
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 02 September 2020
Sec. Comparative Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.02030
This article is part of the Research TopicNovel Insights into Insect Antiviral ImmunityView all 16 articles
Antimicrobial peptides (AMPs) with antiviral activity (antiviral peptides: AVPs) have become a research hotspot and already show immense potential to become pharmaceutically available antiviral drugs. AVPs have exhibited huge potential in inhibiting viruses by targeting various stages of their life cycle. Insects are the most speciose group of animals that inhabit almost all ecosystems and habitats on the land and are a rich source of natural AMPs. However, insect AVP mining, functional research, and drug development are still in their infancy. This review aims to summarize the currently validated insect AVPs, explore potential new insect AVPs and to discuss their possible mechanism of synthesis and action, with a view to providing clues to unravel the mechanisms of insect antiviral immunity and to develop insect AVP-derived antiviral drugs.
The role that insects have played as models in innate immunity research is unquestionable. Since the 1990's, the fruit fly Drosophila melanogaster emerged as an important paradigm of genetic analysis of innate immunity. Outstanding pioneering achievements were awarded the Nobel Prize, which has since greatly stimulated interest in this field (1, 2). Studies in insects initially focused on resistance to bacteria and fungi, and later slowly expanded into antiviral immunity. However, besides the discovery that RNA interference (RNAi) is crucial in insect antiviral immunity, knowledge of other antiviral pathways and antiviral factors is very limited (3–7). In contrast, in mammals, a diverse series of antiviral immune responses including virus recognition, downstream cascade reactions, and production of effectors were gradually unveiled (8–10). In particular, hundreds of interferon-stimulated genes (ISGs), which exert numerous antiviral effector functions, have been identified in multiple vertebrate species (11–15). This raises the question whether antiviral host factors, similar to interferon-stimulated effectors in mammals, also exist in insects.
In insects, antimicrobial peptides (AMPs) are a group of immune proteins that mainly function against bacteria and fungi (16, 17). A considerable number of AMP genes have been identified in Drosophila, the honey bee Apis cerana and the silkworm Bombyx mori (18–20). However, two antiviral screening experiments failed to show that AMPs are a class of antiviral factors in Drosophila (21, 22). Intriguingly, other data in the literature have indicated that AMPs have antiviral function in Drosophila and B. mori (23, 24). On the other hand, it should be kept in mind that the interaction between host and virus is a complex process in which the immune response of the host is counteracted by the immune escape mechanisms of the virus. A recent study found that Kallithea virus (DNA virus of D. melanogaster) gp83 inhibits Toll signaling through the regulation of NF-κB transcription factors (25). The immunosuppression by Kallithea virus infection is also accompanied by the general down-regulation of AMP gene expression (25). Because the action of AMPs may be neutralized by the virus, simple tests cannot decide or exclude whether AMPs have antiviral activity. In fact, AMPs with antiviral activity (antiviral peptides: AVPs) have become a research hotspot and already show considerable potential to become pharmaceutically available antiviral drugs (26). AMPs and AVPs are usually derived from natural sources but they can be readily modified by adding non-natural amino acids or chemical groups to further enhance their stability and activity (27). Insects are an extremely successful and diverse group of animals that produce a wide range of AMPs which also could display potent antiviral activity. Accordingly, a review of insect antibacterial peptides with antiviral activity is considered timely to provide an assessment of the current knowledge as well as to stimulate efforts for the identification of additional insect-derived antiviral AMPs.
Herein, we will summarize the AMPs with antiviral activity reported in the database and literature and we will predict the antiviral activity of insect AMPs through AVP prediction software. This article aims to compile relevant information from insect AVPs as important components of insect antiviral innate immunity and to inspire the development of effective antiviral drugs.
AVPs are considered as a subset of AMPs which act as the first line of defense in many organisms as an innate immune response to viral infection. Compared to a hot field such as the development of antiviral and antitumor drugs in human medicine, the concept of AVP has not appeared often in the field of insect research, although the idea appeared more than 10 years ago (28, 29). With increasing interest for natural AMPs as potential new drugs, many databases, such as APD (30), AVPdb (31) and ParaPep (32), have been developed to centralize information about AMPs. Among AMP databases, a few databases integrate the AMPs with antiviral activity such as APD (30), AVPdb (31), DRAMP 2.0 (33), and dbAMP (34). The information incorporated in DRAMP 2.0 and dbAMP is relatively new and complete. The advantage of AVPdb is that it summarizes AVPs according to various anti-virus mechanisms. In addition, software for AVP prediction has been developed, e.g., AVPpred (35), AntiVPP 1.0 (36), and Meta-iAVP (37). Based on a series of concepts relevant to insect AVP research, we have cataloged several user-friendly and recently released databases and websites that are suitable for insect AVP research (Table 1). The data of known AVPs and prediction methods in this article also come from these databases and websites.
The dbAMP was recently created as a useful resource for accumulating synthetic and natural AMPs from public AMP databases and scientific literature (34). In the dbAMP database, a total of 305 AVPs and 596 insect AMPs are collected (Figure 1A). Nine insect AVPs were obtained from the intersection of these two data sets in the dbAMP (Figure 1A). DRAMP 2.0 is an open-access comprehensive database containing general, patented and clinical AMPs (33). From this database, we identified 8 insect AVPs from a total 214 AVPs (Figure 1B). Integrating the insect AVPs information from the dbAMP and DRAMP 2.0 database, we obtained a total of 13 insect AVPs, which are shown in Figure 1C. Among hundreds of insect AMPs in the database, only 13 were associated with antiviral activity, which suggests that the research on insect AVP is still in in its infancy and requires more data. It can be assumed that many insect AMPs need to be explored for potential antiviral activity. Thus, the 596 insect AMPs in dbAMP database were further used to predict antiviral activity using Meta-iAVP (37). Unexpectedly, 392 insect AMPs were predicted as AVPs (predicted value >0.5) (Supplementary File 1). These predicted insect AVPs originated from B. mori, Galleria mellonella, Aedes aegypti, Pachycondyla goeldii (Ponerine ant), Manduca sexta, D. melanogaster, Danaus plexippus, Anopheles gambiae, Apis mellifera and others (Figure 1D). Based on this evidence, we have reason to believe that insect AMPs are a potential source for identification of AVPs, which is worthy of more in-depth study. However, at present, there is no special insect AMP database that can incorporate the latest review articles of insect AVPs. The existing databases continue to have omissions unless the information also becomes curated by professional insect researchers.
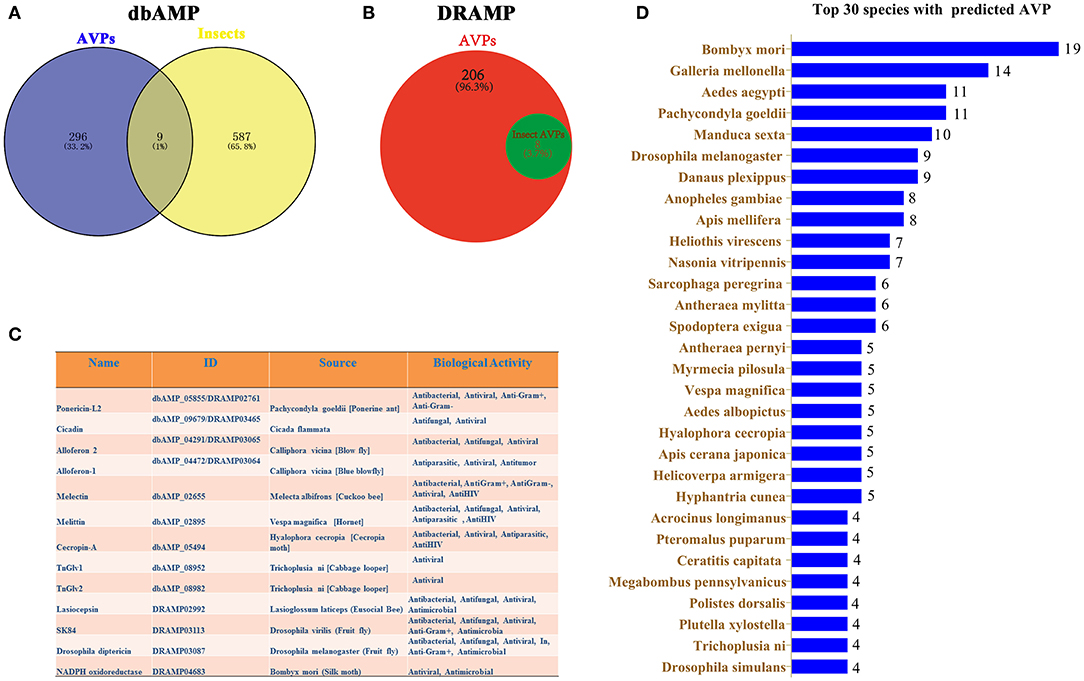
Figure 1. Prediction of insect AVPs from published databases. (A) The intersection between AMPs with potential antiviral activity and insect AMPs in the dbAMP database. (B) Insect AMPs with predicted antiviral activity in the DRAMP 2.0 database. (C) Integration of information on predicted insect AVPs from the dbAMP and DRAMP 2.0 databases. (D) Top 30 insects with predicted AVPs that were identified using Meta-iAVP.
Although the study of insect AVP as an important part of insect antiviral research was promoted more than 10 years ago (29), the available literature is still very limited. Surprisingly, until recently, few insect-derived AMPs were reported with documented antiviral activity. As shown in Table 2, ten insect AVPs were found to be involved in the antiviral response and the antiviral action was directed against both mammalian and insect viruses.
Cecropin-A was one of the first animal antimicrobial peptides to be isolated and fully characterized from the hemolymph of the moth Hyalophora cecropia (43, 44). Subsequent research confirmed that Cecropin-A has inhibitory activity against human immunodefciency virus 1 (HIV-1; Retroviridae), herpes simplex virus 1 and 2 (HSV; Herpesviridae) and against the arenavirus Junin virus (JV) (39, 40).
Melittin belongs to the class of bee venom-derived AVPs and was isolated from the honeybee A. mellifera (45). This AVP was also tested against HSV, HIV-1 and JV, showing inhibition of viral replication for all tested viruses (40, 46). In addition, melittin also curbs infectivity of a diverse array of viruses including Coxsackie Virus and other enteroviruses (Picornaviridae), Influenza A viruses (Orthomyxoviridae), Respiratory Syncytial Virus (RSV; Pneumoviridae), Vesicular Stomatitis Virus (VSV; Rhabdoviridae) and the plant virus tobacco mosaic virus (TMV; Virgaviridae) (47). More information about the antiviral activity of melittin can be found in a review by Memariani et al. (47).
The insect AMP alloferon 1 and 2, derived from the hemolymph of blow fly Calliphora vicina, showed antiviral activity against influenza virus A and influenza virus B (28). Additional research also found that alloferon 1 inhibits human herpes virus type 1 (HHV-1; Herpesviridae) and analogs were active against coxsackievirus in vitro using cell lines (48, 49). Despite the mechanism of antiviral activity of alloferon is still unknown, Alloferon 1 and its analogs are considered as promising candidates for the design of new AVPs (50).
The antiviral compound N-myristoylated-peptide containing only six amino acids with molecular weight of 916 Da was purified from larval hemolymph of the tobacco budworm Heliothis virescens (41). Insect myristoylated-peptide has been confirmed to be effective against HIV-1 and HSV-1 (41). The N-terminus of N-myristoylated-peptide contains the fatty acid myristoyl and the C-terminus contains histidine with two methyl groups giving the histidine a permanent positive charge (41). The structure of the antiviral compound resembles the “myristate plus basic” motif present in particular viral proteins for binding to the cytoplasmic side of the plasma membrane to initiate virus assembly and budding from a host cell (41). It is speculated that the N-myristoylated-peptide is therefore able to specifically block or inhibit viruses like HIV-1 and HSV-1 that use this motif for exit from a host cell (41, 51).
Gloverin, a small cationic antibacterial protein, has been isolated from the hemolymph of various insects such as the giant silk moth Hyalophora (52) and the cabbage looper Trichoplusia ni (53). Two T. ni gloverin peptides named TnGlv1 and TnGlv2 showed resistance to the budded virus (BV) of Autographa californica multiple nucleopolyhedrovirus (AcMNPV; Baculoviridae) (42). The antiviral mechanism was speculated to be based on the accumulation of gloverin on the surface of BVs that may cause membrane strain or formation of pores that disrupt the BV envelope (42).
Two Drosophila AMP coding genes, diptericin B (dptB) and attacin C (attC), are upregulated in transgenic flies expressing a Sindbis virus (SINV) replicon. Silencing their expression led to a significant increase in SINV titers, suggesting that dptB and attC involved in Drosophila antiviral response to SINV (23). However, their mechanism of action remains to be elucidated.
Lysozyme is a ubiquitous peptide that is widely distributed in animals, plants, bacteria and viruses (54). The antibacterial, immunomodulatory and antiviral functions of lysozyme are well-known in vertebrates (55–57). More than fifty lysozyme genes have been identified from several insects (58), but the antiviral activity of insect lysozymes has not been widely investigated. In a recent study, the overexpression of C-lysozyme of B. mori could reduce B. mori nucleopolyhedrovirus (BmNPV) production and progeny virus virulence in vivo and in vitro (24). Further research is required to elucidate the antiviral mechanism of lysozyme peptides.
Insects are the most speciose group of animals that inhabit almost all ecosystems and habitats on the land (17, 59). Although insects are a rich source of natural AMPs (17), only few insect AMPs have been confirmed with antiviral activity (Figure 1C, Table 2). In this study we have predicted 392 potential AVPs from 596 insect AMPs in the dbAMP database (Figure 1D, Supplementary File 1). This information may stimulate researchers to carry out in-depth and extensive research on the activity of the predicted insect AVPs. Insects, especially D. melanogaster, has been widely used as model for the study of innate immunity and microbial pathogens and for assessing the in vivo efficacy of antimicrobial agents (60). The silkworm and honeybee are well-known representative economic insects. In the following section, we will elaborate on potential AVPs in the fruit fly D. melanogaster, the two honeybee species A. mellifera and A. cerana and the silkworm B. mori.
In general, seven well-characterized families including 21 inducible AMP/AMP-like genes have been identified in Drosophila (61, 62). The functions of Drosophila AMPs are not only involved in host defense, but expand also to gut microbiota homeostasis, tumor control, lifespan regulation and neurological processes (62, 63). However, to our knowledge, only two Drosophila AMPs, attC and dptB, have been reported to have antiviral function (23). Since the first animal AMP was discovered in insects (44), D. melanogaster has emerged as a powerful model for their characterization. Unfortunately, the research on antiviral immunity involving Drosophila AMPs has not received enough attention. After downloading the latest updated Drosophila AMP/AMP-like genes (including lysozyme) and their corresponding peptides from the NCBI database, their antiviral activity was predicted using Meta-iAVP (37). For AMP genes for which the mature peptide sequence was not determined, SignalP-5.0 was employed to predict the signal peptide and mature peptide (38).
Following this procedure, as shown in Table 3, a total of 23 potential AVPs were identified in D. melanogaster. We further analyzed these potential AVPs for their induction by viral infection in published transcriptome studies. Expression of Defensin, Cecropin A1, Cecropin B, Andropin, Drosocin, Drosomycin, Metchnikowin, Lysozyme S, Attacin-B, Attacin-C, Diptericin A, and Lysozyme X was found to be induced after viral infection in cell lines or adult flies (Table 3). Screening of transcriptome data for identification of key viral host factors is based on this concept (13). However, viruses may also interfere with the expression of antiviral factors as an immune escape strategy. Determination of antiviral activity based by induction of expression during viral infection is only indicative and cannot be considered as conclusive. But for screening of antiviral genes it can turn out to be a simple and effective method. Therefore, AMPs/AVPs that are up-regulated by a specific virus may be relatively reliable candidate host antiviral factors, for which further verification experiments have to be performed. It should also be noted that dptB has been shown to inhibit SINV replication (23), but it is not among the predicted candidate AVPs (Table 3). Thus, a strategy that screens virus-inducible genes clearly will not identify all potential AVPs.
In addition, some non-classical AMPs such as Bomanins (72), Daishos (73) and Listericin (74) in Drosophila have also attracted our attention. An effector peptide family encoded by twelve Bomanin (Bom) genes has been found to be essential for effective Drosophila Toll-mediated immune responses (72). Daisho peptides, a new class of innate immune effectors in Drosophila, were recently found to have humoral activity against a set of filamentous fungi (73). Currently, these Drosophila peptides have not been confirmed to have antiviral activity. Using Meta-iAVP (37) prediction, we found that BomS1, BomS4, BomS6, BomT1, BomBc2, and Listericin have potential AVPs activity (Supplementary File 2).
Honeybees are important plant pollinators in both natural and agricultural ecosystems (75). Through pollination of flowering plants, honeybees do not only help to maintain biodiversity but in addition they also supply commodities such as honey, royal jelly, propolis (bee glue), pollen and wax. Viruses are significant threats to the health and well-being of the honeybee (76). Due to the abundance and economic importance of the honeybee, research on the interaction with bee viruses has received a lot of research interest. Honeybee antiviral defense mechanisms include RNAi, endocytosis, melanization, encapsulation, autophagy, pathogen-associated molecular pattern (PAMP)-triggered signal transduction cascades, and generation of reactive oxygen species (7, 77). There is currently no evidence that AMPs are involved in the antiviral response of honeybees (7, 77). However, melittin, the principal constituent in the venom of A. mellifera, has been demonstrated to be effective against the infectivity of a diverse array of mammalian viruses such as HIV and HSV (47). Venom-derived AMPs may not play a role in the antiviral response of its host, but the results of the antiviral experiments in vitro are an important reference of which the significance is not clear yet.
Following infection by pathogens, AMPs of four families comprising apidaecins (78), abaecins (79), hymenoptaecins (80), and defensins (81) are synthesized, representing a broad spectrum of antimicrobial activity in the haemolymph. Detailed comparison of these four AMP gene families between A. mellifera and A. cerana revealed that there are many similarities in the number and amino acid composition of the peptides in the abaecin, defensing, and apidaecin families, while many more hymenoptaecin peptides are found in A. cerana than in A. mellifera (19). Compared to A. mellifera that has a longer history of domestication, selection on A. cerana has favored the generation of more variable AMPs as protection against pathogens (19).
Using the predictive tools of Meta-iAVP (37), a total of 7 and 16 AVPs were obtained from A. mellifera and A. cerana, respectively (Table 4). Potential AVP genes of A. mellifera such as defensin 1, defensin 2, abaecin, apisimin, hymenoptaecin, and lysozyme 3 were found to be up-regulated after infection with viruses such as Deformed wing virus (DWV), Sacbrood virus (SBV), black queen cell virus (BQCV), and Israeli acute paralysis virus (IAPV) in transcriptome data (Table 4). Almost all honeybee transcriptome studies that analyze virus infection are restricted to A. mellifera while little related research has been conducted on A. cerana. Recent research found that in A. cerana the predicted AVP genes abaecin and hymenoptaecin were significantly upregulated by Chinese Sacbrood virus (CSBV) infection (85). These potential AVPs, which are up-regulated by a specific honeybee virus, are important leads for future research on the antiviral immunity of honeybee AMPs.
The domestic silkworm B. mori, is an important lepidopteran insect of high scientific and economic value (86). Like in apiculture, the viral disease can cause enormous economic loss in sericulture (87). For viral diseases of silkworm, currently there is no effective treatment. Although there exist specific strains of silkworm that are resistant to some viruses, the specific mechanism is unclear (88–90). Like other insects, RNAi was considered as the major defense strategy against viral infections in B. mori (91). However, the antiviral innate immune response of silkworm has not been systematically studied although specific antiviral molecules such as PP2A (92), BmSTING (93), BmAtlastin-n (94), BmNOX (95), Bmlipase-1 (96), were identified. In a review article the involvement of AMPs in the antiviral response of silkworm was claimed (6), but in fact very few specific cases of antiviral activity of silkworm AMPs are known, an exception being a recent article on inhibition of BmNPV by lysozyme (24). Interestingly, a study reported that B. mori peptidoglycan recognition protein S2 (BmPGRP-S2) overexpression could activate the Imd pathway and induce AMP upregulation, enhancing silkworm antiviral resistance (97).
Following the publication of the genome of the silkworm (86), 35 silkworm AMP genes were identified based on the silkworm genome sequence and expressed sequence tags databases (20). These silkworm AMP genes belong to six families including cecropins, moricins, gloverins, attacins, enbocins, and lebocin (20). Following analysis of updated AMP gene data in the NCBI database, 21 potential silkworm AVPs (Table 5) were obtained using Meta-iAVP prediction (37). Among these potential AVP genes, gloverin-2, gloverin-3, lebocin, attacin 1, and lysozyme have been found to be induced by BmNPV infection in both resistant and susceptible silkworms (98, 100). It is worth noting that the expression of the potential AVP gene gloverin-4 was significantly up-regulated only in BmNPV-infected resistant silkworm, while no changes were found in the BmNPV-infected susceptible silkworm and BmN cells, further suggesting that gloverin-4 is an AVP against BmNPV infection (98). The expression of the potential AVP gene cecropin A and cecropin B also tended to be up-regulated during infection with B. mori cytoplasmic polyhedrosis virus (BmCPV), but expression levels were too low to be considered as biologically important (99). Moreover, it is curious that although many omics data related to silkworm virus infection have been published, no more clues were obtained about the involvement of AMPs in the defense against B. mori bidensovirus (BmBDV), BmNPV and BmCPV infection (101–105).
Universally, after the virus invades the host, the host will initiate a recognition mechanism and induce a downstream antiviral cascade reaction. In vertebrates, during various viral infections, virus-associated PAMPs are recognized by pathogen recognition receptors (PRRs) such as Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), NOD-like receptors (12), interferon-γ-inducible protein 16 (IFI16), AIM2 (absent in melanoma 2) and cyclic GMP-AMP synthase (cGAS) that subsequently lead to the activation of inflammatory cytokines and chemokines as well as interferon (IFN) and ISG production through a cascade reaction (106). However, similar antiviral response systems have not been systematically studied in insects. At present, we have very limited knowledge of how insects recognize virus invasion and initiate cascade reactions to exert antiviral functions.
In insects, a number of actual and potential PRRs such as TLRs, peptidoglycan recognition proteins (PGRPs), Gram-negative bacteria-binding proteins (GNBPs), scavenger receptors (SRs), thioester-containing proteins (TEPs) and lectins have been identified (107, 108). Unfortunately, there is currently no evidence that any of the above-mentioned PRRs are involved in insect virus recognition, with the exception of the nucleic acid sensor Dicer-2 that can act as a PRR of double-stranded RNA in parallel to the RNAi pathway (107). Recently, B. mori cGAMP and PGRP2 were confirmed to be involved in host responses to BmNPV (93, 109). In Drosophila, Toll, IMD and JAK/STAT pathway may be involved in antiviral immunity (4, 65, 110). In addition, JAK/STAT pathway could also be activated by challenge with BmNPV and BmBDV (111). The classical innate immune pathways are also transcriptionally induced during pathogenic infection of Bm5 cells with RNA virus (112). However, the insect PRRs for viral recognition and signaling pathway activation have not been fully elucidated. Thus, there is currently no exact mechanism identified for the generation of AVPs and more in-depth research is needed. Based on evidence obtained in vertebrate (mammalian) systems, we can make the hypothesis that insect viral PAMPs are recognized by specific PRRs located in the cell membrane or cytoplasm of hemocytes, epithelia or fat body which then triggers downstream signaling cascades for the production of AVPs (Figure 2).
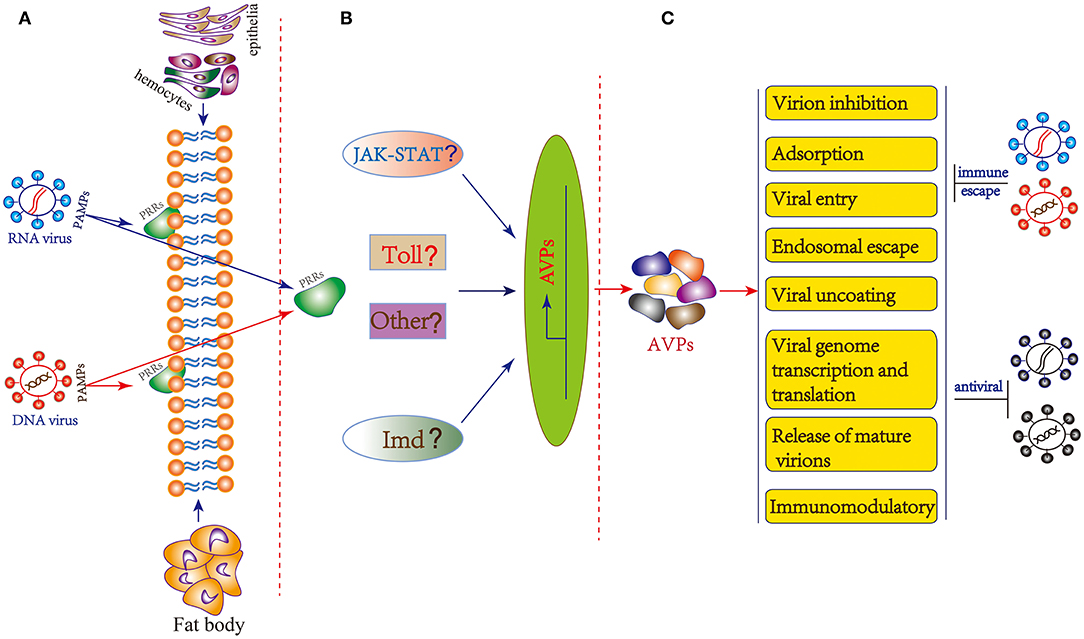
Figure 2. General hypothesis of AVP synthesis and possible mechanism of action in insects. (A) Immune recognition of insect viruses. The PAMPs of insect RNA and DNA viruses are recognized by specific PRRs located in the cell membrane or cytoplasm of hemocytes, epithelia or fat body. (B) Potential downstream signaling cascade reactions including JAK-STAT, Toll, Imd, and other pathway to produce AVPs. (C) The mechanism of action of AVPs covers stages in almost the entire life cycle of the virus: virion inhibition; adsorption; viral entry; endosomal escape; viral uncoating; viral genome transcription and translation, and release of mature virions. Additionally, AVPs may inhibit viral infection by regulating the host immune system. As a counterdefense, insect viruses may employ several strategies to escape the antiviral effect of AVPs.
The AVPs possess diverse structures as well as might act according to different mechanisms. Based on the antiviral peptide database AVPdb (Table 1), a total of 45 virus targeting strategies employed by AVPs can be distinguished such as “Virus entry,” “Virucidal on progeny virions,” “Viral assembly,” “Release,” “Transcription,” “Translation,” “Transport,” and “Replication” (31). The mechanism of action of AVPs summarized in the AVPdb database covers almost the entire life cycle of the virus (Figure 2). Additionally, AVPs may act against viral infection by regulating the host immune system (Figure 2). For instance AVP like alloferons from the blow fly are able to stimulate natural killer cells (NK) activity and interferon synthesis in animal and human models (28).
Many scientific questions about the identities of insect AVPs and their modes of action remain unresolved. Besides, viruses are the causative agents of various dreadful diseases in humans and animals. Recently, the testing and discovery of AVPs was accelerated because extraordinary advantages. Insects are considered an important source of natural AMPs, and their potential to act as AVPs is worthy of in-depth studies. In future research, the research on insect AVPs can mainly focus on the following key issues: (1) Identification of insect AVPs; (2) Recognition by PRRs and downstream cascade reactions involved in insect AVPs production; (3) Molecular mechanism of action of AVPs against insect viruses and vertebrate viruses; (4) AVP counter defense (immune escape) mechanisms by viruses; (5) Evaluation and application of insect AVPs as antiviral drugs.
MF participated in the design, collected and analyzed data, and drafted the manuscript. SF and JX helped with data collection. VL, LS, and JS participated in the design and coordination of the study, and revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31872426); Guangdong Natural Science Foundation (2018A030310210); the project Target Identification and Development of Novel Approaches for Health and Environmental Applications (MIS 5002514) which is implemented under the Action for the Strategic Development on the Research and Technological Sectors, funded by the Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020), and co-financed by Greece and the European Union (European Regional Development Fund); and Guangdong Provincial Promotion Project on Preservation and Utilization of Local Breed of Livestock and Poultry (No.2018-143).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.02030/full#supplementary-material
Supplementary File 1. Antiviral activity prediction of all insect AMPs in the dbAMP database.
Supplementary File 2. Antiviral activity prediction of Bomanins, Daishos, and Listericin in Drosophila.
1. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. (1996) 86:973–83. doi: 10.1016/S0092-8674(00)80172-5
2. Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. (1999) 284:1313–8. doi: 10.1126/science.284.5418.1313
3. Mussabekova A, Daeffler L, Imler JL. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol Life Sci. (2017) 74:2039–54. doi: 10.1007/s00018-017-2453-9
4. Xu J, Cherry S. Viruses and antiviral immunity in Drosophila. Dev Comp Immunol. (2014) 42:67–84. doi: 10.1016/j.dci.2013.05.002
5. Swevers L, Liu J, Smagghe G. Defense mechanisms against viral infection in Drosophila: RNAi and Non-RNAi. Viruses. (2018) 10:230. doi: 10.3390/v10050230
6. Lu P, Pan Y, Yang Y, Zhu F, Li C, Guo Z, et al. Discovery of anti-viral molecules and their vital functions in Bombyx mori. J Invertebr Pathol. (2018) 154:12–8. doi: 10.1016/j.jip.2018.02.012
7. McMenamin AJ, Daughenbaugh KF, Parekh F, Pizzorno MC, Flenniken ML. Honey bee and bumble bee antiviral defense. Viruses. (2018) 10:395. doi: 10.3390/v10080395
8. Chen H, Jiang Z. The essential adaptors of innate immune signaling. Protein Cell. (2013) 4:27–39. doi: 10.1007/s13238-012-2063-0
9. Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. (2012) 13:214–22. doi: 10.1038/ni.2229
10. Schoggins JW. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol. (2014) 6:40–6. doi: 10.1016/j.coviro.2014.03.006
11. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. (2011) 472:481–5. doi: 10.1038/nature09907
12. Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA. (2012) 109:4239–44. doi: 10.1073/pnas.1114981109
13. Xie T, Feng M, Dai M, Mo G, Ruan Z, Wang G, et al. Cholesterol-25-hydroxylase is a chicken ISG that restricts ALV-J infection by producing 25-hydroxycholesterol. Viruses. (2019) 11:498. doi: 10.3390/v11060498
14. Wang X, Li Y, Li LF, Shen L, Zhang L, Yu J, et al. RNA interference screening of interferon-stimulated genes with antiviral activities against classical swine fever virus using a reporter virus. Antiviral Res. (2016) 128:49–56. doi: 10.1016/j.antiviral.2016.02.001
15. Levraud JP, Jouneau L, Briolat V, Laghi V, Boudinot P. IFN-stimulated genes in zebrafish and humans define an ancient arsenal of antiviral immunity. J Immunol. (2019) 203:3361–73. doi: 10.4049/jimmunol.1900804
16. Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. (2014) 98:5807–22. doi: 10.1007/s00253-014-5792-6
17. Sheehan G, Farrell G, Kavanagh K. Immune priming: the secret weapon of the insect world. Virulence. (2020) 11:238–46. doi: 10.1080/21505594.2020.1731137
18. Imler JL, Bulet P. Antimicrobial peptides in Drosophila: structures, activities and gene regulation. Chem Immunol Allergy. (2005) 86:1–21. doi: 10.1159/000086648
19. Xu P, Shi M, Chen XX. Antimicrobial peptide evolution in the Asiatic honey bee Apis cerana. PLoS ONE. (2009) 4:e4239. doi: 10.1371/journal.pone.0004239
20. Cheng T, Zhao P, Liu C, Xu P, Gao Z, Xia Q, et al. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics. (2006) 87:356–65. doi: 10.1016/j.ygeno.2005.11.018
21. Liao JF, Wu CP, Tang CK, Tsai CW, Rouhova L, Wu YL. Identification of regulatory host genes involved in sigma virus replication using RNAi knockdown in Drosophila. Insects. (2019) 10:339. doi: 10.3390/insects10100339
22. Panda D, Rose PP, Hanna SL, Gold B, Hopkins KC, Lyde RB, et al. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep. (2013) 5:1737–48. doi: 10.1016/j.celrep.2013.11.028
23. Huang Z, Kingsolver MB, Avadhanula V, Hardy RW. An antiviral role for antimicrobial peptides during the arthropod response to alphavirus replication. J Virol. (2013) 87:4272–80. doi: 10.1128/JVI.03360-12
24. Chen TT, Tan LR, Hu N, Dong ZQ, Hu ZG, Jiang YM, et al. C-lysozyme contributes to antiviral immunity in Bombyx mori against nucleopolyhedrovirus infection. J Insect Physiol. (2018) 108:54–60. doi: 10.1016/j.jinsphys.2018.05.005
25. Palmer WH, Joosten J, Overheul GJ, Jansen PW, Vermeulen M, Obbard DJ, et al. Induction and suppression of NF-kappaB signalling by a DNA virus of Drosophila. J Virol. (2019) 93:e01443–18. doi: 10.1128/JVI.01443-18
26. Vilas Boas LCP, Campos ML, Berlanda RLA, de Carvalho Neves N, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci. (2019) 76:3525–42. doi: 10.1007/s00018-019-03138-w
27. Gentilucci L, De Marco R, Cerisoli L. Chemical modifications designed to improve peptide stability: incorporation of non-natural amino acids, pseudo-peptide bonds, and cyclization. Curr Pharm Des. (2010) 16:3185–203. doi: 10.2174/138161210793292555
28. Chernysh S, Kim SI, Bekker G, Pleskach VA, Filatova NA, Anikin VB, et al. Antiviral and antitumor peptides from insects. Proc Natl Acad Sci USA. (2002) 99:12628–32. doi: 10.1073/pnas.192301899
29. Slocinska M, Marciniak P, Rosinski G. Insects antiviral and anticancer peptides: new leads for the future? Protein Pept Lett. (2008) 15:578–85. doi: 10.2174/092986608784966912
30. Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. (2016) 44:D1087–93. doi: 10.1093/nar/gkv1278
31. Qureshi A, Thakur N, Tandon H, Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. (2014) 42:D1147–53. doi: 10.1093/nar/gkt1191
32. Mehta D, Anand P, Kumar V, Joshi A, Mathur D, Singh S, et al. ParaPep: a web resource for experimentally validated antiparasitic peptide sequences and their structures. Database. (2014) 2014:bau051. doi: 10.1093/database/bau051
33. Kang X, Dong F, Shi C, Liu S, Sun J, Chen J, et al. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci Data. (2019) 6:148. doi: 10.1038/s41597-019-0154-y
34. Jhong JH, Chi YH, Li WC, Lin TH, Huang KY, Lee TY. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. (2019) 47:D285–97. doi: 10.1093/nar/gky1030
35. Thakur N, Qureshi A, Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. (2012) 40:W199–204. doi: 10.1093/nar/gks450
36. Beltran Lissabet JF, Belen LH, Farias JG. AntiVPP 1.0: a portable tool for prediction of antiviral peptides. Comput Biol Med. (2019) 107:127–30. doi: 10.1016/j.compbiomed.2019.02.011
37. Schaduangrat N, Nantasenamat C, Prachayasittikul V, Shoombuatong W. Meta-iAVP: a sequence-based meta-predictor for improving the prediction of antiviral peptides using effective feature representation. Int J Mol Sci. (2019) 20:5743. doi: 10.3390/ijms20225743
38. Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. (2019) 37:420–3. doi: 10.1038/s41587-019-0036-z
39. Wachinger M, Kleinschmidt A, Winder D, von Pechmann N, Ludvigsen A, Neumann M, et al. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J Gen Virol. (1998) 79:731–40. doi: 10.1099/0022-1317-79-4-731
40. Albiol Matanic VC, Castilla V. Antiviral activity of antimicrobial cationic peptides against Junin virus and herpes simplex virus. Int J Antimicrob Agents. (2004) 23:382–9. doi: 10.1016/j.ijantimicag.2003.07.022
41. Ourth DD. Antiviral activity against human immunodeficiency virus-1 in vitro by myristoylated-peptide from Heliothis virescens. Biochem Biophys Res Commun. (2004) 320:190–6. doi: 10.1016/j.bbrc.2004.05.137
42. Moreno-Habel DA, Biglang-awa IM, Dulce A, Luu DD, Garcia P, Weers PM, et al. Inactivation of the budded virus of Autographa californica M nucleopolyhedrovirus by gloverin. J Invertebr Pathol. (2012) 110:92–101. doi: 10.1016/j.jip.2012.02.007
43. Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. (1980) 106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x
44. Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. (1981) 292:246–8. doi: 10.1038/292246a0
45. Bazzo R, Tappin MJ, Pastore A, Harvey TS, Carver JA, Campbell ID. The structure of melittin. A 1H-NMR study in methanol. Eur J Biochem. (1988) 173:139–46. doi: 10.1111/j.1432-1033.1988.tb13977.x
46. Wachinger M, Saermark T, Erfle V. Influence of amphipathic peptides on the HIV-1 production in persistently infected T lymphoma cells. FEBS Lett. (1992) 309:235–41. doi: 10.1016/0014-5793(92)80780-K
47. Memariani H, Memariani M, Moravvej H, Shahidi-Dadras M. Melittin: a venom-derived peptide with promising anti-viral properties. Eur J Clin Microbiol Infect Dis. (2020) 39:5–17. doi: 10.1007/s10096-019-03674-0
48. Kuczer M, Dziubasik K, Midak-Siewirska A, Zahorska R, Luczak M, Konopinska D. Studies of insect peptides alloferon, Any-GS and their analogues. Synthesis and antiherpes activity. J Pept Sci. (2010) 16:186–9. doi: 10.1002/psc.1219
49. Kuczer M, Midak-Siewirska A, Zahorska R, Luczak M, Konopinska D. Further studies on the antiviral activity of alloferon and its analogues. J Pept Sci. (2011) 17:715–19. doi: 10.1002/psc.1388
50. Kuczer M, Majewska A, Zahorska R. New alloferon analogues: synthesis and antiviral properties. Chem Biol Drug Des. (2013) 81:302–9. doi: 10.1111/cbdd.12020
51. Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta. (1999) 1451:1–16. doi: 10.1016/S0167-4889(99)00075-0
52. Axen A, Carlsson A, Engstrom A, Bennich H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur J Biochem. (1997) 247:614–9. doi: 10.1111/j.1432-1033.1997.00614.x
53. Lundstrom A, Liu G, Kang D, Berzins K, Steiner H. Trichoplusia ni gloverin, an inducible immune gene encoding an antibacterial insect protein. Insect Biochem Mol Biol. (2002) 32:795–801. doi: 10.1016/S0965-1748(01)00162-X
54. Gajda E, Bugla-Ploskonska G. Lysozyme–occurrence in nature, biological properties and possible applications. Postepy Hig Med Dosw. (2014) 68:1501–15. doi: 10.5604/17322693.1133100
55. Ragland SA, Criss AK. From bacterial killing to immune modulation: recent insights into the functions of lysozyme. PLoS Pathog. (2017) 13:e1006512. doi: 10.1371/journal.ppat.1006512
56. Singh IP, Bodiwala HS. Recent advances in anti-HIV natural products. Nat Prod Rep. (2010) 27:1781–800. doi: 10.1039/c0np00025f
57. Villa TG, Feijoo-Siota L, Rama JLR, Ageitos JM. Antivirals against animal viruses. Biochem Pharmacol. (2017) 133:97–116. doi: 10.1016/j.bcp.2016.09.029
58. Mohamed AA, Zhang L, Dorrah MA, Elmogy M, Yousef HA, Bassal TT, et al. Molecular characterization of a c-type lysozyme from the desert locust, Schistocerca gregaria (Orthoptera: acrididae). Dev Comp Immunol. (2016) 61:60–9. doi: 10.1016/j.dci.2016.03.018
59. Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. (2014) 346:763–7. doi: 10.1126/science.1257570
60. Yamaguchi M, Yoshida H. Drosophila as a model organism. Adv Exp Med Biol. (2018) 1076:1–10. doi: 10.1007/978-981-13-0529-0_1
61. Hetru C, Troxler L, Hoffmann JA. Drosophila melanogaster antimicrobial defense. J Infect Dis. (2003) 187(Suppl. 2):S327–34. doi: 10.1086/374758
62. Hanson MA, Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr Opin Immunol. (2020) 62:22–30. doi: 10.1016/j.coi.2019.11.008
63. Loch G, Zinke I, Mori T, Carrera P, Schroer J, Takeyama H, et al. Antimicrobial peptides extend lifespan in Drosophila. PLoS ONE. (2017) 12:e0176689. doi: 10.1371/journal.pone.0176689
64. Tsai CW, McGraw EA, Ammar ED, Dietzgen RG, Hogenhout SA. Drosophila melanogaster mounts a unique immune response to the Rhabdovirus sigma virus. Appl Environ Microbiol. (2008) 74:3251–6. doi: 10.1128/AEM.02248-07
65. Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. (2005) 6:946–53. doi: 10.1038/ni1237
66. Roxstrom-Lindquist K, Terenius O, Faye I. Parasite-specific immune response in adult Drosophila melanogaster: a genomic study. EMBO Rep. (2004) 5:207–12. doi: 10.1038/sj.embor.7400073
67. Zhu F, Ding H, Zhu B. Transcriptional profiling of Drosophila S2 cells in early response to Drosophila C virus. Virol J. (2013) 10:210. doi: 10.1186/1743-422X-10-210
68. Merkling SH, Overheul GJ, van Mierlo JT, Arends D, Gilissen C, van Rij RP. The heat shock response restricts virus infection in Drosophila. Sci Rep. (2015) 5:12758. doi: 10.1038/srep12758
69. Castorena KM, Stapleford KA, Miller DJ. Complementary transcriptomic, lipidomic, and targeted functional genetic analyses in cultured Drosophila cells highlight the role of glycerophospholipid metabolism in flock house virus RNA replication. BMC Genom. (2010) 11:183. doi: 10.1186/1471-2164-11-183
70. Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci USA. (2005) 102:7257–62. doi: 10.1073/pnas.0409181102
71. Kemp C, Mueller S, Goto A, Barbier V, Paro S, Bonnay F, et al. Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J Immunol. (2013) 190:650–8. doi: 10.4049/jimmunol.1102486
72. Clemmons AW, Lindsay SA, Wasserman SA. An effector peptide family required for Drosophila toll-mediated immunity. PLoS Pathog. (2015) 11:e1004876. doi: 10.1371/journal.ppat.1004876
73. Cohen LB, Lindsay SA, Xu Y, Lin SJH, Wasserman SA. The daisho peptides mediate Drosophila defense against a subset of filamentous fungi. Front. Immunol. (2020) 11:9. doi: 10.3389/fimmu.2020.00009
74. Goto A, Yano T, Terashima J, Iwashita S, Oshima Y, Kurata S. Cooperative regulation of the induction of the novel antibacterial listericin by peptidoglycan recognition protein LE and the JAK-STAT pathway. J Biol Chem. (2010) 285:15731–8. doi: 10.1074/jbc.M109.082115
75. Calderone NW. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992–2009. PLoS ONE. (2012) 7:e37235. doi: 10.1371/journal.pone.0037235
76. Chen YP, Siede R. Honey bee viruses. Adv Virus Res. (2007) 70:33–80. doi: 10.1016/S0065-3527(07)70002-7
77. Brutscher LM, Daughenbaugh KF, Flenniken ML. Antiviral defense mechanisms in honey bees. Curr Opin Insect Sci. (2015) 10:71–82. doi: 10.1016/j.cois.2015.04.016
78. Casteels P, Ampe C, Jacobs F, Vaeck M, Tempst P. Apidaecins: antibacterial peptides from honeybees. EMBO J. (1989) 8:2387–91. doi: 10.1002/j.1460-2075.1989.tb08368.x
79. Casteels P, Ampe C, Riviere L, Van Damme J, Elicone C, Fleming M, et al. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera). Eur J Biochem. (1990) 187:381–6. doi: 10.1111/j.1432-1033.1990.tb15315.x
80. Casteels P, Ampe C, Jacobs F, Tempst P. Functional and chemical characterization of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J. Biol. Chem. (1993) 268:7044–54.
81. Casteels-Josson K, Zhang W, Capaci T, Casteels P, Tempst P. Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures. J Biol Chem. (1994) 269:28569–75.
82. Ryabov EV, Fannon JM, Moore JD, Wood GR, Evans DJ. The iflaviruses sacbrood virus and deformed wing virus evoke different transcriptional responses in the honeybee which may facilitate their horizontal or vertical transmission. PeerJ. (2016) 4:e1591. doi: 10.7717/peerj.1591
83. Doublet V, Paxton RJ, McDonnell CM, Dubois E, Nidelet S, Moritz RF, et al. Brain transcriptomes of honey bees (Apis mellifera) experimentally infected by two pathogens: black queen cell virus and Nosema ceranae. Genom. Data. (2016) 10:79–82. doi: 10.1016/j.gdata.2016.09.010
84. Galbraith DA, Yang X, Nino EL, Yi S, Grozinger C. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog. (2015) 11:e1004713. doi: 10.1371/journal.ppat.1004713
85. Shan L, Liuhao W, Jun G, Yujie T, Yanping C, Jie W, et al. Chinese sacbrood virus infection in Asian honey bees (Apis cerana cerana) and host immune responses to the virus infection. J Invertebr Pathol. (2017) 150:63–9. doi: 10.1016/j.jip.2017.09.006
86. Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. (2004) 306:1937–40. doi: 10.1126/science.1102210
87. Swevers L, Feng M, Ren F, Sun J. Antiviral defense against Cypovirus 1 (Reoviridae) infection in the silkworm, Bombyx mori. Arch. Insect Biochem Physiol. (2020) 103:e21616. doi: 10.1002/arch.21616
88. Li G, Zhou K, Zhao G, Qian H, Xu A. Transcriptome-wide analysis of the difference of alternative splicing in susceptible and resistant silkworm strains after BmNPV infection. 3 Biotech. (2019) 9:152. doi: 10.1007/s13205-019-1669-9
89. Li G, Qian H, Luo X, Xu P, Yang J, Liu M, et al. Transcriptomic analysis of resistant and susceptible Bombyx mori strains following BmNPV infection provides insights into the antiviral mechanisms. Int J Genomics. (2016) 2016:2086346. doi: 10.1155/2016/2086346
90. Guo R, Wang S, Xue R, Cao G, Hu X, Huang M, et al. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl Microbiol Biotechnol. (2015) 99:5175–87. doi: 10.1007/s00253-015-6634-x
91. Zografidis A, Van Nieuwerburgh F, Kolliopoulou A, Apostolou-Karampelis K, Head SR, Deforce D, et al. Viral Small-RNA analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. J Virol. (2015) 89:11473–86. doi: 10.1128/JVI.01695-15
92. Hu ZG, Dong ZQ, Dong FF, Zhu Y, Chen P, Lu C, et al. Identification of a PP2A gene in Bombyx mori with antiviral function against B. mori nucleopolyhedrovirus. Insect Sci. (2019) 27:687–96. doi: 10.1111/1744-7917
93. Hua X, Li B, Song L, Hu C, Li X, Wang D, et al. Stimulator of interferon genes (STING) provides insect antiviral immunity by promoting dredd caspase-mediated NF-kappaB activation. J Biol Chem. (2018) 293:11878–90. doi: 10.1074/jbc.RA117.000194
94. Liu TH, Dong XL, Pan CX, Du GY, Wu YF, Yang JG, et al. A newly discovered member of the Atlastin family, BmAtlastin-n, has an antiviral effect against BmNPV in Bombyx mori. Sci Rep. (2016) 6:28946. doi: 10.1038/srep28946
95. Selot R, Kumar V, Shukla S, Chandrakuntal K, Brahmaraju M, Dandin SB, et al. Identification of a soluble NADPH oxidoreductase (BmNOX) with antiviral activities in the gut juice of Bombyx mori. Biosci Biotechnol Biochem. (2007) 71:200–5. doi: 10.1271/bbb.60450
96. Ponnuvel KM, Nakazawa H, Furukawa S, Asaoka A, Ishibashi J, Tanaka H, et al. A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus. J. Virol. (2003) 77:10725–9. doi: 10.1128/JVI.77.19.10725-10729.2003
97. Zhao P, Xia F, Jiang L, Guo H, Xu G, Sun Q, et al. Enhanced antiviral immunity against Bombyx mori cytoplasmic polyhedrosis virus via overexpression of peptidoglycan recognition protein S2 in transgenic silkworms. Dev Comp Immunol. (2018) 87:84–9. doi: 10.1016/j.dci.2018.05.021
98. Bao YY, Tang XD, Lv ZY, Wang XY, Tian CH, Xu YP, et al. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics. (2009) 94:138–45. doi: 10.1016/j.ygeno.2009.04.003
99. Kolliopoulou A, Van Nieuwerburgh F, Stravopodis DJ, Deforce D, Swevers L, Smagghe G. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS ONE. (2015) 10:e0121447. doi: 10.1371/journal.pone.0121447
100. Bao YY, Lv ZY, Liu ZB, Xue J, Xu YP, Zhang CX. Comparative analysis of Bombyx mori nucleopolyhedrovirus responsive genes in fat body and haemocyte of B. mori resistant and susceptible strains. Insect Mol Biol. (2010) 19:347–58. doi: 10.1111/j.1365-2583.2010.00993.x
101. Sun Q, Guo H, Xia Q, Jiang L, Zhao P. Transcriptome analysis of the immune response of silkworm at the early stage of Bombyx mori bidensovirus infection. Dev Comp Immunol. (2020) 106:103601. doi: 10.1016/j.dci.2019.103601
102. Xue J, Qiao N, Zhang W, Cheng RL, Zhang XQ, Bao YY, et al. Dynamic interactions between Bombyx mori nucleopolyhedrovirus and its host cells revealed by transcriptome analysis. J Virol. (2012) 86:7345–7359. doi: 10.1128/JVI.07217-12
103. Sagisaka A, Fujita K, Nakamura Y, Ishibashi J, Noda H, Imanishi S, et al. Genome-wide analysis of host gene expression in the silkworm cells infected with Bombyx mori nucleopolyhedrovirus. Virus Res. (2010) 147:166–75. doi: 10.1016/j.virusres.2009.10.015
104. Gao K, Deng XY, Qian HY, Qin G, Guo XJ. Digital gene expression analysis in the midgut of 4008 silkworm strain infected with cytoplasmic polyhedrosis virus. J. Invertebr Pathol. (2014) 115:8–13. doi: 10.1016/j.jip.2013.10.016
105. Jiang L, Peng Z, Guo Y, Cheng T, Guo H, Sun Q, et al. Transcriptome analysis of interactions between silkworm and cytoplasmic polyhedrosis virus. Sci Rep. (2016) 6:24894. doi: 10.1038/srep24894
106. Feng M, Zhang X. Immunity to Avian leukosis virus: where are we now and what should we do? Front Immunol. (2016) 7:624. doi: 10.3389/fimmu.2016.00624
107. Wang X, Zhang Y, Zhang R, Zhang J. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr Opin Insect Sci. (2019) 33:105–10. doi: 10.1016/j.cois.2019.05.004
108. Lu Y, Su F, Li Q, Zhang J, Li Y, Tang T, et al. Pattern recognition receptors in Drosophila immune responses. Dev Comp Immunol. (2020) 102:103468. doi: 10.1016/j.dci.2019.103468
109. Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, et al. Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front Immunol. (2019) 10:776. doi: 10.3389/fimmu.2019.00776
110. Lamiable O, Imler JL. Induced antiviral innate immunity in Drosophila. Curr Opin Microbiol. (2014) 20:62–8. doi: 10.1016/j.mib.2014.05.006
111. Liu W, Liu J, Lu Y, Gong Y, Zhu M, Chen F, et al. Immune signaling pathways activated in response to different pathogenic micro-organisms in Bombyx mori. Mol Immunol. (2015) 65:391–7. doi: 10.1016/j.molimm.2015.02.018
Keywords: antiviral peptides, antimicrobial peptides, insect, viruses, antiviral drugs
Citation: Feng M, Fei S, Xia J, Labropoulou V, Swevers L and Sun J (2020) Antimicrobial Peptides as Potential Antiviral Factors in Insect Antiviral Immune Response. Front. Immunol. 11:2030. doi: 10.3389/fimmu.2020.02030
Received: 16 June 2020; Accepted: 27 July 2020;
Published: 02 September 2020.
Edited by:
Katherine Buckley, Auburn University, United StatesReviewed by:
Robert Unckless, University of Kansas, United StatesCopyright © 2020 Feng, Fei, Xia, Labropoulou, Swevers and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luc Swevers, c3dldmVyc0BiaW8uZGVtb2tyaXRvcy5ncg==; Jingchen Sun, Y3lmekBzY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.