- 1Department of Rheumatology, Endocrinology and Nephrology, Faculty of Medicine, Hokkaido University, Sapporo, Japan
- 2Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
T cell metabolism is central to cell proliferation, survival, differentiation, and aberrations have been linked to the pathophysiology of systemic autoimmune diseases. Besides glycolysis and fatty acid oxidation/synthesis, amino acid metabolism is also crucial in T cell metabolism. It appears that each T cell subset favors a unique metabolic process and that metabolic reprogramming changes cell fate. Here, we review the mechanisms whereby amino acid transport and metabolism affects T cell activation, differentiation and function in T cells in the prototype systemic autoimmune disease systemic lupus erythematosus. New insights in amino acid handling by T cells should guide approaches to correct T cell abnormalities and disease pathology.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by autoantibody production, immune complex deposition, tissue inflammation and damage of multiple organs (1). SLE can affect practically all organs, including skin, kidney, and central nerve system (2–4). The etiology of SLE is multifactorial and includes contributions from genetic, environmental, hormonal and epigenetic factors (2). These factors, acting serially or simultaneously, lead to generalize breakdown of tolerance to self-antigens, which results in autoantibody production and tissue inflammation (5). T cells have a vital role in the pathogenesis of SLE. Many subsets of T cells, especially Th1, Th17, regulatory T (Treg) cells, and double-negative (CD4-CD8-) T cells, are involved through distinct mechanisms in the development of organ inflammation in SLE (6). Since helper T cells can activate B cells to secrete antibodies, which are also involved in the lupus pathogenesis, T cells have earned claim as main therapeutic targets in patients with SLE (7).
Recent studies have shown that the differentiation and function of each T cell subset is controlled by intracellular metabolic processes (8–10). Cell metabolism operates mainly through glycolysis, fatty acid oxidation and amino acid metabolism including glutaminolysis (8–11). Amino acids are classified as essential (leucine, isoleucine, lysine, histidine, valine, threonine, phenylalanine, tryptophan, and methionine), conditionally essential (glutamine, arginine, cysteine, glycine, proline, and tyrosine), or non-essential (alanine, glutamate, serine, asparagine, and aspartate) (12). Essential amino acids cannot be synthesized within the body and must be supplied through dietary intake. Amino acid metabolism is used in many processes that are involved in cell proliferation, growth and cell function. Furthermore, amino acids are also critical for the biosynthesis of nucleotides (13). It has been documented that some amino acids such as leucine, methionine, glutamine, arginine, and alanine, are more essential than other amino acids during T cell activation and expansion or in determining distinct T cell fates (14, 15). The importance of glycolysis, and fatty acid oxidation/synthesis in lupus T cells has been extensively reviewed elsewhere (8–10, 16–18). Here we summarize amino acid metabolism in mice and people with SLE with a focus on T cells.
Amino Acid Transporters
Amino acid transporters are important in transporting amino acids from the environment into the cell (19). T-cell receptor (TCR) stimulation triggers dramatic metabolic changes including increased glycolysis, pentose phosphate pathway activity, and glutaminolysis (19, 20). SLC7A5, known as large neutral amino acid transporter 1 (LAT-1), is a transporter dedicated to the transport of essential amino acids (21). SLC3A2, also known as CD98, is a transmembrane protein, which chaperones amino-acid transporters, including SLC7A5 SLC7A6, SLC7A7, SLC7A8, SLC7A10, and SLC7A11 (12), and enables them to execute their function. The LAT-1/CD98 heterodimer transports large hydrophobic amino acids, including the seven essential amino acids leucine, isoleucine, histidine, valine, phenylalanine, tryptophan, and methionine. Notably, the expression of LAT-1 and CD98 in T cells is induced after activation (19). Slc7a5-/- CD4+ T cells cannot respond to antigen, undergo clonal expansion or effector cell differentiation. Although Slc7a5-/- CD4+ T cells do not differentiate into Th1 and Th17 cells, differentiation into iTreg is not affected (22). LAT-1 deletion or inhibition blocks the expansion of IL-17 secreting γδ and CD4+ T cells in both human cells and imiquimod (a TLR7 agonist)-induced lupus and psoriasis-like animal models (Figure 1). The heterodimer comprising CD98 and SLC7A7 transports among other amino acids lysine, arginine, methionine, leucine, alanine, and cysteine (12). Interestingly, whole-exome sequencing in patients with childhood-onset SLE identified a SLC7A7 mutation to be linked to disease expression (23).
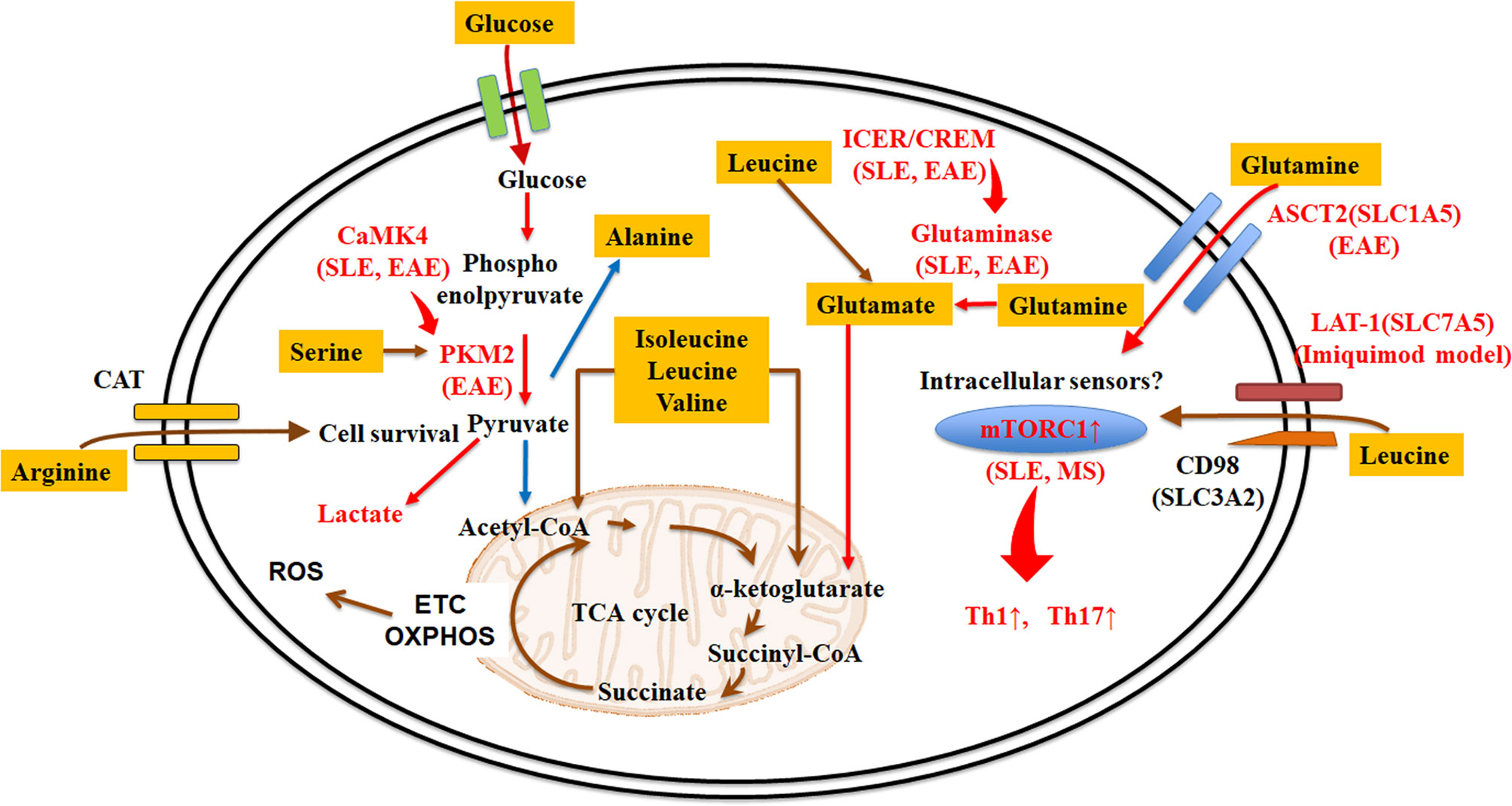
Figure 1 Amino acid transporters and metabolism in lupus T cells. Amino acid acquisition is crucial for cell function. Amino acid transporters play central roles in acquiring amino acids from the external environment. Some amino acids (e.g. leucine, methionine, glutamine, arginine, and alanine) are more essential than other amino acids in during T cell activation and expansion, or in determining different T cell fates in autoimmune diseases. Red arrows or letters indicate “enhance or active”, whereas blue arrows indicate “inhibit or inactivate”. ASCT2, alanine-serine-cysteine transporter 2; CaMK4, calcium/calmodulin–dependent protein kinase IV; CAT, cationic amino acid transporters; CREM, cAMP response element modulator; EAE, experimental autoimmune encephalomyelitis; ETC, electron transport chain; ICER, inducible cAMP early repressor; LAT-1, large neutral amino acid transporter 1; mTORC, mammalian target of rapamycin complex; OXPHOS, oxidative phosphorylation; PKM2, pyruvate kinase muscle isozyme 2; ROS, reactive oxygen species; SLE, systemic lupus erythematosus; TCA cycle, tricarboxylic acid cycle.
Alanine is also important in T cell activation. It is transported through SLC38A1 in CD4+ T cells and TCR stimulation induces its expression (12). Alanine deprivation impairs naïve and memory T cell activations, but it does not affect T cell effector functions (24). Although alanine can be made from pyruvate by a single transamination, extracellular alanine is used mainly for protein synthesis (12, 24).
Glutamine is the most abundant amino acid in the serum (25, 26). T cell stimulation promotes a rapid increase of glutamine uptake and activated T cells need more glutamine than naïve T cells (27). SLC1A5, known as alanine-serine-cysteine transporter 2 (ASCT2), is a transporter of neutral amino acids including glutamine (28). Although Slc1a5-/- CD4+ T cells do not affect TCR-mediated activation, deletion of Slc1a5 impaired Th1 and Th17 cell differentiation (Figure 1) (27).
Arginine is transported through cationic amino acid transporters (CAT) (29), which are shared by lysine and ornithine. Elevation of arginine levels induces metabolic changes including a shift from glycolysis to oxidative phosphorylation in activated T cells and promotes the generation of central memory-like cells (30). Arginine and the transporter CAT-1 (SLC7A1) are also requisite for human T cell survival (31).
These findings demonstrate distinct roles for amino acid transporters in TCR/CD3-mediated T cell stimulation, differentiation, and function and indicate that manipulation of these transporters could serve therapeutic approaches for autoimmune diseases including SLE (Figure 1). Because several other amino acid transporters have not been studied carefully in T cells, further research is needed.
Amino Acid Sensors
Although multiple mechanisms are involved in sensing amino acids within the intracellular space, it has been well established that the presence or absence of amino acids is sensed by distinct signaling pathways which involve the mechanistic target of rapamycin (mTOR) or the general control nonderepressible 2 (GCN2) (32, 33).
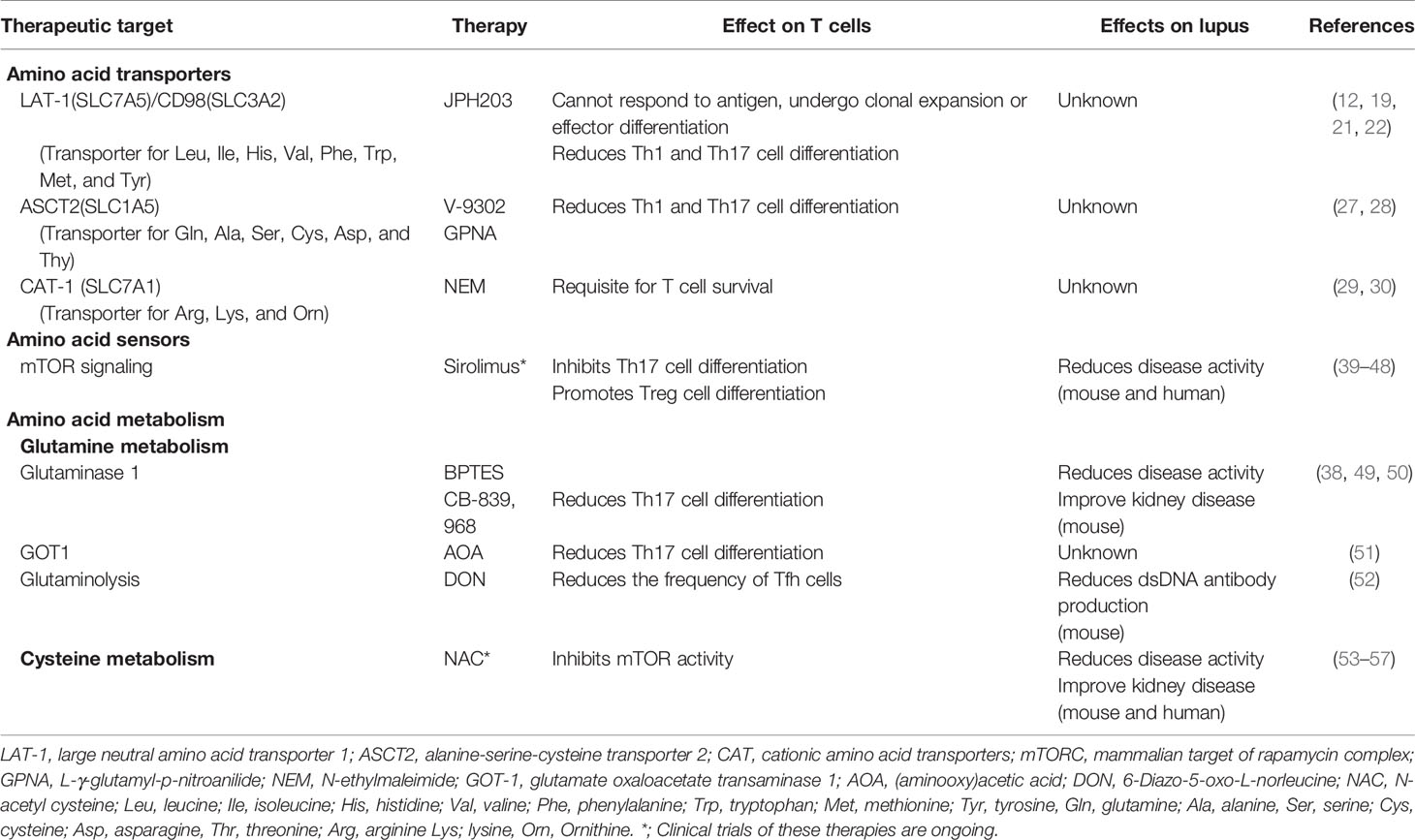
mTOR activity is regulated by amino acid availability, energy levels, and growth factors (34). In mammalian cells mTOR forms two distinct complexes: the mTORC complex 1 (mTORC1) and mTORC2. In fact, mTORC1 senses various stress signals, including the accumulation of amino acids such as leucine, isoleucine, kynurenine, and glutamine (35, 36). Glutamine activates mTORC1 via its metabolic product α-ketoglutarate which is generated during glutaminolysis (37). Inhibition of the first enzyme of glutaminolysis, glutaminase 1, reduces the activity of mTORC1 under Th17-polarized conditions (38). mTORC activity is enhanced in Th17 cells and IL-4-producing double negative T cells resulting in the proinflammatory profile recorded in patients with SLE (39). During Th17 cell differentiation, mTOR is required for the induction of hypoxia-inducible factor 1α (HIF1α) which enhances glycolysis (40). In Th1 and Th17 cells, mTORC1 activity, and glycolysis are increased compared with Tregs and Tfh cells (40, 41). Sirolimus, a mTOR inhibitor, was reported to improve disease activity in patients with refractory SLE in a single-arm, open-label, phase I/II trial (42), and other non-randomized controlled studies have reported that sirolimus is efficacious in patients with SLE (Table 1) (43). Sirolimus normalized Th17/Treg balance and TCR-induced Ca2+ fluxing in patients with SLE (44, 45). Besides the effect on T cells, inhibition of mTOR in plasmacytoid dendritic cells reduced the production of type I interferons (58) and B cell stimulating factor BAFF-mediated B cell activation (59, 60). These results indicate that sirolimus can modify T, B, and plasmacytoid dendritic cell function (46). Further randomized controlled trials are needed to prove the efficacy and record the side effects of sirolimus in patients with SLE (47).
GCN2, a serine/threonine-protein kinase, also senses amino acid starvation by detecting uncharged transfer RNA (33, 61). It plays a vital role in the control of amino acid metabolism as a response to nutrient deprivation. Gcn2 deficiency significantly inhibits in vitro differentiation of Th9 cells but not Th1, Th2, and Treg cells in mouse model, and it ameliorated allergic airway inflammation in mice (62). On the other hand, myeloid cell deletion of Gcn2 in lupus-prone mice resulted in increased immune cell activation, humoral autoimmunity, renal pathology, and mortality (63). These results suggest that therapeutic inhibition of GCN2 should not be considered to treat SLE.
Glutamine Metabolism
Glutaminolysis has a vital role in energy production in proliferating cells including T cells. Because of the indispensable roles of glutaminolysis in the generation of pro-inflammatory effector T cells Th1 and Th17 cells, enzymes involved in glutaminolysis have been studied extensively.
Glutaminase, in charge of converting glutamine to glutamate, promotes Th17 cells through distinct mechanisms (38, 49). Glutaminase expression is controlled by the transcription factor inducible cAMP early repressor (ICER)/cAMP response element modulator (CREM) (38), which is known to be overexpressed in T cells both from patients with SLE or MRL/lpr lupus-prone mice (64, 65). The glutaminase 1 inhibitor Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (BPTES) reduces Th17 cell differentiation and disease activity in animals subjected to experimental autoimmune encephalomyelitis (EAE) (38). BPTES also ameliorates disease activity in MRL/lpr mice (50). Glutamate oxaloacetate transaminase 1 (GOT1), which converts glutamate to α-ketoglutarate, an intermediate of the TCA cycle, also contributes to enhance Th17 cell differentiation through epigenetic processes (51). Selective inhibition of GOT1 with aminooxy acetic acid (AOA) treatment or short hairpin RNA (shRNA) silencing markedly decreased Th17 differentiation of murine T cells (51). Systemic AOA treatment or adoptive transfer of Got1 knockdown Th17-polarized T cells ameliorated EAE (51). Furthermore, inhibition of glutaminolysis with the glutamine analog 6-Diazo-5-oxo-L-norleucine (DON) reduces the frequency of Tfh cells, exogenous antigen-specific germinal center responses, and the production of dsDNA antibody in lupus-prone B6.Sle1.Sle2.Sle3 mice after T cell-dependent immunization (52).
Branched-Chain Amino Acid Metabolism
The branched-chain amino acids (BCAAs) include leucine, isoleucine, and valine. As the most abundant of essential amino acids, BCAAs are not only the substrates for synthesis of nitrogenous compounds, but they also serve as signaling molecules regulating the metabolism of glucose, lipid, and protein synthesis, intestinal health, and immunity through special signaling networks, especially the phosphoinositide 3-kinase/protein kinase B/mTOR (PI3K/AKT/mTOR) signal pathway. The leucine antagonist N-acetyl-leucine amide (NALA) inhibits mTORC1 activity and T cells function, impairs IL-2 and IFNγ production in in vitro Th1 polarized murine T cells (66). Leucine is also essential for Treg cell function. Leucine promotes mTORC1 activity in Treg cells via the small G proteins RagA/B and Rheb1/2 to drive their suppressive activity by inducing the expression of inducible T cell costimulator (ICOS) and CTLA4. Mice bearing RagA-RagB- or Rheb1-Rheb2-deficient Treg cells developed a Scurfy-like autoimmune disease and have reduced effector Treg cell accumulation and function (48).
Unlike most other essential amino acids, BCAAs catabolism is initially catalyzed either by transamination by branched-chain amino acid aminotransferases (BCAT) or decarboxylation by branched-chain α-keto acid dehydrogenase enzyme complex (BCKDC). After these reactions BCAA metabolites are further converted to acetyl-CoA and succinyl-CoA and participate in the TCA cycle (67). In CD4+ T cells, BCAT negatively regulates mTOR and glycolysis. Activated T cells from cytosolic branched chain aminotransferase (BCATc)-deficient mice show increased mTORC1 activation compared to T cells from control mice. Furthermore, T cells from Bcatc-/- mice display higher rates of glycolysis (68). In another study, the oral administration of a leucine analogue, ERG240, selectively inhibited the activity of BCAT1, reduced the severity of collagen-induced arthritis in mice, and crescentic glomerulonephritis in rats (69).
Serine Metabolism
Serine is used in proliferating cells for protein synthesis as well as the synthesis of other amino acids, such as glycine and cysteine (70). Serine-derived glycine is used in nucleotide synthesis. Moreover, serine is also a precursor for the synthesis of lipids, such as phosphatidylserine and sphingolipids, which have central roles in apoptotic cell clearance and immune cell activation, respectively (71, 72). A key molecule which is associated with serine is the M2 isoform of pyruvate kinase (PKM2) because it ligates and allosterically activates its activity (73). Even in the absence of exogenous serine, PKM2 expression contributes to endogenous serine synthesis and to the maintenance of mTORC1 activity (74).
Upon T cell activation, upregulated enzymes of the serine, glycine, one-carbon (SGOC) metabolic network, increase processing of serine into one-carbon metabolism. Extracellular serine is required for optimal T cell proliferation both in vitro and in vivo. Shortage of dietary serine impairs pathogen-driven expansion of T cells in vivo. Serine supplies glycine and one-carbon units for de novo nucleotide biosynthesis in proliferating T cells, and one-carbon units from formate can rescue T cells from serine deprivation (75).
We previously reported that calcium/calmodulin–dependent protein kinase IV (CaMK4) binds to PKM2 and promotes pyruvate kinase activity. Activated PKM2 is requisite for the Th1 and Th17 differentiation (76). Because inhibition of CaMK4 ameliorates pathogenesis of SLE though a Th17 cell manner (77, 78), the serine/PKM2 metabolism axis represents a hub of abnormal T cells in autoimmunity and needs further attention.
Serine also supports mitochondrial metabolism. In Jurkat cells, the catabolic enzyme serine hydroxymethyltransferase (SHMT2) is required for mitochondrial and respiratory activity (79). It has been also shown that SHMT2 promotes inflammatory cytokine signaling, including that of type I interferons, by interacting with the deubiquitylating BRCC36 isopeptidase complex (BRISC) (80). Since it has been recently shown that an inactive form of SHMT2 dimer has the capacity to bind and inhibit BRISC (80), control of the SHMT2-BRISC interaction may represent a new target to control autoimmune diseases.
Glutathione/Cysteine Metabolism
Glutathione is made from three amino acids: cysteine, glutamate, and glycine. Glutathione is important in the antioxidant defense, nutrient metabolism, and regulation of cellular events including gene expression, DNA and protein synthesis, cell proliferation and apoptosis, signal transduction, cytokine production and protein glutathionylation (81). Glutathione reduces intracellular reactive oxygen species (ROS) levels and inhibits Th17 cell differentiation (49, 82). Glutathione is reported to be decreased in the peripheral blood of patients with SLE (83). Glutathione regulates the elevation of mitochondrial transmembrane potential, which in turn activates mTOR in T cells from patients with SLE (53, 84). To date, N-acetylcysteine (NAC) has been used to correct glutathione levels because NAC is the cell-permeable precursor of cysteine which is the rate-limiting constituent of de novo reduced glutathione (53–55). Administration of NAC improves lupus disease activity and ameliorates organ damage mainly by blocking the mTOR pathway in T cells in humans and mice with SLE (56, 57).
Because cysteine contains sulfur, cysteine supports sulfur-dependent metabolism. As discussed above, cysteine is a key amino acid for glutathione function, as it supplies the sulfur necessary for the formation of the disulfide bridge in the glutathione disulfide (13), but its roles extend beyond glutathione synthesis. In humans, naïve T cells express none or very low levels of cystine and cysteine transporters. Thus, early T cells activation does not require cystine and cysteine. However, upon activation, T cells rapidly upregulate the expression of cystine and cysteine transporters and display dependency on exogenous supply of cystine/cysteine for their proliferation (85).
Metabolism of Other Amino Acids
Tryptophan, an essential amino acid used for the biosynthesis of crucial compounds, including 5-hydroxytryptamine (5-HT, serotonin) and kynurenine, is important in T cell function. Indoleamine-2,3-dioxygenase 1 (IDO-1) catabolizes tryptophan to kynurenine and T cells require tryptophan for proliferation and activation (13). Accordingly, IDO-1 inhibits T cell activation and Treg cell differentiation of human and murine T cells (86–89). The dysbiotic gut microbiota of lupus-prone mice which is characterized by altered distribution of tryptophan metabolites in the feces of the mice, including an increase in kynurenine levels, has been linked to the production of autoantibodies and autoimmune pathology (90). Low dietary tryptophan prevents disease activity of the lupus-prone mice, whereas high dietary tryptophan has the opposite effect (90).
Methionine can affect the epigenetic reprogramming in CD4+ T cells (91). Activated T cells transport methionine via SLC7A5 (92). Methionine serves as the major substrate for the biosynthesis of S-adenosyl-L-methionine (SAM) (91, 93). SAM functions as a substrate for epigenetic modifications. Methionine restriction reduces histone H3K4 methylation at promoter regions of genes associated with Th17 cell proliferation and cytokine production in murine T cells (91).
Conclusions
During the last decade great progress has been achieved in the field of immunometabolism. It has now been established that T cell metabolism controls the fate and function of T cells. Amino acids are also crucial in T cell survival, function and differentiation. Besides glycolysis, amino acid metabolism is also involved in the pathogenesis in SLE and by inference to other autoimmune diseases. Although 2-deoxy-d-glucose monotherapy has partial efficacy in improving disease in lupus-prone mice, when combined with metformin, a mitochondrial electron transport chain complex I inhibitor, it leads to normalization of T cell metabolism and reversal of disease activity (94). These results revealed that monotherapy targeting only glycolysis is not sufficient to treat lupus-prone mice. Thus, the focus of research on T cell metabolism in lupus is expanding our understanding of amino acid metabolism.
Although many reports have shown that some metabolic pathways involving amino acids including glutamine, tryptophan, and cysteine can serve as therapeutic targets in lupus-prone mice, the tentative therapeutic targeting of metabolic pathways of other amino acids remains unclear. Sirolimus and NAC are undergoing rigorous clinical trials in patients with SLE (42, 43, 80) and they may end up serving as significant entries in the list of available therapeutic tools for these patients. There are though several challenges to overcome in order to exploit additional amino acid-related treatment targets. Although many studies using mouse models have revealed potential therapeutic targets in amino acid metabolism, further insights are needed from the ex vivo study of immune cells from patients with SLE. Such studies should be followed by properly designed clinical trials in patients with SLE and probably other autoimmune diseases. As all drugs display invariably side effects, cell/tissue targeted delivery should be considered (73, 95, 96).
In this brief review we presented evidence that amino acids are important in T cell function and aberrant metabolism may be linked to autoimmunity and related pathology. It appears that their central role in the control of the immune response is underwritten by being indispensable for the generation of building blocks needed for cell proliferation, the generation of energy by controlling metabolic pathways, the control of epigenetic pathways, the production of phospholipids and the control of oxidative stress.
Amino acids and products of metabolic processes dictate the effector function of T cells and determine whether they will serve as regulators, instigators of inflammation or effectors of cytotoxicity. Alterations of the levels of metabolites within immune cells can be achieved by simply changing their levels in the environment or modulating the activity of transporters and intracellular metabolic enzymes. Drugs altering metabolism or supplementation of amino acids or metabolites or their precursors may prove of great value as modulators of T cell functions in the treatment and well-being of patients with autoimmune disease.
Author Contributions
MK, NY, and GT conceptualized the article, reviewed the literature, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the United States National Institutes of Health grants R01AR064350, R37 AI 49954 (to GT), AMED under Grant Number JP 20ek0410078, a SENSHIN Medical Research Foundation grant (to MK), and Gilead Sciences research scholars program in rheumatology (to NY).
Conflict of Interest
MK reports grants from GlaxoSmithKline plc, Mitsubishi Tanabe, Astellas, Sanofi, Taisho Pharmaceutical, NIPPON SHINYAKU CO., LTD., and Taiju Life Social Welfare Foundation, outside the submitted work. GT reports consultancies, speaking fees, or honoraria from Janssen, Novartis, ABPRO, Silicon Therapeutics, A2 Thera.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with the authors GT and NY.
References
1. Tsokos GC, Lo MS, Costa Reis P, Sullivan KE. New insights into the immunopathogenesis of systemic lupus erythematosus. Nat Rev Rheumatol (2016) 12:716–30. doi: 10.1038/nrrheum.2016.186
2. Tsokos GC. Systemic lupus erythematosus. N Engl J Med (2011) 365:2110–21. doi: 10.1056/NEJMra1100359
3. Kono M, Yasuda S, Kato M, Kanetsuka Y, Kurita T, Fujieda Y, et al. Long-term outcome in Japanese patients with lupus nephritis. Lupus (2014) 23:1124–32. doi: 10.1177/0961203314536246
4. Aso K, Kono M, Kono M, Watanabe T, Shimizu Y, Ogata Y, et al. Low C4 as a risk factor for severe neuropsychiatric flare in patients with systemic lupus erythematosus. Lupus (2020) 29:1238–47. doi: 10.1177/0961203320938453
5. Moulton VR, Suarez-Fueyo A, Meidan E, Li H, Mizui M, Tsokos GC. Pathogenesis of Human Systemic Lupus Erythematosus: A Cellular Perspective. Trends Mol Med (2017) 23:615–35. doi: 10.1016/j.molmed.2017.05.006
6. Katsuyama T, Tsokos GC, Moulton VR. Aberrant T Cell Signaling and Subsets in Systemic Lupus Erythematosus. Front Immunol (2018) 9:1088. doi: 10.3389/fimmu.2018.01088
7. Moulton VR, Tsokos GC. T cell signaling abnormalities contribute to aberrant immune cell function and autoimmunity. J Clin Invest (2015) 125:2220–7. doi: 10.1172/JCI78087
8. Sharabi A, Tsokos GC. T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol (2020) 16:100–12. doi: 10.1038/s41584-019-0356-x
9. Kono M, Yoshida N, Tsokos GC. Metabolic control of T cells in autoimmunity. Curr Opin Rheumatol (2020) 32:192–9. doi: 10.1097/BOR.0000000000000685
10. Vukelic M, Kono M, Tsokos GC. T cell Metabolism in Lupus. Immunometabolism (2020) 2:e200009. doi: 10.20900/immunometab20200009
11. Kono M, Yoshida N, Maeda K, Skinner NE, Pan W, Kyttaris VC, et al. Pyruvate dehydrogenase phosphatase catalytic subunit 2 limits Th17 differentiation. Proc Natl Acad Sci U S A (2018) 115:9288–93. doi: 10.1073/pnas.1805717115
12. Wang W, Zou W. Amino Acids and Their Transporters in T Cell Immunity and Cancer Therapy. Mol Cell (2020) 80:384–95. doi: 10.1016/j.molcel.2020.09.006
13. Kelly B, Pearce EL. Amino Assets: How Amino Acids Support Immunity. Cell Metab (2020) 32:154–75. doi: 10.1016/j.cmet.2020.06.010
14. Wei J, Raynor J, Nguyen TL, Chi H. Nutrient and Metabolic Sensing in T Cell Responses. Front Immunol (2017) 8:247. doi: 10.3389/fimmu.2017.00247
15. Shyer JA, Flavell RA, Bailis W. Metabolic signaling in T cells. Cell Res (2020) 30:649–59. doi: 10.1038/s41422-020-0379-5
16. Teng X, Cornaby C, Li W, Morel L. Metabolic regulation of pathogenic autoimmunity: therapeutic targeting. Curr Opin Immunol (2019) 61:10–6. doi: 10.1016/j.coi.2019.07.001
17. Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol (2017) 13:280–90. doi: 10.1038/nrrheum.2017.43
18. Shan J, Jin H, Xu Y. T Cell Metabolism: A New Perspective on Th17/Treg Cell Imbalance in Systemic Lupus Erythematosus. Front Immunol (2020) 11:1027. doi: 10.3389/fimmu.2020.01027
19. Ren W, Liu G, Yin J, Tan B, Wu G, Bazer FW, et al. Amino-acid transporters in T-cell activation and differentiation. Cell Death Dis (2017) 8:e2757. doi: 10.1038/cddis.2017.207
20. Menk AV, Scharping NE, Moreci RS, Zeng X, Guy C, Salvatore S, et al. Early TCR Signaling Induces Rapid Aerobic Glycolysis Enabling Distinct Acute T Cell Effector Functions. Cell Rep (2018) 22:1509–21. doi: 10.1016/j.celrep.2018.01.040
21. Cibrian D, Castillo-Gonzalez R, Fernandez-Gallego N, de la Fuente H, Jorge I, Saiz ML, et al. Targeting L-type amino acid transporter 1 in innate and adaptive T cells efficiently controls skin inflammation. J Allergy Clin Immunol (2020) 145:199–214.e111. doi: 10.1016/j.jaci.2019.09.025
22. Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol (2013) 14:500–8. doi: 10.1038/ni.2556
23. Tirosh I, Spielman S, Barel O, Ram R, Stauber T, Paret G, et al. Whole exome sequencing in childhood-onset lupus frequently detects single gene etiologies. Pediatr Rheumatol Online J (2019) 17:52. doi: 10.1186/s12969-019-0349-y
24. Ron-Harel N, Ghergurovich JM, Notarangelo G, LaFleur MW, Tsubosaka Y, Sharpe AH, et al. T Cell Activation Depends on Extracellular Alanine. Cell Rep (2019) 28:3011–21.e3014. doi: 10.1016/j.celrep.2019.08.034
25. Karinch AM, Pan M, Lin CM, Strange R, Souba WW. Glutamine metabolism in sepsis and infection. J Nutr (2001) 131:2535S–8S; discussion 2550S-2531S. doi: 10.1093/jn/131.9.2535S
26. Newsholme P. Why is L-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J Nutr (2001) 131:2515S–22S; discussion 2523S-2514S. doi: 10.1093/jn/131.9.2515S
27. Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity (2014) 40:692–705. doi: 10.1016/j.immuni.2014.04.007
28. Jiang H, Zhang N, Tang T, Feng F, Sun H, Qu W. Target the human Alanine/Serine/Cysteine Transporter 2(ASCT2): Achievement and Future for Novel Cancer Therapy. Pharmacol Res (2020) 158:104844. doi: 10.1016/j.phrs.2020.104844
29. Beyer SR, Mallmann RT, Jaenecke I, Habermeier A, Boissel JP, Closs EI. Identification of cysteine residues in human cationic amino acid transporter hCAT-2A that are targets for inhibition by N-ethylmaleimide. J Biol Chem (2013) 288:30411–9. doi: 10.1074/jbc.M113.490698
30. Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell (2016) 167:829–42.e813. doi: 10.1016/j.cell.2016.09.031
31. Werner A, Amann E, Schnitzius V, Habermeier A, Luckner-Minden C, Leuchtner N, et al. Induced arginine transport via cationic amino acid transporter-1 is necessary for human T-cell proliferation. Eur J Immunol (2016) 46:92–103. doi: 10.1002/eji.201546047
32. Gonzalez A, Hall MN. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J (2017) 36:397–408. doi: 10.15252/embj.201696010
33. Battu S, Minhas G, Mishra A, Khan N. Amino Acid Sensing via General Control Nonderepressible-2 Kinase and Immunological Programming. Front Immunol (2017) 8:1719. doi: 10.3389/fimmu.2017.01719
34. Takahara T, Amemiya Y, Sugiyama R, Maki M, Shibata H. Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes. J BioMed Sci (2020) 27:87. doi: 10.1186/s12929-020-00679-2
35. Perl A. Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases. Nat Rev Rheumatol (2016) 12:169–82. doi: 10.1038/nrrheum.2015.172
36. Wyman B, Perl A. Metabolic pathways mediate pathogenesis and offer targets for treatment in rheumatic diseases. Curr Opin Rheumatol (2020) 32:184–91. doi: 10.1097/BOR.0000000000000687
37. Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell (2012) 47:349–58. doi: 10.1016/j.molcel.2012.05.043
38. Kono M, Yoshida N, Maeda K, Tsokos GC. Transcriptional factor ICER promotes glutaminolysis and the generation of Th17 cells. Proc Natl Acad Sci U S A (2018) 115:2478–83. doi: 10.1073/pnas.1714717115
39. Kato H, Perl A. Mechanistic target of rapamycin complex 1 expands Th17 and IL-4+ CD4-CD8- double-negative T cells and contracts regulatory T cells in systemic lupus erythematosus. J Immunol (2014) 192:4134–44. doi: 10.4049/jimmunol.1301859
40. Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med (2011) 208:1367–76. doi: 10.1084/jem.20110278
41. Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, et al. The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity (2015) 43:690–702. doi: 10.1016/j.immuni.2015.08.017
42. Lai ZW, Kelly R, Winans T, Marchena I, Shadakshari A, Yu J, et al. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet (2018) 391:1186–96. doi: 10.1016/S0140-6736(18)30485-9
43. Eriksson P, Wallin P, Sjowall C. Clinical Experience of Sirolimus Regarding Efficacy and Safety in Systemic Lupus Erythematosus. Front Pharmacol (2019) 10:82. doi: 10.3389/fphar.2019.00082
44. Fernandez D, Bonilla E, Mirza N, Niland B, Perl A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheumatol (2006) 54:2983–8. doi: 10.1002/art.22085
45. Chu Y, Zhao C, Zhang B, Wang X, Wang Y, An J, et al. Restoring T-helper 17 cell/regulatory T-cell balance and decreasing disease activity by rapamycin and all-trans retinoic acid in patients with systemic lupus erythematosus. Lupus (2019) 28:1397–406. doi: 10.1177/0961203319877239
46. Oaks Z, Winans T, Huang N, Banki K, Perl A. Activation of the Mechanistic Target of Rapamycin in SLE: Explosion of Evidence in the Last Five Years. Curr Rheumatol Rep (2016) 18:73. doi: 10.1007/s11926-016-0622-8
47. Ji L, Xie W, Zhang Z. Efficacy and safety of sirolimus in patients with systemic lupus erythematosus: A systematic review and meta-analysis. Semin Arthritis Rheumatol (2020) 50:1073–80. doi: 10.1016/j.semarthrit.2020.07.006
48. Shi H, Chapman NM, Wen J, Guy C, Long L, Dhungana Y, et al. Amino Acids License Kinase mTORC1 Activity and Treg Cell Function via Small G Proteins Rag and Rheb. Immunity (2019) 51:1012–27.e1017. doi: 10.1016/j.immuni.2019.10.001
49. Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, et al. Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell (2018) 175:1780–95.e1719. doi: 10.1016/j.cell.2018.10.001
50. Kono M, Yoshida N, Maeda K, Suarez-Fueyo A, Kyttaris VC, Tsokos GC. Glutaminase 1 Inhibition Reduces Glycolysis and Ameliorates Lupus-like Disease in MRL/lpr Mice and Experimental Autoimmune Encephalomyelitis. Arthritis Rheumatol (2019) 71:1869–78. doi: 10.1002/art.41019
51. Xu T, Stewart KM, Wang X, Liu K, Xie M, Kyu Ryu J, et al. Metabolic control of TH17 and induced Treg cell balance by an epigenetic mechanism. Nature (2017) 548:228–33. doi: 10.1038/nature23475
52. Choi SC, Titov AA, Abboud G, Seay HR, Brusko TM, Roopenian DC, et al. Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells. Nat Commun (2018) 9:4369. doi: 10.1038/s41467-018-06686-0
53. Lai ZW, Hanczko R, Bonilla E, Caza TN, Clair B, Bartos A, et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthritis Rheumatol (2012) 64:2937–46. doi: 10.1002/art.34502
54. Suwannaroj S, Lagoo A, Keisler D, McMurray RW. Antioxidants suppress mortality in the female NZB x NZW F1 mouse model of systemic lupus erythematosus (SLE). Lupus (2001) 10:258–65. doi: 10.1191/096120301680416940
55. Piranavan P, Bhamra M, Perl A. Metabolic Targets for Treatment of Autoimmune Diseases. Immunometabolism (2020) 2:e200012. doi: 10.20900/immunometab20200012
56. Shah D, Sah S, Nath SK. Interaction between glutathione and apoptosis in systemic lupus erythematosus. Autoimmun Rev (2013) 12:741–51. doi: 10.1016/j.autrev.2012.12.007
57. Perl A. Review: Metabolic Control of Immune System Activation in Rheumatic Diseases. Arthritis Rheumatol (2017) 69:2259–70. doi: 10.1002/art.40223
58. Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, Murthy N, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol (2008) 9:1157–64. doi: 10.1038/ni.1645
59. Ge F, Wang F, Yan X, Li Z, Wang X. Association of BAFF with PI3K/Akt/mTOR signaling in lupus nephritis. Mol Med Rep (2017) 16:5793–8. doi: 10.3892/mmr.2017.7367
60. Ke Z, Liang D, Zeng Q, Ren Q, Ma H, Gui L, et al. hsBAFF promotes proliferation and survival in cultured B lymphocytes via calcium signaling activation of mTOR pathway. Cytokine (2013) 62:310–21. doi: 10.1016/j.cyto.2013.03.011
61. Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, Anthony TG. GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. J Biol Chem (2009) 284:32742–9. doi: 10.1074/jbc.M109.047910
62. Wang P, Xu Y, Zhang J, Shi L, Lei T, Hou Y, et al. The amino acid sensor general control nonderepressible 2 (GCN2) controls TH9 cells and allergic airway inflammation. J Allergy Clin Immunol (2019) 144:1091–105. doi: 10.1016/j.jaci.2019.04.028
63. Ravishankar B, Liu H, Shinde R, Chaudhary K, Xiao W, Bradley J, et al. The amino acid sensor GCN2 inhibits inflammatory responses to apoptotic cells promoting tolerance and suppressing systemic autoimmunity. Proc Natl Acad Sci U S A (2015) 112:10774–9. doi: 10.1073/pnas.1504276112
64. Tenbrock K, Juang YT, Gourley MF, Nambiar MP, Tsokos GC. Antisense cyclic adenosine 5’-monophosphate response element modulator up-regulates IL-2 in T cells from patients with systemic lupus erythematosus. J Immunol (2002) 169:4147–52. doi: 10.4049/jimmunol.169.8.4147
65. Yoshida N, Comte D, Mizui M, Otomo K, Rosetti F, Mayadas TN, et al. ICER is requisite for Th17 differentiation. Nat Commun (2016) 7:12993. doi: 10.1038/ncomms12993
66. Zheng Y, Delgoffe GM, Meyer CF, Chan W, Powell JD. Anergic T cells are metabolically anergic. J Immunol (2009) 183:6095–101. doi: 10.4049/jimmunol.0803510
67. Nie C, He T, Zhang W, Zhang G, Ma X. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int J Mol Sci (2018) 19:954. doi: 10.3390/ijms19040954
68. Ananieva EA, Patel CH, Drake CH, Powell JD, Hutson SM. Cytosolic branched chain aminotransferase (BCATc) regulates mTORC1 signaling and glycolytic metabolism in CD4+ T cells. J Biol Chem (2014) 289:18793–804. doi: 10.1074/jbc.M114.554113
69. Papathanassiu AE, Ko JH, Imprialou M, Bagnati M, Srivastava PK, Vu HA, et al. BCAT1 controls metabolic reprogramming in activated human macrophages and is associated with inflammatory diseases. Nat Commun (2017) 8:16040. doi: 10.1038/ncomms16040
70. Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci (2014) 39:191–8. doi: 10.1016/j.tibs.2014.02.004
71. Birge RB, Boeltz S, Kumar S, Carlson J, Wanderley J, Calianese D, et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ (2016) 23:962–78. doi: 10.1038/cdd.2016.11
72. Bartke N, Hannun YA. Bioactive sphingolipids: metabolism and function. J Lipid Res (2009) 50 Suppl:S91–96. doi: 10.1194/jlr.R800080-JLR200
73. Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature (2012) 491:458–62. doi: 10.1038/nature11540
74. Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A (2012) 109:6904–9. doi: 10.1073/pnas.1204176109
75. Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, et al. Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab (2017) 25:345–57. doi: 10.1016/j.cmet.2016.12.011
76. Kono M, Maeda K, Stocton-Gavanescu I, Pan W, Umeda M, Katsuyama E, et al. Pyruvate kinase M2 is requisite for Th1 and Th17 differentiation. JCI Insight (2019) 4:e127395. doi: 10.1172/jci.insight.127395
77. Koga T, Ichinose K, Mizui M, Crispin JC, Tsokos GC. Calcium/calmodulin-dependent protein kinase IV suppresses IL-2 production and regulatory T cell activity in lupus. J Immunol (2012) 189:3490–6. doi: 10.4049/jimmunol.1201785
78. Maeda K, Otomo K, Yoshida N, Abu-Asab MS, Ichinose K, Nishino T, et al. CaMK4 compromises podocyte function in autoimmune and nonautoimmune kidney disease. J Clin Invest (2018) 128:3445–59. doi: 10.1172/JCI99507
79. Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW, et al. Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol Cell (2018) 69:610–21.e615. doi: 10.1016/j.molcel.2018.01.024
80. Walden M, Tian L, Ross RL, Sykora UM, Byrne DP, Hesketh EL, et al. Metabolic control of BRISC-SHMT2 assembly regulates immune signalling. Nature (2019) 570:194–9. doi: 10.1038/s41586-019-1232-1
81. Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr (2004) 134:489–92. doi: 10.1093/jn/134.3.489
82. Gerriets VA, Kishton RJ, Nichols AG, Macintyre AN, Inoue M, Ilkayeva O, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest (2015) 125:194–207. doi: 10.1172/JCI76012
83. Gergely P Jr, Grossman C, Niland B, Puskas F, Neupane H, Allam F, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheumatol (2002) 46:175–90. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H
84. Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol (2009) 182:2063–73. doi: 10.4049/jimmunol.0803600
85. Levring TB, Hansen AK, Nielsen BL, Kongsbak M, von Essen MR, Woetmann A, et al. Activated human CD4+ T cells express transporters for both cysteine and cystine. Sci Rep (2012) 2:266. doi: 10.1038/srep00266
86. Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med (1999) 189:1363–72. doi: 10.1084/jem.189.9.1363
87. Mellor AL, Keskin DB, Johnson T, Chandler P, Munn DH. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J Immunol (2002) 168:3771–6. doi: 10.4049/jimmunol.168.8.3771
88. Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol (2009) 183:2475–83. doi: 10.4049/jimmunol.0900986
89. Chen W, Liang X, Peterson AJ, Munn DH, Blazar BR. The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J Immunol (2008) 181:5396–404. doi: 10.4049/jimmunol.181.8.5396
90. Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med (2020) 12:eaax2220. doi: 10.1126/scitranslmed.aax2220
91. Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, et al. Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metab (2020) 31:250–66.e259. doi: 10.1016/j.cmet.2020.01.006
92. Klein Geltink RI, Pearce EL. The importance of methionine metabolism. Elife (2019) 8:e47221. doi: 10.7554/eLife.47221
93. Sinclair LV, Howden AJ, Brenes A, Spinelli L, Hukelmann JL, Macintyre AN, et al. Antigen receptor control of methionine metabolism in T cells. Elife (2019) 8:e44210. doi: 10.7554/eLife.44210
94. Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, et al. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med (2015) 7:274ra218. doi: 10.1126/scitranslmed.aaa0835
95. Otomo K, Koga T, Mizui M, Yoshida N, Kriegel C, Bickerton S, et al. Cutting Edge: Nanogel-Based Delivery of an Inhibitor of CaMK4 to CD4+ T Cells Suppresses Experimental Autoimmune Encephalomyelitis and Lupus-like Disease in Mice. J Immunol (2015) 195:5533–7. doi: 10.4049/jimmunol.1501603
Keywords: cell metabolism, amino acid, T cell, systemic lupus erythematosus, amino acid transporters
Citation: Kono M, Yoshida N and Tsokos GC (2021) Amino Acid Metabolism in Lupus. Front. Immunol. 12:623844. doi: 10.3389/fimmu.2021.623844
Received: 30 October 2020; Accepted: 07 January 2021;
Published: 22 February 2021.
Edited by:
Masayuki Mizui, Osaka University, JapanReviewed by:
Damian Maseda, University of Pennsylvania, United StatesSun Jung Kim, Northwell Health, United States
Copyright © 2021 Kono, Yoshida and Tsokos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michihito Kono, bS1rb25vQGhva3VkYWkuYWMuanA=; Nobuya Yoshida, bnlvc2hpZGFAYmlkbWMuaGFydmFyZC5lZHU=; George C. Tsokos, Z3Rzb2tvc0BiaWRtYy5oYXJ2YXJkLmVkdQ==
†These authors have contributed equally to this work
 Michihito Kono
Michihito Kono Nobuya Yoshida
Nobuya Yoshida George C. Tsokos
George C. Tsokos