
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 11 September 2020
Sec. T Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.02187
This article is part of the Research TopicThe Role of Biomembranes and Biophysics in Immune Cell SignalingView all 11 articles
 Ron Orbach1
Ron Orbach1 Xiaolei Su1,2*
Xiaolei Su1,2*Microvilli are finger-like membrane protrusions, supported by the actin cytoskeleton, and found on almost all cell types. A growing body of evidence suggests that the dynamic lymphocyte microvilli, with their highly curved membranes, play an important role in signal transduction leading to immune responses. Nevertheless, challenges in modulating local membrane curvature and monitoring the high dynamicity of microvilli hampered the investigation of the curvature-generation mechanism and its functional consequences in signaling. These technical barriers have been partially overcome by recent advancements in adapted super-resolution microscopy. Here, we review the up-to-date progress in understanding the mechanisms and functional consequences of microvillus formation in T cell signaling. We discuss how the deformation of local membranes could potentially affect the organization of signaling proteins and their biochemical activities. We propose that curved membranes, together with the underlying cytoskeleton, shape microvilli into a unique compartment that sense and process signals leading to lymphocyte activation.
Sea looks calm miles away but wavy inches ahead; same applies to the plasma membrane. A variety of membrane protrusions have been identified on the cell surface, including microvilli, filopodia, lamellipodia, and cilia (see Table 1). Those structures play a classical function in sensing the environmental cues as well as facilitating cell migration. Meanwhile, accumulating evidence suggests that membrane protrusions also play an active role in regulating biochemical reactions that transduce membrane-proximal signaling (1–3), and dysregulation of their formation has been associated with diseases like Huntington’s disease, PAPA syndrome, Wiskott–Aldrich syndrome (WAS), and renal dysfunction (4–6).
In the immune system, microvilli are among the most common types of membrane protrusions found on lymphocytes. Although they have been well-described by electron microscopy (EM) studies (7), the biochemical and signaling functions of microvilli remained neglected until recently. In this review, we discuss the potential of physical feature of microvilli in regulating chemical reactions that transduce membrane-proximal signaling. We also summarize the development of new techniques for imaging cell surface topography at high spatial or temporal resolutions, and for modulating membrane curvature in a precision manner, which could provide powerful tools for investigating the signaling function of microvilli.
Microvilli are thin finger-like membrane protrusions that are found on the surface of a wide variety of cell types (8), including intestinal epithelial cells (9), dendritic cells (10), and neurons (11). They are supported by actin filaments (F-actin) that are organized in parallel bundles of 10–30 filaments (12, 13), which resemble the actin network that constitutes filopodia (14). However, filopodia often protrude from the lamellipodial and lamellar actin network (15), while the microvilli actin network does not (16, 17). In the case of lymphocytes, EM studies showed the presence of microvilli on the surface of both T cells and B cells (18). The diameter of the microvilli ranges from 50 to 550 nm, as revealed by EM and fluorescence microscopy studies, while their length varies between 100 nm to several microns (Figure 1) (7, 13, 19, 20). Thus, microvilli dramatically increase the cell surface area, while having a negligible effect on the cytosolic volume. Furthermore, actin depolymerizing toxin Latrunculin A (LatA) eliminates most microvilli within 1 min in a reversible manner, suggesting that microvilli are highly dynamic structures (13). Recent technological advances in lattice light-sheet microscopy allow 3D real-time tracing of such dynamic microvilli (20). It was discovered that microvilli move laterally on the plasma membrane and survey antigen-presenting cells (APCs) within 1 min, which is, coincidently or not, the half-life of T cell–APC contact duration in vivo (20). Therefore, the dynamics of microvilli fits well into their function in searching antigens.
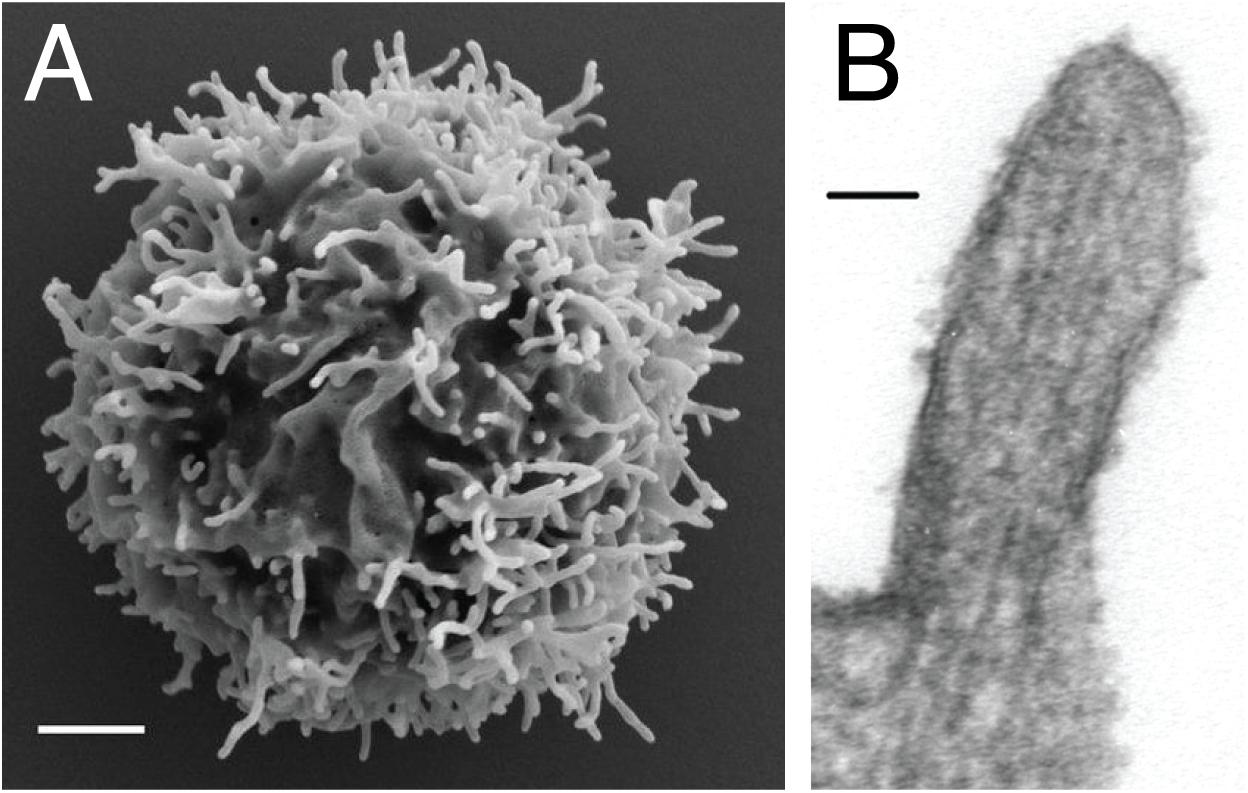
Figure 1. Microvilli decorating the plasma membrane of lymphocytes. (A) Scanning electron microscopy micrograph showing microvilli that protrude from the cell surface of resting peripheral blood human T cells. Scale bar: 1 μm. Reproduced from Jung et al. (19). Copyright 2016 National Academy of Sciences. (B) Transmission electron microscopy micrograph showing the parallel arrangement of F-actin within the microvilli of 300.19 cell (Abelson-transformed murine pre-B lymphoma) Scale bar: 50 nm. Republished with permission of ASH from Ref. (13).
Despite a handful of studies on microvilli morphology, our knowledge on the regulatory mechanism of microvilli size, structure, and dynamics is still limited. Evidently, their fate following the formation of immunological synapse is still a matter of debate. Cai et al. (20) demonstrated that there is no change in the microvilli density before and after the immunological synapse is formed. In contrast, Kim et al. (21) showed that at an early stage of synapse development microvilli polarize toward the synapse, but as the synapse matures, most of the microvilli disappear. The later was further supported by a recent study by Ghosh et al. (22), showing the loss of the microvilli after T cell receptor (TCR) stimulation.
The microvilli are also regulated by cytokines and chemokines. Westerberg et al. (23) found that CD40 antibody together with IL-4 induces microvilli on the surface of B cells. On the other hand, the chemokines stromal derived factor 1α (SDF-1α) and B lymphocyte chemokine (BLC) induce resorption of microvilli (24, 25), which promotes B cell homing by transition from rolling adhesion to integrin-mediated adhesion. Not surprisingly, members of the ezrin-radixin-moesin (ERM) family, which link the cortical F-actin cytoskeleton to the plasma membrane, were found to regulate microvilli assembly, namely, dephosphorylation of ERM proteins (ezrin, T567; radixin, T564, moesin, T558), induced by chemokines, results in resorption of microvilli within a few seconds (25, 26). Because ERM dephosphorylation can be triggered by TCR activation (27), ERM could mediate TCR-induced microvilli resorption (22).
Interestingly, changes in the microvilli shape and density are also linked to several diseases. Uneven distribution of long microvilli was observed on B cells from hairy cell leukemia patients (28–31). Moreover, changes in microvilli morphology were observed in WAS, a severe immunodeficiency disorder that is caused by defective or missing Wiskott–Aldrich syndrome protein (WASp). WASp activates Arp2/3 complex by inducing a conformational change of Arp2/3 and by delivering the first actin monomer of the nascent filament (32–36). On the other hand, WASp could directly promote actin polymerization independently of Arp2/3 (37). It has been demonstrated that lymphocytes derived from patients with WAS, either in resting or activated states, exhibit various microvillar morphological abnormalities. These abnormalities include a decrease in microvilli density and length, as well as formation of dysmorphic structures (5, 24, 38–42). However, knockdown of Arp2 in Jurkat T cells caused no significant effect on microvilli assembly (43), which suggests that WASp might regulate microvilli formation independent of Arp2/3. Meanwhile, further studies are required to confirm the Arp2 phenotype in primary T cells.
The notion that microvilli could serve as a signaling center was primed by studies showing that certain signaling proteins are enriched in microvilli. Immunogold EM studies demonstrated the enrichment of various receptors and adhesion molecules on the microvilli, including insulin receptors, selectin, integrin, and the T cell co-receptor CD4 (44–49). Mass-spectrometry analysis was also implemented to compare the protein composition between isolated microvilli and whole cell, from both human peripheral blood T-lymphocytes and a mouse pre-B lymphocyte line (50). It revealed that microvilli are enriched of GTP-binding proteins, cytoskeletal proteins, and transmembrane proteins as compared to the cell body (after removing the nucleus). This study provides the first global mapping of the microvilli proteome. However, it should be noted that using the cell body as a control could lead to the identification of membrane-associated proteins rather than microvilli-specific proteins, because the surface-to-volume ratio is much higher in microvilli as compared to the cytoplasm. It should be also noted that the identification of membrane proteins by mass-spectrometry remains as a challenge because of the proteins limited solubility in aqueous buffer (51). Therefore, certain hits might be missing in the dataset. Thus, orthogonal approaches will be needed to verify the microvilli-enriched proteins.
For many years the dynamic nature of microvilli together with their small dimensions have hindered the structural and functional characterization of microvilli. Although EM provides high spatial resolutions, understanding the signaling function of microvilli requires characterizing microvilli morphology and signaling protein localization with high temporal resolutions. The advancement of fluorescence microscopy techniques in the past decades has enabled an investigation of membrane morphology and protein localization in microvilli at either high temporal or spatial resolutions, though the combination of both is still technically challenging (19–22, 52, 53).
The Haran group comprehensively characterized the localization of TCR signaling proteins on the microvilli by a unique imaging technique that allows accurate mapping of membrane protein localization. By combining variable-angle total internal reflection microscopy and stochastic localization nanoscopy, the authors reconstructed 3D topographical maps of T cells (19, 22). They have shown that TCR, co-receptor CD4, kinase Lck, adaptor LAT, and adhesion receptor CD2 are highly enriched in the microvilli. On the sub-microvilli scale, the Jun group, using Total Internal Reflection Fluorescence (TIRF) microscopy, showed that TCR is specifically enriched on the microvilli tip (Figure 2) (21). Thus, the tip localization of TCR could promote searching of antigens and establishing contacts with APCs (20). The Haran group also found that treatment with LatA or expressing a dominant negative form of ezrin, both of which reduce microvilli, leads to a random distribution of TCR throughout the plasma membrane. Intriguingly, the authors showed that following TCR stimulation, T cells lose their microvilli, which consequently leads to an even distribution of TCRαβ throughout the plasma membrane (22). This result suggests that microvilli-dependent TCR enrichment could be regulated by TCR triggering.
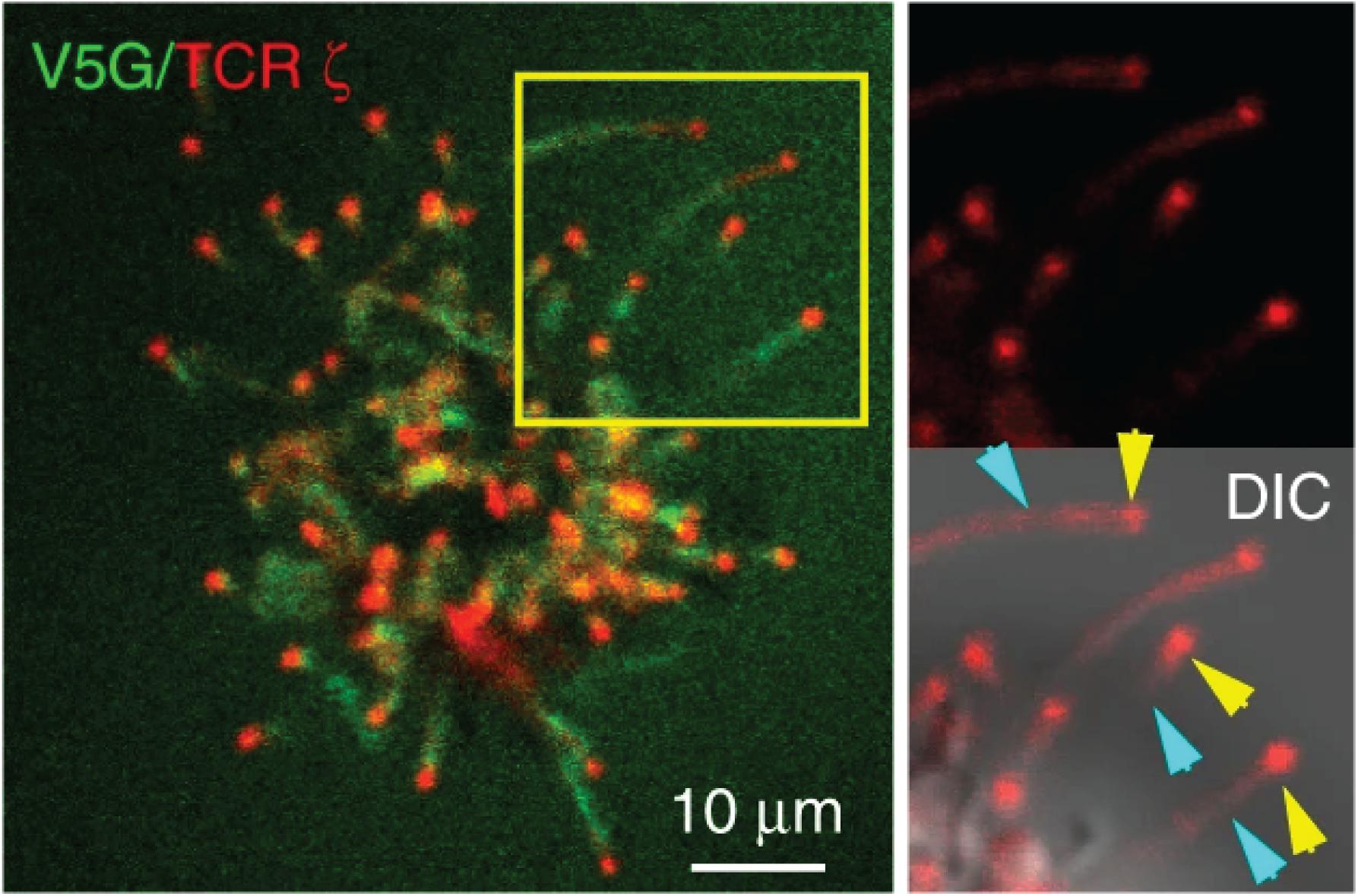
Figure 2. TCR localizes to the microvilli tip. Jurkat T cells expressing GFP-V5G and TCRζ-tdTomato were imaged at the terminal stage of T cell activation. Reproduced from Kim et al. (21) licensed under Creative Commons (CC BY 4.0).
The “kinetic-segregation” model serves as one of the prevalent mechanisms explaining TCR triggering (54–57). A key part to this model is the segregation of the large tyrosine phosphatase CD45 from the TCR-pMHC contact zone. Therefore, multiple groups have investigated the localization of CD45 in the context of microvilli and showed that the segregation between TCR and CD45 occurs a few seconds after contacts are established (58–61). Interestingly, although it has been assumed and supported by experimental data that CD45 is evenly distributed on the cell surface in resting T cells (19, 60), a new study revealed, using expansion microscopy, that CD45 is excluded from the microvilli tip even before contacts are established with APC (53). These discrepancies could be caused by differences in T cell subtypes, activation methods, and resolution of individual imaging techniques.
Summarizing localization studies above, key components mediating TCR-proximal signaling reside in microvilli. These include TCR itself, kinase Lck, and adaptor LAT. It is expected that cytosolic proteins that are associated with these membrane proteins, including ZAP70, Grb2, Sos1, PLCγ1, Gads, and SLP76, are likely to be enriched in microvilli as well. The physical proximity of these molecules could increase the rate of chemical reactions and efficiency of signal transduction. Microvilli, therefore, could serve as a compartment to enrich signaling proteins to promote TCR signaling (Figure 3). In this regard, T cell microclusters are another entity that has been proposed for promoting TCR signaling (62, 63). Because both microvilli-localized proteins and T cell microcluster components display a puncta-like structure on the cell membrane, it raises an interesting question on the relationship between the two. Current evidence suggests that these two entities are different but related structures. The microvilli-enriched proteins are mostly characterized in resting T cells and microvilli disappear, at least in some studies, after TCR activation (22). In contrast, T cell microclusters are formed after TCR activation (62, 63). They also displayed limited mobility as compared to the highly mobile microvilli (20). Meanwhile, the “pre-enrichment” of signaling components in microvilli could facilitate T microcluster formation upon TCR activation.
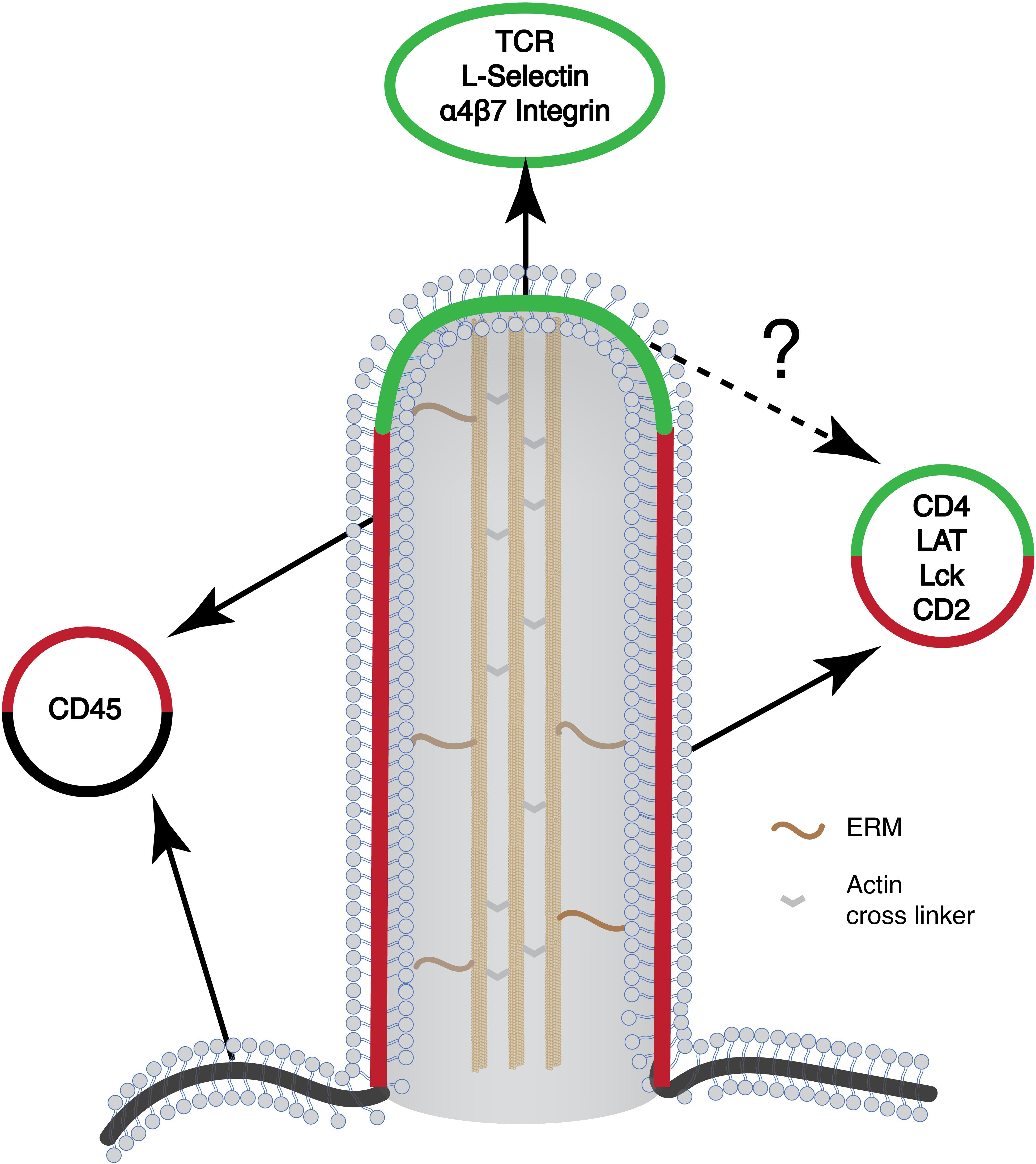
Figure 3. Schematic of the microvilli and the organization of different signaling proteins on the microvilli. Many of the signaling molecules that are involved in T cell activation preferentially localize to the microvilli. Yet, their organization within the microvilli is not known (marked in question mark). Green, microvilli tip region; red, microvilli body; black, plasma membrane.
Besides a potential signaling function in T cells, microvilli have also been proposed by the Jun Lab to serve as precursors for generating TCR-enriched extracellular vesicles (or synaptosomes) that activate dendritic cells (21). It remains to be determined whether the synaptosomes are similar or different to other TCR-enriched microvesicles (synaptic ectosomes) that were described by the Dustin Lab (64). Ectosomes are generated by the ESCRT complex, of which TSG101 facilitates the sorting of TCR into the ectosomes whereas Vps4 facilitates the scission of ectosomes from the plasma membrane. Interestingly, CD40L, a key effector delivered by helper T cells to activate APC, is also enriched in the microvilli and ectosomes, though CD40L is spatially segregated from TCR in ectosomes (65). This probably suggests that TCR and CD40L are independently sorted into microvilli, whereas the exact mechanism needs to be determined. It also remains as an intriguing question on how many microvilli and their associated TCRs end up in synaptosomes or ectosomes. Is ESCRT, a general membrane-shaping machinery, involved in microvilli dynamics regulation and resorption? Answering these questions will help to generate a complete picture of the microvilli life cycle during T cell activation (65).
Highly curved membranes represent a unique feature of microvilli, which could also serve as a platform for enriching proteins in microvilli. Several mechanisms have been proposed for generating curved membranes. Polymerization of actin filaments drives membrane protrusion; in parallel, membrane-associated proteins can also induce membrane curvature by the insertion of conical transmembrane proteins or hydrophobic protein domains into the membrane (66, 67). Intriguingly, intrinsically disordered domains, when attached to membranes, can drive membrane protrusion either with a positive or negative curvature (68–70).
The Bin-Amphiphysin-Rvs (BAR) superfamily is a key player involved in regulation, formation, and detection of cell membrane curvature (71). In this superfamily, the N-BAR and the F-BAR are associated with positive curvatures (e.g., membrane invagination or endocytic pits). In contrast, the I-BAR subfamily of proteins is associated with negative curvatures as found in various membrane protrusions (72). Many members of the BAR superfamily contain the structurally conserved SH2 or SH3 domains that recruit their binding partners to the curved membranes (73). One particular interesting example is the I-BAR protein IRSp53 (also known as BAIAP2) that binds cytoskeletal effectors such as N-WASP through its SH3 domain (74). In an in vitro biochemical assay, IRSp53 induces tubular membrane protrusions with similar dimensions of microvilli (75). IRSp53 is localized to filopodia when ectopically expressed in neuronal NSC34 cells (76) and regulates filopodia dynamics (77). Interestingly, IRSp53 is also expressed in T cells (78), raising its possible role in regulating microvilli formation. Surprisingly, the N-BAR protein sorting nexin 9 (SNX9), which is expected to recognize positive curvatures, is involved the biogenesis of filopodia (79). In a cell-free system for reconstituting actin bundles of filopodia (80), immunodepletion of SNX9 resulted in shorter actin bundles. Moreover, SNX9 localizes to the filopodial tip and shaft in RPE-1 cells (79). The positive role of SNX9 in filopodia formation is dependent on its activity in stimulating N-WASp and Arp2/3 (81), which probably overrides the curvature-sensing function of the N-BAR domain. In terms of the function of SNX9 in T cells, SNX9 was found to interact with WASp, p85, and CD28 to form a signaling complex on endocytic vesicles when T cells are activated by soluble CD3/CD28 antibodies (82). It remains to be determined if SNX9 can regulate microvilli when T cells are activated by surface or bilayer-presented stimuli because stimuli with a physical support could cause different outcomes as compared to those in a soluble format. Previous reports showed that TCR is internalized through endocytosis when T cells are treated with soluble MHC tetramer (83), whereas TCR is sorted into extracellular microvesicles when T cells are activated by supported lipid bilayer (SLB)-presented pMHC (64).
While the BAR proteins sense membrane curvature within the nanometer scale, there are proteins that can also sense a larger length scale. Septins and stage V sporulation protein M (SpoVM) sense positive micron-scale curvatures (84, 85). It was suggested that these nanometer-sized proteins sense micron-scale curvature by polymerization into micro-scale filaments. In contrast, the protein machinery that directly senses micron- or submicron-scale negative curvatures remains to be determined.
What is the cellular function of membrane deformation? Various studies have highlighted the role of membrane curvature in regulating sorting of transmembrane proteins (86–89). Using patterned nanostructure surfaces, Zhao et al. (90) found that the protein machinery mediating clathrin-mediated endocytosis prefers a positive curvature with a radius below 200 nm. Liang et al. (91) discovered that small GTPase Ras senses membrane curvatures in an isoform-dependent manner. One isoform binds to membranes with low curvatures, whereas the other binds to membranes with high curvatures. The effect of local membrane curvature may also influence cell polarization. For many years it had been assumed that cell polarization is induced exclusively by a gradient of a chemoattractant. A recent report revealed that chemical signaling is not sufficient for inducing cell polarization of neutrophils and CD8+ T cells (92). Instead, the authors found that polarization initiates with the formation of curved membranes, which recruits BAR domain protein SRGAP2, activates PI4KA, and results in PtdIns4P polarization. Furthermore, to understand the mechanism by which membrane curvature affects actin-dependent processes, such as endocytosis, focal adhesion maturation, and stress fiber organization, Lou et al. (93) have used patterned nanostructure surfaces to study actin rearrangement. Intriguingly, the authors found that the actin nucleator Arp2/3 and its regulators N-WASP and cortactin are recruited by BAR proteins to membranes with positive curvatures (with radii <200 nm). Consequently, branched actin networks assemble around curved membranes, depleting the monomeric actin pool for assembling stress fibers and mature focal adhesions. Interestingly, members of the formin family, which promote the polymerization of linear F-actin, showed no preferential localization to curved membranes.
The lipid composition of the plasma membrane also influences membrane geometry and protein localization. The size of the lipid headgroups, their charge, as well as the saturation state of acyl chains determines lipid shapes, and consequently the local membrane curvature (66, 94). Lipids with small headgroups such as cardiolipin, phosphatidylethanolamine, ceramide, diacylglycerol, and phosphatic acid induce negative membrane curvatures, whereas lipids with large headgroups like lysophosphatidylcholine and phosphatidylinositol phosphate induce positive curvatures (95). Some of these lipids can also recruit proteins to the membrane. For example, the negatively charged lipids phosphatidylserine and phosphatidylinositol 4,5-bisphosphate recruit positively charged proteins by electrostatic interactions (96–98). Sphingomyelin was found to selectively localize to the microvilli of epithelial cells and to induce microvilli formation through the indirect recruitment of ERM proteins (99). Whether this is also the case in lymphocytes and what the role of sphingomyelinase is in regulating microvilli formation need to be further explored. On the other hand, lipids can mediate the exclusion of proteins from curved membranes. A recent intriguing study from Jung et al. showed that CD45 is excluded out of the tip of microvilli in a cholesterol-dependent manner (53). Although the localization of cholesterol needs to be determined in the context of microvilli, cholesterol was previously reported to be enriched in the negative curved membranes in vitro or in silico (100, 101), where it, together with sphingomyelin, also thickens the membrane (102, 103). The thickened membrane caused by the accumulation of cholesterol was suggested to exclude CD45 of which the transmembrane domain is not long enough to be integrated into the thickened membrane. Depletion of cholesterol by cyclodextrin reduced the exclusion of CD45 from the tip, accompanied by a decrease in the membrane thickness and number of microvilli (53, 104). Besides the contribution from individual lipids, membrane tension, by serving as a physical barrier, can antagonize actin based-protrusion (105).
The composition and organization of the glycocalyx layer, which covers the outer leaflet of the plasma membrane, also contributes to cell morphology and membrane protrusions (106). Mucins are flexible transmembrane glycoprotein polymers within the glycocalyx that are enriched on the surface of many membrane protrusions, such as epithelial microvilli (107). A recent study has demonstrated the role of the mucins in generating forces driving the tubularization of the plasma membrane (108). In contrast, rigid glycoproteins have not shown similar phenomenon as the mucins. In the case of T cells, many cell surface proteins are highly glycosylated, among which CD43 and CD45 are the most abundant glycoproteins (109, 110). Notably, different isoforms of CD45 are expressed at different T cell development stages, and these isoforms differ significantly in their extracellular domain sizes (58, 111). It remains as an interesting question whether these isoforms contribute differently to microvilli formation.
Investigation of the microvilli function can be extremely challenging due to limited tools to specifically manipulate them in cells without perturbing other actin-based processes. Moreover, their small dimensions (r < 200 nm) and unique architecture (i.e., negative membrane curvature viewed from inside of microvilli) hamper the application of traditional in vitro reconstitution approach to the study of microvilli. Nevertheless, methods for studying filopodia, which present similar structural properties as the microvilli, as well as other recent technological advances that enable the accurate shaping of membranes, can be implemented to interrogate microvilli.
In microvilli the actin filaments are organized in parallel bundles (13). Various crosslinkers such as fascin, fimbrin, and espin promote the formation of actin bundles (112–115). While direct evidence is required, these actin crosslinkers can be attractive targets for specifically modulating microvilli shape and density, as demonstrated in filopodia (113). The ERM family, another component involved in microvilli formation, plays a major role in connecting actin cytoskeleton to the cell membrane. Overexpressing a dominant-negative form of ezrin dramatically reduces microvilli formation (24). Yet, a recent finding suggests that the enrichment of ezrin around membrane protrusions is facilitated by I-BAR-domain proteins (116).
Complementing genetic manipulations, pharmacological treatments perturbing membrane composition or cytoskeleton can modulate microvilli in a rapid fashion. Greicius et al. (104) reported a decrease in microvilli density by depleting cholesterol using cyclodextrin. A similar effect could be achieved by using the actin depolymerizing toxin LatA (13, 19). However, both drugs are expected to affect the whole membrane structure, actin network, and surface presence of many signaling proteins, all of which may complicate the interpretation of the results regarding microvilli-specific functions. Recently, an inhibitor of the crosslinker protein fascin has been identified, which could be potentially used to manipulate microvilli. This inhibitor blocks the activity of fascin to bundle actin filaments in vitro, and filopodial formation in multiple cell lines. Furthermore, it blocks cancer cell metastasis, potentially by inhibiting filopodia formation (117).
In vitro assays have been developed to isolate the effect of membrane curvature from complex cellular environment. Yet, while methods to generate positive membrane curvature are well established, it is not the case with negative membrane curvatures, especially in the range of microvillus sizes (r < 200 nm). In one approach, a giant unilamellar vesicle (GUV) is held by a micropipette at one side and pulled, on the other side, by a polystyrene bead holding by optical traps (Figure 4A). A membrane nanotube can be generated with controlled radii, ranging from 7 to 100 nm, by adjusting the micropipette pressure (118). Similarly, an optical trap has been used to pull short tethers (r < 100 nm) from the cell membrane (119). Meanwhile, these manipulations are technically challenging and may be time-consuming. GUVs are also sensitive to osmotic changes and therefore can bring difficulty to long-duration experiments. Alternative approaches to study negative curvature employ substrates that serve as a mold to induce membrane curvature on artificial membranes and cells (Figure 4B). Focused ion beam has been applied to etch an array of invaginations with a radius of 100 nm on a glass surface (120). The fabricated substrate can then be covered with SLB to mimic the highly curved membranes in microvilli. Another promising means to induce membrane curvature is by using nanofabricated substrates (90, 121). Yet, the diffusion of membrane components may be affected by the substrate (122, 123), which should be carefully evaluated beforehand.
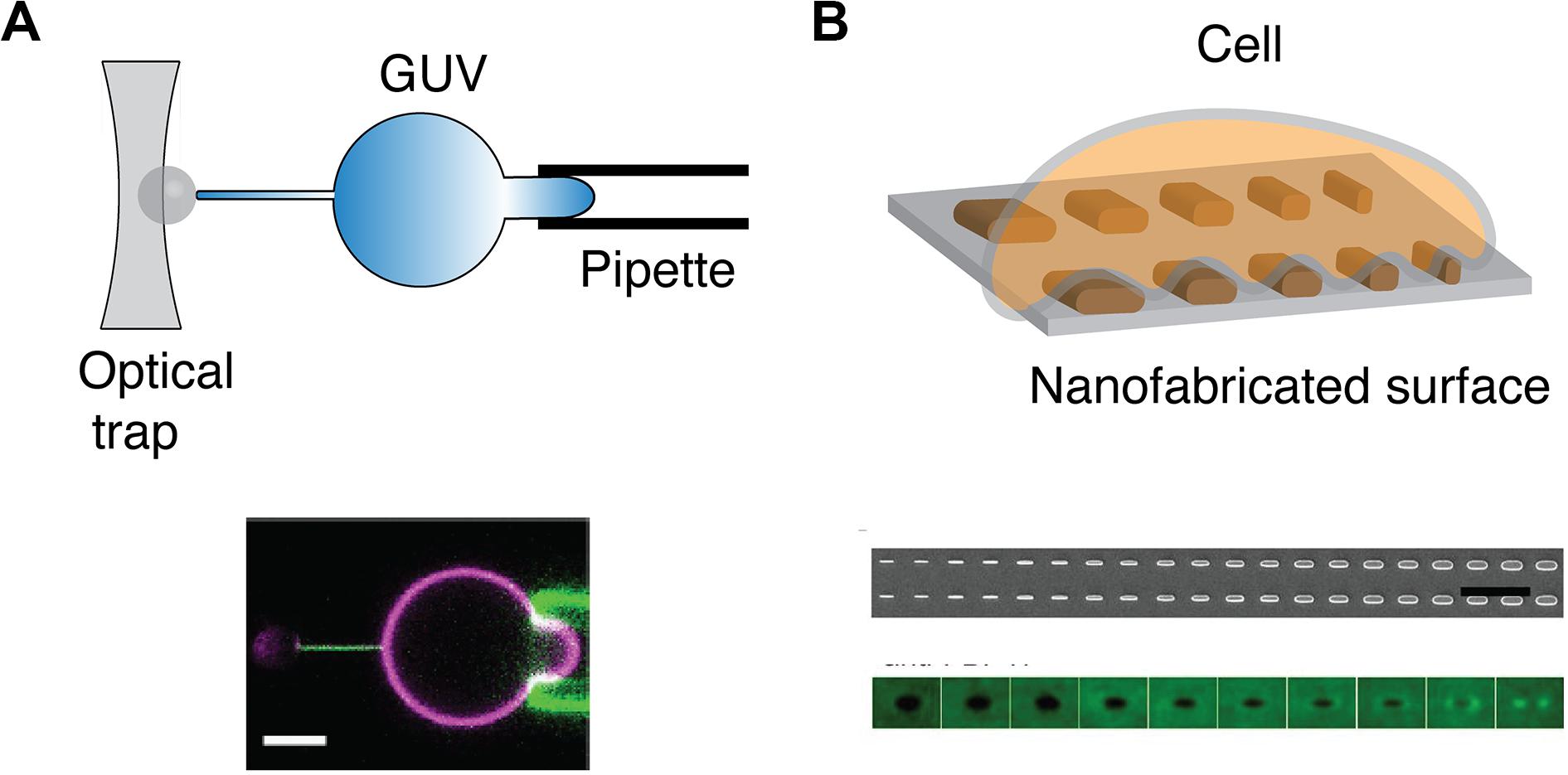
Figure 4. Methods to physically manipulate membrane curvature. (A) Schematic of optical trap that is used to pull a thin nanotube from GUV held by a pipette (top). Confocal microscopy reveals that GFP-IRSp53 BAR protein localizes to nanotube pulled from GUV (magenta) that is held by a pipette (bottom). Scale bar: 5 μm. Reproduced from Prévost et al. (118) licensed under Creative Commons (CC BY 4.0). (B) Schematic of a cell on a nanofabricated surface with structures of different radii (top). Scanning electron microscopy microfabricated of nanofabricated chip with a gradient nanobar array with a variable width from 100 to 1,000 nm (100 nm increment; bar length: 2 μm) (middle). The averaged nanobar images of anti-FBP17 immunostaining for 10 different nanobar widths. FBP17 localizes to positively curved structures with a width <400 nm (bottom). Reproduced from (93). Copyright 2019 National Academy of Sciences.
Looking forward, significant questions remain to be addressed in terms of the mechanism and signaling function of T cell microvilli:
(1) What are the mechanisms that regulate microvilli formation and dynamics? Besides identifying the key protein and lipid components regulating microvilli, it will also be necessary to understand the relationship between microvilli and other membrane structures, for example, the recently identified CD2 Corolla which seems to be devoid of microvilli (124).
(2) How do microvilli regulate the localization and oligomerization state of proteins and lipids? Phase separation, or the formation of liquid-like microclusters, emerges as a new principle in regulating TCR signaling (125, 126). The unique membrane topology in microvilli could play an important role in regulating the assembly of signaling microclusters. On the other hand, microvilli could bring proteins physically close even if there are no direct interactions between those proteins. Future co-localization studies should be best performed in the context of microvilli to understand the exact nature of the entities that are examined. In addition, many other tiny and transient proteo-lipid nanodomains have been identified on the plasma membrane (127–130). It remains as an open question on the relationship between these structures and the microvilli-localized proteins.
(3) How do microvilli modulate chemical reactions? The src-family kinase Lck has been shown to be enriched in microvilli (22). Future studies are expected to reveal the microvilli localization or even sub-microvilli localization (tip, side, or base) of other enzymes in the TCR signaling (e.g., PLCγ1, ZAP70, SHP1, CBL), together with their corresponding substrates. Elegant reconstitution approaches will be needed to recapitulate the essential physical and chemical environment of microvilli to understand how curved membranes in microvilli affect the specific activity of kinases, phosphatases, and lipases.
(4) How do microvilli regulate TCR signal transduction? As protrusive structures that search the surrounding space and make contacts with the APCs, microvilli are constantly experiencing mechanical forces from the environment (131). These forces may be involved in the regulation of cell recognition and calcium flux, as found in the microvillar photoreceptor cells (132). Moreover, TCRs on the tip of the microvilli receive stimuli from APCs. Meanwhile, what happens after antigen recognition remains unclear. How is signal transduced from the tip of microvilli to the cell body? Do microvilli participate in kinetic proof-reading since microvilli are enriched with proteins mediating multiple steps along the TCR pathway? Do microvilli serve as a signaling unit that integrates signals from TCR and co-receptors first before sending them to the cell body, or are different signals transduced individually across the microvilli? With these questions being addressed, the current map of T cell membrane signaling is likely to be significantly expanded from a 2D surface to a 3D world.
RO and XS conceived and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
XS was supported by the American Cancer Society Institutional Research Grant, the Charles H. Hood Foundation Child Health Research Awards, the Andrew McDonough B+ Foundation Research Grant, the Gilead Sciences Research Scholars Program in Hematology/Oncology, and the Rally Foundation a Collaborative Pediatric Cancer Research Awards Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Su lab for fruitful discussions.
1. Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem Sci. (2012) 37:526–33. doi: 10.1016/j.tibs.2012.09.001
2. Jarsch IK, Daste F, Gallop JL. Membrane curvature in cell biology: an integration of molecular mechanisms. J Cell Biol. (2016) 214:375–87. doi: 10.1083/jcb.201604003
3. Pettmann J, Santos AM, Dushek O, Davis SJ. Membrane ultrastructure and T cell activation. Front Immunol. (2018) 9:1263. doi: 10.3389/fimmu.2018.02152
4. Liu S, Xiong X, Zhao X, Yang X, Wang H. F-BAR family proteins, emerging regulators for cell membrane dynamic changes-from structure to human diseases. J Hematol Oncol. (2015) 8:47. doi: 10.1186/s13045-015-0144-2
5. Molina IJ, Kenney DM, Rosen FS, Remold-O’Donnell E. T cell lines characterize events in the pathogenesis of the Wiskott-Aldrich syndrome. J Exp Med. (1992) 176:867–74. doi: 10.1084/jem.176.3.867
6. Varkey J, Isas JM, Mizuno N, Jensen MB, Bhatia VK, Jao CC, et al. Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J Biol Chem. (2010) 285:32486–93. doi: 10.1074/jbc.m110.139576
7. Polliack A, Lampen N, Clarkson BD, Harven ED, Bentwich Z, Siegal FP, et al. Identification of human B and T lymphocytes by scanning electron microscopy. J Exp Med. (1973) 138:607–24. doi: 10.1084/jem.138.3.607
8. Gorelik J, Shevchuk AI, Frolenkov GI, Diakonov IA, Lab MJ, Kros CJ, et al. Dynamic assembly of surface structures in living cells. Proc Natl Acad Sci USA. (2003) 100:5819–22. doi: 10.1073/pnas.1030502100
9. Sauvanet C, Wayt J, Pelaseyed T, Bretscher A. Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu Rev Cell Dev Biol. (2015) 31:593–621. doi: 10.1146/annurev-cellbio-100814-125234
10. Fisher PJ, Bulur PA, Vuk-Pavlovic S, Prendergast FG, Dietz AB. Dendritic cell microvilli: a novel membrane structure associated with the multifocal synapse and T-cell clustering. Blood. (2008) 112:5037–45. doi: 10.1182/blood-2008-04-149526
11. Elsaesser R, Paysan J. The sense of smell, its signalling pathways, and the dichotomy of cilia and microvilli in olfactory sensory cells. BMC neuroscience. (2007) 8(Suppl. 3):S1. doi: 10.1186/1471-2202-8-s3-s1
12. Mooseker MS, Tilney LG. Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol. (1975) 67:725–43. doi: 10.1083/jcb.67.3.725
13. Majstoravich S, Zhang J, Nicholson-Dykstra S, Linder S, Friedrich W, Siminovitch KA, et al. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require Wiskott-Aldrich syndrome protein (WASp) for their morphology. Blood. (2004) 104:1396–403. doi: 10.1182/blood-2004-02-0437
14. Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. (1992) 119:1219–43. doi: 10.1083/jcb.119.5.1219
15. Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. (2003) 160:409–21. doi: 10.1083/jcb.200210174
16. Hirokawa N, Tilney LG, Fujiwara K, Heuser JE. Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol. (1982) 94:425–43. doi: 10.1083/jcb.94.2.425
17. Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. (2007) 9:1110–21. doi: 10.1038/ncb1007-1110
18. Alexander E, Sanders S, Braylan R. Purported difference between human T-and B-cell surface morphology is an artefact. Nature. (1976) 261:239–41. doi: 10.1038/261239a0
19. Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci USA. (2016) 113:E5916–24. doi: 10.1073/pnas.1605399113
20. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science. (2017) 356:eaal3118. doi: 10.1126/science.aal3118
21. Kim H-R, Mun Y, Lee K-S, Park Y-J, Park J-S, Park J-H, et al. T cell microvilli constitute immunological synaptosomes that carry messages to antigen-presenting cells. Nat Commun. (2018) 9:3630. doi: 10.1038/s41467-018-06090-8
22. Ghosh S, Bartolo VD, Tubul L, Shimoni E, Kartvelishvily E, Dadosh T, et al. Dependent assembly of T cell receptor signaling and co-stimulatory molecules on microvilli prior to activation. Cell Rep. (2020) 30:3434–47.e6. doi: 10.1016/j.celrep.2020.02.069
23. Westerberg L, Greicius G, Snapper SB, Aspenström P, Severinson E. Cdc42, Rac1, and the Wiskott-Aldrich syndrome protein are involved in the cytoskeletal regulation of B lymphocytes. Blood. (2001) 98:1086–94. doi: 10.1182/blood.v98.4.1086
24. Brown MJ, Nijhara R, Hallam JA, Gignac M, Yamada KM, Erlandsen SL, et al. Chemokine stimulation of human peripheral blood T lymphocytes induces rapid dephosphorylation of ERM proteins, which facilitates loss of microvilli and polarization. Blood. (2003) 102:3890–9. doi: 10.1182/blood-2002-12-3807
25. Parameswaran N, Matsui K, Gupta N. Conformational switching in ezrin regulates morphological and cytoskeletal changes required for B cell chemotaxis. J Immunol. (2011) 186:4088–97. doi: 10.4049/jimmunol.1001139
26. Nijhara R, van Hennik PB, Gignac ML, Kruhlak MJ, Hordijk PL, Delon J, et al. Rac1 mediates collapse of microvilli on chemokine-activated T lymphocytes. J Immunol. (2004) 173:4985–93. doi: 10.4049/jimmunol.173.8.4985
27. Faure S, Salazar-Fontana LI, Semichon M, Tybulewicz VLJ, Bismuth G, Trautmann A, et al. ERM proteins regulate cytoskeleton relaxation promoting T cell-APC conjugation. Nat Immunol. (2004) 5:272–9. doi: 10.1038/ni1039
28. Sasaki M, Sugimoto K, Mori T, Karasawa K, Oshimi K. Effective treatment of a refractory hairy cell leukemia variant with splenic pre-irradiation and alemtuzumab. Acta Haematol. (2008) 119:48–53. doi: 10.1159/000115785
29. Tabata R, Tabata C, Iwama H, Yasumizu R, Kojima M. CD27-positive hairy cell leukemia-Japanese variant. Virchows Arch. (2016) 468:375–9. doi: 10.1007/s00428-015-1881-x
30. Machii T, Yamaguchi M, Inoue R, Tokumine Y, Kuratsune H, Nagai H, et al. Polyclonal B-cell lymphocytosis with features resembling hairy cell leukemia-Japanese variant. Blood. (1997) 89:2008–14.
31. Polliack A, Tadmor T. Surface topography of hairy cell leukemia cells compared to other leukemias as seen by scanning electron microscopy. Leuk Lymphoma. (2011) 52(Suppl. 2):14–7. doi: 10.3109/10428194.2011.565095
32. Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. (1998) 8:1347–56. doi: 10.1016/s0960-9822(98)00015-3
33. Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. (2001) 3:76–82. doi: 10.1038/35050590
34. Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. (2005) 12:26–31. doi: 10.1038/nsmb870
35. Dayel MJ, Mullins RD. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. (2004) 2:E91. doi: 10.1371/journal.pbio.0020091
36. Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA. (2011) 108:E472–9. doi: 10.1073/pnas.1100236108
37. Urbanek AN, Smith AP, Allwood EG, Booth WI, Ayscough KR. A novel actin-binding motif in Las17/WASP nucleates actin filaments independently of Arp2/3. Curr Biol. (2013) 23:196–203. doi: 10.1016/j.cub.2012.12.024
38. Kenney D, Cairns L, Remold-O’Donnell E, Peterson J, Rosen FS, Parkman R. Morphological abnormalities in the lymphocytes of patients with the Wiskott-Aldrich syndrome. Blood. (1986) 68:1329–32.
39. Gallego MD, Santamaría M, Peña J, Molina IJ. Defective actin reorganization and polymerization of Wiskott-Aldrich T cells in response to CD3-mediated stimulation. Blood. (1997) 90:3089–97.
40. Facchetti F, Blanzuoli L, Vermi W, Notarangelo LD, Giliani S, Fiorini M, et al. Defective actin polymerization in EBV-transformed B-cell lines from patients with the Wiskott-Aldrich syndrome. J Pathol. (1998) 185:99–107. doi: 10.1002/(sici)1096-9896(199805)185:13.0.co;2-l
41. Burns SO, Killock DJ, Moulding DA, Metelo J, Nunes J, Taylor RR, et al. A congenital activating mutant of WASp causes altered plasma membrane topography and adhesion under flow in lymphocytes. Blood. (2010) 115:5355–65. doi: 10.1182/blood-2009-08-236174
42. Westerberg L, Larsson M, Hardy SJ, Fernández C, Thrasher AJ, Severinson E. Wiskott-Aldrich syndrome protein deficiency leads to reduced B-cell adhesion, migration, and homing, and a delayed humoral immune response. Blood. (2005) 105:1144–52. doi: 10.1182/blood-2004-03-1003
43. Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. (2008) 65:904–22. doi: 10.1002/cm.20312
44. Carpentier JL, Obberghen EV, Gorden P, Orci L. Surface redistribution of 125I-insulin in cultured human lymphocytes. J Cell Biol. (1981) 91:17–25. doi: 10.1083/jcb.91.1.17
45. Hasslen SR, von Andrian UH, Butcher EC, Nelson RD, Erlandsen SL. Spatial distribution of L-selectin (CD62L) on human lymphocytes and transfected murine L1-2 cells. Histochem J. (1995) 27:547–54.
46. von Andrian UH, Hasslen SR, Nelson RD, Erlandsen SL, Butcher EC. A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell. (1995) 82:989–99. doi: 10.1016/0092-8674(95)90278-3
47. Berlin C, Bargatze RF, Campbell JJ, von Andrian UH, Szabo MC, Hasslen SR, et al. alpha 4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell. (1995) 80:413–22. doi: 10.1016/0092-8674(95)90491-3
48. Singer II, Scott S, Kawka DW, Chin J, Daugherty BL, DeMartino JA, et al. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J Virol. (2001) 75:3779–90. doi: 10.1128/jvi.75.8.3779-3790.2001
49. Foti M, Phelouzat M-A, Holm A, Rasmusson BJ, Carpentier J-L. p56Lck anchors CD4 to distinct microdomains on microvilli. Proc Natl Acad Sci USA. (2002) 99:2008–13. doi: 10.1073/pnas.042689099
50. Hao J-J, Wang G, Pisitkun T, Patino-Lopez G, Nagashima K, Knepper MA, et al. Enrichment of distinct microfilament-associated and GTP-binding-proteins in membrane/microvilli fractions from lymphoid cells. J Proteome Res. (2008) 7:2911–27. doi: 10.1021/pr800016a
51. Alfonso-Garrido J, Garcia-Calvo E, Luque-Garcia JL. Sample preparation strategies for improving the identification of membrane proteins by mass spectrometry. Anal Bioanal Chem. (2015) 407:4893–905. doi: 10.1007/s00216-015-8732-0
52. Razvag Y, Neve-Oz Y, Sajman J, Yakovian O, Reches M, Sherman E. T Cell activation through isolated tight contacts. Cell Rep. (2019) 29:3506–21.e6. doi: 10.1016/j.celrep.2019.11.022
53. Jung Y, Wen L, Altman A, Ley K. CD45 pre-exclusion from the tips of microvilli establishes a phosphatase-free zone for early TCR triggering. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.05.21.109074
54. Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol. (2006) 7:803–9. doi: 10.1038/ni1369
55. van der Merwe PA, Davis SJ, Shaw AS, Dustin ML. Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin Immunol. (2000) 12:5–21. doi: 10.1006/smim.2000.0203
56. Davis SJ, van der Merwe PA. The structure and ligand interactions of CD2: implications for T-cell function. Immunol Today. (1996) 17:177–87. doi: 10.1016/0167-5699(96)80617-7
57. Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. (2005) 436:578–82. doi: 10.1038/nature03843
58. Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, McColl J, et al. Initiation of T cell signaling by CD45 segregation at “close contacts”. Nat Immunol. (2016) 17:574–82. doi: 10.1038/ni.3392
59. Razvag Y, Neve-Oz Y, Sajman J, Reches M, Sherman E. Nanoscale kinetic segregation of TCR and CD45 in engaged microvilli facilitates early T cell activation. Nat Commun. (2018) 9:732. doi: 10.1038/s41467-018-03127-w
60. Fernandes RA, Ganzinger KA, Tzou JC, Jönsson P, Lee SF, Palayret M, et al. A cell topography-based mechanism for ligand discrimination by the T cell receptor. Proc Natl Acad Sci USA. (2019) 116:14002–10. doi: 10.1073/pnas.1817255116
61. Bakalar MH, Joffe AM, Schmid EM, Son S, Podolski M, Fletcher DA. Size-dependent segregation controls macrophage phagocytosis of antibody-opsonized targets. Cell. (2018) 174:131–42.e13. doi: 10.1016/j.cell.2018.05.059
62. Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol. (2002) 158:1263–75. doi: 10.1083/jcb.200203043
63. Campi G, Varma R, Dustin ML. Actin and agonist MHC–peptide complex–dependent T cell receptor microclusters as scaffolds for signaling. J Exp Med. (2005) 202:1031–6. doi: 10.1084/jem.20051182
64. Choudhuri K, Llodrá J, Roth EW, Tsai J, Gordo S, Wucherpfennig KW, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. (2014) 507:118–23. doi: 10.1038/nature12951
65. Saliba DG, Céspedes-Donoso PF, Bálint Š, Compeer EB, Korobchevskaya K, Valvo S, et al. Composition and structure of synaptic ectosomes exporting antigen receptor linked to functional CD40 ligand from helper T cells. eLife. (2019) 8:247. doi: 10.7554/elife.47528
66. McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. (2005) 438:590–6. doi: 10.1038/nature04396
67. Wolf D, Hofbrucker-MacKenzie SA, Izadi M, Seemann E, Steiniger F, Schwintzer L, et al. Ankyrin repeat-containing N-Ank proteins shape cellular membranes. Nat Cell Biol. (2019) 21:1191–205. doi: 10.1038/s41556-019-0381-7
68. Snead WT, Zeno WF, Kago G, Perkins RW, Richter JB, Zhao C, et al. BAR scaffolds drive membrane fission by crowding disordered domains. J Cell Biol. (2019) 218:664–82. doi: 10.1083/jcb.201807119
69. Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun. (2015) 6:7875. doi: 10.1038/ncomms8875
70. Yuan F, Alimohamadi H, Bakka B, Trementozzi AN, Fawzi NL, Rangamani P, et al. Membrane bending by protein phase separation. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.05.21.109751
71. Frost A, Unger VM, Camilli PD. The BAR domain superfamily: membrane-molding macromolecules. Cell. (2009) 137:191–6. doi: 10.1016/j.cell.2009.04.010
72. Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PKJ, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. (2009) 19:95–107. doi: 10.1016/j.cub.2008.12.029
73. Carman PJ, Dominguez R. BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys Rev. (2018) 10:1587–604. doi: 10.1007/s12551-018-0467-7
74. Lim KB, Bu W, Goh WI, Koh E, Ong SH, Pawson T, et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J Biol Chem. (2008) 283:20454–72. doi: 10.1074/jbc.m710185200
75. Mattila PK, Pykäläinen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. (2007) 176:953–64. doi: 10.1083/jcb.200609176
76. Crespi A, Ferrari I, Lonati P, Disanza A, Fornasari D, Scita G, et al. LIN7 regulates the filopodium- and neurite-promoting activity of IRSp53. J Cell Sci. (2012) 125:4543–54. doi: 10.1242/jcs.106484
77. Kast DJ, Dominguez R. IRSp53 coordinates AMPK and 14-3-3 signaling to regulate filopodia dynamics and directed cell migration. Mol Biol Cell. (2019) 30:1285–97. doi: 10.1091/mbc.e18-09-0600
78. Thomas A, Mariani-Floderer C, López-Huertas MR, Gros N, Hamard-Péron E, Favard C, et al. Involvement of the Rac1-IRSp53-Wave2-Arp2/3 Signaling Pathway in HIV-1 Gag particle release in CD4 T Cells. J Virol. (2015) 89:8162–81. doi: 10.1128/jvi.00469-15
79. Jarsch IK, Gadsby JR, Nuccitelli A, Mason J, Shimo H, Pilloux L, et al. A direct role for SNX9 in the biogenesis of filopodia. J Cell Biol. (2020) 219:931. doi: 10.1083/jcb.201909178
80. Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science. (2010) 329:1341–5. doi: 10.1126/science.1191710
81. Yarar D, Waterman-Storer CM, Schmid SL. SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell. (2007) 13:43–56. doi: 10.1016/j.devcel.2007.04.014
82. Badour K, McGavin MKH, Zhang J, Freeman S, Vieira C, Filipp D, et al. Interaction of the Wiskott-Aldrich syndrome protein with sorting nexin 9 is required for CD28 endocytosis and cosignaling in T cells. Proc Natl Acad Sci USA. (2007) 104:1593–8. doi: 10.1073/pnas.0610543104
83. Weder P, Schumacher TNM, Spits H, Luiten RM. Testing for HLA/peptide tetramer-binding to the T cell receptor complex on human T lymphocytes. Results Immunol. (2012) 2:88–96. doi: 10.1016/j.rinim.2012.04.001
84. Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. (2009) 323:1354–7. doi: 10.1126/science.1169218
85. Bridges AA, Jentzsch MS, Oakes PW, Occhipinti P, Gladfelter AS. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J Cell Biol. (2016) 213:23–32. doi: 10.1083/jcb.201512029
86. Hägerstrand H, Mrówczyńska L, Salzer U, Prohaska R, Michelsen KA, Kralj-Igliè V, et al. Curvature-dependent lateral distribution of raft markers in the human erythrocyte membrane. Mol Membr Biol. (2009) 23:277–88. doi: 10.1080/09687860600682536
87. Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GES, Bassereau P. Membrane shape modulates transmembrane protein distribution. Dev Cell. (2014) 28:212–8. doi: 10.1016/j.devcel.2013.12.012
88. Strahl H, Ronneau S, González BS, Klutsch D, Schaffner-Barbero C, Hamoen LW. Transmembrane protein sorting driven by membrane curvature. Nat Commun. (2015) 6:1–9. doi: 10.1038/ncomms9728
89. Rosholm KR, Leijnse N, Mantsiou A, Tkach V, Pedersen SL, Wirth VF, et al. Membrane curvature regulates ligand-specific membrane sorting of GPCRs in living cells. Nat Chem Biol. (2017) 13:724–9. doi: 10.1038/nchembio.2372
90. Zhao W, Hanson L, Lou, H-Y, Akamatsu M, Chowdary PD, Santoro F, et al. Nanoscale manipulation of membrane curvature for probing endocytosis in live cells. Nat Nanotechnol. (2017) 12:750–6. doi: 10.1038/nnano.2017.98
91. Liang H, Mu H, Jean-Francois F, Lakshman B, Sarkar-Banerjee S, Zhuang Y, et al. Membrane curvature sensing of the lipid-anchored K-Ras small GTPase. Life Sci Alliance. (2019) 2:e201900343. doi: 10.26508/lsa.201900343
92. Ren C, Yuan Q, Braun M, Zhang X, Petri B, Zhang J, et al. Leukocyte cytoskeleton polarization is initiated by plasma membrane curvature from cell attachment. Dev Cell. (2019) 49:206–19.e7. doi: 10.1016/j.devcel.2019.02.023
93. Lou H-Y, Zhao W, Li X, Duan L, Powers A, Akamatsu M, et al. Membrane curvature underlies actin reorganization in response to nanoscale surface topography. Proc Natl Acad Sci USA. (2019) 116:23143–51. doi: 10.1073/pnas.1910166116
94. Hirama T, Lu SM, Kay JG, Maekawa M, Kozlov MM, Grinstein S, et al. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat Commun. (2017) 8:1393. doi: 10.1038/s41467-017-01554-9
95. Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. (2006) 7:9–19. doi: 10.1038/nrm1784
96. Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. (2006) 314:1458–61. doi: 10.1126/science.1134389
97. Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. (2008) 319:210–3. doi: 10.1126/science.1152066
98. Fairn GD, Hermansson M, Somerharju P, Grinstein S. Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat Cell Biol. (2011) 13:1424–30. doi: 10.1038/ncb2351
99. Ikenouchi J, Hirata M, Yonemura S, Umeda M. Sphingomyelin clustering is essential for the formation of microvilli. J Cell Sci. (2013) 126:3585–92. doi: 10.1242/jcs.122325
100. Wang W, Yang L, Huang HW. Evidence of cholesterol accumulated in high curvature regions: implications to the curvature elastic enregy for lipid mixtures. Biophys J. (2007) 92:2319–30. doi: 10.1529/biophysj.106097923
101. Koldsø H, Shorthouse D, Hélie J, Sansom MSP. Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput. Biol. (2014) 10:e1003911. doi: 10.1371/journal.pcbi.1003911
102. Favela-Rosales F, Galván-Hernández A, Hernández-Cobos J, Kobayashi N, Carbajal-Tinoco MD, Nakabayashi S, et al. A molecular dynamics study proposing the existence of statistical structural heterogeneity due to chain orientation in the POPC-cholesterol bilayer. Biophys Chem. (2020) 257:106275. doi: 10.1016/j.bpc.2019.106275
103. Smondyrev AM, Berkowitz ML. Structure of dipalmitoylphosphatidylcholine/cholesterol bilayer at low and high cholesterol concentrations: molecular dynamics simulation. Biophys J. (1999) 77:2075–89. doi: 10.1016/s0006-3495(99)77049-9
104. Greicius G, Westerberg L, Davey EJ, Buentke E, Scheynius A, Thyberg J, et al. Microvilli structures on B lymphocytes: inducible functional domains? Int Immunol. (2004) 16:353–64. doi: 10.1093/intimm/dxh031
105. Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, et al. Mechanism of shape determination in motile cells. Nature. (2008) 453:475–80. doi: 10.1038/nature06952
106. Möckl L. The emerging role of the mammalian glycocalyx in functional membrane organization and immune system regulation. Front Cell Dev Biol. (2020) 8:253. doi: 10.3389/fcell.2020.00253
107. Kesimer M, Ehre C, Burns KA, Davis CW, Sheehan JK, Pickles RJ. Molecular organization of the mucins and glycocalyx underlying mucus transport over mucosal surfaces of the airways. Mucosal Immunol. (2013) 6:379–92. doi: 10.1038/mi.2012.81
108. Shurer CR, Kuo JC-H, Roberts LM, Gandhi JG, Colville MJ, Enoki TA, et al. Physical principles of membrane shape regulation by the glycocalyx. Cell. (2019) 177:1757–70.e21. doi: 10.1016/j.cell.2019.04.017
109. Clark MC, Baum LG. T cells modulate glycans on CD43 and CD45 during development and activation, signal regulation, and survival. Ann N Y Acad Sci USA. (2012) 1253:58–67. doi: 10.1111/j.1749-6632.2011.06304.x
110. Pereira MS, Alves I, Vicente M, Campar A, Silva MC, Padrão NA, et al. Glycans as key checkpoints of T cell activity and function. Front Immunol. (2018) 9:2754. doi: 10.3389/fimmu.2018.02754
111. McCall MN, Shotton DM, Barclay AN. Expression of soluble isoforms of rat CD45. Analysis by electron microscopy and use in epitope mapping of anti-CD45R monoclonal antibodies. Immunology. (1992) 76: 310–7.
112. Yamashiro-Matsumura S, Matsumura F. Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J Biol Chem. (1985) 260:5087–97.
113. Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. (2006) 174:863–75. doi: 10.1083/jcb.200603013
114. Loomis PA, Zheng L, Sekerková G, Changyaleket B, Mugnaini E, Bartles JR. Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J Cell Biol. (2003) 163:1045–55. doi: 10.1083/jcb.200309093
115. Tilney MS, Tilney LG, Stephens RE, Merte C, Drenckhahn D, Cotanche DA, et al. Preliminary biochemical characterization of the stereocilia and cuticular plate of hair cells of the chick cochlea. J Cell Biol. (1989) 109:1711–23. doi: 10.1083/jcb.109.4.1711
116. Tsai F-C, Bertin A, Bousquet H, Manzi J, Senju Y, Tsai M-C, et al. Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner. eLife. (2018) 7:129. doi: 10.7554/elife.37262
117. Huang F-K, Han S, Xing B, Huang J, Liu B, Bordeleau F, et al. Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat Commun. (2015) 6:7465. doi: 10.1038/ncomms8465
118. Prévost C, Zhao H, Manzi J, Lemichez E, Lappalainen P, Callan-Jones A, et al. IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun. (2015) 6:8529. doi: 10.1038/ncomms9529
119. Shi Z, Graber ZT, Baumgart T, Stone HA, Cohen AE. Cell membranes resist flow. Cell. (2018) 175:1769–79.e13. doi: 10.1016/j.cell.2018.09.054
120. Lee I-H, Kai H, Carlson L-A, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci USA. (2015) 112:15892–7. doi: 10.1073/pnas.1518765113
121. Li X, Matino L, Zhang W, Klausen L, McGuire AF, Lubrano C, et al. A nanostructure platform for live-cell manipulation of membrane curvature. Nat Protoc. (2019) 14:1772–802. doi: 10.1038/s41596-019-0161-7
122. Przybylo M, Sıkora J, Humpolíckova J, Benda A, Zan A, Hof M. Lipid diffusion in giant unilamellar vesicles is more than 2 times faster than in supported phospholipid bilayers under identical conditions. Langmuir. (2006) 22:9096–9. doi: 10.1021/la061934p
123. Hsieh C-L, Spindler S, Ehrig J, Sandoghdar V. Tracking single particles on supported lipid membranes: multimobility diffusion and nanoscopic confinement. J Phys Chem B. (2014) 118:1545–54. doi: 10.1021/jp412203t
124. Demetriou P, Abu-Shah E, McCuaig S, Mayya V, Valvo S, Korobchevskaya K, et al. CD2 expression acts as a quantitative checkpoint for immunological synapse structure and T-cell activation. Biorxiv [Preprint]. (2019). doi: 10.1101/589440
125. Lillemeier BF, Mörtelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. (2010) 11:90–6. doi: 10.1038/ni.1832
126. Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. (2016) 352:595–9. doi: 10.1126/science.aad9964
127. Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, Polyakova S, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. (2008) 457:1159–62. doi: 10.1038/nature07596
128. Honigmann A, Mueller V, Ta H, Schoenle A, Sezgin E, Hell SW, et al. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat Commun. (2014) 5:5412. doi: 10.1038/ncomms6412
129. Brameshuber M, Kellner F, Rossboth BK, Ta H, Alge K, Sevcsik E, et al. Monomeric TCRs drive T cell antigen recognition. Nat Immunol. (2018) 19:487–96. doi: 10.1038/s41590-018-0092-4
130. Reth M. Matching cellular dimensions with molecular sizes. Nat Immunol. (2013) 14:765–7. doi: 10.1038/ni.2621
131. Harrison DL, Fang Y, Huang J. T-cell mechanobiology: force sensation, potentiation, and translation. Front Phys. (2019) 7:835. doi: 10.3389/fphy.2019.00045
Keywords: microvilli, actin, membrane curvature, BAR protein, WASp, TCR, super-resolution microscopy, T-cell signaling
Citation: Orbach R and Su X (2020) Surfing on Membrane Waves: Microvilli, Curved Membranes, and Immune Signaling. Front. Immunol. 11:2187. doi: 10.3389/fimmu.2020.02187
Received: 30 May 2020; Accepted: 11 August 2020;
Published: 11 September 2020.
Edited by:
Erdinc Sezgin, Karolinska Institutet (KI), SwedenReviewed by:
Pablo F. Céspedes-Donoso, University of Oxford, United KingdomCopyright © 2020 Orbach and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolei Su, eGlhb2xlaS5zdUB5YWxlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.