- 1Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, United States
- 2Department of Medicine, Indiana University School of Medicine, Indianapolis, IN, United States
- 3Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, IN, United States
The obligate human pathogen Haemophilus ducreyi causes both cutaneous ulcers in children and sexually transmitted genital ulcers (chancroid) in adults. Pathogenesis is dependent on avoiding phagocytosis and exploiting the suppurative granuloma-like niche, which contains a myriad of innate immune cells and memory T cells. Despite this immune infiltrate, long-lived immune protection does not develop against repeated H. ducreyi infections—even with the same strain. Most of what we know about infectious skin diseases comes from naturally occurring infections and/or animal models; however, for H. ducreyi, this information comes from an experimental model of infection in human volunteers that was developed nearly three decades ago. The model mirrors the progression of natural disease and serves as a valuable tool to determine the composition of the immune cell infiltrate early in disease and to identify host and bacterial factors that are required for the establishment of infection and disease progression. Most recently, holistic investigation of the experimentally infected skin microenvironment using multiple “omics” techniques has revealed that non-canonical bacterial virulence factors, such as genes involved in central metabolism, may be relevant to disease progression. Thus, the immune system not only defends the host against H. ducreyi, but also dictates the nutrient availability for the invading bacteria, which must adapt their gene expression to exploit the inflammatory metabolic niche. These findings have broadened our view of the host-pathogen interaction network from considering only classical, effector-based virulence paradigms to include adaptations to the metabolic environment. How both host and bacterial factors interact to determine infection outcome is a current focus in the field. Here, we review what we have learned from experimental H. ducreyi infection about host-pathogen interactions, make comparisons to what is known for other skin pathogens, and discuss how novel technologies will deepen our understanding of this infection.
Introduction
As a primary barrier to infection, the skin possesses multiple defense mechanisms including antimicrobial peptides, immune cells, and the commensal microbiome. These factors, among others, contribute to inter-person variability in response to infection. Studies investigating natural infection outcomes have been very useful for determining pathology later in infection (1), but are limited in their ability to inform what happens early in infection, since patients generally do not seek care at these times. Differences in host factors as well as different strains of a given pathogen, which may have varying degrees of virulence, further confound understanding how the immune system responds to infection.
The skin immune response differs among pathogens despite the general conservation of cell types that either reside in and patrol the skin or that are recruited to combat the infectious agent. Much of what we know about skin immunology has been inferred from murine infection models or by comparing datasets from convenience human skin samples obtained from plastic surgery (2) and from biopsy of diseased skin (3, 4). Both strategies have elucidated important players in the skin immune response, but have notable caveats. While animal models allow for better controlled studies, the skin of mice differs from human skin in terms of thickness, time to healing, microbiome, and immune response, which can all be attributed to differences in both genetics and environments. Thus, interpretations of animal studies are limited to pathogenesis and immune mechanisms shared by both the animal model and humans. Extrapolation of differential gene expression between diseased and either healthy or reference datasets from different anatomical locales, such as convenience human tissue samples, can have similar limitations.
To circumvent these limitations and further our understanding of how human skin responds to infection in a controlled fashion, we have used experimental skin infection of human volunteers with the extracellular, Gram-negative bacterium Haemophilus ducreyi for nearly thirty years (5, 6). H. ducreyi is the causative agent of two diseases: the sexually transmitted genital ulcer (GU) disease chancroid and cutaneous ulcers (CU) in children (7). Chancroid is a sexually transmitted genital ulcer disease and facilitates the transmission of HIV-1 (8). Due to syndromic management, defined as the provision of treatment without diagnostic testing, the epidemiology of chancroid is not well understood, but chancroid remains endemic in developing countries via reservoirs of infected commercial sex workers (7). Recently, in tropical or equatorial regions with high prevalence of CU caused by Treponema pallidum subsp. pertenue (yaws), H. ducreyi was found to be the etiological agent in ~40-60% of lesions (9–11). For both yaws and H. ducreyi, the ulcers are primarily found on the lower leg and affect approximately 4-10% of children in endemic regions. While the prevalence of H. ducreyi-associated CU initially decreased following mass drug administration of azithromycin, the disease was not eliminated due to environmental reservoirs (12, 13). Thus, there is a continuing need to understand H. ducreyi pathogenesis and the host response to this infection.
There are two classes of H. ducreyi isolates; class I and II strains can be differentiated based on several variable extracellular or secreted proteins (DsrA, NcaA, DltA, LspA1, LspA2, and others), and lipooligosaccharide (LOS) structure (14), but are otherwise highly conserved, clinically indistinguishable, and found in significant proportions of both CU and GU (12, 15–17). Although the majority of CU are typically caused by a single genotype, coinfections with both classes are common in CU (12); such studies have not been done in GU. Most in vitro work and experimental human infection utilizes H. ducreyi 35000HP, a human-passaged class I GU strain. By whole genome sequencing, ~70% of CU strains are nearly identical to 35000HP, indicating that our model is highly relevant to both GU and CU (15, 16).
Although both class I and II strains cause CU and GU, whether individual isolates can cause both diseases within a population is unknown due to a lack of surveillance of GU in CU endemic areas and vice versa. In addition, the number of HD isolates that have been sequenced is quite limited, and syndromic management of GU precludes our understanding of how prevalent each class is in the population. Whether there are differences in infectivity or the ability each class to cause disease in different anatomical locales in natural infection is unknown. To date, 35000HP is the only strain used in experimental human infection. Since 35000HP is a GU isolate and able to infect nongenital skin following the same disease kinetics and pathology seen in GU, it appears that its pathogenic potential is similar in genital and nongenital skin.
Here, we highlight and discuss the recent advances made in understanding H. ducreyi pathogenesis via mutant versus parent comparison trials, the immune response to H. ducreyi infection, and how H. ducreyi may be exploiting the immune system to promote its pathogenesis.
Overview of the Human Challenge Model
The H. ducreyi experimental infection model is arguably one of the best studied human infection model systems. It is also a powerful model for investigating bacterial pathogenesis due to FDA approval for not only experimental infection of wild type and mutant bacterial strains but also the ability to perform punch biopsies of infected sites. In this model, as few as one bacterium introduced into wounded skin on the upper arm is sufficient to cause infection (18). This suggests that experimental human infection mimics natural infection—especially considering that infection of animal models requires 3–6 log-fold higher inocula. Although most individuals experimentally infected with wild type H. ducreyi form papules within 24 hours of infection, only ~70% of individuals progress to the pustular stage; the other ~30% spontaneously resolve the infection within 2 to 5 days (5) (Figure 1). The clinical endpoints are resolution of infection or development of a painful pustule; volunteers with pustules (N=231) are infected for 8.1 ± 2.4 (mean ± SD) days. As subject safety considerations preclude progression of experimental infection to the ulcerative stage, experimental infection mirrors natural disease only until the pustular stage. Pustule formation is dependent on host effects and gender, as men are twice as likely to form pustules as women (5). Re-infection of individuals from past H. ducreyi challenge studies demonstrated that individuals who initially formed pustules tend to form pustules and individuals who resolved infection the first time tend to resolve infection a second time at different statistically significant rates (19). This was not attributable to serum bactericidal or opsonizing antibodies. Similarly, in patients with natural chancroid, there is no apparent protection to re-infection. Although the underlying molecular mechanisms leading to pustule formation or resolution are unknown, varying degrees of immune tolerance—particularly with regard to dendritic cells (DCs)—are hypothesized to play a role.
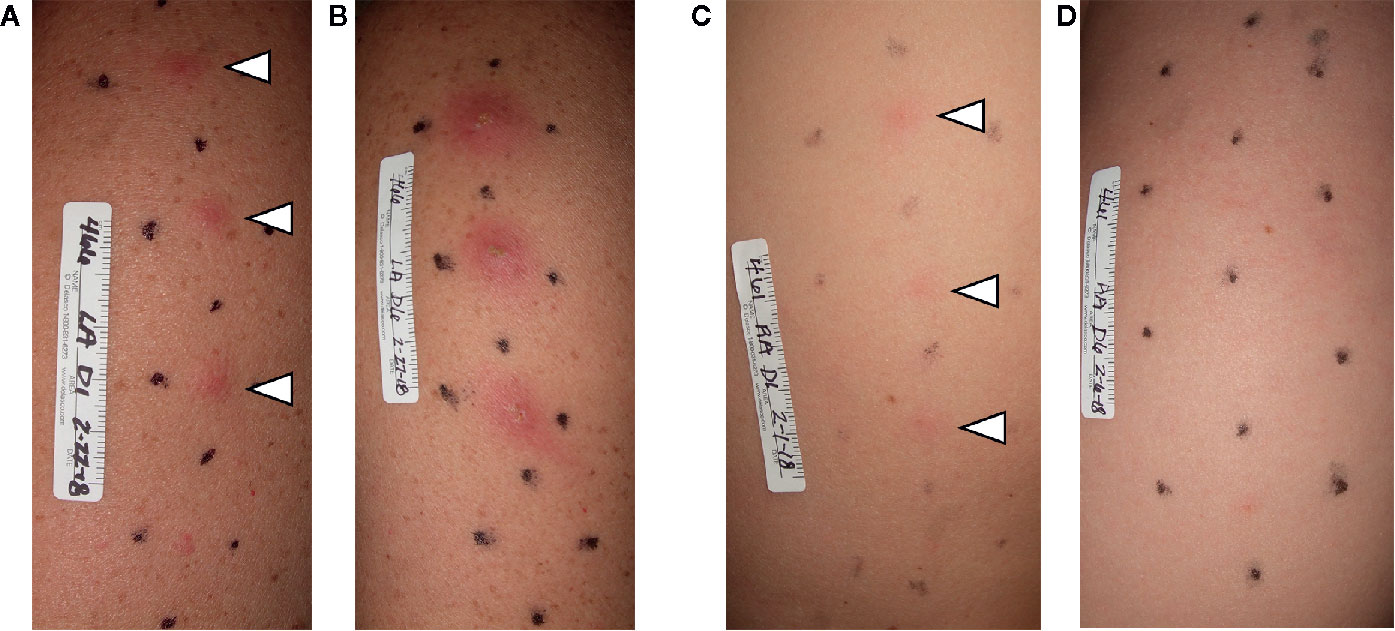
Figure 1 Photographs of clinical outcomes in the H. ducreyi human challenge model. Volunteers were infected in three sites and mock infected in the fourth, bottom-most site. On day 1 (24 hpi), both volunteers had papules at all three infected sites (A, C; white arrowheads). On day 6 of infection, the volunteer in (A) had developed pustules at all infected sites (B). In contrast, by day 6, the volunteer in (C) had resolved infection at all three infected sites (D).
To date, 34 mutants have been tested in the human model to define which virulence factors are required for infection (Table 1). The methods for these double-blinded, multi-stage, dose ranging studies have been reviewed previously (5). Mutants are categorized as virulent (form pustules at doses similar to the parent), partially attenuated (form pustules at doses 2- or 3-fold that of the parent, but not at doses equivalent to the parent) or as attenuated (unable to form pustules even at doses 10-fold that of the parent). Of the 34 mutants, 10 were attenuated, 9 were partially attenuated, and 15 were virulent. The trials have shown the importance of evasion of phagocytosis, resistance to complement mediated killing and antimicrobial peptides, the ability to form aggregates, quorum sensing, adherence to collagen and fibrin, nutrient uptake, and adaptation to starvation as pathogenic mechanisms for disease progression. The interaction of some of these virulence determinants with the host immune response will be discussed in detail below.
To our knowledge, the only other active human challenge model using a skin pathogen is the Bacille Calmette-Guérin (BCG) model, which was developed about twelve years ago to assess BCG vaccine efficiency against BCG infection (52). Later the BCG vaccine challenge model was used to evaluate if specific Mycobacterium tuberculosis (53) and M. leprae (54) proteins were cross-reactive. This model injects BCG into the forearm skin and has also been instrumental in defining the immune infiltrate and cytokines released into the microenvironment (52).
The Role of the Host During H. ducreyi Infection
Histopathology of the Cutaneous Ulcer
Our understanding of the role of the host during disease has been aided by histological examination of both experimental and natural infections (1, 55). A schematic of the progression of disease from the papular to pustular stage is found in Figure 2. Infection is dependent on entry through wounds in the skin; intact skin is resistant to infection. H. ducreyi first encounters keratinocytes, fibroblasts, tissue resident and memory T cells, and tissue resident DCs in healthy skin but remains extracellular throughout infection (1, 18, 55). Collagen and fibrin are deposited in the wounds, and innate immune cells are recruited to the site of infection (55, 57). Neutrophils initially surround and attempt to phagocytose the bacteria, forming a microabscess in the epidermis and dermis (55, 57). Macrophages concurrently migrate into the tissue and form a “collar” around the base of the abscess (1, 57). Immature DCs, natural killer (NK) cells, T cells, and a few B cells are simultaneously recruited and localize primarily below the macrophage collar (58–61). If the phagocytic response fails to clear the organism, the papule evolves into a pustule that resembles a suppurative granuloma and eventually erodes into a painful ulcer (1).
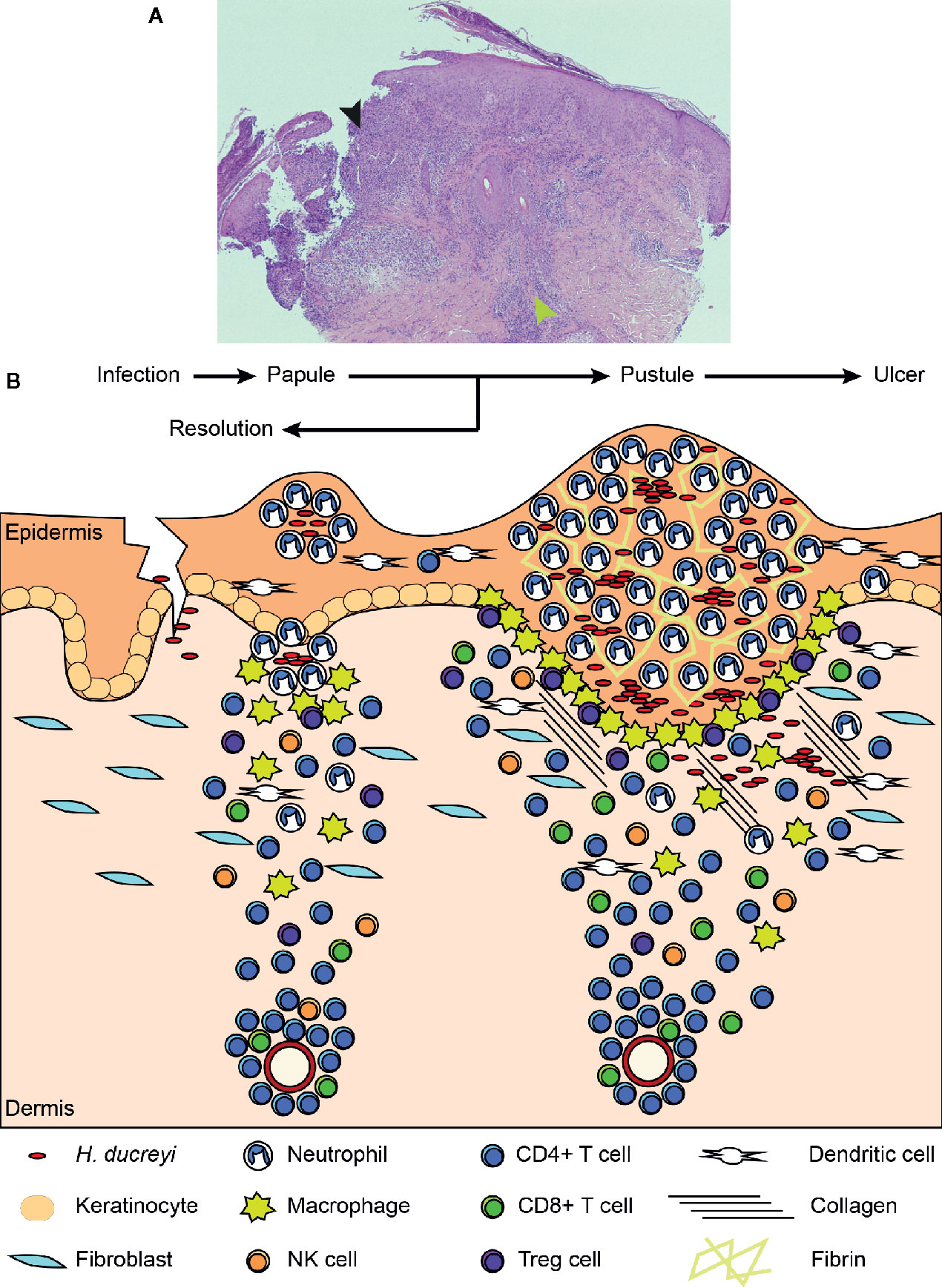
Figure 2 Histology of an experimental lesion and the evolution of disease progression. (A) H & E staining of an endpoint pustule. The black arrowhead indicates the site of the pustule, which has eroded through the epidermis. The green arrowhead indicates the mononuclear cell infiltrate in the dermis. (B) Schematic of disease progression from time of infection to papule and pustule formation. Details of this process appear in the text. Adapted from Atlas of Sexually Transmitted Diseases and AIDS, 4th edition (56).
Immunocytochemistry of papules and pustules revealed that the bacteria are mainly confined to the upper levels of the pustule, where they are surrounded by neutrophils (1, 55, 57). In experimental infection, H. ducreyi is localized to the epidermis and upper dermis, which contrasts to the localization of other skin pathogens that form granulomas in the deep dermis. For instance, Mycobacterium leprae is an intracellular skin pathogen that lives in macrophages. Similar to H. ducreyi, the immune response appears to involve walling off the infection, so the infected macrophages are surrounded by T cells whose subtype determines whether a pro- or anti-inflammatory environment is created (62, 63). Another mycobacterium, M. ulcerans remains extracellular due to secretion of mycolactone and, ideally, is prevented from spreading further into the tissue by macrophages like H. ducreyi, but ulcers can progress to the deep dermis if the infection cannot be controlled (64).
When considering other genital skin infections, chancroidal ulcers can be macroscopically similar to syphilitic chancres caused by T. pallidum. However, in syphilis, bacterial replication is typically in the deep dermis and involves the vasculature (65). Like chancroid, neutrophils are first recruited, but macrophages and the T cells required for infection control are not recruited until late in the second week of infection with T. pallidum (65).
How immune cells are recruited to the site of infection and how H. ducreyi responds to the immune system is discussed below and summarized in Figure 3.
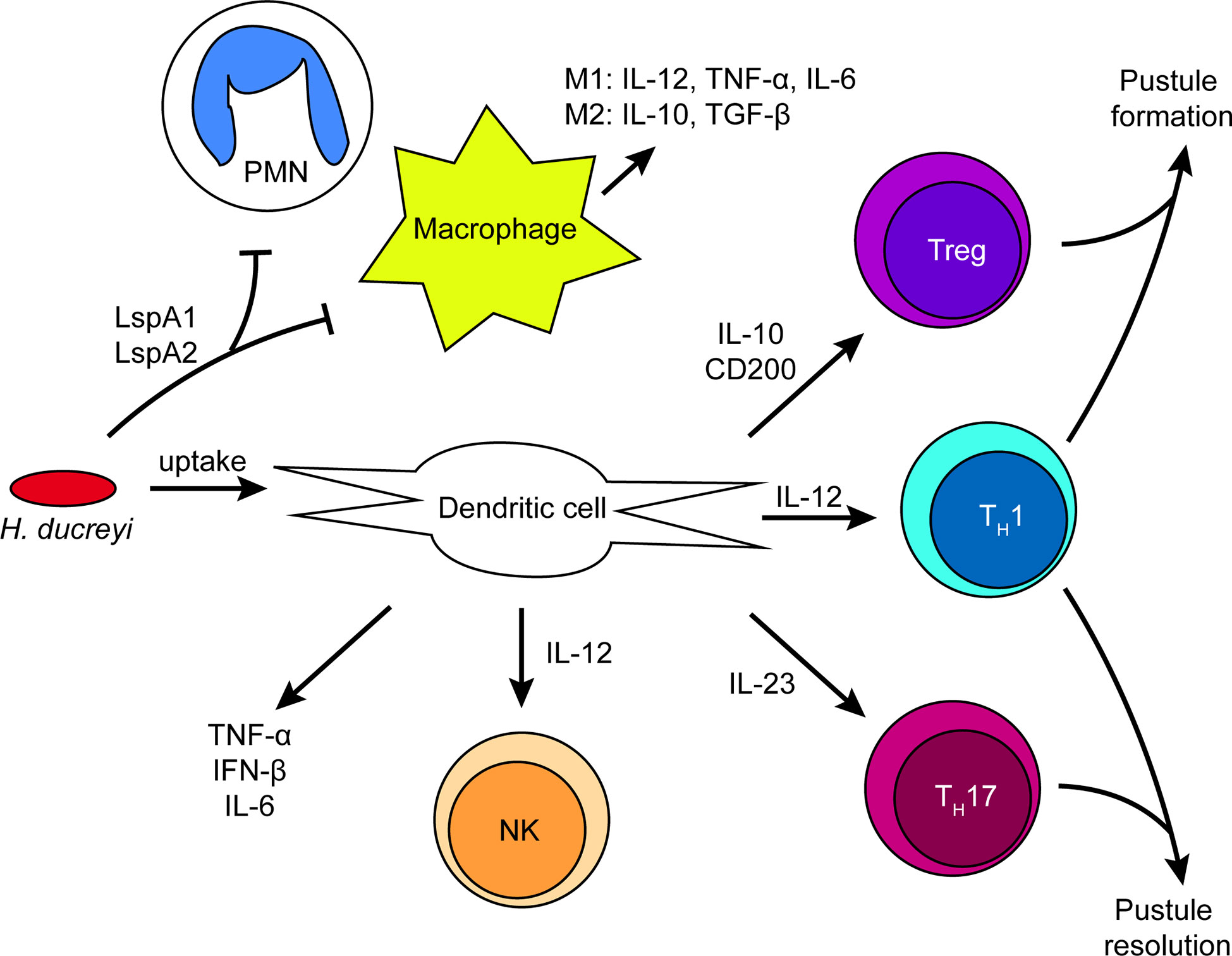
Figure 3 Summary of immune cell activation by H. ducreyi in a pustule. Schematic of immune cell interactions and the cytokine environment in experimental infection based on transcriptomics and in vitro data. H. ducreyi secretes Large supernatant protein (Lsp)A1 and LspA2 to inhibit phagocytosis by neutrophils and macrophages. However, H. ducreyi is directly taken up by dendritic cells; ingested bacteria and/or soluble antigens from lysed bacteria such as lipooligosaccharide (LOS) result in dendritic cell activation. Dendritic cells subsequently secrete the indicated cytokines that lead to activation of NK cells and T cell subsets. The ratio of different T cell subsets in the lesion may be correlated with disease outcome. Further details appear in the text.
Antigen Presentation and Cytokine Secretion by Dendritic Cells
Myeloid DCs are among the most important antigen presenting cells recruited to sites of H. ducreyi infection (58). DCs have both surface and intracellular pattern recognition receptors (PRRs) that recognize foreign molecules or molecules from damaged cells. Ligand binding to PRRs activates DCs, leading to phagocytosis and the transcription of pro- and anti-inflammatory immune response genes that influence the subsequent T cell response. While large supernatant protein (Lsp) A1 and LspA2 have been shown to inhibit phagocytosis of H. ducreyi by neutrophils and macrophages (66), their expression does not impact DC uptake of H. ducreyi (58). Whether this is because DCs are not as sensitive to phagocytic inhibition by LspA1 and LspA2 or whether DCs use a different mechanism to phagocytose H. ducreyi compared to neutrophils and macrophages remains to be determined. DCs can also be activated by H. ducreyi LOS in a non-contact-dependent manner (67), indicating that phagocytosis of H. ducreyi by DCs is sufficient but not necessary to promote DC activitation. H. ducreyi is able to partially inhibit DC activation and maturation through an unknown mechanism; however, DCs are still able to secrete high levels of IL-6 and TNF-α in response to live H. ducreyi (58). Secretion of high levels of IL-6 along with partially activated or immature DCs are hypothesized to be mechanisms by which H. ducreyi avoids a full adaptive immune response and survives immune onslaught.
DCs present antigen to activate T cells and subsequently secrete cytokines that polarize T cells; however, H. ducreyi also partially inhibits DC activation and maturation (58). Despite this, H. ducreyi-specific T cell lines have been isolated from pustules of volunteers who have not been previously exposed to H. ducreyi, which suggests that DCs are able to present antigen to naïve T cells that differentiate into memory cells (68). Differences in pathogen detection and cytokine production that could influence T cell responses have been examined in experimental H. ducreyi infection. Single-nucleotide polymorphisms in Toll-like receptor 9 and interleukin-10 alleles have been identified and are predictive of pustule formation or resolution in experimental infection (69). Microarray analysis of infected and mock infected skin obtained from pustule formers and resolvers who were re-infected for 48 hours suggested that the two groups had both core and distinct tissue transcriptional responses to infection (19). Compared to resolvers, pustule formers had a hyperinflammatory, dysregulated response in the tissue. Monocyte-derived myeloid DCs obtained from resolvers and pustule formers who were re-infected with H. ducreyi also shared a core transcriptional response that should promote Th1 responses. However, resolver DC transcriptomes also suggested the promotion of a Th17 response and the upregulation of IL-23, whereas pustule former DCs promoted a Treg response and the upregulation of IL-10 and CD200. LOS binding to TLR4 on DCs leads to production of TNF-α and IFN-β, which induces indolamine 2,3-dioxygenase (IDO) expression, and IDO induction in DCs may explain the increased number of Tregs found in pustules (67). IDO expression in DCs can promote conversion of naïve T cells to Tregs, prevent Treg conversion to effector T cells, inhibit T cell activation/proliferation, and promote T cell death through tryptophan depletion and production of pro-apoptotic metabolites (70–72), which would result in a decrease in the number of effector T cells and an increase in Tregs. These data suggest that H. ducreyi infection simultaneously increases both pro-inflammatory and anti-inflammatory cytokines and that the balance between the pro-inflammatory and anti-inflammatory signals in different hosts may dictate whether a pustule forms or resolves.
Along with T cells, DCs also activate NK cells (60). H. ducreyi-infected DCs activate NK cells in vitro through production of IL-12 and subsequently lead to NK cell secretion of IFN-γ. IFN-γ secretion, primarily originating from NK cells early in infection, could increase immune-mediated phagocytosis of H. ducreyi.
T Cell Response in Infection
In experimental lesions, CD4+ T cells are primarily recruited and localize beneath the macrophage collar; the average CD4:CD8 ratio is 3:1 (61). T cells comprise 50% of leukocytes in papules at two days post infection. Approximately 70% of T cells collected from papules are CD45RO+ memory T cells, which are likely a mix of skin-resident memory T cells and circulating central memory T cells that responded to chemokines released at the infected site. Whether the T cells in the papule can respond to H. ducreyi has not been tested. We previously observed that DCs in pustule formers and resolvers upregulate different transcripts; these transcripts suggest that the DCs in resolvers promote a type 1 and type 17 response while the DCs in pustule formers promote a type 1 and regulatory response (19). Therefore, we hypothesize that the T cells in the skin of resolvers and pustule formers may similarly differ. In the skin, CD49a (integrin α1) expression has been used to differentiate IFN-γ- versus IL-17-expressing CD8+ cells, which are CD49a+ and CD49a-, respectively (73). Although the differences between CD49a+ and CD49a- CD4 T cells have yet to be determined, CD4 T cells in diseased skin can also express CD49a+ (74). If the T cells identified in papules do respond to H. ducreyi either directly or through an unknown cross-reactive memory T cell response, then pustule formers may have more CD49a+ T cells and resolvers may have more CD49a- T cells. Alternatively, if the T cells are not responding to H. ducreyi, then the T cells that were observed in papules are likely representative of the basal level of T cells in the skin and of non-specific memory T cells that may be following chemotactic gradients.
A hallmark of the adaptive immune system is the recall response. T cell lines have been derived from pustules that were biopsied 6-14 days after experimental infection (68). Treatment of the T cell lines with H. ducreyi antigen in the presence of autologous peripheral blood mononuclear cells (PBMCs), but not irradiated allogeneic PBMCs, led to proliferation and cytokine production from T cells, indicating that the response is MHC restricted. The T cell lines produced minimal responses to related members of the Pasteurellaceae family, demonstrating that the T cell lines are H. ducreyi antigen-specific. Interestingly, this antigen-specific response does not seem to confer protection against re-challenge, as 88% of pustule formers form pustules when re-challenged (75, 76). The reason for the lack of immunity is unclear, but it may in part be due to the lack of antibody responses during experimental infection. Antibody responses are made late in the ulcerative stage of natural infection, but these antibodies are not bactericidal (77).
Avoiding Phagocytosis by Macrophages and Neutrophils
H. ducreyi remains extracellular and prevents its uptake by both neutrophils and macrophages primarily by the secretion of two proteins: LspA1 and LspA2 (66). LspA1 and LspA2 are the largest proteins encoded by the H. ducreyi 35000HP genome, have proportionately higher numbers of SNPs compared to most H. ducreyi genes (16), and are secreted by the LspB transporter. Secreted LspA1 and LspA2 are taken up by neutrophils and macrophages by an unknown mechanism, bind to and are phosphorylated by the human C-terminal tyrosine kinase Csk, and increase Csk activity via a positive feedback loop (78). This increased Csk activity prevents human Src kinases from inducing the actin rearrangements required for FcγR-dependent phagocytosis. Phagocytes with inhibited FcγR-dependent phagocytosis are unable to engulf opsonized bacteria. Ducreyi serum resistance protein A (DsrA) is an outer membrane protein that inhibits IgM from binding to the bacterial surface thereby preventing complement deposition and serum killing (79, 80); theoretically, DsrA could also inhibit complement-mediated phagocytosis. Both DsrA and fibrinogen binding protein A (FgbA) are adhesins that bind fibrinogen, similar to the M proteins of Streptococcus pyogenes (37, 81). Although the role of FgbA and DsrA in resistance to phagocytosis has not been established, binding to fibrinogen in other bacteria is thought to help prevent phagocytosis through adhesion to host cells, steric hindrance, and shielding pathogen associated molecular patterns (PAMPs) that would be recognized by phagocytes (82, 83). Deletion mutants of lspA1 and lspA2, dsrA, or fgbA are attenuated for pustule formation in humans (26, 31, 37). Thus, despite appearing somewhat redundant, they are all required to establish productive infection and, given their posited in vitro functions, to avoid phagocytosis.
With so many mechanisms to prevent phagocytosis, how does the immune response manage to help kill the bacteria and resolve infection? Characterization of macrophages from biopsies of H. ducreyi infected pustules and from in vitro H. ducreyi -infected monocyte-derived macrophages has shown that infection produces a mixed M1/M2 macrophage population (84). The M1 phenotype is characterized by being microbicidal, producing high levels of IL-12, TNF-α, IL-6, and reactive nitrogen and oxygen intermediates; the M2 phenotype is associated with tissue repair and reducing inflammation, producing IL-10 and TGF-β (85). M2 macrophages are also better at phagocytosis than M1 macrophages, especially M2c macrophages, which are polarized by IL-10 and are efficient at phagocytosing H. ducreyi via class A scavenger receptors (84). The proportion of M1 versus M2 macrophages may play an important role in resolving H. ducreyi infection, but is yet to be studied in the context of pustule formation and resolution. Given that resolvers who are re-infected upregulate IL-10 transcripts in tissue compared to pustule formers (19), we hypothesize that resolvers may have more M2c macrophages than pustule formers and are therefore better able to phagocytose and clear H. ducreyi infection via class A scavenger receptor uptake.
The ratio of type 1 and type 2 immune cells has been instrumental in predicting disease progression for bacterial pathogens. M. leprae infection is dynamic. Type 1 and type 17 cells dominate in tuberculoid lesions and type 2 and regulatory cells dominate in lepromatous lesions; oscillation between these two states during natural disease correlates with changes of immune cell phenotypes (62). Similarly, prevention of Staphylococcus aureus and Streptococcus pyogenes skin infections requires a predominantly type 1 immune response rather than a type 2 immune response (86–89). Interestingly, the immune cell typing that controls infection of M. leprae (90), S. aureus (86), or S. pyogenes (89) favors skewing towards a predominately type 1 response, whereas the immune cell typing that controls infection of H. ducreyi may favor a mixed type 1 and type 17 response (19). Important distinctions between these pathogens may account for this difference. For instance, M2 macrophages express an M. leprae entry receptor, CD163 (91), which would explain why leprosy lesions worsen with increased numbers of M2 macrophages. The skewing of macrophage typing towards an M2 response leads to their release of cytokines that can also push the T cells toward a type 2 response.
Defensin and Cathelicidin Resistance
In addition to avoiding phagocytosis, H. ducreyi also resists antimicrobial peptides that are secreted by phagocytes and keratinocytes. H. ducreyi encodes two transporters that confer resistance to some human defensins and the cathelicidin LL-37. A proton motive force-dependent transmembrane efflux pump, Mtr, confers resistance to LL-37 and β-defensins—especially HBD-3. A mtrC deletion mutant had decreased expression of lspB and dsrA, indicating that multiple virulence factors are impacted by the absence of MtrC (92). Another transporter system, Sap, also ameliorates LL-37 activity (93). SapB and SapC form a heterodimeric transporter that transports cargo bound by the periplasmic chaperone SapA. A sapA deletion mutant is 25-50% more sensitive to killing by LL-37 in vitro and has an approximately 50% reduction in pustule formation in the human infection model (39). However, deletion of sapBC results in a mutant that is fully attenuated for pustule formation in the human infection model, suggesting that SapBC may be secreting additional SapA-like protein(s) that serve redundant functions (43).
The positive charge of the H. ducreyi outer membrane was also hypothesized to repulse antimicrobial peptide binding (49). The outer membrane of most Gram-negative bacteria contains lipopolysaccharide (LPS); however, H. ducreyi has a similar, less decorated version—LOS (94). Class I strains have a sialylated nonasaccharide (95), while Class II stains have a non-sialylated pentasaccharide (96). Deletion of either the sialyltransferase lst or the N-acylneuraminate cytidylyltransferase neuA eliminates LOS sialylation in the Class I strain H. ducreyi 35000HP (44). Both mutants are fully virulent and form pustules in human volunteers. This contrasts to the pathogenic Neisseria spp. in which sialylation of LOS is a major virulence determinant (97). H. ducreyi LOS and core oligosaccharide each contain a positively charged phosphoethanolamine (PEA) group (96, 98), which helps to neutralize the otherwise negatively charged LOS and was hypothesized to repel positively charged LL-37. However, deletion of lptA, which attaches PEA to the LOS, did not affect pustule formation (49). Overall, cathelicidin resistance is primarily mediated by the Sap and Mtr transporters rather than membrane charge or sialylation (43, 92).
Interaction of the Host With H. ducreyi
While cytokines and redox species certainly shape the lesion environment, what the microenvironment contains on a molecular level is a current focus of investigation. The molecular composition of the lesion bridges bacterial growth and the recruitment of immune cells. Transcriptomic data were used to first investigate how H. ducreyi alters its gene expression from in vitro growth to the environment of a pustule (99). Comparison of biopsy samples to historic mid-log, transition, and stationary phase in vitro data sets showed that the in vivo transcriptome of H. ducreyi was unique (99). The genes that consistently differed between all growth stages and the biopsies are involved in essential growth pathways and suggest a shift towards anaerobiosis and the use of alternative carbon sources, such as ascorbic acid and citrate, as well as increased amino acid and metal transport, protein folding chaperones, growth arrest, and DNA replication and repair. Given that the in vitro and skin environments are metabolically distinct, a differential growth signature is unsurprising.
A limitation of differential gene expression studies is that only differences at the transcript level are detected and, depending on the gene, may or may not accurately reflect the other biology at the site such as post-transcriptional regulatory mechanisms or the metabolome. Since many terminally differentiated cells, such as neutrophils, generally do not contain high levels of mRNA, genes that are not well-expressed or that are post-transcriptionally regulated may not be detected in RNA-seq datasets, but their metabolic products may be detected by metabolomics. Expanding on the previous study, the H. ducreyi mid-log phase inoculum used to infect volunteers and biopsies of pustules along with their matched wounded skin controls were subjected to unbiased bulk RNA-seq and metabolic profiling (100). The most differentially regulated host genes in the lesion indicated a robust immune response. This metabolomics data showed that there are elevated levels of ascorbic acid as well as the presence of prostaglandins, glutamate, linoleate, and glycosphingolipids in pustules versus sham inoculated skin (100).
The matched metabolomes and transcriptomes support a richer understanding of the lesion. The elevated levels of ascorbic acid in the pustule coupled with the upregulation of H. ducreyi genes that metabolize ascorbic acid suggests that H. ducreyi is using ascorbic acid as an alternative carbon source (99, 100). Although the definitive cellular source of ascorbic acid (and other) metabolites are unknown, neutrophils take up and sequester ascorbic acid and likely release it into the abscess as they become necrotic (101). Thus, this data set helps to generate hypotheses about how different carbon sources are used by H. ducreyi and what host cells may be responsible for producing these carbon sources.
Since spatial information is lost during sample processing, a limitation of tissue metabolomics is that whether the metabolite is sequestered in host cells or freely available extracellularly cannot be determined. Therefore, the outstanding questions involve both sides of the host-pathogen interaction. In addition, because of the extreme difference in the amount of host material compared to the bacteria, the metabolome overwhelmingly reflects host components. For H. ducreyi, we hypothesize that, given the upregulation of genes to metabolize a given compound, such as ascorbic acid, coupled with the inability to synthesize the input metabolite of the pathway, H. ducreyi is using these metabolites as energy sources (99, 100). Generation of gene deletion mutant strains and subsequent experimental infection with these strains will confirm whether these are important carbon sources for H. ducreyi.
Because both the host and bacterial transcripts present in the pustule were captured from the infected samples, it was possible to use bioinformatics to generate an interaction network of host-bacterial gene clusters in an unbiased manner that were either positively or negatively correlated. Notably, genes involved in anaerobiosis were correlated with several pro-inflammatory host genes that are upregulated during infection (100). A similar strategy was used to determine an interaction network for naturally occurring lepromatous M. leprae biopsies; an interactome for tuberculoid M. leprae infection could not be established, since there were too few bacterial reads in the biopsies (4). The M. leprae interaction network confirmed that the type I interferon response is upregulated and that lepromatous lesions have increased bacterial metabolism (4). This was correlated to class switching in B cells (4). Application of these techniques to biopsies of lesions caused by other organisms will also elucidate whether immune system metabolism is altered along a common pathway(s) or whether infections differentially influence host metabolism. Thus, dual RNA-seq provides a tool to examine interaction networks that may identify therapeutic targets.
Toward Uncovering Metabolic Shifts in Specific Cell Types
Understanding how the host contributes to these differences is more complicated; central carbon metabolites—namely citrate, α-ketogluterate, and succinate—have been shown to have alternative uses outside of central metabolism during inflammation (102). These metabolites can be transported out of the mitochondria to form antimicrobial metabolites, protect against oxidation, and alter protein functions through post-translational modification (102). Furthermore, whether the differences observed in H. ducreyi pustules are representative of changes in local cellular metabolism (e.g., from keratinocytes and fibroblasts), the infiltration of specific cell types that are high in a given metabolite, or both is not known. As such, determining whether these compounds become available to H. ducreyi through diffusion or secretion from live host cells or become available after host cell death and lysis is an outstanding question. Utilization of emerging spatial metabolomics approaches in the future will further advance our understanding of the lesion on the molecular level (103).
Emerging technologies are providing new insights into specific host cell contributions to disease. Instead of restricting lesional cell characterization by using pre-selected phenotypic markers for flow cytometry, next generation sequencing provides the possibility of identifying the source of transcripts at the single cell level as well as their physical location within a lesion using spatial transcriptomics. This will allow us to better correlate the proportion of different cell types observed in previous studies (57–61); determine the cellular source(s) of differentially expressed genes observed in bulk RNA-seq studies (99, 100); and better estimate the effectiveness of the immune response in individual volunteers. In preliminary experiments, we have generated single cell suspensions and performed droplet single cell RNA sequencing on biopsies of pustules. Using Seurat (104) to cluster cells, macrophages, monocytes, DCs, T cells, NK cells, as well as endothelial cells, fibroblasts, and keratinocytes were identified in the biopsies (Figure 4). Interestingly, neutrophils were not detected perhaps because they are terminally differentiated and do not have high levels of mRNA. Nevertheless, the results of single cell RNA-seq studies can then be compared to our bulk transcriptomics data sets and to other publicly available single cell data sets that are available from normal and diseased human skin (2).
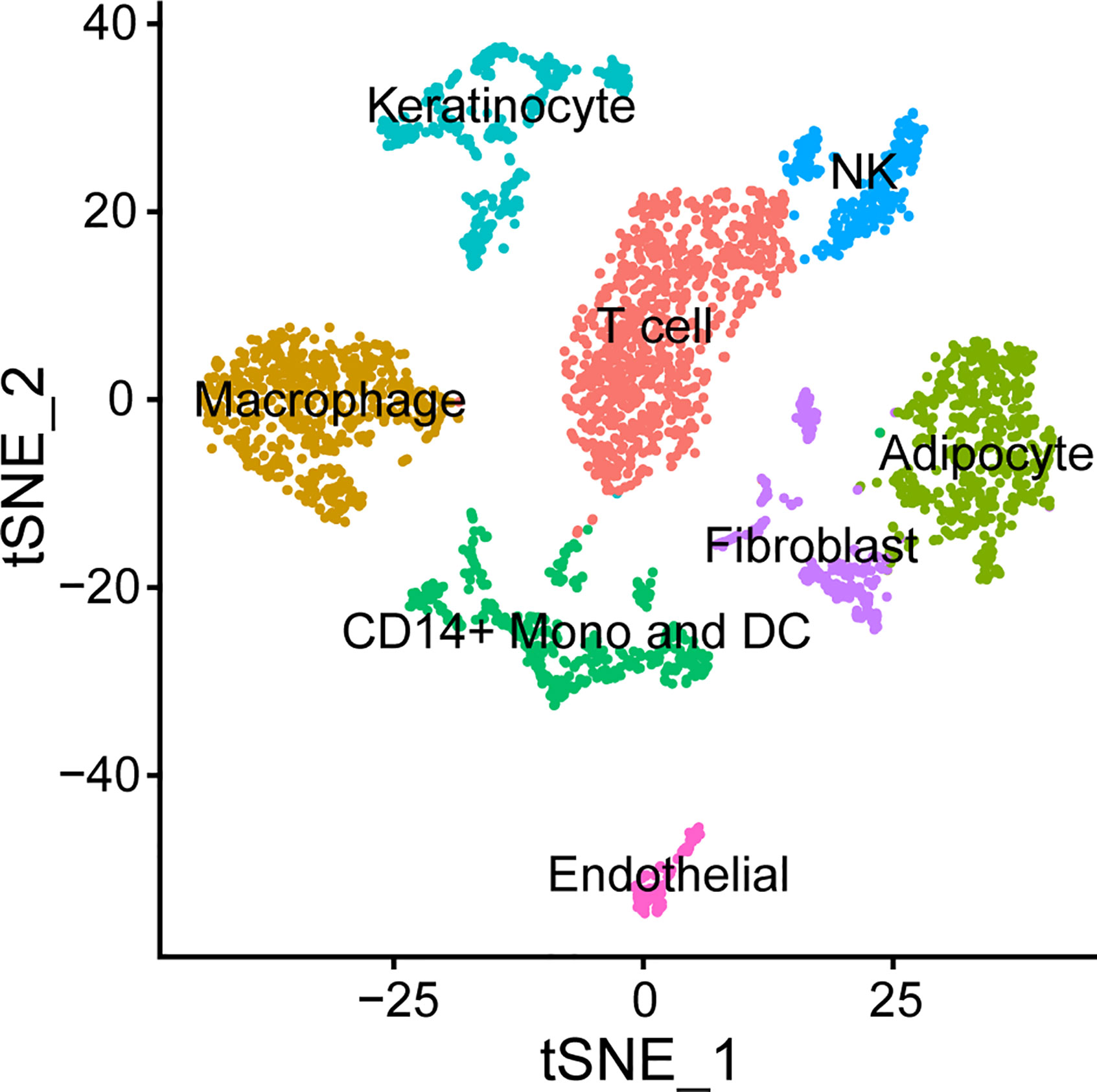
Figure 4 Cell types found in the H. ducreyi pustule by single cell RNA-seq. Cells from a pustule were dissociated and single cell sequencing was performed. Cells were clustered in Seurat (104) and identified based off the unique, abundant transcripts in each cluster.
In previous studies, we showed that papules of pustule formers and resolvers who were re-infected for 48 hours had distinct transcription profiles that correlated with disease progression versus resolution (19). Due to the limitations of technology available at that time, we were not able to identify the cells responsible for differential transcript expression. Although we have shown that women are less susceptible to pustule formation than men, we have not been able to identify the cause(s) of the gender difference. A similar gender difference has been observed for progression from tuberculoid to lepromatous lesions for M. leprae, where more men progress to lepromatous lesions (90). Interestingly, some immunometabolites that are known to be regulated by sex hormones, such as prostaglandins, are upregulated in pustules (19, 100) and prostaglandin E2 has been shown to increase cytokine secretion in a monocyte-derived DC model (67). During infection, these cytokines are predicted to promote inflammatory responses in leukocytes and tissue repair in keratinocytes and fibroblasts. Single cell RNA-seq and metabolomics have the potential to unravel the differences in the tissues of pustule formers and resolvers who are re-infected and of men and women.
The Future of Understanding Host-Pathogen Interaction in the Skin
A full understanding of how an immune response is mounted in infectious human skin diseases will require determining the metabolic environment during infection and the cell-cell signaling involved as the lesion is first formed. Because of the difficulty of obtaining sufficient numbers of cells responding to invading pathogens early in infection, the first cells to initiate the immune response is largely unknown. Thus, we do not know the in vivo signal(s) received by the immune system that begins the recruitment of additional leukocytes. We anticipate that new “omics” technologies will help close this knowledge gap and help the field to generate a more holistic model of skin infection.
In addition to the interaction between the pathogen and host, other environmental factors, such as the skin microbiome, may play a role in determining infection outcome. In a prospective study, the pre-infection microbiomes of sites of people who resolved experimental infection clustered together while those of pustule formers were more diverse (105). In the course of experimental infection, the microbiomes of resolvers did not change, while the microbiomes of pustule formers were driven to a similar composition; in the latter, H. ducreyi dominates the lesion, and the proportions of the other genera decrease substantially during disease progression (105). The pre-infection microbiomes of resolvers tend to have higher levels of Actinobacteria, Firmicutes, and Bacteroidetes and lower levels of Proteobacteria compared to pustule formers, which had higher levels of micrococci and Staphylococcus. Whether these microbiome differences are because the former genera actively outcompete H. ducreyi (e.g., secrete toxins or metabolites that H. ducreyi is sensitive to), are a product of environmental factors (e.g., different hygiene practices and/or products), or underlying immune responsiveness is not known. However, the data suggest that the environment of a pustule drives the microbiome to a more uniform composition.
Future of Treatment Options for Skin Pathogens
H. ducreyi is currently only reliably susceptible to macrolides, quinolones, and third generation cephalosporins. For chancroid, the preferred Center for Diseases Control and Prevention regimens include a single dose of oral azithromycin, a single dose of intramuscular ceftriaxone or a 3 day course of oral ciprofloxacin (8). For CU, a single oral dose of azithromycin has been the only regimen that has been studied and is highly effective (106). Further emergence of antibiotic resistance in endemic regions may require alternative treatment strategies. The higher differential expression of genes involved in carbon source switching and hypoxia/anaerobiosis in vivo indicates that interfering with bacterial nutrition may be a viable treatment tactic (99, 100).
If H. ducreyi is found to be dependent on “unique” metabolite(s), it may be possible to develop inhibitors to enzymes used in its metabolism in H. ducreyi. This strategy would require that host enzymes—and potentially those of other members of the healthy skin microbiome—are not impacted by the inhibitor. If the metabolite is only used by H. ducreyi, then it may also be possible to prevent its uptake and essentially starve the bacteria of a carbon source. Given the emergence of (multiple) drug resistant skin pathogens such as S. aureus or sexually transmitted bacteria such as N. gonorrhoeae, limiting broad spectrum antibiotics may help to decrease the prevalence of drug resistant strains in the population. Because H. ducreyi infects the upper layers of the skin, topical treatments could also be developed.
Finally, experimental human infection with deletion of candidate gene targets has been used to define essential virulence genes. Identification of vaccine antigens that produce high titers of opsonizing antibody have been complicated by the low tendency of H. ducreyi antigens to elicit long lasting immune responses. Evaluating vaccine efficacy in animal models is challenging because animal models lead to pathogen clearance and/or require very high dosages for infection. Nevertheless, two vaccine candidates have been studied in the porcine ear model. They are Class I specific and are most likely ineffective against Class II strains because the evaluated genes are among the genes with the most sequence diversity between Class I and Class II strains (107, 108). Therefore, these results are not wholly unanticipated, but suggest that targeting essential specific outer membrane proteins may not be as fruitful of a strategy as originally thought.
Conclusions
The ability to infect humans with H. ducreyi has been instrumental in exploring host-pathogen interaction at the tissue level. Nearly thirty years of human challenge experiments are helping to confirm previous studies as well as guide us toward new biological insights for future study on the cellular and molecular levels. This model is beneficial for both the skin immunology community, since it is one of the few human experimental infection models that captures the cells and metabolites of the lesion microenvironment. Thus, a rich understanding of cell types in the lesion, immune activation, and lesion architecture is achieved.
Advancements in “omics” technologies afford researchers a more holistic view of host-pathogen adaptions during infection. Bacterial virulence has expanded from primarily being thought of as the result of extracellular and secreted bacterial effectors to including how metabolism—both in the host and the bacteria—influence infection outcomes. Increased awareness of differences in microbiomes (105) and nutrient availability in vivo (99, 100) has highlighted that bacteria differentially regulate genes in the context of the host response and alterations in nutrient availability.
As we investigate skin inflammatory responses, we must also acknowledge that responses can differ not only by pathogen but also potentially by skin site. For other inflammatory conditions such as psoriasis, single cell RNA-seq revealed that while most dysregulated genes in psoriasis are independent of location on the body, some differentially expressed genes are unique to where psoriatic lesions are located (2). Most notably, different T cell subsets are activated based on location, which may influence therapeutic intervention strategies and outcomes (2). Similarly, location of disease may be of particular interest in H. ducreyi infection, since it infects both cutaneous lower leg and arm skin as well as genital skin. How or if the immune response significantly differs in these two locations has not been studied; however, the relationships between H. ducreyi and host cells are similar in experimental and natural infection (1). The use of clinical samples in infectious disease will be instrumental in elucidating the mechanisms behind various skin pathologies and in appreciating both inter-person and intra-person response diversity. As more data become available for different diseases, we anticipate that common immune response pathways will emerge, giving us a better understanding of how skin infections and the immune system interact.
Author Contributions
JB, BG, and SS conceptualized the manuscript. JB and BG wrote the original draft. LC analyzed transcriptomic data. JB, BG, LC, and SS revised the final draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
JB and BG were supported by T32AI007637. This work was supported by grant UL RR052761 from the Indiana Clinical and Translational Sciences Institute and the Indiana Clinical Research Center and by R01AI134727 and R01AI137116 from the National Institute of Allergy and Infectious Diseases to SS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Drs. Byron Batteiger and Margaret Bauer for critiquing this manuscript. We are also grateful to all the volunteers that have participated in the human challenge model.
References
1. Bauer ME, Townsend CA, Ronald AR, Spinola SM. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microbes Infect (2006) 8(9-10):2465–8. doi: 10.1016/j.micinf.2006.06.001
2. Cheng JB, Sedgewick AJ, Finnegan A II, Harirchian P, Lee J, Kwon S, et al. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep (2018) 25(4):871–83. doi: 10.1016/j.celrep.2018.09.006
3. Zhu J, Koelle DM, Cao J, Vazquez J, Huang ML, Hladik F, et al. Virus-specific CD8+ T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J Exp Med (2007) 204(3):595–603. doi: 10.1084/jem.20061792
4. Montoya DJ, Andrade P, Silva BJA, Teles RMB, Ma F, Bryson B, et al. Dual RNA-Seq of Human Leprosy Lesions Identifies Bacterial Determinants Linked to Host Immune Response. Cell Rep (2019) 26(13):3574–85.e3. doi: 10.1016/j.celrep.2019.02.109
5. Janowicz DM, Ofner S, Katz BP, Spinola SM. Experimental infection of human volunteers with Haemophilus ducreyi: fifteen years of clinical data and experience. J Infect Dis (2009) 199(11):1671–9. doi: 10.1086/598966
6. Spinola SM, Wild LM, Apicella MA, Gaspari AA, Campagnari AA. Experimental human infection with Haemophilus ducreyi. J Infect Dis (1994) 169(5):1146–50. doi: 10.1093/infdis/169.5.1146
7. Gonzalez-Beiras C, Marks M, Chen CY, Roberts S, Mitja O. Epidemiology of Haemophilus ducreyi Infections. Emerg Infect Dis (2016) 22(1):1–8. doi: 10.3201/eid2201.150425
8. Spinola SM. Chancroid and Haemophilus ducreyi. In: Sparling PF, Holmes KK, Stamm WE, Piot P, Wasserheit JN, Corey L, Cohen MS, Watts DH, editors. Sexually Transmitted Diseases. New York: McGraw-Hill (2008).
9. Mitja O, Godornes C, Houinei W, Kapa A, Paru R, Abel H, et al. Re-emergence of yaws after single mass azithromycin treatment followed by targeted treatment: a longitudinal study. Lancet (2018) 391(10130):1599–607. doi: 10.1016/S0140-6736(18)30204-6
10. Ghinai R, El-Duah P, Chi KH, Pillay A, Solomon AW, Bailey RL, et al. A cross-sectional study of ‘yaws’ in districts of Ghana which have previously undertaken azithromycin mass drug administration for trachoma control. PLoS Negl Trop Dis (2015) 9(1):e0003496. doi: 10.1371/journal.pntd.0003496
11. Marks M, Chi KH, Vahi V, Pillay A, Sokana O, Pavluck A, et al. Haemophilus ducreyi associated with skin ulcers among children, Solomon Islands. Emerg Infect Dis (2014) 20(10):1705–7. doi: 10.3201/eid2010.140573
12. Grant JC, Gonzalez-Beiras C, Amick KM, Fortney KR, Gangaiah D, Humphreys TL, et al. and Class II Haemophilus ducreyi Strains Cause Cutaneous Ulcers in Children on an Endemic Island. Clin Infect Dis (2018) 67(11):1768–74. doi: 10.1093/cid/ciy343
13. Houinei W, Godornes C, Kapa A, Knauf S, Mooring EQ, Gonzalez-Beiras C, et al. Haemophilus ducreyi DNA is detectable on the skin of asymptomatic children, flies and fomites in villages of Papua New Guinea. PLoS Negl Trop Dis (2017) 11(5):e0004958. doi: 10.1371/journal.pntd.0004958
14. White CD, Leduc I, Olsen B, Jeter C, Harris C, Elkins C. Haemophilus ducreyi Outer membrane determinants, including DsrA, define two clonal populations. Infect Immun (2005) 73(4):2387–99. doi: 10.1128/IAI.73.4.2387-2399.2005
15. Gangaiah D, Spinola SM. Haemophilus ducreyi Cutaneous Ulcer Strains Diverged from Both Class I and Class II Genital Ulcer Strains: Implications for Epidemiological Studies. PLoS Negl Trop Dis (2016) 10(12):e0005259. doi: 10.1371/journal.pntd.0005259
16. Gangaiah D, Webb KM, Humphreys TL, Fortney KR, Toh E, Tai A, et al. Haemophilus ducreyi Cutaneous Ulcer Strains Are Nearly Identical to Class I Genital Ulcer Strains. PLoS Negl Trop Dis (2015) 9(7):e0003918. doi: 10.1371/journal.pntd.0003918
17. Marks M, Fookes M, Wagner J, Ghinai R, Sokana O, Sarkodie YA, et al. Direct Whole-Genome Sequencing of Cutaneous Strains of Haemophilus ducreyi. Emerg Infect Dis (2018) 24(4):786–9. doi: 10.3201/eid2404.171726
18. Spinola SM, Bauer ME, Munson RS Jr. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect Immun (2002) 70(4):1667–76. doi: 10.1128/iai.70.4.1667-1676.2002
19. Humphreys TL, Li L, Li X, Janowicz DM, Fortney KR, Zhao Q, et al. Dysregulated immune profiles for skin and dendritic cells are associated with increased host susceptibility to Haemophilus ducreyi infection in human volunteers. Infect Immun (2007) 75(12):5686–97. doi: 10.1128/IAI.00777-07
20. Palmer KL, Thornton AC, Fortney KR, Hood AF, Munson RS Jr, Spinola SM. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis (1998) 178(1):191–9. doi: 10.1086/515617
21. Young RS, Fortney K, Haley JC, Hood AF, Campagnari AA, Wang J, et al. Expression of sialylated or paragloboside-like lipooligosaccharides are not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect Immun (1999) 67(12):6335–40. doi: 10.1128/IAI.67.12.6335-6340.1999
22. Al-Tawfiq JA, Bauer ME, Fortney KR, Katz BP, Hood AF, Ketterer M, et al. A pilus-deficient mutant of Haemophilus ducreyi is virulent in the human model of experimental infection. J Infect Dis (2000) 181(3):1176–9. doi: 10.1086/315310
23. Al-Tawfiq JA, Fortney KR, Katz BP, Hood AF, Elkins C, Spinola SM. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J Infect Dis (2000) 181(3):1049–54. doi: 10.1086/315309
24. Throm RE, Al-Tawfiq JA, Fortney KR, Katz BP, Hood AF, Slaughter CA, et al. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect Immun (2000) 68(5):2602–7. doi: 10.1128/iai.68.5.2602-2607.2000
25. Fortney KR, Young RS, Bauer ME, Katz BP, Hood AF, Munson RS Jr, et al. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect Immun (2000) 68(11):6441–8. doi: 10.1128/iai.68.11.6441-6448.2000
26. Bong CT, Throm RE, Fortney KR, Katz BP, Hood AF, Elkins C, et al. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect Immun (2001) 69(3):1488–91. doi: 10.1128/IAI.69.3.1488-1491.2001
27. Young RS, Fortney KR, Gelfanova V, Phillips CL, Katz BP, Hood AF, et al. Expression of cytolethal distending toxin and hemolysin is not required for pustule formation by Haemophilus ducreyi in human volunteers. Infect Immun (2001) 69(3):1938–42. doi: 10.1128/IAI.69.3.1938-1942.2001
28. Young RS, Filiatrault MJ, Fortney KR, Hood AF, Katz BP, Munson RS Jr, et al. Haemophilus ducreyi lipooligosaccharide mutant defective in expression of beta-1,4-glucosyltransferase is virulent in humans. Infect Immun (2001) 69(6):4180–4. doi: 10.1128/IAI.69.6.4180-4184.2001
29. Bong CT, Fortney KR, Katz BP, Hood AF, San Mateo LR, Kawula TH, et al. A superoxide dismutase C mutant of Haemophilus ducreyi is virulent in human volunteers. Infect Immun (2002) 70(3):1367–71. doi: 10.1128/iai.70.3.1367-1371.2002
30. Spinola SM, Fortney KR, Katz BP, Latimer JL, Mock JR, Vakevainen M, et al. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect Immun (2003) 71(12):7178–82. doi: 10.1128/iai.71.12.7178-7182.2003
31. Janowicz DM, Fortney KR, Katz BP, Latimer JL, Deng K, Hansen EJ, et al. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect Immun (2004) 72(8):4528–33. doi: 10.1128/IAI.72.8.4528-4533.2004
32. Janowicz D, Luke NR, Fortney KR, Katz BP, Campagnari AA, Spinola SM. Expression of OmpP2A and OmpP2B is not required for pustule formation by Haemophilus ducreyi in human volunteers. Microb Pathog (2006) 40(3):110–5. doi: 10.1016/j.micpath.2005.11.005
33. Janowicz D, Leduc I, Fortney KR, Katz BP, Elkins C, Spinola SM. A DltA mutant of Haemophilus ducreyi Is partially attenuated in its ability to cause pustules in human volunteers. Infect Immun (2006) 74(2):1394–7. doi: 10.1128/IAI.74.2.1394-1397.2006
34. Fulcher RA, Cole LE, Janowicz DM, Toffer KL, Fortney KR, Katz BP, et al. Expression of Haemophilus ducreyi collagen binding outer membrane protein NcaA is required for virulence in swine and human challenge models of chancroid. Infect Immun (2006) 74(5):2651–8. doi: 10.1128/IAI.74.5.2651-2658.2006
35. Banks KE, Fortney KR, Baker B, Billings SD, Katz BP, Munson RS Jr, et al. The enterobacterial common antigen-like gene cluster of Haemophilus ducreyi contributes to virulence in humans. J Infect Dis (2008) 197(11):1531–6. doi: 10.1086/588001
36. Leduc I, Banks KE, Fortney KR, Patterson KB, Billings SD, Katz BP, et al. Evaluation of the repertoire of the TonB-dependent receptors of Haemophilus ducreyi for their role in virulence in humans. J Infect Dis (2008) 197(8):1103–9. doi: 10.1086/586901
37. Bauer ME, Townsend CA, Doster RS, Fortney KR, Zwickl BW, Katz BP, et al. A fibrinogen-binding lipoprotein contributes to the virulence of Haemophilus ducreyi in humans. J Infect Dis (2009) 199(5):684–92. doi: 10.1086/596656
38. Labandeira-Rey M, Janowicz DM, Blick RJ, Fortney KR, Zwickl B, Katz BP, et al. Inactivation of the Haemophilus ducreyi luxS gene affects the virulence of this pathogen in human subjects. J Infect Dis (2009) 200(3):409–16. doi: 10.1086/600142
39. Mount KL, Townsend CA, Rinker SD, Gu X, Fortney KR, Zwickl BW, et al. Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect Immun (2010) 78(3):1176–84. doi: 10.1128/IAI.01014-09
40. Spinola SM, Fortney KR, Baker B, Janowicz DM, Zwickl B, Katz BP, et al. Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect Immun (2010) 78(9):3898–904. doi: 10.1128/IAI.00432-10
41. Janowicz DM, Cooney SA, Walsh J, Baker B, Katz BP, Fortney KR, et al. Expression of the Flp proteins by Haemophilus ducreyi is necessary for virulence in human volunteers. BMC Microbiol (2011) 11:208. doi: 10.1186/1471-2180-11-208
42. Labandeira-Rey M, Dodd D, Fortney KR, Zwickl B, Katz BP, Janowicz DM, et al. A Haemophilus ducreyi CpxR deletion mutant is virulent in human volunteers. J Infect Dis (2011) 203(12):1859–65. doi: 10.1093/infdis/jir190
43. Rinker SD, Gu X, Fortney KR, Zwickl BW, Katz BP, Janowicz DM, et al. Permeases of the sap transporter are required for cathelicidin resistance and virulence of Haemophilus ducreyi in humans. J Infect Dis (2012) 206(9):1407–14. doi: 10.1093/infdis/jis525
44. Spinola SM, Li W, Fortney KR, Janowicz DM, Zwickl B, Katz BP, et al. Sialylation of lipooligosaccharides is dispensable for the virulence of Haemophilus ducreyi in humans. Infect Immun (2012) 80(2):679–87. doi: 10.1128/IAI.05826-11
45. Gangaiah D, Li W, Fortney KR, Janowicz DM, Ellinger S, Zwickl B, et al. Carbon storage regulator A contributes to the virulence of Haemophilus ducreyi in humans by multiple mechanisms. Infect Immun (2013) 81(2):608–17. doi: 10.1128/IAI.01239-12
46. Janowicz DM, Zwickl BW, Fortney KR, Katz BP, Bauer ME. Outer membrane protein P4 is not required for virulence in the human challenge model of Haemophilus ducreyi infection. BMC Microbiol (2014) 14:166. doi: 10.1186/1471-2180-14-166
47. Gangaiah D, Labandeira-Rey M, Zhang X, Fortney KR, Ellinger S, Zwickl B, et al. Haemophilus ducreyi Hfq contributes to virulence gene regulation as cells enter stationary phase. mBio (2014) 5(1):e01081–13. doi: 10.1128/mBio.01081-13
48. Holley C, Gangaiah D, Li W, Fortney KR, Janowicz DM, Ellinger S, et al. A (p)ppGpp-null mutant of Haemophilus ducreyi is partially attenuated in humans due to multiple conflicting phenotypes. Infect Immun (2014) 82(8):3492–502. doi: 10.1128/IAI.01994-14
49. Trombley MP, Post DM, Rinker SD, Reinders LM, Fortney KR, Zwickl BW, et al. Phosphoethanolamine Transferase LptA in Haemophilus ducreyi Modifies Lipid A and Contributes to Human Defensin Resistance In Vitro. PLoS One (2015) 10(4):e0124373. doi: 10.1371/journal.pone.0124373
50. Holley CL, Zhang X, Fortney KR, Ellinger S, Johnson P, Baker B, et al. DksA and (p)ppGpp have unique and overlapping contributions to Haemophilus ducreyi pathogenesis in humans. Infect Immun (2015) 83(8):3281–92. doi: 10.1128/IAI.00692-15
51. Leduc I, Fortney KR, Janowicz DM, Zwickl B, Ellinger S, Katz BP, et al. A Class I Haemophilus ducreyi Strain Containing a Class II hgbA Allele Is Partially Attenuated in Humans: Implications for HgbA Vaccine Efficacy Trials. Infect Immun (2019) 87(7):e00112-19. doi: 10.1128/IAI.00112-19
52. Minassian AM, Satti I, Poulton ID, Meyer J, Hill AV, McShane H. A human challenge model for Mycobacterium tuberculosis using Mycobacterium bovis bacille Calmette-Guerin. J Infect Dis (2012) 205(7):1035–42. doi: 10.1093/infdis/jis012
53. Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet (2013) 381(9871):1021–8. doi: 10.1016/S0140-6736(13)60177-4
54. Duthie MS, Saunderson P, Reed SG. The potential for vaccination in leprosy elimination: new tools for targeted interventions. Mem Inst Oswaldo Cruz (2012) 107 Suppl 1:190–6. doi: 10.1590/s0074-02762012000900027
55. Bauer ME, Spinola SM. Localization of Haemophilus ducreyi at the pustular stage of disease in the human model of infection. Infect Immun (2000) 68(4):2309–14. doi: 10.1128/iai.68.4.2309-2314.2000
56. Spinola SM, Ballard R. Chancroid. In: Morse SA, Ballard RC, Holmes KK, Moreland AA, editors. Atlas of Sexually Transmitted Diseases and AIDS, 4th ed. London, UK: Elsevier Ltd (2010).
57. Bauer ME, Goheen MP, Townsend CA, Spinola SM. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect Immun (2001) 69(4):2549–57. doi: 10.1128/IAI.69.4.2549-2557.2001
58. Banks KE, Humphreys TL, Li W, Katz BP, Wilkes DS, Spinola SM. Haemophilus ducreyi partially activates human myeloid dendritic cells. Infect Immun (2007) 75(12):5678–85. doi: 10.1128/IAI.00702-07
59. Humphreys TL, Baldridge LA, Billings SD, Campbell JJ, Spinola SM. Trafficking pathways and characterization of CD4 and CD8 cells recruited to the skin of humans experimentally infected with Haemophilus ducreyi. Infect Immun (2005) 73(7):3896–902. doi: 10.1128/IAI.73.7.3896-3902.2005
60. Li W, Janowicz DM, Fortney KR, Katz BP, Spinola SM. Mechanism of human natural killer cell activation by Haemophilus ducreyi. J Infect Dis (2009) 200(4):590–8. doi: 10.1086/600123
61. Palmer KL, Schnizlein-Bick CT, Orazi A, John K, Chen CY, Hood AF, et al. The immune response to Haemophilus ducreyi resembles a delayed-type hypersensitivity reaction throughout experimental infection of human subjects. J Infect Dis (1998) 178(6):1688–97. doi: 10.1086/314489
62. de Sousa JR, Sotto MN, Simões Quaresma JA. Leprosy As a Complex Infection: Breakdown of the Th1 and Th2 Immune Paradigm in the Immunopathogenesis of the Disease. Front Immunol (2017) 8:1635. doi: 10.3389/fimmu.2017.01635
63. Pinheiro RO, Schmitz V, Silva BJA, Dias AA, de Souza BJ, de Mattos Barbosa MG, et al. Innate Immune Responses in Leprosy. Front Immunol (2018) 9:518. doi: 10.3389/fimmu.2018.00518
64. Roltgen K, Pluschke G. Buruli ulcer: The Efficacy of Innate Immune Defense May Be a Key Determinant for the Outcome of Infection With Mycobacterium ulcerans. Front Microbiol (2020) 11:1018. doi: 10.3389/fmicb.2020.01018
65. Carlson JA, Dabiri G, Cribier B, Sell S. The immunopathobiology of syphilis: the manifestations and course of syphilis are determined by the level of delayed-type hypersensitivity. Am J Dermatopathol (2011) 33(5):433–60. doi: 10.1097/DAD.0b013e3181e8b587
66. Vakevainen M, Greenberg S, Hansen EJ. Inhibition of phagocytosis by Haemophilus ducreyi requires expression of the LspA1 and LspA2 proteins. Infect Immun (2003) 71:5994–6003. doi: 10.1128/iai.71.10.5994-6003.2003
67. Li W, Katz BP, Spinola SM. Haemophilus ducreyi lipooligosaccharides induce expression of the immunosuppressive enzyme indoleamine 2,3-dioxygenase via type I interferons and tumor necrosis factor alpha in human dendritic cells. Infect Immun (2011) 79(8):3338–47. doi: 10.1128/IAI.05021-11
68. Gelfanova V, Humphreys TL, Spinola SM. Characterization of Haemophilus ducreyi-specific T cell lines from lesions of experimentally infected human subjects. Infect Immun (2001) 69:4224–31. doi: 10.1128/IAI.69.7.4224-4231.2001
69. Singer M, Li W, Morre SA, Ouburg S, Spinola SM. Host Polymorphisms in TLR9 and IL10 Are Associated With the Outcomes of Experimental Haemophilus ducreyi Infection in Human Volunteers. J Infect Dis (2016) 214(3):489–95. doi: 10.1093/infdis/jiw164
70. Baban B, Chandler PR, Sharma MD, Pihkala J, Koni PA, Munn DH, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J Immunol (2009) 183(4):2475–83. doi: 10.4049/jimmunol.0900986
71. Mellor AL, Baban B, Chandler P, Jhaver K, Hansen A, Koni PA, et al. Cutting edge: induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J Immunol (2003) 171:1652–5. doi: 10.4049/jimmunol.171.4.1652
72. Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, et al. T cell apoptosis by tryptophan catabolism. Cell Death Differ (2002) 9(10):1069–77. doi: 10.1038/sj.cdd.4401073
73. Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, et al. CD49a Expression Defines Tissue-Resident CD8(+) T Cells Poised for Cytotoxic Function in Human Skin. Immunity (2017) 46(2):287–300. doi: 10.1016/j.immuni.2017.01.009
74. Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med (2007) 13(7):836–42. doi: 10.1038/nm1605
75. Al-Tawfiq JA, Palmer KL, Chen C-Y, Haley JC, Katz BP, Hood AF, et al. Experimental infection of human volunteers with Haemophilus ducreyi does not confer protection against subsequent challenge. J Infect Dis (1999) 179:1283–7. doi: 10.1086/314732
76. Spinola SM, Bong CTH, Faber AL, Fortney KR, Bennett SL, Townsend CA, et al. Differences in host susceptibility to disease progression in the human challenge model of Haemophilus ducreyi infection. Infect Immun (2003) 71:6658–63. doi: 10.1128/iai.71.11.6658-6663.2003
77. Chen CY, Mertz KJ, Spinola SM, Morse SA. Comparison of enzyme immunoassays for antibodies to Haemophilus ducreyi in a community outbreak of chancroid in the United States. J Infect Dis (1997) 175:1390–5. doi: 10.1086/516471
78. Dodd DA, Worth RG, Rosen MK, Grinstein S, van Oers NS, Hansen EJ. The Haemophilus ducreyi LspA1 Protein Inhibits Phagocytosis By Using a New Mechanism Involving Activation of C-Terminal Src Kinase. MBio (2014) 5(3):1–11. doi: 10.1128/mBio.01178-14
79. Elkins C, Morrow KJ, Olsen B. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect Immun (2000) 68:1608–19. doi: 10.1128/iai.68.3.1608-1619.2000
80. Abdullah M, Nepluev I, Afonina G, Ram S, Rice P, Cade W, et al. Killing of dsrA mutants of Haemophilus ducreyi by normal human serum occurs via the classical complement pathway and is initiated by immunoglobulin M binding. Infect Immun (2005) 73:3431–9. doi: 10.1128/IAI.73.6.3431-3439.2005
81. Herwald H, Cramer H, Mörgelin M, Russell W, Sollenberg U, Norrby-Teglund A, et al. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell (2004) 116(3):367–79. doi: 10.1016/s0092-8674(04)00057-1
82. Berends ET, Kuipers A, Ravesloot MM, Urbanus RT, Rooijakkers SH. Bacteria under stress by complement and coagulation. FEMS Microbiol Rev (2014) 38(6):1146–71. doi: 10.1111/1574-6976.12080
83. Vaca DJ, Thibau A, Schutz M, Kraiczy P, Happonen L, Malmstrom J, et al. Interaction with the host: the role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med Microbiol Immunol (2020) 209(3):277–99. doi: 10.1007/s00430-019-00644-3
84. Li W, Katz BP, Spinola SM. Haemophilus ducreyi-induced IL-10 promotes a mixed M1 and M2 activation program in human macrophages. Infect Immun (2012) 80(12):4426–34. doi: 10.1128/IAI.00912-12
85. Atri C, Guerfali FZ, Laouini D. Role of Human Macrophage Polarization in Inflammation during Infectious Diseases. Int J Mol Sci (2018) 19(6):1–15. doi: 10.3390/ijms19061801
86. Brown AF, Murphy AG, Lalor SJ, Leech JM, O’Keeffe KM, Mac Aogáin M, et al. Memory Th1 Cells Are Protective in Invasive Staphylococcus aureus Infection. PLoS Pathog (2015) 11(11):e1005226. doi: 10.1371/journal.ppat.1005226
87. Travers JB. Toxic interaction between Th2 cytokines and Staphylococcus aureus in atopic dermatitis. J Invest Dermatol (2014) 134(8):2069–71. doi: 10.1038/jid.2014.122
88. Veckman V, Miettinen M, Pirhonen J, Sirén J, Matikainen S, Julkunen I. Streptococcus pyogenes and Lactobacillus rhamnosus differentially induce maturation and production of Th1-type cytokines and chemokines in human monocyte-derived dendritic cells. J Leukoc Biol (2004) 75(5):764–71. doi: 10.1189/jlb.1003461
89. Mortensen R, Nissen TN, Blauenfeldt T, Christensen JP, Andersen P, Dietrich J. Adaptive Immunity against Streptococcus pyogenes in Adults Involves Increased IFN-γ and IgG3 Responses Compared with Children. J Immunol (2015) 195(4):1657–64. doi: 10.4049/jimmunol.1500804
90. Bezerra-Santos M, do Vale-Simon M, Barreto AS, Cazzaniga RA, de Oliveira DT, Barrios MR, et al. Mycobacterium leprae Recombinant Antigen Induces High Expression of Multifunction T Lymphocytes and Is Promising as a Specific Vaccine for Leprosy. Front Immunol (2018) 9:2920. doi: 10.3389/fimmu.2018.02920
91. Moura DF, de Mattos KA, Amadeu TP, Andrade PR, Sales JS, Schmitz V, et al. CD163 favors Mycobacterium leprae survival and persistence by promoting anti-inflammatory pathways in lepromatous macrophages. Eur J Immunol (2012) 42(11):2925–36. doi: 10.1002/eji.201142198
92. Rinker SD, Trombley MP, Gu X, Fortney KR, Bauer ME. Deletion of mtrC in Haemophilus ducreyi increases sensitivity to human antimicrobial peptides and activates the CpxRA regulon. Infect Immun (2011) 79:2324–34. doi: 10.1128/IAI.01316-10
93. Groisman EA, Parra-Lopez C, Salcedo M, Lipps CJ, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci U S A (1992) 89:11939–43. doi: 10.1073/pnas.89.24.11939
94. Campagnari AA, Spinola SM, Lesse AJ, Abu Kwaik Y, Mandrell RE, Apicella MA. Lipooligosaccharide epitopes shared among gram-negative non-enteric mucosal pathogens. Microb Pathog (1990) 8:353–62. doi: 10.1016/0882-4010(90)90094-7
95. Melaugh W, Campagnari AA, Gibson BW. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J Bacteriol (1996) 178:564–70. doi: 10.1128/jb.178.2.564-570.1996
96. Post DMB, Munson RS Jr, Baker B, Zhong H, Bozue JA, Gibson BW. Identification of genes involved in the expression of atypical lipooligosaccharide structures from a second class of Haemophilus ducreyi. Infect Immun (2007) 75:113–21. doi: 10.1128/IAI.01016-06
97. Vogel U, Frosch M. Mechanisms of neisserial serum resistance. Mol Microbiol (1999) 32:1133–9. doi: 10.1046/j.1365-2958.1999.01469.x
98. Melaugh W, Phillips NJ, Campagnari AA, Tullius MV, Gibson BW. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry (1994) 33:13070–8. doi: 10.1021/bi00248a016
99. Gangaiah D, Zhang X, Baker B, Fortney KR, Gao H, Holley CL, et al. Haemophilus ducreyi Seeks Alternative Carbon Sources and Adapts to Nutrient Stress and Anaerobiosis during Experimental Infection of Human Volunteers. Infect Immun (2016) 84(5):1514–25. doi: 10.1128/IAI.00048-16
100. Griesenauer B, Tran TM, Fortney KR, Janowicz DM, Johnson P, Gao H, et al. Determination of an Interaction Network between an Extracellular Bacterial Pathogen and the Human Host. MBio (2019) 10(3):1–15. doi: 10.1128/mBio.01193-19
101. Wang Y, Russo TA, Kwon O, Chanock S, Rumsey SC, Levine M. Ascorbate recycling in human neutrophils: induction by bacteria. Proc Natl Acad Sci U S A (1997) 94(25):13816–9. doi: 10.1073/pnas.94.25.13816
102. Diskin C, Palsson-McDermott EM. Metabolic Modulation in Macrophage Effector Function. Front Immunol (2018) 9:270. doi: 10.3389/fimmu.2018.00270
103. Geier B, Sogin EM, Michellod D, Janda M, Kompauer M, Spengler B, et al. Spatial metabolomics of in situ host-microbe interactions at the micrometre scale. Nat Microbiol (2020) 5(3):498–510. doi: 10.1038/s41564-019-0664-6
104. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell (2019) 177(7):1888–902.e21. doi: 10.1016/j.cell.2019.05.031
105. van Rensburg JJ, Lin H, Gao X, Toh E, Fortney KR, Ellinger S, et al. The Human Skin Microbiome Associates with the Outcome of and Is Influenced by Bacterial Infection. mBio (2015) 6(5):e01315–15. doi: 10.1128/mBio.01315-15
106. Gonzalez-Beiras C, Kapa A, Vall-Mayans M, Paru R, Gavilan S, Houinei W, et al. Single-Dose Azithromycin for the Treatment of Haemophilus ducreyi Skin Ulcers in Papua New Guinea. Clin Infect Dis (2017) 65(12):2085–90. doi: 10.1093/cid/cix723
107. Fusco WG, Afonina G, Nepluev I, Cholon DM, Choudhary N, Routh PA, et al. Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA with adjuvant monophosphoryl lipid A protects swine from a homologous but not a heterologous challenge. Infect Immun (2010) 78(9):3763–72. doi: 10.1128/IAI.00217-10
Keywords: Haemophilus ducreyi, interactome, metabolome, skin, immune response
Citation: Brothwell JA, Griesenauer B, Chen L and Spinola SM (2021) Interactions of the Skin Pathogen Haemophilus ducreyi With the Human Host. Front. Immunol. 11:615402. doi: 10.3389/fimmu.2020.615402
Received: 09 October 2020; Accepted: 21 December 2020;
Published: 03 February 2021.
Edited by:
Roberta Olmo Pinheiro, Oswaldo Cruz Foundation, BrazilReviewed by:
Michael Marks, University of London, United KingdomYu-Ching Su, Lund University, Sweden
Kristian Riesbeck, Lund University, Sweden
Copyright © 2021 Brothwell, Griesenauer, Chen and Spinola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie A. Brothwell, amJyb3Rod2VAaXUuZWR1
 Julie A. Brothwell
Julie A. Brothwell Brad Griesenauer
Brad Griesenauer Li Chen
Li Chen Stanley M. Spinola
Stanley M. Spinola