- 1College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2Key Laboratory of Aquatic, Livestock and Poultry Feed Science and Technology in South China, Ministry of Agriculture, Zhanjiang, China
- 3Aquatic Animals Precision Nutrition and High Efficiency Feed Engineering Research Center of Guangdong Province, Zhanjiang, China
- 4Department of Medical Laboratories, Faculty of Medical and Health Science, Taiz University/AL-Turba Branch, Taiz, Yemen
- 5Guangdong Provincial Key Laboratory of Marine Resources and Coastal Engineering, Guangzhou, China
Glycogen synthase kinase-3 (GSK3), a cytoplasmic serine/threonine-protein kinase involved in a large number of key cellular processes, is a little-known signaling molecule in virus study. In this study, a GSK3 protein which was highly similar to GSK3β homologs from other species in Litopenaeus vannamei (designated as LvGSK3β) was obtained. LvGSK3β was expressed constitutively in the healthy L. vannamei, at the highest level in the intestine and the lowest level in the eyestalk. White spot syndrome virus (WSSV) reduced LvGSK3β expression was in immune tissues including the hemocyte, intestine, gill and hepatopancreas. The inhibition of LvGSK3β resulted in significantly higher survival rates of L. vannamei during WSSV infection than the control group, and significantly lower WSSV viral loads in LvGSK3β-inhibited L. vannamei were observed. Knockdown of LvGSK3β by RNAi resulted in increases in the expression of LvDorsal and several NF-κB driven antimicrobial peptide (AMP) genes (including ALF, PEN and crustin), but a decrease in LvCactus expression. Accordingly, overexpression of LvGSK3β could reduce the promoter activity of LvDorsal and several AMPs, while the promoter activity of LvCactus was increased. Electrophoretic mobility shift assays (EMSA) showed that LvDorsal could bind to the promoter of LvGSK3β. The interaction between LvGSK3β and LvDorsal or LvCactus was confirmed using co-immunoprecipitation (Co-IP) assays. In addition, the expression of LvGSK3β was dramatically reduced by knockdown of LvDorsal. In summary, the results presented in this study indicated that LvGSK3β had a negative effect on L. vannamei by mediating a feedback regulation of the NF-κB pathway when it is infected by WSSV.
Introduction
Shrimp is one of the main internationally traded aquatic products, which has important economic value. As an important aquaculture shrimp species, the produce of Litopenaeus vannamei accounts for nearly two-thirds of the total output of shrimp worldwild (1). In recent years, emerging diseases caused by various pathogens have been the main menace for sustainable development of the shrimp industry worldwide. Among these pathogens, white spot syndrome virus (WSSV) has always been a viral agent of greatest concern, although other viruses such as Taura syndrome virus (TSV), infectious hypodermal and hematopoietic necrosis virus (IHHNV) and decapod iridescent virus 1 (DIV1) were reported to cause losses in farmed shrimp (2–5). As an invertebrate, the innate immunity is the first defense line against viral infections in shrimp (6). Although a considerable number of scientific studies have focused on this for several years, knowledge of the host-virus interaction is quite insufficient. Thus, it is in urgent need to investigate the mechanisms of immune defence for disease controling in shrimp culture.
Glycogen synthase kinase 3 (GSK3), a multifunctional serine/threonine-protein kinase, initially identified as a key regulator of glycogen metabolism and insulin signaling, has been verified as being involved in various physiological and pathological processes, such as protein synthesis, signal transduction, cell proliferation and differentiation, immune response, inflammation and tumorigenesis, and so on (7, 8). GSK3 is highly conserved in species from the amoeba, Dictyostelium discoideum, to humans. In vertebrates, there are two isoforms of GSK3 (GSK3α and GSK3β) known, which share a highly conserved catalytic domain but differ at both termini (9). GSK3α and GSK3β perform different functions in many biological processes. GSK3α is mainly involved in glycogen metabolism, while GSK3β functions as a convergence point for multiple signaling pathways, including nuclear factor κB (NF-κB), Wnt/β-catenin, phosphatidylinositide 3-kinase/protein kinase B (PI3K/Akt), Hedgehog and STAT signaling pathways (10–12). Recent studies have unearthed GSK3β’s role in viral infections through its alteration of viral entry, replication and egress. GSK3β is known as an AKT substrate which plays a crucial role in influenza viral entry (13, 14). GSK3β silence could decrease the hepatitis C virus (HCV) replication and the production of infectious particle while GSK3β overexpression enhanced HCV replication (15). Studies in vitro displayed that specific inhibitors (small molecules) of GSK3β significantly decreased Tat-dependent replication of human immunodeficiency virus 1 (HIV1) (16). In addition, GSK3β inhibitors could effectively prevent the assembly and release of HCV because of the key role in HCV virion assembly and release GSK3β played, through inhibiting apolipoprotein synthesis (17). But overall, studying on the function of GSK3β and its related signaling pathways in the research field of virology is still in its initial stage, especially in invertebrates.
GSK3β has been demonstrated to play a role in the interaction between host and virus in shrimp, but studies on its specific regulation mechanism during the anti-viral immune reaction in shrimp is required, as GSK3β is a cross point in several signal pathways (18). An important pro-inflammatory pathway promoted by GSK3β is the activation of NF-κB (19). GSK3β plays a vital role in gene transcription by the way of regulating the promoter-specific recruitment of NF-κB (20). Through restraining the transcriptional activation of NF-κB, GSK3β inhibitors could lessen the inflammatory cytokine production of the TLR pathway (21). Consistently, activation of the PI3K/Akt/GSK3β signaling pathway reduced the NF-κB nuclear translocation and the pathway acts as the influencing factor in inflammatory protection (22). NF-κB plays an essential role in the antiviral immunity and is an obstacle to the survival of virus (23). The Toll and immune deficiency (IMD) pathways are Two NF-κB related signaling pathways which are considered as the main regulatory factors of the anti-viral immune response in shrimp (24). Building on the current research, the mechanism behind GSK3β mediation of NF-κB during WSSV infection in L. vannamei was studied in this paper. The results showed that L. vannamei GSK3β (LvGSK3β) has a negative effect for the shrimp during WSSV infection by regulating NF-κB activity, which is beneficial for the study on the host-virus interactional mechanisms and may offer potential strategies for disease prevention in shrimp industry.
Materials and Methods
Shrimp and WSSV
L. vannamei (approximately 4–6 g each) were purchased from a local shrimp farm in Zhanjiang City, Guangdong Province, China. As our previous study (25) described, L. vannamei were cultured in a recirculating water tank system for over 7 days before the experiments with water salinity and temperature maintained at 27‰ salinity at 25–27 °C. The shrimp were fed with a commercial shrimp pellet diet (Haid Group, Guangzhou, China) twice daily. At the beginning of all experimental treatments, shrimp (5% of the total) were analyzed and ensured to be free of WSSV via PCR method performed according to published standard operating procedures. The WSSV (Chinese strain, AF332093) was extracted from WSSV-infected shrimp muscle tissue and stored at −80 °C. Before injection, the muscle tissue was homogenised and prepared as a WSSV inoculum with approximately 1×105 copies/µl, as previously described (26).
Cloning the cDNA and Genome of LvGSK3β
Total RNA and the genomic DNA were extracted from the shrimp tissues using the RNeasy Mini kit (Qiagen, Hilden, Germany) and TIANGEN Marine Animals DNA Kit (TIANGEN, Guangzhou, China), respectively. First-strand cDNA synthesis was performed using a cDNA Synthesis Kit (Takara, Dalian, China) following the manufacturer’s instructions. The partial cDNA sequence of LvGSK3β was obtained from transcriptomic sequencing of L. vannamei (27) and its full-length cDNA sequence was cloned by a rapid amplification of cDNA ends (RACE) PCR using the SMARTer™ RACE cDNA Amplification kit (Clontech, Japan), following the user manual and the specific primers used are listed in Table 1. The genomic DNA sequences of LvGSK3β were obtained via PCR, according to a previously published method (28) and were based on the genome sequences of Penaeus vannamei breed Kehai No.1 (GenBank No. NW_020869560). The PCR products were cloned into the pMD-20 vector (Takara, Japan) and sequenced. The gene sequences obtained in this study have been deposited in the NCBI GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
Genomic Walking
The 5’ flanking regulatory regions of LvGSK3β were cloned by genome walking PCR amplification via the GenomeWalker Universal Kit (Clontech, Dalian, China), according to the methods of a previous paper (29). Two pairs of primers, AP1/LvGSK3β-R1 and AP2/LvGSK3β-R2, were used to perform the primary and nested genome walking PCR amplification. The PCR products were cloned into the pMD-20T vector (Takara, Dalian, China) and sequenced. Primers are listed in Table 1.
Bioinformatics Analyses
The protein domain topologies and the genome sequences were analysed using the SMART program (http://smart.embl-heidelberg.de/) and Splign program (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi). Protein sequences of GSK3β homologues from other species were selected from the NCBI databases. Multiple sequence alignments were performed using the ClustalX 2.0 program (http://www.ebi.ac.uk/tools/clustalw2) and visualized using GeneDoc. The similarities among the GSK3β proteins were calculated in GeneDoc. The neighbor-joining phylogenetic trees were constructed based on the amino acid sequences using MEGA 5.0 (http://www.megasoft-ware.net/download_form), applying the Poisson distribution substitution and bootstrapping procedure with a minimum of 1,000 bootstraps. The LvGSK3β promoter sequence was analyzed using Consite (http://consite.genereg.net/).
Tissue Distribution, Immune Challenge, and Gene Expression Analyses
Eleven tissues, including the hemocytes, hepatopancreas, gill, heart, stomach, pyloric caecum, nerve, epithelium, eyestalk, intestine and muscle, were obtained from 10 healthy L. vannamei for RNA extraction. For the challenge experiments, 60 healthy L. vannamei divided into two groups (n = 30) were intramuscularly injected with WSSV (106 copies/g) in the third abdominal segment. L. vannamei injected with PBS were used as controls. At 0, 4, 8, 12, 24, 48, and 72 h post-injection (hpi), three animals from each group were randomly sampled for hemocyte, hepatopancreas, gill and intestine collection. Total RNA extraction, reverse transcription and real-time PCR (qPCR) analysis were performed as detailed in previous research (30). The qPCR was performed in a Roche Light Cycler480 thermal cycler (Roche Applied Science, Germany). Elongation factor 1α (EF1α, GenBank accession No. GU136229) was used as the internal control. The expression level of LvGSK3β was calculated using the Livak(2−ΔΔCt) method after normalization to LvEF1α. All samples were tested in triplicate. Primer sequences are listed in Table 1.
RNA Interference
As described in our previous studies (31, 32), the double-stranded RNAs (dsRNAs) specific to LvGSK3β, LvDorsal and GFP (as control) were synthesized using an in vitro transcription method, by using the T7 RiboMAX™ Express RNAi System (Promega, USA), and the quality and amount of dsRNA were checked after annealing via gel electrophoresis. For the RNA interference efficiency determination, healthy L. vannamei were injected with 50 µl PBS containing 5 µg LvGSK3β, LvDorsal or GFP dsRNA. At 24 and 48 hpi, the hemocytes from each group were sampled for qPCR to detect the RNAi efficiency, the effects of LvGSK3β on the expression of LvDorsal, LvCactus and nine antimicrobial peptides (AMP) genes and the effects of LvDorsal on the expression of LvGSK3β in L. vannamei. Primer sequences are listed in Table 1.
WSSV Challenge Experiments in LvGSK3β-Inhibited L. vannamei
LvGSK3β was inhibited both on the enzyme activity and mRNA expression levels by injecting with the specific inhibitor and dsRNA. Specific inhibitor of LvGSK3β, named TWS-119 (Selleck Chemicals, USA) was used with the dose refer to the Inhibitory concentration 50 (IC50) (33). At 4, 8 and 12 hpi, the hemocytes from each group were sampled for the detection of inhibition efficiency of LvGSK3β activities, using the GSK3β Kinase Activity Quantitative Detection Kit (Genmed, China). The expression of LvGSK3β was inhibited by RNAi as described above in Section RNA Interference. For the mortality test, L. vannamei were injected with 50 µl PBS containing or not containing 106 WSSV after 8 h of TWS-119 injection or 48 h of dsRNA injection. Cumulative mortality was recorded every 4 h and the differences between groups were analysed using the Mantel–Cox (log-rank χ2 test) method with the software GraphPad Prism. To investigate the genome copies of WSSV, total DNA was extracted from muscle at 48, 72 and 96 hpi and absolute real-time PCR was performed using primers WSSV32678-F/WSSV32753-R and a Taq Man fluorogenic probe, as in a previous study (34).
Dual-Luciferase Reporter Assays
The gene-specific primers were designed to amplify the ORF of LvGSK3β without introducing a stop codon. The amplified product was cloned into a pAc5.1/V5-His A vector (Invitrogen) to allow subsequent expression of V5-tagged LvGSK3β proteins. All of the vectors with reporter genes used in this paper, including Dorsal, Cactus and AMPs in shrimp or Drosophila, were obtained from our previous studies (30, 35). Since no permanent shrimp cell line was available, Drosophila Schneider 2 (S2) cells were cultured for promoter activity analysis at 28 °C in Schneider’s Insect Medium (Sigma, USA) containing 10% fetal bovine serum (Gibco, USA) for the in vitro experiments. After 24 h of culturing in a 96-well plate, the S2 cells of each well were transfected with 0.05 μg of firefly luciferase reporter-gene plasmids, 0.01 μg of pRL-TK renilla luciferase plasmid (Promega, as an internal control) and 0.05 μg of (otherwise indicated) either pAc-LvGSK3β-V5 plasmids or pAc5.1-Basic plasmids (control). Each experiment was repeated in 6 wells. 48 hours post-transfection, the activity of the luciferases was detected in order to calculate the relative ratios of firefly and renilla luciferase activity using the Dual-Glo® Luciferase Assay System kit (Promega, USA), according to the manufacturer’s instructions.
Electrophoretic Mobility Shift Assay
An electrophoretic mobility shift assay (EMSA) was performed using a Light Shift Chemiluminescent EMSA kit (Thermo Fisher Scientific, Waltham, MA, USA) according to a previously published method (36). Briefly, the biotin-labeled or unbiotin-labeled probes were designed using the Dorsal binding motif sequence. The mutant probe was designed by deleting the Dorsal-binding motif sequence. All of the probes were synthesized by Life Technologies (Shanghai, China) and the sequences are listed in Table 1. RHD domain of Dorsal (Dorsal-RHD) was cloned into the modified pGEX-4T plasmid to get rDorsal-RHD-GST. GST tag of rDorsal-RHD-GST was then removed using rPorcine Enterokinase from Glutathione Resin Kit. The purification productions of rDorsal-RHD-GST and rDorsal-RHD were analyzed using SDS-PAGE and stained with Coomassie blue. Purified rDorsal-RHD protein (10 μg) was incubated with 20 fmol of the probes for the binding reactions. The reactions were separated on a 5% native PAGE gel, transferred to positively charged nylon membranes (Roche, Germany) and cross-linked by UV light. Then, the biotin-labeled DNA on the membrane was detected by chemiluminescence and developed on X-ray film, followed by enhanced chemiluminescence (ECL) visualisation (Tanon, Shanghai, China).
Co-Immunoprecipitation Assays
Co-immunoprecipitation (Co-IP) assays were performed as described in the previous study (36). To investigate the interaction of LvGSK3β with LvDorsal or LvCactus, pAc-LvGSK3β-GFP or pAc5.1-GFP (control) were co-transfected with individual plasmids along with pAc-LvDorsal-HA or pAc-LvCactus-HA into the S2 cells. Forty eight hours post transfection, cells were harvested and washed with ice-cold PBS three times, and then lysed in IP Lysis Buffer with a protease inhibitor cocktail (Sigma, USA). Both Co-IP and reciprocal Co-IP experiments were carried out using anti-HA affinity gel (Sigma, USA) and samples were subjected to SDS-PAGE assay. The precipitated protein was examined using western-blots, with rabbit anti-GFP antibody as the primary antibody, and alkaline phosphatase-conjugated goat anti-rabbit as the secondary antibody (Sigma, USA). A standardized aliquot (5%) of each total input cell lysates was also examined as control.
Statistical Analyses
The relative gene expression data were statistically analysed and all data were presented as mean ± SD. Student’s t-tests were used to make comparisons among groups. For the mortality analysis, the data were statistically analysed using GraphPad Prism (Graphpad, San Diego, CA, USA) to obtain Kaplan–Meier plots (log-rank χ2 test).
Results
Bioinformatics Analyses of LvGSK3β
As shown in Figure 1A, the transcript of LvGSK3β was 2,382 bp in length, with a 286 bp 5'-untranslated region (UTR), an 863 bp 3'-UTR containing a poly (A) tail and a 1233 bp open reading frame (ORF) encoded a protein of 410 amino acids at the calculated molecular weight of 46,098.78 Da and an estimated pI of 8.70 (GenBank No. KU641425). Additionally, a protein kinase ATP-binding region signature (between amino acids 61 and 85), a serine/threonine protein kinases active site signature (between amino acids 176 and 188) and two candidate regulatory residues at positions 9 (serine) and 216 (tyrosine) were identified. The genomic sequence of LvGSK3β (GenBank No. MT891030) was also obtained, showing that the LvGSK3β gene consists of eight exons and seven introns (Figure 1B). Analysis of the genome sequences showed that all of the exon-intron boundaries in LvMyD88 conform to the consensus GT/AG rule for splicing (37). SMART analysis revealed that the LvGSK3β protein contained one protein kinase domain named STKc_GSK3 between residues 54 and 338 (Figure 1C).

Figure 1 The sequence analysis of LvGSKβ. (A) The full-length cDNA sequence and deduced amino acid sequences of LvGSKβ. The ORF of the nucleotide sequence was shown in upper-case letters, while the 5’ and 3’-UTR sequences were shown in lowercase. The nucleotide (lower row) and deduced amino acid (upper row) sequences are shown and numbered on the left. The translation initiation codon (ATG) and stop codon (TAA) are in bold. The STKc_GSK3 domain is shaded and protein kinase ATP-binding region, serine/threonine protein kinases active site and two candidate regulatory residues (Ser9 and Tyr216) are marked out. The poly A signal (aataaa) are underlined. (B) Gene structure of LvGSKβ. Boxes in represent exons, and lines in represent introns. The numbers indicate the length (bp) of the exons and introns. (C) Structural features of LvGSKβ. Conserved domains of LvGSKβ are predicted by SMART program.
Multiple sequence alignment indicated that LvGSK3β shares a high similarity of more than 80% with GSK3β proteins from invertebrates to vertebrates, with the lowest homology (80%) to GSK3β from Aplysia californica and the highest homology (100%) to GSK3β from Penaeus monodon (Figure 2A), which is also a shrimp species. The neighbour-joining (NJ) phylogenetic tree showed that GSK3α and GSK3β were closely related to their own homologues from other species. All of the GSK3β proteins used in this study were separated into five clades including the crustacea, insecta, arachnids, molluscs and vertebrates groups (Figure 2B). GSK3β from L. vannamei, P. monodon, Daphnia pulex and Eriocheir sinensis were clustered together in the crustacea group. L. vannamei GSK3β and P. monodon GSK3β showed the closest evolutionary relationship. As shown in Figure 3A, the promoter region of LvGSK3β (1,169 bp in length) contained two potential Dorsal binding sites (−700 to −711 bp and −962 to −973 bp) (Figure 3B).
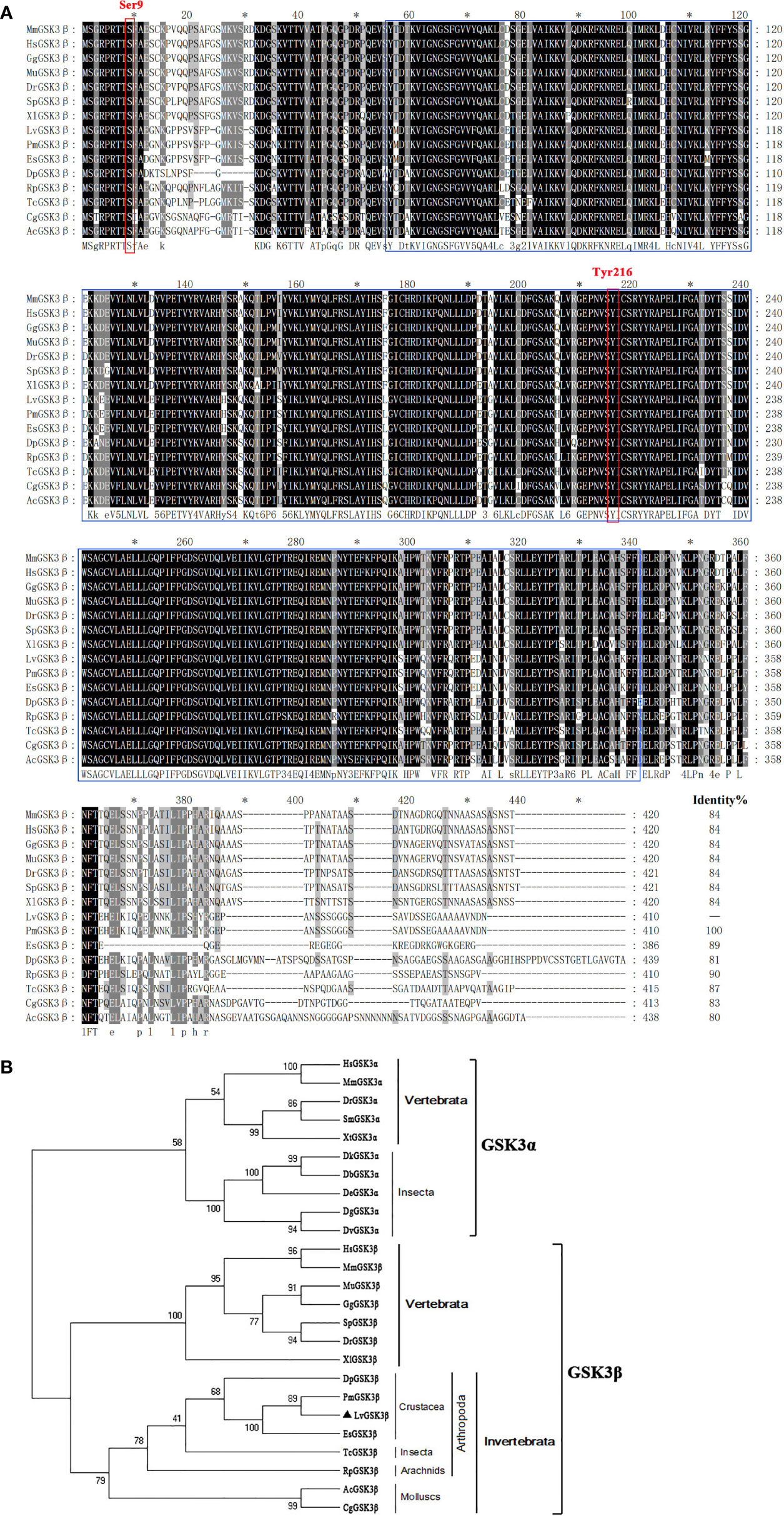
Figure 2 Multiple sequence alignment and phylogenetic tree analysis of GSK3β from L. vannamei and other species. (A) Multiple sequence alignment of GSK3β proteins. The amino acid sequences of GSK3β from typical organisms were aligned using the ClustalX 2.0 program. The black shade represent 100% identity, dark gray represented 80% identity, light gray represented 60% identity. The region of STKc_GSK3 domain was boxed in blue and two candidate regulatory residues Ser9 and Tyr216 were boxed in red. (B) Neighborjoining phylogenetic tree analysis. A rooted tree was constructed via the neighbor-joining method and was bootstrapped 1000 times using MEGA 5.0 software (http://www.megasoftware.net/index.html). LvGSK3β is denoted by ▲. The genes used are listed as follows, GSK3α isoform: HsGSK3α, Homo sapiens GSK3α (Accession No. NP_001139628); MmGSK3α, Mus muscμlus GSK3α (Accession No. AAD39258); DrGSK3α, Danio rerio GSK3α (Accession No. NP_571465); DrGSK3α, Scophthalmus maximus (Accession No. AWP21281); DrGSK3α, Xenopus tropicalis (Accession No. XP_002936450); DbGSK3α, Drosophila bipectinata (Accession No. XP_017108632); DeGSK3α, Drosophila elegans (Accession No. XP_017115500); DtGSK3α, Drosophila takahashii (Accession No. 017013230); DvGSK3α, Drosophila virilis (Accession No. XP_002051677); DgGSK3α, Drosophila grimshawi (Accession No. XP_001988855); GSK3β isoform: LvGSK3β, L. vannamei GSK3β (Accession No. KU641425); HsGSK3β, Homo sapiens GSK3β (Accession No. AAH00251); MmGSK3β, Mus muscμlus GSK3β (Accession No. AAD39258); MuGSK3β, Melopsittacus undulatus GSK3β (Accession No. XP_005143085); GgGSK3β, Gallus gallus GSK3β (Accession No. XP_004938236); SpGSK3β, Schizothorax prenanti GSK3β (Accession No. AKA09693); DrGSK3β, Danio rerio GSK3β (Accession No. NP_571456); XlGSK3β, Xenopus laevis GSK3β (Accession No. AAA84444); DpGSK3β, Daphnia pulex GSK3β (Accession No. EFX72595); PmGSK3β, Penaeus monodon GSK3β (Accession No. QIK00339); EsGSK3β, Eriocheir sinensis GSK3β (Accession No. ANZ22981); TcGSK3β, Tribolium castaneum GSK3β (Accession No. XP_008192199); RpGSK3β, Rhipicephalus pulchellus GSK3β (Accession No. JAA60305); AcGSK3β, Aplysia californica GSK3β (Accession No. XP_005107895); CgGSK3β, Crassostrea gigas GSK3β (Accession No. EKC35379).

Figure 3 The sequence analysis of the LvGSK3β promoter. (A) The promoter sequence of LvGSK3β. The predicted TATA-box was boxed and the putative Dorsal binding sites were underlined. (B) Prediction of the Dorsal binding sites sequence in LvGSK3β promoter using Consite software.
Expression Profiles of LvGSK3β in Healthy and WSSV Challenged L. vannamei
LvGSK3β was detected in all of the examined tissues (Figure 4). The relative expression levels of LvGSK3β in the other tissues were normalised to that in eyestalk, which was the lowest and set as the baseline (1.0). LvGSK3β was expressed highest in the intestine, in which the expression level of LvGSK3β was 14.07-fold over the baseline. As shown in Figure 4, LvGSK3β expression in the gill, muscle, nerve, heart, hepatopancreas and hemocyte were all nearly 5.00-fold over the baseline and not significantly different from each other.
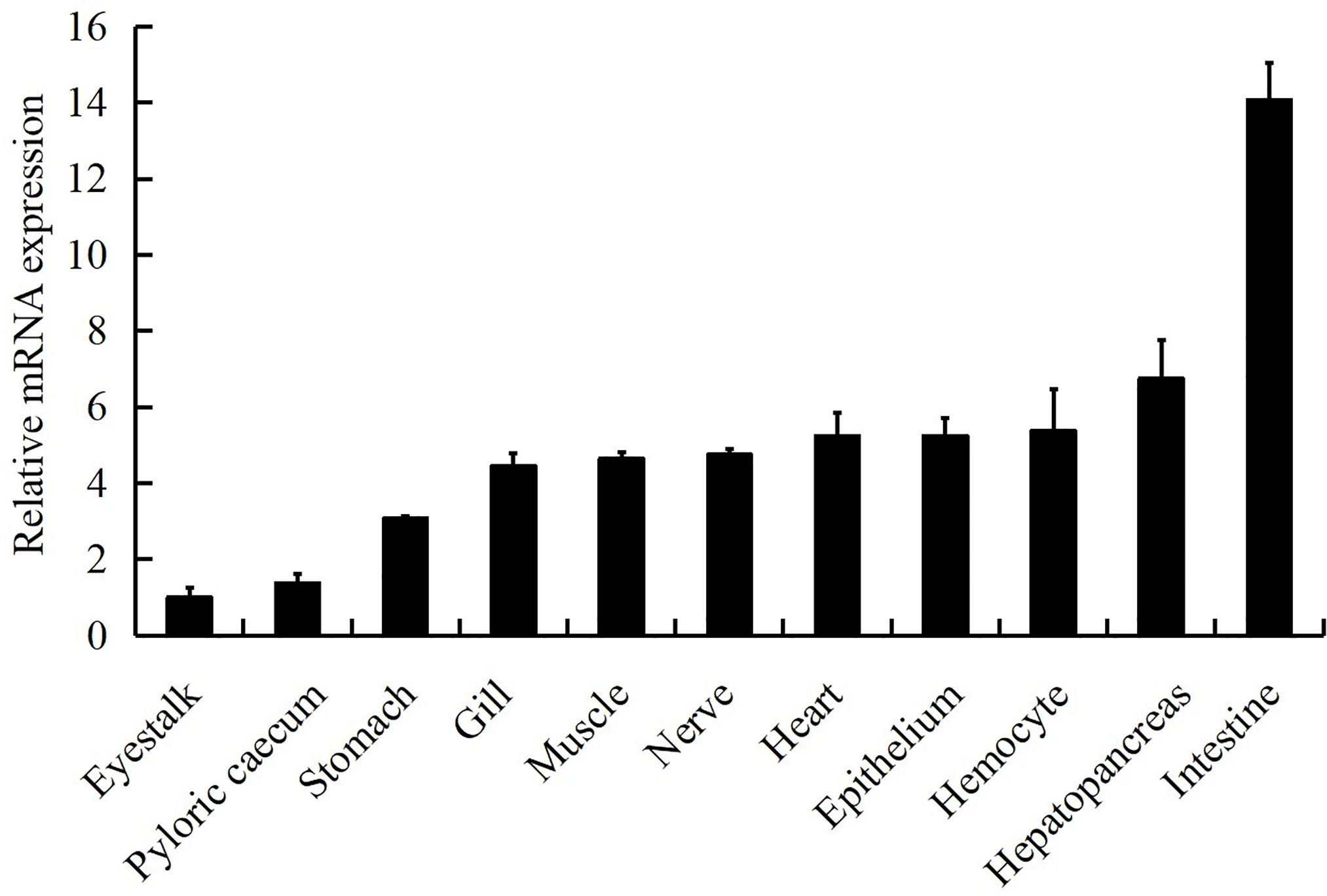
Figure 4 Tissue distribution of LvGSK3β expression in healthy L. vannamei. Ten animals were used for tissue sampling. The expression of LvGSK3β in eyestalk was set as 1.0. LvEF1α was used as the internal control to normalize the cDNA template used for qPCR analysis. The results were based on three independent experiments and expressed as mean values ± SD.
Using the four important immune tissues, including hemocytes, hepatopancreas, intestine and gill, LvGSK3β expression after WSSV challenge was investigated using qPCR (Figure 5). The expression of LvGSK3β was reduced to different degrees by WSSV challenge in the haemocytes, intestine and gill but not in the hepatopancreas. In the hepatopancreas there was no significant difference in LvGSK3β expression between the infection group and the control group at all of the sampled timepoints (Figure 5B). In the hemocyte, LvGSK3β expression was significantly down-regulated starting at 4 hpi, reaching a low-point at 12 hpi at 25.21% of the control. At 72 hpi, LvGSK3β expression recovered to a level not perceptibly different from the control (Figure 5A). In the intestine, the expression of LvGSK3β in the infection group was notably lower than the control group at 4, 8 and 72 hpi, with the lowest level at 8 hpi at 21.00% of the control (Figure 5C). When compared with the control group, LvGSK3β expression in the gill of the infection group was significantly decreased at 12, 24 and 48 hpi by 72.02%, 54.78% and 65.16%, respectively (Figure 5D).
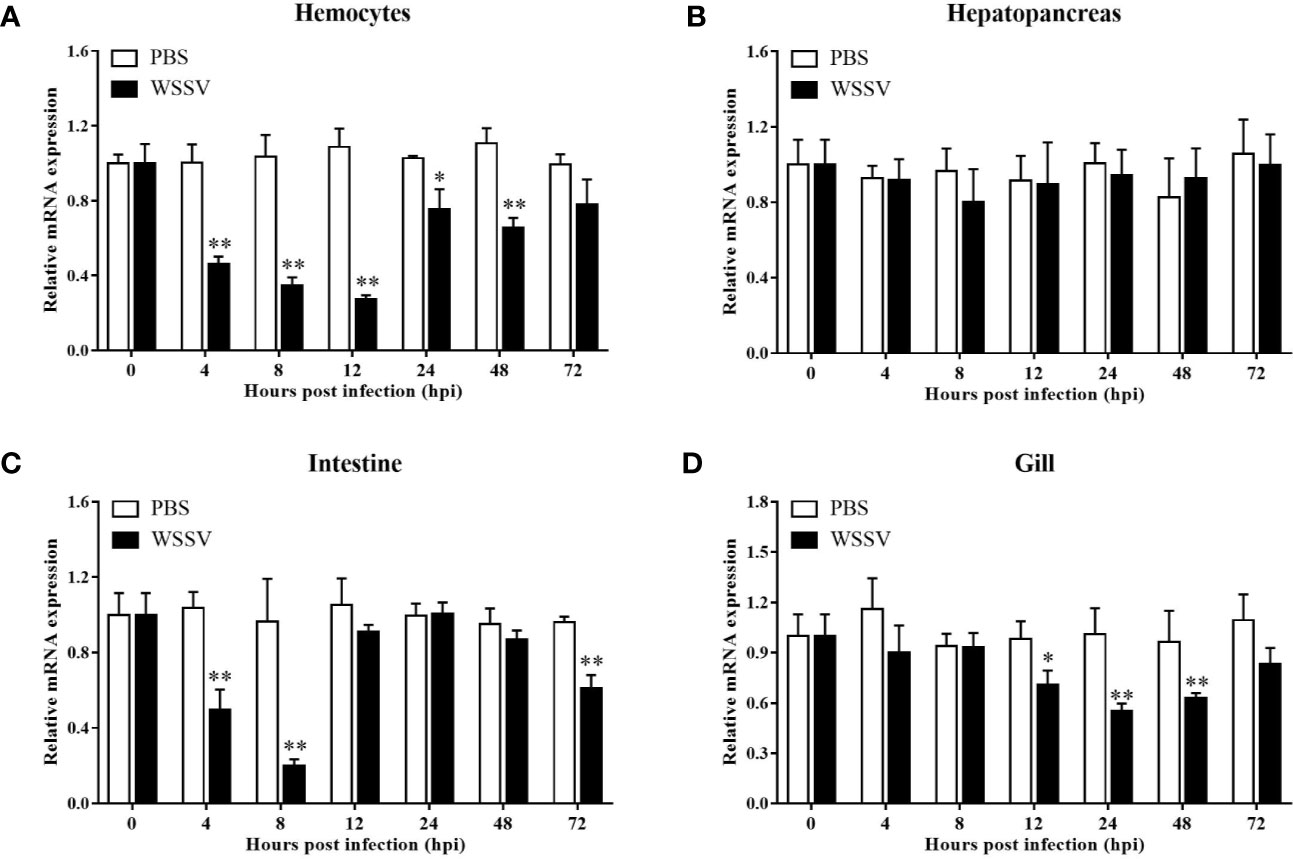
Figure 5 Expression profiles of LvGSK3β in haemocytes (A), hepatopancreas (B), gill (C) and intestine (D) from WSSV or PBS challenged L. vannamei. qPCR was performed in triplicate for each sample. Expression values were normalized to those of LvEF1α using the Livak (2−ΔΔCt) method and the data were provided as the means ± SD of triplicate assays. The statistical significance was calculated using Student’s t-test (*p < 0.05, **p < 0.01).
Function of LvGSK3β in WSSV Infection
Refer to IC50 of TWS-119 in animals (33), a dose of 5 µg/g TWS-119 per shrimp was chosen for the LvGSK3β-inhibition test. As Figure 6A shows, the enzyme activity of LvGSK3β was significantly reduced at 4, 8 and 12 hpi, with the lowest level at 8 hpi, which was the timepoint chosen for the following WSSV challenge experiment. In the TWS-119 + WSSV group, the survival rates of L. vannamei were notably higher than that of the PBS + WSSV group starting at 56 hpi (Figure 6B). At 124 hpi, the survival rate of L. vannamei in the TWS-119 + WSSV group was 43.60% but the L. vannamei in the PBS+WSSV group had all died. The final survival rate of L. vannamei at 144 hpi was 10.30% for the TWS-119 + WSSV group. Consistently, the WSSV genome copies in the muscle of the TWS-119 injected L. vannamei at 48, 72 and 96 hpi were significantly less than those in the PBS injected L. vannamei (Figure 6C).
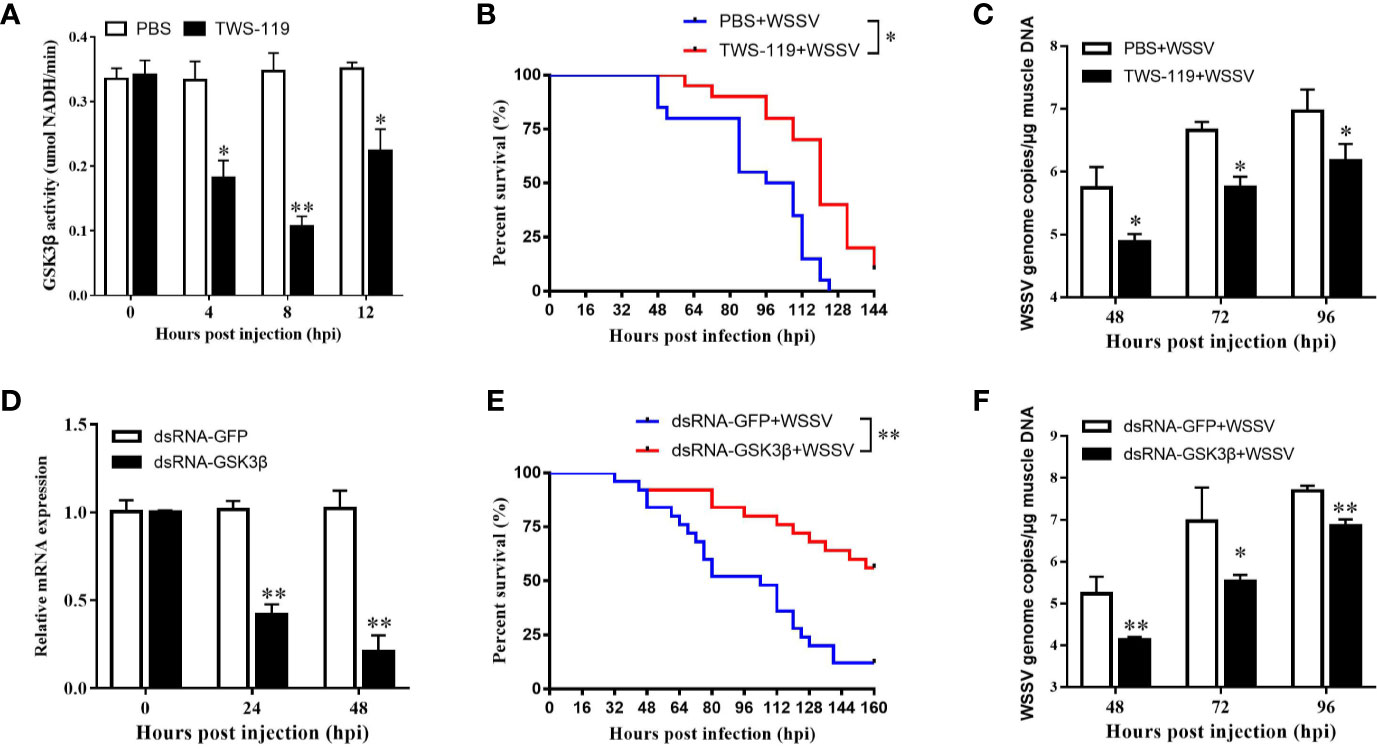
Figure 6 Functional analysis of LvGSK3β in WSSV infection. The inhibiting efficiencies of LvGSK3β by TWS-119 (A) or dsRNA-LvGSK3β (C) injection. The detection of LvGSK3β activities and expression was performed in triplicate for each sample. Expression values were normalized to those of LvEF1α using the Livak (2−ΔΔCt) method and the data were provided as the means ± SD of triplicate assays. Cumulative mortalities of TWS-119 (B) and dsRNA-LvGSK3β (D) treated L. vannamei during WSSV infection. The experiments were performed two times with identical results. Differences in cumulative mortality levels between treatments were analyzed by using the log-rank χ2 test (*p <0.05, **p <0.01). WSSV genome copies in muscle tissue of TWS-119 (C) and dsRNA-LvGSK3β (E) treated L. vannamei post WSSV infection. Absolute real-time PCR was performed using primers WSSV32678-F/WSSV32753-R and a Taq Man fluorogenic probe. Each bar represented the mean ± SD of three samples. Statistically significant differences were represented with asterisks (**p < 0.01 and * p < 0.05).
RNAi was also performed to investigate the role of LvGSK3β during WSSV infection. qPCR was used to check the silencing efficiency of LvGSK3β. As shown in Figure 6D, at 48 h after injection of LvGSK3β dsRNA, the expression levels of LvGSK3β at 24 and 48 hpi were significantly down-regulated to ~0.43-fold and ~0.21-fold of the GFP dsRNA injection group, respectively. Similarly as for the results when LvGSK3β were inhibited by TWS-119, the survival rates of L. vannamei in the LvGSK3β dsRNA + WSSV group were also significantly higher than that of the GFP dsRNA + WSSV group, and the final survival rate of L. vannamei at 144 hpi was 53.85 and 14.29% for the TWS-119 + WSSV group and GFP dsRNA + WSSV group, respectively (Figure 6E). Furthermore, the viral loads of the LvGSK3β dsRNA + WSSV group were significantly lower than those of the GFP dsRNA + WSSV control group, with a ~15.52-fold, ~40.78-fold and~7.06-fold decrease at 48, 72 and 96 h, respectively (Figure 6F).
Influence of LvGSK3β on Expression of NF-κB Related Genes in L. vannamei
NF-κB has important roles in the regulation of AMP genes in shrimp (2). Considering the fact that the promoter of LvGSK3β contained two potential Dorsal binding sites, the expression of NF-κB related genes (Dorsal, Cactus and AMP genes) in LvGSK3β dsRNA injected shrimp were investigated to primarily explore the underlying mechanism of LvGSK3β during WSSV infection. When compared with the control GFP dsRNA, the LvGSK3β dsRNA significantly increased LvDorsal expression but decreased LvCactus expression in the haemocyte of L. vannamei. Consistent with this, the expression of several AMP genes, including three antilipopolysaccharide factors (LvALF1, LvALF2, LvALF3), two penaeidins (LvPEN3, LvPEN4) and one crustin (LvCRU1) were significantly up-regulated at 48 h after injection with LvGSK3β dsRNA (Figure 7A).
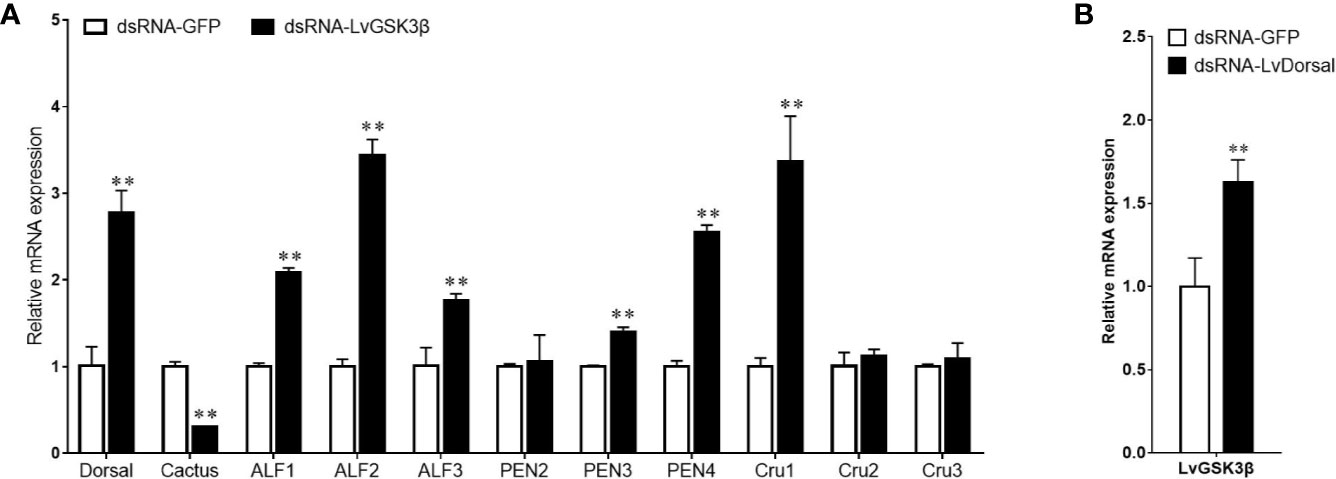
Figure 7 Influence of LvGSK3β on expression of NF-κB related genes (A) and LvDorsal on expression of LvGSK3β (B) in L. vannamei The mRNA levels of NF-κB related genes and LvGSK3β in L. vannamei at 48 h post dsRNA injection were detected using qPCR with LvEF1α gene as internal control. For each gene, the value in the dsRNA-GFP injected sample was set as the baseline (1.0) for comparison (**p < 0.01).
Influence of LvDorsal on Expression of LvGSK3β in L. vannamei
The expression of LvGSK3β in LvDorsal dsRNA injected shrimp was also investigated. When LvDorsal was knocked down by dsRNA injection, the expression of LvGSK3β in the hemocyte of L. vannamei was significantly higher than the control GFP dsRNA injected group (Figure 7B).
Regulatory Effects of LvGSK3β on the Promoter Activities of NF-κB Related Genes
Previous results in vivo showed that LvGSK3β could regulate the expression of NF-κB related genes, so the effects of LvGSK3β on the promoter activities of NF-κB related genes were further explored in vitro using dual-luciferase reporter assays. As shown in Figure 8, overexpression of LvGSK3β dramatically down-regulated the promoter activities of LvDorsal (56.61%) and the AMP genes including the L. vannamei AMPs LvALF2 (65.80%), LvPEN2 (28.29%), LvPEN3 (41.18%), LvCru1 (65.96%), LvCru2 (59.04%) and LvCru3 (64.23%), the Penaeus monodon AMP PmPEN536 (66.05%), and the Drosophila AMPs, DmCecA (44.37%), DmDpt (49.16%), DmDrs (65.76%) and DmDef (21.23%). However, the promoter activity of LvCactus was significantly induced by ~1.88 fold.
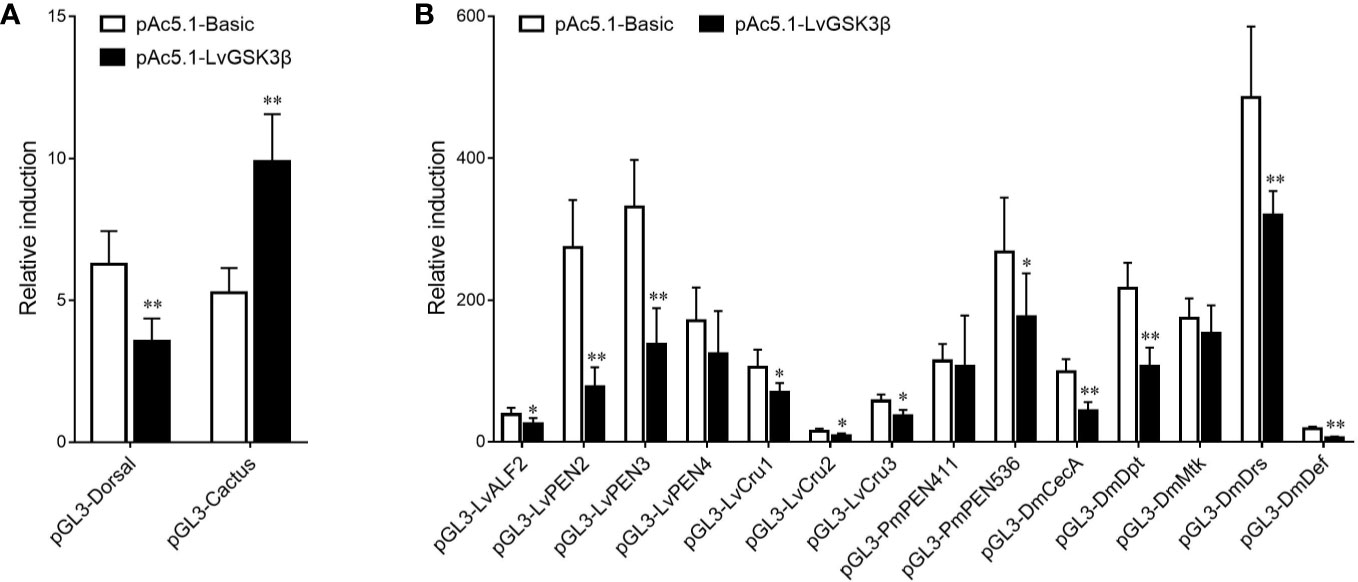
Figure 8 Effects of LvGSK3β on the promoter activities of NF-κB related genes in L. vannamei. (A) LvDorsal and LvCactus promoters; (B) AMPs promoters. Drosophila S2 cells were transfected with the protein expression vectors (LvGSK3β and the pAc5.1 empty vector as a control), the reporter gene plasmid, and the pRL-TK Renilla luciferase plasmid (as an internal control). After 48 h, the cells were harvested for measurement of luciferase activity using the dualluciferase reporter assay system (Promega). The bars indicated the mean ± SD of the luciferase activity (n = 6). The statistical significance was calculated using Student’s t-test (*p < 0.05, **p < 0.01).
Interaction Between LvDorsal and the LvGSK3β Promoter
To further investigate the interaction between LvDorsal and the LvGSK3β promoter, EMSA was performed using the purified 6His-tagged RHD domain of Dorsal protein (rDorsal-RHD) expressed in E. coli cells. As shown in Figure 9, rLvDorsal-RHD, but not the control rTrx, effectively retarded the mobility of the bio-labeled probes 1 and 2. The DNA/protein complex was markedly reduced by the competitive unlabeled probe 1 at the level of 10×, 50× and 100×. Contrarily, the DNA/protein complex was only faintly reduced by the competitive 10× and 50× unlabeled probe 2 but markedly reduced by the competitive 100× unlabeled probe 2. These results indicate the specificity of the interaction between rLvDorsal-RHD and the two probes.
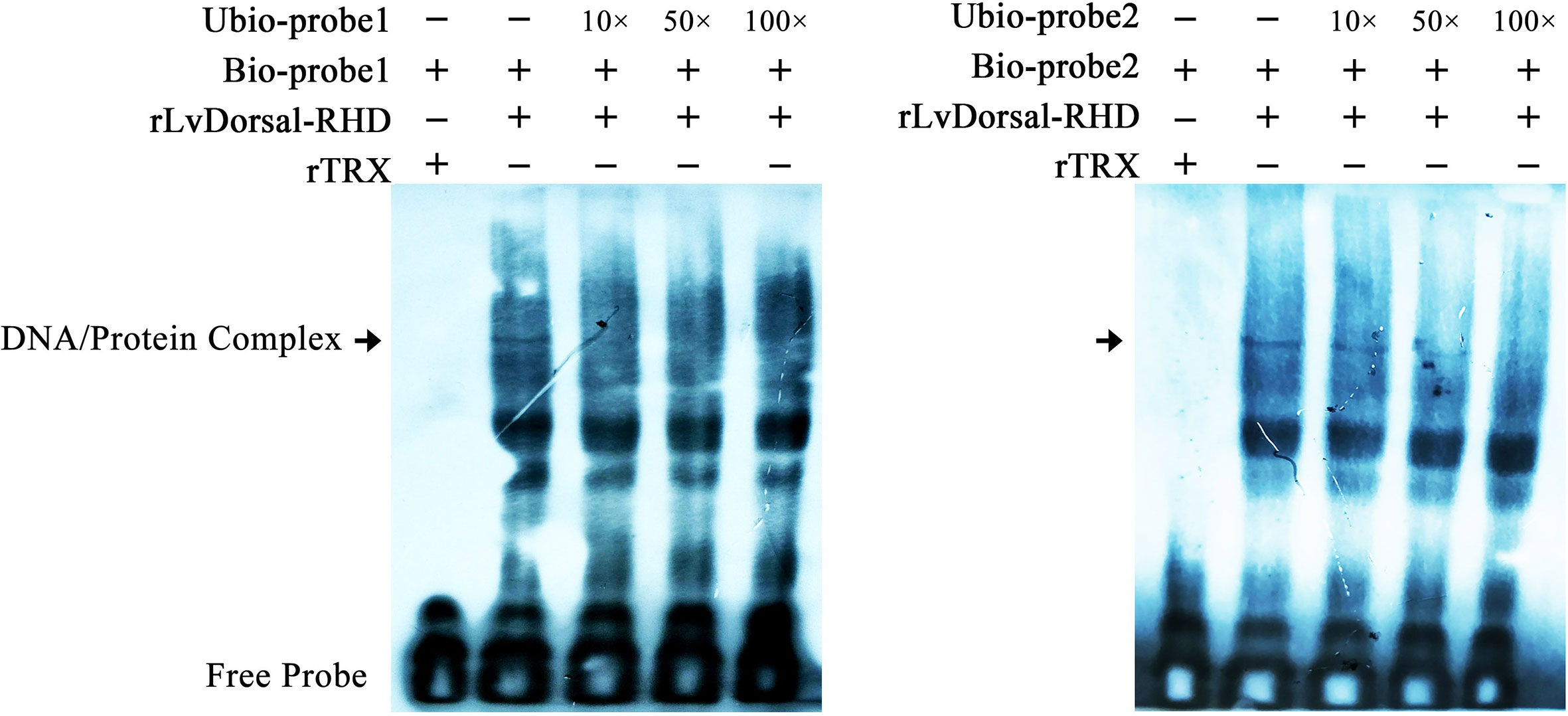
Figure 9 Dorsal interacted with the predicted binding motifs of LvGSK3β promoter in vitro. An EMSA was performed using biotin-labeled (Bio-) or unlabeled (Unbio-) probes containing or not containing the Dorsal binding motifs of LvGSK3β. Biotin-labeled or mutated biotin-labeled dsDNA probes were incubated with 10 μg of purified rLvDorsal-RHD protein. Unlabeled probe was added to compete with binding, and an rTrx protein was used as a control. Probe1 and probe 2 referred to the Dorsal binding motif 1 and 2, respectively. All experiments were performed three times with similar results.
Interaction of LvGSK3β With LvDorsal and LvCactus
To explore whether the interaction of LvGSK3β with LvDorsal or LvCactus occurs in L. vannamei, Co-IP analyses were carried out by using the S2 cell line. GFP-tagged LvGSK3β or GFP (as a control) were co-expressed with HA-tagged LvDorsal or HA-tagged LvCactus, respectively. The results showed that both LvDorsal-HA and LvCactus-HA co-precipitated with GFP-tagged LvGSK3β but not the control GFP (Figures 10A, B), indicating an interaction of LvGSK3β with LvDorsal and LvCactus.
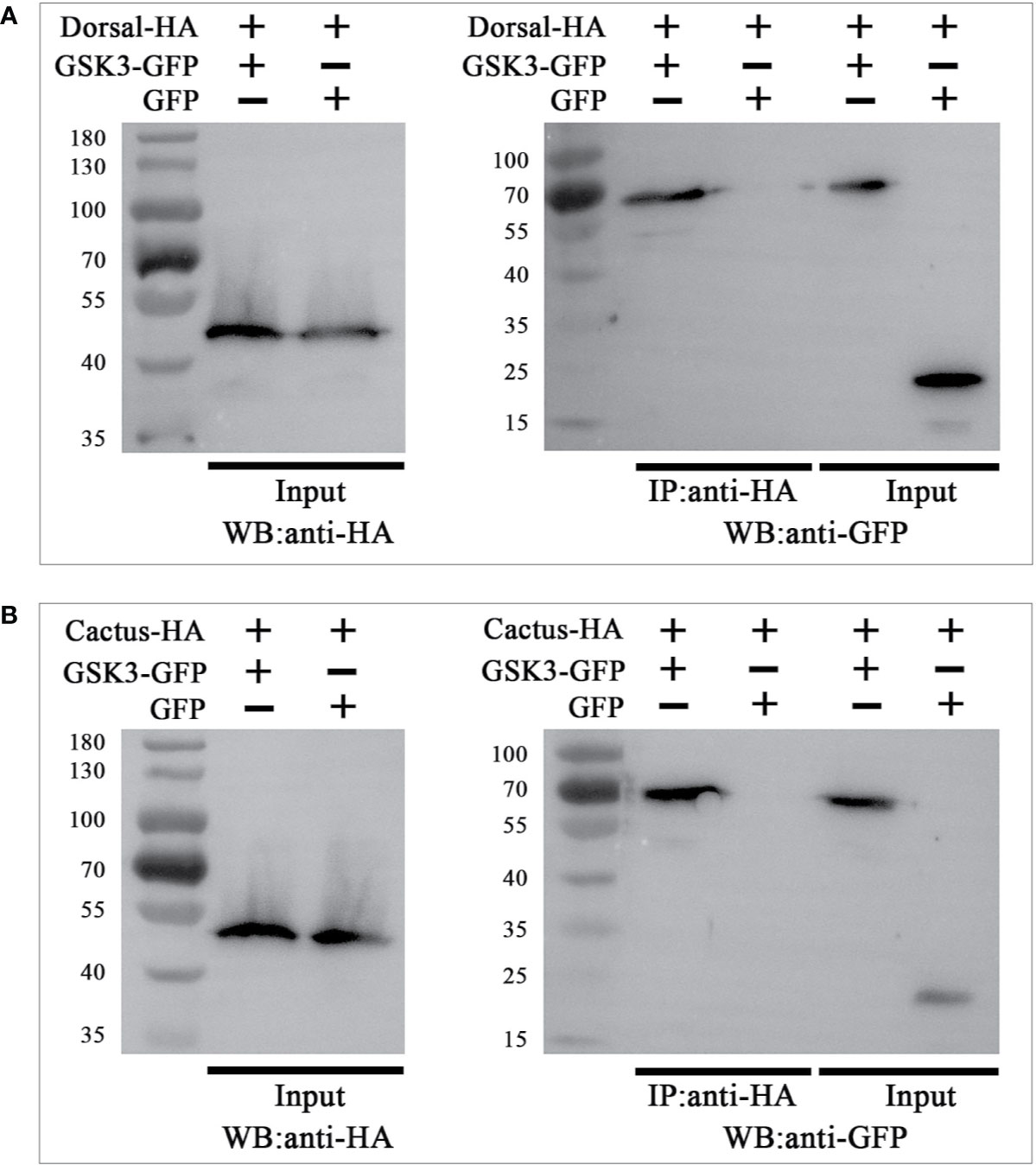
Figure 10 LvGSK3β interacted with LvDorsal (A) and LvCactus (B). Co-immunoprecipitation (Co-IP) assays showed that the GFP-tagged LvGSK3β but not the control GFP protein can be co-precipitated by HA-tagged LvDorsal or LvCactus. Input: western-blotting analysis of the input cell lysates (5%) before immunoprecipitation.
Discussion
Previous studies on GSK3, concentrating on vertebrates, have shown that it is a multifunctional enzyme involved in glycogen metabolism, insulin signaling, inflammatory response and innate immunity (38). However, the function of GSK3 in invertebrates has rarely been reported. In the present study, a GSK3β homolog was cloned from L. vannamei (LvGSK3β) and was found to be strongly downregulated in vivo after WSSV infection, while the inhibition of LvGSK3β significantly increased the survival rate of L. vannamei in response to WSSV infection. The effects of inhibition and overexpression of LvGSK3β on NF-κB related genes were the opposite. In addition, interaction between LvDorsal and the LvGSK3β promoter, and the interaction of LvGSK3β with LvDorsal/LvCactus were revealed. All of the results indicated that LvGSK3β has a negative effect on the shrimp by negatively regulating NF-κB activity in L. vannamei when it is infected by WSSV.
In vertebrates, GSK3 comprises two homologues, GSK3α and GSK3β, encoded by two similar genes, which share similar Serine/Threonine kinase domains but differ substantially in their termini (39). Unique to the GSK3α is an N-terminal domain consisting of 63 residues that is glycine-rich. Resembling the previously identified GSK3β, L. vannamei GSK3β has Ser9 and Tyr216 phosphorylation sites, a STKc_GSK3 domain but no glycine-rich extension at the N-terminus, and the NJ phylogenetic tree showed that LvGSK3β belonged to the cluster of GSK3β homologues from other species, which suggested that LvGSK3β was a member of the β isoform of the GSK3 family in L. vannamei. Based on the information on NCBI, most of the GSK3 genes identified were GSK3β in invertebrates including P. monodon, D. pulex and E. sinensis, and GSK3α was only reported in Drosophila species. In fact, only GSK3β could be found in L. vannamei through scanning the transcriptome and genomic data published on NCBI. The reason why GSK3β but not GSK3α is common in invertebrates has not yet been revealed.
In E. sinensis, GSK3β regulated hemocyte phagocytosis in the immune response (40). A previous study reported that LvGSK3β played an important role in shrimp immunomodulation, and shrimp might promote the apoptosis to restrain WSSV infection by inhibition of LvGSK3β (18). Similarly, it was also found that LvGSK3β expression decreased with WSSV challenge, and LvGSK3β inhibition by injection with a specific inhibitor or RNAi reduced the WSSV loads in L. vannamei in this study. More intuitively, our study showed that the survival rates of LvGSK3β inhibited L. vannamei after the WSSV challenge was greater. All of the results further verified the close relationship between LvGSK3β and WSSV infection, implying the positive function of LvGSK3β on WSSV replication. To our knowledge, this study was a rare report about the role GSK3β plays during viral infection in invertebrates. Comparatively speaking, many more studies about the effect of GSK3β on viral replication have been performed on vertebrates. In humans, there was a significant drop in GSK3β expression in response to Japanese encephalitis virus (JEV) infection (41), and the replication of HCV (15), HIV-1 (16), varicella zoster virus (VZV) (42) and severe acute respiratory syndrome coronavirus (SARS-CoV) (43) was decreased by silencing GSK3β. In addition, GSK3β functioned in inhibiting porcine epidemic diarrhoea virus (PEDV) replication in vitro (44). Thus, it can be seen that the effect of GSK3β on viral replication is consistent in vertebrates and invertebrates, indicating the functional conservatism of GSK3β in different animal species.
As an intersection of multiple cell signaling pathways such as Wnt/β-catenin, NF-κB and PI3K/AKT, the function of GSK3β in proliferation, differentiation, apoptosis, and immune responses could not be ignored (10–12). This raises difficulties for clarifying the role of GSK3β and the associated regulatory mechanisms regarding virology. Nevertheless, great progress has been made in humans. For example, the downregulation of GSK3β resulted in a long viral persistence of JEV by enhancing the stability of cyclin D1 (39). For the herpes simplex virus type 1 (HSV-1) -induced Kaposi's sarcoma-associated herpesvirus (KSHV) reactivation, the activation of the MAPK and the PTEN/PI3K/AKT/GSK3β pathways was required (45). VZV benefited its own survival and replication through activating AKT signaling that induces the phosphorylation of GSK3β, which in turn blocks Bad's pro-apoptotic pathway (40). In addition, the decrease of GSK3β expression can also inhibit replication of the influenza virus by positively regulating the virus-induced host cell apoptosis (46). Similar results were presented in the functional study of GSK3β in shrimp. LvGSK3β play an important role in WSSV clearance by mediating apoptosis (18). Consistently, this study found that the expression of LvGSK3β was inhibited in L. vannamei upon WSSV challenge and the mortality rates of LvGSK3β-inhibited L. vannamei in response to WSSV infection were significantly reduced when compares with the control group. Besides, LvGSK3β expression has a great impact on the expression and transcriptional regulation of LvDorsal, LvCactus and AMP genes, and LvGSK3β interacted with LvDorsal and LvCactus. Furthermore, knockdown of LvDorsal significantly reduced the expression of LvGSK3β. All these results implied that LvGSK3β plays a role during virus infection by regulating NF-κB activity in L. vannamei and there was a feedback regulatory loop existed. Briefly speaking, the silence of LvGSK3β by dsRNA resulted in upregulation of LvDorsal and downregulation of LvCactus. The downregulation of LvCactus could indirectly increase the expression of LvDorsal, followed by the increased expression of AMP genes which acted as important effectors of host defense against WSSV (24). At the same time, the increase of LvDorsal brought about the downregulation of LvGSK3β could enhance the effect. When suffer WSSV infection, the inhibition of LvGSK3β expression was also strengthened. All the series of reactions led to the final result was that the survival rates of LvGSK3β-inhibited L. vannamei significantly increased when suffer WSSV infection. To our knowledge, although the role of the GSK3β or NF-κB related signaling pathways in viral infection and the control of NF-κB signaling activity by GSKβ has been confirmed separately (18–23, 37), no previous studies have explored the functional mechanisms of GSKβ in viral infection from the perspective of the regulation of NF-κB; therefore, this study is the first such report.
In conclusion, this study revealed that LvGSK3β has a negative effect on L. vannamei by negatively regulating NF-κB activity when it is infected by WSSV. The results detail the immune mechanisms and provide a theoretical basis for enhancing the immunity and improving the disease resistance of shrimp, which lays the foundation for the establishment of effective prevention and control of WSS.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
The experiments were conceived and designed by SZ and LS. SZ, L-LZ, CH, HY, and SY collected samples and performed the experiments. MD, SZ, and LS analyzed the data and completed the writing of the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2019YFD0900200), National Natural Science Foundation of China (Grant Nos. 31702377 and 32072988), General Program of Natural Science Foundation of Guangdong Province, China (Grant Nos. 2018A030313963 and 2020A1515010319) and Research Fund Program of Guangdong Provincial Key Laboratory of Marine Resources and Coastal Engineering.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge NATIVE English Editing (www.nativeee.com) for its linguistic assistance during the preparation of this manuscript.
References
1. Alday-Sanz V, Brock J, Flegel TW, Mcintosh R, Bondad-Reantaso M, Salazar M, et al. Facts, truths and myths about spf shrimp in aquaculture. Rev Aquaculture (2018) 1:1–9. doi: 10.1111/raq.12305
2. Li CZ, Weng SP, He JG. WSSV-host interaction: Host response and immune evasion. Fish Shellfish Immunol (2019) 84:558–71. doi: 10.1016/j.fsi.2018.10.043
3. Thitamadee S, Prachumwat A, Srisala J, Jaroenlak P, Salachan PV, Sritunyalucksana K, et al. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture (2016) 452:69–87. doi: 10.1016/j.aquaculture.2015.10.028
4. Qiu L, Chen MM, Wan XY, Li C, Zhang QL, Wang RY, et al. Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei). Sci Rep (2017) 7(1):11834. doi: 10.1038/s41598-017-10738-8
5. Liao XZ, Wang CG, Wang B, Qin HP, Hu SK, Zhao JC, et al. Research into the hemocyte immune response of Fenneropenaeus merguiensis under decapod iridescent virus 1 (DIV1) challenge using transcriptome analysis. Fish Shellfish Immunol (2020) 104:8–17. doi: 10.1016/j.fsi.2020.05.053
6. Flegel TW, Sritunyalucksana K. Shrimp molecular responses to viral pathogens. Marine Biotechnol (NewYork NY) (2011) 13(4):587–607. doi: 10.1007/s10126-010-9287-x
7. Larner J, Villar-Palasi C, Goldberg ND, Bishop JS, Huijing F, Wenger JI, et al. Hormonal and non-hormonal control of glycogen synthesis-control of transferase phosphatase and transferase I kinase. Adv Enzyme Regul (1968) 6:409–23. doi: 10.1016/0065-2571(68)90025-3
8. Patel P, Woodgett JR. Glycogen Synthase Kinase 3: A Kinase for All Pathways? Curr Topics Dev Biol (2017) 123:277–302. doi: 10.1016/bs.ctdb.2016.11.011
9. Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J (1990) 9(8):2431–8. doi: 10.1002/j.1460-2075.1990.tb07419.x
10. Lin J, Song T, Li C, Mao W. GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim Biophys Acta (BBA) - Mol Cell Res (2020) 1867(5):118659. doi: 10.1016/j.bbamcr.2020.118659
11. Zhang S, Chen C, Ying J, Wei C, Qi F. Alda-1, an Aldehyde Dehydrogenase 2 Agonist, Improves Cutaneous Wound Healing by Activating Epidermal Keratinocytes via Akt/GSK-3β/β-Catenin Pathway. Aesthetic Plast Surg (2020) 44(3):993–1005. doi: 10.1007/s00266-020-01614-4
12. Terzioglu-Usak S, Nalli A, Elibol B, Ozek E, Hatiboglu MA. Anvirzel™ regulates cell death through inhibiting GSK-3 activity in human U87 glioma cells. Neurol Res (2020) 42(1):68–75. doi: 10.1080/01616412.2019.1709744
13. Müller KH, Kakkola L, Nagaraj AS, Cheltsov AV, Anastasina M, Kainov DE. Emerging cellular targets for influenza antiviral agents. Trends Pharmacol ences (2012) 33(2):89–99. doi: 10.1016/j.tips.2011.10.004
14. Hirata N, Suizu F, Matsuda-Lennikov M, Edamura T, Bala J, Noguchi M. Inhibition of Akt kinase activity suppresses entry and replication of influenza virus. Biochem Biophys Res Commun (2014) 450(1):891–8. doi: 10.1016/j.bbrc.2014.06.077
15. Saleh M, Rüschenbaum S, Welsch C, Zeuzem S, Moradpour D, Gouttenoire J, et al. Glycogen Synthase Kinase 3β Enhances Hepatitis C Virus Replication by Supporting miR-122. Front Microbiol (2018) 9:2949. doi: 10.3389/fmicb.2018.02949
16. Guendel I, Iordanskiy S, Van DR, Kylene KH, Mohammed S, Ravi D, et al. Novel neuroprotective GSK-3β inhibitor restricts Tat-mediated HIV-1 replication. J Virol (2014) 88(2):1189–208. doi: 10.1128/JVI.01940-13
17. Sarhan MA, Abdel-Hakeem MS, Mason AL, Tyrrell DL, Houghton M. Glycogen synthase kinase 3β inhibitors prevent hepatitis C virus release/assembly through perturbation of lipid metabolism. Sentific Rep (2017) 7(1):2495. doi: 10.1038/s41598-017-02648-6
18. Ruan LW, Liu HC, Shi H. Characterization and function of GSK3β from Litopenaeus vannamei in WSSV infection. Fish Shellfish Immunol (2018) 82:220–8. doi: 10.1016/j.fsi.2018.08.032
19. Cortés-Vieyra R, Bravo-Patiño A, Valdez-Alarcón JJ, Juárez MC, Finlay BB, Baizabal-Aguirre VM. Role of glycogen synthase kinase-3 beta in the inflammatory response caused by bacterial pathogens. J Inflammation (London England) (2012) 9(1):23. doi: 10.1186/1476-9255-9-23
20. Steinbrecher KA, Wilson W, Cogswell PC, Baldwin AS. Glycogen synthase kinase 3beta functions to specify gene-specific, NF-kappaB-dependent transcription. Mol Cell Biol (2005) 25(19):8444–55. doi: 10.1128/MCB.25.19.8444-8455.2005
21. Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol (2005) 6(8):777–84. doi: 10.1038/ni1221
22. Choi JY, Park HJ, Lee YJ, Byun J, Youn YS, Choi JH, et al. Upregulation of Mer receptor tyrosine kinase signaling attenuated lipopolysaccharide-induced lung inflammation. J Pharmacol Exp Ther (2013) 344(2):447–58. doi: 10.1124/jpet.112.199778
23. Deng LY, Zeng QR, Wang MS, Cheng AC, Jia RY, Chen S, et al. Suppression of NF-κB Activity: A Viral Immune Evasion Mechanism. Viruses (2018) 10(8):409. doi: 10.3390/v10080409
24. Li CZ, Wang S, He JG. The Two NF-κB Pathways Regulating Bacterial and WSSV Infection of Shrimp. Front Immunol (2019) 10:1785. doi: 10.3389/fimmu.2019.01785
25. Panichareon B, Khawsak P, Deesukon W, Sukhumsirichart W. Multiplex real-time PCR and high-resolution melting analysis for detection of white spot syndrome virus, yellow-head virus, and Penaeus monodon densovirus in penaeid shrimp. J Virol Methods (2011) 178(1-2):16–21. doi: 10.1016/j.jviromet.2011.07.010
26. Zhang S, Li CZ, Yang QH, Dong XH, Chi SY, Liu HY, et al. Molecular cloning, characterization and expression analysis of Wnt4, Wnt5, Wnt6, Wnt7, Wnt10 and Wnt16 from Litopenaeus vannamei. Fish Shellfish Immunol (2016) 54:445–55. doi: 10.1016/j.fsi.2016.04.028
27. Liao XZ, Wang CG, Wang B, Qin HP, Hu SK, Wang P, et al. Comparative Transcriptome Analysis of Litopenaeus vannamei Reveals That Triosephosphate Isomerase-Like Genes Play an Important Role During Decapod Iridescent Virus 1 Infection. Front Immunol (2020) 11:1–17. doi: 10.3389/fimmu.2020.01904
28. Zuo HL, Gao JF, Yuan J, Deng HW, Yang LW, Weng SP, et al. Fatty acid synthase plays a positive role in shrimp immune responses against Vibrio parahaemolyticus infection. Fish Shellfish Immunol (2017) 60:282–8. doi: 10.1016/j.fsi.2016.11.054
29. Zhang S, Li CZ, Yan H, Qiu W, Chen YG, Wang PH, et al. Identification and function of myeloid differentiation factor 88 (MyD88) in Litopenaeus vannamei. PloS One (2012) 7:e47038. doi: 10.1371/journal.pone.0047038
30. Zhang S, Shi LL, Lǚ K, Li HY, Wang SP, He JG, et al. Cloning, identification and functional analysis of a β-catenin homologue from Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol (2016) 54:411–8. doi: 10.1016/j.fsi.2016.03.162
31. Shi LL, Chan SM, Li CZ, Zhang S. Identification and characterization of a laccase from Litopenaeus vannamei involved in anti-bacterial host defense. Fish Shellfish Immunol (2017) 66:1–10. doi: 10.1016/j.fsi.2017.04.026
32. Li HY, Wang S, Qian Z, Wu ZZ, Lǚ K, Weng SP, et al. MKK6 from pacific white shrimp Litopenaeus vannamei is responsive to bacterial and WSSV infection. Mol Immunol (2016) 70:72–83. doi: 10.1016/j.molimm.2015.12.011
33. Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med (2009) 15:808–13. doi: 10.1038/nm.1982
34. Li CZ, Chen YH, Weng S, Li SD, Zuo HL, Yu XQ, et al. Presence of Tube isoforms in Litopenaeus vannamei suggests various regulatory patterns of signal transduction in invertebrate NF-κB pathway. Dev Comp Immunol (2014) 42:174–85. doi: 10.1016/j.dci.2013.08.012
35. Qiu W, He JH, Zuo HL, Niu SW, Xu XP. Identification, characterization, and function analysis of the NF-κB repressing factor (NKRF) gene from Litopenaeus vannamei. Dev Comp Immunol (2017) 76:83–92. doi: 10.1016/j.dci.2017.05.020
36. Li CZ, Li HY, Chen YX, Chen YG, Wang S, Weng SP, et al. Activation of Vago by interferon regulatory factor (IRF) suggests an interferon system-like antiviral mechanism in shrimp. Sci Rep (2015) 5:15078. doi: 10.1038/srep15078
37. Sharp PA, Burge CB. Classification of introns: U2-type or U12-type. Cell (1997) 91(7):875–9. doi: 10.1016/s0092-8674(00)80479-1
38. Alfhili MA, Alsughayyir J, McCubrey JA, Akula SM. GSK-3-associated signaling is crucial to virus infection of cells. Biochim Biophys Acta (BBA) - Mol Cell Res (2020) 1867:118767. doi: 10.1016/j.bbamcr.2020.118767
39. Chen X, Wang RZ, Liu X, Wu YM, Zhou T, Yang YJ, et al. A Chemical-Genetic Approach Reveals the Distinct Roles of GSK3α and GSK3β in Regulating Embryonic Stem Cell Fate. Dev Cell (2017) 43(5):563–76.e4. doi: 10.1016/j.devcel.2017.11.007
40. Li XW, Jia ZH, Wang WL, Wang LL, Liu ZQ, Yang B, et al. Glycogen synthase kinase-3 (GSK3) regulates TNF production and haemocyte phagocytosis in the immune response of Chinese mitten crab Eriocheir sinensis. Dev Comp Immunol (2017) 73:144–55. doi: 10.1016/j.dci.2017.03.022
41. Kim JY, Park SY, Lyoo HR, Koo ES, Kim MS, Jeong YS. Extended stability of cyclin D1 contributes to limited cell cycle arrest at G1-phase in BHK-21 cells with Japanese encephalitis virus persistent infection. J Microbiol (2015) 53:77–83. doi: 10.1007/s12275-015-4661-z
42. Rahaus M, Desloges N, Wolff MH. Varicella-zoster virus requires a functional PI3K/Akt/GSK-3α/β signaling cascade for efficient replication. Cell Signal (2007) 19(2):312–20. doi: 10.1016/j.cellsig.2006.07.003
43. Wu CH, Yeh SH, Tsay YG, Shieh YH, Kao CL, Chen YS, et al. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem (2009) 284(8):5229–39. doi: 10.1074/jbc.M805747200
44. Ning K, Wu Y, Meng Q, Wang Z, Zuo Y, Xi P, et al. Suppression of Virulent Porcine Epidemic Diarrhea Virus Proliferation by the PI3K/Akt/GSK-3α/β Pathway. PloS One (2016) 11:e0161508. doi: 10.1371/journal.pone.0161508
45. Qin D, Feng N, Fan W, Ma X, Yan Q, Lv Z, et al. Activation of PI3K/AKT and ERK MAPK signal pathways is required for the induction of lytic cycle replication of Kaposi’s Sarcoma-associated herpesvirus by herpes simplex virus type 1. BMC Microbiol (2011) 11:240. doi: 10.1186/1471-2180-11-240
Keywords: glycogen synthase kinase 3β, nuclear factor κB, negative regulation, white spot syndrome virus, Litopenaeus vannamei
Citation: Zhang S, Zhu L, Hou C, Yuan H, Yang S, Dehwah MAS and Shi L (2020) GSK3β Plays a Negative Role During White Spot Syndrome Virus (WSSV) Infection by Regulating NF-κB Activity in Shrimp Litopenaeus vannamei. Front. Immunol. 11:607543. doi: 10.3389/fimmu.2020.607543
Received: 17 September 2020; Accepted: 30 October 2020;
Published: 26 November 2020.
Edited by:
Chaozheng Li, Sun Yat-sen University, ChinaReviewed by:
Hongliang Zuo, Sun Yat-sen University, ChinaMuting Yan, South China Agricultural University, China
Copyright © 2020 Zhang, Zhu, Hou, Yuan, Yang, Dehwah and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Shi, c2hpbGxAZ2RvdS5lZHUuY24=
 Shuang Zhang
Shuang Zhang Lulu Zhu1
Lulu Zhu1