
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 22 June 2021
Sec. Inflammation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.624753
The pathological mechanism underlying heat stroke (HS) is associated with the dysbalanced inflammation and coagulation cascade. Cell-derived circulating extracellular vesicles (EVs), as a novel pathway mediating intercellular communication, are associated with the immune response and inflammation in critical inflammatory syndromes, such as sepsis. Although these vesicles contain genetic material correlated with their biological function, their molecular cargo during HS remains unknown. In this study, we evaluate the presence of microRNAs (miRNAs) and messenger RNAs (mRNAs) associated with inflammatory responses and coagulation cascade in exosomes of patients with HS. Blood samples were collected from three patients with HS at the time of admission to the intensive care unit; three healthy volunteers were selected as control. Exosomes were isolated using ultracentrifugation, and their miRNA content was profiled using next-generation sequencing; mRNA content was evaluated using qPCR array. Compared with those from healthy volunteers, exosomes from patients with HS showed substantial changes in the expression of 202 exosomal miRNAs (154 upregulated and 48 downregulated miRNAs). The most upregulated miRNAs included miR-511-3p, miR-122-5p, miR-155-3p, miR-1290, and let7-5p, whereas the most downregulated ones included miR-150-3p, 146a-5p, and 151a-3p. Gene ontology enrichment of the miRNAs of patients with HS compared with control subjects were associated mostly with inflammatory response, including T cell activation, B cell receptor signaling, dendritic cell chemotaxis and leukocyte migration, and platelet activation and blood coagulation. The identified miRNAs were primarily enriched to the signal transduction pathways namely, T cell receptor signaling, Ras signaling, chemokine signaling, platelet activation, and leukocyte transendothelial migration, all of which are associated with inflammation and hemostasis. Multiple targeted mRNAs associated with the inflammatory response, blood coagulation, and platelet activation were further verified in serum exosomes. Exosomes from patients with HS convey miRNAs and mRNAs associated with pathogenic pathways, including inflammatory response and coagulation cascade. Exosomes may represent a novel mechanism for intercellular communication during HS.
Heatstroke (HS) is a severe condition with limited treatment options that can even be fatal. Recently, HS incidence has increased due to the growing frequency of heatwaves; the resultant mortality rates have also increased, particularly HS-induced multiple organ failure, with a mortality rate of 40–60% (1, 2).
The molecular mechanism underlying HS pathogenesis is complicated and poorly investigated (3). Evidence has suggested that the HS mechanism is associated with dysbalanced inflammation and coagulation cascade (4). Cell-derived circulating extracellular vesicles (EVs) shed from different cell types during acute inflammatory syndromes such as sepsis. The roles of EVs in disease pathophysiology have been extensively investigated, especially those associated with immune responses and inflammation (5, 6). In addition, we have previously demonstrated that hepatocyte-derived EVs from patients with HS correlate with liver dysfunction, which is associated with activation of EV-induced inflammatory NOD-like receptor pathway and necrosis in hepatocytes (7, 8).
EVs can potentially mediate cell communication under normal physical conditions or disease state via functional microRNAs (miRNAs) and messenger RNA (mRNA). A pilot study has suggested, using pathway enrichment analysis, that the miRNA profile in serum EVs from sepsis patients might have had significant roles during sepsis (9). The authors also reported that the differential expression pattern of EV-miRNA in sepsis modulates the immune system and cell cycle. However, no existing study has yet examined the genetic exosomal content in HS.
This study analyzes the changes in the miRNA profiles in serum EVs and their potential biological functions in patients with HS. The results of this study provide insights into the pathophysiology of HS and the development of novel treatment strategies.
From June 2019 to June 2020, six patients admitted within the first 24 h of severe HS onset into the ICU at the General Hospital of South Theatre of People’s Liberation Army (PLA) of China, a tertiary hospital, were retrospectively enrolled in this study. HS was diagnosed according to the Expert Consensus on Diagnosis and Treatment of Heat Stroke in China (10) released by the Expert Group on Prevention and Treatment of Heat Stroke and Critical Care Committee of PLA of China. Patients with active malignant tumors, chronic liver and kidney diseases, chronic cardiac insufficiency (New York grades 3–4), chronic pulmonary insufficiency, underlying central nervous system (CNS) disease, metabolic disorders, or those using heparin or any other medications, were eliminated from this study. Additionally, three healthy individuals from the Physical Examination Center were enrolled as the control group. Baseline demographic characteristics were recorded upon admission to ICU (day 1). Blood samples and organ function data were collected on day 1; besides, the sequential organ failure assessment (SOFA) and the age and chronic health evaluation (APACHE II) scores were determined to evaluate the disease severity. Patients or the corresponding family members provided informed consent. Our study protocol was approved by the Medical Ethics Review Committee of General Hospital of Southern Theater of PLA of China.
A total of 30 ml peripheral venous blood sample was collected from each patient using the ethylenediaminetetraacetic acid (EDTA) anticoagulant tubes (the first few 5 ml was discarded). Next, the blood collection tubes were left standing vertically for 30 min at 22–27°C. The plasma was obtained from whole blood following centrifugation for 10 min at 4°C and 2,500 × g. After the addition of protease inhibitors (supplemented with 3 mM phenylmethylsulfonyl fluoride, 1 µg/ml pepstatin, as well as 1 µg/ml aprotinin), the plasma was stored at −80°C until further use. When processing, plasma was diluted with equivalent volume of phosphate-buffered saline (PBS) and centrifuged at 4°C (30 min at 2,500 × g, 45 min at 12,000 × g, and 2 h at 110,000 × g, SW 28 Ti Rotor, Optima L-90K Ultracentrifuge, Beckman Coulter, Fullerton, USA). Finally, 50–200 µl PBS was used to resuspend the precipitate (exosomes) and was stored in −80°C.
Exosome samples were collected from a healthy individual, a patient with mild HS, and a patient with severe HS (randomly selected) and prepared for TEM evaluation. Firstly, exosomes were dehydrated in 2% formalin. After adding 5 ml exosome suspension into the copper grid coated with formvar (Mecalab, QC, Canada), the sample was incubated for 30 min, at 4°C, washed with 100 µl PBS, fixed with 2% paraformaldehyde for 10 min, and stained for 15 min with 2% uranyl acetate (dissolved into 50% ethanol). Thereafter, the Philips CM10 transmission electron microscope (JEM-2100F, Netherlands) was utilized for sample visualization.
For detecting exosome content and size distribution, NTA was performed using the NanoSight NS3000 (Malvern Instruments, Worcestershire, UK). In brief, exosome sample was diluted with sterile PBS at 1:5,000, and the NanoSight LM10 and NTA software (NanoSight Ltd, Amesbury, UK) was employed for sample analysis, thrice (60 s each).
Proteins were extracted from 1 ml ultracentrifuged plasma (120,000 × g for 2.5 h) using the DE buffer (consisting of 20 mM Tris-HCL, 12 mM 2-mercaptoethanol, 1 mM EGTA, 1 mM EDTA, 1% Triton-X 100, together with 10% glycerol) that contained the protease inhibitor mix (GE Healthcare, Uppsala, Sweden). Total proteins were quantified using Bradford, then 5 µg protein from each sample was separated on a 12% SDS-PAGE. The protein bands were then transferred onto nitrocellulose membranes and blotted using polyclonal antibodies against CD9 (1:1,000 diluted, Abcam ab10895, Cambridge, UK), CD63 (1:1,000 diluted, Abcam ab92726, Cambridge, UK), and Tsg-101 (1:1,000 diluted, Abcam ab30871, Cambridge, UK) at 4°C, overnight. GAPDH was used as a loading control. The target protein expression level was quantified by densitometry analysis using the ImageJ software.
TRIzol reagent was used to extract the RNA from exosomes.
The NEB SmallRNA Library kit was adopted for library construction, whereas Illumina HiSeq was applied for sequencing, with a data volume of 20 M reads per sample.
Clean data were filtered from raw reads after sequencing in accordance with the following criteria: a) 30% base quality <20; b) read length <17 bp; c) adaptor sequence. Clean data were then mapped to the Danio Rerio miRNA database (miRBase v21.0) together with the Danio Rerio genome (Zv10, NCBI) using the BWA software, whereas the unmapped reads were used to map the rat, mouse, and human miRNA database (miRBase v21.0) to achieve the expression profiles of miRNAs.
The novel miRNA prediction was applied using mirdeep based on the reads mapped to Zv10 genome but unmapped against the Danio Rerio miRNA database (miRBase v21.0) to identify new miRNA. The BWA software mapping was applied in measuring the expression of new miRNA (11).
Pair-end read mapping. Before read mapping, adaptor sequences, low-quality reads with over 20% bases that had <20 quality and those having over 5% ambiguous bases (N) were eliminated from raw reads to obtain clean reads. The zebrafish genome (version: Zv10) was used to align clean reads by the Tophat program (12).
The DESeq2 package was adopted to identify miRNA and mRNA with differential expression according to the numbers of miRNA and mRNA, FDR, and P-value (13), with the following selection criteria: i) Fold-Change (FC) >2 or <0.5; ii) P-value <0.05 and FDR <0.05.
Miranda target analysis (14) was adopted for predicting the miRNA target in the mRNA of zebrafish. To identify the possible miRNA target, targets with no negative association of mRNA with miRNA were excluded.
GO analysis was conducted to identify the roles of specific genes in typical or significant differentially expressed gene (DEG) profiles (15). The GO annotations were first downloaded from Gene Ontology (http://www.geneontology.org/), UniProt (http://www.uniprot.org/), along with NCBI (http://www.ncbi.nlm.nih.gov/) databases. Thereafter, significant GO categories were identified using Fisher’s exact test, whereas P-values were corrected using FDR.
Pathway enrichment analysis was also applied to identify DEGs based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Significant pathways were selected using Fisher’s exact test, with FDR and P-value satisfying significance thresholds (16).
The Statminer v5 software (Integromics, Granada, Spain) was used to analyze the cycle threshold (Ct) data. According to the manufacturer’s instructions, Ct values ≥32 were excluded from immune and miRNA studies. For the OS studies, the cutoff value was set at 35 in line with the manufacturer’s instructions. For the miRNA analysis, the optimal reference was identified using the geNorm approach, and the median miR17, miR20a, and miR106a were applied to normalize the data. The 18S rRNA was used to normalize the immune gene expression, and beta-actin was used to normalize OS gene expression. Finally, the 2DDCt method was adopted to calculate the relative gene expression.
The data normality was assessed using the Kurtosis test. Data with normal distribution were expressed as mean ± standard deviation (SD), whereas those with abnormal distribution were expressed as median ± interquartile range. The two-tailed Student’s t-test was used to compare two groups, whereas one-way analysis of variance followed by post hoc test was used to compare multiple groups. A P-value of <0.05 (two-tailed) was considered statistically significant, whereas GraphPad version 7.0 (La Jolla, CA, USA) was used for all tests.
All patients and healthy controls were males with low comorbidities. Ages were comparable between healthy controls and patients with HS (25 ± 6 vs. 21 ± 4 years, respectively). No participant has previously used any medication. Clinical and laboratory data are summarized in Table 1. Core body temperatures on admission were higher in patients with HS (39.00 ± 1.94 vs. 36.42 ± 0.46°C, P < 0.001). In addition, three out of the six patients with HS had died during the ICU hospitalization. The patients remained at ICU for 5–10 (median, 7) days.
All severe HS cases were complicated with multiple organ dysfunction. There was significant organ support when patients were enrolled because 67% of patients received mechanical ventilation, whereas 50% received norepinephrine. The overall disease severity, as assessed using the APACHE II and SOFA scores, was 17.05 ± 4.35 and 10.95 ± 5.64, respectively (Table 2). None of the enrolled patients received prophylactic heparin, or transfusion of red blood cells/platelet/fresh frozen plasma, or antibiotics on admission before the collection of study samples.
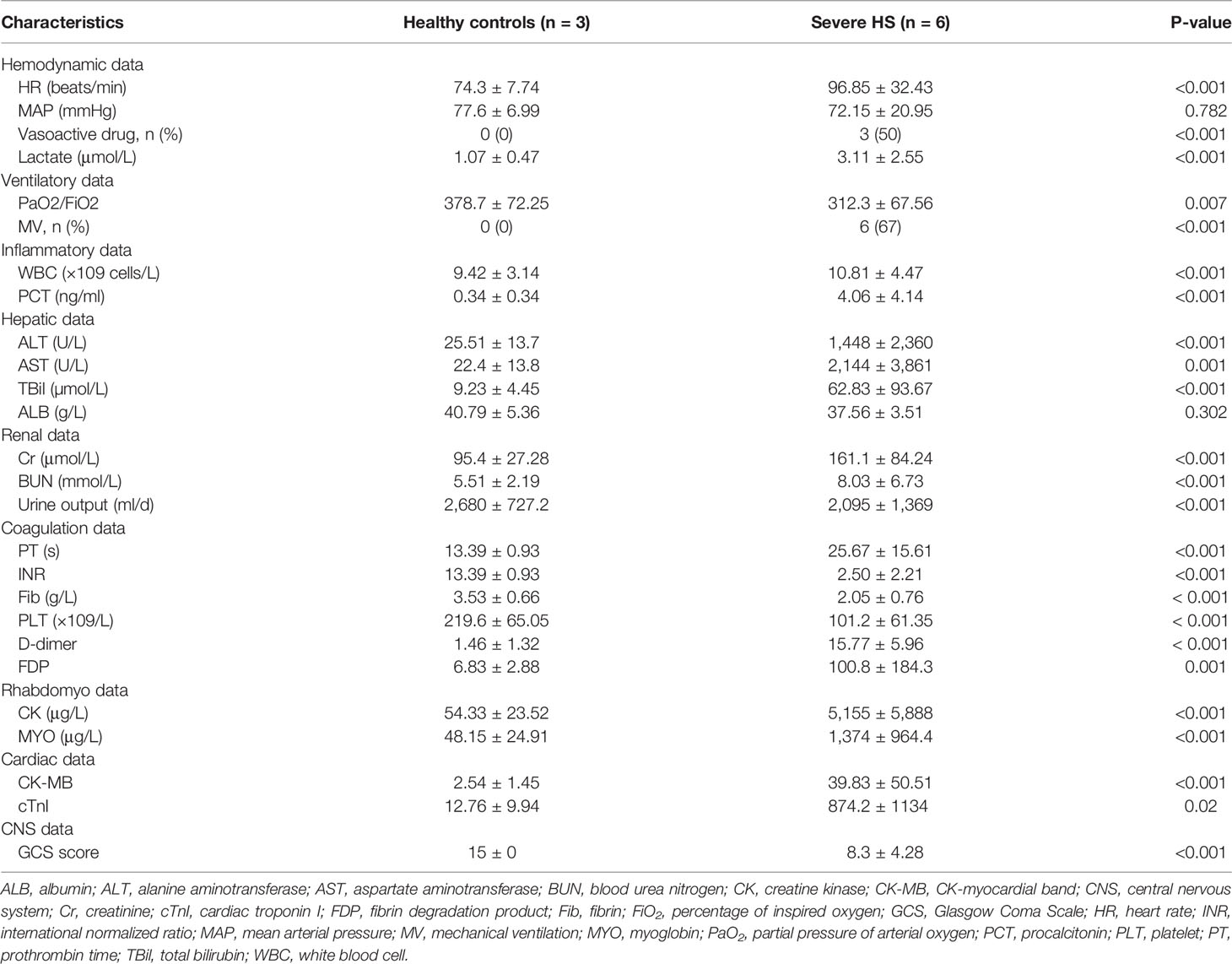
Table 2 Comparison of clinical and laboratory indices of patients with heat stroke and healthy controls according to the day of admission and outcome status.
TEM showed that the separated preparation from the plasma of healthy controls and patients with HS exhibited a double-membrane vesicle-like structure approximately 100 nm in diameter (Figure 1A). Besides, the NTA results indicated that the plasma exosome concentration of the HS group increased compared with that of the control group (5.67 ± 2.34 × 109 vs. 0.47 ± 0.15 × 109 particles/ml, respectively; Figure 1B). Finally, western blotting analysis showed the positive expression of the membrane-associated proteins related to endocytosis, as well as the exosomal markers (CD9, CD63, and Tsg-101) in both control and HS groups (Figure 1C). Taken together, the extracted vesicles were mostly consistent with exosomes.
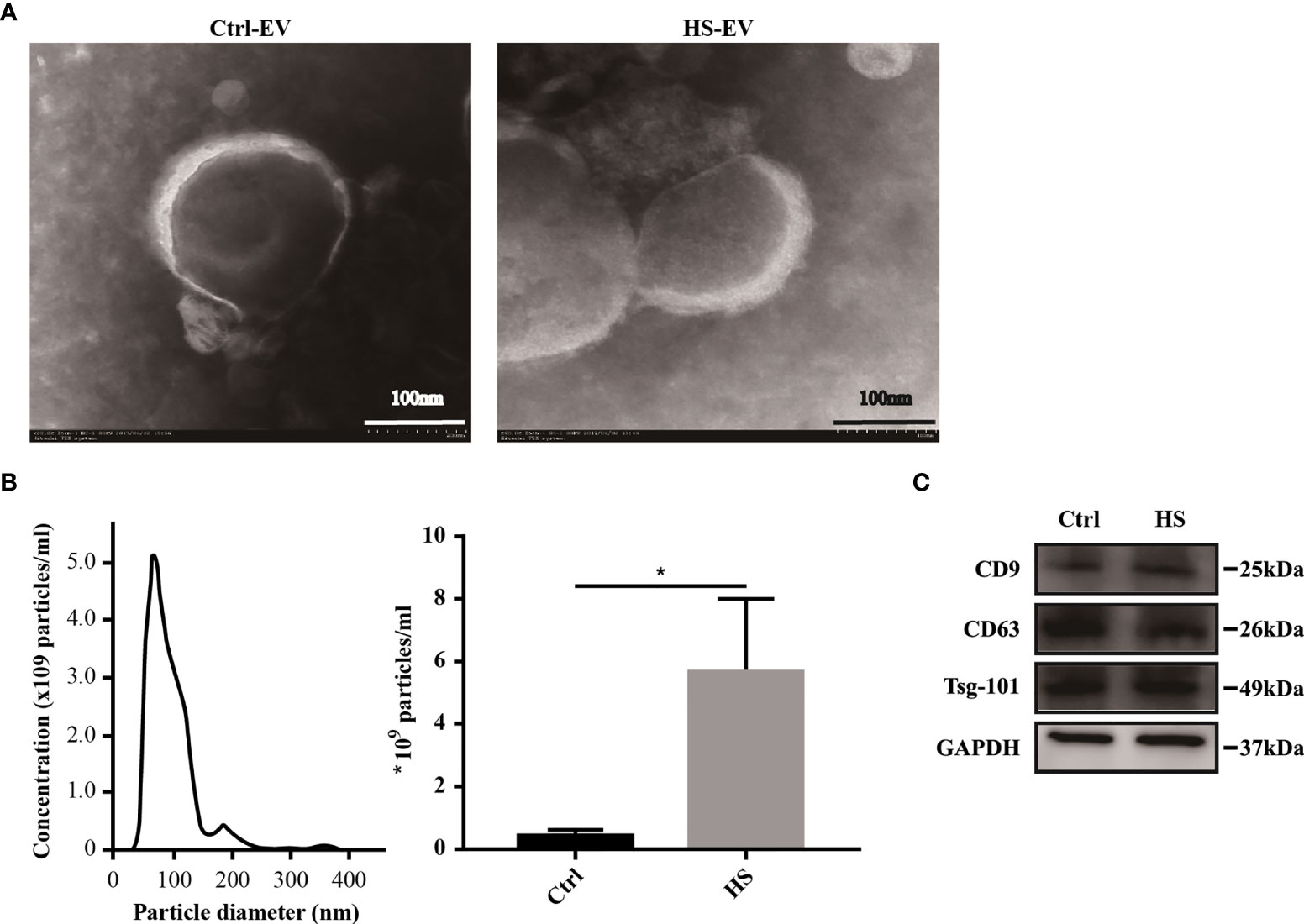
Figure 1 Characterization of plasma exosomes in both healthy controls and HS patients. (A) Morphology of plasma exosomes visualized under TEM. Bar, 100 nm. (B) Exosome size distribution examined through NTA. (*) p < 0.05. (C) Levels of representative exosomal surface markers (CD9, CD63 along with Tsg-101) using western blot analysis. All experiments were repeated thrice.
To determine whether HS changed exosomal miRNA expression, the miRNA profiling analysis was conducted firstly among the HS cases admitted to ICU, and the results were compared against the healthy controls. HS had no impact on 90.72% of miRNA accumulation (Figure 2A). Figure 2B shows that differences in most identified miRNAs were not significant due to the close abundance ratio of HS versus control EVs. Nonetheless, in patients with HS, 202 miRNAs (9.28%) were identified as differentially expressed compared with controls [154 upregulated (7.08%) and 48 downregulated (2.21%)] (Figure 2A). The miRNAs expression FC in HS versus control EVs (ratio of HS to control group <0.5 or >2), along with the P-values are summarized in Table 1 and displayed in Figure 2B.
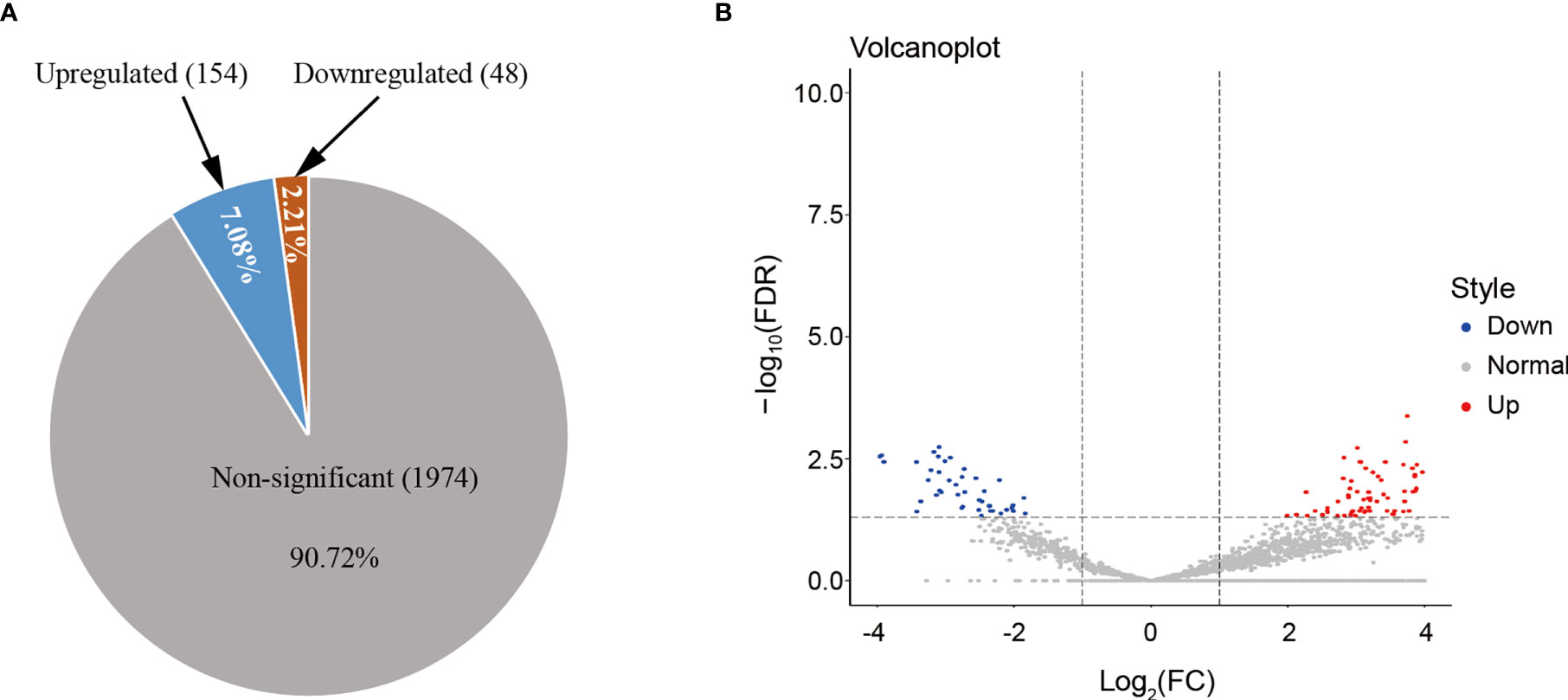
Figure 2 (A) HS altered the miRNA expression patterns of hepatocyte-derived EVs, as analyzed using NGS. The number and proportion of differentially regulated (upregulated, downregulated, and non-significant) EV miRNAs isolated from HS compared with the control (n = 3 samples/group). Numbers indicate proportions. Data base: Homo Sapiens. (B) Volcano plot representing the miRNA fold-change and RNA intensity in the HS and control hepatocyte EVs. X-axis shows miRNA fold-change (HS/Control, log2), whereas Y-axis depicted the summary of RNA intensity (LFQ intensity, lg).
Additionally, hierarchical cluster analysis was conducted on 202 differentially expressed miRNAs based on samples or stimulants to examine the creditability of those selected target miRNAs (Figure 3). There was significant differential expression in HS patients versus controls, with low SD in three samples, which suggested that our results were highly repeatable. Some specific miRNAs with significant elevation within HS EVs relative to controls were identified, including miR-511-3p, miR-122-5p, miR-155-3p, miR-1290, and let7-5p. The most downregulated miRNAs included miR-150-3p, 146a-5p, and 151a-3p. The ACC IDs of all the dis-regulated miRNAs and the corresponding FCs are presented in Table S1. The results suggested that, HS changed certain miRNA levels within plasma EVs, and this was correlated with the biological effect.
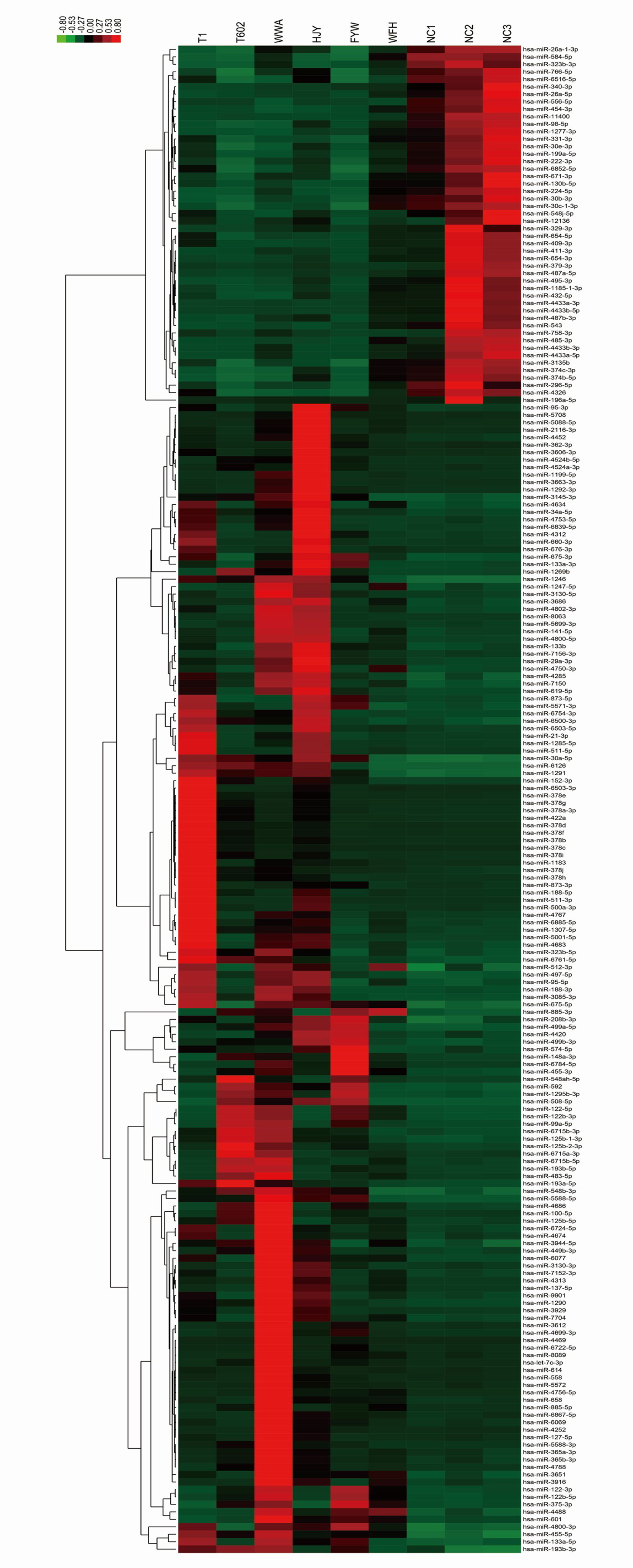
Figure 3 Cluster analysis of the dysregulated miRNA components from HS EVs. The list of all 202 significantly differentially expressed miRNAs (HS-EV group/control EV group) has been shown in heat map.
Thereafter, each EV miRNA identified using NGS was classified to determine the specific biological role using GO functional annotation clustering approach. Most identified EV miRNAs were involved in regulating the signaling and molecular functions. Therefore, the targeted miRNAs may influence the EVs’ biological activity.
The 20 most significant biological terms related to the upregulated or downregulated miRNAs in HS EVs, as well as the miRNA enrichment significance determined for all clusters, are illustrated in Figure 4. Particularly, inflammatory responses including T cell activation, B cell receptor signaling, dendritic cell chemotaxis and leukocyte migration, and platelet activation and blood coagulation, as well as cell responses to various stimuli including cytokines, regulation of cell death and cell adhesion/migration, and cell cycle were the most significantly upregulated clusters and were all involved in the mechanism of HS.
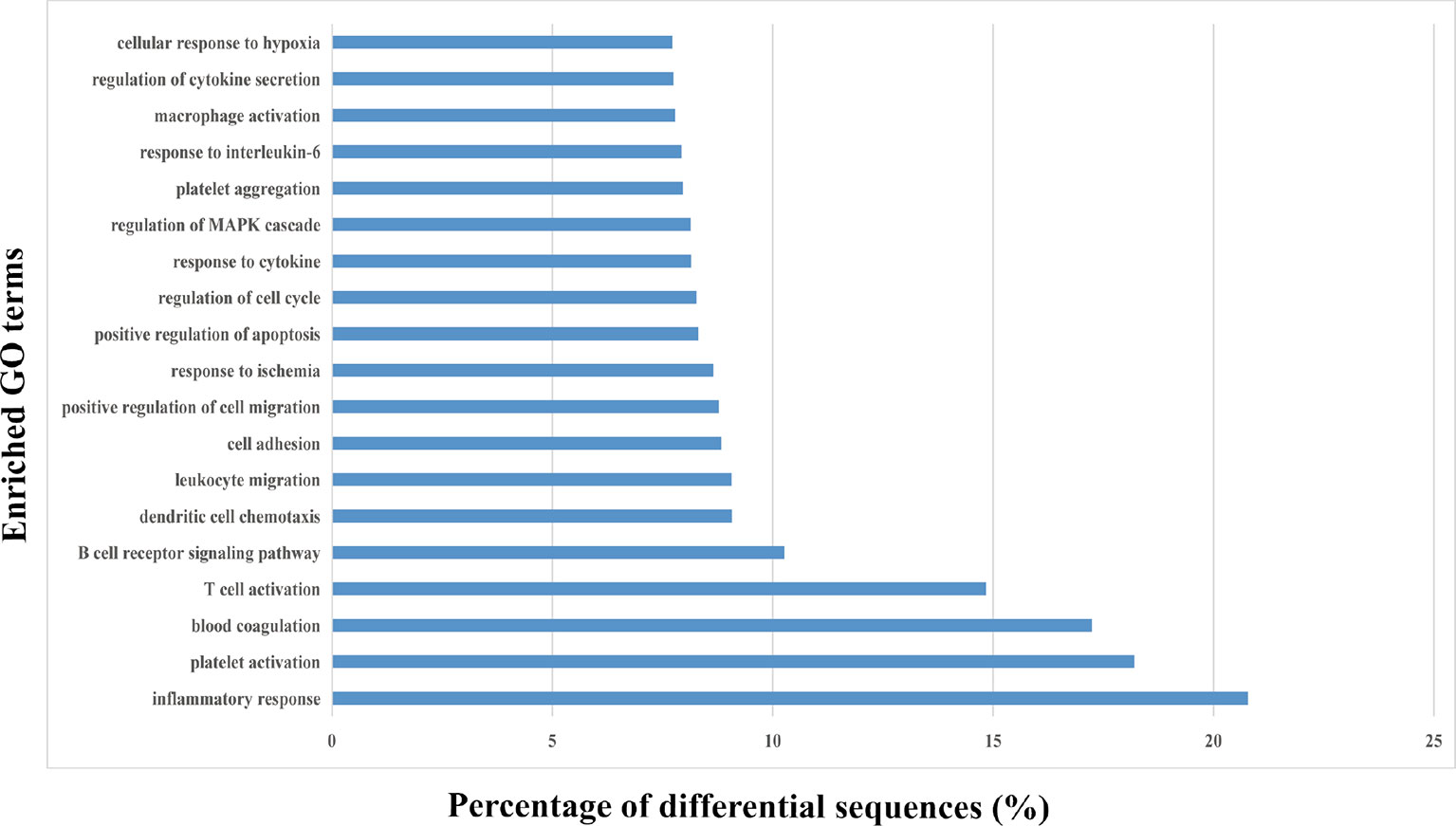
Figure 4 Gene ontology (GO) enrichment analysis of the differentially regulated proteins. The top 20 enriched terms according to the GO functional annotation clustering of the 202 differentially expressed miRNAs in the heat-stroked hepatocyte-derived EVs. Percentages of the sequences involved are shown.
KEGG enrichment helps to determine the miRNA enrichment significance of every pathway. In this study, KEGG analysis indicated that those identified miRNAs were primarily enriched to the signal transduction pathways namely, T cell receptor signaling, Ras signaling, chemokine signaling, platelet activation, and leukocyte transendothelial migration (Figure 5). These pathways are mainly associated with inflammation and hemostasis. Thus, circulating EVs may be associated with HS-induced injury via the activation of these signal transduction pathways within target cells.
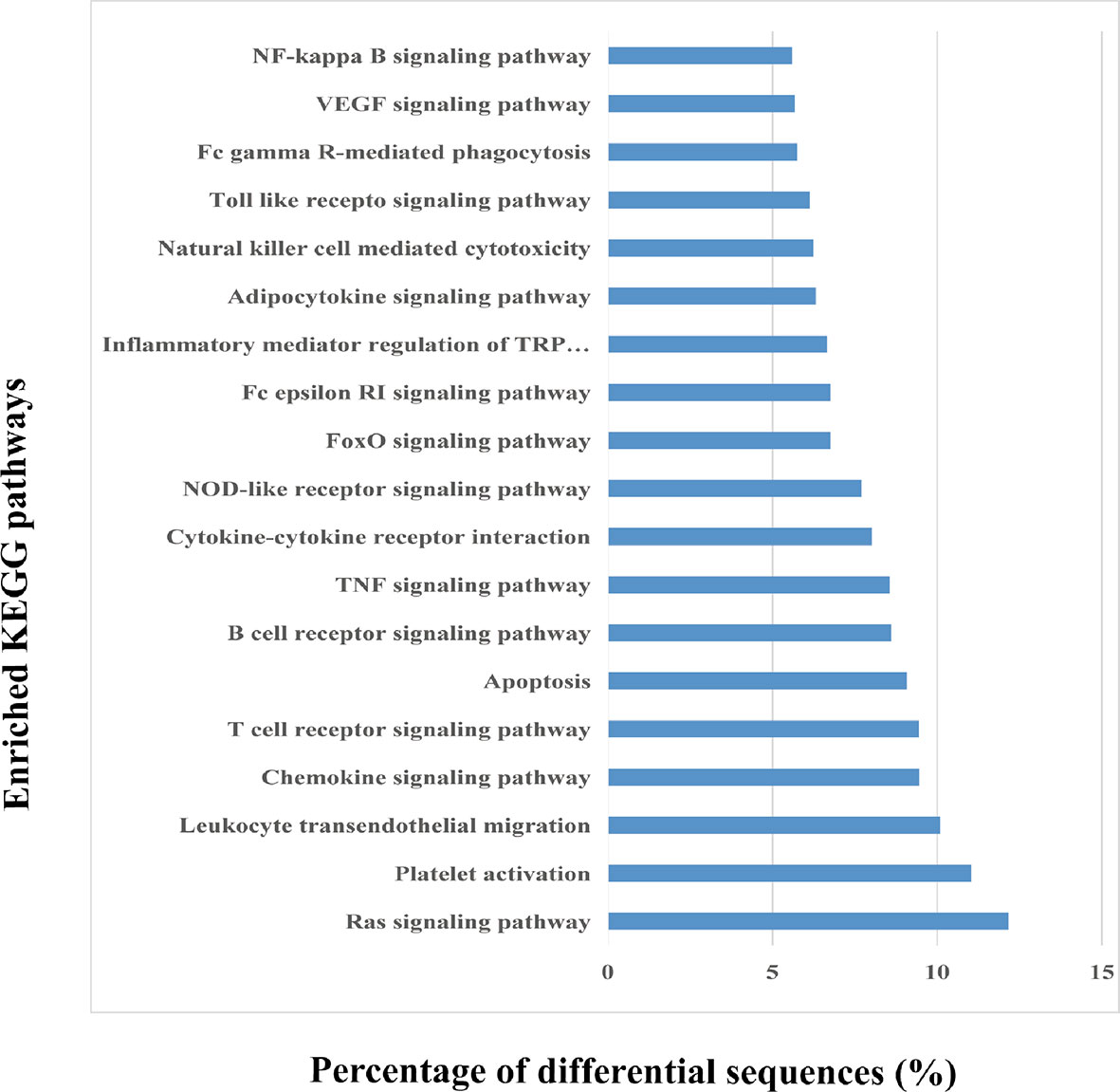
Figure 5 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the differentially expressed EV miRNAs. The top 20 enriched KEGG pathways in the heat-stroked hepatocyte-derived EV-miRNA and the percentages of the sequences involved in each pathway are shown.
Tables S2–S4 list the targeted mRNAs associated with the top three upregulated clusters, inflammatory response, blood coagulation, and platelet activation.
For assessing the presence or absence of immune response-related mRNAs within exosomes, qPCR was performed to compare the inflammation/immune-related gene expression levels based on RNA extracted from patients with HS and healthy individuals. A total of 18 genes, including PRKD1, SP100, IL25, CRLF2, NLRP2, AOAN, and CXCR1 were significantly upregulated in patients with HS (P < 0.05; Figure 6A).
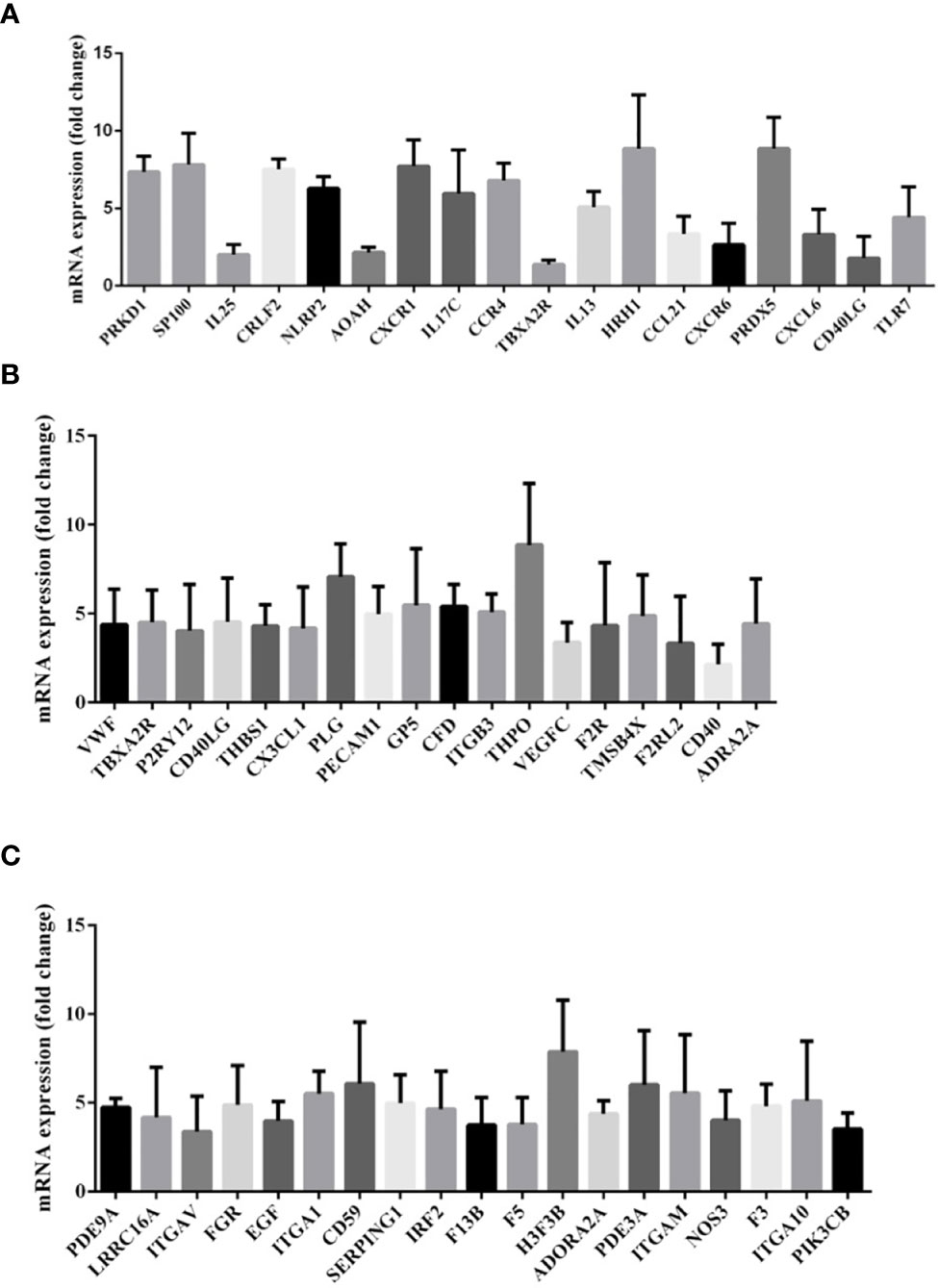
Figure 6 (A) Gene expression of messenger RNA (mRNA) related to inflammatory response in the comparison between patients with heat stroke versus healthy control subjects. (B) Gene expression of messenger RNA (mRNA) related to platelet activation in the comparison between patients with heat stroke versus healthy control subjects. (C) Gene expression of messenger RNA (mRNA) related to blood coagulation in the comparison between patients with heat stroke versus healthy control subjects.
As illustrated in Figure 6B, the platelet activation-related mRNA expression levels in exosomes like VWF, TBXA2R, P2RY12, CD40LG, and THBS1 in patients with HS were also upregulated in comparison with those in healthy controls.
We also investigated the expression of mRNAs associated with blood coagulation and demonstrated an increased expression of exosomal mRNAs of PDE9A, LRRC16A, ITGAV, FCR, EGF, ITGA1, CD59, SERPING1, and IRF2 (Figure 6C).
Putative pathways, including chemokine signaling, platelet activation, T cell receptor signaling, and leukocyte transendothelial migration, were constructed based on KEGG mapping (Figure 7).
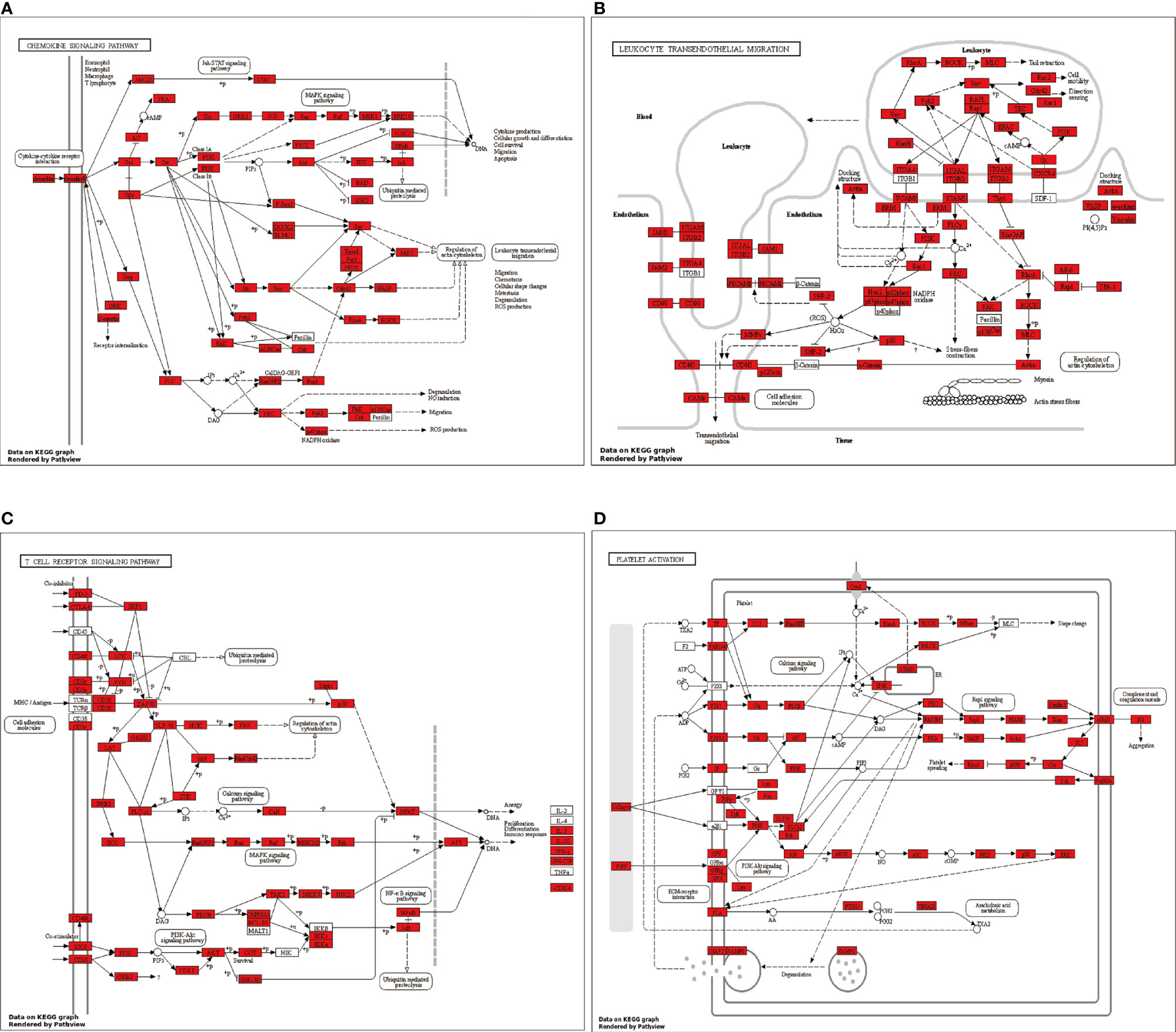
Figure 7 Putative (A) chemokine signaling pathway, (B) platelet activation pathway, (C) T cell receptor signaling pathway, and (D) leukocyte transendothelial migration pathway were constructed based on KEGG mapping. Red squares indicate proteins identified as differentially expressed miRNAs; white circles indicate miRNAs not identified as differentially expressed in our study.
In the present study, the different circulating exosome-miRNA expression profiles with the corresponding targets involved in the immune and coagulation system were firstly identified in patients with HS.
The organ injury mechanism during HS remains unknown. The current consensus is that an inflammatory and coagulation response initiated by the hyperthermal injury may play a more important role than the direct physical damage from heat exposure (3). The pathological findings of HS (core temperature attained 42.5–43°C) baboon models include massive transmural leukocyte migration, widespread microthrombosis, microvascular endothelium injury, endothelial leukocyte–platelet interaction, and extensive apoptosis in multiple organs including the spleen, gut, liver, kidney, and lung (4).
As demonstrated in the present study, the enriched miRNAs conveyed in circulating exosomes may have participated in the inflammatory response and coagulation cascade, suggesting that exosomal miRNAs may potentially mediate HS-associated damage. Data on the presence of miRNAs in exosomes during HS are limited. We have previously studied exosomes derived from human vascular endothelial cells exposed to 41°C hyperthermal stress (17). Gene expression profiles in the lung tissue of an HS rat model showed that the miRNAs associated with inflammatory/immune responses, including leukocyte migration, lipopolysaccharide response, NIK/NF-kappaB signaling, and response to reactive oxygen species, were upregulated, which is in agreement with our findings (18). Whole blood mRNA and microRNA have been identified as biomarkers of tissue damage and immune function resulting from HS (19).
In other acute critical inflammatory diseases, miRNAs are involved in the immune/inflammatory responses during sepsis (20). In one study, miRNA profiling analysis on ten subjects with polytrauma, sepsis, and septic shock identified specific differentially expressed miRNAs (21). Furthermore, Real et al. (9) enrolled 24 patients with septic shock and reported that the circulating exosomal miRNAs were significantly altered, and the same expression kinetics was maintained during the course of the disease. These results suggest similar physiological responses as those in severe inflammatory injury.
miR-150 is one of the earliest detected miRNAs in critically ill patients or those with sepsis. Through microarray gene expression profiling, miR-150 has been reported as a part of the miRNA panels with abnormal expression within leukocytes/PBMC obtained from sepsis patients compared to normal subjects (22–24). Such alterations are reflected by consistent changes in the serum miR-150 expression. For instance, Vasilescu et al. (22) discovered that miR-150 expression was downregulated in 16 cases with abdominal sepsis. The reduced miR-150 expression in serum was associated with increased SOFA scores and sepsis severity. Furthermore, Ma et al. (25) discovered that miR-150 expression was downregulated in sepsis patients compared with that with non-infectious systemic inflammatory response syndrome or normal subjects.
miRNA-122 was one of the most significantly enriched miRNAs in our profiling. Changes in its serum concentration significantly increase systemic inflammatory diseases such as sepsis, which is associated with the inflammatory response level. Therefore, miRNA-22 is a reliable biomarker for early disease stratification and prognostic evaluation (26). The level of miRNA-122 can predict the degree of inflammation and organ injury, such as acute respiratory distress syndrome (27) and coagulation disorders (28). According to the longitudinal samples obtained from sepsis patients, the miR-122 level was significantly increased on day 14 upon ICU admission, which strongly correlated with antithrombin III (R = 0.913, P < 0.001) (26). To determine the direct or indirect impacts of miRNAs on coagulation, the crosstalk between cytokines, miRNAs dysregulation, and thrombocyte synthesis/apoptosis should be further investigated.
miRNA-155 was also among the most upregulated miRNAs in our study. In a heat stress cell model, microRNA-155 promoted heat stress-induced inflammatory responses in the microglia by facilitating inflammatory factor expression by increasing NF-κB pathway activation via targeting liver X receptor α (29).
Little is known regarding the interactions between exosomal miRNAs and the corresponding targets. It has been proposed that exosomes can act as effective carriers of genetic materials and proteins to the surrounding or distant, cells. Nonetheless, the exosome–recipient cell interaction mechanism remains unknown. Such interaction generally begins with the internalization of exosomes through various pathways, such as phagocytosis, clathrin-mediated endocytosis, as well as macropinocytosis (30). The exosomal miRNAs and mRNAs in recipient cells have specific functions and may interact with related targets to synthesize novel proteins or modulating gene expression levels (31). However, the interaction of exosomal genetic contents with the target cells in HS remains speculative since most data available are obtained in vitro.
This study has some limitations. First, the number of patients enrolled was small, and the results obtained were interpreted as hypothesis-generating. The incidence of severe HS is relatively low though is increasing due to the climate change. Actually, only about 10 patients with severe HS was admitted into our ICU per year. In the further study, we will consider to expand the sample size. Additionally, only cases at the first HS episode were enrolled in this study to prevent subjects with immunosuppression or chronic inflammation from possibly affecting our results, and hence may limit the generalizability of our results. The plasma exosome counts were not determined either in patients with HS or normal controls; thus, it was impossible to estimate the impacts of HS on exosome number. The contents of exosomal miRNAs and mRNAs were not normalized relative to the plasma exosome amount. Moreover, only mRNAs associated with coagulation activation and inflammatory response were assessed, since they have been suggested by previous studies to regulate exosomes in HS. Using such method, it was feasible to assess additional pathophysiological targets for exosomal mRNAs. To compare the effects between infectious and non-infectious injuries, the control group comprising infected ICU cases was not enrolled in this study. Finally, no functional or mechanistic miRNA studies were taken. We did not perform additional experiments to identify the specific roles of these differentially expressed miRNAs; thus, these markers need to be further explored in the future. Any of these miRNAs may be a potential target in the treatment of HS-induced injury. Time-course and thoroughly step-by-step pathway study can be employed to clarify these issues.
Exosomes from patients with HS convey genetic material that may be associated with key pathways in the pathogenesis of HS, including inflammatory response, blood coagulation, and platelet activation. Further functional studies are warranted to identify the exact contribution of these vesicles to the exchange of genetic material and intercellular communication during HS.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/ (accession number: GSE174224).
All animal experiments were conducted in compliance with the criteria outlined in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 86-23, 1985 revision) and were approved by the Animal Care and Use Committee of the General Hospital of Guangzhou Military Command. Blood sample collection from HS patients was approved by the Ethics Committee of the General Hospital of Guangzhou Military Command. Informed consent was obtained from the patients or their representatives.
HT supervised the complete study. YL designed the complete study and performed the NTA, western blotting, statistical analysis, as well as manuscript writing. QW, BL, and HL performed the isolation of exosomes and data collection. XW and HC performed the TEM. LS performed the clinical data and blood sample collection. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (grant No. 81671896), Military Medical Innovation Project (18CXZ032), and Natural Science Foundation of Guangdong Province (2019A1515012088).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Mr. Panweilun for the technical assistance of NTA. We would like to also thank the Guangzhou Branch of Chinese Academy of Life Sciences for technical assistance of TEM.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.624753/full#supplementary-material
1. Hifumi T, Kondo Y, Shimizu K, Miyake Y. Heat Stroke. J Intensive Care (2018) 6:30. doi: 10.1186/s40560-018-0298-4
2. Epstein Y, Yanovich R. Heatstroke. N Engl J Med (2019) 380(25):2449–59. doi: 10.1056/NEJMra1810762
3. Bouchama A, Roberts G, Al Mohanna F, El-Sayed R, Lach B, Chollet-Martin S, et al. Inflammatory, Hemostatic, and Clinical Changes in a Baboon Experimental Model for Heatstroke. J Appl Physiol (2005) 98(2):697–705. doi: 10.1152/japplphysiol.00461.2004
4. Roberts GT, Ghebeh H, Chishti MA, Al-Mohanna F, El-Sayed R, Al-Mohanna F, et al. Microvascular Injury, Thrombosis, Inflammation, and Apoptosis in the Pathogenesis of Heatstroke: A Study in Baboon Model. Arterioscler Thromb Vasc Biol (2008) 28(6):1130–6. doi: 10.1161/ATVBAHA.107.158709
5. Reid VL, Webster NR. Role of Microparticles in Sepsis. Br J Anaesth (2012) 109(4):503–13. doi: 10.1093/bja/aes321
6. Terrasini N, Lionetti V. Exosomes in Critical Illness. Crit Care Med (2017) 45(6):1054–60. doi: 10.1097/CCM.0000000000002328
7. Li Y, Zhu X, Zhang M, Tong H, Su L. Heatstroke-Induced Hepatocyte Exosomes Promote Liver Injury by Activating the NOD-Like Receptor Signaling Pathway in Mice. PeerJ (2019) 7:e8216. doi: 10.7717/peerj.8216
8. Li Y, Zhu X, Wang G, Tong H, Su L, Li X. Proteomic Analysis of Extracellular Vesicles Released From Heat-Stroked Hepatocytes Reveals Promotion of Programmed Cell Death Pathway. BioMed Pharmacother (2020) 129:110489. doi: 10.1016/j.biopha.2020.110489
9. Real JM, Ferreira LRP, Esteves GH, Koyama FC, Dias MVS, Bezerra-Neto JE, et al. Exosomes From Patients With Septic Shock Convey miRNAs Related to Inflammation and Cell Cycle Regulation: New Signaling Pathways in Sepsis? Crit Care (2018) 22(1):68. doi: 10.1186/s13054-018-2003-3
10. Expert Group on Prevention and Treatment of Heat Stroke and Critical Care Committee of PLA of China. Expert Consensus on Diagnosis and Treatment of Heat Stroke in China. Med J Chin People’s Liberation Army (2019) 44:181–97. doi: 10.1186/s40779-019-0229-2
11. An J, Lai J, Lehman ML, Nelson CC. miRDeep*: An Integrated Application Tool for miRNA Identification From RNA Sequencing Data. Nucl Acids Res (2013) 41(2):727–37. doi: 10.1093/nar/gks1187
12. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments With TopHat and Cufflinks. Nat Protoc (2012) 7(3):562–78. doi: 10.1038/nprot.2012.016
13. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the False Discovery Rate in Behavior Genetics Research. Behav Brain Res (2001) 125(1–2):279–84. doi: 10.1016/s0166-4328(01)00297-2
14. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA Targets. PloS Biol (2004) 2(11):e363. doi: 10.1371/journal.pbio.0020363
15. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat Genet (2000) 25(1):25–9. doi: 10.1038/75556
16. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A Systems Biology Approach for Pathway Level Analysis. Genome Res (2007) 17(10):1537–45. doi: 10.1101/gr.6202607
17. Chen HS, Tong HS, Zhao Y, Hong CY, Bin JP, Su L. Differential Expression Pattern of Exosome Long Non-Coding RNAs (lncRNAs) and microRNAs (miRNAs) in Vascular Endothelial Cells Under Heat Stroke. Med Sci Monit (2018) 24:7965–74. doi: 10.12659/MSM.909983
18. Permenter MG, McDyre BC, Ippolito DL, Stallings JD. Alterations in Tissue microRNA After Heat Stress in the Conscious Rat: Potential Biomarkers of Organ-Specific Injury. BMC Genomics (2019) 20(1):141. doi: 10.1186/s12864-019-5515-6
19. Camacho L, Silva CS, Hanig JP, Schleimer RP, George NI, Bowyer JF. Identification of Whole Blood mRNA and microRNA Biomarkers of Tissue Damage and Immune Function Resulting From Amphetamine Exposure or Heat Stroke in Adult Male Rats. PloS One (2019) 14(2):e0210273. doi: 10.1371/journal.pone.0210273
20. Essandoh K, Fan GC. Role of Extracellular and Intracellular microRNAs in Sepsis. Biochim Biophys Acta Mol Basis Dis (2014) 1842(11):2155–62. doi: 10.1016/j.bbadis.2014.07.021
21. Uhlich RM, Konie JA, Davis JW, Misfeldt ML, Nelson C, Calaluce R, et al. Novel microRNA Correlations in the Severely Injured. Surgery (2014) 156(4):834–40. doi: 10.1016/j.surg.2014.06.017
22. Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, et al. MicroRNA Fingerprints Identify miR-150 as a Plasma Prognostic Marker in Patients With Sepsis. PloS One (2009) 4(10):e7405. doi: 10.1371/journal.pone.0007405
23. How CK, Hou SK, Shih HC, Huang MS, Chiou SH, Lee CH, et al. Expression Profile of microRNAs in Gram-Negative Bacterial Sepsis. Shock (2015) 43(2):121–7. doi: 10.1097/SHK.0000000000000282
24. Zhou J, Chaudhry H, Zhong Y, Ali MM, Perkins LA, Owens WB, et al. Dysregulation in microRNA Expression in Peripheral Blood Mononuclear Cells of Sepsis Patients Is Associated With Immunopathology. Cytokine (2015) 71(1):89–100. doi: 10.1016/j.cyto.2014.09.003
25. Ma Y, Vilanova D, Atalar K, Delfour O, Edgeworth J, Ostermann M, et al. Genome-Wide Sequencing of Cellular microRNAs Identifies a Combinatorial Expression Signature Diagnostic of Sepsis. PloS One (2013) 8(10):e75918. doi: 10.1371/journal.pone.0075918
26. Wang H, Yu B, Deng J, Jin Y, Xie L. Serum miR-122 Correlates With Short-Term Mortality in Sepsis Patients. Crit Care (2014) 18(6):704. doi: 10.1186/s13054-014-0704-9
27. Rahmel T, Schäfer ST, Frey UH, Adamzik M, Peters J. Increased Circulating microRNA-122 Is a Biomarker for Discrimination and Risk Stratification in Patients Defined by Sepsis-3 Criteria. PloS One (2018) 13(5):e0197637. doi: 10.1371/journal.pone.0197637
28. Wang HJ, Deng J, Wang JY, Zhang PJ, Xin Z, Xiao K, et al. Serum miR-122 Levels Are Related to Coagulation Disorders in Sepsis Patients. Clin Chem Lab Med (2014) 52(6):927–33. doi: 10.1515/cclm-2013-0899
29. Li P, Wang G, Zhang XL, He GL, Luo X, Yang J, et al. MicroRNA-155 Promotes Heat Stress-Induced Inflammation Via Targeting Liver X Receptor α in Microglia. Front Cell Neurosci (2019) 13:12. doi: 10.3389/fncel.2019.00012
30. Fernández-Messina L, Gutiérrez-Vázquez C, Rivas-García E, Sánchez-Madrid F, de la Fuente H. Immunomodulatory Role of microRNAs Transferred by Extracellular Vesicles. Biol Cell (2015) 107(3):61–77. doi: 10.1111/boc.201400081
Keywords: heat stroke, exosome, miRNA, next-generation sequencing, inflammation, coagulation
Citation: Li Y, Wen Q, Chen H, Wu X, Liu B, Li H, Su L and Tong H (2021) Exosomes Derived From Heat Stroke Cases Carry miRNAs Associated With Inflammation and Coagulation Cascade. Front. Immunol. 12:624753. doi: 10.3389/fimmu.2021.624753
Received: 03 November 2020; Accepted: 19 May 2021;
Published: 22 June 2021.
Edited by:
Beate E. Kehrel, University Hospital Münster, GermanyReviewed by:
Philip Brandon Busbee, University of South Carolina, United StatesCopyright © 2021 Li, Wen, Chen, Wu, Liu, Li, Su and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huasheng Tong, ZmltbXV0aHNAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.