Corrigendum: A Detailed View of KIR Haplotype Structures and Gene Families as Provided by a New Motif-Based Multiple Sequence Alignment
- 1Bioinformatics and Computational Biology, University of Minnesota, Rochester, MN, United States
- 2Center for International Blood and Marrow Transplant Research, Minneapolis, MN, United States
- 3Clinical Research Division, Fred Hutchinson Cancer Research Center, Seattle WA, United States
- 4Department of Computer Science and Engineering, University of Minnesota, Minneapolis, MN, United States
Human chromosome 19q13.4 contains genes encoding killer-cell immunoglobulin-like receptors (KIR). Reported haplotype lengths range from 67 to 269 kb and contain 4 to 18 genes. The region has certain properties such as single nucleotide variation, structural variation, homology, and repetitive elements that make it hard to align accurately beyond single gene alleles. To the best of our knowledge, a multiple sequence alignment of KIR haplotypes has never been published or presented. Such an alignment would be useful to precisely define KIR haplotypes and loci, provide context for assigning alleles (especially fusion alleles) to genes, infer evolutionary history, impute alleles, interpret and predict co-expression, and generate markers. In order to extend the framework of KIR haplotype sequences in the human genome reference, 27 new sequences were generated including 24 haplotypes from 12 individuals of African American ancestry that were selected for genotypic diversity and novelty to the reference, to bring the total to 68 full length genomic KIR haplotype sequences. We leveraged these data and tools from our long-read KIR haplotype assembly algorithm to define and align KIR haplotypes at <5 kb resolution on average. We then used a standard alignment algorithm to refine that alignment down to single base resolution. This processing demonstrated that the high-level alignment recapitulates human-curated annotation of the human haplotypes as well as a chimpanzee haplotype. Further, assignments and alignments of gene alleles were consistent with their human curation in haplotype and allele databases. These results define KIR haplotypes as 14 loci containing 9 genes. The multiple sequence alignments have been applied in two software packages as probes to capture and annotate KIR haplotypes and as markers to genotype KIR from WGS.
Introduction
The proteins of the killer-cell immunoglobulin-like receptor (KIR) family are important to human health. Whether or not these transmembrane receptors on natural killer (NK) cells bind with peptide-bound human leukocyte antigen (HLA) class I molecules determines how they help educate, activate, and inhibit NK cell functionality, including cytotoxicity and cytokine release. The accumulated evidence seems to indicate that KIR evolved with HLA to balance its pathogen defense effects with its effects on reproduction via the embryo-uterus interface (1–3). KIR effects are generally tissue-specific and tissue-variable. Within a given NK cell, KIR expression is stochastic, depending on the epigenetic profiles, inter- and intra-genic content of the haplotypes, binding alleles or lack thereof, methylation status, alternative splicing, and randomness (4–6). Besides investigating KIR in the context of viruses and pregnancy, medicine is also studying its effects in cancer, hematopoietic stem cells transplants, various autoimmune diseases, and immunotherapy (7–9).
KIR gene names reflect their protein structures (10). First the prefix “KIR,” followed by the number of extracellular domains (“2D” or “3D”), followed by a short or long intracellular domain (“S,” L”), followed by an index. KIR2DS1, for example, has two extracellular domains (“2D”) and a short intracellular domain (“S”); it is the first gene named with that structure (“1”), and KIR2DS2 is the second. The Immuno Polymorphism Database for KIR (IPD-KIR) names KIR gene alleles, records their DNA-RNA-protein relationships, and annotates each gene’s alleles in a multiple sequence alignment (11). It contains over 300 full-length DNA and almost 1,000 protein reference alleles in the latest release, 2.9.0.
There is no equivalent of IPD’s allele annotation for haplotypes. KIR haplotypes have no official nomenclature, although the majority of contributions to the human genome reference use a convention set by Pyo et al. in 2010 (12) and 2013 (13). The haplotype names reflect the two-part structure of the region: a proximal centromeric (“c”) region is paired with (“~”) a distal telomeric (“t”) region. Each region is a variant of a “A” haplotype or “B” haplotype family, followed by a two-digit index. The haplotype named “cB02~tA01,” for example, is comprised of the second centromeric B region (“cB02”) in cis with (“~”) the first telomeric A region (“tA01”).
Although there are many publications that analyze gene or intergene allele alignments, none report full haplotype multiple sequence alignments (MSAs), and therefore the haplotype nomenclature has not been formalized. Allele gene assignments are evaluated largely by amplification primers or sequence similarly to other alleles, as opposed to location in its haplotype. To help solve these issues, we present a simple bioinformatics approach to represent KIR haplotypes as a string of alignment-based motifs. We applied it by creating a full-haplotype DNA MSA for all 68 human haplotype sequences in the human genome project and a chimp haplotype for an outlier.
In our recent preprint describing a KIR long-read assembler (14), we show that 18 120 bp sequences can be used to capture full haplotype KIR DNA from PacBio circular consensus sequencing (CCS) reads. In this manuscript, we show when those probe sequences are aligned to assembled haplotypes, the pattern of the probes (“motifs”) provides a structural annotation. These alignment locations of the motifs allow the haplotype sequences to be represented accurately and efficiently at the structural level. We leveraged that annotation to align the 68 published human KIR haplotypes and one chimpanzee that haplotype to within an average of 2,398 bases, and then we micro aligned each locus for precise full haplotype multiple sequence alignment. The results are concordant with the annotation in the human genome reference and reveal 13 structural haplotypes for the 68 human haplotype sequences. The MSA also shows the haplotypes have 14 loci containing 9 genes. KIR2DS4 shares a locus with KIR2DS3 and KIR2DS5, as opposed to KIR2DS1 or by itself. Sharing a gene motif with locus KIR2DS3/KIR2DS4/KIR2DS5 is the shared locus KIR2DL2/KIR2DL3. KIR2DL1, KIR2DS1, and KIR2DS2 share a gene motif at different loci, as do KIR2DL5A and KIR2DL5B.
The MSA includes this study’s contribution of 27 new haplotypes to the human genome project, including 24 haplotypes from 12 individuals of African American ancestry.
Materials and Methods
Source Sequences and Annotation
The source sequences consisted of two newly assembled haplotypes from an Ashkenazim individual (from the assembler preprint), along with all 66 full-sequence alternative haplotypes in the human genome reference (12, 13, 15, 16) and a chimpanzee haplotype (GenBank accession AC155174.2) (17). The two Ashkenazim haplotypes were assembled with PacBio reads obtained from the Genome In a Bottle (GIAB) consortium (18) and have not yet been submitted to genetic databases. All other haplotypes were generated by physically separating chromosomes via fosmid cloning and then sequencing and assembling fragments into full haplotype sequences. The 68 include 27 new sequences first described in this manuscript, including 24 haplotypes from 12 individuals of African American ancestry that were selected by genotypic diversity and/or lack of representation in the human genome reference. An additional three haplotypes (one Asian, two European) of opportunity from two individuals were contributed by Scisco Genetics, who sequenced all non-GIAB haplotypes following previously reported protocols (12, 13). The GenBank entries also provided structural and allelic annotation of all the human haplotypes, except for the two Ashkenazim haplotypes. Broken down by population, the haplotype counts include 31 African or African American, 3 Asian, 2 Ashkenazim, 24 European, 2 Guarani South Amerindian, 2 Romani in Spain, and 4 unknowns. The structures are depicted schematically in Figure 1, excluding cA01~tB04, which contains a large insertion and was excluded for compact visualization.
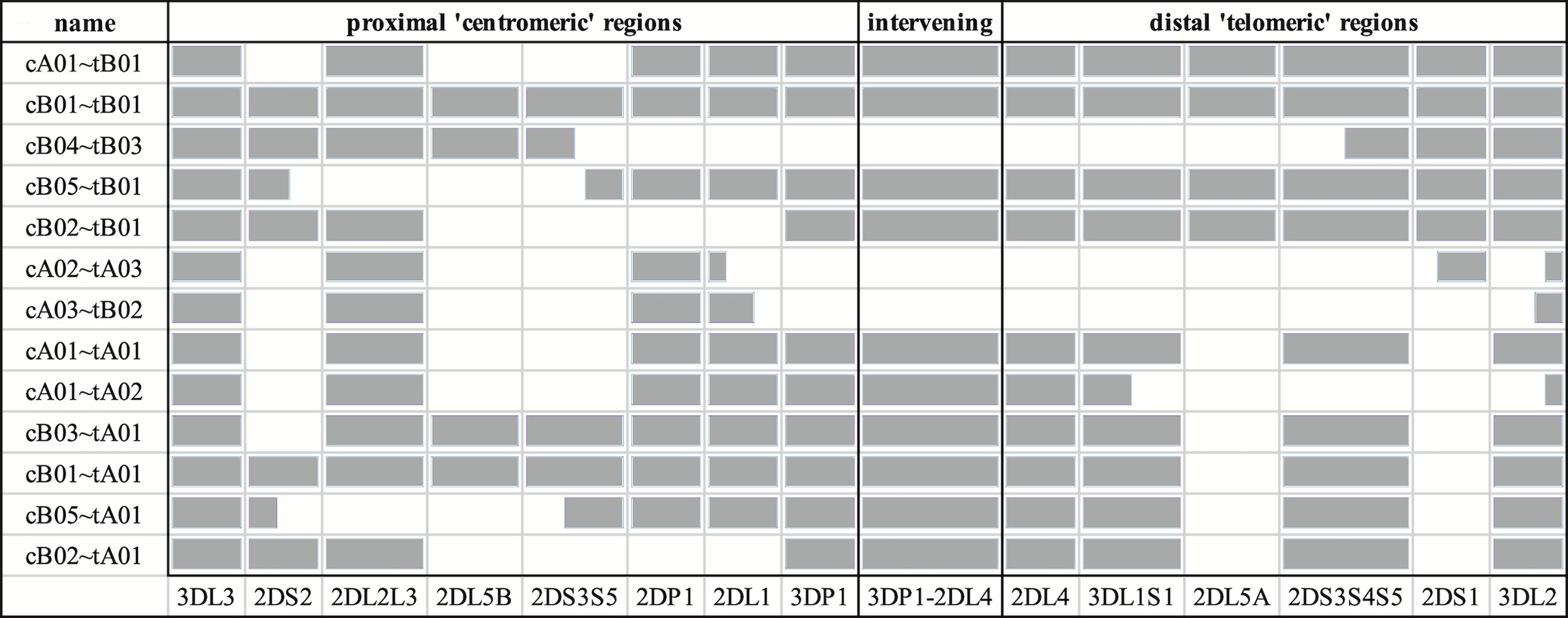
Figure 1 Schematic representation of the informal names and definitions of 13 human haplotype structures. The informal names of the haplotypes are in the first column. The definitions of the centromeric and telomeric regions are in the top row. The abbreviated names of the genes are in the bottom row; the intergenic region between KIR3DP1 and KIR2DL4 is represented as “3DP1-2DL4.” Gray cells indicates the presence of alleles for that gene; white indicates absence of an allele. Some cells are partially colored to indicate two-gene fusion alleles. The names of the haplotypes, as well as the gene content and locations are taken from the human curated annotations in the human genome reference. Haplotype cA01~tB04 contains a large insertion and was excluded for visualization.
Workflow Overview
The result of the workflow was a multiple sequence alignment of 68 human and 1 chimp haplotypes. As shown in Figure 2, the first step was to align the 18 capture probes to the haplotype sequences (Figure 2A). Next, the sequence between each pair of probes (e.g., probe 4 to probe 3) was assigned a letter (e.g., C) (Figure 2B). In this way, each haplotype can be represented by a string (“motifs,” e.g., “…CIJKL…”) whose length is 105 characters or less. These haplotype motifs were aligned to produce a high-level MSA (Figure 2C). The motifs allowed loci to be located in the haplotype sequences. The sequences for all alleles of each locus was aligned separately, and then the MSAs were concatenated to create the full haplotype DNA MSA (Figure 2D). In this two-alignment approach, the motif alignment provides structural annotation and the locus-specific DNA alignment provides base-level accuracy. The following paragraphs detail each step.
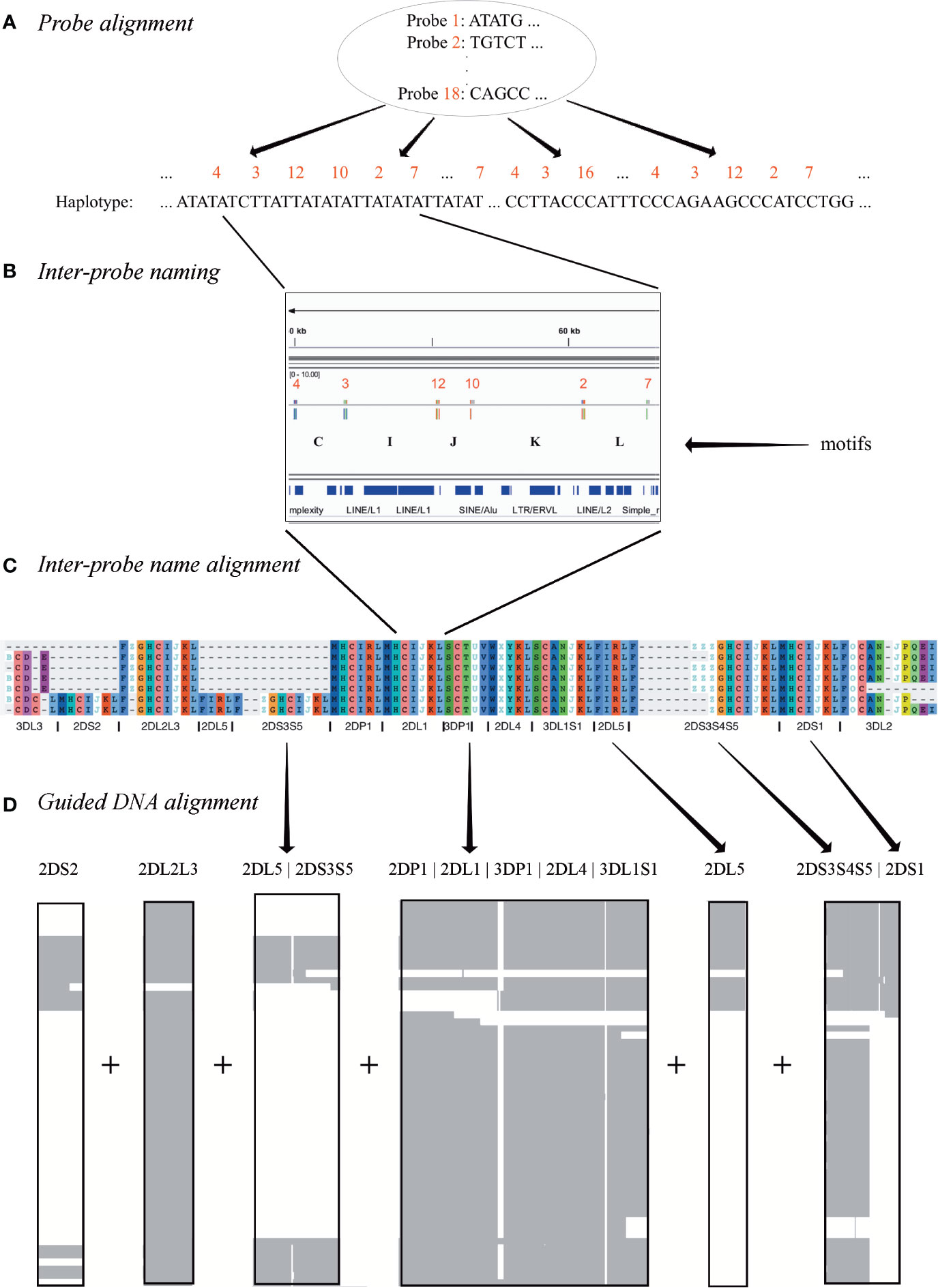
Figure 2 Multiple sequence alignment (MSA) workflow. Step (A) depicts the alignment of 18 120 base probes to each haplotype sequence. Step (B) shows how each ordered pair of probes in the alignment is assigned a letter. For example, the letter “I” represents the sequence between the alignment of probe 3 and probe 12. Step (C) shows how each haplotype is depicted by a string of letters, how a haplotype motif MSA is generated from them, and how loci are defined in the MSA. Step (D) shows how the DNA in the motif-defined loci were separately aligned and then joined to create the final MSA.
Probe Alignment and Inter-Probe Naming
The assembler preprint describes a method that uses 18 120 bp probes to capture KIR PacBio long-read sequences and assemble them into haplotypes with an average of 97% coverage and 99.7% concordance compared with reference sequences. When the probes are aligned to the haplotypes (Figure 2A), they align every 2,398 bases on average. Probe locations are discovered by alignment with bowtie2 with options “-a –end-to-end –rdg 3,3 –rfg 3,3.” The alignment order of the 18 probes across a haplotype sequence allows that haplotype structure to be succinctly annotated as sequences of probe pairs (Figure 2B). For example, assume the alignment of probe 4 followed by probe 3 is called “C,” probe 3 followed by probe 12 is called “I,” and probe 12 followed by probe 10 is called “J.” Then the region “probe 4 to probe 3 to probe 12 to probe 10” can be called “CIJ.” Probe 1 repeating once in the alignment is “Z” and twice is “ZZ,” etc. There are 42 such probe pairs in this collection of 69 haplotypes. In this way, haplotypes can be briefly annotated as strings of a 42-character alphabet. See Figure 3 for an example of KP420440, whose full motif is MHCIJKLFGHCIJKLAIRLFZGHCIJKLMHCIRLMHCIJKLSCTUVWXYKLSCNOJKLAIRLFZGHCIJKLMHCIJKLFPCNOJQ. The motif pairs are defined in Supplementary Table 1; the probe sequences are defined in the assembly manuscript and are also available via GitHub at https://github.com/droeatumn/kpi/tree/master/input.
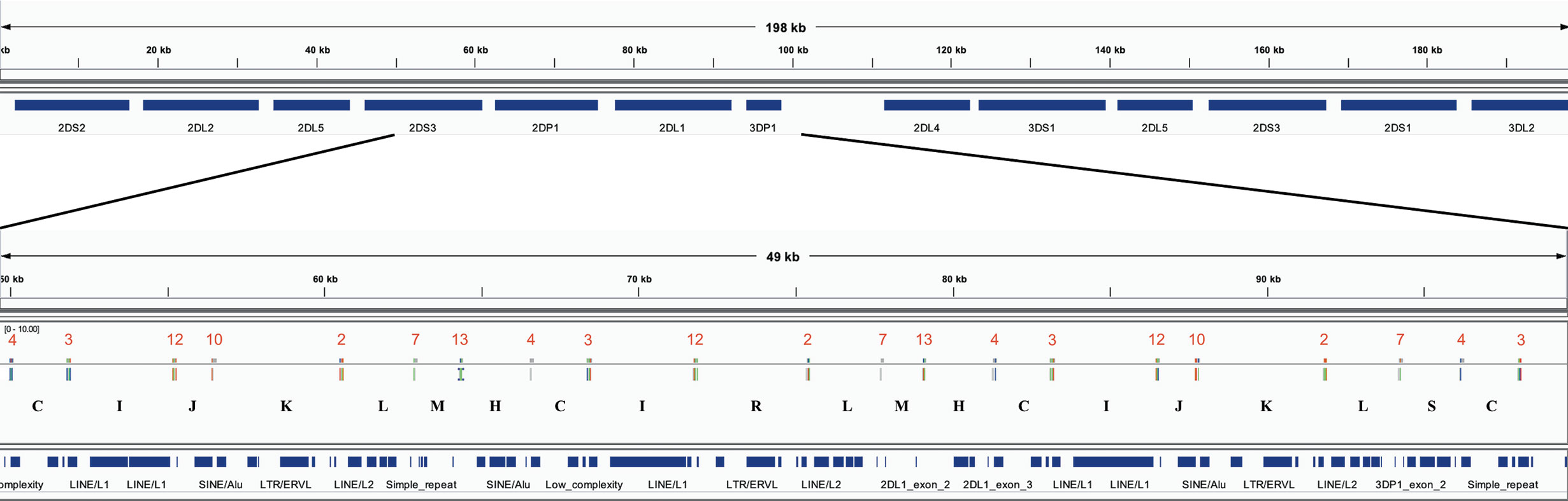
Figure 3 Example of the motif-generating alignment. The top frame shows the annotation for 198 kb haplotype KP420440 (cB01~tB01). The bottom frame zooms in to a 49 kb region depicting the alignment of the probes. In the middle of the bottom track, the locations of the probes are displayed by the vertical ticks with the red numbers above them and black letters below them. The red numbers label each probe, and the black letters label each probe pair (i.e., motif). The locations of exons and repeat elements (horizontal blue bars) are on the bottom track. Seven distinct probes align in this window. The motif pattern CIJKL occurs in two locations: 50–63kb and 81–94kb; the variation CIRL occurs 66–79kb.
Inter-Probe Name Alignment
A full-haplotype multiple sequence alignment of the probe motifs was generated for the 68 human haplotypes plus the chimpanzee as an outgroup (Figure 2C). Alignment of the motifs was created with MAFFT (19), but mostly aligned manually with Aliview 1.2.6; manual alignment was required to resolve ambiguities, as the motif alphabet is not directly supported by DNA or protein aligners. Ambiguous alignments were resolved by following the human curation of the reference haplotypes.
DNA Alignment
MAFFT was used to merge or add these haplotypes in the order cB02~tA01, cA01~tA01, cA02~tA01, cB02~tB01, cA0X~tB0X, cB0X~tA01, and cB0X~tB0X, where “X” a general number not already used. Then, the full-length DNA sequences were separated into sets as defined by their motif structures (Figure 2D). Each set was aligned separately with MAFFT. The alignment was then edited manually by adding or deleting gaps to conform to locations in the probe motif MSA. Using the loci defined in the motifs, and the motif locations defined in the DNA, the alignment was refined for all alleles in each locus with MUSCLE in Aliview. A high-level depiction of the alignment was created with Jalview 2.11.0’s “Overview Window” functionality and NCBI’s Multiple Sequence Alignment Viewer 1.13.1.
The MSA was validated at the structural level by showing that it recapitulates the human curation of the reference haplotypes in GenBank and the human genome reference. The alignment was validated at the allele level by showing each allele is assigned and annotated as expected with respect to IPD-KIR classifications.
The ability of existing software to align the 69 haplotypes was also evaluated with MAFFT stand-alone (–thread 19 –threadtb 10 –threadit 0 –reorder –adjustdirection –anysymbol –leavegappyregion –kimura 1 –auto), PASTA v1.8.5 (docker run –rm -it -v $PWD:/opt/droe smirarab/pasta run_pasta.py), Clustal Omega v1.2.4 (clustalo –threads=29), M-Coffee metaserver 13.41.0.28, MUSCLE v3.8.31 (default parameters), and WebPrank (Updated 8 October, 2017). Implementation was conducted on the web servers when possible or a server running Ubuntu 18.04.4 LTS with 32 AMD Opteron (TM) Processor 6220 and 200 GB RAM and maximum java heap space set to -Xmx100G.
Results
Haplotype References
The African American haplotype sequences MN167504-MN167530 were deposited in GenBank; Supplementary Table 2 contains details, including allele calls as IPD-KIR names for all 68 haplotypes in the human genome reference. Structural annotation for the new sequences was confirmed by the workflow by Pyo et al. for annotation of the previously-submitted reference haplotypes. The 27 haplotypes include 8 cA01~tA01, 4 cA01~tA02, 4 cB01~tA01, 3 cB01~tB01, 3 cB03~tA01, 2 cA01~tB01, 2 cB02~tA01, and 1 cB01~tB01. Haplotype MN167506 (cB02~tB01), presumed to be of Asian ancestry, contains a KIR2DL5B allele (KIR2DL5B*00804) at the KIR2DL5A locus. Four haplotypes (MN167507, MN167509, MN167516, MN167530) with a KIR3DL1/KIR3DL2 fusion (10) have been deposited in the human genome reference for the first time; they have been labeled as telomeric region “tA02.” Some of the diversity of structures in the African American cohort are depicted visually in Figure 4. In total, the 68 human haplotype structures consist of 27 cA01~tA01, 7 cB01~tA01, 6 cB01~tB01, 6 cB02~tA01, 5 cA01~tB01, 4 cA01~tA02, 4 cB03~tA01, 3 cB02~tB01, and 1 each for cA01~tB04, cA02~tA03, cA03~tB02, cB04~tB03, cB05~tA01, cB05~tB01.
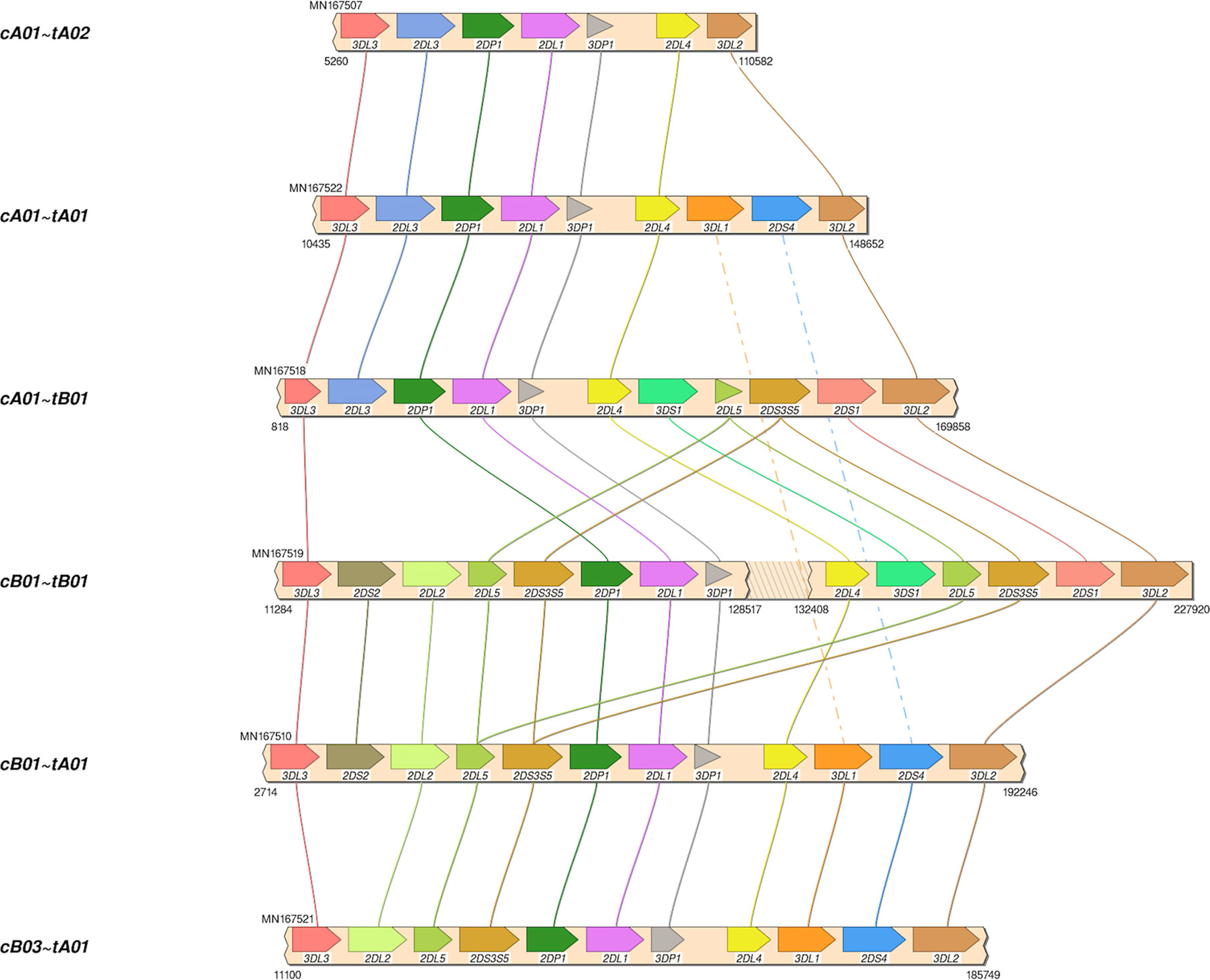
Figure 4 Haplotype structures in African American cohort. Shown are six of the eight structures in the African American cohort, which was added to the human genome project as part of this study. Not shown are cB02~tA01, and 1 cB01~tB01.
Validation of the Multiple Sequence Alignment by Annotation in Human Genome Reference
Figure 5 shows a multiple sequence alignment of the probe motifs for the human haplotypes, excluding insertion-containing cA01~tB04 for space considerations. It recapitulates the human-annotated structures in Figure 1 with 105 positions. The common ~140 kb cA01~tA01 haplotypes are marked up in ~63 motif characters, and the ~220 kb cB01~tB01 haplotypes are marked up in ~94 motif characters. The average distance between probes is 2,398 bases; the maximum distance for non-KIR3DL3 genes is ~5,700 bases and for KIR3DL3 it is ~7,800 bases. The haplotype motifs subdivide into 14 genic and 1 intergenic (KIR3DP1-KIR2DL4) loci. From 5’ (left) to 3’ (right) at the bottom of Figure 5, the 15 abbreviated loci are: 3DL3, 2DS2, 2DL2L3, 2DL5, 2DS3S5, 2DP1, 2DL1, 3DP1, 3DP1-2DL4, 2DL4, 3DL1S1, 2DL5, 2DS3S4S5, 2DS1, and 3DL2. The 14 genic loci consist of 9 distinct motifs: 2DL1, 2DS1, 2DS2 share MHCIJ; 2DL5A and 2DL5B share FIRL; 2DS3, 2DS4, and 2DS5 share FZ+GHCIJKL. The genes can be summarized as a short motif, or in some cases a regular expression. For example, FZ+GHCIJKL means: a F followed by one-or-more Zs followed by GHCIJKL. Supplementary Data Sheet 1 contains the motif MSA from Figure 5. Supplementary Data Sheet 2 adds cA01~tB04 (KU645196) and the chimpanzee haplotype to the MSA. The chimp haplotype contains 55 motif characters, encoding genes that align to the human KIR3DL3~KIR2DS2~KIR2DP1~KIR2DL1~KIR3DP1~KIR2DL4~KIR3DL1S1~KIR3DL2.
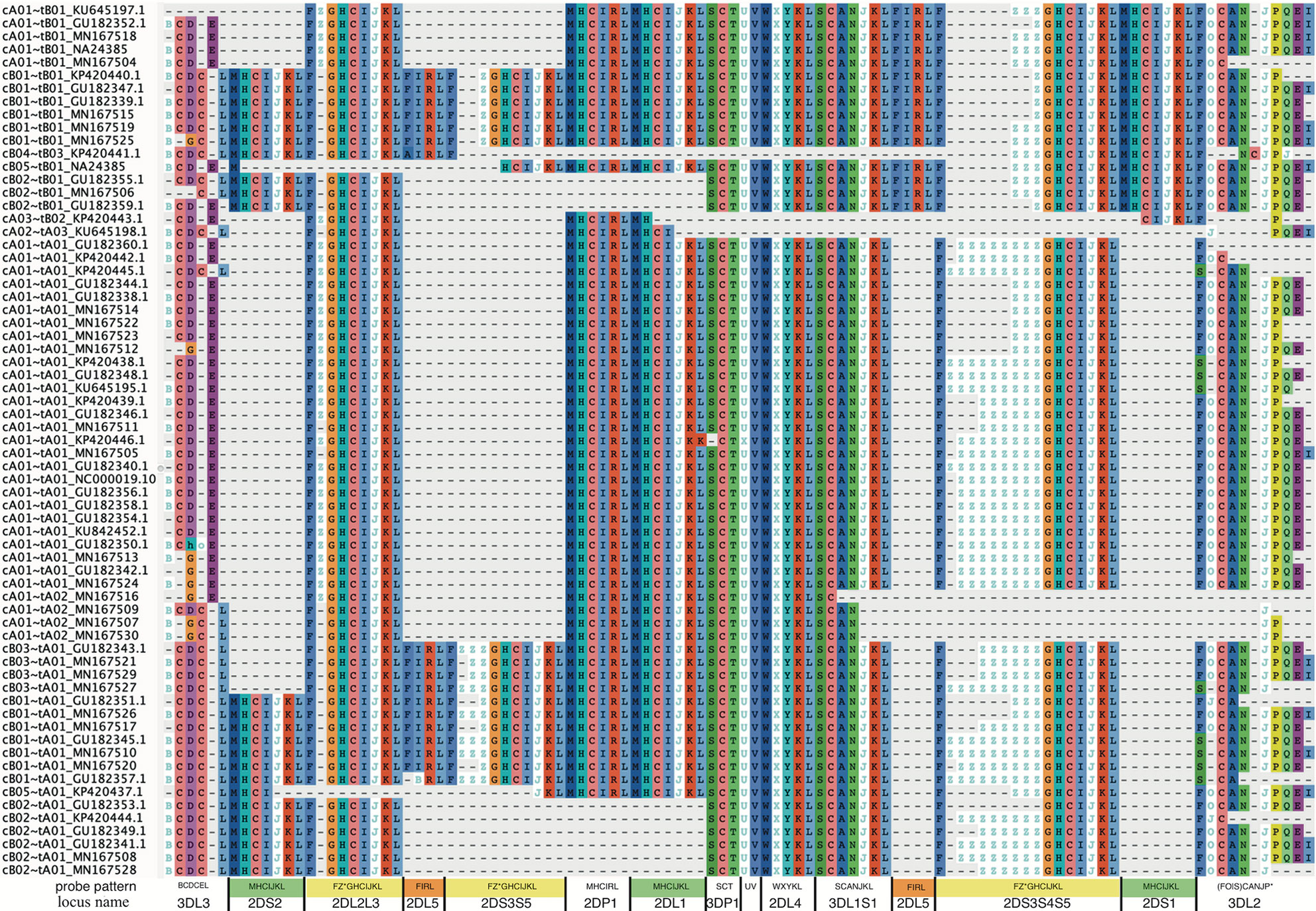
Figure 5 Probe motif multiple sequence alignment of the human haplotypes. The informal haplotype name and GenBank accession number are in the first column. The consensus motif pattern with each abbreviated locus name is described in the bottom row. Some loci share motifs: KIR2DL1, KIR2DS1, KIR2DS2 (green); KIR2DL5A and KIR2DL5B (orange); KIR2DS3, KIR2DS4, and KIR2DS5 (yellow). Some gene names are combined when they align at the same locus: KIR2DL2 and KIR2DL3 (2DL2L3), KIR2DS3 and KIR2DS5 centromeric side (2DS3S5), KIR2DS3, and KIR2DS4, and KIR2DS5 telomeric side (2DS3S4S5), and KIR3DL1 and KIR3DS1 (3DL1S1). Haplotype cA01~tB04, which contains a 106 kb insertion, was omitted for visualization.
Haplotype sizes by number of loci (not including the KIR3DP1-KIR2DL4 intergenic region) range from four loci (cA04~tA03) to 18 (cA01~tB04). Figure 6 displays a phylogenetic tree from the motifs of the 69 haplotypes, including cA01~tB04 and the chimp haplotype; the clusters recapitulate the their human-annotated names shown on the right part of the figure.
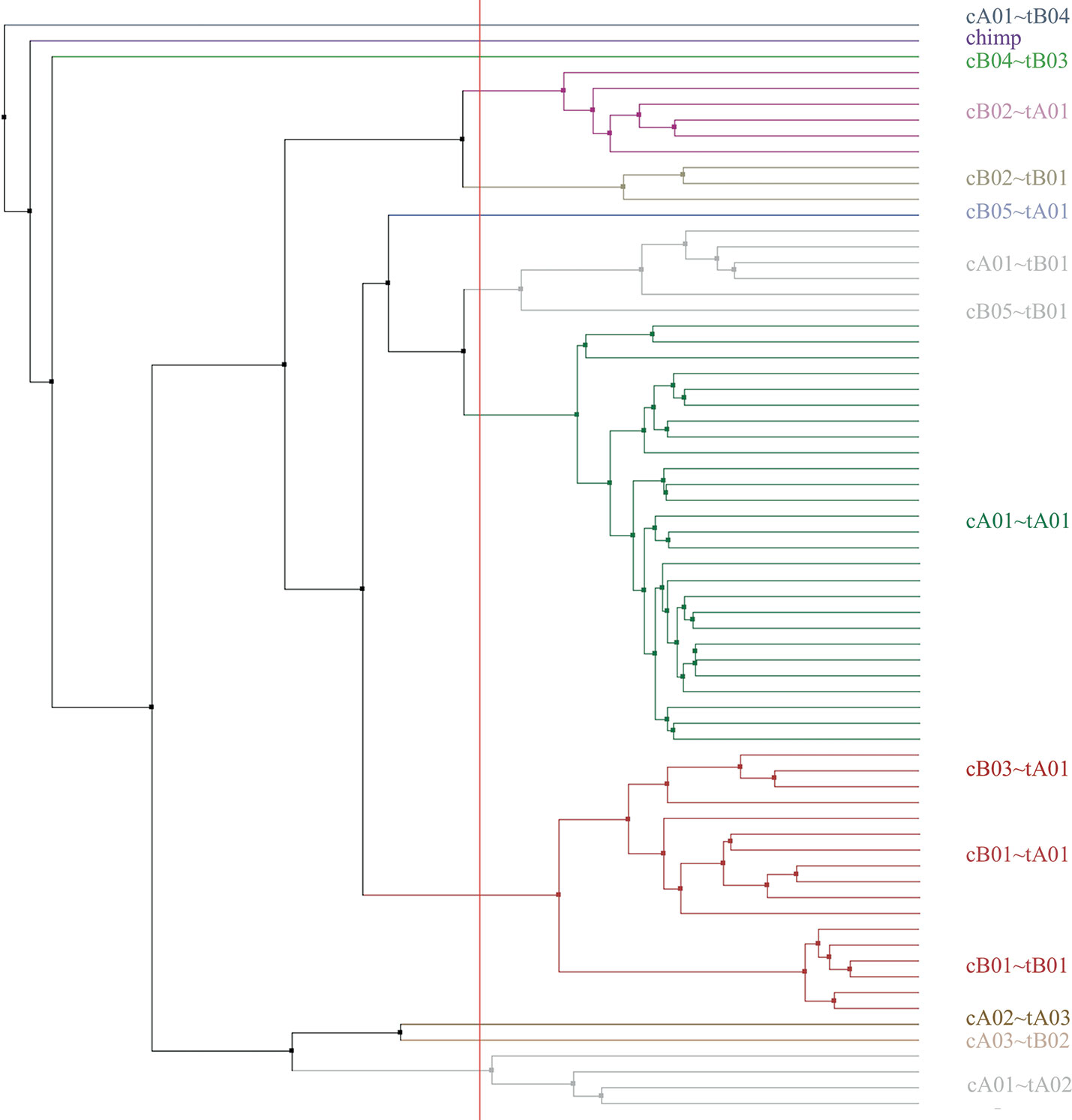
Figure 6 Phylogenetic tree made from the multiple sequence alignment (MSA) from the motifs of all the human haplotypes plus the chimpanzee (n=69). The leaf descriptions on the right include the informal name of the structural haplotype as documented in GenBank. The leaves are colored by their alignment order and the grouping defined by the red vertical line. The tree may not reflect the evolutionary history.
The DNA alignment of the 67 human haplotypes (minus cA01~tB04) has 263,556 positions; it is included in Supplementary Data Sheet 3. Figure 7B shows an overview of the DNA haplotype alignment as generated by Jalview’s “Overview Window” function; the names of the haplotypes were added after exporting the overview image from Jalview. The pattern of white (gaps) and gray (aligned sequences) mirror the patterns in the schematic depictions of the human curated haplotypes in Figure 1 (recapitulated in Figure 7A). This evidence shows the DNA alignment matches the human curation at a structural level.
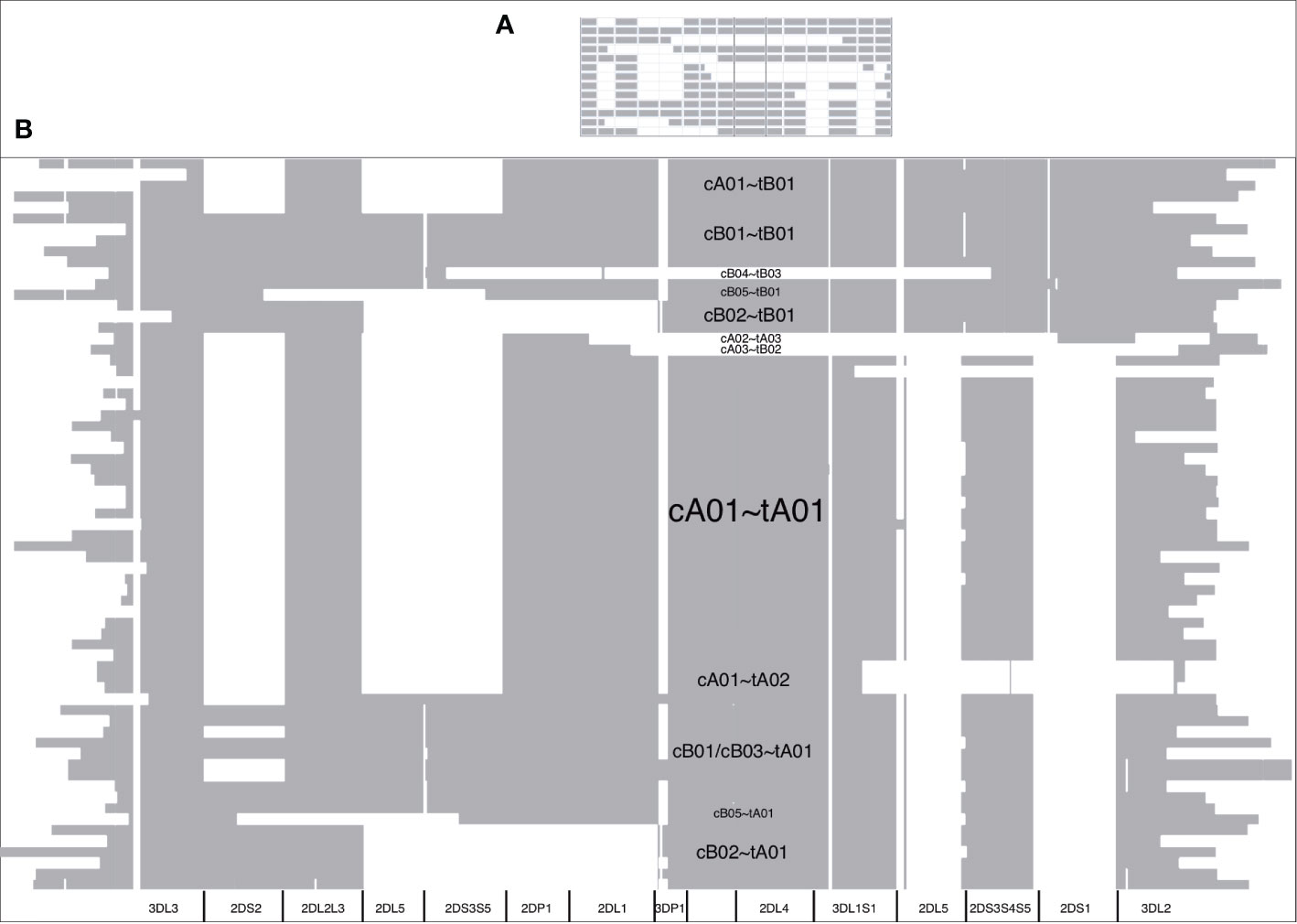
Figure 7 DNA multiple sequence alignment (MSA) overview. The cartoon of the GenBank annotations from Figure 1 is in (A) and is included to compare with the Jalview overview of the DNA full-haplotype MSA from the 67 haplotypes in (B). The gray denotes alignment of the DNA bases. White denotes gaps between the sequences. cA01~tB04 is excluded for better visualization.
Figure 8 shows a view of the alignment of the 67 haplotypes using NCBI’s MSA Viewer. Positions that are in an agreement with consensus are colored in gray, positions that are not in an agreement with consensus are colored in red. In this tool, consensus includes gaps (lack of an allele at that position). For KIR, this causes the regions specific to B haplotypes to show as pure red, and the other regions to show as mixture of red and gray.
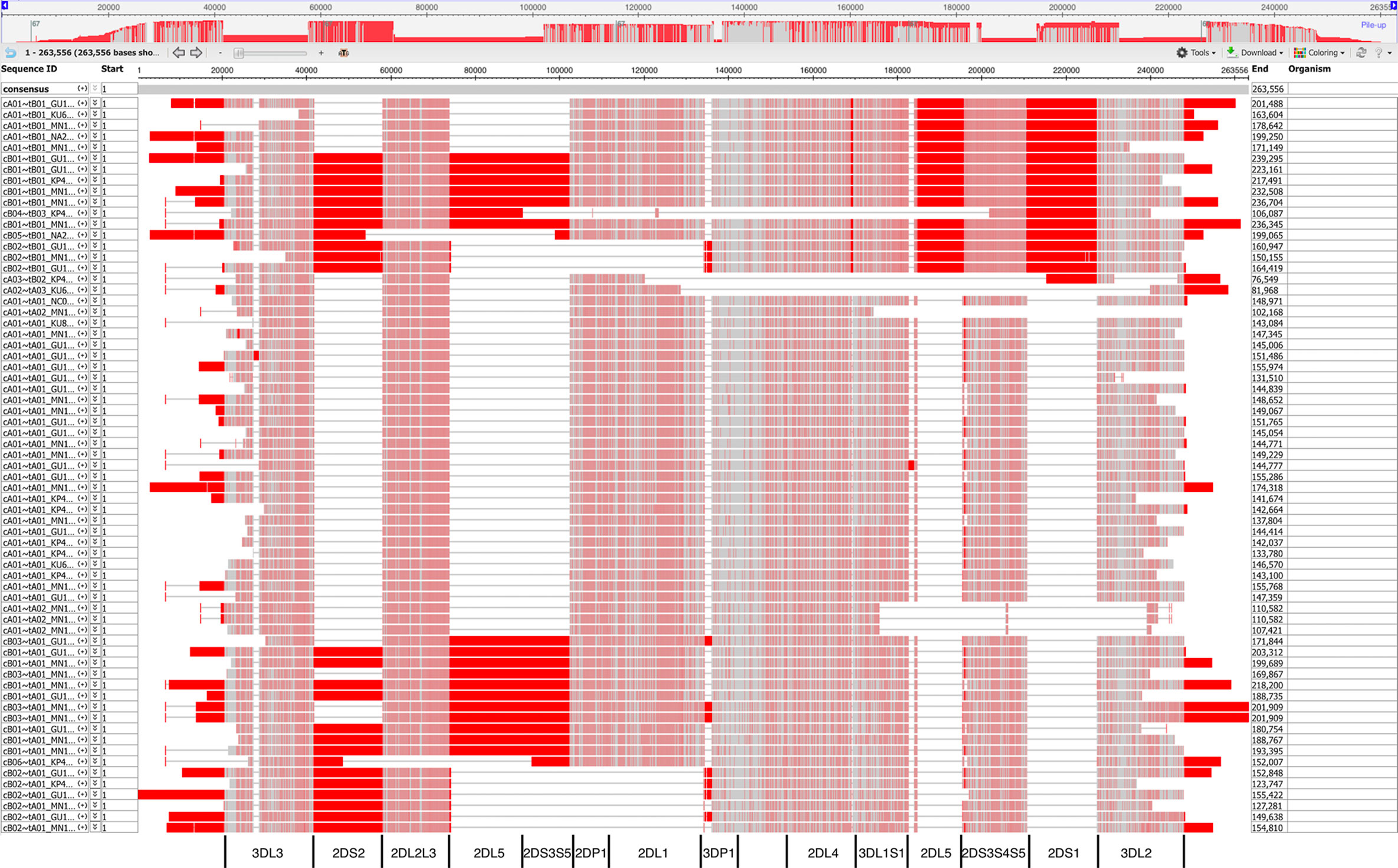
Figure 8 Multiple sequence alignment (MSA) Viewer depiction of the DNA alignment of 67 human haplotypes. Positions that are in an agreement with consensus are colored in gray, positions that are not in an agreement with consensus are colored in red. The location and borders of the genes have been added at the bottom.
Validation of the Multiple Sequence Alignment-Defined Allele Assignments by Immuno Polymorphism Database for KIR Annotation
The haplotypes are annotated by the names and orders of their alleles in Supplementary Table 2. Of the 647 alleles in the 68 haplotypes, 556 (86%) can be assigned names via IPD-KIR 2.9.0, at least at protein resolution. Unnamed alleles occur either because they have not yet been included in the IPD-KIR database, or they are partial alleles in the case of KIR3DL3 and KIR3DL2. 97% of alleles can be named excluding KIR3DL3, KIR2DP1, and KIR3DL2. 39 KIR2DP1 alleles are unnamed. To check the gene assignment for the 91 alleles that could not be named, the sequences were aligned to a set of 17 full gene alleles, each the first named allele for its assigned gene in IPD-KIR. Each allele being evaluated was considered to be correctly annotated if the allele sequence aligned closet to the IPD-KIR reference to which it was assigned by the motifs. Those results show that every unnamed allele aligned to the reference allele predicted by its motif assignment. The only exception was the GU182360 KIR3DL2, which aligns closest to KIR3DP1; however, this haplotype sequence is incomplete on the 3’ end, and the KIR3DL2 allele only contains the 2,221 5’-end sequences. Alleles that are a fusion of KIR3DL1 and KIR3DL2 (e.g., KIR3DL1 alleles 059-061) are classified as KIR3DL2 alleles using motifs but are classified as KIR3DL1 alleles in IPD-KIR.
Comparison with Existing Multiple Sequence Alignment Software
Of the existing alignment software that was evaluated on the 67 human haplotypes, only stand-alone MAFFT was able to generate an alignment, using the FFT-NS-2 strategy. The input size was too large for Clustal Omega local server, M-Coffee web server, MUSCLE local server, and WebPrank web server, and the other algorithms ran out of memory. The MAFFT alignment, output, and overview are included in Supplementary Data Sheet 4 and Supplementary Image 2. Except for large portions of KIR3DL3, distal KIR2DL3/KIR2DS4/2DS3S5, and KIR3DL2, the MAFFT alignment does not recapitulate human-curated KIR haplotypes or gene alleles. It is 75% larger than our motif-guided alignment (960,221 vs. 263,556 positions), and its overview shows the alignment columns are mostly gaps compared with the more expected block shaped column in Figure 7.
Discussion
Although our evaluation of existing MSA software methods was not exhaustive, we believe it is unlikely that any current general-purpose alignment software can align all human KIR haplotype sequences consistent with the human curation. Alignment of the 68 sequences, each 67–269 kb, each homologous with itself and every other haplotype over lengths of 15 kb is very challenging without prior knowledge. Scoring matrices and algorithms like dynamic programming do not generally support sequences of this size under conditions where relatively young gene regions have duplicated both within and between haplotypes. To the best of our knowledge, a MSA of KIR haplotypes has never been published, despite the fact that dozens of haplotype sequences have been public for years.
Figures 5 and 6 demonstrate that the capture probe alignment motifs alone can annotate KIR haplotype sequences in a way that recreates the human curated annotation in GenBank. Figures 7 and 8 similarly show the accuracy of the DNA alignment; they demonstrate that the DNA annotated by the motifs recreates the assignment of the alleles in IPD-KIR. Also, for two practical examples, the capture probes have demonstrated the ability to assemble haplotypes with an average of 97% coverage (preprint), and the DNA MSA was used to discover gene markers that can genotype whole genome sequences (WGS) with almost perfect accuracy (11) (preprint). The combination of multi-resolution validation and software applications demonstrates that both the motif and DNA alignments are likely to be accurate.
Figure 5 (alignment in Supplementary Data Sheet 1 suggest 14 human KIR loci and 1 intergenic locus. The genic loci are abbreviated and labeled as 3DL3, 2DS2, 2DL2L3, 2DL5B, 2DS3S5, 2DP1, 2DL1, 3DP1, 2DL4, 3DL1S1, 2DS3S4S5, 2DS1, 3DL2, and the intergenic locus is between 3DP1 and 2DL4 (3DP1-2DL4). There are nine motif-defined genes in the 14 genic loci (Figure 5). Loci abbreviated 2DL1, 2DS1, and 2DS2 share motif MHCIJKL. Loci 2DL2L3, 2DS3S5, and 2DS3S4S5 share variations of the FZ*GHCIJKL motif. Loci 2DL5A and 2DL5B share variations of FIRL. The motif MSA suggests KIR2DS4 on the A haplotype shares a locus with KIR2DS3 and KIR2DS5 (not KIR2DS1) on the B haplotypes. The MSA similarly suggests KIR2DL2 and KIR2DL3 share a locus as do KIR3DL1 and KIR3DS1. The Z character is unique to centromeric 2DS3S5 and telomeric 2DS3S4S5 loci and marks the alleles to a certain extent. In both, the KIR2DS3 alleles have 1 Z in their motifs, KIR2DS5 has 2 or 3, and KIR2DS4 alleles are more variable.
Although the motifs correctly annotate the haplotype structures, they do not always do so unambiguously, nor do they always agree with the existing nomenclature as utilized in IPD-KIR. In the current gene nomenclature, there are 16 KIR genes. Some researchers have previously considered KIR2DL2 and KIR2DL3 to occupy the same locus as well as KIR2DS3 and KIR2DS5 (20, 21). Conversely, KIR2DL5 was once considered one gene but was split into KIR2DL5A for alleles in the centromeric locus and KIR2DL5B for alleles in the telomeric locus, creating new “A” and “B” designations that are independent from the “A” and “B” haplotype classifications (22). The motif MSA suggests the KIR region has 14 loci in 9 gene motifs. Under the existing gene nomenclature, KIR3DL1 and KIR3DL2 differ only by an index, since they are both three domain (“3D”) long tail (“L”) genes; since the extracellular domains of KIR3DL1/KIR3DL2 fusion alleles are from KIR3DL1, those fusion alleles are labelled in IPD-KIR as KIR3DL1. They are considered KIR3DL2 alleles under the motif structure because the proximal portions of KIR3DL1 and KIR3DL2 share variations of SCANJ, but the distal portions are different; since the fusions are comprised of proximal KIR3DL1 and distal KIR3DL2, they share a partial proximal motif with KIR3DL1 and a full motif with KIR3DL2. The motif ambiguity between the KIR3DL1/KIR3DL2 fusion and KIR3DL2 can be resolved by linkage disequilibrium with KIR2DS4, since the fusion lacks KIR2DS4 and the haplotypes with KIR2DS4 cannot contain the fusion. Both systems consider the cB05 KIR2DS2/KIR2DS3 fusion to be KIR2DS2 (KIR2DS2*005 in IPD-KIR).
A total of 27 new haplotypes were added to the human genome reference as part of this study (accessions MN167504-30), 24 of which are from African Americans. The haplotypes cluster by structure, not population. Haplotypes from Africans or African Americans now constitute 47% of the KIR alternate references in the human genome project, and the human genome project contains more than three times as many alternative references for KIR than any full chromosome. These new haplotypes contain the first deposits of unusual linkages such as KIR2DL5B*00804 in the KIR2DL5A locus (MN167506), KIR3DL1 or KIR3DL2 (fusion) without KIR2DS4 (tA02), KIR2DL2 without KIR2DS2 (cA03), and a KIR2DS2 (fusion) without KIR2DL2 (cB05). MN167526’s KIR2DS2 allele has a one base deletion after 285th base in the CDS sequence, which leads to a premature stop code at position 435 in the CDS.
Scisco Genetics’ contribution to the human genome project of the KIR haplotypes has—like the human genome project itself—been an important advancement and will support the downstream discovery of the human immune system. Building on that work, the motifs and alignments presented here provide a means to help unify interpretation of the entire KIR region. They can be used to precisely define KIR haplotypes and loci, provide context for assigning alleles (especially fusion alleles) to genes, improve evolutionary inferences, improve imputation, interpret co-expression, and generate markers. The motif probes have been applied to a workflow to capture, assemble, and annotate KIR haplotypes at https://github.com/droeatumn/kass; it includes the ability to annotate KIR contigs/haplotypes as a separate workflow. The DNA alignments have been applied to discover markers used in a workflow to genotype KIR presence/absence from WGS at https://github.com/droeatumn/kpi.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by National Marrow Donor Program. The patients/participants provided their written informed consent to participate in this study. No animal studies are presented in this manuscript. No potentially identifiable human images or data is presented in this study.
Author Contributions
DR and RK designed the experiments. DR performed the in silico experiments and wrote the majority of the manuscript. C-WP and DG carried out KIR haplotype sequencing. All authors contributed to the article and approved the submitted version.
Funding
Supported by grants N00014-18-2888 and N00014-20-1-2705 from the Department of the Navy, Office of Naval Research. The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); U24HL138660 from NHLBI and NCI; R21HL140314, and U01HL128568 from the NHLBI; HHSH250201700006C, SC1MC31881-01-00, and HHSH250201700007C from the Health Resources and Services Administration (HRSA); and N00014-18-1-2850, N00014-18-1-2888, and N00014-20-1-2705 from the Office of Naval Research. Additional federal support is provided by P01CA111412, R01CA152108, R01CA215134, R01CA218285, R01CA231141, R01AI128775, R01HL129472, R01HL130388, R01HL131731, U01AI069197, U01AI126612, and BARDA. The views expressed in this article do not reflect the official policy or position of the Department of the Navy, the Department of Defense or any other agency of the U.S. Government.
Conflict of Interest
C-WP and DG are employees of Scisco Genetics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.4
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.585731/full#supplementary-material
Supplementary Table 1 | Motif assignments. The first column lists the alignment order of a pair of probes. The second column assigns a character to the pair.
Supplementary Table 2 | Allelic haplotypes. Each row represents a haplotype and columns D-X contain the presence/absence of the gene and allele names from IPD-KIR 2.9.0. Column A contains and ID consisting of the haplotype structure and GenBank accession. Column B contains the population of the individual. Column C contains the GenBank accession of the haplotype.
Supplementary Figure 1 | Phylogenetic tree made from the phylogenetic tree of the probe motifs of 68 human haplotypes and 1 chimpanzee in Supplemental Data Sheet 2. The relatively short chimpanzee haplotype is an outlier on the bottom of the tree and the relatively long cA01~tB04 is an outlier on the top.
Supplementary Figure 2 | Jalview overview of the MAFFT-generated multiple sequence alignment of the DNA of 68 human haplotypes plus 1 chimpanzee (Supplementary Data Sheet 4).
Supplementary Data Sheet 1 | Zipped fasta file of the multiple sequence alignment of the probe motifs of 67 human haplotypes. Human haplotype cA01~tB04 is not included for purposes of visualization in the manuscript.
Supplementary Data Sheet 2 | Zipped fasta file of the multiple sequence alignment of the probe motifs of 68 human haplotypes and 1 chimpanzee. The phylogenetic tree is in Supplementary Image 1.
Supplementary Data Sheet 3 | Multiple sequence alignment of the DNA of 67 human haplotypes. Human haplotype cA01~tB04 is not included for purposes of visualization in the manuscript.
Supplementary Data Sheet 4 | MAFFT-generated multiple sequence alignment of the DNA of 68 human haplotypes plus 1 chimpanzee. The overview image is in Supplementary Image 2.
References
1. Wroblewski EE, Parham P, Guethlein LA. Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC. Front Immunol (2019) 10:177. doi: 10.3389/fimmu.2019.00177
2. Bastidas-Legarda LY, Khakoo SI. Conserved and variable natural killer cell receptors: diverse approaches to viral infections. Immunology (2019) 156:319–28. doi: 10.1111/imm.13039
3. Díaz-Peña R, de los Santos MJ, Lucia A, Castro-Santos P. Understanding the role of killer cell immunoglobulin-like receptors in pregnancy complications. J Assist Reprod Genet (2019) 36:827–35. doi: 10.1007/s10815-019-01426-9
4. Lau CM, Adams NM, Geary CD, Weizman O-E, Rapp M, Pritykin Y, et al. Epigenetic control of innate and adaptive immune memory. Nat Immunol (2018) 19:963–72. doi: 10.1038/s41590-018-0176-1
5. Parham P, Guethlein LA. Genetics of Natural Killer Cells in Human Health, Disease, and Survival. Annu Rev Immunol (2018) 36:519–48. doi: 10.1146/annurev-immunol-042617-053149
6. Bruijnesteijn J, van der Wiel MKH, de Groot N, Otting N, de Vos-Rouweler AJM, Lardy NM, et al. Extensive Alternative Splicing of KIR Transcripts. Front Immunol (2018) 9:2846. doi: 10.3389/fimmu.2018.02846
7. Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front Immunol (2019) 10:1179. doi: 10.3389/fimmu.2019.01179
8. Agrawal S, Prakash S. Significance of KIR like natural killer cell receptors in autoimmune disorders. Clin Immunol (2020) 216:108449. doi: 10.1016/j.clim.2020.108449
9. Chauhan SKS, Koehl U, Kloess S. Harnessing NK Cell Checkpoint-Modulating Immunotherapies. Cancers (2020) 12:1807. doi: 10.3390/cancers12071807
10. Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics (2003) 55:220–6. doi: 10.1007/s00251-003-0571-z
11. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SGE. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res (2014) 43(D1):D423–431. doi: 10.1093/nar/gku1161
12. Pyo C-W, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different Patterns of Evolution in the Centromeric and Telomeric Regions of Group A and B Haplotypes of the Human Killer Cell Ig-Like Receptor Locus. PLoS One (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
13. Pyo C-W, Wang R, Vu Q, Cereb N, Yang SY, Duh F-M, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics (2013) 14:89. doi: 10.1186/1471-2164-14-89
14. Roe D, Williams J, Ivery K, Brouckaert J, Downey N, Locklear C, et al. Efficient Sequencing, Assembly, and Annotation of Human KIR Haplotypes. bioRxiv (2020). doi: 10.1101/2020.07.12.199570
15. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci (2000) 97:4778–83. doi: 10.1073/pnas.080588597
16. Roe D, Vierra-Green C, Pyo C-W, Eng K, Hall R, Kuang R, et al. Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun (2017) 18:127–34. doi: 10.1038/gene.2017.10
17. Carrillo-Bustamante P, Keşmir C, de Boer RJ. The evolution of natural killer cell receptors. Immunogenetics (2016) 68:3–18. doi: 10.1007/s00251-015-0869-7
18. Wenger AM, Peluso P, Rowell WJ, Chang P-C, Hall RJ, Concepcion GT, et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol (2019) 34:1155–62. doi: 10.1038/s41587-019-0217-9
19. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf (2017) 20(4):1160–6. doi: 10.1093/bib/bbx108
20. Falco M, Moretta L, Moretta A, Bottino C. KIR and KIR ligand polymorphism: a new area for clinical applications?: Relevance of KIR and KIR-L polymorphism. Tissue Antigens (2013) 82:363–73. doi: 10.1111/tan.12262
21. Ordóñez D, Meenagh A, Gómez-Lozano N, Castaño J, Middleton D, Vilches C. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. Genes Immun (2008) 9:431–7. doi: 10.1038/gene.2008.34
Keywords: killer-cell immunoglobulin-like receptors (KIR), alignment, DNA, haplotype, nomenclature, natural killer
Citation: Roe D, Vierra-Green C, Pyo C-W, Geraghty DE, Spellman SR, Maiers M and Kuang R (2020) A Detailed View of KIR Haplotype Structures and Gene Families as Provided by a New Motif-Based Multiple Sequence Alignment. Front. Immunol. 11:585731. doi: 10.3389/fimmu.2020.585731
Received: 21 July 2020; Accepted: 20 October 2020;
Published: 18 November 2020.
Edited by:
Eleanor Riley, University of Edinburgh, United KingdomReviewed by:
Hisashi Arase, Osaka University, JapanBen Roediger, Novartis Institutes for BioMedical Research, Switzerland
Michael G. Brown, University of Virginia, United States
Copyright © 2020 Roe, Vierra-Green, Pyo, Geraghty, Spellman, Maiers and Kuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Roe, roex0050@umn.edu
 David Roe
David Roe Cynthia Vierra-Green2
Cynthia Vierra-Green2 Chul-Woo Pyo
Chul-Woo Pyo Daniel E. Geraghty
Daniel E. Geraghty Stephen R. Spellman
Stephen R. Spellman Martin Maiers
Martin Maiers