
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 24 September 2020
Sec. Microbial Immunology
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01980
This article is part of the Research Topic Host Biomarkers as Specific or General Indicators for Infections View all 8 articles
 Dao Huy Manh1,2‡§
Dao Huy Manh1,2‡§ Lan Nguyen Weiss3*†‡§
Lan Nguyen Weiss3*†‡§ Nguyen Van Thuong3‡
Nguyen Van Thuong3‡ Shusaku Mizukami1,4‡§
Shusaku Mizukami1,4‡§ Shyam Prakash Dumre1§
Shyam Prakash Dumre1§ Quang Chan Luong5
Quang Chan Luong5 Le Chi Thanh6
Le Chi Thanh6 Cao Minh Thang3
Cao Minh Thang3 Pham Thanh Huu7
Pham Thanh Huu7 Le Hong Phuc7
Le Hong Phuc7 Cao Thi Hong Nhung3
Cao Thi Hong Nhung3 Nguyen Thi Mai3
Nguyen Thi Mai3 Nguyen Quang Truong3
Nguyen Quang Truong3 Vu Thien Thu Ngu3
Vu Thien Thu Ngu3 Do Kien Quoc5
Do Kien Quoc5 Tran Thi Ngoc Ha1
Tran Thi Ngoc Ha1 Tran Ton6
Tran Ton6 Tran Van An7
Tran Van An7 Oday Halhouli8,9
Oday Halhouli8,9 Le Nhat Quynh9,10
Le Nhat Quynh9,10 Mohamed Gomaa Kamel9,11§
Mohamed Gomaa Kamel9,11§ Juntra Karbwang4
Juntra Karbwang4 Vu Thi Que Huong3#
Vu Thi Que Huong3# Nguyen Tien Huy4,9§
Nguyen Tien Huy4,9§ Kenji Hirayama1,2*§
Kenji Hirayama1,2*§Background: The protective or pathogenic role of T lymphocytes during the acute phase of dengue virus (DENV) infection has not been fully understood despite its importance in immunity and vaccine development.
Objectives: This study aimed to clarify the kinetics of T lymphocyte subsets during the clinical course of acute dengue patients.
Study design: In this hospital-based cohort study, 59 eligible Vietnamese dengue patients were recruited and admitted. They were investigated and monitored for T cell subsets and a panel of clinical and laboratory parameters every day until discharged and at post-discharge from the hospital.
Results: We described for the first time the kinetics of T cell response during the clinical course of DENV infection. Severe cases showed significantly lower levels of effector CD8+ T cells compared to mild cases at day −1 (p = 0.017) and day 0 (p = 0.033) of defervescence. After defervescence, these cell counts in severe cases increased rapidly to equalize with the levels of mild cases. Our results also showed a decline in total CD4+ T, Th1, Th1/17 cells during febrile phase of dengue patients compared to normal controls or convalescent phase. On the other hand, Th2 cells increased during DENV infection until convalescent phase. Cytokines such as interferon-γ, IL-12p70, IL-5, IL-23, IL-17A showed tendency to decrease on day 0 and 1 compared with convalescence and only IL-5 showed significance indicating the production during acute phase was not systemic.
Conclusion: With a rigorous study design, we uncovered the kinetics of T cells in natural DENV infection. Decreased number of effector CD8+ T cells in the early phase of infection and subsequent increment after defervescence day probably associated with the T cell migration in DENV infection.
Dengue has emerged as a major mosquito-borne disease affecting over 100 countries that results in 96 million symptomatic cases, including 500,000 severe cases and 22,000 deaths annually (1–3). Human infection with any serotype of DENV (DENV-1 to –4) produces a broad range of clinical manifestations from mild fever to severe dengue involving hypovolemic shock characterized by plasma leakage (increased vascular permeability), multi-organ failure, and severe bleeding (1, 3). Unfortunately, no specific treatment for dengue is available. The recently licensed dengue vaccine based on humoral immunity induced high levels of neutralizing antibody against four DENV serotypes but showed lower efficacy for DENV-2 (4), which also suggests the potential protective function of T cells during DENV infection (5).
One of the interesting clinical features of DENV infection is that only a small proportion of the dengue patients progress to severe forms following defervescence, while the rest gradually recover (6). This implies the existence of different immune responses upon DENV infection characterized by their outcomes as protection or pathogenesis (7, 8). Several hypotheses including antibody-dependent enhancement (ADE) (7, 9) and T cell mediated immune response have been put forward, although the latter remained more controversial (7, 8). Secondary infection with heterologous DENV serotype induces cross-reactive T cells that lack killing capacity but release pro-inflammatory cytokines making the condition more severe (8, 10). Moreover, cellular immunity (protective and pathogenic) in an acute phase of other viral infections has been well-characterized utilizing the flow cytometry-based analyses (11–13). Thus, there is a need of further understanding in potential dual roles of T cells during DENV infection.
Several studies on animal models have reported the protective roles of T cells (both CD4+ and CD8+) during DENV infection (14–16). While these observations in animal models are certainly crucial in suggesting the roles of T cell in DENV infection or developing hypotheses, these findings are difficult to extrapolate in actual human infection, which is a major limitation of animal studies. Therefore, a rigorous study design is necessary to uncover the T cell phenomenon in natural human DENV infection, which often requires tremendous efforts. As a consequence, limited information known about T cell roles in human DENV infection except for a few reports that suggested cytotoxic CD4+ T cells contribute in controlling DENV infection (17) while others suggested the roles of both CD8+ cytotoxic and CD4+ helper T cells in DENV infection (7–9, 18). Detailed knowledge of the kinetics of T cell subsets in dengue patients will contribute to improve clinical management and vaccine strategy.
Considering the knowledge gap in differential T cell response during natural DENV infection in human, we carried out a hospital-based prospective study to observe the precise kinetics of T cells during the entire course of illness in mild and severe cases of dengue.
This study was approved by Institutional Review Boards of the Pasteur Institute in Ho Chi Minh City (PIHCM) (No. 602/QD-Pas 27/12/10) and Institute of Tropical Medicine (NEKKEN), Nagasaki University (No. 11063072). The study was conducted according to the Declaration of Helsinki. Written informed consent was obtained from all participants and/or their parent/legal guardian (in the case of minors).
This was a hospital-based prospective study conducted at Nguyen Dinh Chieu Hospital, Ben Tre province, Vietnam, from July 2011 to May 2013. The inclusion and exclusion criteria have been described elsewhere in our previous report (19) and provided in the Supplementary Materials. In our design, after the identification of dengue patients who met the inclusion criteria, they were required to be hospitalized despite having no severe manifestations at the time of recruitment. This was done to ensure stringent monitoring and to eliminate potential biases which could otherwise occur. All the patients were rigorously investigated and monitored by an experienced physician for a panel of clinical and laboratory parameters every day until discharged from the hospital (Supplementary Figure 1). All patients were treated according to the guidelines of the Ministry of Health, Vietnam.
Dengue was screened by DENV NS1 antigen ELISA kit (Bio-Rad Laboratories, Inc., Marnes-la-Coquette, France) and confirmed by either DENV RNA RT-PCR or MAC-ELISA as described elsewhere (19, 20). In-house IgM/IgG capture ELISA assays (PIHCM, Vietnam) in paired sera (acute and convalescent) were performed to identify primary and secondary dengue infections where the secondary was defined as IgM/IgG ratio <1.8 and ≥4-fold rise of the IgG, respectively (20, 21).
Blood samples were collected each day from the enrolled patients until discharge. One additional sample was also taken two weeks after discharge (post-discharge) from each patient (Supplementary Figure 1). Whole blood was used for T cell subset analysis and routine hematological parameters, while the plasma (multiple aliquots) was used for other laboratory profiles. All samples were transferred to PIHCM within 24 h after collection.
Control samples were taken from nine healthy Vietnamese donors, from a highly endemic area and the same Kinh ethnic group, without current or recent history of fever or any other disease symptoms. To rule out DENV infection, DENV NS1 antigen and DENV-specific IgM antibody in the plasma were tested by ELISA as described above.
Each patient was monitored daily as in-patient and all the clinical data were duly recorded which include, but are not limited to, blood collection time, clinical manifestations [vomiting, hemorrhagic tendencies (such as mucosal, gastrointestinal, menstruation, nosebleed etc.), liver enlargement, and progression to severe syndromes, e.g., shock], treatment history, and laboratory parameters (hematocrit level, platelet count, leukocyte count, etc.). Clinical outcome was then linked back to the patient data. Clinical outcome was classified based on the established criteria as described previously (22–29) and compatible with the WHO 2009 classification for severity grading (1). For T cell kinetic analysis, the day of defervescence was designated as day 0, which was defined as the first time when a patient's body temperature dropped below 38°C for a 24 h period. One day before defervescence was assigned as day −1, while 1 day after defervescence was assigned as day 1 and so on. Post-discharge indicated 15–33 days after defervescence (Supplementary Figure 1).
The absolute number of CD4+ and CD8+ T cells were measured using TBNK Multi 6-colors BD kit (BD, San Diego, CA) according to the manufacturer's protocol (Supplementary Figure 2). To identify T cell subsets, these surface markers were analyzed using the following antibodies: CD3-APC/Cy7 (SK7), CD4-PerCP/Cy5.5 (RPA-T4), CD8-PerCP/Cy5.5 (RPA-T8), CD45RA-FITC (5H9), CD45RO-FITC (UCHL1), CXCR3-APC (1C6), CCR6-PE (11A9), CCR7-PE (150503), and CD62L-APC (DREG-56) (BD). Briefly, whole blood was stained with these fluorochrome-conjugated antibodies for 30 min and treated with lysis solution (BD) for 20 min at room temperature. After rinsing with phosphate buffer (pH 7.2–7.4), cells were acquired by FACS Canto II (BD). The gating strategy for flow cytometric analysis has been given in Supplementary Figures 2, 3. Analysis of flow cytometry data was accomplished with FlowJo version 10.0 (FLOWJO, LCC, Ashland, OR). Based on the cell surface markers, percentage of Th1 (CD3+, CD4+, CD45RA−, CXCR3+CCR6−), Th2 (CD3+, CD4+, CD45RA−, CXCR3−CCR6−), Th1/17 (CD3+, CD4+, CD45RA−, CXCR3+CCR6+), Th17 (CD3+, CD4+, CD45RA−, CXCR3−CCR6+) (17, 30, 31), and effector CD8+ T cell (CD3+, CD8+, CD45RO−, CCR7−CD62L−) (32– 34) were calculated (Supplementary Figures 2, 3).
The absolute cell number in each T cell subset was calculated by multiplying the percentage of each subset in activated CD4+ or effector CD8+ with the absolute number of CD4+ and CD8+ cell, respectively, which were counted with TBNK Multi 6-colors BD kit.
For the quantification of plasma cytokines, MILLIPLEX MAP kits (Human Cytokine/Chemokine Magnetic Beads Panel and Human Th17 Magnetic Beads Panel, Millipore) were used. Frozen plasma samples were measured as indicated in instructions of the kits. Data were analyzed with LABScan 100 (Luminex).
Data were analyzed with R version 3.3.2. The two groups were compared using Fisher's exact or Chi-square test for categorical variables, and Wilcoxon rank sum test for continuous variable (T cell data). Significant difference was determined when p < 0.05. Graphs were presented as line and LOESS curve fitting (35). Post-discharge data were presented as dot blot with the median bar while control data were presented as solid triangle with median bar.
To understand the kinetics of T cell in DENV infection, a total of 175 samples (115 samples from mild and 60 from severe patients) collected every day from day −4 before defervescence to post-discharge day were analyzed by flow cytometry. Additionally, nine healthy controls samples from the epidemic area were also examined (Figure 1, Supplementary Table 1). There were no significant differences for participant age, gender, and hemorrhagic tendencies between the three groups (mild dengue, severe dengue, and healthy control). The severe dengue group had significantly higher increase in hematocrit levels (p < 0.05). Only two patients had liver enlargement (>2 cm) and both were from the severe dengue group. Other clinical features such as primary and secondary infection or thrombocytopenia were not different between mild and severe patients, and so was the distribution of different serotypes. DENV-2 accounted for 40% of severe cases compared to 28.2% in mild cases (non-significant, Table 1). Samples from a total of 59 patients were applied to T cell analysis.
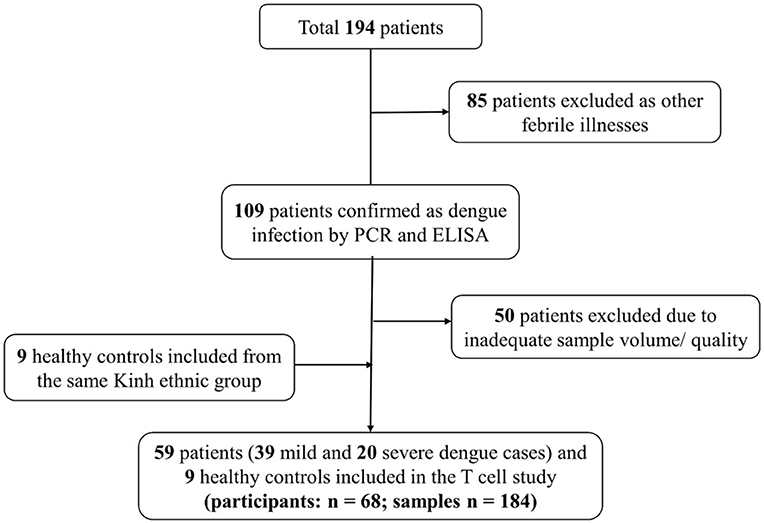
Figure 1. Flow diagram illustrating patient selection and sample investigation. Fifty-nine symptomatic dengue patients were enrolled. Blood samples were collected each day from the enrolment until discharge. One sample was also obtained 2 weeks after discharge as convalescent sample. T cell subsets were analyzed using these samples (n = 184).
We conducted kinetic T cell subset analysis from day −4 to 5 and during the convalescent phase of acute DENV infection. Number of samples examined each day varied from 1 to 22, as shown in Supplementary Table 1 and each figure. CD8+ T cells generally contribute to the antiviral response by directly killing infected cells. The kinetic profiles of CD8+ and effector CD8+ T cells during the course of DENV infection showed diminished level of absolute total CD8+ counts compared to healthy controls in both mild and severe cases during the febrile phase, which continued to incline, reaching normal level around day 0 and further increased to remain higher than normal levels until the convalescent phase (Figure 2, Supplementary Table 1). CD8+ T cell counts at post-discharge period were significantly higher in both mild (p = 0.009) and severe (p = 0.0496) groups compared to healthy controls. Effector CD8+ cells were significantly lower in severe than mild cases at day −1 (p = 0.017) and day 0 (p = 0.033). However, after defervescence, effector CD8+ T cell counts in severe cases equalized with the levels of mild cases, and high levels of the effector CD8+ count remained until convalescence (Figure 2, Supplementary Table 1).
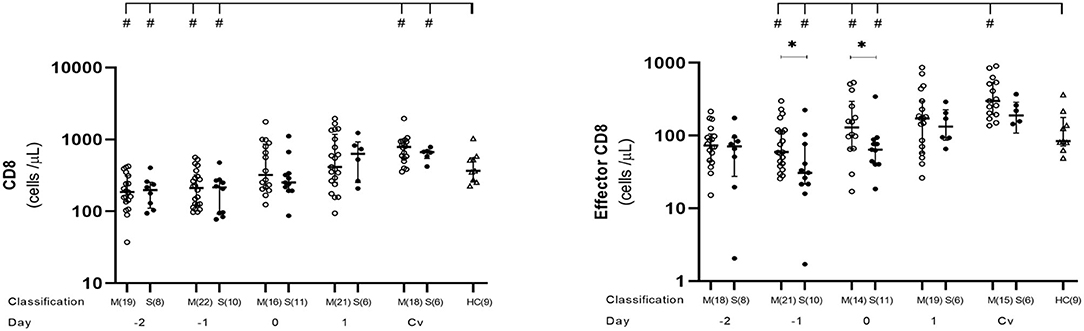
Figure 2. Kinetics of CD8+ and effector CD8+ T cells in mild and severe dengue infection. Number of CD8+ and effector CD8+ T cells from dengue patients was monitored in acute and convalescent phase. Open and closed circles show number of cell count from mild dengue patients and severe dengue patients, respectively. Open triangle shows data from healthy control. Asterisks (*) indicate significant difference (p < 0.05) between mild and severe group on a single day. Hash (#) indicates significant difference (p < 0.05) between healthy control and other groups. M and S indicate mild and severe dengue patients, respectively. Number in parentheses indicates number of samples in each time point. Day 0 denotes defervescence day. Cv and HC indicate convalescent phase and healthy control, respectively.
We observed here that absolute number of activated CD4+ as well as Th1 and Th17 cells behaved in the same trend. Overall, the number of these cells were significantly lower in dengue cases than that in the healthy controls before day 0 (Figure 3, Supplementary Table 1), which then reached normal levels at day 1 and remained stable until convalescent phase. At day −1, severe cases showed significantly lower activated Th1 counts than in the mild cases (p = 0.0498) (Figure 3, Supplementary Table 1).
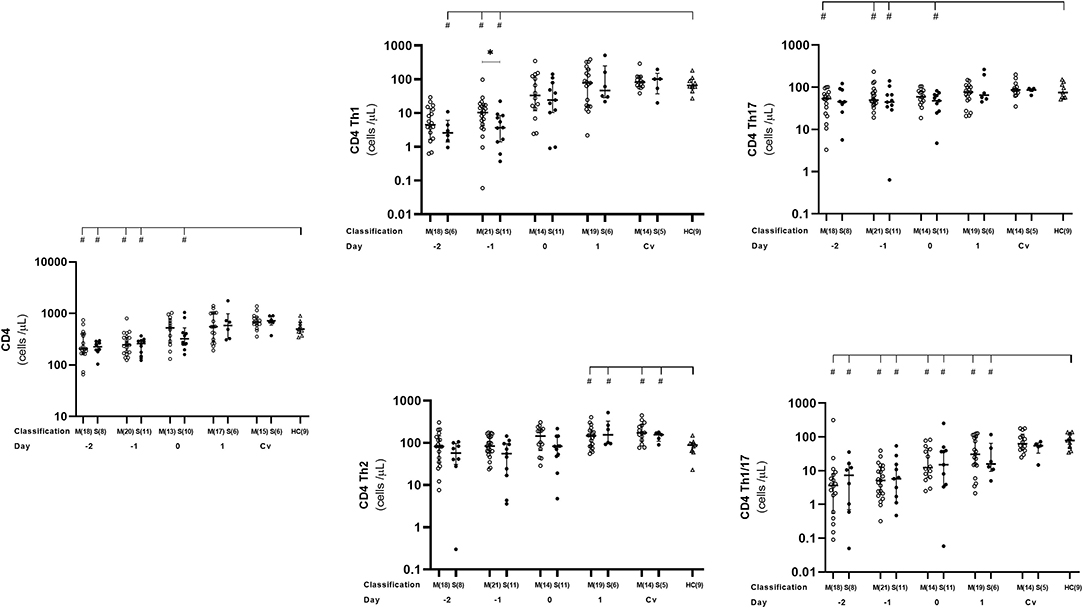
Figure 3. Kinetics of CD4+, Th1, Th2, Th1/17, and Th17 T cells in mild and severe dengue infection. Number of CD4+, Th1, Th2, Th1/17, and Th17 T cells from dengue patients was monitored in acute and convalescent phase. Open and closed circles show number of cell count from mild dengue patients and severe dengue patients, respectively. Open triangle shows data from healthy control. Asterisks (*) indicates significant difference (p < 0.05) between mild and severe group on a day. Hash (#) indicates significant difference (p < 0.05) between healthy control and other groups. M and S indicate mild and severe dengue patients, respectively. Number in parentheses indicates number of samples in each time point. Day 0 denotes defervescence day. Cv and HC indicate convalescent phase and healthy control, respectively.
We also analyzed Th1/17 (CCR6+CXCR3+) cells which were originally related to autoimmunity, but recently turned out to be significant in microorganism infections including dengue virus (17, 35, 36). We noticed that kinetics of Th1/17 cells was comparable to Th1 and Th17, with lower counts in dengue patients compared to that in healthy controls during the early phase of infection from day −2 until 1 (severe cases) or day −4 (mild cases). Interestingly, mild patients had significantly lower number of Th1/17 cells than severe ones on day 3 (p = 0.03; Figure 3, Supplementary Table 1).
While overall CD4+ T cell subsets were lower at the early phase of dengue infection, then increased toward normal level after defeverescence and maintained until convalescence, Th2 cells behaved differently. Dengue infection cases had significantly higher levels of Th2 cells than normal at day 1 and remained high until convalesce (p = 0.036 and 0.019, respectively).
Plasma level of five cytokines (IFN-γ, IL-5, IL-10, IL-12p70, IL-17A, and IL-23) were estimated. All cytokines except IL-10 showed a similar tendency with the plasma level decreasing from day −2 to 1, with day 1 as the lowest level. After that, the plasma level started to increase and became similar to or higher than day −2 at convalescent phase. On the other hand, IL-10 did not show such a trend, and the plasma levels were almost similar on every time point. When compared between mild and severe cases, IL-5 plasma levels was statistically significant difference (p < 0.05) at day 0 and 1 (Figure 4).
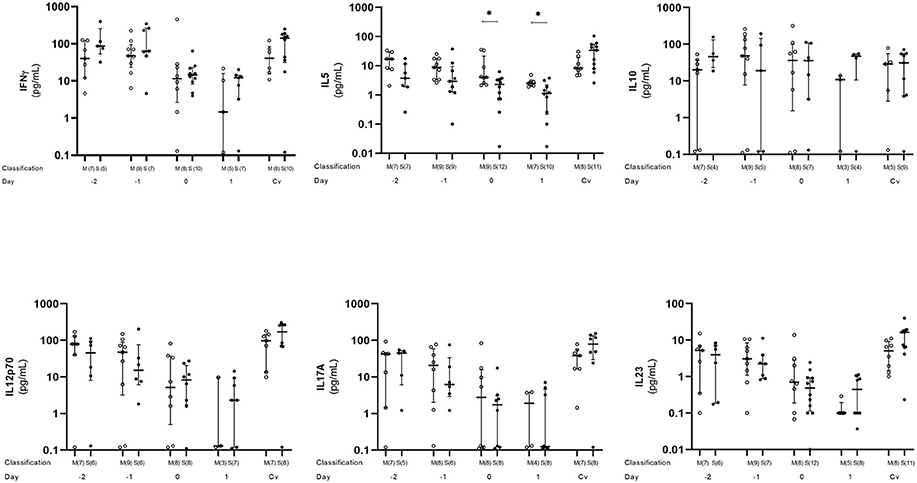
Figure 4. Kinetics of plasma cytokine in dengue infection. Cytokines from dengue patients' plasma were monitored in acute and convalescent phase. Open and closed circles show number of cell count from mild dengue patients and severe dengue patients, respectively. Open triangle shows data from healthy control. Asterisks (*) indicates significant difference (p < 0.05) between mild and severe group on a single day. M and S indicate mild and severe dengue patients, respectively. Number in parentheses indicates number of samples in each time point. Day 0 denotes defervescence day. Cv and HC indicate convalescent phase and healthy control, respectively.
Significant effort has been invested over the last two decades in order to understand the input of DENV-specific T cells in protection or enhanced immunopathology. In general, CD8+ T cells can eliminate viral infection by generating effector cells, induce cytotoxicity, IFN-γ, and TNFα (37). CD4+ T cells indirectly control infection by enhancing B and CD8+ T cell activation, memory to control recurring pathogen, and production of inflammatory and anti-viral cytokines (38).
HLA restricted CD4+ and CD8+ T cells are activated upon viral infection and several epitopes have been identified (39). Multiple lines of evidence suggest that CD4+ and specially CD8+ T cells importantly contributed against DENV infection in mouse models (15, 16) and in humans (39–41). In many recent human studies (39–41), analysis was conducted on blood collected from a healthy donor in the endemic area. Here, we characterized for the first time the precise kinetics of T cell subsets in the meticulously monitored cohorts of dengue patients.
In this cohort, CD8+ and CD4+ T cells showed similar kinetic patterns. All DENV infection cases had a tendency of lower number of total CD8+, effector CD8+, Th1, and Th1/17 cells than those in normal control before the defervescence day which then started to increase till post-discharge period and, interestingly, the increase seems to be faster in severe than mild patients. This emphasized the protective role of the CD8+ and CD4+ T cells in natural human dengue infection. After activation, CD8+ cells possibly migrate to sites of infection and kill infected cells. While CD4+ cells provide help to innate immune responses, B cells, and CD8+ cells contribute to eliminate DENV from infected tissues (42).
Multiple studies discovered that not only CD8+ but also CD4+ T cells appeared to contribute in protecting DENV infection through cytotoxic functions such as antigen-specific killing targeting non-structural proteins (43). Additionally, CD4+ T cells interfere the lysis of non-antigen-presenting bystander target cells (16) via two main pathways: perforin-dependent and FAS/FAS ligand (44). Cytotoxic activity and protecting role of DENV specific CD4+ T cells could strengthen the finding from our laboratory and others the association of HLA class II molecules DRB1*09:01 (Vietnamese) and DRB1*04:01 (Sri Lankan) (45, 46). Th1/17 clone helped antibody production by B cells and to display cytotoxic activity (47). In our patients, a remarkable significant movement of Th1/17 cells during acute DENV infection was observed highlighting their protective role. This finding opens an important area of research as previous studies reported. Th1/17 cells, together with Th1 and other Th1-like cells, showed to be significantly activated (17, 36). As they are the INF-γ producers, their activation could lead to cytokine storm and severity (36).
Contrary to Th1, Th17, and Th1/17 cells, Th2 cells increased from defervescence day until convalescence and were significantly higher than normal levels in both mild and severe cases (Figure 3). A study by Chng et al. (36) had detected this difference in Th2 frequency which was an upward trend during the recovery phase. The increased level of Th2 cells during defervescence period in dengue cases is correlated with our previous finding of elevated levels of plasma mast cell granule-derived factors. Increased vascular permeability, the hallmark of severe dengue, is thought to be provoked by mast cells (48). We and others have previously shown the important role of mast cell-derived mediators, including vascular endothelial growth factor (VEGF), tryptase, chymase, in vascular permeability in dengue severity (19, 48, 49). However, it is still unknown why the onset of vascular leakage occurs during defervescence.
Higher levels of inflammatory cytokines including IFN-γ in acute phase than in defervescence phase were reported (36, 50). Because our study focuses on T cells and the subsets, inflammatory cytokines (e.g., IL-1β, TNF-α) were not well-covered in analysis. We revealed a similar tendency on a major CD8+ T cell cytokine IFN-γ, and cytokines represent several helper subsets (Th1, Th2, and Th17) (Figure 4). Decrease of plasma level between day −2 and 1 as a common trend for cytokines except IL-10 and no significant cytokine surge was observed. Our observation strongly suggested that the T cell mediated immunological reaction provoked by dengue virus infection is highly limited in the lesion and is difficult to see from systemic circulating blood levels. To have a clear conclusion, further study will be required.
Looking into the differences between clinical groups, we observed significant lower levels of effector CD8+ (at days −1 and 0) (Figure 2), as well as CD4+ Th1 (at days −1) in severe group than in mild group (Figure 3). CD4+ Th2 cells also behaved the same way between the two groups even though it was not statistically significant (Figure 3). Clinically, the critical phase of dengue begins at day of defervescence (6–9). It is reasonable to speculate that around day-1 is critical time of kinetic T cells which in turns could possibly control the clinical outcome of the infection.
In both groups, effector CD8+ increased after day 0, probably due to the migration of these cells to infected tissues and organs before day 0, as seen with the Th1 population. In a study on T cell epitope reactivity against DENV proteome, specific CD4+ and CD8+ T cells recognized distinct viral proteins confirming the crucial roles of CD4+ Th1 in INF-γ production (18). CD4+ Th1 subset was claimed to have a positive interaction with naïve CD8+ to become effectors (51, 52).
The combination of total CD8+, effector CD8+, Th1, Th1/17 subsets during the early stage is important to inhibit viral replication in the host cells of tissue sites. The later increment of these cells in the peripheral circulation after defervescence might indicate their return from the infected tissues. There are reports of T cell homing to the skin during the early phase of DENV (53) and HSV-2 infections (54, 55). Our observation of early phase reduction and delayed regain of these cell levels may also be explained by the same phenomenon of committed T cells homing markers to the original skin tissue or inflammation places such as liver, although there is a report that CD8+ T cell activation does not enhance prior to the commencement of resolution of viremia or hemo-concentration (39, 53, 56–61).
Secondary heterotypic dengue infection has been associated with severity due to the ineffective viral control of preexisting cross-reactive and low-affinity memory T cells (62). However, the risk of severe dengue in secondary heterotypic infection was not high in our studies (45, 49) including the current study and some others (62). We have also analyzed T cell kinetics in the context of primary and secondary infection (Supplementary Figures 4A, B). At convalescent phase across all studied groups, there was a trend of higher levels of dengue specific or memory T cells in dengue patients compared with healthy controls (Supplementary Figure 4B). The significantly higher activated CD4+ and Th17 cells in secondary infection compared with primary infection was revealed at convalescent time (Supplement Figure 4B). This difference occurred by second simulation may be due to some activated memory. The same tendency was also observed in Th1 cells. Th2 cells, however, in both primary and secondary infection cases were higher when compared with that in healthy controls from day 1 until convalescence (Supplementary Figure 4B). As both types of infections similarly increase, perhaps a memory independent response may have occurred. More focused study on the T cell epitopes that are critical for the repeated heterologous infection could reveal the difference (39).
Amino acid differences between DENV serotypes can range from 14 to 67% (63). Recognized T-cell epitopes in the case of sequential heterologous serotype infection can interfere with the outcome from being protective to pathogenic response (63). Out of four serotypes (DENV 1–4), DENV-2 had more severe clinical manifestations than other serotypes in South East Asian and South American populations (45). In this study, although not significant, DENV-2 was still predominant in severe cases while DENV-4 was predominant in the mild cases (Table 1). Engagingly, patients with DENV-4 had the highest CD8+ and effector CD8+ T cells in convalescent time. It is noteworthy that there were the clusters of severe cases with DENV-2 which had the lowest CD8+ (84 and 97 at day −1), as well as effector CD8+ T cell count (26, 33, 34 at day −1, and 40, 63, 76 at day 0; Supplementary Figure 5A). Additionally, DENV-2 subjects were found to have the significant higher cell counts (p < 0.05) in convalescence across Th1, Th2, and Th1/17 populations (Supplementary Figure 5B).
We observed CD4+ and CD8+ T cell subset kinetics during the course of natural infection in dengue patients; however, we did not distinguish DENV specific T cell populations that could be analyzed by tetramers (36). Recent findings of Chng et al. showed the full activation and proliferation of all immune subsets, except for B cells in the acute phase of dengue infection. Moreover, by using the strategy of 430 multiplexed peptide-HLA staining, the adaption of T cell during dengue infection revealed specifically to DENV antigen against EBV and influenza's epitopes (36). Additionally, DENV has the T-cell cross-reactivity with zika virus, the other species of Flavivirus (56). We believe that the kinetics of T cells showed here have the specification to DENV, at least with other no relative viruses. We consider these limitations to instruct for our comprehensive future studies. Alternatively, comprehensive data from Weiskopf et al. in the general population from Sri Lanka using multiple peptide technology showed CD8+ T cells targeted at non-structural protein epitopes, dominantly at NS3 protein. Meanwhile CD4+ T cell epitopes identified at capsid, envelop, and non-structural proteins, thus contributing to protective immunity to DENV infection. Moreover, CD4 + and CD8 + T cells increased after a single dose of a tetravalent live-attenuated dengue vaccine were directed to the same region of non-structural protein antigens as T cells from a naturally infected human (17). Those comprehensive HLA reactive T epitope analysis will enhance the availability of dominant tetramers that can be applied to our further analysis.
With a rigorous study design, we have uncovered the kinetics of T cell in natural dengue infection in our cohorts which, we believe, aids in the understanding of the cellular immune response in dengue. Moreover, the significantly lower number of the effector CD8+ cells we observed in severe patients during the febrile phase is probably related to the movement of T cell upon DENV infection (as we hypothesized), which warrants further investigations.
All datasets generated for this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Institutional Review Boards of the Pasteur Institute in Ho Chi Minh City (PIHCM) (No. 602/QD-Pas 27/12/10) and Ethical Review Committee of the Institute of Tropical Medicine (NEKKEN), Nagasaki University (No. 11063072). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
LW, VH, SM, NH, and KH conceived and designed the experiments. DM, LW, NVT, SM, SD, CT, CN, NM, NQT, VN, TH, and TT performed the experiments. CN, PH, LP, TA, LW, VH, NH, and KH performed clinical management. DM, LW, QL, LT, NVT, SM, VH, OH, LQ, MK, NH, DQ, SM, SD, KH, and JK analyzed and interpreted the data. All authors wrote and approved the final manuscript.
This work was supported by Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) (Grant No. 16fm0108001h0002 to KH). This fund supported clinical and laboratory work at the hospital and the research institute in Vietnam and Japan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors thank Dr. Lbachir BenMohamed, Laboratory of Cellular and Molecular Immunology, University of California Irvine, Irvine, CA, for his critical review comments and edits.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01980/full#supplementary-material
Supplementary Figure 1. Time frame of blood sample collection. Blood samples were collected each day from the enrolment until discharge. One additional sample was also taken two weeks after discharge (convalescent phase) from each patient.
Supplementary Figure 2. Absolute T cell count using TBNK + TruC Kit. TBNK Multi 6- colours BD- Kit was used to stain whole blood. Major cell populations were defined as CD4+ (CD3+CD4+), CD8+ (CD3+CD8+), B (CD19+), and NK (CD16+CD56+). The FACSCanto II automatically calculated the absolute number of each cell type from number of beads run together with cells during flow cytometry.
Supplementary Figure 3. Gating strategy to identify CD4+ and CD8+ T cell subsets. Whole blood was stained by fluorochrome-conjugated antibodies. After that the cells were acquired by FACSCanto II followed by the analysis of data using FlowJo v 10.0. T cell subsets were defined (based on cell surface markers) as: CD8 T cell effector subset (CD3+, CD8+, CD45RO−, CCR7−CD62L−) (A), and CD4 T cell subsets, Th1 (CD3+, CD4+, CD45RA−, CXCR3+CCR6−), Th2 (CD3+, CD4+, CD45RA−, CXCR3−CCR6−), Th1/17 (CD3+, CD4+, CD45RA−, CXCR3+CCR6+), Th17 (CD3+, CD4+, CD45RA−, CXCR3−CCR6+) (B). Percentage of each subset was used to calculate absolute number of that subset.
Supplementary Figure 4. T cells kinetics in primary and secondary dengue infection. Number of CD8+, effector CD8+ T cells (A) and CD4+ and CD4+ T cell subsets (B) from primary and secondary dengue patients was monitored in acute and convalescent phase. Open and closed circles show number of cell count from primary infection (PI) and secondary infection (SI), respectively. Triangle show data from healthy control. Number in parentheses indicates number of samples in each time point. Day 0 denotes defervescence day. Cv and HC indicate convalescent phase and healthy control, respectively. Asterisks (*) indicates significant difference between primary and secondary infection group on a single day. Hash (#) indicates significant difference (p < 0.05) between healthy control and other groups (# p < 0.05, ## p < 0.01, ### p < 0.005, #### p < 0.0001).
Supplementary Figure 5. T cells kinetics by virus serotype. Number of CD8+, effector CD8+ T cells (A) and CD4+ and CD4+ T cell subsets (B) from patient infected with dengue virus 1, 2, 3, 4 serotypes was monitored in acute and convalescent phase. Open circle, triangle, diamond and closed circle show number of cell count from dengue virus 1, 2, 3, 4 serotype, respectively. Asterisks (*) indicates significant difference (p < 0.05) between dengue serotypes on a single day. Day indicates day from defervescence day. n indicates number of samples each time point. Cv denotes samples from convalescent phase.
Supplementary Table 1. Absolute T cell count in dengue patients and healthy control.
1. Dengue W. Guidelines for Diagnosis, Treatment. Prevention and Control Geneva: World Health Organization (2009).
2. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. (2013) 496:504–07. doi: 10.1038/nature12060
3. Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB. Dengue infection. Nat Rev Dis Primers. (2016) 2:16055. doi: 10.1038/nrdp.2016.55
4. Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. (2012) 380:1559–67. doi: 10.1016/S0140-6736(12)61428-7
5. Guy B, Jackson N. Dengue vaccine: hypotheses to understand CYD-TDV-induced protection. Nat Rev Microbiol. (2016) 14:45–54. doi: 10.1038/nrmicro.2015.2
6. Pang T, Mak TK, Gubler DJ. Prevention and control of dengue-the light at the end of the tunnel. Lancet Infect Dis. (2017) 17:e79–e87. doi: 10.1016/S1473-3099(16)30471-6
7. Screaton G, Mongkolsapaya J, Yacoub S, Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol. (2015) 15:745–59. doi: 10.1038/nri3916
8. Mongkolsapaya J, Dejnirattisai W, Xu X-n, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. (2003) 9:921. doi: 10.1038/nm887
9. Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol. (2011) 11:532–43. doi: 10.1038/nri3014
10. Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. (2006) 176:3821–29. doi: 10.4049/jimmunol.176.6.3821
11. Sitati EM, Diamond MS. CD4(+) T-cell responses are required for clearance of West Nile virus from the central nervous system. J Virol. (2006) 80:12060–69. doi: 10.1128/JVI.01650-06
12. Schmidt J, Blum HE, Thimme R. T-cell responses in hepatitis B and C virus infection: similarities and differences. Emer Microbes Infect. (2013) 2:e15. doi: 10.1038/emi.2013.14
13. Rabenstein H, Behrendt AC, Ellwart JW, Naumann R, Horsch M, Beckers J, et al. Differential kinetics of antigen dependency of CD4+ and CD8+ T cells. J Immunol. (2014) 192:3507–17. doi: 10.4049/jimmunol.1302725
14. Wan SW, Chen PW, Chen CY, Lai YC, Chu YT, Hung CY, et al. Therapeutic effects of monoclonal antibody against dengue virus NS1 in a STAT1 knockout mouse model of dengue infection. J Immunol. (2017) 199:2834–44. doi: 10.4049/jimmunol.1601523
15. Hertz T, Beatty PR, MacMillen Z, Killingbeck SS, Wang C, Harris E, et al. Antibody epitopes identified in critical regions of dengue virus nonstructural 1 protein in mouse vaccination and natural human infections. J Immunol. (2017) 198:4025–35. doi: 10.4049/jimmunol.1700029
16. Kao YS, Yu CY, Huang HJ, Tien SM, Wang WY, Yang M, et al. Combination of modified NS1 and NS3 as a novel vaccine strategy against dengue virus infection. J Immunol. (2019) 203:1909–17. doi: 10.4049/jimmunol.1900136
17. Weiskopf D, Bangs DJ, Sidney J, Kolla RV, De Silva AD, de Silva AM, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc Natl Acad Sci U S A. (2015) 112:E4256–E63. doi: 10.1073/pnas.1505956112
18. Rivino L, Kumaran EA, Jovanovic V, Nadua K, Teo EW, Pang SW, et al. Differential targeting of viral components by CD4+ versus CD8+ T lymphocytes in dengue virus infection. J Virol. (2013) 87:2693–706. doi: 10.1128/JVI.02675-12
19. Inokuchi M, Dumre SP, Mizukami S, Tun MMN, Kamel MG, Manh DH, et al. Association between dengue severity and plasma levels of dengue-specific IgE and chymase. Arch Virol. (2018) 163:2337–47. doi: 10.1007/s00705-018-3849-z
20. Innis B, Nisalak A, Nimmannitya S, Kusalerdchariya S, Chongswasdi V, Suntayakorn S, et al. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am J Trop Med Hygiene. (1989) 40:418–27. doi: 10.4269/ajtmh.1989.40.418
21. Ha TTN, Huy NT, Murao LA, Lan NTP, Thuy TT, Tuan HM, et al. Elevated levels of cell-free circulating DNA in patients with acute dengue virus infection. PLoS One. (2011) 6:e25969. doi: 10.1371/journal.pone.0025969
22. Alexander N, Balmaseda A, Coelho IC, Dimaano E, Hien TT, Hung NT, et al. Multicentre prospective study on dengue classification in four South-east Asian and three Latin American countries. Trop Med Int Health. (2011) 16:936–48. doi: 10.1111/j.1365-3156.2011.02793.x
23. Pozo-Aguilar JO, Monroy-Martínez V, Díaz D, Barrios-Palacios J, Ramos C, Ulloa-García A, et al. Evaluation of host and viral factors associated with severe dengue based on the 2009 WHO classification. Parasites Vectors. (2014) 7:1. doi: 10.1186/s13071-014-0590-7
24. Van De Weg CA, van Gorp EC, Supriatna M, Soemantri A, Osterhaus AD, Martina BE. Evaluation of the 2009 WHO dengue case classification in an Indonesian pediatric cohort. Am J Trop Med Hygiene. (2012) 86:166–70. doi: 10.4269/ajtmh.2012.11-0491
25. Narvaez F, Gutierrez G, Pérez MA, Elizondo D, Nuñez A, Balmaseda A, et al. Evaluation of the traditional and revised WHO classifications of dengue disease severity. PLoS Negl Trop Dis. (2011) 5:e1397. doi: 10.1371/journal.pntd.0001397
26. Basuki PS, Puspitasari BD, Husada D, Darmowandowo W, Soegijanto S, Yamanaka A. Application of revised dengue classification criteria as a severity marker of dengue viral infection in Indonesia. Southeast Asian J Trop Med Public Health. (2010) 41:1088–94.
27. Srikiatkhachorn A, Gibbons RV, Green S, Libraty DH, Thomas SJ, Endy TP, et al. Dengue hemorrhagic fever: the sensitivity and specificity of the world health organization definition for identification of severe cases of dengue in Thailand, 1994-2005. Clin Infect Dis. (2010) 50:1135–43. doi: 10.1086/651268
28. Potts JA, Gibbons RV, Rothman AL, Srikiatkhachorn A, Thomas SJ, Supradish P-o, et al. Prediction of dengue disease severity among pediatric Thai patients using early clinical laboratory indicators. PLoS Negl Trop Dis. (2010) 4:e769. doi: 10.1371/journal.pntd.0000769
29. Potts JA, Thomas SJ, Srikiatkhachorn A, Supradish P-o, Li W, Nisalak A, et al. Classification of dengue illness based on readily available laboratory data. Am J Trop Med Hygiene. (2010) 83:781–88. doi: 10.4269/ajtmh.2010.10-0135
30. Lubberts E. The IL-23-IL-17 axis in inflammatory arthritis. Nat Rev Rheumatol. (2015) 11:415–29. doi: 10.1038/nrrheum.2015.53
31. Wacleche VS, Goulet JP, Gosselin A, Monteiro P, Soudeyns H, Fromentin R, et al. New insights into the heterogeneity of Th17 subsets contributing to HIV-1 persistence during antiretroviral therapy. Retrovirology. (2016) 13:59. doi: 10.1186/s12977-016-0293-6
32. Kuss I, Donnenberg AD, Gooding W, Whiteside TL. Effector CD8+CD45RO-CD27-T cells have signalling defects in patients with squamous cell carcinoma of the head and neck. Br J Cancer. (2003) 88:223–30. doi: 10.1038/sj.bjc.6600694
33. Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. (2002) 2:251–62. doi: 10.1038/nri778
34. Prado-Garcia H, Aguilar-Cazares D, Flores-Vergara H, Mandoki JJ, Lopez-Gonzalez JS. Effector, memory and naïve CD8+ T cells in peripheral blood and pleural effusion from lung adenocarcinoma patients. Lung Cancer. (2005) 47:361–71. doi: 10.1016/j.lungcan.2004.07.046
35. Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. (2007) 8:639–46. doi: 10.1038/ni1467
36. Chng MHY, Lim MQ, Rouers A, Becht E, Lee B, MacAry PA, et al. Large-scale HLA tetramer tracking of T cells during dengue infection reveals broad acute activation and differentiation into two memory cell fates. Immunity. (2019) 51:1119–35.e5. doi: 10.1016/j.immuni.2019.10.007
37. Remakus S, Sigal LJ. Memory CD8+ T cell protection. Adv Exp Med Biol. (2013) 785:77–86. doi: 10.1007/978-1-4614-6217-0_9
38. Sant AJ, McMichael A. Revealing the role of CD4+ T cells in viral immunity. J Exp Med. (2012) 209:1391–5. doi: 10.1084/jem.20121517
39. Tian Y, Grifoni A, Sette A, Weiskopf D. Human T cell response to dengue virus infection. Front. Immunol. (2019) 10:2125. doi: 10.3389/fimmu.2019.02125
40. Grifoni A, Angelo MA, Lopez B, O'Rourke PH, Sidney J, Cerpas C, et al. Global assessment of dengue virus-specific CD4+ T cell responses in dengue-endemic areas. Front Immunol. (2017) 8:1309. doi: 10.3389/fimmu.2017.01309
41. Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc Natl Acad Sci U S A. (2013) 110:E2046–53. doi: 10.1073/pnas.1305227110
42. Tian Y, Sette A, Weiskopf D. Cytotoxic CD4 T cells: differentiation, function, and application to dengue virus infection. Front Immunol. (2016) 7:531. doi: 10.3389/fimmu.2016.00531
43. Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. (1998) 72:3999–4004. doi: 10.1128/JVI.72.5.3999-4004.1998
44. Gagnon SJ, Ennis FA, Rothman AL. Bystander target cell lysis and cytokine production by dengue virus-specific human CD4+ cytotoxic T-lymphocyte clones. J Virol. (1999) 73:3623–9. doi: 10.1128/JVI.73.5.3623-3629.1999
45. Lan NTP, Kikuchi M, Huong VTQ, Ha DQ, Thuy TT, Tham VD, et al. Protective and enhancing H2LA alleles, HLA-DRB1*0901 and HLA-A*24, for severe forms of dengue virus infection, dengue hemorrhagic fever and dengue shock syndrome. PLoS Negl Trop Dis. (2008) 2:e304. doi: 10.1371/journal.pntd.0000304
46. Weiskopf D, Angelo A, Grifoni A, O'Rourke PH, Sidney J, Paul S, et al. HLA-DRB1 alleles are associated with different magnitudes of dengue virus-specific CD4+ T-cell responses. J Infect Dis. (2016) 214:1117–24. doi: 10.1093/infdis/jiw309
47. Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. (2007) 204:1849–61. doi: 10.1084/jem.20070663
48. St. John AL, Rathore AP, Raghavan B, Ng ML, Abraham SN. Contributions of mast cells and vasoactive products, leukotrienes and chymase, to dengue virus-induced vascular leakage. Elife. (2013) 2:e00481. doi: 10.7554/eLife.00481.001
49. Furuta T, Murao LA, Lan NT, Huy NT, Huong VT, Thuy TT, et al. Association of mast cell-derived VEGF and proteases in Dengue shock syndrome. PLoS Negl Trop Dis. (2012) 6:e1505. doi: 10.1371/journal.pntd.0001505
50. Imad HA, Phumratanaprapin W, Phonrat B, Chotivanich K, Chotivanich P, Muangnoicharoen S, et al. Cytokine expression in dengue fever and dengue hemorrhagic fever patients with bleeding and severe hepatitis. Am J Trop Med Hyg. (2020) 102:943–50. doi: 10.4269/ajtmh.19-0487
51. Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4+ T cells in immunity to viruses. Nat Rev Immunol. (2012) 12:136–48. doi: 10.1038/nri3152
52. Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. (2003) 4:835–42. doi: 10.1038/ni969
53. Rivino L, Kumaran EA, Thein T-L, Too CT, Gan VCH, Hanson BJ, et al. Virus-specific T lymphocytes home to the skin during natural dengue infection. Sci Transl Med. (2015) 7:278ra35–78ra35. doi: 10.1126/scitranslmed.aaa0526
54. Koelle DM, Liu Z, McClurkan CM, Topp MS, Riddell SR, Pamer EG, et al. Expression of cutaneous lymphocyte-associated antigen by CD8+ T cells specific for a skin-tropic virus. J Clin Invest. (2002) 110:537–48. doi: 10.1172/JCI0215537
55. Zhu J, Koelle DM, Cao J, Vazquez J, Huang ML, Hladik F, et al. Virus-specific CD8+T cells accumulate near sensory nerve endings in genital skin during subclinical HSV-2 reactivation. J Exp Med. (2007) 204:595–603. doi: 10.1084/jem.20061792
56. Rivino L Lim NQ. CD4+ and CD8+ T-cell immunity to Dengue - lessons for the study of Zika virus. Immunology. (2016) 150:146–54. doi: 10.1111/imm.12681
57. Kangwanpong D, Bhamarapravati N, Lucia HL. Diagnosing dengue virus infection in archived autopsy tissues by means of the in situ PCR method: a case report. Clin Diagn Virol. (1995) 3:165–72. doi: 10.1016/0928-0197(94)00032-P
58. Liu L, Zhong Q, Tian T, Dubin K, Athale SK, Kupper TS. Epidermal injury and infection during poxvirus immunization is crucial for the generation of highly protective T cell-mediated immunity. Nat Med. (2010) 16:224–7. doi: 10.1038/nm.2078
59. Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature. (2012) 483:227–31. doi: 10.1038/nature10851
60. Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. (2009) 10:524–30. doi: 10.1038/ni.1718
61. Dung NTP, Le Duyen HT, Van Thuy NT, Van Ngoc T, Chau NVV, Hien TT, et al. Timing of CD8+ T cell responses in relation to commencement of capillary leakage in children with dengue. J Immunol. (2010) 184:7281–87. doi: 10.4049/jimmunol.0903262
62. Halstead S. Recent advances in understanding dengue [version 1; peer review: 2 approved]. F1000Research. (2019) 8:1279. doi: 10.12688/f1000research.19197.1
Keywords: dengue, T helper cells (Th cells), cytokine, T cell subsets, cytoxic T cells
Citation: Manh DH, Weiss LN, Thuong NV, Mizukami S, Dumre SP, Luong QC, Thanh LC, Thang CM, Huu PT, Phuc LH, Nhung CTH, Mai NT, Truong NQ, Ngu VTT, Quoc DK, Ha TTN, Ton T, An TV, Halhouli O, Quynh LN, Kamel MG, Karbwang J, Huong VTQ, Huy NT and Hirayama K (2020) Kinetics of CD4+ T Helper and CD8+ Effector T Cell Responses in Acute Dengue Patients. Front. Immunol. 11:1980. doi: 10.3389/fimmu.2020.01980
Received: 08 March 2020; Accepted: 22 July 2020;
Published: 24 September 2020.
Edited by:
Hyundoo Hwang, BBB Inc., South KoreaReviewed by:
Leticia Cedillo-Barron, Instituto Politécnico Nacional de México (CINVESTAV), MexicoCopyright © 2020 Manh, Weiss, Thuong, Mizukami, Dumre, Luong, Thanh, Thang, Huu, Phuc, Nhung, Mai, Truong, Ngu, Quoc, Ha, Ton, An, Halhouli, Quynh, Kamel, Karbwang, Huong, Huy and Hirayama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lan Nguyen Weiss, bG53ZWlzc0B1Y2kuZWR1; bmd1LnZ1MDFAZ21haWwuY29t; Kenji Hirayama, aGlyYWtlbkBuYWdhc2FraS11LmFjLmpw
†Present address: Lan Nguyen Weiss, Division of Genetic and Genomic Medicine, Department of Pediatrics, University of California Irvine, Irvine, CA, United States
‡These authors have contributed equally to this work
§ORCID: Dao Huy Manh orcid.org/0000-0003-3874-5051
Lan Nguyen Weiss orcid.org/0000-0003-4130-8725
Shusaku Mizukami orcid.org/0000-0002-7722-4884
Shyam Prakash Dumre orcid.org/0000-0002-4072-0745
Mohamed Gomaa Kamel orcid.org/0000-0001-9848-2965
Nguyen Tien Huy orcid.org/0000-0002-9543-9440
Kenji Hirayama orcid.org/0000-0001-9467-1777
#Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.