
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 07 October 2020
Sec. Nutritional Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.584288
This article is part of the Research Topic Immunity in Infancy and Impact on Immediate and Long-Term Health View all 16 articles
 Sabine Pirr1,2*
Sabine Pirr1,2* Dorothee Viemann1,2,3
Dorothee Viemann1,2,3Gut microbial colonization starts with birth and initiates a complex process between the host and the microbiota. Successful co-development of both establishes a symbiotic mutual relationship and functional homeostasis, while alterations thereof predispose the individual life-long to inflammatory and metabolic diseases. Multiple data have been provided how colonizing microbes induce a reprogramming and maturation of immunity by providing crucial instructing information to the newborn immune system. Less is known about what host factors have influence on the interplay between intestinal immunity and the composition of the gut microbial ecology. Here we review existing evidence regarding host factors that contribute to a favorable development of the gut microbiome and thereby successful maturation of gut mucosal immunity.
Several hundred bacterial species colonize the human gastrointestinal tract and live in a mutual relationship with the host and its immune system (1, 2). A healthy adult gut microbiota is stable and aids in digestion, detoxification and as a barrier against pathogens (3, 4). Importantly, the development of a health gut microbiota keeps the host immune system in a balanced state of tolerance and defense activity (Figure 1A). Though still ill defined, dysbiotic microbiome states are characterized by an altered microbiota composition of loss of diversity resulting in a bloom of pathobionts, loss of commensals, shift in metabolic capacities, and associate with the development of highly prevalent chronic inflammatory and metabolic diseases and/or an increased susceptibility to infections (5–9).
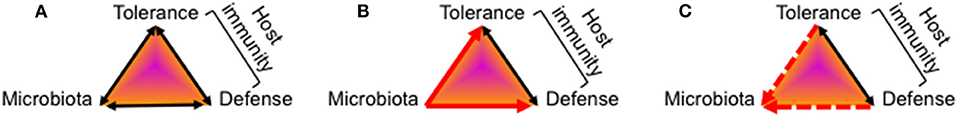
Figure 1. Mutual relationship between the host's immune system and the gut microbiota. The function of the host's gut mucosal immune system toward colonizing microbes is determined by the net sum of tolerance and defense activities which is depicted in a model of a triangular relationship. All elements of the triangle can be impacted by diverse environmental factors not included in this model. (A) Healthy gut colonization and immune adaptation mutually co-develops into a stable, energy-efficient state of homeostasis between the host's immune system and the microbiota. (B) Multiple evidence exists that after birth colonizing microbiota induce maturation of gut mucosal immunity by activation of tolerance-mediating signaling pathways as well as antimicrobial defense programs. (C) The knowledge on how the host's immune system at the beginning of life impacts on the kind of colonizing microbes and the further development of the microbiota composition is still fragmentary.
The postnatal period is particularly critical for the establishment of a healthy gut microbiota composition that promotes the postnatal maturation of gut mucosal and extra-intestinal immunity (10–15). The character of resulting host–microbe interactions determines what degree of gut homeostasis is achievable. Hornef and Torow recently proposed to implement the concept of the “neonatal window of opportunity” into the model of “layered immunity” as a timed succession of non-redundant phases during postnatal immune system maturation and establishment of host-microbial immune homeostasis (16). However, it often remains elusive whether a host factor or a microbiota state evolves as a result from the impact of the respective other feature or whether it represents a true driving force in this deeply interwoven developmental process. Clarification of this question is challenging and requires studies focusing on the beginning life with a high and longitudinal time resolution of effect sizes and directions.
Multiple data, especially those from experiments involving fecal transplants, demonstrated how colonizing microbes induce a reprogramming and maturation of immunity by providing crucial instructing information to the newborn immune system (10, 17–20) (Figure 1B). The importance of intestinal bacterial colonization for normal immune system development became particularly obvious by studies in germ-free mice that are lacking several immune functions (21–23) or the comparison of wild and laboratory mice (24). A surrogate in humans is antibiotic treatment that was shown to affect microbiome development of neonates and infants and translates into short- and long-term disorders characterized by dysregulated or impaired immune functions (25–33).
More challenging is the elucidation of the role of the host's state of immunity for the development of the gut microbiome (Figure 1C), especially that of infant-derived immune factors compared to maternally provided immune factors that supplement or impact on the infant's immune system. Moreover, the mode of action is often not clearly detangled in terms of microbiota-shaping by direct antimicrobial activity or indirectly via priming or regulation of neonatal mucosal immunity which then in turn impacts on the colonization or expansion of specific microbial communities. Finally, many studies have identified factors with immunoregulatory functions for the newborn's immune system but without further demonstration of a link to the microbiome development (those works will therefore not be discussed here).
Of course, exogenous factors such as the mode of delivery, infant feeding patterns, maternal diet, and environmental factors including drugs are also well-known to impact on the microbiome development either directly or indirectly by modulating the host's state of immunity. While these important microbiota-shaping exogenous directors have been extensively discussed elsewhere (34–36), we will focus in this review on endogenous host factors. Maternal (Table 1) and infant-derived (Table 2) host factors will be highlighted and evaluated regarding the present level of evidence on their role in shaping the development of the gut microbiome in humans and animals and thereby the maturation of gut mucosal immunity.
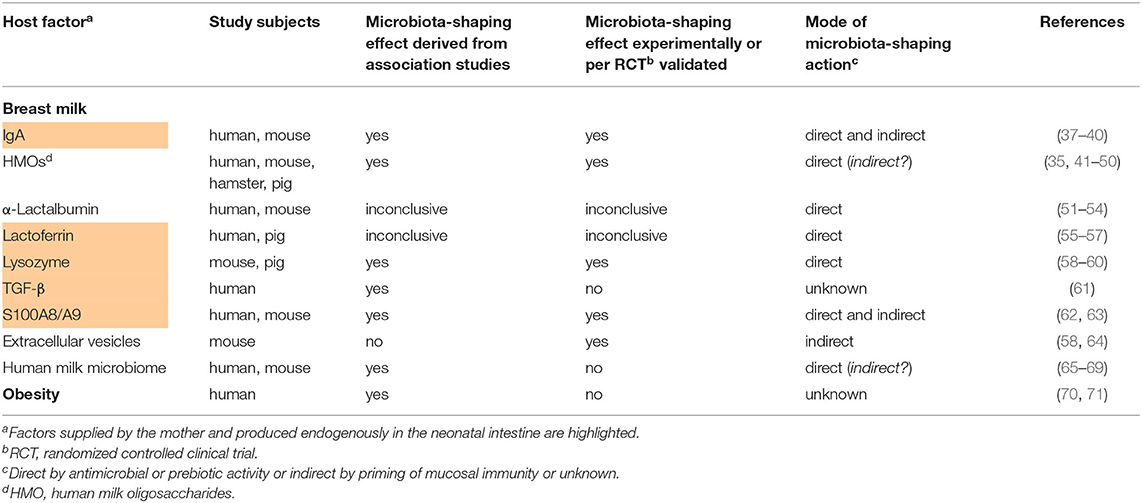
Table 1. Studies providing evidence for the influence of maternal host factors on the postnatal development of the infant's gut microbiome.
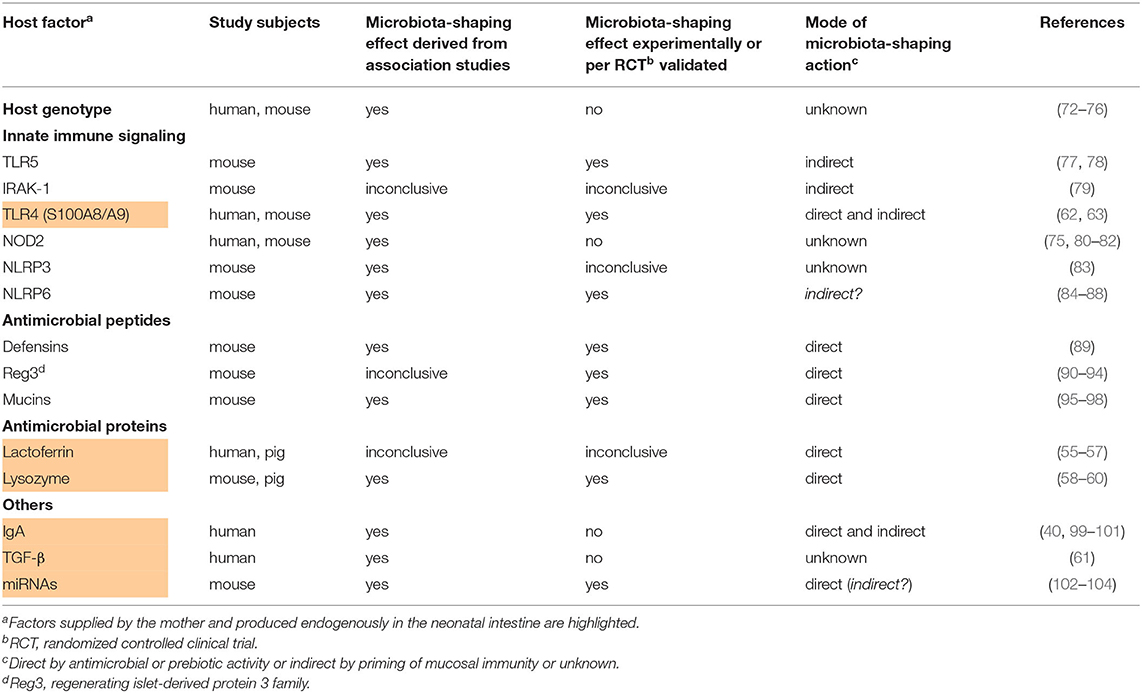
Table 2. Studies providing evidence for the influence of infant-derived host factors on the postnatal development of the infant's gut microbiome.
Human breast milk is the ideal nourishment for infants and has been shaped by evolution to contain essential nutrients and immunological components necessary for the neonate during its initial phase of life. Numerous studies indicate that breastfed neonates are better protected against infections, inflammatory disorders and allergies and show improved cognitive development compared to infants fed with formula (105). In the following we report human breast milk components of different substance classes with the strongest evidence for a capacity to modulate the developing microbiota either directly via antimicrobial or prebiotic activities or indirectly by modulating immune functions of the host.
During the first months of life, breast milk is the predominant source of IgA as B cells generally do not populate the intestine until about 4 weeks of age (37, 106). In mice, early exposure to maternal soluble IgA via breast milk prevented the translocation of aerobic bacteria from the neonatal gut into draining lymph nodes (38). In human preterm infants, a high ratio of IgA-coated Enterobacteriaceae to uncoated Enterobacteriaceae was associated with a lower risk of NEC while a mouse model confirmed the NEC-protective effect of breast milk IgA (37). It is speculated that secretory IgA contributes to the creation of temporal and spatial niches and exerts a strong selection pressure on colonization along the gastrointestinal tract. IgA levels are particularly high in crypts. Agglutination and immune exclusion might primarily act against highly prevalent bacteria and thus favor colonization by minorities and microbial diversity (107). In addition, secretory IgA binds to components of the mucus (108) and co-localizes specifically with gut bacteria in the outer mucus layer (109). The outer mucus layer forms a distinct microbial niche colonized by bacteria with mucolytic capability (110). IgA-coated bacteria might be retained in the mucus layer, which reduces their shedding and promotes diverse colonization (107). In neonatal mice, secretory IgA dampened T cell-dependent immune responses against commensal bacteria (39) and was involved in the selective suppression of Proteobacteria during establishment of the early gut flora (40). A reduced IgA supply via breast milk during infancy came along with an altered gene expression profile of intestinal epithelial cells characterized by an increased expression of genes associated with intestinal inflammatory diseases in humans (38). By the age of weaning, the induction of a Gammaproteobacteria-specific IgA response partially contributes to the transition from a neonatal to a mature microbiota (40). Experiments in germfree mice showed that colonization with microbes from mice lacking IgA have persistent increased colonization with Gammaproteobacteria that results in sustained intestinal inflammation and increased susceptibility to neonatal and adult models of intestinal injury (40). Collectively, there is clear evidence in mice and humans that that IgA supplied via breast milk has direct and indirect beneficial effects on the developing gut microbiota.
HMOs are the third largest solid component of breast milk after lipids and lactose and known to have a bifidogenic effect on the infant's microbiome. HMOs promoted the growth of specific Bifidobacteria, supporting an early Bifidobacteria-dominated gut microbiome (35, 41, 42). The structure of some HMOs resembles that of epithelial pathogen receptors, enabling them to serve as a decoy receptor to prevent pathogen binding and enhance pathogen clearance (42). A mother's secretor status is genetically defined and determines the HMO concentration and profile in her breast milk (111–113). The abundance of Bifidobacteria in the intestinal microbiome of breastfed infants of non-secretor women was lower than in that of children of secretor women (43, 44). In preterm infants, colonization with Bifidobacterium breve was shown to depend on the composition of HMOs but not the mother's secretor status (45). Infant fecal HMO concentrations change over time and correlate with a shift in the fecal microbiota from a non-saccharolytic population dominated by commensals of the birth canal to a population dominated by saccharolytic microbes (46) Several randomized controlled trials showed that infants fed with a formula supplemented with the HMOs 2′-FL ± lacto-N-neotetraose or galacto-oligosaccharides develop a distinctive stool bacterial population compared to infants fed a control formula which is more similar to that of breastfed infants (increased colonization with Bifidobacteria and Lactobacillus and decreased colonization with pathobionts) and was accompanied by a lower plasma inflammatory cytokine concentrations (47–50). In vitro, HMOs prevented the intestinal epithelial attachment of enteropathogenic Escherichia (E.) coli but whether this was a direct or indirect effect remained elusive (114) With respect to the mode of action, HMOs were shown to modulate also toll-like receptor (TLR)-dependent proinflammatory signaling, while elevating cytokines involved in tissue repair and homeostasis (115–117). However, these effects could not be linked to the gut microbiota-shaping effect so far. Further mechanistic studies are required to understand how HMOs influence the development of the gut microbiota.
α-Lactalbumin is the major whey protein in breast milk and is digested in the small intestine (118). Several peptides released from α-lactalbumin during digestion have been proposed to elicit biological effects. In vitro studies demonstrated their antimicrobial activity toward potential pathogens as E. coli, Klebsiella pneumoniae, Staphylococcus aureus, Staphylococcus epidermis, Streptococci, and Candida albicans as well as their prebiotic activity by stimulating the growth of Bifidobacteria (51–53). A study in mice showed significantly higher proportions of Lactobacillus, Parabacteroides and Bifidobacterium in animals fed a high-fat diet supplemented with α-lactalbumin compared to a control diet (54). However, only a few data have been published on the effects of α-Lactalbumin on developing gut microbiota communities in humans and remain inconclusive (119, 120).
Lactoferrin is a non-heme iron binding protein with direct microbial as well as immunomodulatory activities. Lactoferrin is digested to some extend in the infant's intestine yielding bioactive peptides (121). Degradation products of lactoferrin such as lactoferricin (122–125) or Lf(1–11) and lactoferrampin (126, 127) also showed strong bactericidal activity against various kinds of both Gram-positive and Gram-negative bacteria and might contribute to the following net effect of lactoferrin. Piglets fed recombinant human lactoferrin had a greater abundance of Lactobacillus and lower abundances of Veillonella and Escherichia-Shigella in the jejunum and of Actinobacillus in the ileum at 1 week resp. 3 weeks of life (55). Several in vitro studies have shown that human lactoferrin is able to stimulate the growth of Bifidobacteria (56, 57). In vivo, a bifidogenic effect of lactoferrin has also been suggested, but only for bovine and not for human lactoferrin (128), or based on a non-confounder-controlled association between fecal lactoferrin levels on day 3 of life with the amount of fecal Bifidobacteria and Lactobacilli in human infants (129). Clinical trials on the microbiota-shaping effect of the nutritional supplementation of newborn infants with bovine lactoferrin have reported conflicting results with no significant impact on the developing fecal microflora (130, 131) or reduction of pathobionts in a very small cohort of 21 very low birth weight preterm infants (132) or a minor contribution to the total Bifidobacterium population in 202 healthy term infants (133). Human lactoferrin exerts direct antibacterial and antiviral activities through iron depletion (134), interaction with microbial and target host cell surfaces (135) and by preventing the entry of viral particles into the host cells, either by blocking cellular receptors or by directly binding to the viral particles (136). Furthermore, lactoferrin modulates innate and adaptive immune responses thereby controlling the expression of different anti- and pro-inflammatory cytokines and chemokines [e.g., interleukin (IL)-4, IL-10, IL-1, IL-6, IL-12, tumor necrosis factor-α] which impacts on the growth, differentiation, activation and function of immune cells (136–138). Whether and how the immunomodulatory effects contribute to the microbiota-shaping effect or whether the latter primarily results from the antimicrobial activity of lactoferrin remains to be shown.
Lysozyme is an antimicrobial protein secreted by glandular epithelial cells that functions by cleaving bacterial peptidoglycan, causing a loss of cellular membrane integrity and cell lysis. In adult sows, feeding of high amounts of lysozyme reduced the intestinal richness of E. coli and increased abundance of Lactobacillus amylovorus (58). Recombinant human lysozyme expressed at high level in milk of transgenic pigs inhibited the growth of E. coli in the duodenum of piglets (59). In transgenic mice, expression of recombinant human lysozyme promoted the growth of Bifidobacterium and inhibited the growth of Salmonella in the intestine (60). In humans, the role of lysozyme for a developing gut microbiome is unknown.
There is only one study showing that breast milk cytokines TGF-β1 and TGF-β2 partially explains variation in gut microbial composition among human breast-fed neonates. Especially, high concentrations of TGF-β2 were associated with a richer microbiome with an increased abundance of several bacteria, including members of Streptococcaceae and Ruminococcaceae, and lower relative abundance of distinct Staphylococcaceae taxa in neonatal gut composition (61). The exact mode of the microbiota-shaping effect has not been clarified and experimentally validated yet.
The alarmins S100A8 and S100A9 are myeloid-related proteins. Upon extracellular release they typically form the heterodimer complex S100A8/A9, also known as calprotectin (139). Human breast milk contains exceeding high amounts of S100A8/A9 with the highest levels found after the vaginal delivery of a full-term neonate compared to birth of a preterm infant per cesarean section (62). S100A8/A9 proved to be an important mediator of the antimicrobial activity of breast milk, especially inhibiting the growth of manganese sensitive bacteria such as Staphylococcus aureus and group B streptococci (62). Willers et al. found that human infants with reduced levels of fecal S100A8/A9 during the first year of life had an altered development of the gut microbiome characterized by less abundant Actinobacteria including Bifidobacteriaceae but dominant colonization with Gammaproteobacteria, particularly opportunistic Enterobacteriaceae (63). This gut microbial state in turn translated into a higher risk of neonatal sepsis and development of obesity (33, 63). It could furthermore be demonstrated in mice that the high enteral supply of S100A8/A9 after birth primes a regulatory phenotype in lamina propria macrophages with a high expression of Cx3cr1, Il-10, and Tgf-β, that is followed by a stronger expansion of regulatory T cells in intestinal tissues and a restricted expansion of Enterobacteriaceae. A single feeding of S100A8 after birth prevented the excess expansion of Enterobacteriaceae as well as fatal sepsis in neonatal mice (63).
Extracellular vesicles in breast milk may influence the establishment of the intestinal microbiota by altering the local immune response toward challenges from microbial colonization (105). Breast milk exosomes contain proteins, RNA, microRNA and long non-coding RNA (140–142). Profiling studies found these microRNAs to be related to multiple biological functions of a wide range of immunological pathways (143, 144). Likewise, proteomic analyses revealed that human breast milk extracellular vesicles include proteins involved in regulation of cell growth and controlling inflammatory signaling pathways (142). In addition to vesicles released by maternal cells (exosomes), a recent study revealed the presence of bacteria-derived extracellular vesicles in breast milk (145). Their vertical transfer or indirect involvement into the development of the infant's gut microbiota is possible but has not been shown yet. The presence of serum-derived extracellular vesicles impacted on epithelial and immune cell responses to the gut microbes Lactobacillus or Bifidobacterium and enhanced their aggregation and phagocytosis and modulated TLR-induced cellular responses (64). Whether breast milk extracellular vesicles have similar effects is not known yet. Feeding of bovine milk exosomes to young adult mice altered bacterial communities in the murine cecum (higher abundance of Tenericutes and Lachnospiraceae, lower abundance of Verrucomicrobiaceae) (58). Data on the influence of milk extracellular vesicles and their cargo on human infant's gut microbial communities are still lacking.
Although not in a strict sense an endogenous maternal factor, the mother the human milk microbiota is at least mother-specific and represents therefore for the infant an endogenous factor in the broader sense. Human milk contains ~106 bacterial cells/mL. Commonly isolated microbiota in human milk are Staphylococcus, Streptococcus, Propionibacterium, Bifidobacterium, and Lactobacillus among others (146–150). The composition and variation of the human milk microbiota are associated with the maternal body mass index, parity, glucose tolerance status, mode of delivery, ethnicity, breast feeding practices, and other milk components (151, 152). The origin of bacteria in human milk is not well-established. Two different routes, which are probably not mutually exclusive, have been proposed: (i) surface skin contamination and retrograde flow during breastfeeding and (ii) translocation through a more speculative gut-mammary route (153). The assumption that microbes in breast milk contribute to the establishment of the infant intestinal microbiome is supported by several studies in mother-infant pairs using sequencing techniques and demonstrating that breast milk and term infants' feces share specific microbial strains of Bifidobacterium, Lactobacillus, Enterococcus, and Staphylococcus (65–69). In vitro and mouse studies demonstrated that several Lactobacillus strains isolated from human milk exert immune-modulatory effects on bone marrow-derived macrophages as well as antibacterial properties against pathogenic bacteria (149, 150). However, clear evidence for a colonization of human milk microbiota in the infant's intestine or an indirect impact of human milk microbiota on the developing intestinal microbiome has not been provided yet. Kim and Yi recently raised the question whether extracellular vesicles derived from the human milk microbiota are involved in the vertical transfer from mothers to their progeny (145), which remains unanswered to date.
Neonates born to mothers with obesity showed a 50% reduction in the intestinal abundance of Gammaproteobacteria at 2 weeks of age compared with infants of normal-weight mothers (70). Furthermore, infants born to overweight mothers were found to have an increased abundance of Bacteroides and Staphylococcus at 1 and 6 months of age compared to infants of normal-weight mothers (71). Reasons might be the different and less diverse bacterial community in breast milk from mothers with obesity compared to normal-weight mothers, including higher levels of Staphylococcus and Akkermansia muciniphila and lower levels of Bifidobacterium (154, 155). Differences in the composition of hormones, cytokines and HMOs might also play a microbiota-shaping role (156, 157). These findings might even challenge the beneficial value of breast milk feeding in case of overweight mothers. This concern is corroborated by the data that antibiotic use in infants born to mothers with obesity reduced the risk of obesity, whereas antibiotic use in infants born to normal-weight mothers increased the risk of obesity (158). Another reason for the altered microbiome composition in infants born to obese mothers might be iron deficiency (157, 159, 160). Mothers with obesity and those with excess gestational weight gain as well as their infants are significantly more often iron deficient (161, 162). Several clinical and in vitro studies have demonstrated the impact of iron deficiency and supplementation on the gut microbiota composition and function and intestinal inflammation in infants and children (163–168). However, whether iron deficiency is the causative mechanism altering the postnatal gut microbiota colonization in infants of mothers with obesity needs to be further explored.
There is growing scientific evidence indicating that host genetics influence the acquisition and development of the infant gut microbiota (72–74). Studies in mice revealed that host genetics in the interplay with the diet shape the intestinal microbiota composition with individual genes and regulatory circuits only beginning to be identified (72, 73). Benson et al. found 18 quantitative trait loci in mice that show significant or suggestive genome-wide linkage with the relative abundances of specific microbial taxa (72). A study using three large human cohorts established a clear association between the host genotype and the gut microbiota state in adulthood. In this analysis, 58 single nucleotide polymorphisms (SNPs) at nine loci were associated with microbial taxa and a total of 33 loci were associated with functional units, with 21 loci associated with metabolic pathways and 12 loci associated with gene ontogeny categories. For example, variants of the LCT locus responsible for the human lactase production linked to the abundance of Bifidobacteria, while variants of the NOD2 locus had an impact on the pathway abundance of enterobactin biosynthesis which in turn strongly correlated with the abundance of E. coli (75). Another study in a large human twin cohort uncovered associations between heritable taxa and genes related to diet, metabolism, olfaction, barrier defense, and self/non-self-recognition (76). Summarized, linkage studies on associations between genetic variants and gut microbiota states point to possible important host factors that are crucial for the establishment of a healthy gut microbiome. For most of presently identified genetic trait loci further research is needed to confirm the relevance of these host factors for the development of the gut microbiome experimentally and in human infants.
Age-dependent differences exist for the intestinal expression of individual TLR receptors in mice and humans (77, 169). This fact already suggests TLR signaling playing a role for the postnatal development of the gut microbiome and evidence for this specific relationship is accumulating. Several mouse models with knock-downs or genetic variants targeting TLR-signaling pathways at different levels (MyD88, Tlr2, Tlr4, Tlr5, Tlr9, Trif , NF-kB-pathway) are characterized by spontaneous or permissive colitis (20, 170–181). We learned from these mouse models how critical commensal-mediated TLR-signaling is for the training and maintenance of gut mucosal immunity and integrity. The host's primary phenotype and programming of TLR-signaling is conversely probably likewise important for the development of the gut microbiome. However, this reverse effect order has been clarified only for a few TLR-related host factors and barely in humans.
For the flagellin receptor TLR5 it has been clearly shown that its transient expression on the gut epithelium of neonatal mice contributes to the establishment of a favorable gut microbiota that lowers the risk of intestinal inflammatory injuries (77, 78). Fulde et al. demonstrated in mice that a TLR5-mediated epithelial production of the regenerating islet-derived protein 3 γ (REG3γ) is critical to restrict the colonizing of flagellated bacteria. This host-mediated regulatory circuit of bacterial colonization acts solely during the early neonatal period but influences life-long microbiota composition. The relevance of TLR5-signaling on the gut microbiota development in human infants has not been demonstrated yet.
Chassin et al. demonstrated that the microRNA-146a-mediated translational repression and proteolytic degradation of the essential TLR signaling molecule interleukin 1 receptor associated kinase 1 (IRAK1), an integral element of MyD88-dependent TLR-signaling pathways, induces intestinal epithelial innate immune tolerance toward lipopolysaccharide (LPS) of Gram-negative bacteria. It furthermore protects murine neonates against epithelial damage from oral challenge with E. coli (79), suggesting that IRAK1 has a beneficial impact on the established microbiota. In contrast, however, 16S rRNA profiling of stool samples revealed miR-146a-deficient mice having a rather favorable gut microbiota composition protecting against Listeria monocytogenes infection compared to wild-type mice with decreased levels of the Proteobacteria phylum, Prevotellaceae family, and Parasutterella genus, and significantly increased short-chain fatty acid producing bacteria, including the genera Alistipes, Blautia, Coprococcus_1, and Ruminococcus_1 (102). Thus, the role of IRAK1 for the development of the gut microbiota remains to be clarified and also elucidate in humans.
The alarmins S100A8 and S100A9 are endogenous TLR4 ligands (182). Next to their high supply via breast milk (62), S100A8 and S100A9 are also endogenously produced in intestinal tissues by lamina propria macrophages, in infants at significantly higher levels than in intestinal tissues from adults (63). Studies in S100a9 knock-out mice and cross-fostering settings delineated the mode of action. S100A8/A9-mediated TLR4-signaling during the neonatal period tolerized the inflammatory responsivity of lamina propria macrophages and increased the expression tonus of regulatory genes like IL-10 and transforming growth factor-β. This regulatory macrophage phenotype impacted on the developing microbiota in mouse and humans as outlined above. To achieve full effects regarding immunoregulation and modulation of gut colonization the additional supply of S100A8/A9 via breast milk was mandatory (63).
Similar as in the case of TLRs, murine NLR knock-out models (Nod2, Nlrp3, Nlrp6, Nlrc4, caspase 1) with increased (albeit partly controversial) susceptibility in different colitis models (183–188) point to NLRs as important host factors for the establishment of a protective gut microbiota. However, for most factors this assumption awaits experimental approval.
Mutations in the peptidoglycan sensor NOD2 locus represent one of the most important genetic risk factors for inflammatory bowel disease (IBD) (189, 190). As mentioned above, genetic variants of the NOD2 locus in humans correlated with the intestinal abundance of E. coli (75). In two cohorts of adult patients with IBD, a significant positive correlation was found between the number of IBD risk alleles in the NOD2 exon and the relative abundance of Enterobacteriaceae (80). Two recent studies in mice demonstrated that NOD2 influences microbial resilience with prolonged alterations of the microbiota in Nod2-deficient mice after antibiotic treatment, in case of neonatally treated mice resulting in an increased susceptibility to colitis in adulthood (81, 82). However, experimental evidence for NOD2 shaping the primary development of the gut microbiota in neonates and infants and the related mode of action is still lacking.
For NLRP3 it could recently been shown that the compositions of the gut microbiota in Nlrp3 knock-out mice and wild-type mice were significantly different (83). At the genus level, NLRP3 deficiency decreased the relative abundance of Bacteroides but increased the abundance of Desulfovibrio, Mucispirillum, Oscillospira, and Ruminococcus. Surprisingly, fecal microbiota transplantation from Nlrp3 knock-out mice significantly ameliorated depressive-like behaviors of recipient wild-type mice in a chronic stress model suggesting that restriction of NLRP3-dependent inflammasome signaling is important to ensure healthy gut microbiota development. For a final evaluation of the role and mode of action of NLRP3 regarding its impact on developing gut microbiota compositions further clarifying data in mice as well as humans is desirable.
Elinav et al. were the first group demonstrating that the lack of intestinal epithelial NLRP6-dependent inflammasome activity results in an expansion of the bacterial phyla Bacteroidetes and TM7 and that this microbiota state exacerbates chemical colitis (84). This microbiota-shaping effect was later ascribed, but not directly linked, to a regulating impact of NLRP6 on mucus secretion by goblet cells (85). However, littermate-controlled mouse studies then dismissed the impact of NLRP6-dependent inflammasomes on the composition of the commensal gut microbiota (86, 87). This conflict could be detangled by showing that NLRP6 matters in dependence of the microbial community structure. In pathobiont-free conditions WT and Nlrp6-deficient mice had a similar microbiota while upon introduction of pathobionts a high prevalence, specifically of Prevotellaceae, Verrucomicrobiaceae, and Helicobacteraceae, in Nlrp6-deficient mice became obvious (88). This finding was also largely in line with the previous findings (84). Demonstration of the biological relevance of NLRP6 shaping the microbiota development in humans would now be convincing.
The anatomical distribution of AMPs in mice and humans shows age-dependent differences (77, 191). In fetal intestine AMPs are already expressed prenatally and levels increase with gestational age (192, 193). Major AMP families are cathelicidins, α-defensins, and β-defensins, lectins, and mucins (194, 195). The peptides are stored in vacuoles of granulocytes and/or epithelial cells ready to be secreted upon microbial stimulation. In addition to their ability to kill bacteria by disrupting critical membrane functions, many AMPs have chemotactic and immunomodulatory activities (194, 196). It is evident that induced expression of AMPs is frequently an integral part of a response to a microbial challenge, reflecting how colonizing microbes train the host's immunity (197, 198). Conversely, though not in humans but at least in mice, there is also evidence that the levels and composition of AMPs in the gut can regulate the development of the microbiota.
In human α-defensin 5 (HD-5) transgenic mice significant changes in the microbiota composition were observed compared to wild-type littermate controls, specifically a lower relative abundance of Firmicutes and higher percentages of Bacteroidetes (89). Human REG3α and its murine homolog REG3γ are C-type soluble, carbohydrate-binding lectins found throughout the small intestinal epithelium, and in the large intestine in response to infection and inflammation (199). Bacterial binding (90, 91) and membrane permeabilization (92) are proposed to be important for the bactericidal activity of REG3. Hitherto shown effects of REG3α resp. REG3γ on the gut microbiota are not consistent, most likely due to diverse confounders like REG3 expression levels, age, and initial health condition. In a murine model of alcoholic steatohepatitis, intestinal REG3γ was protective by reducing the mucosa-associated microbiota and preventing bacterial translocation (93). On the other side, the fecal microbiota of mice that express human REG3α in hepatocytes (hREG3α travels via the bile to the intestinal lumen) showed a significant shift in microbiota composition compared to control mice, with an enrichment of Clostrididales (Ruminococcaceae, Lachnospiraceae) and depletion of Bacteroidetes (Prevotellaceae) (94). These mice were less sensitive to colitis, suggesting that increased intestinal levels of REG3α might impair the development of a healthy gut microbiota state.
Mucins are the main structural components of mucus, the biopolymer found throughout the gastrointestinal tract (199). Some commensal intestinal bacteria, including Bacteroidetes, are mucolytic and use mucin glycoproteins as an energy source (95, 96). Therefore, mucins have been proposed to play an important role in shaping microbial communities at the intestinal luminal surface. Recent studies in mice suggest a correlation between changes in mucin glycosylation profiles and deviations of overall microbial community ecology as well as altered abundances of specific microbes (97, 98). Mice lacking terminal fucose residues in their distal gut mucins showed significantly higher abundance of members of the genera Parabacteroides, Eubacterium, Parasutterella, Bacteroides, and family Lachnospiraceae and lower abundance of an unclassified genus within the order Clostridiales (97). Changes in mucus glycosylation due to loss of core 1-derived O-glycans in mice led to inverse shifts in the abundance of the phyla Bacteroidetes and Firmicutes (98).
Summarized, further supporting studies in mice and particularly first evidences in humans are warranted to verify the host-microbiome direction of the AMP-microbiota axis.
Antimicrobial proteins play important roles in the first line of defense, by either killing pathogens directly, or by contributing to an efficient immune response to eliminate pathogens. Most abundant antimicrobial proteins in humans are lactoferrin and lysozyme, both of which are also contained in breast milk. Their potential to influence the postnatal development of the gut microbiome composition has been discussed above. Whether the endogenous production of these proteins is important for the putative microbiota-shaping effect or to what extent the endogenous production contributes to the effect of the maternal supply of these factors has not been clarified so far. However, the lack of considering also the endogenous production and levels of these factors might have hampered the significance of effect sizes of the exogenous supply in previous studies.
The knowledge on how IgA impacts on the developing gut microbiota is mainly based on studies with a focus on the IgA supply via breast milk and has been outlined above. However, also the endogenous IgA production by the infant is of relevance in this context. Patients with common variable immunodeficiency (CVID) showed a large shift in the microbiota (reduced abundance of Actinobacteria and Firmicutes and increased abundance of Gammaproteobacteria) which was particularly pronounced in patients with undetectable serum IgA and also found in patients with selective IgA deficiency (99–101). In patients with selective IgA deficiency, metagenomic analyses revealed pathobiont expansion (e.g., Gammaproteobacteria including E. coli) and depletion in some typically beneficial symbionts belonging to the Bacteroidetes and Firmicutes phylum (100, 101, 200, 201).
TGF-β is a regulatory cytokine ubiquitously produced by immune cells and epithelial cells in intestinal tissues. While breast milk levels of TGF-β correlated with the neonate's microbiota composition (61), the role endogenously produced TGF-β plays in this context has not been explored so far.
MicroRNAs are single-stranded non-coding RNA molecules with a length varying between 18 and 23 nucleotides that operate in post-transcriptional regulation of gene expression (202). miRNAs are found in various body fluids as plasma, urine and breast milk as well as in human feces (103, 143, 203, 204). Fecal miRNAs are secreted by intestinal epithelial cells (IEC), particularly homeodomain-only protein homeobox (HOPX)-positive cells, such as goblet and Paneth cells (103). In vitro studies revealed that specific fecal miRNAs were able to enter Fusobacterium (F.) nucleatum and E. coli, regulate bacterial gene transcripts, and affect bacterial growth. Furthermore, IEC-miRNA deficient mice showed uncontrolled gut microbiota compared to wild-type mice and exacerbated colitis. Wild-type fecal miRNA transplantation restored fecal microbes and ameliorated colitis (103). Moreover, the abundances of the phyla Bacteroidetes, Firmicutes, Actinobacteria, Bacteroidetes, Cyanobacteria, and Proteobacteria correlated with distinct murine miRNA levels (104). Another murine model revealed differing gut microbiota compositions with decreased levels of the Proteobacteria phylum, Prevotellaceae family, and Parasutterella genus, and significantly increased short-chain fatty acid producing bacteria, including the genera Alistipes, Blautia, Coprococcus_1, and Ruminococcus_1 in miR-146a-deficient mice relative to wild-type mice (102). Whether this is an indirect effect linked to the miR-146a-mediated induction of intestinal epithelial innate immune tolerance (79, 205) or due to a direct microbial activity of miR-146a has not been detangled yet. Ji et al. demonstrated that human fecal miRNAs associated with inflammatory bowel disease have diverse effects on the proliferative activity of the intestinal bacteria F. nucleatum, E. coli and segmental filamentous bacteria following the uptake of these miRNAs by the microorganisms (206). Of note, data on the influence of fecal miRNAs on the developing intestinal microbiome in neonates and infants are missing to date. Moreover, for miRNAs found in breast milk (143) neither a microbiota-shaping effect nor correspondences with microbiota-shaping fecal miRNAs could be demonstrated so far (wherefore they have not been listed in Table 1 but highlighted in Table 2 as factors supplied by the mother and produced endogenously in the neonatal intestine).
Acknowledging that the establishment of a diverse and stable gut microbiota during the first years of life is crucial for life-long immune homeostasis, acquisition of tolerance, resistance against pathogens, and overall health fueled recent research work for factors that regulate the development of a health-promoting host-microbiota relationship. Present studies proposing host factors possibly important in this process of gut microbiome development differ regarding the therefore provided level of evidence. The spectrum of proof ranges from sole assumptions based on known biological functions, e.g., factors with antimicrobial activity, through association studies of host factor levels or genetic variants with specific microbiota states up to the experimental confirmation of the microbiota-shaping function, at best including delineation of the related mode of action. Moreover, the studies that provide functional data are mostly restricted to investigations in the mouse but lack demonstration of the biological relevance of the respective host factor in humans. Only if the latter condition is met together with a high grade of functional evidence for a beneficial microbiota-shaping effect this would allow and encourage translation of the concept into a clinical trial. Currently, only a few factors (IgA, HMOs, and S100A8/A9) meet these criteria. Further studies are needed to advance existing data or to unravel novel host factors important for a favorable postnatal development of the gut microbiome. Unveiling such endogenous mechanisms of microbiota regulation is important as they are highly attractive candidates for exploiting nature's principles for effective intervention strategies to ensure the development of health.
SP and DV equally conceptualized and composed the manuscript. All authors contributed to the article and approved the submitted version.
SP was supported by the Ellen-Schmidt-Stipend and a HiLF-grant of Hannover Medical School. The Volkswagen Foundation (Az 90 005), the Appenrodt Foundation and the Deutsche Forschungsgemeinschaft (DFG) (VI 538/6-1), and the DFG under Germany's Excellence Strategy-EXC 2155 RESIST-Project ID 39087428 supported research in this area.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Round JL, Mazmanian SK. The gut microbiome shapes intestinal immune responses during health and disease. Nat Rev Immunol. (2009) 9:313–23. doi: 10.1038/nri2515
2. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7. doi: 10.1126/science.1223813
3. Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. (2005) 307:1915–20. doi: 10.1126/science.1104816
4. Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. (2017) 66:515–22. doi: 10.1016/j.alit.2017.07.010
5. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. doi: 10.1038/nature05414
6. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490:55–60. doi: 10.1038/nature11450
7. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. (2007) 104:13780–5. doi: 10.1073/pnas.0706625104
8. Gensollen T, Blumberg RS. Correlation between early-life regulation of the immune system by microbiota and allergy development. J Allergy Clin Immunol. (2017) 139:1084–91. doi: 10.1016/j.jaci.2017.02.011
9. Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. (2008) 359:61–73. doi: 10.1056/NEJMra0708473
10. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
11. Macpherson AJ, Agüero MG de, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol. (2017) 17:508–17. doi: 10.1038/nri.2017.58
12. Torow N, Marsland BJ, Hornef MW, Gollwitzer ES. Neonatal mucosal immunology. Mucosal Immunol. (2017) 10:5–17. doi: 10.1038/mi.2016.81
13. Menckeberg CL, Hol J, Simons-Oosterhuis Y, Raatgeep HR, de Ruiter LF, Lindenbergh-Kortleve DJ, et al. Human buccal epithelium acquires microbial hyporesponsiveness at birth, a role for secretory leukocyte protease inhibitor. Gut. (2015) 64:884–93. doi: 10.1136/gutjnl-2013-306149
14. Claud EC, Lu L, Anton PM, Savidge T, Walker WA, Cherayil BJ. Developmentally regulated IkappaB expression in intestinal epithelium and susceptibility to flagellin-induced inflammation. Proc Natl Acad Sci USA. (2004) 101:7404–8. doi: 10.1073/pnas.0401710101
15. Kasahara K, Matsumura Y, Ui K, Kasahara K, Komatsu Y, Mikasa K, et al. Intranasal priming of newborn mice with microbial extracts increases opsonic factors and mature CD11c+ cells in the airway. Am J Physiol Lung Cell Mol Physiol. (2012) 303:L834–43. doi: 10.1152/ajplung.00031.2012
16. Hornef MW, Torow N. 'Layered immunity' and the 'neonatal window of opportunity' - timed succession of non-redundant phases to establish mucosal host-microbial homeostasis after birth. Immunology. (2020) 159:15–25. doi: 10.1111/imm.13149
17. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. (2010) 10:159–69. doi: 10.1038/nri2710
18. Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351:1296–302. doi: 10.1126/science.aad2571
19. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. (2017) 46:562–76. doi: 10.1016/j.immuni.2017.04.008
20. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. (2004) 118:229–41. doi: 10.1016/j.cell.2004.07.002
21. Purchiaroni F, Tortora A, Gabrielli M, Bertucci F, Gigante G, Ianiro G, et al. The role of intestinal microbiota and the immune system. Eur Rev Med Pharmacol Sci. (2013) 17:323–33.
22. Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. (2004) 4:478–85. doi: 10.1038/nri1373
23. Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. (2008) 455:808–12. doi: 10.1038/nature07240
24. Abolins S, King EC, Lazarou L, Weldon L, Hughes L, Drescher P, et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat Commun. (2017) 8:14811. doi: 10.1038/ncomms14811
25. Zeissig S, Blumberg RS. Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat Immunol. (2014) 15:307–10. doi: 10.1038/ni.2847
26. Gibson MK, Wang B, Ahmadi S, Burnham CA, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. (2016) 1:16024. doi: 10.1038/nmicrobiol.2016.24
27. Cotten CM, Taylor S, Stoll B, et al. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. (2009) 123:58–66. doi: 10.1542/peds.2007-3423
28. Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. J Pediatr. (2011) 159:720–5. doi: 10.1016/j.jpeds.2011.05.033
29. Alexander VN, Northrup V, Bizzarro MJ. Antibiotic exposure in the newborn intensive care unit and the risk of necrotizing enterocolitis. J Pediatr. (2011) 159:392–7. doi: 10.1016/j.jpeds.2011.02.035
30. Hoskin-Parr L, Teyhan A, Blocker A, Henderson AJ. Antibiotic exposure in the first two years of life and development of asthma and other allergic diseases by 7.5 yr: a dose-dependent relationship. Pediatr Allergy Immunol. (2013) 24:762–71. doi: 10.1111/pai.12153
31. Lamont RF, Møller Luef B, Stener Jørgensen J. Childhood inflammatory and metabolic disease following exposure to antibiotics in pregnancy, antenatally, intrapartum and neonatally. F1000Res. (2020) 9:F1000. doi: 10.12688/f1000research.19954.1
32. Oosterloo BC, Van't Land B, de Jager W, Rutten NB, Klöpping M, Garssen J, et al. Neonatal antibiotic treatment is associated with an altered circulating immune marker profile at year of age. Front Immunol. (2020) 10:2939. doi: 10.3389/fimmu.2019.02939
33. Graspeuntner S, Waschina S, Künzel S, Twisselmann N, Rausch TK, Cloppenborg-Schmidt K, et al. Gut dysbiosis with bacilli dominance and accumulation of fermentation products precedes late-onset sepsis in preterm infants. Clin Infect Dis. (2019) 69:268–77. doi: 10.1093/cid/ciy882
34. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036–17. doi: 10.1128/MMBR.00036-17
35. Vandenplas Y, Carnielli VP, Ksiazyk J, Sanches Luna M, Migacheva N, Mosselmans JM, et al. Factors affecting early-life intestinal microbiota development. Nutrition. (2020) 78:110812. doi: 10.1016/j.nut.2020.110812
36. Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW; GI-MDH consortium, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. (2020) 158:1584–96. doi: 10.1053/j.gastro.2020.01.024
37. Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, et al. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med. (2019) 25:1110–5. doi: 10.1038/s41591-019-0480-9
38. Rogier EW, Frantz AL, Bruno MEC, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci USA. (2014) 111:3074–9. doi: 10.1073/pnas.1315792111
39. Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, et al. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. (2016) 165:827–41. doi: 10.1016/j.cell.2016.04.055
40. Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. (2014) 5:28–39. doi: 10.4161/gmic.26489
41. Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr Res. (2015) 77:229–35. doi: 10.1038/pr.2014.156
42. Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagström H, Sanchez Luna M, et al. Human milk oligosaccharides: 2'-fucosyllactose. (2'-FL) and lacto-n-neotetraose. (LNnT) in infant formula. Nutrients. (2018) 10:1161. doi: 10.3390/nu10091161
43. Lewis ZT, Totten MS, Smilowitz JT, Popovic M, Parker E, Lemay DG, et al. Maternal fucosyltransverase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. (2015) 3:13. doi: 10.1186/s40168-015-0071-z
44. Smith-Brown P, Morrison M, Krause L, Davies PS. Mothers secretor status affects development of childrens microbiota composition and function: a pilot study. PLoS ONE. (2016) 11:e0161211. doi: 10.1371/journal.pone.0161211
45. Underwood MA, Davis JCC, Kalanetra KM, Gehlot S, Patole S, Tancredi DJ, et al. Digestion of human milk oligosaccharides by bifidobacterium breve in the premature infant. J Pediatr Gastroenterol Nutr. (2017) 65:449–55. doi: 10.1097/MPG.0000000000001590
46. De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, et al. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. (2015) 14:491–502. doi: 10.1021/pr500759e
47. Puccio G, Alliet P, Cajozzo C, Janssens E, Corsello G, Sprenger N, et al. Effects of infant formula with human milk oligosaccharides on growth and morbidity: a randomized multicenter trial. J Pediatr Gastroenterol Nutr. (2017) 64:624–31. doi: 10.1097/MPG.0000000000001520
48. Steenhout P, Sperisen P, Martin FP, Sprenger N, Wernimont S, Pecquet S, et al. Term infant formula supplemented with human milk oligosaccharides. (2′Fucosyllactose and Lacto-N-neotetraose) shifts stool microbiota and metabolic signatures closer to that of breastfed infants. FASEB J. (2016) 30:S1. doi: 10.1096/fasebj.30.1_supplement.275.7
49. Goehring KC, Marriage BJ, Oliver JS, Wilder JA, Barrett EG, Buck RH. Similar to those who are breastfed, infants fed a formula containing 2'-fucosyllactose have lower inflammatory cytokines in a randomized controlled trial. J Nutr. (2016) 146:2559–66. doi: 10.3945/jn.116.236919
50. Giovannini M, Verduci E, Gregori D, Ballali S, Soldi S, Ghisleni D, et al. PLAGOS Trial Study Group. Prebiotic effect of an infant formula supplemented with galacto-oligosaccharides: randomized multicenter trial. J Am Coll Nutr. (2014) 33:385–93. doi: 10.1080/07315724.2013.878232
51. Brück WM, Graverholt G, Gibson GR. Use of batch culture and a two-stage continuous culture system to study the effect of supplemental α-lactalbumin and glycomacropeptide on mixed populations of human gut bacteria. FEMS Microbiol Ecol. (2002) 41:231–7. doi: 10.1016/S0168-6496(02)00296-9
52. Brück WM, Graverholt G, Gibson GR. A two-stage continuous culture system to study the effect of supplemental α-lactalbumin and glycomacropeptide on mixed cultures of human gut bacteria with entero-pathogenic Escherichia coli and salmonella serotype typhimurium. J Appl Microbiol. (2003) 95:44–53. doi: 10.1046/j.1365-2672.2003.01959.x
53. Pellegrini A, Thomas U, Bramaz N, Hunziker P, von Fellenberg R. Isolation and identification of three bactericidal domains in the bovine a-lactalbumin molecule. Biochim Biophys Acta. (1999) 1426:439–48. doi: 10.1016/S0304-4165(98)00165-2
54. Boscaini S, Cabrera-Rubio R, Speakman JR, Cotter PD, Cryan JF, Nilaweera KN. Dietary α-lactalbumin alters energy balance, gut microbiota composition and intestinal nutrient transporter expression in high-fat diet-fed mice. Br J Nutr. (2019) 121:1097–107. doi: 10.1017/S0007114519000461
55. Hu P, Zhao F, Zhu W, Wang J. Effects of early-life lactoferrin intervention on growth performance, small intestinal function and gut microbiota in suckling piglets. Food Funct. (2019) 10:5361–73. doi: 10.1039/C9FO00676A
56. Kim WS, Tanaka T, Kumura H, Kim GY, Kwon IK, Goh JS, et al. Growth promoting effects of lactoferrin on L. acidophilus and Bifidobacterium spp. Biometals. (2004) 17:279–83. doi: 10.1023/B:BIOM.0000027705.57430.f1
57. Liepke C, Adermann K, Raida M, Magert HJ, Forssmann WG, Zucht HD. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. (2002) 269:712–8. doi: 10.1046/j.0014-2956.2001.02712.x
58. Zhou F, Paz HA, Sadri M, Cui J, Kachman SD, Fernando SC, et al. Dietary bovine milk exosomes elicit changes in bacterial communities in C57BL/6 mice. Am J Physiol Gastrointest Liver Physiol. (2019) 317:G618–24. doi: 10.1152/ajpgi.00160.2019
59. Lu D, Li Q, Wu Z, Shang S, Liu S, Wen X, et al. High-level recombinant human lysozyme expressed in milk of transgenic pigs can inhibit the growth of Escherichia coli in the duodenum and influence intestinal morphology of sucking pigs. PLoS ONE. (2014) 9:e89130. doi: 10.1371/journal.pone.0089130
60. Dan L, Liu S, Shang S, Zhang H, Zhang R, Li N. Expression of recombinant human lysozyme in bacterial artificial chromosome transgenic mice promotes the growth of Bifidobacterium and inhibits the growth of Salmonella in the intestine. J Biotechnol. (2018) 272–3:33–39. doi: 10.1016/j.jbiotec.2018.03.005
61. Sitarik AR, Bobbitt KR, Havstad SL, Fujimura KE, Levin AM, Zoratti EM, et al. Breast milk transforming growth factor β is associated with neonatal gut microbial composition. Pediatr Gastroenterol Nutr. (2017) 65:e60–7. doi: 10.1097/MPG.0000000000001585
62. Pirr S, Richter M, Fehlhaber B, Pagel J, Härtel C, Roth J, et al. High amounts of S100-alarmins confer antimicrobial activity on human breast milk targeting pathogens relevant in neonatal sepsis. Front Immunol. (2017) 8:1822. doi: 10.3389/fimmu.2017.01822
63. Willers M, Ulas T, Völlger L, Vogl T, Heinemann AS, Pirr S, et al. S100A8 and S100A9 are important for postnatal development of gut microbiota and immune system in mice and infants. Gastroenterology. (2020) S0016-5085(20)35058-7. doi: 10.1053/j.gastro.2020.08.019
64. van Bergenhenegouwen J, Kraneveld AD, Rutten L, Kettelarij N, Garssen J, Vos AP. Extracellular vesicles modulate host-microbe responses by altering TLR2 activity and phagocytosis. PLoS ONE. (2014) 9:e89121. doi: 10.1371/journal.pone.0089121
65. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. (2018) 24:133–45.e5. doi: 10.1016/j.chom.2018.06.005
66. Murphy K, Curley D, O'Callaghan TF, O'Shea CA, Dempsey EM, O'Toole PW, et al. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. (2017) 7:40597. doi: 10.1038/srep40597
67. Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. (2017) 2:e00164–16. doi: 10.1128/mSystems.00164-16
68. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
69. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. (2014) 16:2891–904. doi: 10.1111/1462-2920.12238
70. Lemas DJ, Young BE, Baker PR II, Tomczik AC, Soderborg TK, Hernandez TL, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. Am J Clin Nutr. (2016) 103:1291–300. doi: 10.3945/ajcn.115.126375
71. Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother's weight on infant's microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. (2010) 92:1023–30. doi: 10.3945/ajcn.2010.29877
72. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. (2010) 107:18933–8. doi: 10.1073/pnas.1007028107
73. Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. (2014) 15:552. doi: 10.1186/s13059-014-0552-6
74. Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. (2016) 48:1413–7. doi: 10.1038/ng.3693
75. Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. (2016) 48:1407–12. doi: 10.1038/ng.3663
76. Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. (2016) 19:731–43. doi: 10.1016/j.chom.2016.04.017
77. Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, et al. Neonatal selection by toll-like receptor 5 influences long-term gut microbiota composition. Nature. (2018) 560:489–93. doi: 10.1038/s41586-018-0395-5
78. Chassaing B, Ley RE, Gewirtz AT. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. (2014) 147:1363–77.e17. doi: 10.1053/j.gastro.2014.08.033
79. Chassin C, Kocur M, Pott J, Duerr CU, Gütle D, Lotz M, et al. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. (2010) 8:358–68. doi: 10.1016/j.chom.2010.09.005
80. Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, Huang H, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. (2014) 6:107. doi: 10.1186/s13073-014-0107-1
81. Goethel A, Turpin W, Rouquier S, Zanello G, Robertson SJ, Streutker CJ, et al. Nod2 influences microbial resilience and susceptibility to colitis following antibiotic exposure. Mucosal Immunol. (2019) 12:720–32. doi: 10.1038/s41385-018-0128-y
82. Moltzau Anderson J, Lipinski S, Sommer F, Pan WH, Boulard O, Rehman A, et al. NOD influences trajectories of intestinal microbiota recovery after antibiotic perturbation. Cell Mol Gastroenterol Hepatol. (2020) 10:365–89. doi: 10.1016/j.jcmgh.2020.03.008
83. Zhang Y, Huang R, Cheng M, Wang L, Chao J, Li J, et al. Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome. (2019) 7:116. doi: 10.1186/s40168-019-0733-3
84. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. (2011) 145:745–57. doi: 10.1016/j.cell.2011.04.022
85. Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. (2014) 156:1045–59. doi: 10.1016/j.cell.2014.01.026
86. Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira-Silva S, Yilmaz B, Martens L, et al. Nlrp6- and ASC-dependent inflammasomes do not shape the commensal gut microbiota composition. Immunity. (2017) 47:339–48.e4. doi: 10.1016/j.immuni.2017.07.011
87. Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, et al. The NLR protein NLRP does not impact gut microbiota composition. Cell Rep. (2017) 21:3653–61. doi: 10.1016/j.celrep.2017.12.026
88. Gálvez EJC, Iljazovic A, Gronow A, Flavell R, Strowig T. Shaping of intestinal microbiota in Nlrp6- and Rag2-deficient mice depends on community structure. Cell Rep. (2017) 21:3914–26. doi: 10.1016/j.celrep.2017.12.027
89. Salzman NH, Hung K, Haribhai D, Chu H, Karsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. (2010) 11:76–83. doi: 10.1038/ni.1825
90. Lehotzky RE, Partch CL, Mukherjee S, Cash HL, Goldman WE, Gardner KH, et al. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci USA. (2010) 107:7722–7. doi: 10.1073/pnas.0909449107
91. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. (2006) 313:1126–30. doi: 10.1126/science.1127119
92. Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, et al. Antibacterial membrane attack by a pore-forming intestinal C- type lectin. Nature. (2014) 505:103–7. doi: 10.1038/nature12729
93. Wang L, Fouts DE, Stärkel P, Hartmann P, Chen P, Llorente C, et al. Intestinal REG lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. (2016) 19:227–39. doi: 10.1016/j.chom.2016.01.003
94. Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology. (2018) 154:1009–23.e14. doi: 10.1053/j.gastro.2017.11.003
95. Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. (2005) 307:1955–9. doi: 10.1126/science.1109051
96. Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. (2010) 105:2420–8. doi: 10.1038/ajg.2010.281
97. Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, et al. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci USA. (2013) 110:17059–64. doi: 10.1073/pnas.1306070110
98. Sommer F, Adam N, Johansson MEV, Xia L, Hansson GC, Bäckhed F. Altered mucus glycosylation in core O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS ONE. (2014) 9:e85254. doi: 10.1371/journal.pone.0085254
99. Jørgensen SF, Troseid M, Kummen M, Anmarkrud JA, Michelsen AE, Osnes LT, et al. Altered gut microbiota profile in common variable immunodeficiency associates with levels of lipopolysaccharide and markers of systemic immune activation. Mucosal Immunol. (2016) 9:1455–65. doi: 10.1038/mi.2016.18
100. Jørgensen SF, Holm K, Macpherson ME, Storm-Larsen C, Kummen M, Fevang B, et al. Selective IgA deficiency in humans is associated with reduced gut microbial diversity. J Allergy Clin Immunol. (2019) 143:1969–71.e11. doi: 10.1016/j.jaci.2019.01.019
101. Sterlin D, Fieschi C, Malphettes M, Larsen M, Gorochov G, Fadlallah J. Immune/microbial interface perturbation in human IgA deficiency. Gut Microbes. (2019) 10:429–33. doi: 10.1080/19490976.2018.1546520
102. Du CT, Gao W, Ma K, Yu SX, Li N, Yan SQ, et al. MicroRNA-146a deficiency protects against listeria monocytogenes infection by modulating the gut microbiota. Int J Mol Sci. (2018) 19:993. doi: 10.3390/ijms19040993
103. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. (2016) 19:32–43. doi: 10.1016/j.chom.2015.12.005
104. Moloney GM, Viola MF, Hoban AE, Dinan TG, Cryan JF. Faecal microRNAs: indicators of imbalance at the host-microbe interface? Benef Microbes. (2018) 9:175–83. doi: 10.3920/BM2017.0013
105. Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother's milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol. (2018) 9:361. doi: 10.3389/fimmu.2018.00361
106. Rognum TO, Thrane RS, Stoltenberg L, Vege Å, Brandtzaeg P. Development of intestinal mucosal immunity in fetal life and the first postnatal months. Pediatr Res. (1992) 32:145–8. doi: 10.1203/00006450-199208000-00003
107. Pabst O, Cerovic V, Hornef M. Secretory IgA in the coordination of establishment and maintenance of the microbiota. Trends Immunol. (2016) 37:287–96. doi: 10.1016/j.it.2016.03.002
108. Phalipon A, Cardona A, Kraehenbuhl JP, Edelman L, Sansonetti PJ, Corthésy B. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity. (2002) 17:107–15. doi: 10.1016/S1074-7613(02)00341-2
109. Rogier EW, Frantz AL, Bruno ME, Kaetzel CS. Secretory IgA is concentrated in the outer layer of colonic mucus along with gut bacteria. Pathogens. (2014) 3:390–403. doi: 10.3390/pathogens3020390
110. Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. (2015) 6:8292. doi: 10.1038/ncomms9292
111. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. (2012) 22:1147–62. doi: 10.1093/glycob/cws074
112. Kunz C, Meyer C, Collado MC, Geiger L, García-Mantrana I, Bertua-Ríos B, et al. Influence of gestational age, secretor, and Lewis blood group status on the oligosaccharide content of human milk. J Pediatr Gastroenterol Nutr. (2017) 64:789–98. doi: 10.1097/MPG.0000000000001402
113. Cabrera-Rubio R, Kunz C, Rudloff S, García-Mantrana I, Crehuá-Gaudiza E, Martínez-Costa C, et al. Association of maternal secretor status and human milk oligosaccharides with milk microbiota: an observational pilot study. J Pediatr Gastroenterol Nutr. (2019) 68:256–63. doi: 10.1097/MPG.0000000000002216
114. Manthey CF, Autran CA, Eckmann L, Bode L. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr. (2014) 58:165–8. doi: 10.1097/MPG.0000000000000172
115. He Y, Liu S, Leone S, Newburg DS. Human colostrum oligosaccharides modulate major immunologic pathways of immature human intestine. Mucosal Immunol. (2014) 7:1326–39. doi: 10.1038/mi.2014.20
116. He Y, Liu S, Kling DE, Leone S, Lawlor NT, Huang Y, et al. The human milk oligosaccharide 2'-fucosyllactose modulates CD14 expression in human enterocytes, thereby attenuating LPS-induced inflammation. Gut. (2016) 65:33–46. doi: 10.1136/gutjnl-2014-307544
117. Sodhi CP, Wipf P, Yamaguchi Y, Fulton WB, Kovler M, Niño DF, et al. The human milk oligosaccharides 2'-fucosyllactose and 6'-sialyllactose protect against the development of necrotizing enterocolitis by inhibiting toll-like receptor 4 signaling. Pediatr Res. (2020). doi: 10.1038/s41390-020-0852-3. [Epub ahead of print].
118. Layman DK, Lönnerdal B, Fernstrom JD. Applications for α-lactalbumin in human nutrition. Nutr Rev. (2018) 76:444–60. doi: 10.1093/nutrit/nuy004
119. Brück WM, Redgrave M, Tuohy KM, Lönnerdal B, Graverholt G, Hernell O, et al. Effects of bovine alpha-lactalbumin and casein glycomacropeptide-enriched infant formulae on faecal microbiota in healthy term infants. J Pediatr Gastroenterol Nutr. (2006) 43:673–9. doi: 10.1097/01.mpg.0000232019.79025.8f
120. Rozé JC, Barbarot S, Butel MJ, Kapel N, Waligora-Dupriet AJ, De Montgolfier I, et al. An α-lactalbumin-enriched and symbiotic-supplemented v. a standard infant formula: a multicentre, double-blind, randomised trial. Br J Nutr. (2012) 107:1616–22. doi: 10.1017/S000711451100479X
121. Davidson LA, Lönnerdal B. Persistence of human milk proteins in the breast-fed infant. Acta Paediatr Scand. (1987) 76:733–40. doi: 10.1111/j.1651-2227.1987.tb10557.x
122. Lönnerdal B. Bioactive proteins in breast milk. J Paediatr Child Health. (2013) 49 (Suppl. 1):1–7. doi: 10.1111/jpc.12104
123. Wakabayashi H, Yamauchi K, Takase M. Inhibitory effects of bovine lactoferrin and lactoferricin B on Enterobacter sakazakii. Biocontrol Sci. (2008) 13:29–32. doi: 10.4265/bio.13.29
124. Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. (1992) 1121:130–6. doi: 10.1016/0167-4838(92)90346-F
125. Ulvatne H, Samuelsen Ø, Haukland HH, Krämer M, Vorland LH. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol Lett. (2004) 237:377–84. doi: 10.1111/j.1574-6968.2004.tb09720.x
126. Bruni N, Capucchio MT, Biasibetti E, Pessione E, Cirrincione S, Giraudo L, et al. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules. (2016) 21:752. doi: 10.3390/molecules21060752
127. Brouwer CPJM, Rahman M, Welling MM. Discovery and development of a synthetic peptide derived from lactoferrin for clinical use. Peptides. (2011) 32:1953–63. doi: 10.1016/j.peptides.2011.07.017
128. Hentges DJ, Marsh WW, Petschow BW, Thal WR, Carter MK. Influence of infant diets on the ecology of the intestinal tract of human flora-associated mice. J Pediatr Gastroenterol Nutr. (1992) 14:146–52. doi: 10.1097/00005176-199202000-00005
129. Mastromarino P, Capobianco D, Campagna G, Laforgia N, Drimaco P, Dileone A, et al. Correlation between lactoferrin and beneficial microbiota in breast milk and infant's feces. Biometals. (2014) 27:1077–86. doi: 10.1007/s10534-014-9762-3
130. Balmer SE, Scott PH, Wharton BA. Diet and faecal flora in the newborn: lactoferrin. Arch Dis Child. (1989) 64:1685–90. doi: 10.1136/adc.64.12.1685
131. Roberts AK, Chierici R, Sawatzki G, Hill MJ, Volpato S, Vigi V. Supplementation of an adapted formula with bovine lactoferrin: I. effect on the faecal flora. Acta Paediatr. (1992) 81:119–24. doi: 10.1111/j.1651-2227.1992.tb12186.x
132. Sherman MP, Sherman J, Arcinue R, Niklas V. Randomized control trial of human recombinant lactoferrin: a substudy reveals effects on the fecal microbiome of very low birth weight infants. J Pediatr. (2016) 173:S37–42. doi: 10.1016/j.jpeds.2016.02.074
133. Castanet M, Costalos C, Haiden N, Hascoet JM, Berger B, Sprenger N, et al. Early effect of supplemented infant formulae on intestinal biomarkers and microbiota: a randomized clinical trial. Nutrients. (2020) 12:1481. doi: 10.3390/nu12051481
134. Griffiths EA, Duffy LC, Schanbacher FL, Dryja D, Leavens A, Neiswander RL, et al. In vitro growth responses of bifidobacteria and enteropathogens to bovine and human lactoferrin. Dig Dis Sci. (2003) 48:1324–32. doi: 10.1023/A:1024111310345
135. Wada Y, Lönnerdal B. Bioactive peptides derived from human milk proteins - mechanisms of action. J Nutr Biochem. (2014) 25:503–14. doi: 10.1016/j.jnutbio.2013.10.012
136. Mancinelli R, Rosa L, Cutone A, Lepanto MS, Franchitto A, Onori P, et al. Viral hepatitis and iron dysregulation: molecular pathways and the role of lactoferrin. Molecules. (2020) 25:1997. doi: 10.3390/molecules25081997
137. Liao Y, Jiang R, Lönnerdal B. Biochemical and molecular impacts of lactoferrin on small intestinal growth and development during early life. Biochem Cell Biol. (2012) 90:476–84. doi: 10.1139/o11-075
138. Legrand D. Overview of lactoferrin as a natural immune modulator. J Pediatr. (2016) 173:S10–5. doi: 10.1016/j.jpeds.2016.02.071
139. Vogl T, Leukert N, Barczyk K, Strupat K, Roth J. Biophysical characterization of S100A8 and S100A9 in the absence and presence of bivalent cations. Biochim Biophys Acta. (2006) 1763:1298–306. doi: 10.1016/j.bbamcr.2006.08.028
140. Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. (2011) 9:9. doi: 10.1186/1479-5876-9-9
141. Karlsson O, Rodosthenous RS, Jara C, Brennan KJ, Wright RO, Baccarelli AA, et al. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: implications for early child development. Epigenetics. (2016) 11:721–9. doi: 10.1080/15592294.2016.1216285
142. van Herwijnen MJ, Zonneveld MI, Goerdayal S, Nolte-'t Hoen EN, Garssen J, Stahl B, et al. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol Cell Proteomics. (2016) 15:3412–23. doi: 10.1074/mcp.M116.060426
143. Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. (2010) 1:7. doi: 10.1186/1758-907X-1-7
144. Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. (2012) 8:118–23. doi: 10.7150/ijbs.8.118
145. Kim SY, Yi DY. Analysis of the human breast milk microbiome and bacterial extracellular vesicles in healthy mothers. Exp Mol Med. (2020) 52:1288–97. doi: 10.20944/preprints202004.0404.v1
146. Fitzstevens JL, Smith KC, Hagadorn JI, Caimano MJ, Matson AP, Brownell EA. Systematic review of the human milk microbiota. Nutr Clin Pract. (2017) 32:354–64. doi: 10.1177/0884533616670150
147. Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr. (2012) 55:321–7. doi: 10.1097/MPG.0b013e31824fb899
148. Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. (2010) 18:298–307. doi: 10.1016/j.tim.2010.03.008
149. Díaz-Ropero MP, Martín R, Sierra S, Lara-Villoslada F, Rodríguez JM, Xaus J, et al. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol. (2007) 102:337–43. doi: 10.1111/j.1365-2672.2006.03102.x
150. Olivares M, Díaz-Ropero MP, Martín R, Rodríguez JM, Xaus J. Antimicrobial potential of four Lactobacillus strains isolated from breast milk. J Appl Microbiol. (2006) 101:72–9. doi: 10.1111/j.1365-2672.2006.02981.x
151. Moossavi S, Sepehri S, Robertson B, Bode L, Goruk S, Field CJ, et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe. (2019) 25:324–35.e4. doi: 10.1016/j.chom.2019.01.011
152. LeMay-Nedjelski L, Butcher J, Ley SH, Asbury MR, Hanley AJ, Kiss A, et al. Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum. BMC Microbiol. (2020) 20:219. doi: 10.1186/s12866-020-01901-9
153. Beghetti I, Biagi E, Martini S, Brigidi P, Corvaglia L, Aceti A. Human milk's hidden gift: implications of the milk microbiome for preterm infants' health. Nutrients. (2019) 11:2944. doi: 10.3390/nu11122944
154. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. (2012) 96:544–51. doi: 10.3945/ajcn.112.037382
155. Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. (2012) 72:77–85. doi: 10.1038/pr.2012.42
156. Andreas NJ, Hyde MJ, Gale C, Parkinson JR, Jeffries S, Holmes E, et al. Effect of maternal body mass index on hormones in breast milk: a systematic review. PLoS ONE. (2014) 9:e115043. doi: 10.1371/journal.pone.0115043
157. Mulligan CM, Friedman JE. Maternal modifiers of the infant gut microbiota: metabolic consequences. J Endocrinol. (2017) 235:R1–12. doi: 10.1530/JOE-17-0303
158. Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes. (2011) 35:522–9. doi: 10.1038/ijo.2011.27
159. Tang M, Frank DN, Sherlock L, Ir D, Robertson CE, Krebs NF. Effect of vitamin E with therapeutic iron supplementation on iron repletion and gut microbiome in US iron deficient infants and toddlers. J Pediatr Gastroenterol Nutr. (2016) 63:379–85. doi: 10.1097/MPG.0000000000001154
160. Knight LC, Wang M, Donovan SM, Dilger RN. Early-life iron deficiency and subsequent repletion alters development of the colonic microbiota in the pig. Front Nutr. (2019) 6:120. doi: 10.3389/fnut.2019.00120
161. Dosch NC, Guslits EF, Weber MB, Murray SE, Ha B, Coe CL, et al. Maternal obesity affects inflammatory and iron indices in umbilical cord blood. J Pediatr. (2016) 172:20–8. doi: 10.1016/j.jpeds.2016.02.023
162. Jones AD, Zhao G, Jiang YP, Zhou M, Xu G, Kaciroti N, et al. Maternal obesity during pregnancy is negatively associated with maternal and neonatal iron status. Eur J Clin Nutr. (2016) 70:918–24. doi: 10.1038/ejcn.2015.229
163. Simonyté Sjödin K, Domellöf M, Lagerqvist C, Hernell O, Lönnerdal B, Szymlek-Gay EA, et al. Administration of ferrous sulfate drops has significant effects on the gut microbiota of iron-sufficient infants: a randomised controlled study. Gut. (2019) 68:2095–7. doi: 10.1136/gutjnl-2018-316988
164. Dostal A, Fehlbaum S, Chassard C, Zimmermann MB, Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol Ecol. (2013) 83:161–75. doi: 10.1111/j.1574-6941.2012.01461.x
165. Dostal A, Baumgartner J, Riesen N, Chassard C, Smuts CM, Zimmermann MB, et al. Effects of iron supplementation on dominant bacterial groups in the gut, faecal SCFA and gut inflammation: a randomised, placebo-controlled intervention trial in South African children. Br J Nutr. (2014) 112:547–56. doi: 10.1017/S0007114514001160
166. Dostal A, Lacroix C, Bircher L, Pham VT, Follador R, Zimmermann MB, et al. Iron modulates butyrate production by a child gut microbiota in vitro. mBio. (2015) 6:e01453–15. doi: 10.1128/mBio.01453-15
167. Forgie AJ, Drall KM, Bourque SL, Field CJ, Kozyrskyj AL, Willing BP. The impact of maternal and early life malnutrition on health: a diet-microbe perspective. BMC Med. (2020) 18:135. doi: 10.1186/s12916-020-01584-z
168. Rusu IG, Suharoschi R, Vodnar DC, Pop CR, Socaci SA, Vulturar R, et al. Iron supplementation influence on the gut microbiota and probiotic intake effect in iron deficiency-a literature-based review. Nutrients. (2020) 12:E1993. doi: 10.3390/nu12071993
169. Iram N, Mildner, Prior M, Petzelbauer P, Fiala C, Hacker S, et al. Age-related changes in expression and function of Toll-like receptors in human skin. Development. (2012) 139:4210–9. doi: 10.1242/dev.083477
170. Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, Rudensky B, et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. (2004) 126:520–8. doi: 10.1053/j.gastro.2003.11.019
171. Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol. (2005) 288:G1055–65. doi: 10.1152/ajpgi.00328.2004
172. Jilling T, Simon D, Lu J, Meng FJ, Li D, Schy R, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol. (2006) 177:3273–82. doi: 10.4049/jimmunol.177.5.3273
173. Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. (2007) 446:552–6. doi: 10.1038/nature05590
174. Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. (2007) 117:3909–21. doi: 10.1172/JCI33084
175. Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. (2007) 446:557–61. doi: 10.1038/nature05698
176. Leaphart CL, Cavallo J, Gribar SC, Cetin S, Li J, Branca MF, et al. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. Immunol. (2007) 179:4808–20. doi: 10.4049/jimmunol.179.7.4808
177. Gribar SC, Sodhi CP, Richardson WM, Anand RJ, Gittes GK, Branca MF, et al. Reciprocal expression and signaling of TLR4 and TLR9 in the pathogenesis and treatment of necrotizing enterocolitis. J Immunol. (2009) 182:636–46. doi: 10.4049/jimmunol.182.1.636
178. Asquith MJ, Boulard O, Powrie F, Maloy KJ. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. (2010) 139:519–29, 529.e1–2. doi: 10.1053/j.gastro.2010.04.045
179. Zhou W, Cao Q, Peng Y, Zhang QJ, Castrillon DH, DePinho RA, et al. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology. (2009) 137:1403–14. doi: 10.1053/j.gastro.2009.06.049
180. Stockinger S, Duerr CU, Fulde M, Dolowschiak T, Pott J, Yang I, et al. TRIF signaling drives homeostatic intestinal epithelial antimicrobial peptide expression. J Immunol. (2014) 193:4223–34. doi: 10.4049/jimmunol.1302708
181. Kanagavelu S, Flores C, Termini JM, Riveron R, Romero L, Chung K, et al. TIR-domain-containing adapter-inducing interferon-β. (TRIF) regulates Th17-mediated intestinal immunopathology in colitis. Mucosal Immunol. (2015) 8:296–306. doi: 10.1038/mi.2014.67
182. Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen, et al. Mrp8 and Mrp14 are endogenous activators of toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. (2007) 13:1042–9. doi: 10.1038/nm1638
183. Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, et al. Colitis induced in mice with dextran sulfate sodium. (DSS) is mediated by the NLRP3 inflammasome. Gut. (2010) 59:1192–9. doi: 10.1136/gut.2009.197822
184. Siegmund B, Lehr HA, Fantuzzi G, Dinarello C A. IL-1 beta -converting enzyme. (caspase-1) in intestinal inflammation. Proc Natl Acad Sci USA. (2001) 98:13249–54. doi: 10.1073/pnas.231473998
185. Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. (2010) 32:379–91. doi: 10.1016/j.immuni.2010.03.003
186. Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. (2010) 207:1745–55. doi: 10.1084/jem.20100257
187. Hirota SA, Ng J, Lueng A, Khajah M, Parhar K, Li Y, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. (2011) 17:1359–72. doi: 10.1002/ibd.21478
188. Kim YG, Kamada N, Shaw MH, Warner N, Chen GY, Franchi L, et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. (2011) 34:769–80. doi: 10.1016/j.immuni.2011.04.013
189. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. (2001) 411:603–6. doi: 10.1038/35079114
190. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. (2001) 411:599–603. doi: 10.1038/35079107
191. Mallow EB, Harris A, Salzman N, Russell JP, DeBerardinis RJ, Ruchelli E, et al. Human enteric defensins. gene structure and developmental expression. J Biol Chem. (1996) 271:4038–45. doi: 10.1074/jbc.271.8.4038
192. Ouellette AJ, Greco RM, James M, Frederick D, Naftilan J, Fallon JT. Developmental regulation of cryptdin, a corticostatin/defensin precursor mRNA in mouse small intestinal crypt epithelium. J Cell Biol. (1989) 108:1687–95. doi: 10.1083/jcb.108.5.1687
193. Cunliffe RN, Mahida YR. Expression and regulation of antimicrobial peptides in the gastrointestinal tract. J Leukoc Biol. (2004) 75:49–58. doi: 10.1189/jlb.0503249
194. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. (2003) 3:710–20. doi: 10.1038/nri1180
195. Hazlett L, Wu M. Defensins in innate immunity. Cell Tissue Res. (2011) 343:175–88. doi: 10.1007/s00441-010-1022-4
196. Blyth GAD, Connors L, Fodor C, Cobo ER. The network of colonic host defense peptides as an innate immune defense against enteropathogenic bacteria. Front Immunol. (2020) 11:965. doi: 10.3389/fimmu.2020.00965
197. Schauber J, Svanholm C, Termen S, Iffland K, Menzel T, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. (2003) 52:735–41. doi: 10.1136/gut.52.5.735
198. O'Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. (1999) 163:6718–24.
199. Chairatana P, Nolan EM. Defensins, lectins, mucins, and secretory immunoglobulin A: microbe-binding biomolecules that contribute to mucosal immunity in the human gut. Crit Rev Biochem Mol Biol. (2017) 52:45–56. doi: 10.1080/10409238.2016.1243654
200. Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. (2018) 10:eaan1217. doi: 10.1126/scitranslmed.aan1217
201. Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep. (2019) 9:13574. doi: 10.1038/s41598-019-49923-2
202. Sarshar M, Scribano D, Ambrosi C, Palamara AT, Masotti A. Fecal microRNAs as innovative biomarkers of intestinal diseases and effective players in host-microbiome interactions. Cancers. (2020) 12:E2174. doi: 10.3390/cancers12082174
203. Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. (2016) 7:11106. doi: 10.1038/ncomms11902
204. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. (2004) 101:13368–73. doi: 10.1073/pnas.0403453101
205. Chassin C, Hempel C, Stockinger S, Dupont A, Kübler JF, Wedemeyer J, et al. MicroRNA-146a-mediated downregulation of IRAK1 protects mouse and human small intestine against ischemia/reperfusion injury. EMBO Mol Med. (2012) 4:1308–19. doi: 10.1002/emmm.201201298
Keywords: neonatal mucosal immunity, developing microbiota, shaping of microbiota, gut microbial colonization, host factors
Citation: Pirr S and Viemann D (2020) Host Factors of Favorable Intestinal Microbial Colonization. Front. Immunol. 11:584288. doi: 10.3389/fimmu.2020.584288
Received: 17 July 2020; Accepted: 09 September 2020;
Published: 07 October 2020.
Edited by:
Philipp Henneke, University of Freiburg, GermanyCopyright © 2020 Pirr and Viemann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabine Pirr, UGlyci5TYWJpbmVAbWgtaGFubm92ZXIuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.