- 1State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China
- 2Biological Science Research Center, Southwest University, Chongqing, China
- 3Department of Biological Sciences, University of Rhode Island, Kingston, RI, United States
Insects are the largest group of animals. Nearly all organisms, including insects, have viral pathogens. An important domesticated economic insect is the silkworm moth Bombyx mori. B. mori nucleopolyhedrovirus (BmNPV) is a typical baculovirus and a primary silkworm pathogen. It causes major economic losses in sericulture. Baculoviruses are used in biological pest control and as a bioreactor. Silkworm and baculovirus comprise a well-established model of insect–virus interactions. Several recent studies have focused on this model and provided novel insights into viral infections and host defense. Here, we focus on baculovirus invasion, silkworm immune response, baculovirus evasion of host immunity, and enhancement of antiviral efficacy. We also discuss major issues remaining and future directions of research on silkworm antiviral immunity. Elucidation of the interaction between silkworm and baculovirus furnishes a theoretical basis for targeted pest control, enhanced pathogen resistance in economically important insects, and bioreactor improvement.
Introduction
Insects are globally distributed and play vital roles in the biosphere. Lepidoptera is a major insect taxon with an estimated 150,000 to 180,000 described species (1, 2). Many lepidopterans are pests that adversely affect agricultural production. However, the silkworm moth Bombyx mori, the only fully domesticated insect, is an economically important lepidopteran used for silk production in many developing countries (3, 4). China is the largest producer of silkworm cocoons, with an annual value for the output of the silk industry of about 200 billion Yuan (about 30 billion USD) (3). Pathogenic viruses are severe threats to all organisms and silkworm viruses cause losses of almost 16% of potential cocoon production each year. Bombyx mori nucleopolyhedrovirus (BmNPV) is a primary silkworm pathogen. This typical baculovirus causes major economic losses in sericulture (3). Baculovirus is also used as a biological control agent against insect pests and as a bioreactor. The Silkworm Genome Project was completed >10 years ago (5–8) and promoted B. mori to model insect status in basic and applied research (9). Here, we present a broad overview of silkworm–baculovirus interactions. We also discuss the major challenges and future directions of research in silkworm antiviral immunity.
Baculovirus Host Invasion Mechanism
Baculovirus consists of a circular double-stranded DNA genome that combines with capsid proteins to form an enveloped nucleocapsid (3, 10). Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is a close relative of BmNPV and the most well studied baculovirus (11, 12). Both NPVs are models for basic molecular research which have been used to elucidate the baculovirus infection cycle. The baculovirus replication cycle includes two virion phenotypes, an occlusion-derived virus (ODV) and a budded virus (BV). ODVs are packaged in occlusion bodies and induce host infection, whereas BVs spread throughout the host after infection (12, 13). ODVs and BVs have the same nucleocapsids but different envelopes. BVs mature early during infection and acquire their envelopes from modified host cell membranes. In contrast, ODVs mature late in infection and form their envelopes within host nuclei (14, 15). BVs and ODVs interact differently with host cells: ODVs fuse with the midgut epithelial cell membrane, whereas BVs are internalized by adsorptive endocytosis (15).
Baculovirus occurs in the environment in the form of occlusion bodies. For infection, it invades insect larvae mainly by ingestion (3). ODVs are released after these occlusion bodies dissociate in the alkaline environment of larval gut juice. They pass through the peritrophic membrane, invade the midgut, and cause primary infection (Figure 1) (11–13). Several envelope proteins known as per os infectivity factors (PIFs) are unique to ODVs. They mediate specific ODV binding to midgut columnar epithelial cells and initiate oral infection by binding to receptors (16–19), after which nucleocapsids enter the epithelial cells via envelope-mediated membrane fusion (3, 11). Viral DNA is then released from the nucleocapsids and used as a template to generate new DNA and mRNA (3, 11).
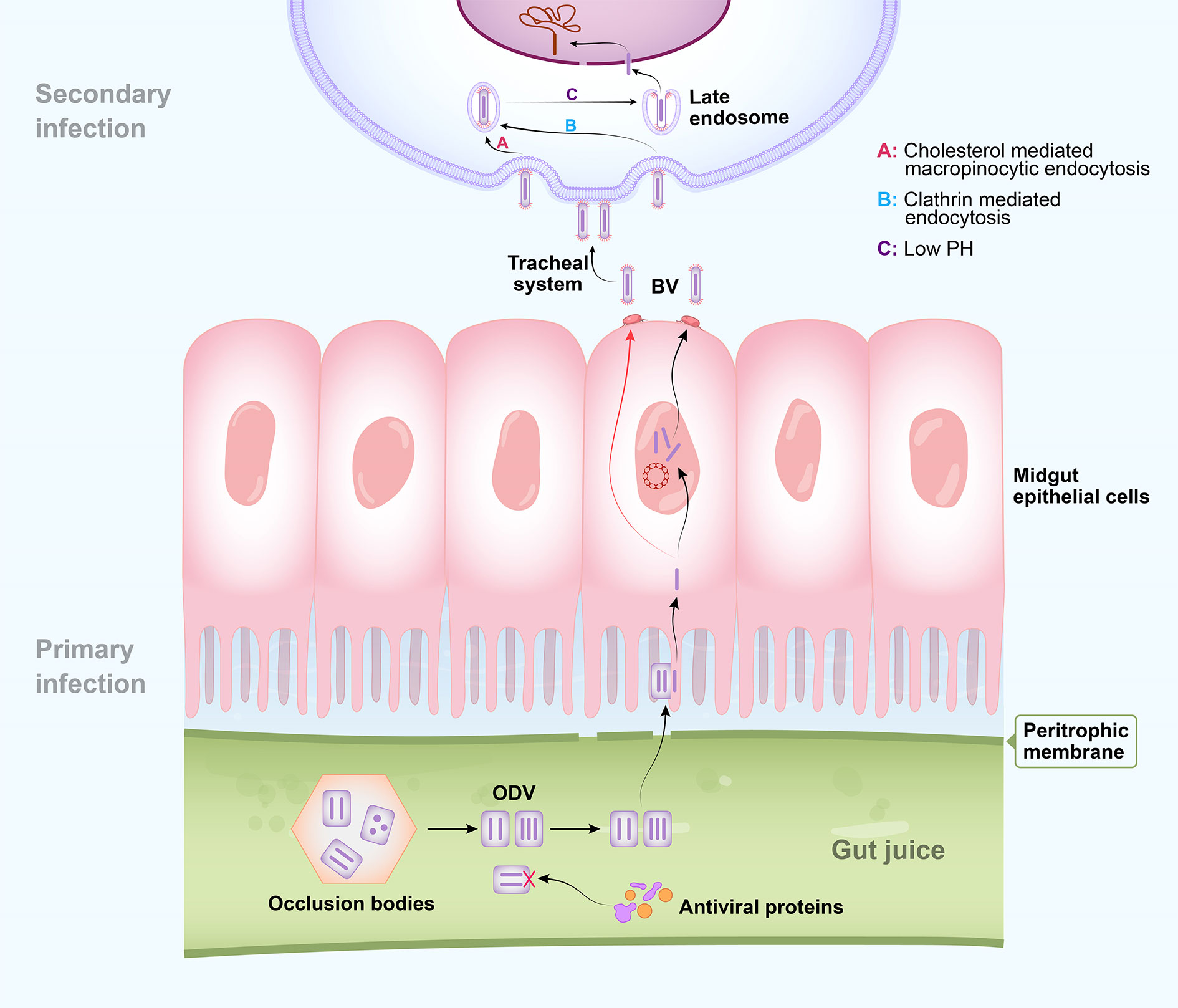
Figure 1 Schematic diagram of baculovirus entry. Occlusion-derived viruses (ODVs) are released from occlusion bodies in the alkaline environment of larval gut juice after ingestion. Several insect gut juice proteins have strong antiviral capacity against ODVs. Intact ODVs pass through the peritrophic membrane and nucleocapsids enter the midgut epithelial cells via envelope-mediated membrane fusion to cause primary infection. Progeny budded viruses (BVs) spread through the host via the tracheal system to cause secondary infection. Binding and penetration into host cells by BV of both Bombyx mori nucleopolyhedrovirus (BmNPV) and Autographa californica multiple nucleopolyhedrovirus (AcMNPV) are mediated by the GP64 envelope glycoprotein which is specific to BV. BmNPV BV penetrates nonmidgut host cells by multiple strategies, including clathrin-independent macropinocytic endocytosis mediated by cholesterol on the cell membrane and clathrin- and dynamin-dependent endocytosis pathways. Successful BV entry also requires low pH. Nucleocapsid uncoating in the nucleus results in the subsequent virus infection process.
Baculoviral gene expression occurs in four phases: immediate early, delayed early, late, and very late. In an infected cell, viral DNA replication starts at 8 h post infection (hpi) and represents the transition from the early stage to the late stage (20, 21). During early infection, host RNA polymerase transcribes the viral DNA and produces the elements required for its replication (15). Viral DNA replication and transcription then form nucleocapsid progeny that acquire envelopes by budding from the host cell membrane. The latter is modified mainly by virally encoded fusion protein GP64 to generate a BV, which causes systemic infection via the host tracheal system (13, 22, 23). At the latter infection stages, progeny ODVs acquire envelopes in the nucleus, possibly derived from nuclear membranes modified by several viral proteins (24), are subsequently assembled into occlusion bodies and released into the environment after host disintegration (11, 22).
BmNPV BV utilizes multiple strategies to invade host cells (Figure 1). Binding and penetration into host cells by BV of both BmNPV and AcMNPV are mediated by the GP64 envelope glycoprotein which is specific to BV (12, 25, 26). GP64 contains a cholesterol recognition amino acid consensus (CRAC) domain which is known to be essential for fusion between the BV envelope and mammalian cell membrane (26, 27). The BmN and BmE cell lines are derived from the ovary and embryonic cells of silkworm, respectively. Various endocytic inhibitor assays disclosed that BmNPV BV penetrates BmN cells by clathrin-independent macropinocytic endocytosis mediated by cholesterol on the cell membrane (28). The cholesterol transporter BmNPC1 interacts with GP64. Its deficiency inhibits viral penetration rather than viral binding to BmE cells (29). In contrast, BmNPV BV uses clathrin- and dynamin-dependent endocytosis pathways to penetrate BmN cells. Successful BV entry also requires low pH (25). A number of studies were performed to identify the host receptor of GP64 (12). The membrane protein BmREEPa is not a direct NPV receptor but interacts with GP64 and may participate in BV attachment or binding (30). Yeast two-hybrid and coimmunoprecipitation (Co-IP) assays demonstrated that the silkworm protein SINAL10 binds GP64, is concentrated near the cell membrane, and stimulates BmNPV proliferation in BmN cells (14). Nevertheless, to date, unequivocal identification of a receptor for GP64 remains elusive (12).
Baculovirus encodes some auxiliary genes to enhance its infection in insect larvae, including viral fibroblast growth factor (vfgf), ecdysteroid (UDP)-glucosyltransferase (egt), and p35 (31). Horizontal gene transfer (HGT) between host and pathogen might augment pathogen survival and propagation. Several BmNPV auxiliary genes were acquired from the silkworm genome via HGT. These include egt, vfgf, and protein tyrosine phosphatase (ptp) (32). BmNPV PTP is a virus-associated structural protein which might have originated from insect ptp-h (32). Deleting it reduces production of progeny in larval silkworm hosts; moreover, the mutation can be rescued by inserting Bmptp-h into BmNPV ptp-deleted virus (33), and overexpression of Bmptp-h accelerates BmNPV multiplication in BmE host cells (34). Other experiments involving deletion and insertion of ptp and egt (34, 35) showed that HGT-derived genes are dispensable for virus production in certain cell lines but affect progeny contents and may control host physiology.
Silkworm Immune Response to Baculovirus
Innate immune responses in insects control and clear pathogens following infection (36, 37). Lepidopteran insects have several antiviral immune responses which they use against baculovirus infections. These include global protein synthesis shutdown, rRNA degradation, inactivation by gut juice antiviral proteins, melanization, apoptosis, RNAi-based antiviral response, and host gene-encoded resistance (Figure 2) (3, 36–39). Among these immune responses, there are relatively few studies on the mechanisms of the first two processes. After AcMNPV infection of B. mori cells, rRNA degradation is triggered by six amino acid residues (positions 514 and 599) of viral protein P143 as a primary antiviral response. Global protein synthesis shutdown then follows viral DNA replication, resulting in abortive infection (38, 40). The latter processes are more clearly delineated, and each process is described in turn here.
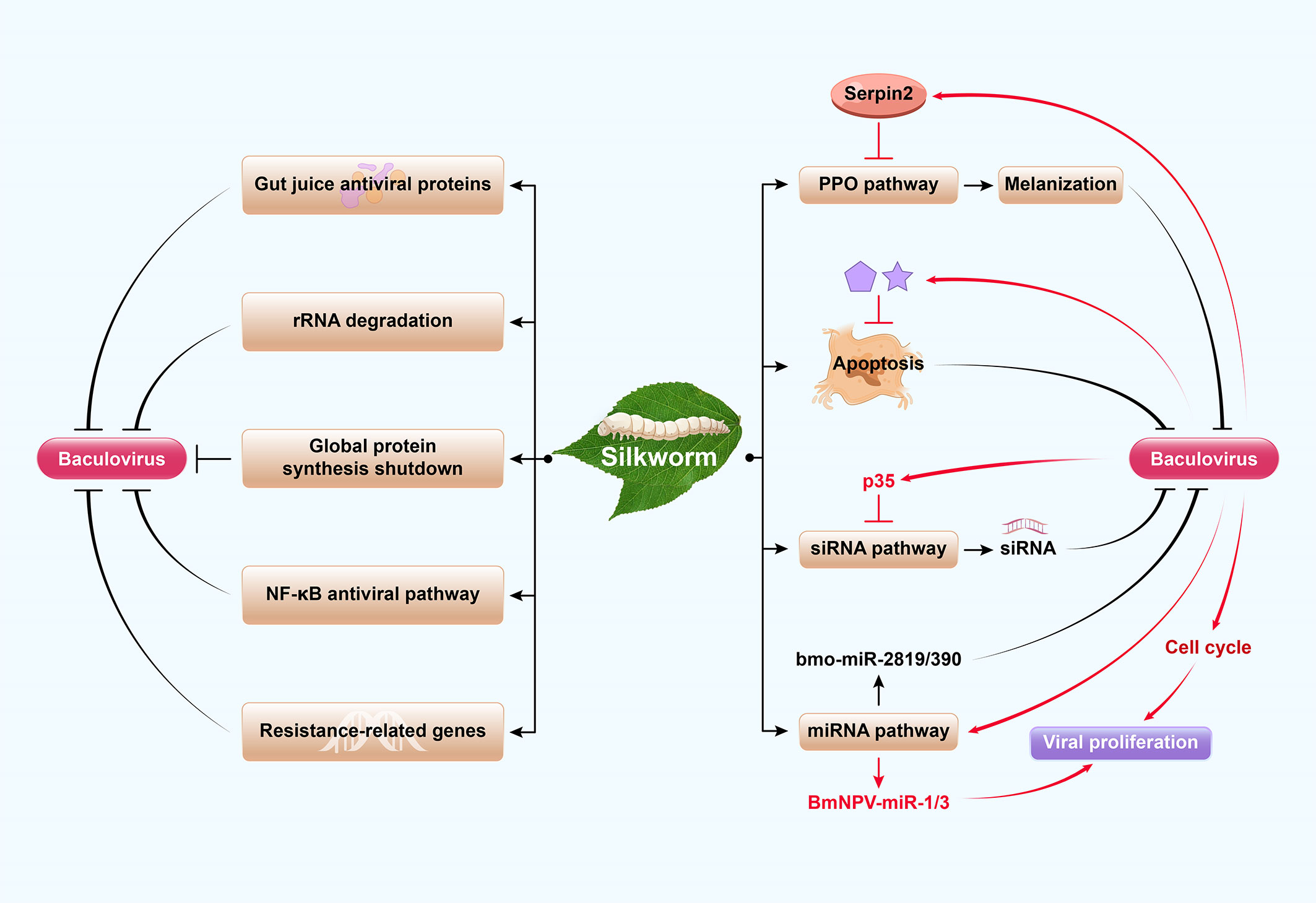
Figure 2 Model of the arms race of silkworm and baculovirus. Silkworms have several antiviral immune responses which they use against baculovirus infections. These include global protein synthesis shutdown, rRNA degradation, inactivation by gut juice antiviral proteins, host gene-encoded resistance, NF-κB antiviral pathway, apoptosis, melanization, and RNAi-based antiviral response. The prophenoloxidase (PPO) activation cascade causes melanization to block baculovirus infection, which is negatively regulated by serpins. RNAi antiviral defense of insects includes the major mechanism of the siRNA pathway and the minor contribution of the miRNA pathway. The silkworm-encoded miRNA bmo-miR-2819 and bmo-miRNA-390 inhibit BmNPV proliferation by downregulating viral genes. As a confrontation, baculovirus have developed several strategies to escape host immunity and promote their own replication and proliferation, including inhibition of antiviral apoptosis, melanization, RNAi and regulation of the cell cycle. For example, Bombyx mori nucleopolyhedrovirus (BmNPV) induces Bmserpin2 to inhibit host melanization. Meanwhile, Autographa californica multiple nucleopolyhedrovirus (AcMNPV) p35 inhibits siRNA pathway. Additionally, baculoviruses exploit the miRNA pathway to encode their own miRNAs (such as BmNPV-miR-1 and BmNPV-miR-3) for viral propagation.
The insect midgut is the first tissue to be infected after baculovirus ingestion. Hence, it is an important immune organ which acts as a first line of defense against pathogens (41, 42). Several insect gut juice proteins secreted from the midgut have strong antiviral capacity. The antiviral proteins Bmlipase-1 (43), BmSP-2 (44), BmNOX (45), red fluorescent proteins (RFPs) (46), Bmtryp (47), and BmLHA (48) have been isolated from silkworm larva gut juice, which inhibit BmNPV at an initial infection stage. The activation of energy synthesis by adenosine signaling following baculovirus infection is a physiological response in the silkworm that supports its innate immunity (49). Melanization is a prominent humoral response in insects. It consists of a cascade of clip-domain serine proteases (cSPs) that converts zymogen prophenoloxidase (PPO) into active phenoloxidase (PO), which is negatively regulated by serpins. PO catalyzes melanin formation to encapsulate and kill invading pathogens (50, 51). Baculovirus infection is efficiently blocked by the PPO activation cascade (50). Bmserpin2 knockdown increases PO activity and decreases viral DNA content in silkworm haemolymph infection with BmNPV (52). The stage of infection at which melanization inhibits baculovirus infection needs further exploration.
Apoptosis is a genetically controlled process that removes unwanted or damaged cells. It serves as an important antiviral defense mechanism in insects (15, 37, 53–55). The apoptotic caspase cascade comprises upstream initiator caspases (ICs) and downstream effector caspases (ECs) (15, 53) (Figure 3). To initiate apoptosis, ECs are activated by ICs, and then cleave other signaling proteins (56). In lepidopterans, caspase-1, caspase-2, and caspase-3 are ECs while caspase-5 (Dronc) and caspase-6 (Dredd) are ICs (57). A cellular inhibitor of apoptosis (IAP) binds caspases, blocks their function, and prevents apoptosis activation in normal cells (15, 58). In BmN cells, B. mori iap1 (BmIAP1) interacts with BmDronc and Bmcaspase-1 and downregulates apoptosis (58). Apoptotic signaling, which is initiated upon baculovirus infection, promotes iap-antagonist (iap-A) binding to cellular IAP and releases free caspases to facilitate apoptosis (15, 53). The host p53 protein is pro-apoptotic and triggers antiviral apoptosis upon viral DNA replication. It elevates caspase-3-like protease activity and enhances BmDronc processing in BmN cells after BmNPV infection (53) (Figure 3). Nevertheless, a DNA damage response, which is elicited upon viral DNA replication, depletes cellular IAP protein, activates apoptosis, and promotes baculovirus multiplication in infected cells (59–61). Although apoptotic pathways and their associated viral and cellular factors play important roles in regulating the outcome of baculovirus infection in insect cells, their mechanisms and interactions are complex and remain to be fully elucidated.
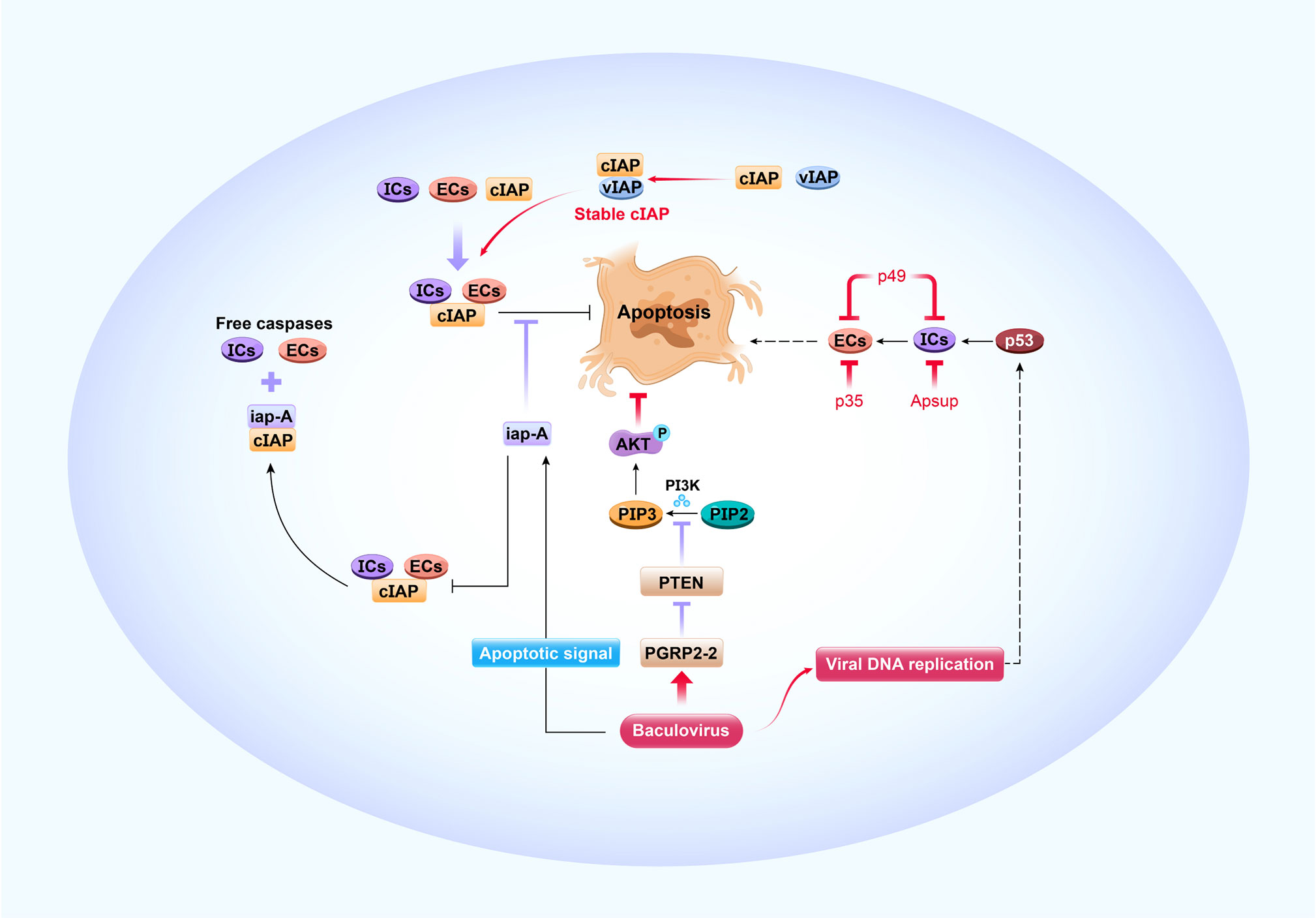
Figure 3 Antiviral apoptosis and its modification by baculoviruses. The apoptotic caspase cascade comprises upstream initiator caspases (ICs) and downstream effector caspases (ECs). The cellular inhibitor of apoptosis (cIAP) binds caspases and blocks apoptosis in normal cells. Apoptotic signaling is initiated upon baculovirus infection, which causes iap-Antagonist (iap-A) to bind cIAP and release free caspases that facilitate apoptosis. Viral DNA replication triggers host p53 pro-apoptosis, which accelerates IC and EC activity. Progression of antiviral apoptotic signaling cascades is prevented by baculovirus-encoded apoptosis suppressors such as viral IAP (vIAP), p35, p49, and Apsup. When the apoptotic signal is initiated, vIAP blocks apoptosis by interacting with unstable cIAP such that the cIAP levels and antiapoptotic activity are maintained. Viral p35 binds ECs and p49 binds ICs and ECs to block apoptosis. Apsup inhibits apoptosis by preventing IC activity. BmNPV induces the pattern recognition receptor protein PGRP2-2 to suppress PTEN and prevent it from inhibiting PI3K/Akt signaling and activating p-Akt. In this manner, cell apoptosis is inhibited. The resultant increase in cell survival is conducive to NPV proliferation.
RNA interference (RNAi) is an ancient post-transcriptional antiviral regulatory process in insects (36, 62) whereby the host RNAi response degrades baculovirus transcripts (63). In this process, viral infections generate dsRNAs that trigger the RNAi machinery and process them into viral short interfering RNAs (vsiRNAs) that target viral RNA sequences and inhibit viral proliferation (64). Another RNAi response involves the microRNA (miRNA) pathway in which precursor miRNA (pre-miRNA) is cleaved into mature miRNA that regulates gene expression by targeting specific mRNAs (65). Cellular miRNAs also affect viral infections and play important roles in host–pathogen interactions. The silkworm-encoded miRNA bmo-miR-2819 is upregulated at the delayed early stage in infection, and its overexpression inhibits BmNPV proliferation by downregulating viral ie-1 (66). Similarly, bmo-miRNA-390 downregulates the expression of BmNPV-cg30 (67). The PIWI-associated RNA (piRNA) pathway is also involved in an antiviral response but little information is reported in silkworm (68). Results from published reports reveal that the siRNA pathway is the major mechanism, whereas the contribution of the miRNA pathway is minor in RNAi antiviral defense of insects (Figure 2).
Innate immune signaling pathways and resistance-related genes play an important role in antiviral defense. The Imd and Toll signaling pathways participate in the antiviral immune response (36, 54) but do not seem to play roles in the silkworm BmNPV response. BmNPV infection induces cGAMP production in BmE cells and BmSTING responds to the cGAMP and activated Dredd caspase-mediated NF-κB antiviral signaling pathways (69). Antimicrobial peptides (AMPs), humoral immunity, and reactive oxygen species (ROS) may also be involved in the antiviral response (45, 70). Dozens of candidate genes regulating the silkworm immune response to baculovirus have been screened via multi-omics using various resistant hosts. However, the functions of only a few of them are verified in cells or individuals. For example, the BmLHA level in the digestive juice of resistant silkworm strains is relatively higher than that of susceptible silkworms, and recombinant BmLHA inhibits BmNPV proliferation in silkworm larvae (48). Similarly, BmAtlastin-n is highly expressed in resistant BmE-SWU2 cells but not in BmE-SWU1 cells susceptible to BmNPV, and BmAtlastin-n overexpression inhibits BmNPV reproduction in BmE-SWU1 cells and transgenic silkworms (71). Additionally, B. mori heat shock protein 19.9 (Bmhsp19.9) is upregulated at the late stage after BmNPV challenge in BmE cells and silkworms, and its overexpression markedly inhibits BmNPV proliferation in the hosts (72). Finally, overexpression of lysozyme BmC-LZM, which is upregulated at the very late stage of BmNPV infection in BmE cells, inhibits BmNPV virus in BmE cells but does not decrease mortality in silkworm larvae (73). The anti-BmNPV mechanisms of the aforementioned resistance-related genes are unclear and merit further investigation.
Viral Immune Evasion Mechanism
Viruses have developed several strategies to escape host immunity and promote their own replication and proliferation, including inhibition of antiviral melanization, autophagy, apoptosis, RNAi and regulation of the cell cycle (Figure 2). Baculoviruses can suppress host melanization so that they can proliferate. Several SPs (serine proteases) and their homologs are upregulated in response to bacterial or fungal challenge but downregulated in response to baculovirus infection (50, 51). For example, when serpins 5 and 9 are induced by HearNPV in Helicoverpa armigera, they inhibit SPs and melanization and promote viral infection (51). Similarly, Bmserpin2 is upregulated and PO activity is diminished in haemolymph following BmNPV infection in silkworm. Hence, BmNPV inhibits host melanization by regulating Bmserpin2 expression (Figure 2) (52). Additionally, several potential resistance-related genes such as BmPP2A (74) and BmPEPCK-2 (75) are downregulated by BmNPV to allow robust viral proliferation.
Autophagy is a catabolic biological process in the body, which has antiviral efficacy by targeting viruses and sending them to the lysosome for phagocytosis and degradation. At the same time, viruses can also use autophagy to enhance their own replication (76). However, little is known about the association between BmNPV and autophagy. Atg 6, Atg 7, Atg 8, and Atg 13, proteins involved in various stages of autophagy, are all upregulated in BmN-SWU1 cells (77) but downregulated in BmE cells (75) following BmNPV infection, possibly because of the relative differences among cell lines and internal reference genes used in these experiments. Understanding the roles and mechanisms of such immuno-suppressive processes during BmNPV infection is clearly important for future applications to enhance their impact (for pests) or protecting their hosts (for beneficials) and merit further examination.
Baculoviruses can inhibit host antiviral apoptosis through a variety of strategies (Figure 3). The progression of apoptotic signaling cascades is prevented by virus-encoded apoptosis suppressors such as viral IAPs, p35, p49, and Apsup (55, 78, 79). Six IAPs (iap1-6) have been identified in baculoviruses that inhibit apoptosis in insects (78–80). Unlike their cellular counterparts, they lack an N-terminal instability motif (81) and stabilize cellular IAPs (82). In a model mechanism, Op-IAP3 derived from OpMNPV blocks apoptosis by interacting with an unstable auto-ubiquitinating host IAP such that cellular IAP levels and antiapoptotic activity are maintained (82). Similarly, IAP1 and IAP2 from BmNPV interact with BmIAP, and both BmIAP and viral IAPs increase BmNPV proliferation in infected silkworm cells (80). Numerous studies have shown that viral protein p35 blocks apoptosis by binding ECs (79, 83, 84), and p49 protein binds ICs and ECs and blocks apoptosis (85, 86). Additionally, Apsup from LdMNPV inhibits apoptosis by preventing proteolytic Dronc (IC) processing (87). Recently, our research demonstrated that peptidoglycan recognition protein (PGRP) is regulated by virus to inhibit host antiviral apoptosis, which is well known to recognize invading bacteria and fungi to activate host immune defenses (54). For example, BmNPV induces BmPGRP2-2 to suppress PTEN and the inhibition of PI3K/Akt signaling, increase p-Akt production and activation, and inhibit cell apoptosis (54). Clearly, enhanced host cell survival is beneficial for viral proliferation (Figure 3).
Viruses have evolved strategies to circumvent host antiviral RNAi (siRNA and miRNA pathways). Almost all plant viruses and some insect viruses encode viral suppressors of RNAi (VSRs) to counteract the host siRNA pathway and inhibit vsiRNA production (88, 89). AcMNPV p35 is responsible for the suppression of RNAi in various insect cells; its VSR activity acts downstream in the RNAi pathway and is not associated with its antiapoptotic activity (89). The identification of BmNPV VSRs and clarification of their modes of action require further research. On the other hand, it is evident that baculoviruses exploit the miRNA pathway for their own propagation, suppress cellular miRNAs after infection, encode their own miRNAs, and disrupt host defense mechanisms that interfere with viral propagation (90–92). For example, BmNPV-miR-1 suppresses host miRNA biogenesis by regulating the exportin-5 cofactor Ran and enhancing viral multiplication (92). Simultaneously, BmNPV-miR-3 facilitates viral infection by modulating the expression of P6.9 and other late BmNPV genes (91) (Figure 2). Several miRNAs have been predicted in the BmNPV genome; however, only four miRNAs (BmNPV-miR-1, BmNPV-miR-2, BmNPV-miR-3, and BmNPV-miR-4) have been empirically identified (90) and biological functions of only two miRNAs have been uncovered thus far. Deciphering viral miRNA targets and functions remains a challenging task.
Virus regulation of the host cell cycle might be an important immune evasion strategy and could promote its proliferation. The normal insect cell life cycle is characterized by a complex series of events ranging from cell growth to replication, but this process is disrupted during infection (15). Baculovirus infection arrests the cell cycle at S or G2/M. The AcMNPV protein EC27 arrests the host cell cycle in the G2/M phase, and this arrest enables ODV maturation (93, 94). ERK regulates cell proliferation, differentiation, and apoptosis and is conserved among different species (95). The ERK signaling pathway is activated during the late phase of BmNPV infection via the B. mori epidermal growth factor receptor (BmEGFR). The latter inhibits cell proliferation and increases viral replication by increasing the G2/M phases of the cell cycle (96). BmSpry is a negative feedback regulator of the BmEGFR-ERK cascade; its inhibitory activity is upstream of ERK. It is downregulated by BmNPV to elevate ERK phosphorylation (p-ERK), thereby enhancing viral reproduction (95, 97). The modification mechanisms of cell cycle phases during baculovirus infection are only partially elucidated and need more experimentation.
Enhancement of Host Antiviral Capacity
No fundamental strategies have been established to cope with BmNPV during sericulture; instead, this industry mainly relies on thorough disinfection and strict breeding operation techniques to prevent virus infectivity. Breeding resistant host insect strains would help contend with baculovirus infection in sericulture (3, 98, 99). However, enhancing pathogen resistance in the host is usually accomplished at the expense of economically important traits, which is a major constraint in traditional silkworm breeding methods. This compromise may be avoided by applying transgenic and gene editing techniques (3). The antiviral capacity of transgenic silkworms could be enhanced using strategies based on the BmNPV infection process such as inhibiting BmNPV at the initial infection stage via Bmlipase-1 overexpression (100), targeting BmNPV mRNA with RNAi (21), inhibiting BmNPV protein synthesis by hycu-ep32 overexpression (101), and suppressing BmNPV by regulating the host immune pathway (54). Antiviral capacity could be further increased by optimizing and integrating the aforementioned anti-BmNPV strategies (41, 42, 102). Transgenic CRISPR/Cas9 system-mediated mutagenesis randomly targeting and inactivating the viral genome has been studied as a potential approach against BmNPV infection in silkworm (103). Theoretically the inhibitory effect of the CRISPR/Cas9 system (knock out) on the virus should be higher than that of the RNAi system (knock down) when targeting the same viral genes. However, silkworms with inserted DNA fragments expressing dsRNA (21) or gRNA (103) are all transgenic strains and security assessment is an unavoidable challenge under the conditions of mass rearing practiced in sericulture.
Several drugs have been evaluated for their antiviral activity against BmNPV. The bacterial secondary metabolite prodigiosin inhibits BmNPV in BmN cells and is a potential antiviral compound (104). However, its antiviral efficacy must be tested in insect larvae. The single-crystal compound seselin extracted from Aegle marmelos (a kind of citrus fruit) shows antiviral activity against BmNPV in silkworm larvae (105). AZD8835, AMG319, HS173, AS605240, GDC0941, BEZ235 are PI3K inhibitors and afuresertib is an Akt inhibitor. These seven drugs target the PI3K/Akt pathway to decrease p-Akt and all inhibit BmNPV in BmE cells; nevertheless, only AMG319 and AZD8835 inhibit viral proliferation in silkworm larvae. Of these two, AZD8835 exhibits a stronger antiviral efficacy which might be due to lower drug toxicity in larvae and stronger inhibition of p-Akt (106). The development of drugs with high antiviral capacity in silkworms could decrease mortality in sericulture. However, their absorption and utilization efficiency, inhibitory efficacy, and cost-effectiveness must be increased while their cytotoxicity is decreased (106).
Major Issues in Silkworm Antiviral Studies
Several conflicting results have been reported for the same genes in previous studies on the interaction between silkworm and baculovirus. These discrepancies may be explained by the use of different silkworm strains and cell lines as well as inappropriate internal reference genes (RGs). RGs must not be affected by experimental conditions and should be expressed at the same constant level in all samples. Unsuitable RGs lead to the incorrect interpretation of gene expression patterns and functions (107). As a widely used example, actin participates in baculovirus proliferation and expression after viral infection in silkworms (107). Hence, actin cannot serve as the RG for mRNA and protein detection in studies involving the interaction between silkworms and viruses. In contrast, TIF-4A is an appropriate RG for gene expression analysis (107) and GAPDH (54) is an appropriate internal reference for protein content measurements following viral challenges in silkworms.
Transgenic silkworms with high antiviral capacity have been constructed (102, 103, 108, 109). Nevertheless, their commercial application still faces great challenges. Security assessment must be performed on transgenic silkworms before they are commercialized (3). There are operational guidelines for safety assessments of genetically modified (GM) vertebrates and plants but not for insects, including silkworms. Thus, safety evaluations are difficult to execute on transgenic silkworms. Based on GM animal safety assessment guidelines, we conducted a preliminary evaluation of transgenic silkworms in our laboratory. A classical genetic analysis and molecular characterization of 11 successive generations showed that an inserted foreign DNA fragment was stably inherited in transgenic silkworms (110). The disposition of the inserted DNA in transgenic silkworms fed to chickens was also examined, with no apparent transfer of transgenic DNA from silkworms to chickens (111). A subacute toxicity test comprising a 28 d feeding study in rats showed that transgenic silkworms are toxicologically equivalent to normal silkworms and are safe for rats (112). Transgenic silkworms are unable to survive and reproduce in the field and would not cause environmental risks of competition with other insects, and no interspecific hybridization of transgenic silkworms and Bombyx mandarina was observed in nature, so transgenic silkworms have no risks to biodiversity (113). The transgenic silkworms that produce green fluorescent silk have been reared in a sericulture farm in Japan since 2017 (113). Nevertheless, the design of safety assessment procedures and identification of transgenic antiviral silkworm indicators are urgently required as they cannot be the same as those already implemented for GM vertebrates. A notable difference in appropriate safety assessment design is that although GM vertebrates are used for food and feed, transgenic silkworms are used only in silk production.
Future Directions of Silkworm Antivirus Research
Current research on the mechanisms by which baculovirus penetrates its host has focused mainly on BVs and insect cell lines (25, 28–30). Some of the constraints of investigations into the interactions between individual insects and baculovirus include limitations in insect genetic manipulation, long experimental periods, and intensive labor. The PIFs of ODV envelopes form complexes that mediate viral invasion in the insect midgut (16–19). The receptors involved in ODV invasion may also be part of a complex. Screening and identifying ODV receptor genes in the silkworm midgut are difficult exercises. The process of ODV entry must first be clarified in order to develop methods to block BmNPV infection in silkworm. Earlier studies reported that the resistance of silkworms to BmNPV is controlled by major genes and modified by minor genes (98); however, a major resistance gene has not yet been identified despite numerous attempts using various methods. Identification of resistance genes and analysis of silkworm antiviral mechanisms against BmNPV merit further investigation. In future experiments, we will screen for negative regulatory factors in the immune pathway using genome-wide CRISPR (114) and identify the host proteins that bind the virus by use of inhibitors. The target genes will be knocked out via gene editing to improve silkworm resistance. Immune priming is a new strategy to increase host antiviral capacity (115, 116) and we will clarify its mechanism of action in silkworm. The influences of gut microbes, heat shock response, and DNA methylation on viral silkworm infections will also be evaluated.
The baculovirus expression vector system (BEVS) is a bioreactor for the production of recombinant proteins and vaccines. Several vaccines produced by BEVS have been approved for human and/or veterinary use (15, 117, 118). The BEVS was invented using AcMNPV in combination with an insect cell system (117). However, the cost of silkworm rearing is much lower than that of insect cell culture, promoting the use of BmNPV to generate foreign proteins using silkworm larvae as bioreactors. Understanding the baculovirus infection mechanism including modification of host and viral proteins will facilitate application of a combined BmNPV-silkworm system in production of high value-added medical proteins. Explorations of the silkworm immune response to baculovirus will help construct silkworms less sensitive to BmNPV by inhibiting the host immune system and resistance genes, and in combination with BmNPV with attenuated virulence, further reduce the costs of foreign protein fabrication.
Baculoviruses have been applied worldwide as biopesticides for the control of various insect pests (119, 120). Compared to chemical pesticides, baculoviruses are environmentally safe. Nevertheless, their killing rates are low, and their host range is narrow (15, 31, 119). In the future, baculovirus should be modified to expand its target pest host range. Its antagonism against the host immune defense must be strengthened by accentuating viral host immune evasion mechanisms which will enable use of lower viral titers to kill pests faster. Less sensitive insect bioreactors for baculovirus-based biopesticides should be designed to reduce production costs. Further investigations into silkworm antiviral mechanisms will provide a reverse theoretical basis and reference for biological insect pest control.
Conclusion
Viruses exert strong selection pressure on their hosts to evolve resistance pathways. In turn, these genetic modifications enable viruses to escape host antiviral mechanisms. This arms race favors host defense diversification and the development of viral escape mechanisms (37). Several factors contribute to viral coevolution with its natural host. A complete elucidation of antiviral immunity and immune evasion is challenging as numerous complex pathways are involved (37). Hence, BmNPV research should focus on actual silkworms rather than cell lines and novel technologies such as gene editing and value-added protein biosynthesis. Studies involving the silkworm–baculovirus model are highly informative as they disclose original antiviral strategies, immune evasion mechanisms, and weaknesses of viruses. In this way, genetic antiviral improvement of silkworms may be achieved along with the development of more effective approaches to control lepidopteran and other insect pests. These applications, along with the realization of more productive and efficient bioreactors for novel baculovirus-insect-derived products, are promising applications for the future.
Author Contributions
LJ: analyzed data, drew figure, drafted the article, and supervision. MG: review and editing. QX: supervision. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Zhihong Hu for critical reading of the manuscript. This work was funded by the National Natural Science Foundation of China (no. 31501875), the Fundamental Research Funds for the Central Universities (SWU120029, XDJK2020C006), and the Venture & Innovation Support Program for Chongqing Overseas Returnees (cx2019152).
Abbreviations
AcMNPV, Autographa californica multiple nucleopolyhedrovirus; AMP, antimicrobial peptide; B. mori, Bombyx mori; BEVS, baculovirus expression vector system; BmEGFR, B. mori epidermal growth factor receptor; Bmhsp19.9, B. mori heat shock protein 19.9; BmNPV, Bombyx mori nucleopolyhedrovirus; BV, budded virus; Co-IP, coimmunoprecipitation; CRAC, cholesterol recognition amino acid consensus; cSPs, clip-domain serine proteases; ECs, effector caspases; egt, ecdysteroid (UDP)-glucosyltransferase; GM, genetically modified; HGT, horizontal gene transfer; hpi, h post infection; IAP, inhibitor of apoptosis; iap-A, iap-antagonist; ICs, initiator caspases; miRNA, microRNA; ODV, occlusion-derived virus; PGRP, peptidoglycan recognition protein; PIF, per os infectivity factor; piRNA, PIWI-associated RNA; PO, phenoloxidase; PPO, prophenoloxidase; pre-miRNA, precursor miRNA; ptp, protein tyrosine phosphatase; RFPs, red fluorescent proteins; RGs, reference genes; RNAi, RNA interference; ROS, reactive oxygen species; siRNAs, short interfering RNAs; SPs, serine proteases; vfgf, viral fibroblast growth factor; VSRs, viral suppressors of RNAi.
References
1. Kawahara AY, Plotkin D, Espeland M, Meusemann K, Toussaint EFA, Donath A, et al. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc Natl Acad Sci U S A (2019) 116(45):22657–63. doi: 10.1073/pnas.1907847116
2. Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science (2014) 346(6210):763–7. doi: 10.1126/science.1257570
3. Jiang L, Xia Q. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem Mol Biol (2014) 48:1–7. doi: 10.1016/j.ibmb.2014.02.003
4. Goldsmith MR, Shimada T, Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol (2005) 50:71–100. doi: 10.1146/annurev.ento.50.071803.130456
5. Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science (2004) 306(5703):1937–40. doi: 10.1126/science.1102210
6. Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, et al. The genome sequence of silkworm, Bombyx mori. DNA Res An Int J Rapid Publ Rep Genes Genomes (2004) 11(1):27–35. doi: 10.1093/dnares/11.1.27
7. International Silkworm Genome, C. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol (2008) 38(12):1036–45. doi: 10.1016/j.ibmb.2008.11.004
8. Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science (2009) 326(5951):433–6. doi: 10.1126/science.1176620
9. Xia Q, Li S, Feng Q. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu Rev Entomol (2014) 59:513–36. doi: 10.1146/annurev-ento-011613-161940
10. Gomi S, Majima K, Maeda S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Virol (1999) 80( Pt 5):1323–37. doi: 10.1099/0022-1317-80-5-1323
11. Keddie BA, Aponte GW, Volkman LE. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science (1989) 243(4899):1728–30. doi: 10.1126/science.2648574
12. Blissard GW, Theilmann DA. Baculovirus Entry and Egress from Insect Cells. Annu Rev Virol (2018) 5:113–39. doi: 10.1146/annurev-virology-092917-043356
13. Rahman MM, Gopinathan KP. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res (2004) 101(2):109–18. doi: 10.1016/j.virusres.2003.12.027
14. Feng M, Kong X, Zhang J, Xu W, Wu X. Identification of a novel host protein SINAL10 interacting with GP64 and its role in Bombyx mori nucleopolyhedrovirus infection. Virus Res (2018) 247:102–10. doi: 10.1016/j.virusres.2018.02.005
15. Saxena A, Byram PK, Singh SK, Chakraborty J, Murhammer D, Giri L. A structured review of baculovirus infection process: integration of mathematical models and biomolecular information on cell-virus interaction. J Gen Virol (2018) 99(9):1151–71. doi: 10.1099/jgv.0.001108
16. Xiang X, Chen L, Guo A, Yu S, Yang R, Wu X. The Bombyx mori nucleopolyhedrovirus (BmNPV) ODV-E56 envelope protein is also a per os infectivity factor. Virus Res (2011) 155(1):69–75. doi: 10.1016/j.virusres.2010.08.024
17. Boogaard B, van Lent JWM, Theilmann DA, Erlandson MA, van Oers MM. Baculoviruses require an intact ODV entry-complex to resist proteolytic degradation of per os infectivity factors by co-occluded proteases from the larval host. J Gen Virol (2017) 98(12):3101–10. doi: 10.1099/jgv.0.000974
18. Wang X, Shang Y, Chen C, Liu SR, Chang M, Zhang N, et al. Baculovirus Per Os Infectivity Factor Complex: Components and Assembly. J Virol (2019) 93(6):e02053-18. doi: 10.1128/JVI.02053-18
19. Boogaard B, van Oers MM, van Lent JWM. An Advanced View on Baculovirus per Os Infectivity Factors. Insects (2018) 9(3):84. doi: 10.3390/insects9030084
20. Huh NE, Weaver RF. Categorizing Some Early and Late Transcripts Directed by the Autographa-Californica Nuclear Polyhedrosis-Virus. J Gen Virol (1990) 71:2195–200. doi: 10.1099/0022-1317-71-9-2195
21. Jiang L, Zhao P, Wang GH, Cheng TC, Yang Q, Jin SK, et al. Comparison of factors that may affect the inhibitory efficacy of transgenic RNAi targeting of baculoviral genes in silkworm, Bombyx mori. Antivir Res (2013) 97(3):255–63. doi: 10.1016/j.antiviral.2012.12.020
22. Slack J, Arif BM. The baculoviruses occlusion-derived virus: Virion structure and function. Adv Virus Res (2007) 69:99–165. doi: 10.1016/S0065-3527(06)69003-9
23. Engelhard EK, Kammorgan LNW, Washburn JO, Volkman LE. The Insect Tracheal System - a Conduit for the Systemic Spread of Autographa-Californica-M Nuclear Polyhedrosis-Virus. Proc Natl Acad Sci U S A (1994) 91(8):3224–7. doi: 10.1073/pnas.91.8.3224
24. Braunagel SC, Summers MD. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr Drug Targets (2007) 8(10):1084–95. doi: 10.2174/138945007782151315
25. Feng M, Zhang JJ, Xu WF, Wang HP, Kong XS, Wu XF. Bombyx mori nucleopolyhedrovirus utilizes a clathrin and dynamin dependent endocytosis entry pathway into BmN cells. Virus Res (2018) 253:12–9. doi: 10.1016/j.virusres.2018.05.020
26. Kataoka C, Kaname Y, Taguwa S, Abe T, Fukuhara T, Tani H, et al. Baculovirus GP64-Mediated Entry into Mammalian Cells. J Virol (2012) 86(5):2610–20. doi: 10.1128/JVI.06704-11
27. Luz-Madrigal A, Asanov A, Camacho-Zarco AR, Sampieri A, Vaca L. A Cholesterol Recognition Amino Acid Consensus Domain in GP64 Fusion Protein Facilitates Anchoring of Baculovirus to Mammalian Cells. J Virol (2013) 87(21):11894–907. doi: 10.1128/JVI.01356-13
28. Huang JS, Hao BF, Cheng C, Liang F, Shen XJ, Cheng XW. Entry of Bombyx mori nucleopolyhedrovirus into BmN cells by cholesterol-dependent macropinocytic endocytosis. Biochem Bioph Res Co (2014) 453(1):166–71. doi: 10.1016/j.bbrc.2014.09.073
29. Li ZH, Fan YP, Wei JH, Mei XOG, He Q, Zhang YH, et al. Baculovirus Utilizes Cholesterol Transporter NIEMANN-Pick C1 for Host Cell Entry. Front Microbiol (2019) 10:2825. doi: 10.3389/fmicb.2019.02825
30. Dong XL, Liu TH, Wang W, Pan CX, Wu YF, Du GY, et al. BmREEPa Is a Novel Gene that Facilitates BmNPV Entry into Silkworm Cells. PLoS One (2015) 10(12):e0144575. doi: 10.1371/journal.pone.0144575
31. Wang ML, Hu ZH. Cross-talking between baculoviruses and host insects towards a successful infection. Philos T R Soc B (2019) 374(1767):20180324. doi: 10.1098/rstb.2018.0324
32. Katsuma S, Kawaoka S, Mita K, Shimada T. Genome-wide survey for baculoviral host homologs using the Bombyx genome sequence. Insect Biochem Mol Biol (2008) 38(12):1080–6. doi: 10.1016/j.ibmb.2008.05.008
33. Katsuma S. Phosphatase activity of Bombyx mori nucleopolyhedrovirus PTP is dispensable for enhanced locomotory activity in B. mori larvae. J Invertebr Pathol (2015) 132:228–32. doi: 10.1016/j.jip.2015.11.002
34. Wang F, Xue RJ, Li XY, Hu CM, Xia QY. Characterization of a protein tyrosine phosphatase as a host factor promoting baculovirus replication in silkworm, Bombyx mori. Dev Comp Immunol (2016) 57:31–7. doi: 10.1016/j.dci.2015.12.002
35. Zhang X, Xue R, Cao G, Hu X, Wang X, Pan Z, et al. Effects of egt gene transfer on the development of Bombyx mori. Gene (2012) 491(2):272–7. doi: 10.1016/j.gene.2011.09.026
36. Kingsolver MB, Huang ZJ, Hardy RW. Insect Antiviral Innate Immunity: Pathways, Effectors, and Connections. J Mol Biol (2013) 425(24):4921–36. doi: 10.1016/j.jmb.2013.10.006
37. Marques JT, Imler JL. The diversity of insect antiviral immunity: insights from viruses. Curr Opin Microbiol (2016) 32:71–6. doi: 10.1016/j.mib.2016.05.002
38. Hamajima R, Saito A, Makino S, Kobayashi M, Ikeda M. Antiviral immune responses of Bombyx mori cells during abortive infection with Autographa californica multiple nucleopolyhedrovirus. Virus Res (2018) 258:28–38. doi: 10.1016/j.virusres.2018.09.014
39. Ikeda M, Yamada H, Hamajima R, Kobayashi M. Baculovirus genes modulating intracellular innate antiviral immunity of lepidopteran insect cells. Virology (2013) 435(1):1–13. doi: 10.1016/j.virol.2012.10.016
40. Hamajima R, Kobayashi M, Ikeda M. Identification of amino acid residues of AcMNPV P143 protein involved in rRNA degradation and restricted viral replication in BM-N cells from the silkworm Bombyx mori. Virology (2015) 485:244–51. doi: 10.1016/j.virol.2015.08.008
41. Jiang L, Cheng TC, Dang YH, Peng ZW, Zhao P, Liu SP, et al. Identification of a midgut-specific promoter in the silkworm Bombyx mori. Biochem Bioph Res Co (2013) 433(4):542–6. doi: 10.1016/j.bbrc.2013.03.019
42. Jiang L, Huang CL, Sun Q, Guo HZ, Cheng TC, Peng ZW, et al. The 5 ‘-UTR intron of the midgut-specific BmAPN4 gene affects the level and location of expression in transgenic silkworms. Insect Biochem Mol Biol (2015) 63:1–6. doi: 10.1016/j.ibmb.2015.05.005
43. Ponnuvel KM, Nakazawa H, Furukawa S, Asaoka A, Ishibashi J, Tanaka H, et al. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J Virol (2003) 77(19):10725–9. doi: 10.1128/JVI.77.19.10725-10729.2003
44. Nakazawa H, Tsuneishi E, Ponnuvel KM, Furukawa S, Asaoka A, Tanaka H, et al. Antiviral activity of a serine protease from the digestive juice of Bombyx mori larvae against nucleopolyhedrovirus. Virology (2004) 321(1):154–62. doi: 10.1016/j.virol.2003.12.011
45. Selot R, Kumar V, Shukla S, Chandrakuntal K, Brahmaraju M, Dandin SB, et al. Identification of a soluble NADPH oxidoreductase (BmNOX) with antiviral activities in the gut juice of Bombyx mori. Biosci Biotechnol Biochem (2007) 71(1):200–5. doi: 10.1271/bbb.60450
46. Sunagar SG, Savanurmath CJ, Hinchigeri SB. The profiles of red fluorescent proteins with antinucleopolyhedrovirus activity in races of the silkworm Bombyx mori. J Insect Physiol (2011) 57(12):1707–14. doi: 10.1016/j.jinsphys.2011.09.009
47. Ponnuvel KM, Nithya K, Sirigineedi S, Awasthi AK, Yamakawa M. In Vitro Antiviral Activity of an Alkaline Trypsin from the Digestive Juice of Bombyx Mori Larvae against Nucleopolyhedrovirus. Arch Insect Biochem (2012) 81(2):90–104. doi: 10.1002/arch.21046
48. Zhang SZ, Zhu LB, You LL, Wang J, Cao HH, Liu YX, et al. A Novel Digestive Proteinase Lipase Member H-A in Bombyx mori Contributes to Digestive Juice Antiviral Activity Against B. mori Nucleopolyhedrovirus. Insects (2020) 11(3):154. doi: 10.3390/insects11030154
49. Lin YH, Tai CC, Broz V, Tang CK, Chen P, Wu CP, et al. Adenosine Receptor Modulates Permissiveness of Baculovirus (Budded Virus) Infection via Regulation of Energy Metabolism in Bombyx mori. Front Immunol (2020) 11:763. doi: 10.3389/fimmu.2020.00763
50. Wang Q, Yin M, Yuan C, Liu X, Hu Z, Zou Z, et al. Identification of a Conserved Prophenoloxidase Activation Pathway in Cotton Bollworm Helicoverpa armigera. Front Immunol (2020) 11:785. doi: 10.3389/fimmu.2020.00785
51. Yuan CF, Xing LS, Wang ML, Wang X, Yin MY, Wang QR, et al. Inhibition of melanization by serpin-5 and serpin-9 promotes baculovirus infection in cotton bollworm Helicoverpa armigera. PLoS Pathog (2017) 13(9):e1006645. doi: 10.1371/journal.ppat.1006645
52. Toufeeq S, Wang J, Zhang SZ, Li B, Hu P, Zhu LB, et al. Bmserpin2 Is Involved in BmNPV Infection by Suppressing Melanization in Bombyx mori. Insects (2019) 10(11):399. doi: 10.3390/insects10110399
53. Makino S, Hamajima R, Saito A, Tomizaki M, Iwamoto A, Kobayashi M, et al. Bombyx mori homolog of tumor suppressor p53 is involved in apoptosis-mediated antiviral immunity of B-mori cells infected with nucleopolyhedrovirus. Dev Comp Immunol (2018) 84:133–41. doi: 10.1016/j.dci.2018.02.009
54. Jiang L, Liu WQ, Guo HZ, Dang YH, Cheng TC, Yang WY, et al. Distinct Functions of Bombyx mori Peptidoglycan Recognition Protein 2 in Immune Responses to Bacteria and Viruses. Front Immunol (2019) 10:776. doi: 10.3389/fimmu.2019.00776
55. Clem RJ. The role of apoptosis in defense against baculovirus infection in insects. Curr Topics Microbiol Immunol (2005) 289:113–29. doi: 10.1007/3-540-27320-4_5
56. Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ (2007) 14(1):66–72. doi: 10.1038/sj.cdd.4402059
57. Courtiade J, Pauchet Y, Vogel H, Heckel DG. A comprehensive characterization of the caspase gene family in insects from the order Lepidoptera. BMC Genomics (2011) 12:357. doi: 10.1186/1471-2164-12-357
58. Hamajima R, Iwamoto A, Tomizaki M, Suganuma I, Kitaguchi K, Kobayashi M, et al. Functional analysis of inhibitor of apoptosis 1 of the silkworm Bombyx mori. Insect Biochem Mol Biol (2016) 79:97–107. doi: 10.1016/j.ibmb.2016.10.012
59. Huang N, Wu WB, Yang K, Passarelli AL, Rohrmann GF, Clem RJ. Baculovirus Infection Induces a DNA Damage Response That Is Required for Efficient Viral Replication. J Virol (2011) 85(23):12547–56. doi: 10.1128/JVI.05766-11
60. Mitchell JK, Friesen PD. Baculoviruses Modulate a Proapoptotic DNA Damage Response To Promote Virus Multiplication. J Virol (2012) 86(24):13542–53. doi: 10.1128/JVI.02246-12
61. Vandergaast R, Schultz KLW, Cerio RJ, Friesen PD. Active Depletion of Host Cell Inhibitor-of-Apoptosis Proteins Triggers Apoptosis upon Baculovirus DNA Replication. J Virol (2011) 85(16):8348–58. doi: 10.1128/JVI.00667-11
62. Vogel E, Santos D, Mingels L, Verdonckt TW, Vanden Broeck J. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front Physiol (2019) 9:1912. doi: 10.3389/fphys.2018.01912
63. Jayachandran B, Hussain M, Asgari S. RNA Interference as a Cellular Defense Mechanism against the DNA Virus Baculovirus. J Virol (2012) 86(24):13729–34. doi: 10.1128/JVI.02041-12
64. van Mierlo JT, van Cleef KW, van Rij RP. Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol Biol (2011) 721:3–22. doi: 10.1007/978-1-61779-037-9_1
65. Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5’ precision of both MicroRNAs and their miRNA* strands in flies. Curr Biol (2008) 18(2):147–51. doi: 10.1016/j.cub.2007.12.049
66. Wu P, Shang Q, Dweteh OA, Huang HL, Zhang SL, Zhong JB, et al. Over expression of bmo-miR-2819 suppresses BmNPV replication by regulating the BmNPV ie-1 gene in Bombyx mori. Mol Immunol (2019) 109:134–9. doi: 10.1016/j.molimm.2019.03.013
67. Kang LQ, Wang ML, Cao XL, Tang SM, Xia DG, Shen XJ, et al. Inhibition of expression of BmNPV cg30 by bmo-miRNA-390 is a host response to baculovirus invasion. Arch Virol (2018) 163(10):2719–25. doi: 10.1007/s00705-018-3912-9
68. Wang GH, Jiang L, Zhu L, Cheng TC, Niu WH, Yan YF, et al. Characterization of Argonaute family members in the silkworm, Bombyx mori. Insect Sci (2013) 20(1):78–91. doi: 10.1111/j.1744-7917.2012.01555.x
69. Hua X, Li B, Song L, Hu C, Li X, Wang D, et al. Stimulator of interferon genes (STING) provides insect antiviral immunity by promoting Dredd caspase-mediated NF-kappaB activation. J Biol Chem (2018) 293(30):11878–90. doi: 10.1074/jbc.RA117.000194
70. Lu P, Pan Y, Yang YH, Zhu FF, Li CJ, Guo ZJ, et al. Discovery of anti-viral molecules and their vital functions in Bombyx mori. J Invertebr Pathol (2018) 154:12–8. doi: 10.1016/j.jip.2018.02.012
71. Liu TH, Dong XL, Pan CX, Du GY, Wu YF, Yang JG, et al. A newly discovered member of the Atlastin family, BmAtlastin-n, has an antiviral effect against BmNPV in Bombyx mori. Sci Rep-Uk (2016) 6:28946. doi: 10.1038/srep28946
72. Jiang L, Xie EY, Guo HZ, Sun Q, Liuli HY, Wang YM, et al. Heat shock protein 19.9 (Hsp19.9) from Bombyx mori is involved in host protection against viral infection. Dev Comp Immunol (2021) 114:103790. doi: 10.1016/j.dci.2020.103790
73. Chen TT, Tan LR, Hu N, Dong ZQ, Hu ZG, Jiang YM, et al. C-lysozyme contributes to antiviral immunity in Bombyx mori against nucleopolyhedrovirus infection. J Insect Physiol (2018) 108:54–60. doi: 10.1016/j.jinsphys.2018.05.005
74. Hu ZG, Dong ZQ, Dong FF, Zhu Y, Chen P, Lu C, et al. Identification of a PP2A gene in Bombyx mori with antiviral function against B. mori nucleopolyhedrovirus. Insect Sci (2020) 27(4):687–96. doi: 10.1111/1744-7917.12678
75. Guo HZ, Xu GW, Wang BB, Xia F, Sun Q, Wang YM, et al. Phosphoenolpyruvate carboxykinase is involved in antiviral immunity against Bombyx mori nucleopolyhedrovirus. Dev Comp Immunol (2019) 92:193–8. doi: 10.1016/j.dci.2018.11.015
76. Yin HC, Shao SL, Jiang XJ, Xie PY, Sun WS, Yu TF. Interactions between Autophagy and DNA Viruses. Viruses-Basel (2019) 11(9):776. doi: 10.3390/v11090776
77. Wang L, Xiao Q, Zhou XL, Zhu Y, Dong ZQ, Chen P, et al. Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection. Viruses-Basel (2018) 10(1):14. doi: 10.3390/v10010014
78. Clem RJ. Baculoviruses and apoptosis: the good, the bad, and the ugly. Cell Death Differ (2001) 8(2):137–43. doi: 10.1038/sj.cdd.4400821
79. Clem RJ. Baculoviruses and apoptosis: a diversity of genes and responses. Curr Drug Targets (2007) 8(10):1069–74. doi: 10.2174/138945007782151405
80. Chen P, Kang TT, Bao XY, Dong ZQ, Zhu Y, Xiao WF, et al. Evolutionary and functional analyses of the interaction between the Bombyx mori inhibitor of apoptosis (IAP) and nucleopolyhedrovirus IAPs. Insect Sci (2020) 27(3):463–74. doi: 10.1111/1744-7917.12664
81. Vandergaast R, Mitchell JK, Byers NM, Friesen PD. Insect Inhibitor-of-Apoptosis (IAP) Proteins Are Negatively Regulated by Signal-Induced N-Terminal Degrons Absent within Viral IAP Proteins. J Virol (2015) 89(8):4481–93. doi: 10.1128/JVI.03659-14
82. Byers NM, Vandergaast RL, Friesen PD. Baculovirus Inhibitor-of-Apoptosis Op-IAP3 Blocks Apoptosis by Interaction with and Stabilization of a Host Insect Cellular IAP. J Virol (2016) 90(1):533–44. doi: 10.1128/JVI.02320-15
83. Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, et al. Inhibition of Ice Family Proteases by Baculovirus Antiapoptotic Protein P35. Science (1995) 269(5232):1885–8. doi: 10.1126/science.7569933
84. Nakanishi T, Shimada T, Katsuma S. Characterization of a Bombyx mori nucleopolyhedrovirus mutant lacking both fp25K and p35. Virus Genes (2010) 41(1):144–8. doi: 10.1007/s11262-010-0492-5
85. Jabbour AM, Ekert PG, Coulson EJ, Knight MJ, Ashley DM, Hawkins CJ. The p35 relative, p49, inhibits mammalian and Drosophila caspases including DRONC and protects against apoptosis. Cell Death Differ (2002) 9(12):1311–20. doi: 10.1038/sj.cdd.4401135
86. Guy MP, Friesen PD. Reactive-site cleavage residues confer target specificity to baculovirus P49, a dimeric member of the P35 family of caspase inhibitors. J Virol (2008) 82(15):7504–14. doi: 10.1128/JVI.00231-08
87. Yamada H, Kitaguchi K, Hamajima R, Kobayashi M, Ikeda M. Novel Apoptosis Suppressor Apsup from the Baculovirus Lymantria dispar Multiple Nucleopolyhedrovirus Precludes Apoptosis by Preventing Proteolytic Processing of Initiator Caspase Dronc. J Virol (2013) 87(23):12925–34. doi: 10.1128/JVI.02065-13
88. Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell (2007) 130(3):413–26. doi: 10.1016/j.cell.2007.07.039
89. Mehrabadi M, Hussain M, Matindoost L, Asgari S. The Baculovirus Antiapoptotic p35 Protein Functions as an Inhibitor of the Host RNA Interference Antiviral Response. J Virol (2015) 89(16):8182–92. doi: 10.1128/JVI.00802-15
90. Singh J, Singh CP, Bhavani A, Nagaraju J. Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology (2010) 407(1):120–8. doi: 10.1016/j.virol.2010.07.033
91. Singh CP, Singh J, Nagaraju J. bmnpv-miR-3 facilitates BmNPV infection by modulating the expression of viral P6.9 and other late genes in Bombyx mori. Insect Biochem Mol Biol (2014) 49:59–69. doi: 10.1016/j.ibmb.2014.03.008
92. Singh CP, Singh J, Nagaraju J. A baculovirus-encoded MicroRNA (miRNA) suppresses its host miRNA biogenesis by regulating the exportin-5 cofactor Ran. J Virol (2012) 86(15):7867–79. doi: 10.1128/JVI.00064-12
93. Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology (2007) 368(2):219–26. doi: 10.1016/j.virol.2007.05.043
94. Belyavskyi M, Braunagel SC, Summers MD. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci U S A (1998) 95(19):11205–10. doi: 10.1073/pnas.95.19.11205
95. Guo HZ, Sun Q, Wang BB, Wang YM, Xie EY, Xia QY, et al. Spry is downregulated by multiple viruses to elevate ERK signaling and ensure viral reproduction in silkworm. Dev Comp Immunol (2019) 98:1–5. doi: 10.1016/j.dci.2019.04.001
96. Jin S, Cheng T, Guo Y, Lin P, Zhao P, Liu C, et al. Bombyx mori epidermal growth factor receptor is required for nucleopolyhedrovirus replication. Insect Mol Biol (2018) 27(4):464–77. doi: 10.1111/imb.12386
97. Jin SK, Cheng TC, Jiang L, Lin P, Yang Q, Xiao Y, et al. Identification of a New Sprouty Protein Responsible for the Inhibition of the Bombyx mori Nucleopolyhedrovirus Reproduction. PLoS One (2014) 9(6):e99200. doi: 10.1371/journal.pone.0099200
98. Cheng Y, Wang XY, Du C, Gao J, Xu JP. Expression analysis of several antiviral related genes to BmNPV in different resistant strains of silkworm, Bombyx mori. J Insect Sci (2014) 14:76. doi: 10.1673/031.014.76
99. Chen TT, Hu N, Tan LR, Xiao Q, Dong ZQ, Chen P, et al. Resistant silkworm strain block viral infection independent of melanization. Pestic Biochem Phys (2019) 154:88–96. doi: 10.1016/j.pestbp.2018.12.012
100. Jiang L, Wang GH, Cheng TC, Yang Q, Jin SK, Lu G, et al. Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch Virol (2012) 157(7):1323–8. doi: 10.1007/s00705-012-1309-8
101. Jiang L, Cheng TC, Zhao P, Yang Q, Wang GH, Jin SK, et al. Resistance to BmNPV via Overexpression of an Exogenous Gene Controlled by an Inducible Promoter and Enhancer in Transgenic Silkworm, Bombyx mori. PLoS One (2012) 7(8):e41838. doi: 10.1371/journal.pone.0041838
102. Jiang L, Zhao P, Cheng TC, Sun Q, Peng ZW, Dang YH, et al. A transgenic animal with antiviral properties that might inhibit multiple stages of infection. Antivir Res (2013) 98(2):171–3. doi: 10.1016/j.antiviral.2013.02.015
103. Chen SQ, Hou CX, Bi HL, Wang YQ, Xu J, Li MW, et al. Transgenic Clustered Regularly Interspaced Short Palindromic Repeat/Cas9-Mediated Viral Gene Targeting for Antiviral Therapy of Bombyx mori Nucleopolyhedrovirus. J Virol (2017) 91(8):e02465-16. doi: 10.1128/JVI.02465-16
104. Zhou W, Zeng C, Liu RH, Chen J, Li R, Wang XY, et al. Antiviral activity and specific modes of action of bacterial prodigiosin against Bombyx mori nucleopolyhedrovirus in vitro. Appl Microbiol Biot (2016) 100(9):3979–88. doi: 10.1007/s00253-015-7242-5
105. Somu C, Karuppiah H, Sundaram J. Antiviral activity of seselin from Aegle marmelos against nuclear polyhedrosis virus infection in the larvae of silkworm, Bombyx mori. J Ethnopharmacol (2019) 245:112155. doi: 10.1016/j.jep.2019.112155
106. Wang BB, Jiang L, Guo HZ, Sun Q, Wang YM, Xie EY, et al. Screening of PI3K-Akt-targeting Drugs for Silkworm against Bombyx mori Nucleopolyhedrovirus. Molecules (2019) 24(7):1260. doi: 10.3390/molecules24071260
107. Guo HZ, Jiang L, Xia QY. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol Genet Genomics (2016) 291(2):999–1004. doi: 10.1007/s00438-015-1125-4
108. Jiang L, Peng ZW, Guo HZ, Sun JC, Sun Q, Xia F, et al. Enhancement of antiviral capacity of transgenic silkworms against cytoplasmic polyhedrosis virus via knockdown of multiple viral genes. Dev Comp Immunol (2017) 77:138–40. doi: 10.1016/j.dci.2017.07.020
109. Sun Q, Jiang L, Guo HZ, Xia F, Wang BB, Wang YM, et al. Increased antiviral capacity of transgenic silkworm via knockdown of multiple genes on Bombyx mori bidensovirus. Dev Comp Immunol (2018) 87:188–92. doi: 10.1016/j.dci.2018.06.002
110. Jiang L, Sun Q, Liu WQ, Guo HZ, Peng ZW, Dang YH, et al. Postintegration stability of the silkworm piggyBac transposon. Insect Biochem Mol Biol (2014) 50:18–23. doi: 10.1016/j.ibmb.2014.03.006
111. Wang YM, Wang ZL, Guo HZ, Huang J, Li XY, Sun Q, et al. Potential of transferring transgenic DNA from silkworm to chicken. Int J Biol Macromol (2020) 142:311–9. doi: 10.1016/j.ijbiomac.2019.09.102
112. Jiang L, Wang YM, Guo HZ, Sun Q, Xie EY, Liuli HY, et al. Toxicological evaluation of transgenic silkworms. Toxicol Res (2020) 9(6):845–53. doi: 10.1093/toxres/tfaa089
113. Kômoto N, Tomita S. Risk Assessment of Transgenic Silkworms. GMOs Topics Biodiversity Conserv 19. (2020). doi: 10.1007/978-3-030-53183-6_10
114. Chang J, Wang R, Yu K, Zhang T, Chen X, Liu Y, et al. Genome-wide CRISPR screening reveals genes essential for cell viability and resistance to abiotic and biotic stresses in Bombyx mori. Genome Res (2020) 30(5):757–67. doi: 10.1101/gr.249045.119
115. Valdez A, Yepiz-Plascencia G, Ricca E, Olmos J. First Litopenaeus vannamei WSSV 100% oral vaccination protection using CotC::Vp26 fusion protein displayed on Bacillus subtilis spores surface. J Appl Microbiol (2014) 117(2):347–57. doi: 10.1111/jam.12550
116. Tidbury HJ, Pedersen AB, Boots M. Within and transgenerational immune priming in an insect to a DNA virus. P Roy Soc B-Biol Sci (2011) 278(1707):871–6. doi: 10.1098/rspb.2010.1517
117. Smith GE, Summers MD, Fraser MJ. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol (1983) 3(12):2156–65. doi: 10.1128/MCB.3.12.2156
118. van Oers MM, Pijlman GP, Vlak JM. Thirty years of baculovirus-insect cell protein expression: from dark horse to mainstream technology. J Gen Virol (2015) 96(Pt 1):6–23. doi: 10.1099/vir.0.067108-0
119. Moscardi F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu Rev Entomol (1999) 44:257–89. doi: 10.1146/annurev.ento.44.1.257
Keywords: antiviral immunity, baculovirus, Bombyx mori nucleopolyhedrovirus, immune evasion, silkworm
Citation: Jiang L, Goldsmith MR and Xia Q (2021) Advances in the Arms Race Between Silkworm and Baculovirus. Front. Immunol. 12:628151. doi: 10.3389/fimmu.2021.628151
Received: 11 November 2020; Accepted: 04 January 2021;
Published: 09 February 2021.
Edited by:
Humberto Lanz-Mendoza, National Institute of Public Health, MexicoReviewed by:
Jorge Cime-Castillo, National Institute of Public Health, MexicoJorge Contreras-Garduño, National Autonomous University of Mexico, Mexico
Jiaping Xu, Anhui Agricultural University, China
Copyright © 2021 Jiang, Goldsmith and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Jiang, amlhbmdsaWFuZ0Bzd3UuZWR1LmNu
 Liang Jiang
Liang Jiang Marian R. Goldsmith
Marian R. Goldsmith Qingyou Xia1,2
Qingyou Xia1,2