
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 21 July 2021
Sec. Viral Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.632798
This article is part of the Research Topic Updates on Immunity to Influenza A Virus in Humans and Animals View all 5 articles
Influenza A virus (IAV) has a higher genetic variation, leading to the poor efficiency of traditional vaccine and antiviral strategies targeting viral proteins. Therefore, developing broad-spectrum antiviral treatments is particularly important. Host responses to IAV infection provide a promising approach to identify antiviral factors involved in virus infection as potential molecular drug targets. In this study, in order to better illustrate the molecular mechanism of host responses to IAV and develop broad-spectrum antiviral drugs, we systematically analyzed mRNA expression profiles of host genes in a variety of human cells, including transformed and primary epithelial cells infected with different subtypes of IAV by mining 35 microarray datasets from the GEO database. The transcriptomic results showed that IAV infection resulted in the difference in expression of amounts of host genes in all cell types, especially those genes participating in immune defense and antiviral response. In addition, following the criteria of P<0.05 and |logFC|≥1.5, we found that some difference expression genes were overlapped in different cell types under IAV infection via integrative gene network analysis. IFI6, IFIT2, ISG15, HERC5, RSAD2, GBP1, IFIT3, IFITM1, LAMP3, USP18, and CXCL10 might act as key antiviral factors in alveolar basal epithelial cells against IAV infection, while BATF2, CXCL10, IFI44L, IL6, and OAS2 played important roles in airway epithelial cells in response to different subtypes of IAV infection. Additionally, we also revealed that some overlaps (BATF2, IFI44L, IFI44, HERC5, CXCL10, OAS2, IFIT3, USP18, OAS1, IFIT2) were commonly upregulated in human primary epithelial cells infected with high or low pathogenicity IAV. Moreover, there were similar defense responses activated by IAV infection, including the interferon-regulated signaling pathway in different phagocyte types, although the differentially expressed genes in different phagocyte types showed a great difference. Taken together, our findings will help better understand the fundamental patterns of molecular responses induced by highly or lowly pathogenic IAV, and the overlapped genes upregulated by IAV in different cell types may act as early detection markers or broad-spectrum antiviral targets.
Influenza A virus (IAV) infection causes severe respiratory symptoms and persistent morbidity as well as mortality during annual seasonal or pandemic outbreaks, resulting in a severe threat to public health and safety, and even huge economic burden (1). Over the past decade, influenza outbreaks and pandemics have been caused by different subtypes of IAV, including H1N1, H3N2, swine-origin H1N1, and highly pathogenic avian influenza viruses (2–4), suggesting that the deeper biologic and epidemiologic mechanisms should be revealed to confidently and accurately predict the next influenza outbreak. Accumulative evidence has shown that IAV is capable of eliciting cellular immune response thought changing the expression of multiple genes in diverse types of cells, which in turn inhibit IAV infection. Airway epithelial cells are the preferred location for IAV replication and dissemination, and IAV infection induced toll-like receptors (TLRs)-related genes expression in responses to the pathogen (5). Moreover, other cell types, including endothelial cells, macrophages, monocytes, dendritic cells, and neutrophils, play important roles in response to IAV infection (6–10). During IAV infection, interferon, interferon-stimulated genes, and cytokines were secreted and activated in epithelial cells and immune cell types such as macrophages, monocytes, and neutrophils to facilitate antiviral responses. However, IAV could also elicit inflammation and cause various disorders of the respiratory system. Therefore, the systematic comparison of host responses of various types of cells to a range of strains of IAV still need to be further investigated.
Microarray technology with maturity is a powerful tool for the global view of gene expression levels, and enormous amounts of genome-wide gene expression microarray studies were distributed and archived in the gene expression omnibus (GEO) repository at the National Centre for Biotechnology Information (NCBI) in the last few decades, providing the chance for investigators revisiting these data to solve scientific questions. In this current study, we collected various transcriptomic datasets that were involved in diverse types of cells infected with subtypes of IAV, in order to examine common aspects of host cell responses to various subtypes of IAV infection. By integrating the global gene expression data, our results suggested that although the differentially expressed genes involved in host responses might not conform, the similar immune responses of diverse cell types were triggered by the infection of different subtypes of IAV.
The public gene expression resource of human cells infected with different IAV subtypes are mainly collected from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Totally, after searching for keywords related to IAV, we selected 35 data series about IAV for this research (Tables 1–3, and 5). The normalization of data of target profiles from the GEO database was performed using the limma package (30) to detect differentially expressed genes (DEGs). Then, significant DEGs were obtained using this set of parameters: P value < 0.05 and abs [log fold change (logFC)] > 1.5.
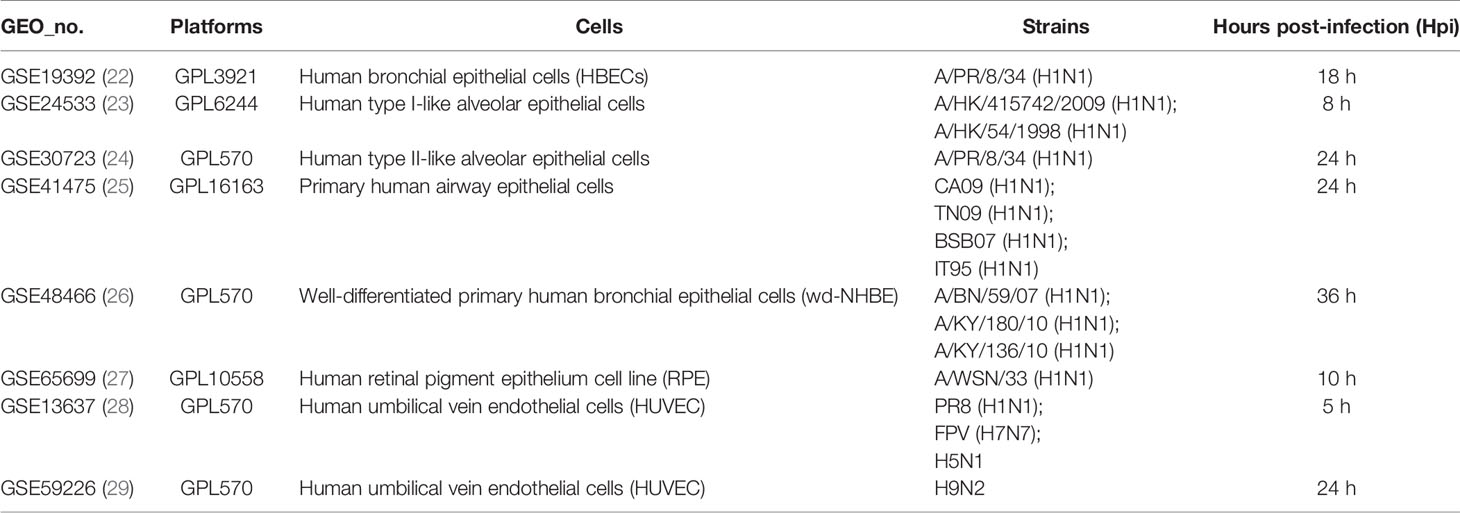
Table 3 The details of gene expression profiles on epithelial and endothelial cells from the GEO database.
DEGs with abs (logFC) > 1.5 and P value < 0.05 from each data series were obtained to analyze the overlap genes. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway analyses were performed to predict the functionalities of differentially expressed overlap genes using the R package clusterProfiler (31). In this study, a Venn diagram, heatmap, and volcano plot were constructed using R language and R packages, including VennDiagram (32), ggplot2 (33), and pheatmap (34).
To illustrate host cell response to influenza virus infection, the global gene expression profiles from four cell-based time-series gene expression datasets in A549, a lung epithelial cell known to be highly susceptible to IAV, were analyzed (Table 1) (11–15). Matched with the criteria of P<0.05 and |logFC|≥1.5, the Venn diagram showed that 17 genes (IFI6, IFIT2, HERC5, ISG15, RSAD2, GBP1, IFIT3, IFNB1, IFITM1, LAMP3, USP18, CXCL10, IER5L, HAMP, MX2, FERMT3, and ZC3HAV1) were differentially expressed genes (DEGs) and overlapped in these different databases (Figure 1A). Then, to further identify whether the mRNA expression of these genes had the same expression pattern in different subtypes of influenza virus infection, the databases with the other subtypes of influenza virus were analyzed and the result showed that the mRNA expression of 11 overlapped genes (IFI6, IFIT2, ISG15, HERC5, RSAD2, GBP1, IFIT3, IFITM1, LAMP3, USP18, and CXCL10) were also remarkably upregulated in A549 cells infected with A/Puerto Rico/8/1934 (H1N1), H5N2, H5N3, and H9N2 (Figure 1B) compared with mock, respectively. Overall, these results indicated that IFI6, IFIT2, ISG15, HERC5, RSAD2, GBP1, IFIT3, IFITM1, LAMP3, USP18, and CXCL10 might play key roles in IAV infection.
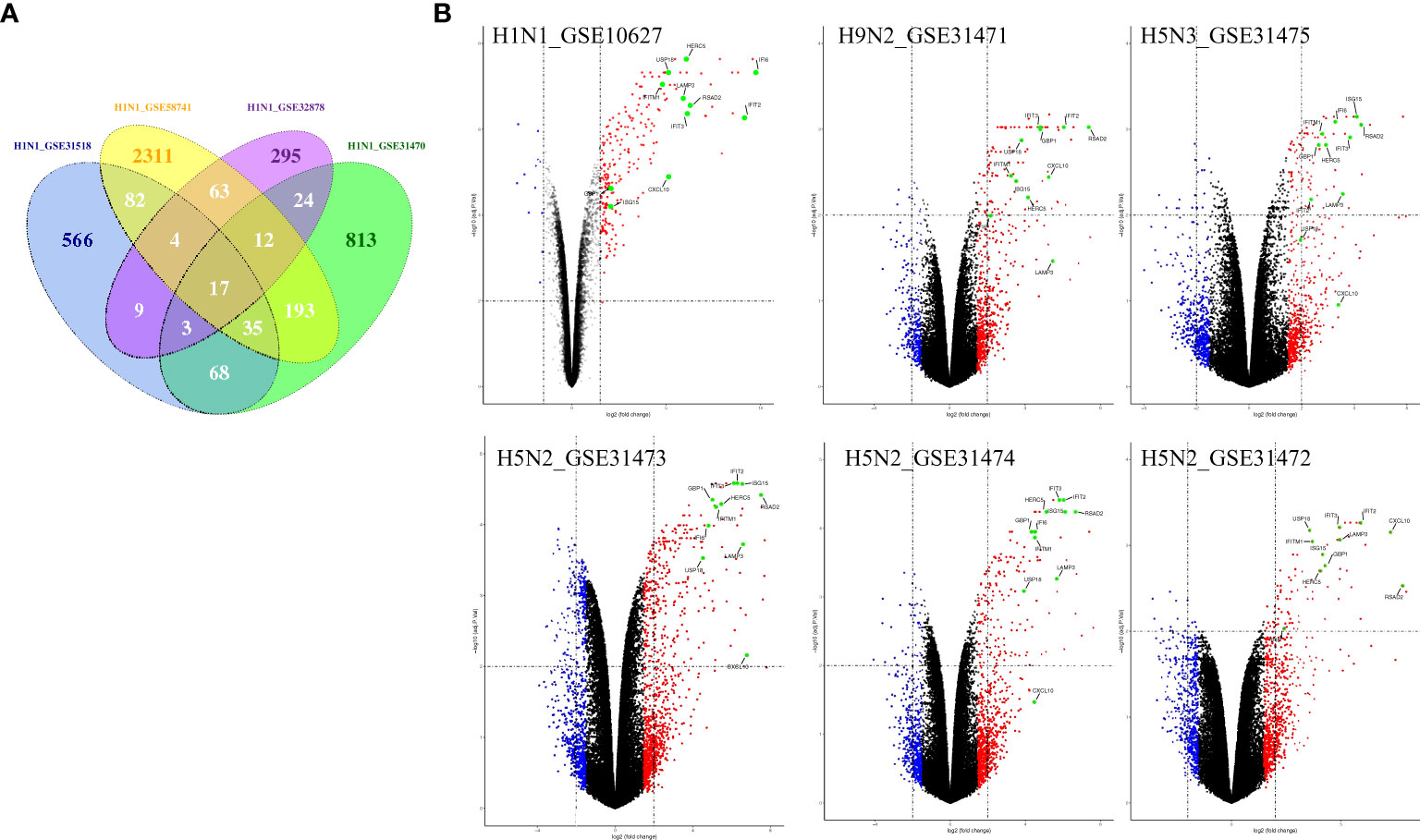
Figure 1 The common gene expression in A549 cells infected with IAV. (A) The Venn diagram shows the overlapping genes in 4 H1N1-infected datasets, 17 genes were common to the 4 H1N1-infected groups; (B) Volcano plots showing differentially expressed genes for H1N1, H9N2, H5N3, and H5N2-infected A549 cells. The vertical dashed lines correspond to 1.5-fold up and down expression, and the horizontal dashed line represents a p-value of 0.01. Blue and red dots indicate significantly downregulated and upregulated genes, respectively. Genes that were not classified as differentially expressed are plotted in black. The 11 overlapped genes identified in different IAV subtype infection are exhibited in the Volcano plots.
Airway epithelial cells are key to regulate pulmonary inflammatory and immune responses against influenza virus challenges. To acquire the knowledge on how airway epithelial cells contribute to the defense against IAV infection, the data from Calu-3 cells infected with influenza virus strains (H1N1, H3N2, H5N1, H7N7, H7N9, and H11N9) were collected and analyzed (Table 2) (16–21). In the Calu-3 cells infected with H1N1, the results from four transcriptomic datasets (GSE80697, GSE40844_CA, GSE40844_NL, and GSE37571) showed that twelve overlapping DEGs containing USP18, RSAD2, IFIT2, OAS1, MX1, IFI44, MX2, IFIT1, DHX58, IFITM1, IFI44L, and FOLR2 were identified to have a higher expression compared with the uninfected cells at 12 h post-infection (hpi). In addition, at 24 hpi, there were 66 overlapping DEGs (Supplementary File 1) significantly regulated by IAV (Figure 2A), according to the criteria of P<0.05 and |logFC|≥1.5. Interestingly, all of those 12 common DEGs at 12 hpi were also significantly upregulated by IAV infection at 24 hpi. In addition, hierarchical clustering analysis was used to assess the expression profiling of the common DEGs in different samples with different infection time-points in four H1N1 infection data series, and heatmap diagrams revealed that a large proportion of the overlapping DEGs mRNA expression exhibited an infection time-dependent upregulation (Figure 2B). Subsequently, host transcriptome profiles in Calu-3 cells infected with H5N1 from GSE28166, GSE33142, and GSE49840 were analyzed. The results showed that a total of 158 overlapping DEGs were identified in Calu-3 cells at 12 hpi, while there were 2233 overlapping DEGs at 24 hpi upon H5N1 strains infection following the criteria of P<0.05 and |logFC|≥1.5 (Figures 3A, B). In addition, KEGG pathway analysis showed that the overlapping genes were mainly concentrated on type I interferon-mediated signaling and host response to the virus at 12 hpi, while the genes related with the steroid biosynthetic process and adaptive immune response were enriched in H5N1-infected Calu-3 cells at 24 hpi (Figures 3A, B). Moreover, hierarchical clustering analysis showed that a large proportion of the overlapping DEGs mRNA expression exhibited an infection time-dependent upregulation in three H5N1 infection data series (Figure 3C).Additionally, H3N2, H7N7, H7N9, and H11N9-induced gene expression profiles at 24 hpi were overlapped to identify 15 upregulated genes (IFNB1, IL6, CMPK2, IL28A, IFIT3, IL29, RSAD2, MX2, IFI44, IFIT2, IFIH1, IFIT1, IFI44L, CXCL10, BATF2) following the criteria of P<0.05 and |logFC|≥1.5 (Figures 4A–E). Furthermore, integration of the overlapping DEGs regulated with the six subtypes of influenza virus at 24 hpi identified eight overlapped genes (BATF2, IFNB1, IL28A, IL29, IFIT2, CXCL10, IFI44L, and IL6) that were significantly overexpressed in IAV-infected Calu-3 cells.
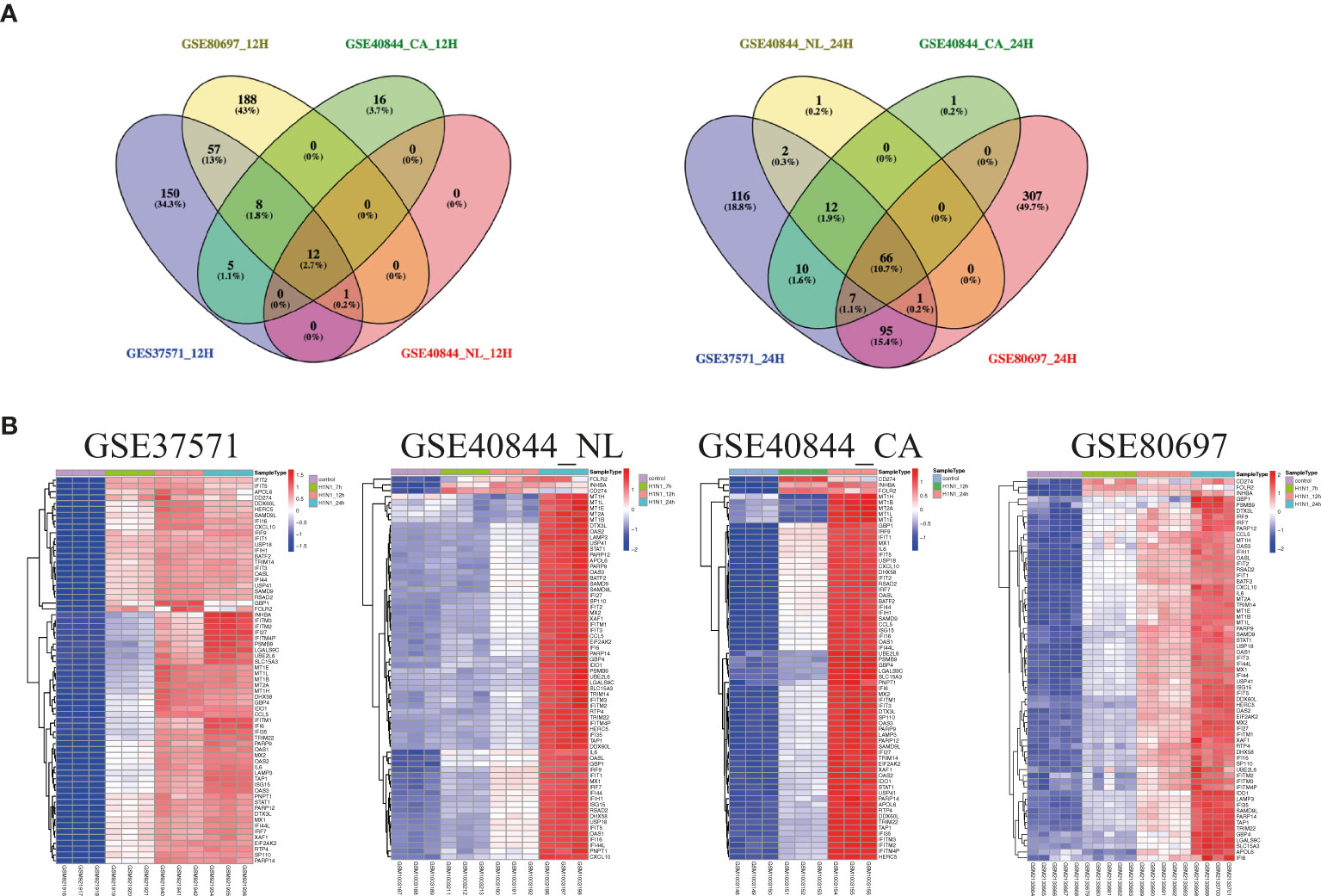
Figure 2 The expression profiling of common genes from four different data series infected with H1N1 in Calu-3 cells. (A) Venn diagram of the overlap genes between different data with 12 hpi (left) or 24 hpi (right). Twelve genes were common to the four H1N1-infected Calu-3 cells at 12 hpi, while 66 overlapped genes were found at 24 hpi. (B) Heatmap of differentially expressed patterns of the overlap genes in IAV-infected Calu-3 cells. Row represents the overlap genes and each column corresponds to a sample. NL: A/Netherland/219/2003, CA: A/California/04/2009.
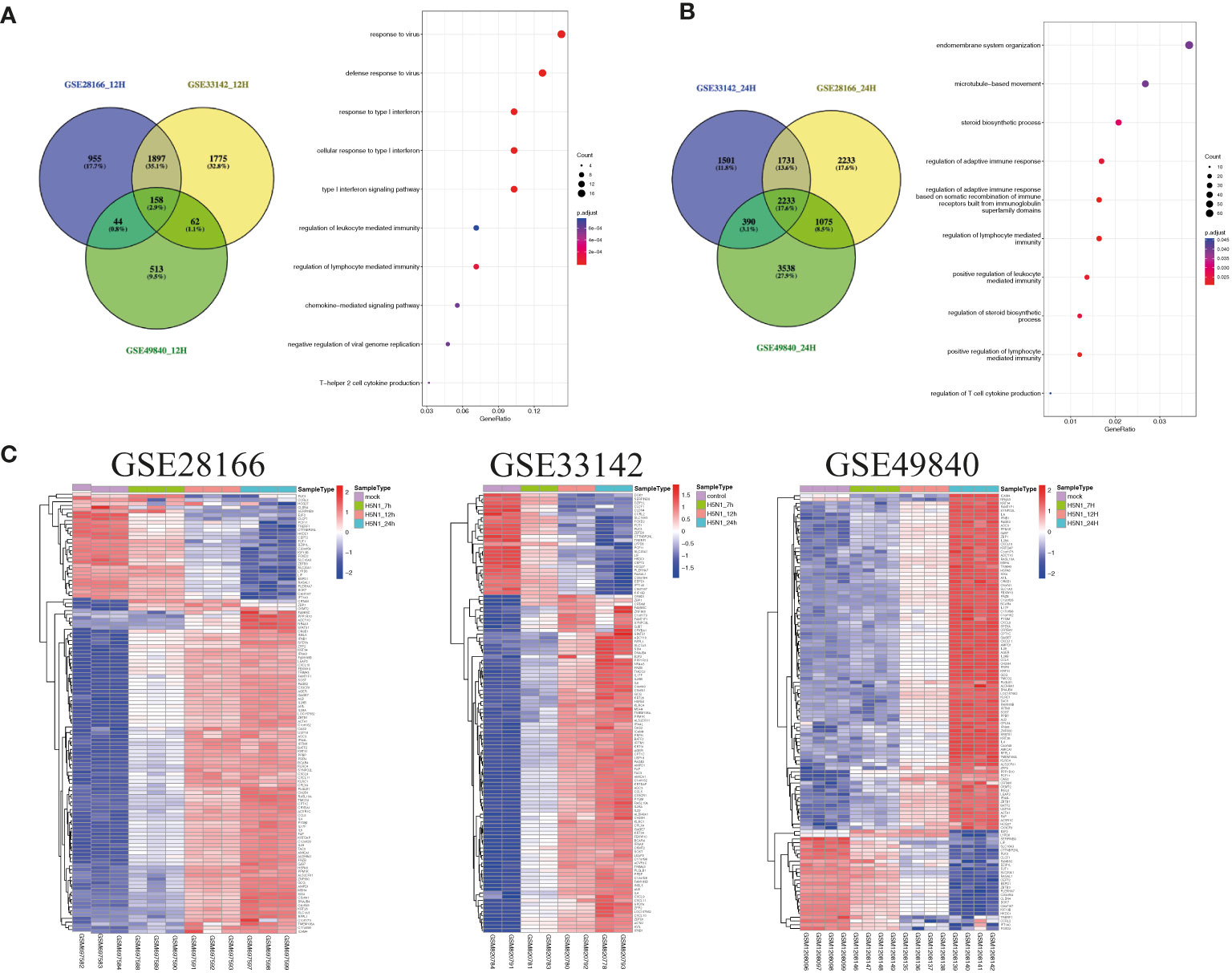
Figure 3 The common gene expression from three different data series in Calu-3 cells infected with H5N1. (A) A Venn diagram of the overlap genes between different data with 12 hpi. A total of 158 genes were common to the four H5N1-infected Calu-3 cells at 12 hpi; KEGG pathways of differentially expressed overlap genes. The vertical axis shows the number of genes, and the horizontal axis shows the GeneRatio. (B) A Venn diagram of the overlap genes between different data with 24 hpi. In total, 2233 overlapped genes were common to the four H5N1-infected Calu-3 cells at 24 hpi. KEGG pathways of differentially expressed overlap genes. The vertical axis shows the number of genes, and the horizontal axis shows the GeneRatio. (C) Heatmap of differentially expressed patterns of the overlap genes in IAV-infected Calu-3 cells. Row represents the overlap genes and each column corresponds to a sample.
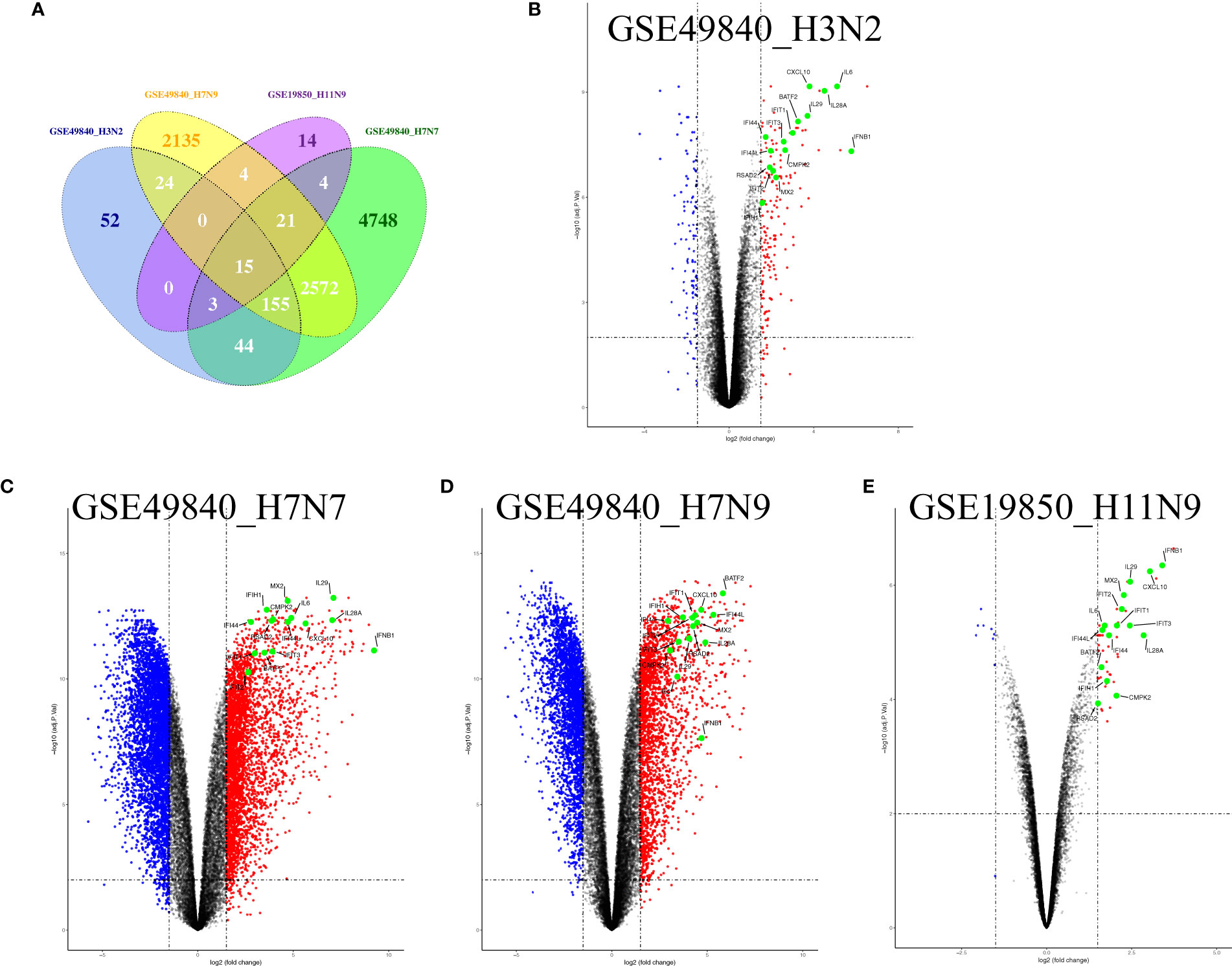
Figure 4 Fifteen overlap genes were identified to significantly upregulate in H3N2, H7N7, H7N9, and H11N9-infected Calu-3 cells. (A) Venn diagram of the overlap genes; (B) Volcano plots of the overlaps between mock and H3N2-infected cells; blue and red dots indicate significantly downregulated and upregulated genes, respectively. (C) Volcano plots of the overlaps between mock and H7N7-infected cells; blue and red dots indicate significantly downregulated and upregulated genes, respectively. (D) Volcano plots of the overlaps between mock and H7N9-infected cells; blue and red dots indicate significantly downregulated and upregulated genes, respectively. (E) Volcano plots of the overlaps between mock and H11N9-infected cells. The vertical dashed lines correspond to 1.5-fold up and down expression and the horizontal dashed line represents a p-value of 0.01. Blue and red dots indicate significantly downregulated and upregulated genes, respectively. The 15 overlapped genes identified in different IAV subtype infection are exhibited in the Volcano plots.
To further illustrate the effect of transcriptome alteration in influenza virus infection on human respiratory tract epithelial cells, we analyzed gene expression profiles of influenza virus-infected primary epithelial cells (Table 3) (22–29), including primary human bronchial epithelial cells (HBEC), well-differentiated human bronchial epithelial cells (wd-NHBE), human primary airway epithelial cell, human type I-like alveolar epithelial cells, and human type II-like alveolar epithelial cells, all of which were infected by H1N1 influenza. As above, matching the criteria of P<0.05 and |logFC|≥1.5, in total 17 overlapping genes (IFI44L, IFI44, HERC5, OASL, CXCL10, MX2, CXCL11, OAS2, XAF1, IFIT3, USP18, IFIH1, OAS1, DDX58, MX1, IFIT2, and IFIT1) were identified to be significantly increased with influenza virus infection in all human primary epithelial cells (Figures 5A–D) and these commonly responsive genes were largely related with interferon-stimulated response. To identify whether other strains of influenza virus also induced the same response in other epithelium and endothelial cells lines, human retinal pigment epithelium cell line (RPE) and human umbilical vein endothelial cells (HUVEC) were infected with other subtypes of influenza virus (H7N7, H5N1, and H9N2) and the genome-wide gene expression pattern of infected cells was analyzed. The results showed that 14 of 17 overlapping genes (IFI44L, IFI44, HERC5, OASL, CXCL10, MX2, OAS2, XAF1, IFIT3, USP18, IFIH1, OAS1, IFIT2, and IFIT1) in primary epithelial cells were remarkably upregulated in H1N1, H7N7, H5N1, and H9N2-infected RPE or HUVEC (Figures 6A–C), indicating that these overlapped DEGs may play key roles in regulating epithelium cells response against IAV infection. Moreover, we found that BATF2 could be significantly upregulated with H1N1 and other strain infections in all collected datasets of human primary epithelial cells and endothelial cells, expect one dataset (GSE19392) where the BATF2 gene was not included in the microarray platform (Table 4). Overall, our results suggested that a similar host response was induced by different influenza strains in human epithelial and endothelial cells, regardless of high or low pathogenicity.
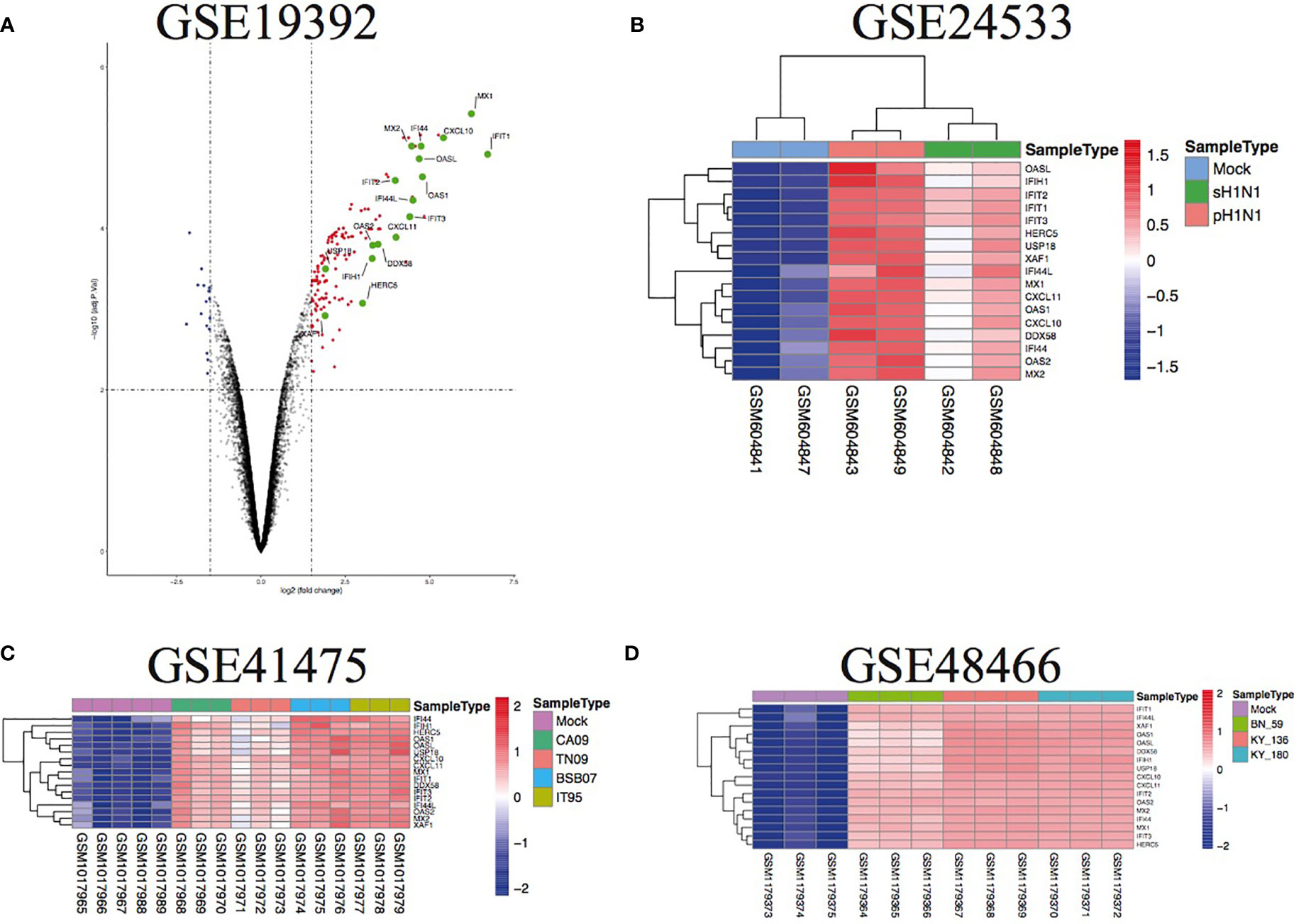
Figure 5 The overlap regulation gene expression of diverse primary epithelial cells in response to H1N1 infection. (A) Volcano plots of the overlaps between mock and H1N1-infected human bronchial epithelial cells. The vertical dashed lines correspond to 1.5-fold up and down expression and the horizontal dashed line represents a p-value of 0.01. Blue and red dots indicate significantly downregulated and upregulated genes, respectively. The 17 overlapped genes identified in different IAV subtype infections are exhibited in the Volcano plots. (B) Heatmap of the overlaps in mock or H1N1-infected human type I-like alveolar epithelial cells. Row represents the overlap genes and each column corresponds to a sample; (C) heatmap of the overlaps in mock or H1N1-infected primary human airway epithelial cells. Row represents the overlap genes and each column corresponds to a sample; (D) heatmap of the overlaps in mock or H1N1-infected well-differentiated primary human bronchial epithelial cells. Row represents the overlap genes and each column corresponds to a sample.
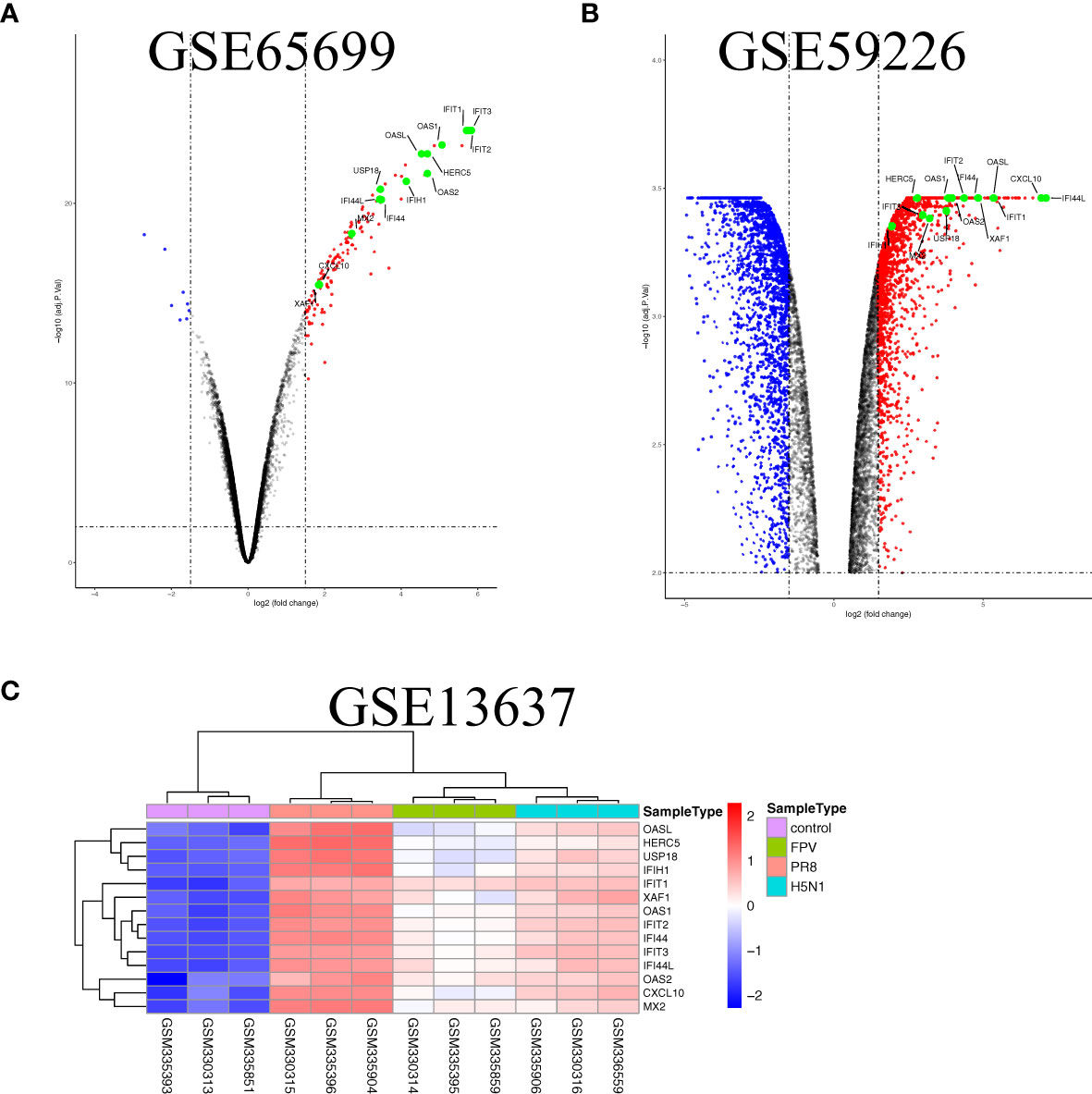
Figure 6 The overlap regulation gene expression of epithelium and endothelial cells lines in response to IAV infection. (A) Volcano plots of the overlaps between mock and the H1N1-infected human retinal pigment epithelium cell line. The vertical dashed lines correspond to 1.5-fold up and down expression and the horizontal dashed line represents a p-value of 0.01. Blue and red dots indicate significantly downregulated and upregulated genes, respectively. The 13 overlapped genes identified in different IAV subtype infections are exhibited in the Volcano plots. (B) Volcano plots of the overlaps between mock and H9N2-infected human umbilical vein endothelial cells. The vertical dashed lines correspond to 1.5-fold up and down expression and the horizontal dashed line represents a p-value of 0.01. Blue and red dots indicate significantly downregulated and upregulated genes, respectively. The 13 overlapped genes identified in different IAV subtype infections are exhibited in the Volcano plots. (C) Heatmap of the overlaps in mock or H1N1, H5N1, and H7N7-infected human umbilical vein endothelial cells. Row represents the overlap genes and each column corresponds to a sample.
Phagocytes containing monocytes, macrophages, dendritic cells, and neutrophils are critical in the recognition, engulfment, and destruction of invading pathogens. To uncover the antiviral activities of phagocytes against different subtypes of influenza viruses, the transcriptome analysis of gene expression profiles was performed in human phagocytic cells (PBMC, monocytes, macrophages, pDC, and neutrophils) that were infected with different subtypes of influenza viruses, including H1N1, H5N1, and H7N7 (Table 5) (24, 27, 35–41). Following the criteria of P<0.05 and |logFC|≥1.5, 12 genes (C19orf66, IL6, HESX1, IFNB1, IFITM3, ISG20, ISG15, HERC5, IFIT1, CCL8, IFIT2, and CXCL10) were overlapped and upregulated in H1N1-infected monocyte-derived macrophages and primary alveolar macrophages (Figure 7A). Moreover, BAFT2 was also significantly increased in multiple data series of macrophages, except GSE27702 where the BATF2 gene was not included in the microarray platform. On the side, 29, 54, and 35 overlapped DEGs were significantly regulated in H1N1-infected monocytes, monocyte-derived DCs, and neutrophils, respectively (Supplementary File 2), and further Gene Ontology (GO) and Venn diagram analysis showed that H1N1 infection triggered a stronger host response with an elevated expression of cytokines and interferons signaling molecules in phagocytic cells, which in turn blocked IAV infection (Figures 7B–D). Five common genes (OASL, IFIT3, RSAD2, HERC5, and IFIT1) identified in macrophages were also found to be upregulated in monocytes, monocyte-derived DCs, and neutrophils infected with H1N1.
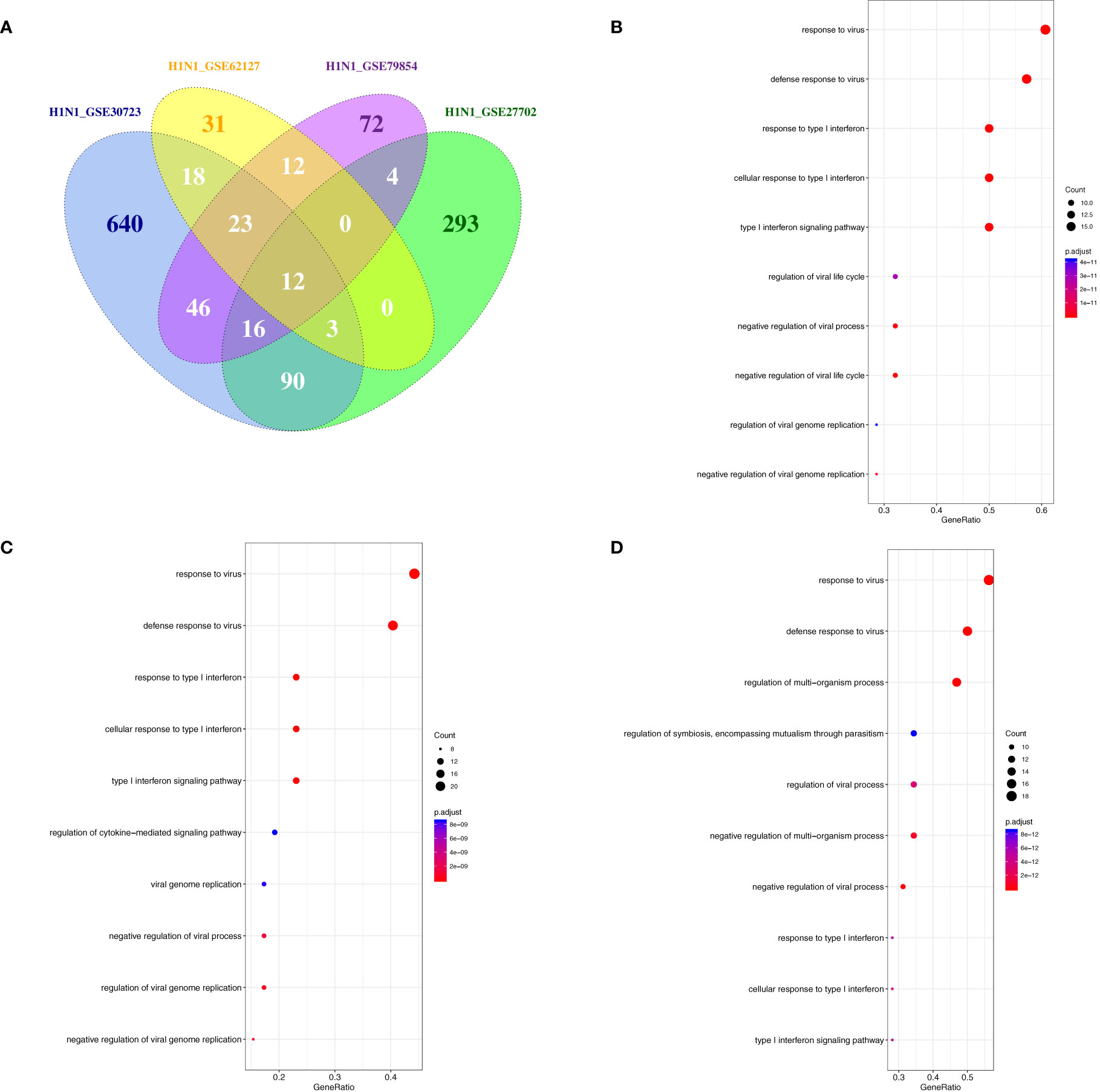
Figure 7 The overlap regulation gene expression of macrophages in response to H1N1 infection. (A) A Venn diagram of the overlap genes in H1N1-infected macrophages, 12 genes were common to the two infected groups. (B) GO biological processes of differentially expressed overlap in H1N1-infected monocytes. The vertical axis shows the functional classification, and the horizontal axis shows the GeneRatio; (C) GO biological processes of differentially expressed overlap in H1N1-infected monocyte-derived DCs. The vertical axis shows the functional classification, and the horizontal axis shows the GeneRatio; (D) GO biological processes of differentially expressed overlap in H1N1-infected neutrophils. The vertical axis shows the functional classification, and the horizontal axis shows the GeneRatio.
In addition, to illustrate whether there was a similar response of phagocytes to other subtypes of influenza A virus infection, the globe gene expression profiles of avian H5N1 and H7N7-infceted monocytes and monocyte-derived macrophages were analyzed. The Venn diagram revealed that a total of 125 different expression genes were found to overlap in the avian flu-infected cells (Figure 8A). Moreover, the results of GO and KEGG pathway analysis showed that the overlap genes were mainly concentrated on cytokine-mediated signaling, type I interferon-mediated signaling, defense response to virus, and necroptosis (Figures 8B, C). A similar antiviral response, including the type I interferon-mediated signaling pathway, cytokine signaling pathway, and antiviral defense in H1N1-infceted phagocytes was also induced in human phagocytes during highly or lowly pathogenic influenza virus infection, although an overlapped gene was not found in H1N1, H5N1, and H7N7-infceted phagocytes.
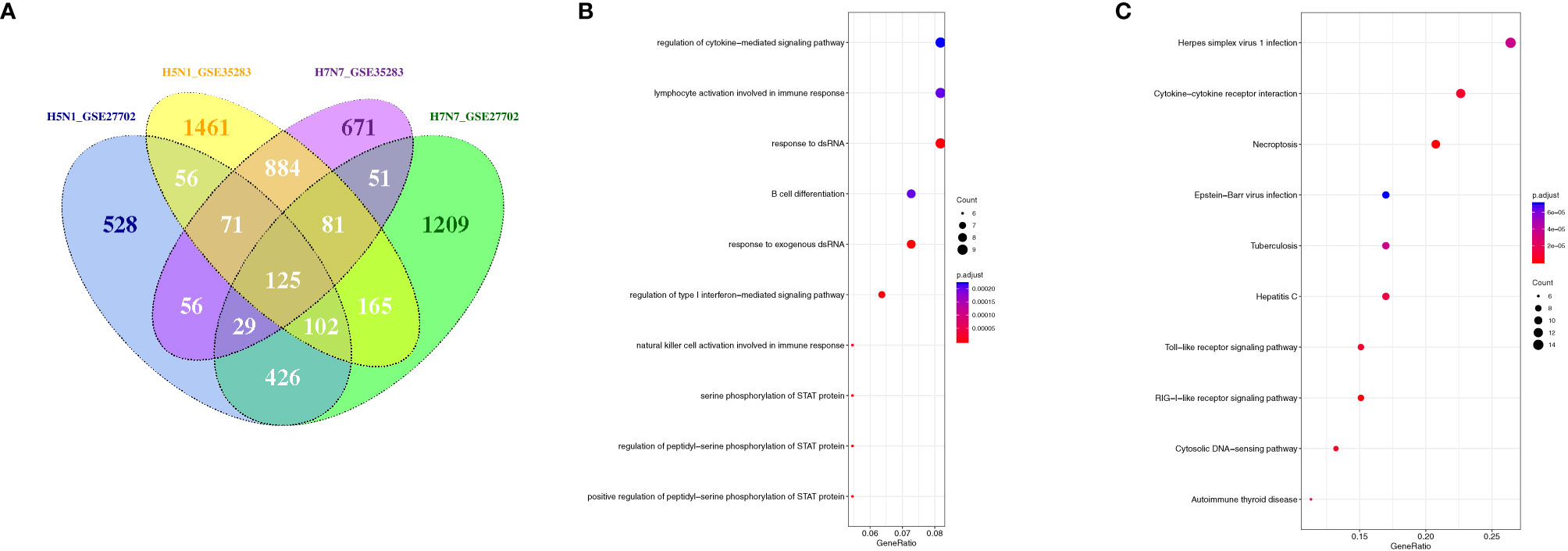
Figure 8 The overlapped regulation gene expression of monocytes and macrophages in response to avian influenza virus infection. (A) A Venn diagram of the overlap genes between H5N1 and H7N7-infected monocytes and monocyte-derived macrophages; (B) GO biological processes of differentially expressed overlap. The vertical axis shows the functional classification, and the horizontal axis shows the GeneRatio; (C) KEGG pathways of differentially expressed overlap genes. The vertical axis shows the number of genes, and the horizontal axis shows the GeneRatio.
Host defense responses elicited by IAV are critical to protect the host against IAV. However, the mechanisms underlying how the host response is activated among IAVs was not fully understood. In this study, we collected and analyzed a large amount of gene expression data of different host cells infected with different subtypes of IAV. We found that IAV infection could affect host response and generate special differentially expressed genes in the distinct cell lines. And then we identified some upregulated genes (IFIT2, HERC5, and BATF2) that may exhibit antiviral activities in different cell types against IAV infection. Moreover, with the available data collections from distinct types of cells with different strains of IAV infection, we compared gene expression difference and the potential response difference of distinct types of cells to IAV stains.
Epithelial cells are the primary targets of IAV infection and can produce a protective environment by continuously secreting antiviral substances to initiate defense responses against infection. Many studies have shown that epithelial cells could produce many host cellular restriction factors in response to IAV infection (42). We summarized gene expression profiles of different human epithelial cells including human lung epithelial cells (A549), human airway epithelial cells (Calu-3), primary human bronchial epithelial cells (HBEC), well-differentiated human bronchial epithelial cells (wd-NHBE), human primary airway epithelial cells, human type I-like alveolar epithelial cells, and human type II-like alveolar epithelial cells. The results showed that various subtypes of IAV infection affected the amount of differentially expression genes, mainly involved in the interferon signaling pathway and cytokine and chemokine signaling pathway in different epithelial cells. But some overlapped genes (IFIT2, HERC5, and BATF2) were found to upregulate during different subtypes of IAV infection.
Interferon-induced protein with tetratricopeptide repeats 2 (IFIT2) is an interferon-stimulated gene (ISG) with a possible RNA-binding capacity and acts as an important restriction factor for many viruses, including rabies virus, Sendai virus, mouse hepatitis virus, hepatitis B virus, West Nile virus, and influenza virus (43–48). However, recent genome-wide knockout screens provided a novel proviral function of IFIT2, and knockout of IFIT2 remarkedly reduced diverse influenza viruses infection by decreasing the translational efficiency for viral mRNA and IFIT2-bound mRNAs. The influenza virus hijacked IFIT2 to preferentially bind viral mRNAs and prevent ribosome pausing for increasing viral replication (49).
HERC5, an interferon-induced HECT E3 enzyme was identified to upregulate in distinct cell types infected with different subtypes of IAV and might potentially play an important role for IAV replication. Existing research has revealed the antiviral activity of HERC5 against IAV; knockdown of HERC5 weakens IFN-beta-induced antiviral activities against IAV. HERC5 activated the ISGylation system by catalyzing conjugation of ISG15 onto IAV-NS1 proteins, leading to ISG15 modification of NS1 protein and blocking of the nuclear import of the NS1A protein (50, 51). In addition, our result that HERC5 acts as a key antiviral factor in IAV infection was in accordance with the result of a previous report, which identified HERC5 as a potential novel biomarker for the treatment of IAV by employing weighted gene co-expression network analysis (WGCNA) (52). Moreover, we found that although some microarray platforms did not include the BATF2 gene owing to the design of the array, the mRNA expression of BATF2, a member of AP-1 family transcription factor (53), was still significantly upregulated in multiple cells infected with different subtypes of IAV. BATF2 was proved as an antibacterial gene and was able to induce inflammatory responses in lipopolysaccharides and mycobacterium tuberculosis infection (54). IFNγ induced high levels of BATF2 mRNA expression to downregulate trypanosoma cruzi-induced IL-23 production in innate immune cells by blocking the recruitment of the c-JUN-ATF-2 heterodimer to the IL23a promoter and preventing the formation of the c-JUN-ATF-2 complex, and IFN-γ–induced BATF2 expression plays a key role for controlling Th17-mediagted immune responses during trypanosoma cruzi infection (54). In addition, BATF2 was broadly highly expressed in multiple tissues, including the spleen, lung, small intestine, cecum, and large intestine, and IFN-γ–induced BATF2 also disturbed T cell-mediated intestinal inflammation through the regulation of the IL-23/IL-17 axis that was associated with intestine inflammation (55). Previous studies reported that BATF2 was a proapoptotic gene and overexpression of BATF2 could lead to inhibition of DNA binding activation protein (AP1), which causes growth inhibition and induces apoptosis particularly in cancerous cells (53). Additionally, BATF2 could dephosphorylate phosphor-STAT3 to promote DUSP2 expression and upregulate of NF-κB activity, and could also be modified by N6-methyladenosine (m6A) to suppress its expression (56, 57). During feline infectious peritonitis virus (FIPV) infection, BATF2 showed continuous high expression and might be an important regulation factor of the death stages of infected cells (58). Our transcriptome analysis also showed that BATF2 was significantly increased in distinct cell types with IAV stains infection, indicating that it will be increasingly interesting to illustrate the role of BATF2 during IAV infection.
In conclusion, we demonstrated the gene expression pattern and molecular responses of distinct cells types among different subtypes of IAV infection. In general, IAV strains triggered a similar defense response among distinct cells types via the production of various antiviral cytokines and interferon-related genes, although few overlapped genes were present in distinct cells types. We identified that IFIT2, HERC5, and BATF2 might act as key antiviral factors to regulate IAV infection, but the molecular regulatory mechanisms of IFIT2, HERC5, and BATF2 involved in IAV infection still need to be validated.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All authors contributed to the article and approved the submitted version. AZ designed this project, analyzed and interpreted data, and contributed to the writing of the manuscript. XD collected the data and modified the manuscript. BT analyzed and interpreted the data. ML drew the figures. BT and AZ supervised the financial support.
This work was supported by the Key Lab of Process Analysis and Control of Sichuan Universities (No. 2018001). The project was sponsored by Sichuan Province for ROCS (0903/00021728) and NSFC (81902073).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.632798/full#supplementary-material
1. Florian K, Smith GJD, Fouchier RAM, Peiris M, Kedzierska K, Doherty PC, et al. Influenza. Nat Rev Dis Primers (2018) 4(1):3. doi: 10.1038/s41572-018-0002-y
2. Cook PW, Stark T, Jones J, Kondor R, Zanders N, Benfer J, et al. Detection and Characterization of Swine-Origin Influenza A(H1N1) Pandemic 2009 Viruses in Humans Following Zoonotic Transmission. J Virol (2020) 95(2):e01066-20. doi: 10.1128/JVI.01066-20
3. Barbara JJ, Uyeki TM, Jernigan DB. Fifty Years of Influenza A(H3N2) Following the Pandemic of 1968. Am J Public Health (2020) 110(5):669–76. doi: 10.2105/AJPH.2019.305557
4. Uyeki TM, Peiris M. Novel Avian Influenza A Virus Infections of Humans. Infect Dis Clin North Am (2019) 33(4):907–32. doi: 10.1016/j.idc.2019.07.003
5. Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, et al. Influenza Virus Receptor Specificity and Cell Tropism in Mouse and Human Airway Epithelial Cells. J Virol (2006) 80(15):7469–80. doi: 10.1128/JVI.02677-05
6. Feng B, Zhang Q, Wang J, Dong H, Mu X, Hu G, et al. IFIT1 Expression Patterns Induced by H9N2 Virus and Inactivated Viral Particle in Human Umbilical Vein Endothelial Cells and Bronchus Epithelial Cells. Mol Cells (2018) Apr 3041(4):271–81. doi: 10.14348/molcells.2018.2091
7. Tumpey TM, García-Sastre A, Taubenberger JK, Palese P, Swayne DE, Pantin-Jackwood MJ, et al. Pathogenicity of Influenza Viruses With Genes From the 1918 Pandemic Virus: Functional Roles of Alveolar Macrophages and Neutrophils in Limiting Virus Replication and Mortality in Mice. J Virol (2005) 79:14933–44. doi: 10.1128/JVI.79.23.14933-14944.2005
8. Bussfeld D, Kaufmann A, Meyer RG, Gemsa D, Sprenger H. Differential Mononuclear Leukocyte Attracting Chemokine Production After Stimulation With Ac- Tive and Inactivated Influenza A Virus. Cell Immunol (1998) 186:1–7. doi: 10.1006/cimm.1998.1295
9. Hao X, Kim TS, Braciale TJ. Differential Response of Respiratory Dendritic Cell Subsets to Influenza Virus Infection. J Virol (2008) 82:4908–19. doi: 10.1128/JVI.02367-07
10. Wang JP, Bowen GN, Padden C, Cerny A, Finberg RW, Newburger PE, et al. Toll-Like Receptor-Mediated Activation of Neutrophils by Influenza A Virus. Blood (2008) 112:2028–34. doi: 10.1182/blood-2008-01-132860
11. Sutejo R, Yeo DS, Zu Myaing M, Hui C, Xia J, Ko D, et al. Activation of Type I and III Interferon Signalling Pathways Occurs in Lung Epithelial Cells Infected With Low Pathogenic Avian Influenza Viruses. PloS One (2012) 7(3):e33732. doi: 10.1371/journal.pone.0033732
12. Taye B, Yeo D, Chuen Lee RT, Huan Tan B, Sugrue RJ, Maurer-Stroh S. Inter-Species Host Gene Expression Differences in Response to Human and Avian Influenza A Virus Strains. Int J Mol Sci (2017) 18(11):2295. doi: 10.3390/ijms18112295
13. Song W, Li H, Chen Y, Wei H, Gao GF, Liu H, et al. Transport of Influenza Virus Neuraminidase (NA) to Host Cell Surface Is Regulated by ARHGAP21 and Cdc42 Proteins. J Biol Chem (2012) 287(13):9804–16. doi: 10.1074/jbc.M111.312959
14. Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, et al. Robust Expression of Vault RNAs Induced by Influenza A Virus Plays a Critical Role in Suppression of PKR-Mediated Innate Immunity. Nucleic Acids Res (2015) 43(21):10321–37. doi: 10.1093/nar/gkv1078
15. Wang B, Lam TH, Soh MK, Ye Z, Chen J, Chee Ren EE. Influenza A Virus Facilitates Its Infectivity by Activating P53 to Inhibit the Expression of Interferon-Induced Transmembrane Proteins. Front Immunol (2018) 9:1193. doi: 10.3389/fimmu.2018.01193
16. Chen-Yu Hsu A, Barr I, Hansbro PM, Wark PA. Human Influenza is More Effective Than Avian Influenza at Antiviral Suppression in Airway Cells. Am J Respir Cell Mol Biol (2011) 44(6):906–13. doi: 10.1165/rcmb.2010-0157OC
17. Li C, Bankhead A 3rd, Eisfeld AJ, Hatta Y, Jeng S, Chang JH, et al. Host Regulatory Network Response to Infection With Highly Pathogenic H5N1 Avian Influenza Virus. J Virol (2011) 85(21):10955–67. doi: 10.1128/JVI.05792-11
18. McDermott JE, Shankaran H, Eisfeld AJ, Belisle SE, Neuman G, Li C, et al. Conserved Host Response to Highly Pathogenic Avian Influenza Virus Infection in Human Cell Culture, Mouse and Macaque Model Systems. BMC Syst Biol (2011) 5:190. doi: 10.1186/1752-0509-5-190
19. Mitchell HD, Eisfeld AJ, Sims AC, McDermott JE, Matzke MM, Webb-Robertson B-JM, et al. A Network Integration Approach to Predict Conserved Regulators Related to Pathogenicity of Influenza and SARS-CoV Respiratory Viruses. PloS One (2013) 8(7):e69374. doi: 10.1371/journal.pone.0069374
20. Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. Transcriptomic Characterization of the Novel Avian-Origin Influenza A (H7N9) Virus: Specific Host Response and Responses Intermediate Between Avian (H5N1 and H7N7) and Human (H3N2) Viruses and Implications for Treatment Options. mBio (2014) 5(1):e01102–13. doi: 10.1128/mBio.01102-13
21. Burnum-Johnson KE, Kyle JE, Eisfeld AJ, Casey CP, Stratton KG, Gonzalez JF, et al. MPLEx: A Method for Simultaneous Pathogen Inactivation and Extraction of Samples for Multi-Omics Profiling. Analyst (2017) 142(3):442–8. doi: 10.1039/c6an02486f
22. Shapira SD, Gat-Viks I, Shum BOV, Dricot A, de Grace MM, Wu L, et al. A Physical and Regulatory Map of Host-Influenza Interactions Reveals Pathways in H1N1 Infection. Cell (2009) 139(7):1255–67. doi: 10.1016/j.cell.2009.12.018
23. Lee SMY, Chan RWY, Gardy JL, Lo C-K, Sihoe ADL, Kang SSR, et al. Systems-Level Comparison of Host Responses Induced by Pandemic and Seasonal Influenza A H1N1 Viruses in Primary Human Type I-Like Alveolar Epithelial Cells In Vitro. Respir Res (2010) 11(1):147. doi: 10.1186/1465-9921-11-147
24. Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, et al. Innate Immune Response to Influenza A Virus in Differentiated Human Alveolar Type II Cells. Am J Respir Cell Mol Biol (2011) 45(3):582–91. doi: 10.1165/rcmb.2010-0108OC
25. Gold ES, Diercks AH, Podolsky I, Podyminogin RL, Askovich PS, Treuting PM, et al. 25-Hydroxycholesterol Acts as an Amplifier of Inflammatory Signalling. Proc Natl Acad Sci USA (2014) 111(29):10666–71. doi: 10.1073/pnas.1404271111
26. Gerlach RL, Camp JV, Chu Y-K, Jonsson CB. Early Host Responses of Seasonal and Pandemic Influenza A Viruses in Primary Well-Differentiated Human Lung Epithelial Cells. PloS One (2013) 8(11):e78912. doi: 10.1371/journal.pone.0078912
27. Anastasina M, Le May N, Bugai A, Fu Y, Söderholm S, Gaelings L, et al. Influenza Virus NS1 Protein Binds Cellular DNA to Block Transcription of Antiviral Genes. Biochim Biophys Acta (2016) 1859(11):1440–8. doi: 10.1016/j.bbagrm.2016.09.005
28. Viemann D, Schmolke M, Lueken A, Boergeling Y, Friesenhagen J, Wittkowski H, et al. H5N1 Virus Activates Signaling Pathways in Human Endothelial Cells Resulting in a Specific Imbalanced Inflammatory Response. J Immunol (2011) 186(1):164–73. doi: 10.4049/jimmunol.0904170
29. Wang W, Mu X, Zhao L, Wang J, Chu Y, Feng X, et al. Transcriptional Response of Human Umbilical Vein Endothelial Cell to H9N2 Influenza Virus Infection. Virology (2015) 482:117–27. doi: 10.1016/j.virol.2015.03.037
30. Smyth G. Limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer (2005). p. 397–420.
31. Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics: J Integr Biol (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
32. Chen H, Boutros PC. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinf (2011) 12:35. doi: 10.1186/1471-2105-12-35
33. Wickham H. ggplot2: Elegant Graphics for Data Analysis . New York: Springer-Verlag (2016). doi: 10.1007/978-3-319-24277-4
34. Kolde R. Pheatmap: Pretty Heatmaps, R Package Version 1.0.8. 2015. Available at: http://CRAN.Rproject.org/package=pheatmap.
35. Friesenhagen J, Boergeling Y, Hrincius E, Ludwig S, Roth J, Viemann D. Highly Pathogenic Avian Influenza Viruses Inhibit Effective Immune Responses of Human Blood-Derived Macrophages. J Leukoc Biol (2012) 92(1):11–20. doi: 10.1189/jlb.0911479
36. Fu Y, Gaelings L, Söderholm S, Belanov S, Nandania J, Sampsa Matikainen TAN, et al. JNJ872 Inhibits Influenza A Virus Replication Without Altering Cellular Antiviral Responses. Antiviral Res (2016) 133:23–31. doi: 10.1016/j.antiviral.2016.07.008
37. Hou W, Gibbs JS, Lu X, Brooke CB, Roy D, Modlin RL, et al. Viral Infection Triggers Rapid Differentiation of Human Blood Monocytes Into Dendritic Cells. Blood (2012) 119(13):3128–31. doi: 10.1182/blood-2011-09-379479
38. Zaslavsky E, Nudelman G, Marquez S, Hershberg U, Hartmann BM, Thakar J, et al. Reconstruction of Regulatory Networks Through Temporal Enrichment Profiling and its Application to H1N1 Influenza Viral Infection. BMC Bioinf (2013) 14 Suppl 6(Suppl 6):S1. doi: 10.1186/1471-2105-14-S6-S1
39. Ciancanelli MJ, Huang SXL, Luthra P, Garner H, Itan Y, Volpi S, et al. Infectious Disease. Life-Threatening Influenza and Impaired Interferon Amplification in Human IRF7 Deficiency. Science (2015) 348(6233):448–53. doi: 10.1126/science.aaa1578
40. Bajwa G, DeBerardinis RJ, Shao B, Hall B, Farrar JD, Gill MA. Cutting Edge: Critical Role of Glycolysis in Human Plasmacytoid Dendritic Cell Antiviral Responses. J Immunol (2016) 196(5):2004–9. doi: 10.4049/jimmunol.1501557
41. Malachowa N, Freedman B, Sturdevant DE, Kobayashi SD, Nair 3 V, Feldmann F, et al. Differential Ability of Pandemic and Seasonal H1N1 Influenza A Viruses To Alter the Function of Human Neutrophils. mSphere (2018) 3(1):e00567–17. doi: 10.1128/mSphereDirect.00567-17
42. Villalón-Letelier F, Brooks AG, Saunders PM, Londrigan SL, Reading PC. Host Cell Restriction Factors That Limit Influenza A Infection. Viruses (2017) 9(12):376. doi: 10.3390/v9120376
43. Davis BM, Fensterl V, Lawrence TM, Hudacek AW, Sen GC, Schnell MJ. Ifit2 Is a Restriction Factor in Rabies Virus Pathogenicity. J Virol (2017) 91(17):e00889–17. doi: 10.1128/JVI.00889-17
44. Wetzel JL, Fensterl V, Sen GC. Sendai Virus Pathogenesis in Mice is Prevented by Ifit2 and Exacerbated by Interferon. J Virol (2014) 88(23):13593–601. doi: 10.1128/JVI.02201-14
45. Butchi NB, Hinton DR, Stohlman SA, Kapil P, Fensterl V, Sen GC, et al. Ifit2 Deficiency Results in Uncontrolled Neurotropic Coronavirus Replication and Enhanced Encephalitis via Impaired Alpha/Beta Interferon Induction in Macrophages. J Virol (2014) 88(2):1051–64. doi: 10.1128/JVI.02272-13
46. Pei R, Qin B, Zhang X, Zhu W, Kemper T, Ma Z, et al. Interferon-Induced Proteins With Tetratricopeptide Repeats 1 and 2 Are Cellular Factors That Limit Hepatitis B Virus Replication. J Innate Immun (2014) 6(2):182–91. doi: 10.1159/000353220
47. Cho H, Shrestha B, Sen GC, Diamond MS. A Role for Ifit2 in Restricting West Nile Virus Infection in the Brain. J Virol (2013) 87(15):8363–71. doi: 10.1128/JVI.01097-13
48. Hou L, Li J, Qu H, Yang L, Chen Y, Du Q, et al. Inhibition of Replication and Transcription of WSN Influenza A Virus by IFIT Family Genes. Sheng Wu Gong Cheng Xue Bao (2015) 31(1):123–34. doi: 10.13345/j.cjb.140160-en
49. Tran V, Ledwith MP, Thamamongood T, Higgins CA, Tripathi S, Chang MW, et al. Influenza Virus Repurposes the Antiviral Protein IFIT2 to Promote Translation of Viral mRNAs. Nat Microbiol (2020) 5(12):1490–503. doi: 10.1038/s41564-020-0778-x
50. Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, et al. Herc5 Attenuates Influenza A Virus by Catalyzing ISGylation of Viral NS1 Protein. J Immunol (2010) 184(10):5777–90. doi: 10.4049/jimmunol.0903588
51. Zhao C, Hsiang T-Y, Kuo R-L, Krug RM. ISG15 Conjugation System Targets the Viral NS1 Protein in Influenza A Virus-Infected Cells. Proc Natl Acad Sci USA (2010) 107(5):2253–8. doi: 10.1073/pnas.0909144107
52. Zarei Ghobadi M, Mozhgani S-H, Farzanehpour M, Behzadian F. Identifying Novel Biomarkers of the Pediatric Influenza Infection by Weighted Co-Expression Network Analysis. Virol J (2019) 16(1):124. doi: 10.1186/s12985-019-1231-8
53. Su ZZ, Lee SG, Emdad L, Lebdeva IV, Gupta P, Valerie K, et al. Cloning and Characterization of SARI (Suppressor of AP-1, Regulated by IFN). Proc Natl Acad Sci USA (2008) 105(52):20906–11. doi: 10.1073/pnas.0807975106
54. Kitada S, Kayama H, Okuzaki D, Koga R, Kobayashi M, Arima Y, et al. BATF2 Inhibits Immunopathological Th17 Responses by Suppressing Il23a Expression During Trypanosoma Cruzi Infection. J Exp Med (2017) 214(5):1313–31. doi: 10.1084/jem.20161076
55. Kayama H, Tani H, Kitada S, Opasawatchai A, Okumura R, Motooka D, et al. BATF2 Prevents T-Cell-Mediated Intestinal Inflammation Through Regulation of the IL-23/IL-17 Pathway. Int Immunol (2019) 31(6):371–83. doi: 10.1093/intimm/dxz014
56. Kanemaru H, Yamane F, Tanaka H, Maeda K, Satoh T, Akira S. BATF2 Activates DUSP2 Gene Expression and Up-Regulates NF-κb Activity via Phospho-STAT3 Dephosphorylation. Int Immunol (2018) 30(6):255–65. doi: 10.1093/intimm/dxy023
57. Xie J-W, Huang X-B, Chen Q-Y, Ma Y-B, Zhao Y-J, Liu L-C, et al. M 6 A Modification-Mediated BATF2 Acts as a Tumor Suppressor in Gastric Cancer Through Inhibition of ERK Signalling. Mol Cancer (2020) 19(1):114. doi: 10.1186/s12943-020-01223-4
Keywords: gene express profile, Influenza A virus, immune response, BATF2, HERC5
Citation: Zhou A, Dong X, Liu M and Tang B (2021) Comprehensive Transcriptomic Analysis Identifies Novel Antiviral Factors Against Influenza A Virus Infection. Front. Immunol. 12:632798. doi: 10.3389/fimmu.2021.632798
Received: 25 November 2020; Accepted: 04 June 2021;
Published: 21 July 2021.
Edited by:
Aitor Nogales, Centro de Investigación en Sanidad Animal (CISA), SpainReviewed by:
Xavier Saelens, Ghent University, BelgiumCopyright © 2021 Zhou, Dong, Liu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Dong, ZHh0b25nMjAyMEB3aHB1LmVkdS5jbg==; Bin Tang, dGJAc3dtdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.