
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 08 January 2021
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.613957
This article is part of the Research TopicNovel Insights into Insect Antiviral ImmunityView all 16 articles
 Yu-Juan He1,2†
Yu-Juan He1,2† Gang Lu2†
Gang Lu2† Yu-Hua Qi2
Yu-Hua Qi2 Yan Zhang2
Yan Zhang2 Xiao-Di Zhang2
Xiao-Di Zhang2 Hai-Jian Huang2
Hai-Jian Huang2 Ji-Chong Zhuo2
Ji-Chong Zhuo2 Zong-Tao Sun2
Zong-Tao Sun2 Fei Yan2
Fei Yan2 Jian-Ping Chen1,2*
Jian-Ping Chen1,2* Chuan-Xi Zhang2*
Chuan-Xi Zhang2* Jun-Min Li2*
Jun-Min Li2*The Toll pathway plays an important role in defense against infection of various pathogenic microorganisms, including viruses. However, current understanding of Toll pathway was mainly restricted in mammal and some model insects such as Drosophila and mosquitoes. Whether plant viruses can also activate the Toll signaling pathway in vector insects is still unknown. In this study, using rice stripe virus (RSV) and its insect vector (small brown planthopper, Laodelphax striatellus) as a model, we found that the Toll pathway was activated upon RSV infection. In comparison of viruliferous and non-viruliferous planthoppers, we found that four Toll pathway core genes (Toll, Tube, MyD88, and Dorsal) were upregulated in viruliferous planthoppers. When the planthoppers infected with RSV, the expressions of Toll and MyD88 were rapidly upregulated at the early stage (1 and 3 days post-infection), whereas Dorsal was upregulated at the late stage (9 days post-infection). Furthermore, induction of Toll pathway was initiated by interaction between a Toll receptor and RSV nucleocapsid protein (NP). Knockdown of Toll increased the proliferation of RSV in vector insect, and the dsToll-treated insects exhibited higher mortality than that of dsGFP-treated ones. Our results provide the first evidence that the Toll signaling pathway of an insect vector is potentially activated through the direct interaction between Toll receptor and a protein encoded by a plant virus, indicating that Toll immune pathway is an important strategy against plant virus infection in an insect vector.
In invertebrates, host defense against pathogens, including bacteria, fungi, and viruses, is known to rely on innate immunity, while in vertebrates, the innate immune system provides the first defense line against pathogens before activation of acquired immune response (1). In insects, various evolutionarily conserved signaling pathways mediate antiviral immunity, including small RNA interference (RNAi), Toll, the immune deficiency (IMD), and JAK-STAT (2, 3). These pathways mainly rely on different pattern recognition receptors (PRRs), which recognize signature molecules of pathogens, known as pathogen associated molecular patterns (PAMPs) and induce downstream effectors against viral infection (4, 5). Toll receptor superfamily, including invertebrate Tolls and vertebrate Toll-like receptors (TLRs), is important class of PRRs and the primary sensor of pathogens in all metazoans (6). The activation of Toll pathway in vertebrate is initiated by TLRs binding to various PAMPs, whereas in invertebrate, it is activated indirectly by Toll receptors binding to the cytokine-like molecule Spätzle (Spz) (7). Tolls and TLRs are characterized by an extracellular domain containing leucine-rich repeats (LRRs), a transmembrane domain, and a cytoplasmic tail that contains a conserved region called the Toll/IL-1 receptor (TIR) domain (8). The first identified Toll (Toll1) is the receptor of the Toll pathway, and to date, nine Toll genes have been identified in Drosophila (8). In invertebrate, pathogen infection is censored by extracellular recognition and the inactive precursor of the Spz is cleaved to active form. Then the activated Spz binds to Toll receptor and a cassette of proteins (MyD88, Tube and Pelle) are recruited to assemble a receptor-proximal oligomeric complex (9–11). In Drosophila, the complex further trigger the phosphorylation and degradation of Cactus, freeing Dorsal or Dif (Dorsal-related immunity factor) to transfer from the cytoplasm into the nucleus for the regulation of different antibacterial peptides (AMPs) expressions (12).
Although the importance of the Toll pathway against bacteria and fungi has been well demonstrated, accumulated evidences suggested that it also plays essential antiviral roles in invertebrate, such as the fly (Drosophila) and mosquitoes (Culex, Aedes, and Anopheles) (1, 13). The importance of Toll pathway against virus was firstly reported in Drosophila when challenged with Drosophila X virus (DXV) infection (1). Further studies indicated that the Toll pathway also mediate resistance to other RNA viruses including Drosophila C virus, cricket paralysis virus, flock house virus, and norovirus (14). In mosquito, Toll immune pathway was activated upon viral infection, and they controlled the conserved anti-dengue defenses across diverse Aegypti strains and against multiple dengue virus serotypes (13, 15). Interestingly, recent studies found that several members of Toll receptors can also act as PRRs analogous to the TLRs in mammal, triggering conventional or non-conventional Toll-Dorsal pathway. RNAi screening suggested that Toll-4 might be one of upstream PRR to detect white spot syndrome virus (WSSV) infection in shrimp, and thereby leading to conventional Toll-Dorsal pathway (16). Another example is three shrimp Tolls (Toll1-3) directly bind to PAMPs from bacterial infection, resulting in Dorsal translocation into nucleus to regulate the expression of different AMPs (17). In contrast, Drosophila Toll-7 can also act as a PRR and directly interact with vesicular stomatitis virus (VSV) at the plasma membrane, but induces antiviral autophagy independent of the canonical Toll-Dorsal signaling pathway (2).
Rice stripe virus (RSV) is a filamentous, negative-strand RNA virus of the genus Tenuivirus that causes rice stripe disease, one of the most severe rice diseases in East Asia (18–20). RSV is transmitted by the vector insect, small brown planthopper (SBPH, Laodelphax striatellus), in a persistent-propagative manner. RSV can replicate in L. striatellus, and can be transmitted to the progeny of the planthopper through infection of the embryos or germ cells in the female insects (21). The viral genome of RSV consists of four single-stranded RNA segments: RNA1-RNA4. RNA1 is negative-sense RNA and encodes a 337-kDa protein referred to as RNA-dependent RNA polymerase (22). The other three genomic segments exhibit ambisense coding features and each RNA encodes two proteins. Sense and antisense strands of RNA2 encode RNA silencing suppressor NS2 and the putative membrane glycoprotein NSvc2, respectively (23–25). RNA3 encodes a second viral suppressor NS3 (26), and complementary sense RNA3 (vcRNA3) encodes the nucleocapsid protein (NP) (27, 28). RNA4 encodes the disease-specific protein NS4 (29), and vcRNA4 encodes the movement protein (MP) (30, 31). Previous studies suggested the induced active response of L. striatellus during RSV infection. For example, analysis of viral-derived small interfering RNAs (siRNAs) revealed that RNAi-mediated antiviral response can successfully be induced by the infection of RSV and another reovirus, rice black-streaked dwarf virus (32). Activation of c-Jun N-terminal kinase (JNK) promoted RSV replication in L. striatellus, whereas JNK inhibition caused a significant reduction in virus production and thus delayed disease incidence in plants (33). In addition, silencing of the autophagy-related-8 (Atg8) expression of L. striatellus significantly decreased the phosphorylation of JNK in the midgut of the planthoppers, suggesting that ATG8 might activate the JNK machinery (34). Nevertheless, to date, whether the classical Toll-Dorsal pathway involved in antiviral response of L. striatellus or other plant virus vectors have never been investigated.
In this study, open reading frames (ORFs) of four core components from Toll pathway, including Toll, Tube, MyD88, and Dorsal, were identified from L. striatellus and their potential antiviral roles were further explored. Our results revealed that the Toll signaling pathway in L. striatellus is potentially induced through the direct interaction between Toll and RSV-NP. Knockdown of Toll increased the replication of RSV, indicating that Toll in insect vectors might act as PRR in perceiving plant viruses, similar to that of TLRs in mammalian.
The planthopper populations are maintained on susceptive japonica rice seedlings (cv Wuyujing No. 3) in a temperature-controlled room at 25 ± 1°C, with 70–80% relative humidity, and a light/dark photoperiod of 14/10 h. The infection ratio of the viruliferous planthopper population (RSV-infected) was around 80% and monitored every 3–4 generations by reverse transcription polymerase chain reaction (RT-PCR) as described previously (32).
Four Toll pathway core genes (Toll, Tube, MyD88, and Dorsal) were obtained from transcriptome of L. striatellus (Accession Number: SRR4020768) by homology search based on the corresponding genes of Nilaparvata lugens as query sequences (nlToll, XP_022198839; nlTube, XP_022207725; nlMyD88, XP_022187892; nlDorsal, XP_022195378). Conserved protein domains were predicted using NCBI conserved domains database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Phylogenetic trees were constructed based on the deduced amino-acid sequences in MEGA 6.0 using the maximum likelihood (ML) algorithm with 1,000 bootstrap replications. The full-length ORFs of the four identified genes were amplified with the respective primer pairs (Supplementary Table 1) from planthoppers using RT-PCR and further confirmed by Sanger sequencing (Sangon, China).
For the yeast two-hybrid assay (Y2H) interaction assay, the full-length of RSV NS2, NSvc2-C (C-terminal of glycoprotein), NSvc2-N (N-terminal of glycoprotein), NS3, NP, NS4, and MP were cloned into the DNA-binding domain of the vector pGBK-T7 to create bait plasmids. The full-length ORF of Toll was cloned into the activation domain of the yeast vector pGAD-T7. Yeast cells (AH109) were co-transformed with RSV protein libraries and pGAD-T7-Toll. Positive clones were selected on quadruple dropout medium (SD/-Leu/-Trp/-His/-Ade).
To further confirm the protein interactions, the full-length genes of RSV proteins NS2, NSvc2-C, NSvc2-N, NS3, NP, NS4, and MP were amplified and cloned into pCV-nYFP expression vector, respectively. The full-length ORF of Toll was cloned into pCV-cYFP expression vector. Constructed vector pCV-cYFP-Toll were then transformed into Agrobacterium tumefaciens GV3101 by heat transfer method, and co-transformed with pCV-nYFP-NS2, pCV-nYFP-NSvc2-C, pCV-nYFP-NSvc2-N, pCV-nYFP-NS3, pCV-nYFP-NP, pCV-nYFP-NS4, and pCV-nYFP-MP, into Nicotiana benthamiana, respectively. YFP fluorescence signal was observed under Nikon confocal (Nikon, Japan).
The total RNA was extracted using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. The concentration and quality of total RNA was determined using a NanoDrop spectrophotometer (Thermo Scientific, USA). The first strand of complementary DNA (cDNA) from 1,000 ng of total RNA was synthesized with HiScript ®II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China) following the manufacturer’s protocol. In brief, quantitative real-time PCR (qPCR) was performed in 10 µl-reaction agent containing 0.5 µl of template cDNA and 5 µl of Hieff ® qPCR SYBR Green PCR Master Mix (YESEN, China), 0.2 µl of 1 µM forward and reverse primers, and 4.1 µl of ddH2O on LightCycler® 480 II (Roche, Switzerland). The thermal cycling conditions were 95°C for 5 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 70°C for 30 s, followed by melting curve analysis. The data were analyzed using the 2−ΔΔCT method and statistically significant differences at P < 0.05 (*) and P < 0.01 (**) level are indicated according to one-way analysis of variance (ANOVA) test.
Planthopper samples of different developmental stages (eggs, 1st to 5th instar nymphs, female adults, and male adults) and various tissues (salivary gland, gut, ovary of female adult, epidermis, hemolymph, fat body, and testis of male adult) from non-viruliferous L. striatellus were collected. For the collection of hemolymph and fat body, the PBS solution after the dissect of planthoppers was centrifuged at 5,000 × g for 5 min at 4°C, and the hemolymph in supernatant and fat body in precipitate were separately collected, respectively. Five independent replicates were used in this experiment. For the collection of different developmental stages, various numbers of insects were obtained according to the sample size for each replicate. While for tissues, each replicate contains different tissues derived from approximately 40–50 individual adult planthoppers.
To determine the expression of Toll, Tube, MyD88, and Dorsal in response to RSV infection, approximately 20 adult planthoppers from non-viruliferous and viruliferous cultures were collected for RNA extraction individually. In addition, to further investigate the response of Toll pathway core gene expressions during the whole process of RSV infection, approximately 1,000 2nd instar nymphs of non-viruliferous planthoppers were transferred and feeding onto RSV-infected rice seedlings for 2 days. Then the planthoppers were transferred to healthy rice seedlings and collected at various time points (1, 3, 6, 9, or 12 days post-infection). About 20 planthoppers were collected at each time point and expressions of Toll pathway genes were determined from individual insect by qPCR as described above.
Toll, Tube, MyD88, and Dorsal fragments of L. striatellus were amplified using gene-specific primer ligated with a T7-promoter sequence, and the green fluorescent proteins (GFP) fragment was used as a negative control. The primers used for the amplification were listed in Supplementary Table 1. The double-stranded RNA (dsRNA) was synthesized using the T7 RiboMAX Express RNAi System (Promega, USA) following the manufacturer’s instructions. The quality of synthesized dsRNA was evaluated using agarose gel electrophoresis. Each planthopper was injected with 40 nl of dsRNA into the insect ventral thorax with a glass needle (35).
Adult planthoppers from viruliferous culture were collected and injected with dsToll, dsTube, dsMyD88, and dsDorsal, respectively. dsGFP was used as a negative control. Each of the injected planthopper was used for RNA extraction and the RNAi efficiency was determined at 48 h post-injection using qPCR. The accumulation level of RSV was quantified by qPCR as described above with specific primer pairs for RSV-NP.
Second instar planthoppers from non-viruliferous culture were injected with dsToll individually and maintained in healthy rice seedling for 2 days. The injected planthoppers were then transferred onto RSV-infected rice seedlings for another 2 days for virus acquisition. Finally, the planthoppers were moved to healthy rice seedling again and collected at various time points for the detection of virus accumulation. dsGFP was used as a negative control. Each of the injected planthopper was used for RNA extraction at 0, 1, 3, 9 days post-RSV acquisition. The expression of RSV-NP was measured by qPCR after silencing of Toll. Approximately 20 planthoppers were used for the detection at each time point. Meanwhile, the mortality rate of dsToll injected planthoppers was investigated. Three biological replicates were performed for each treatment in this experiment.
To explore the potential antiviral roles of classic Toll pathway in L. striatellus, full ORF of Toll pathway core genes (Toll, Tube, MyD88, and Dorsal) were identified and cloned. The ORF of Toll consists of 3336 bp nucleotides encoding a predicted protein of 1,111 amino-acid residues with a calculated molecular mass of 125.82 kDa. The predicted Toll protein contains five conserved domains including a PRK15370 super family (type III secretion system effector E3 ubiquitin transferase SlrP), a LRR_8 (Leucine rich repeat), a PCC super family (polycystin cation channel protein), a LRR, and a TIR (Toll—interleukin 1—resistance) (Figure 1A); Tube contains a 1,500 bp ORF, encoding a predicted protein of 499 amino-acid residues with a calculated molecular mass of 55.58 kDa. The putative Tube protein contains a Death_Tube domain and a PKc (protein kinases) domain (Figure 1B); MyD88 consists of 1,230 bp and encodes a predicted protein of 409 amino acids. The putative MyD88 protein contains two conserved domains Death_MyD88 and TIR_2 (a family of bacterial TLRs) (Figure 1C). Dorsal contains a continuous 2,712 bp ORF, encoding a predicted protein of 903 amino-acid residues. The putative Dorsal protein contains domains including a Dorsal_Dif, a RHD_dimer (Rel homology dimerization), and an AidA superfamily (Figure 1D). The full ORF sequences of Toll, Tube, MyD88, and Dorsal were submitted to GenBank with the accession numbers of MW048393, MW048395, MW048396, and MW048394.
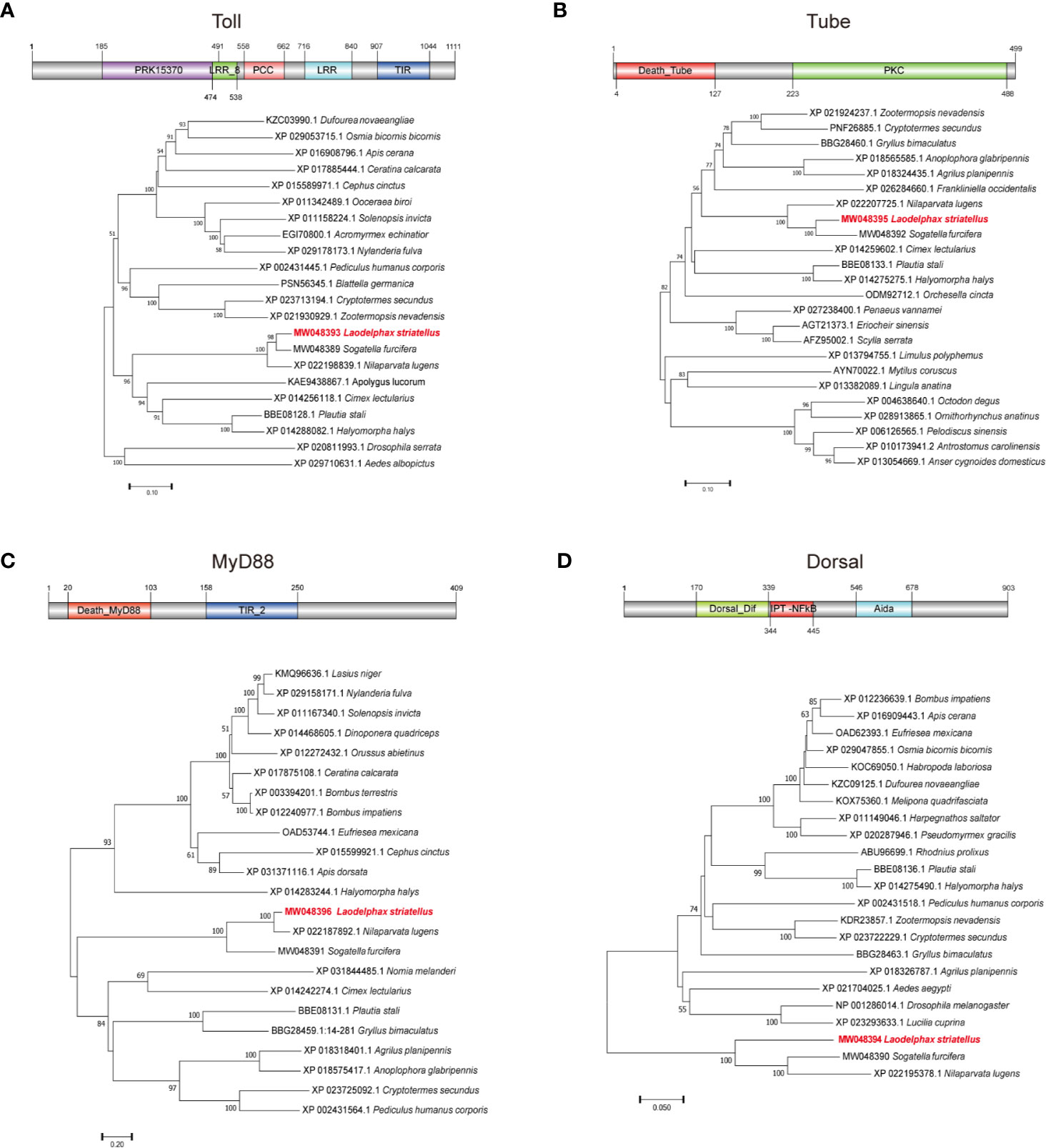
Figure 1 The architecture and phylogenetic analysis of Toll, Tube, MyD88, and Dorsal. (A) Toll contains five conserved domains: PRK15370, TIR, LRR_8, leucine-rich repeat (LRR), and PCC. (B) Tube contains two conserved domains: Death and S_TKC. (C) MyD88 contains two conserved domains: Death_MyD88 and TIR_2. (D) Dorsal contains three conserved domains: Dorsal_Dif, RHD_dimer, and AidA. Phylogenetic tree analysis with the maximum likelihood method was based on homologous amino-acid sequences of Laodelphax striatellus and other insects.
Homology analysis showed that the predicted amino acids of the four Toll pathway proteins of L. striatellus share highest homologies to the other two rice planthoppers, N. lugens and Sogatella furcifera, with identities of 88.44 and 88.07% for Toll, 70.18 and 60.39% for Tube, 77.64 and 31.05% for MyD88, 65.11 and 49.77% for Dorsal, respectively. Phylogenetic analysis based on the putative amino-acid sequences suggested the four proteins of L. striatellus clustered together with the other two planthoppers (N. lugens and S. furcifera) with high strap value support (Figure 1).
To investigate the potential interaction between Toll and RSV proteins, seven viral proteins (NS2, NSvc2-C, NSvc2-N, NS3, NP, NS4, and MP) were used as baits to screen against the L. striatellus Toll. We found that Toll interacted with RSV-NP protein, but not the other six RSV proteins, and similar results were found when Toll was used as a bait and RSV-NP as a prey (Figure 2A, and Figure S1). In addition, yeast two-hybrid assay result showed that Toll could not interact directly with SPZ family including SPZ1, SPZ2, SPZ3, SPZ4, SPZ5, and SPZ6 proteins in SD/–Leu/–Trp/–His/–Ade medium (Figure S2). To confirm the interaction between planthopper Toll and RSV-NP, bimolecular fluorescence complementation (BiFC) assays were further performed in N. benthamiana. When pCV-cYFP-Toll and pCV-nYFP-NP were transiently co-expressed in N. benthamiana leaves, strong YFP fluorescence signals were observed in the cytomembrane, whereas no visible signal was detected in the negative control of pCV-cYFP–Toll and pCV–nYFP (Figure 2B). Similar results were found when pCV-cYFP-NP and pCV-nYFP-Toll were transiently co-expressed (Figure 2B). These results indicated that Toll and RSV-NP proteins interact directly and the L. striatellus Toll might act as PRR in recognizing signaling molecules of pathogen.
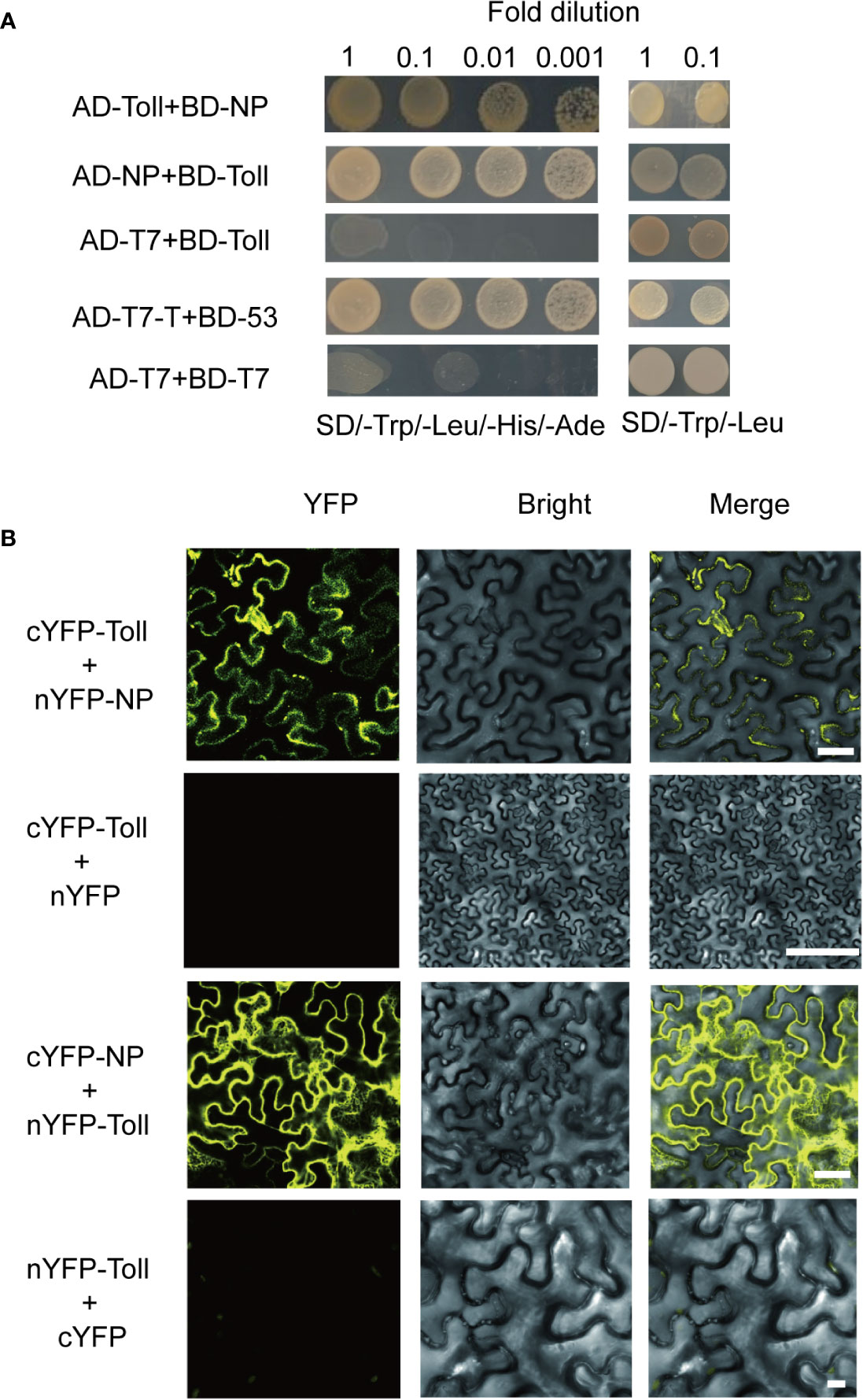
Figure 2 Protein-protein interaction analysis of Toll and rice stripe virus (RSV)-NP. (A) Yeast two-hybrid assay result showed that Toll interacted with RSV-NP protein in SD/–Leu/–Trp/–His/–Ade medium. (B) Bimolecular fluorescence complementation assays showed that pCV-cYFP–Toll and pCV–nYFP–RSV-NP, pCV–cYFP–RSV–NP and pCV–nYFP–Toll fluorescent strong YFP signals in the cytomembrane but there were no detectable signals in the negative control combinations pCV-cYFP–Toll and pCV–nYFP, pCV–cYFP, and pCV–nYFP–Toll. Bars, 50 µm.
To explore the expression pattern of Toll receptors, non-viruliferous planthopper samples from eight developmental stages and seven tissues were collected and quantified by qPCR. The results showed that Toll was ubiquitously expressed in all collected developmental stages and tissues of L. striatellus (Figures 3A, B). Messenger RNA (mRNA) of Toll was most abundant in the first instar nymphs of non-viruliferous planthopper, followed by eggs (Figure 3A). Furthermore, highest expression of Toll was observed in salivary glands compared with the other tissues of non-viruliferous planthoppers (Figure 3B).
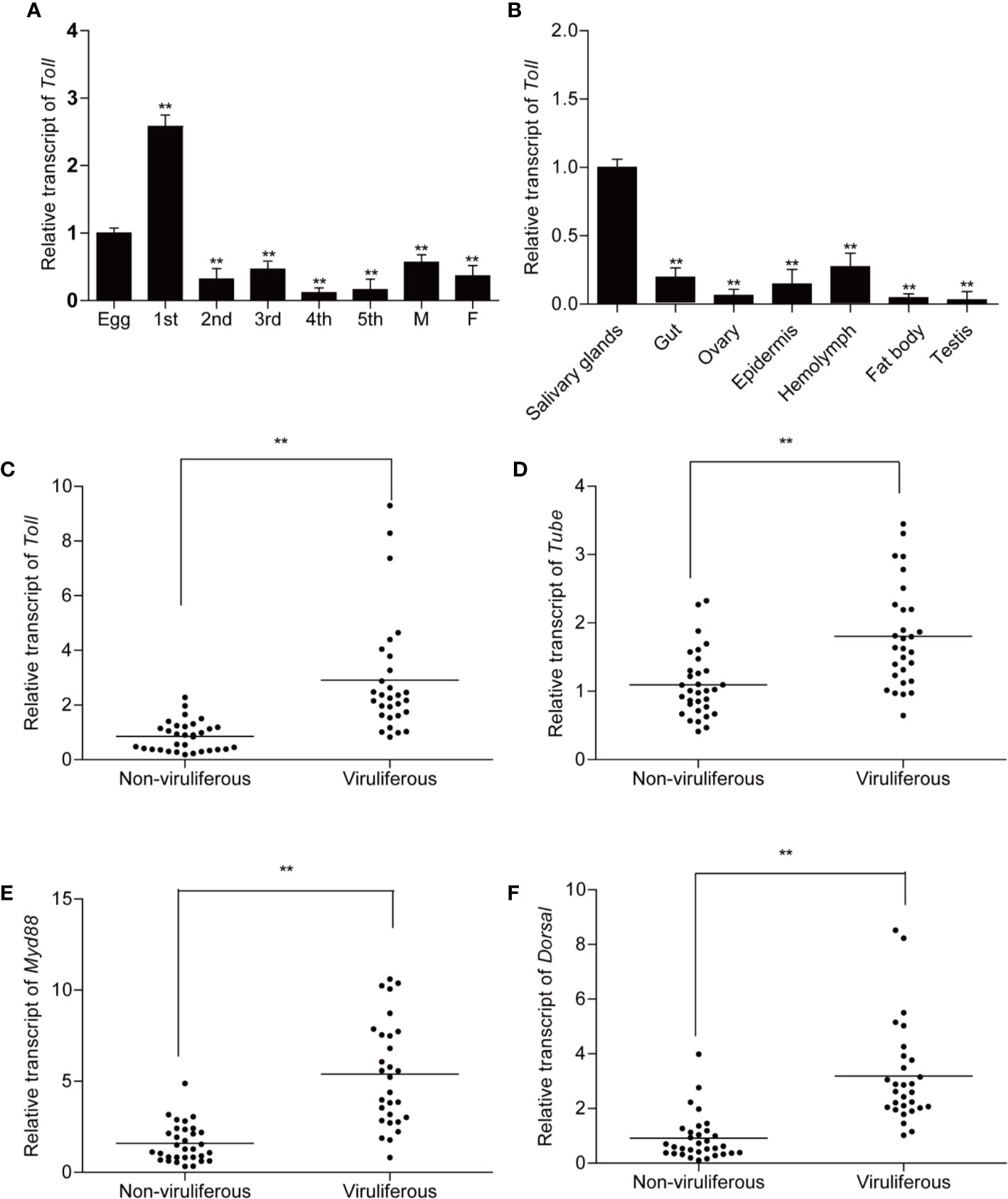
Figure 3 Expression patterns of Toll and relative transcript levels of Toll, Tube, MyD88, and Dorsal in non-viruliferous and viruliferous planthopper. For qPCR detection of Toll, samples from different developmental stages (eggs, nymphs from 1st to 5th instars, female and male adults) (A) and different tissues (salivary gland, gut, ovary, epidermis, hemolymph, fat body, and testicle) (B) were collected from non-viruliferous planthoppers. Five biological replicates were performed. For analysis of relative transcript levels of Toll (C), Tube (D, E) MyD88 (E), and Dorsal (F), non-viruliferous and viruliferous planthopper samples were collected individually. Actin gene was used as housekeeping gene. Each point represents a biological replicate. Statistically significant differences at P < 0.01 (**) level are indicated according to one-way analysis of variance (ANOVA) test.
To illustrate the potential roles of Toll signaling pathway in RSV infection, the expressions of Toll, Tube, MyD88, and Dorsal were compared between viruliferous planthopper and non-viruliferous planthopper. Significantly increased expressions of all the four genes were observed in viruliferous planthopper population (Figures 3C–F), suggesting that Toll pathway might be actively involved in the stable maintenance of RSV in planthopper.
Moreover, previous studies demonstrated that virus infection activated the Toll pathway within a short period (16). Considering the remarkable upregulation of Toll, Tube, MyD88, and Dorsal in viruliferous planthopper, how the Toll pathway of non-viruliferous planthoppers responded to RSV infection is of interest. As a result, Toll, Tube, and MyD88, but not Dorsal, were actively responded during early stage of RSV infection (Figure 4). The expressions of MyD88 and Toll were significantly increased after 1 and 3 days post-infection (dpi) (Figures 4A, C), whereas Tube was notably decreased after 1 dpi compare to that of the control (0 dpi) (Figure 4B). No significant change was detected after 6 dpi of RSV infection for both Toll, Tube, and MyD88. However, all of the four Toll pathway core genes were up-regulated after 9 and 12 dpi (at the late stage) of RSV infection (Figure 4). These dynamic expressions of Toll, Tube, MyD88, and dorsal imply the active and complexed involvement of the canonical Toll signaling pathway in response to the infection process of RSV.

Figure 4 The expression pattern of Toll, Tube, MyD88, and Dorsal when non-viruliferous planthoppers were infected with rice stripe virus (RSV). Non-viruliferous planthoppers were fed on RSV-infected rice seedlings, and the samples were collected at 1, 3, 6, 9, and 12 days post-infection (dpi). Relative transcript levels of Toll (A), Tube (B), MyD88 (C), and Dorsal (D) at the indicated time points were analyzed by qPCR. The non-viruliferous planthopper that did not contact with RSV was used as a control. Actin gene was used as housekeeping gene. Each point represents a biological replicate. Statistically significant differences at P < 0.05 (*) and P < 0.01 (**) level are indicated according to one-way ANOVA test.
To investigate the potential roles of Toll, Tube, MyD88, and Dorsal in RSV infected planthoppers, dsRNA fragments corresponding to these four genes were synthesized and injected into the viruliferous planthoppers. Assessment of silencing efficient indicated the significant transcripts reduction (70%) for all of the four genes after 2 dpi (Figures 5A, C, E, G). Meanwhile, significant increase in titer of RSV was observed in dsToll- and dsDorsal-treated planthoppers (Figures 5B, H), whereas RNA level of RSV-NP was notably reduced for dsTube-injected insects when compared with that of the control (dsGFP) (Figure 5D). In contrast, no significant difference was detected in RSV-NP between dsMyD88 and dsGFP-treated planthoppers (Figure 5F).
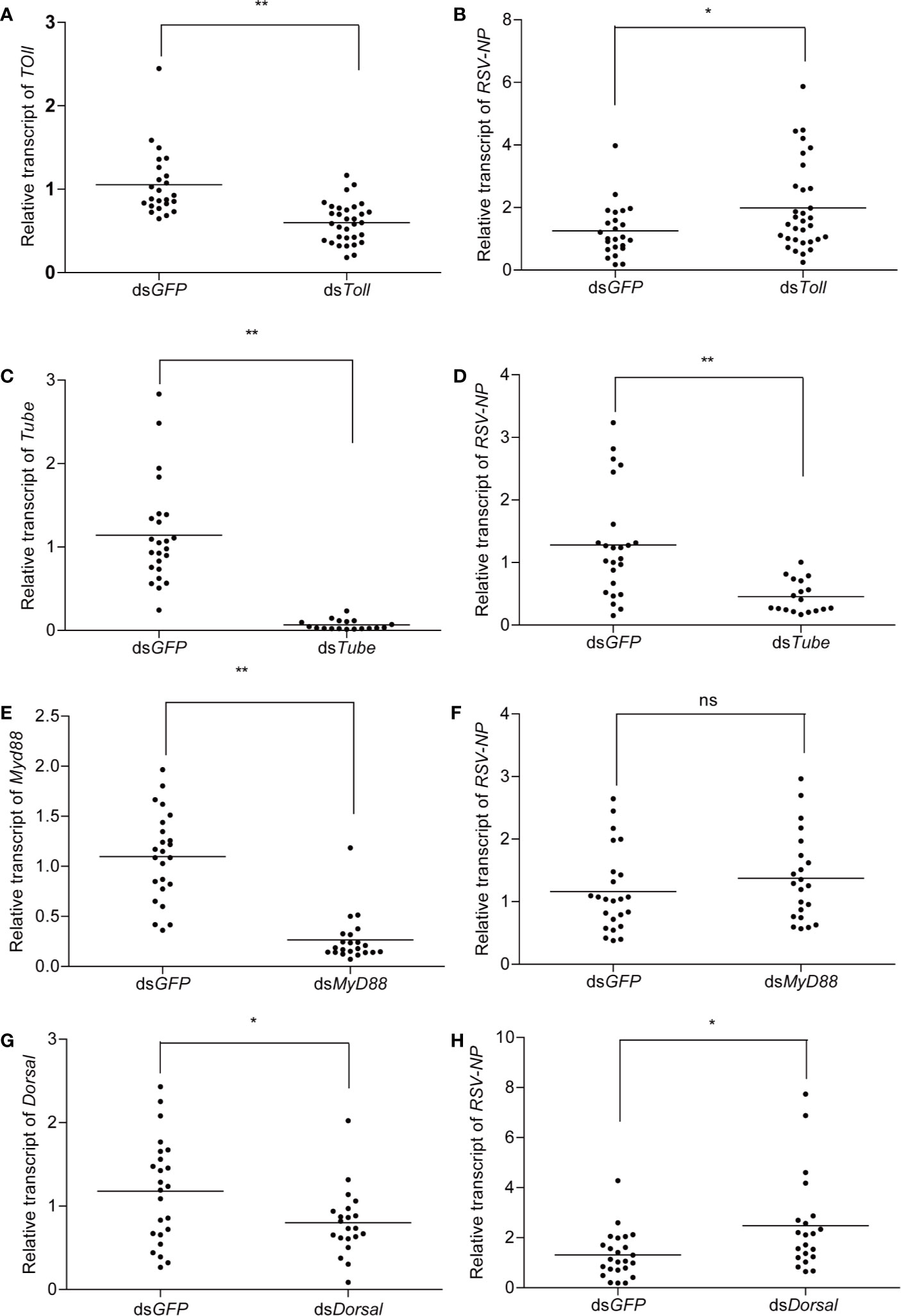
Figure 5 The influence of double-stranded RNA (dsRNA) treatment on viruliferous planthoppers. Approximately 25 viruliferous planthopper were injected with dsToll, dsTube, dsMyD88, and dsDorsal. The silencing efficiency of Toll (A), Tube (C), MyD88 (E), and Dorsal (G) were determined. Meanwhile, the relative transcript level of RSV-NP after silencing of Toll (B), Tube (D), MyD88 (F), and Dorsal (H) were analyzed by qPCR. Planthopers treated with dsGFP were used as a negative control. Each point represents a biological replicate. Statistically significant differences at P < 0.05 (*) and P < 0.01 (**) level are indicated according to one-way ANOVA test. ns, not significant.
In view of the direct interaction between Toll and RSV-NP, the accumulation of RSV-NP transcripts was further examined in dsToll-treated planthoppers when non-viruliferous planthoppers were infected by RSV. Injection of dsToll successfully and stably inhibits the Toll expression in planthoppers after RSV acquisition for various days (Figure 6A). The effects of dsToll-injection on the RSV proliferation in planthoppers were further determined. Significant increase in RSV-NP transcripts were observed in dsToll injected planthopper compared to that of the control (dsGFP) at various infected time point (1, 3, and 9 dpi) (Figure 6B). Additionally, the mortality of RSV-infected planthopper was significantly higher after dsToll treatment in 3 and 9 dpi than that of dsGFP control (Figure 6C). These data suggested that Toll might play an essential role in restricting RSV proliferation.
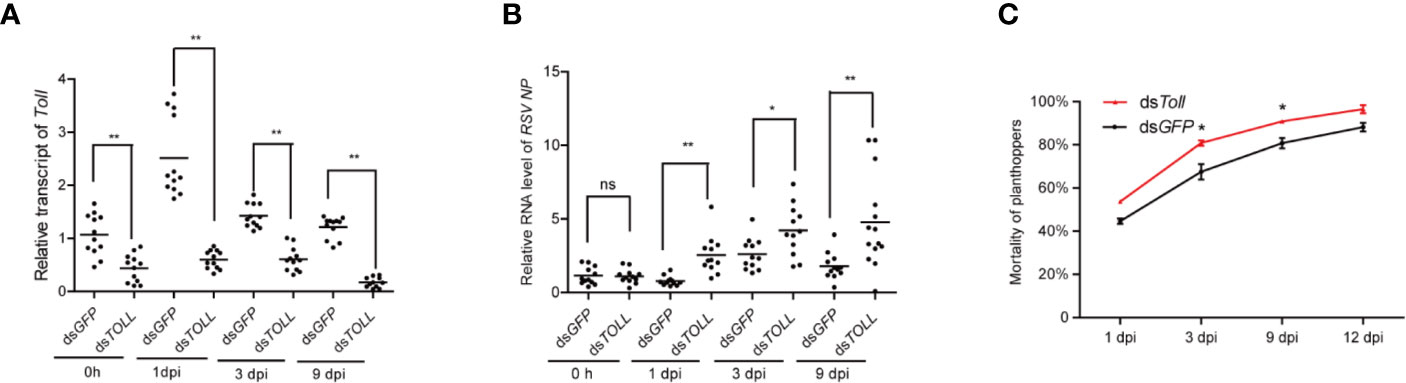
Figure 6 The influence of double-stranded RNA (dsRNA) treatment on non-viruliferous planthoppers that orally infected with rice stripe virus (RSV). The non-viruliferous planthopper were treated with dsToll or dsGFP. Two days later, the insects were provided with RSV-infected rice seedlings, and the RNAi efficiency (A) and the amount of RSV in vector insects (B) were determined by qPCR at the indicated time point. Each point represents a biological replicate. (C) The mortality of RSV-infected planthopper after dsToll and dsGFP treatments. Three biological replicates were performed. Statistically significant differences at P < 0.05 (*) and P < 0.01 (**) level are indicated according to one-way ANOVA test. ns, not significant.
Accumulated evidence demonstrated that the innate immune system plays an important role in defense against viruses in mammal and some model insects such as Drosophila and mosquitoes (1, 13–16). However, whether the canonical pathway of vector insects also involved in defense against plant viruses remained unknown. In this study, we found that RSV activated the Toll immune pathway of L. striatellus through direct interaction between Toll protein and RSV-NP. Knockdown of Toll significantly increased the proliferation of RSV in vector insect, and the dsToll-treated insects exhibited higher mortality than that of dsGFP-treated ones. Our results suggested a potential role of Toll pathway in restrict plant virus infection.
Activation of immune pathways relies on an array of PRRs to recognize the PAMPs, and subsequently induce an appropriate effector response to clear the infection (36). For Toll pathway, this process was mainly accomplished by Toll, which is the upstream receptor of this pathway. In Drosophila, Toll-7 is a PRR that interacted with VSV at the plasma membrane and induced antiviral autophagy (2). In shrimp, knockdown of Toll4 results in elevated viral loads and renders shrimp more susceptible to WSSV infection. Furthermore, Toll4 could act as an upstream PRR to detect WSSV, and lead to nuclear translocation and phosphorylation of Dorsal for the trigger of AMP production against the virus (16). Our study identified a strong interaction between Toll and RSV-NP, indicating that Toll in L. striatellus might be an upstream receptor to recognize RSV, and Toll pathway was associated with plant virus infection in insect vector.
Toll pathways is the major constitute of insect immune pathways that activate a battery of immune proteins in response to various microorganism invasion. Remarkable upregulation in Toll pathway genes were reported in Aedes aegypti challenged with Plasmodium gallinaceum (37), Drosophila challenged with Vesicular stomatitis virus (2), and Litopenaeus vannamei challenged with WSSV (16). Our study demonstrated that Toll, Tube, and MyD88 were actively responded during early stage of RSV oral infection (Figure 4), in accordance of previous work. For the transcription factor Dorsal, it is stable expressed at the early stage of viral infection, but significantly upregulated at the late stage (Figure 4). Since Toll, Tube, MyD88, and Dorsal are the four core genes of the canonical Toll-Dorsal signaling pathway, the upregulation of these four genes at various stages during RSV infection (Figure 4), the interaction between Toll and RSV-NP (Figure 2), as well as the increased viral titers observed in dsToll-treated planthoppers (Figure 6), implying that this classical pathway was actively involved in response to RSV infection. Nevertheless, more studies are needed to further investigate on the detail of downstream antiviral response, such as how does Dorsal translocate from cytoplasm into the nucleus, and which downstream effectors are regulated by Dorsal induced with RSV infection. In addition, for viruliferous planthopper, they can harbor the viruses for several generations, and no significant phenotype can be found in the RSV-infected insects. In this study, it is interesting to find that the expression level of four Toll pathway core genes were significantly higher in viruliferous planthopper than that in non-viruliferous one (Figure 3). Sustained activation of defense pathway inevitably consumes extra resources, which is detrimental to insects (38). We presumed that it might be more important for planthoppers to restrict RSV infection than other physiological metabolisms. Interestingly, higher mortality rate was recognized in dsToll-treated viruliferous planthoppers (Figure 6C), suggesting that dsToll-treatment might interfere with the established delicate balance between innate immunity of planthopper and persistent RSV infection, as described in mosquitoes (39). Our results also consist with the previous report that TLR4 knockdown mice exhibited greater viral replication (Vaccinia virus) and mortality compared to the wild-type mice following respiratory infection (40), indicating that the Toll signaling pathway of the host might be essential for the virus persistent infection.
Involvement of Toll pathway in restrict virus infection has been well documented in previous work. In Drosophila, Toll and Dif mutant lines showed increased susceptibility to Drosophila X virus (1), and Toll-7 depletion promoted vesicular stomatitis virus replication (2). In shrimps, silencing of Toll-4 resulted in high WSSV titers, with the average viral DNA burden approximately 150 times higher than that of the control (16). In A. aegypti, silencing of MyD88 led to a significant increase in dengue virus titers, demonstrating the importance of this innate immune pathway in the defense against different dengue virus serotypes at the early stages of infection (13). Our study demonstrated that Toll-inhibition and Dorsal-inhibition significantly increased the RSV titer, suggesting the potential antiviral roles of Toll pathway against plant virus. However, for dsMyD88-treatment viruliferous planthoppers, no significant change in RSV titer was observed when compared to the control (dsGFP) (Figure 5F), which is inconsistent with the previous studies in mosquitoes (13) and mice (41). Considering the increased expression of MyD88 in response to RSV infection (Figure 4C), we presume that MyD88 might play more important roles during the process of RSV infection, rather than the maintenance of RSV persistent infection in planthoppers. Furthermore, unexpectedly, RSV titer in dsTube-treatment was significantly decreased compare to control (dsGFP) (Figure 5D). It will be interesting to further explore the possibility that whether Tube can interact directly with the protein of RSV and might be hijacked by the virus to promote its proliferation.
In summary, we found that Toll pathway was activated upon RSV infection, and the viruliferous planthopper exhibited higher level of Toll, Tube, MyD88, and Dorsal. More intriguing, unlike the classical Toll signaling pathway which rely on the Spz binding to the Toll receptor, our study provide the first evidence that the antiviral Toll signaling pathway of L. striatellus is potentially activated through the direct interaction between Toll receptor and PAMPs (RSV-NP), suggesting that Toll immune pathway is an important strategy against plant viruses in insect vectors.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW048393 https://www.ncbi.nlm.nih.gov/genbank/, MW048395 https://www.ncbi.nlm.nih.gov/genbank/, MW048396 https://www.ncbi.nlm.nih.gov/genbank/, MW048394.
J-ML, J-PC, and C-XZ conceived the study and designed the project. Y-JH, GL, YZ, Y-HQ, Z-TS, X-DZ, and J-CZ performed the experiment, analyzed the data, and drafted the manuscript. H-JH, FY, and J-ML helped to revise the manuscript. All authors contributed to the article and approved the submitted version.
Present research was financially supported by the National Natural Science Foundation of China (32000121), Ningbo Science and Technology Innovation 2025 Major Project (2019B10004) and Commonweal Project (202002N3004, 202002N3008). This work was sponsored by K.C. Wong Magna Fund in Ningbo University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.613957/full#supplementary-material
Supplementary Figure 1 | Protein-protein interaction between Toll and RSV NS2, NSvc2-C, NSvc2-N, NS3, NS4, and MP.
Supplementary Figure 2 | Protein-protein interaction between Toll and SPZ family of planthopper.
1. Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci U S A (2005) 102(20):7257–62. doi: 10.1073/pnas.0409181102
2. Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, et al. Virus Recognition by Toll-7 Activates Antiviral Autophagy in Drosophila. Immunity (2012) 36(4):658–67. doi: 10.1016/j.immuni.2012.03.003
3. Janeway CA Jr, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
4. Palmer WH, Varghese FS, van Rij RP. Natural Variation in Resistance to Virus Infection in Dipteran Insects. Viruses (2018) 10(3):118. doi: 10.3390/v10030118
5. Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun (2009) 388(4):621–5. doi: 10.1016/j.bbrc.2009.08.062
6. Chtarbanova S, Imler JL. Microbial Sensing by Toll Receptors - A Historical Perspective. Arterioscler Thromb Vasc Biol (2011) 31(8):1734–8. doi: 10.1161/atvbaha.108.179523
7. Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat Immunol (2003) 4(8):794–800. doi: 10.1038/ni955
8. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34(5):637–50. doi: 10.1016/j.immuni.2011.05.006
9. Schneider DS, Jin Y, Morisato D, Anderson KV. A processed form of the Spätzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development (1994) 120(5):1243–50.
10. Horng T, Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc Natl Acad Sci U S A (2001) 98(22):12654–8. doi: 10.1073/pnas.231471798
11. Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A (2002) 99(20):12871–6. doi: 10.1073/pnas.202396399
12. Towb P, Bergmann A, Wasserman SA. The protein kinase Pelle mediates feedback regulation in the Drosophila Toll signaling pathway. Development (2001) 128(23):4729–36.
13. Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev Comp Immunol (2010) 34(6):625–9. doi: 10.1016/j.dci.2010.01.006
14. Ferreira ÁG, Naylor H, Esteves SS, Pais IS, Martins NE, Teixeira L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog (2014) 10(12):e1004507. doi: 10.1371/journal.ppat.1004507
15. Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog (2008) 4(7):e1000098. doi: 10.1371/journal.ppat.1000098
16. Li H, Yin B, Wang S, Fu Q, Xiao B, Lǚ K, et al. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PLoS Pathog (2018) 14(9):e1007109. doi: 10.1371/journal.ppat.1007109
17. Sun JJ, Xu S, He ZH, Shi XZ, Zhao XF, Wang JX. Activation of Toll Pathway Is Different between Kuruma Shrimp and Drosophila. Front Immunol (2017) 8:1151. doi: 10.3389/fimmu.2017.01151
18. Falk BW, Tsai JH. Biology and molecular biology of viruses in the genus Tenuivirus. Annu Rev Phytopathol (1998) 36:139–63. doi: 10.1146/annurev.phyto.36.1.139
19. S Toriyama. Rice stripe virus: prototype of a new group of viruses that replicate in plants and insects. Microbiol Sci (1986) 3(11):347–51. doi: 10.1016/0166-0934(86)90035-2
20. Wei TY, Yang JG, Liao FL, Gao FL, Lu LM, Zhang XT, et al. Genetic diversity and population structure of rice stripe virus in China. J Gen Virol (2009) 90(Pt 4):1025–34. doi: 10.1099/vir.0.006858-0
21. Hogenhout SA, Ammar E-D, Whitfield AE, Redinbaugh MG. Insect Vector Interactions with Persistently Transmitted Viruses. Annu Rev Phytopathol (2008) 46(1):327–59. doi: 10.1146/annurev.phyto.022508.092135
22. Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A. Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J Gen Virol (1994) 75(Pt 12):3569–79. doi: 10.1099/0022-1317-75-12-3569
23. Takahashi M, Toriyama S, Hamamatsu C, Ishihama A. Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J Gen Virol (1993) 74(Pt 4):769–73. doi: 10.1099/0022-1317-74-4-769
24. Yao M, Liu X, Li S, Xu Y, Zhou Y, Zhou X, et al. Rice stripe tenuivirus NSvc2 glycoproteins targeted to the golgi body by the N-terminal transmembrane domain and adjacent cytosolic 24 amino acids via the COP I- and COP II-dependent secretion pathway. J Virol (2014) 88(6):3223–34. doi: 10.1128/JVI.03006-13
25. Zheng L, Du Z, Lin C, Mao Q, Wu K, Wu J, et al. Rice stripe tenuivirus p2 may recruit or manipulate nucleolar functions through an interaction with fibrillarin to promote virus systemic movement. Mol Plant Pathol (2015) 16(9):921–30. doi: 10.1111/mpp.12220
26. Wu G, Wang J, Yang Y, Dong B, Wang Y, Sun G, et al. Transgenic rice expressing rice stripe virus NS3 protein, a suppressor of RNA silencing, shows resistance to rice blast disease. Virus Genes (2014) 48(3):566–9. doi: 10.1007/s11262-014-1051-2
27. Hayano Y, Kakutani T, Hayashi T, Minobe Y. Coding strategy of rice stripe virus major nonstructural protein is encoded in viral RNA segment 4 and coat protein in RNA complementary to segment 3. Virology (1990) 177(1):372–4. doi: 10.1016/0042-6822(90)90493-b
28. Kakutani T, Hayano Y, Hayashi T, Minobe Y. Ambisense segment 3 of rice stripe virus the first instance of a virus containing two ambisense segments. J Gen Virol (1991) 72(Pt 2):465–8. doi: 10.1099/0022-1317-72-2-465
29. Kong L, Wu J, Lu L, Xu Y, Zhou X. Interaction between Rice stripe virus disease-specific protein and host PsbP enhances virus symptoms. Mol Plant (2014) 7(4):691–708. doi: 10.1093/mp/sst158
30. Kakutani T, Hayano Y, Hayashi T, Minobe Y. Ambisense segment 4 of rice stripe virus possible evolutionary relationship with phleboviruses and uukuviruses (Bunyaviridae). J Gen Virol (1990) 71(Pt 7):1427–32. doi: 10.1099/0022-1317-71-7-1427
31. Xiong R, Wu J, Zhou Y, Zhou X. Identification of a movement protein of the tenuivirus rice stripe virus. J Virol (2008) 82(24):12304–11. doi: 10.1128/JVI.01696-08
32. Li J, Andika IB, Shen J, Lv Y, Ji Y, Sun L, et al. Characterization of Rice Black-Streaked Dwarf Virus- and Rice Stripe Virus-Derived siRNAs in Singly and Doubly Infected Insect Vector Laodelphax striatellus. PLoS One (2013) 8(6):e66007. doi: 10.1371/journal.pone.0066007
33. Wang W, Zhao W, Li J, Luo L, Kang L, Cui F. The c-Jun N-terminal kinase pathway of a vector insect is activated by virus capsid protein and promotes viral replication. Elife (2017) 6(e26591). doi: 10.7554/eLife.26591
34. Yu YL, Zhang MT, Huo Y, Tang JL, Liu Q, Chen XY, et al. Laodelphax striatellus Atg8 facilitates Rice stripe virus infection in an autophagy-independent manner. Insect Sci (2020) 00:1–15. doi: 10.1111/1744-7917.12771
35. Xu HJ, Xue J, Lu B, Zhang XC, Zhuo JC, He SF, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature (2015) 519(7544):464–7. doi: 10.1038/nature14286
36. Wang XL, Zhang YQ, Zhang R, Zhang JH. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr Opin Insect Sci (2019) 33:105–10. doi: 10.1016/j.cois.2019.05.004
37. Zou Z, Souza-Neto J, Xi Z, Kokoza V, Shin SW, Dimopoulos G, et al. Transcriptome Analysis of Aedes aegypti Transgenic Mosquitoes with Altered Immunity. PLoS Pathog (2011) 7(11):e1002394. doi: 10.1371/journal.ppat.1002394
38. Huang HJ, Cui JR, Hong XY. Comparative analysis of diet-associated responses in two rice planthopper species. BMC Genomics (2020) 21:565. doi: 10.1186/s12864-020-06976-2
39. Lee WS, Webster JA, Madzokere ET, Stephenson EB, Herrero LJ. Mosquito antiviral defense mechanisms: a delicate balance between innate immunity and persistent viral infection. Parasit Vectors (2019) 12(1). doi: 10.1186/s13071-019-3433-8
40. Hutchens MA, Luker KE, Sonstein J, Núñez G, Curtis JL, Luker GD. Protective Effect of Toll-like Receptor 4 in Pulmonary Vaccinia Infection. PLoS Pathog (2008) 4(9):e1000153. doi: 10.1371/journal.ppat.1000153
Keywords: Toll pathway, rice stripe virus, small brown planthopper, immune perception, protein interaction
Citation: He Y-J, Lu G, Qi Y-H, Zhang Y, Zhang X-D, Huang H-J, Zhuo J-C, Sun Z-T, Yan F, Chen J-P, Zhang C-X and Li J-M (2021) Activation of Toll Immune Pathway in an Insect Vector Induced by a Plant Virus. Front. Immunol. 11:613957. doi: 10.3389/fimmu.2020.613957
Received: 04 October 2020; Accepted: 01 December 2020;
Published: 08 January 2021.
Edited by:
Xiao-Qiang Yu, University of Missouri–Kansas City, United StatesReviewed by:
Christine A. Jansen, Utrecht University, NetherlandsCopyright © 2021 He, Lu, Qi, Zhang, Zhang, Huang, Zhuo, Sun, Yan, Chen, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Ping Chen, anBjaGVuMjAwMUAxMjYuY29t; Chuan-Xi Zhang, Y2h4emhhbmdAemp1LmVkdS5jbg==; Jun-Min Li, bGlqdW5taW5AbmJ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.