- 1Department of Epidemiology and Biostatistics, Key Laboratory of Infectious Diseases, School of Public Health, Nanjing Medical University, Nanjing, China
- 2Department of Critical Care Medicine, The Affiliated Yixing Hospital of Jiangsu University, Yixing, China
- 3Department of Epidemiology and Statistics, School of Public Health, Medical College of Soochow University, Suzhou, China
- 4Institute of Epidemiology and Microbiology, Eastern Theater Command Centers for Disease Control and Prevention, Nanjing, China
- 5Department of Epidemiology, Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing, China
- 6Department of Digestive Medicine, Third Affiliated Hospital of Nantong University, Nantong, China
- 7Department of General Surgery, Third Affiliated Hospital of Nantong University, Nantong, China
- 8Department of Information, First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 9Department of Infectious Diseases, First Affiliated Hospital of Nanjing Medical University, Nanjing, China
Background: KIR/HLA-C signaling pathway influences the innate immune response which is the first defense to hepatitis C virus (HCV) infection. The aim of this study was to determine the association between the genetic polymorphisms of KIR/HLA-C genes and the outcomes of HCV infection in a high-risk Chinese population.
Methods: In this case-control study, four single nucleotide polymorphisms (SNPs) of KIR/HLA-C genes (KIR2DS4/KIR2DS1/KIR2DL1 rs35440472, HLA-C rs2308557, HLA-C rs1130838, and HLA-C rs2524094) were genotyped by TaqMan assay among drug users and hemodialysis (HD) patients including 1,378 uninfected control cases, 307 subjects with spontaneous viral clearance, and 217 patients with persistent HCV infection. Bioinformatics analysis was used to functionally annotate the SNPs.
Results: After logistic regression analysis, the rs35440472-A and rs1130838-A alleles were found to be associated with a significantly elevated risk of HCV infection (OR = 1.562, 95% CI: 1.229–1.987, P < 0.001; OR = 2.134, 95% CI: 1.180–3.858, P = 0.012, respectively), which remained significant after Bonferroni correction (0.05/4). The combined effect of their risk alleles and risk genotypes (rs35440472-AA and rs1130838-AA) were linked to the increased risk of HCV infection in a locus-dosage manner (all Ptrend < 0.001). Based on the SNPinfo web server, rs35440472 was predicted to be a transcription factor binding site (TFBS) while rs1130838 was predicted to have a splicing (ESE or ESS) function.
Conclusion: KIR2DS4/KIR2DS1/KIR2DL1 rs35440472-A and HLA-C rs1130838-A variants are associated with increased susceptibility to HCV infection in a high-risk Chinese population.
Introduction
Hepatitis C virus (HCV) infection is a global health problem, affecting more than 185 million individuals worldwide (1, 2). Among them, approximately 71 million people have progressed to chronic HCV infection and over 10 million are in China (3). Without effective and timely treatment, chronic HCV infection can induce decompensated cirrhosis and hepatocellular carcinoma (HCC), resulting in significant socioeconomic burden (4). Despite the high cure rates of direct-acting antivirals (DAAs), HCV elimination continues to be a challenge due to frequent re-infection in the high-risk population (5). Elucidating the mechanisms underlying HCV infection and development is essential to conducting precision prevention for the high-risk population and eliminating HCV.
The initiation and progression of HCV infection depend on a complex interplay of pathogenic and host factors. As we all know, immune responses affect disease development and innate immunity is the first line of defense against HCV. As a crucial member of the innate immune system, natural killer cells (NK cells), are well known for their rapid and efficient immune responses against HCV infection (6, 7). Furthermore, the functional activation of NK cells can affect HCV RNA replication as well as the expression of HCV proteins (8). The NK cell response and host susceptibility to infection are tightly regulated by specific immune receptors and their corresponding ligands (9–11).
Killer cell immunoglobulin-like receptors (KIRs), an important receptor of NK cells, are type I transmembrane glycoproteins belonging to the immunoglobulin superfamily (12). KIRs can activate or inhibit NK cells by recognizing class I major histocompatibility complex (MHC) molecules (13–15). Generally, the nucleated cells normally express inhibitory ligands, such as human leukocyte antigen class I (HLA-I), on their surface that keep the NK cells in an inactivated state by engaging with the KIRs (7). In contrast, virus-infected cells might reduce the expression of MHC molecules and present activating ligands on their surfaces. NK cells would be elicited and exert their antiviral effector functions to participate in immune responses (16, 17). However, aberrant activation of NK cells resulting in excessive or low immune responses may lead to persistent infection (18, 19). Not surprisingly, the exertion of NK cell function is directly impacted by the genetic variations of KIRs and HLA-I molecules (12, 20).
The KIR gene cluster is located on chromosome 19q13.4 within the 150 kb long leukocyte receptor complex region (21, 22). HLA-I proteins are encoded by the MHC genes that span a region of about 3.6 Mb on chromosome 6p21.3 (23, 24). The independent segregation of unlinked KIR and HLA-I genes both feature significant variations, which could affect the health and disease status in different individuals (25). Recent studies have shown that the diversity in the number and type of KIR-HLA gene combinations are associated with infections, cancer, and reproductive disorders (26–29). Dring et al. reported an association between KIR2DS3 and chronic HCV infection, and Nozawa et al. found that the presence of KIR2DL3 and HLA-C1 was associated with both treatment-induced and spontaneous clearance of HCV infection in the Caucasian population (30, 31). Recently, exciting researches have shown that KIR2DS4 selectively binds to HLA-C (32). This finding may be important for the host NK cell response and the outcome of viral infection, including the susceptibility to HIV and hepatitis B virus-associated hepatocellular carcinoma (33, 34). It is interesting that rs35440472 is attributed to KIR2DS4, KIR2DS1, and KIR2DL1 according to the NCBI dbSNP. KIR2DS4/KIR2DS1/KIR2DL1 rs35440472 was abbreviated as KIR2DS4 rs35440472 when appropriate in this study. In addition, even a single nucleotide polymorphism (SNP) also can markedly alter KIRs expression, and the specificity and activity of HLA-I (12, 35–38). However, little is known regarding the association between KIR/HLA-C SNPs and the outcome of HCV infection. Given all of the above, we hypothesized that SNPs of these signaling pathway genes may influence the outcomes of HCV infection. Therefore, the current study was to explore the associations between four potentially functional SNPs in the genes of the KIR/HLA-C and outcomes of HCV infection in a high-risk Chinese population, including KIR2DS4/KIR2DS1/KIR2DL1 rs35440472 G>A, HLA-C rs1130838 G>A, HLA-C rs2524094 A>G, and HLA-C rs2308557 A>G.
Method
Ethics Statement
This study was conducted according to the national legislation and the ethical guidelines of the Declaration of Helsinki (39), and approved by the Institutional Review Board of Nanjing Medical University. Written consent was obtained from all individuals before participating in this study and sampling.
Subjects
A total of 1,902 high-risk subjects, including 1,153 drug users from two compulsory detoxification centers (Nanjing and Yixing, China) and 749 hemodialysis (HD) patients from nine hospital-based hemodialysis centers (Jiangsu, China), were consecutively recruited from 2008 to 2018. The exclusion criteria were as follows (1): age below 18 years or over 80 years, (2) history of any anti-HCV treatment, (3) co-infection with human immunodeficiency virus (HIV) or other hepatitis viruses, and (4) any autoimmune, alcoholic, or metabolic liver diseases. The participants were categorized into the following groups: (1) Group A—uninfected controls who were seronegative for both anti-HCV antibodies and HCV RNA; (2) Group B—spontaneous HCV clearance, seropositive for anti-HCV antibodies and seronegative for HCV RNA; (3) Group C—persistent HCV infection and seropositive for both anti-HCV antibodies and HCV RNA. All subjects came from high-risk populations and had high-risk behaviors. Therefore, all three groups were exposed to HCV. The serological tests were conducted at least three times during the 6-month follow-up period to confirm the infection status. The demographic data and history of HCV exposure were recorded by interviewing each participant with a standard questionnaire.
Serological Testing
Blood samples (5–10 ml) were collected from all subjects and centrifuged to separate the plasma fractions, which were then stored at −20°C. Anti-HCV antibodies were detected through the qualitative third-generation enzyme-linked immunosorbent assay (ELISA) (Diagnostic Kit for Antibody to HCV 3.0 ELISA, Intec Products Inc, Xiamen, China) which is a qualitative experiment. Trizol LS reagent (Takara Biotech, Tokyo, Japan) was used to extract HCV RNA from the sera. The HCV RNA load was measured by real time fluorescent quantitative polymerase chain reaction with the detection limit of 1 × 10^3 IU/ml (Promotor® Hepatitis C Virus Quantitative PCR Test Kits; ACON Biotech Co., Ltd., Hangzhou, China). HCV genotypes were determined using the murex HCV serotype ELISA kit (Abbott, Wiesbaden, Germany) based on type-specific antibodies. Genomic DNA was extracted from the white blood cells (WBCs) using protease K digestion, followed by phenol-chloroform purification and ethanol precipitation. All serological tests were performed with the same analytical systems.
SNP Selection and Genotyping
The selection of candidate genes is based on the importance and the frequencies of KIR/HLA-C genes among the Chinese population. Candidate Tag SNPs of these genes were selected by searching the 1000 Genomes database (37.0 version, https://www.internationalgenome.org/home) and the HaploReg database Version 4.1 (http://pubs.broadinstitute.org/mammals/haploreg/haploreg.php). The selected SNPs were filtered using the following criteria: (1) minor allele frequency (MAF) >5% in the Chinese Han population and (2) pairwise linkage disequilibrium (LD) r2 ≥ 0.8. The above data can be acquired from Haploview software (version 4.2; Broad Institute, Cambridge, MA, USA). The immune-related KIR2DS4/HLA-C SNPs reported in previous studies were also considered. Finally, four SNP candidates (KIR2DS4/KIR2DS1/KIR2DL1 rs35440472, HLA-C rs2308557, HLA-C rs1130838, and HLA-C rs2524094) were chosen for genotyping. It should be mentioned that, according to the NCBI dbSNP, rs35440472 is shown to belong to KIR2DS4, KIR2DS1, and KIR2DL1. Not surprisingly, rs35440472 is located in the overlapping region of three genes. Rs35440472 is presented in these three genes, which all encode receptors for HLA-C. For simplicity, KIR2DS4/KIR2DS1/KIR2DL1 rs35440472 was abbreviated as KIR2DS4 rs35440472.
Genotyping of the four SNPs was conducted with a TaqMan allelic discrimination assay on the LightCycler® 480 IIReal-Time PCR System (Roche, Switzerland). The primers and probes are summarized in Table S1. The SNPs were genotyped in a blinded manner, and the success rate was >90%. Furthermore, 10% random samples were re-genotyped and the accordance rates were 100%. Besides, all tests were carried out in accordance with the manufacturer’s instructions. The genotyping experiments of all samples used identical procedures and instruments.
In Silico Analysis
The functions of the selected SNPs were predicted using the HaploReg database Version 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) and the SNPinfo Web Server (https://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi). The RNAfold Web Server (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi) was applied to detect mRNA secondary structures. The expression quantitative trait loci (eQTL) analysis was conducted with the Genotype-Tissue Expression (GTEx) database (https://www.gtexportal.org/home/snp/). The UCSC Genome Browser (http://genome.ucsc.edu/) was used to assess the potential biological function. The H3K4Me1 histone marker expression data in the GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, NHLF, and HepG2 cell lines were analyzed with the Encyclopedia of DNA Elements (ENCODE).
Statistical Analysis
The demographic, clinical, virological data were compared using the chi-square (χ2) test, one-way analysis of variance (ANOVA), or Kruskal-Wallis test as appropriate. Hardy-Weinberg equilibrium (HWE) was assessed in the control group by goodness-of-fit χ2 test. The co-dominant model, dominant, additive, and recessive genetic models were used to explore the association between each SNP and the outcomes of HCV infection. Logistic regression adjusted for gender, age, ALT, AST, IL28B-rs12979860, IL28B-rs8099917, and route of infection was performed to analyze the relationship between these SNPs and HCV infection outcome by calculating odds ratio (OR) and 95% confidence interval (CI). Stratified analysis was used to further reduce the bias of confounding factors. All statistical analyses were two-sided with a significance level of P < 0.05 was considered statistically significant. Bonferroni correction was applied to correct for multiple comparisons and the P-value was adjusted to 0.0125 (0.05/4) (40). All statistical analyses were performed using SPSS (version 22.0; SPSS Institute, Chicago, IL, USA) and Stata (version14.0; STATA Corp, College Station, TX, USA).
Results
Demographic and Clinical Characteristics
The demographic and clinical characteristics of all subjects are summarized in Table 1. Group A consisted of 1,378 individuals (1,003 males and 375 females; mean age 42.92 ± 12.91 years), Group B included 307 subjects (218 males and 89 females; mean age 41.61 ± 13.10 years), and Group C included 217 cases (154 males and 63 females; mean age 39.44 ± 11.25 years). There were no significant differences in the distribution of gender and the frequency of IL28B-rs12979860 among the three groups (P = 0.737 and P = 0.439, respectively). However, age, alanine aminotransferase (ALT), aspartate aminotransferase (AST), routes of infection, HCV genotype, and the frequency of IL28B-rs8099917 showed significant differences (all P < 0.001). Furthermore, the demographic and clinical characteristics of the HD and IVDU patients are shown in Tables S1 and S2, respectively. The genotype distributions in the control group were in accordance with the Hardy-Weinberg Equilibrium for all six SNPs (all P>0.05) (Table S3).
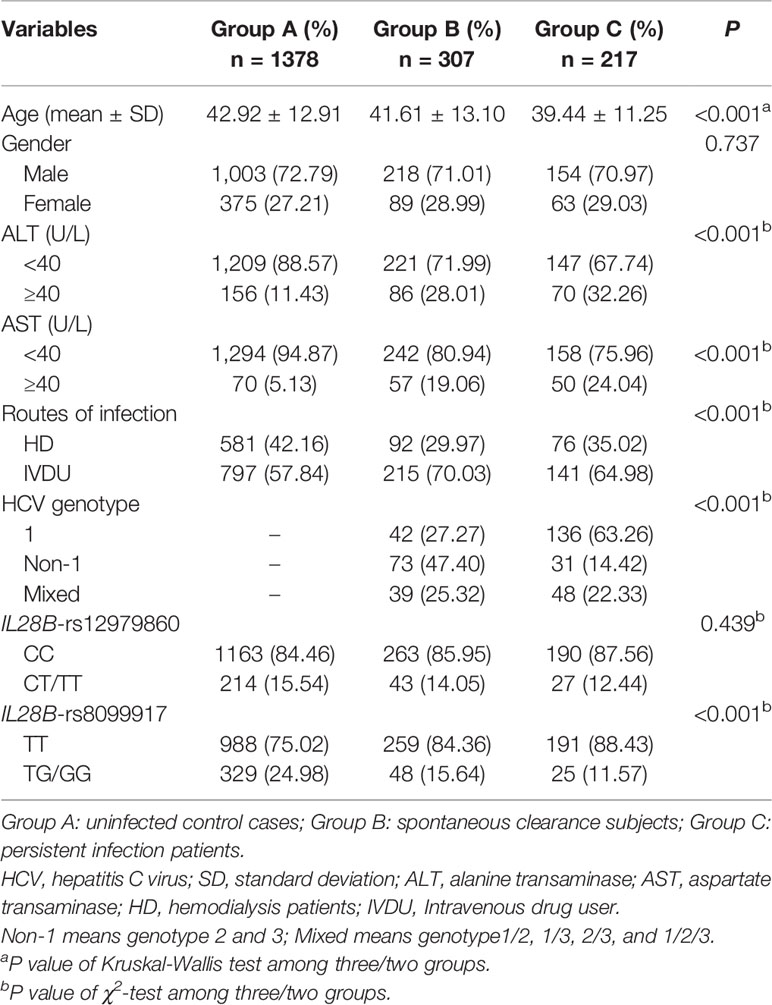
Table 1 Demographic and clinical characteristics among HCV control, spontaneous clearance, and persistent infection groups.
Association of Candidate SNPs With the Susceptibility and Chronicity of HCV Infection
The genotype distribution of rs35440472, rs1130838, rs2524094, and rs2308557 in the three groups is shown in Table 2. A representative TaqMan allelic discrimination assay result of rs35440472 is shown in Figure S1. To analyze the association between these SNPs and susceptibility to HCV infection, Group B and Group C patients were combined into the HCV-infected group and compared them with the control Group A. After adjusting for gender, age, ALT, AST, IL28B-rs12979860, IL28B-rs8099917, and route of infection, the results of logistic regression analysis revealed that KIR2DS4-rs35440472 and HLA-C-rs1130838 were associated with susceptibility to HCV infection. Furthermore, the frequency of the rs35440472-A allele was significantly higher in the infected versus the uninfected groups compared to that of the wild type rs35440472-G allele (dominant model: adjusted OR = 1.562, 95% CI = 1.229–1.987, P < 0.001; recessive model: adjusted OR = 1.460, 95% CI = 1.108–1.924, P = 0.007; additive model: adjusted OR = 1.361, 95% CI = 1.165–1.589, P < 0.001). Compared to rs1130838-GG genotype, the AA genotype correlated to a significantly increased risk of HCV infection (co-dominant model: adjusted OR = 2.134, 95% CI = 1.180–3.858, P = 0.012). Furthermore, the association between the selected SNPs and HCV infection chronicity was compared among the persistent infection subjects (Group C) and the spontaneous infection subjects (Group B). However, no significant association was observed in the logistic regression analyses between these four SNPs and HCV infection chronicity (all P > 0.05/4) (Table 2).
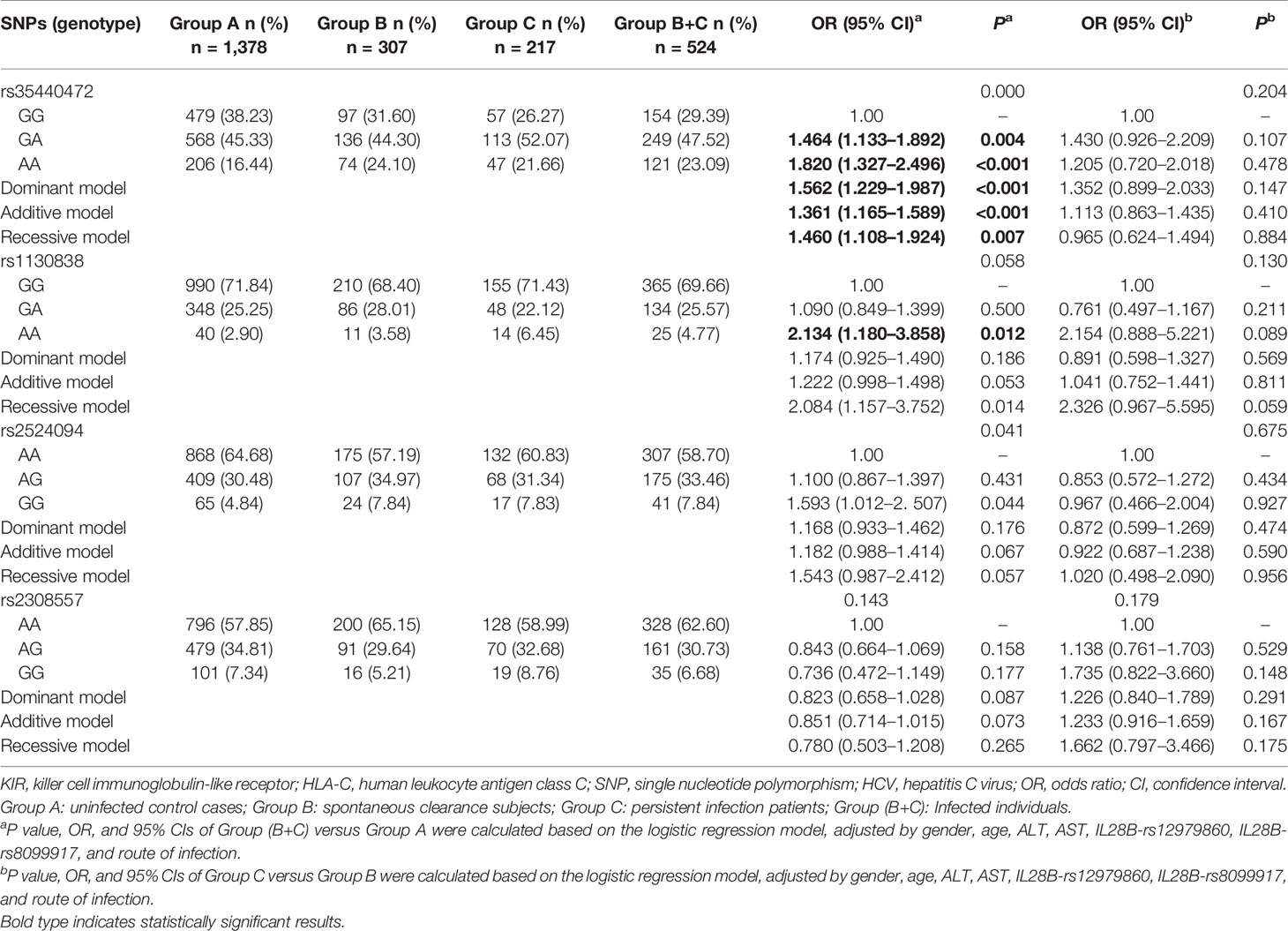
Table 2 Genotypes distributions of KIR/HLA-C genes among HCV control, spontaneous clearance, and persistent infection groups.
A stratified analysis was conducted to decrease the bias of gender, age, ALT, AST, and route of infection. Compared to the GG genotype of rs35440472, a significantly higher of infection was associated with the A allele in the following subgroups: age <50 years (adjusted OR: 1.417, 95% CI: 1.181–1.699, P<0.001), the male subgroup (adjusted OR: 1.378, 95% CI: 1.144–1.659, P = 0.001), ALT < 40 (adjusted OR: 1.348, 95% CI: 1.135–1.602, P = 0.001), ALT ≥ 40 (adjusted OR: 1.503, 95% CI: 1.028–2.197, P = 0.035), AST < 40 (adjusted OR: 1.392, 95% CI: 1.180–1.641, P<0.001), and IVDU (adjusted OR: 1.399, 95% CI: 1.145–1.710, P = 0.008). In addition, rs1130838 variant genotypes were significantly associated with an increased risk of HCV infection in the age ≥50 years (adjusted OR: 4.542, 95% CI: 1.492–13.830, P = 0.008), AST < 40 (adjusted OR: 2.397, 95% CI: 1.261–4.557, P = 0.008), and HD (adjusted OR: 4.027, 95% CI: 1.534–10.576, P = 0.005) subgroups (Table S4).
Combined Effects Analysis
The combined effects of KIR2DS4 rs35440472 and HLA-C rs1130838 on susceptibility to HCV infection were calculated by counting the number of their risk alleles (rs35440472-A and rs1130838-A) and risk genotypes (rs35440472-AA and rs1130838-AA) respectively. The risk for HCV infection increased with the presence of more unfavorable alleles (PTrend < 0.001), and carrying all four unfavorable alleles correlated to the highest risk (adjusted OR = 3.629, 95% CI = 1.174–11.213, P = 0.025) (Table 3). Similarly, the more unfavorable alleles the subjects carried, the more likely they were to be infected with HCV (PTrend<0.001). Compared with those carrying the rs35440472-GG/GA or rs1130838-GG/GA genotypes, subjects with rs35440472-AA or rs1130838-AA (one risk genotype: adjusted OR = 1.476, 95% CI = 1.120–1.945, P = 0.006; two risk genotypes: adjusted OR = 1.793, 95% CI = 1.278–2.516, P = 0.001) were more susceptible to HCV infection (Table 3).
Bioinformatics Analysis
Since rs35440472 is located in the overlapping region of KIR2DS4, KIR2DS1, and KIR2DL1, the bioinformatics analysis results can be applied to all three genes. Rs35440472 was predicted to be a transcription factor binding site (TFBS) by using the SNPinfo web server. The RNAfold web server showed the impact of this mutation on the mRNA secondary structure of gene. The local structural changes are shown in Figure 1, which indicate a difference in the lowest free energy between rs35440472-A and -G alleles (−10.70 vs. −11.70 kcal/mol). Furthermore, the eQTL analysis with the GTEx database showed that the expression of rs35440472-AA was lower than that of rs35440472-AG and GG in the liver tissue (P < 0.001) (Figure S2).
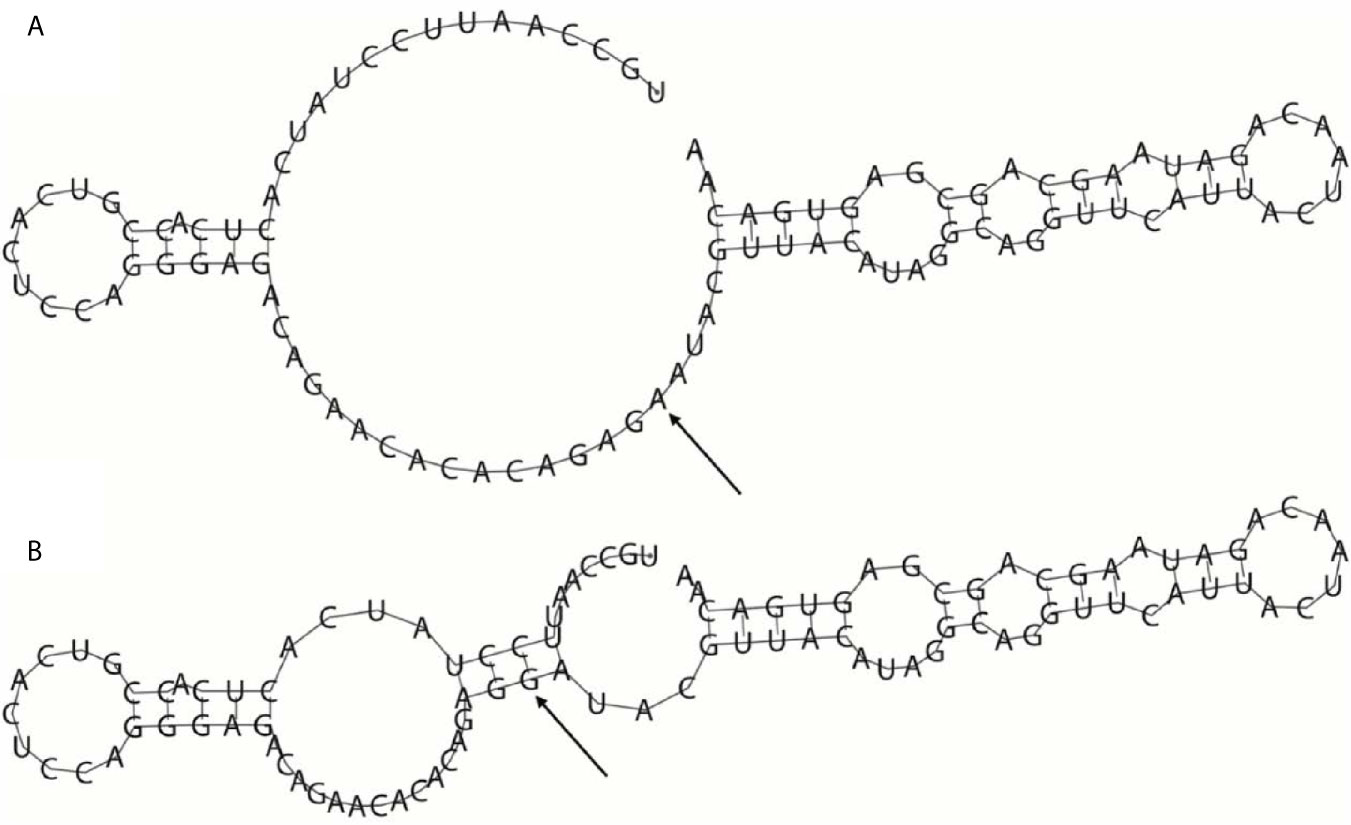
Figure 1 The influence of rs35440472 on KIR2DS4 mRNA optimal secondary structures. (A) The minimum free energy of the mRNA optimal secondary structure with a minimum free energy of −10.70 kcal/mol for rs35440472-A; (B) The minimum free energy of the mRNA optimal secondary structure with a minimum free energy of −11.70 kcal/mol for rs35440472-G. Changes in the local structure were illustrated by the RNAfold Web Server (Data available: http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi). The arrow marks the position of the mutation (50 bases upstream and 50 bases downstream from the mutation).
Based on the HaploReg database, rs1130838 is located in the functional regions of enhancer histone marks, DNase, and PLZF Motifs. In addition, there are 10 SNPs in linkage disequilibrium (r2 = 1.0) with rs1130838 in the Asian population (Table S5). According to the SNPinfo web server and UCSC, rs1130838 was predicted to have a splicing (ESE or ESS) function and located on the high peak of the histone H3 at lysine 4 (H3K4Me1) marker in seven cell lines as well as in the HepG2 liver cell line from Encyclopedia of DNA Elements (ENCODE) (Figure S3).
Discussion
This study systematically explored the association between SNPs in KIR/HLA-C genes and HCV infection. Our results indicate that the rs35440472-A and HLA-C rs1130838-A alleles are associated with a significantly higher probability of HCV infection in two high-risk Chinese populations, including drug users and hemodialysis patients. The unfavorable effects of both alleles were also confirmed by stratified analysis. The KIR/HLA-C genes may influence NK cell effector functions by regulating signal transduction via the activator and inhibitor receptors, which in turn affect the outcomes of HCV infection (41). The KIRs modulate NK cell stimulation through their signaling strength and specificity of interaction. Besides, strongly inhibitory KIR/HLA combinations display a higher threshold for cell activation than those with weak inhibitory action (12).
In this study, the selection of candidate genes is based on the importance of haplotypes of complex KIR/HLA genes and the gene frequency in the Chinese population. Therefore, the KIR2DS4 gene was chosen because it was the only active gene in haplotype A of the KIR gene cluster that encodes inhibitory receptors, and it is highly prevalent in the Chinese Han population. The frequency of KIR2DS4 in the Chinese, Japanese, Caucasian, and African American populations are 92.7, 87, 94.9, and 100%, respectively (25, 42–44). Consistent with this, the frequency of KIR2DS4 in this study was around 93%. Furthermore, KIR2DS4 binds with multiple HLA ligands encoded by the HLA-C1, HLA-C2, HLA-A*11:02 (45), and HLA-F (46). However, a recent study showed that KIR2DS4 exhibits peptide selective binding to HLA-C which might be significant for the NK cell response to human bacterial infections (32). Apart from bacterial infections, KIR2DS4 may be associated with the outcome of viral infections. It is associated with susceptibility to HIV and hepatitis B virus-associated hepatocellular carcinoma (33, 34, 47). Therefore, KIR2DS4 and HLA-C were selected as candidate genes, followed by the adoption of a screening strategy to select candidate SNP. In addition, this study is a part of our research series for KIR/HLA, and more KIR/HLA genes will be explored in further researches.
The rs35440472-A allele (chromosome position: 54846254) in particular was correlated to a higher susceptibility to HCV infection. Interestingly, the NCBI dbSNP shows that rs35440472 belongs to KIR2DS4, KIR2DS1, and KIR2DL1. All of these three genes encode receptors for HLA-C, which also justify this study on HLA-C as a ligand of rs35440472-possessing KIRs. The “multiple attribution” of rs35440472 is reasonable because this site is located in the overlapping region of three genes. It is due to the complex splicing mechanism of KIR gene cluster (48). A kind of splicing event defined as “cryptic exon” is involved. For simplicity, this site is called KIR2DS4 rs35440472 in most of the text. Rs35440472 doesn’t have a direct protein-coding function because it is located on these three genes’ intron region. Rs35440472 is a potential TFBS with homotypic clusters that are a key component of transcriptional promoters and enhancers (49) and is therefore biologically relevant on basis of the SNPinfo web server. Beyond that, according to predictions about this SNP location in the KIR2DS4/KIR2DS1/KIR2DL1 genes, the optimal mRNA secondary structure has changed and the minimum free energy for the A allele is higher than the G allele. The eQTL analysis further revealed that the genetic variation of rs35440472 affected the expression of the KIR2DS4/KIR2DS1/KIR2DL1 genes and the rs35440472-A allele reduced the gene expression. Given that KIR2DS4/KIR2DS1 are activating receptors of NK cells (45), rs35440472-A may result in lower activating signals due to decreased gene expression levels. However, not only other activating KIR, KIR2DS1, but also the inhibitory KIR2DL1 possess this SNP and that inhibitory KIRs exert stronger effect than activating ones. Thus, the variation of rs35440472 may affect NK cell effector functions, which ultimately influence the innate immune responses and susceptibility to HCV infection. However, no significant association was found between rs35440472 and the chronicity of HCV infection. These results are consistent with the findings of De Re et al. and Li et al., who also excluded the association between KIR2DS4 and chronic HCV infection in the Chinese Han or Italian populations, although the grouping was slightly different (50, 51).
The HLA-C rs1130838-A allele was also linked to an increased risk of HCV infection. Rs1130838 (chromosome position: 31269347) maps to the exon region of HLA-C and its allele changes create missense mutations resulting in residue changes (Thr > Pro, Thr > Ala or Thr > Ser). Studies show that missense mutations are often severely detrimental because they cause complete loss of function (52). Not surprisingly, variations in rs1130838 have been associated with psoriasis and Behcet’s disease (BD) (53). Additionally, rs1130838 has a splicing (ESE or ESS) function and is located on the high peak of the H3K4Me1 marker in different cell lines, including the hepatic carcinoma HepG2 cells. Recent studies show that H3K4me1 affects enhancers by promoting binding of the BAF complex and possibly other chromatin regulators (54, 55). The accumulating evidence implied that the genetic or structural disruption of enhancer function is a major cause of human diseases (56). Therefore, we inferred rs1130838 may affect the susceptibility to HVC by regulating the transcription and translation of HLA-C.
In the combined effects analyses of rs35440472 and HLA-C rs1130838, it is revealed that the more unfavorable alleles (rs35440472-A and rs1130838-A) or genotypes (rs35440472-AA and rs1130838-AA) patients carried, the more susceptible to HCV they were. The activating receptor KIR2DS4 recognizes unique, selective HLA-C molecules and six HLA-C allotypes (three carrying the C1 epitope and three carrying the C2 epitope) can reliably bind KIR2DS4 (45). The activating KIR2DS4/KIR2DS1 or the inhibitory KIR2DL1, which exerts stronger effect than activating ones, has to interact with HLA-C presenting on healthy or diseased cells to affect NK cell activity (19). As mentioned above, the risk alleles of rs35440472 and rs1130838 could affect the expression of three KIR genes as well as HLA-C, respectively. Taken together, the variation of these two SNPs not only may affect the activation of KIRs but also the KIR receptors binding with HLA-C ligands because of the incorrect expression of HLA-C gene, as the risk alleles increase. Hence, this may be the reason why the SNPs affect NK cell functions, and ultimately associated with the susceptibility to HCV infection. However, the explanations above were based on bioinformatics analysis and further studies are warranted to confirm the functional contribution of rs35440472-A and rs1130838-A in NK cell response to HCV infection.
In our research, some issues need to be mentioned. To minimize inherent selection bias, we adjusted for confounding factors such as gender, age, ALT, AST, and route of infection. Moreover, the IL28B-rs12979860 and IL28B-rs8099917 were also included because both are significantly associated with both spontaneous virus clearance and response to peg-IFN-α/RBV treatment (57–60). Therefore, the two SNPs were also included as the adjusted factors in logistic regression. However, all subjects came from China and more large survey samples of other nations and ethnic populations are needed to confirm the findings. Regrettably, this study couldn’t extract more data of some subjects including the duration of HD and the number of blood transfusions due to the privacy protection policy.
Nevertheless, there are several limitations in this study that ought to be considered. Firstly, there is a single 22 bp deletion in exon 5 of KIR2DS4, which can form a new truncated soluble KIR2DS4 protein. It is defined as KIR1D protein devoid of transmembrane region (45). The effect of KIR1D must be stronger than a presumable effect of SNPs increasing or decreasing receptor expression. And no linkage disequilibrium between the deletion mutation and KIR2DS4 rs35440472 was found. However, even if it is in LD in KIR2DS4 itself, it cannot generally display LD because it is present also in two other KIRs not possessing this deletion. Since the KIR2DS4-del variant may have influenced our findings, the association between KIR2DS4/KIR1D and the outcomes of HCV infection should be explored next. Secondly, KIR2DS4 may bind not only HLA-C, but also HLA-A*11 (45) and HLA-F (61). As HLA-F is only marginally polymorphic and has a limited tissue distribution, it should not play important role here. However, HLA-A is at least as polymorphic as HLA-C, and is generally regarded as more important in antigen recognition by T cells (although much less important for the NK cells). Therefore, the possible association between the genetic polymorphisms within KIR2DS4/HLA-A*11 and the outcomes of HCV infection should also be explored further. Finally, further genetic and functional studies are warranted to assure the mechanism of KIR/HLA-C genetic variation in the course of HCV infection.
Conclusion
In summary, our results revealed that polymorphisms within the KIR/HLA-C pathway genes are associated with HCV susceptibility in a high-risk Chinese population. KIR2DS4/KIR2DS1/KIR2DL1 rs35440472 and HLA-C rs1130838 might serve as potential biomarkers and own potential functions of the risk and progression of HCV infection.
Data Availability Statement
According to national legislation/guidelines, specifically the Administrative Regulations of the People’s Republic of China on Human Genetic Resources (http://www.gov.cn/zhengce/content/2019-06/10/content_5398829.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm, http://english.www.gov.cn/policies/latest_releases/2019/06/10/content_281476708945462.htm), no additional raw data is available at this time. Data of this project can be accessed after an approval application to the China National Genebank (CNGB, https://db.cngb.org/cnsa/). Please refer to https://db.cngb.org/, or email: Q05HQmRiQGNuZ2Iub3Jn for detailed application guidance. The accession code CNP0001926 should be included in the application.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Ethics Review Committee of Nanjing Medical University (Nanjing, China). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CS, MY, and ZG participated in the design of the study. CS, ZG, WC, JS, PH, and WC carried out the surveys and experiments. CS, HF, and CD performed the statistical analysis. YZ, JL, and MY contributed to analysis. CS, ZG, and MY wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Science Foundation for Distinguished Young Scholars of Jiangsu Province (BK20190106), the National Natural Science Foundation of China (81773499), Clinical Research Center for emerging respiratory infectious diseases (HS2020002), Key Project of Natural Science Foundation of Yunnan Province (2019FA005), Jiangsu Program for Young Medical Talents (QNRC2016616), and Natural Science Foundation of Jiangsu Province (No. BK20171054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.632353/full#supplementary-material
References
1. Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global Epidemiology and Genotype Distribution of the Hepatitis C Virus Infection. J Hepatol (2014) 61(1 Suppl):S45–57. doi: 10.1016/j.jhep.2014.07.027
2. Xiao J, Wang F, Wong NK, He J, Zhang R, Sun R, et al. Global Liver Disease Burdens and Research Trends: Analysis From a Chinese Perspective. J Hepatol (2019) 71(1):212–21. doi: 10.1016/j.jhep.2019.03.004
3. Lim SG, Aghemo A, Chen PJ, Dan YY, Gane E, Gani R, et al. Management of Hepatitis C Virus Infection in the Asia-Pacific Region: An Update. Lancet Gastroenterol Hepatol (2017) 2(1):52–62. doi: 10.1016/s2468-1253(16)30080-2
4. Awan AA, Jadoul M, Martin P. Hepatitis C in Chronic Kidney Disease: An Overview of the KDIGO Guideline. Clin gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc (2020) 18(10):2158–67. doi: 10.1016/j.cgh.2019.07.050
5. Bailey JR, Barnes E, Cox AL. Approaches, Progress, and Challenges to Hepatitis C Vaccine Development. Gastroenterology (2019) 156(2):418–30. doi: 10.1053/j.gastro.2018.08.060
6. Shi J, Li Y, Chang W, Zhang X, Wang FS. Current Progress in Host Innate and Adaptive Immunity Against Hepatitis C Virus Infection. Hepatol Int (2017) 11(4):374–83. doi: 10.1007/s12072-017-9805-2
7. Yoon JC, Yang CM, Song Y, Lee JM. Natural Killer Cells in Hepatitis C: Current Progress. World J gastroenterol (2016) 22(4):1449–60. doi: 10.3748/wjg.v22.i4.1449
8. Cho YJ, Lee HH, Kang H, Cho H. Cytokine-Modulated Natural Killer Cells Differentially Regulate the Activity of the Hepatitis C Virus. Int J Mol Sci (2018) 19(9):2771. doi: 10.3390/ijms19092771
9. Díaz-Peña R, Blanco-Gelaz MA, López-Larrea C. Kir Genes and Their Role in Spondyloarthropathies. Adv Exp Med Biol (2009) 649:286–99. doi: 10.1007/978-1-4419-0298-6_22
10. Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA Compound Genotypes Affect the Kinetics of Human Antiviral Natural Killer Cell Responses. J Clin Invest (2008) 118(3):1017–26. doi: 10.1172/jci32400
11. Martínez-Sánchez MV, Periago A, Legaz I, Gimeno L, Mrowiec A, Montes-Barqueros NR, et al. Overexpression of KIR Inhibitory Ligands (HLA-I) Determines That Immunosurveillance of Myeloma Depends on Diverse and Strong Nk Cell Licensing. Oncoimmunology (2016) 5(4):e1093721. doi: 10.1080/2162402x.2015.1093721
12. Béziat V, Hilton HG, Norman PJ, Traherne JA. Deciphering the Killer-Cell Immunoglobulin-Like Receptor System at Super-Resolution for Natural Killer and T-Cell Biology. Immunology (2017) 150(3):248–64. doi: 10.1111/imm.12684
13. Parham P, Guethlein LA. Genetics of Natural Killer Cells in Human Health, Disease, and Survival. Annu Rev Immunol (2018) 36:519–48. doi: 10.1146/annurev-immunol-042617-053149
14. Zeng J, Tang SY, Toh LL, Wang S. Generation of “Off-the-Shelf” Natural Killer Cells From Peripheral Blood Cell-Derived Induced Pluripotent Stem Cells. Stem Cell Rep (2017) 9(6):1796–812. doi: 10.1016/j.stemcr.2017.10.020
15. Thielens A, Vivier E, Romagné F. Nk Cell Mhc Class I Specific Receptors (Kir): From Biology to Clinical Intervention. Curr Opin Immunol (2012) 24(2):239–45. doi: 10.1016/j.coi.2012.01.001
16. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or Adaptive Immunity? The Example of Natural Killer Cells. Sci (New York NY) (2011) 331(6013):44–9. doi: 10.1126/science.1198687
17. Vivier E TE, Baratin M, Walzer T, Ugolini S. Functions of Natural Killer Cells. Nat Immunol (2008) 9):503–10. doi: 10.1038/ni1582
18. Peng H, Tian Z. Nk Cells in Liver Homeostasis and Viral Hepatitis. Sci China Life Sci (2018) 61(12):1477–85. doi: 10.1007/s11427-018-9407-2
19. Njiomegnie GF, Read SA, Fewings N, George J, McKay F, Ahlenstiel G. Immunomodulation of the Natural Killer Cell Phenotype and Response During HCV Infection. J Clin Med (2020) 9(4):1030. doi: 10.3390/jcm9041030
20. Hilton HG, Parham P. Missing or Altered Self: Human Nk Cell Receptors That Recognize HLA-C. Immunogenetics (2017) 69(8-9):567–79. doi: 10.1007/s00251-017-1001-y
21. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the Organization and Sequences of Human Kir/Ilt Gene Families. Proc Natl Acad Sci USA (2000) 97(9):4778–83. doi: 10.1073/pnas.080588597
22. Trowsdale J. Genetic and Functional Relationships Between MHC and NK Receptor Genes. Immunity (2001) 15(3):363–74. doi: 10.1016/s1074-7613(01)00197-2
23. Klein J, Sato A. The HLA System. First of Two Parts. N Engl J Med (2000) 343(10):702–9. doi: 10.1056/nejm200009073431006
24. Klein J, Sato A. The HLA System. Second of Two Parts. N Engl J Med (2000) 343(11):782–6. doi: 10.1056/nejm200009143431106
25. Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-Ligand Analyses Define Minimal Killer Cell Ig-Like Receptor (KIR) in Humans. Immunogenetics (2007) 59(1):1–15. doi: 10.1007/s00251-006-0168-4
26. Khakoo SI, Carrington M. KIR and Disease: A Model System or System of Models? Immunol Rev (2006) 214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x
27. Bao X, Hanson AL, Madeleine MM, Wang SS, Schwartz SM, Newell F, et al. HLA and KIR Associations of Cervical Neoplasia. J Infect Dis (2018) 218(12):2006–15. doi: 10.1093/infdis/jiy483
28. Moffett A, Chazara O, Colucci F, Johnson MH. Variation of Maternal KIR and Fetal HLA-C Genes in Reproductive Failure: Too Early for Clinical Intervention. Reprod biomed Online (2016) 33(6):763–9. doi: 10.1016/j.rbmo.2016.08.019
29. Stelma F, Jansen L, Sinnige MJ, van Dort KA, Takkenberg RB, Janssen HL, et al. HLA-C and KIR Combined Genotype as New Response Marker for HBeAg-Positive Chronic Hepatitis B Patients Treated With Interferon-Based Combination Therapy. J Viral Hepatitis (2016) 23(8):652–9. doi: 10.1111/jvh.12525
30. Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, O’Farrelly C, et al. Innate Immune Genes Synergize to Predict Increased Risk of Chronic Disease in Hepatitis C Virus Infection. Proc Natl Acad Sci USA (2011) 108(14):5736–41. doi: 10.1073/pnas.1016358108
31. Nozawa Y, Umemura T, Joshita S, Katsuyama Y, Shibata S, Kimura T, et al. Kir, HLA, and IL28B Variant Predict Response to Antiviral Therapy in Genotype 1 Chronic Hepatitis C Patients in Japan. PloS One (2013) 8(12):e83381. doi: 10.1371/journal.pone.0083381
32. Sim MJW, Rajagopalan S, Altmann DM, Boyton RJ, Sun PD, Long EO. Human NK Cell Receptor Kir2ds4 Detects a Conserved Bacterial Epitope Presented by HLA-C. Proc Natl Acad Sci USA (2019) 116(26):12964–73. doi: 10.1073/pnas.1903781116
33. Merino AM, Dugast AS, Wilson CM, Goepfert PA, Alter G, Kaslow RA, et al. Kir2ds4 Promotes Hiv-1 Pathogenesis: New Evidence From Analyses of Immunogenetic Data and Natural Killer Cell Function. PloS One (2014) 9(6):e99353. doi: 10.1371/journal.pone.0099353
34. Pan N, Jiang W, Sun H, Miao F, Qiu J, Jin H, et al. KIR and HLA Loci Are Associated With Hepatocellular Carcinoma Development in Patients With Hepatitis B Virus Infection: A Case-Control Study. PloS One (2011) 6(10):e25682. doi: 10.1371/journal.pone.0025682
35. VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A Single Polymorphism Disrupts the Killer Ig-Like Receptor 2dl2/2dl3 D1 Domain. J Immunol (Baltimore Md 1950) (2006) 177(8):5347–57. doi: 10.4049/jimmunol.177.8.5347
36. Mulrooney TJ, Hou L, Steiner NK, Chen M, Belle I, Ng J, et al. Promoter Variants of KIR2DL5 Add to Diversity and May Impact Gene Expression. Immunogenetics (2008) 60(6):287–94. doi: 10.1007/s00251-008-0273-7
37. Snyder GA, Brooks AG, Sun PD. Crystal Structure of the HLA-Cw3 Allotype-Specific Killer Cell Inhibitory Receptor Kir2dl2. Proc Natl Acad Sci USA (1999) 96(7):3864–9. doi: 10.1073/pnas.96.7.3864
38. Saulquin X, Gastinel LN, Vivier E. Crystal Structure of the Human Natural Killer Cell Activating Receptor KIR2DS2 (Cd158j). J Exp Med (2003) 197(7):933–8. doi: 10.1084/jem.20021624
39. World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. Jama (2013) 310(20):2191–4. doi: 10.1001/jama.2013.281053
40. Ranstam J. Multiple P-values and Bonferroni Correction. Osteoarthritis cartilage (2016) 24(5):763–4. doi: 10.1016/j.joca.2016.01.008
41. Cheent K, Khakoo SI. Natural Killer Cells: Integrating Diversity With Function. Immunology (2009) 126(4):449–57. doi: 10.1111/j.1365-2567.2009.03045.x
42. Bao X, Hou L, Sun A, Qiu Q, Yuan X, Chen M, et al. Distribution of Killer Cell Immunoglobulin-Like Receptor Genes and 2DS4 Alleles in the Chinese Han Population. Hum Immunol (2010) 71(3):289–92. doi: 10.1016/j.humimm.2009.12.004
43. Miyashita R, Tsuchiya N, Yabe T, Kobayashi S, Hashimoto H, Ozaki S, et al. Association of Killer Cell Immunoglobulin-Like Receptor Genotypes With Microscopic Polyangiitis. Arthritis Rheumat (2006) 54(3):992–7. doi: 10.1002/art.21653
44. Middleton D, Menchaca L, Rood H, Komerofsky R. New Allele Frequency Database: Http://Www.Allelefrequencies.Net. Tissue Antigens (2003) 61(5):403–7. doi: 10.1034/j.1399-0039.2003.00062.x
45. Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 Is a Product of Gene Conversion With Kir3dl2 That Introduced Specificity for HLA-A*11 While Diminishing Avidity for HLA-C. J Exp Med (2009) 206(11):2557–72. doi: 10.1084/jem.20091010
46. Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC Class I Open Conformers Are Ligands for NK Cell Ig-Like Receptors. J Immunol (Baltimore Md 1950) (2013) 191(7):3553–62. doi: 10.4049/jimmunol.1300081
47. Olvera A, Pérez-Álvarez S, Ibarrondo J, Ganoza C, Lama JR, Lucchetti A, et al. The HLA-C*04: 01/Kir2ds4 Gene Combination and Human Leukocyte Antigen Alleles With High Population Frequency Drive Rate of HIV Disease Progression. AIDS (London England) (2015) 29(5):507–17. doi: 10.1097/qad.0000000000000574
48. Bruijnesteijn J, van der Wiel MKH, de Groot N, Otting N, de Vos-Rouweler AJM, Lardy NM, et al. Extensive Alternative Splicing of KIR Transcripts. Front Immunol (2018) 9:2846. doi: 10.3389/fimmu.2018.02846
49. Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic Clusters of Transcription Factor Binding Sites Are a Key Component of Human Promoters and Enhancers. Genome Res (2010) 20(5):565–77. doi: 10.1101/gr.104471.109
50. De Re V, Caggiari L, De Zorzi M, Repetto O, Zignego AL, Izzo F, et al. Genetic Diversity of the KIR/HLA System and Susceptibility to Hepatitis C Virus-Related Diseases. PloS One (2015) 10(2):e0117420. doi: 10.1371/journal.pone.0117420
51. Li Q, Liu S, Zhang S, Liu C, Sun M, Li C, et al. Human Leucocyte Antigen But Not KIR Alleles and Haplotypes Associated With Chronic HCV Infection in a Chinese Han Population. Int J Immunogenet (2019) 46(4):263–73. doi: 10.1111/iji.12425
52. Stein A, Fowler DM, Hartmann-Petersen R, Lindorff-Larsen K. Biophysical and Mechanistic Models for Disease-Causing Protein Variants. Trends Biochem Sci (2019) 44(7):575–88. doi: 10.1016/j.tibs.2019.01.003
53. Lee YH, Choi SJ, Ji JD, Song GG. Genome-Wide Pathway Analysis of a Genome-Wide Association Study on Psoriasis and Behcet’s Disease. Mol Biol Rep (2012) 39(5):5953–9. doi: 10.1007/s11033-011-1407-9
54. Local A, Huang H, Albuquerque CP, Singh N, Lee AY, Wang W, et al. Identification of H3K4me1-Associated Proteins at Mammalian Enhancers. Nat Genet (2018) 50(1):73–82. doi: 10.1038/s41588-017-0015-6
55. Rada-Iglesias A. Is H3K4me1 at Enhancers Correlative or Causative? Nat Genet (2018) 50(1):4–5. doi: 10.1038/s41588-017-0018-3
56. Smith E, Shilatifard A. Enhancer Biology and Enhanceropathies. Nat Struct Mol Biol (2014) 21(3):210–9. doi: 10.1038/nsmb.2784
57. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, et al. Genetic Variation in IL28B and Spontaneous Clearance of Hepatitis C Virus. Nature (2009) 461(7265):798–801. doi: 10.1038/nature08463
58. Montes-Cano MA, García-Lozano JR, Abad-Molina C, Romero-Gómez M, Barroso N, Aguilar-Reina J, et al. Interleukin-28B Genetic Variants and Hepatitis Virus Infection by Different Viral Genotypes. Hepatol (Baltimore Md) (2010) 52(1):33–7. doi: 10.1002/hep.23624
59. Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, et al. Genetic Variation in IL28B Is Associated With Chronic Hepatitis C and Treatment Failure: A Genome-Wide Association Study. Gastroenterology (2010) 138(4):1338–45.e1-7. doi: 10.1053/j.gastro.2009.12.056
60. Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV–response to Infection and Treatment. Nat Rev Gastroenterol Hepatol (2012) 9(7):406–17. doi: 10.1038/nrgastro.2012.101
Keywords: hepatitis C virus, killer cell immunoglobulin-like receptors, polymorphism, human leukocyte antigen, infection
Citation: Shen C, Ge Z, Dong C, Wang C, Shao J, Cai W, Huang P, Fan H, Li J, Zhang Y and Yue M (2021) Genetic Variants in KIR/HLA-C Genes Are Associated With the Susceptibility to HCV Infection in a High-Risk Chinese Population. Front. Immunol. 12:632353. doi: 10.3389/fimmu.2021.632353
Received: 25 November 2020; Accepted: 31 May 2021;
Published: 18 June 2021.
Edited by:
Pascal Pineau, Institut Pasteur, FranceReviewed by:
Nadia Bouzgarrou-Debbabi, Higher Institute of Biotechnology of Monastir, TunisiaPiotr Kusnierczyk, Polish Academy of Sciences, Poland
Copyright © 2021 Shen, Ge, Dong, Wang, Shao, Cai, Huang, Fan, Li, Zhang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Yue, eXVlbWluZ0Buam11LmVkdS5jbg==
†These authors have contributed equally to this work
 Chao Shen1†
Chao Shen1† Peng Huang
Peng Huang Haozhi Fan
Haozhi Fan Jun Li
Jun Li Ming Yue
Ming Yue