
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Immunol. , 29 September 2020
Sec. Vaccines and Molecular Therapeutics
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.561851
Many vaccines are under clinical trials to fight against pandemic COVID-19. Here, we briefly reviewed vaccines under trial and their limitations, with a possible alternate solution to overcome these limitations. In most cases, vaccine targets are all viral proteins or a specific protein, mainly spike protein. As shown in previous studies on coronavirus strains, these vaccine targets may cause a wide range of side effects. These include the induction of cytokine storms (i.e., IL6), lung immunopathology, hepatitis, hemotoxicity, cytotoxicity, cross-reactive antibodies, allergenicity. Minimizing the size of vaccine candidates from protein to epitope/peptide will overcome the side effects. In our opinion, there is a need to utilize computer-aided techniques for the identification of potential vaccine candidates to fight against COVID-19.
The pandemic COVID-19, a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) disease, emerged as the most recent significant challenge to global health and prosperity since World War II. WHO has reported more than 26 million cases, with 0.87 million deaths up to September 6, 2020 (1). Lack of proper medication forced many countries to opt for complete lockdown, which caused chaos in the economy (2). COVID-19 posed a threat not only to lives but also to freedom, it threatened to tear the world into boundaries and lock people into confined regions. An exponential increase in the infected cases forced the researcher to look up for the complete cure as early as possible. Thus, the COVID-19 vaccine has become a holy grail. At the start of May 2020, more than 120 vaccines were in the pipeline throughout the world, and at least six groups had started injecting the formulation into volunteers (3). According to WHO, 34 vaccine candidates were in clinical evaluation, among which eight candidates entered into phase 3 of the clinical trial as of September 3, 2020. Besides this, about 142 vaccines are in preclinical evaluation (4).
Ad5-nCoV, developed by CanSino Biologics and an arm of the People’s Liberation Army, was the first candidate to enter human trials and is currently in clinical phase 2. It is a viral vector vaccine and uses non-replicating adenovirus type 5 vector, to transport the DNA of spike proteins of SARS-CoV-2 (3, 5). The concept is to transform the spike proteins within the body, ultimately leading to the activation of the immune system. Using the same technology, researchers from Oxford University developed ChAdOx1, which will eventually produce spike proteins within the human body, leading to immune system activation. ChAdOx1 has already started phase 3 clinical trials, after showing an acceptable safety profile in single blind, randomized phase 1/2 clinical trials (6). Inovio Pharmaceuticals, a US-based biotechnology firm, has developed INO-4800, a DNA-based vaccine, utilizing a relatively newer vaccine technique. Here, DNA containing the genetic code of SARS-CoV-2 spike proteins will be injected into human cells by electroporation devices, and the transformed spike proteins will activate the immune system. PiCoVacc, currently in phase 3 trial, is an inactivated vaccine developed by Sinovac, a Chinese private biopharma company, based on the traditional fact that exposure to an inactivated virus will eventually lead up to the immune response (7). The Beijing Institute of Biological Products/Wuhan Institute of Biological Products is working on few inactivated vaccines, which are in the clinical phases 2/3. However, they are relatively unadvertised outside Chinese media (8). Besides this, two RNA vaccines are also in the pipeline, named as mRNA-1273 by US biotech firm Moderna/NIAID and BNT-162 jointly by German company BioNTech and US pharma giant Pfizer. Both these vaccines follow the concept of delivering information molecules to instruct human body cells to produce the spike proteins of SARS-CoV-2. This information molecule is mRNA in the case of BNT-162, whereas LNP-encapsulated mRNA in mRNA-1273 (9, 10) (Figure 1).
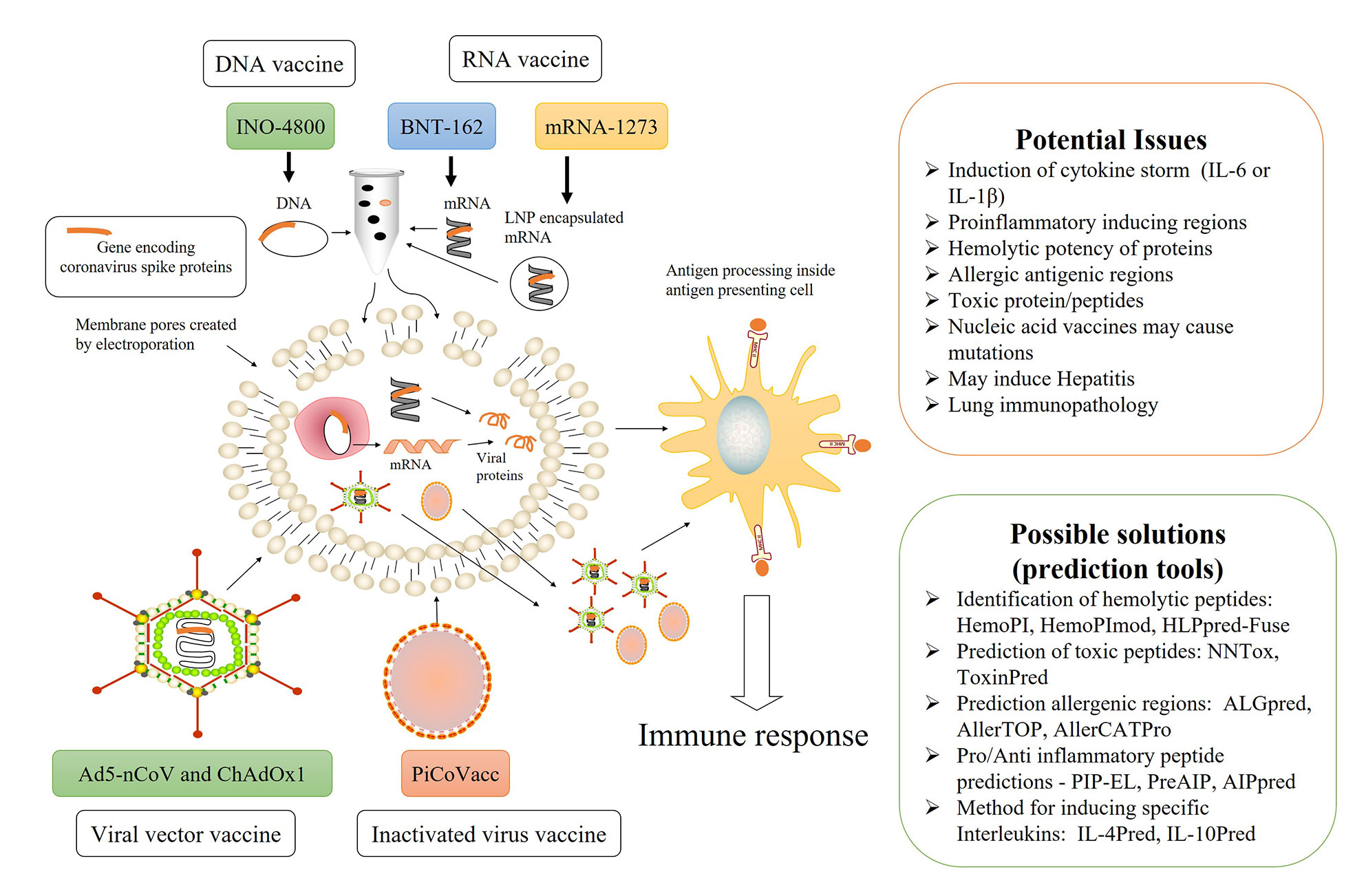
Figure 1 Overview of current SARS-CoV-2 vaccines under trial, their possible limitations, and computer-aided solutions.
The current distressing worldwide scenario demands a vaccine at the earliest, causing a race among pharmaceutical industries. This accelerated version of vaccine development implements parallel steps like animal testing, clinical phase 1, or simultaneous phase 1 and 2 clinical trials. By involving several manufacturers, the developers of ChAdOx1 have already planned to produce a million-dose by September 2020 (11). However, the question is whether speeding the trail is safe. What are the possibilities of getting a vaccine with the highest efficacy concerning diverse ages and ethnicity? What is the financial risk in scaling up production capacity, even before sufficient efficacy data, like in the case of ChAdOx1? These questions make sense if we re-evaluate recent virus outbreaks, including Ebola, Zika, or earlier coronaviruses, i.e., SARS and MERS. The Ebola and Zika epidemics ended before vaccine development, leaving the manufacturer with financial loss due to the reallocation of funds by federal agencies (12). The SARS (severe acute respiratory syndrome) coronavirus emerged in 2002-03 and affected 26 countries, whereas the MERS (Middle East respiratory syndrome) coronavirus emerged in 2012 and affected 27 countries (13). However, the astonishing fact is that no vaccine has been commercially available for these coronaviruses until now (14). Inactivated SARS virus (15), live attenuated/host-adapted SARS virus (16), replicating and non-replicating viral vector (17, 18), DNA based vaccine (19), soluble proteins/adjuvants (20), virus-like peptide/adjuvants (21), and a combination of vaccines approach (22, 23) have been used to develop a vaccine against SARS-CoV, but to the best of our knowledge only inactivated SARS virus, DNA and soluble SARS S-glycoprotein reached the clinical phase 1 trials (14). Similarly, for MERS, several vaccines have been developed, but only a DNA based vaccine, targeting S-glycoproteins and subunits, is in a clinical phase 1 trial (24). The vaccine development might have been slowed by several reasons such as a lack of suitable animal models, as they exhibited limited viral replication and clinical manifestations (25) or geographically centralized cases might have declined the interest of pharmaceutical companies. Whatever the reason for the lack of vaccine or financial losses, there is a need to revisit these cases while investing in the development of the SARS-CoV-2 vaccine.
The nature of COVID-19 transmission makes it more devastating, as the median incubation time from infection to symptoms ranges from 4-7 days. Besides this, many infected patients remain asymptomatic but prone to transmit the virus (26). It is quite evident that both innate and adaptive immunity plays a role during the COVID-19 infection, but how their interaction mediates viral control as well as host toxicity, is not very clear yet. Based on pieces of evidence, it has been suggested that an innate immune-mediated ‘cytokine storm’ is responsible for toxicity and organ damage in a subgroup of patients with a severe COVID-19 infection (27). Extended populations of IL-6 and IL-1β secreting monocytes have been found in COVID-19 infected patients, resulting in elevated serum IL-6 and lactate dehydrogenase, a marker of pyroptosis, a highly inflammatory form of programmed cell death (28, 29). Similarly, an elevated level of ferritin and IL-6 in 150 multicenter COVID-19 confirmed cases suggested that virally driven hyper-inflammation might be the reason for mortality (30). It is a known fact that virally derived pathogen and damage-associated molecular patterns (PAMP and DAMP) activate macrophages, resulting in the downstream production of IL-6 and IL-1β, which further recruits neutrophils and CD8+ T cells, thus controlling viral growth. However, within the lung parenchyma, neutrophils also induce tissue damage by releasing leukotrienes and reactive oxygen species, giving passage to alveolar flooding and fibrosis. In severe infection, persistent IL-6 elevation leads to constant neutrophil-mediated alveolar damage, resulting in the need for mechanical ventilation and, ultimately, mortality (31). We all know that an early innate and adaptive immune response leads to the suppression of acute viral infection. However, chronic viral infection causes T cell depletion and exhaustion, leaving persistent innate activation, triggering inflammation, and cytokine toxicity. An intact T cell-mediated immune response is the key contributor in clearing and maintaining long term suppression (32). It is a growing possibility that immunosuppression due to the depletion and exhaustion of T cells contribute to COVID-19 persistence and mortality (27, 33).
There are several studies showing a paradoxical phenomenon, in which vaccinated animals or people exhibit a more severe disease while exposed to the virus than non-activated fellows (34–36). This immune backfiring is known as antibody dependent enhancement (ADE), in which the virus influences antibodies for its own benefit and enhances the infection or immune enhancement, consisting of allergic inflammation due to Th2 immunopathology. ADE has been observed with dengue, HIV, flavivirus, as well as Zika virus (37–40). The hunt for a SARS and MERS vaccine has been stymied by ADE. It has been shown that, antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins (41, 42). ADE or eosinophil-mediated immunopathology has been observed while immunizing mice with inactivated whole SARS-CoV (43) or DNA vaccine encoding full length S-protein (44) as well as immunizing macaque models with MVA encoded S protein (45). There have been various recently published articles concerning the potential danger of suboptimal antibody responses or ADE of SARS-CoV-2, providing an insightful discussion (46–49).
It is obvious to consider the fact that both an innate and adaptive immunity participates in COVID-19 mediated toxicity while designing vaccines. There is always a possibility of mutation within a human genome by nucleic acid vaccines (50). What if the spike proteins contain the region which could elicit the IL-6 or IL-1β level? What if the nucleic acid fragment delivered within the body transforms into a region of spike proteins that are proinflammatory or have any other interleukin-inducing peptides? There are significant possibilities of having toxic peptide fragments in COVID-19 spike proteins. The peptide region may have hemolytic potential, resulting in hemolysis as a severe side effect. Besides this, the coronavirus spike protein may contain the regions prone to allergy, and their processing inside the cell may lead to allergic antigens. Have they been ruled out, while delivering spike protein regions into the body? Bioinformatics might provide an answer to these queries to some extent. There are several in silico tools to predict interleukin-inducing properties, pro/anti- inflammatory as well as the toxicity of peptides (51–60). Besides this, many in silico tools are available for designing subunit vaccines and immunotherapeutics (61, 62). There are 244,682 proteins and 22,892 nucleotide sequence information about SARS-CoV-2 available in the NCBI virus datahub, as of September 8, 2020 (63). It is obvious that all 244,682 proteins cannot be a vaccine candidate, so we can use various in silico tools to identify potential vaccine candidates. These candidates must be further scrutinized for potential epitopes, which can activate the desired arm of the immune system. The half-life, toxicity, and unwanted properties like in SARS-CoV-2 cases such as IL-6 inducing potential, immunosuppressive property, hemolysis, and allergy must also be checked. We have enlisted some useful in silico tools in Figure 1. Various researchers have already identified potential B and T cell epitopes, HLA susceptibility mapping as well as candidate targets by applying a bioinformatic approach (64–69). In the cat and mouse game of a viral pandemic and vaccine development, bioinformatic resources are beneficial and will cut down the chances of failure, a lesson by previous virus endemics. There are significant chances of severe side effects, as the frenetic race against time forced the vaccine trials on a fast track. Due to emergency and the worldwide crisis, there is a possibility of accelerating the trial mechanism by reducing numbers and waving off some of the population, as reported in several news agencies (70, 71). We hope that the devastating worldwide scenario will end up with a successful vaccine candidate soon.
SU wrote the manuscript while GR conceived the idea as well as edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. World Health Organization. “Coronavirus Disease (COVID-19)”. In: Weekly Epidemiological Update. WHO (2020a). Accessed on Sep 8, 2020.
2. Lora Jones, Palumbo D, Brown D. Coronavirus: A visual guide to the economic impact. BBC NEWS (2020). Available at: https://www.bbc.com/news/business-51706225 Acessed on Jul 10, 2020.
3. Callaway E. Scores of coronavirus vaccines are in competition - how will scientists choose the best? Nature (2020). Accessed on May 1, 2020. doi: 10.1038/d41586-020-01247-2
4. World Health Organization. “Draft landscape of COVID-19 candidate vaccines”. WHO (2020b). Accessed on Sep 8, 2020.
5. Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, et al. A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Int J Mol Sci (2020) 21(7):2657. doi: 10.3390/ijms21072657
6. Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet (2020) 396(10249):467–78. doi: 10.1016/S0140-6736(20)31604-4
7. Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science (2020) 369(6499):77–81. doi: 10.1126/science.abc1932
8. Xinhua. “China Focus: China’s inactivated COVID-19 vaccine starts phase-3 clinical trial internationally”, (ed.) huaxia.) XINHUANET (2020). Accessed on Sep 8, 2020.
9. Shi Y, Wang N, Zou QM. [Progress and challenge of vaccine development against 2019 novel coronaviru-nCoV)]. Zhonghua Yu Fang Yi Xue Za Zhi (2020) 54(0):E029. doi: 10.3760/cma.j.cn112150-20200317-00366
10. Yang L, Tian D, Liu W. [Strategies for vaccine development of COVID-19]. Sheng Wu Gong Cheng Xue Bao (2020) 36(4):593–604. doi: 10.13345/j.cjb.200094
11. Rees V. “UK COVID-19 vaccine candidate to be manufactured at seven outsourced sites”. Eur Pharm Rev (2020) https://www.europeanpharmaceuticalreview.com/news/117636/uk-covid-19-vaccine-candidate-to-be-manufactured-at-seven-outsourced-sites/ Acessed on May 5, 2020.
12. Lurie N, Saville M, Hatchett R, Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N Engl J Med (2020) 382:1969–73. doi: 10.1056/NEJMp2005630
13. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol (2016) 14(8):523–34. doi: 10.1038/nrmicro.2016.81
14. Padron-Regalado E. Vaccines for SARS-CoV-2: Lessons from Other Coronavirus Strains. Infect Dis Ther (2020) 9(2):255–74. doi: 10.1007/s40121-020-00300-x
15. Lin JT, Zhang JS, Su N, Xu JG, Wang N, Chen JT, et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir Ther (2007) 12(7):1107–13.
16. Netland J, DeDiego ML, Zhao J, Fett C, Alvarez E, Nieto-Torres JL, et al. Immunization with an attenuated severe acute respiratory syndrome coronavirus deleted in E protein protects against lethal respiratory disease. Virology (2010) 399(1):120–8. doi: 10.1016/j.virol.2010.01.004
17. Czub M, Weingartl H, Czub S, He R, Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine (2005) 23(17-18):2273–9. doi: 10.1016/j.vaccine.2005.01.033
18. See RH, Petric M, Lawrence DJ, Mok CPY, Rowe T, Zitzow LA, et al. Severe acute respiratory syndrome vaccine efficacy in ferrets: whole killed virus and adenovirus-vectored vaccines. J Gen Virol (2008) 89(Pt 9):2136–46. doi: 10.1099/vir.0.2008/001891-0
19. Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, et al. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature (2004) 428(6982):561–4. doi: 10.1038/nature02463
20. Zhou Z, Post P, Chubet R, Holtz K, McPherson C, Petric M, et al. A recombinant baculovirus-expressed S glycoprotein vaccine elicits high titers of SARS-associated coronavirus (SARS-CoV) neutralizing antibodies in mice. Vaccine (2006) 24(17):3624–31. doi: 10.1016/j.vaccine.2006.01.059
21. Tseng CT, Sbrana E, Iwata-Yoshikawa N, Newman PC, Garron T, Atmar RL, et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PloS One (2012) 7(4):e35421. doi: 10.1371/journal.pone.0035421
22. Huang J, Cao Y, Du J, Bu X, Ma R, Wu C. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine (2007) 25(39-40):6981–91. doi: 10.1016/j.vaccine.2007.06.047
23. Du L, Zhao G, Lin Y, Chan C, He Y, Jiang S, et al. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine (2008) 26(13):1644–51. doi: 10.1016/j.vaccine.2008.01.025
24. Modjarrad K, Roberts CC, Mills KT, Castellano AR, Paolino K, Muthumani K, et al. Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect Dis (2019) 19(9):1013–22. doi: 10.1016/S1473-3099(19)30266-X
25. Sutton TC, Subbarao K. Development of animal models against emerging coronaviruses: From SARS to MERS coronavirus. Virology (2015) 479–80:247–58. doi: 10.1016/j.virol.2015.02.030
26. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (2020) 395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5
27. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (2020) 395(10229):1033–4. doi: 10.1016/s0140-6736(20)30628-0
28. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery (2020) 6:31. doi: 10.1038/s41421-020-0168-9
29. Zhang D, Guo R, Lei L, Liu H, Wang Y, Wang Y, et al. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv (2020), 2020.2003.2024.20042655. doi: 10.1101/2020.03.24.20042655
30. Ruan Q, Yang K, Wang W, Jiang L, Song J. Correction to: Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intens Care Med (2020) 46(6):1294–7. doi: 10.1007/s00134-020-06028-z
31. Vardhana SA, Wolchok JD. The many faces of the anti-COVID immune response. J Exp Med (2020) 217(6):e20200678. doi: 10.1084/jem.20200678
32. Yoo JK, Kim TS, Hufford MM, Braciale TJ. Viral infection of the lung: host response and sequelae. J Allergy Clin Immunol (2013) 132(6):1263. doi: 10.1016/j.jaci.2013.06.006
33. Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, et al. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin Exp Rheumatol (2020) 38(2):337–42.
34. Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis (1979) 140(4):527–33. doi: 10.1093/infdis/140.4.527
35. Yoon KJ, Wu LL, Zimmerman JJ, Hill HT, Platt KB. Antibody-dependent enhancement (ADE) of porcine reproductive and respiratory syndrome virus (PRRSV) infection in pigs. Viral Immunol (1996) 9(1):51–63. doi: 10.1089/vim.1996.9.51
36. Tirado SM, Yoon KJ. Antibody-dependent enhancement of virus infection and disease. Viral Immunol (2003) 16(1):69–86. doi: 10.1089/088282403763635465
37. Gorlani A, Forthal DN. Antibody-dependent enhancement and the risk of HIV infection. Curr HIV Res (2013) 11(5):421–6. doi: 10.2174/1570162x113116660062
38. Taylor A, Foo SS, Bruzzone R, Dinh LV, King NJ, Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol Rev (2015) 268(1):340–64. doi: 10.1111/imr.12367
39. Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science (2017) 358(6365):929–32. doi: 10.1126/science.aan6836
40. Martin-Acebes MA, Saiz JC, Jimenez de Oya N. Antibody-Dependent Enhancement and Zika: Real Threat or Phantom Menace? Front Cell Infect Microbiol (2018) 8:44. doi: 10.3389/fcimb.2018.00044
41. Jaume M, Yip MS, Cheung CY, Leung HL, Li PH, Kien F, et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcgammaR pathway. J Virol (2011) 85(20):10582–97. doi: 10.1128/JVI.00671-11
42. Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun (2014) 451(2):208–14. doi: 10.1016/j.bbrc.2014.07.090
43. Bolles M, Deming D, Long K, Agnihothram S, Whitmore A, Ferris M, et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol (2011) 85(23):12201–15. doi: 10.1128/JVI.06048-11
44. Yang ZY, Werner HC, Kong WP, Leung K, Traggiai E, Lanzavecchia A, et al. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc Natl Acad Sci USA (2005) 102(3):797–801. doi: 10.1073/pnas.0409065102
45. Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight (2019) 4(4):e123158. doi: 10.1172/jci.insight.123158
46. Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature (2020) 584(7821):353–63. doi: 10.1038/s41586-020-2538-8
47. Fierz W, Walz B. Antibody Dependent Enhancement Due to Original Antigenic Sin and the Development of SARS. Front Immunol (2020) 11:1120. doi: 10.3389/fimmu.2020.01120
48. Hotez PJ, Corry DB, Bottazzi ME. COVID-19 vaccine design: the Janus face of immune enhancement. Nat Rev Immunol (2020) 20(6):347–8. doi: 10.1038/s41577-020-0323-4
49. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol (2020) 20(6):339–41. doi: 10.1038/s41577-020-0321-6
50. Chary MA, Barbuto AF, Izadmehr S, Hayes BD, Burns MM. COVID-19: Therapeutics and Their Toxicities. J Med Toxicol (2020) 16(3):284–94. doi: 10.1007/s13181-020-00777-5
51. Saha S, Raghava GP. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res (2006) 34(Web Server issue):W202–209. doi: 10.1093/nar/gkl343
52. Dhanda SK, Gupta S, Vir P, Raghava GP. Prediction of IL4 inducing peptides. Clin Dev Immunol (2013) 2013:263952. doi: 10.1155/2013/263952
53. Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP. In silico approach for predicting toxicity of peptides and proteins. PloS One (2013) 8(9):e73957. doi: 10.1371/journal.pone.0073957
54. Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v.2–a server for in silico prediction of allergens. J Mol Model (2014) 20(6):2278. doi: 10.1007/s00894-014-2278-5
55. Chaudhary K, Kumar R, Singh S, Tuknait A, Gautam A, Mathur D, et al. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci Rep (2016) 6:22843. doi: 10.1038/srep22843
56. Nagpal G, Usmani SS, Dhanda SK, Kaur H, Singh S, Sharma M, et al. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Sci Rep (2017) 7:42851. doi: 10.1038/srep42851
57. Jain A, Kihara D. NNTox: Gene Ontology-Based Protein Toxicity Prediction Using Neural Network. Sci Rep (2019) 9(1):17923. doi: 10.1038/s41598-019-54405-6
58. Maurer-Stroh S, Krutz NL, Kern PS, Gunalan V, Nguyen MN, Limviphuvadh V, et al. AllerCatPro-prediction of protein allergenicity potential from the protein sequence. Bioinformatics (2019) 35(17):3020–7. doi: 10.1093/bioinformatics/btz029
59. Hasan MM, Schaduangrat N, Basith S, Lee G, Shoombuatong W, Manavalan B. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics (2020) 36(11):3350–6. doi: 10.1093/bioinformatics/btaa160
60. Kumar V, Kumar R, Agrawal P, Patiyal S, Raghava GPS. A Method for Predicting Hemolytic Potency of Chemically Modified Peptides From Its Structure. Front Pharmacol (2020) 11:54. doi: 10.3389/fphar.2020.00054
61. Dhanda SK, Usmani SS, Agrawal P, Nagpal G, Gautam A, Raghava GPS. Novel in silico tools for designing peptide-based subunit vaccines and immunotherapeutics. Brief Bioinform (2017) 18(3):467–78. doi: 10.1093/bib/bbw025
62. Usmani SS, Kumar R, Bhalla S, Kumar V, Raghava GPS. In Silico Tools and Databases for Designing Peptide-Based Vaccine and Drugs. Adv Protein Chem Struct Biol (2018) 112:221–63. doi: 10.1016/bs.apcsb.2018.01.006
63. NCBI. (2020). “Severe acute respiratory syndrome coronavirus 2 data hub”. NCBI, Accessed on Sep 8, 2020.
64. Ahmed SF, Quadeer AA, McKay MR. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses (2020) 12(3):254. doi: 10.3390/v12030254
65. Baruah V, Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol (2020) 92(5):495–500. doi: 10.1002/jmv.25698
66. Fast E, Altman RB, Chen B. Potential T-cell and B-cell Epitopes of 2019-nCoV. bioRxiv (2020) 2020.02.19.955484. doi: 10.1101/2020.02.19.955484
67. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe (2020) 27671-680(4):e672. doi: 10.1016/j.chom.2020.03.002
68. Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, et al. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J Virol (2020) 94(13):e00510-20. doi: 10.1128/JVI.00510-20
69. Patiyal S, Kaur D, Kaur H, Shrama N, Dhall A, Sahai S, et al. A web-based platform on COVID-19 to maintain Predicted Diagnostic, Drug and Vaccine candidates. OSF Prepr (2020). doi: 10.31219/osf.io/xegzu
70. Cohen J. (2020). Speed coronavirus vaccine testing by deliberately infecting volunteers? Not so fast, some scientists warn. Science. doi: 10.1126/science.abc0006
Keywords: coronavirus, COVID-19, SARS-CoV-2, vaccine candidates, immunoinformatic
Citation: Usmani SS and Raghava GPS (2020) Potential Challenges for Coronavirus (SARS-CoV-2) Vaccines Under Trial. Front. Immunol. 11:561851. doi: 10.3389/fimmu.2020.561851
Received: 13 May 2020; Accepted: 14 September 2020;
Published: 29 September 2020.
Edited by:
Rashika El Ridi, Cairo University, EgyptReviewed by:
Arun Kumar, Coalition for Epidemic Preparedness Innovations (CEPI), NorwayCopyright © 2020 Usmani and Raghava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gajendra P. S. Raghava, cmFnaGF2YUBpaWl0ZC5hYy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.