Corrigendum: Assessing the Function of the ZFP90 Variant rs1170426 in SLE and the Association Between SLE Drug Target and Susceptibility Genes
- 1Department of Dermatology, The First Affiliated Hospital, Anhui Medical University, Hefei, China
- 2Department of Rheumatology and Immunology, Arthritis Research Institute, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 3Department of Dermatology, Lu’an People’s Hospital, Lu’an, China
- 4Department of Dermatology, The Second Affiliated Hospital, Hainan Medical University, Haikou, China
- 5Clinical Research Centre, Zhujiang Hospital, Southern Medical University, Zhujiang, China
- 6Menzies Institute for Medical Research, University of Tasmania, Hobart, TAS, Australia
- 7Department of Medical and Molecular Genetics, King’s College London, London, United Kingdom
A genome-wide association study (GWAS) has discovered that a polymorphism in the ZFP90 gene is associated with systemic lupus erythematosus (SLE). In this study, we explored the candidate function of a ZFP90 variant (rs1170426) in the context of SLE and detected the relationship between SLE susceptible genes and SLE drug target genes. First, we investigated the regulatory role of rs1170426 on ZFP90 expression by expression quantitative trait loci (eQTL) analysis in peripheral blood mononuclear cells (PBMCs), T, B, and monocytes cells and annotated the regulatory function of rs1170426 using bioinformatic databases. Second, we compared the case-control difference in ZFP90 expression levels. Third, we analyzed the association of genotype and ZFP90 expression levels with SLE clinical characters. Last, we showed the interaction of SLE susceptibility genes with SLE drug target genes. Subjects with the risk allele “C” of rs1170426 had lower expression levels of ZFP90 in PBMCs (P = 0.006) and CD8+ T cells (P = 0.003) from controls. SLE cases also had lower expression levels compared with controls (P = 2.78E-9). After correction for multiple testing, the ZFP90 expression levels were related to serositis (FDR p = 0.004), arthritis (FDR p = 0.020), hematological involvement (FDR p = 0.021), and increased C-reactive protein (CRP) (FDR p = 0.005) in cases. Furthermore, the SLE susceptible genes and the recognized SLE drug target genes were more likely to act upon each other compared with non-SLE genetic genes (OR = 2.701, P = 1.80E-5). These findings suggest that ZFP90 might play a role in the pathogenesis of SLE, and SLE genetics would contribute to therapeutic drug discovery.
Introduction
Systemic lupus erythematosus (SLE) is a chronic multi-system autoimmune disease that mostly affects women of childbearing age (1). Though the pathogenesis of SLE is still unclear, inherited susceptibility is an important aspect (2). The risk allele “C” of rs1170426 was first identified to be significantly associated with SLE in Han Chinese and European populations (Supplementary Table 1) (3).
Zinc finger proteins (ZFP) are a diversified family of proteins performing numerous biological functions (4), such as regulating gene expression in many tissues (5). The expression of ZFP90 is involved in various diseases, for example, colorectal cancer (6), cardiac dysfunction (7), intellectual disability (8), and obesity (9). In this study, we aimed to determine if rs1170426 was associated with SLE pathogenesis by affecting the expression of ZFP90. Previous studies have proved that the discoveries from GWAS play a vital role in pharmaceutical development and drug repositioning (10, 11). To assess the function of SLE genes in pharmaceutical development, we also aimed to map the protein-protein interaction (PPI) network between SLE susceptibility genes and recognized SLE drug target genes, using the drug database.
Methods
Subjects
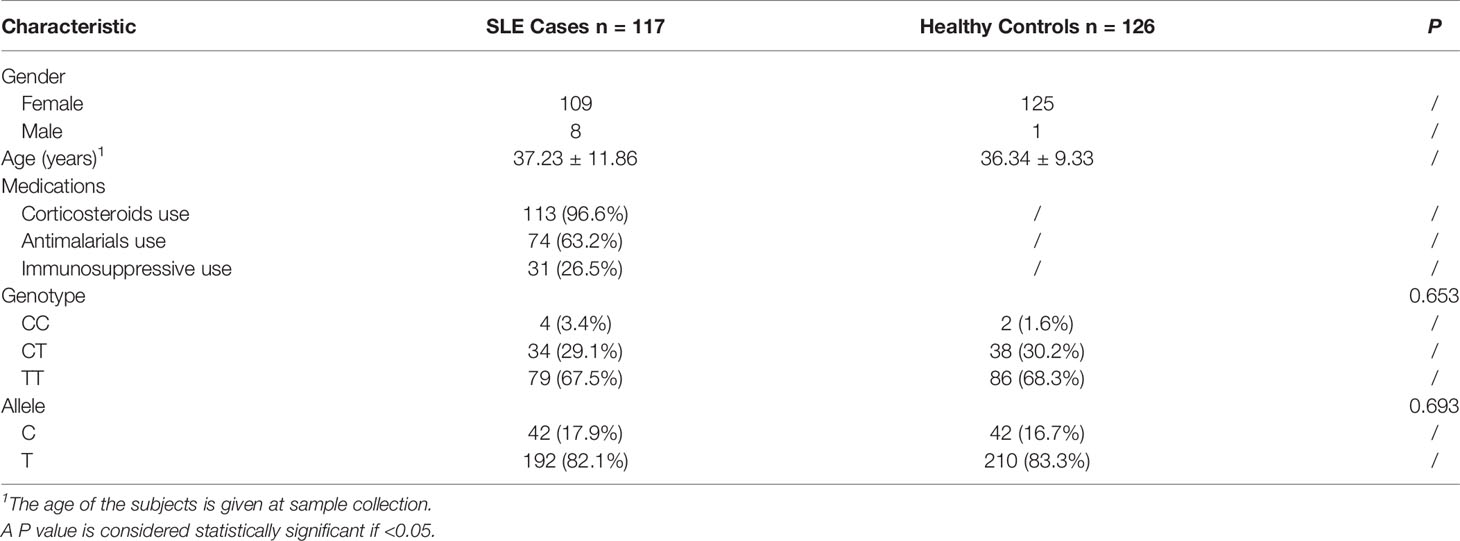
To test the mRNA expression level of ZFP90, we recruited 135 SLE cases and 130 healthy controls. The demographic characteristics of the samples included in the analysis were summarized in Table 1. The genotyping and gene expression study of four immune cells was conducted in 116 other healthy controls. All subjects were Han Chinese and enrolled from the First Affiliated Hospital of Anhui Medical University, Hefei, Anhui Province, China. SLE cases were diagnosed by two or more consultants in the Rheumatology or Dermatology Department according to the revised American College of Rheumatology (ACR) SLE classification criteria (12). All clinical phenotypes of SLE cases were defined by the ACR SLE classification criteria. The clinically verified healthy controls had no history of SLE, family history of SLE, or any other autoimmune diseases. Each subject signed informed consent to participate in this study, which was performed following the 1964 Declaration of Helsinki and approved by the Ethical Committee of Anhui Medical University.
Isolation of PBMCs, T Cells, B Cells, and Monocytes
The whole blood was collected from the above subjects using an anti-coagulation tube. We removed the plasma after centrifugal isolation (RT, 400 × g, 5 min) and added an equal volume of phosphate-buffered saline (PBS) to replace the removed plasma. The diluted blood was layered onto Ficoll-Hypaque Solution. The PBMCs were isolated by density-gradient centrifugation. The fresh PBMCs of 116 healthy controls were incubated for 15 min at 4°C with the following fluorescent conjugated monoclonal antibodies: anti-CD3-FITC, anti-CD14-PE, anti-CD19-APC, anti-CD4-PERCP-CY5.5, anti-CD8-PE-CY7. Stained cells were sorted on a BD FACSAria III. CD4+T, CD8+T, monocytes, and B cells were identified as CD4+/CD3+, CD8+/CD3+, CD14+/CD3−, and CD19+/CD3−, respectively. All stained cells were sorted to >98% purity.
Total RNA Isolation and qPCR
We extracted total mRNA from PBMCs and immune cell subsets using TRIzol Reagent. After reverse transcription of RNA (400 ng of each sample, using the PrimeScript RT reagent Kit) isolated from the samples, we used qPCR assays to confirm the relative expression of ZFP90 mRNA in collected PBMCs. The glyceraldehyde phosphate dehydrogenase (GAPDH) acted as the internal control. Below primers were employed in the qPCR: ZFP90 primers: forward 5′- CGCCCCAGGAATCAGTGACA -3′ and reverse 5′- GCTATAGTTCTCCAGCATCACATCC-3′. The experiment was conducted using the ViiA 7 Real-Time PCR System. The level of ZFP90 mRNA expression was computed using the 2-ΔΔCt method (13).
Genotyping
We extracted the genomic DNA using a Wizard Genomic DNA Purification Kit (QIAGEN, Germany). For 135 SLE cases and 130 healthy controls, we genotyped rs1170426 of each sample using the improved multiplex ligation detection reaction (iMLDR) technology. For 116 other healthy controls, we genotyped each sample using Sanger sequencing. The sequencing products were conducted on the ABI3730XL automatic sequencer (Applied Biosystems).
Functional Annotation and Bioinformatic Insights
We used HaploRegv4.1 (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php) (14) and rSNPBase (http://rsnp.psych.ac.cn/) (15) databases to annotate the functional elements containing rs1170426. We evaluated the effect of rs1170426 on ZFP90 expression in SLE implicated tissues or organs by using the Genotype-Tissue Expression (GTEx) Analysis Release V8 (http://www.gtexportal.org/home/) (16). In addition, we downloaded the RNA-seq data of EBV-transformed lymphocytes from the website (http://ebv-b.helmholtz-muenchen.de/) to address the influence of EBV infection on ZFP90 expression (17). We used the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/) (18) to discover the epigenomic regulatory potential of rs1170426.
The Association Between SLE Susceptibility Genes and Drug Target Genes
We got the target genes of SLE therapeutic drugs by consulting the DrugBank database (https://www.drugbank.ca/) (19) and Therapeutic target database (http://db.idrblab.org/ttd/) (20). We extracted PPI information from InWeb (21) to evaluate the potential relationship between SLE risk genes and drug target genes. Furthermore, we obtained the PPI network of ZFP90 with drug target genes by using the BioGRID database (https://thebiogrid.org/) (22).
Statistical Analysis
We used one-way analysis of variance to evaluate the cis-eQTL regulatory effect of rs1170426 on ZFP90 and compared ZFP90 mRNA expression levels in four kinds of immune cells. Mann-Whitney U test was used to detect the case-control difference in ZFP90 mRNA expression level, the correlation between ZFP90 mRNA expression level and clinical characteristics, the effect of treatment on ZFP90 expression, as well as the difference in ZFP90 mRNA expression level between cases and controls in each rs1170426 genotype group, which did not conform to the normal distribution. Chi-square test was applied to conduct association analysis in our cohort, genotype-phenotype analysis, and to analyze the difference of interaction probability between SLE risk genes and non-risk genes with drug targets. The PPI network diagram was plotted by Cytoscape software (version v3.8.0). SPSS (version 26.0) software and GraphPad Prism (version 8.01) were used for other data analysis and plotting. For multiple testing, a false discovery rate (FDR) p-value of 0.05 was used. The P-value (2 tails) of less than 0.05 indicated statistical significance.
Results
Association of rs1170426 With ZFP90 mRNA Expression
To predict the regulatory function of rs1170426 on ZFP90 mRNA expression, we performed eQTL analysis in PBMCs, CD4+ T, CD8+ T, CD14+ monocytes, and CD19+ B cells. As shown in Figure 1A, the risk allele “C” of SNP rs1170426 was correlated with lower expression levels of ZFP90 in PBMCs extracted from healthy controls (P = 0.006). However, there was no significant association in SLE cases (P = 0.548) (Figure 1B). Considering the medicine would affect eQTL, we grouped the patients by medicine type (corticosteroids, antimalarials, and immunosuppressive) and conducted eQTL analysis in each group. The results are all negative (Supplementary Table 2). In the four kinds of immune cells, we only found rs1170426 was an eQTL for ZFP90 in CD8+ T cells (P = 0.003) (Figures 1C–F).
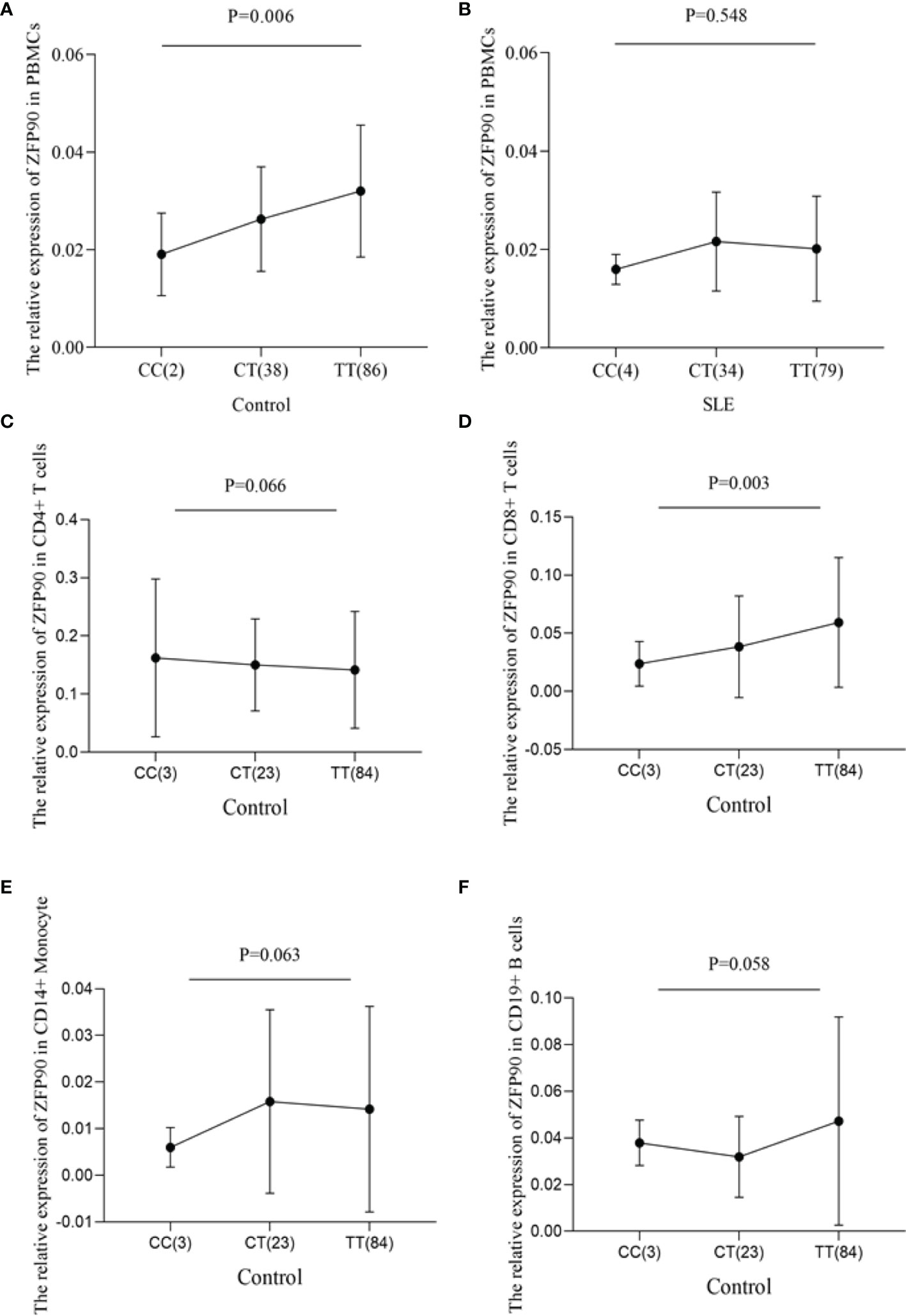
Figure 1 (A) The effect of rs1170426 on ZFP90 mRNA expression levels in PBMCs from healthy controls. Of the 126 controls, 2 individuals with CC, 38 with CT, and 86 with TT were analyzed. The group with “CC” homozygous has the lowest expression levels (P = 0.006). (B) The effect of rs1170426 on ZFP90 mRNA expression levels in PBMCs from SLE cases. Of the 117 cases, 4 individuals with CC, 34 with CT, and 79 with TT were analyzed. The expression did not significantly correlate with genotype of rs1170426 (P = 0.548). (C–F) The effect of rs1170426 on ZFP90 mRNA expression levels in CD4+ T cells, CD8+ T cells, CD19+ B cells, and CD14+ monocytes from other 110 healthy controls. Of the 110 controls, 3 individuals with CC, 23 with CT, and 84 with TT were analyzed. ZFP90 expression levels of samples with risk allele “C” of rs1170426 were significantly decreased in CD8+ T cells (P = 0.003).
Detection of rs1170426 Regulatory Effects
The HaploReg v4.1 database showed rs1170426 located in the intron of ZFP90 and overlapped with the binding site motifs of the transcription factor CCCTC-Binding Factor (CTCF) and Estrogen Receptor in T cells and monocytes (Table 2). It also played the role of a regulatory element in T cells and monocytes, i.e., gene enhancers marked by histone modifications histone H3 lysine 4 trimethylation (H3K4me3). We further found 158 SNPs in strong linkage disequilibrium (r2 > 0.8) with rs1170426 in the Asian population. Among them, 135 SNPs were regarded as regulatory SNPs (rSNPs) from SNPBase database. According to the GTEx Portal (version v8), rs1170426 is a cis-eQTL impacting on the expression of ZFP90 in EBV-transformed cells, sun-exposed skin, not sun-exposed skin, and whole blood (P = 6.90E-8, P = 7.91E-2, P = 1.60E-4, P = 1.13E-116, respectively) (Figure 2). By analyzing the processed RNA-seq data of EBV transfected lymphocytes in controls, we found that EBV transformation does not affect ZFP90 expression (Supplementary Figure 1). The epigenome annotation results show that the variant rs1170426 is located in the enhancer region of T cells and PBMCs respectively marked by H3 lysine 4 monomethylation (H3K4me1), H3K4me3, H3 lysine 9 trimethylation (H3K9me3), and H3 lysine 27 trimethylation (H3K27me3). However, we did not observe the signal in B cells (Figure 3A).
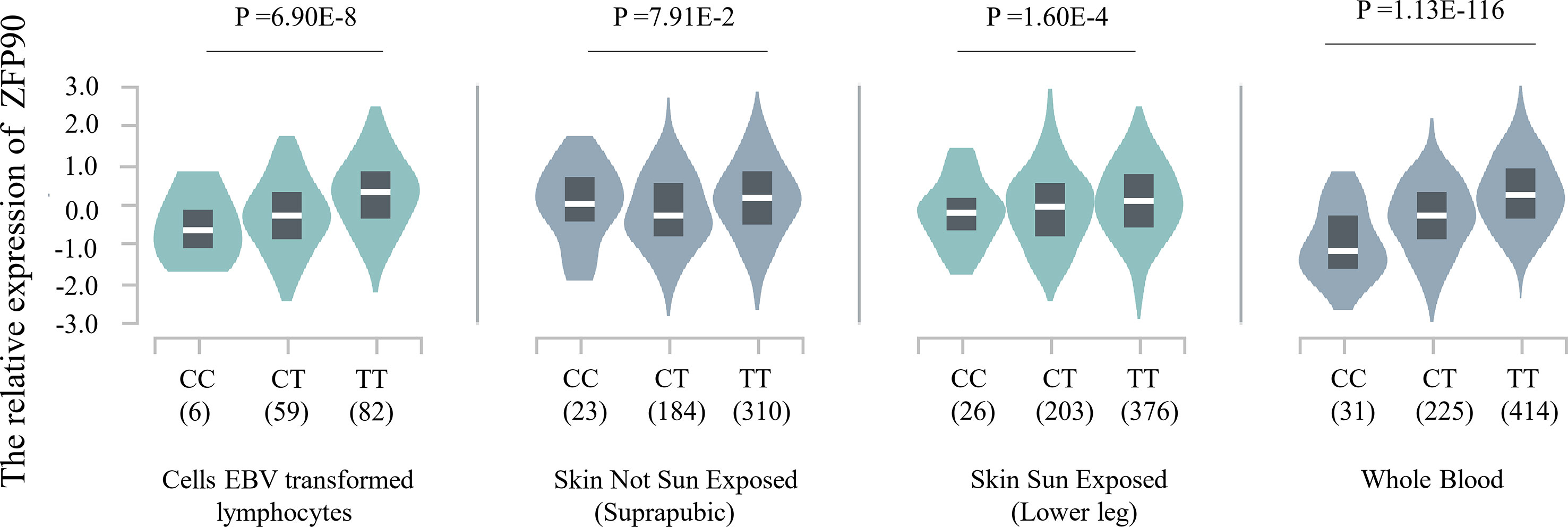
Figure 2 Plot showing the results of cis-eQTL analysis for SNP rs1170426 in EBV-transformed cells, sun-exposed skin, not sun-exposed skin, and whole blood (P = 6.90E-8, P = 7.91E-2, P = 1.60E-4, P = 1.13E-116, respectively).
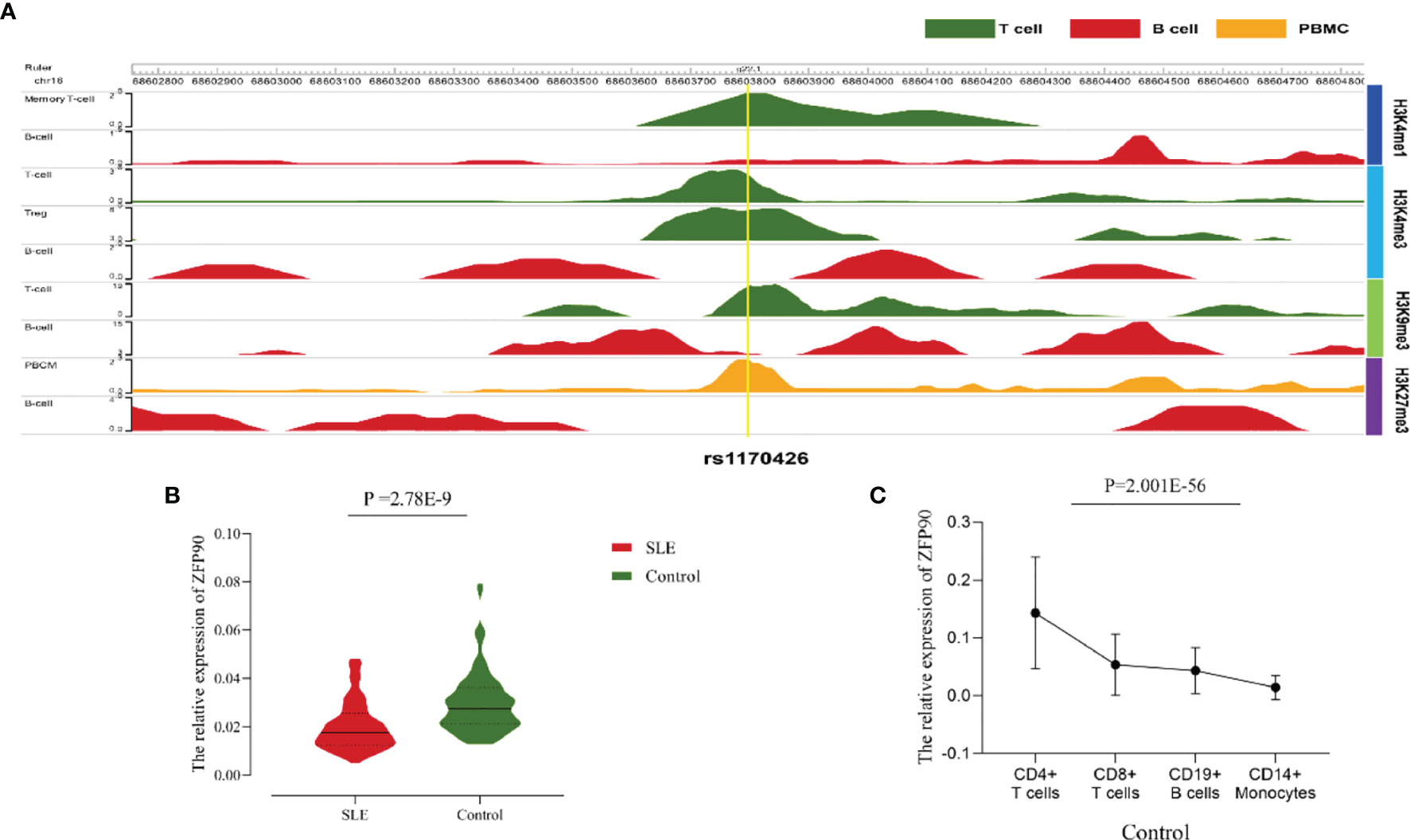
Figure 3 (A)The epigenome annotation results of rs1170426 in T cells (green), B cells (red), and PBMCs (orange). The signal can be found in T cells and PBMCs, while not in B cells. (B) Compared the ZFP90 mRNA expression levels between SLE cases (n = 117, red) and healthy controls (n = 126, green) in PBMCs. The expression levels were lower in cases than in healthy controls (P =2.78E-9). (C) The ZFP90 mRNA expression levels in four kinds of immune cells were remarkably different (P = 2.001E-56) and were higher in T cells.
Difference in ZFP90 mRNA Expression Levels
A total of 135 cases and 130 controls were collected for qPCR, 117 SLE cases and 126 healthy controls passed quality control for inclusion in the subsequent analysis. The median level of ZFP90 mRNA expression in SLE cases [Q50 = 0.018 (Q25 = 0.013, Q75 = 0.027)] was 1.5 times lower than in healthy controls [0.027 (0.021,0.036)]. Accordingly, the mRNA expression level of ZFP90 was substantially decreased in SLE cases compared to healthy controls (P = 2.78E-9) (Figure 3B). Then, we divided the samples into three groups by rs1170426 genotype and compared the case-control ZFP90 mRNA expression levels in each group, respectively. The results showed that the ZFP90 mRNA expression levels of the “CT” and “TT” groups were significantly decreased in cases (P = 0.004 and 1.836E-9, respectively). However, there is no statistical significance in the “CC” group (P = 0.800) (Supplementary Figure 2). In addition, we compared the ZFP90 expression level between patients with and without treatment by a certain class of drug (corticosteroids, antimalarials, and immunosuppressive). The results were not statistically significant (Supplementary Table 2).
At last, we found the ZFP90 mRNA expression levels was remarkably different among four kinds of immune cells (P = 2.001E-56), including CD4+ T, CD8+ T, CD14+ monocytes, and CD19+ B cells. Compared to other two types of cells, it especially expressed higher in T cells (Figure 3C).
The Correlation Between ZFP90 mRNA Expression and Clinical Characteristics
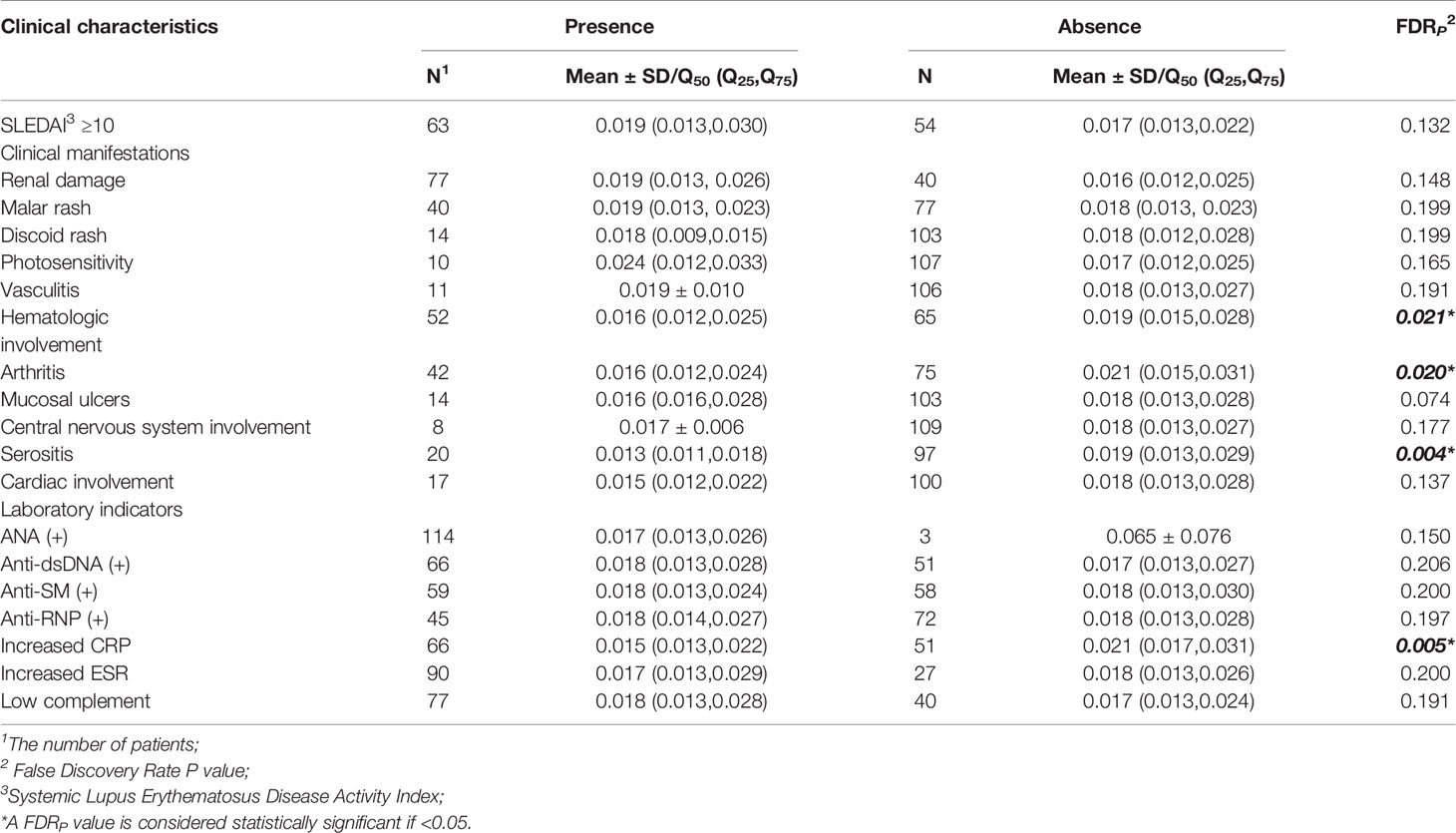
Compared with SLE cases without serositis, arthritis, or hematological involvement, cases with these symptoms had much lower levels of ZFP90 mRNA expression (FDR p = 0.004, FDR p = 0.020, FDR p = 0.021, respectively, Figure 4, Table 3). Moreover, the expression level was also significantly lower in cases with increased CRP than cases without a change in CRP (FDR p = 0.005).
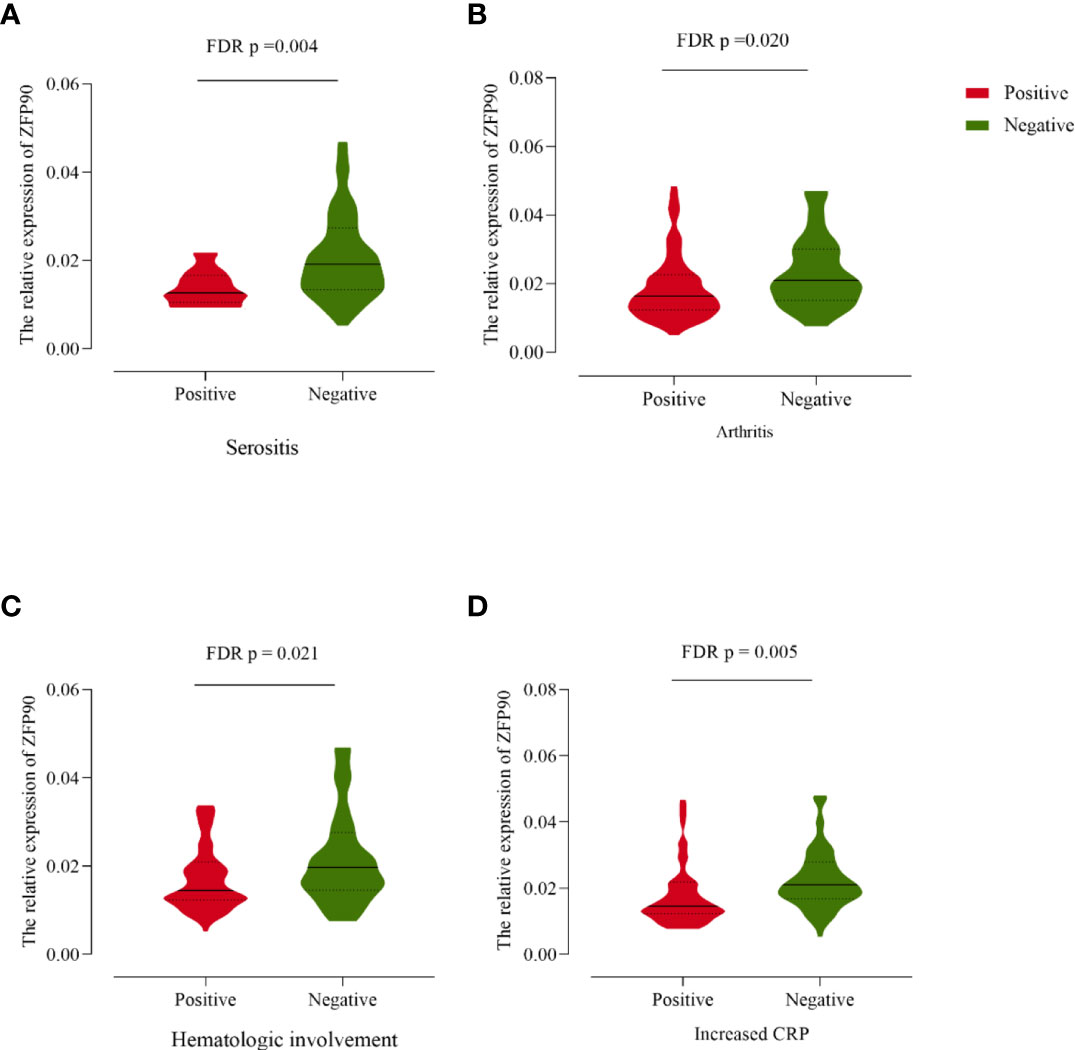
Figure 4 (A) The ZFP90 mRNA expression levels were lower (FDR p = 0.004) in cases with serositis than those without. (B) The levels were lower (FDR p = 0.020) in cases with arthritis than those without. (C) The levels were lower (FDR p = 0.021) in cases with hematologic involvement than those without. (D) The levels were lower (FDR p = 0.005) in cases with increased CRP than those without.
Association Study and Genotype-Phenotype Analysis of rs1170426 and SLE
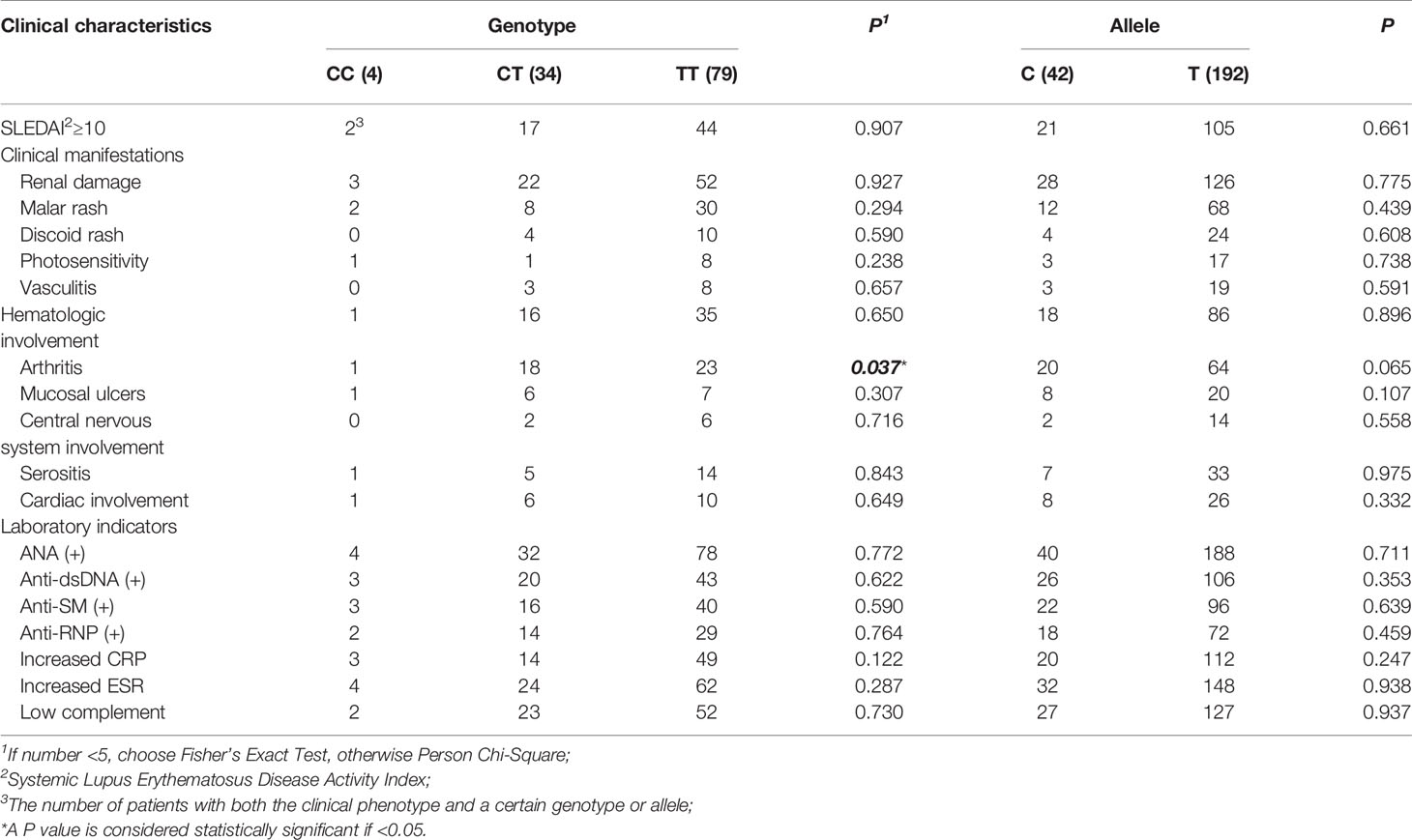
We found the frequency of the risk allele “C” in SLE cases was higher than in healthy controls, however, there is no statistical significance (Table 1). Then, we performed the genotype-phenotype analysis in two steps. First, we analyzed the correlation between genotype frequency and clinical character. Second, we conducted it for allele frequency. We only discovered that rs1170426 was statistically significant with arthritis involved (P = 0.037) (Table 4).
The PPI Information for SLE Susceptibility Genes and Drug Target Genes
We summarized 86 published and putative SLE risk genes that achieved genome-wide significance (P < 5.0E-8) (23, 24) (Supplementary Table 3). We then discovered 41 SLE drug target genes by the DrugBank database and Therapeutic target database (Supplementary Table 4). From the Inweb database, 72 SLE susceptibility genes (without ZFP90) were found to have PPI information with other genes, of which 29 were found to have protein interactions with SLE drug-targeted genes (Figure 5A). However, only 2,496 out of 12,491 SLE unassociated genes had PPI information with SLE drug-targeted genes in the Inweb database. Although only NFKBIA was mapped as both SLE risk genes and SLE drug target genes, the probability of SLE drug-targeted genes interaction with SLE susceptibility genes was much higher than unassociated genes (OR = 2.701, P = 1.80E-5) (Figure 5A). In the BioGRID database, we further discovered protein interaction between ZFP90 and PRKAB2, a drug target gene of SLE (Figure 5B).
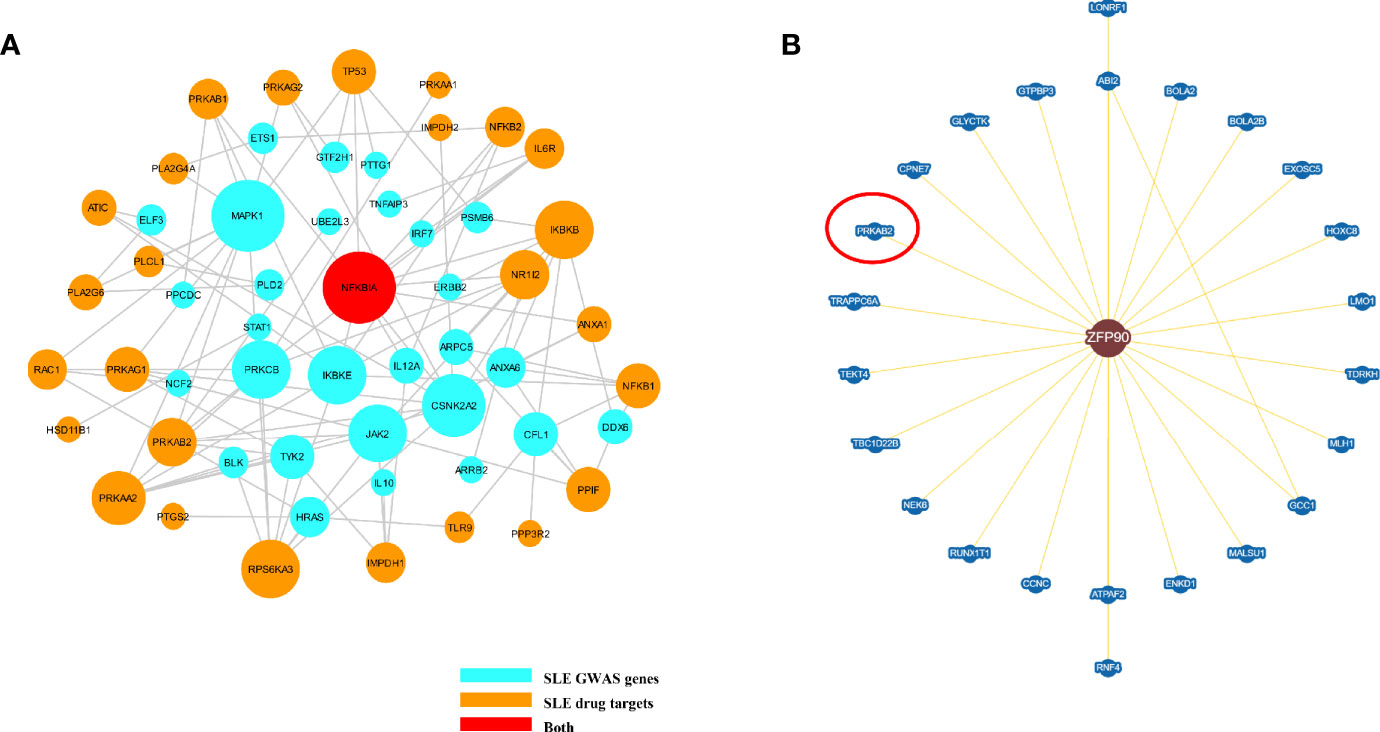
Figure 5 (A) The plot shows the PPI network of putative SLE risk genes and SLE drug target genes. SLE risk genes are shown as light blue nodes and SLE drugs target genes are shown as orange nodes. NFKBIA, which was shown as red node, is both SLE risk genes and SLE drug target genes. (B) This figure plots the protein interaction between ZFP90 and drug-targeted genes. PRKAB2 is a SLE drug-targeted gene (circled in red).
Discussion
SLE is a chronic autoimmune disease that often involves multiple organs, and its pathogenesis is related to numerous signaling pathways. GWAS is an efficient strategy to define disease susceptibility variants (25, 26). Our team has detected that rs1170426 located in the ZFP90 region is strongly linked to SLE by a trans-ancestral GWAS meta-analysis (3). Nonetheless, its potential causal effect and clinical relevance remain to be investigated in SLE. In this study, we found rs1170426 was a cis-eQTL for ZFP90 in PBMCs and primary CD8+T cells extracted from healthy controls. SLE cases expressed markedly lower transcriptional levels of ZFP90 compared with controls. After correction for multiple testing, the ZFP90 expression levels were still correlated with some clinical characters in cases. Importantly, we mapped the protein-protein interaction (PPI) network between SLE susceptibility genes and recognized SLE drug target genes.
ZFP90 is a zinc-finger protein that contains 13 zinc finger domains and a KRAB domain (27). The dysregulation of ZFP90 was associated with some autoimmune diseases, including inflammatory bowel disease, vitiligo, and multiple sclerosis (3). According to the expression data from GTEx, ZFP90 is widely expressed in various tissues such as skin and whole blood. We also confirmed it abundantly expressed in PBMCs and especially in T cells. Through functional annotation using multiple databases, we revealed that the variant rs1170426 had a regulatory function on ZFP90. The SNP rs1170426 of ZFP90 is predicted to span the binding site motifs of the transcription factor CTCF and Estrogen Receptor in T cells and monocytes. We also discovered rs1170426 was a cis-eQTL for ZFP90 in healthy PBMCs and primary CD8+T cells. The CTCF has been reported to take part in regulating the aberrant gene expression in SLE T cells (28). The estrogen contributes to immune cell trafficking and inflammation (29), which advances the understanding of significant gender bias in SLE cases. These findings provide essential clues to explore the relationship between SLE and the variant rs1170426 of ZFP90.
ZFP90 has been reported to regulate the activation of Ca2+ channels to a certain extent (7). An increased and prolonged Ca2+ influx was also observed in T cells of SLE patients (30). Altered Ca2+ flux can cause mitochondrial dysfunction and predispose to necrotic cell death (31). Increased cell necrosis may trigger the pro-inflammatory status resulting from the production of pro-inflammatory cytokines [such as TNF-α, interleukin-6 (IL-6) and IL-8] and interferon (32).
We also found ZFP90 mRNA expression level was strongly linked with clinical characteristics. It was significantly lowered in SLE cases with serositis than those without. Serositis is one of the manifestations of lupus, which is often accompanied by the presence of infections. The exact pathogenesis of SLE serositis is still elusive. A study showed abnormal Ca2+ influx could lead to an inflammatory response associated with the onset of serositis (33). Our results also displayed that ZFP90 mRNA expression level was lower in cases with arthritis than without. A previous study has shown that arthritis affects no less than 90% of SLE cases at some point during the course of the disease (34). It has been reported that IL-6, related to ZFP90, can promote arthritis and joint deformation in SLE cases (35). Consequently, we can reasonably suspect that the low expression of ZFP90 promotes the production of pro-inflammatory factors leading to the onset of arthritis. Besides, some studies have shown that pro-inflammatory factors can stimulate the production of CRP (36, 37). Hence, in this study, SLE cases with a lower ZFP90 mRNA expression level had an elevated CRP level. As we all know, infection is a common and leading cause of morbidity and mortality in SLE patients (38). CRP is generally associated with infection. So we would imagine the lower expression of ZFP90 was associated with SLE through the infection pathway.
Hematopoietic stem cells (HSCs), which take on the potency to self-renew and differentiate to all lineage blood cells, play an essential role in hematopoiesis (39, 40). Studies have shown that ZFP90 can be used as a transcription factor to participate in the regulation of HSC self-renewal and repopulation potential in vivo (41). Besides, excessive activation of the Wnt/β-catenin signaling pathway, an important signal pathway to participate in SLE (42), facilitates HSCs to lose the abilities of differentiation (43, 44). SLE cases with low expression of ZFP90 are more likely to have a hematologic abnormality, perhaps due to the loss of the balance between proliferation and differentiation in HSCs.
In the PPI network of ZFP90 and SLE drug target genes, ZFP90 interacts with PRKAB2. PRKAB2 can activate AMP-activated protein kinase (AMPK) in response to Ca2+ increase (45). ZFP90 also participates in the regulation of Ca2+ channels and affects the flow of Ca2+ as mentioned above. This may contribute to explain the relationship between ZFP90 and PRKAB2. PRKAB2 is also one of the target genes of Fostamatinib that is a drug approved by the US Food and Drug Administration (FDA) in 2018 for the treatment of idiopathic thrombocytopenic purpura. Fostamatinib has been confirmed to suppress the development of lupus skin and kidney disease in lupus-prone mice (46). This find might promote that Fostamatinib would be used for SLE patients in the future. Choosing genetically supported targets can significantly increase the success rate of clinical drug development (11), which is supported by our analysis of the relationship between SLE susceptibility genes and drug target genes.
The current study still has some caveats. The limited sample size may weak the statistical power. Besides, as the real target could be mapped far away from the associated variants (47), lists of the causal genes for disease associations can be incomplete, and inaccurate at times.
In summary, we have dissected the possible pathogenic role of ZFP90 in SLE and its impact on related clinical characteristics. Given the clinical characteristics, bioinformatics, and functional significance of genetic variation and the defined functional target gene, ZFP90, we draw the conclusion that the risk SNP locus of rs1170426 and its associated pathways might be meaningful for SLE, and targeting this pathway may be vital in the prevention or treatment of SLE. In addition, we provide evidence that the SLE genetics play important role in therapeutic drug discovery. However, further studies will be needed to elucidate this further.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
LL conceptualized and supervised this study. CD, JL, and XZ contributed to the study design. TZ conducted data analysis and wrote the manuscript. LL polished and revised the manuscript. TZ and YH performed functional annotation and qPCR experiments. YH and DQ sorted the cells and extracted PBMC, DNA, and RNA. YS performed genotyping. CZ and SC conducted sample recruitment. HZ and HW collected clinical information. All authors contributed to the article and approved the submitted version.
Funding
This study was supported financially by First batch of construction fund for innovation Capacity “135” Project (4601011226), Innovation research Group recipient cultivation Fund (4601011223), and Young Program (82003328) of National Natural Science Foundation of China (NSFC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YS declared a shared affiliation with several of the authors, TZ, YH, YS, CZ, SC, HZ, HW, XZ, CD, and LL, to the handling editor at the time of review.
The handling editor declared a shared affiliation, though no other collaboration, with one of the authors LL.
Acknowledgments
We thank all the study participants and all the volunteers who have so willingly participated in this subject. We also appreciate the Genotype-Tissue Expression (GTEx) Project, which was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.611515/full#supplementary-material
References
1. Parikh SV, Almaani S, Brodsky S, Rovin BH. Update on Lupus Nephritis: Core Curriculum 2020. Am J Kidney Dis (2020) 762:265–81. doi: 10.1053/j.ajkd.2019.10.017
2. Kuo CF, Grainge MJ, Valdes AM, See LC, Luo SF, Yu KH, et al. Familial Aggregation of Systemic Lupus Erythematosus and Coaggregation of Autoimmune Diseases in Affected Families. JAMA Intern Med (2015) 1759:1518–26. doi: 10.1001/jamainternmed.2015.3528
3. Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet (2016) 488:940–6. doi: 10.1038/ng.3603
4. Laity JH, Lee B M, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol (2001) 111:39–46. doi: 10.1016/s0959-440x(00)00167-6
5. Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, et al. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell (2016) 366:611–23. doi: 10.1016/j.devcel.2016.02.024
6. Yu CY, Han JX, Zhang J, Jiang P, Shen C, Guo F, et al. A 16q22.1 variant confers susceptibility to colorectal cancer as a distal regulator of ZFP90. Oncogene (2020) 396:1347–60. doi: 10.1038/s41388-019-1055-4
7. Hata L, Murakami M, Kuwahara K, Nakagawa Y, Kinoshita H, Usami S, et al. Zinc-finger protein 90 negatively regulates neuron-restrictive silencer factor-mediated transcriptional repression of fetal cardiac genes. J Mol Cell Cardiol (2011) 506:972–81. doi: 10.1016/j.yjmcc.2011.01.017
8. Palka Bayard de Volo C, Alfonsi M, Gatta V, Novelli A, Bernardini L, Fantasia D, et al. 16q22.1 microdeletion detected by array-CGH in a family with mental retardation and lobular breast cancer. Gene (2012) 4982:328–31. doi: 10.1016/j.gene.2012.01.028
9. Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet (2005) 377:710–7. doi: 10.1038/ng1589
10. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature (2014) 5067488:376–81. doi: 10.1038/nature12873
11. Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, et al. The support of human genetic evidence for approved drug indications. Nat Genet (2015) 478:856–60. doi: 10.1038/ng.3314
12. Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum (1997) 409:1725. doi: 10.1002/art.1780400928
13. Livak K J, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 254:402–8. doi: 10.1006/meth.2001.1262
14. Ward L D, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res (2012) 40Database issue:D930–4. doi: 10.1093/nar/gkr917
15. Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature (2012) 4917422:119–24. doi: 10.1038/nature11582
16. Gibson G. Human genetics. GTEx detects genetic effects. Science (2015) 3486235:640–1. doi: 10.1126/science.aab3002
17. Mrozek-Gorska P, Buschle A, Pich D, Schwarzmayr T, Fechtner R, Scialdone A, et al. Epstein-Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc Natl Acad Sci U S A (2019) 11632:16046–55. doi: 10.1073/pnas.1901314116
18. Li D, Hsu S, Purushotham D, Sears R L, Wang T. WashU Epigenome Browser update 2019. Nucleic Acids Res (2019) 47W1:W158–65. doi: 10.1093/nar/gkz348
19. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res (2018) 46D1:D1074–82. doi: 10.1093/nar/gkx1037
20. Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res (2020) 48D1:D1031–41. doi: 10.1093/nar/gkz981
21. Lage K, Karlberg EO, Storling ZM, Olason PI, Pedersen AG, Rigina O, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol (2007) 253:309–16. doi: 10.1038/nbt1295
22. Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res (2019) 47D1:D529–41. doi: 10.1093/nar/gky1079
23. Chen L, Morris D L, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol (2017) 295:423–33. doi: 10.1097/BOR.0000000000000411
24. Yin X, Kim K, Suetsugu H, Bang SY, Wen L, Koido M, et al. Meta-analysis of 208370 East Asians identifies 113 susceptibility loci for systemic lupus erythematosus. Ann Rheum Dis (2020) annrheumdis-2020-219209. doi: 10.1136/annrheumdis-2020-219209
25. Couzin J, Kaiser J. Genome-wide association. Closing the net on common disease genes. Science (2007) 3165826:820–2. doi: 10.1126/science.316.5826.820
26. Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med (2010) 3632:166–76. doi: 10.1056/NEJMra0905980
27. Lange R, Christoph A, Thiesen HJ, Vopper G, Johnson KR, Lemaire L, et al. Developmentally regulated mouse gene NK10 encodes a zinc finger repressor protein with differential DNA-binding domains. DNA Cell Biol (1995) 1411:971–81. doi: 10.1089/dna.1995.14.971
28. Zhao M, Wang J, Liao W, Li D, Li M, Wu H, et al. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J Autoimmun (2016) 69:64–73. doi: 10.1016/j.jaut.2016.03.001
29. Moulton VR. Sex Hormones in Acquired Immunity and Autoimmune Disease. Front Immunol (2018) 9:2279. doi: 10.3389/fimmu.2018.02279
30. Kyttaris VC, Wang Y, Juang YT, Weinstein A, Tsokos GC. Increased levels of NF-ATc2 differentially regulate CD154 and IL-2 genes in T cells from patients with systemic lupus erythematosus. J Immunol (2007) 1783:1960–6. doi: 10.4049/jimmunol.178.3.1960
31. Gergely P Jr, Grossman C, Niland B, Puskas F, Neupane H, Allam F, et al. Mitochondrial hyperpolarization and ATP depletion in patients with systemic lupus erythematosus. Arthritis Rheum (2002) 461:175–90. doi: 10.1002/1529-0131(200201)46:1<175::AID-ART10015>3.0.CO;2-H
32. Crow M K, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol (2004) 165:541–7. doi: 10.1097/01.bor.0000135453.70424.1b
33. Andrews BS, Arora NS, Shadforth MF, Goldberg S K, Davis JST. The role of immune complexes in the pathogenesis of pleural effusions. Am Rev Respir Dis (1981) 1242:115–20. doi: 10.1164/arrd.1981.124.2.115
34. Esdaile JM, Danoff D, Rosenthall L, Gutkowski A. Deforming arthritis in systemic lupus erythematosus. Ann Rheum Dis (1981) 402:124–6. doi: 10.1136/ard.40.2.124
35. Eilertsen GO, Nikolaisen C, Becker-Merok A, Nossent JC. Interleukin-6 promotes arthritis and joint deformation in patients with systemic lupus erythematosus. Lupus (2011) 206:607–13. doi: 10.1177/0961203310392432
36. Volanakis JE. Human C-reactive protein: expression, structure, and function. Mol Immunol (2001) 382-3:189–97. doi: 10.1016/s0161-5890(01)00042-6
37. Wlodarczyk M, Ciebiera M, Nowicka G. TNF-alpha G-308A genetic variants, serum CRP-hs concentration and DNA damage in obese women. Mol Biol Rep (2020) 472:855–66. doi: 10.1007/s11033-019-04764-0
38. Doaty S, Agrawal H, Bauer E, Furst DE. Infection and Lupus: Which Causes Which? Curr Rheumatol Rep (2016) 183:13. doi: 10.1007/s11926-016-0561-4
39. Orkin S H, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell (2008) 1324:631–44. doi: 10.1016/j.cell.2008.01.025
40. Rowe RG, Mandelbaum J, Zon L I, Daley GQ. Engineering Hematopoietic Stem Cells: Lessons from Development. Cell Stem Cell (2016) 186:707–20. doi: 10.1016/j.stem.2016.05.016
41. Liu T, Kong WX, Tang XY, Xu M, Wang QH, Zhang B, et al. The transcription factor Zfp90 regulates the self-renewal and differentiation of hematopoietic stem cells. Cell Death Dis (2018) 96:677. doi: 10.1038/s41419-018-0721-8
42. Olferiev M, Jacek E, Kirou K A, Crow MK. Novel molecular signatures in mononuclear cell populations from patients with systemic lupus erythematosus. Clin Immunol (2016) 172:34–43. doi: 10.1016/j.clim.2016.08.018
43. Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol (2006) 710:1037–47. doi: 10.1038/ni1387
44. Kirstetter P, Anderson K, Porse BT, Jacobsen S E, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat Immunol (2006) 710:1048–56. doi: 10.1038/ni1381
45. Mihaylova M M, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol (2011) 139:1016–23. doi: 10.1038/ncb2329
46. Lucas MC, Goldstein DM, Hermann JC, Kuglstatter A, Liu W, Luk KC, et al. Rational design of highly selective spleen tyrosine kinase inhibitors. J Med Chem (2012) 5523:10414–23. doi: 10.1021/jm301367c
Keywords: ZFP90, single nucleotide polymorphism, systemic lupus erythematosus, eQTL, SLE drug target genes
Citation: Zhu T, Huang Y, Qian D, Sheng Y, Zhang C, Chen S, Zhang H, Wang H, Zhang X, Liu J, Ding C and Liu L (2021) Assessing the Function of the ZFP90 Variant rs1170426 in SLE and the Association Between SLE Drug Target and Susceptibility Genes. Front. Immunol. 12:611515. doi: 10.3389/fimmu.2021.611515
Received: 29 September 2020; Accepted: 25 February 2021;
Published: 16 March 2021.
Edited by:
Deborah Cunninghame Graham, King’s College London, United KingdomReviewed by:
Zhimin Lu, Affiliated Hospital of Nantong University, ChinaKenneth Kaufman, Cincinnati Children’s Hospital Medical Center, United States
Copyright © 2021 Zhu, Huang, Qian, Sheng, Zhang, Chen, Zhang, Wang, Zhang, Liu, Ding and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Liu, bGl1bHU4ODg3QDE2My5jb20=; bHUuMi5saXVAa2NsLmFjLnVr; Changhai Ding, Y2hhbmdoYWkuZGluZ0B1dGFzLmVkdS5hdQ==; Junlin Liu, bGl1anVubGluMDc1OUAxNjMuY29t
†These authors have contributed equally to this work
 Tingting Zhu
Tingting Zhu Yuandi Huang
Yuandi Huang Danfeng Qian
Danfeng Qian Yuming Sheng
Yuming Sheng Chaowen Zhang
Chaowen Zhang Shirui Chen
Shirui Chen Hui Zhang
Hui Zhang Hui Wang
Hui Wang Xuejun Zhang
Xuejun Zhang Junlin Liu
Junlin Liu Changhai Ding
Changhai Ding Lu Liu
Lu Liu