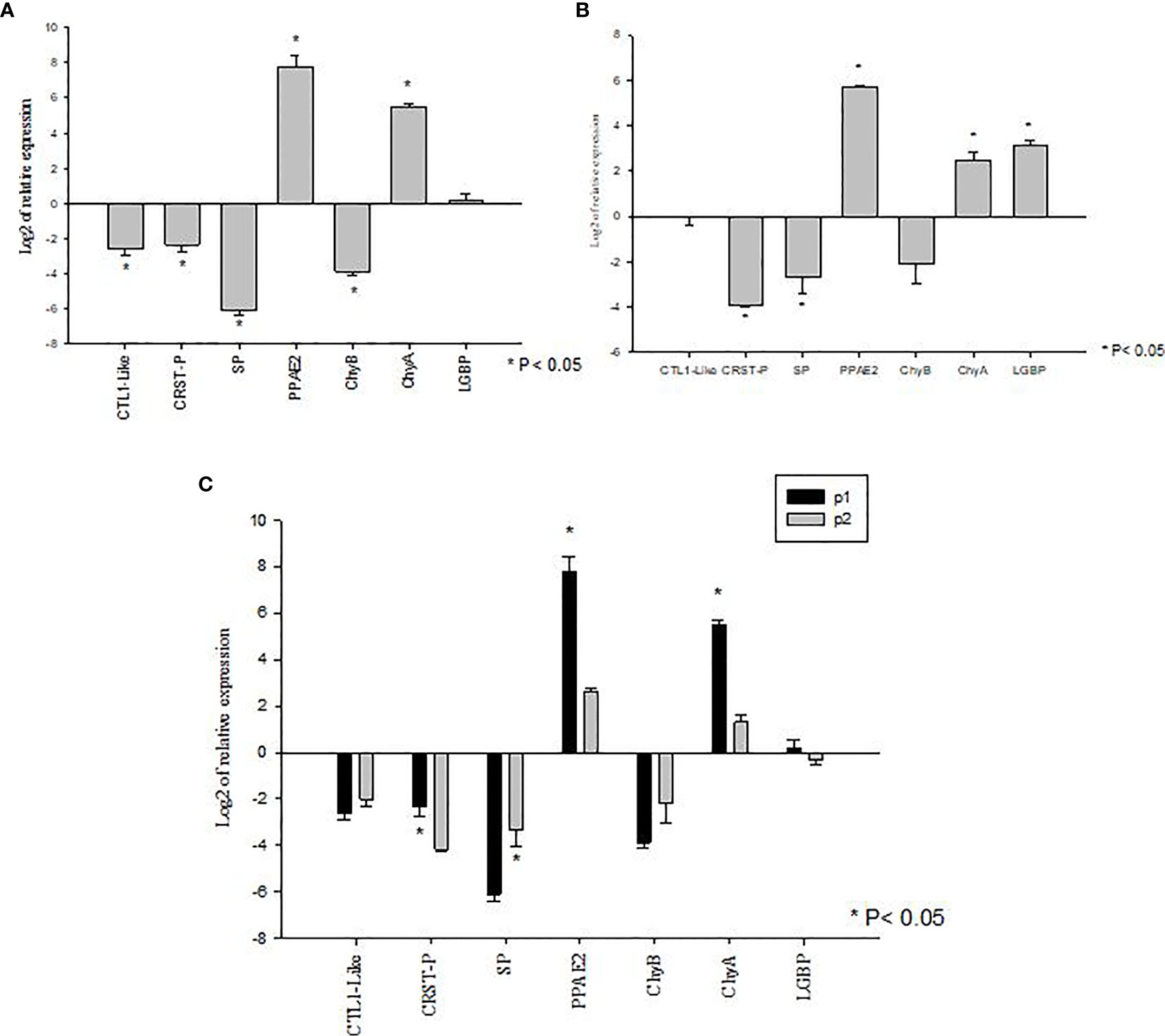
Figure 4 Validation of gene expression profiles of metabolic and immune genes in Penaeus vannamei from Bioassay 3 (A) The mRNA expression profile in P. vannamei AHPND susceptible (P1) population (Panel A) and AHPND resistant/tolerant (P2) population (Panel B) following experimental challenge. Expression level of each gene is shown relative to the expression in correspond control treatment (C) Comparison of gene expression profile from AHPND susceptible (P1) vs AHPND resistant/tolerant (P2) population. Expression levels of each gene are shown relative to the expression in P1 negative control treatment. The data are presented as log2 of relative expression ± SD. Statistical significance between control and challenged animals for each of the candidate gene was determined using Student’s t-test. *P<0.05. CRST P, Crustin P; CTL, C-type lectin 1-like; LGBP, Lipopolysaccharide and β-1,3-glucan-binding protein; PPAE2, Prophenol oxidase activation system 2; SP, Serine protease; CHYA, Chymotrypsin A; CHYB, Chymotrypsin B.
In P2 population, PPAE2, ChyA and LGBP expression levels were significantly up regulated (P<0.05). In contrast, the expression levels of CRSTP and SP were down regulated (P<0.05) (Figure 4B). CTL1-like and ChyB expression were not significantly different during the experiment (P>0.05) (Figure 4B).
By comparison, P1 and P2 challenged animals showed the same expression pattern in both Bioassay 2 and Bioassay 3. The expression levels of CRSTP, PPAE2 and ChyA were significantly higher in P1 population (P<0.05). LGBP expression level was higher in P1 population but not significantly different (P>0.05). Meanwhile, SP had a significant higher expression level in P2 population than P1 population (P<0.05) (Figure 4C). ChyB expression level was higher in P2 population than P1 population even though there was no significant difference observed during the experiment (P>0.05) (Figure 4C) (Supplementary Table S3).
Discussion
Bacterial pathogenesis in crustaceans is well studied and genes involved in humoral and cellular immunity are known. We decided to take advantage of this background knowledge by measuring the expression of genes that are well known to be involved in defense and metabolic responses during bacterial infections in shrimp. Coincidentally, we had access to AHPND-tolerant lines of P. vannamei and considering the lethal nature of AHPND-causing V. parahaemolyticus we explored if genes known to be involved in other bacterial pathogenesis in shrimp are also involved in AHPND pathogenesis. To our knowledge, a recently published paper is the first report of the development of AHPND resistant/tolerant lines of P. vannamei, Aranguren Caro et al. (15), and as of today, there is no report of looking into the gene expression profiles of AHPND-tolerant vs. susceptible lines.
It is now widely accepted that the etiology of AHPND is the insecticidal binary toxin-like genes carried by plasmid DNA in Vibrio spp (11, 12, 27). An AHPND resistant/tolerant shrimp line would be ideal in controlling the disease in shrimp aquaculture. Tinwongger and colleagues (16) showed that shrimp exposed to formalin killed cells (FKC) of AHPND causing V. parahaemolyticus can survive upon AHPND challenge, and anti-lipopolysaccharide factor AV-R isoform (LvALF AV-R) showed significantly higher expression in the hepatopancreas from the survivor. Interestingly, only four out of two hundred shrimp (2%) survived after feeding with FKC diet, and only three shrimp from the survivor group were used for gene expression analysis. Despite examining a limited number of animals, the authors were successful in identifying genes that could be potentially involved in AHPND pathogenesis. In this study, three bioassays were performed. Bioassay 1 was performed to identify an AHPND tolerant line, P2, using mortality and histopathology as end point data of the bioassay. The P2 population suffered 28% mortality compared to the susceptible line P1 that experienced 87.5% mortality. The bioassay was repeated using the same AHPND-susceptible (P1) and tolerant line (P2) (i.e. Bioassay 2) to examine the mRNA expression of twelve candidate immune and metabolic genes. When the expression of these genes were compared before and after challenge within a population, P1 and P2, a number of genes showed differential expression. However, when the expression profiles were compared between P1 and P2 populations after AHPND-challenge, seven candidate genes showed differential expression. These genes are likely involved in AHPND pathogenesis. In order to further validate the expression of these seven genes, a third bioassay was conducted (i.e. Bioassay 3) and samples were collected for the gene expression validation. The data shed light on the molecular basis of AHPND pathogenesis and enabled to identify potential markers for AHPND tolerance/susceptibility, as discussed below.
Bioassay 1
The mortality data in the Bioassay 1 showed that the survival rate of the P2 population was over five times higher than P1 population indicating that P2 population is indeed an AHPND tolerant line. The mortality data was consistent with the histopathology findings that cellular damage in the hepatopancreas was far greater in animals from the P1 compared to the P2 population. For example, sloughing of the epithelial cells in the hepatopancreatic tubules followed by massive infiltration of bacterial cells that are considered pathognomonic for AHPND was clearly evident by H&E histology in animals from P1 population (Figure 1), whereas in the P2 population, the lesions present in the hepatopancreas resembled more of a chronic infection as seen during SHPN. The differences in mortality and histopathology data led to examining the expression profiles of twelve metabolic and immune-related genes in these two populations. Interestingly, some genes in the tolerant shrimp such as CRST-P, SP, ChyB and LGBP showed the discrepancies in the trend of expression between Bioassay 2 and Bioassay 3. However, only LGBP expression significantly down-regulated in tolerant shrimp from Bioassay 2 meanwhile LGBP expression in tolerant shrimp from Bioassay 3 showed significantly up-regulated. The reasons for discrepancies could be different animals reacting differently to the same pathogen although they are similar genetic line. That would be the reason we had to do validation test with larger number animals.
Gene Expression in P. vannamei From Bioassay 2 vs. Bioassay 3
The expression of twelve candidate genes known to be involved in other bacterial diseases were examined to determine if similar genes are involved in AHPND pathogenesis. It was interesting to note that there was no significant difference in the expression levels of pirA or pirB toxin genes between the P1 and P2 populations suggesting that the animals from the two populations were exposed to equivalent levels of toxin. Thus, the difference in tolerance was most likely due to the difference in immune response between the two populations.
It is known that shrimp, like other invertebrates, elicit cellular and humoral immune responses when exposed to microbes or non-self-protein containing pathogen associated molecular patterns (PAMP) (28, 29). PAMP is easily recognized by pattern recognition proteins including BGBP, LGBP and CTL (19, 30–33). Several studies indicate that the expression of BGBP, LGBP and CTL are up-regulated in shrimp challenged with pathogens such as bacteria, viruses and fungi (34–37).
In Bioassay 2, when the mRNA expressions of BGBP, LGBP and CTL1-like genes were compared between AHPND-susceptible P1 and AHPND-tolerant P2 populations, LGBP and CTL1-like genes were found to be upregulated in P1 compared to P2 population. However, there was no difference in BGBP expression between the two populations (Figure 3). Interestingly, in Bioassay 3, although LGBP expression was higher in P1 than P2 populations as in Bioassay 2 samples, the difference in expression was not statistically significant (p=0.365). In contrary, CTL1-like gene did not show any differential expression between the two populations (Figure 4), as observed in samples derived from Bioassay 2 (Figure 3). These discrepancies highlight two important facts: (i) it is critical to validate gene expression data with biological samples derived from independent bioassays, and (ii) it is important to validate initial findings with larger sample sets, as in Bioassay 3 compared to Bioassay 2. These gives further credence to the mRNA expression findings reported in this study.
In shrimp, upon microbial infection pattern recognition protein(s) circulating in the hemolymph triggers immune response by activating proPO cascade and releasing anti-microbial peptides such as PEN and CRSTP to eliminate the invading pathogen(s) (20). Although PEN and CRSTP were shown to have antimicrobial activities against Vibrio spp. and Gram positive bacteria in penaeid shrimp (21, 23, 38), neither of these genes were significantly upregulated in the challenged animals in Bioassay 2. In shrimp, PEN2 is mostly detected in hemocytes and to a lesser level in hepatopancreas. Since we examined the gene expression in hepatopancreas tissue, it is possible that for this reason PEN2 was not found to be differentially expressed. Interestingly, CRSTP which is also predominantly expressed in hemocytes and less in hepatopancreas showed significantly higher expression in AHPND susceptible P1 compared to AHPND tolerant P2 populations in both Bioassays 2 and 3. It remains to be determined if higher CRSTP expression in the susceptible population is due to increased bacterial cell deaths and consequently the release of PirABVP toxin in the infected animals.
The proPO activation is an important event in crustacean immunity to eliminate pathogens from the circulatory system (39, 40). The final event in the proPO activation process is the conversion of proPO to phenoloxidase (PO) by the PPAE (26, 41, 42). In both Bioassays 2 and 3, PPAE2 showed significantly higher expression after AHPND challenge in P1 compared to P2 populations. The finding is consistent with a previously published report in P. monodon where PPAE2 expression in the stomach showed significant upregulation at 24 hour post-challenge with AHPND causing V. parahaemolyticus (43). In a separate study, Apitanyasai et al. (44) suggested an overreaction of the proPO cascade causes damage to host cells during AHPND infection and leads to higher mortality. The higher expression of PPAE2 in AHPND susceptible P1 population supports this observation. Taken together, this evidence suggests that upon infection with VpAHPND, not only the expression of antimicrobial peptide like CRSTP but also the genes involved in the proPO pathway are elevated in AHPND susceptible compared to tolerant animals.
It is known that the activation of proPO leads to production of quinones and other intermediate reactions which polymerizes quinones to melanin resulting in pathogen capsulation (45). Quinone, however, also causes cell death by inducing reactive oxygen species (ROS) production (46–48). In crustacean, ROS can be scavenged by an anti-oxidant system involving enzymes of the SOD family (49–51). The EC-SOD expression in the animals studied was not significantly modulated upon AHPND challenge. The proPO cascade is also modulated by a series of protease inhibitors such as KPI and SEP8 to prevent the over activation (52, 53). Again, both KPI and SEP8 mRNA levels in animals from the P1 and P2 populations were not significantly different. It is tempting to speculate that for the lack of modulation of efforts in EC-SOD, proPO cascade activation led to cell toxicity more in P1 compared to P2 population. It is also possible that killing of VpAHPND cells leads to further release of PirAB toxin from the inactivated bacterial cells exerting lethal effects in genetically susceptible animals.
The involvement of PO activity in the susceptibility of invertebrates due to bacterial toxins has been well documented in many insect species. For example, cabbage loopers (Trichoplusia ni) is inherently susceptible to the Cry toxin secreted from Bacillus thuringiensis. However, the Cry toxin resistance is negatively correlated with the PO activity in B. thuringiensis challenged T.ni (54). In addition, the effectiveness of biological insecticide is also higher in lepidopteran species that show high PO activity (55). Recently, it has been reported that during VPAHPND infection resulting in AHPND, reduction in activation of the proPO system by a serine protease inhibitor, LvSerpin7, results in reduction of the toxic effects compared to an unregulated activation of the PO cascade (44).
Apart from the difference in immune gene expression, the expression of metabolic genes also showed differences between healthy and AHPND-challenged animals in each population and between challenged animals in populations P1 vs P2. The expression of ChyB and SP were down regulated whereas ChyA expression was up regulated in AHPND challenged shrimp in both P1 and P2 populations and in both Bioassays 2 and 3. These results are consistent with the findings from a recent study in which SP was shown to be upregulated in AHPND tolerant P. monodon (18). In addition, Chymotrypsin is upregulated in AHPND susceptible P. vannamei after 24 hour of infection (17) which is also in agreement with our findings.
Recently, the crystal structure of PirABVP toxin shows homology to the structure of the Cry toxin released by B. thuringiensis (56), and it is known that the Cry protein is activated by protease enzymes (56–58). The tertiary structure of PirABVP involves a heterotetrameric interaction between two PirAVP and two PirBVP. It has been hypothesized that PirAVP plays a role in receptor binding while PirBVP is involved in pore formation in the cell membrane (59). In silico analysis (https://web.expasy.org/peptide_cutter/) showed that the PirAVP toxin contains twelve and nine putative sites that are likely to be cleaved by chymotrypsin and trypsin, respectively (Supplementary Figure S2). Meanwhile, PirBVP toxin contains 53 and 36 sites cleaved by chymotrypsin and trypsin, respectively. Multiple alignments between Cry toxin and PirAVP showed that 3 out of 12 and 3 out of 9 cleaved sites for chymotrypsin and trypsin were conserved. Meanwhile, the multiple alignment between Cry toxin and PirBVP showed that 9 out of 53 and 1 out of 36 cleaved sites for chymotrypsin and trypsin were conserved (Supplementary Figure S2). It is interesting to note that SP expression showed higher levels in the AHPND tolerant than AHPND susceptible P. vannamei in this study and this enzyme belongs to trypsin family (60). It remains to be determined if SP is involved in cleaving PirABVP toxins to de-activate the toxin. If so, a higher expression of this enzyme in AHPND tolerant population (P2) may prevent the activation of toxin from exerting a lethal effect.
To summarize, we compared the gene expression profiles of two populations of P. vannamei that differ in susceptibility to AHPND. The two populations showed a major difference in survival upon experimental challenge. The difference in susceptibility was further evidenced by the differences observed by histopathology. In order to understand the molecular mechanisms governing tolerance and susceptibility, a set of candidate genes that are known to be involved in bacterial pathogenesis and in metabolism in crustaceans were evaluated. Seven genes that showed differential expression in Bioassay 2 were further evaluated in a follow-up challenge, Bioassay 3. The pattern of differential expression between the susceptible (P1) and tolerant (P2) population in Bioassays 2 and 3 were in agreement. Despite the fact that the mRNA expression of only handful genes were measured and a limited number of animals were screened, the information, albeit limited, provides valuable insight in V. parahaemolyticus pathogenesis and sheds light on how susceptible and tolerant populations of P. vannamei respond differently to VpAHPND. To our knowledge, this is the first report looking into the differences in gene expression profiles between AHPND tolerant and susceptible lines in P. vannamei.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AD conceived the idea. HM and AD designed the study. HM and RC-F performed gene expression analyses. HM wrote the manuscript. LA performed histopathological examine. BN carried out experimental challenge experiment. HM, RC-F, LA, and AD reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding for this research was provided by Aquaculture Pathology Laboratory Diagnostic Fund. Part of this work is also supported by the USDA National Institute of Food and Agriculture, Hatch/Multistate project 1018120.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Mr. Paul Schofield and Mr. Tanner Padilla for their support in conducting the AHPND experimental challenges and Ms. Jasmine Millabas for preparing the histological slides.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.634152/full#supplementary-material
References
1. De La Peña LLD, Cabillon NARN, Catedral DDD, Amar EEC, Usero RRC, Monotilla WWD, et al. Acute Hepatopancreatic Necrosis Disease (AHPND) Outbreaks in Penaeus Vannamei and P. Monodon Cultured in the Philippines. Dis Aquat Organ (2015) 116:251–4. doi: 10.3354/dao02919
2. Dhar AK, Piamsomboon P, Aranguren Caro LFL, Kanrar S, Adami R, Juan Y-SY. First Report of Acute Hepatopancreatic Necrosis Disease (AHPND) Occurring in the USA. Dis Aquat Organ (2019) 132:241–7. doi: 10.3354/dao03330
3. Han J, Choi S, Han S, Chan S, Jin H, Lee C, et al. Genomic and Histopathological Characteristics of Vibrio Parahaemolyticus Isolated From an Acute Hepatopancreatic Necrosis Disease Outbreak in Paci Fi C White Shrimp (Penaeus Vannamei ) Cultured in Korea. Aquaculture (2020) 524:735284. doi: 10.1016/j.aquaculture.2020.735284
4. Joshi J, Srisala J, Truong VH, Chen IT, Nuangsaeng B, Suthienkul O, et al. Variation in Vibrio parahaemolyticus Isolates From a Single Thai Shrimp Farm Experiencing an Outbreak of Acute Hepatopancreatic Necrosis Disease (AHPND). Aquaculture (2014) 428–429:297–302. doi: 10.1016/j.aquaculture.2014.03.030
5. Nunan L, Lightner D, Pantoja C, Gomez-jimenez S. Detection of Acute Hepatopancreatic Necrosis Disease (AHPND ) in Mexico. Dis Aquat Organ (2014) 111:81–6. doi: 10.3354/dao02776
6. Thitamadee S, Prachumwat A, Srisala J, Jaroenlak P, Salachan PV, Sritunyalucksana K, et al. Review of Current Disease Threats for Cultivated Penaeid Shrimp in Asia. Aquaculture (2016) 452:69–87. doi: 10.1016/j.aquaculture.2015.10.028
7. MME E, MM A, NJ P, MK B, Rahman M. Molecular Identification of AHPND Positive Vibrio Parahaemolyticus Causing an Outbreak in South-West Shrimp Farming Regions of Bangladesh. J Bangladesh Acad Sci (2017) 41:127–35. doi: 10.3329/jbas.v41i2.35492
8. Han JE, Tang KFJ, Aranguren LF, Piamsomboon P. Characterization and Pathogenicity of Acute Hepatopancreatic Necrosis Disease Natural Mutants, Pirabvp(-) V. Parahaemolyticus, and Pirabvp(+) V. Campbellii Strains. Aquaculture (2017) 470:84–90. doi: 10.1016/j.aquaculture.2016.12.022
9. Tran L, Nunan L, Redman RRMR, Mohney LLL, Pantoja CCR, Fitzsimmons K, et al. Determination of the Infectious Nature of the Agent of Acute Hepatopancreatic Necrosis Syndrome Affecting Penaeid Shrimp. Dis Aquat Organ (2013) 105:45–55. doi: 10.3354/dao02621
10. Liu L, Ge M, Zheng X, Tao Z, Zhou S, Wang G. Investigation of Vibrio Alginolyticus, V. Harveyi, and V. Parahaemolyticus in Large Yellow Croaker, Pseudosciaena Crocea (Richardson) Reared in Xiangshan Bay, China. Aquac Rep (2016) 3:220–4. doi: 10.1016/j.aqrep.2016.04.004
11. Jesús M, Ana DAV, Francisco J, Fabiola ZM, Manuel J, Norberto AJG. Pir A - and Pirb - Like Gene Identification in Micrococcus Luteus Strains in Mexico. J Fish Dis (2018) 11:1667–73. doi: 10.1111/jfd.12874
12. Han JE, Tang KFJ, Tran LH, Lightner DV. Photorhabdus Insect-Related (Pir ) Toxin-Like Genes in a Plasmid of Vibrio Parahaemolyticus, the Causative Agent of Acute Hepatopancreatic Necrosis Disease (AHPND ) of Shrimp. Dis Asian Aquac (2015) 113:33–40. doi: 10.3354/dao02830
13. Soto-Rodriguez SA, Gomez-Gil B, Lozano-Olvera R, Betancourt-Lozano M, Morales-Covarrubias MS. Field and Experimental Evidence of Vibrio Parahaemolyticus as the Causative Agent of Acute Hepatopancreatic Necrosis Disease of Cultured Shrimp (Litopenaeus Vannamei) in Northwestern Mexico. Appl Environ Microbiol (2015) 81:1689–99. doi: 10.1128/AEM.03610-14
14. Lai H, Hann T, Ando M, Lee C, Chen I, Chiang Y, et al. Fish & Shell Fi Sh Immunology Pathogenesis of Acute Hepatopancreatic Necrosis Disease (AHPND) in Shrimp. Fish Shellfish Immunol (2015) 47:1006–14. doi: 10.1016/j.fsi.2015.11.008
15. Fernando L, Caro A, Mai HN, Noble B, Dhar AK. Acute Hepatopancreatic Necrosis Disease (VP AHPND ), a Chronic Disease in Shrimp Penaeus Vannamei Population Raised in Latin America. J Invertebr Pathol (2020) 174:107424. doi: 10.1016/j.jip.2020.107424
16. Tinwongger S, Thawonsuwan J, Kondo H, Hirono I. Fish and Shell Fi Sh Immunology Identi Fi Cation of an Anti-Lipopolysaccharide Factor AV-R Isoform (Lv Alf Av- R) Related to Vp _ PirAB-like Toxin Resistance in Litopenaeus Vannamei. Fish Shellfish Immunol (2019) 84:178–88. doi: 10.1016/j.fsi.2018.10.005
17. Vela E, Jua L, Id ISR, Id HV, Valdes-lopez O, Luna-gonza A. Transcriptomic Analysis of Pacific White Shrimp (Litopenaeus Vannamei, Boone 1931) in Response to Acute Hepatopancreatic Necrosis Disease Caused by Vibrio Parahaemolyticus. PloS One (2019) 14:1–28, e0220993. doi: 10.1371/journal.pone.0220993
18. Soo C, Chiew T, Devadas S, Shariff M, Din M, Bhassu S. Differential Transcriptome Analysis of the Disease Tolerant Madagascar – Malaysia Crossbred Black Tiger Shrimp, Penaeus Monodon Hepatopancreas in Response to Acute Hepatopancreatic Necrosis Disease (AHPND ) Infection : Inference on Immune Gene Response. Gut Pathog (2019) 11:1–13. doi: 10.1186/s13099-019-0319-4
19. Lee SY, Wang R, So K. A Lipopolysaccharide- and -1, 3-Glucan-Binding Protein From Hemocytes of the Freshwater Crayfish Pacifastacus Leniusculus. J Biol Chem (2000) 275:1337–43. doi: 10.1074/jbc.275.2.1337
20. Bernard D, Söderhäll K. β-1, 3-Glucan-Binding Proteins From Plasma of the Fresh-Water Crayfishes Astacus Astacus and Procambarus Clarkii Author (s): Bernard Duvic and Kenneth Söderhäll Published by: Oxford University Press on Behalf of The Crustacean Society Stable URL: Htt. J Crustac Biol (1993) 13:403–8. doi: 10.2307/1548783
21. Shockey JE, O’Leary NA, de la Vega E, Browdy CL, Baatz JE, Gross PS. The Role of Crustins in Litopenaeus Vannamei in Response to Infection With Shrimp Pathogens: An In Vivo Approach. Dev Comp Immunol (2009) 33:668–73. doi: 10.1016/j.dci.2008.11.010
22. Amparyup P, Charoensapsri W, Tassanakajon A. Fish & Shell Fi Sh Immunology Prophenoloxidase System and its Role in Shrimp Immune Responses Against Major Pathogens. Fish Shellfish Immunol (2013) 34:990–1001. doi: 10.1016/j.fsi.2012.08.019
23. Vandenbulcke F, Saulnier D, Bache E, Mun M. Expression and Distribution of Penaeidin Antimicrobial Peptides are Regulated by Haemocyte Reactions in Microbial Challenged Shrimp. Eur J Biochem (2002) 2689:2678–89. doi: 10.1046/j.1432-1033.2002.02934.x
24. Charoensapsri W, Amparyup P, Hirono I, Aoki T, Tassanakajon A. Pm PPAE2, a New Class of Crustacean Prophenoloxidase (proPO ) -Activating Enzyme and its Role in PO Activation. Dev Comp Immunol (2011) 35:115–24. doi: 10.1016/j.dci.2010.09.002
25. Aranguren LF, Mai HN, Noble B, Dhar AK. Acute Hepatopancreatic Necrosis Disease (VPAHPND), a Chronic Disease in Shrimp (Penaeus Vannamei) Population Raised in Latin America. J Invertebr Pathol (2020) 174:107424. doi: 10.1016/j.jip.2020.107424
26. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real- Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 408:402–8. doi: 10.1006/meth.2001.1262
27. Kondo H, Van PT, Dang LT. Draft Genome Sequence of Non- Vibrio Parahaemolyticus Acute Diseased Shrimp in Vietnam. Genome Announc (2015) 3:2014–5. doi: 10.1128/genomeA.00978-15
28. Hauton C. The Scope of the Crustacean Immune System for Disease Control. J Invertebrate Pathol (2012) 110:251–60. doi: 10.1016/j.jip.2012.03.005
29. Jiravanichpaisal P, Lee BL, Söderhäll K. Cell-Mediated Immunity in Arthropods: Hematopoiesis, Coagulation, Melanization and Opsonization. Immunobiology (2006) 211:213–36. doi: 10.1016/j.imbio.2005.10.015
30. Wang X, Wang J. Fish & Shell Fi Sh Immunology Pattern Recognition Receptors Acting in Innate Immune System of Shrimp Against Pathogen Infections. Fish Shellfish Immunol (2013) 34:981–9. doi: 10.1016/j.fsi.2012.08.008
31. Yu X, Kanost MR. Immulectin-2, a Pattern Recognition Receptor That Stimulates Hemocyte Encapsulation and Melanization in the Tobacco Hornworm, Manduca Sexta. Dev Comp Immunol (2004) 28:891–900. doi: 10.1016/j.dci.2004.02.005
32. Beschin A, Bilej M, Hanssens F, Raymakers J, Van Dyck E, Revets H, et al. Identification and Cloning of a Glucan- and Lipopolysaccharide-Binding Protein From Eisenia Foetida Earthworm Involved in the Activation of Prophenoloxidase Cascade. J Biol Chem (1998) 273:24948–54. doi: 10.1074/jbc.273.38.24948
33. Kim Y, Ryu J-H, Han S-J, Choi K-H, Nam K-B, Jang I-H, et al. Gram-Negative Bacteria-binding Protein, a Pattern Recognition Receptor for Lipopolysaccharide and B -1, 3-Glucan That Mediates the Signaling for the Induction of Innate Immune Genes in Drosophila Melanogaster Cells. J Biol Chem (2000) 275:32721–7. doi: 10.1074/jbc.M003934200
34. Roux MM, Pain A, Klimpel KR, Dhar AK. The Lipopolysaccharide and B-1, 3-Glucan Binding Protein Gene is Upregulated in White Spot Virus-Infected Shrimp (Penaeus Stylirostris). J Virol (2002) 76:7140–9. doi: 10.1128/JVI.76.14.7140-7149.2002
35. Zhao Z-Y, Yin Z-X, Xu X-P, Weng S-P, Rao X-Y, Dai Z-X, et al. A Novel C-Type Lectin From the Shrimp Litopenaeus Vannamei Possesses Anti-White Spot Syndrome Virus Activity. J Virol (2009) 83:347–56. doi: 10.1128/JVI.00707-08
36. Cheng W, Liu C, Tsai C, Chen J. Molecular Cloning and Characterisation of a Pattern Recognition Molecule, Lipopolysaccharide- and B -1, 3-Glucan Binding Protein (LGBP ) From the White Shrimp Litopenaeus Vannamei. Fish Shellfish Immunol (2005) 18:297–310. doi: 10.1016/j.fsi.2004.08.002
37. Amparyup P, Sutthangkul J, Charoensapsri W, Tassanakajon A. Pattern Recognition Protein Binds to Lipopolysaccharide and β-1,3-glucan and Activates Shrimp Prophenoloxidase System. J Biol Chem (2012) 287:10060–9. doi: 10.1074/jbc.M111.294744
38. Destoumieux D, Bulet P, Strub J, Van Dorsselaer A, Bache E. Recombinant Expression and Range of Activity of Penaeidins, Antimicrobial Peptides From Penaeid Shrimp. Enviromental Microbiol (1999) 346:335–46. doi: 10.1046/j.1432-1327.1999.00855.x
39. Sritunyalucksana K, Söderhäll K. The proPO and Clotting System in Crustaceans. Aquaculture (2000) 191:53–69. doi: 10.1016/S0044-8486(00)00411-7
40. Cerenius L, Kawabata S, Lee BL, Nonaka M. Proteolytic Cascades and Their Involvement in Invertebrate Immunity. Trends Biochem Sci (2010) 35:575–83. doi: 10.1016/j.tibs.2010.04.006
41. Wang R, Lee SY, Cerenius L, So K. Properties of the Prophenoloxidase Activating Enzyme of the Freshwater Crayfish, Pacifastacus Leniusculus. Eur J Biochem (2001) 902:895–902. doi: 10.1046/j.1432-1327.2001.01945.x
42. Charoensapsri W, Amparyup P, Hirono I, Aoki T, Tassanakajon A. Gene Silencing of a Prophenoloxidase Activating Enzyme in the Shrimp, Penaeus Monodon, Increases Susceptibility to Vibrio Harveyi Infection. Dev Comp Immunol (2009) 33:811–20. doi: 10.1016/j.dci.2009.01.006
43. Soonthornchai W, Chaiyapechara S, Klinbunga S. Differentially Expressed Transcripts in Stomach of Penaeus Monodon in Response to AHPND Infection. Dev Comp Immunol (2016) 65:53–63. doi: 10.1016/j.dci.2016.06.013
44. Apitanyasai K, Chang C, Hann T, Siong Y, Liou J. Penaeus Vannamei Serine Proteinase Inhibitor 7 (Lv Serpin7 ) Acts as an Immune Brake by Regulating the proPO System in AHPND-a Ff Ected Shrimp. Dev Comp Immunol (2020) 106:103600. doi: 10.1016/j.dci.2019.103600
45. Cerenius L, Lee BL, So K. The proPO-system: Pros and Cons for its Role in Invertebrate Immunity. Trends Immunol (2008) 29:263–71. doi: 10.1016/j.it.2008.02.009
46. Hergenhahn H, Aspant A, Soderhalltt K. Purification and Characterization of a high-MA Proteinase Inhibitor of Pro-Phenol Oxidase Activation From Crayfish Plasma. Biochem J (1987) 248:223–8. doi: 10.1042/bj2480223
47. Kan H, Kim C-H, Kwon H-M, Park J-W, Roh K-B, Lee H, et al. Molecular Control of Phenoloxidase-induced Melanin Synthesis in an Insect. J Biol Chem (2008) 283:25316–23. doi: 10.1074/jbc.M804364200
48. Saibu M, Sagar S, Green I, Ameer F, Meyer M. Evaluating the Cytotoxic Effects of Novel Quinone Compounds. Anticancer Res (2014) 34:4077–86.
49. Fridovich I. Superoxide Radical and Superoxide Dismutases. Annu Rev Biochem (1995) 64:97–112. doi: 10.1146/annurev.bi.64.070195.000525
50. Hart PJ, Balbirnie MM, Ogihara NL, Nersissian AM, Weiss MS, Valentine JS, et al. A Structure-Based Mechanism for Copper - Zinc Superoxide Dismutase †,‡. Biochemistry (1999) 38:2167–78. doi: 10.1021/bi982284u
51. Chakravarthy N, Aravindan K, Kalaimani N. Intracellular Copper Zinc Superoxide Dismutase (icCuZnSOD) From Asian Seabass (Lates Calcarifer): Molecular Cloning, Characterization and Gene Expression With Reference to Vibrio Anguillarum Infection. Dev Comp Immunol (2012) 36:751–5. doi: 10.1016/j.dci.2011.11.002
52. Wedde M, Weise C, Kopacek P, Franke P, Vilcinskas A. Purification and Characterization of an Inducible Metalloprotease Inhibitor From the Hemolymph of Greater Wax Moth Larvae, Galleria Mellonella. Eur J Biochem (1998) 225:535–43. doi: 10.1046/j.1432-1327.1998.2550535.x
53. Kanost MR. Serine Proteinase Inhibitors in Arthropod Immunity. Dev Comp Immunol (1999) 23:291–301. doi: 10.1016/S0145-305X(99)00012-9
54. Ericsson JD, Janmaat AF, Lowenberger C, Myers JH. Is Decreased Generalized Immunity a Cost of Bt Resistance in Cabbage Loopers Trichoplusia N? J Invertebr Pathol (2009) 100:61–7. doi: 10.1016/j.jip.2008.10.007
55. Damas G, Oppert B. Comparative Evaluation of Phenoloxidase Activity in Different Larval Stages of Four Lepidopteran Pests After Exposure to Bacillus Thuringiensis. J Insect Sci (2012) 12:1–11. doi: 10.1673/031.012.8001
56. Lin S, Hsu K, Wang H. Structural Insights Into the Cytotoxic Mechanism of Vibrio Parahaemolyticus PirA Vp and PirB Vp Toxins. Mar Drugs (2017) 15:9–12. doi: 10.3390/md15120373
57. Loseva O, Ibrahim M, Candas M, Koller CN, Bauer LS, Bulla LA Jr. Changes in Protease Activity and Cry3Aa Toxin Binding in the Colorado Potato Beetle: Implications for Insect Resistance to Bacillus Thuringiensis Toxins. Insect Biochem Mol Biol (2002) 32:567–77. doi: 10.1016/S0965-1748(01)00137-0
58. Sa J, Real MD, Bravo A. Role of Toxin Activation on Binding and Pore Formation Activity of the Bacillus Thuringiensis Cry3 Toxins in Membranes of Leptinotarsa Decemlineata (Say ). Biochim Biophys Acta (2004) 1660:99–105. doi: 10.1016/j.bbamem.2003.11.004
59. Lin SJ, Chen Y-F, Hsu K-C, Chen Y-L, Ko T-P, Lo C-F, et al. Structural Insights to the Heterotetrameric Interaction Between the Vibrio Parahaemolyticus PirA Vp and PirB Vp Toxins and Activation of the Cry-Like Pore-Forming Domain. Toxins (Basel) (2019) 11. doi: 10.3390/toxins11040233
Keywords: AHPND, Penaeus vannamei, AHPND tolerant P vannamei, shrimp immunity, immune genes
Citation: Mai HN, Caro LFA, Cruz-Flores R, White BN and Dhar AK (2021) Differentially Expressed Genes in Hepatopancreas of Acute Hepatopancreatic Necrosis Disease Tolerant and Susceptible Shrimp (Penaeus vannamei). Front. Immunol. 12:634152. doi: 10.3389/fimmu.2021.634152
Received: 27 November 2020; Accepted: 15 April 2021;
Published: 13 May 2021.
Edited by:
Kunlaya Somboonwiwat, Chulalongkorn University, ThailandReviewed by:
Ikuo Hirono, Tokyo University of Marine Science and Technology, JapanYeong Yik Sung, University of Malaysia Terengganu, Malaysia
Parin Chaivisuthangkura, Srinakharinwirot University, Thailand
Copyright © 2021 Mai, Caro, Cruz-Flores, White and Dhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arun K. Dhar, YWRoYXJAZW1haWwuYXJpem9uYS5lZHU=
 Hung N Mai
Hung N Mai Luis Fernando Aranguren Caro
Luis Fernando Aranguren Caro Brenda Noble White
Brenda Noble White Arun K. Dhar
Arun K. Dhar