
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 30 November 2020
Sec. Molecular Innate Immunity
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.573915
Cytosolic DNA sensing is a fundamental mechanism by which organisms handle various stresses, including infection and genotoxicity. The hematopoietic system is sensitive to stresses, and hematopoietic changes are often rapid and the first response to stresses. Based on the transcriptome database, cytosolic DNA sensing pathways are widely expressed in the hematopoietic system, and components of these pathways may be expressed at even higher levels in hematopoietic stem and progenitor cells (HSPCs) than in their certain progeny immune cells. Recent studies have described a previously unrecognized role for cytosolic DNA sensing pathways in the regulation of hematopoiesis under both homeostatic and stress conditions. In particular, the recently discovered cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) pathway is a critical modulator of hematopoiesis. Perturbation of the cGAS-STING pathway in HSPCs may be involved in the pathogenesis of hematopoietic disorders, autoimmune diseases, and inflammation-related diseases and may be candidate therapeutic targets. In this review, we focus on the recent findings of the cGAS-STING pathway in the regulation of hematopoiesis, and its physiopathological significance including its implications in diseases and therapeutic potential.
Hematopoietic stem and progenitor cells (HSPCs) in the bone marrow (BM) are responsible for the maintenance of both the homeostatic and stressed hematopoiesis. By modulating the balance between self-renewal and multilineage differentiation, HSPCs give rise to all mature hematopoietic cell types within the blood and immune system, including lymphoid cells, granulocytes, monocytes, dendritic cells (DCs), erythrocytes, and platelets (1). Granulocytes, monocytes, erythrocytes, and platelets are collectively called myeloid cells. Under homeostatic conditions, HSPCs are maintained in a quiescent state in the BM and undergo balanced differentiation into lymphoid and myeloid cells. However, the hematopoietic system is sensitive to stresses, such as infections and genotoxicity, and hematopoietic changes are often the rapid and the first response to stress. Under stress conditions, the quiescent state is disrupted, and HSPCs will biasedly differentiate into specific hematopoietic populations. During these processes, stresses are directly sensed by HSPCs or by HSPC progeny cells and niche cells, which in turn modulate hematopoiesis (2). Therefore, HSPCs serve as integrative hubs for the conversion of stress signals into demand-adapted hematopoiesis.
The mechanisms by which HSPCs sense stress have not been completely elucidated; however, it is well-recognized that the recognition of stress-associated molecular patterns and triggering of the immune response are fundamental functions of the immune system (3). Among these pathways, aberrant nucleic acid recognition has been regarded as a key mechanism. The pathways involved in RNA recognition have been reviewed elsewhere (4–6) and will not be covered here. Regarding DNA recognition, Toll-like receptor 9 (TLR9), absent in melanoma 2 (AIM2) and cyclic GMP-AMP synthase (cGAS), are the main specialized receptors that initiate DNA-driven immune responses in mammalian cells (7). The detection of cytosolic DNA activates the stimulator of interferon genes (STING)-dependent type I interferon (IFN-I)-stimulatory pathway, which is essential for antiviral responses. Generally, the cGAS-STING pathway is required for the IFN-I response triggered by various types of exogenous pathogenic DNA. Loss of the compartmentalization of endogenous DNA, including nuclear DNA (nDNA) and mitochondrial DNA (mtDNA), as well as disturbances in endogenous DNA metabolism also result in cytosolic self-DNA accumulation, which induces immune responses via the cGAS-STING pathway (8–10). Emerging evidence has indicated roles for the cGAS-STING pathway in modulating antitumor immunity (8, 9). Therefore, cellular and organismal homeostasis may be achieved through the recognition of aberrant cytosolic DNA and activation of the cGAS-STING pathway.
Interestingly, based on the transcriptome database, cGAS may be expressed at high levels in HSPCs and even exceed the levels in their certain progeny cells (11). In this review, we primarily focus on the role of the cGAS-STING pathway in hematopoietic regulation and its physiopathological significance. We envision that a better understanding of the stress sensing in HSPCs mediated by the cGAS-STING pathway will help refine potential targets for interventions in patients with hematopoiesis-related diseases.
cGAS is a recently discovered DNA-binding protein that condenses ATP and GTP to synthesize 2’3’-cyclic GMP-AMP (2’3’-cGAMP), which functions as a second messenger that activates STING (12) (Figure 1). In fact, cGAS is not a cytosolic protein but is instead localized to the plasma membrane via the interaction of an N-terminal phosphoinositide-binding domain with phosphatidylinositol 4,5-bisphosphate. The location of cGAS at the plasma membrane ensures the efficient discrimination between self- and non-self-DNA, as cGAS mutants defective in lipid binding induce potent IFN-I responses to genotoxic stress (self-DNA) but weaker responses to viral infection (non-self-DNA) (13). cGAS binds double-stranded DNA (dsDNA) and sometimes single-stranded DNA (ssDNA) via a positively charged surface and its zinc thumb to interact with the DNA (14). The activation of cGAS is also modulated by circular RNA (15), protein interactions (16–18), and posttranslational modifications, including glutamylation, ubiquitination, acetylation, and sumoylation (19–23).
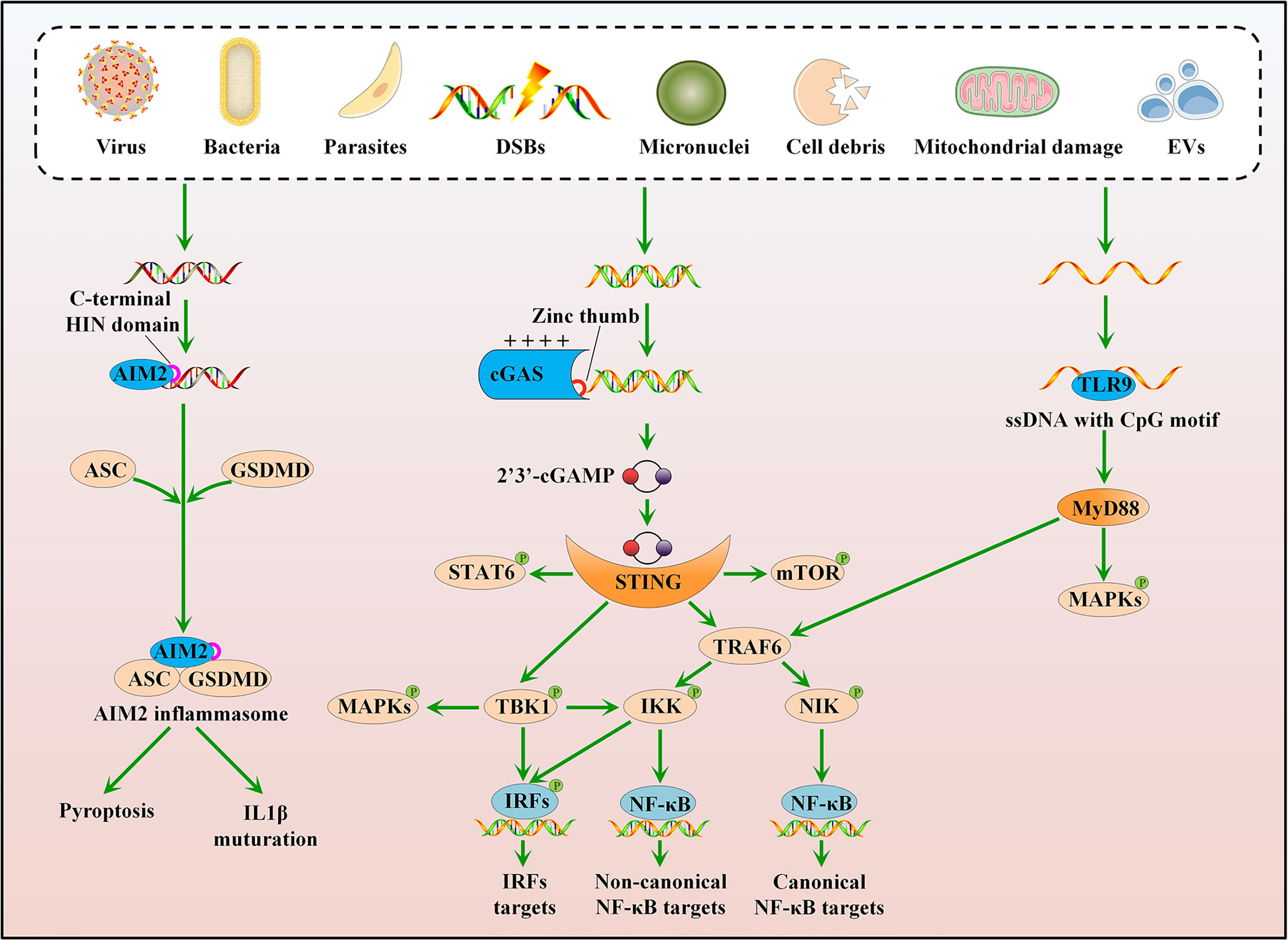
Figure 1 Cytosolic DNA sensing pathways. cGAS can sense various stress signals of cytosolic DNA and trigger a STING-dependent response. Upon activation, STING mainly recruits and activates three kinases: TBK1, IKK, and NIK. Besides, the cGAS-STING pathway can directly or indirectly activate IRF1, IRF5, IRF7, mTOR, STAT6, and MAPKs. TLR9 and AIM2 also recognize cytosolic DNA. The ligated TLR9 interacts with MyD88 to activate TRAF6 and MAPKs, whereas activated AIM2 associates with ASC and GSDMD to form an inflammasome complex.
Upon activation, STING translocates from the endoplasmic reticulum (ER) membrane to the Golgi and primarily recruits and activates three kinases: TANK-binding kinase 1 (TBK1), which phosphorylates interferon regulatory factor 3 (IRF3); IκB kinase (IKK), which is recruited by TNF receptor-associated factor 6 (TRAF6) and subsequently activates canonical nuclear factor-κB (NF-κB) signaling; and mitogen-activated protein kinase kinase kinase 14 (MAP3K14, best known as NIK), which activates noncanonical NF-κB signaling (Figure 1) (24). TBK1 acts redundantly with IKK to drive NF-κB signaling, and IKKα/IKKβ also activates IRF3 (Figure 1) (25, 26). The cGAS-STING pathway directly or indirectly activates other signaling molecules, such as IRF1, IRF5, IRF7, mammalian target of the rapamycin (mTOR), signal transducer and activator of transcription 6 (STAT6), and mitogen-activated protein kinases (MAPKs), including p38, extracellular signal-regulated kinases 1/2 (ERK1/2), and c-Jun-N-terminal kinase (JNK) (Figure 1) (27–33). Consequently, the activated cGAS-STING pathway induces the production of IFN-I and other cytokines involved in the immune response and inflammation. The cGAS-STING pathway is also implicated in the regulation of other important cellular processes, such as autophagy, cell cycle, cell death, and chromosomal stability, to eliminate pathogens, infected cells, or malignantly transformed cells (34–38).
Other sensors, including TLR9 and AIM2, also recognize cytosolic DNA. TLR9 plays a role in DNA sensing in the endosomal compartment and preferentially binds to unmethylated CpG motifs of ssDNA (39), whereas AIM2 senses cytosolic DNA via its C-terminal HIN domain in a sequence-independent manner (Figure 1) (40). Consequently, the ligated TLR9 interacts with its proximal adaptor protein, myeloid differentiation primary response 88 (MyD88), to activate TRAF6 and MAPKs (41); activated AIM2 associates with the adaptor molecule apoptosis-associated speck-like protein containing caspase recruitment domain (ASC), to form an inflammasome complex, which activates gasdermin D (GSDMD)-mediated pyroptosis and caspase 1 (CASP1)-dependent maturation of interleukin 1β (IL1β) (Figure 1) (40, 42). However, both TLR9 and AIM2 are dispensable for IFN-I induction, suggesting a central role of the cGAS-STING pathway in cytosolic DNA sensing.
Upon stresses, such as infections and genotoxicity, a considerable number of hematopoietic cells, especially myeloid cells, are depleted. The hematopoietic system needs to respond rapidly to replenish myeloid cells via a process called “emergency myelopoiesis” (43). According to recent studies, HSPCs may be the central hub of stress responses (43–48). HSPCs are able to skip several steps in the traditional tree-like hematopoietic hierarchy and directly differentiate into myeloid progenitor cells (49–51). Meanwhile, hematopoietic stem cells (HSCs) are heterogeneous, and myeloid-biased HSC subtypes may selectively expand in response to stress (52). Through these mechanisms, the hematopoietic system rapidly satisfies the increased demand for myeloid cells during stress. Therefore, the rapid myeloid-biased differentiation of HSPCs is required for the hematopoietic system to adapt to stresses. Although the cGAS-STING pathway is essential for sensing and defending against stresses, its role in regulation hematopoiesis has rarely been examined. The present review summarizes the role of the cGAS-STING pathway to obtain a better understanding of the mechanism regulating hematopoiesis (Figure 2).
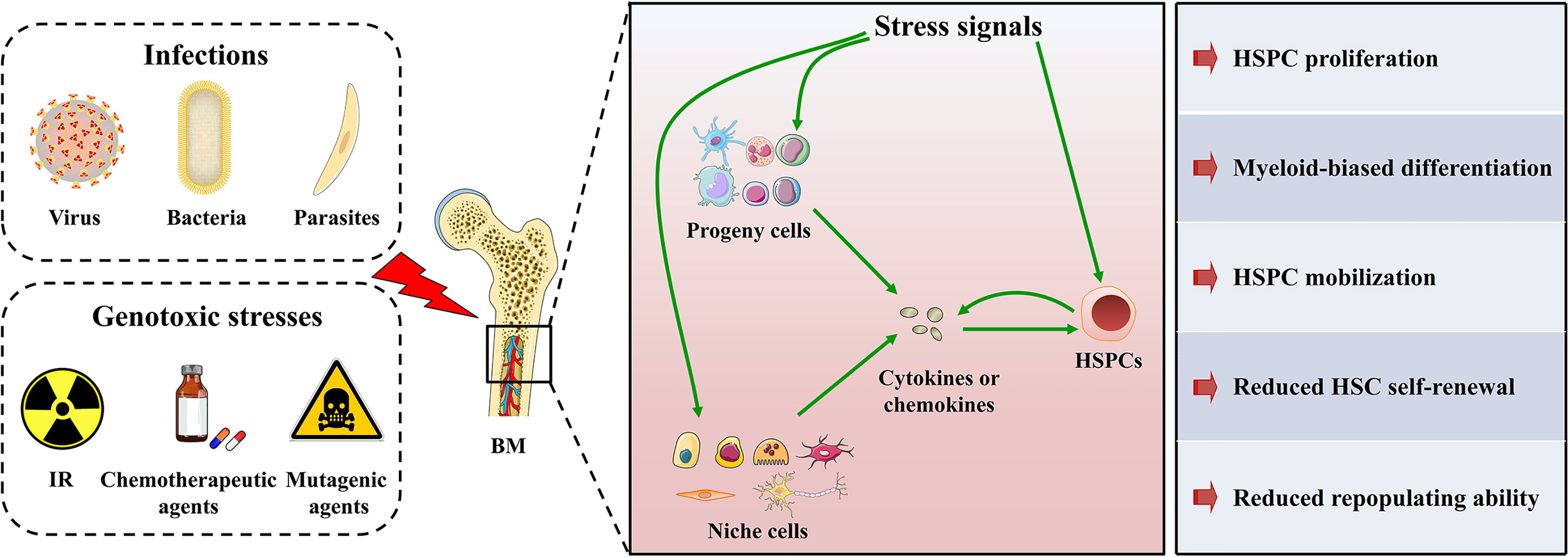
Figure 2 HSPC maintenance under stress conditions. HSPCs in BM are sensitive to various stresses. On one hand, HSPCs can directly sense and respond to stress signals through specialized receptors. On the other hand, HSPC progeny cells and niche cells in the BM microenvironment can sense stress signals and convert the stress signals into the secretion of cytokines or chemokines, which in turn modulate HSPC maintenance. In addition, HSPCs themselves possess cytokine production ability that even overmatches that of their progeny cells in both magnitude and breadth. Therefore, HSPCs can also convert stress signals into cytokine or chemokine signals and modulate their maintenance in an autocrine manner. Consequently, HSPCs will undergo mobilization, proliferation, and myeloid-biased differentiation to rapidly produce myeloid cells, but their self-renewal and repopulating ability will be mitigated.
A circular RNA named cia-cGAS was recently reported to be expressed at high levels in the nuclei of HSCs. Cia-cGAS displays a stronger binding affinity for cGAS than self-DNA. Cia-cGAS binds cGAS in the nucleus to block its synthase activity under steady-state condition, while a cia-cGAS deficiency will lead to increased expression of IFN-I in the BM but decreased numbers of dormant HSCs. Therefore, cia-cGAS suppresses the cGAS-mediated production of IFN-I in HSCs and the exhaustion of dormant HSCs (Table 1) (15). R-loops, the nucleic acid structures consisting of displaced ssDNAs and RNA : DNA hybrids, also activate cGAS-STING pathway in HSPCs. DEAD-box Helicase 41 (DDX41) binds R-loops and acts as a suppressor of R-loop accumulation. DDX41 insufficiency will result in excessive R-loop accumulation, which subsequently activate the cGAS-STING-NF-κB pathway to promote HSPC expansion via secreting inflammatory cytokines (Table 1) (53). Meanwhile, BAK/BAX-mediated apoptosis will trigger the release of mtDNA, which is recognized by the cGAS-STING pathway. The subsequent production of IFN-I in hematopoietic cells will expand HSCs but suppress their self-renewal capacity (Table 1). However, caspase activation following BAK/BAX activation will attenuate the IFN-I response possibly through global cellular demolition (54). In addition, gain-of-function STING mutation-induced constitutive activation of the cGAS-STING pathway causes myeloid cell expansion and lymphoid cell cytopenia in an IRF3-independent manner (75, 76). Of note, in this case, the HSC pool is only moderately affected, whether HSC function such as self-renewal capacity is influenced remains unknown (76).
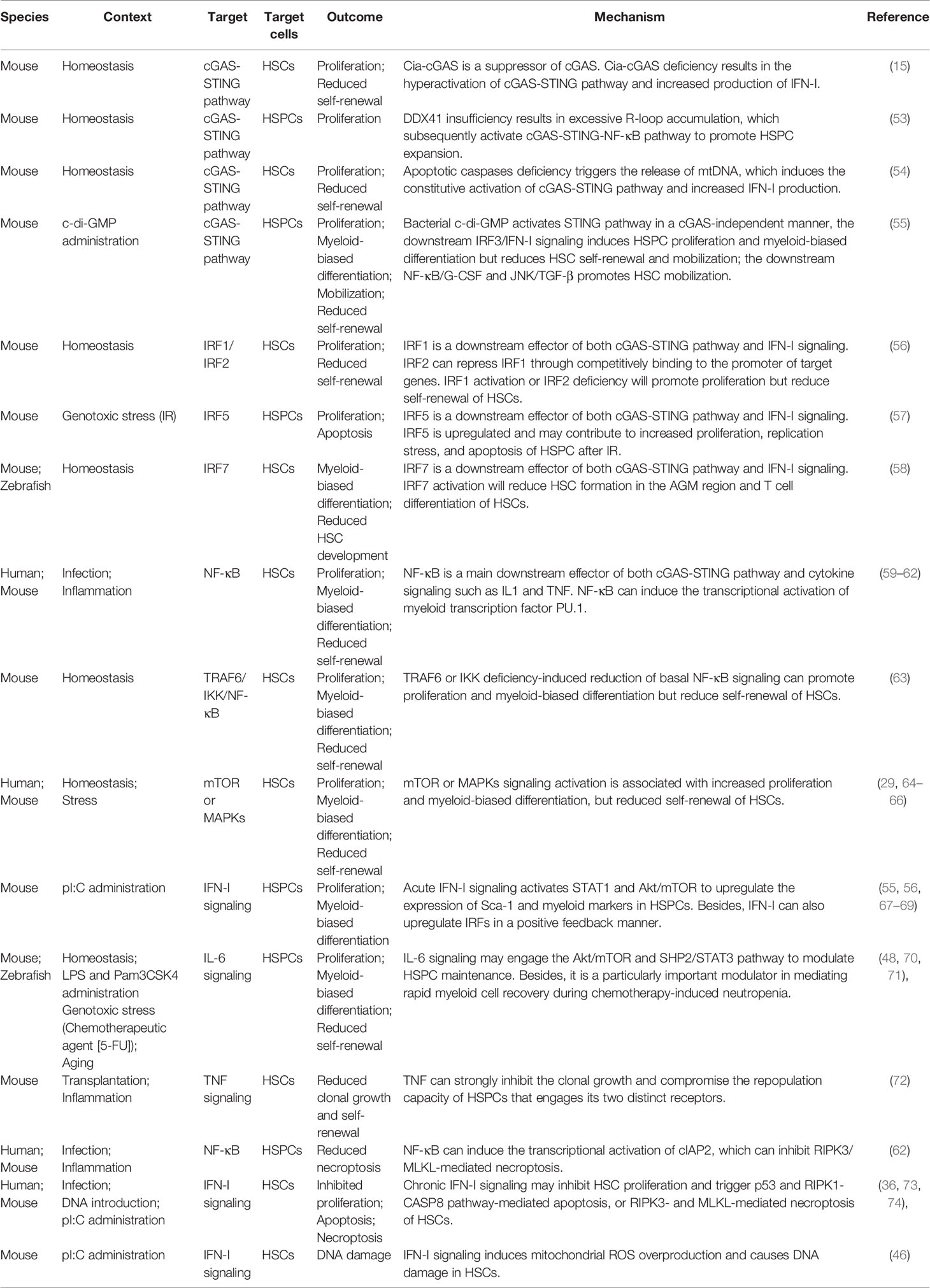
Table 1 Roles of stress-related pathways including the cGAS-STING pathway in regulating HSPC maintenance.
The cGAS-STING pathway directly and indirectly regulates cell activities, including proliferation, differentiation, cell death, autophagy, senescence, and metabolism, through the activation of its downstream effectors or the secretion of cytokines. However, evidence for whether and how this pathway regulates HSPC maintenance is scarce. We discuss the recently discovered functions of the cGAS-STING pathway and its potential roles in regulating HSPC maintenance.
Downstream effectors of the cGAS-STING pathway, including IRFs, NF-κB, mTOR, and MAPKs, are closely associated with hematopoiesis. The roles of mTOR and MAPKs in HSPC maintenance are well-defined (Table 1) (29, 64–66). IRFs and NF-κB are transcription factors, and their activation directly modulates hematopoiesis at the genetic level. IRF1, IRF3, IRF5, and IRF7 act downstream of the cGAS-STING pathway (31–33, 55, 77). IRF2 represses IRF1 by competitively binding to the promoter of target genes. IRF1 activation and IRF2 deficiency promote the proliferation of HSCs but reduce their self-renewal capacity (Table 1) (56). IRF3 is implicated in infection-associated HSPC proliferation and a reduction in self-renewal (55). IRF5 may participate in the mechanism regulating HSPC survival, because a deletion of IRF5 protects HSPCs from DNA damage-induced apoptosis (Table 1) (57). IRF7 activation reduces HSC populations in the aorta-gonad-mesonephros (AGM) region and the T cell differentiation of HSCs, suggesting roles for IRF7 in HSC development and specification (Table 1) (58). These data suggest distinct functions of IRFs in regulating HSC maintenance. NF-κB promotes the myeloid differentiation of HSCs through the transcriptional activation of the myeloid transcription factor PU.1 (Table 1) (59). However, a reduction in basal NF-κB signaling induced by a TRAF6 or IKK deficiency promotes proliferation and myeloid-biased differentiation but reduces the self-renewal of HSCs under homeostatic conditions (Table 1) (63).
The most important function of the cGAS-STING pathway is the production of various cytokines and chemokines. HSPCs and their progeny and niche cells contribute to cytokine production during stress. Notably, the ability of HSPCs to produce cytokines exceeds their progeny cells in both magnitude and breadth (48). Given the high expression of cytokine and chemokine receptors, cytokines and chemokines may help HSPCs rapidly adapt to stresses in an autocrine manner. Here, we discuss the most common cytokines downstream of the cGAS-STING pathway, including IFN-I, Interleukin-1 (IL-1), IL-6, and TNF.
IFN-I is the hallmark cytokine produced following the activation of the cGAS-STING pathway. Acute IFN-I signaling promotes HSC proliferation but reduces the self-renewal capacity and mobilization of HSCs (Table 1) (55, 56, 67, 68). IFN-I signaling increases the phosphorylation of STAT1 and Akt/mTOR and upregulates the expression of stem cell antigen-1 (Sca-1) and myeloid markers, such as CD41. Conversely, inhibition of STAT1, Akt/mTOR, or Sca-1 will cause HSCs to become insensitive to IFN-I stimulation, indicating that STAT1, Akt/mTOR, and Sca-1 mediate the effects of IFN-I (67, 68). IFN-I also upregulates IRFs through a positive feedback mechanism (69), which may in turn regulate HSPC maintenance, as described above. However, chronic IFN-I signaling inhibits HSC proliferation and triggers apoptosis mediated by p53 and the receptor-interacting protein kinase-1 (RIPK1)-CASP8 pathway-mediated or RIPK3- and mixed-lineage kinase domain-like protein (MLKL)-mediated necroptosis of HSCs, leading to HSC exhaustion (Table 1) (36, 73, 74, 78). Notably, IFN-I signaling-induced mitochondrial reactive oxygen species (ROS) overproduction causes DNA damage in HSCs (Table 1) (46), which provides a novel connection between stress hematopoiesis and the occurrence of DNA damage and decreased function of adult HSCs.
IL-1 promotes the proliferation and myeloid-biased differentiation of HSCs and prevents chemotherapy-mediated myelosuppression by activating the NF-κB-PU.1 axis, but it mitigates the self-renewal capacity of HSCs (Table 1) (59–61). Similarly, IL-6 also promotes HSPC proliferation and myeloid-biased differentiation in an auto- or paracrine manner (Table 1) (48, 70, 71). In particular, Zhao et al. identified IL-6 as a particularly important modulator that mediates rapid myeloid cell recovery during chemotherapy-induced neutropenia (48).
TNF overproduction is closely associated with the BM hematopoietic failure observed in many patients with acute or chronic inflammatory diseases. TNF strongly inhibits clonal growth and compromises the repopulation capacity of HSCs (72). Notably, actively cycling rather than quiescent HSCs are the primary targets of TNF-mediated hematopoietic suppression (Table 1) (72). The latest research has revealed different effects of TNF on HSCs, and myeloid progenitors are actually different. TNF prevents HSC necroptosis and promotes myeloid-biased differentiation via the NF-κB-dependent transcriptional activation of cellular inhibitor of apoptosis protein 2 (cIAP2) and PU.1, respectively, whereas it induces myeloid progenitor apoptosis. Consequently, TNF-α prevents the necroptosis, but not apoptosis, of HSPCs and poises HSPCs for myeloid cell production but kills other blood cells, which facilitates hematopoietic clearance and promotes regeneration in response to inflammatory stress (Table 1) (62).
The cGAS-STING pathway protects against infections with viruses, bacteria, and protozoan parasites by sensing pathogenic DNA, infection-induced mitochondrial damage, bacterial proteins, virulence factors and metabolites, and bacteria-derived cyclic dinucleotides (CDNs) (79–84). Meanwhile, infected cells transactivate the cGAS-STING pathway in bystander cells via the cell-to-cell transmission of cGAMP or the release of extracellular vesicles (EVs) (85–89). Based on accumulating evidence, HSPCs, and not only their progeny immune cells, are able to directly sense and respond to infections. For example, HSPCs directly sense a mimic of viral nucleic acid, polyinosinic:polycytidylic acid (pI:C), to elicit IFN-I production through a process independent of TLR signaling (68). Meanwhile, HSPCs were recently shown to sense one of the CDNs, c-di-GMP, via STING. Upon c-di-GMP exposure, STING activation in HSPCs induces HSC entry into the cell cycle and promotes HSPC proliferation and mobilization, but it decreases the number and repopulation capacity of LT-HSC (55). On the other hand, HSPCs indirectly recognize the acute or chronic infectious state by sensing the extrinsic signals, such as the cytokines or chemokines released by their progeny immune cells, niche cells, or themselves (Figure 2). In particular, the abundance of these receptors on HSPCs increases significantly during systemic infections (48).
Genotoxic stress is derived from a wide variety of physical and chemical sources, such as ionizing radiation, chemotherapeutic agents, and mutagenic agents. In fact, the long-living property of HSCs makes these cells particularly susceptible to genotoxic stress. HSCs are thought to be the gatekeeper of genomic integrity of the entire hematopoietic system, because an unrepaired genetic lesion in a single HSC may spread throughout the HSC pool and propagate to all of the hematological compartments via self-renewing division and differentiation (90). DNA damage occurs when cells are exposed to genotoxic stress. In fact, HSPCs are sensitive to genotoxic stress-induced DNA damage, including DSBs and micronuclei formation (91). Recent advances suggest that the cGAS-STING pathway may play an essential role in the genotoxic stress of HSPCs by sensing DSBs and micronuclei (91–94). Therefore, reacquainting the role of the cGAS-STING pathway in HSPC (Table 1) maintenance under genotoxic stress is of great value to understand the pathogenesis of some hematopoietic disorders.
Numerous functions of the cGAS-STING pathway have been established recently, and many of these functions may be extrapolated to HSPC maintenance. First, the activation of the cGAS-STING pathway may promote or inhibit HSC death, depending on the context or HSC subtype. Activation of the cGAS-STING pathway in mouse myeloid cells allows them to resist apoptosis, but activation is amplified in mouse lymphoid cells and results in apoptosis (95). Activation of the cGAS-STING pathway in human myeloid cells also elicits lysosomal cell death with apoptotic-like features (35). Besides, activation of cGAS-STING pathway induces apoptosis by disrupting calcium homeostasis and/or suppressing of B-cell lymphoma-extra large (Bcl-xL) expression (17). A positive feedback loop of death signaling may also exist because PUMA induction during cell death may promote the cytosolic release of mtDNA and subsequent activation of the cGAS-STING pathway (96). Autophagy, which is closely associated with HSC maintenance (97), is a primordial function of the cGAS-STING pathway. The cGAS-STING pathway induces autophagy by facilitating the lipidation of microtubule-associated protein 1 light chain 3 (LC3) or inactivating mTOR. However, the cytosolic chromatin fragments that result from replicative crisis-induced telomeric DNA damage activate the cGAS-STING pathway and engage the autophagic cell death to restrict chromosomal instability and prevent tumorigenesis (98). cGAS is also recruited to DSBs in the nucleus and interacts with poly(ADP-ribose) polymerase 1 (PARP1) to suppress homologous recombination by impeding the formation of the PARP1-Timeless complex, which may aggravate genomic instability and subsequently induce malignant transformation or cell death (99, 100).
An understanding of the role of the cGAS-STING pathway in regulating hematopoiesis provides new insights into the pathogenesis of and interventions for hematopoietic disorders. Gain-of-function mutations or aberrant activation of the cGAS-STING pathway may participate in the pathogenesis of myelopoiesis-associated clonal hematopoiesis and myeloid malignancies (75, 101, 102). Recent advances spotlighted on the role of leukemia stem cells (LSCs) in the driving of persistent disease and the relapse after robust antitumor therapy (103). Given the many common characteristics shared between LSCs and HSCs, aberrant activation of the cGAS-STING pathway may be involved in the maintenance of myeloid LSCs through promoting malignant reprogramming or self-renewal (104, 105). On the other hand, the activation of a cGAS-STING pathway may improve the therapeutic effect of myeloid leukemia via inducing cell death or differentiation of myeloid LSCs (95, 106, 107).
The cGAS-STING pathway in HSPCs may be employed as a therapeutic target or to develop vaccine adjuvants for chronic infections. Activation of the cGAS-STING pathway may promote the myeloid-biased differentiation of HSPCs to enhance the anti-infection reservoir of the immune system. However, it should bear in mind that the activation of the cGAS-STING pathway may also increase the vulnerability to or severity of infections through dampening adaptive immunity, exacerbating inflammation or facilitating pathogen replication, survival, and infection (81, 108–113).
HSPC maintenance is closely associated with the pathogenesis of autoimmune diseases. HSPCs confer protection to secondary infection, in a phenomenon termed “trained innate immunity” or “innate immune memory,” which contributes to autoimmune diseases through exaggerating immune responses (114–116). Meanwhile, the myeloid progenitors released from the BM may infiltrate peripheral tissues and produce primed neutrophils in situ to exacerbate inflammatory responses within the affected tissues (117, 118). On the other hand, patients with autoimmune diseases may have a poor response to infectious challenges due to the deficiency of lymphoid cells (119). During these processes, the cGAS-STING pathway in HSPCs may play an important role.
Myelopoiesis or lymphocytopenia are closely associated with inflammation-related diseases including gain-of-function STING mutation-induced vasculopathy, cardiovascular diseases (CVD), and metabolic diseases (75, 120–126). Conversely, the modulation of HSPC maintenance that reduces myeloid cell production can protect against CVD (127, 128). Therefore, the cGAS-STING pathway in HSPCs may be a promising target to provide new insights into the pathogenesis and treatment of inflammation-related diseases.
The cGAS-STING pathway has been considered as a promising immunotherapy target in recent years (8). Synergetic effects can be achieved in multiple tumors using the combination immunotherapies with cGAS-STING pathway agonists (129, 130). However, the activated cGAS-STING pathway may promote growth or metastasis of certain tumors (131), and its chronic activation paradoxically induces an immune-suppressive tumor microenvironment by promoting the myeloid-biased differentiation of HSPCs and apoptosis of T cells (95). Individualized or specific cell-targeted application of cGAS-STING pathway agonists or antagonists will be feasible in the future.
Although the immunomodulatory effects of the cGAS-STING pathway in hematopoietic cells have been extensively studies, little attention has been paid to its roles in hematopoiesis. The regulation of HSPC maintenance is an attractive field to understand hematopoiesis under homeostatic and stress conditions. Components of the cGAS-STING pathway may be expressed at high levels in HSPCs, which may allow HSPCs to rapidly sense and respond to stresses, including infection and genotoxicity, by promoting HSPC proliferation, mobilization, and myeloid-biased differentiation (Figure 2). Moreover, this rapid response may also facilitate the elimination of damaged HSPCs or malignantly transformed HSPCs via the induction of cell death to maintain the fitness of the HSPC pool. On the other hand, modulation of the cGAS-STING pathway in HSPCs may boost anti-infection or antitumor immunity and suppress autoimmunity and peripheral inflammation. However, multiple obstacles must be addressed before its clinical application. For example, chronic activation of the cGAS-STING pathway may promote the pathogenesis of myeloid malignancies or induce an adaptive immunodeficiency. Future studies aiming to discover the distinct functions of the cGAS-STING pathway in HSPC maintenance and develop modulators targeting specific cell types will be meaningful.
CD and JW decided on the topics. WL and CD wrote the manuscript and prepared the figures and the table. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of China (No. 81725019, 81930090, 81874256).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AIM2 absent in melanoma 2 CASP1 caspase 1; CDNs: cyclic dinucleotides; cGAS: cyclic GMP-AMP synthase; CMPs: common myeloid progenitors; DCs: dendritic cells; DDR: DNA damage response; GMPs: granulocyte-monocyte progenitors; GSDMD: gasdermin D; HSCs: hematopoietic stem cells; HSPCs: hematopoietic stem and progenitor cells; IFN-I: type I interferon; IL1β: interleukin 1β; IKK: IkB kinase; JNK: c-Jun-N-terminal kinase; MDS: myelodysplastic syndromes; MDPs: monocyte-dendritic cell progenitors; MyD88: myeloid differentiation primary response 88; NF-κB: nuclear factor-κB; NLRP3: NOD-, LRR-, and pyrin domain-containing protein 3; STING: stimulator of interferon genes; TBK1: TANK-binding kinase 1; TLR9: Toll-like receptor 9; TRAF6: TNF receptor associated factor 6; PARP1: poly (ADP–ribose):;: polymerase 1; PUMA: p53 upregulated modulator of apoptosis.
1. Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol (2019) 20(5):303–20. doi: 10.1038/s41580-019-0103-9
2. Chavakis T, Mitroulis I, Hajishengallis G. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat Immunol (2019) 20(7):802–11. doi: 10.1038/s41590-019-0402-5
3. Taguchi T, Mukai K. Innate immunity signalling and membrane trafficking. Curr Opin Cell Biol (2019) 59:1–7. doi: 10.1016/j.ceb.2019.02.002
4. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol (2020) 20(9):537–51. doi: 10.1038/s41577-020-0288-3
5. Ablasser A, Hur S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat Immunol (2020) 21(1):17–29. doi: 10.1038/s41590-019-0556-1
6. Tatematsu M, Funami K, Seya T, Matsumoto M. Extracellular RNA Sensing by Pattern Recognition Receptors. J Innate Immun (2018) 10(5-6):398–406. doi: 10.1159/000494034
7. Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi L. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell (2018) 34(3):361–78. doi: 10.1016/j.ccell.2018.05.013
8. Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet (2019) 20(11):657–74. doi: 10.1038/s41576-019-0151-1
9. Ablasser A, Chen ZJ. cGAS in action: Expanding roles in immunity and inflammation. Science (2019) 363(6431):eaat8657. doi: 10.1126/science.aat8657
10. Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Sakar MS, et al. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Sci (N Y NY) (2020) 369(6505):823–8. doi: 10.1126/science.aaw6421
11. Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, et al. The Dlk1-Gtl2 Locus Preserves LT-HSC Function by Inhibiting the PI3K-mTOR Pathway to Restrict Mitochondrial Metabolism. Cell Stem Cell (2016) 18(2):214–28. doi: 10.1016/j.stem.2015.11.001
12. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (2013) 339(6121):786–91. doi: 10.1126/science.1232458
13. Barnett KC, Coronas-Serna JM, Zhou W, Ernandes MJ, Cao A, Kranzusch PJ, et al. Phosphoinositide Interactions Position cGAS at the Plasma Membrane to Ensure Efficient Distinction between Self- and Viral DNA. Cell (2019) 176(6):1432–46.e11. doi: 10.1016/j.cell.2019.01.049
14. Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell (2014) 54(2):289–96. doi: 10.1016/j.molcel.2014.03.040
15. Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, et al. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity (2018) 48(4):688–701.e7. doi: 10.1016/j.immuni.2018.03.016
16. Liu ZS, Cai H, Xue W, Wang M, Xia T, Li WJ, et al. G3BP1 promotes DNA binding and activation of cGAS. Nat Immunol (2019) 20(1):18–28. doi: 10.1038/s41590-018-0262-4
17. Zierhut C, Yamaguchi N, Paredes M, Luo JD, Carroll T, Funabiki H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell (2019) 178(2):302–15.e23. doi: 10.1016/j.cell.2019.05.035
18. Srikanth S, Woo JS, Wu B, El-Sherbiny YM, Leung J, Chupradit K, et al. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat Immunol (2019) 20(2):152–62. doi: 10.1038/s41590-018-0287-8
19. Guo Y, Jiang F, Kong L, Li B, Yang Y, Zhang L, et al. Cutting Edge: USP27X Deubiquitinates and Stabilizes the DNA Sensor cGAS to Regulate Cytosolic DNA-Mediated Signaling. J Immunol (2019) 203(8):2049–54. doi: 10.4049/jimmunol.1900514
20. Chen X, Chen Y. Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochem Biophys Res Commun (2019) 514(3):659–64. doi: 10.1016/j.bbrc.2019.05.022
21. Dai J, Huang YJ, He X, Zhao M, Wang X, Liu ZS, et al. Acetylation Blocks cGAS Activity and Inhibits Self-DNA-Induced Autoimmunity. Cell (2019) 176(6):1447–60.e14. doi: 10.1016/j.cell.2019.01.016
22. Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, et al. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity (2016) 45(3):555–69. doi: 10.1016/j.immuni.2016.08.014
23. Xia P, Ye B, Wang S, Zhu X, Du Y, Xiong Z, et al. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol (2016) 17(4):369–78. doi: 10.1038/ni.3356
24. Galluzzi L, Vanpouille-Box C, Bakhoum SF, Demaria S. SnapShot: CGAS-STING Signaling. Cell (2018) 173(1):276–.e1. doi: 10.1016/j.cell.2018.03.015
25. Balka KR, Louis C, Saunders TL, Smith AM, Calleja DJ, D’Silva DB, et al. TBK1 and IKKϵ Act Redundantly to Mediate STING-Induced NF-κB Responses in Myeloid Cells. Cell Rep (2020) 31(1):107492. doi: 10.1016/j.celrep.2020.03.056
26. Yu Q, Katlinskaya YV, Carbone CJ, Zhao B, Katlinski KV, Zheng H, et al. DNA-damage-induced type I interferon promotes senescence and inhibits stem cell function. Cell Rep (2015) 11(5):785–97. doi: 10.1016/j.celrep.2015.03.069
27. Bodur C, Kazyken D, Huang K, Ekim Ustunel B, Siroky KA, Tooley AS, et al. The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J (2018) 37(1):19–38. doi: 10.15252/embj.201696164
28. Chen H, Sun H, You F, Sun W, Zhou X, Chen L, et al. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell (2011) 147(2):436–46. doi: 10.1016/j.cell.2011.09.022
29. Wang YY, Shen D, Zhao LJ, Zeng N, Hu TH. Sting is a critical regulator of spinal cord injury by regulating microglial inflammation via interacting with TBK1 in mice. Biochem Biophys Res Commun (2019) 517(4):741–8. doi: 10.1016/j.bbrc.2019.07.125
30. Luo X, Li H, Ma L, Zhou J, Guo X, Woo SL, et al. Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology (2018) 155(6):1971–84.e4. doi: 10.1053/j.gastro.2018.09.010
31. Park SM, Omatsu T, Zhao Y, Yoshida N, Shah P, Zagani R, et al. T cell fate following Salmonella infection is determined by a STING-IRF1 signaling axis in mice. Commun Biol (2019) 2:464. doi: 10.1038/s42003-019-0701-2
32. Andzinski L, Spanier J, Kasnitz N, Kröger A, Jin L, Brinkmann MM, et al. Growing tumors induce a local STING dependent Type I IFN response in dendritic cells. Int J Cancer (2016) 139(6):1350–7. doi: 10.1002/ijc.30159
33. Suschak JJ, Wang S, Fitzgerald KA, Lu S. A cGAS-Independent STING/IRF7 Pathway Mediates the Immunogenicity of DNA Vaccines. J Immunol (2016) 196(1):310–6. doi: 10.4049/jimmunol.1501836
34. Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature (2019) 567(7747):262–6. doi: 10.1038/s41586-019-1006-9
35. Gaidt MM, Ebert TS, Chauhan D, Ramshorn K, Pinci F, Zuber S, et al. The DNA Inflammasome in Human Myeloid Cells Is Initiated by a STING-Cell Death Program Upstream of NLRP3. Cell (2017) 171(5):1110–24.e18. doi: 10.1016/j.cell.2017.09.039
36. Brault M, Olsen TM, Martinez J, Stetson DB, Oberst A. Intracellular Nucleic Acid Sensing Triggers Necroptosis through Synergistic Type I IFN and TNF Signaling. J Immunol (Baltimore Md 1950) (2018) 200(8):2748–56. doi: 10.4049/jimmunol.1701492
37. Ranoa DRE, Widau RC, Mallon S, Parekh AD, Nicolae CM, Huang X, et al. STING Promotes Homeostasis via Regulation of Cell Proliferation and Chromosomal Stability. Cancer Res (2019) 79(7):1465–79. doi: 10.1158/0008-5472.Can-18-1972
38. Basit A, Cho MG, Kim EY, Kwon D, Kang SJ, Lee JH. The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp Mol Med (2020) 52(4):643–57. doi: 10.1038/s12276-020-0416-y
39. Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature (1995) 374(6522):546–9. doi: 10.1038/374546a0
40. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature (2009) 458(7237):509–13. doi: 10.1038/nature07710
41. Marongiu L, Gornati L, Artuso I, Zanoni I, Granucci F. Below the surface: The inner lives of TLR4 and TLR9. J Leukoc Biol (2019) 106(1):147–60. doi: 10.1002/jlb.3mir1218-483rr
42. Banerjee I, Behl B, Mendonca M, Shrivastava G, Russo AJ, Menoret A, et al. Gasdermin D Restrains Type I Interferon Response to Cytosolic DNA by Disrupting Ionic Homeostasis. Immunity (2018) 49(3):413–26.e5. doi: 10.1016/j.immuni.2018.07.006
43. Schultze JL, Mass E, Schlitzer A. Emerging Principles in Myelopoiesis at Homeostasis and during Infection and Inflammation. Immunity (2019) 50(2):288–301. doi: 10.1016/j.immuni.2019.01.019
44. Hérault A, Binnewies M, Leong S, Calero-Nieto FJ, Zhang SY, Kang YA, et al. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature (2017) 544(7648):53–8. doi: 10.1038/nature21693
45. Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell (2015) 17(1):35–46. doi: 10.1016/j.stem.2015.05.003
46. Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature (2015) 520(7548):549–52. doi: 10.1038/nature14131
47. Takizawa H, Fritsch K, Kovtonyuk LV, Saito Y, Yakkala C, Jacobs K, et al. Pathogen-Induced TLR4-TRIF Innate Immune Signaling in Hematopoietic Stem Cells Promotes Proliferation but Reduces Competitive Fitness. Cell Stem Cell (2017) 21(2):225–40.e5. doi: 10.1016/j.stem.2017.06.013
48. Zhao JL, Ma C, O’Connell RM, Mehta A, DiLoreto R, Heath JR, et al. Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell (2014) 14(4):445–59. doi: 10.1016/j.stem.2014.01.007
49. Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell (2013) 154(5):1112–26. doi: 10.1016/j.cell.2013.08.007
50. Carrelha J, Meng Y, Kettyle LM, Luis TC, Norfo R, Alcolea V, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature (2018) 554(7690):106–11. doi: 10.1038/nature25455
51. Grinenko T, Eugster A, Thielecke L, Ramasz B, Krüger A, Dietz S, et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat Commun (2018) 9(1):1898. doi: 10.1038/s41467-018-04188-7
52. Haas S, Trumpp A, Milsom MD. Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell (2018) 22(5):627–38. doi: 10.1016/j.stem.2018.04.003
53. Weinreb J, Gupta V, Bowman TV. Excessive R-Loops Trigger an Inflammatory Cascade Leading to Aberrant Hematopoietic Stem and Progenitor Cell Expansion. Blood (2019) 134(Supplement_1):772–. doi: 10.1182/blood-2019-124888
54. White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell (2014) 159(7):1549–62. doi: 10.1016/j.cell.2014.11.036
55. Kobayashi H, Kobayashi CI, Nakamura-Ishizu A, Karigane D, Haeno H, Yamamoto KN, et al. Bacterial c-di-GMP affects hematopoietic stem/progenitors and their niches through STING. Cell Rep (2015) 11(1):71–84. doi: 10.1016/j.celrep.2015.02.066
56. Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat Med (2009) 15(6):696–700. doi: 10.1038/nm.1973
57. Bi X, Feng D, Korczeniewska J, Alper N, Hu G, Barnes BJ. Deletion of Irf5 protects hematopoietic stem cells from DNA damage-induced apoptosis and suppresses γ-irradiation-induced thymic lymphomagenesis. Oncogene (2014) 33(25):3288–97. doi: 10.1038/onc.2013.295
58. Lu X, Li X, He Q, Gao J, Gao Y, Liu B, et al. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res (2013) 23(12):1356–68. doi: 10.1038/cr.2013.145
59. Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol (2016) 18(6):607–18. doi: 10.1038/ncb3346
60. Frisch BJ, Hoffman CM, Latchney SE, LaMere MW, Myers J, Ashton J, et al. Aged marrow macrophages expand platelet-biased hematopoietic stem cells via Interleukin1B. JCI Insight (2019) 5(10):e124213. doi: 10.1172/jci.insight.124213
61. Hemmati S, Sinclair T, Tong M, Bartholdy B, Okabe RO, Ames K, et al. PI3 kinase alpha and delta promote hematopoietic stem cell activation. JCI Insight (2019) 5(13):e125832. doi: 10.1172/jci.insight.125832
62. Yamashita M, Passegué E. TNF-α Coordinates Hematopoietic Stem Cell Survival and Myeloid Regeneration. Cell Stem Cell (2019) 25(3):357–72.e7. doi: 10.1016/j.stem.2019.05.019
63. Fang J, Muto T, Kleppe M, Bolanos LC, Hueneman KM, Walker CS, et al. TRAF6 Mediates Basal Activation of NF-κB Necessary for Hematopoietic Stem Cell Homeostasis. Cell Rep (2018) 22(5):1250–62. doi: 10.1016/j.celrep.2018.01.013
64. Oburoglu L, Romano M, Taylor N, Kinet S. Metabolic regulation of hematopoietic stem cell commitment and erythroid differentiation. Curr Opin Hematol (2016) 23(3):198–205. doi: 10.1097/moh.0000000000000234
65. Li J, Zhang L, Yin L, Ma N, Wang T, Wu Y, et al. In Vitro Expansion of Hematopoietic Stem Cells by Inhibition of Both GSK3 and p38 Signaling. Stem Cells Dev (2019) 28(22):1486–97. doi: 10.1089/scd.2019.0119
66. Baumgartner C, Toifl S, Farlik M, Halbritter F, Scheicher R, Fischer I, et al. An ERK-Dependent Feedback Mechanism Prevents Hematopoietic Stem Cell Exhaustion. Cell Stem Cell (2018) 22(6):879–92.e6. doi: 10.1016/j.stem.2018.05.003
67. Essers MA, Offner S, Blanco-Bose WE, Waibler Z, Kalinke U, Duchosal MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature (2009) 458(7240):904–8. doi: 10.1038/nature07815
68. Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell (2015) 17(4):422–34. doi: 10.1016/j.stem.2015.07.007
69. Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses. Front Immunol (2018) 9:1135. doi: 10.3389/fimmu.2018.01135
70. Ho YH, Del Toro R, Rivera-Torres J, Rak J, Korn C, García-García A, et al. Remodeling of Bone Marrow Hematopoietic Stem Cell Niches Promotes Myeloid Cell Expansion during Premature or Physiological Aging. Cell Stem Cell (2019) 25(3):407–18.e6. doi: 10.1016/j.stem.2019.06.007
71. Tie R, Li H, Cai S, Liang Z, Shan W, Wang B, et al. Interleukin-6 signaling regulates hematopoietic stem cell emergence. Exp Mol Med (2019) 51(10):1–12. doi: 10.1038/s12276-019-0320-5
72. Pronk CJ, Veiby OP, Bryder D, Jacobsen SE. Tumor necrosis factor restricts hematopoietic stem cell activity in mice: involvement of two distinct receptors. J Exp Med (2011) 208(8):1563–70. doi: 10.1084/jem.20110752
73. Smith JNP, Zhang Y, Li JJ, McCabe A, Jo HJ, Maloney J, et al. Type I IFNs drive hematopoietic stem and progenitor cell collapse via impaired proliferation and increased RIPK1-dependent cell death during shock-like ehrlichial infection. PloS Pathog (2018) 14(8):e1007234. doi: 10.1371/journal.ppat.1007234
74. Pietras EM, Lakshminarasimhan R, Techner JM, Fong S, Flach J, Binnewies M, et al. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med (2014) 211(2):245–62. doi: 10.1084/jem.20131043
75. Warner JD, Irizarry-Caro RA, Bennion BG, Ai TL, Smith AM, Miner CA, et al. STING-associated vasculopathy develops independently of IRF3 in mice. J Exp Med (2017) 214(11):3279–92. doi: 10.1084/jem.20171351
76. Bouis D, Kirstetter P, Arbogast F, Lamon D, Delgado V, Jung S, et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J Allergy Clin Immunol (2019) 143(2):712–25.e5. doi: 10.1016/j.jaci.2018.04.034
77. Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol (2018) 18(5):309–24. doi: 10.1038/nri.2017.142
78. Sarhan J, Liu BC, Muendlein HI, Weindel CG, Smirnova I, Tang AY, et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Different (2019) 26(2):332–47. doi: 10.1038/s41418-018-0122-7
79. Lahaye X, Gentili M, Silvin A, Conrad C, Picard L, Jouve M, et al. NONO Detects the Nuclear HIV Capsid to Promote cGAS-Mediated Innate Immune Activation. Cell (2018) 175(2):488–501.e22. doi: 10.1016/j.cell.2018.08.062
80. Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, et al. Opposing roles of Toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PloS Pathog (2017) 13(7):e1006496. doi: 10.1371/journal.ppat.1006496
81. Hu X, Peng X, Lu C, Zhang X, Gan L, Gao Y, et al. Type I IFN expression is stimulated by cytosolic MtDNA released from pneumolysin-damaged mitochondria via the STING signaling pathway in macrophages. FEBS J (2019) 286(23):4754–68. doi: 10.1111/febs.15001
82. Das S, Kumar A, Mandal A, Abhishek K, Verma S, Kumar A, et al. Nucleic acid sensing activates the innate cytosolic surveillance pathway and promotes parasite survival in visceral leishmaniasis. Sci Rep (2019) 9(1):9825. doi: 10.1038/s41598-019-45800-0
83. Cheng Y, Schorey JS. Mycobacterium tuberculosis-induced IFN-β production requires cytosolic DNA and RNA sensing pathways. J Exp Med (2018) 215(11):2919–35. doi: 10.1084/jem.20180508
84. Kim BR, Kim BJ, Kook YH, Kim BJ. Mycobacterium abscessus infection leads to enhanced production of type 1 interferon and NLRP3 inflammasome activation in murine macrophages via mitochondrial oxidative stress. PloS Pathog (2020) 16(3):e1008294. doi: 10.1371/journal.ppat.1008294
85. Nandakumar R, Tschismarov R, Meissner F, Prabakaran T, Krissanaprasit A, Farahani E, et al. Intracellular bacteria engage a STING-TBK1-MVB12b pathway to enable paracrine cGAS-STING signalling. Nat Microbiol (2019) 4(4):701–13. doi: 10.1038/s41564-019-0367-z
86. Deschamps T, Kalamvoki M. Extracellular Vesicles Released by Herpes Simplex Virus 1-Infected Cells Block Virus Replication in Recipient Cells in a STING-Dependent Manner. J Virol (2018) 92(18):e01102–18. doi: 10.1128/jvi.01102-18
87. Zhou C, Chen X, Planells-Cases R, Chu J, Wang L, Cao L, et al. Transfer of cGAMP into Bystander Cells via LRRC8 Volume-Regulated Anion Channels Augments STING-Mediated Interferon Responses and Anti-viral Immunity. Immunity (2020) 52(5):767–81. doi: 10.1016/j.immuni.2020.03.016
88. Pépin G, De Nardo D, Rootes CL, Ullah TR, Al-Asmari SS, Balka KR, et al. Connexin-Dependent Transfer of cGAMP to Phagocytes Modulates Antiviral Responses. mBio (2020) 11(1):e03187–19. doi: 10.1128/mBio.03187-19
89. Choudhuri S, Garg NJ. PARP1-cGAS-NF-κB pathway of proinflammatory macrophage activation by extracellular vesicles released during Trypanosoma cruzi infection and Chagas disease. PloS Pathog (2020) 16(4):e1008474. doi: 10.1371/journal.ppat.1008474
90. Mandal PK, Blanpain C, Rossi DJ. DNA damage response in adult stem cells: pathways and consequences. Nat Rev Mol Cell Biol (2011) 12(3):198–202. doi: 10.1038/nrm3060
91. Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature (2018) 553(7687):171–7. doi: 10.1038/nature25154
92. Shao L, Luo Y, Zhou D. Hematopoietic stem cell injury induced by ionizing radiation. Antioxid Redox Signaling (2014) 20(9):1447–62. doi: 10.1089/ars.2013.5635
93. Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature (2017) 548(7668):466–70. doi: 10.1038/nature23470
94. Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature (2017) 548(7668):461–5. doi: 10.1038/nature23449
95. Gulen MF, Koch U, Haag SM, Schuler F, Apetoh L, Villunger A, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun (2017) 8(1):427. doi: 10.1038/s41467-017-00573-w
96. Chen D, Tong J, Yang L, Wei L, Stolz DB, Yu J, et al. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc Natl Acad Sci U States America (2018) 115(15):3930–5. doi: 10.1073/pnas.1717190115
97. Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature (2017) 543(7644):205–10. doi: 10.1038/nature21388
98. Nassour J, Radford R, Correia A, Fusté JM, Schoell B, Jauch A, et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature (2019) 565(7741):659–63. doi: 10.1038/s41586-019-0885-0
99. Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature (2018) 563(7729):131–6. doi: 10.1038/s41586-018-0629-6
100. Jiang H, Xue X, Panda S, Kawale A, Hooy RM, Liang F, et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J (2019) 38(21):e102718. doi: 10.15252/embj.2019102718
101. Méndez-Ferrer S, Bonnet D, Steensma DP, Hasserjian RP, Ghobrial IM, Gribben JG, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer (2020) 20(5):285–98. doi: 10.1038/s41568-020-0245-2
102. Cai Z, Kotzin JJ, Ramdas B, Chen S, Nelanuthala S, Palam LR, et al. Inhibition of Inflammatory Signaling in Tet2 Mutant Preleukemic Cells Mitigates Stress-Induced Abnormalities and Clonal Hematopoiesis. Cell Stem Cell (2018) 23(6):833–49.e5. doi: 10.1016/j.stem.2018.10.013
103. Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer (2020) 20(3):158–73. doi: 10.1038/s41568-019-0230-9
104. Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U.S.A. (2013) 110(3):1041–6. doi: 10.1073/pnas.1213021110
105. Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell (2016) 19(2):177–91. doi: 10.1016/j.stem.2016.05.004
106. Tang CH, Zundell JA, Ranatunga S, Lin C, Nefedova Y, Del Valle JR, et al. Agonist-Mediated Activation of STING Induces Apoptosis in Malignant B Cells. Cancer Res (2016) 76(8):2137–52. doi: 10.1158/0008-5472.Can-15-1885
107. Preudhomme C, Guilhot J, Nicolini FE, Guerci-Bresler A, Rigal-Huguet F, Maloisel F, et al. Imatinib plus peginterferon alfa-2a in chronic myeloid leukemia. N Engl J Med (2010) 363(26):2511–21. doi: 10.1056/NEJMoa1004095
108. Zhang H, Zeng L, Xie M, Liu J, Zhou B, Wu R, et al. TMEM173 Drives Lethal Coagulation in Sepsis. Cell Host Microbe (2020) 27(4):556–70.e6. doi: 10.1016/j.chom.2020.02.004
109. Martin GR, Blomquist CM, Henare KL, Jirik FR. Stimulator of interferon genes (STING) activation exacerbates experimental colitis in mice. Sci Rep (2019) 9(1):14281. doi: 10.1038/s41598-019-50656-5
110. Heipertz EL, Harper J, Walker WE. STING and TRIF Contribute to Mouse Sepsis, Depending on Severity of the Disease Model. Shock (2017) 47(5):621–31. doi: 10.1097/shk.0000000000000771
111. Bennion BG, Ingle H, Ai TL, Miner CA, Platt DJ, Smith AM, et al. A Human Gain-of-Function STING Mutation Causes Immunodeficiency and Gammaherpesvirus-Induced Pulmonary Fibrosis in Mice. J Virol (2019) 93(4) 142(3):e01806–18. doi: 10.1128/jvi.01806-18
112. Majumdar T, Chattopadhyay S, Ozhegov E, Dhar J, Goswami R, Sen GC, et al. Induction of interferon-stimulated genes by IRF3 promotes replication of Toxoplasma gondii. PloS Pathog (2015) 11(3):e1004779. doi: 10.1371/journal.ppat.1004779
113. Guimarães ES, Gomes MTR, Campos PC, Mansur DS, Dos Santos AA, Harms J, et al. Brucella abortus Cyclic Dinucleotides Trigger STING-Dependent Unfolded Protein Response That Favors Bacterial Replication. J Immunol (2019) 202(9):2671–81. doi: 10.4049/jimmunol.1801233
114. Fermaintt CS, Sano K, Liu Z, Ishii N, Seino J, Dobbs N, et al. A bioactive mammalian disaccharide associated with autoimmunity activates STING-TBK1-dependent immune response. Nat Commun (2019) 10(1):2377. doi: 10.1038/s41467-019-10319-5
115. Kumar V. A STING to inflammation and autoimmunity. J Leukoc Biol (2019) 106(1):171–85. doi: 10.1002/jlb.4mir1018-397rr
116. Vanpouille-Box C, Hoffmann JA, Galluzzi L. Pharmacological modulation of nucleic acid sensors - therapeutic potential and persisting obstacles. Nat Rev Drug Discovery (2019) 18(11):845–67. doi: 10.1038/s41573-019-0043-2
117. Cairns AP, Crockard AD, McConnell JR, Courtney PA, Bell AL. Reduced expression of CD44 on monocytes and neutrophils in systemic lupus erythematosus: relations with apoptotic neutrophils and disease activity. Ann Rheum Dis (2001) 60(10):950–5. doi: 10.1136/ard.60.10.950
118. Orr Y, Taylor JM, Bannon PG, Geczy C, Kritharides L. Circulating CD10-/CD16low neutrophils provide a quantitative index of active bone marrow neutrophil release. Br J Haematol (2005) 131(4):508–19. doi: 10.1111/j.1365-2141.2005.05794.x
119. Bach JF. The hygiene hypothesis in autoimmunity: the role of pathogens and commensals. Nat Rev Immunol (2018) 18(2):105–20. doi: 10.1038/nri.2017.111
120. Luksch H, Stinson WA, Platt DJ, Qian W, Kalugotla G, Miner CA, et al. STING-associated lung disease in mice relies on T cells but not type I interferon. J Allergy Clin Immunol (2019) 144(1):254–66.e8. doi: 10.1016/j.jaci.2019.01.044
121. Quail DF, Olson OC, Bhardwaj P, Walsh LA, Akkari L, Quick ML, et al. Obesity alters the lung myeloid cell landscape to enhance breast cancer metastasis through IL5 and GM-CSF. Nat Cell Biol (2017) 19(8):974–87. doi: 10.1038/ncb3578
122. Barrett TJ, Distel E, Murphy AJ, Hu J, Garshick MS, Ogando Y, et al. Apolipoprotein AI) Promotes Atherosclerosis Regression in Diabetic Mice by Suppressing Myelopoiesis and Plaque Inflammation. Circulation (2019) 140(14):1170–84. doi: 10.1161/circulationaha.119.039476
123. Nahrendorf M. Myeloid cell contributions to cardiovascular health and disease. Nat Med (2018) 24(6):711–20. doi: 10.1038/s41591-018-0064-0
124. Murphy AJ, Tall AR. Disordered haematopoiesis and athero-thrombosis. Eur Heart J (2016) 37(14):1113–21. doi: 10.1093/eurheartj/ehv718
125. Hoyer FF, Zhang X, Coppin E, Vasamsetti SB, Modugu G, Schloss MJ, et al. Bone Marrow Endothelial Cells Regulate Myelopoiesis in Diabetes. Circulation (2020) 142(3):244–58. doi: 10.1161/circulationaha.120.046038
126. Sreejit G, Abdel-Latif A, Athmanathan B, Annabathula R, Dhyani A, Noothi SK, et al. Neutrophil-Derived S100A8/A9 Amplify Granulopoiesis After Myocardial Infarction. Circulation (2020) 141(13):1080–94. doi: 10.1161/circulationaha.119.043833
127. McAlpine CS, Kiss MG, Rattik S, He S, Vassalli A, Valet C, et al. Sleep modulates haematopoiesis and protects against atherosclerosis. Nature (2019) 566(7744):383–7. doi: 10.1038/s41586-019-0948-2
128. Frodermann V, Rohde D, Courties G, Severe N, Schloss MJ, Amatullah H, et al. Exercise reduces inflammatory cell production and cardiovascular inflammation via instruction of hematopoietic progenitor cells. Nat Med (2019) 25(11):1761–71. doi: 10.1038/s41591-019-0633-x
129. Berger G, Marloye M, Lawler SE. Pharmacological Modulation of the STING Pathway for Cancer Immunotherapy. Trends Mol Med (2019) 25(5):412–27. doi: 10.1016/j.molmed.2019.02.007
130. Saeed A, Ruan X, Guan H, Su J, Ouyang S. Regulation of cGAS-Mediated Immune Responses and Immunotherapy. Adv Sci (Weinh) (2020) 7(6):1902599. doi: 10.1002/advs.201902599
Keywords: cGAS-STING pathway, Hematopoiesis, Hematopoietic stem and progenitor cells, Cytosolic DNA sensing, Innate Immunity and inflammation
Citation: Liao W, Du C and Wang J (2020) The cGAS-STING Pathway in Hematopoiesis and Its Physiopathological Significance. Front. Immunol. 11:573915. doi: 10.3389/fimmu.2020.573915
Received: 18 June 2020; Accepted: 29 October 2020;
Published: 30 November 2020.
Edited by:
Jagadeesh Bayry, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Marieke Essers, German Cancer Research Center (DKFZ), GermanyCopyright © 2020 Liao, Du and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junping Wang, d2FuZ2p1bnBpbmdAdG1tdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.