
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 09 March 2021
Sec. Vaccines and Molecular Therapeutics
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.627688
This article is part of the Research Topic Development of Vaccines Against Emerging Pathogens View all 16 articles
 Hadar Marcus
Hadar Marcus Emily Thompson
Emily Thompson Yan Zhou
Yan Zhou Michael Bailey
Michael Bailey Mitzi M. Donaldson
Mitzi M. Donaldson Daphne A. Stanley
Daphne A. Stanley Clement Asiedu
Clement Asiedu Kathryn E. Foulds
Kathryn E. Foulds Mario Roederer
Mario Roederer Juan I. Moliva†
Juan I. Moliva† Nancy J. Sullivan*†
Nancy J. Sullivan*†Heterologous prime-boost immunization regimens are a common strategy for many vaccines. DNA prime rAd5-GP boost immunization has been demonstrated to protect non-human primates against a lethal challenge of Ebola virus, a pathogen that causes fatal hemorrhagic disease in humans. This protection correlates with antibody responses and is also associated with IFNγ+ TNFα+ double positive CD8+ T-cells. In this study, we compared single DNA vs. multiple DNA prime immunizations, and short vs. long time intervals between the DNA prime and the rAd5 boost to evaluate the impact of these different prime-boost strategies on vaccine-induced humoral and cellular responses in non-human primates. We demonstrated that DNA/rAd5 prime-boost strategies can be tailored to induce either CD4+ T-cell or CD8+ T-cell dominant responses while maintaining a high magnitude antibody response. Additionally, a single DNA prime immunization generated a stable memory response that could be boosted by rAd5 3 years later. These results suggest DNA/rAd5 prime-boost provides a flexible platform that can be fine-tuned to generate desirable T-cell memory responses.
DNA vaccines have been demonstrated to induce both durable cellular and humoral responses; however, these responses, while broad, were weak in primates when DNA was given alone without an additional vaccine (1–8). Both humoral and cellular immune responses can be significantly enhanced by combining a priming immunization with a heterologous boost vaccination (2, 6, 9–19). Homologous prime-boost immunization regimens, where the initial vaccine agent is re-administered via the same immunization route, have been used since the beginning of vaccine development and are common strategies for many licensed vaccines. The benefit of homologous prime-boost immunization is the efficient boosting of the humoral response and relative simplicity. In the case of DNA vaccines against HIV, three immunizations with DNA generated a superior humoral response than two immunizations, and this humoral response was maintained over time (20–22). However, the memory CD8+ T-cell response was relatively weak (20).
In contrast to homologous prime-boost regimens, heterologous prime-boost vaccinations are more effective in generating a memory CD8+ T-cell response of higher quality and magnitude (2, 9, 11–16, 23–26), and can bypass anti-vector immunity (27, 28). DNA prime followed by a protein or vector-based boost generates strong T-cell responses in addition to a potent antibody response (7, 29, 30). Various strategies consisting of a single or multiple DNA vaccines followed by recombinant adenovirus serotype 5 (rAd5) boost have been tested (31–35). While several of these regimens were reported to efficiently induce multifunctional CD4+ and CD8+ T-cell responses (35, 36), other strategies failed to induce strong CD8+ T-cell responses. For example, three DNA prime immunizations followed by rAd5 boost 1 month later elicited a high frequency of SIV-specific CD4+ T-cells but failed to induce CD8+ T-cells against some subdominant epitopes (37). Immunogenicity data obtained from clinical trials using HIV DNA prime, NYVAC, or rAd boost have demonstrated a high magnitude T-cell response with CD4+ T-cells observed more frequently than CD8+ T-cells (38). It is known that DNA immunization could skew the cellular immune response toward CD4+ T-cell responses (33, 39) but how variation in DNA/Ad5 prime-boost strategies affect memory humoral and cellular responses has not been systematically examined.
In this study, we evaluated the impact of multiple DNA vaccination and the time intervals between DNA prime and rAd5 boost on Ebola vaccine immunogenicity in non-human primates. Ebola virus (EBOV) is a member of the Filovirus family that causes hemorrhagic fever with a 32–89% fatality rate in humans (40). We have previously demonstrated that the combination of three immunizations with a DNA plasmid encoding Ebola glycoprotein (GP) followed by a rAd5-GP boost, as well as a single rAd5-GP immunization, protected 100% of non-human primates (NHP) against lethal EBOV challenge (41, 42), and antigen-specific IgG antibody level is a correlate of protection (43). At the same time, CD8+ cellular immunity is required for uniform protection (42). By changing the frequency of DNA vaccination and prime-boost interval, we found that all regimens induced high and durable antibody responses. However, in contrast to the CD8+ T-cell dominated response generated after a single DNA prime-rAd5-GP boost, multiple DNA primes resulted in higher CD4+ T-cell magnitude and reduced CD8+ T-cell responses. Extending the time interval between the multiple DNA primes and the Ad5-GP boost reversed the CD4+ T-cell dominancy. Importantly, CD8+ effector memory cells expressing both IFNγ and TNFα, a phenotypic quality associated with Ebola vaccine protection (44), could be preferentially expanded by modifying vaccine component order, frequency, or time interval. Our data demonstrate the importance of fine-tuning the immunization regimens according to the desired immune responses.
To assess the impact of DNA immunization frequency on the immunogenicity of DNA prime/rAd5-GP boost regimen, we immunized groups of four macaques with single or multiple doses of DNA vaccine before boosting them with rAd5-GP (Figure 1). Anti-GP ELISA IgG specific responses were measured 2 weeks after the last DNA immunization (Figure 2). Single DNA prime induced modest plasma antibody titers with an average effective concentration (EC90) of 807. Subsequent additional DNA immunization increased GP-specific antibody titer to 1,410 and 4,996 with 2x DNA and 3x DNA, respectively. However, even with 3x DNA primes, the GP-specific IgG titer was significantly lower than that induced by a single rAd5 immunization (p = 0.026, Figure 2), which is consistent with the hypothesis that DNA is weaker for antibody induction than rAd5.
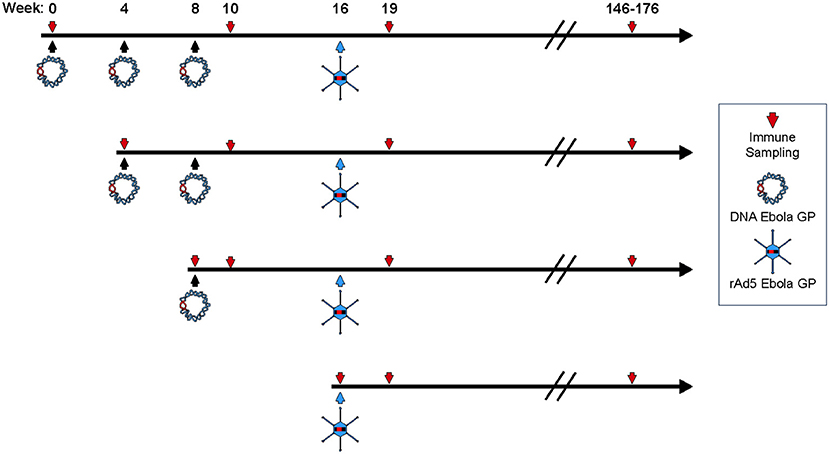
Figure 1. Study design. Four female cynomolgus macaques in each group were primed with plasmid vectors encoding GP(Z) and GP(S/G) 1, 2, or 3 times. Eight weeks after the last DNA immunization some NHP's were boosted with 1011 PFU of rAd5-GP. Blood sampling for anti-GP antibody and T-cell analysis were collected as indicated by the downward arrows.
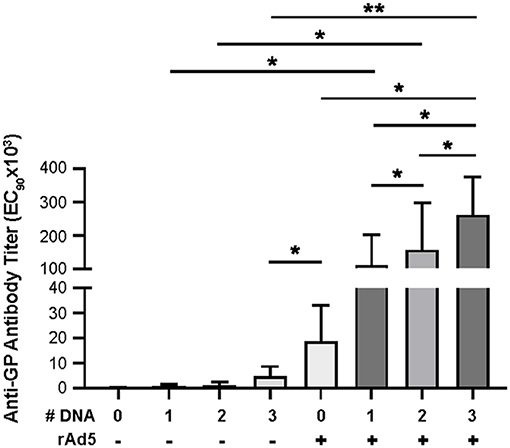
Figure 2. Anti-GP antibody secretion. Anti-GP specific antibodies were detected in immunized NHP sera using ELISA. Sera were diluted from 1:50 to 1:50,000 in half-log increments. Goat anti-human IgG HRP 1:5,000 was used as secondary antibody. ELISA titers are expressed as EC90 values, the dilution at which there is a 90% reduction in antigen binding. Statistical analysis between more than two groups was performed using a one-way ANOVA with Tukey's post hoc test; *p < 0.05; **p < 0.01; mean ± standard deviation shown.
Following boost immunization with rAd5-GP, GP-specific IgG titers measured at week three post boost were one to two orders of magnitude higher than those induced by DNA immunization (Figure 2). A significant increase in GP-specific IgG titers was observed in all DNA immunization groups after the rAd5 boost compared to DNA immunization alone (p = 0.026, 0.038, and 0.002 for groups with 1x, 2x, or 3x DNA vaccines, respectively). Furthermore, consistent with higher GP-specific IgG titers induced by multiple DNA immunization, we observed a trend of higher post-boost titers being associated with an increased number of DNA primes. The average post-boost titer of the 3x DNA prime group was significantly higher than the titer of the single DNA prime group (p = 0.04, Figure 2), which in turn was significantly higher than the titer induced by single rAd5 immunization (p = 0.039, Figure 2). These results suggest that DNA prime imprints a humoral response even when the titers are moderate after the prime, and allows rAd5 boost to elicit a stronger humoral response than the rAd5 prime alone.
Having shown that the DNA prime strategy impacts the magnitude of humoral response after the rAd5 boost, we next evaluated the influence of the number of DNA prime doses on the cellular immune response. We used intracellular cytokine staining to measure the antigen-specific T-cell magnitude, defined as the frequency of memory T-cells expressing any one of three cytokines, IFNγ, TNFα, and IL-2 (Figure 3A, see Supplementary Figure 1 for cytokine gating strategy). At 3 weeks post rAd5-GP boost, the 1x DNA prime/rAd5 boost regimen generated CD4+ and CD8+ T-cell responses with magnitudes similar to those induced by a single immunization with rAd5-GP (Figure 3B). In both cases, average CD8+ T-cell responses were stronger than CD4+ T-cell responses, accounting for 90% of the total T-cell response (Figure 3C). In contrast to a single DNA prime, multiple DNA primes were associated with an increase in the magnitude of CD4+ T-cell response, but not in the magnitude of CD8+ T-cell response. The proportion of the CD4+ T-cell response among total T-cell responses post boost increased from 11% in 1x DNA prime group to 60% in the 2x DNA prime group, and further to 90% in the 3x DNA prime group (Figures 3B,C). Thus, multiple DNA primes favor the development of CD4+ T-cell responses, and as a result, a CD4+ dominant memory T-cell response was generated after the rAd5 boost.
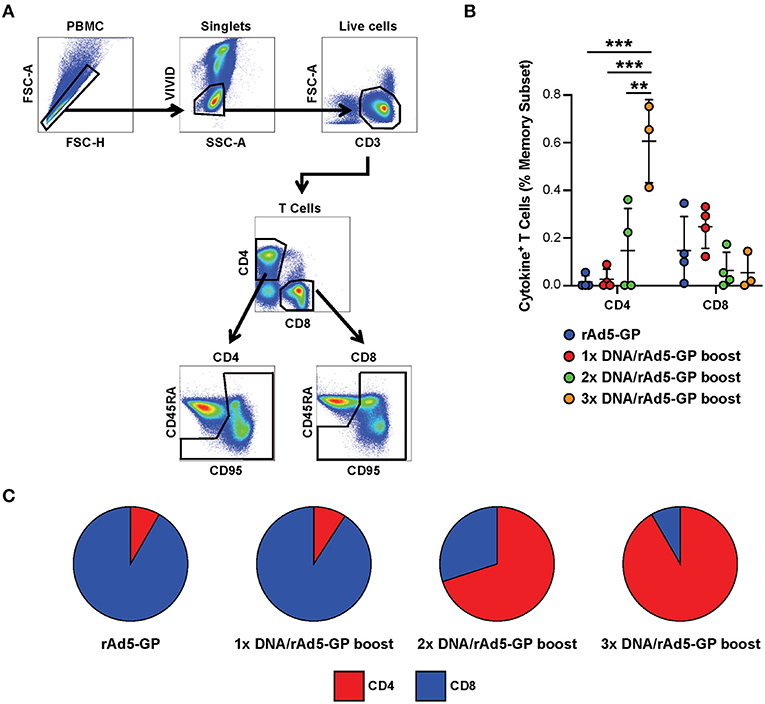
Figure 3. Reduction of DNA priming reverses the CD4/CD8 ratio in heterologous DNA prime rAd5 boost vaccination. Ebola GP reactive CD4+ and CD8+ T-cells secreting any of the three cytokines measured (IFNg, IL-2, or TNFa) were detected in the memory (CD95+CD45RAhi and CD95+CD45RAlo) subset of the NHP's PBMCs following 6 h in vitro stimulation with EBOV GP peptide pool or DMSO control. (A) T-cell gating tree. (B) Magntitude of cytokine positive T-cells in animals vaccinated with: rAd5 GP (blue), 1x DNA prime-rAd5-GP boost (red), 2x DNA prime-rAd5-GP boost (green), and 3x DNA prime-rAd5-GP boost (orange). (C) The relative proportion of CD4+ or CD8+ T-cell responses in each group. Statistical analysis between more than two groups was performed using a one-way ANOVA with Tukey's post hoc test; **p < 0.01, ***p < 0.001; mean ± standard deviation shown.
In addition to the magnitude of T-cell response, T-cell quality, defined as the frequency of T-cells with certain combinations of effector functions, is important for vaccine-induced protection (45, 46). We tested if the DNA prime frequency also affects post-boost T-cell quality. Quality analysis of the CD4+ T-cell response revealed a significant increase in the frequency of IFNγ+IL2+TNFα+ CD4+ T-cells with the addition of each DNA prime immunization (Figure 4A). In contrast, the magnitude of IFNγ+TNFα+ CD8+ T-cells, a subset that is associated with rAd5 vaccine-induced protection against Ebola virus infection (47), was significantly reduced by 3- and 4.5-fold with the addition of the second and third DNA primes, respectively (Figure 4B). Thus, increasing the number of DNA primes may not only affect the magnitude but also the quality of the T-cell response.
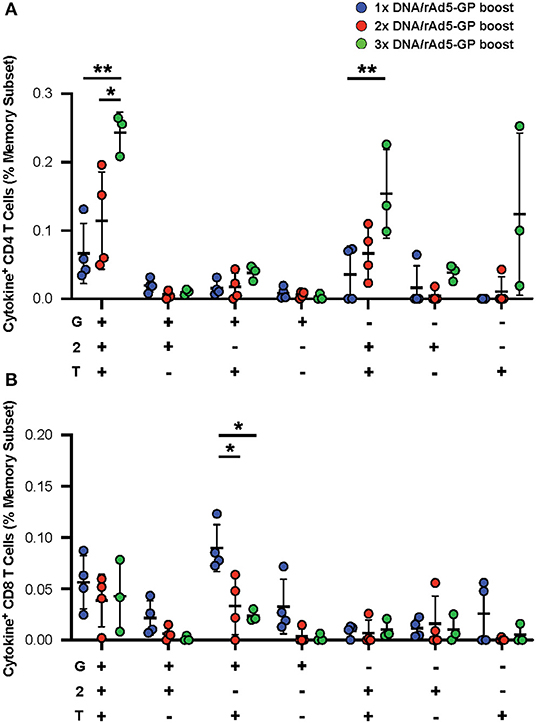
Figure 4. GP specific polyfunctional T-cell. PBMCs were stimulated with EBOV GP peptides. The functionality of antigen-specific CD4+ (A) and CD8+ (B) T-cell responses were assessed in DNA prime rAd5 boost NHP's by analyzing the individual cytokine (G:IFNγ, T:TNFa, and 2:IL-2) response pattern. Statistical analysis between more than two groups was performed using a one-way ANOVA with Tukey's post hoc test; *p < 0.05, **p < 0.01; mean ± standard deviation shown.
To assess the durability of the T-cell response following DNA prime and rAd5-GP boost, the frequencies of antigen-specific CD4+ and CD8+ T-cells were analyzed 130–160 weeks after the rAd5-GP boost (Figure 1, first and third regimens). Single or multiple DNA primes followed by rAd5-GP boost 8 weeks later resulted in sustained CD4+ T-cell responses that lasted for at least 2.5 years. The magnitude of the CD4+ response 2.5 years after the boost was found to be similar between the 1x DNA/rAd5 and the 3x DNA/rAd5 groups (Figure 5A). This response was higher than the CD4+ response that was measured 3 weeks post the rAd boost in the case of single DNA prime immunization but lower in the case of 3x DNA prime immunization (Figure 3C). In contrast, post rAd5 boost, specific CD8+ T-cells could be detected mainly in animals that received a single DNA prime immunization and were barely detected in the multiple DNA primed animals (Figure 5C). Quality analysis of the CD4+ T-cells 2.5 years after boost revealed the dominancy of TNFα+ single positive cells (Figure 5B). The CD8+ T-cell dominance in the single DNA prime rAd5-GP consisted mainly of IFNγ+IL2+TNFα+ and IFNγ+IL2−TNFα+ secreting cells, indicating a memory phenotype (Figure 5D) (48). Anti-Ebola GP IgG could also be detected 130–160 weeks after the rAd5 boost of either single or multiple DNA primed groups. Anti-GP IgG titers were significantly higher in NHPs that received a single DNA prime/rAd5 boost than NHPs that were boosted after multiple DNA primes (Figure 5E).
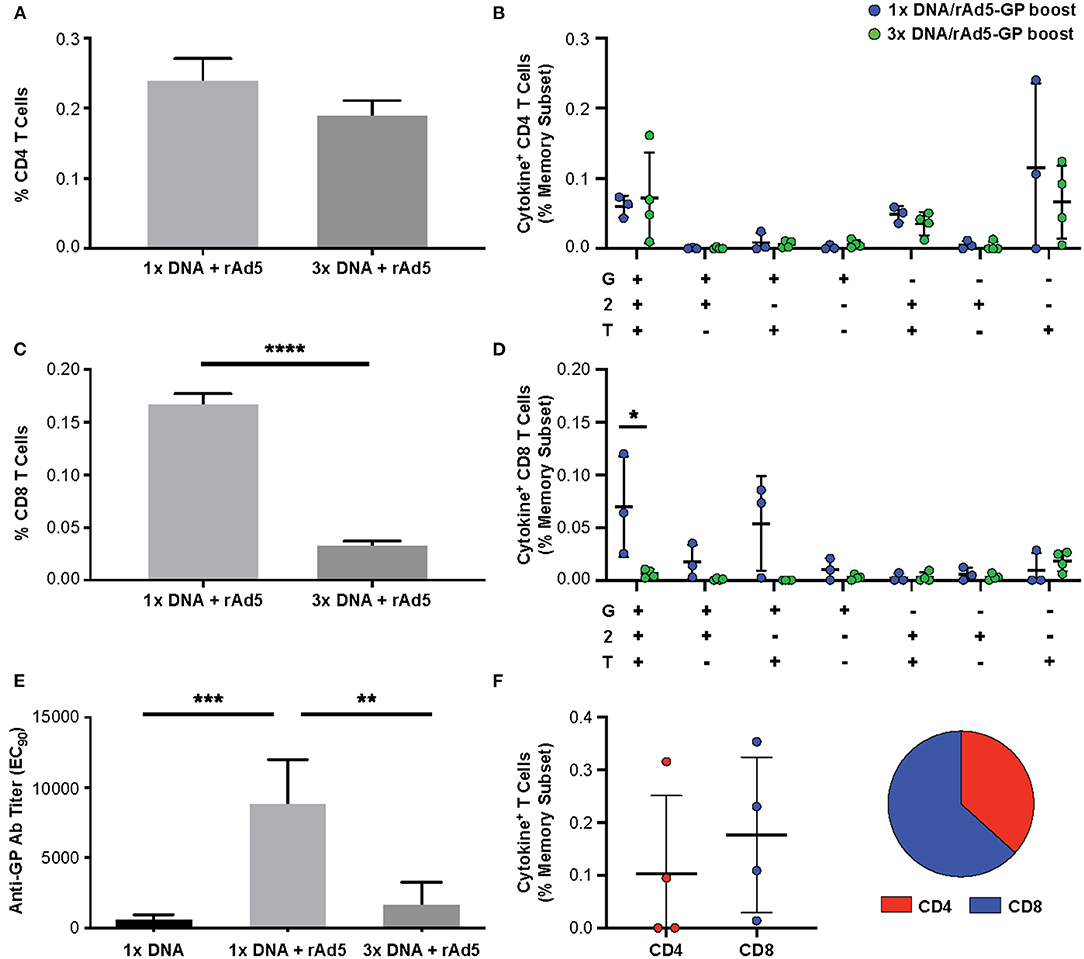
Figure 5. Durability of the immune response following DNA prime rAd5-GP boost. Cynomolgus macaques were primed with plasmid vectors encoding GP(Z) and GP(S/G) one or three times. Eight weeks after the last DNA immunization 4 NHP's from each group were boosted with 1011 PFU of rAd5-GP vaccine. Blood sampling for anti-GP antibody and T-cell analysis were collected 130–160 weeks after the rAd5-GP boost, and at 159 weeks from four animals which received only a single DNA prime immunization without a boost. PBMCs were stimulated with EBOV GP peptides. The magnitude of antigen-specific cytokine response and pattern of CD4+ (A,B) and CD8+ (C,D) T-cells (G:IFNγ, T:TNFa, and 2:IL-2). (E) Specific anti-GP antibody titers were measured using ELISA. (F) Ebola GP specific CD4+ and CD8+ T-cells after rAd5 GP boost that was given 159 weeks after a single DNA prime. Statistical analysis between two groups was performed using an unpaired student's t-test, comparison between more than two groups were performed using a one-way ANOVA with Tukey's post hoc test; *p < 0.05, **p < 0.01, ***p < 0.001,****p < 0.0001; mean ± standard deviation shown.
Interestingly, 3 years after a single DNA immunization with no additional boost, low yet measurable CD4+ T-cells (data not shown), as well as anti-Ebola GP antibody could be detected (Figure 5E). To assess the immunological memory established by a single DNA prime long-term, this group was boosted with rAd5-GP 3 years later. High titers of anti-GP antibody (200,373 ± 127,700) were detected (data not shown). We found that previously undetected CD8+ T-cell responses could be boosted after 3 years, and shifted the memory phenotype to one that slightly favored CD8+ T-cells (Figure 5F). These data indicate that durable and stable immunological memory can be established after a single DNA prime and leaves a large window of time for a subsequent boost with rAd5.
To assess whether the long time interval following prime could improve CD8+ T-cell responses in the multiple DNA prime/rAd5 strategy, we boosted four animals with rAd5 1 year after multiple DNA immunizations (Figure 6A). In contrast to CD4+ T-cell dominant responses observed when the boost immunization was given 8 weeks post-prime (Figure 3B), a long time interval between the DNA prime and rAd5-GP boost resulted in high magnitudes of both CD4+ and CD8+ T-cell responses, with CD8+ T-cell responses dominating (Figure 6B). Furthermore, boosting with rAd5 1 year after multiple DNA immunizations yielded the same high levels of GP-specific IgG titers as boosting with rAd5 8 weeks after multiple DNA immunizations (240,000 ± 186,000 and 262,000 ± 111,000, respectively) (Figure 6C).

Figure 6. Effect of time interval between prime and boost on GP responses. (A) Study design: Four cynomolgus macaques were primed with multiple plasmids encoding Ebola GP(Z) and GP(S/G). Eight or 54 weeks after the last DNA immunization NHP's were boosted with 1011 PFU of rAd5-GP vaccine. Blood sampling for anti-GP antibody and T-cell analysis was taken before and after the rAd5 boost. (B) Antigen-specific CD4+ and CD8+ T-cells analyed 4 weeks after the boost in the short interval or 4 weeks after the boost in the long interval. (C) Anti-GP antibody titers, pre boost (black), post boost (gray). Statistical analysis between two groups was performed using an unpaired student's t-test; **p < 0.01, ***p < 0.001; mean ± standard deviation.
Following vaccination with rAd5 Ebola GP, high quality Ebola-specific memory CD8+ T-cells secreting multiple cytokines are needed to confer protection against lethal challenge, and moreover, the protection in this vaccination regimen was found to be correlated specifically with IFNγ+TNFα+ CD8+ T-cells (42). In addition, a high antibody titer was found to be a quantitative predictor for rAd5 Ebola GP vaccine efficacy (43). Therefore, generating a large population of this specific high-quality memory CD8+ T-cell population is an essential goal. Based upon experience with licensed vaccines, multiple vaccinations as a prime-boost regimen is a feasible approach to rapidly generate a large population of memory CD8+ T-cells.
The initial immunization has a major influence on the nature of the immune response that follows the boost. One important variable that impacts how strongly the CD8+ T-cell population can be boosted is the length of time separating the primary and secondary antigen administrations. A time period of at least 40–60 days is required before optimal boosting of the CD8+ T-cell population is possible (49). A single DNA prime immunization before the rAd5 boost 8 weeks later resulted in the expansion of the CD8+ T-cell population (mainly IFNγ+TNFα+ cells that are known to correlate with protection against Ebola infection), while the CD4+ T-cell population was very small. In contrast, after three prime immunizations with DNA, we could not detect expansion of the CD8+ T-cells after the rAd5 GP boost, while the expansion of the CD4+ T-cell population could be observed. The reduction in the CD8+ T-cells observed after multiple DNA prime immunizations were mainly in the IFNγ+TNFα+ and IFNγ+ secreting cells. The increase in the CD4+ T-cell population after a single DNA prime rAd boost was mainly in the polyfunctional memory T-cells. The induction of high magnitude, high quality CD8+ T-cells is likely beneficial in the control of viral replication at its initiation and therefore might confer protection against lethal challenge. Thus, a single DNA prime followed by a rAd5-GP boost 8 weeks later is beneficial for the generation of CD8+ T-cells in a shorter time period than multiple DNA prime immunizations.
As shown in our work, DNA prime immunization resulted in low anti-GP antibody titers and tracked with the number of DNA primes. As a high antibody titer was found to be a surrogate marker for vaccine efficacy, this regimen by itself may not be sufficient for protection against Ebola. However, this prime generates humoral immunological memory that lasts for at least 3 years, even after a single DNA prime, and that memory could be activated by the rAd5-GP boost at any time. Thus, prime immunization with a single DNA vaccine followed by a rAd5-GP boost given when needed might be a useful approach for generating a polyfunctional effector CD8+ T-cell population for the rapid development of protective immunity that is mediated by CD8+ T-cells and antibodies.
The impact of the prime frequencies on the durable antibody response is interesting and is likely associated with memory B-cells. It was not surprising to observe a drop in Ag-specific IgG levels 130–160 weeks after the boost given that there was no additional exposure to antigen. It was, however, interesting that we observed a higher Ag-specific antibody response in animals vaccinated with a single DNA plus rAd5-GP vs. 3x DNA plus rAd5-GP 130–160 weeks post boost. The low Ag-specific IgG titers in the 3x DNA group could be explained by our observation that increasing time after multiple DNA primes favors CD8+ T-cell responses. In contrast, a single DNA plus rAd5-GP over time may favor CD4+ T-cells. Thus, since CD4+ T-cells support B-cell function, this may be the obvious explanation. Furthermore, the presence of specific anti-GP antibodies years after immunization is probably due to long lived plasma cells in survival niches. It might be that multiple DNA primes followed by rAd5-GP boost leads to a very high Ag-specific antibody response that results in B-cell exhaustion and elimination. The outcome might be a decreased number of long lived plasma cells leading to low levels of anti-GP antibodies. A single DNA immunization is probably insufficient for the generation of a substantial antibody response and long-lived plasma cells.
The CD8+ T-cells from all treatment groups showed dominance of IFNγ+TNFα+ and IFNγ+ IL2+TNFα+, with similar polyfunctional profiles and the major difference being found in the magnitude of the specific T-cell subpopulations. Thus, DNA prime regimens may not influence the maturation of the T-cells, but instead affect the expansion of the T-cell population, with the CD4+ T-cells expanded following multiple DNA primes at the expense of the CD8+ T-cells. The effect of the number of DNA primes on cellular and humoral immune responses suggests a unique and complex immune mechanism rather than simply impairing the expression of the GP protein by the rAd5-GP boost (Figure 7A). Our results revealed that compared to multiple DNA immunizations, a single DNA prime skewed toward CD8+ T-cell responses after the rAd boost. This observation is likely explained by the fact that adenoviral vectors are strong inducers of CD8+ T-cell responses, while a single DNA vaccination alone generates relatively weaker responses. Therefore, it is not surprising that a single DNA plus rAd5-GP boost is very similar in T-cell phenotype to that of a single rAd5-GP prime without a boost. Similarly, a long-time interval (1 year) between multiple DNA primes and the rAd5-GP boost allowed the robust rAd-induced CD8+ T-cells to dominate the DNA-induced memory CD4+ T-cells, and thus reversed the CD4+/CD8+ T-cell ratio observed following multiple DNA primes, with a short time interval before rAd5-GP boost (8 weeks), from a CD4+ T-cell dominance to that of a CD8+ T-cell dominance (Figure 7B). These observations could be explained by the immune response generated following the prime. If the immune response after the prime was not fully contracted, the response generated by the boost will be influenced by the prime. On the other hand, if the response generated by the prime was fully contracted and low, the boost may be efficiently boosting the primed (memory) response, and instead could more closely resemble a primary response from the “boost.” In essence, the strength and nature of the primary immunization could imprint particular qualities on the memory T-cells irrespective of the boosting agent. Additionally, the magnitude of T-cell reactivation could also have an impact on the quantity and quality of the memory T-cell response after the boost. There are several studies in which a prolonged time interval between the prime and the boost was found to be critical for the establishment of a memory response (50–53). Since CD8+ cells are important in the context of rAd5-GP-induced immune protection against Ebola virus infection, an immunization regimen that induces robust CD8+ cells may be beneficial.
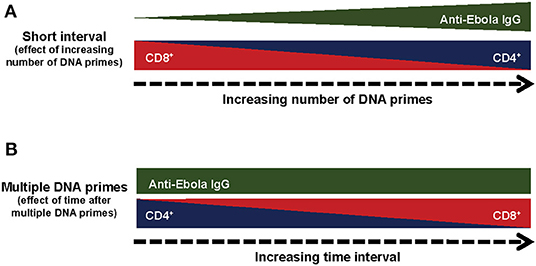
Figure 7. Summary of the immunological responses to different regiments of DNA prime rAd5-GP Boost. (A) Effect of the number of prime immunizations. (B) Effect of time after multiple DNA prime immunizations.
The data presented herein suggest it is possible to tailor immune responses and provide a framework for testing the relative contribution of skewed or balanced immune responses to immune protection against Ebola or other pathogens using infectious challenge models. Our study indicates that the DNA prime regimen, when reduced to a smaller number of injections, can contribute to the expansion of the CD8+ T-cells without changing their polyfunctional properties. The benefits of such a prime-boost regimen are the induction of a high magnitude antibody response, without sacrificing the CD8+ T-cell advantage induced after the rAd5-GP immunization, and in a much shorter time than the three DNA prime-rAd5 boost regimen. Thus, when considering the DNA prime rAd5 boost regimen, a single DNA prime will allow for the establishment of a robust and durable immunological response in a shorter period of time with the benefit of high magnitude antibody responses and CD8+ T-cell responses. The same approach could be applied for a condition in which the rapid generation of both antibodies and polyfunctional CD8+ T-cell responses are desired, either for vaccine design or treatment of human diseases.
While these studies were focused on Ebola vaccines, the results show that vaccine platforms can drive the relative balance of immune responses. Therefore, similar CD4+ to CD8+ phenotypes would be expected with a similar DNA/rAd5 vaccination strategy expressing a different protein, although this would have to be systematically evaluated. A vaccination strategy requiring four administrations to drive specific immune response ratios has pragmatic limitations, but the data provide proof of principle that vaccination strategies can be tailored to induce CD4+ or CD8+ T-cell responses depending on the desired outcome.
The vaccine vectors used in this study have been described previously (41). DNA and replication-defective rAd5 GP vectors were cloned and purified as described previously (54).
Animal experiments were conducted in full compliance with all relevant federal guidelines and NIH policies. All animal experiments were conducted under protocols approved by NIH Animal Care and Use committees. Female cynomolgus macaques (Macaca fascicularis) 3–5 years of age and weighing between 2–3 kg were obtained from Covance for all studies. Monkeys were housed individually and given enrichment regularly as recommended by the Guide for the Care and Use of Laboratory Animals (DHEW number NIH 86-23). Animals were anesthetized with ketamine prior to blood sampling or vaccination. Each vaccination group in this study contained three or four cynomolgus macaques.
DNA immunizations were administered to cynomolgus macaques to both deltoids by Biojector intramuscular injection with a mixture of two milligrams each of two plasmid vectors encoding the glycoprotein from Zaire ebolavirus Mayinga strain, GP(Z), and the glycoprotein from Sudan ebolavirus Gulu strain, GP(S/G). One to four DNA immunizations were administered as indicated in each experiment. Following the final DNA priming immunization, at a time indicated for each group subjects received a boost immunization with 1011 particle units (PU) of rAd5 encoding the glycoprotein from Zaire ebolavirus Mayinga strain, GP(Z). The boost was performed by intramuscular injection in the deltoids by needle and syringe containing the vectors.
Anti-EBOV GP IgG ELISA titers were measured as described previously (55). Transmembrane-deleted EBOV GP (EBOV GPΔTM) was generated by calcium phosphate-mediated transient transfection of 293T cells using the Promega ProFection® Mammalian Transfection System and the plasmid VRC6008 [pVR1012-GP(Z) delta Tm]. 293T cells were plated in complete media [high glucose DMEM (ThermoFisher Scientific, Waltham, MA) containing 10% fetal bovine serum (Gemini Bio, Sacramento, CA)] 14–18 h prior to transfection. Two to three hours before transfection the media was removed and replaced with fresh complete media. The cells were transfected at 30–60% confluency. Eight hours post transfection, the media was once again replaced with complete media. Twenty-four hours post transfection, the media was replaced with serum-free media (DMEM without FBS). The cell culture supernatant was harvested 36 h later, centrifuged, and filtered using 0.22 mm filters. This cell culture supernatant served as the antigen. Polyvinyl chloride enzyme-linked immunosorbent assay (ELISA) plates (Nunc, Rochester, NY) were coated with 100 μl of antigen per well and incubated at 4°C overnight. Subsequent incubations were performed at room temperature. Plates were washed with PBS containing Tween 20 after antigen coating. Test sera were serially diluted and added to the antigen-coated wells for 60 min. The plates were washed followed by incubation with the detection antibody, goat anti-human IgG (H+L; SouthernBiotech/Millipore, Billerica, MA) conjugated to horseradish peroxide. SigmaFast o-phenylenediamine dihydrochloride (Sigma, St. Louis, MO) substrate was added to the wells, and the optical density was determined (450 nm). A positive control serum sample from a single animal with a known EBOV GP IgG response was run every time the assay was performed. Background-subtracted ELISA titers are expressed as EC90, reciprocal optical density values, which represent the dilution at which there is a 90% decrease in antigen binding.
Intracellular cytokine staining was performed as described previously (44). After the 6 h stimulation with EBOV GP peptides or DMSO control, PBMC were stained with a mixture of antibodies against lineage markers [CD3-Cy7-APC clone SP34-2 (BD Biosciences), CD4-QD605 clone M-T477 (BD Biosciences), CD8-PerCP Cy5.5 clone RPA-T8, CD95 Cy5-PE clone DX2 (BD Biosciences), CD28 Alexa 488 clone 28.2 (Biolegend), CD45RA QD655 clone 5H3] at room temperature for 20 min. After two washes the cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) followed by staining with antibodies against cytokines: tumor necrosis factor (TNF)α-APC clone MAb11 (BD Biosciences), interleukin (IL)-2 PE clone MQ17H12 (BD Biosciences), and IFNγ PE-Cy7 clone B27. The viability dye ViViD (Invitrogen) was included to allow discrimination between live and dead cells (56). EBOV-specific cytokine positive cells were defined as a percentage within CD4+ and CD8+ T-cell memory subsets (CD95+CD45RAhi and CD95+CD45RAlo) secreting any of the three cytokines measured (IFNγ IL-2 or TNFα) above DMSO background. Cells were acquired on BD-LSR II cytometer collecting up to 1,000,000 total events, resulting in >100,000 events in the CD4/CD8 T-cell gates. Typically, this yields >10 total events in the cytokine gates of the antigen-stimualted samples. Samples were analyzed using FACSDiva and analyzed with FlowJo 9.4.9 (Tree Star, Inc.) and SPICE software (57).
Comparison of anti-GP ELISA IgG titers and intracellular cytokine production by T-cell memory subsets was performed using an unpaired two-tailed Student's t-test for comparisons between two groups or one-way ANOVA with Tukey's post hoc test for comparisons between more than two groups in GraphPad Prism version 9 software.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by VRC ACUC.
HM and NS designed these studies. HM, ET, CA, MD, KF, and DS executed experiments. NS and CA wrote animal study protocols. MB and CA executed vaccination. HM, YZ, JM, and NS wrote the manuscript. MR provided scientific support. All authors contributed to the article and approved the submitted version.
This work was supported by the Intramural Research Program of the Vaccine Research Center, the National Institute of Allergy and Infectious Diseases, and the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the Vaccine Research Center's Non-human Primate Immunogenicity Core and the Flow Cytometry Core for NHP sample processing. We thank Ruth Hunegnaw, Ph.D. for her careful review of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.627688/full#supplementary-material
1. Barouch DH, Craiu A, Santra S, Egan MA, Schmitz JE, Kuroda MJ, et al. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J Virol. (2001) 75:2462–7. doi: 10.1128/JVI.75.5.2462-2467.2001
2. Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. (2000) 290:486–92. doi: 10.1126/science.290.5491.486
3. Doria-Rose NA, Ohlen C, Polacino P, Pierce CC, Hensel MT, Kuller L, et al. Multigene DNA priming-boosting vaccines protect macaques from acute CD4+-T-cell depletion after simian-human immunodeficiency virus SHIV89.6P mucosal challenge. J Virol. (2003) 77:11563–77. doi: 10.1128/JVI.77.21.11563-11577.2003
4. Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. (2000) 18:927–74. doi: 10.1146/annurev.immunol.18.1.927
5. Kong WP, Huang Y, Yang ZY, Chakrabarti BK, Moodie Z, Nabel GJ. Immunogenicity of multiple gene and clade human immunodeficiency virus type 1 DNA vaccines. J Virol. (2003) 77:12764–72. doi: 10.1128/JVI.77.23.12764-12772.2003
6. Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev. (2011) 239:62–84. doi: 10.1111/j.1600-065X.2010.00980.x
7. Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines. (2012) 11:189–209. doi: 10.1586/erv.11.188
8. Zhou F, Wang G, Buchy P, Cai Z, Chen H, Chen Z, et al. A tri-clade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of HPAI H5N1 viruses. J Virol. (2012) 86:6970–8. doi: 10.1128/JVI.06930-11
9. Amara RR, Smith JM, Staprans SI, Montefiori DC, Villinger F, Altman JD, et al. Critical role for Env as well as Gag-pol in control of a simian-human immunodeficiency virus 89.6P challenge by a DNA prime/recombinant modified vaccinia virus Ankara vaccine. J Virol. (2002) 76:6138–46. doi: 10.1128/JVI.76.12.6138-6146.2002
10. Amara RR, Villinger F, Altman JD, Lydy SL, O'Neil SP, Staprans SI, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. (2001) 292:69–74. doi: 10.1126/science.1058915
11. Chege GK, Thomas R, Shephard EG, Meyers A, Bourn W, Williamson C, et al. A prime-boost immunisation regimen using recombinant BCG and Pr55(gag) virus-like particle vaccines based on HIV type 1 subtype C successfully elicits Gag-specific responses in baboons. Vaccine. (2009) 27:4857–66. doi: 10.1016/j.vaccine.2009.05.064
12. Hanke T, Samuel RV, Blanchard TJ, Neumann VC, Allen TM, Boyson JE, et al. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J Virol. (1999) 73:7524–32. doi: 10.1128/JVI.73.9.7524-7532.1999
13. Subbramanian RA, Kuroda MJ, Charini WA, Barouch DH, Costantino C, Santra S, et al. Magnitude and diversity of cytotoxic-T-lymphocyte responses elicited by multiepitope DNA vaccination in rhesus monkeys. J Virol. (2003) 77:10113–8. doi: 10.1128/JVI.77.18.10113-10118.2003
14. Vuola JM, Keating S, Webster DP, Berthoud T, Dunachie S, Gilbert SC, et al. Differential immunogenicity of various heterologous prime-boost vaccine regimens using DNA and viral vectors in healthy volunteers. J Immunol. (2005) 174:449–55. doi: 10.4049/jimmunol.174.1.449
15. Wilks AB, Christian EC, Seaman MS, Sircar P, Carville A, Gomez CE, et al. Robust vaccine-elicited cellular immune responses in breast milk following systemic simian immunodeficiency virus DNA prime and live virus vector boost vaccination of lactating rhesus monkeys. J Immunol. (2010) 185:7097–106. doi: 10.4049/jimmunol.1002751
16. Yang ZY, Wyatt LS, Kong WP, Moodie Z, Moss B, Nabel GJ. Overcoming immunity to a viral vaccine by DNA priming before vector boosting. J Virol. (2003) 77:799–803. doi: 10.1128/JVI.77.1.799-803.2003
17. Fiedler M, Kosinska A, Schumann A, Brovko O, Walker A, Lu M, et al. Prime/boost immunization with DNA and adenoviral vectors protects from hepatitis D virus (HDV) infection after simultaneous infection with HDV and woodchuck hepatitis virus. J Virol. (2013) 87:7708–16. doi: 10.1128/JVI.00645-13
18. Hayes P, Gilmour J, von Lieven A, Gill D, Clark L, Kopycinski J, et al. Safety and immunogenicity of DNA prime and modified vaccinia ankara virus-HIV subtype C vaccine boost in healthy adults. Clin Vaccine Immunol. (2013) 20:397–408. doi: 10.1128/CVI.00637-12
19. Lin SC, Lin YF, Chong P, Wu SC. Broader neutralizing antibodies against H5N1 viruses using prime-boost immunization of hyperglycosylated hemagglutinin DNA and virus-like particles. PLoS ONE. (2012) 7:e39075. doi: 10.1371/journal.pone.0039075
20. Bakari M, Aboud S, Nilsson C, Francis J, Buma D, Moshiro C, et al. Broad and potent immune responses to a low dose intradermal HIV-1 DNA boosted with HIV-1 recombinant MVA among healthy adults in Tanzania. Vaccine. (2011) 29:8417–28. doi: 10.1016/j.vaccine.2011.08.001
21. Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204). PLoS ONE. (2011) 6:e21225. doi: 10.1371/journal.pone.0021225
22. Koblin BA, Casapia M, Morgan C, Qin L, Wang ZM, Defawe OD, et al. Safety and immunogenicity of an HIV adenoviral vector boost after DNA plasmid vaccine prime by route of administration: a randomized clinical trial. PLoS ONE. (2011) 6:e24517. doi: 10.1371/journal.pone.0024517
23. Chege GK, Burgers WA, Stutz H, Meyers AE, Chapman R, Kiravu A, et al. Robust immunity to an auxotrophic Mycobacterium bovis BCG-VLP prime-boost HIV vaccine candidate in a nonhuman primate model. J Virol. (2013) 87:5151–60. doi: 10.1128/JVI.03178-12
24. Lin SC, Liu WC, Lin YF, Huang YH, Liu JH, Wu SC. Heterologous prime-boost immunization regimens using adenovirus vector and virus-like particles induce broadly neutralizing antibodies against H5N1 avian influenza viruses. Biotechnol J. (2013) 8:1315–22. doi: 10.1002/biot.201300116
25. Lorin C, Vanloubbeeck Y, Baudart S, Ska M, Bayat B, Brauers G, et al. Heterologous prime-boost regimens with a recombinant chimpanzee adenoviral vector and adjuvanted F4 protein elicit polyfunctional HIV-1-specific T-Cell responses in macaques. PLoS ONE. (2015) 10:e0122835. doi: 10.1371/journal.pone.0122835
26. Swadling L, Capone S, Antrobus RD, Brown A, Richardson R, Newell EW, et al. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci Transl Med. (2014) 6:261ra153. doi: 10.1126/scitranslmed.3009185
27. Delany I, Rappuoli R, De Gregorio E. Vaccines for the 21st century. EMBO Mol Med. (2014) 6:708–20. doi: 10.1002/emmm.201403876
28. Pinschewer DD. Virally vectored vaccine delivery: medical needs, mechanisms, advantages and challenges. Swiss Med Wkly. (2017) 147:w14465. doi: 10.4414/smw.2017.14465
29. Gudmundsdotter L, Nilsson C, Brave A, Hejdeman B, Earl P, Moss B, et al. Recombinant modified vaccinia Ankara (MVA) effectively boosts DNA-primed HIV-specific immune responses in humans despite pre-existing vaccinia immunity. Vaccine. (2009) 27:4468–74. doi: 10.1016/j.vaccine.2009.05.018
30. Kibuuka H, Kimutai R, Maboko L, Sawe F, Schunk MS, Kroidl A, et al. A phase 1/2 study of a multiclade HIV-1 DNA plasmid prime and recombinant adenovirus serotype 5 boost vaccine in HIV-Uninfected East Africans (RV 172). J Infect Dis. (2010) 201:600–7. doi: 10.1086/650299
31. Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. (2003) 77:6305–13. doi: 10.1128/JVI.77.11.6305-6313.2003
32. Casimiro DR, Wang F, Schleif WA, Liang X, Zhang ZQ, Tobery TW, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with dna and recombinant adenoviral vaccine vectors expressing Gag. J Virol. (2005) 79:15547–55. doi: 10.1128/JVI.79.24.15547-15555.2005
33. Geisbert TW, Bailey M, Geisbert JB, Asiedu C, Roederer M, Grazia-Pau M, et al. Vector choice determines immunogenicity and potency of genetic vaccines against Angola Marburg virus in nonhuman primates. J Virol. (2010) 84:10386–94. doi: 10.1128/JVI.00594-10
34. Jaoko W, Karita E, Kayitenkore K, Omosa-Manyonyi G, Allen S, Than S, et al. Safety and immunogenicity study of multiclade HIV-1 adenoviral vector vaccine alone or as boost following a multiclade HIV-1 DNA vaccine in Africa. PLoS ONE. (2010) 5:e12873. doi: 10.1371/journal.pone.0012873
35. Koup RA, Roederer M, Lamoreaux L, Fischer J, Novik L, Nason MC, et al. Priming immunization with DNA augments immunogenicity of recombinant adenoviral vectors for both HIV-1 specific antibody and T-cell responses. PLoS ONE. (2010) 5:e9015. doi: 10.1371/journal.pone.0009015
36. De Rosa SC, Thomas EP, Bui JY, Huang deCamp A, Morgan C, Kalams SA, et al. HIV-DNA priming alters T cell responses to HIV-adenovirus vaccine even when responses to DNA are undetectable. J Immunol. (2011) 187:3391–401. doi: 10.4049/jimmunol.1101421
37. Vojnov L, Bean AT, Peterson EJ, Chiuchiolo MJ, Sacha JB, Denes FS, et al. DNA/Ad5 vaccination with SIV epitopes induced epitope-specific CD4 T cells, but few subdominant epitope-specific CD8 T cells. Vaccine. (2011) 29:7483–90. doi: 10.1016/j.vaccine.2011.07.048
38. Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, et al. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis. (2011) 203:610–9. doi: 10.1093/infdis/jiq105
39. Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, et al. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine. (2012) 30:1737–45. doi: 10.1016/j.vaccine.2011.12.026
40. Centers for Disease Control and Prevention. Outbreaks Chronology: Ebola Virus Disease. Atlanta, GA (2018).
41. Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. Development of a preventive vaccine for Ebola virus infection in primates. Nature. (2000) 408:605–9. doi: 10.1038/35046108
42. Sullivan NJ, Hensley L, Asiedu C, Geisbert TW, Stanley D, Johnson J, et al. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat Med. (2011) 17:1128–31. doi: 10.1038/nm.2447
43. Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat Rev Microbiol. (2009) 7:393–400. doi: 10.1038/nrmicro2129
44. Stanley DA, Honko AN, Asiedu C, Trefry JC, Lau-Kilby AW, Johnson JC, et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nat Med. (2014) 20:1126–9. doi: 10.1038/nm.3702
45. Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. (2008) 8:247–58. doi: 10.1038/nri2274
46. Zhou Y, Sullivan NJ. Immunology and evolvement of the adenovirus prime, MVA boost Ebola virus vaccine. Curr Opin Immunol. (2015) 35:131–6. doi: 10.1016/j.coi.2015.06.006
47. Hensley LE, Mulangu S, Asiedu C, Johnson J, Honko AN, Stanley D, et al. Demonstration of cross-protective vaccine immunity against an emerging pathogenic Ebolavirus species. PLoS Pathog. (2010) 6:e1000904. doi: 10.1371/journal.ppat.1000904
48. Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. (2007) 13:843–50. doi: 10.1038/nm1592
49. Nolz JC, Harty JT. Strategies and implications for prime-boost vaccination to generate memory CD8 T cells. Adv Exp Med Biol. (2011) 780:69–83. doi: 10.1007/978-1-4419-5632-3_7
50. Fournillier, Frelin L, Jacquier E, Ahlén G, Brass A, Gerossier E, et al. A heterologous prime/boost vaccination strategy enhances the immunogenicity of therapeutic vaccines for hepatitis C virus. J Infect Dis. (2013) 208:1008–19. doi: 10.1093/infdis/jit267
51. Khurana S, Wu J, Dimitrova M, King LR, Manischewitz J, Graham BS, et al. DNA priming prior to inactivated influenza A(H5N1) vaccination expands the antibody epitope repertoire and increases affinity maturation in a boost-interval-dependent manner in adults. JID. (2013) 208:413–17. doi: 10.1093/infdis/jit178
52. Ledgerwood JE, Wei CJ, Hu Z, Gordon IJ, Enama ME, Hendel CS, et al. DNA priming and influenza vaccine immunogenicity: two phase 1 open label randomised clinical trials. Lancet Infect Dis. (2011) 11:916–24. doi: 10.1016/S1473-3099(11)70240-7
53. Ledgerwood JE, Zephir K, Hu Z, Wei CJ, Chang L, Enama ME, et al. Prime-boost interval matters: a randomized phase 1 study to identify the minimum interval necessary to observe the H5 DNA influenza vaccine priming effect. J Infect Dis. (2013) 208:418–22. doi: 10.1093/infdis/jit180
54. Aoki K, Barker C, Danthinne X, Imperiale MJ, Nabel GJ. Efficient generation of recombinant adenoviral vectors by Cre-lox recombination in vitro. Mol Med. (1999) 5:224–31. doi: 10.1007/BF03402119
55. Geisbert TW, Bailey M, Hensley L, Asiedu C, Geisbert J, Stanley D, et al. Recombinant adenovirus serotype 26 (Ad26) and Ad35 vaccine vectors bypass immunity to Ad5 and protect nonhuman primates against ebolavirus challenge. J Virol. (2011) 85:4222–33. doi: 10.1128/JVI.02407-10
56. Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. (2006) 313:199–208. doi: 10.1016/j.jim.2006.04.007
Keywords: vaccine, Ebola, prime, boost, CD8+, CD4+, T-cell, non-human primates
Citation: Marcus H, Thompson E, Zhou Y, Bailey M, Donaldson MM, Stanley DA, Asiedu C, Foulds KE, Roederer M, Moliva JI and Sullivan NJ (2021) Ebola-GP DNA Prime rAd5-GP Boost: Influence of Prime Frequency and Prime/Boost Time Interval on the Immune Response in Non-human Primates. Front. Immunol. 12:627688. doi: 10.3389/fimmu.2021.627688
Received: 10 November 2020; Accepted: 19 February 2021;
Published: 09 March 2021.
Edited by:
Neeltje van Doremalen, Rocky Mountain Laboratories (NIAID), United StatesReviewed by:
Axel T. Lehrer, University of Hawaii at Manoa, United StatesCopyright © 2021 Marcus, Thompson, Zhou, Bailey, Donaldson, Stanley, Asiedu, Foulds, Roederer, Moliva and Sullivan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nancy J. Sullivan, bnN1bGxpdmFuQG5paC5nb3Y=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.