
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol. , 02 February 2021
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.623993
This article is part of the Research Topic New Insights into the Complexity of Tumor Immunology in B-cell Malignancies: Prognostic and Predictive Biomarkers and Therapy View all 11 articles
The monocyte/macrophage lineage has been shown to be involved in the promotion of a protumoral tumor microenvironment and resistance to treatment in B cell lymphomas. However, it is still poorly described at the single cell level, and tissue samples are not easily accessible. Thus, a detailed analysis of the circulating myeloid cell compartment in the different B lymphomas is needed to better understand the mechanisms of resistance to treatment and identify at risk patients. In this Perspective, we review current knowledge on the phenotypic and functional description of the circulating monocytic lineage in B cell lymphomas and provide first insights into the heterogeneity of these cell populations in health and lymphoma, using mass cytometry. Indeed, the monocytic compartment is a continuum more than distinct subpopulations, as demonstrated by our high-resolution approach, explaining the sometimes confusing and contradictory conclusions on the prognostic impact of the different populations, including monocytes and monocytic myeloid derived suppressor cells (M-MDSC). By identifying S100A9high monocytic cells as a potential biomarker in diffuse large B cell lymphoma (DLBCL) in this proof-of-concept preliminary study including a limited number of samples, we underline the potential of circulating myeloid regulatory cells as diagnostic and prognostic biomarkers in B-cell lymphomas.
Lymphomas are malignancies that arise from the lymphoid system with an involvement of the B lineage in 90% of cases (1). The lymphoma nomenclature, based on clinical, pathological, genetic, and molecular description (2), is constantly evolving. Different subtypes are described, depending on the stage of maturation from which the tumoral B cell derives. Beside the first historically described Hodgkin lymphoma (HL), non-Hodgkin lymphomas (NHL) include essentially mantle cell lymphoma (MCL), follicular lymphoma (FL), diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma, and chronic lymphoid leukemia (CLL). As in many cancers, the importance of the tumor microenvironment (TME) in the development of the disease and response to treatment has been emphasized in B-cell lymphoma, in particular, lymphomas can be defined by their dependence to the TME (1, 3–6). The TME is in particular composed of immune cells including T cell subsets (follicular helper T cells [Tfh], regulatory T cells [Treg]), stromal cells, dendritic cells, and myeloid cells (monocytes, macrophages, myeloid derived suppressor cells [MDSC]) and is affected by the tumor localization (e.g. lymph node and bone marrow in FL or blood in CLL). The myeloid cell compartment has lately gained great interest and is extensively studied in several cancer types, including B lymphoma, due to its numerous key and ambivalent roles in pro-tumoral immune suppression, anti-tumor immunity, immunotherapy efficacy through antibody-dependent cellular phagocytosis (ADCP), or as a therapeutic target in immune-checkpoint blockade immunotherapy (1, 7). The monocyte/macrophage lineage has been shown to interact with tumoral and non-tumor B cells in lymphoma, and to be involved in the promotion of protumoral TME and resistance to treatment (8). However, the TME in lymphoma is heterogenous and still poorly described at the single cell level (9, 10). Moreover, there is currently an overall lack of biological material due to the popularity of the fine-needle aspiration for routine diagnosis. Thus, peripheral blood remains of interest for the analysis of immune cells and a fine knowledge of the circulating myeloid cell compartment in the different B lymphomas is needed to better understand the mechanisms of resistance to treatment and identify at risk patients. Herein, we propose to review current advances in the high-resolution description and functional role understanding of circulating monocytic lineage in B cell lymphomas and to discuss future perspectives.
Myeloid-cell related prognostic signatures have been demonstrated in situ in secondary lymphoid organs of DLBCL, FL, and HL by gene expression profiling studies (4–6), and several studies have shown an association between macrophage infiltration in lymphoma tissues and prognosis (5, 11–15), but the prognostic impact of myeloid cells in the blood is less clear in these pathologies. Nevertheless, soluble prognostic factors related to the biology of myeloid cells have been proposed to be of prognostic interest in these pathologies, including soluble PD-L1 (16, 17), soluble CD163 (18), CXCL10 (19), and IL-10 (19).
More recently, the circulating myeloid compartment, including in particular monocytes and MDSC, has been evaluated as prognostic biomarkers in B lymphoma. An increased monocyte count has been associated with poor prognosis in DLBCL (20–22), HL (23), MCL (24–26), and FL (27). A slightly different approach, with the lymphocyte/monocyte ratio, gives the same results in HL (23, 28, 29) and DLBCL (30). This could be linked to an increase in monocytic myeloid derived suppressor cells (M-MDSC), as it has been suggested in DLBCL (20, 31) and CLL, in which M-MDSC levels correlate with response to treatment (32–35). Concerning the granulocytic lineage, polymorphonuclear MDSC (PMN-MDSC) have been proposed as a marker of poor prognosis in DLBCL (36), but other works did not find any association between PMN-MDSC and DLBCL prognosis (31, 37). In HL, the presence of a CD34pos MDSC subtype in the blood has been associated with worsened prognosis (38).
Overall, there is ample evidence of the clinical relevance of the monocytic lineage, and in particular of circulating suppressor myeloid cells, in the prognosis of patients diagnosed with B lymphomas. However, the existence of conflicting data, largely due to inconsistent phenotype, highlights the need for an in-depth phenotypic study of these populations in order to better characterize them and evaluate their role in these pathologies.
To fully understand the role of the monocytic lineage in the physiopathology of B-cell lymphomas, it is first necessary to clearly define these cell subsets. Unfortunately, this raises a pitfall in the different data available on the circulating myeloid compartment in B-cell lymphomas, indeed there is no consensus on the definition of M-MDSC and PMN-MDSC, and monocyte subsets are defined with a continuum of markers (CD14 and CD16). Nevertheless, some low-resolution phenotypical investigations have been performed on these cell subsets in B-cell lymphomas, using various and sometimes overlapping phenotypical definitions.
MDSC is a heterogeneous population of myeloid regulatory cells derived from polymorphonuclear cells (PMN-MDSC) and monocytes (M-MDSC) and defined by their immunosuppressive functions (39, 40). Their existence in inflammatory and cancerous diseases could reshape the TME or more distant sites. Since these cells were first described in mice and defined by their immunosuppressive functions, their identification and study in humans are challenging and discussed. In blood, PMN-MDSC (historically referred to as granulocytic MDSC [G-MDSC]) are classically identified as CD11bpos CD14neg CD15pos or CD11bpos CD14neg CD66bpos, and M-MDSCs as CD11bpos CD14pos HLA-DRneg/low CD15neg. It should be noted that these phenotypical definitions do not discriminate PMN-MDSC from normal neutrophils, as a consequence it is recommended to evaluate PMN-MDSC in the low density fraction after ficoll (39). Linneg HLA-DRneg CD33pos CD123neg cells contain more immature progenitors named early-stage MDSC (e-MDSC) (39–41). Additional markers, including CD116, CD124, VEGF-R, CD11c, CD11b, PD-L1 are commonly used, and various phenotypes have been described in tumors (39, 40, 42).
In human B-cell lymphoma, various PMN-MDSC phenotypes were described. In particular, circulating Linneg/low HLA-DRneg CD11bpos CD33pos cells were increased in HL, FL, DLBCL, and MCL, compared to healthy donors blood (31, 37, 38), whereas CD66bpos CD33dim HLA-DRneg were more abundant in the blood of HL, and indolent or aggressive NHL B-cell lymphomas (36). Finally, immature MDSC defined as CD11bpos CD33pos CD14neg CD34pos HLA‐DRneg were more abundant in HL compared to blood from healthy donors (38). M-MDSC phenotype is more consistent and CD14pos HLA-DRlow/neg cells are accumulated in peripheral blood from FL, DLBCL, MCL, CLL, and HL when compared to healthy samples (20, 31–33, 43, 44). However, HLA-DR is expressed as a continuum and thus the M-MDSC identification remains subjective and will benefit from additional markers and high-resolution analysis.
Overall, comparing all these data is challenging due to the use of different and few phenotypic criteria to assess the myeloid compartment. To overcome these difficulties, high resolution tools, such as mass cytometry, could be of use to fully decipher at the phenotypic level these complex populations in tissue and in blood (45). Deep phenotyping of the circulating myeloid compartment in human blood in both healthy and B-cell lymphoma context is needed. Such data already exist for healthy donor blood, but have not been compared with blood samples obtained from lymphoma patients (9), and a clear landscape of blood mononuclear phagocytes in human health and B-cell lymphoma would allow relevant and reproducible functional experiments.
We propose here such analysis on blood monocytic cells from FL (n=3), DLBCL (n=5), and CLL patients (n=3), compared to healthy donors (n=3) (Table 1), using a previously described mass cytometry (CyTOF) panel dedicated to regulatory myeloid cell exploration (9, 46) (Figure 1). With one exception, all samples were obtained at diagnosis, before any treatment. Our analysis was performed on cryopreserved peripheral blood mononuclear cells (PBMC), thus allowing the exploration of monocytes and M-MDSC, but not PMN-MDSC. Using an unsupervised approach, we realized a dimension reduction followed by clustering, defining 8 monocytic clusters (Figure 1A). The hSNE representation shows all the CD14pos myeloid cells compartment as a continuum, underlining the difficulty to clearly and consistently distinguish different subsets. Nevertheless, based on canonical markers expression, we can identify non-classical CD14dim CD16pos monocytes in cluster 1, classical CD14pos CD16neg HLA-DRpos monocytes in clusters 4, 6, and 8, and putative CD14pos HLA-DRlow M-MDSC in clusters 2, 3, 5, and 7 (Figures 1A, B). Interestingly, the hierarchical clustering of all 8 clusters do not delineate these 3 clusters groups, emphasizing the importance of other less classical markers (Figure 1B). In particular, clusters 2, 3, and 4 display a strong expression of S100A9, a protein which is involved in MDSC metabolism (49). These three clusters are mainly found in DLBCL, the most aggressive disease of our panel, in which they represent up to more than 90% of the monocytic population (Figure 1B). Of note, the cells in cluster 4 highly express HLA-DR, and would usually be included in the classical monocytes subset, even though their phenotype and presence in DLBCL patients may indicate a regulatory M-MDSC-like function. If clusters 1, 5, and 6 are found in all sample types, half of the cells present in cluster 5 come from FL patients. This cluster exhibits an interesting CD14pos HLA-DRlow CD36pos S100A9high M-MDSC like phenotype (Figure 1B). Surprisingly, the cells in cluster 6 display heterogenous and partly low levels of HLA-DR, even though they highly originate from healthy donors. This intriguing result emphasizes the need to not reduce M-MDSC evaluation to HLA-DR expression. Finally, an unsupervised clustering can hide some residual heterogeneity inside the clusters, and thus mask disparities between sample types in a given cluster. For example, the cluster 1 gathers all non-classical monocytes, but looking more precisely at their phenotype across the different sample types, we can see that the cells from DLBCL patients express less HLA-DR and CD36, or that FL cells express more CD32 (Figure 1C). Of course, this preliminary study requires further investigations with a larger number of samples to overcome the limitations due to inter-sample variability, as underlined by the peculiar profiles displayed by one of the HD and one of the CLL samples. It is also difficult to conclude from these findings what impact these phenotypic differences have on cell function, since, for example, the CD32 subtype, which could be an activator or inhibitor, is not known. Linking these phenotypes to cell functionality and pathogeny would therefore require further functional studies.
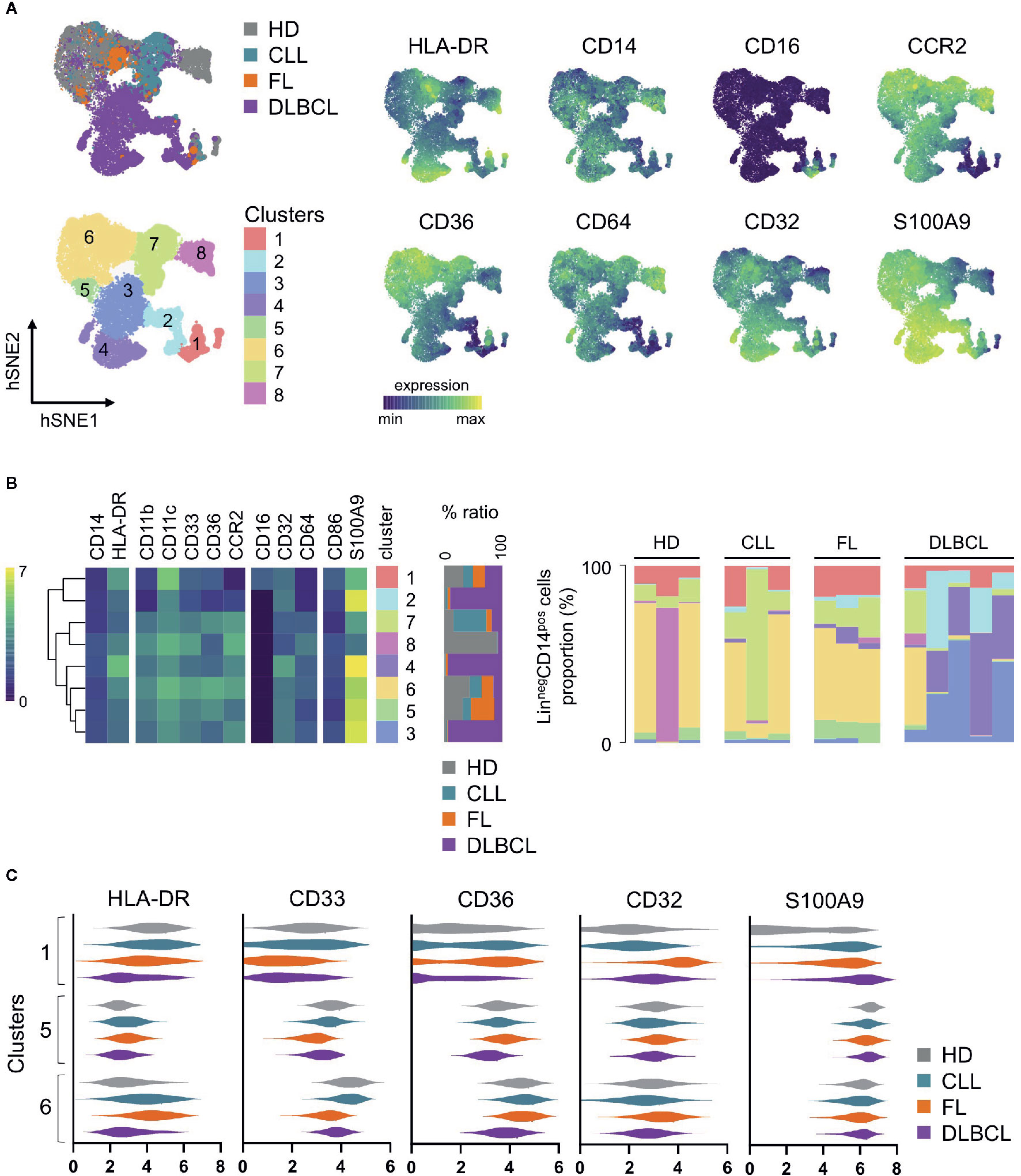
Figure 1 Peripheral blood mononuclear cells (PBMC) from healthy donors (HD, n = 3), follicular lymphoma (FL, n = 3), diffuse large B-cell lymphoma (DLBCL, n = 5) and chronic lymphocytic leukemia (CLL, n = 3) patients were stained with a CyTOF panel already published and dedicated to regulatory myeloid cells assessment (9, 46). LinnegCD14pos viable cells were selected by manual gating on hSNE for each sample, using all channels and the default settings on Cytosplore (47, 48). Then, a hSNE algorithm followed by clustering was performed on all pooled selected cells (n=79,760) with Cytosplore (47, 48) using the following channels: CD11b, CD64, CD36, CCR2, CD45RA, CD123, CD86, CD33, CD11c, CD14, CD32, HLA-DR, CD16, and S100A9 and the default settings. Eight clusters were identified. (A) hSNE plot of CD14pos myeloid cells, colored by sample type, clusters, and selected marker expression. (B) Heatmaps of selected markers in each cluster (left), relative distribution of the cells contained in each cluster per sample type (middle), and relative proportion of each cluster in each sample (right). (C) Violin plots of selected markers on clusters 1, 5, and 6 for each sample type. Expression values are arc-sinh transformed. Hierarchical clustering, heatmaps and violin plots were generated with R v3.6.3, using Rstudio v1.2.5033 and the pheatmap package, and Graphpad Prism 8.4.3.
Altogether, these results suggest that circulating monocytic cells display diverse and sometimes specific phenotypes depending on the physiological or lymphomatous context, and that these phenotypes may be related to cell functionality and/or severity of the disease.
Even if their definition remains quite elusive, particularly at the phenotypic level, it is now clear that MDSC are important players in B-cell lymphoma pathogenesis. Since the characterization of MDSC, although challenging, is classically based on functional assays (39), we have data on their functional role in tumors, including lymphoma.
In cancer, induction and expansion of MDSC can be induced by factors produced by the tumour or the TME cells (T-cells, macrophages, stromal cells), such as VEGF, GM-CSF, M-CSF, S100A8/9, IL-4, IL-6, or IL-10 (49). MDSC could derive from myeloid progenitors from the bone marrow or the spleen, or arise from monocytes (M-MDSC) or activated mature or immature PMN (PMN-MDSC) (50), after the activation of genes implicated in myeloid differentiation blockade or immune regulation. The main signaling mechanism involved seems to be the phosphorylation of transcription factors known as signal transducer and activator of transcription (STAT) protein family members like STAT3, STAT6, STAT1, STAT5 (40, 51). The fate of these circulating myeloid cells is not completely elucidated. MDSC can differentiate into macrophages and DC (52, 53), especially under hypoxia (54). It has been demonstrated in murine models of solid tumors that MDSC can differentiate into TAM at the tumor site (51, 55–57). MDSC are defined by their immunosuppressive capabilities. They can impair effector T- and NK-cell functions and polarize macrophages into « M2 » phenotype via the expansion of regulatory T cells (Treg), amino acids depletion through arginase-1 (Arg1) and Indoleamine 2,3-dioxygenase (IDO), ROS production through NOX2, expressions of immunoregulatory proteins PDL1, TGFβ, IL-10, and S100A12 (42, 58, 59). The crosstalk between MDSC, macrophages, and DC could enhance the tumor-induced immune suppression (60). MDSC could induce a systemic immune suppression by affecting trafficking of T- and B-cells and reducing antigen response outside tumor sites (61).
Some of these immunosuppressive mechanisms have been demonstrated in B-cell lymphomas. The role of arginine metabolism has been demonstrated in lymphoma mice models (54, 62), where an upregulation of inducible nitric oxide synthase (iNOS) and Arg1 is mediated in MDSC by hypoxia-inducible factor-1a (HIF1a) (54). Another mechanism identified in mice in the upregulation of Arg1 in both PMN-MDSCs and M-MDSC is an increase in the microRNA miR-30a expression, which leads to a decrease in SOCS3 mRNA and the activation of the Janus kinase/STAT3 pathway (63). It has also been demonstrated that MDSC arising from the bone marrow are recruited to the tumor through blood circulation, and that tumor infiltrating M-MDSC mediate the recruitment of Treg in tumor-bearing mice via CCL4 and CCL5 production (64). High levels of ROS were also reported in PMN-MDSCs from the blood of this lymphoma murine model (64). The role of MDSC in IL-10 production is suggested by results in B-cell NHL patients and lymphoma murine models (65), but serum IL-10 could also be produced by lymphoma cells and contribute to an increased number of M-MDSC (66). In DLBCL patients, the release of IL-10, together with S100A12 and an increased PD-L1 expression, could explain the M-MDSC mediated T-cell immunosuppression (31). Accordingly, monocytes from NHL patients (including DLBCL, FL, MCL) induce less T-cell proliferation in vitro than HD monocytes, and the removal of these monocytes restore T cell functions (44). The role of arginine metabolism in the suppressive activity of MDSC from the blood of NHL patients is debated (31, 36, 44), as well as PMN-MDSC pathogeny. Even though no immunosuppressive effect of PMN-MDSC has been reported in a study including only DLBCL patients (31), another study including both HL and B-cell NHL patients showed that in vitro depletion of lymphoma PMN-MDSC restored the proliferation of autologous T cells (36). This could be due to the different phenotypic definition of the cells studied, or to the pathologies included. In CLL, the efficacy of the depletion of monocytes to reduce tumorigenesis has been shown in mice models (67). CLL cells induce IDOhigh MDSC in vitro, and CLL patients display an increase in M-MDSC, with suppressing T cell activity, via IDO and Treg expansion (33). Accordingly, monocytes from CLL patients inhibit T cell proliferation, an effect abrogated by anti-TGFβ, anti-IL10 antibodies and IDO inhibitor, and support Treg expansion (68).
Overall, the sometimes conflicting results of the different works focusing on explaining the functional roles of myeloid cells in B-cell lymphomas highlight the need for a better and clearer definition of these different cells subsets.
Over the last few years, an increasing interest has been shown in the myeloid regulatory compartment in B cell lymphomas, and notably the circulating monocytic population. MDSC in particular were shown to be involved in disease prognosis and could have a pathogenic role. However, there is still no consensus on the phenotypic definition of the different circulating myeloid populations. Moreover, the monocytic compartment is more of a continuum than constituted of distinct subtypes, as highlighted by our results, resulting in sometimes contradictory conclusions on the prognostic impact of the different MDSC populations. We confirm here previous results, such as HLA-DR down regulation in B-cell lymphomas, and provide a first exhaustive phenotypic evaluation of the monocytic compartment in different B-cell lymphomas, identifying for example S100A9high monocytic cells as a potential biomarker in DLBCL. This proof-of-concept preliminary study underlines the need to further assess the circulating myeloid compartment at the phenotypic level, on a larger panel of patients and possibly with more markers, to validate clinically relevant prognostic signatures.
The therapeutic management of lymphomas today largely relies on immunotherapy, including rituximab and immune checkpoint blockade. The efficacy of immunotherapy could be modulated by MDSC, as it has been shown in TAM, and their monitoring in the blood could help evaluate response to treatment. However, the characterization of MDSC is currently based on functional assays (39), preventing their monitoring in personalized medicine. Linking easily accessible and monitorable surface phenotypic markers to the functionality of these cells would help overcome this issue. A refined characterization through deep phenotyping of the myeloid compartment could also help identify potential new therapeutic targets (69) and study the immune response to treatment (70).
Altogether, the observations discussed in this article support the idea that circulating myeloid regulatory cells in B-cell lymphomas are promising diagnostic and prognostic biomarkers, and that a comprehensive phenotypic evaluation could serve as a surrogate biomarker of their pathological activity.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by IRB Rennes University Hospital. The patients/participants provided their written informed consent to participate in this study.
JF analyzed data and wrote the manuscript; FL, SLG performed experiments; JMI analyzed data; MR designed and supervised research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a fellowship from the Nuovo-Soldati Foundation (Switzerland) [M.R.], from the FHU CAMIn (Federation Hospitalo-Universitaire Cancer Microenvironnement et Innovation) [J.F.], from the comité de la recherche clinique et translationnelle (CORECT), CHU of Rennes [F.L.], and from the association pour le développement de l’hématologie oncologie (ADHO) (F.L.).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are indebted to the clinicians of the BREHAT network and to the French Blood Bank (EFS) of Rennes for providing samples. The authors acknowledge the Centre de Ressources Biologiques (CRB-santé) of Rennes (BB-0033-00056, http://www.crbsante-rennes.com) for managing samples.
1. Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer (2014) 14:517. doi: 10.1038/nrc3774
2. Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood (2016) 127:2375–90. doi: 10.1182/blood-2016-01-643569
3. Amé-Thomas P, Tarte K. The yin and the yang of follicular lymphoma cell niches: Role of microenvironment heterogeneity and plasticity. Semin Cancer Biol (2014) 24:23–32. doi: 10.1016/j.semcancer.2013.08.001
4. Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of Survival in Follicular Lymphoma Based on Molecular Features of Tumor-Infiltrating Immune Cells. N Engl J Med (2004) 351:2159–69. doi: 10.1056/NEJMoa041869
5. Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med (2008) 359:2313–23. doi: 10.1056/NEJMoa0802885
6. Steidl C, Lee T, Shah SP, Farinha P, Han G, Nayar T, et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N Engl J Med (2010) 362:875–85. doi: 10.1056/NEJMoa0905680
7. Nicholas NS, Apollonio B, Ramsay AG. Tumor microenvironment (TME)-driven immune suppression in B cell malignancy. Biochim Biophys Acta (2016) 1863:471–82. doi: 10.1016/j.bbamcr.2015.11.003
8. Pham LV, Pogue E, Ford RJ. The Role of Macrophage/B-Cell Interactions in the Pathophysiology of B-Cell Lymphomas. Front Oncol (2018) 8:147. doi: 10.3389/fonc.2018.00147
9. Roussel M, Ferrell PB, Greenplate AR, Lhomme F, Gallou SL, Diggins KE, et al. Mass cytometry deep phenotyping of human mononuclear phagocytes and myeloid-derived suppressor cells from human blood and bone marrow. J Leukoc Biol (2017) 102:437–47. doi: 10.1189/jlb.5MA1116-457R
10. Roussel M, Irish JM, Menard C, Lhomme F, Tarte K, Fest T. Regulatory myeloid cells: an underexplored continent in B-cell lymphomas. Cancer Immunol Immunother (2017) 66:1103–11. doi: 10.1007/s00262-017-2036-5
11. Connors JM. Risk assessment in the management of newly diagnosed classical Hodgkin lymphoma. Blood (2015) 125:1693–702. doi: 10.1182/blood-2014-07-537480
12. Keane C, Vari F, Hertzberg M, Cao K-AL, Green MR, Han E, et al. Ratios of T-cell immune effectors and checkpoint molecules as prognostic biomarkers in diffuse large B-cell lymphoma: a population-based study. Lancet Haematol (2015) 2:e445–55. doi: 10.1016/S2352-3026(15)00150-7
13. Marchesi F, Cirillo M, Bianchi A, Gately M, Olimpieri OM, Cerchiara E, et al. High density of CD68+/CD163+ tumour-associated macrophages (M2-TAM) at diagnosis is significantly correlated to unfavorable prognostic factors and to poor clinical outcomes in patients with diffuse large B-cell lymphoma. Hematol Oncol (2015) 33:110–2. doi: 10.1002/hon.2142
14. Riihijärvi S, Fiskvik I, Taskinen M, Vajavaara H, Tikkala M, Yri O, et al. Prognostic influence of macrophages in patients with diffuse large B-cell lymphoma: a correlative study from a Nordic phase II trial. Haematologica (2015) 100:238–45. doi: 10.3324/haematol.2014.113472
15. Vaidya R, Witzig TE. Prognostic factors for diffuse large B-cell lymphoma in the R(X)CHOP era. Ann Oncol Off J Eur Soc Med Oncol (2014) 25:2124–33. doi: 10.1093/annonc/mdu109
16. Rossille D, Gressier M, Damotte D, Maucort-Boulch D, Pangault C, Semana G, et al. High level of soluble programmed cell death ligand 1 in blood impacts overall survival in aggressive diffuse large B-Cell lymphoma: results from a French multicenter clinical trial. Leukemia (2014) 28:2367–75. doi: 10.1038/leu.2014.137
17. Rossille D, Azzaoui I, Feldman AL, Maurer MJ, Labouré G, Parrens M, et al. Soluble programmed death-ligand 1 as a prognostic biomarker for overall survival in patients with diffuse large B-cell lymphoma: a replication study and combined analysis of 508 patients. Leukemia (2017) 31:988–91. doi: 10.1038/leu.2016.385
18. Jones K, Vari F, Keane C, Crooks P, Nourse JP, Seymour LA, et al. Serum CD163 and TARC as disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res Off J Am Assoc Cancer Res (2013) 19:731–42. doi: 10.1158/1078-0432.CCR-12-2693
19. Charbonneau B, Maurer MJ, Ansell SM, Slager SL, Fredericksen ZS, Ziesmer SC, et al. Pretreatment circulating serum cytokines associated with follicular and diffuse large B-cell lymphoma: a clinic-based case-control study. Cytokine (2012) 60:882–9. doi: 10.1016/j.cyto.2012.08.028
20. Tadmor T, Fell R, Polliack A, Attias D. Absolute monocytosis at diagnosis correlates with survival in diffuse large B-cell lymphoma-possible link with monocytic myeloid-derived suppressor cells. Hematol Oncol (2013) 31:65–71. doi: 10.1002/hon.2019
21. Tadmor T, Bari A, Sacchi S, Marcheselli L, Liardo EV, Avivi I, et al. Monocyte count at diagnosis is a prognostic parameter in diffuse large B-cell lymphoma: results from a large multicenter study involving 1191 patients in the pre- and post-rituximab era. Haematologica (2014) 99:125–30. doi: 10.3324/haematol.2013.088161
22. Wu C, Wu X, Liu X, Yang P, Xu J, Chai Y, et al. Prognostic Significance of Monocytes and Monocytic Myeloid-Derived Suppressor Cells in Diffuse Large B-Cell Lymphoma Treated with R-CHOP. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2016) 39:521–30. doi: 10.1159/000445644
23. Tadmor T, Bari A, Marcheselli L, Sacchi S, Aviv A, Baldini L, et al. Absolute Monocyte Count and Lymphocyte-Monocyte Ratio Predict Outcome in Nodular Sclerosis Hodgkin Lymphoma: Evaluation Based on Data From 1450 Patients. Mayo Clin Proc (2015) 90:756–64. doi: 10.1016/j.mayocp.2015.03.025
24. von Hohenstaufen KA, Conconi A, de Campos CP, Franceschetti S, Bertoni F, Margiotta Casaluci G, et al. Prognostic impact of monocyte count at presentation in mantle cell lymphoma. Br J Haematol (2013) 162:465–73. doi: 10.1111/bjh.12409
25. Koh YW, Shin S-J, Park C, Yoon DH, Suh C, Huh J. Absolute monocyte count predicts overall survival in mantle cell lymphomas: correlation with tumour-associated macrophages. Hematol Oncol (2014) 32:178–86. doi: 10.1002/hon.2106
26. Porrata LF, Ristow K, Markovic SN. Absolute monocyte count at diagnosis and survival in mantle cell lymphoma. Br J Haematol (2013) 163:545–7. doi: 10.1111/bjh.12531
27. Wilcox RA, Ristow K, Habermann TM, Inwards DJ, Micallef INM, Johnston PB, et al. The absolute monocyte count is associated with overall survival in patients newly diagnosed with follicular lymphoma. Leuk Lymphoma (2012) 53:575–80. doi: 10.3109/10428194.2011.637211
28. Porrata LF, Ristow K, Colgan JP, Habermann TM, Witzig TE, Inwards DJ, et al. Peripheral blood lymphocyte/monocyte ratio at diagnosis and survival in classical Hodgkin’s lymphoma. Haematologica (2012) 97:262–9. doi: 10.3324/haematol.2011.050138
29. Wilcox RA, Ristow K, Habermann TM, Inwards DJ, Micallef INM, Johnston PB, et al. The absolute monocyte and lymphocyte prognostic score predicts survival and identifies high-risk patients in diffuse large-B-cell lymphoma. Leukemia (2011) 25:1502–9. doi: 10.1038/leu.2011.112
30. Wang J, Gao K, Lei W, Dong L, Xuan Q, Feng M, et al. Lymphocyte-to-monocyte ratio is associated with prognosis of diffuse large B-cell lymphoma: correlation with CD163 positive M2 type tumor-associated macrophages, not PD-1 positive tumor-infiltrating lymphocytes. Oncotarget (2017) 8:5414–25. doi: 10.18632/oncotarget.14289
31. Azzaoui I, Uhel F, Rossille D, Pangault C, Dulong J, Le Priol J, et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood (2016) 128:1081–92. doi: 10.1182/blood-2015-08-662783
32. Gustafson MP, Abraham RS, Lin Y, Wu W, Gastineau DA, Zent CS, et al. Association of an increased frequency of CD14+ HLA-DR lo/neg monocytes with decreased time to progression in chronic lymphocytic leukaemia (CLL). Br J Haematol (2012) 156:674–6. doi: 10.1111/j.1365-2141.2011.08902.x
33. Jitschin R, Braun M, Büttner M, Dettmer-Wilde K, Bricks J, Berger J, et al. CLL-cells induce IDOhi CD14+HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs. Blood (2014) 124:750–60. doi: 10.1182/blood-2013-12-546416
34. Liu J, Zhou Y, Huang Q, Qiu L. CD14+HLA-DRlow/- expression: A novel prognostic factor in chronic lymphocytic leukemia. Oncol Lett (2015) 9:1167–72. doi: 10.3892/ol.2014.2808
35. Zarobkiewicz M, Kowalska W, Chocholska S, Tomczak W, Szymańska A, Morawska I, et al. High M-MDSC Percentage as a Negative Prognostic Factor in Chronic Lymphocytic Leukaemia. Cancers (2020) 12:2614. doi: 10.3390/cancers12092614
36. Marini O, Spina C, Mimiola E, Cassaro A, Malerba G, Todeschini G, et al. Identification of granulocytic myeloid-derived suppressor cells (G-MDSCs) in the peripheral blood of Hodgkin and non-Hodgkin lymphoma patients. Oncotarget (2016) 7:27676–88. doi: 10.18632/oncotarget.8507
37. Amini R-M, Enblad G, Hollander P, Laszlo S, Eriksson E, Ayoola Gustafsson K, et al. Altered profile of immune regulatory cells in the peripheral blood of lymphoma patients. BMC Cancer (2019) 19:316. doi: 10.1186/s12885-019-5529-0
38. Romano A, Parrinello NL, Vetro C, Forte S, Chiarenza A, Figuera A, et al. Circulating myeloid-derived suppressor cells correlate with clinical outcome in Hodgkin Lymphoma patients treated up-front with a risk-adapted strategy. Br J Haematol (2015) 168:689–700. doi: 10.1111/bjh.13198
39. Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun (2016) 7:12150. doi: 10.1038/ncomms12150
40. Gabrilovich DI. Myeloid-Derived Suppressor Cells. Cancer Immunol Res (2017) 5:3–8. doi: 10.1158/2326-6066.CIR-16-0297
41. Uhel F, Azzaoui I, Le Gallou S, Fest T, Tarte K, Roussel M. Early-stage myeloid-derived suppressor cell count: Basophil exclusion matters. J Allergy Clin Immunol (2019) 144:1125–7. doi: 10.1016/j.jaci.2019.06.027
42. Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv Cancer Res (2015) 128:95–139. doi: 10.1016/bs.acr.2015.04.002
43. Gustafson MP, Lin Y, Maas ML, Van Keulen VP, Johnston PB, Peikert T, et al. A method for identification and analysis of non-overlapping myeloid immunophenotypes in humans. PloS One (2015) 10:e0121546. doi: 10.1371/journal.pone.0121546
44. Lin Y, Gustafson MP, Bulur PA, Gastineau DA, Witzig TE, Dietz AB. Immunosuppressive CD14+HLA-DR(low)/- monocytes in B-cell non-Hodgkin lymphoma. Blood (2011) 117:872–81. doi: 10.1182/blood-2010-05-283820
45. Ferrant J, Le Gallou S, Manson G, Genebrier S, Mourcin F, Tarte K, Roussel M. High-Dimensional Phenotyping of Human Myeloid-Derived Suppressor Cells/Tumor-Associated Macrophages in Tissue by Mass Cytometry. Methods Mol Biol (2021) 2236:57–66. doi: 10.1007/978-1-0716-1060-2
46. Roussel M, Lhomme F, Roe CE, Bartkowiak T, Gravelle P, Laurent C, et al. Mass cytometry defines distinct immune profile in germinal center B-cell lymphomas. Cancer Immunol Immunother CII (2020) 69:407–20. doi: 10.1007/s00262-019-02464-z
47. Höllt T, Pezzotti N, van UV, Koning F, Eisemann E, Lelieveldt B, et al. Cytosplore: Interactive Immune Cell Phenotyping for Large Single-Cell Datasets. Comput Graph Forum (2016) 35:171–80. doi: 10.1111/cgf.12893
48. van Unen V, Höllt T, Pezzotti N, Li N, Reinders MJT, Eisemann E, et al. Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun (2017) 8:1740. doi: 10.1038/s41467-017-01689-9
49. Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol (2009) 9:162–74. doi: 10.1038/nri2506
50. Bizymi N, Bjelica S, Kittang AO, Mojsilovic S, Velegraki M, Pontikoglou C, et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. HemaSphere (2019) 3:e168. doi: 10.1097/HS9.0000000000000168
51. Kumar V, Cheng P, Condamine T, Mony S, Languino LR, McCaffrey JC, et al. CD45 Phosphatase Inhibits STAT3 Transcription Factor Activity in Myeloid Cells and Promotes Tumor-Associated Macrophage Differentiation. Immunity (2016) 44:303–15. doi: 10.1016/j.immuni.2016.01.014
52. Kusmartsev S, Gabrilovich DI. Inhibition of myeloid cell differentiation in cancer: the role of reactive oxygen species. J Leukoc Biol (2003) 74:186–96. doi: 10.1189/jlb.0103010
53. Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol Baltim Md 1950 (2005) 174:4880–91. doi: 10.4049/jimmunol.174.8.4880
54. Corzo CA, Condamine T, Lu L, Cotter MJ, Youn J-I, Cheng P, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med (2010) 207:2439–53. doi: 10.1084/jem.20100587
55. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol (2016) 37:208–20. doi: 10.1016/j.it.2016.01.004
56. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol (2017) 14:399. doi: 10.1038/nrclinonc.2016.217
57. Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest (2015) 125:3365–76. doi: 10.1172/JCI80006
58. Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol (2012) 12:253–68. doi: 10.1038/nri3175
59. Nagaraj S, Schrum AG, Cho H-I, Celis E, Gabrilovich DI. Mechanism of T cell tolerance induced by myeloid-derived suppressor cells. J Immunol Baltim Md 1950 (2010) 184:3106–16. doi: 10.4049/jimmunol.0902661
60. Ostrand-Rosenberg S, Sinha P, Beury DW, Clements VK. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol (2012) 22:275–81. doi: 10.1016/j.semcancer.2012.01.011
61. Ku AW, Muhitch JB, Powers CA, Diehl M, Kim M, Fisher DT, et al. Tumor-induced MDSC act via remote control to inhibit L-selectin-dependent adaptive immunity in lymph nodes. eLife (2016) 5:e17375. doi: 10.7554/eLife.17375
62. Serafini P, Mgebroff S, Noonan K, Borrello I. Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res (2008) 68:5439–49. doi: 10.1158/0008-5472.CAN-07-6621
63. Xu Z, Ji J, Xu J, Li D, Shi G, Liu F, et al. MiR-30a increases MDSC differentiation and immunosuppressive function by targeting SOCS3 in mice with B-cell lymphoma. FEBS J (2017) 284:2410–24. doi: 10.1111/febs.14133
64. Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol Baltim Md 1950 (2012) 189:5602–11. doi: 10.4049/jimmunol.1201018
65. Sato Y, Shimizu K, Shinga J, Hidaka M, Kawano F, Kakimi K, et al. Characterization of the myeloid-derived suppressor cell subset regulated by NK cells in malignant lymphoma. Oncoimmunology (2015) 4:e995541. doi: 10.1080/2162402X.2014.995541
66. Xiu B, Lin Y, Grote DM, Ziesmer SC, Gustafson MP, Maas ML, et al. IL-10 induces the development of immunosuppressive CD14(+)HLA-DR(low/-) monocytes in B-cell non-Hodgkin lymphoma. Blood Cancer J (2015) 5:e328. doi: 10.1038/bcj.2015.56
67. Hanna BS, McClanahan F, Yazdanparast H, Zaborsky N, Kalter V, Rößner PM, et al. Depletion of CLL-associated patrolling monocytes and macrophages controls disease development and repairs immune dysfunction in vivo. Leukemia (2016) 30:570–9. doi: 10.1038/leu.2015.305
68. Giannoni P, Pietra G, Travaini G, Quarto R, Shyti G, Benelli R, et al. Chronic lymphocytic leukemia nurse-like cells express hepatocyte growth factor receptor (c-MET) and indoleamine 2,3-dioxygenase and display features of immunosuppressive type 2 skewed macrophages. Haematologica (2014) 99:1078–87. doi: 10.3324/haematol.2013.091405
69. Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer (2016) 16:447–62. doi: 10.1038/nrc.2016.54
Keywords: monocyte, myeloid-derived suppressor cells, lymphoma, biomarker, mass cytometry
Citation: Ferrant J, Lhomme F, Le Gallou S, Irish JM and Roussel M (2021) Circulating Myeloid Regulatory Cells: Promising Biomarkers in B-Cell Lymphomas. Front. Immunol. 11:623993. doi: 10.3389/fimmu.2020.623993
Received: 30 October 2020; Accepted: 14 December 2020;
Published: 02 February 2021.
Edited by:
Martina Seiffert, German Cancer Research Center (DKFZ), GermanyReviewed by:
Selcen Öztürk, German Cancer Research Center (DKFZ), GermanyCopyright © 2021 Ferrant, Lhomme, Le Gallou, Irish and Roussel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mikael Roussel, bWlrYWVsLnJvdXNzZWxAY2h1LXJlbm5lcy5mcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.