
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 25 September 2020
Sec. Inflammation
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.550145
This article is part of the Research TopicMitochondria at the Crossroads of Immunity and Inflammatory Tissue DamageView all 17 articles
 Samantha E. C. Wolff1,2,3
Samantha E. C. Wolff1,2,3 Xiao-Lan Wang1,2,4
Xiao-Lan Wang1,2,4 Han Jiao3
Han Jiao3 Jia Sun3
Jia Sun3 Andries Kalsbeek1,2,5
Andries Kalsbeek1,2,5 Chun-Xia Yi1,2,5*
Chun-Xia Yi1,2,5* Yuanqing Gao3*
Yuanqing Gao3*Microglia are the immune cells of the brain. Hyperactivation of microglia contributes to the pathology of metabolic and neuroinflammatory diseases. Evidence has emerged that links the circadian clock, cellular metabolism, and immune activity in microglia. Rev-erb nuclear receptors are known for their regulatory role in both the molecular clock and cell metabolism, and have recently been found to play an important role in neuroinflammation. The Rev-erbα agonist SR9011 disrupts circadian rhythm by altering intracellular clock machinery. However, the exact role of Rev-erbα in microglial immunometabolism remains to be elucidated. In the current study, we explored whether SR9011 also had such a detrimental impact on microglial immunometabolic functions. Primary microglia were isolated from 1–3 days old Sprague-Dawley rat pups. The expression of clock genes, cytokines and metabolic genes was evaluated using RT-PCR and rhythmic expression was analyzed. Phagocytic activity was determined by the uptake capacity of fluorescent microspheres. Mitochondria function was evaluated by measuring oxygen consumption rate and extracellular acidification rate. We found that key cytokines and metabolic genes are rhythmically expressed in microglia. SR9011 disturbed rhythmic expression of clock genes in microglia. Pro-inflammatory cytokine expression was attenuated by SR9011 during an immune challenge by TNFα, while expression of the anti-inflammatory cytokine Il10 was stimulated. Moreover, SR9011 decreased phagocytic activity, mitochondrial respiration, ATP production, and metabolic gene expression. Our study highlights the link between the intrinsic clock and immunometabolism of microglia. We show that Rev-erbα is implicated in both metabolic homeostasis and the inflammatory responses in microglia, which has important implications for the treatment of metabolic and neuroinflammatory diseases.
The circadian brain clock orchestrates physiological and metabolic processes to prepare the body for environmental changes and to optimize energy metabolism (1). Cells in peripheral tissues, including metabolic tissues such as liver, pancreas, and kidney, also display rhythms in activity that are regulated by an intrinsic clock machinery. Since metabolic events are tightly regulated by the circadian clock, disruptions of the circadian timing system can result in metabolic dysfunctions and contribute to obesity and type 2 diabetes (2–10). Thus, the intracellular clock system is tightly involved in the control of metabolic rhythms that are essential for metabolic homeostasis.
The nuclear receptor reverse viral erythroblastosis oncogene product alpha (Rev-erbα) plays a crucial role in both the molecular clock of the circadian timing system and the regulation of metabolism. Rev-erbα stabilizes circadian rhythms by inhibiting the expression of the core clock genes brain and muscle ARNT-Like 1 (Bmal1) and circadian locomotor output cycles kaput (Clock) (11–13). The known endogenous ligand of Rev-erbα is heme, which plays an important role in mitochondrial respiration, and is required for the repressive activity of Rev-erbα/β on target genes (14–17). Additionally, nuclear receptors are sensors for dietary lipids and lipid-soluble hormones, thus rhythmic expression of Rev-erbα/β is also involved in regulating metabolic processes in a circadian manner (18, 19). Intriguingly, a recent Rev-erbα knock-out study proposed an important role for Rev-erbα in regulating neuroinflammation (20). In previous research, Rev-erbα agonists (GSK4112 and SR9011), and antagonists (SR8278) have provided us with insights about the function of Rev-erbα (21–24). For example, systemic administration of the Rev-erbα agonist SR9011 altered clock genes expression and disrupted the circadian behavior of mice leading to loss of locomotor activity during the active phase (dark phase), while vehicle administration caused no disruption (23). These results show that Rev-erb nuclear receptors have profound effects on circadian rhythm, metabolism and neuroinflammation, and possibly are eligible targets for treating metabolic diseases.
Recent research has demonstrated that a high fat, high sugar diet is associated with chronic activation of microglia, also known as microgliosis, which contributes to disturbed energy homeostasis and diet-induced obesity (DIO) (25–29). Additionally, our group recently reported that microglia activity shows a clear day/night rhythm when animals are fed a regular diet, but this rhythm is disrupted in DIO (30, 31). These findings suggest that the daily microglial rhythm is important for its normal activity and disturbance may result in metabolic diseases. However, little is known about the mechanism behind the intrinsic clock and the function of microglia. There is also a complex interplay between metabolic processes and immune responses, known as immunometabolism, in which metabolic reprogramming underlies the inflammatory state of microglia (32, 33). Therefore, considering the important role of Rev-erbα in the molecular clock machinery, neuroinflammation, and metabolism, in the current study we used the Rev-erbα agonist SR9011 to investigate the role of Rev-erbα in microglial immunometabolism.
Primary microglia cultures were prepared from brains of newborn, male Sprague Dawley rat pups (1–3 days old; Nanjing Medical University, China). After removal of the meninges, the brains were dissected and homogenized. Tissue lysates were centrifuged and suspended in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, United States; C11885500BT) containing 1 g/L D-Glucose, L-Glutamine, 110 mg/L sodium pyruvate, and supplemented with 10% Fetal Bovine Serum (FBS; Gibco, United States; 10099141) and 1% Penicillin/Streptomycin (Gibco, United States; 15140122). Mixed glia cells were plated at a density of one brain per T75 flask or three brains per T175 flask, and incubated at 37°C in a humid atmosphere with 5% CO2. Culture medium was refreshed every 3 days. After the astrocyte layer reached confluency (8–14 days), the microglial cells were collected by shaking the flasks 150 rpm/min for 1 h at 37°C. Cells were seeded in 96-well plates (30k–50k cells/well) for the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay, 12-well plates (250k–300k cells/well) for qPCR experiments, or coverslipped in a 24-well plate (100k–150k cells/well) for phagocytosis experiments, or the Seahorse XF96 Cell Culture Microplate (Agilent, United States; 50k cells/well) for Seahorse experiments. For palmitic acid study, the palmitic – BSA solution was prepared 5:1 molar ratio palmitate: BSA in Krebs ringer buffer. All plates were coated with poly-L-lysine hydrobromide (MDBio, China). Dexamethasone (Sigma-Aldrich, United States; D4902), SR9011 (Sigma-Aldrich, United States; SML2067), DMSO (MDBio, China; D015), Fatty acids free BSA (Sigma A7030), Sodium Palmitate (Sigma-Aldrich, United States; P9767), TNFα (Peprotech, United States; 315-01A), and 0.1% BSA (Beyotime, China; ST023) were used to treat the cells during the experiments.
Cell viability was determined with the CellTiter 96 Aqueous assay (Promega, United States; G3582). Cells were treated with dexamethasone for 2 h, followed by a 24-h treatment with 5 μM SR9011 (or DMSO for control). Afterward, MTS solution was added followed by incubation for 3 h at 37°C, and the absorbance at 490 nm was measured on a SpectraMax M2 microplate reader (Molecular Devices).
For gene expression analysis, total RNA was extracted from primary microglia using the RNAeasyTM kit (Beyotime, China; R0026) according to the manufacturer’s protocol. RNA concentration was measured with the NanoDrop Onec Spectrophotometer (Thermo Scientific, United States) and reverse transcribed into cDNA using the HiScript II qRT Supermix (Vazyme Biotech; R222-01). Gene expression levels of Bmal1, Clock, Nr1d1 (Rev-erbα), Per2, Cry1, Il1β, Il4 Il6, Il10, Tnfα, Ccl2, Gm-csf, Tgfβ, CD36, CD68, Cpt1, Pdk1, Hk2, Fasn, Glut5, and Hprt (housekeeping gene; see Supplementary Table 1 for primer sequences) were measured using real-time quantitative PCR on a QuantStudio 5 (Applied Biosystems Thermo Scientific), using the AceQ qPCR SYBR Green Master Mix (Vazyme Biotech; Q131-02). Gene expression levels were normalized to the housekeeping gene. Primers were designed using the Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI) and purchased from GeneRay Biotech.
After treatment with SR9011 or DMSO for 12 h, microglia were incubated with 0.05% fluorescent latex microspheres (Sigma, United States; L1030-1 ml) in DMEM containing 0.25% FBS for 1 h. Subsequently, the cells were fixed with 4% paraformaldehyde for 30 min and a fluorescence staining of microglia was performed with Rabbit Anti-Iba1 (Wako, Japan; 019-19741) and the nuclei were counterstained with 4’,6-diamidino-2-phenylindole (DAPI; Bioprox, France). Images were captured with an Olympus BX53F microscope equipped with an Olympus U-HGLGPS light source, and analyzed with ImageJ software (version 1.48). The relative fluorescent intensity of the beads was determined by dividing the total fluorescent intensity of the beads by the number of DAPI-stained nuclei.
Microglia were treated with 5 μM SR9011 or DMSO for 4 h prior to placement in the Seahorse XFe96 Analyzer (Agilent, United States). During the run, oxygen consumption rate (OCR), and extracellular acidification rate (ECAR) were measured and the wells were injected with modulators from the Agilent Seahorse XF Mito Stress Kit to determine parameters of mitochondrial function. After measuring basal levels of OCR and ECAR, oligomycin was injected to block ATP synthase, which decreases mitochondrial respiration. The second injection was carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), which disrupts the mitochondrial membrane potential and maximizes oxygen consumption. The third injection was a mixture of rotenone and antimycin A, which block complex I and III (respectively) and shut down mitochondrial respiration. Lastly, 2-deoxy-D-glucose (2-DG) was added to inhibit glycolysis, leading to a decrease in ECAR. Cellular respiration, mitochondrial respiration, cellular acidification, maximum substrate utilization, and maximum glycolytic capacity were calculated as previously described (34). ATP production was calculated from mitochondrial respiration by using a phosphate/oxygen ratio of 2.3 (35). After the measurements, cells were fixed with 4% paraformaldehyde and attached cells were quantified using a crystal violet assay by measuring absorbance at 590 nm with a SpectraMax M2 microplate reader (Molecular Devices). Data obtained from the Seahorse XF96 Analyzer was normalized to cell quantity per well.
All results are expressed as mean ± SEM. Statistical analysis was performed using Graph-Pad PRISM (version 7.00), ImageJ software (version 1.48), and JTK_Cycle software (36). Two-way ANOVA and Bonferroni’s post hoc test was used to assess the SR9011 effects on clock genes within 24 h after synchronization. Unpaired t-tests were used to evaluate the differences between DMSO and SR9011 groups in the rest of the study. ImageJ software was used for the analysis of phagocytosis assays. JTK_Cycle software (36) was used to identify rhythmic components in circadian PCR data.
The intrinsic clock machinery is present in microglia with clock genes being rhythmically expressed (30, 37). It has been well documented that cytokines and metabolic genes exhibit rhythmic expression (23, 38). To confirm whether cytokines and metabolic genes were also temporally regulated in primary microglia, we investigated the expression of three key pro-inflammatory cytokines – interleukin-1 beta (Il1β), interleukin-6 (Il6), and tumor necrosis factor-alpha (Tnfα), as well as three important metabolic genes – carnitine palmitoyltransferase 1 (Cpt1), hexokinase 2 (Hk2), and pyruvate dehydrogenase kinase 1 (Pdk1). Primary microglia were synchronized by 10 nM dexamethasone for 2 h as reported before (39) and cultured for 0, 6, 12, 18, or 24 h before being harvested. Gene expression of the cytokines and metabolic genes was determined over these time points [time post-synchronization, from now on referred to as “Time (T)” in hours] and rhythmicity was analyzed by JTK_Cycle software (Figure 1). Tnfα, Il6, Pdk1, and Cpt1 showed a clear rhythmic expression, while Il1β and Hk2 were not rhythmically expressed (Figure 1). The acrophase of the curves was estimated at T6, for Il6, Pdk1 and Cpt1, and T12 for Tnfα. These data show that Il6, Tnfα, Pdk1, and Cpt1 expression are likely orchestrated by the circadian timing system in primary microglia.
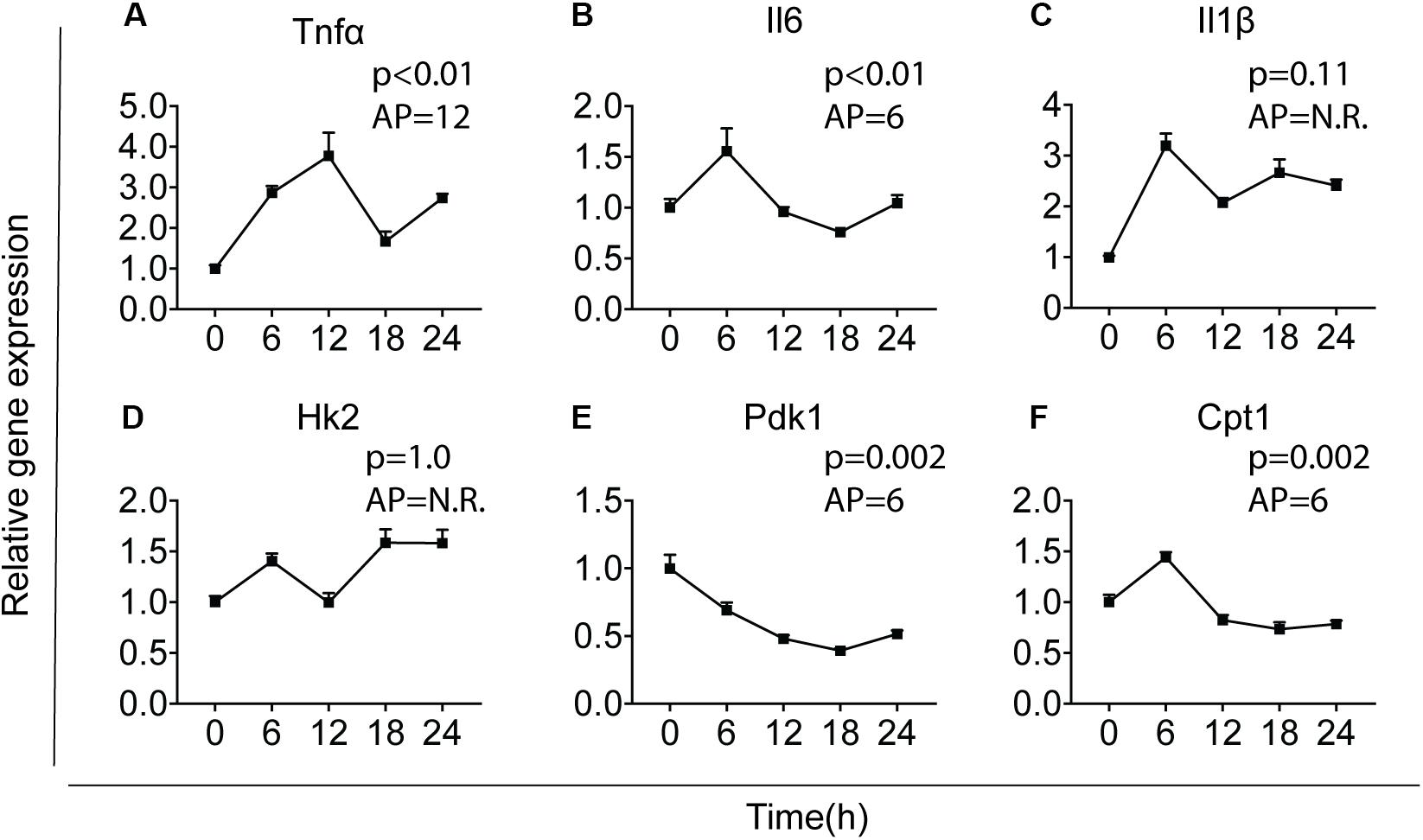
Figure 1. Major cytokines and metabolic genes are rhythmically expressed in microglia. Relative gene expression of major cytokines and metabolic genes was measured in primary microglia. After synchronization by dexamethasone for 2 h, cells were cultured and harvested at the indicated post-synchronization time points (T; n = 6 per group, per time point). Rhythmic expression is signified by a p-value of less than 0.05 and can be found for Tnf (A), Il6 (B), Pdk1 (E), and Cpt1 (F). The expression of Il1 (C) and Hk2 (D) was not rhythmic. The acrophase of the curve is not relevant (N.R.) if data does not fit a curve. Data are presented as means ± SEM and statistical significance was determined using JTK_Cycle software.
We first determined whether SR9011 enhances Rev-erbα activity and disrupts the clock machinery in primary microglia. For this, we performed a dose-response study with 1, 5, 10, and 50 μM SR9011 (data not shown) and decided to use 5 μM for this study. To test the efficiency of SR9011, we first analyzed the effects of SR9011 on Rev-erb targets Serpine1, Cyp7a1, and Srebf1 and found significant reductions in their expression compared to control, which confirms that SR9011 acts through Rev-erb (Supplementary Figure 1). When using SR9011 as a pretreatment before synchronization with dexamethasone, we found that the effect of SR9011 pretreatment did not persist after removing SR9011 from the culture (Supplementary Figure 2 and Supplementary Table 2), contrary to the result from Nakazato et al. (39). Therefore, the following design was used in this study: primary rat microglia were exposed to dexamethasone for 2 h to synchronize the cells, followed by treatment with. 5 μm SR9011 or DMSO for 0, 6, 12, 18, or 24 h. Rhythmic expression of Bmal1, Clock, period 2 (Per2), cryptochrome 1 (Cry1), period 1 (Per1) and nuclear receptor subfamily 1 group d member 1 (Nr1d1 or Rev-erbα) was found in primary microglia treated with DMSO, as evaluated with Two-way ANOVA (Table 1) and JTK_Cycle Software (Figure 2 and Table 2). SR9011 exerted an inhibitory effect on Bmal1 and Per2 expression, as analyzed by Two way ANOVA (Table 1). An interactive effect of SR9011 and Time was found on Bmal1, Clock, and Nr1d1 (=Rev-erbα, see Table 1). The impact of SR9011 at each time point was analyzed by multiple comparison (Figures 2A–F). Notably, SR9011 disrupted the rhythmic expression of Bmal1 and Clock, and caused a shift in the acrophase of Per2 rhythm, as analyzed by JTK_Cycle (Table 2). SR9011 in combination with dexamethasone had no impact on cell viability (Figure 2G), indicating that a decrease in gene expression could not be attributed to cell death. These results show that SR9011 disrupted the rhythm of clock gene expression, pertaining to both amplitude and phase depending on the clock gene, which means that rhythmic expression of clock gene expression became non-rhythmic after adding SR9011. SR9011 had the strongest impact on Bmal1 and Clock, while the effect on Per2 and Cry1 was less potent. This is probably due to the direct inhibitory effect of Rev-erbα on the BMAL1 and CLOCK complex, which indirectly influences Per2 and Cry1 expression.
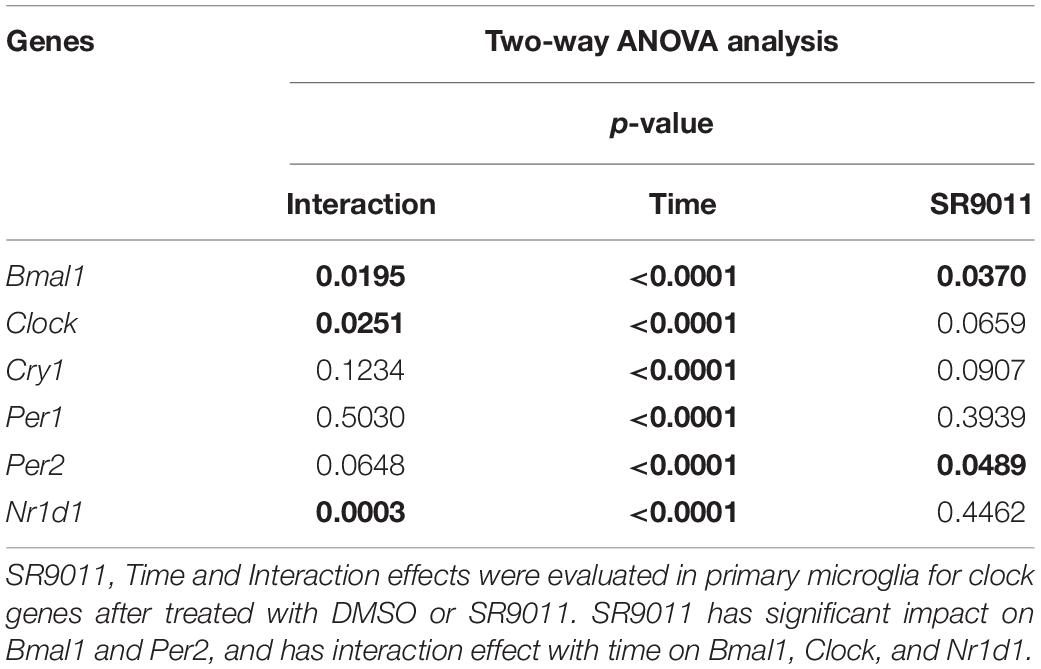
Table 1. Two-way ANOVA assessment of effect of Time, SR9011, and Interaction in clock genes in primary microglia with SR9011.
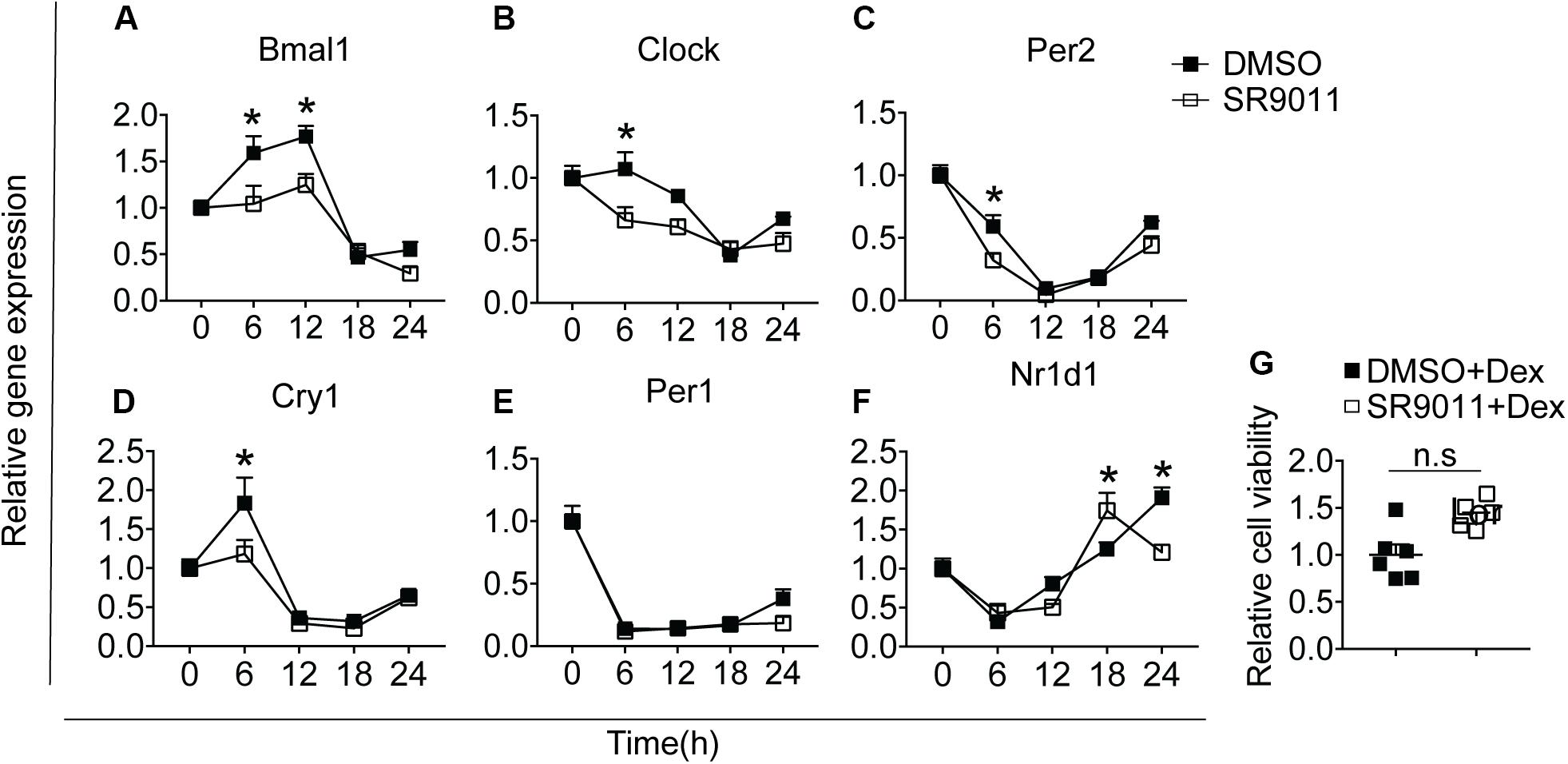
Figure 2. SR9011 disrupts clock gene expression in microglia. Rhythmic and relative gene expression of clock genes in primary microglia. Cells were treated with dexamethasone for 2 h followed by SR9011 or DMSO and were harvested at the indicated time points (n = 4 per group, per time point). (A–F) Clock gene expression is altered by SR9011 treatment compared to DMSO (control). SR9011 disrupted the rhythmic expression of Bmal1 and Clock, and caused a shift in the acrophase of Per2 rhythm, as analyzed by JTK_Cycle. (G) Cell viability of primary microglia was not affected by the 24-h SR9011 treatment with dexamethasone. Data are presented as means ± SEM. p < 0.05* vs. DMSO group was determined using Two-way ANOVA followed by Bonferroni’s post hoc test and multiple comparison.
Cytokine secretion constitutes an essential part of microglial immune function. Rev-erbα has very recently been reported to regulate neuroinflammation (20). To evaluate the role of Rev-erbα in regulating cytokine expression, we stimulated the cells with TNFα to resemble an immune challenge and analyzed the effect of SR9011. Primary microglia were exposed to 5 μM SR9011 (or DMSO for control) for 12 h, followed by 100 ng/ml TNFα treatment (or BSA for control) for 12 h in combination with SR9011 or DMSO. TNFα significantly increased the expression of the pro-inflammatory cytokines tumor necrosis factor-alpha (Tnfα), interleukin-6 (Il6), interleukin-1 beta (Il1β), and C-C Motif Chemokine Ligand (Ccl2; Figures 3A–E). TNFα also significantly increased the regulatory cytokine granulocyte-macrophage colony-stimulating factor (Gm-csf; Figure 3F). In all cases, the increase in gene expression by TNFα treatment was attenuated after treatment with SR9011. Different results were found for anti-inflammatory cytokines. TNFα had no effect on the regulatory cytokine transforming growth factor-beta (Tgfβ) expression, however, SR9011 treatment decreased Tgfβ expression (Figure 3G). No changes were found in interleukin-4 (Il4) expression (Figure 3H). Interestingly, while TNFα had no effect on the expression of anti-inflammatory cytokine interleukin-10 (Il10), SR9011 stimulated the expression of Il10 (Figure 3I). These results suggest that SR9011 attenuates the pro-inflammatory response in primary microglia in the context of an immune challenge, while stimulating the expression of the anti-inflammatory cytokine Il10.
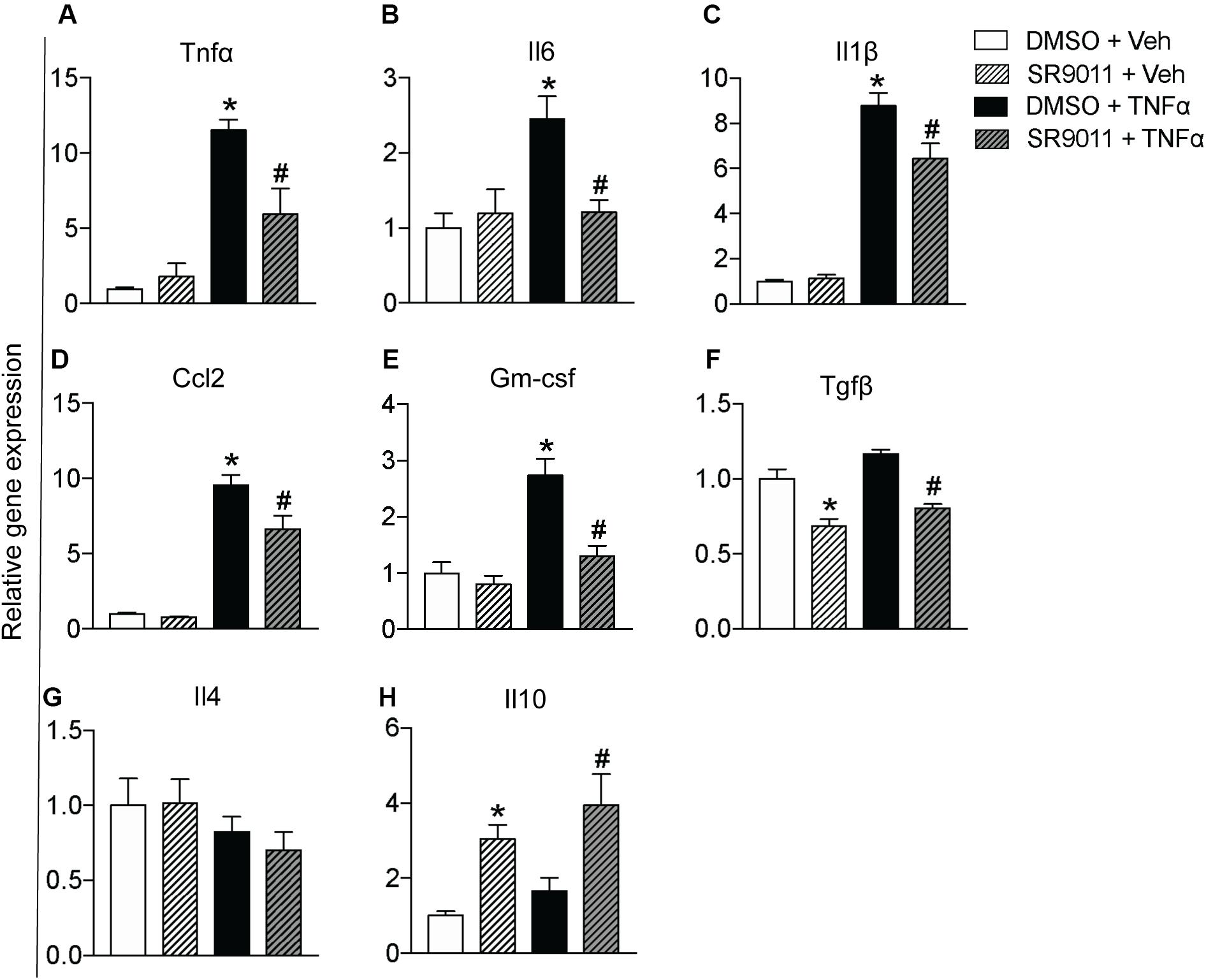
Figure 3. The effect of SR9011 on cytokine expression in microglia. Relative gene expression of cytokines in primary microglia treated with SR9011 or DMSO for 12 h followed by TNFα or BSA treatment for 12 h (n = 6 per group). This figure shows the effect of TNFα and SR9011 on pro-inflammatory cytokine expression (A–E), regulatory cytokine expression (F), and anti-inflammatory cytokine expression (G–H). TNFα increases the gene expression of Tnfα, Il6, Il1β, Ccl2, and Gm-csf. SR9011 attenuates the expression of Tnfα, Il6, Il1β, Ccl2, and Gm-csf compared to treatment with TNFα alone. Conversely, SR9011 increases Il10 expression after TNFα stimulation. Data are presented as means ± SEM and statistical significance was determined using Unpaired t-test in all experiments. p < 0.05* vs DMSO + BSA and p < 0.05# vs DMSO + TNFα.
To further test whether SR9011 has a potential therapeutic effect for overnutrition-induced neuroinflammation, we stimulated the microglia with palmitic acid (PA) to resemble a pro-inflammatory stimulus on high-fat diet feeding. Primary microglia were exposed to 5 μM SR9011 (or DMSO for control) for 12 h, followed by 50 μM palmitic acid (or BSA for control) for 12 h in combination with SR9011 or DMSO. Palmitic acid significantly increased the expression of pro-inflammatory cytokines Il6 and Il1β. which were attenuated by SR9011 (Figures 4A,B). Palmitic acid also significantly stimulated Gm-csf expression, while SR9011 had a profound inhibitory effect on Gm-csf expression (Figure 4C). Tnfα and Ccl2 expression was not enhanced by palmitic acid treatment (Figures 4D,E), which is a slightly different outcome compared to the TNFα stimulus. In addition, palmitic acid treatment exerted impacts on the expression of core clock genes Nr1d1 and Per1 after 12h incubation (Supplementary Figure 3), which indicates palmitic acid treatment might also influence circadian rhythms. This is consistent with our previous observation that circadian rhythms of microglia are disrupted in DIO animals (30). Thus, these data suggest that SR9011 attenuates the pro-inflammatory response in primary microglia upon palmitic acid treatment, which may be beneficial for DIO-induced neuroinflammation.
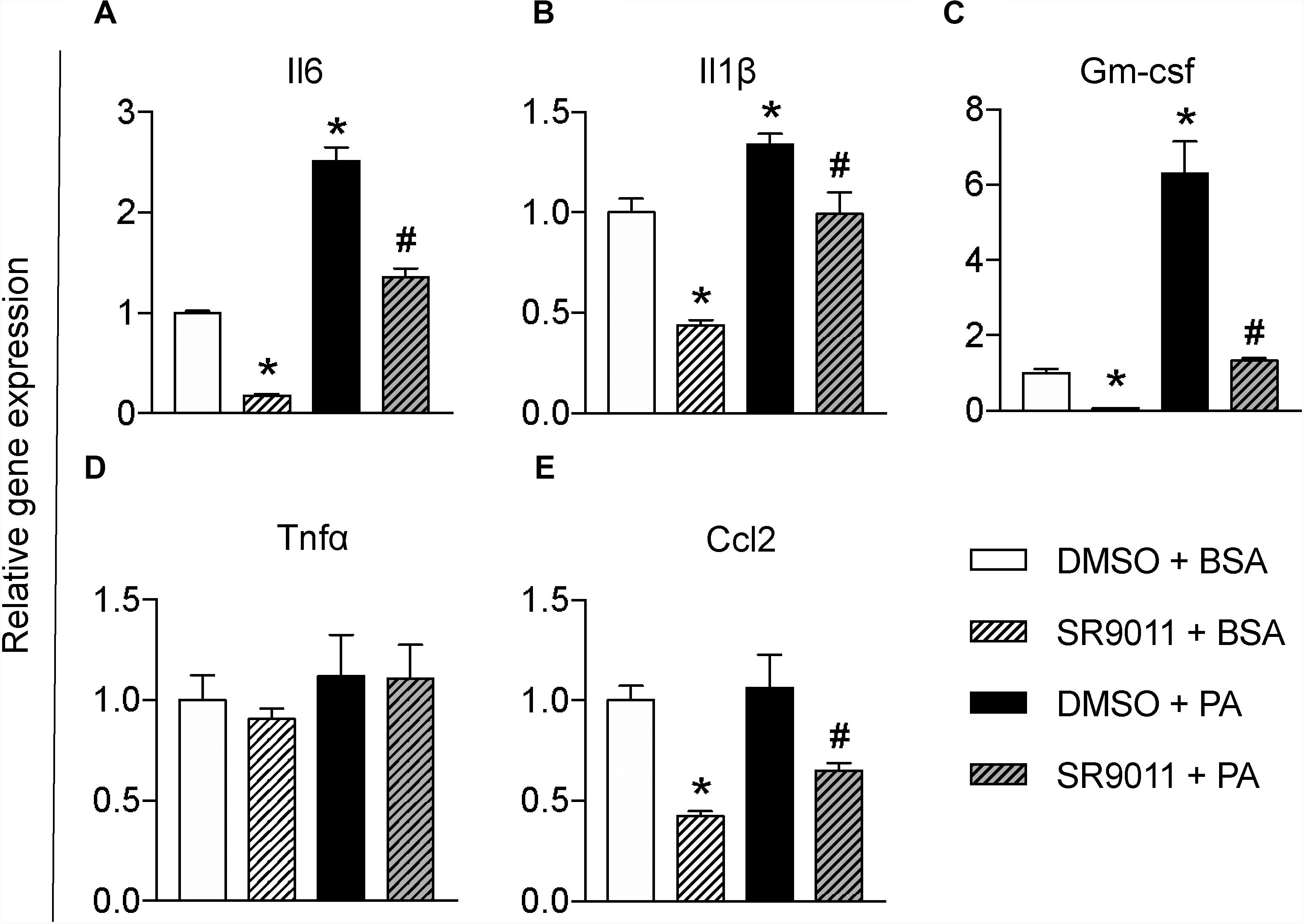
Figure 4. The effect of SR9011 on palmitic acid induced inflammatory responses in microglia. Relative gene expression of cytokines in primary microglia treated with SR9011 or DMSO for 12 h followed by 50 μM palmitic acid or BSA treatment for 12 h (n = 4 per group). Palmitic acid treatment increases Il6 (A), Il1β (B), and Gm-csf (C), which are inhibited by SR9011. Palmitic acid treatment has no profound effect on the expression of Tnfα (D) and Ccl2 (E), while SR9011 decreased Ccl2 expression with and without palmitic acid treatment (E). p < 0.05* vs DMSO + BSA and p < 0.05# vs DMSO + PA.
One of the main neuroprotective functions of microglia is phagocytosis, so we evaluated the effect of SR9011 on phagocytic activity. Primary microglia were treated with 5 μM SR9011 (or DMSO) for 12 h and subsequently exposed to 0.05% fluorescent beads for 1 h. A fluorescence staining was performed for IBA1 (microglia marker) and DAPI, after which the intensity of fluorescent beads per cell was determined (Figure 5A). The uptake of fluorescent beads was decreased in primary microglia treated with SR9011 (Figure 5B). Additionally, the expression of cluster of differentiation 68 (CD68), a phagocytic marker, was decreased in primary microglia that were starved for 6 h (0% FBS) and subsequently treated with 5 μM SR9011 for 12 h (Figure 5C). The purity of the cultures was tested by staining primary microglia with IBA1 and DAPI and a microglia purity of 98.81% was determined (data not shown). These results indicate that SR9011 decreases phagocytic activity, meaning that Rev-erbα plays an inhibitory role in regulating phagocytosis.
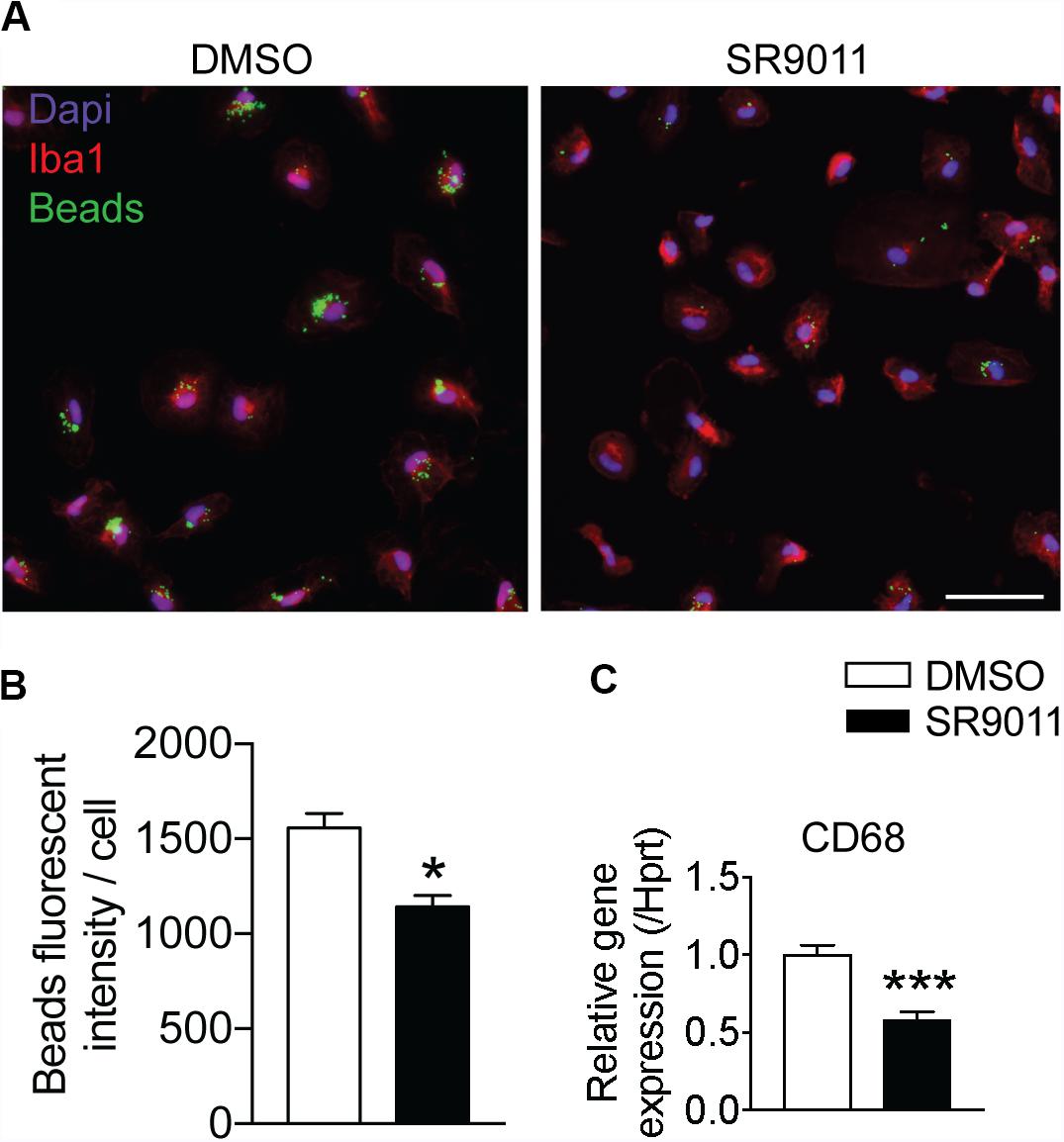
Figure 5. SR9011 decreased phagocytic activity in primary microglia. (A) The uptake of fluorescent beads by primary microglia treated with DMSO or SR9011. (B) Primary microglia treated with SR9011 exhibit decreased phagocytic activity (n = 15). (C) The expression of phagocytic marker CD68 is decreased in primary microglia treated with SR9011 for 12 h (n = 6). Data are presented as means ± SEM and statistical significance was determined using Unpaired t-test in all experiments. p < 0.05* and p < 0.001*** vs. DMSO.
Major metabolic genes are expressed in a rhythmic manner in microglia, as shown in Figure 1. We evaluated metabolism after disrupting the intrinsic microglial clock with SR9011. Primary microglia were treated with 5 μM SR9011 (or DMSO) for 12 h after which cellular respiration and ECAR were measured using the Seahorse XFe96 Analyzer to evaluate mitochondrial respiration and glycolysis. The raw data of OCR and ECAR are shown in Figures 6A,B. Cellular respiration was significantly decreased by SR9011, which was mainly attributed to less ATP-linked mitochondrial respiration, while the H+ proton leak remained unchanged (Figures 6C,D). Consequently, ATP production was greatly reduced (Figure 6E). Furthermore, maximum substrate oxidation was decreased after SR9011 treatment (Figure 6F). No changes were found in cellular acidification (glycolysis) and maximum glycolytic capacity (Figures 6G,H). These results indicate that SR9011 decreases ATP production by inhibiting oxidative phosphorylation. This mitochondrial dysfunction is not compensated with glycolysis. Furthermore, SR9011 inhibits the expression of Hk2, Pdk1, and Cpt1 (Figures 6I–K), which are key enzymes involved in substrate utilization by the citric acid cycle. Taken together, these results suggest an overall decrease in cellular metabolism caused by activation of Rev-erbα by SR9011. Thus, Rev-erbα is a potent inhibitor of cell metabolism in primary microglia.
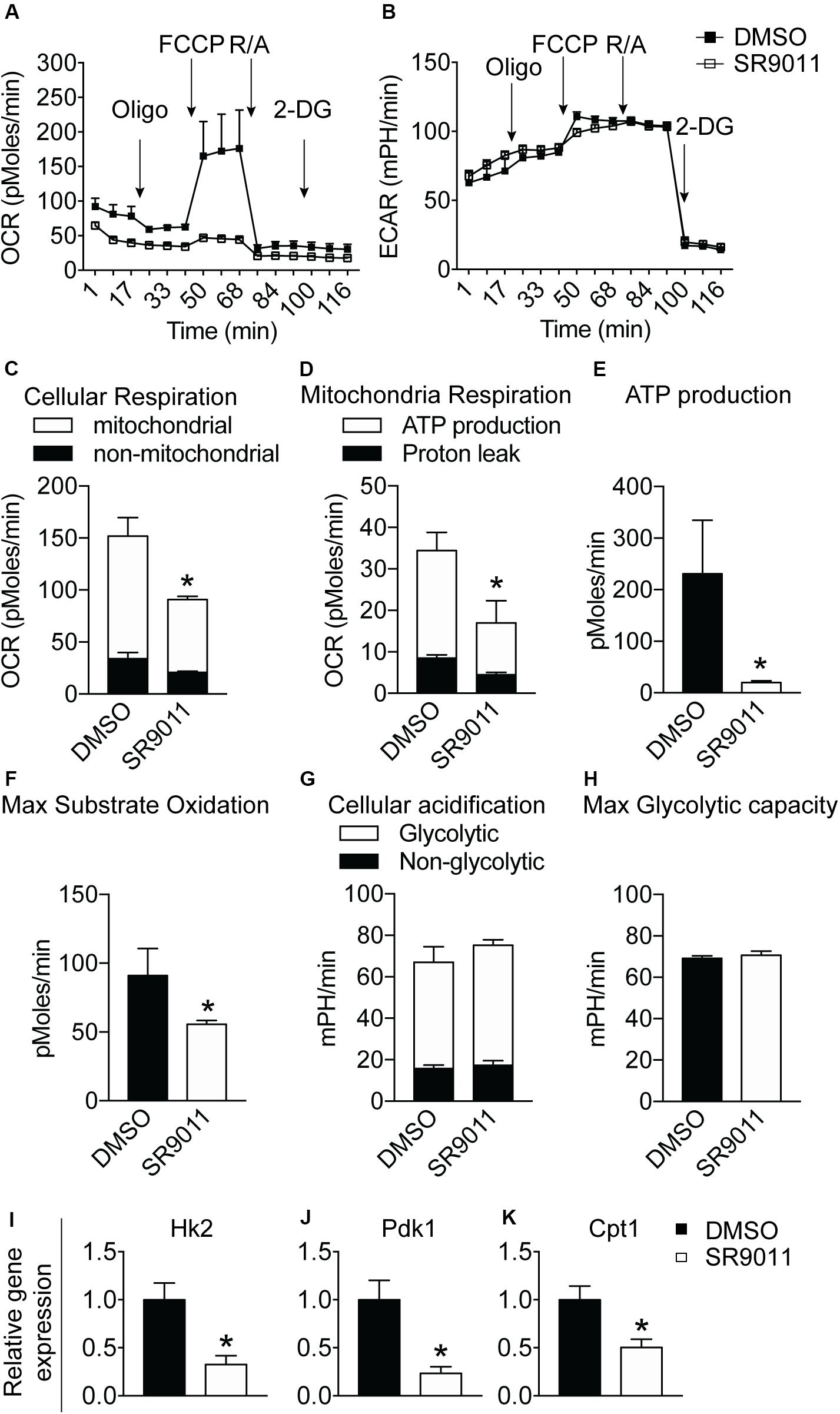
Figure 6. The effect of SR9011 on cellular metabolism in primary microglia. Cellular bioenergetics and metabolic gene expression in primary microglia after SR9011 treatment (n = 4 per group). (A, B) Overview of OCR (A) and ECAR (B) of primary microglia treated with DMSO or SR9011 for 12 h. Indicated are the times on which oligomycin, FCCP, rotenone/antimycin A (R/A) and 2DG were administered. (C–H) SR9011 decreased cellular respiration (C), which was caused by a decline in ATP-linked mitochondrial respiration (D). Consequently, ATP production was significantly decreased in response to SR9011 (E). SR9011 also caused a decrease in maximum substrate oxidation (F). No changes were found in cellular acidification (G) and maximum glycolytic capacity (H). (I–K) SR9011 decreased the expression of metabolic genes Hk2 (I), Pdk1 (J), and Cpt1 (K) in primary microglia treated by dexamethasone for 2 h and SR9011 for 6 h. Data are presented as means ± SEM and statistical significance was determined using Unpaired t-test in all experiments. p < 0.05*.
The aim of the current study was to determine the role of Rev-erbα in microglial immunometabolism. We show that key cytokine and metabolic genes are rhythmically expressed in primary microglia. Activation of Rev-erbα with SR9011 disrupted the intrinsic microglial clock and attenuated the phagocytosis and the pro-inflammatory response. SR9011 also decreased mitochondrial respiration and metabolic gene expression in microglia. These findings shed new light on the link between the circadian clock, cell metabolism and immune function in microglia and identify Rev-erbα as a possible important therapeutic target for treatment of neuroinflammatory diseases.
The rhythmic expression of major cytokines and metabolic genes in primary microglia observed in the current study supports our previous idea that in vivo the intrinsic clock is coupled with cellular metabolism (30). Such rhythmicity of cytokines may have physiological relevance in vivo – we reported before that microglia activity displays a day and night difference and is accompanied by a change in TNFα expression in the medial basal hypothalamus, which in turn affected the mitochondria function of nearby neurons (31). A growing body of literature reports that an obesogenic diet is associated with constant activation of microglia, leading to chronic hypothalamic inflammation, which contributes to the pathology observed (26, 27, 31, 40). We also reported that microglial circadian rhythmicity is disrupted in DIO animals (27). In this study, we found that SR9011 prevents the microglia pro-inflammatory responses usually observed during an inflammatory challenge and an obesogenic metabolic challenge. The mechanism behind the metabolic improvements observed in DIO mice can partially be explained by the mitigating effect of SR9011 on hyperactive microglia, thereby attenuating inflammation. This finding can have major implications for reducing microgliosis caused by obesity or neuroinflammatory diseases. A better ex vivo experimental setting to mimic the in vivo situation under a chronic high-fat diet challenge would be helpful to further elucidate these mechanisms.
The current study examined the immune function of microglia upon SR9011-associated Rev-erbα activation. Similar topics have been discussed before. In studies on knock-out mice or mice pharmacologically administered with SR9011, microglia activation and Il6 expression was downregulated upon LPS exposure (39, 41). In another Rev-erbα deletion study, mice displayed microgliosis, increased CD68, and neuroinflammation in the hippocampus (20). Consistent with these results, we enhanced Rev-erbα activity with SR9011 and found a decreased pro-inflammatory response and phagocytosis. A recent study describes a regulatory role of Bmal1 in macrophage motility and phagocytosis (42), which may indicate a possible interplay between Rev-erb and Bmal1 in the regulation of phagocytosis in microglia – an interesting topic for future research. Additionally, SR9011 stimulated the expression of the anti-inflammatory cytokine Il10. This suggests Rev-erbα not only inhibits pro-inflammatory cytokines, but also promotes the anti-inflammatory response. Moreover, to more accurately mimic microglia’s immune response, we used TNFα as an innate immune challenge, which is more physiologically relevant compared to lipopolysaccharide (LPS) used in the studies mentioned above. These findings suggest that the molecular clock components Bmal1 and Rev-erbα are closely related to the microglial immune response, and that Rev-erbα activity can regulate the inflammatory state in microglia under physiological and pathological conditions.
Rev-erbα nuclear receptors were reported to regulate metabolic homeostasis in cancer cells, hepatic cells and hepatoma cells (17, 18, 43). The findings of the current study show that Rev-erbα nuclear receptors regulate cell metabolism in primary microglia, the innate immune cells, as evidenced by the decrease in mitochondrial respiration and ATP production, as well as the decreased expression of rate-limiting enzymes such as Cpt1, Pdk1, and Hk2 in response to SR9011. Our study dissected how the Rev-erbα-associated clock machinery interacts with immunometabolism in microglia. It is generally assumed that immune function is tightly linked to cell metabolism due to the metabolic demand of an inflammatory response (44). One explanation for the decline in energy metabolism is that it is a consequential phenomenon caused by less energy demand due to the attenuated pro-inflammatory response. Another possibility is a direct effect of Rev-erbα on cellular metabolism. Rev-erbα nuclear receptors, when bound to their ligand, repress the transcription of not only clock genes like Bmal1, but also repress metabolic genes and are involved in mitochondrial respiration (14, 15, 45, 46). The inflammatory response of microglia also involves metabolic reprogramming, which also might explain the attenuation of pro-inflammatory cytokines (32, 33). Another speculation would be that the intrinsic molecular clock orchestrates both microglial metabolism and immune response, and that it can also change the activation state of microglia. The latter would mean that SR9011-induced enhanced Rev-erbα activity is associated with disturbed microglial circadian rhythms, which consequently changed the cellular metabolism and immune response. Indeed, the in vivo situation might be far more complicated, because it is unknown what occurs first in the development of metabolic disorders: the disrupted circadian rhythmicity, the alterations in intracellular metabolism, the changes in immune response, or any other unknown factors. In our opinion, the disrupted circadian rhythmicity is very likely to be the driving force of the other two. In addition, the appropriate immune response of microglia is required to defend the CNS under normal conditions, so attenuated pro-inflammatory responses might not be always beneficial for the body. Further research is needed to elucidate the causal relation among the intracellular clock, cellular metabolism and immune response in microglia under physiological condition and metabolic challenges. It is interesting to note that SR9009, a REV-ERB agonist similar to SR9011, has been reported to exert Rev-erbα independent side-effects on cell proliferation and metabolism (47). Whether SR9011 has the same side-effects is currently unknown, and requires further research.
Future research should also investigate the significance of the microglial intrinsic clock in vivo. It is likely that the exact consequences of disturbing the microglial clock depend on the brain regions involved. Microgliosis in the hypothalamus is associated with metabolic disorders, while microglia in other brain regions will have other functions, depending on the micro-environment. On the other hand, the role of Rev-erbα should also be investigated in other immune cells and their related immune-diseases.
Our study demonstrates that disturbing circadian rhythmicity by SR9011-induced Rev-erbα activation attenuates pro-inflammatory cytokine expression and reduces cellular metabolism in microglia. In addition, expression of the anti-inflammatory cytokine Il10 is stimulated, emphasizing the mitigating effect of Rev-erbα activation on the pro-inflammatory response. The findings of this study identify an interconnected role of Rev-erbα in the circadian rhythmicity, cell metabolism and inflammatory response of microglia and may have important implications for the employment of Rev-erbα as a potential therapeutic target for the treatment of neuroinflammatory diseases, however, further studies are required to achieve this goal.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The experiments were approved by the Animal Care and Use Committee of Nanjing Medical University, and the experiments were performed according to the Guide for the Care and Use of Laboratory Animals of China.
SW, JS, HJ, and YG performed the experiments. C-XY and YG designed the study. SW, X-LW, and AK analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
This work was funded by the international student fellowship from Amsterdam Gastroenterology & Metabolism (AG&M), grants from the National Natural Science Foundation of China 81873654 and 31800971, and Jiangsu Province Science Foundation for Youths BK20180684, the Department of Cardiovascular and Cerebrovascular Medicine, Nanjing Medical University (YG Lab); and AMC fellowship, Amsterdam University Medical Center (2014; C-XY lab); and the doctoral fellowship from the ‘NeuroTime” Erasmus+ Program (X-LW).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank AG&M for granting SW the international student fellowship to conduct this valuable research at Nanjing Medical University under the supervision of YG.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.550145/full#supplementary-material
1. Kalsbeek A, Palm IF, La Fleur SE, Scheer FAJL, Perreau-Lenz S, Ruiter M, et al. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. (2006) 21:458–69. doi: 10.1177/0748730406293854
2. Boden G, Chen X, Polansky M. Disruption of circadian insulin secretion is associated with reduced glucose uptake in first-degree relatives of patients with type 2 diabetes. Diabetes. (1999) 48:2182–8. doi: 10.2337/diabetes.48.11.2182
3. Depner CM, Stothard ER, Wright KP. Metabolic consequences of sleep and circadian disorders. Curr Diab Rep. (2014) 14:507. doi: 10.1007/s11892-014-0507-z
4. McGinnis GR, Young ME. Circadian regulation of metabolic homeostasis: causes and consequences. Nat Sci Sleep. (2016) 8:163–80. doi: 10.2147/NSS.S78946
5. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian clock mutant nice. Science. (2005) 308:1043–5. doi: 10.1126/science.1108750
6. Ma D, Liu T, Chang L, Rui C, Xiao Y, Li S, et al. The liver clock controls cholesterol homeostasis through trib1 protein-mediated regulation of PCSK9/Low density lipoprotein receptor (LDLR) axis. J Biol Chem. (2015) 290:31003–12. doi: 10.1074/jbc.M115.685982
7. Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. (2010) 466:627–31. doi: 10.1038/nature09253
8. Inoue I, Shinoda Y, Ikeda M, Hayashi K, Kanazawa K, Nomura M, et al. CLOCK/BMAL1 is involved in lipid metabolism via transactivation of the peroxisome proliferator-activated receptor (PPAR) response element. J Atheroscler Thromb. (2005) 12:169–74. doi: 10.5551/jat.12.169
9. Sato F, Kohsaka A, Bhawal UK, Muragaki Y. Potential roles of dec and bmal1 genes in interconnecting circadian clock and energy metabolism. Int J Mol Sci. (2018) 19:781. doi: 10.3390/ijms19030781
10. Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, et al. Deficient of a clock gene, brain and muscle arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. (2011) 6:e25231. doi: 10.1371/journal.pone.0025231
11. Crumbley C, Burris TP. Direct regulation of CLOCK expression by REV-ERB. PLoS One. (2011) 6:e17290. doi: 10.1371/journal.pone.0017290
12. Gaucher J, Montellier E, Sassone-Corsi P. Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol. (2018) 28:368–79. doi: 10.1016/j.tcb.2018.01.006
13. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. (2002) 110:251–60. doi: 10.1016/S0092-8674(02)00825-5
14. Padmanaban G, Venkateswar V, Rangarajan PN. Haem as a multifunctional regulator. Trends Biochem Sci. (1989) 14:492–6. doi: 10.1016/0968-0004(89)90182-5
15. Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. (2007) 14:1207–13. doi: 10.1038/nsmb1344
16. Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. (2009) 23:2201–9. doi: 10.1101/gad.1825809
17. Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. (2007) 318:1786–9. doi: 10.1126/science.1150179
18. Duez H, Staels B. Rev-erbα gives a time cue to metabolism. FEBS Lett. (2008) 582:19–25. doi: 10.1016/j.febslet.2007.08.032
19. Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. (2006) 126:801–10. doi: 10.1016/j.cell.2006.06.050
20. Griffin P, Dimitry JM, Sheehan PW, Lananna BV, Guo C, Robinette ML, et al. Circadian clock protein Rev-erbα regulates neuroinflammation. Proc Natl Acad Sci USA. (2019) 116:5102–7. doi: 10.1073/pnas.1812405116
21. Grant D, Yin L, Collins JL, Parks DJ, Orband-Miller LA, Wisely GB, et al. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor rev-erbα. ACS Chem Biol. (2010) 5:925–32. doi: 10.1021/cb100141y
22. Chen H, Chu G, Zhao L, Yamauchi N, Shigeyoshi Y, Hashimoto S, et al. Rev-erbα regulates circadian rhythms and StAR expression in rat granulosa cells as identified by the agonist GSK4112. Biochem Biophys Res Commun. (2012) 420:374–9. doi: 10.1016/j.bbrc.2012.02.164
23. Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. (2012) 485:62–8. doi: 10.1038/nature11030
24. Kojetin D, Wang Y, Kamenecka TM, Burris TP. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol. (2011) 6:131–4. doi: 10.1021/cb1002575
25. Dragano NRV, Solon C, Ramalho AF, de Moura RF, Razolli DS, Christiansen E, et al. Polyunsaturated fatty acid receptors, GPR40 and GPR120, are expressed in the hypothalamus and control energy homeostasis and inflammation. J Neuroinflammation. (2017) 14:91. doi: 10.1186/s12974-017-0869-7
26. Gao Y, Bielohuby M, Fleming T, Grabner GF, Foppen E, Bernhard W, et al. Dietary sugars, not lipids, drive hypothalamic inflammation. Mol Metab. (2017) 6:897–908. doi: 10.1016/j.molmet.2017.06.008
27. Maldonado-Ruiz R, Montalvo-Martínez L, Fuentes-Mera L, Camacho A. Microglia activation due to obesity programs metabolic failure leading to type two diabetes. Nutr Diabetes. (2017) 7:e254. doi: 10.1038/nutd.2017.10
28. Paolicelli RC, Angiari S. Microglia immunometabolism: from metabolic disorders to single cell metabolism. Semin Cell Dev Biol. (2019) 94:129–37. doi: 10.1016/j.semcdb.2019.03.012
29. Valdearcos M, Robblee MM, Benjamin DI, Nomura DK, Xu AW, Koliwad SK. Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep. (2014) 9:2124–38. doi: 10.1016/j.celrep.2014.11.018
30. Milanova IV, Kalsbeek MJT, Wang XL, Korpel NL, Stenvers DJ, Wolff SEC, et al. Diet-induced obesity disturbs microglial immunometabolism in a time-of-day manner. Front Endocrinol. (2019) 10:424. doi: 10.3389/fendo.2019.00424
31. Yi CX, Walter M, Gao Y, Pitra S, Legutko B, Kälin S, et al. TNFα drives mitochondrial stress in POMC neurons in obesity. Nat Commun. (2017) 8:15143. doi: 10.1038/ncomms15143
32. Orihuela R, McPherson CA, Harry GJ. Microglial M1/M2 polarization and metabolic states. Br J Pharmacol. (2016) 173:649–65. doi: 10.1111/bph.13139
33. Wang A, Luan HH, Medzhitov R. An evolutionary perspective on immunometabolism. Science. (2019) 363:eaar3932. doi: 10.1126/science.aar3932
34. Keuper M, Jastroch M, Yi CX, Fischer-Posovszky P, Wabitsch M, Tschöp MH, et al. Spare mitochondrial respiratory capacity permits human adipocytes to maintain ATP homeostasis under hypoglycemic conditions. FASEB J. (2014) 28:761–70. doi: 10.1096/fj.13-238725
35. Brand M. The efficiency and plasticity of mitochondrial energy transduction. Biochem Soc Trans. (2005) 33(Pt 5):897–904. doi: 10.1042/BST20050897
36. Hughes ME, Hogenesch JB, Kornacker K. JTK-CYCLE an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. (2010) 25:372–80. doi: 10.1177/0748730410379711
37. Nakazato R, Takarada T, Yamamoto T, Hotta S, Hinoi E, Yoneda Y. Selective upregulation of Per1 mRNA expression by ATP through activation of P2X7 purinergic receptors expressed in microglial cells. J Pharmacol Sci. (2011) 116:350–61. doi: 10.1254/jphs.11069FP
38. Logan RW, Sarkar DK. Circadian nature of immune function. Mol Cell Endocrinol. (2012) 349:82–90. doi: 10.1016/j.mce.2011.06.039
39. Nakazato R, Hotta S, Yamada D, Kou M, Nakamura S, Takahata Y, et al. The intrinsic microglial clock system regulates interleukin-6 expression. Glia. (2017) 65:198–208. doi: 10.1002/glia.23087
40. Gao Y, Ottaway N, Schriever SC, Legutko B, García-Cáceres C, de la Fuente E, et al. Hormones and diet, but not body weight, control hypothalamic microglial activity. Glia. (2014) 62:17–25. doi: 10.1002/glia.22580
41. Guo D, Zhu Y, Sun H, Xu X, Zhang S, Hao Z, et al. Pharmacological activation of REV-ERBα represses LPS-induced microglial activation through the NF-κB pathway. Acta Pharmacol Sin. (2019) 40:26–34. doi: 10.1038/s41401-018-0064-0
42. Kitchen GB, Cunningham PS, Poolman TM, Iqbal M, Maidstone R, Baxter M, et al. The clock gene Bmal1 inhibits macrophage motility, phagocytosis, and impairs defense against pneumonia. Proc Natl Acad Sci USA. (2020) 117:1543–51. doi: 10.1073/pnas.1915932117
43. Sulli G, Rommel A, Wang X, Kolar MJ, Puca F, Saghatelian A, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature. (2018) 553:351–5. doi: 10.1038/nature25170
44. Borst K, Schwabenland M, Prinz M. Microglia metabolism in health and disease. Neurochem Int. (2019) 130:104331. doi: 10.1016/j.neuint.2018.11.006
45. Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, et al. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. (2012) 26:657–67. doi: 10.1101/gad.186858.112
46. Everett LJ, Lazar MA. Nuclear receptor Rev-erbα: up, down, and all around. Trends Endocrinol Metab. (2014) 25:586–92. doi: 10.1016/j.tem.2014.06.011
Keywords: clock genes, Innate immunity, microglia, immunometabolism, neuroinflammation, cytokines, phagocytosis
Citation: Wolff SEC, Wang X-L, Jiao H, Sun J, Kalsbeek A, Yi C-X and Gao Y (2020) The Effect of Rev-erbα Agonist SR9011 on the Immune Response and Cell Metabolism of Microglia. Front. Immunol. 11:550145. doi: 10.3389/fimmu.2020.550145
Received: 09 April 2020; Accepted: 04 September 2020;
Published: 25 September 2020.
Edited by:
Pedro Manoel Mendes Moraes Vieira, Campinas State University, BrazilReviewed by:
Jennifer Lee, Beth Israel Deaconess Medical Center and Harvard Medical School, United StatesCopyright © 2020 Wolff, Wang, Jiao, Sun, Kalsbeek, Yi and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanqing Gao, eXVhbnFpbmdnYW9AbmptdS5lZHUuY24=; Chun-Xia Yi, Yy55aUBhbXN0ZXJkYW11bWMubmw=; Yy55aUBhbWMudXZhLm5s
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.