- 1Biological Resource Center, TaiZhou Hospital of Zhejiang Province, Taizhou Enze Medical Center (Group), LinHai, China
- 2Medical Research Center, TaiZhou Hospital of Zhejiang Province, Taizhou Enze Medical Center (Group), LinHai, China
Signaling pathway between human leukocyte antigen (HLA)-G and immune inhibitory receptors immunoglobulin-like transcript (ILT)-2/4 has been acknowledged as one of immune checkpoints, and as a potential target for cancer immunotherapy. Like other immune checkpoints, inter- and even intratumor heterogeneity of HLA-G could render a rather complexity for HLA-G-target immunotherapy. However, little information for intratumor heterogeneity of HLA-G is available. In this study, HLA-G expression in a serial section of colorectal cancer (CRC) lesions from three CRC patients (each sample with serial section of 50 slides, 10 randomized slides for each antibody), three different locations within a same sample (five CRC), and three case-matched blocks that each includes 36 esophageal cancer samples, were evaluated with immunohistochemistry using anti-HLA-G antibodies (mAbs 4H84, MEM-G/1 and MEM-G/2 probing for all denatured HLA-G isoforms, 5A6G7, and 2A12 probing for denatured HLA-G5 and HLA-G6 isoforms). Our results revealed that, in addition to the frequently observed inter-tumor heterogeneity, intratumor heterogeneous expression of HLA-G is common in different areas within a tumor in CRC and esophageal cancer samples included in this study. Moreover, percentage of HLA-G expression probed with different anti-HLA-G antibodies also varies dramatically within a tumor. Given HLA-G has been considered as an important immune checkpoint, intratumor heterogeneity of HLA-G expression, and different specificity of anti-HLA-G antibodies being used among studies, interpretation and clinical significance of HLA-G expression in cancers should be with caution.
Introduction
Immune suppressive functions induced by the interaction between human leukocyte antigen-G (HLA-G) and its immune inhibitory receptors, the immunoglobulin-like transcripts (ILTs), have been widely acknowledged (1). Receptors ILT-2 and ILT-4 express on various immune cells, the immune tolerogenic effects induced by HLA-G are comprehensive (2). Due to alternative splicing of its primary transcripts, seven confirmed HLA-G isoforms (HLA-G1~HLA-G7), and recently predicted novel isoforms such as lacking a transmembrane region and α1 domain have been reported (3).
In the context of cancers, different degree of inter-tumor HLA-G expression has been observed in most histological types of cancers studied, and the significance of HLA-G/ILTs signaling pathway as an immune checkpoint in cancer biology has been highlighted (4). Look back to its expression firstly observed in cancer, the melanoma lesions in 1998 (5), immune tolerance induced by HLA-G has been solidified by large numbers of studies both in vitro and in vivo preclinical experimental animal models (6–8).
HLA-G/ILTs binding can inhibit the proliferation of natural killer cells (NK), T and B lymphocytes and maturation and antigen presentation of dendritic cells (DC), suppress NK and T cell’s cytotoxic function, B cell’s immunoglobulin production and neutrophils’ reactive oxygen species production and phagocytosis capability (9–11). To the contrary, HLA-G/ILTs binding can promote myeloid-derived suppressor cells (MDSC) proliferation and polarize M1 macrophages towards to M2 type (12, 13). Moreover, immune tolerance can be induced by HLA-G-bearing exosomes between cells at long-distance, and by cellular membrane fragments containing HLA-G through trogocytosis in a close cell-to-cell contact manner (14, 15). In preclinical murine models, HLA-G could promote tumor immune escape and growth through murine MDSC proliferation and Th2 cytokine production, or reduce T and B cell tumor infiltrate, impair B cell immune responses in immunocompetent mice (8, 16). Findings also revealed that HLA-G expression in ovarian cancer cells could enhance the tumor cell migration and metastasis in tumor-bearing immunodeficient nude mice through induction of matrix metalloproteinase-15 (MMP-15) expression (7, 17). Moreover, a recent study showed that depletion of CD4lowHLA-G+ T cells could favor the castration-resistance prostate cancer therapy (18). Echoing the above mentioned in vitro and in vivo preclinical experimental observations, lesion HLA-G expression was observed to be closely associated with tumor metastasis, poor tumor cell differentiation, advanced disease stage and worse survival in a variety of cancers in clinical settings (14).
Inter- and intratumor heterogeneity of immune checkpoints is the main obstacle for immune checkpoint inhibitor (ICI) immunotherapy. Consequently, the benefits of the ICI therapy varies dramatically among patients (19). As a new immune checkpoint, the inter-tumor heterogeneous pattern of HLA-G expression is well evidenced; however, information for the intratumor heterogeneity of HLA-G is very limited. Previous studies revealed that the degree of HLA-G or its receptors ILT2/4 expression varies markedly among different locations in a primary renal cell cancer tumor lesion, indicating the complexity of intratumor heterogeneity of HLA-G and its receptor expression (3, 20).
In this study, inter- and intratumor heterogeneity of HLA-G expression was evaluated with immunohistochemistry using a panel of anti-HLA-G antibodies (mAbs 4H84, MEM-G/1 and MEM-G/2 probing for all denatured HLA-G isoforms, 5A6G7 and 2A12 probing for denatured HLA-G5 and HLA-G6 isoforms) in a serial section of colorectal cancer lesions from three CRC patients, three different locations within a same sample from five CRC patients, and three case-match blocks that each includes 36 esophageal cancer samples, and our findings solidify the heterogeneity of HLA-G in cancers.
Materials and Methods
Tumor Lesion Specimen
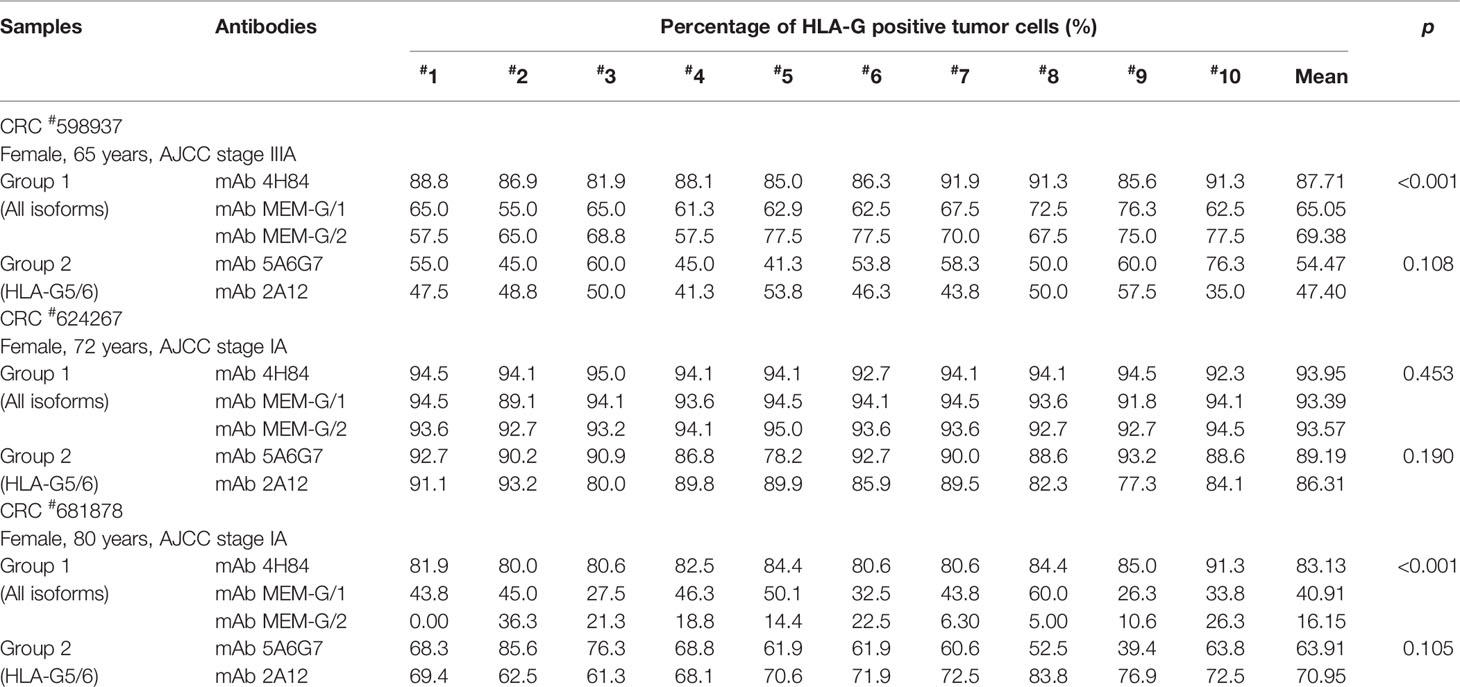
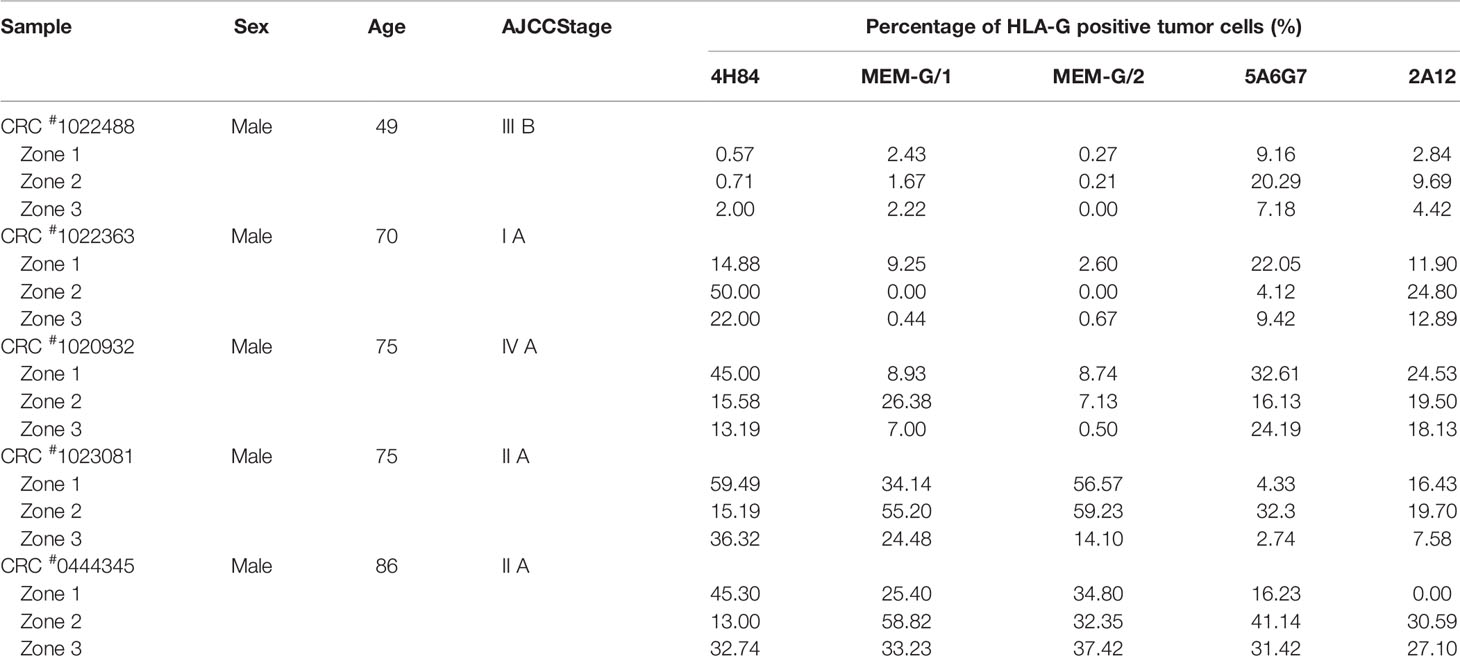
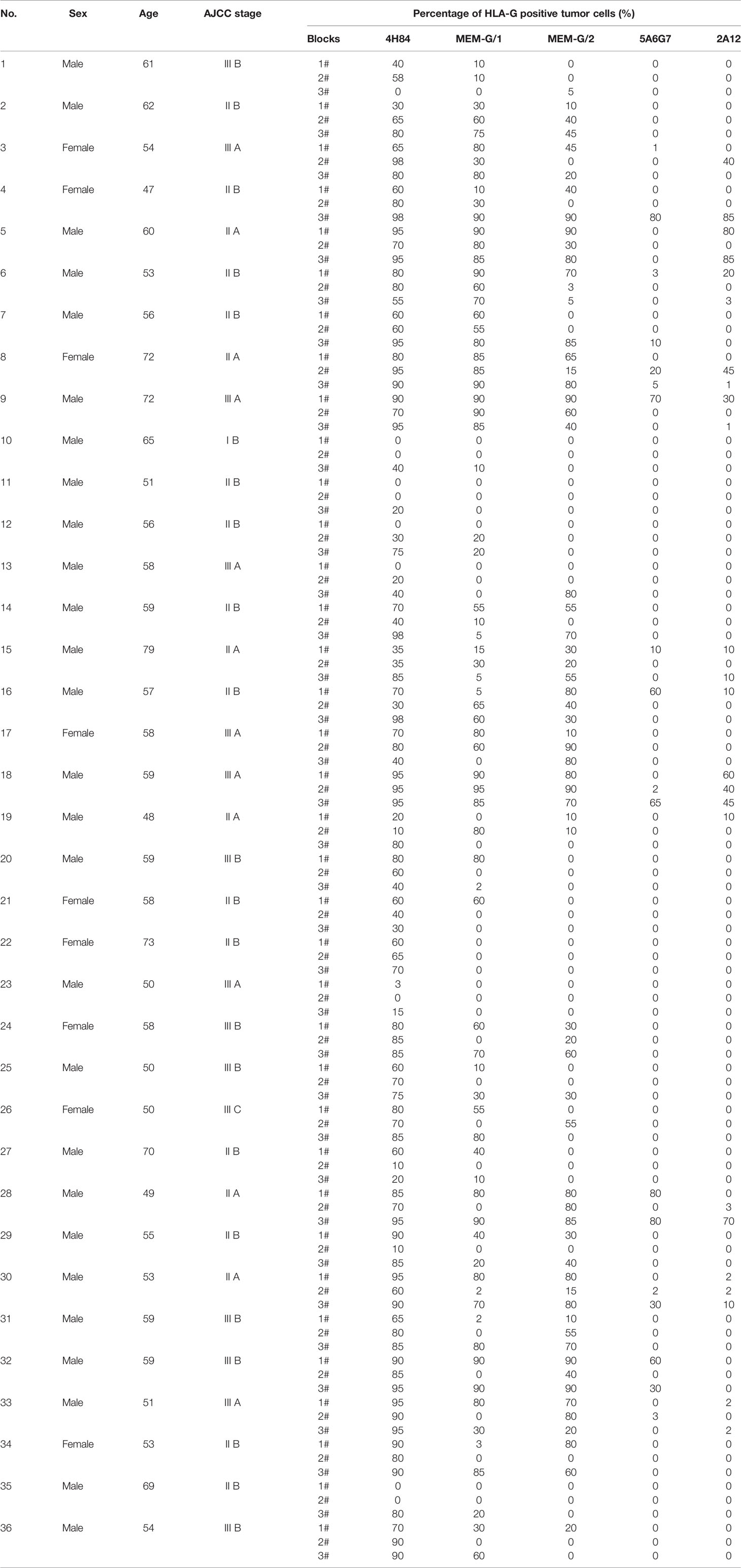
Tumor lesion specimen and clinical records were retrospectively reviewed. In this study, three CRC lesions #598937 (Female, 65 years, AJCC stage IIIA), #624267 (Female, 72 years, AJCC stage I A) and #681878 (Female, 80 years, AJCC stage I A; Table 1), and each sample was serially sectioned for 50 slides. Slides from three different locations within a same sample from another five CRC samples were obtained #1022488 (Male, 49 years, AJCC stage III B), #1022363 (Male, 70 years, AJCC stage I A), #1020932 (Male, 75 years, AJCC stage IV A), #1023081 (Male, 75 years, AJCC stage II A) and #444345 (Male, 86 years, AJCC stage II A; Table 2). Furthermore, slides from three case-matched blocks that each includes 36 esophageal squamous cell carcinoma (ESCC) samples were included in the study. Among 36 ESCC patients (27 male and nine female; median age: 58 years; range from 47 to 79 years), there were one patient with stage I B, six patients with II A, 14 patients with II B, seven patients with III A, seven patients with III B, and one patient with III C. The detailed clinical information was shown in Table 3. The clinicopathological findings were determined according to 7th American Joint Committee on Cancer (AJCC) Tumor-Node-Metastasis (TNM) staging system (21). None of them received radiotherapy, chemotherapy, or other medical interventions before the study. All these patients were diagnosed and treated at Taizhou Hospital of Zhejiang Province, China, and samples were retrieved by Biological Resource Center, Taizhou Hospital of Zhejiang Province (National Human Genetic Resources Platform of China YCZYPT [2017]02). Written informed consent was obtained from each participant before the surgical operation, and this study was approved by Medical Ethics Review Board of Taizhou Hospital of Zhejiang Province.
HLA-G Antibodies and Immunohistochemistry
Five anti-HLA-G murine antibodies were used in this study. mAbs 4H84 (dilution 1:200), MEM-G/1 (dilution 1:100) and MEM-G/2 (dilution 1:100), IgG1 antibodies detect denatured heavy chain of all HLA-G isoforms (Exbio, Prague, Czech Republic); mAbs 5A6G7 and 2A12, IgG1 antibodies probe denaturized heavy chain of HLA-G5/HLA-G6 isoforms (dilution 1:100; Exbio, Prague, Czech Republic). Immunohistochemistry assay was performed on 4-μm-thick, formalin-fixed and paraffin-embedded tumor lesion sections. Details of the protocols was according to our previous study (22). Immunohistochemistry staining was visualized with a Dako EnVison kit (Dako, Glostrup, Denmark). The percentage of HLA-G positive tumor cells was determined by presence of HLA-G staining while irrespective of staining intensity. HLA-G staining was evaluated by two reviewers who were blind to the patient clinicopathological information. Membrane or/and cytoplasmic expression of HLA-G were interpreted as positive. Percentage of HLA-G-positive tumor cells was determined by each observer, and the average of scores was calculated.
Statistical Analysis
Statistical analysis was performed with the SPSS 13.0 statistical software package (SPSS, Inc., Chicago, IL, USA). Comparison between groups was analyzed with non-parametric Mann-Whitney U or Kruskal-Wallis H test. p<0.05 (two-tailed) was considered statistically significant.
Results
To evaluate the heterogeneity of HLA-G expression in cancers, three different types of tumor tissue samples were prepared. a) For three CRC tissue samples (#598937, #624267 and #681878), 50 slides was serially sectioned for each sample. Among 50 slides, 10 randomized slides for each antibody probing. b) Slides from three different zones within a same sample from another five CRC samples (#1022488, #1022363, #1020932, #1023081, and #444345), and c) slides from three case-matched blocks that each includes 36 esophageal cancer samples. These slides were probed with five different anti-HLA-G antibodies. Anti-HLA-G antibodies were divided into two groups according to the specificity of these antibodies. Group 1: mAbs 4H84, MEM-G/1 and MEM-G/2, which detect denatured heavy chain of all HLA-G isoforms; Group 2: mAbs 5A6G7 and 2A12, which detect denaturized heavy chain of HLA-G5/HLA-G6 isoforms. The representative immunohistochemistry HLA-G staining patterns of CRC and ESCC were shown in Figure 1.
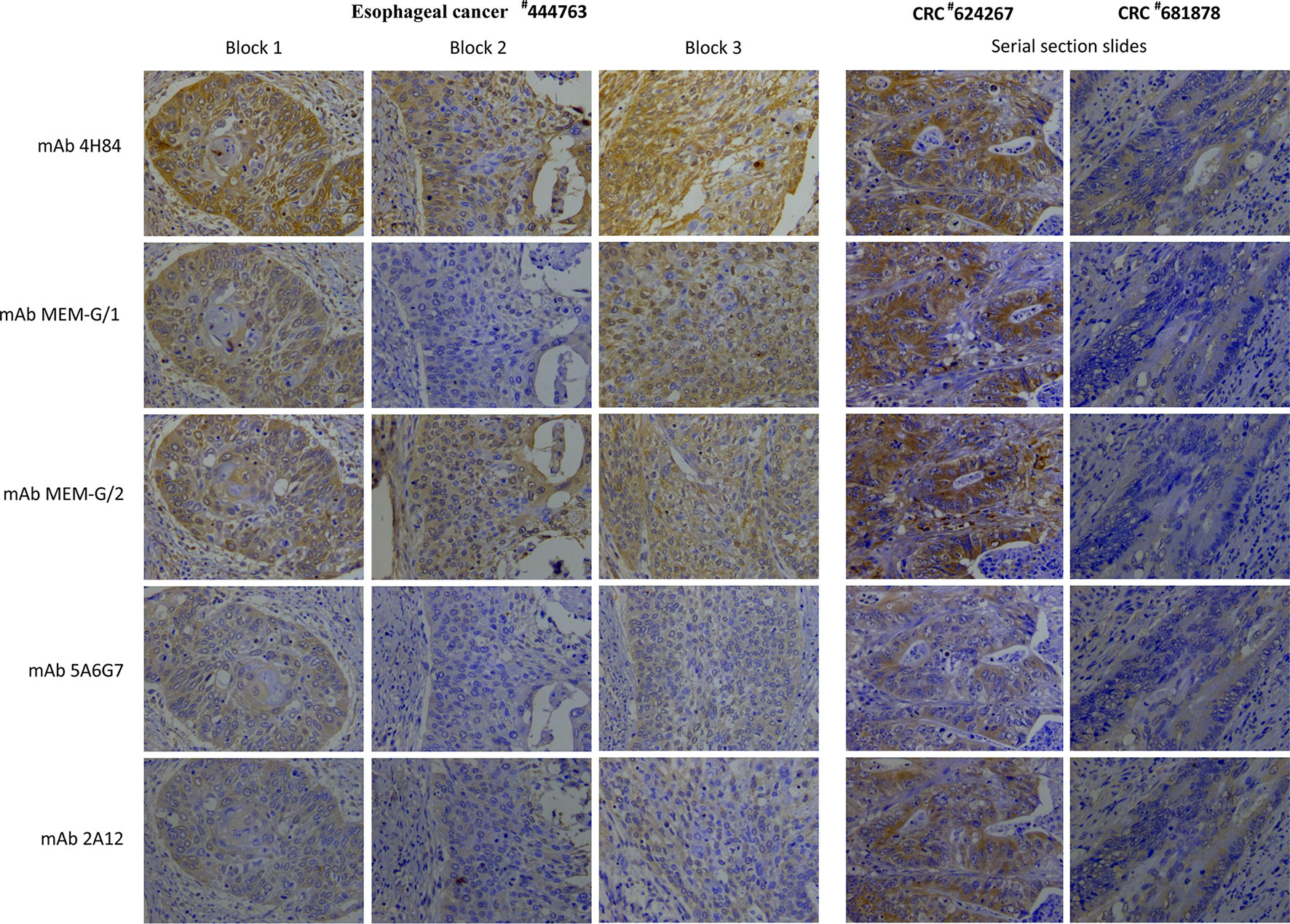
Figure 1 Representatives of intratumor heterogeneous staining of HLA-G expression with different anti-HLA-G antibodies in three blocks of a esophageal squamous cell carcinoma sample (#444763), and serial sections of two colorectal cancer samples (#624267 and #681878) (400×).
Intratumor Heterogeneity of HLA-G
Intratumor heterogeneous expression of HLA-G was observed among different sections and antibodies used in three CRC tissue samples (#598937, #624267, and #681878).
For the Group 1 antibodies (mAbs 4H84, MEM-G/1, and MEM-G/2), HLA-G expression was dramatically different in samples CRC#598937 (p<0.001) and CRC#681878 (p<0.001), while comparable degree of HLA-G expression was observed in sample CRC#624267 (p=0.453). Among these samples, no significant variation of HLA-G expression was found for the Group 2 antibodies (mAbs 5A6G7 and 2A12; Table 1).
Moreover, HLA-G expression in samples from different zones of a same tumor also varied significantly when detected with a distinct anti-HLA-G antibody. CRC#1022488 for an example, the percentage of HLA-G expression detected with mAbs 5A6G7 and 2A12 are much higher than that probed with mAbs 4H84, MEM-G/1 and MEM-G/2. Zone 2 particularly, percentage of HLA-G expression detected by mAb 5A6G7 is 20.29% while HLA-G is nearly negative detected by mAb 4H84 (0.71%). In CRC#1022363, the degree of HLA-G detected by mAb 4H84 was 14.88%, 50.0% and 22.0% in zone 1, zone 2, and zone 3, respectively. HLA-G expression in zone 2 and 3 was almost undetectable, while HLA-G was positive in zone 1 when detected by mAbs MEM-G/1 and MEM-G/2. Moreover, HLA-G expression was observed in all three zones when detected with mAb 5A6G7 and mAb 2A12, respectively (Table 2). Similarly, intratumor heterogeneity of HLA-G expression was also found in case-matched esophageal cancer blocks (Table 3).
Intratumor Heterogeneity of HLA-G Isoforms
Distinct pattern and variation of HLA-G expression was also observed for each antibody for HLA-G detection among 10 randomized slides from a same tumor sample. No significant variation of HLA-G expression was observed when detected by mAb 2A12 in CRC#598937 (p=0.1151), mAb 4H84 in CRC#681878 (p=0.154), and mAbs MEM-G/1 (p=0.203) and MEM-G/2 (p=0.386) in CRC#624267. HLA-G expression was found varied dramatically among 10 slides when probed with a distinct anti-HLA-G antibody (Figure 2A). To be noted, previously considered as unexpected immunohistochemistry staining patters such as mAb 4H84neg mAb 5A6G7pos was observed in this study (Table 2). In CRC#1022488, HLA-G expression is low/negative stained with mAbs 4H84, MEM-G/1, and MEM-G/2, while HLA-G is positive when stained with mAbs 5A6G7 and 2A12. This staining pattern now could be explained by the findings that novel HLA-G isoforms such as lacking the α1 domain was depicted by Tronik-Le Roux et al. (3) in a renal cancer study. Similar data were also observed in slides from three different zones within a same sample from another five CRC samples (#1022488, #1022363, #1020932, #1023081, and #444345; Figure 2B).
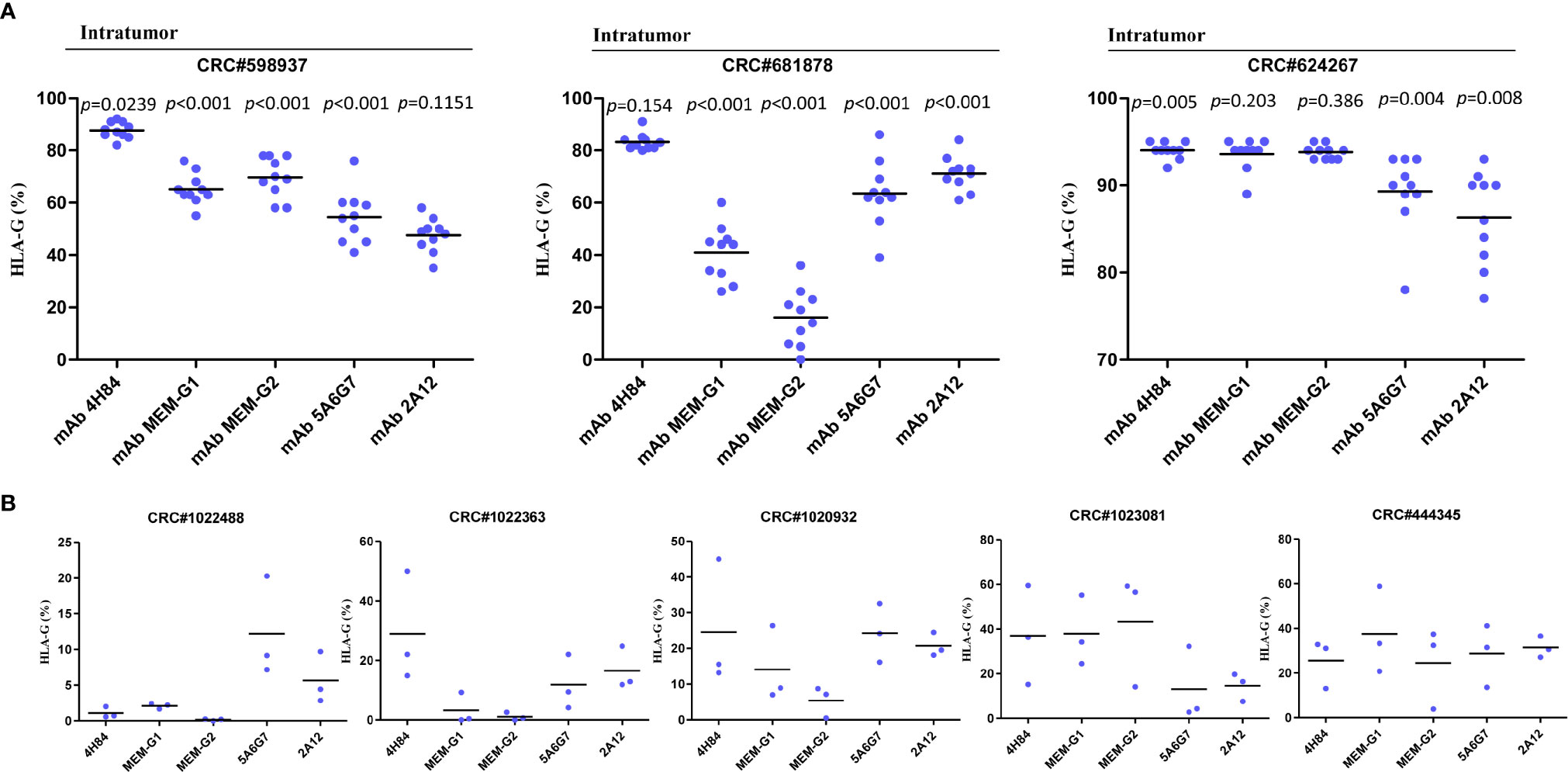
Figure 2 Intratumor heterogeneous staining of HLA-G expression with different anti-HLA-G antibodies in (A) serial sections of three colorectal cancer samples (each sample with 50 serial sections, 10 randomized slides for each antibody). Dot represents each section. Black line represents median. Comparison among the sections was analyzed with analyzed with Kruskal-Wallis H test. (B) three different zones within a same sample (five CRC samples). Dot represents each zone. Black line represents median.
Among 36 ESCC samples, HLA-G expression could be detected by mAbs 4H84, MEM-G/1 and MEM-G/2, while HLA-G expression is negative detected by mAbs 5A6G7 and 2A12 in most cases. Moreover, the staining pattern for mAbs 4H84 and 5A6G7 seems more consistent according to their recognizing epitope in the HLA-G heavy chain, that no mAbs 4H84neg5A6G7pos was observed (Table 3).
Discussion
Inter-tumor HLA-G expression in various types of tumor tissues has been widely investigated and its clinical significance has been well acknowledged. A large body of studies have evidenced that higher degree of HLA-G expression in cancers is related to disease progression and worse clinical outcome (14). Based on the signaling pathway of HLA-G/ILTs and its clinical relevance, HLA-G as a potential immune checkpoints is expected (1). Though ICIs such as targeting the PD-L/PD-L1 is certainly an effective and promising strategic regime for cancer immunotherapy, limited effects of the ICIs therapy resulted from inter- and intratumor heterogeneous expression of immune checkpoints is gaining concern (19).
Indeed, the degree and percentage of HLA-G in cancers varies significantly among different types of cancers which have been observed to be negative in uveal melanoma to totally positive in hydatidiform moles (23, 24), and inconsistent HLA-G findings among different cohorts or laboratories existed in most cases even on a same type of cancer such as breast cancer (25–27) and CRC (22, 28–30). These controversies might be raised by the different specificities of HLA-G monoclonal antibodies, varied laboratory technical procedures, or different composition and HLA-G genetic backgrounds of the included cohorts (14, 31). In line with this, our data showed that different staining pattern of HLA-G expression has been observed between the CRC and ESCC, where HLA-G is almost negative in ESCC but positive in CRC samples when detected by mAbs 5A6G7 and 2A12. This finding indicated that HLA-G isoforms could be differentially regulated among different types of cancers. Moreover, mechanisms involved in regulation of HLA-G expression are complex. In addition to the HLA-G genetic variations both in 5′-upstream regulatory region and in 3′-untranslated region which comprise binding sites for transcription factor and microRNAs and epigenetic modifications (32), other environmental factor such as hypoxia, cytokines, hormones, and even immunotherapy chemicals and radiation have been acknowledged to be related to the regulation of HLA-G expression (33–35).
Intratumor heterogeneity of HLA-G expression has been firstly detailed in 19 primary renal cell cancer (RCC) tumor tissues. HLA-G expression was sharply differed either between samples or inside a tumor tissue (20). In that study, with mAb 4H84, authors revealed that various degree of HLA-G expression exists among different areas (zones) as they illustrated in sample RCC#2 (70% in area T1, 37% in T2, 58% in T3 and T4, respectively), while no HLA-G expression was observed in the T1 or T2 areas in sample RCC#10. In line with their findings, as our data in this study revealed that intratumor heterogeneous expression of HLA-G is a common phenomenon among different zones within a sample in CRC and ESCCs. According to these results, similar findings that intratumor HLA-G heterogeneity could be expected in other malignancies. Shortly afterwards, with transcriptome analysis in RCC samples, they further depicted that, besides the already identified HLA-G1~HLA-G7 isoforms, novel HLA-G isoforms without an α1 domain and transmembrane region could be existed (3). This important finding do explain previously unexpected immunohistochemistry staining patters such as mAb 4H84neg mAb 5A6G7pos, which was observed in our study such as the CRC#1022488 and other samples. In this context, in an our previous study, we found 44 out of 379 (11.6%) CRC patients were with the staining pattern of mAbs 4H84neg 5A6G7pos, and CRC patients with the patterns of mAbs 4H84neg 5A6G7pos had a longer survival time than those with the pattern of mAbs 4H84pos5A6G7neg (36). However, future investigations for the biological functions and clinical significance of novel HLA-G isoforms with mAbs 4H84neg 5A6G7pos are extremely necessary.
However, our study have notable limitations. First, this study is based on a very limited size of patients and types of cancers, the real-world of the heterogeneity of HLA-G expression in more different types of cancers and in larger cohorts of cancer patients remain to be explored. Second, being the very limited size of the patients included, clinical significance of the heterogeneity of HLA-G and HLA-G isoform expression in cancers is still unknown. Third, potential mechanisms underlying the heterogeneity of HLA-G in cancers remain to be uncovered. Finally, more specific antibodies for HLA-G isoforms are needed to define the clinical significance of a particular HLA-G isoforms.
In summary, our study revealed a rather high degree of intratumor heterogeneity of HLA-G expression in cancers, and degree of HLA-G expression is also varied among anti-HLA-G antibodies with different specificities. Therefore, to evaluate the clinical significance of HLA-G expression in cancers, important issues including location of the tumor tissues isolated, HLA-G isoforms and specificity of the anti-HLA-G antibodies should be concerned.
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical Ethics Review Board of Taizhou Hospital of Zhejiang Province. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
W-HY: study design. XZ, JG, and AL: performed experiments. J-GZ, Q-YH, Q-YC, Y-HY, W-JZ, and H-HX: material support and data acquisition. W-HY: performed statistical analysis and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from National Natural Science Foundation of China (81901625) and Science and Technology Bureau of Taizhou (1901ky01; 1901ky04, 1901ky05, 1901ky09).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: An Immune Checkpoint Molecule. Adv Immunol (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
2. Amiot L, Vu N, Samson M. Biology of the immunomodulatory molecule HLA-G in human liver diseases. J Hepatol (2015) 62:1430–7. doi: 10.1016/j.jhep.2015.03.007
3. Tronik-Le Roux D, Renard J, Vérine J, Renault V, Tubacher E, LeMaoult J, et al. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol (2017) 11:1561–78. doi: 10.1002/1878-0261.12119
4. Carosella ED, Ploussard G, LeMaoult J, Desgrandchamps FA. Systematic Review of Immunotherapy in Urologic Cancer: Evolving Roles for Targeting of CTLA-4, PD-1/PD-L1, and HLA-G. Eur Urol (2015) 68:267–79. doi: 10.1016/j.eururo.2015.02.032
5. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci U S A (1998) 95:4510–5. doi: 10.1073/pnas.95.8.4510
6. Chen BG, Xu DP, Lin A, Yan WH. NK cytolysis is dependent on the proportion of HLA-G expression. Hum Immunol (2013) 74:286–9. doi: 10.1016/j.humimm.2012.12.005
7. Lin A, Zhang X, Xu HH, Xu DP, Ruan YY, Yan WH. HLA-G expression is associated with metastasis and poor survival in the Balb/c nu/nu murine tumor model with ovarian cancer. Int J Cancer (2012) 131:150–7. doi: 10.1002/ijc.26375
8. Loumagne L, Baudhuin J, Favier B, Montespan F, Carosella ED, Rouas-Freiss N. In vivo evidence that secretion of HLA-G by immunogenic tumor cells allows their evasion from immunosurveillance. Int J Cancer (2014) 135:2107–17. doi: 10.1002/ijc.28845
9. Lesport E, Baudhuin J, Sousa S, LeMaoult J, Zamborlini A, Rouas-Freiss N, et al. Inhibition of human gamma delta [corrected] T-cell antitumoral activity through HLA-G: implications for immunotherapy of cancer. Cell Mol Life Sci (2011) 68:3385–99. doi: 10.1007/s00018-011-0632-7
10. Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol (2014) 192:1536–46. doi: 10.4049/jimmunol.1300438
11. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6–STAT3 signaling pathway. Proc Natl Acad Sci U S A (2008) 105:8357–62. doi: 10.1073/pnas.0803341105
12. Lee CL, Guo Y, So KH, Vijayan M, Guo Y, Wong VH, et al. Soluble human leukocyte antigen G5 polarizes differentiation of macrophages toward a decidual macrophage-like phenotype. Hum Reprod (2015) 30:2263–74. doi: 10.1093/humrep/dev196
13. Köstlin N, Ostermeir AL, Spring B, Schwarz J, Marmé A, Walter CB, et al. HLA-G promotes myeloid-derived suppressor cell accumulation and suppressive activity during human pregnancy through engagement of the receptor ILT4. Eur J Immunol (2017) 47:374–84. doi: 10.1002/eji.201646564
14. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
15. Lin A, Yan WH. Intercellular transfer of HLA-G: its potential in cancer immunology. Clin Transl Immunol (2019) 8:e1077. doi: 10.1002/cti2.1077
16. Agaugué S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood (2011) 117:7021–31. doi: 10.1182/blood-2010-07-294389
17. Lin A, Xu HH, Xu DP, Zhang X, Wang Q, Yan WH. Multiple steps of HLA-G in ovarian carcinoma metastasis: alter NK cytotoxicity and induce matrix metalloproteinase-15 (MMP-15) expression. Hum Immunol (2013) 74:439–46. doi: 10.1016/j.humimm.2012.11.021
18. Wang C, Chen J, Zhang Q, Li W, Zhang S, Xu Y, et al. Elimination of CD4(low)HLA-G(+) T cells overcomes castration-resistance in prostate cancer therapy. Cell Res (2018) 28:1103–17. doi: 10.1038/s41422-018-0089-4
19. Jiang Y, Zhao X, Fu J, Wang H. Progress and Challenges in Precise Treatment of Tumors With PD-1/PD-L1 Blockade. Front Immunol (2020) 11:339. doi: 10.3389/fimmu.2020.00339
20. Rouas-Freiss N, LeMaoult J, Verine J, Tronik-Le Roux D, Culine S, Hennequin C, et al. Intratumor heterogeneity of immune checkpoints in primary renal cell cancer: Focus on HLA-G/ILT2/ILT4. Oncoimmunology (2017) 6:e1342023. doi: 10.1080/2162402X.2017.1342023
21. Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol (2010) 17:1471–4. doi: 10.1245/s10434-010-0985-4
22. Zhang RL, Zhang X, Dong SS, Hu B, Han QY, Zhang JG, et al. Predictive value of different proportion of lesion HLA-G expression in colorectal cancer. Oncotarget (2017) 8:107441–51. doi: 10.18632/oncotarget.22487
23. Anastassiou G, Rebmann V, Wagner S, Bornfeld N, Grosse-Wilde H. Expression of classic and nonclassic HLA class I antigens in uveal melanoma. Invest Ophthalmol Vis Sci (2003) 44:2016–9. doi: 10.1167/iovs.02-0810
24. Goldman-Wohl D, Ariel I, Greenfield C, Hochner-Celnikier D, Lavy Y, Yagel S. A study of human leukocyte antigen G expression in hydatidiform moles. Am J Obstet Gynecol (2001) 185:476–80. doi: 10.1067/mob.2001.115994
25. Chen HX, Lin A, Shen CJ, Zhen R, Chen BG, Zhang X, et al. Upregulation of human leukocyte antigen-G expression and its clinical significance in ductal breast cancer. Hum Immunol (2010) 71:892–8. doi: 10.1016/j.humimm.2010.06.009
26. He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, et al. HLA-G expression in human breast cancer: implications for diagnosis and prognosis, and effect on allocytotoxic lymphocyte response after hormone treatment in vitro. Ann Surg Oncol (2010) 17:1459–69. doi: 10.1245/s10434-009-0891-9
27. da Silva GB, Silva TG, Duarte RA, Neto NL, Carrara HH, Donadi EA, et al. Expression of the Classical and Nonclassical HLA Molecules in Breast Cancer. Int J Breast Cancer (2013) 2013:250435. doi: 10.1155/2013/250435
28. Swets M, König MH, Zaalberg A, Dekker-Ensink NG, Gelderblom H, van de Velde CJ, et al. HLA-G and classical HLA class I expression in primary colorectal cancer and associated liver metastases. Hum Immunol (2016) 77:773–9. doi: 10.1016/j.humimm.2016.03.001
29. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human leukocyte antigen G expression: as a significant prognostic indicator for patients with colorectal cancer. Mod Pathol (2007) 20:375–83. doi: 10.1038/modpathol.3800751
30. Guo ZY, Lv YG, Wang L, Shi SJ, Yang F, Zheng GX, et al. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol (2015) 293:10–6. doi: 10.1016/j.cellimm.2014.10.003
31. Swets M, Wouters A, Krijgsman D, van Vlierberghe RLP, Boot A, van Eendenburg JD, et al. HLA-G protein expression in colorectal cancer evaluated by immunohistochemistry and western blot analysis: Its expression characteristics remain enigmatic. Clin Immunol (2018) 194:80–6. doi: 10.1016/j.clim.2018.07.005
32. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res (2014) 2014:734068. doi: 10.1155/2014/734068
33. Ziliotto M, Rodrigues RM, Chies JAB. Controlled hypobaric hypoxia increases immunological tolerance by modifying HLA-G expression, a potential therapy to inflammatory diseases. Med Hypotheses (2020) 140:109664. doi: 10.1016/j.mehy.2020.109664
34. Persson G, Bork JBS, Isgaard C, Larsen TG, Bordoy AM, Bengtsson MS, et al. Cytokine stimulation of the choriocarcinoma cell line JEG-3 leads to alterations in the HLA-G expression profile. Cell Immunol (2020) 352:104110. doi: 10.1016/j.cellimm.2020.104110
35. Ishikawa M, Brooks AJ, Fernández-Rojo MA, Medina J, Chhabra Y, Minami S, et al. Growth hormone stops excessive inflammation after partial hepatectomy allowing liver regeneration and survival via induction of H2-Bl/HLA-G. Hepatology (2020). doi: 10.1002/hep.31297
Keywords: HLA-G, tumor, heterogeneity, isoform, antibody, colorectal cancer, esophageal cancer
Citation: Zhang X, Lin A, Han Q-Y, Zhang J-G, Chen Q-Y, Ye Y-H, Zhou W-J, Xu H-H, Gan J and Yan W-H (2020) Intratumor Heterogeneity of HLA-G Expression in Cancer Lesions. Front. Immunol. 11:565759. doi: 10.3389/fimmu.2020.565759
Received: 03 June 2020; Accepted: 27 October 2020;
Published: 19 November 2020.
Edited by:
Brian Duncan Tait, Australian Red Cross Blood Service, AustraliaReviewed by:
Uma Kanga, All India Institute of Medical Sciences, IndiaStanislaw Stepkowski, University of Toledo, United States
Vera Rebmann, University of Duisburg-Essen, Germany
Copyright © 2020 Zhang, Lin, Han, Zhang, Chen, Ye, Zhou, Xu, Gan and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Hua Yan, eWFud2hjb21AeWFob28uY29t
 Xia Zhang1
Xia Zhang1 Aifen Lin
Aifen Lin Hui-Hui Xu
Hui-Hui Xu Wei-Hua Yan
Wei-Hua Yan