- 1Department of Allergy, Beijing Shijitan Hospital, Capital Medical University, Beijing, China
- 2Research Division of Clinical Pharmacology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 3Department of Allergy, Duolun People’s Hospital, Duolun, China
- 4School of Basic Medicine and Clinical Pharmacy, China Pharmaceutical University, Nanjing, China
- 5Department of Allergy, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing Key Laboratory of Precision Medicine for Diagnosis and Treatment on Allergic Diseases, Beijing, China
- 6Department of Allergy, General Hospital of Northern Theater Command, Shenyang, China
- 7Department of Otolaryngology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
Background: Allergic rhinitis is a common disorder that affects 10% to 40% of the population worldwide. Allergen immunotherapy (AIT) represents the only therapy that has the potential to resolve clinical symptoms of allergic rhinitis. However, up to 30% of patients do not respond to AIT. Biomarkers predicting the clinical efficacy of AIT as early as possible would significantly improve the patient selection and reduce unnecessary societal costs.
Methods: Artemisia pollen allergic patients who received at least 1-year AIT were enrolled. Clinical responses before and after 1-year AIT were evaluated to determine AIT responders. Artemisia specific IgE and IgG4 levels were measured by using ImmunoCAP and enzyme-linked immunosorbent assay (ELISA) separately. Stepwise regression analysis was performed to identify which rhinitis-relevant parameters explained the most variability in AIT results. Liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomics was applied to identify the potential candidate biomarkers in the sera of responders and non-responders collected before and after 1-year therapy. The diagnostic performance of the potential biomarkers was then assessed using enzyme-linked immunosorbent assay (ELISA) in 30 responders and 15 non-responders.
Results: Artemisia specific IgE and IgG4 levels were elevated only in the responders. Regression analysis of allergic rhinitis-relevant parameters provided a robust model that included two most significant variables (sneeze and nasal congestion). Thirteen candidate biomarkers were identified for predicting AIT outcomes. Based on their association with allergy and protein fold change (more than 1.1 or less than 0.9), four proteins were identified to be potential biomarkers for predicting effective AIT. However, further ELISA revealed that only leukotriene A4 hydrolase (LTA4H) was consistent with the proteomics data. The LTA4H level in responders increased significantly (P < 0.001) after 1-year therapy, while that of non-responders remained unchanged. Assessment of LTA4H generated area under curve (AUC) value of 0.844 (95% confidence interval: 0.727 to 0.962; P < 0.05) in distinguishing responders from the non-responders, suggesting that serum LTA4H might be a potential biomarker for predicting the efficiency of AIT.
Conclusion: Serum LTA4H may be a potential biomarker for early prediction of an effective AIT.
Introduction
Allergic rhinitis is a common disorder that affects 10% to 40% of the population worldwide (1). It is defined as symptoms of sneezing, rhinorrhea, nasal pruritus and airflow obstruction (2). Allergic rhinitis may occur at any age, peaking in the teenage years (3). It also underlies many complications and is a major risk factor for poor asthma control (4–6). Uncontrolled allergic rhinitis has a negative impact on social life, work productivity and school performance, particularly in patients with severe symptoms. However, allergic rhinitis is often ignored, misdiagnosed, and mistreated, which can be detrimental to health and increases societal costs (3, 7).
Pollens are the major cause of seasonal allergic rhinitis. In northern China, over 50% patients with respiratory allergies are sensitized to Artemisia pollen (8). Many classes of drug are available to relieve allergy symptoms, including intranasal corticosteroids and antihistamines. However, even with the best pharmacotherapy, 20% affected individuals remain highly symptomatic (3). Allergen immunotherapy (AIT) represents the only available cause-oriented therapy so far that has the potential to alter the natural history of allergic rhinitis (9–12). AIT is defined as the repeated administration of specific allergens to patients with IgE-mediated allergies in order to provide protection against allergies and inflammatory reactions associated with natural exposure to the same allergens. It is often accompanied by an initial early increase in allergen-specific IgE (sIgE) and allergen-specific (sIgG4) levels (13). The exact mechanism by which AIT conveys a clinical benefit is unknown. Although efficacy of AIT has been demonstrated against highly prevalent allergens such as pollens and house dust mites, up to 30% of patients do not respond to it (14). No predictive biomarkers or diagnostic tests have been developed for its efficacy (15). The unpredictable outcome and a long course (up to 5 years) (16) of AIT imposes a high or even unnecessary cost to the health system especially for people who turn out to be non-responder. Therefore, early prediction of AIT efficacy would greatly reduce the overall cost associated with the treatment.
Clinical analysis of serum is the most widespread diagnostic procedure in medicine, and serum biomarkers are usually used to categorize patients and to support treatment decisions (17). Due to its availability and stability, serum represents valuable resource for proteome analysis and biomarker discovery (18, 19). This biological specimen is considered a key source of physiological information about the overall status of each tissue, and it reflects the status of several pathological conditions. The assessment of the circulating molecules, e.g., proteins, in serum may present a potential approach to help predict the outcome of AIT (12, 20).
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic analysis can measure global protein abundance and post-translational modifications to provide additional biological insights (21) and rapidly identify candidate biomarkers in clinical samples (22). In this study, we used LC-MS/MS to profile the blood samples from patients after AIT to identify potential biomarkers and further validated the results using enzyme-linked immunosorbent assay (ELISA) (Figure 1). The outcomes of this work might help develop serum-based early assay for AIT efficacy using validated biomarkers and may help reduce the overall therapeutic cost and improve the outcomes.
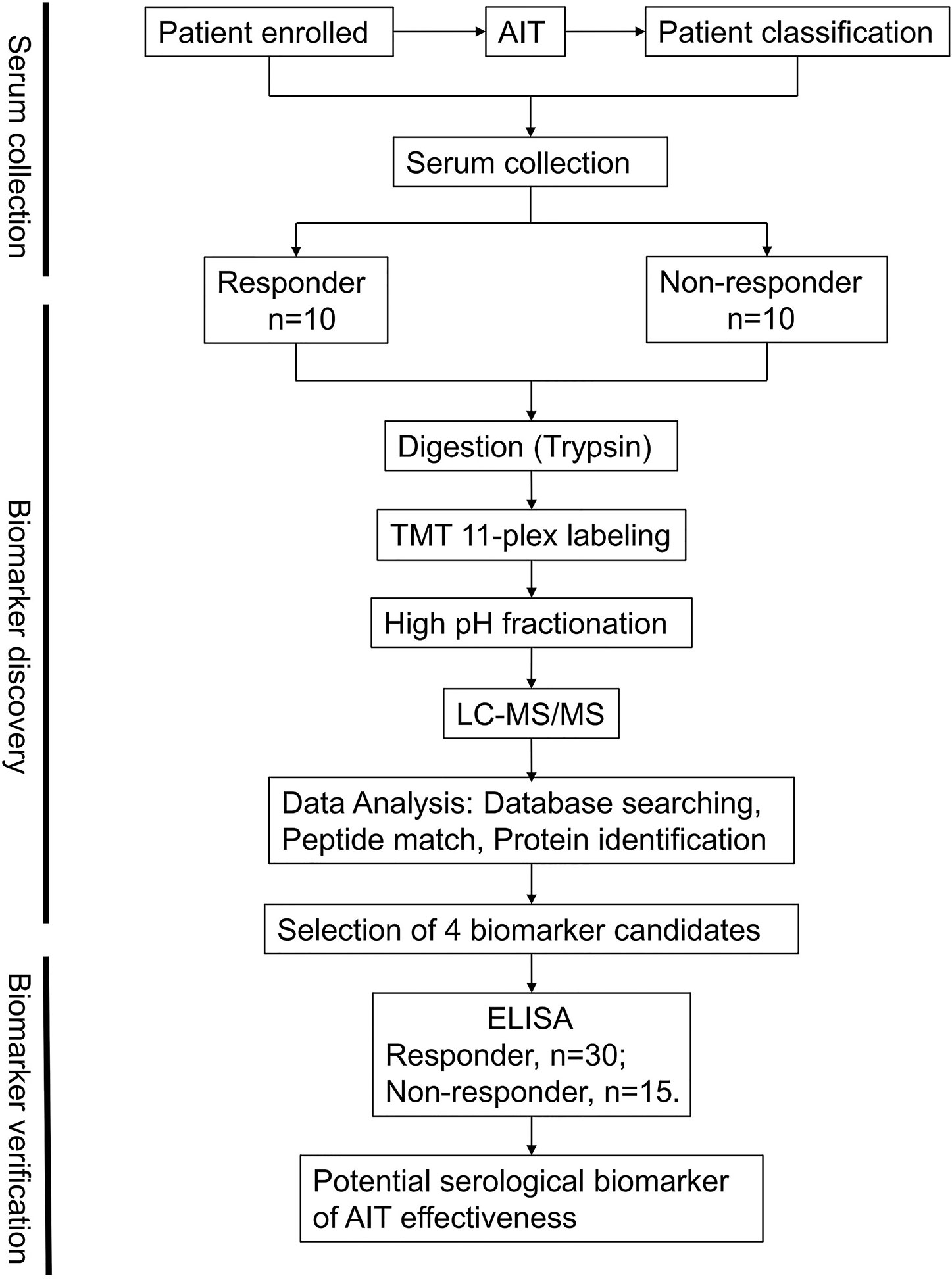
Figure 1 Schematic workflow. An overview of the workflow used for the identification of allergen immunotherapy (AIT) efficacy biomarkers.
Methods
Patients and Samples
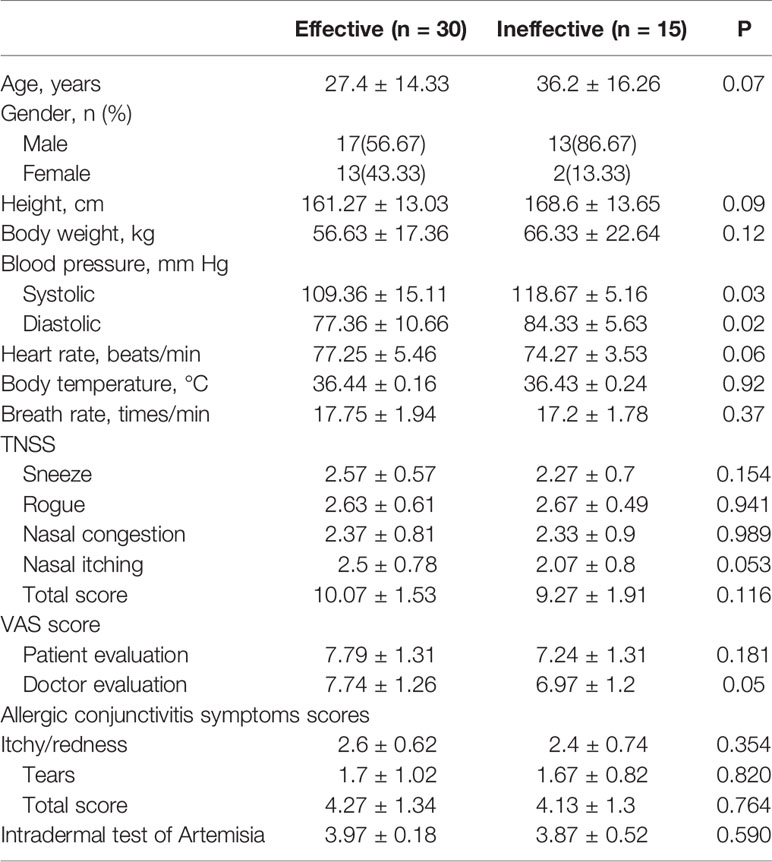
Patients from 6 to 60 years of age with Artemisia pollen allergic rhinitis were recruited at Beijing Shijitan Hospital, affiliated to Capital Medical University on 28-31 July 2016 with a course of disease more than 12 months. Clinical responses were evaluated before and after AIT, including total nasal symptoms scores (TNSS), rhinoconjunctivitis quality of life questionnaires (RQLQ), VAS and allergic conjunctivitis symptoms. According to the treatment results after one year, patients were classified as responders (n = 30) and non-responders (n = 15). The curative effect was evaluated by symptoms and sign scores. The curative effect index (%) = (pretreatment total score − posttreatment total score)/pre-treatment total score × 100%. Patients with ineffective therapeutic index (≤25%, n = 15) were included the ineffective group. And those with effective therapeutic index (≥66%, n = 30) were classified as the effective group. Patients with not significantly effective or ineffective results (>25%, <66%, n=3) were not included in the analysis because of unclear treatment results.
Ten ml fresh blood was collected from the patients before AIT, immediately centrifuged at 1500 rpm at 4°C for 10 min and stored at −80°C for use in subsequent LC-MS/MS and ELISA. Then, serum was collected and processed in the same way before the last injection of 1-year AIT. Serum samples from 20 patients (10 AIT responders, and 10 AIT non-responders) with Artemisia pollen allergic rhinitis were used for comprehensive proteome profiling. For further development of potential biomarkers, ELISA verification was performed on an independent set of 45 pairs of serum samples (30 AIT responders, and 15 AIT non-responders). Characteristics of enrolled patients were summarized in Table 1. The study was conducted under the guidance of the Helsinki Declaration and approved by the Institutional Review Board of Beijing Shijitan Hospital, affiliated to Capital Medical University, Beijing, China. Written informed consent was obtained from every participant in this study.
Inclusion Criteria
The inclusion criteria were as follows: (1) age between 6 and 60, (2) course of disease more than 1 year, (3) typical symptoms appearing in the summer and autumn pollen season, and asymptomatic or mild symptoms in the non-pollen season (non-pollen season VAS <3), (4) intradermal test of Artemisia pollen allergen was ≥+++, and sIgE was ≥II (based on UniCAP allergen-specific IgE detection system), (5) other types of allergen skin test was negative, or “+” and above (including “+”) but specific IgE was <II level. Other pollen allergen specific IgE (including pollens of ragweed, Humulus, Chenopodium, sunflower and oilseed rape) were at least two grades below the Artemisia pollen test. More specifically, if a wheal was 1/3 times as large as the histamine positive control, the result was defined as +; if a wheal was 2/3 times as large as the histamine positive control, the result was defined as ++; if a wheal was as large as the histamine positive control, the result was defined as +++; if a wheal was larger than the histamine positive control, the result was defined as ++++. The 0.7–3.5 kU/L sIgE was regarded as II level.
Exclusion Criteria
The exclusion criteria were as follows: (1) simple rhinoconjunctivitis with TNSS<6, (2) forced expiratory volume in 1 s <75% during non-pollen season, (3) with nasal polyps, significant vasomotor rhinitis, severe asthma, malignancy, severe cardiovascular diseases and impaired renal function, etc., (4) having accepted AIT, (5) be using β-blockers, (6) cannot accept regular follow-up, (7) smokers of 30 packs-per-year or more, (8)be pregnant or plan to become pregnant in the next two years.
Allergen Immunotherapy
AIT was initiated after confirmation of Artemisia sensitivity. At the build-up phase, patients were injected standardized Artemisia sieversiana pollen allergen extract with 1:1012 dilution (purchased from Beijing MacroUnion Pharmaceutical Limited Corporation, Beijing, China, batch number: S20130001, total protein content 1.75 mg/5 ml) for 10 times starting from 0.1 ml at the first time with an increment of 0.1 ml per time. The concentration of allergen extract was increased by 10 times each time, from 1:1012 dilution to 1:100 dilution of the original concentration, then reaching the maintenance dose. In the build-up phase, subjects received each concentration 10 times for 5 weeks from 0.1 to 1 ml with an increment of 0.1 ml per time. During the maintenance phase, the concentration was no longer increased, 0.5 ml was injected twice a week. Patients received AIT during the pollen season.
Serum Immunoglobulin Analysis
Before AIT initiation and after 1-year therapy, Artemisia-sIgE levels were measured by using ImmunoCAP (Thermo Fisher Scientific, Uppsala, Sweden) according to the manufacturer’s protocols. The concentrations of Artemisia-sIgG4 in sera were also measured by a sandwich ELISA. Briefly, 96-well plates (467320, Nunc, Denmark) were coated overnight at 4°C with 100 μl of allergen extracts (XP61D3A2.5, Stallergenes Greer, USA) at a 1:1,000 dilution. Test wells were incubated with 10 μl of the sera sample and 40 μl 1% BSA/PBS (bovine serum albumin dissolved in phosphate-buffered saline at the concentration of 1%), while control wells were incubated with 50 μl 1% BSA/PBS. After adding secondary antibody (Mouse Anti-Human IgG4 pFc, 9190-05, Southern Biotech, USA)at a 1:4,000 dilution and 3,3′,5,5′-tetramethylbenzidine (TMB, PR1210, Solarbio, China), sIgG4 was determined at 450 nm using an ELISA plate reader (SPARK 10 M, Tecan, Switzerland). Antibody levels in the sera were quantified by extrapolation against the standard curve.
Filter-Aided Sample Preparation and Digestion
For protein extraction, the sodium dodecyl sulfate (SDS) lysis buffer was added into the samples. Proteins were separated and digested by SDS polyacrylamide gel electrophoresis. However, SDS removal is critical before mass spectrometry analysis due to its ability to contaminate liquid chromatography systems and dominate mass spectra. Filter-aided sample preparation (FASP) is the most popular method for detergent removal and protein digestion in recent years (23). FASP was performed according to a previously described method with slight modifications (24). Briefly, 100 µg proteins were moved into a EP tube and centrifuged at 12,000×g for 30 min. One microliter M dithiothreitol (DTT) was added and the sample was incubated for 2-3 h at 37°C. The digested sample was loaded to a 30 k Microcon filtration device (Millipore, Darmstadt, Germany) and centrifuged for 40 min at 12,000×g. The concentrate was then mixed with 100 µl 50 mM iodoacetamide (IAA) and incubated for 30 min in darkness at room temperature. IAA was removed by centrifugation at 12,000×g for 40 min. The sample was washed with 100 µl triethyl ammonium bicarbonate (TEAB) for four times and digested with trypsin (Promega) at a trypsin: protein ratio (w/w) of 1:50 in 50 µl TEAB overnight and followed by another 4 h digestion at 37°C. The tryptic peptides were collected by centrifugation at 12,000×g and desalted using StageTipC18 purification system.
Isobaric Tandem Mass Tags Labeling and High pH Fractionation
Peptides were reconstituted in 100 mM TEAB and concentration was determined by the bicinchoninic acid (BCA) assay. Total of 40 samples were divided into 4 groups. Each group contained 10 samples (5 pairs) and 1 internal reference. For the ‘‘internal reference’’ mixed sample used in tandem mass tags (TMT) labeling, all the samples in same group were mixed in equal protein amount, and labeled using TMT 10 isobaric label reagent and TMT11-131C label reagent (Thermo Fisher Scientific). Each sample containing 30 μg peptide in 50 μl TEAB buffer was combined with 41 μl of respective TMT-labeled reagent and this mixture incubated for 1 h at room temperature. 8 μl of 5% hydroxylamine was then added to the sample and incubated for 15 min to quench the reaction. After that, 11 TMT-labeled samples in the same group were pooled and lyophilized. Half of the TMT-labeled peptide mixture was fractionated using a Waters XBridge BEH130 C18 3.5 μm 2.1 × 150 mm column on an Agilent 1260 HPLC operating at 0.2 ml/min. Buffer A consisted of 10 mM ammonium formate and buffer B consisted of 10 mM ammonium formate with 90% acetonitrile; both buffers were adjusted to pH 10 with ammonium hydroxide as described previously (25). A CBS-B programed multifunction automatic fraction collecting instrument (Huxi instrument, Shanghai, China) was coupled to the HPLC to collect eluted peptides.
Nanoflow LC-MS/MS
An online LC-MS/MS setup consisting of an EasynanoLC system and a Q-Exactive mass spectrometer (Thermo, Bremen, Germany) equipped with a nano-electrospray ion source was used for LC-MS/MS experiments. The tryptic digested peptides were loaded on a 75 μm × 200 mm fused silica column packed in-house with 3 μm ReproSil-Pur C18 beads (Dr. Maisch GmbH, Ammerbuch, Germany) and separated with a 120-min gradient at a flow rate of 300 nl/min. Solvent A contained 100% H2O and 0.1% formic acid; Solvent B contained 100% acetonitrile and 0.1% formic acid. The gradient was 3%−5% B, 2 min; 5%−28% B, 93 min; 28%−40% B, 16 min; 40%–90% B, 2 min; 90% B, 7 min. The mass spectrometry instrument was operated with the temperature of the heated capillary set at 320°C and the source voltage set at 2.0 kV, MS1 full scan resolution, 70,000 at m/z 200; automatic gain control target, 3 × 106; maximum injection time, 50 ms. MS2 scan resolution 70,000 at m/z 200; automatic gain control target, 2 × 105; maximum injection time, 120 ms. The precursor ions were fragmented by higher energy collisional dissociation (HCD) with a normalized collision energy of 33%.
Proteomics Analysis
Raw MS data were searched using MaxQuant (version 1.5.1.0) software (26) against a human database downloaded from UniProt (UP000005640; containing 74470 entries). The search criteria were set as the following: Trypsin/P was selected as the digestive enzyme with two potential missed cleavages. Carbamidomethyl (C) was set as a fixed modification, while acetyl (protein N-term) and oxidation (M) were set as variable modifications. The false discovery rates (FDR) of peptides and proteins were both set at 1%.
Enzyme-Linked Immunosorbent Assay
Leukotriene A4 hydrolase (LTA4H) (abx572445, Abbexa Ltd, Cambridge, UK), Mucin 5 subtype B (MUC5B) (CSB-E11201h; CUSABIO, Hubei, China), lipopolysaccharide binding protein (LBP) (DY870-05, R&D Systems Inc) and C4b-binding protein (C4BPB) (CSB-EL003954HU; CUSABIO, Hubei, China) from serum samples were measured using ELISA according to the manufacturer’s protocols. Absorbance was measured on a plate reader at 450 nm. Serum concentration of the protein of interest was quantified by extrapolation against the standard curve.
Statistical Analysis
We used t-test and Wilcoxon signed rank test to determine significant differences of values between groups. The stepwise regression analysis was performed to identify which rhinitis-relevant parameters explained the most variability in AIT results. We applied the receiver-operator characteristic (ROC) curve and area under the curve (AUC) analyses to determine the accuracy of using LTA4H as a potential biomarker in a given specimen. The ROC curve is a fundamental tool for diagnostic test evaluation. AUC is defined as the area enclosed by the coordinate axis under the ROC curve. The closer the AUC value is to 1.0, the higher the authenticity of the detection method is. All P values shown were two sided, and a P value of <0.05 was considered statistically significant.
Results
Serum Immunoglobulin Level
Serum immunoglobulin levels were detected in both groups before and after 1-year therapy. Artemisia-sIgE and Artemisia-sIgG4 were elevated only in the responders (P<0.01) (Figure 2). Furthermore, we applied the ROC curve to determine the accuracy of using Artemisia-sIgE and Artemisia-sIgG4 as potential biomarkers in our specimen. The assessment of Artemisia-sIgE generated an AUC value of 0.620 (95% confidence interval: 0.438 to 0.802; P=0.194) in distinguishing responders from the non-responders (Figure 2B). The assessment of Artemisia-sIgG4 generated an AUC value of 0.712 (95% confidence interval: 0.550 to 0.875; P=0.021) in distinguishing responders from the non-responders (Figure 2D).
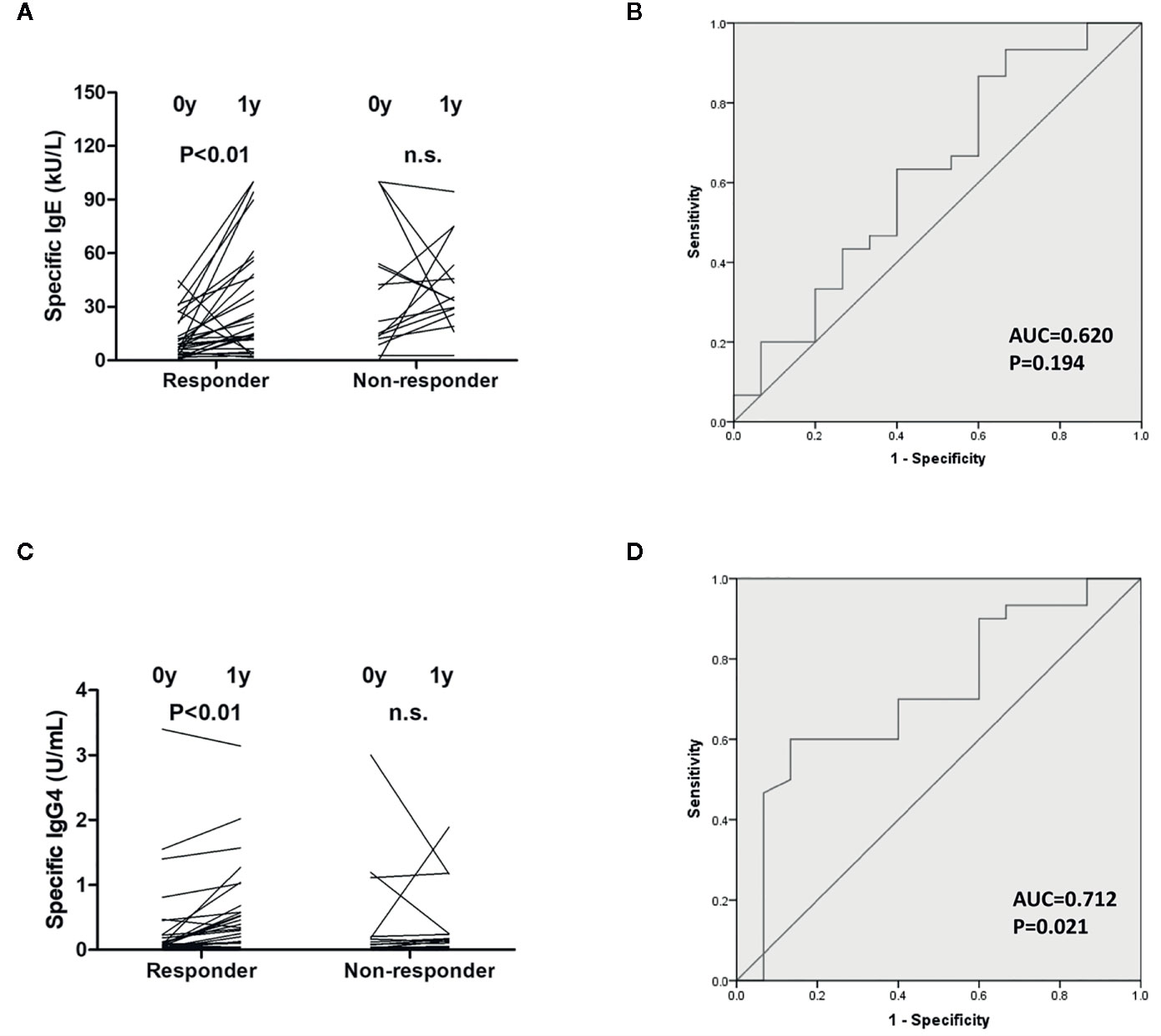
Figure 2 Measurements of serum immunoglobulin level. Artemisia-sIgE (A) and Artemisia-sIgG4 (C) in the sera of responders and non-responders before and after 1-year allergen immunotherapy (AIT) were measured as described in the Methods section. Receiver-operator characteristic (ROC) curve of Artemisia-sIgE (B) and Artemisia-sIgG4 (D) in the sera of responders and non-responders. n.s., not significant.
Stepwise Regression Analysis
A stepwise regression analysis, with AIT results as the dependent variable and changes of those allergic rhinitis-relevant parameters listed in Table 1 as the independent variables, was performed to identify which rhinitis-relevant parameters explained the most variability in AIT results. As shown in Table 2, the final step of the regression analyses retained two of the eight predictor variables: sneeze and nasal congestion. Taken together, these two variables explained approximately 84.1% of the variability in AIT results.
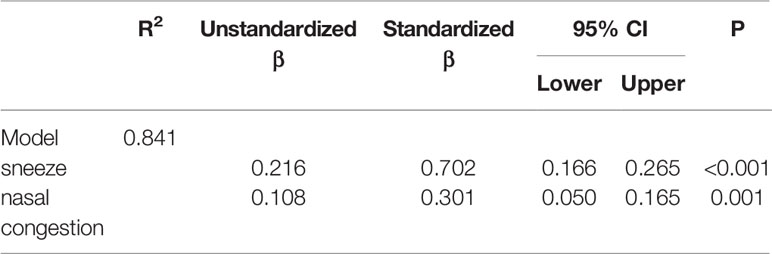
Table 2 Regression statistics for a stepwise regression of allergic rhinitis-relevant predictors on allergen immunotherapy (AIT) results, final model.
Characteristics of Serum Proteins by Quantitative LC-MS/MS
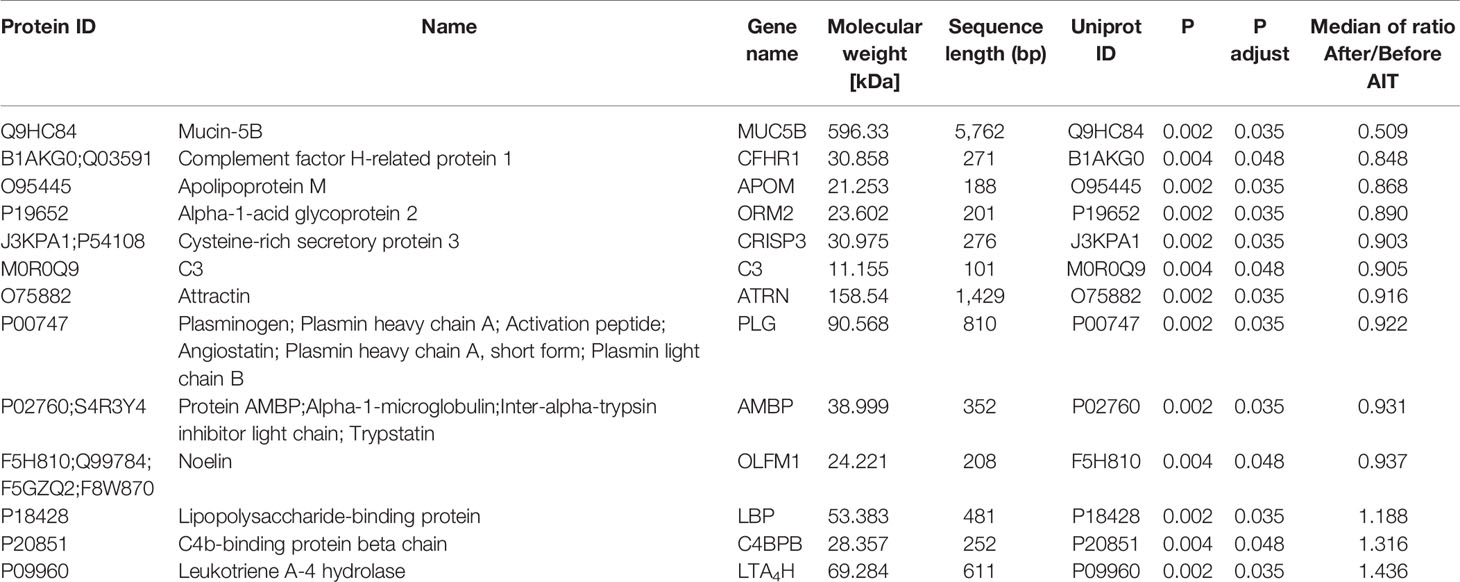
To identify potential biomarkers, proteomics of 10 AIT responders and 10 non-responders before and after 1-year AIT were compared. The unimodal distributions of the ratios of sample to mixture suggested no obvious degradation in serum samples (Figure 3B). Principal-component analysis clearly separated the serum samples before and after 1-year treatment in non-responder cohort and responder cohort based on TMT global data, and no batch effects were observed (Figures 3C, D). Among 573 proteins evaluated, 47 proteins were differentially expressed before and after 1-year AIT in responders and 143 proteins were identified expressed differentially in non-responders (Figures 3E, F and Supplementary Table S1) by differential analysis with P-adjust value < 0.05. The biological processes associated with those proteins were shown in Figures 2G, H. 13 proteins were found to have statistical difference in expression level after 1-year therapy in AIT responder cohort (P values less than 0.01) (Table 3 and Figure 3A), while in non-responders their level remained unchanged. Based on their association with allergy and the protein fold change (more than 1.1 or less than 0.9) between ATI responders and non-responders, four potential genes (MUC5B, LBP, C4BPB, LTA4H) were identified as potential biomarkers that indicate an effective AIT. Of the four proteins, MUC5B displayed a low level of expression, while the other three (LBP, C4BPB, LTA4H) showed high serum levels in the AIT responders after 1-year treatment as compared with the levels before the treatment (Figure 4).
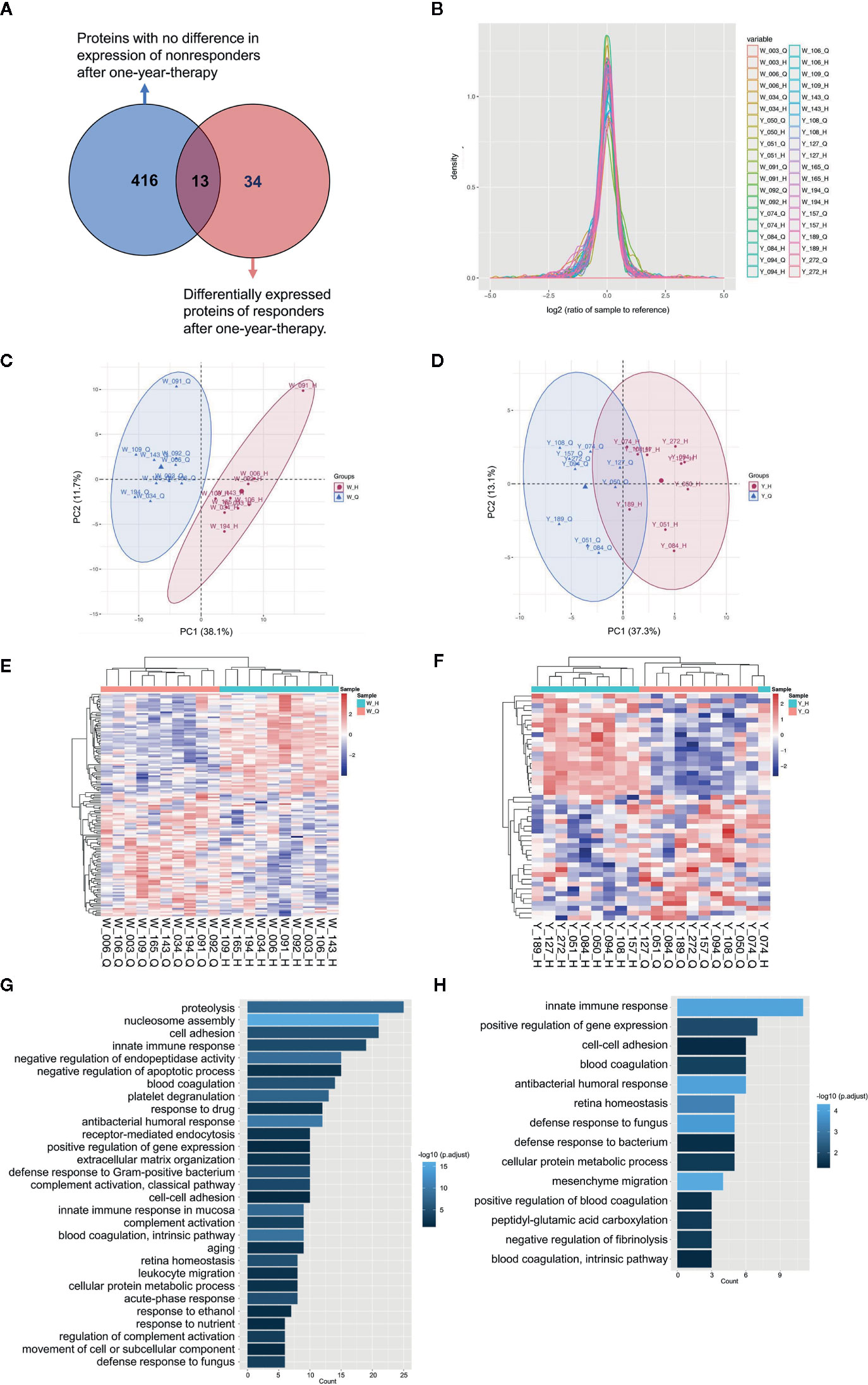
Figure 3 Characterization of serum proteins using quantitative MS. (A) Venn diagram depicts the overlap between proteins with no difference in expression of non-responders and differentially expressed proteins of responders after 1-year therapy. (B) The unimodal distributions of the ratios of samples to the internal reference. Principal-component analysis of serum proteins before and after 1-year allergen immunotherapy (AIT) in non-responder cohort (C) and responder cohort (D) based on tandem mass tags (TMT) global data. The heat map and Gene Ontology biological processes enrichment of the significantly differentially expressed proteins (P-adjusted < 0.05, t test) in AIT non-responders (E, G) and responders (F, H) after 1-year therapy. *W_Q: non-responder, before AIT; W_H: non-responder, after AIT; Y_Q: responder, before AIT; Y_H: responder, after AIT.
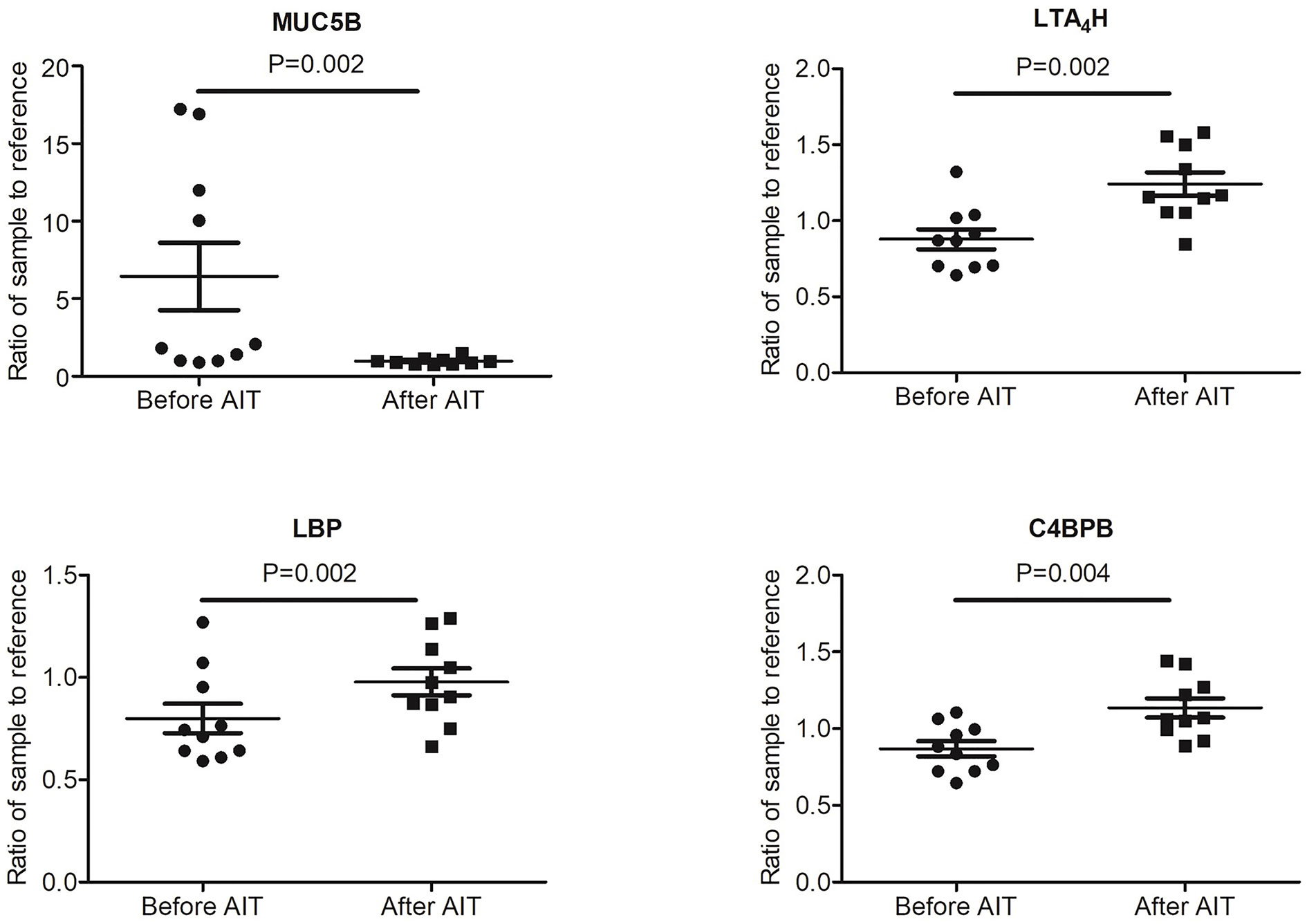
Figure 4 Expression level of potential biomarkers (MUC5B, LBP, C4BPB and LTA4H) of allergen immunotherapy (AIT) efficacy identified by MS-based proteomics in AIT responder cohort.
Validation of LC-MS/MS Data by ELISA
To evaluate the diagnostic value of the candidate four proteins, we measured their expression levels in 30 AIT responders and 15 AIT non-responders (Table 1), which were also included in the quantitative MS. The clinical responses of AIT responders and non-responders were not statistically different before AIT, enabling to control many potential confounders. Among the four candidate proteins, only LTA4H showed were consistent results with the proteomics data (Figure 5). The LTA4H level in the responders increased significantly (mean 1.802 ng/ml, P<0.001) after 1-year therapy, while that of non-responders remained unchanged (Figures 5A, B). Furthermore, we applied the ROC curve to determine the accuracy of using LTA4H as a potential biomarker in our specimen. The assessment of LTA4H generated an AUC value of 0.844 (95% confidence interval: 0.727 to 0.962; P< 0.05) in distinguishing responders from the non-responders (Figure 5C). The AUC value of LTA4H was larger than those of Artemisia-sIgE and Artemisia-sIgG4, indicating that LTA4H might be superior as biomarker to Artemisia-sIgE and Artemisia-sIgG4 in predicting response after 1 year of AIT. These findings imply that the serum LTA4H might be potentially useful in estimating the efficiency of AIT.
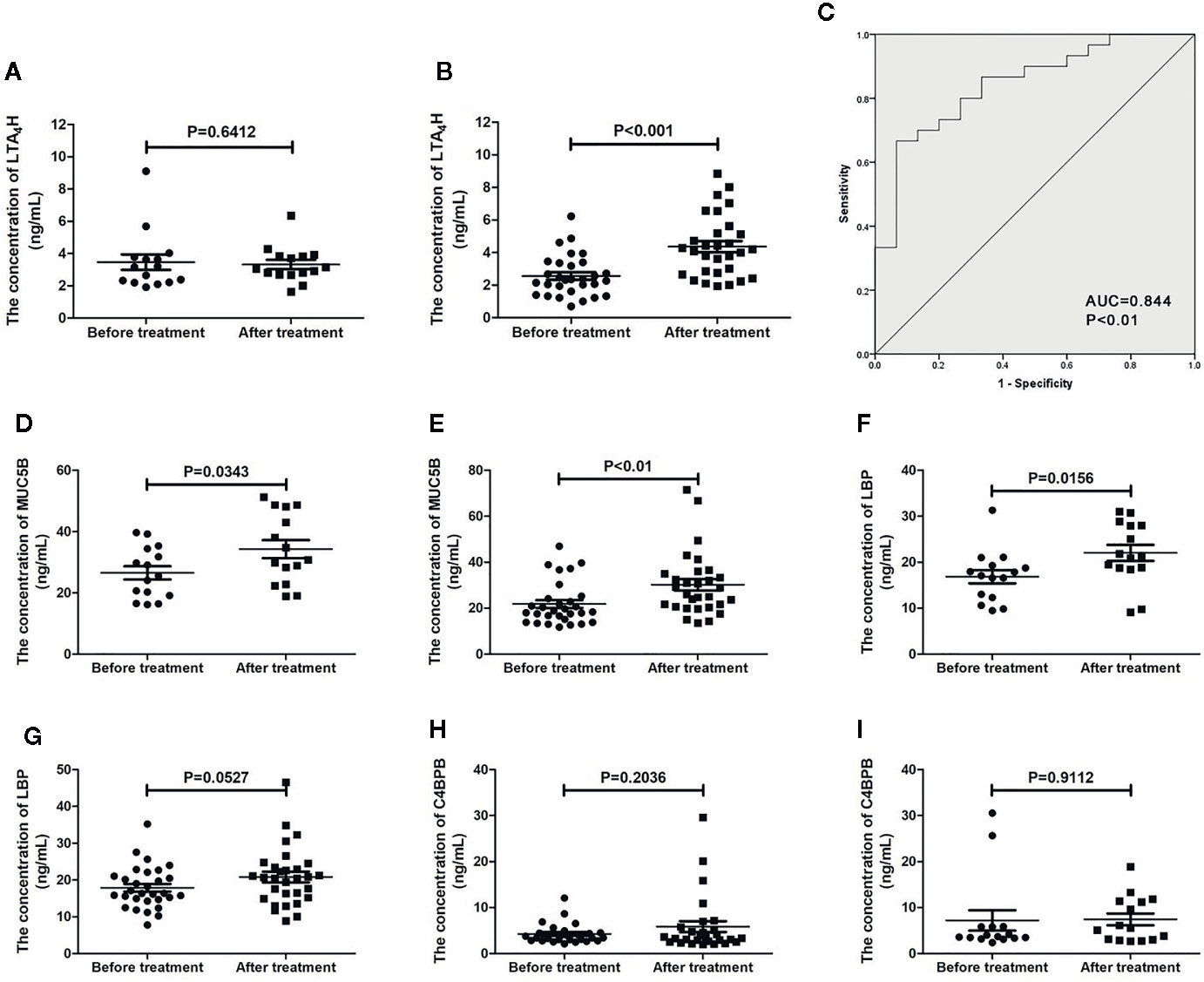
Figure 5 Validation of proteomic prognostic biomarkers using enzyme-linked immunosorbent assay (ELISA). The serum level of LTA4H in allergen immunotherapy (AIT) non-responders (A) and AIT responders (B). (C) Receiver-operator characteristic (ROC) curve of LTA4H measured using ELISA. The serum level of MUC5B in AIT non-responders (D) and responders (E). The serum level of lipopolysaccharide binding protein (LBP) in AIT non-responders (F) and responders (G). The serum level of C4BPB in AIT non-responders (H) and responders (I).
Discussion
AIT is a well-established treatment scheme for IgE-mediated respiratory allergies, inducing a long-lasting reduction of symptoms in association with a down-regulation of allergen-specific TH2 responses (27–29). However, less than one third of patients are offered this therapy and up to 30% of patients do not respond to it (30, 31). Patient selection and compliance might be improved if predictive biological markers were available to identify those individuals more likely to benefit from AIT.
To address this issue, a total of 45 patients with Artemisia pollen allergic rhinitis were included in this study. Clinical responses and immunoglobulin levels were evaluated before and after AIT. A success AIT significantly increased Artemisia-sIgE and the Artemisia-sIgG4, which was in line with previous reports (13, 32). Regression analysis of allergic rhinitis-relevant parameters collected in this study provided a robust model that included two most significant variables (sneeze and nasal congestion) that explain 84.1% of the variability in AIT results. Allergic rhinitis-relevant parameters were based on subjective feelings of patients. Single subjective index is difficult to standardize and is easily affected by many factors. Thus, it should be used combined with objective biomarkers to predict the efficacy of AIT. This is also the reason why we want to look for serum objective biomarkers of AIT efficacy. Nasal secretion is also a good sample type to mirror the pathological status of allergic rhinitis. However, nasal secretion is less stable and easy to be polluted by the external environment, which may affect the results. The clinical collection procedure of nasal secretions is more complicated than that of blood, which may decrease the practical utility of biomarkers. Serum represents valuable resource for proteome analysis and biomarker discovery due to its availability and stability. Thus, we chose blood instead of nasal secretions as study samples.
Our study used a LC-MS/MS-based proteomics approach followed by ELISA validation to identify and confirm proteins associated with AIT results. First, we performed a comparative proteomic analysis on sera obtained from 10 AIT responders and 10 non-responders before and after 1-year treatment to select candidate biomarkers. Then we used sera from 30 AIT responders and 15 AIT non-responders to validate their diagnostic usefulness. Finally, we found that LTA4H is up-regulated only in AIT-responders. The ROC assessment of LTA4H generated an AUC value of 0.844 (95% confidence interval: 0.727 to 0.962; P<0.05) in distinguishing responders from the non-responders. The AUC value of LTA4H was larger than those of Artemisia-sIgE and Artemisia-sIgG4, demonstrating that LTA4H may have considerable accuracy in predicting AIT results. Taken together, these findings suggest the high potential value of LTA4H as a marker of effective AIT after the treatment of the first year. On other hand, the ELISA results of LBP, C4BPB and MUC5B are inconsistent with that of MS-based proteomics. However, they may be worthy further study for their potential as predictive markers. A previous study showed that the existence of isoforms might reduce the reliability of protein detection by antibody probes (33), suggesting there might be room to improve the quantification of these proteins with better ELISA, which may subsequently improve the consistence with MS-based proteomics.
LTA4H is responsible for the synthesis of Leukotrienes (LTs). LTs have been identified as central mediators in asthma and allergy (34). But we did not find significantly changes in their serum level before and after 1-year AIT through MS analysis. We speculated that their serum levels might not be influenced by AIT. Moreover, no studies have shown that they could be reliably used as a predictor of AIT efficacy. Thus, we did not consider these mediators in this study. LTA4H plays important roles in the recruitment of neutrophils, with disparate functions in generating one neutrophil chemoattractant (LTB4), while degrading another (Pro-Gly-Pro, PGP) (35, 36). Neutrophils are defined as important cells in the development of allergic sensitization and inflammation (37). They are one of the first innate immune cells recruited into the lungs during specific asthma-related events such as allergenic challenges (38). Uncontrolled allergic rhinitis and asthma are associated with elevated numbers of neutrophils (39, 40). Activated neutrophils can lead to allergic inflammation in allergic rhinitis by priming T cells and attracting eosinophils (40). Clinical data showed increased levels of neutrophils in nasal biopsies, nasal lavage fluid (NAL) and peripheral blood from patients with allergic rhinitis during the pollen season compared to healthy controls (40–42). The aminopeptidase activity of LTA4H with PGP as substrate is augmented in the presence of albumin (43) and chloride ions (44). Due to the disparate concentrations of chloride ions and albumin intracellularly and extracellularly, the epoxide hydrolase capacity of LTA4H occurs intracellularly and the aminopeptidase activity operates extracellularly. Thus, we thought serum LTA4H mainly operated the aminopeptidase capacity, which degraded PGP, and then restricting recruitment of neutrophils and facilitating the resolution of inflammation. The elevation of serum LTA4H in responders can better inhibit the progress of neutrophil driven inflammation by degrading PGP, which may be the reason for the good efficacy of AIT (45).
AIT candidate biomarkers proposed so far could be grouped into six domains: (i) antibodies, including IgE and IgG4, (ii) serum inhibitory activity for IgE, including IgE-FAB and IgE-BF, (iii) basophil activation, including CD63 and CD203c, (iv) cytokines and chemokines; (v) cellular markers and (vi) in vivo biomarkers (13, 46). Due to the differences in treatment course, desensitization vaccine type and allergic disease type between our studies and previous studies, we did not find the above biomarkers in our results. Previous studies often proposed some targeted candidate markers at first, and then determined the predictive value of these candidate biomarkers by single detective methods, including immunoCAP, ELISA, real-time PCR or flow cytometry (13, 46–49). In contrast, our study used a LC-MS/MS-based proteomics approach to screen candidate biomarkers in whole serum without presetting any target. Then, ELISA was performed to validate the results of proteomics, ensuring the accuracy.
Our study had a number of notable strengths. First, we had a complete set of allergic rhinitis control symptoms scoring system. All patients were evaluated on a uniform standard, enabling controls of many potential confounders. Second, our incorporation of an ELISA validation based on the initial proteomics approach allowed for confirmation of initially identified proteins and verified the robustness of our findings. This repeated analysis study design provided protection against the detection of false positive markers. Third, we found that in patients undergoing AIT, the increase of LTA4H level was associated with a more effective AIT. LTA4H level in serum may identify patients at-risk for AIT failure, enabling proactive and targeted interventions, and serum LTA4H level might provide a means of monitoring course of AIT. Last, these findings have the potential to lead to new studies to design and target intervention strategies that modulate the LTA4H level to achieve an effective AIT.
The limitations of the study were as follows: (i) it was a single-center study with relatively small sample size; (ii) the biomarker was estimated after 1-year treatment and therefore might be not able to predict the outcomes of AIT at the very beginning; (iii) due to the high drop-off rate of AIT, this study was not include placebo control and subjects were not blinded; (iv) this study could not identify late responders and sustained unresponsiveness following discontinuation of AIT; (v) it was also possible that some proteins might have degraded before analysis and would have reduced our ability to detect them in this analysis. We will continue to track the follow-up treatment of all patients.
In conclusion: using proteomics together with ELISA, we have identified serum LTA4H level as a diagnostic candidate biomarker for predicting AIT results. Further efforts are required to elucidate its biological functions in AIT and to confirm its diagnostic values in larger cohorts in clinic setting. Future validation of other candidates defined by proteomics in a large cohort study may help develop additional biomarkers that can be added to LTA4H to have more diagnostic efficacy for the successful AIT.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD022130.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of Beijing Shijitan Hospital, Affiliated to Capital Medical University. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
J-FW, X-YW, and J-SY supervised the project. T-TM, J-LS and H-YS collected the sera. R-LY, W-JY collected important background information. M-DC and CP carried out the study and analyzed the data. J-GL, Q-YW, and D-YW provided assistance for data acquisition, data analysis, and statistical analysis. M-DC and T-TM wrote the manuscript. M-DC, T-TM and J-FW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was partially supported by Beijing Municipal Science & Technology Commission (Grant No. Z161100000516006), Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (Grant No. ZYLX201826), Science and technology research and development project of China National Railway Group Co (Grant No. J2019Z603) and the fund of Beijing Shijitan Hospital (Grant No. 2018 q-16).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.559746/full#supplementary-material
References
1. Brozek JL, Bousquet J, Agache I, Agarwal A, Bachert C, Bosnic-Anticevich S, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol (2017) 140:950–8. doi: 10.1016/j.jaci.2017.03.050
2. Dumitru AF, Shamji M, Wagenmann M, Hindersin S, Scheckenbach K, Greve J, et al. Petasol butenoate complex (Ze 339) relieves allergic rhinitis-induced nasal obstruction more effectively than desloratadine. J Allergy Clin Immunol (2011) 127:1515–21.e6. doi: 10.1016/j.jaci.2011.02.045
3. Greiner AN, Hellings PW, Rotiroti G, Scadding GK. Allergic rhinitis. Lancet (2011) 378:2112–22. doi: 10.1016/S0140-6736(11)60130-X
4. Bousquet J, Van Cauwenberge P, Khaltaev N, Aria Workshop G, World Health O. Allergic rhinitis and its impact on asthma. J Allergy Clin Immunol (2001) 108:S147–334. doi: 10.1067/mai.2001.118891
5. Deliu M, Belgrave D, Simpson A, Murray CS, Kerry G, Custovic A. Impact of rhinitis on asthma severity in school-age children. Allergy (2014) 69:1515–21. doi: 10.1111/all.12467
6. Ponte EV, Franco R, Nascimento HF, Souza-Machado A, Cunha S, Barreto ML, et al. Lack of control of severe asthma is associated with co-existence of moderate-to-severe rhinitis. Allergy (2008) 63:564–9. doi: 10.1111/j.1398-9995.2007.01624.x
7. Colas C, Brosa M, Anton E, Montoro J, Navarro A, Dordal MT, et al. Estimate of the total costs of allergic rhinitis in specialized care based on real-world data: the FERIN Study. Allergy (2017) 72:959–66. doi: 10.1111/all.13099
8. Gao Z, Fu WY, Sun Y, Gao B, Wang HY, Liu M, et al. Artemisia pollen allergy in China: Component-resolved diagnosis reveals allergic asthma patients have significant multiple allergen sensitization. Allergy (2019) 74:284–93. doi: 10.1111/all.13597
9. Larenas-Linnemann D. Allergen immunotherapy: an update on protocols of administration. Curr Opin Allergy Clin Immunol (2015) 15:556–67. doi: 10.1097/aci.0000000000000220
10. Klimek L, Pfaar O, Bousquet J, Senti G, Kundig T. Allergen immunotherapy in allergic rhinitis: current use and future trends. Expert Rev Clin Immunol (2017) 13:897–906. doi: 10.1080/1744666x.2017.1333423
11. Pohlit H, Bellinghausen I. Recent advances in the use of nanoparticles for allergen-specific immunotherapy. Allergy (2017) 72:1461–74. doi: 10.1111/all.13199
12. Shi H-Y, Pan C, Ma T-T, Chen Y-L, Yan W-J, Liu J-G, et al. Clinical Efficacy Evaluation of 1-Year Subcutaneous Immunotherapy for Artemisia sieversiana Pollen Allergic Rhinitis by Serum Metabolomics. Front Pharmacol (2020) 11:305. doi: 10.3389/fphar.2020.00305
13. Shamji MH, Durham SR. Mechanisms of allergen immunotherapy for inhaled allergens and predictive biomarkers. J Allergy Clin Immunol (2017) 140:1485–98. doi: 10.1016/j.jaci.2017.10.010
14. Fernandez TD, Gomez E, Dona I, Campo P, Rondon C, Gonzalez M, et al. Differential Plasma-cell evolution is linked with Dermatophagoides pteronyssinus immunotherapy response. Sci Rep (2015) 5:14482. doi: 10.1038/srep14482
15. Gotoh M, Kaminuma O, Nakaya A, Katayama K, Motoi Y, Watanabe N, et al. Identification of biomarker sets for predicting the efficacy of sublingual immunotherapy against pollen-induced allergic rhinitis. Int Immunol (2017) 29:291–300. doi: 10.1093/intimm/dxx034
17. Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol (2017) 13:942. doi: 10.15252/msb.20156297
18. Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Sys (2016) 2:185–95. doi: 10.1016/j.cels.2016.02.015
19. Pieragostino D, Petrucci F, Del Boccio P, Mantini D, Lugaresi A, Tiberio S, et al. Pre-analytical factors in clinical proteomics investigations: impact of ex vivo protein modifications for multiple sclerosis biomarker discovery. J Proteomics (2010) 73:579–92. doi: 10.1016/j.jprot.2009.07.014
20. Greco V, Piras C, Pieroni L, Urbani A. Direct Assessment of Plasma/Serum Sample Quality for Proteomics Biomarker Investigation. Methods Mol Biol (2017) 1619:3–21. doi: 10.1007/978-1-4939-7057-5_1
21. Gao Q, Zhu H, Dong L, Shi W, Chen R, Song Z, et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell (2019) 179:561–77.e22. doi: 10.1016/j.cell.2019.08.052
22. Yu L, Shen J, Mannoor K, Guarnera M, Jiang F. Identification of ENO1 as a potential sputum biomarker for early-stage lung cancer by shotgun proteomics. Clin Lung Cancer (2014) 15:372–8.e1. doi: 10.1016/j.cllc.2014.05.003
23. Ludwig KR, Schroll MM, Hummon AB. Comparison of In-Solution, FASP, and S-Trap Based Digestion Methods for Bottom-Up Proteomic Studies. J Proteome Res (2018) 17:2480–90. doi: 10.1021/acs.jproteome.8b00235
24. Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods (2009) 6:359–62. doi: 10.1038/nmeth.1322
25. Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics (2011) 11:2019–26. doi: 10.1002/pmic.201000722
26. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol (2008) 26:1367–72. doi: 10.1038/nbt.1511
27. Akdis CA, Akdis M. Mechanisms of allergen-specific immunotherapy. J Allergy Clin Immunol (2011) 127:18–27; quiz 8-9. doi: 10.1016/j.jaci.2010.11.030
28. Globinska A, Boonpiyathad T, Satitsuksanoa P, Kleuskens M, van de Veen W, Sokolowska M, et al. Mechanisms of allergen-specific immunotherapy: Diverse mechanisms of immune tolerance to allergens. Ann Allergy Asthma Immunol (2018) 121:306–12. doi: 10.1016/j.anai.2018.06.026
29. Shamji MH, Durham SR. Mechanisms of immunotherapy to aeroallergens. Clin Exp Allergy (2011) 41:1235–46. doi: 10.1111/j.1365-2222.2011.03804.x
30. Caillot N, Bouley J, Jain K, Mariano S, Luce S, Horiot S, et al. Sialylated Fetuin-A as a candidate predictive biomarker for successful grass pollen allergen immunotherapy. J Allergy Clin Immunol (2017) 140:759–70.e13. doi: 10.1016/j.jaci.2016.10.036
31. Weberschock T, Schaefer I, Heigel H, Valesky E, Augustin M, Schmitt J. Use of specific immunotherapy–a survey of 15 164 employed persons in Germany. J der Deutschen Dermatol Gesellschaft = J German Soc Dermatol JDDG (2014) 12:341–5. doi: 10.1111/ddg.12298
32. Schmid JM, Würtzen PA, Dahl R, Hoffmann HJ. Pretreatment IgE sensitization patterns determine the molecular profile of the IgG4 response during updosing of subcutaneous immunotherapy with timothy grass pollen extract. J Allergy Clin Immunol (2016) 137:562–70. doi: 10.1016/j.jaci.2015.05.023
33. Weiland F, Fritz K, Oehler MK, Hoffmann P. Methods for identification of CA125 from ovarian cancer ascites by high resolution mass spectrometry. Int J Mol Sci (2012) 13:9942–58. doi: 10.3390/ijms13089942
34. Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy (2008) 63:1046–53. doi: 10.1111/j.1398-9995.2008.01667.x
35. Snelgrove RJ, Jackson PL, Hardison MT, Noerager BD, Kinloch A, Gaggar A, et al. A critical role for LTA4H in limiting chronic pulmonary neutrophilic inflammation. Science (2010) 330:90–4. doi: 10.1126/science.1190594
36. Wells JM, O’Reilly PJ, Szul T, Sullivan DI, Handley G, Garrett C, et al. An aberrant leukotriene A4 hydrolase-proline-glycine-proline pathway in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med (2014) 190:51–61. doi: 10.1164/rccm.201401-0145OC
37. Hosoki K, Aguilera-Aguirre L, Brasier AR, Kurosky A, Boldogh I, Sur S. Facilitation of Allergic Sensitization and Allergic Airway Inflammation by Pollen-Induced Innate Neutrophil Recruitment. Am J Respir Cell Mol Biol (2016) 54:81–90. doi: 10.1165/rcmb.2015-0044OC
38. Radermecker C, Louis R, Bureau F, Marichal T. Role of neutrophils in allergic asthma. Curr Opin Immunol (2018) 54:28–34. doi: 10.1016/j.coi.2018.05.006
39. Moore WC, Hastie AT, Li X, Li H, Busse WW, Jarjour NN, et al. Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol (2014) 133:1557–63.e5. doi: 10.1016/j.jaci.2013.10.011
40. Arebro J, Ekstedt S, Hjalmarsson E, Winqvist O, Kumlien Georen S, Cardell LO. A possible role for neutrophils in allergic rhinitis revealed after cellular subclassification. Sci Rep (2017) 7:43568. doi: 10.1038/srep43568
41. Varney VA, Jacobson MR, Sudderick RM, Robinson DS, Irani AM, Schwartz LB, et al. Immunohistology of the nasal mucosa following allergen-induced rhinitis. Identification of activated T lymphocytes, eosinophils, and neutrophils. Am Rev Respir Dis (1992) 146:170–6. doi: 10.1164/ajrccm/146.1.170
42. Fransson M, Benson M, Wennergren G, Cardell LO. A role for neutrophils in intermittent allergic rhinitis. Acta Otolaryngol (2004) 124:616–20. doi: 10.1080/00016480310015173
43. Orning L, Fitzpatrick FA. Albumins activate peptide hydrolysis by the bifunctional enzyme LTA4 hydrolase/aminopeptidase. Biochemistry (1992) 31:4218–23. doi: 10.1021/bi00132a010
44. Wetterholm A, Haeggström JZ. Leukotriene A4 hydrolase: an anion activated peptidase. Biochim Biophys Acta (1992) 1123:275–81. doi: 10.1016/0005-2760(92)90007-i
45. Hosoki K, Itazawa T, Boldogh I, Sur S. Neutrophil recruitment by allergens contribute to allergic sensitization and allergic inflammation. Curr Opin Allergy Clin Immunol (2016) 16:45–50. doi: 10.1097/ACI.0000000000000231
46. Shamji MH, Kappen JH, Akdis M, Jensen-Jarolim E, Knol EF, Kleine-Tebbe J, et al. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy (2017) 72:1156–73. doi: 10.1111/all.13138
47. Di Lorenzo G, Mansueto P, Pacor ML, Rizzo M, Castello F, Martinelli N, et al. Evaluation of serum s-IgE/total IgE ratio in predicting clinical response to allergen-specific immunotherapy. J Allergy Clin Immunol (2009) 123:1103–10, 10.e1-4. doi: 10.1016/j.jaci.2009.02.012
48. Ebo DG, Bridts CH, Mertens CH, Hagendorens MM, Stevens WJ, De Clerck LS. Analyzing histamine release by flow cytometry (HistaFlow): a novel instrument to study the degranulation patterns of basophils. J Immunol Methods (2012) 375:30–8. doi: 10.1016/j.jim.2011.09.003
Keywords: allergic rhinitis, allergen immunotherapy, serum, biomarkers, proteomics, LTA4H
Citation: Ma T-T, Cao M-D, Yu R-L, Shi H-Y, Yan W-J, Liu J-G, Pan C, Sun J, Wei Q-Y, Wang D-Y, Wei J-F, Wang X-Y and Yin J-S (2020) Leukotriene A4 Hydrolase Is a Candidate Predictive Biomarker for Successful Allergen Immunotherapy. Front. Immunol. 11:559746. doi: 10.3389/fimmu.2020.559746
Received: 07 May 2020; Accepted: 22 October 2020;
Published: 24 November 2020.
Edited by:
Peisong Gao, Johns Hopkins University, United StatesReviewed by:
Ping-Chang Yang, Shenzhen University, ChinaEric Oliver, Johns Hopkins University, United States
Copyright © 2020 Ma, Cao, Yu, Shi, Yan, Liu, Pan, Sun, Wei, Wang, Wei, Wang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue-Yan Wang, d2FuZ3h1ZXlhbjIwMThAMTYzLmNvbQ==; Ji-Fu Wei, d2VpamlmdUBob3RtYWlsLmNvbQ==; Jin-Shu Yin, eWluamluc2h1NTVAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
 Ting-Ting Ma
Ting-Ting Ma Meng-Da Cao2†
Meng-Da Cao2† Jinlyu Sun
Jinlyu Sun De-Yun Wang
De-Yun Wang Ji-Fu Wei
Ji-Fu Wei