
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 18 February 2021
Sec. B Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.633705
This article is part of the Research Topic Multi-Dimensional Genomic Structures in the Immune System View all 12 articles
A functional adaptive immune system must generate enormously diverse antigen receptor (AgR) repertoires from a limited number of AgR genes, using a common mechanism, V(D)J recombination. The AgR loci are among the largest in the genome, and individual genes must overcome huge spatial and temporal challenges to co-localize with optimum variability. Our understanding of the complex mechanisms involved has increased enormously, due in part to new technologies for high resolution mapping of AgR structure and dynamic movement, underpinning mechanisms, and resulting repertoires. This review will examine these advances using the paradigm of the mouse immunoglobulin heavy chain (Igh) locus. We will discuss the key regulatory elements implicated in Igh locus structure. Recent next generation repertoire sequencing methods have shown that local chromatin state at V genes contribute to recombination efficiency. Next on the multidimensional scale, we will describe imaging studies that provided the first picture of the large-scale dynamic looping and contraction the Igh locus undergoes during recombination. We will discuss chromosome conformation capture (3C)-based technologies that have provided higher resolution pictures of Igh locus structure, including the different models that have evolved. We will consider the key transcription factors (PAX5, YY1, E2A, Ikaros), and architectural factors, CTCF and cohesin, that regulate these processes. Lastly, we will discuss a plethora of recent exciting mechanistic findings. These include Rag recombinase scanning for convergent RSS sequences within DNA loops; identification of Igh loop extrusion, and its putative role in Rag scanning; the roles of CTCF, cohesin and cohesin loading factor, WAPL therein; a new phase separation model for Igh locus compartmentalization. We will draw these together and conclude with some horizon-scanning and unresolved questions.
V(D)J recombination is catalyzed by the recombinase complex, comprising recombination-activating genes 1 and 2 (RAG1-RAG2), that cleaves recombination signal sequences (RSSs) flanking antigen receptor (AgR) genes (1). Referred to as the 12/23 rule, recombination normally occurs exclusively between a 12-RSS and a 23-RSS, thereby for example enabling recombination of Igh DH genes, flanked on both sides by 12-RSS, with both JH and VH genes, flanked by 23-RSS, and preventing direct recombination between VH and JH genes with identical RSSs. Recombination is focused at recombination centers: regions of highly concentrated RAG binding which orchestrate RSS synapsis (2). V(D)J recombination is tightly regulated: it is lineage- and cell stage-specific and is controlled at three distinct levels. The first is the developmental stage-specific and cell cycle-regulated expression of the RAG recombinase (3–5). Secondly, next generation repertoire sequencing methods have revealed that several mechanisms including transcription factor binding, ATP-dependent chromatin remodeling, histone modifications, DNA methylation and non-coding transcription, regulate the local accessibility of AgR gene RSSs to the RAG recombinase within chromatin (6–8). However changes in local chromatin accessibility alone are insufficient to promote V(D)J recombination of spatially separated V, D and J genes (9). The third level of regulation is the spatial genomic organization of the AgR loci. Understanding the architecture of the AgR loci is an enormous technical challenge: they are large, multimegabase, repetitive loci that, given their biological function, are expected to exhibit multiple conformations. The dynamic 3D conformation of the Igh locus and its contributory mechanisms will be the main focus of this review, in part because studies of AgR conformation have made most progress in this locus, but given the countless similarities among them, the principles discussed here will be a very relevant paradigm for the other AgR loci.
The mouse immunoglobulin heavy chain (Igh) locus comprises 195 variable VH genes in 16 families, 10 diversity DH genes, 4 joining JH genes and 8 constant CH genes (Figure 1A) (15). To generate antibody diversity, the VH, DH, and JH genes undergo V(D)J recombination in a strictly regulated, sequential manner. First, DH-JH recombination commences in early pre-pro-B lymphoid progenitors and is completed by the early pro-B cell stage on both Igh alleles. Subsequently, one Igh allele undergoes VH-DJH recombination in committed pro-B cells. The second allele undergoes allelic exclusion. A successful recombination event is achieved when the newly produced heavy chain polypeptide pairs with a surrogate light chain and is expressed as a pre-B cell receptor (pre-BCR) on the cell surface. If this first VH-DJH recombination event is unsuccessful, then recombination is attempted on the remaining allele. Thereafter the Igκ locus (3Mb; 140 Vκ genes, 5 Jκ genes) undergoes a single recombination event—Vκ-Jκ—in pre-B cells to generate an Igκ polypeptide. Pairing of Igh and Igκ generates the BCR (16).
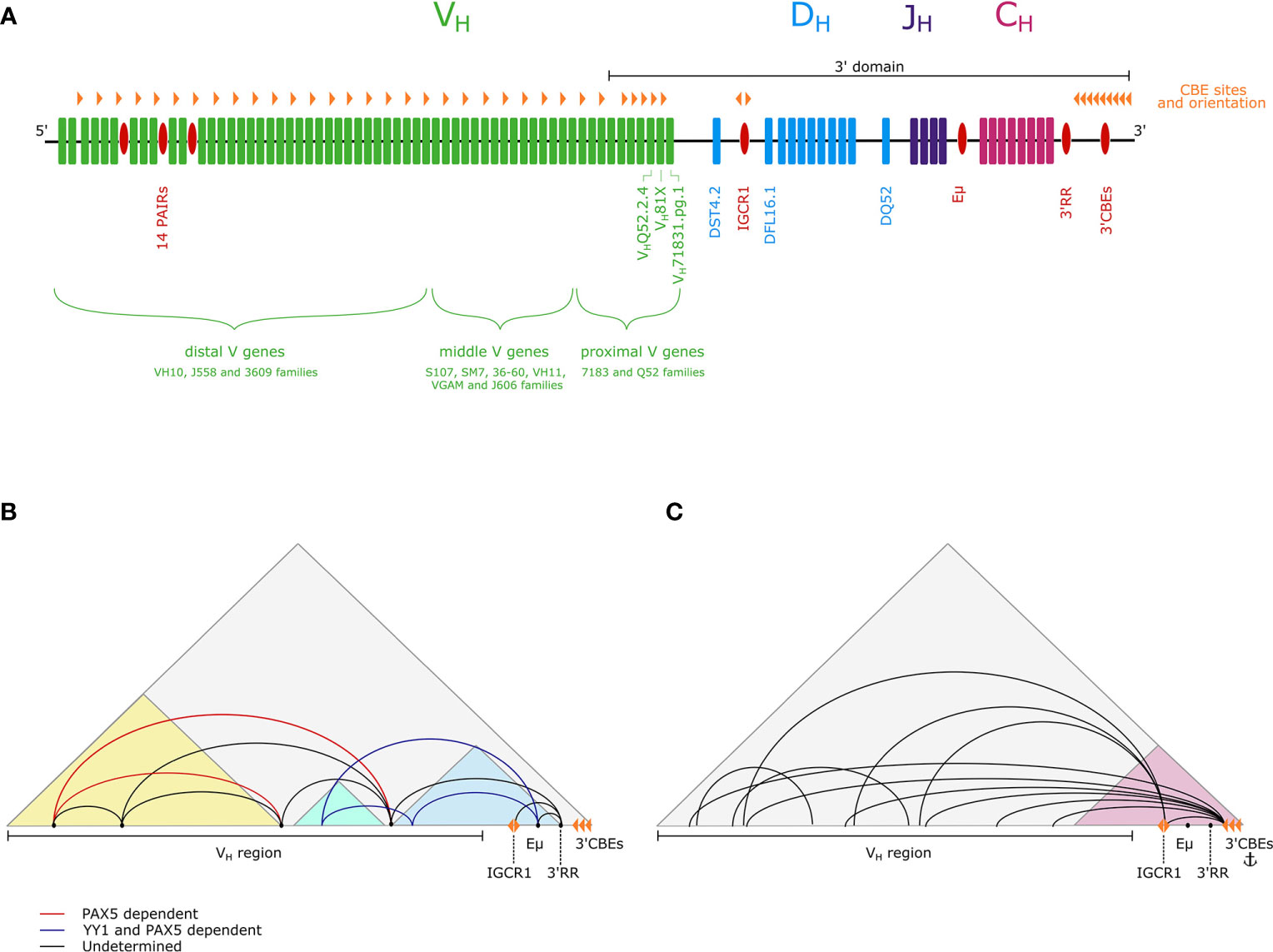
Figure 1 (A) A schematic of the Igh locus showing VH (green), DH (blue), JH (purple) and CH (pink) genes, as well as regulatory elements (red). The positions of CBEs are also marked (orange triangles).The 5’-3’ orientation is indicated and the extent of the 3’domain is depicted by a black line. (B) The three subdomain model proposes that within the Igh TAD (gray) there are three subdomains (light blue, light green, yellow), with extensive looping within them. Interactions between subdomains are mediated by loop anchoring sites (black points). A subset of these loops is PAX5-dependent (red) and PAX5- and YY1-dependent (dark blue) (10–12). (C) The flexible looping model proposes that there is a dynamic continuum of loops from the entire VH region to the 3’ end of the locus, with the 3’ region forming a more structurally invariant subdomain (pink) containing an anchor point for long-range interactions (13, 14).
There are a number of well characterized regulatory elements in the Igh locus that contribute to locus structure (Figure 1A). First, the intronic enhancer Eμ, located between the JH and CH genes (17), is required for effective recombination (18–21). Second, the 3’ regulatory region (3’RR), which contains four DNase I hypersensitive sites (HSs), HS1-4, is a B lineage-specific enhancer (22, 23). Downstream, there is a regulatory element, containing HS5-8 and 10 binding sites for CCCTC-binding factor (CTCF), which is known as the 3’ CTCF binding elements (3’CBEs) and functions as an insulator and a superanchor (13, 24). In addition, the intergenic control region 1 (IGCR1), located just upstream of the DH genes, contains two CBEs, and operates as an insulator to prevent premature VH-DJH recombination (25). Potential regulatory elements have also been discovered in the distal VH region, in the form of 14 tandemly repeating PAX5-activated intergenic repeat (PAIR) elements (26). PAIR elements are bound by several transcription factors and architectural proteins, including the B cell-specific transcription factor PAX5, which mediates non-coding antisense transcription from PAIR elements in pro-B cells (26, 27).
Local chromatin environments within the nucleus can be permissive for or refractory to active nuclear processes. The nuclear periphery is a region of repressive heterochromatin, and in non-B cells the Igh locus is anchored at the nuclear periphery via its VH end (28). FISH studies showed that before V(D)J recombination, the Igh locus is activated by relocation from the nuclear periphery to permissive euchromatin in the nuclear interior (29, 30), dependent on the transcription factors EBF1 and E2A, and on Eμ (10, 29, 31–33). In addition to relocation, the Igh locus undergoes large-scale contraction to bring VH and DH genes in spatial proximity for VH to DJH recombination (29, 30, 34). This was measured by a significant reduction in the distance between FISH probes positioned on opposite ends of the locus in pro-B cells compared with pre-pro-B cells. DNA looping was subsequently visualized with three-color FISH, which showed that distal VH genes frequently loop closer to the CH region than proximal VH genes that are closer in linear sequence (33). These early FISH studies introduced pro-B cells lacking the RAG recombinase as a model to study Igh locus interactions. In Rag-/- pro-B cells, the Igh locus retains its intact germline sequence but is poised for recombination.
Translating individual observations of Igh locus contraction and looping into a comprehensive model of the 3D structure of this enormous locus has been a challenge. The Murre laboratory first visualized a putative overall 3D Igh locus structure using 11 small FISH probes distributed evenly along the Igh locus, combined with mathematical modeling (35). Distal VH probes were more frequently positioned close to the Eμ-JH region in pro-B cells than in pre-pro-B cells, revealing a major conformational change in the Igh locus when it is poised for recombination, suggesting that the distal VH genes are brought close to Eμ-JH region to have equal opportunity with proximal VH genes to recombine. Furthermore, in pro-B cells, the 3’ end of the Igh locus interacted with distal VH genes as frequently as with the proximal VH genes, indicating extensive long-range DNA looping involving this CBE-rich region. This study also reported larger variability of distance measurements in pro-B cells than in pre-pro-B cells, suggesting more variation in the interaction landscape at this stage. Moreover, the locus formed into clusters of loops at both developmental stages.
Subsequently, the Murre lab employed live imaging of fluorescently labeled Igh loci and computer modeling to determine first passage times of interactions between VH and DH genes (36). This is a major advance beyond FISH techniques on fixed cells, giving insights into the real-time dynamics of recombination. This study revealed that a VH gene interacts with the DJH region within minutes of DH-JH recombination, much faster than expected. It was proposed that the DHJH region resides in a cavity surrounded by equally distant VH genes and that the viscous nuclear environment causes VH genes to bounce back and forth rapidly until a specific synapsis is established by the RAG machinery, aligning with the equal opportunity model above (37).
Several mouse models have given insight into the transcription factors required for locus contraction and looping. PAX5, YY1, E2A and Ikaros are essential for contraction and all bind to numerous sites within the Igh locus, but it remains unclear if their roles are direct or indirect (33, 34, 38, 39). CTCF is also implicated in mediating loops in the Igh locus: reduction of CTCF binding decreases locus contraction (40). CTCF binding to the Igh locus is lymphocyte-specific (41). The 10 CBEs in the 3’CBEs (13, 41) are in a convergent orientation to 121 CBEs distributed throughout the VH region (41), likely facilitating looping between the 3’ and 5’ ends of the locus. In addition, there are two CBEs in the IGCR1 that are in divergent orientations: CBE1 can facilitate CTCF-mediated looping between the IGCR1 and the VH region, while CBE2 can facilitate interactions between IGCR1 and the 3’CBEs (13, 14, 42). There is extensive co-localization between CTCF and cohesin binding sites genome-wide (43), and cohesin binds to multiple shared sites within the Igh locus, although its binding is more dynamic, being most enriched at the pro-B stage (44).
Detailed understanding of the 3D DNA interactions governing AgR recombination have benefited enormously from chromosome conformation capture (3C) and its later refinements (4C, 5C, HiC), which have revealed genomic contacts underpinning locus contraction at higher resolution (10s of kb compared with 100s of kb for FISH).
3C-based studies have shown that the Igh locus forms a discrete topologically associated domain (TAD) (11, 13). Furthermore, the 3’ domain of the Igh locus is compacted and highly structured, suggesting little variability between individual B cells. The 3’CBEs, 3’RR and IGCR1 interact with Eμ, in addition to a separate interaction between the 3’ CBEs and IGCR1 (10, 40). These interactions are independent of locus contraction (14). However, more recent 4C and 5C studies of Igh locus conformation suggest that the 3’ domain extends beyond the VH-DH intergenic region, to encompass proximal VH gene segments (11, 14). This is consistent with the observation that proximal VH genes can undergo recombination in the absence of locus contraction in pro-B cells (34, 45).
There are also local interactions within the VH region. A number of these interactions are independent of locus contraction, such as the PAX5-independent interactions of the PAIR elements with other regions in the distal VH (14). Moreover, middle VH36-60 and distal VH10 regions of the locus are involved in local interactions that are CTCF mediated and are present in Eµ deficient cells with impaired contraction (10, 12).
In addition to local looping, there is a requirement for long range contacts between the 5’ and 3’ ends of the locus to facilitate VH-DJH recombination. How VH genes loop to the 3’ region at the pro-B cell stage to enable all VH genes to recombine is an ongoing debate. A 5C and FISH study from the Kenter lab defined loop anchor sites that are key in mediating megabase-scale interactions between subdomains, and demonstrated that these long-range interactions are Eμ independent, with a subset being PAX5-dependent. On this basis they have proposed the three subdomain model, also favored by the Sen lab (11, 12). They propose compaction is achieved by CTCF-dependent intra-subdomain looping, together with long-range looping between these subdomains, mediated by PAX5 and YY1 (Figure 1B) (11, 12, 46).
In contrast, based on observations of interactions from 16 4C-seq viewpoints in the Igh locus, the Busslinger laboratory propose a flexible looping model, with a continuum of dynamic loops from VH gene segments to the more rigid 3’ domain, providing equal opportunity for recombination of all VH genes, aligning with findings from the Murre lab (Figure 1C) (14). The Busslinger study also questioned the importance of Eµ and the IGCR1 as central hubs of intralocus interactions.
Building on FISH and 3C studies that have given a detailed understanding of overall Igh locus conformation and intralocus looping events, recent work has investigated the mechanism of loop formation in the locus. In studies aiming to understand RAG off-target activity in chromosomal translocations, the Alt lab showed that RAG off-target activity is limited to convergent CBE-based loop domains (47). 12-RSS and 23-RSS pairs were inserted at random into the genome of a pro-B cell line to create ectopic recombination centers. RAG mediated recombination occurred within convergent CBE loop domains. Furthermore, deletional recombination of RSSs conforming to the 12/23 rule, and in a convergent orientation was preferred, similar to canonical Igh V(D)J recombination. This indicated that RAG may bind one RSS and then linearly track along chromatin to find a convergent RSS within the same loop domain (47). This model is referred to as the RAG scanning hypothesis (48).
A wealth of evidence demonstrates that TADs and finer-scale loop features are formed through the process of loop extrusion (49–51). Cohesin, a loop extruding complex that holds two strands of DNA together, is loaded onto DNA by Nipbl. Cohesin progressively extrudes larger DNA loops until stalled at boundary elements, principally CTCF bound at convergent CBEs. The enrichment for CBEs at key regulatory regions in the Igh locus suggests a role for CTCF loop domains in organizing locus architecture, and the restriction of off-target RAG scanning to loop domains suggests that scanning could also occur through a loop extrusion related process in the Igh locus (47).
Mutational analysis showed that inversion of DH-12-RSSs impaired DH-JH rearrangement, indicating that the orientation of the JH-23-RSS directs upstream RAG scanning to a convergently orientated DH-12-RSS in the Igh 3’ subdomain (52). Hence in most DH-JH rearrangements, recombination by deletion is favored over recombination by inversion, suggesting a role for RAG scanning by loop extrusion (Figure 2A). However, recombination from the DQ52 gene segment, adjacent to the JH genes (Figure 1A), can be inversional, using a diffusion-based mechanism due to its close spatial proximity to the DJH recombination center.
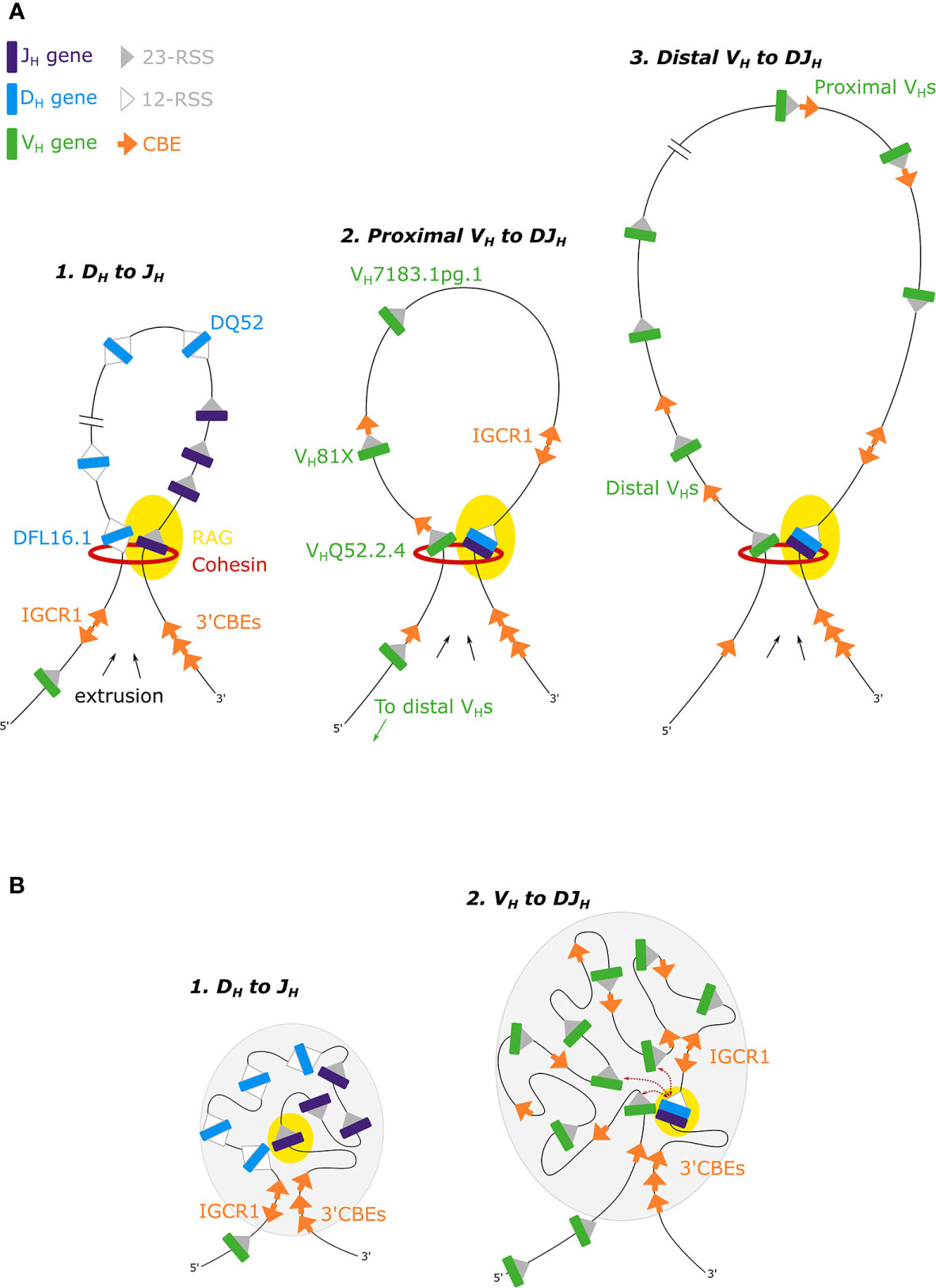
Figure 2 (A) The loop extrusion model for DH-JH, proximal VH-DJH and distal VH-DJH recombination. Cohesin-mediated RAG-scanning leads to the extrusion of progressively larger loops, until loops are stalled by convergent CBEs (53, 54). (B) In the phase separation model for Igh recombination, recombination occurs near the boundary of the gel phase (gray ovals) (55).
A number of recent studies from the Alt and Sen labs have shown that deletion of the IGCR1 leads to increased interactions between the 3’ domain, specifically Eµ, and proximal VH genes, resulting in increased recombination of these proximal genes (56–58). IGCR1 deletion disrupts CBE loop domains within the Igh locus, consequently altering RAG targets (47). Deletion leads to premature VH to germline DH joins, and increased rearrangement of the remote, rarely utilized, DH gene DST4.2 upstream of the IGCR1 (Figure 1A). Thus removing this barrier allows extrusion into the intergenic and proximal VH region (58). The Sen lab has also shown that recombination of VH gene segments to germline DH gene segments can occur by inversion of the DH gene, rather than deletion, on IGCR1-deficient alleles (58). They argue that the mechanism of VH-DJH recombination is distinct from that of DH-JH recombination, and propose that VH-DJH recombination occurs primarily through a diffusion-based mechanism. However, VH-DJH recombination via diffusion has not been shown on wild type alleles, and it is unclear what can be extrapolated from the IGCR1 deletion context.
Other barriers to RAG scanning include active transcription (59), which has been proposed to focus RAG to its target RSSs (52). Transcription impedes RAG scanning in an artificial construct with cryptic scanning across the downstream CH region (52). It will be important to determine whether transcription impedes RAG scanning in canonical locations, for example at the PAIR elements, since this would implicate two bona fide, complementary mechanisms in enabling RAG accumulation at target RSSs.
CTCF binding sites within the VH region exhibit a bipartite pattern. Proximal recombining VH genes are associated with a CBE just 3’ of their RSS but pseudogenes in this region lack adjacent CBEs (37, 44). In contrast, distal VH genes are not closely associated with CBEs, which can either be far upstream of VH genes or intergenic (44, 60). Focusing on the proximal VH genes: mutation of the CBE associated with the first functional VH81X gene (Figure 1A), leads to reduced VH81X gene usage but increased usage of the next upstream gene VHQ52.2.4, alongside a reduced interaction between VH81X and both the IGCR1 and the 3’CBEs (56, 57). Recombination of the VHQ52.2.4 gene is also dependent on its flanking CBE (56). Moreover, repair of a non-functional CBE associated with the first VH gene, pseudogene VH7183.1pg.1, converted it into the most highly rearranging VH gene (56). This work supports a RAG scanning mechanism within the proximal VH genes (Figure 2A), where convergent CBEs impede further scanning, thereby tethering the RAG complex and mediating interactions between VH genes and the 3’ domain to facilitate VH gene recombination.
Two recent studies have extended our knowledge of RAG scanning within the VH domain, suggesting that it is not limited to the proximal VH genes but occurs over the entire multi-megabase VH region. Both the Busslinger and Alt labs generated large inversions in the VH domain of primary pro-B cells. In the first, there was an 890kb inversion in the distal VH region, while in the second, nearly the entire 2.4Mb murine Igh VH region was inverted (53, 61). In both cases, recombination of the inverted VH genes was abrogated. Efficient recombination was dependent on both the orientation of the VH gene and of the associated CBEs (53), and utilization of cryptic RSSs, now in a convergent orientation, was activated (61). Busslinger and colleagues also used 3C-seq to demonstrate that following inversion of the distal domain, interactions across the entire Igh locus were altered (53). Furthermore, insertion of an array of 20 CBEs in the middle of the Igh VH domain in an inverted orientation altered loop domains in the Igh locus and disrupted long range interactions from the 3’ of the locus to the VH domain. These findings implicate a loop extrusion mechanism for recombination across the entire VH domain (Figure 2A), with loops of up to 2.8Mb (53), larger than the average TAD (880kb) (62). However, while recombination to VH genes immediately 5’ of the insertion was reduced, this was limited to a 355kb region, which is not consistent with a single looping event across the VH domain (53). In addition, FISH and 4C studies indicate multiple loops in the distal VH domain (14, 35). It remains unclear how large numbers of CBEs are bypassed to generate loops between distal VH genes and the DJH gene segment. It is also unknown how frequently the >100 CBEs in the VH region are occupied by CTCF. If occupation follows a dynamic pattern on individual Igh alleles, this would facilitate a stochastic pattern of CBE stalling that would enable diverse participation of VH genes in VH-DH recombination. It also remains unclear how loop extrusion across the entire VH domain enables the recombination of all active distal VH genes given that they are not paired with a CBE. Does loop extrusion work in concert with diffusion based mechanisms for distal VH genes, whereby stalling of the loop acts to tether a number of genes in close proximity to the recombination center, obviating the need for juxtaposed CBEs? It is also feasible that other loop impeding mechanisms, such as transcription (59), may contribute to stalling. All of these possibilities require experimental testing.
Clarification of the roles of CTCF and cohesin in RAG scanning by loop extrusion has come from degradation of Rad21 and CTCF in v-Abl-transformed pro-B cell lines, using an auxin-inducible degron system (54). These cell lines have impaired locus contraction and favor proximal over distal VH-DJH recombination compared to primary pro-B cells, despite the distal VH genes being highly transcribed; the underlying defect is not known (54). Rad21 degradation led to: disruption of looping in the 3’ region; severe reduction in DH-JH recombination (DQ52 could still recombine through diffusion based mechanisms); and total abrogation of VH-DJH recombination. This supports a role for cohesin-mediated loop extrusion in both stages of V(D)J recombination, albeit in a system where only the most proximal five VH genes recombine. CTCF degradation reduced both CTCF and Rad21 occupancy in the VH region (54). Strikingly, while CTCF mediated interactions were reduced, interactions of the recombination center with VH regions were increased. Distal VH-DJH recombination was restored, comparable to wild type loci in primary pro-B cells that undergo extensive locus contraction. Bound CTCF impedes RAG scanning at the 3’ VH genes (56), so relief of this impediment through CTCF degradation may promote cohesin-mediated RAG scanning throughout the VH region. However this interesting study raises some puzzling questions. How does restoration of loop extrusion and distal VH gene recombination caused by loss of CTCF binding in a v-Abl-transformed pro-B cell line equate with the wild type in vivo situation where CTCF is bound and recombination to distal VH genes occurs? Loss of CTCF also caused global reorganization of chromatin, thus it cannot be ruled out that the effect of CTCF loss at the Igh locus in the cell lines is an indirect effect of expression of another factor, or of more widespread genome disorganization. CTCF binds the Igh locus earlier in development with cohesin binding more dynamically (41). Moreover, in order to fully understand how diverse antibody repertoires are generated, where loop stalling may be a stochastic process across a population of cells, the occupancy of CTCF and cohesin at a single cell or allele level, the interplay between these two factors, which bind similar motifs, and their binding kinetics need further investigation. Furthermore, in contrast, while beyond the scope of this review, loss of CTCF in vivo leads to impaired recombination of distal Vκ genes (63). Taken together, while it is clear that CTCF plays a pivotal role in organizing Igh (and Igκ) locus structure, how this is achieved requires further investigation.
Reframing the model from a direct to an indirect role has recently occurred for PAX5, a key transcription factor that mediates commitment to the pro-B cell stage, and long implicated in control of the 3D organization of the Igh locus (34). As PAX5 binds to multiple sites in the Igh locus, including the 3’RR, the VH-DH intergenic region (but not at the IGCR1) and the distal VH region at the PAIR elements and at approximately 20 VH gene RSSs (6, 26), it has been hypothesized that its importance in Igh recombination is due to direct regulation of the locus (26, 27). However recent insights from the Busslinger lab indicate that PAX5 regulates Igh recombination, at least in part, indirectly through control of the cohesin DNA release factor WAPL (53). PAX5 represses the transcription of Wapl in both pro- and pre-B cells, through binding to the Wapl promoter and recruiting PRC2. PAX5 downregulation of Wapl increases cohesin residence time, and modulates Igh loop extrusion and recombination. Moreover, Wapl repression by PAX5 controls chromatin architecture at a global scale in pro-B cells. This provides a mechanism for previous reports of a global role for PAX5 in progenitor B cell genome organization that was not dependent on its canonical role as a transcription factor (64).
Interestingly, v-Abl-transformed pro-B cell lines have increased transcription of Wapl (61). Degradation of Wapl in these cell lines restores locus contraction and utilization of distal VH genes in VH-DJH recombination. Taken together, PAX5 repression of Wapl facilitates RAG scanning across the entire VH domain, allowing loops much larger than the average TAD. Lower levels of WAPL enable RAG scanning over longer distances due to increased cohesin residency time on DNA, which, at the Igh locus, results in increased locus contraction and facilitates recombination to distal VH genes. Cohesin and CTCF also play key roles in TCR recombination (65), suggesting that loop extrusion may play a wider role in AgR recombination. However it is unclear whether control of loop extrusion through Wapl regulation can be extended to other AgR loci. Indeed, Wapl expression remains high during T cell development, including during TCRβ and TCRα rearrangement (53), thus different mechanisms may be at play to modulate cohesin dynamics. It will be interesting to see if other lymphocyte specific transcription factors regulate the loop extrusion machinery.
Phase separation describes a concept that a mixture of components can separate based on their physical and chemical properties into two or more distinct phases. It has been proposed that phase separation, driven by cooperative interactions between chromatin associated proteins, underlies A and B compartmental segregation of chromosomal regions (66, 67). Polymer simulations have suggested that the interplay of active chromatin loop extrusion and phase separation mechanisms can explain chromatin organization (66).
Murre and colleagues extended their previous live imaging work, developing a two color labeling strategy to simultaneously track the motion of paired VH and DJH loci (55). Computer modeling where the Igh locus was in a weak gel state, near the sol-gel phase boundary, best reproduced VH-DJH first encounter times (Figure 2B). This phase enables the Igh locus to move flexibly to facilitate rapid and random recombination while providing enough stability to promote ordered DH-JH and then VH-DJH recombination. The authors propose that phase separation acts cooperatively with loop extrusion, employing local chromatin based cross-linking to stabilise the loops. However, this model of restrained motion is rather at odds with earlier studies from the lab which are indicative of rapid, flexible motion (36).
In recent years, enormous progress has been made in three key areas: description of Igh locus conformation; understanding the underlying mechanisms that organize the locus; understanding how the RAG recombinase achieves RSS synapsis. Together, these components provide an extremely intricate and dynamic picture of the role of AgR locus conformation in generation of diverse AgR repertoires. In each sphere, divergent models have emerged and future progress will depend on new methods to resolve their differences. First, convergence of the three subdomain and flexible looping models of conformation could be achieved by high resolution capture Hi-C of the entire Igh locus. Second, of the two rather different mechanisms proposed to organize the locus, loop-extrusion, orchestrated by cohesin, CTCF and Wapl, is supported by numerous genome-wide studies. Phase separation is less established and reflects pioneering work. While the convergence of these two models is not on the immediate horizon, it is conceivable, based on polymer modeling studies that have shown the interplay of these two mechanisms genome-wide (65), that loop extrusion and phase separation act cooperatively to co-orchestrate AgR folding and recombination. Here, we envisage that developments in polymer modeling approaches may resolve the differences between these models. Igh locus conformation per se, and its underlying mechanisms are RAG-independent, and serve to prepare the locus for recombination events. The third piece of the puzzle is RAG targeting. Contrasting models suggest that the RAG complex avails of loop extrusion to scan for convergent RSSs, or brings nearby convergent RSSs together by diffusion. Neither model yet fully explains recombination to distal VH genes. Here too, cooperation between two mechanisms may be at play. Gene-targeting models addressing both may reveal how this may occur.
CR and AC planned the content of the review and wrote the abstract. CR and OM drafted the manuscript, AC edited the manuscript, CR prepared the figures, CR, OM and AC finalized the manuscript. All authors contributed to the article and approved the submitted version.
CR is supported by a PhD studentship (1947339) from the Medical Research Council, UK. OM was supported by a PhD studentship (1426107) from the Medical Research Council, UK. Work in AEC's laboratory is supported by grants from the Biotechnology and Biological Scientific Research Council (BBS/E/B/000C0404, BBS/E/B/000C0405, BBS/E/B/000C0427, BBS/E/B/000C0428). The Babraham Institute provides funds, throough the BBSRC, for open access publication fees.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol (2011) 11:251–63. doi: 10.1038/nri2941
2. Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell (2010) 141:419–31. doi: 10.1016/j.cell.2010.03.010
3. Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19 – early B cell precursors. J Exp Med (2006) 203:675–87. doi: 10.1084/jem.20052444
4. Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity (1996) 5:575–89. doi: 10.1016/S1074-7613(00)80272-1
5. Kuo TC, Schlissel MS. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol (2009) 21:173–8. doi: 10.1016/j.coi.2009.03.008
6. Bolland DJ, Koohy H, Wood AL, Matheson LS, Krueger F, Stubbington MJT, et al. Two Mutually Exclusive Local Chromatin States Drive Efficient V(D)J Recombination. Cell Rep (2016) 15:2475–87. doi: 10.1016/j.celrep.2016.05.020
7. Matheson LS, Bolland DJ, Chovanec P, Krueger F, Andrews S, Koohy H, et al. Local Chromatin Features Including PU.1 and IKAROS Binding and H3K4 Methylation Shape the Repertoire of Immunoglobulin Kappa Genes Chosen for V(D)J Recombination. Front Immunol (2017) 8:1550. doi: 10.3389/fimmu.2017.01550
8. Kumari G, Sen R. Chapter Two - Chromatin Interactions in the Control of Immunoglobulin Heavy Chain Gene Assembly. In: Murre C, editor. Molecular Mechanisms that Orchestrate the Assembly of Antigen Receptor Loci Advances in Immunology. United States: Academic Press (2015). p. 41–92. doi: 10.1016/bs.ai.2015.08.001
9. Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell (1996) 85:887–97. doi: 10.1016/S0092-8674(00)81272-6
10. Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell (2011) 147:332–43. doi: 10.1016/j.cell.2011.08.049
11. Montefiori L, Wuerffel R, Roqueiro D, Lajoie B, Guo C, Gerasimova T, et al. Extremely Long-Range Chromatin Loops Link Topological Domains to Facilitate a Diverse Antibody Repertoire. Cell Rep (2016) 14:896–906. doi: 10.1016/j.celrep.2015.12.083
12. Gerasimova T, Guo C, Ghosh A, Qiu X, Montefiori L, Verma-Gaur J, et al. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev (2015) 29:1683–95. doi: 10.1101/gad.263871.115
13. Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci (2015) 112:12776–81. doi: 10.1073/pnas.1512995112
14. Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity (2013) 39:229–44. doi: 10.1016/j.immuni.2013.08.011
15. Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete Sequence Assembly and Characterization of the C57BL/6 Mouse Ig Heavy Chain V Region. J Immunol (2006) 176:4221–34. doi: 10.4049/jimmunol.176.7.4221
16. Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin Immunol (2010) 22:353–61. doi: 10.1016/j.smim.2010.08.001
17. Banerji J, Olson L. Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell (1983) 33:729–40. doi: 10.1016/0092-8674(83)90015-6
18. Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci (2005) 102:14362–7. doi: 10.1073/pnas.0507090102
19. Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. Regulation of IgH gene assembly: role of the intronic enhancer and 5’DQ52 region in targeting DHJH recombination. J Immunol (2006) 176:2439–47. doi: 10.4049/jimmunol.176.4.2439
20. Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol (2007) 27:5523–33. doi: 10.1128/MCB.02407-06
21. Chakraborty T, Perlot T, Subrahmanyam R, Jani A, Goff PH, Zhang Y, et al. Sen R. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med (2009) 206:1019–27. doi: 10.1084/jem.20081621
22. Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, et al. Localization of the 3′ IgH locus elements that effect long-distance regulation of class switch recombination. Immunity (2001) 15:187–99. doi: 10.1016/S1074-7613(01)00181-9
23. Dunnick WA, Shi J, Zerbato JM, Fontaine CA, Collins JT. Enhancement of Antibody Class-Switch Recombination by the Cumulative Activity of Four Separate Elements. J Immunol (2011) 187:4733–43. doi: 10.4049/jimmunol.1101808
24. Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, et al. Chromatin Architecture near a Potential 3′ End of the Igh Locus Involves Modular Regulation of Histone Modifications during B-Cell Development and In Vivo Occupancy at CTCF Sites. Mol Cell Biol (2005) 25:1511–25. doi: 10.1128/mcb.25.4.1511-1525.2005
25. Lin SG, Guo C, Su A, Zhang Y, Alt FW. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc Natl Acad Sci (2015) 112:1815–20. doi: 10.1073/pnas.1424936112
26. Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, et al. The Distal VH Gene Cluster of the Igh Locus Contains Distinct Regulatory Elements with Pax5 Transcription Factor-Dependent Activity in Pro-B Cells. Immunity (2011) 34:175–87. doi: 10.1016/j.immuni.2011.02.005
27. Verma-Gaur J, Torkamani A, Schaffer L, Head SR, Schork NJ, Feeney AJ. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc Natl Acad Sci (2012) 109:17004–9. doi: 10.1073/pnas.1208398109
28. Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell (2012) 149:1474–87. doi: 10.1016/j.cell.2012.04.035
29. Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science (2002) 296:158–62. doi: 10.1126/science.1068768
30. Rother MB, Palstra RJ, Jhunjhunwala S, Van Kester KAM, Van IJcken WFJ, Hendriks RW, et al. Nuclear positioning rather than contraction controls ordered rearrangements of immunoglobulin loci. Nucleic Acids Res (2016) 44:175–86. doi: 10.1093/nar/gkv928
31. Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev (2010) 237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x
32. Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol (2012) 13:1196–204. doi: 10.1038/ni.2432
33. Sayegh C, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev (2005) 19:322–7. doi: 10.1101/gad.1254305
34. Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev (2004) 18:411–22. doi: 10.1101/gad.291504
35. Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJM, et al. The 3D Structure of the Immunoglobulin Heavy-Chain Locus: Implications for Long-Range Genomic Interactions. Cell (2008) 133:265–79. doi: 10.1016/j.cell.2008.03.024
36. Lucas JS, Zhang Y, Dudko OK, Murre C. 3D trajectories adopted by coding and regulatory DNA elements: First-passage times for genomic interactions. Cell (2014) 158:339–52. doi: 10.1016/j.cell.2014.05.036
37. Lucas JS, Bossen C, Murre C. Transcription and recombination factories: Common features? Curr Opin Cell Biol (2011) 23:318–24. doi: 10.1016/j.ceb.2010.11.007
38. Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev (2007) 21:1179–89. doi: 10.1101/gad.1529307
39. Reynaud D, Demarco I A, Reddy K L, Schjerven H, Bertolino E, Chen Z, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol (2008) 9:927–36. doi: 10.1038/ni.1626
40. Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci (2011) 108:9566–71. doi: 10.1073/pnas.1019391108
41. Loguercio S, Barajas-Mora EM, Shih H-Y, Krangel MS, Feeney AJ. Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci. Front Immunol (2018) 9:425. doi: 10.3389/fimmu.2018.00425
42. Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng H-L, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature (2011) 477:424–30. doi: 10.1038/nature10495
43. Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, et al. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell (2008) 132:422–33. doi: 10.1016/j.cell.2008.01.011
44. Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting Edge: Developmental Stage-Specific Recruitment of Cohesin to CTCF Sites throughout Immunoglobulin Loci during B Lymphocyte Development. J Immunol (2009) 182:44–8. doi: 10.4049/jimmunol.182.1.44
45. Hesslein DGT, Pflugh DL, Chowdhury D, Bothwell ALM, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev (2003) 17:37–42. doi: 10.1101/gad.1031403
46. Kenter AL, Feeney AJ. New insights emerge as antibody repertoire diversification meets chromosome conformation. F1000Research (2019) 8:347. doi: 10.12688/f1000research.17358.1
47. Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, et al. Chromosomal Loop Domains Direct the Recombination of Antigen Receptor Genes. Cell (2015) 163:947–59. doi: 10.1016/j.cell.2015.10.016
48. Lin SG, Ba Z, Alt FW, Zhang Y. RAG Chromatin Scanning During V(D)J Recombination and Chromatin Loop Extrusion are Related Processes. In: Advances in immunology. United States: Elsevier (2018). p. 93–135. doi: 10.1016/bs.ai.2018.07.001
49. Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci (2015) 112:E6456–65. doi: 10.1073/pnas.1518552112
50. Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep (2016) 15:2038–49. doi: 10.1016/j.celrep.2016.04.085
51. Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, et al. Real-time imaging of DNA loop extrusion by condensin. Science (2018) 360:102–5. doi: 10.1126/science.aar7831
52. Zhang Y, Zhang X, Ba Z, Liang Z, Dring EW, Hu H, et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature (2019) 573:600–4. doi: 10.1038/s41586-019-1547-y
53. Hill L, Ebert A, Jaritz M, Wutz G, Nagasaka K, Tagoh H, et al. Wapl repression by Pax5 promotes V gene recombination by Igh loop extrusion. Nature (2020) 584:142–7. doi: 10.1038/s41586-020-2454-y
54. Ba Z, Lou J, Ye AY, Dai H-Q, Dring EW, Lin SG, et al. CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning. Nature (2020) 586:305–10. doi: 10.1038/s41586-020-2578-0
55. Khanna N, Zhang Y, Lucas JS, Dudko OK, Murre C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun (2019) 10:1–13. doi: 10.1038/s41467-019-10628-9
56. Jain S, Ba Z, Zhang Y, Dai H-Q, Alt FW. CTCF-Binding Elements Mediate Accessibility of RAG Substrates During Chromatin Scanning. Cell (2018) 174:1–15. doi: 10.1016/j.cell.2018.04.035
57. Qiu X, Kumari G, Gerasimova T, Du H, Labaran L, Singh A, et al. Sequential Enhancer Sequestration Dysregulates Recombination Center Formation at the IgH Locus. Mol Cell (2018) 70:21–33.e6. doi: 10.1016/j.molcel.2018.02.020
58. Qiu X, Ma F, Zhao M, Cao Y, Shipp L, Liu A, et al. Altered 3D chromatin structure permits inversional recombination at the IgH locus. Sci Adv (2020) 6:eaaz8850. doi: 10.1126/sciadv.aaz8850
59. Brandão HB, Paul P, van den Berg AA, Rudner DZ, Wang X, Mirny LA. RNA polymerases as moving barriers to condensin loop extrusion. Proc Natl Acad Sci USA (2019) 116:20489–99. doi: 10.1073/pnas.1907009116
60. Ciccone DN, Namiki Y, Chen C, Morshead KB, Wood AL, Johnston CM, et al. The murine IgH locus contains a distinct DNA sequence motif for the chromatin regulatory factor CTCF. J Biol Chem (2019) 294:13580–92. doi: 10.1074/jbc.RA118.007348
61. Dai H-Q, Hu H, Lou J, Ye AY, Ba Z, Zhang X, et al. Loop Extrusion Mediates Physiological Locus Contraction for V(D)J Recombination. Nature (2020). doi: 10.1038/s41586-020-03121-7.
62. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature (2012) 485:376–80. doi: 10.1038/nature11082
63. Ribeiro de Almeida C, Stadhouders R, de Bruijn MJW, Bergen IM, Thongjuea S, Lenhard B, et al. The DNA-Binding Protein CTCF Limits Proximal Vκ Recombination and Restricts κ Enhancer Interactions to the Immunoglobulin κ Light Chain Locus. Immunity (2011) 35:501–13. doi: 10.1016/j.immuni.2011.07.014
64. Johanson TM, Lun ATL, Coughlan HD, Tan T, Smyth GK, Nutt SL, et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat Immunol (2018) 19:1257–64. doi: 10.1038/s41590-018-0234-8
65. Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature (2011) 476:467–73. doi: 10.1038/nature10312
66. Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci (2018) 115:E6697–706. doi: 10.1073/pnas.1717730115
Keywords: V(D)J recombination, immunoglobulin, 3D conformation, loop extrusion, RAG scanning, chromatin regulation, genome organization
Citation: Rogers CH, Mielczarek O and Corcoran AE (2021) Dynamic 3D Locus Organization and Its Drivers Underpin Immunoglobulin Recombination. Front. Immunol. 11:633705. doi: 10.3389/fimmu.2020.633705
Received: 26 November 2020; Accepted: 30 December 2020;
Published: 18 February 2021.
Edited by:
Amy L. Kenter, University of Illinois at Chicago, United StatesReviewed by:
Michael Steven Krangel, Duke University Medical Center, United StatesCopyright © 2021 Rogers, Mielczarek and Corcoran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne E. Corcoran, YW5uZS5jb3Jjb3JhbkBiYWJyYWhhbS5hYy51aw==orcid.org/0000-0002-9577-5313
†Present address: Olga Mielczarek, Horizon Discovery, Cambridge, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.